Abstract
Human brain structure changes throughout our lives. Altered brain growth or rates of decline are implicated in a vast range of psychiatric, developmental, and neurodegenerative diseases. Here, we identified common genetic variants that affect rates of brain growth or atrophy, in the first genome-wide association meta-analysis of changes in brain morphology across the lifespan. Longitudinal MRI data from 15,640 individuals were used to compute rates of change for 15 brain structures. The most robustly identified genes GPR139, DACH1 and APOE are associated with metabolic processes. We demonstrate global genetic overlap with depression, schizophrenia, cognitive functioning, insomnia, height, body mass index and smoking. Gene-set findings implicate both early brain development and neurodegenerative processes in the rates of brain changes. Identifying variants involved in structural brain changes may help to determine biological pathways underlying optimal and dysfunctional brain development and ageing.
Under the influence of genes and a varying environment, human brain structure changes throughout the lifespan. Even in adulthood, when the brain seems relatively stable, individuals differ in the profile and rate of brain changes1. Longitudinal studies are crucial to identify genetic and environmental factors that influence the rate of these brain changes throughout development2 and ageing3. Inter-individual differences in brain development are associated with general cognitive function4,5, and risk for psychiatric disorders6,7 and neurological diseases8,9. Genetic factors involved in brain development and ageing overlap with those for cognition10 and risk for neuropsychiatric disorders11. A recent cross-sectional study showed brain age to be advanced in several brain disorders. Brain age is an estimate of biological age based on brain structure, which can deviate from chronological age. Several shared loci were found between the genome wide association study (GWAS) summary statistics for advanced brain age and psychiatric disorders12. However, we still lack information on which genetic variants influence an individual‘s brain changes throughout life, since this requires longitudinal data. Discovering genetic factors that explain variation between individuals in brain structural changes may reveal key biological pathways that drive normal development and ageing, and may contribute to identifying disease risk and resilience: a crucial goal given the urgent need for new treatments for aberrant brain development and ageing worldwide.
As part of the Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) consortium13 the ENIGMA Plasticity Working Group quantified the overall genetic contribution to longitudinal brain changes, by combining evidence from multiple twin cohorts across the world14. Most global and subcortical brain measures showed genetic influences on change over time, with a higher genetic contribution in the elderly (heritability 16 – 42%). Genetic factors that influence longitudinal changes were partially independent of those that influence baseline volumes of brain structures, suggesting that there might be genetic variants that specifically affect the rate of development or ageing. However, the genes involved in these processes are still not known, with only a single, small-scale GWAS performed for longitudinal volume change in gray and white matter of the cerebrum, basal ganglia, and cerebellum15. Here, we set out to find genetic variants that may influence rates of brain changes over time, using genome-wide analysis in individuals scanned with magnetic resonance imaging (MRI) on more than one occasion. We also aimed to identify age-dependent effects of genomic variation on longitudinal brain changes in mostly healthy, but also neurological and psychiatric, populations.
In our GWAS meta-analysis, we sought genetic loci associated with annual change rates in 8 global and 7 subcortical morphological brain measures in a coordinated two-phased analysis using data from 40 longitudinal cohorts (Extended Data Fig 1 and Supplementary Table 1). We extracted global and subcortical brain measures, and assessed annual change rates, using additive genetic association analyses to estimate the effects of genetic variants on the rates of change within each cohort. As brain change is not constant over age1 and gene expression also changes during development and ageing16, we determined whether the estimated genetic variants were age-dependent, i.e., differentially affected rates of brain changes at different stages of life, by using genome-wide meta-regression models with linear or quadratic age effects (Methods). It must be noted that although the cohorts analysed in this study together cover the full lifespan, there is relatively little age overlap between them. This implies that we cannot rule out that cohort-specific characteristics other than age could influence our meta-regression findings.
We employed a rolling cumulative meta-analysis and -regression approach17. In phase 1, for which data collection ended on Feb 1st, 2019, we analysed the cohorts of European descent (N=9,623). We sought replication by adding data from three additional cohorts that became available after our analysis of phase 1: one developmental cohort (average age 10 at baseline) and two in ageing populations (N =5,477; all of European descent; total N=15,100 in phase 2). For all follow-up analyses we used results from phase 2. Finally, we added cohorts of non-European ancestry (total N=15,640).
Longitudinal trajectories
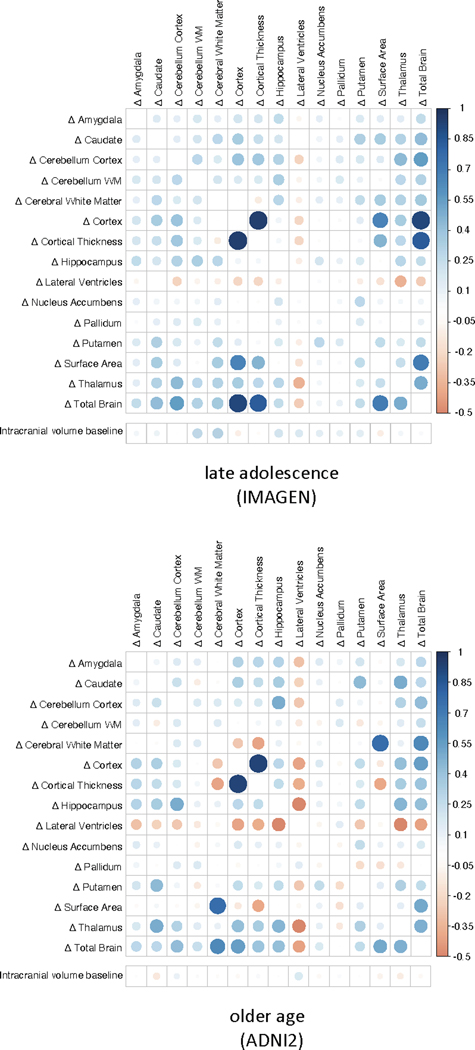
Brain measures showed differing trajectories of change with age (Figures 1,2 and Extended Data Video 1) - either monotonic increases (lateral ventricles), monotonic decreases (cortex volume, cerebellar grey matter volume, cortical thickness, surface area, total brain volume), or increases followed by stabilization and subsequently decreases (cerebral and cerebellar white matter, thalamus, caudate, putamen, nucleus accumbens, pallidum, hippocampus and amygdala volumes). Each brain structure showed a characteristic trajectory of change. Within two of our largest cohorts in phase 1 (one in childhood and one in older age), we computed correlations between the rates of change of all possible pairs of these 15 brain structures. These correlations in both childhood and older age were generally low in our data (Extended Data Fig. 2), except for the correlation between rates of change of cortical thickness and cortex volume. Therefore, we chose to investigate all brain structures separately, maximizing sensitivity of the GWAS to identify region-specific associations of genetic variants. Using the correlation structure, we estimated the effective number of independent variables through matrix spectral decomposition on the rates of change, yielding 14 independent traits for multiple testing corrections (Methods).
Figure 1:
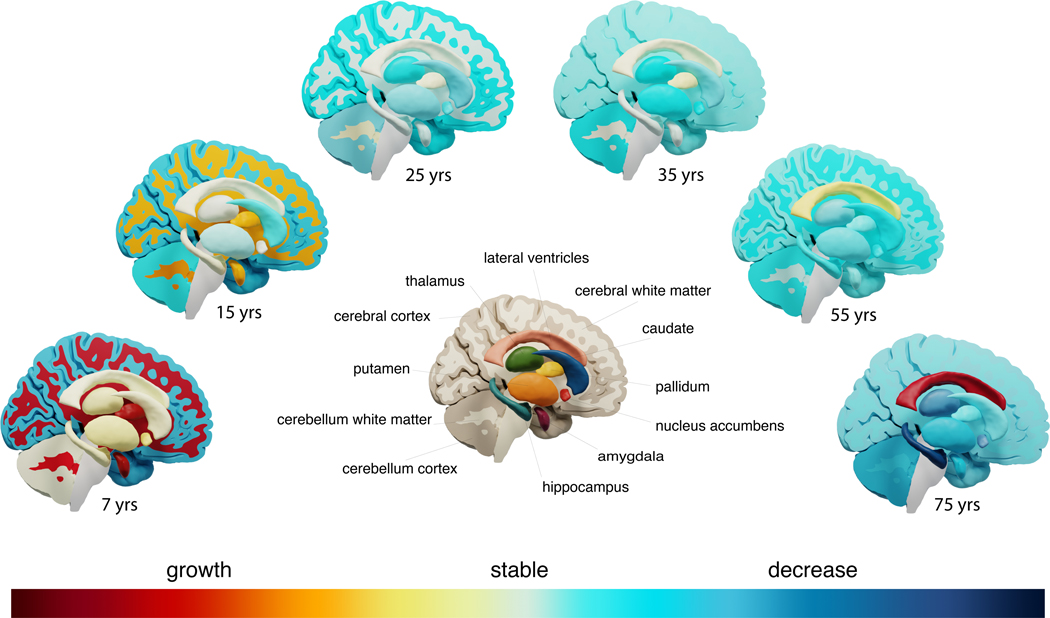
Phenotypic brain changes throughout the lifespan.
Visualization of growth and decline of brain structures throughout the lifespan. The subcortical structures are shown in exploded view.
Figure 2:
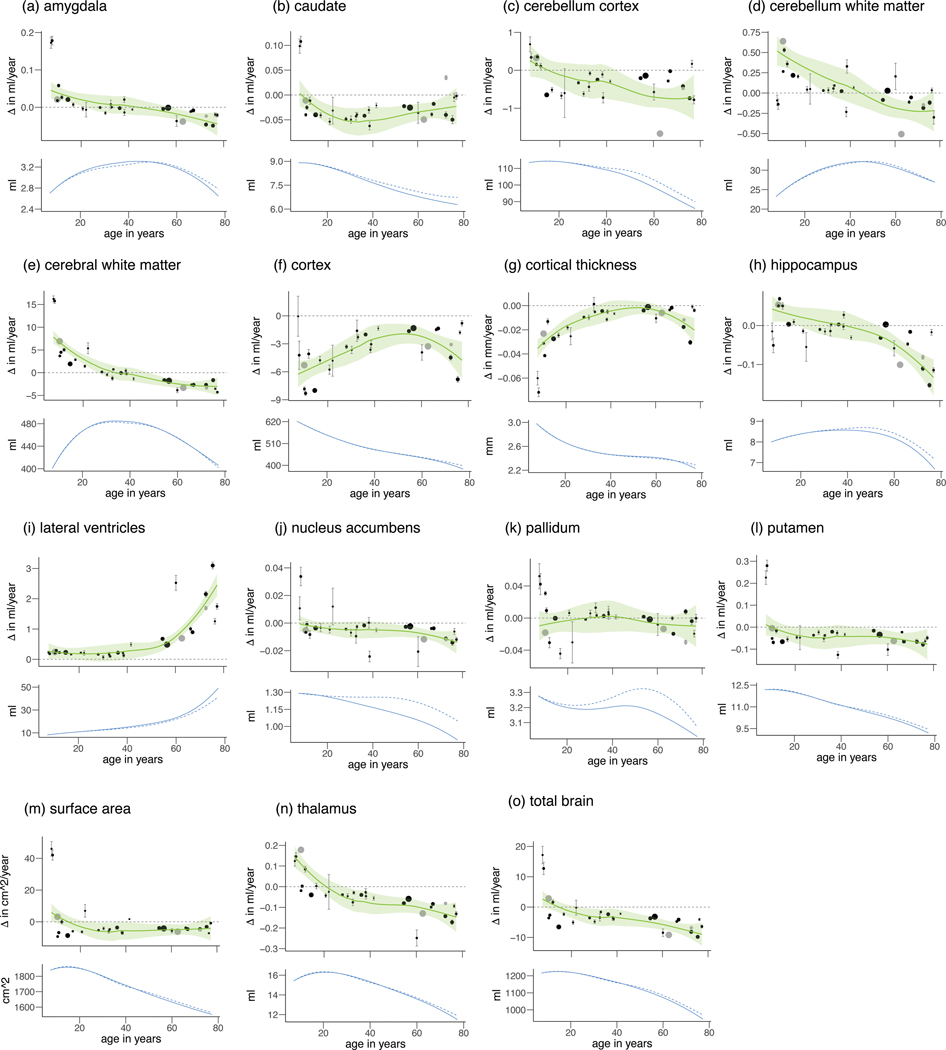
Annual rates of change Δ per cohort for each structure (a-o).
The estimated trajectories with 95% confidence intervals (in green) are displayed in the top row. Mean values of individual cohorts are displayed as points, with error bars representing standard errors displayed in grey. The size of the points represents the relative size of the cohorts, total sample size N=15640. Means and standard deviations are based on raw data – no covariates were included. Cohorts that were added in phase 2 are displayed in grey. Only cohorts that satisfy N>75 and mean interval > 0.5 years are shown. The estimated trajectories of the volumes themselves are displayed in the bottom row, for all subjects (solid line) and for subjects not part of diagnostic groups (dashed line).
Age-independent associations
Two loci showed genome-wide significant effects on the rate of brain change in phase 1, one of which was also genome-wide significant in phase 2 (Figure 3; Supplementary Table 4; p-value replication sample 0.08). This lead SNP, rs72772740 on chromosome 16, is an intronic variant located in the GPR139 gene and was associated with rate of change in lateral ventricle volume (Figure 4). Functional annotation identified numerous significant expression quantitative trait loci (eQTL) associations (FDR < 0.05) in different datasets and highlighted genes by either eQTL mapping (GPRC5B, IQCK, KNOP1, C16orf62) or chromatin interaction mapping (ACSM1, ACSM5, UMOD, GP2). GPR139 is the G-protein-coupling receptor gene 139, which encodes a member of the rhodopsin family of G-protein coupled receptors. The gene is almost exclusively expressed in the central nervous system, with highest expression from 12 to 26 weeks post-conception, and has been suggested as a therapeutic target for metabolic syndromes and motor diseases18. GPR139 may play a role in foetal brain development19. Mice lacking GPR139 exhibited schizophrenia-like behavioural abnormalities20, and functional cell assays showed the inhibitory influence of GPR139 on dopamine receptor 2 (D2R) signalling20. The second lead SNP, rs449998, an intronic variant on chromosome 21 located in the Down Syndrome Cell Adhesion Molecule (DSCAM) gene, was associated with rate of change in nucleus accumbens volume in phase 1, but this association was not significant in the replication sample, or phase 2. Three SNPs were significant in the phase 2 analysis only. These include rs10990953, intergenic on chromosome 9, associated with rate of change in lateral ventricle volume; rs1425034, intergenic and located in long intergenic non-protein coding RNA on chromosome 2, associated with rate of change in pallidum volume; and rs12325429, intron of CDH8 on chromosome 16, associated with rate of change in total brain volume (Supplementary Table 5; Supplementary Figs. 1,2 provide Manhattan plots, QQ plots, locus plots and circos plots). The association of CDH8 with total brain volume rate of change is particularly interesting, since CDH8 has been associated previously with learning disability and autism21. CDH8 is a protein-coding gene and encodes a type II classical cadherin from the cadherin superfamily, integral membrane proteins that mediate calcium-dependent cell–cell adhesion. Genome-wide significant SNPs in phase 1 or phase 2 did not show heterogeneity (I2 < 10.2; p(I2) > 0.31; Supplementary Tables 4,5, Supplementary Fig.3 for forest plots).
Figure 3:
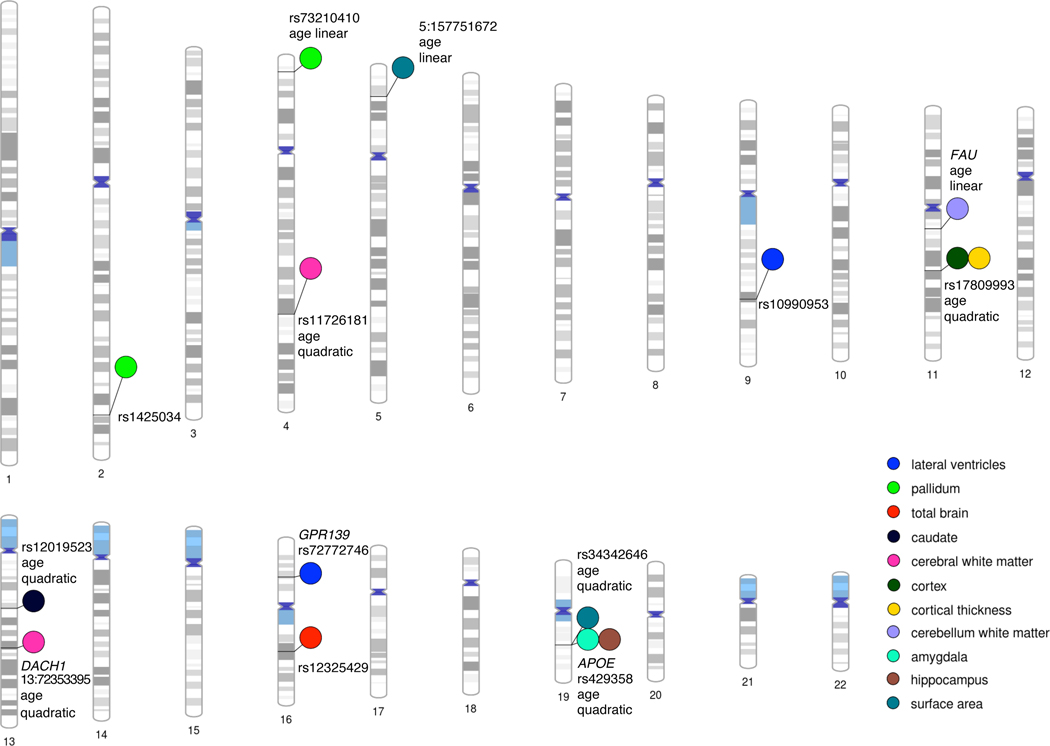
Genetic effects on rates of brain changes throughout the lifespan.
Genome-wide significant SNPs and genes with effects on brain changes at their respective loci across the human genome, from phase 2 (total N=15,100). This plot was created using PhenoGram (http://visualization.ritchielab.org).
Figure 4:
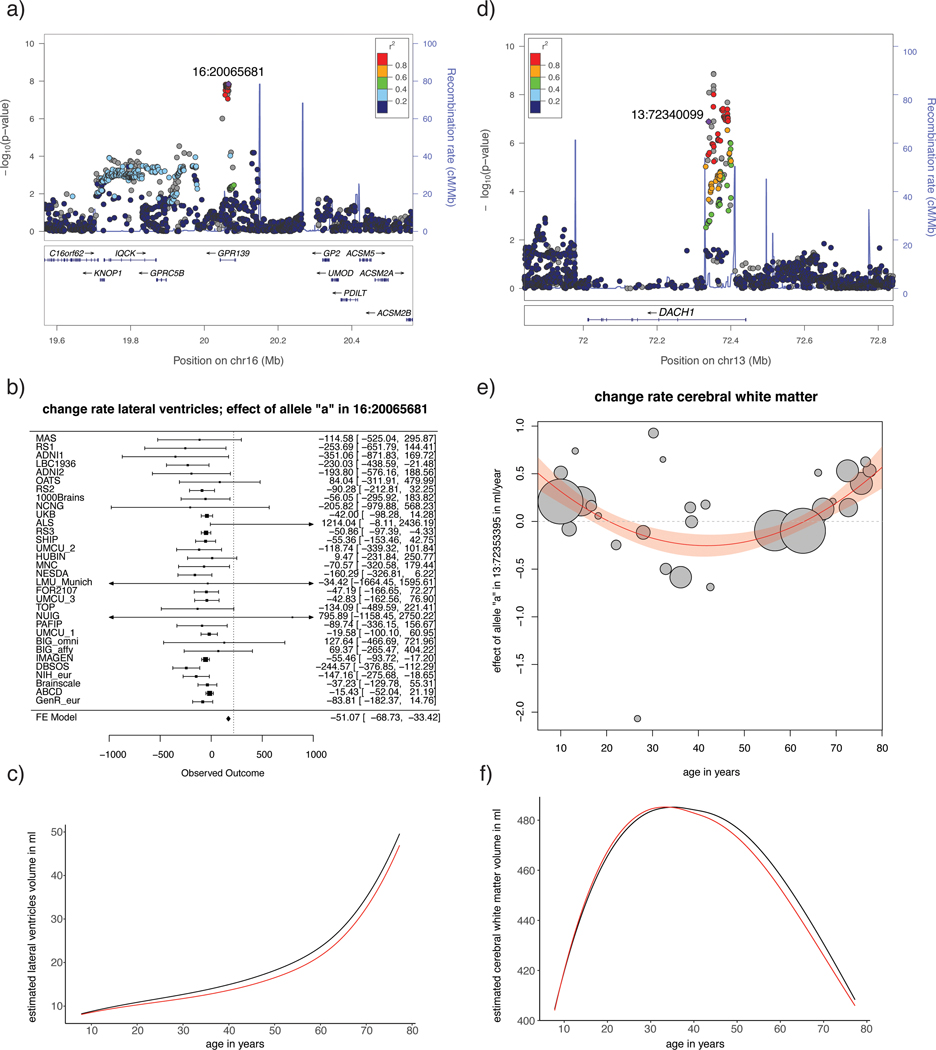
Summary of findings for two top-SNPs.
Shown here is a summary of findings for a top-SNP of an age independent effect (rs72772746; intron to GPR139; associated with rate of change of lateral ventricle volume; left column) and a top-SNP of an age dependent effect (13:72353395; intron to DACH1; associated with rate of change in cerebral white matter volume; right column). Displayed are the locus plots (a) and (d), forest plot (b; total N = 14593, means and 95% confidence intervals are displayed for each cohort; confidence intervals that are outside the axis of the plot are marked with an arrow) and plot of meta-regression (e; total N = 13864, center of the circles represent the effect size of the tested allele for each cohort, radius of the circles are proportional to sample size) and inferred lifespan trajectories for carriers (in red) and non-carriers of the effect allele (in black) (c) and (f). Note that 13:72353395 was not in the reference dataset containing LD structure; the displayed LD structure is based on 13:7234009, R2 = 0.87 with the top-SNP.
Age-dependent associations
Three additional loci had an association with rate of change that was variable across the lifespan in phase 1 (Figure 3; Supplementary Tables 6,8). For two of these, the association remained significant in the phase 2 analysis: rate of change in white matter cerebrum volume was affected by rs573983368 (intronic variant) in the Dachshund Family Transcription Factor 1 (DACH1) gene, and 5:157751672 (intergenic and located in long intergenic non-protein coding RNA LINC02227) on chromosome 5 had an age-dependent effect on the rate of change in surface area (Figure 4; Supplementary Tables 6-9). Rate of change in cerebellar white matter volume was affected by the intronic rs10674957 in the Thyrotropin Releasing Hormone Degrading Enzyme (TRHDE) gene, but this third locus was not significant in phase 2.
The DACH1 locus shows significant chromatin interaction, which can play an important role in gene expression regulation. DACH1 encodes a chromatin-associated protein that associates with DNA-binding transcription factors to regulate gene expression and cell fate determination during development. DACH1 is highly expressed in the proliferating neural progenitor cells of the developing cortical ventricular and subventricular regions, and in the striatum22. We found the effect of DACH1 to have a quadratic age-dependence, with the variant being associated with faster growth in childhood and earlier but slower decline with ageing (Figure 4). The effect of 5:157751672 had a linear age-dependence, with the tested variant being associated with less growth of surface area in childhood, and less decline at older age.
For seven additional loci we found a significant age-dependent association with rate of change only in phase 2 (Supplementary Tables 7,9; Supplementary Figs. 1,2 provide Manhattan plots, QQ plots, locus plots and circos plots). One of these, rs429358, a missense variant of the Alzheimer’s disease (AD)-related23 apolipoprotein E gene (APOE) gene, was associated with change rate in hippocampus, showing prolonged growth into adulthood and faster reductions of volume of the hippocampus for carriers of the AD risk variant. APOE plays a role in maintenance of cellular cholesterol homeostasis by delivering cholesterol to neurons on apoE-containing lipoprotein particles. Cholesterol is important for synapse and dendrite formation, and cholesterol depletion has been shown to cause synaptic and dendritic degeneration24. Other findings include rs12019523, an intronic variant in the CAB39L gene associated with rate of change of the caudate volume; rs34342646, an intronic variant in the NECTIN2 gene associated with rate of change in surface area and rs73210410, an intronic variant in the SORCS2 gene associated with rate of change in pallidum volume.
To visualize the age-dependent effects, we plotted the meta-regression results for the significant loci (Methods, Supplementary Fig. 3). Genome-wide significant SNPs in phase 1 or phase 2 did not show significant residual heterogeneity (p > 0.23; except for the age-dependent effect of rs429358 on hippocampus change rate (p=0.02)). A summary of the genome-wide significant results and the top-10 loci for each phenotype and age model are presented in Supplementary Tables 4-9.
Gene-based analyses
Gene-based associations with all phenotypes were estimated using MAGMA (Methods). We found six genome-wide significant genes influencing structural rates of change in phase 1, four of which were also significant in phase 2 (Supplementary Table 10, 11); among these, DACH1 and GPR139, which were implicated through SNP-based GWAS, also reached genome-wide significance in this gene-based GWAS. In addition, we found APOE to be associated with change rates for both hippocampus and amygdala. The phase 2 analysis showed two new findings: an association of the FAU gene with rate of change in cerebellum white matter volume, and again APOE, associated with rate of change in surface area. Of note, the APOE findings were based on GWAS and subsequent gene analysis, and we did not investigate the classical APOE status, since that is determined by a combination of two SNPs. However, we observed that the effect of APOE on change rate of hippocampus and amygdala was fully driven by rs429358, with the risk variant for AD causing prolonged growth into adulthood and faster decay for both amygdala and hippocampus volumes later in life.
To visualize the age-dependent effects, we plotted the meta-regression results for the top SNP in each of the significant genes (Supplementary Fig. 3). Supplementary Tables 10, 11 display the top-10 genes for each phenotype and each age model. Supplementary Table 12 details putative biological functions of associated genes and genes harbouring genome-wide significant associated loci.
Gene-set analyses
To test whether genetic findings for brain structure change converged onto functional gene sets and pathways, we conducted gene-set analyses using MAGMA (Methods). Competitive testing was used and 10 and 12 genome-wide significant gene sets were found for phase 1 and phase 2, respectively (Supplementary Tables 13, 14 for top-10 gene sets and genes included). Two main themes emerge from this analysis, as biological functions of the gene sets converge onto involvement in early brain development and involvement in neurodegeneration, respectively.
One gene set was significant in both the phase 1 and phase 2 analyses, i.e. GO_neural_nucleus_development. This gene set consists of genes involved in the development of neural nuclei (compact clusters of neurons in the brain) and was associated with rates of change in cerebellar white matter volume in our study. Two other gene sets, significant in phase 1 (GO_substantia_nigra_development associated with rate of change in cerebellum white matter volume) and phase 2 (GO_midbrain_development associated with quadratic age-dependent surface area rates of change) were closely related to neural nucleus development in gene ontology terms.
The most significant gene set was GO_response_to_phorbol_13_acetate_12_myristate (p-value=1.42e-08) in phase 2, related to surface area change. Phorbol 13-acetate 12-myristate is a phorbol ester and an activator of protein kinase C (PKC)25. Two other gene sets, significant in phase 2 (GO_tau_protein_binding and GO_tau_protein_kinase_activity) and both associated with rate of change in caudate volume, imply genes involved in interacting with tau protein. Tau is a microtubule-associated protein, implicated in Alzheimer’s disease, Down Syndrome and amyotrophic lateral sclerosis (ALS).
Follow-up analyses: overlap with cross-sectional findings
SNP-based heritability estimates (h2) of the rates of change based on linkage disequilibrium score regression (LDSC; Methods) were small overall (Supplementary Table 15). For all phenotypes, the h2 z-score was below 4. We thus tested for genetic overlap with cross-sectional brain data and other phenotypes by applying approaches other than LDSC, although these do not provide a measure of genetic correlation. To investigate whether cross-sectional GWAS for brain structure and our GWAS on rates of change identify the same or different genetic variants, we investigated overlap between rate of change and earlier published data on cross-sectional brain structure of the same structure, where available (Methods). Supplementary Fig. 4 displays the number of overlapping genes tested against the expected number of overlapping genes that would occur by chance, in the first 1–1,000 ranked genes. Supplementary Table S11 lists the top-10 gene findings for each of the 15 change-rate phenotypes and compares these with the gene ranks from cross-sectional data. In the top-10 ranked genes, APOE for hippocampus occurred in the top-10 for both cross-sectional data26 and age-dependent effects on rate of change (p=0.006). No overlap was seen for the other measured phenotypes. Extending this search to the top 200 (~1% of genes), we found overlapping genes above chance level for cortical thickness of quadratic age-dependent genes and cross-sectional findings (p = 8.39e-05). In the top 1,000 ranked genes (~5% of genes), further overlapping genes did emerge (Supplementary Fig. 4). Overlapping genes at such a high aggregate level imply that largely different genetic backgrounds underlie changes in brain structure and brain structure per se.
To test for global genomic overlap between our findings and GWAS of cross-sectional volumes we applied independent SNP-Effect Concordance Analyses (iSECA) (Methods) and tested for pleiotropy. We found no significant pleiotropy between longitudinal and cross-sectional results, confirming a largely different genetic background for changes in brain structure and brain structure per se (Figure 5).
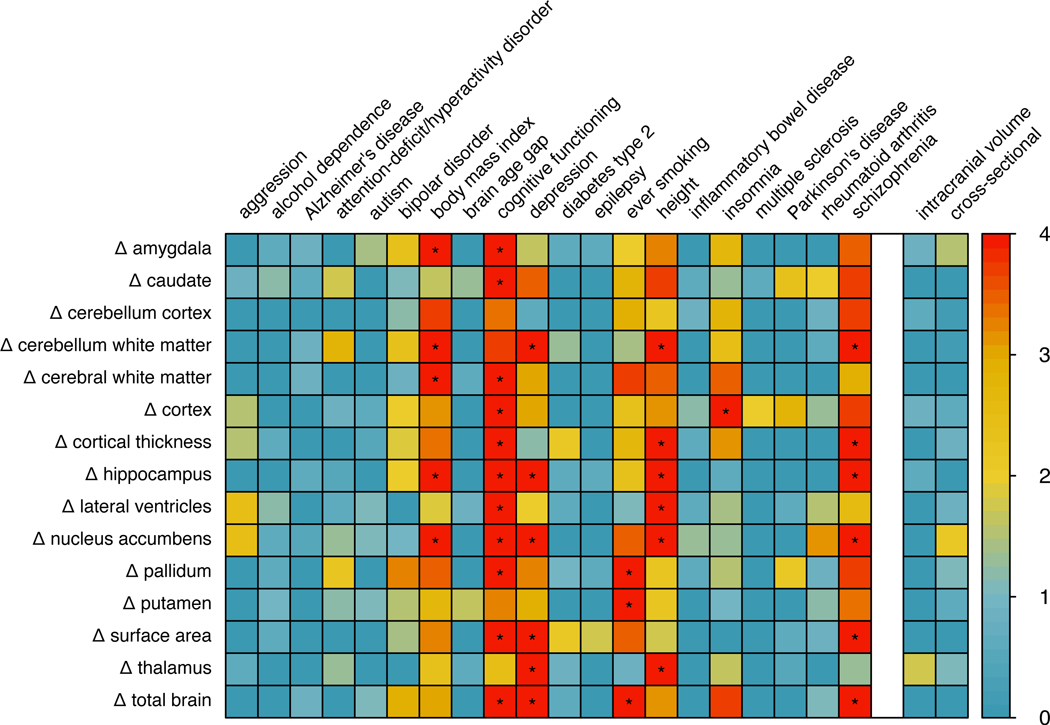
Figure 5:
Genetic overlap with other phenotypes.
P-values for pleiotropy between change rates of structural brain measures (rows, indicated by Δ for change rate) and neuropsychiatric, disease-related and psychological traits (columns on the left). P-values for pleiotropy between change rates of structural brain measures and head size (intracranial volume) and the cross-sectional brain measure are displayed on the right. The colour legend is displayed on the right, indicating the -log10 p-value. Significant overlap (p < 1.6e-04; obtained through permutation testing, two-sided, Bonferroni corrected) is marked with *. P-values underlying this figure can be found in Supplemental Table 16.
Follow-up analyses: overlap with other traits
We applied iSECA for overlap between our age-independent summary statistics for structural brain changes and several neuropsychiatric, neurological, physical, ageing and disease-related phenotypes and psychological traits (Methods). We found significant genomic overlap (p < 1.6e-04) with genetic variants associated with depression27, schizophrenia28, cognitive functioning29, height30, insomnia31, body mass index (BMI)30 and ever-smoking32. Despite significant pleiotropy between rates of change and these traits, we did not find evidence for concordance or discordance of effects (Figure 5, Supplementary Figure 5). For comparison, we computed the genomic overlap between cross-sectional volumes and these phenotypes using the same method. In general, cross-sectional volumes showed overlap for the same traits and several others. Of note, there was also little overlap between the summary statistics for the longitudinal brain measures and summary statistics for the corresponding volumes, based on cross-sectional data. This implies that despite the fact that both cross-sectional brain volume and rates of changes are associated with traits such as schizophrenia or cognitive functioning, these associations are likely not driven by the same genomic locations. Additionally, there was little overlap in the genetic loci associated with the longitudinal brain measures and intracranial volume at baseline, indicating that overall head size did not drive our findings (Figure 5).
Follow-up analyses: gene expression across the lifespan
We determined mRNA expression for genome-wide significant genes and genes associated with genome-wide significant SNPs (Supplementary Tables S5,7,9) in 54 tissue types and in both the developing and adult human brain (Methods). For the prioritized genes, a gene expression heatmap was created, based on GTEx v8 RNAseq data33. This revealed considerable expression levels across several brain tissues for the following genes: APOE, CAB39L, FAU, NECTIN2 (alias PVRL2) and SORCS2, the latter showing higher expression in brain tissue compared to all other tissue types (Supplementary Fig. 6A). These genes show different expression patterns across the lifespan in the BrainSpan data34. DACH1 shows highest expression during early prenatal stages (8–9 post conception weeks), compared to postnatal stages. Several genes demonstrate stable high expression levels throughout development and across the lifespan (APOE, CAB39L, FAU, NECTIN2 (alias PVRL2)). CDH8 shows lower expression in the early prenatal stages and higher expression later in life (Supplementary Fig. 6B).
Follow-up analyses: phenome-wide associations
For the prioritized SNPs and genes (Supplementary Tables 5,7,9,11), exploratory pheWAS (i.e., ‘phenome-wide’) analysis was performed to systematically analyse many phenotypes for association with the genotype and individual genes (Supplementary Table 17). PheWAS was performed using publicly available data from the GWASAtlas32 (https://atlas.ctglab.nl). Gene associations of DACH1, GPR139 and SORCS2 showed pleiotropic effects mainly in the metabolic domain, e.g., with estimated glomerular filtration rate and BMI (Supplementary Table 17, Supplementary Fig. 7). SORCS2 and CDH8 also showed significant associations with psychiatric and cognitive traits. Both APOE and NECTIN2 showed strongest associations with Alzheimer’s disease, cholesterol and lipids (Supplementary Table 17, Supplementary Fig. 7).
Sensitivity analyses
We repeated the SNP and gene analyses in various subgroups: 1) by adding four cohorts of non-European or mixed ancestry (N=540; total N=15,640); 2) by omitting cohorts that did not meet a minimum sample size criterion (N>75) or a minimum scanning interval (> 0.5 years) leaving N=14,601; 3) by excluding diagnostic groups in each cohort, leaving N=13,034, and 4) by including a covariate adjusting for disease status (Supplementary Tables 18,19). In SNP-based and gene-based analyses, effect sizes of SNPs were very similar in all subgroups, suggesting that our results are also applicable for individuals of non-European ancestry (with the caveat that the non-European subgroup was rather small) and were not driven by the smaller cohorts. Findings were also similar in the healthy subgroup and when correcting for disease status, with one notable exception: the association between APOE and rate of volume change in hippocampus and amygdala, with increasing influence of the top SNP with age, was no longer present after correcting for disease (see Supplementary Table 1 for diagnoses). This suggests that these APOE findings were in part driven by the presence of patients in the cohorts and could therefore be explained either by disease-related genes that also influence rates of change or by brain changes occurring as a consequence of the disease.
Given that our main analyses included patients, and iSECA analyses showed several associations with disease, we repeated iSECA analyses excluding diagnostic groups in each cohort. These analyses implicate the same traits, associated with largely the same rates of change of brain measures (Supplementary Fig. 5).
Discussion
Here, we present the first GWAS investigating influences of common genetic variants on brain-structural changes in over 15,000 subjects covering the lifespan. The longitudinal design of our study combined with the large age range assessed provides a flexible framework to detect age-independent and age-dependent effects of genetic variants on rates of structural brain changes. We identified genetic variants for structural brain changes between 4 and 99 years of age. Some of these were independent of age, showing effects that were stable throughout life in terms of strength and direction, suggesting that these genetic variants are equally crucial for early brain development as for brain ageing. In addition, we identified age-dependent genetic variants, suggesting that some genetic variants are predominantly associated with brain development while others are mainly associated with brain ageing.
Amongst our top findings is the APOE gene, a major risk factor for AD23, and specifically a missense variant in that gene, which influences rates of change in amygdala and hippocampus volume with varying and differential effects across the lifespan, with probably most pronounced effects in those affected with brain disorders. While most of the additional genetic loci identified here have not previously been associated with any brain-plasticity-related phenotypes, several others were also linked to brain disorders, including psychiatric (e.g., GPR139 and CDH8) and neurodegenerative disorders (e.g., NECTIN2). Notably, DACH1 and NECTIN2 show increased expression during early development, while other genes’ brain expression patterns are most pronounced during adulthood (e.g., APOE and CDH8), suggesting that these genes may exert specific effects during different developmental periods.
Gene-set analysis also implies a role for both developmental and neurodegenerative processes. We found a gene-set involved in ‘neural nucleus development‘ that influenced rates of change in cerebellar white matter. Other closely related gene ontology terms, ‘development of the substantia nigra and midbrain nuclei’, were associated with rates of change of cerebral white matter volume and surface area. These all implicate the biological process of progression of a neural nucleus, a compact cluster of neurons in the brain, from its initial condition or formation to its mature state. This would also suggest that we observed the influence of genes involved in early developmental mechanisms of (subcortical) nuclei on cortical changes later in life. It is unclear whether this is a direct effect of these gene sets on cortical changes in adulthood, or the consequence of these early developmental pathways. In addition, we found several gene-sets interacting with tau-protein associated with rate of change in caudate volume, and a gene-set associated with rate of change in surface area that implicates phorbol 13-acetate 12-myristate, an activator of protein kinase C (PKC)25. PKC is a family of enzymes whose members transduce a large variety of cellular signals and plays a key role in controlling the balance between cell survival and cell death. Its loss of function is generally associated with cancer, whereas its enhanced activity is associated with neurodegeneration. PKC both directly phosphorylates tau and indirectly causes the dephosphorylation of tau, and has been suggested to play a key role in the pathology of Alzheimer’s disease35. Together these results suggest involvement of genes in ageing and neurodegeneration.
At the global, genome-wide level, we found significant genomic overlap between genetic variants associated with rate of change with genetic variants associated with depression, schizophrenia, cognitive functioning, insomnia, height, body mass index (BMI) and ever-smoking. Several of these traits, such as schizophrenia, smoking, cognitive functioning, and body mass index, have been associated with longitudinal brain-structural changes5,36–38. The global overlap coincides with findings at the individual gene level: several of the identified genetic variants and genes were linked to metabolic processes (APOE, DACH1, GPR139, NECTIN2), cognitive functioning (CDH8), psychiatric traits (GPR139, SORCS2, CDH8) and Alzheimer’s disease (NECTIN2 and APOE) as apparent from the pheWAS results. Despite the pleiotropic effects, concordance of effects was generally null. This is not surprising, as rate-of-change measures for brain structures are not constant and often switch sign over the course of the lifespan1,39, whereas the GWAS for other traits assume stability of both the phenotype and the genetic influences on the phenotype over time. As such, concordance and discordance of effects would not be expected.
The advantage of longitudinal analyses is that each individual acts as their own control, allowing us to separate the genetic effects on volumes in cross-sectional studies from those on the rates of change14. Indeed, we found little overlap between the two: top genes identified in the GWAS on cross-sectional brain structure26,40–42 generally did not overlap with the top genes for the corresponding rates of change. Longitudinal analyses have long been shown to provide different information from cross-sectional approaches. On a phenotypic level, ageing patterns of the hippocampus show different results in cross-sectional studies than in longitudinal studies43. On a genetic level, a study that included a within-sample SNP-by-age interaction in the ADNI cohort showed that the power to detect genetic associations was larger for a longitudinal design than for a cross-sectional analysis44. Of note, that study also identified rs429358 in APOE as being associated with longitudinal hippocampal and amygdala volume change in older age (the ADNI cohort is also included in the current study). Through our meta-regression approach, we now show this variant to exert an effect across the lifespan, with the risk variant for AD causing faster increases in childhood for amygdala volume and faster volume reductions for both amygdala and hippocampus later in life.
Given the dynamics of brain structural changes during the lifespan, we investigated both age-independent and age-dependent genetic effects. The age-independent effects can be interpreted as neurodevelopmental influences that also impact brain structure at older ages45,46, whereas the age-dependent effects can be interpreted as possible changing effects of genes or gene expression during life16. The genome-wide meta-regression approach employed here may enable future GWAS for other phenotypes that change over the human lifespan.
We chose to analyse longitudinal changes for 15 separate brain structures, because we observed generally low correlations between these phenotypic changes. This approach allowed us to find brain-structure-specific associations. However, several longitudinal studies have described phenotypic correlations between structural changes 39,47,48; combining several phenotypes could thus be an alternative approach to identify genetic variants that exert a global effect. Of note, cohort and age are intertwined in our meta-regression analysis. Although the cohorts analysed in this study together cover the full lifespan, there is relatively little age overlap between them; therefore, we cannot be sure that differences between cohorts can be exclusively attributed to age. Mega-analysis would circumvent this problem, but was not feasible in practice. Moreover, we imposed the same stringent criteria of genome-wide significance for the age-independent meta-analysis and age-dependent meta-regression, which renders chance findings equally unlikely in either type of analysis. In addition, residual heterogeneity for the top findings was generally small. That said, our sample size is still relatively modest for GWAS purposes, and replication in larger samples and inclusion of other ancestries is needed once more longitudinal data becomes available.
How exactly variation in these genes impacts brain changes in health and disease cannot be answered based on genome-wide association studies. To this end, our findings may direct future studies into brain development and ageing, and prevention and treatment of brain disorders. For example, biological pathways that guide neural nucleus development in the foetal subcortical brain may be particularly relevant to the cerebral white matter growth and cortical thinning that takes place during childhood and adolescence. Neurodegenerative disorders might be better understood when we identify genetic variants that influence brain atrophy over time, compared with identification of static genetic differences. In conclusion, our study shows that our genetic architecture is associated with the dynamics of human brain structure throughout life.
Methods
Ethical approval
All participants gave written informed consent and all participating sites obtained approval from local research ethics committees/institutional review boards. Ethics approval for meta-analyses within the ENIGMA consortium was granted by the QIMR Berghofer Medical Research Institute Human Research Ethics Committee in Australia (approval: P2204).
Inclusion criteria
Cohorts that had longitudinal magnetic resonance imaging (MRI) data of the brain and genotyped data extracted from blood or saliva available were invited to participate, irrespective of disease status and age. Patients were not excluded as aberrant brain trajectories are often observed and we hypothesize that genetic risk for disease may be associated with genetic influences on rates of change. We included cohorts that had a preferred sample size of at least 75 subjects and a follow up duration (for repeated MRI scans) of at least six months. After quality control of individual subject’s imaging and genotyping data, not all the cohorts could meet these criteria. In total, we included 15,640 subjects aged 4 to 99 (49% female, 14% patients). Please see Extended Data Fig. 1 and Supplementary Table 1 for further description of the cohorts.
Longitudinal imaging
Eight global brain measures (total brain including cerebellum and excluding brainstem, surface area measured at the grey-white matter boundary, average cortical thickness, total lateral ventricle volume, and cortical and cerebellar grey and white matter volume) and seven subcortical structures (thalamus, caudate, putamen, pallidum, hippocampus, amygdala and nucleus accumbens) were extracted from the FreeSurfer processing pipeline49–51; see Supplementary Table 2 for details per cohort). We chose these measures based on the fact that they show generally high test-retest reliability for cross-sectional measures52–54, thereby selecting those measures that would have sufficient signal to noise in change measures. Image processing and quality control were performed at the level of the cohorts, following harmonized protocols (http://enigma.ini.usc.edu/protocols/imaging-protocols/) which included visual inspection of the segmentation. Annual rates of change were computed in each individual for each phenotype by subtracting baseline brain measures from follow up measures and dividing by the number of years of follow-up duration. We chose not to correct for overall head size in the main analysis: while it is common practice to correct for intracranial volume when investigating cross-sectional brain volumes55, the associations between intracranial volume and brain changes over time are small (Extended Data Fig. 2) and GWAS findings are very similar with and without correction (Supplementary Note; Supplementary Figure 8). Distributions of baseline and follow-up measures - as well as annual rates of changes - were visually inspected and change rates were centrally compared for consistency.
Longitudinal trajectories of brain structure rates of change were estimated by applying locally, cohort-size weighted, estimated scatterplot smoothing with a Gaussian kernel, local polynomials of degree 2 and a span of 1 (LOWESS56) implemented in R57. Integrating these trajectories and then fitting these to the baseline values of the phenotypes in the cohorts provides trajectories throughout the lifespan. Trajectories were estimated in the full dataset including patients and by excluding diagnostic groups in each cohort separately.
Genome-wide association analysis
At each participating site, genotypes were imputed using the 1000 Genomes project dataset58 through the Michigan imputation server59 (https://imputationserver.sph.umich.edu/) or the Sanger imputation server60 (Supplementary Table 3). Subsequently, each site ran the same multidimensional scaling (MDS) analysis protocol, computing MDS components from the combination of their cohort’s data with the HapMap3 population61. This ensured that all sites corrected for ancestry in a consistent manner. See http://enigma.ini.usc.edu/protocols/genetics-protocols/ for the imputation and MDS analysis protocol. Within each cohort genome-wide association was conducted using an additive model, modelling change rate as a function of the genetic variant plus covariates age, sex, age*sex, age2, age2*sex and ancestry (the first four MDS components). While it is possible that rates of brain structural changes are different in males and females, we did not have the power to perform analyses separating the sexes. Dummy variables were added where appropriate, e.g., when multiple scanners were used. We re-ran these analyses adding a covariate for disease status if the cohorts contained patients and controls. Most sites used our harmonized GWAS protocol, which used raremetalworker62 for analysis (Supplementary Table 3). Regardless of the study design, a kinship matrix was incorporated in these analyses, accounting for relatedness in family studies, or possible unknown kinship in the other studies.
Given the small sample sizes of the individual cohorts, a stringent cohort level quality control was enforced, to exclude variants with a minor allele frequency (MAF) < 0.05 or variants with imputation R2 / info score < 0.75. Across cohorts and phenotypes, GWAS summary plots (Manhattan plots and QQ plots) were visually inspected at the central site. If a given cohort / trait showed deviation from expectations, sites were asked to re-analyse their data, which usually involved removal of outliers in the phenotypic data. QQ plots per cohort, per phenotype can be found in Supplementary Figure 10.
Meta-analysis and Meta-regression
In the phase 1 cohorts of European ancestry (N=9,604) we aggregated the cohort-level data for each phenotype, using standard-error weighted meta-analysis or meta-regression. We employed a cumulative meta-analysis and meta-regression approach for replication, in phase 2 (N=15,100). The meta-regression could not be performed separately in the three independent cohorts added in phase 2 since a regression line based on three points is prone to overfitting. For age-independent analyses, we list results in the added sample (Supplementary Tables 4 and 10). We tested three models. Under the assumption that effect sizes of single nucleotide polymorphisms (SNPs) were consistent across the lifespan (i.e., a standard meta-analytic approach), where the subscript C denotes a cohort and ε an error term:
-
1
, under the null hypothesis that .
Given that brain changes throughout life are dependent on age, the effects of a genetic variant on brain change are likely to depend on age too. Within cohorts, such an age by SNP effect analysis would not have been feasible since longitudinal cohorts that span the age-range between 4–99 years do not exist. Given the widespread mean age among the cohorts included (Extended Data Fig. 1 and Supplementary Table 1), it was possible to calculate the age-dependent effects across the life span by comparing effects of loci between cohorts, through meta-regression. Meta-regression is a sophisticated tool for addressing heterogeneity between cohorts in meta-analyses when the source of heterogeneity is known (in this case, age)63. We estimated the following model under the assumption that the effects of SNPs may vary in size or direction across the lifespan:
-
2
under the null hypothesis that (1 degree of freedom), and
-
3
under the null hypothesis that (, 2 degrees of freedom).
SNP data were aligned using METAL64 for all three analyses. The age-independent effect of SNPs (model 1) was computed in METAL. For the age-dependent analyses (model 2 for linear age effects and model 3 for quadratic age effects) the aligned data were imported into R52 and fixed effects meta-regression was performed using the R-package metafor65 (version 2.0–0). Results were filtered on SNPs that were present for at least 50% of the cohorts and in at least 50% of the subjects.
Functional mapping
Functional mapping was performed using the FUMA platform designed for prioritization, annotation and interpretation of GWAS results66. As the first step, independent significant SNPs in the individual GWAS meta-analysis summary statistics were identified based on their p-value (p < 5 × 10−8) and independence of each other (r2 < 0.6 in the 1000G phase 3 reference) within a 1Mb window. Thereafter, lead SNPs were identified from independent significant SNPs, which are independent of each other (r2 < 0.1). We used FUMA to annotate lead SNPs in genomic risk loci based on the following functional consequences on genes: eQTL data (GTEx v6 and v767), blood eQTL browser68, BIOS QTL browser69, BRAINEAC70, MuTHER71, xQTLServer72, the CommonMind Consortium73 and 3D chromatin interactions from HI-C experiments of 21 tissues/cell types74. Next for eQTL mapping and chromatin interaction mapping, genes were mapped using positional mapping, which is based on a maximum distance between SNPs (default 10kb) and genes. Chromatin interaction mapping was performed with significant chromatin interactions (defined as FDR < 1 × 10−6). The two ends of significant chromatin interactions were defined as follows: region 1 – a region overlapping with one of the candidate SNPs, and region 2 – another end of the significant interaction, used to map to genes based on overlap with a promoter region (250bp upstream and 50bp downstream of the transcription start site).
Visualization of SNP effects
We visualized the effects of our top SNPs on the lifespan trajectory, assuming no effects of the other SNPs, for easier interpretation of the direction of effect. Similar to the estimation of the lifespan trajectory, we estimated a smoothed version of the phenotypic change rate using LOWESS (see above) and integrated the rate of change. We added the unknown volume C at the start of our age range by fitting the integrated curve to the baseline data. Suppose is the unknown rate of change for non-carriers. The additional change rate for carriers was estimated through the meta-analysis or meta-regression. The full dataset contained a fraction p of the carriers of the tested allele. Assuming . We created a rate of change curve for non-carriers as and a rate of change curve of carriers as . The offset C is potentially different in carriers and non-carriers, so we estimated this difference by taking the effect of the cross-sectional GWAS data (see below) in this SNP, or a proxy SNP in high linkage disequilibrium (LD).
Gene-based and gene-set analyses
Gene-based associations with 15 phenotypes were estimated using MAGMA75 (version 1.09a) using the summary statistics from age-independent and age-dependent GWAS meta-analyses of rate of change of global brain measures. Gene names and locations were based on NCBI 37.3 locations as provided by MAGMA. Association was tested using the SNP-wise mean model, in which the sum of -log(SNP p-value) for SNPs located within the transcribed region was used as the test statistic. LD correction was based on estimates from the 1000 Genomes Project Phase 3 European ancestry samples58. To describe the direction of the age effect for significant genes in the age-dependent analyses, we subsequently identified the SNPs that were used in the gene-based p-value and plotted the age-dependent effect of the top SNP that contributed to the gene-based p-value.
The generated gene-based p-values were used to analyse sets of genes in order to test for association of genes belonging to specific biological pathways or processes. MAGMA applies a competitive test to analyse if the genes of a gene set are more strongly associated with the trait than other genes, while correcting for a series of confounding effects such as gene length and size of the gene set. For gene sets we used 9,975 sets with 10 –1,000 genes from the Gene Ontology sets76 curated from MsigDB 7.077.
Multiple testing corrections
We investigated annual rates of change for 15 brain phenotypes, but these are correlated to some extent (Extended Data Fig. 2). We therefore estimated the effective number of independent variables based on matrix spectral decomposition78 for the largest adolescent cohort (IMAGEN; N=1,068) and for the largest elderly cohort from the phase 1 sample (ADNI2; N=626). The most conservative estimate of the number of independent traits was 13.93. Despite the fact that models 2 and 3 are nested and therefore not independent, we also corrected for performing three analyses per trait. The study-wide significant threshold for the genome was therefore set at p < 1.2e-09 (5e-08/13.93*3). For gene-based significance, we applied a genome-wide significance level of 0.05/17541= 2.85e-06, and a study wide significance of 2.85e-06/(13.93*3), i.e. p < 6.82e-08. For gene-set significance, we applied a genome-wide significance level of 0.05/9,975 = 5.01e-06 and a study-wide significance level of 5.01e-06/(13.93*3), i.e. p < 1.20e-07.
SNP heritability
SNP heritabilities, h2SNP, were estimated by using linkage disequilibrium (LD) score regression79 (LDSR) for the European-ancestry brain change GWASs to ensure matching of population LD structure. For LDSR, we used precomputed LD scores based on the European-ancestry samples of the 1000 Genomes Project58 restricted to HapMap3 SNPs61. The summary statistics with standard LDSC filtering were regressed onto these scores. SNP heritabilities were estimated based on the slope of the LD score regression, with heritabilities on the observed scale calculated. To ensure sufficient power for the genetic correlations, rg was calculated if the Z-score of the h2SNP for the corresponding GWAS was 4 or higher79.
Comparison with cross-sectional results
For the genome-wide significant genes and genes associated with genome-wide significant SNPs, we compared our findings with cross-sectional GWAS summary statistics when available. To this end, datasets26,40–42 were requested and downloaded from http://enigma.ini.usc.edu/research/download-enigma-gwas-results/ and http://big.stats.ox.ac.uk/download_page. Gene-based association analyses for cross-sectional brain GWAS summary statistics were performed using MAGMA (as described above). Additionally, we compared the overlap in the first 1,000 ranked genes to the expected number of overlapping genes based on chance. False discovery rate correction80 was applied to determine over- or under-representation of genes from our longitudinal GWAS to the cross-sectional previously published GWAS26,40–42.
Overlap with cross-sectional results and other traits
To investigate genetic overlap with other traits across the genome we applied an adapted version of iSECA81 (independent SNP effect concordance analysis) which examines pleiotropy and concordance of the direction of effects between two phenotypes by comparing expected and observed overlap in sets of SNPs from both phenotypes that are thresholded at different levels. From the results at each threshold, heatmap plots were generated containing binomial tests for pleiotropy and Fisher’s exact tests for concordance. An empirical p-value for overall pleiotropy and concordance was then generated through permutation testing. Our implementation of iSECA also included a p-value for overall discordance, as we expect some phenotypes to negatively influence brain-structural change rates. P-values were computed using a two-step approach: we first ran 1,000 permutations. If the p-value for pleiotropy was below 0.05/15 we reran the analyses with 10,000 permutations to obtain a more precise p-value. Summary statistics of change rates were first filtered on SNPs for which > 95% of the subjects contributed data to remove the sample size dependency of p-values and subsequently clumped (p=1,kb=1000) to ensure independence of input SNPs.
We investigated the genetic overlap between brain-structural changes and risk for 20 neuropsychiatric, neurological and somatic disorders, and physical and psychological traits. Summary statistics were downloaded or requested for aggression82, alcohol dependence83, Alzheimer’s disease84, attention-deficit/hyperactivity disorder85, autism86, bipolar disorder87, body mass index30, brain age gap12, cognitive functioning29, depression27, diabetes type 288, ever-smoking32, focal epilepsy89, height30, inflammatory bowel disease90, insomnia31, multiple sclerosis91, Parkinson’s disease92, rheumatoid arthritis93 and schizophrenia28. These phenotypes were chosen because of known associations with brain structure or function, and availability of summary statistics based on large GWA-studies. For comparison, we computed the genetic overlap between cross-sectional brain structure and these phenotypes, using the same method.
Apart from these, we also 1) included intracranial volume94 to investigate the effect of overall head size and 2) tested the overlap between each structure’s longitudinal change measure against its cross-sectional brain structure. Pleiotropy, concordance or discordance was considered significant when the p-value was smaller than 0.05/15*22 (#change rates * #phenotypes tested) = 1.5e-04.
Brain gene expression
GENE2FUNC, a core process of FUMA66 (Functional Mapping and Annotation of Genome-wide Association Studies), was employed to analyse gene expression patterns. For this, a set of 8 genes was used as input, including all genome-wide significant genes and genes harbouring genome-wide significant SNPs (compare Supplementary Tables 5,7,9,11). Gene expression heatmap was constructed employing GTEx v833; 54 tissue types) and BrainSpan RNA-seq data across 29 different ages or 11 different developmental stages32. The average of normalized expression per label (zero means across samples) was displayed on the corresponding heatmaps. Expression values are TPM (Transcripts Per Million) for GTEx v8 and RPKM (Read per Kilobase Million) in the case of the BrainSpan data set.
Phenome-wide association studies
To identify phenotypes associated with the candidate SNPs and genes (defined as genome-wide significant SNPs and the genome-wide significant genes and genes associated with genome-wide significant SNPs), a phenome-wide association study (pheWAS) was done for each SNP and/or gene. PheWAS was performed using public data provided by GWASAtlas32(https://atlas.ctglab.nl). To correct for multiple testing, the total number of GWASs (4,756) was considered (including GWASs in which the searched SNP or gene was not tested) and the number of tested SNPs and genes (n=14), resulting in a Bonferroni corrected p-value threshold of 1.05e-05/14, i.e., p < 7.51e-07.
Sensitivity analyses
The phase 2 analyses include available data from all cohorts with European ancestry (N=15,100). The four cohorts of non-European and mixed ancestry together consist of 540 subjects, who are predominantly children and adolescents (Supplementary Table 3). The number of subjects, heterogeneity in ancestry and the age-distribution do not allow for separate meta-analysis or meta-regression. We therefore added the cohorts of non-European ancestry to the original datasets and reran analyses (N=15,640). In a second analysis, we excluded the 9 cohorts that had N < 75 or mean scanning interval < 0.5 years (Supplementary Table 2), leaving N=14,601 subjects. The main analyses include data from all subjects combined, without correction for disease. This approach was chosen because many neurological and neuropsychiatric diseases are characterized by aberrant brain changes over time, and genes involved in the disease may also be involved in these brain changes. To check whether our results were confounded by disease, we repeated the main analyses excluding diagnostic groups of each cohort (N=13,0349) and by correcting for disease status.
Data availability:
This work is a meta-analysis. Upon publication, the meta-analytic results will be made available from the ENIGMA consortium webpage (http://enigma.ini.usc.edu/research/download-enigma-gwas-results). Cohort level data can be shared upon request, after permission of cohort principal investigators. Individual level data can be shared with interested investigators, subject to local and national ethics regulations and legal requirements that respect the informed consent forms and national laws of the country of origin of the persons scanned. Figures that contain cohort level (meta) data: Figures 1, 2, Extended data Figures 1,2, Supplementary Figures 1,3,8,10.
Public data used in this work include the ABCD cohort (data release 3.0, accessible through https://nda.nih.gov/abcd; http://dx.doi.org/10.15154/1519007), ADNI cohort (accessible through adni.loni.usc.edu), and the UK biobank cohort (data request 11559, https://www.ukbiobank.ac.uk).
Extended Data
Extended Data Fig. 1. Demographics and analysis.
Overview of demographics (left). Per cohort, an age distribution is displayed, based on mean and standard deviation of the age at baseline. Cohorts of European ancestry are displayed in green, non-European cohorts are displayed in yellow. On the right, the total number of included subjects is displayed and a pie-chart of the distribution of diagnostic groups (pink) and subjects not belonging to diagnostic groups - often healthy subjects (aqua). Overview of analysis pipeline (right).
Extended Data Fig. 2. Correlations between change rates.
Pearson correlations between rates of change and between baseline intracranial volume and rates of change in the largest adolescent cohort (top, N = 1068) and the largest cohort in older age (bottom, N = 624) in phase 1. The size of the correlations is displayed by color and size of the circles.
Supplementary Material
Supplementary Table 1. Cohort characteristics.
Supplementary Table 2. Description of imaging per study cohort.
Supplementary Table 3. Description of genetics per study cohort.
Supplementary Table 4. Summary of genome-wide significant SNPs and top-10 loci for main effect of genetic variants on brain morphology rates of change in phase 1 + results for same SNPs in replication cohorts and phase 2.
Supplementary Table 5. Summary of genome-wide significant SNPs and top-10 loci for main effect of genetic variants on brain morphology rates of change in phase 2.
Supplementary Table 6. Summary of genome-wide significant SNPs and top-10 loci for linear age effects of genetic variants on brain morphology rates of change in phase 1 + results for same SNPs in phase 2.
Supplementary Table 7. Summary of genome-wide significant SNPs and top-10 loci for linear age effects of genetic variants on brain morphology rates of change in phase 2.
Supplementary Table 8. Summary of genome-wide significant SNPs and top-10 loci for quadratic age effects of genetic variants on brain morphology rates of change in phase 1 + results for same SNPs in phase 2.
Supplementary Table 9. Summary of genome-wide significant SNPs and top-10 loci for quadratic age effects of genetic variants on brain morphology rates of change in phase 2.
Supplementary Table 10. Summary of genome-wide significant genes and top-10 genes for brain morphology rates of change in phase 1 + results for same genes in replication cohorts and phase 2.
Supplementary Table 11. Summary of genome-wide significant genes, top-10 genes for brain morphology rates of change in phase 2 sample, and look-up results for top 10 genes in cross-sectional data.
Supplementary Table 12. Biological functions for top SNPs and genes.
Supplementary Table 13. Summary of genome-wide significant effects and top-10 gene-sets for brain morphology rates of change in phase 1 + results for same gene sets in phase 2.
Supplementary Table 14. Summary of genome-wide significant effects and top-10 gene-sets for brain morphology rates of change in phase 2.
Supplementary Table 15. SNP-based heritabilities as estimated using LDSC.
Supplementary Table 16. P-values and intervals for genetic overlap with cross-sectional volumes, ICV and other traits, underlying Figure 4.
Supplementary Table 17. Phenome-wide association results for genome-wide significant loci and genes.
Supplementary Table 18. Loci for age-(in)dependent effect on longitudinal brain changes in subgroups.
Supplementary Table 19. Genes for age-(in)dependent effect on longitudinal brain changes in subgroups.
Acknowledgements
Data used in preparing this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, many investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators may be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf. A full list of consortium authors can be found in the supplementary information.
BF has received speaking fees from MEDICE Arzneimittel Pütter GmbH & Co. BWJHP has received research funding from Jansen Research and Boehringer Ingelheim. CA has been a consultant to or has received honoraria or grants from Acadia, Angelini, Gedeon Richter, Janssen Cilag, Lundbeck, Minerva, Otsuka, Roche, Sage, Servier, Shire, Schering Plough, Sumitomo Dainippon Pharma, Sunovion and Takeda. CDW is an employee of Biogen Inc. DJS has received research grants and/or consultancy honoraria from Lundbeck and Sun. GJB receives honoraria for teaching from GE Healthcare. HB is on the Advisory Board Nutricia Australia. HEH has received travel fees for membership of the Steering Committee of the Lundbeck Foundation Center for Clinical Intervention and Neuropsychiatric Schizophrenia Research and for two presentations from Philips. These concerned activities unrelated to the submitted work. HJG has received travel grants and speaker’s honoraria from Fresenius Medical Care, Neuraxpharm, Servier and Janssen Cilag as well as research funding from Fresenius Medical Care. LP has served as an advisor or consultant to Shire, Takeda and Roche. She has received speaking fees from Shire and Infectopharm. The present work is unrelated to these relationships. MHJ received grant support from the Brain and behavior Foundation (NARSAD) Independent Investigator grant number 20244. MMN has received fees for memberships in Scientific Advisory Boards from the Lundbeck Foundation and the Robert-Bosch-Stiftung, and for membership in the Medical-Scientific Editorial Office of the Deutsches Ärzteblatt. MMN was reimbursed travel expenses for a conference participation by Shire Deutschland GmbH. MMN receives salary payments from Life & Brain GmbH and holds shares in Life & Brain GmbH. All these concerned activities outside the submitted work. NJ and PMT are MPIs of a research grant from Biogen, Inc (Boston, USA) for work unrelated to the contents of this manuscript. OAA has received Speaker’s honorarium from Lundbeck, Consultant for HealthLytix. PSS reports on-off payment for an advisory board meeting of Biogen. TB served in an advisory or consultancy role for Lundbeck, Medice, Neurim Pharmaceuticals, Oberberg GmbH, Shire, and Infectopharm. He received conference support or speaker’s fee by Lilly, Medice, and Shire. He received royalties from Hogrefe, Kohlhammer, CIP Medien, Oxford University Press; the present work is unrelated to these relationships. TEl has received speaker’s fee from Lundbeck AS. TRM has received honoraria for speaking and chairing engagements from Lundbeck, Janssen and Astellas.
Funding:
The ENIGMA-Plasticity working group is part of the ENIGMA World Aging Center, funded by NIA grants R56 AG058854- and R01 AG058854. The ENIGMA Consortium core funding was supported by NIH Consortium grant U54 EB020403, supported by a cross-NIH alliance that funds Big Data to Knowledge Centers of Excellence.
1000BRAINS:
1000BRAINS is a population-based cohort based on the Heinz-Nixdorf Recall Study and is supported in part by the German National Cohort. We thank the Heinz Nixdorf Foundation (Germany) for their generous support in terms of the Heinz Nixdorf Study. The authors are supported by the Initiative and Networking Fund of the Helmholtz Association (Svenja Caspers) and the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreements 785907 (Human Brain Project SGA2; Svenja Caspers, Sven Cichon, and Katrin Amunts). This work was further supported by the German Federal Ministry of Education and Research (BMBF) through the Integrated Network IntegraMent (Integrated Understanding of Causes and Mechanisms in Mental Disorders) under the auspices of the e:Med Program (grant 01ZX1314A; Sven Cichon), and by the Swiss National Science Foundation (SNSF, grant 156791; Sven Cichon).
ABCD:
Data used in the preparation of this article were obtained from the Adolescent Brain Cognitive DevelopmentSM (ABCD) Study (https://abcdstudy.org), held in the NIMH Data Archive (NDA). This is a multisite, longitudinal study designed to recruit more than 10,000 children age 9–10 and follow them over 10 years into early adulthood. The ABCD Study® is supported by the National Institutes of Health and additional federal partners under award numbers U01DA041048, U01DA050989, U01DA051016, U01DA041022, U01DA051018, U01DA051037, U01DA050987, U01DA041174, U01DA041106, U01DA041117, U01DA041028, U01DA041134, U01DA050988, U01DA051039, U01DA041156, U01DA041025, U01DA041120, U01DA051038, U01DA041148, U01DA041093, U01DA041089, U24DA041123, U24DA041147. A full list of supporters is available at https://abcdstudy.org/federal-partners.html. A listing of participating sites and a complete listing of the study investigators can be found at https://abcdstudy.org/consortium_members/. ABCD consortium investigators designed and implemented the study and/or provided data but did not necessarily participate in the analysis or writing of this report. This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH or ABCD consortium investigators. The ABCD data repository grows and changes over time. The ABCD data used in this report came from Data Release 3.0 (http://dx.doi.org/10.15154/1519007).
ADNI:
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.;Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
ALS Utrecht:
The authors acknowledge grants supporting their work from the European Union’s Horizon 2020 Research and Innovation Programme (H2020/2014-2020) under grant agreements 667302 (CoCA), 728018 (Eat2beNICE), 785907 (HBP SGA2), and 772376 (EScORIAL) and the Netherlands ALS Foundation.
BDC:
Brain Dynamics Centre (BDC), Sydney - cohort is funded by a National Health & Medical Research Council of Australia Project Grant (APP1008080).
BHRCS:
The Brazilian High Risk Cohort Study (BHRCS) was supported by the National Institute of Developmental Psychiatry for Children and Adolescent (INPD) Grant: Fapesp 2014/50917-0 CNPq 465550/2014-2.
BIG:
This study used the BIG database, which was established in Nijmegen in 2007. This resource is now part of Cognomics, a joint initiative by researchers of the Donders Centre for Cognitive Neuroimaging, the Human Genetics and Cognitive Neuroscience departments of the Radboud university medical center, and the Max Planck Institute for Psycholinguistics. The Cognomics Initiative is supported by the participating departments and centres and by external grants, including grants from the Biobanking and Biomolecular Resources Research Infrastructure (Netherlands) (BBMRI-NL) and the Hersenstichting Nederland. In particular, the authors would also like to acknowledge grants supporting their work from the Netherlands Organization for Scientific Research (NWO), i.e., the NWO Brain & Cognition Excellence Program (grant 433-09- 229) and the Vici Innovation Program (grant 016-130-669 to BF). Additional support is received from the European Community’s Seventh Framework Programme (FP7/2007 – 2013) under grant agreements nº 602805 (Aggressotype), nº 603016 (MATRICS), nº 602450 (IMAGEMEND), and nº 278948 (TACTICS), and from the European Community’s Horizon 2020 Programme (H2020/2014 – 2020) under grant agreements nº 643051 (MiND) and nº 667302 (CoCA).
BrainSCALE:
The BrainSCALE study is a collaborative project between Netherlands Twin Register (NTR) at the Vrije Universiteit (VU) Amsterdam and University Medical Center Utrecht (UMCU). The BrainSCALE study was funded by Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO 51.02.061 to H.H., NWO 51.02.062 to DB, NWO-NIHC Programs of excellence 433-09-220 to HEH, NWO-MagW 480-04-004 to DB, and NWO/SPI 56-464-14192 to DB); FP7 Ideas: European Research Council (ERC-230374 to DB), Universiteit Utrecht (High Potential Grant to HEH), Netherlands Twin Registry Repository (NWO-Groot 480-15-001/674 to DB) and Neuroscience Campus Amsterdam (NCA). Biomolecular Resources Research Infrastructure (BBMRI-NL, 184.021.007 and 184.033.111) Developmental trajectories of psychopathology (NIMH 1RC2 MH089995); and the Avera Institute for Human Genetics, Sioux Falls, South Dakota (USA).
Capetown:
The CTAAC study was supported by grant No R01-HD074051.
DBSOS:
The DBSOS study is partially funded by the Brain and behavior Foundation (NARSAD) by an Independent Investigator grant; No 20244. The generation R Study is made possible by financial support from the Erasmus Medical center, Rotterdam and the Netherlands organization for health research and development (ZonMW). The neuroimaging infrastructure is supported by ZonMW TOP (No: 912110210), The NWO Physical Sciences Division, and SURFsara supercomputing center (Cartesius Compute Cluster).
FOR2107:
This work is part of the German multicenter consortium “Neurobiology of Affective Disorders. A translational perspective on brain structure and function”, funded by the German Research Foundation (Deutsche Forschungsgemeinschaft DFG; Forschungsgruppe/Research Unit FOR2107). Grant agreements included the following: FOR2107 DA1151/5-1 and DA1151/5-2 to UD; SFB-TRR58, Projects C09 and Z02 to UD; the Interdisciplinary Center for Clinical Research (IZKF) of the medical faculty of Münster (grant Dan3/012/17 to UD); KR 3822/7-1 and KR 3822/7-2 to AK; KI 588/14-1, KI 588/14-2; NO 246/10-1 and NO 246/10-2 to MMN. AJ was in particular involved as PI in WP6, multi-method data analytics (JA 1890/7-1, JA 1890/7-2). FOR2107 study was also supported by the German Federal Ministry of Education and Research (BMBF), through ERA-NET NEURON, “SynSchiz - Linking synaptic dysfunction to disease mechanisms in schizophrenia - a multilevel investigation” (01EW1810 to MR) and the German Research Foundation (DFG grant FOR2107; RI908/11-2 to MR).
Generation R:
Netherlands Organization for Health Research and Development (ZonMw) TOP project number 91211021. Sophia Children’s Hospital Foundation (Stichting Vrienden van het Sophia) project number S18-68. The Generation R sample further reports the following support: Super computing resources for imaging processing were supported by the NWO Physical Sciences Division (Exacte Wetenschappen) and SURFsara (Cartesius compute cluster, https://www.surf.nl); neuroimaging data analysis was supported in part by Sophia Foundation Project S18-20 and Erasmus University Fellowship awarded to RLM.
HGUGM:
This work was supported by: Spanish Ministry of Science and Innovation, Instituto de Salud Carlos III (SAM16PE07CP1, PI16/02012, PI19/024), co-financed by ERDF Funds from the European Commission, “A way of making Europe”, CIBERSAM; Madrid Regional Government (B2017/BMD-3740 AGES-CM-2), European Union Structural Funds; European Union Seventh Framework Program under grant agreements, FP7-HEALTH-2013-2.2.1-2-603196 (Project PSYSCAN) and European Union H2020 Program under the Innovative Medicines Initiative 2 Joint Undertaking (grant agreement No 115916, Project PRISM, and grant agreement No 777394, Project AIMS-2-TRIALS), Fundación Familia Alonso, Fundación Alicia Koplowitz and Fundación Mutua Madrileña.
HUBIN:
The HUBIN study was funded by: Swedish Research Council (2003-5485, 2006-2992, 2006-986, 2008-2167, K2012-61X-15078-09-3, 521-2011-4622, 521-2014-3487, 2017-00949); regional agreement on medical training and clinical research between Stockholm County Council and the Karolinska Institutet; Knut and Alice Wallenberg Foundation.
IMAGEN:
This work received support from the following sources: the European Union-funded FP6 Integrated Project IMAGEN (Reinforcement-related behaviour in normal brain function and psychopathology) (LSHM-CT-2007-037286), the Horizon 2020 funded ERC Advanced Grant ‘STRATIFY’ (Brain network based stratification of reinforcement-related disorders) (695313), ERANID (Understanding the Interplay between Cultural, Biological and Subjective Factors in Drug Use Pathways) (PR-ST-0416-10004), BRIDGET (JPND: BRain Imaging, cognition Dementia and next generation GEnomics) (MR/N027558/1), Human Brain Project (HBP SGA 2, 785907), the FP7 project MATRICS (603016), the Medical Research Council Grant ‘c-VEDA’ (Consortium on Vulnerability to Externalizing Disorders and Addictions) (MR/N000390/1), the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, the Bundesministeriumfür Bildung und Forschung (BMBF grants 01GS08152; 01EV0711; Forschungsnetz AERIAL 01EE1406A, 01EE1406B), the Deutsche Forschungsgemeinschaft (DFG grants SM 80/7-2, SFB 940, TRR 265, NE 1383/14-1), the Medical Research Foundation and Medical Research Council (grants MR/R00465X/1 and MR/S020306/1), the National Institutes of Health (NIH) funded ENIGMA (grants 5U54EB020403-05 and 1R56AG058854-01). Further support was provided by grants from: - the ANR (ANR-12-SAMA-0004, AAPG2019 - GeBra), the Eranet Neuron (AF12-NEUR0008-01 - WM2NA; and ANR-18-NEUR00002-01 - ADORe), the Fondation de France (00081242), the Fondation pour la Recherche Médicale (DPA20140629802), the Mission Interministérielle de Lutte-contre-les-Drogues-et-les-Conduites-Addictives (MILDECA), the Assistance-Publique-Hôpitaux-de-Paris and INSERM (interface grant), Paris Sud University IDEX 2012, the Fondation de l’Avenir (grant AP-RM-17-013 ), the Fédération pour la Recherche sur le Cerveau; the National Institutes of Health, Science Foundation Ireland (16/ERCD/3797), U.S.A. (Axon, Testosterone and Mental Health during Adolescence; RO1 MH085772-01A1), and by NIH Consortium grant U54 EB020403, supported by a cross-NIH alliance that funds Big Data to Knowledge Centres of Excellence.
LBC1936:
We thank the Lothian Birth Cohort 1936 members who took part in this study, and Lothian Birth Cohort 1936 research team members and radiographers who collected, entered and checked data used in this paper. Magnetic Resonance Image acquisition and analyses were conducted at the Brain Research Imaging Centre, Neuroimaging Sciences, University of Edinburgh (www.bric.ed.ac.uk) which is part of SINAPSE (Scottish Imaging Network—A Platform for Scientific Excellence) collaboration (www.sinapse.ac.uk) funded by the Scottish Funding Council and the Chief Scientist Office. The LBC1936 and this research are supported by Age UK (Disconnected Mind project), the UK Medical Research Council [MRC; G0701120, G1001245, MR/M013111/1, MR/R024065/1], and the University of Edinburgh.
NCNG:
The NCNG sample collection was supported by grants from the Bergen Research Foundation and the University of Bergen, the Dr Einar Martens Fund, the K.G. Jebsen Foundation, the Research Council of Norway, to SLH, VMS and TE.
NESDA:
The infrastructure for the NESDA study (www.nesda.nl) is funded through the Geestkracht program of the Netherlands Organisation for Health Research and Development (ZonMw, grant No 10-000-1002) and financial contributions by participating universities and mental health care organizations (VU University Medical Center, GGZ inGeest, Leiden University Medical Center, Leiden University, GGZ Rivierduinen, University Medical Center Groningen, University of Groningen, Lentis, GGZ Friesland, GGZ Drenthe, Rob Giel Onderzoekscentrum).
NeuroIMAGE:
The NeuroIMAGE study was supported by NIH Grant R01MH62873 (to Stephen V. Faraone), NWO Large Investment Grant 1750102007010 (to Jan Buitelaar), ZonMW grant 60-60600-97-193, NWO grants 056-13-015 and 433-09-242, and matching grants from Radboud University Nijmegen Medical Center, University Medical Center Groningen and Accare, and Vrije Universiteit Amsterdam. The research leading to these results also received support from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement No 278948 (TACTICS), 602805 (Aggressotype), 603016 (MATRICS) and 602450 (Imagemend), and the Innovation Medicine Initiative grants 115300 (EU-AIMS) and 777394 (AIMS-2-TRIALS).
NUIG:
We would like to thank the radiologists at the University Hospital Galway and the participants who generously gave their time to make this study possible. The NUIG sample was supported and funded by the National University of Ireland Galway (NUIG) Millennium Fund and the Health Research Board (HRA_POR/2011/100).
OATS:
We gratefully acknowledge and thank the OATS participants, their supporters and the Research Team. The Older Australian Twin Study (OATS) is supported by the Australian NHMRC/Australian Research Council Strategic Award (Grant 401162) and the NHMRC Project grant 1405325. This study was facilitated through Twins Research Australia, a national resource in part supported by a Centre for Research Excellence from the NHMRC. DNA was extracted by Genetic Repositories Australia (NHMRC Grant 401184). Genome-wide genotyping at the Diamantina Institute, University of Queensland, was partly funded by a CSIRO Flagship Collaboration Fund Grant.
PAFIP:
PAFIP was supported by the Instituto de Salud Carlos III (PI14/00639, PI14/00918 and PI17/01056) and Fundación Instituto de Investigación Marqués de Valdecilla (NCT0235832 and NCT02534363). No pharmaceutical company has financially supported the study.
Rotterdam study:
The GWAS datasets are supported by the Netherlands Organization of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012), the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) Netherlands Consortium for Healthy Aging (NCHA), project no. 050-060-810. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters, MSc, and Carolina Medina-Gomez, MSc, for their help in creating the GWAS database, and Karol Estrada, PhD, Yurii Aulchenko, PhD, and Carolina Medina-Gomez, MSc, for the creation and analysis of imputed data. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists.
SHIP:
The SHIP study is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs and the Social Ministry of the Federal State of Mecklenburg-West Pomerania. MRI scans in SHIP and SHIP-TREND have been supported by a joint grant from Siemens Healthineers, Erlangen, Germany and the Federal State of Mecklenburg-West Pomerania.
Sydney MAS:
We gratefully acknowledge and thank the Sydney MAS participants, their supporters and the Research Team. The Sydney Memory and Ageing Study (MAS) is supported by a National Health & Medical Research Council of Australia Program Grant (Grants 350833, 568969, 109308) and a Capacity Building Grant (Grant 568940). DNA samples were extracted by Genetic Repositories Australia, an Enabling Facility, which is supported by a National Health & Medical Research Council of Australia Grant, 401184.
UK Biobank:
This research has been conducted using the UK Biobank Resource under Application Number ‘11559’.
UMCU:
The UMCU cohort contains a.o. UTWINS and GROUP. UTWINS was funded by the Netherlands Organization for Health Research and Development ZonMw (908.02.123 and 917.46.370 to H.H.), and by the European Union Marie-Curie Research Training Network (MRTN-CT-2006-035987). The GROUP study is partially funded through the Geestkracht programme of the Dutch Health Research Council (Zon-Mw, grant No 10-000-1001), and matching funds from participating pharmaceutical companies (Lundbeck, AstraZeneca, Eli Lilly, Janssen Cilag) and universities and mental health care organizations (Amsterdam: Academic Psychiatric Centre of the Academic Medical Center and the mental health institutions: GGZ Ingeest, Arkin, Dijk en Duin, GGZ Rivierduinen, Erasmus Medical Centre, GGZ Noord Holland Noord. Groningen: University Medical Center Groningen and the mental health institutions: Lentis, GGZ Friesland, GGZ Drenthe, Dimence, Mediant, GGNet Warnsveld, Yulius Dordrecht and Parnassia psycho-medical center The Hague. Maastricht: Maastricht University Medical Centre and the mental health institutions: GGzE, GGZ Breburg, GGZ Oost-Brabant, Vincent van Gogh voor Geestelijke Gezondheid, Mondriaan, Virenze riagg, Zuyderland GGZ, MET ggz, Universitair Centrum Sint-Jozef Kortenberg, CAPRI University of Antwerp, PC Ziekeren Sint-Truiden, PZ Sancta Maria Sint-Truiden, GGZ Overpelt, OPZ Rekem. Utrecht: University Medical Center Utrecht and the mental health institutions Altrecht, GGZ Centraal and Delta.).
UNSW:
The UNSW study was supported by the Australian National Medical and Health Research Council (NHMRC) Program Grant 1037196, Project Grant 1066177, and the Lansdowne Foundation. We gratefully acknowledge the Janette Mary O’Neil Research Fellowship to JMF.
Personal funding:
ALWB received funding from the National Children’s Foundation Tallaght, Ireland. RMB was supported by NIA R56AG058854 and R01AG058854 to the ENIGMA World Aging Center. CD-C was supported by Instituto de Salud Carlos III, Juan Rodés Grant (JR19/00024). CEF was supported by R01 AG050595; R01 AG022381; P01 AG055367; R01R56 AG037985. DAl was supported by South-Eastern Norway Regional Health Authority (2019107). DJS is supported by the SAMRC. DvdM was supported by Research Council of Norway grant No 276082. EGJ was supported by Swedish Research Council (2003-5485, 2006-2992, 2006-986, 2008-2167, K2012-61X-15078-09-3, 521-2011-4622, 521-2014-3487, 2017-00949); regional agreement on medical training and clinical research between Stockholm County Council and the Karolinska Institutet; Knut and Alice Wallenberg Foundation; HUBIN project. ESP is supported by Hypatia Tenure Track Grant (Radboudumc); NARSAD Young Investigator Grant (Brain and Behavior Research Foundation ID:25034); Christine Mohrmann Fellowship. EV was supported by National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. FN was supported by German Research Foundation NE 1383/14-1. HB was supported by NHMRC Australia. GAS was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil; grant No 573974/2008-0), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brazil), the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, Brazil; grant No 2008/57896-8) and the Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS, Brazil). HEH was supported by NIA R56AG058854 and R01AG058854 to the ENIGMA World Aging Center. HJG has received research funding from the EU “Joint Programme Neurodegenerative Disorders” (JPND). HHHA was supported by the Netherlands Organization for Health Research and Development (ZonMW, grant No 916.19.151). IAB was supported by University of Sydney Post-graduate Award. IN was supported by DFG Ne2254/1-2. JBJK was supported by NHMRC Dementia Research Team Grant APP1095127. JH was supported by R21MH107327-01. JLS was supported by grant Nos R01MH118349, R01MH120125. JW was supported by the UK Dementia Research Institute which receives its funding from DRI Ltd, funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK (JW), the Row Fogo Charitable Trust through the Row Fogo Centre for Research into Ageing and the Brain (Ref No: AD.ROW4.35. BRO-D.FID3668413) and the Fondation Leducq Transatlantic Network of Excellence for the Study of Perivascular Spaces in Small Vessel Disease, ref no. 16 CVD 05. KLG was supported by grant No APP1173025. KS was supported by research grants from the National Healthcare Group, Singapore (SIG/05004; SIG/05028; SIG /1103), and the Singapore Bioimaging Consortium (RP C009/2006). LD was supported by R01AG059874 and R01MH117601. JHF was supported by SFB 940/2 and the German Ministry of Education and Research (BMBF Grants 01EV0711 & 01EE1406B). LHvdB was supported by the Netherlands ALS Foundation. OAA was supported by Research Council of Norway (223273), KG Jebsen Stiftelsen, H2020 CoMorMent (847776). LMOL was supported by K99MH116115. LP received funding from the German Research Foundation (DFG), the Ministry of Science and Education (BMBF) and EU. LTW is funded by the European Research Council under the European Union’s Horizon 2020 research and innovation program (ERC Starting Grant 802998), the Research Council of Norway (249795), the South-East Norway Regional Health Authority (2019101), and the Department of Psychology, University of Oslo. MK was supported by funding from the Dutch National Science Agenda NeurolabNL project (grant No 400-17-602). MLPM was supported by the French funding agency ANR (ANR-12-SAMA-0004), the Assistance-Publique-Hôpitaux-de-Paris and INSERM (interface grant), Paris-Descartes-University (collaborative-project-2010), Paris-Sud-University (IDEX-2012). MLS was supported by FAPESP: 2016/13737-0 and 2016/04983-7. MMN was supported by the German Research Foundation (DFG grant FOR2107; NO246/10-2). MNS was supported by the Deutsche Forschungsgemeinschaft (DFG grants TRR 265; SFB 940; SM 80/7-2) and the German Ministry of Education and Research (BMBF grants 01EV0711; 01EE1406B). MR was supported by DFG FOR2107 RI 908/11-1 & RI 908/11-2, BMBF Neuron Eranet Synschiz 01EW1810. MSK was supported by the National Health and Medical Research Council, Australia Project Grant (GNT1008080) and Career Development Fellowship (GNT1090148). MSP was supported by NIA R01AG02238.NJ and LD were supported by R01AG059874 and R01MH117601. PMT and SIT were supported by NIH U54 EB020403, R56AG058854 to the ENIGMA World Aging Center, R01MH116147 and P41EB015922. PRS was supported by National Health and Medical Research Council, Australia grant Nos 1037196, 1063960, 1176716. RA-A is funded by a Miguel Servet contract from the Carlos III Health Institute (CP18/00003), carried out on Fundación Instituto de Investigación Marqués de Valdecilla. PGF received funding from the German Research Foundation, the European Union and the Federeal Ministry of Science. RAB was supported by the European Research Council. SIB was supported by FAPESP 2016/04983-7; FAPESP 2011/50740-5; INCT (CNPq/FAPESP) 2014/50917-0. SEF was supported by the Max Planck Society. SEM was supported by NHMRC APP1103623; APP1172917; APP1158127. SHW was supported by DFG FOR2107 Wi3439/3-2, BMBF Neuron ERANET Synschiz 01EW1810. SLH was supported by the University of Bergen, Trond Mohn Research Foundation, Helse Vest. SRC was supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (Grant Number 221890/Z/20/Z). TEl was funded by the Research Council of Norway, the South-Eastern Norway Regional Health Authority, Oslo University Hospital and a research grant from Mrs. Throne-Holst. TH was supported by grants from the Interdisciplinary Center for Clinical Research (IZKF) of the medical faculty of Münster (grant MzH 3/020/20) and the German Research Foundation (DFG grants HA7070/2-2, HA7070/3, HA7070/4). TJ was supported by National Natural Science Foundation of China (81801773, 81930095, 91630314), the Shanghai Pujiang Project (18PJ1400900), the Key Project of Shanghai Science and Technology Innovation Plan (16JC1420402), the Shanghai Municipal Science and Technology Major Project (No.2018SHZDZX01) and ZHANGJIANG LAB. TRM was supported by Medical Research Council (UK). TW was supported by Netherlands Organization for Health Research and Development (ZonMw) TOP project No 91211021; Sophia Children’s Hospital Foundation (Stichting Vrienden van het Sophia) project No S18-68.VM was supported by CONICYT fellowships 21180871. UFM was supported by the Throne-Holst foundation. VMS was supported by Research Council of Norway (grant No 223273 NORMENT). WSK was supported by NIA grants R01 AG050595, R01 AG022381, R01AG060470, R01 AG054002, and NIAAA grant R01 AA026881.
IMAGEN consortium authors:
Tobias Banaschewski1, Gareth J. Barker2, Arun L.W. Bokde3, Sylvane Desrivières4, Herta Flor5,6, Antoine Grigis7, Hugh Garavan8, Penny Gowland9, Andreas Heinz10, Rüdiger Brühl11, Jean-Luc Martinot12, Marie-Laure Paillère Martinot13, Eric Artiges14, Frauke Nees1,5,23, Dimitri Papadopoulos Orfanos7, Herve Lemaitre7,15, Tomáš Paus16, 17, Luise Poustka18, Sarah Hohmann1, Sabina Millenet1, Juliane H. Fröhner19, Michael N. Smolka19, Henrik Walter10,Robert Whelan20, Gunter Schumann21, 22
1 Department of Child and Adolescent Psychiatry and Psychotherapy, Central Institute of Mental Health, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany.
2 Department of Neuroimaging, Institute of Psychiatry, Psychology & Neuroscience, King’s College London, United Kingdom.
3 Discipline of Psychiatry, School of Medicine and Trinity College Institute of Neuroscience, Trinity College Dublin, Dublin, Ireland.
4 Centre for Population Neuroscience and Precision Medicine (PONS), Institute of Psychiatry, Psychology & Neuroscience, SGDP Centre, King’s College London, United Kingdom.
5 Institute of Cognitive and Clinical Neuroscience, Central Institute of Mental Health, Medical Faculty Mannheim, Heidelberg University, Square J5, Mannheim, Germany.
6 Department of Psychology, School of Social Sciences, University of Mannheim, Mannheim, Germany.
7 NeuroSpin, CEA, Université Paris-Saclay, F-91191 Gif-sur-Yvette, France.
8 Departments of Psychiatry and Psychology, University of Vermont, Burlington, VT, USA.
9 Sir Peter Mansfield Imaging Centre School of Physics and Astronomy, University of Nottingham, University Park, Nottingham, United Kingdom.
10 Department of Psychiatry and Psychotherapy CCM, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin, Humboldt-Universität zu Berlin, and Berlin Institute of Health, Berlin, Germany.
11 Physikalisch-Technische Bundesanstalt (PTB), Braunschweig and Berlin, Germany.
12 Institut National de la Santé et de la Recherche Médicale, INSERM U A10 “Trajectoires développementales en psychiatrie”, Université Paris-Saclay, Ecole Normale supérieure Paris-Saclay, CNRS, Centre Borelli, Gif-sur-Yvette, France.
13 Institut National de la Santé et de la Recherce Médicale, INSERM U A10 “Trajectoires développementales & psychiatrie”, University Paris-Saclay, Ecole Normale Supérieure Paris-Saclay, CNRS; Centre Borelli, Gif-sur-Yvette, France; and AP-HP, Sorbonne Université, Department of Child and Adolescent Psychiatry, Pitié-Salpêtrière Hospital, Paris, France.
14 Institut National de la Santé et de la Recherche Médicale, INSERM U A10 “Trajectoires développementales en psychiatrie”; Université Paris-Saclay, Ecole Normale supérieure Paris-Saclay, CNRS, Centre Borelli, Gif-sur-Yvette; and Psychiatry Department, EPS Barthélémy Durand, Etampes, France.
15 Institut des Maladies Neurodégénératives, UMR 5293, CNRS, CEA, Université de Bordeaux, Bordeaux, France.
16 Department of Psychiatry, Faculty of Medicine and Centre Hospitalier Universitaire Sainte-Justine, University of Montreal, Montreal, QB, Canada.
17 Departments of Psychiatry and Psychology, University of Toronto, Toronto, ON, Canada.
18 Department of Child and Adolescent Psychiatry and Psychotherapy, University Medical Centre Göttingen, Göttingen, Germany.
19 Department of Psychiatry and Neuroimaging Center, Technische Universität Dresden, Dresden, Germany.
20 School of Psychology and Global Brain Health Institute, Trinity College Dublin, Ireland.
21 PONS Centre, Charité Mental Health, Department of Psychiatry and Psychotherapy, Charité Universitätsmedizin Berlin, Berlin, Germany.
22 Centre for Population Neuroscience and Precision Medicine (PONS), Institute for Science and Technology of Brain-inspired Intelligence (ISTBI), Fudan University, Shanghai, China.
Footnotes
Code availability:
The code for processing of individual cohorts (including imaging and QC, imputation and GWAS protocol) can be found on http://enigma.ini.usc.edu/ongoing/enigma-plasticity-working-group/. Code for the meta-regression is available through Github https://github.com/RMBrouwer/GWAS_meta_regression.
Competing interests:
Other authors declare no conflict of interest.
References
- 1.Hedman AM, van Haren NE, Schnack HG, Kahn RS & Hulshoff Pol HE Human brain changes across the life span: a review of 56 longitudinal magnetic resonance imaging studies. Hum Brain Mapp 33, 1987–2002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giedd JN et al. Brain development during childhood and adolescence: a longitudinal MRI study. Nat Neurosci 2, 861–863 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Raz N. et al. Regional brain changes in aging healthy adults: general trends, individual differences and modifiers. Cereb Cortex 15, 1676–1689 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Ramsden S. et al. Verbal and non-verbal intelligence changes in the teenage brain. Nature 0, 6–10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnack HG et al. Changes in thickness and surface area of the human cortex and their relationship with intelligence. Cereb Cortex 25, 1608–1617 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Shaw P. et al. Development of Cortical Asymmetry in Typically Developing Children and its Disruptionin Attention-Deficit/Hyperactivity Disorder. Arch Gen Psychiatry 66, 888–896 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLisi LE, Sakuma M, Maurizio AM, Relja M. & Hoff AL Cerebral ventricular change over the first 10 years after the onset of schizophrenia. Psychiatry Res. - Neuroimaging (2004) doi: 10.1016/j.pscychresns.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Reiter K. et al. Five-Year Longitudinal Brain Volume Change in Healthy Elders at Genetic Risk for Alzheimer’s Disease. J. Alzheimers Dis 55, 1363–1377 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eshaghi A. et al. Deep gray matter volume loss drives disability worsening in multiple sclerosis. Ann Neurol 83, 210–222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brouwer RM et al. Heritability of brain volume change and its relation to intelligence. Neuroimage 100, 676–683 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Brans RGH et al. Heritability of changes in brain volume over time in twin pairs discordant for schizophrenia. Arch. Gen. Psychiatry 65, 1259–1268 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Kaufmann T. et al. Common brain disorders are associated with heritable patterns of apparent aging of the brain. Nat. Neurosci (2019) doi: 10.1038/s41593-019-0471-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson PM et al. ENIGMA and global neuroscience: A decade of large-scale studies of the brain in health and disease across more than 40 countries. Transl. Psychiatry 10, 1–28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brouwer RM et al. Genetic influences on individual differences in longitudinal changes in global and subcortical brain volumes: Results of the ENIGMA plasticity working group. Hum Brain Mapp 38, 4444–4458 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szekely E. et al. Genetic associations with childhood brain growth, defined in two longitudinal cohorts. Genet Epidemiol 42, 405–414 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang H. et al. Spatio-temporal transcriptome of the human brain. Nature 478 (7370), 483–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fletcher SC How (not) to measure replication. Eur. J. Philos. Sci 11, 57 (2021). [Google Scholar]
- 18.Nøhr AC et al. Identification of a novel scaffold for a small molecule GPR139 receptor agonist. Sci. Rep 9, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Süsens U, Hermans-Borgmeyer I, Urny J. & Schaller HC Characterisation and differential expression of two very closely related G-protein-coupled receptors, GPR139 and GPR142, in mouse tissue and during mouse development. Neuropharmacology (2006) doi: 10.1016/j.neuropharm.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Dao M, Stoveken HM, Cao Y. & Martemyanov KA The role of orphan receptor GPR139 in neuropsychiatric behavior. Neuropsychopharmacology (2021) doi: 10.1038/s41386-021-00962-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pagnamenta AT et al. Rare familial 16q21 microdeletions under a linkage peak implicate cadherin 8 (CDH8) in susceptibility to autism and learning disability. J Med Genet 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castiglioni V. et al. Dynamic and Cell-Speci fi c DACH1 Expression in Human Neocortical and Striatal Development. 2115–2124 (2019) doi: 10.1093/cercor/bhy092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolfe CM, Fitz NF, Nam KN, Lefterov I. & Koldamova R. The role of APOE and TREM2 in Alzheimer ′ s disease—Current understanding and perspectives. Int. J. Mol. Sci 20, 65–70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hauser PS, Narayanaswami V. & Ryan RO Apolipoprotein E: From lipid transport to neurobiology. Prog. Lipid Res 50, 62–74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinberg SF Structural Basis of Protein Kinase C Isoform Function. Physiol. Rev 88, 1341–1378 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hibar DP et al. Novel genetic loci associated with hippocampal volume. Nat Commun 8, 13624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard DM et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci 22, 343–352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Psychiatric Genomics Consortium. Biological Insights From 108 Schizophrenia-Associated Genetic Loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savage JE et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet 50, 912–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yengo L. et al. Meta-analysis of genome-wide association studies for height and body mass index in ~700 000 individuals of European ancestry. Hum. Mol. Genet 27, 3641–3649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jansen PR et al. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat. Genet 51, 394–403 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Watanabe K. et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet 51, (2019). [DOI] [PubMed] [Google Scholar]
- 33.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller JA et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Callender JA & Newton AC Conventional protein kinase C in the brain: 40 years later. Neuronal Signal. 1, NS20160005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bobb JF, Schwartz BS, Davatzikos C. & Caffo B. Cross-sectional and longitudinal association of body mass index and brain volume. Hum. Brain Mapp 35, 75–88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim RE et al. Lifestyle-dependent brain change: a longitudinal cohort MRI study. Neurobiol. Aging (2018) doi: 10.1016/j.neurobiolaging.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 38.Hulshoff Pol HE & Kahn RS What happens after the first episode? A review of progressive brain changes in chronically ill patients with schizophrenia. Schizophr Bull 34, 354–366 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fjell AM et al. The genetic organization of longitudinal subcortical volumetric change is stable throughout the lifespan. eLife 10, e66466 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elliott LT et al. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature 562, 210–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satizabal CL et al. Genetic architecture of subcortical brain structures in 38,851 individuals. Nat. Genet 51, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grasby KL et al. The genetic architecture of the human cerebral cortex. Science 367, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfefferbaum A. & Sullivan EV Cross-sectional versus longitudinal estimates of age-related changes in the adult brain: overlaps and discrepancies. Neurobiol Aging 36, 2563–2567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Z, Shen X, Pan W, & for the Alzheimer’s Disease Neuroimaging Initiative. Longitudinal Analysis Is More Powerful than Cross-Sectional Analysis in Detecting Genetic Association with Neuroimaging Phenotypes. PLoS ONE 9, e102312 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fjell AM et al. Development and aging of cortical thickness correspond to genetic organization patterns. Proc Natl Acad Sci U A (2015) doi: 10.1073/pnas.1508831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walhovd KB et al. Neurodevelopmental origins of lifespan changes in brain and cognition. Proc. Natl. Acad. Sci. U. S. A 113, 9357–9362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan EV Differential Rates of Regional Brain Change in Callosal and Ventricular Size: a 4-Year Longitudinal MRI Study of Elderly Men. Cereb. Cortex 12, 438–445 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Storsve AB et al. Differential Longitudinal Changes in Cortical Thickness, Surface Area and Volume across the Adult Life Span: Regions of Accelerating and Decelerating Change. J. Neurosci 34, 8488–8498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 49.Fischl B. et al. Whole Brain Segmentation: Automated Labeling of Neuroanatomical Structures in the Human Brain. Neuron 33, 341–355 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Fischl B. et al. Sequence-independent segmentation of magnetic resonance images. Neuroimage 23 Suppl 1, S69–84 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Reuter M, Schmansky NJ, Rosas HD & Fischl B. Within-Subject Template Estimation for Unbiased Longitudinal Image Analysis. Neuroimage 61, 1402–1418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iscan Z. et al. Test-retest reliability of freesurfer measurements within and between sites: Effects of visual approval process. Hum. Brain Mapp 36, 3472–3485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wonderlick JS et al. Reliability of MRI-derived cortical and subcortical morphometric measures: Effects of pulse sequence, voxel geometry, and parallel imaging. NeuroImage (2009) doi: 10.1016/j.neuroimage.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liem F. et al. Reliability and statistical power analysis of cortical and subcortical FreeSurfer metrics in a large sample of healthy elderly. Neuroimage 108, 95–109 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Voevodskaya O. et al. The effects of intracranial volume adjustment approaches on multiple regional MRI volumes in healthy aging and Alzheimer’s disease. Front. Aging Neurosci 6, 1–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cleveland WS LOWESS : A Program for Smoothing Scatterplots by Robust Locally Weighted Regression. Am. Stat 35, 10–11 (1981). [Google Scholar]
- 57.The R Core Team. R: A language and environment for statistical computing. (2018). [Google Scholar]
- 58.The 1000 Genomes Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Das S. et al. Next-generation genotype imputation service and methods. Nat Genet 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCarthy S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet 48, 1279–1283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.International HapMap Consortium. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng S, Liu D, Zhan X, Wing MK & Abecasis GR RAREMETAL: fast and powerful meta-analysis for rare variants. Bioinformatics 30, 2828–2829 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker WL, Michael White C, Cappelleri JC, Kluger J. & Coleman CI Understanding heterogeneity in meta-analysis: The role of meta-regression. Int. J. Clin. Pract 63, 1426–1434 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Willer CJ, Li Y. & Abecasis GR METAL: fast and efficient meta-analysis of genome-wide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Viechtbauer W. Journal of statistical software. J. Stat. Softw 36, 1–48 (2010). [Google Scholar]
- 66.Watanabe K, Taskesen E, Van Bochoven A. & Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun 8, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lonsdale J. et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Westra HJ et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet 45, 1238–1243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhernakova DV et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet 49, 139–145 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Ramasamy A. et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat. Neurosci 17, 1418–1428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grundberg E. et al. Mapping cis-and trans-regulatory effects across multiple tissues in twins. Nat. Genet 44, 1084–1089 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ng B. et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci 20, 1418–1426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fromer M. et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci 19, 1442–1453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmitt AD et al. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Rep. (2016) doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Leeuw CA, Mooij JM, Heskes T. & Posthuma D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput. Biol 11, 1–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.The Gene Ontology Consortium. Gene Ontology Consortium : going forward. Nucleic Acids Res. 43, 1049–1056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Subramanian A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nyholt DR A Simple Correction for Multiple Testing for Single-Nucleotide Polymorphisms in Linkage Disequilibrium with Each Other. Am J Hum Genet 74, 765–769 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Benjamini Y. & Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc 57, 289–300 (1995). [Google Scholar]
- 81.Nyholt DR SECA: SNP effect concordance analysis using genome-wide association summary results. Bioinformatics 30, 2086–2088 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Pappa I. et al. A genome-wide approach to children’s aggressive behavior: The EAGLE consortium. Am. J. Med. Genet. B Neuropsychiatr. Genet 171, 562–572 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Walters RK et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat. Neurosci 21, 1656–1669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lambert JC et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet 45, 1452–1458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Demontis D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet 51, 63–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Psychiatric Genomics Consortium. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism 8, 21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stahl E. & Bipolar Working Group of the Psychiatric Genomics Consortium. Genome-Wide Association Study Identifies Twenty New Loci Associated With Bipolar Disorder. Eur. Neuropsychopharmacol 29, S816 (2019). [Google Scholar]
- 88.Scott RA et al. An Expanded Genome-Wide Association Study of Type 2 Diabetes in Europeans. Diabetes 66, 2888–2902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.The International League Against Epilepsy Consortium on Complex Epilepsies. Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat. Commun 9, 5269 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu JZ et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet 47, 979–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sawcer S. et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nalls MA et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet 46, 989–993 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Okada Y. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Adams HH et al. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat Neurosci 19, 1569–1582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Cohort characteristics.
Supplementary Table 2. Description of imaging per study cohort.
Supplementary Table 3. Description of genetics per study cohort.
Supplementary Table 4. Summary of genome-wide significant SNPs and top-10 loci for main effect of genetic variants on brain morphology rates of change in phase 1 + results for same SNPs in replication cohorts and phase 2.
Supplementary Table 5. Summary of genome-wide significant SNPs and top-10 loci for main effect of genetic variants on brain morphology rates of change in phase 2.
Supplementary Table 6. Summary of genome-wide significant SNPs and top-10 loci for linear age effects of genetic variants on brain morphology rates of change in phase 1 + results for same SNPs in phase 2.
Supplementary Table 7. Summary of genome-wide significant SNPs and top-10 loci for linear age effects of genetic variants on brain morphology rates of change in phase 2.
Supplementary Table 8. Summary of genome-wide significant SNPs and top-10 loci for quadratic age effects of genetic variants on brain morphology rates of change in phase 1 + results for same SNPs in phase 2.
Supplementary Table 9. Summary of genome-wide significant SNPs and top-10 loci for quadratic age effects of genetic variants on brain morphology rates of change in phase 2.
Supplementary Table 10. Summary of genome-wide significant genes and top-10 genes for brain morphology rates of change in phase 1 + results for same genes in replication cohorts and phase 2.
Supplementary Table 11. Summary of genome-wide significant genes, top-10 genes for brain morphology rates of change in phase 2 sample, and look-up results for top 10 genes in cross-sectional data.
Supplementary Table 12. Biological functions for top SNPs and genes.
Supplementary Table 13. Summary of genome-wide significant effects and top-10 gene-sets for brain morphology rates of change in phase 1 + results for same gene sets in phase 2.
Supplementary Table 14. Summary of genome-wide significant effects and top-10 gene-sets for brain morphology rates of change in phase 2.
Supplementary Table 15. SNP-based heritabilities as estimated using LDSC.
Supplementary Table 16. P-values and intervals for genetic overlap with cross-sectional volumes, ICV and other traits, underlying Figure 4.
Supplementary Table 17. Phenome-wide association results for genome-wide significant loci and genes.
Supplementary Table 18. Loci for age-(in)dependent effect on longitudinal brain changes in subgroups.
Supplementary Table 19. Genes for age-(in)dependent effect on longitudinal brain changes in subgroups.
Data Availability Statement
This work is a meta-analysis. Upon publication, the meta-analytic results will be made available from the ENIGMA consortium webpage (http://enigma.ini.usc.edu/research/download-enigma-gwas-results). Cohort level data can be shared upon request, after permission of cohort principal investigators. Individual level data can be shared with interested investigators, subject to local and national ethics regulations and legal requirements that respect the informed consent forms and national laws of the country of origin of the persons scanned. Figures that contain cohort level (meta) data: Figures 1, 2, Extended data Figures 1,2, Supplementary Figures 1,3,8,10.
Public data used in this work include the ABCD cohort (data release 3.0, accessible through https://nda.nih.gov/abcd; http://dx.doi.org/10.15154/1519007), ADNI cohort (accessible through adni.loni.usc.edu), and the UK biobank cohort (data request 11559, https://www.ukbiobank.ac.uk).