Abstract
Spheroids and organoids are increasingly popular three-dimensional (3D) cell culture models. Spheroid models are more physiologically relevant to a tumor compared to two-dimensional (2D) cultures and organoids are a simplified version of an organ with similar composition. Spheroids are often only formed from a single cell type which does not represent the situation in vivo. However, despite this, both spheroids and organoids can be used in cell migration studies, disease modelling and drug discovery. A drawback of these models is, however, the lack of appropriate analytical tools for high throughput imaging and analysis over a time course. To address this, we have developed an R Shiny app called SpheroidAnalyseR: a simple, fast, effective open-source app that allows the analysis of spheroid or organoid size data generated in a 96-well format. SpheroidAnalyseR processes and analyzes datasets of image measurements that can be obtained via a bespoke software, described herein, that automates spheroid imaging and quantification using the Nikon A1R Confocal Laser Scanning Microscope. However, templates are provided to enable users to input spheroid image measurements obtained by user-preferred methods. SpheroidAnalyseR facilitates outlier identification and removal followed by graphical visualization of spheroid measurements across multiple predefined parameters such as time, cell-type and treatment(s). Spheroid imaging and analysis can, thus, be reduced from hours to minutes, removing the requirement for substantial manual data manipulation in a spreadsheet application. The combination of spheroid generation in 96-well ultra-low attachment microplates, imaging using our bespoke software, and analysis using SpheroidAnalyseR toolkit allows high throughput, longitudinal quantification of 3D spheroid growth whilst minimizing user input and significantly improving the efficiency and reproducibility of data analysis. Our bespoke imaging software is available from https://github.com/GliomaGenomics. SpheroidAnalyseR is available at https://spheroidanalyser.leeds.ac.uk, and the source code found at https://github.com/GliomaGenomics.
Keywords: spheroids, organoids, confocal, R Shiny
BACKGROUND
Spheroids and organoids as 3D cell culture models
The use of three-dimensional (3D) models such as spheroid and organoids has become an intermediate step between two-dimensional (2D) cell culture and in vivo models especially in cancer research. 2D cell culture models have been used since the 19th Century and still play an important role in scientific research due to their ease of use and readily available assays [1]. However, often the results from 2D models do not reflect cellular activity and responses as observed in vivo. Spheroid models are based on the generation of 3D structures of tumor cells and provide a more physiologically relevant environment for cancer cells in comparison to being grown as 2D monolayers; as such they lead to a more clinically relevant recapitulation of the biology or therapeutic response observed in vivo [2] and thus make a case for use as enhanced models in numerous drug discovery and toxicity studies [3]. As miniature versions of organs, organoids, typically grown from stem cells, are often used in disease modelling, drug discovery and cell migration studies [4,5]. Cells contained within a 3D structure have many more physiological relevant features than cells grown as monolayers. For example, in 3D cells display a more natural morphology rather than being elongated, enabling them to grow in aggregates as opposed to a monolayer and they are exposed to varying oxygen and nutrient gradients. This allows them to be in different stages of the cell cycle and have more biologically relevant cell-cell interactions [6,7]. The presence of an oxygen gradient means that tumor spheroids often have a hypoxic core which is a common feature in most solid tumor types such as glioblastoma (GBM) and breast cancer resembling the features of a tissue or tumor in a way that 2D cell cultures cannot. Furthermore, spheroids give a more accurate representation of drug response, and gene and protein expression profiles often show increased similarity to in vivo than profiles from 2D cultures [8]. A sharp increase in the number of publications on 3D cell cultures has been seen over recent years [9]. Pre-clinical in vivo animal models are often used in cancer research to test drug toxicity and new treatment regimens. The development of 3D models supports the reduction in the use of animal models as spheroids and organoids allow scientists to model tumors and organs for pre-clinical large scale treatment screens.
Drawbacks of spheroids and organoids models
Despite the numerous advantages, spheroids and organoids so far are not widely used in biomedical research. As spheroids are often formed from a single cell type and organoids from stem cells, they do not completely reflect the structural complexity of in vivo models. Further adding to this issue is a lack of suitably high-throughput assays and reliable analysis methods meaning that it is difficult to understand how 3D methods reflect the in vivo systems or what is seen in humans [8]. Many spheroid assays are endpoint only, meaning that changes occurring between initial set up and the assay end point are not recorded. Evaluation of spheroid measurement changes over time including parameters such as area, diameter, and circularity could be a valuable tool for many biological studies and help facilitate the acquisition of data to allow the gap between 3D and in vivo or human systems to be bridged. In many cases, obtaining these results involves manual imaging of each spheroid with subsequent manual application of imaging tools to obtain quantification metrics that are individually collated and then laboriously analyzed. This is time-intensive, resulting in fewer technical repeats per condition being studied, and introduces subjectivity and, thus, human error. To address these issues, we have created SpheroidAnalyseR: an R Shiny app that takes spheroid size measurements and associated metadata as input, identifies and optionally removes outliers using a Robust-Z-Score, can merge datasets from multiple plates (including across a time-course) and can plot graphs to enable users to analyze spheroid data quickly and efficiently.
SpheroidAnalyseR identifies outliers in the data, including natural outliers and those caused by erroneous dimensional parameters for example those resulting from incorrect detection of the spheroid by the imaging software, or wells with missing spheroids due to human error. To maintain the quantitative integrity of the measurements it is therefore necessary to identify and remove any outliers from the dataset, which may compromise the validity of the final analysis. A configurable Robust-Z-Score used to measure outlier strength has been incorporated into SpheroidAnalyseR’s outlier detection code using statistical summarization of the parameter data grouped by technical replicates [10].
R Shiny package
The R programming language is a highly regarded open-source platform for data manipulation, visualization and statistical analysis. Despite numerous training initiatives, there is a still a steep learning curve for those without prior programming experience to adopt the language in their daily work. Shiny allows R developers to make their code and analyses accessible to the wider community through a web interface.
PROCEDURE
The starting point for using SpheroidAnalyseR input of size metrics from spheroids/organoids grown in 96-well plates. This requires two prior steps: (1) Growing the spheroids in 96-well plates. This can be done via the user’s preferred method. Herein we used Ultra-low-attachment 96-well plates (Corning®, Corning, NY; Cat# 7007); and (2) Quantification of spheroid images.
There are multiple ways in which to acquire the input data for SpheroidAnalyseR. This includes manual imaging and subsequent measuring for example using an EVOS fluorescent and transmitted light microscope capable of producing high resolution images. ImageJ, a Java-based image processing program, can be used to manually draw around each spheroid [11]. Next, data can then be uploaded into the template spreadsheet provided, and analyzed by SpheroidAnalyseR. Alternatively, published methods such as SpheroidSizer provide a way to calculate the area and other key spheroid measurements from images [12]. In addition, we collaborated with Nikon to create a bespoke JOB (computationally scripted series of commands) for the Confocal Laser Scanning Microscope-Nikon A1R for use with spheroids grown in 96-well plates. This JOB performs spheroid imaging and subsequent measurement of volume, area, diameter, circularity and perimeter automatically using a threshold-based method, with results output to a Microsoft Excel file. A full 96-well plate is imaged, and result outputs are obtained within 2 minutes. The JOB can be accessed via the Glioma Genomic GitHub (https://github.com/GliomaGenomics) with full instructions on how to upload to and run on a Nikon A1R. The resulting Excel spreadsheet can be uploaded directly to SpheroidAnalyseR where users can preview the data, remove outliers, merge result files and plot graphs. The workflow for analyzing the data is as follows:
- 1. Data input SpheroidAnalyseR identifies and visualizes outliers in spheroid data giving the user the option to remove some or all of them. It does this by converting user-defined technical replicate data into Robust-Z-Scores and allowing the user to set a threshold score, with the default set as ±1.96, which equates to a 95% confidence interval. The webpage, after data input, includes graphical visualisation of spheroid measurements across multiple predefined parameters i.e., time, cell-type and treatment(s).
-
1.1. On the Data Input tab, three types of files are required to be uploaded to allow processing (see below). Input file templates that include the specific column headings needed can be downloaded from the Data Input tab.
- 1.1.1. Raw data file—Files can be uploaded either directly from the output file from the bespoke JOB on the confocal (Raw Data Template 1) or, data can be manually inputted into the second template file (Raw Data Template 2). Several parameters can be inputted included treatment date and time, cell line, passage number, drugs and concentrations used, and irradiation dose. Any columns in which no data is present should have “0” for each row. If the user is analyzing multiple plates of data with the same layout, then multiple raw data files can be uploaded simultaneously to allow for faster processing.
- 1.1.2. Plate layout file—This defines the layout of treatments on the 96-well plate for each spheroid or organoid. Each plate can contain spheroids or organoids with multiple different treatments. 1–12 represents columns 1–12 of the 96-well plate and A–H represents rows A–H of a 96-well plate. A number corresponding to the treatment index assigned in the treatment file should be in each cell that a spheroid measurement was for. Only one plate layout can be uploaded at any one time. If a raw data file has a different plate layout to the last file analyzed, then a new plate layout will need to be uploaded.
- 1.1.3. Treatment file—This file defines the treatment index numbers corresponding to the plate layout file. An index denotes a specific treatment, or combination thereof, and cell line as detailed in the corresponding row of the treatment template. If desired, users can input the time and date of treatment, the cell lines used, the passage number, the dose of radiation used and multiple drugs and subsequent concentrations. Multiple wells with the same index constitute technical replicates. Multiple different combinations of treatments can be defined as required.
NOTE: Templates for these files are available to download on the Data Input tab of the SpheroidAnalyseR web page. The column names of any input files must match those given in the supplied templates, and the sheet name must match the template’s sheet name. Four template sheets are provided: (i) Raw Data Template 1—This is the file created after running the bespoke JOB on the Confocal Nikon AR1. It can be uploaded straight to SpheroidAnalyseR; (ii) Raw Data Template 2—This template allows users to input their own spheroid measurements obtained. Users must have the Well.Name column completed and at least one of measurements from the other columns which are: area, perimeter, circularity, count (number of spheroids per well), diameter and volume. If any measurements are not inputted, then a “0” should be inputted in each cell. Optional—The user can input the time and date the spheroids were analyzed, if not required then “0” should be inputted in each cell; (iii) Plate layout template; and (iv) Treatment template.CAUTION: The number of spheroids to be analyzed in the raw data file must match the number of cells filled out in the plate layout or an error will be shown in red on the Outlier Removal tab and the user will not be allowed to process outliers. -
- 2. Previewing files in the Data Input tab
- 2.1. Raw data files can be previewed on the Data Input tab (Fig. 1). A drop-down list enables users to select each raw file if multiple files were uploaded simultaneously. Treatment and layout information based on the templates uploaded can also be reviewed here: the plate layout is shown, and a dropdown list allows the user to color the plate according to each parameter in the treatment file (i.e., cell line, drug used, or time treated).
NOTE: An image of the plate layout along with its designated treatment index is shown. It is recommended to review your uploaded data to ensure that that plate layout and treatment index are correct before proceeding. - 3. Outlier removal Once data has been inputted and reviewed, the user can move to the Outlier Removal tab and determine if any of the data points are spurious and should be removed as an outlier.
- 3.1. On the Outlier Removal tab, choose the raw file to be analyzed from the “Choose a raw file” drop-down list.
- 3.2. A spheroid measurement value (e.g., diameter, area, etc.) for which the outliers should be determined for can be selected from the “Choose a value to plot the outliers” drop-down list.
- 3.3. In the shaded beige/yellow box (Fig. 2) users can adjust the Robust-Z-Score (if required) and choose to apply pre-screen thresholds. These are automatically applied with default settings but can be adjusted if necessary. The pre-screen threshold removes spheroid measurements where the value falls outside the upper or lower limits set by the user. This ensures any wells in which there is an extreme outlier, for example due to an empty well will be removed prior to outlier analysis.
- 3.4. After clicking the “Remove outliers” button, outliers will be removed, and three images will be displayed. The top figure shows the plate layout after wells with spheroids that are determined to be outliers have been removed based on the pre-screen thresholds and Robust-Z-Score limits. Below that, Plot 1 and Plot 2 can be viewed with the selected measurement (e.g., diameter, area, etc.). These plots show the columns of the plate and the measurement each spheroid in these columns has (for example diameter). Plot 2 shows spheroids grouped by technical replicates, i.e., with the same treatment index as defined on the previously inputted treatment file.
HINT: It is advised to apply pre-screen threshold to remove any values that are obvious outliers before running the Robust-Z-Score. This could include measurements that have been taken of empty wells or when the imaging technique failed to recognize and measure a spheroid.- 3.5. Using the “Toggle Cell Status normal/outliers” drop-down list, users can manually override individual results of the outlier process. The user must select a cell or multiple cells and choose the “apply manual adjustment” button. At this point, if the spheroid in that well was determined to be an outlier, it will now be classed as a normal result, and vice-versa. This is to give the user full control over the data and inclusion in subsequent analysis.
- 3.6. The report of the selected file with outliers removed can be downloaded by clicking the “Download” button. This will automatically download and be titled <the file name of the raw file>. The file contains multiple tabs each showing a different measurement (area, diameter etc.) with two plots per tab showing the data for each treatment index with and without outliers. The main dataset tab shows all the data uploaded with new columns added with the outliers removed (OR) (Area_OR etc.). This data can be used by the user to create their own graphs and perform statistical analysis in their preferred method. The downloaded files can also be re-uploaded later to the Merging tab by selecting “Use previous reports” checkbox and using the file selection tool to upload the previously processed files.
- 4. Merging
- 4.1. The Merging tab allows multiple files to be merged into one master file to create plots in the Plotting tab. If the user is only analyzing one file, then the “Merge” button should be chosen and then progress straight to the next tab.
- 4.2. On the main panel on the Merging tab, a table of raw files that have been uploaded on the Data Input tab will be shown. The third column (column name: Processed) shows whether the file has been processed through the outlier removal step on the Outlier Removal tab. The remaining columns show the configuration (e.g., Robust-Z-Score value or Pre-screen thresholds) used in outlier removal so that the user has a record of the parameters should they need to re-analyze a file or analyze future files in the same way later.
- 4.3. Files that have been previously analyzed on the Outlier Removal tab can be uploaded directly to the Merging tab. To do this, select the “Use previous reports” checkbox and browse for the correct files.
- 4.4. The merged file is only available when selected raw files have been processed. Once all the raw files have been processed on the Outlier Removal tab, press the “Merge” button. This will allow plots to be created using the data from multiple files on the Plotting tab. The user can rename (using the “Merged file name” textbox) and download (using “Download the merged file” button) the merged file if required.
- 5. Plotting
- 5.1. Plots of the merged data can be created and viewed on the Plotting tab. SpheroidAnalyseR supports bar charts, point plots, dot plots, and boxplots. The user must select a plot type (bar chart by default) followed by a spheroid/organoid measurement type for the Y-axis (Area by default) and a grouping parameter for the X-axis (Treatment.Label by default) from the drop-down lists. Up to two grouping parameters can be specified to be distinguished by different coloring.
- 5.2. The plot can be named and there is an option to color the plot in black and white. Press the “Plot” button to view the plot on the tab, and the “Add to the report” button to add it to the report which can be downloaded using the “Download the report of plots” button. Multiple plots can be added to the report before downloading.
Figure 1.
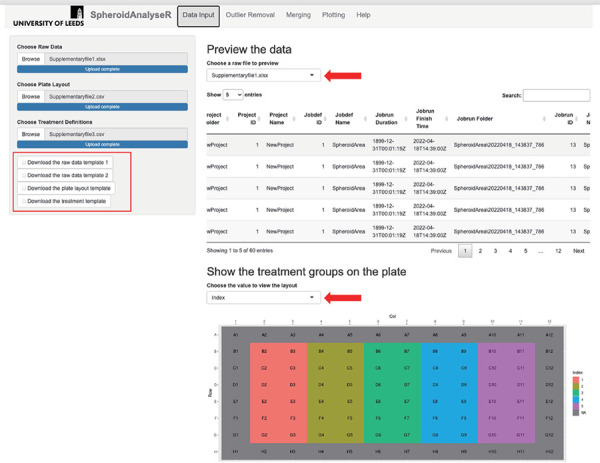
The Data Input tab once raw data has been uploaded. Template files are available to download (see red rectangle). A raw file (File S1), plate layout (File S2) and treatment file (File S3) have been uploaded. A preview of the raw data is shown in the main panel, and a map of the plate colored by treatment index is displayed underneath. Red arrows show the dropdown menus to select a different file to view, and to choose a value to view the layout for.
Figure 2.

The Outlier Removal. A. Inside the beige box are the adjustable outlier settings of Pre-screen-thresholds and Robust-Z-Score. After pressing the “Remove outliers” button, the outliers have been identified on the plate layout seen at the top of the panel. Orange shows outliers removed via pre-screen thresholds and red shows outliers removed after Robust-Z-Score. The results of either of these can be modified using the “Toggle cell status, normal/outlier” drop down menu. The user should select the wells to adjust and press “Apply manual adjustment”. The data can be downloaded as an excel spreadsheet via the Download button. B. The image of the plate of spheroids analyzed.
ANTICIPATED RESULTS
Example work through
A series of screenshots taken from SpheroidAnalyseR highlighting the process of uploading data, data analysis and data presentation is given in Figures 1-4. Key steps are described in the associated figure legends. The example data files are provided in the supplementary materials (raw data is in File S1, plate layout is supplied in File S2 and treatment definitions within File S3).
Figure 3.
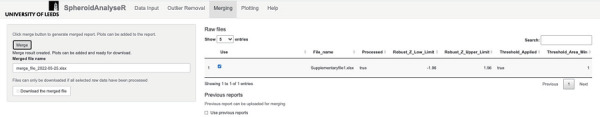
The Merging tab. The status of the file processing is shown “true” or “false” in the “Processed” column. The “Merge” button must be pressed before moving to the next stage even if there is only one file to be analyzed. Files that have been downloaded from the “Outlier Removal” tab can be uploaded here if required.
Figure 4.
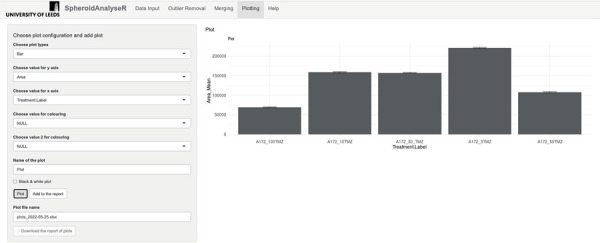
Shows the Plotting tab. The default plot type is a bar plot, but different plot types can be selected in the “Choose plot type” drop-down list. Values to plot on the Y-axis and X-axis can be selected from the drop-down lists. The plot can be displayed in black and white if desired. Plots can be added to the report via the “Add to report” button and then a file of the plots can be downloaded. n = 8 for each treatment group on the bar chart.
TROUBLEHSOOTING
Possible problems and their troubleshooting solutions are listed in Table 1.
Table 1.
Troubleshooting for the SpheroidAnalyseR app
| Step | Problem | Cause | Solution |
|---|---|---|---|
| Outlier removal. 3.4 | Error message—Number of rows of data in raw file does not match the plate setup checksum | The number of rows of data in the raw file is different to that specified in the layout template | Check that the layout template has the correct number of spheroids labelled |
| Merging Tab.4.4 | Error message—Please check all selected files have been processed | Not all files uploaded to the app have been processed on the Outlier Removal tab | Using the select box on the Outlier Removal tab, go through each raw file available and press the remove outlier button for each file |
Supplementary information
File S1. An example raw data file for users to upload to SpheroidAnalyseR for practice using SpheroidAnalyseR.
File S2. An example plate layout corresponding File S1 for users to upload to SpheroidAnalyseR for practice using SpheroidAnalyseR.
File S3. An example treatment file corresponding File S1 for users to upload to SpheroidAnalyseR for practice using SpheroidAnalyseR.
Supplementary information of this article can be found online at
https://polscientific.com/jbm/index.php/jbm/article/view/388/470
https://polscientific.com/jbm/index.php/jbm/article/view/388/471
https://polscientific.com/jbm/index.php/jbm/article/view/388/472.
References
- 1.Ferreira LP, Gaspar VM, Mano JF. Design of spherically structured 3D in vitro tumor models -Advances and prospects. Acta Biomater. 2018. Jul;75:11–34. https://doi.org/10.1016/j.actbio.2018.05.034 10.1016/j.actbio.2018.05.034 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehta G, Hsiao AY, Ingram M, Luker GD, Takayama S. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J Control Release. 2012. Dec;164(2):192–204. https://doi.org/10.1016/j.jconrel.2012.04.045 10.1016/j.jconrel.2012.04.045 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antoni D, Burckel H, Josset E, Noel G. Three-dimensional cell culture: a breakthrough in vivo. Int J Mol Sci. 2015;16(3):5517-27. Epub 2015/03/15. https://doi.org/10.3390/ijms16035517 10.3390/ijms16035517 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Souza N. Organoids. Nature Methods. 2018;15(1):23-. https://doi.org/10.1038/nmeth.4576 10.1038/nmeth.4576 [DOI] [Google Scholar]
- 5.Zanoni M, Cortesi M, Zamagni A, Arienti C, Pignatta S, Tesei A. Modeling neoplastic disease with spheroids and organoids. J Hematol Oncol. 2020;13(1):97. Epub 2020/07/18. https://doi.org/10.1186/s13045-020-00931-0 10.1186/s13045-020-00931-0 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nunes AS, Barros AS, Costa EC, Moreira AF, Correia IJ. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol Bioeng. 2019. Jan;116(1):206–26. https://doi.org/10.1002/bit.26845 10.1002/bit.26845 PMID: [DOI] [PubMed] [Google Scholar]
- 7.Mehta G, Hsiao AY, Ingram M, Luker GD, Takayama S. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J Control Release. 2012;164(2):192-204. Epub 2012/05/23. https://doi.org/10.1016/j.jconrel.2012.04.045 10.1016/j.jconrel.2012.04.045 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katt ME, Placone AL, Wong AD, Xu ZS, Searson PC. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front Bioeng Biotechnol. 2016;4:12. Epub 2016/02/24. https://doi.org/10.3389/fbioe.2016.00012 10.3389/fbioe.2016.00012 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pant D, Narayanan SP, Vijay N, Shukla S. Hypoxia-induced changes in intragenic DNA methylation correlate with alternative splicing in breast cancer. J Biosci. 2020;45(1):3. https://doi.org/10.1007/s12038-019-9977-0 10.1007/s12038-019-9977-0 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006. Feb;24(2):167–75. https://doi.org/10.1038/nbt1186 10.1038/nbt1186 PMID: [DOI] [PubMed] [Google Scholar]
- 11.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671-5. Epub 2012/08/30. https://doi.org/10.1038/nmeth.2089 10.1038/nmeth.2089 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W, Wong C, Vosburgh E, Levine AJ, Foran DJ, Xu EY. High-throughput image analysis of tumor spheroids: a user-friendly software application to measure the size of spheroids automatically and accurately. J Vis Exp. 2014;(89). Epub 2014/07/22. https://doi.org/10.3791/51639 10.3791/51639 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
File S1. An example raw data file for users to upload to SpheroidAnalyseR for practice using SpheroidAnalyseR.
File S2. An example plate layout corresponding File S1 for users to upload to SpheroidAnalyseR for practice using SpheroidAnalyseR.
File S3. An example treatment file corresponding File S1 for users to upload to SpheroidAnalyseR for practice using SpheroidAnalyseR.


