Abstract
Key message
Novel QTLs and candidate genes for Sclerotinia-resistance were identified in B. villosa , a wild Brassica species, which represents a new genetic source for improving oilseed rape resistance to SSR.
Abstract
Sclerotinia stem rot (SSR), caused by Sclerotinia sclerotiorum, is one of the most destructive diseases in oilseed rape growing regions. To date, there is no effective genetic resistance against S. sclerotiorum in the B. napus germplasm and knowledge of the molecular plant–fungal interaction is also limited. To identify new resistance resources, we screened a set of wild Brassica species and identified B. villosa (BRA1896) with a high level of Sclerotinia-resistance. Two segregating F2 populations for Sclerotinia-resistance, generated by interspecific crosses between the resistant B. villosa (BRA1896) and the wild susceptible B. oleracea (BRA1909) were assessed for Sclerotinia-resistance. Genetic mapping using a 15-k Illumina Infinium SNP-array resulted in a high-density genetic map containing 1,118 SNP markers and spanning a total genetic length of 792.2 cM. QTL analysis revealed seven QTLs explaining 3.8% to 16.5% of phenotypic variance. Intriguingly, RNAseq-based transcriptome analysis identified genes and pathways specific to B. villosa, of which a cluster of five genes encoding putative receptor-like kinases (RLKs) and two pathogenesis-related (PR) proteins are co-localized within a QTL on chromosome C07. Furthermore, transcriptomic analysis revealed enhanced ethylene (ET)-activated signaling in the resistant B. villosa, which is associated with a stronger plant immune response, depressed cell death, and enhanced phytoalexin biosynthesis compared to the susceptible B. oleracea. Our data demonstrates that B. villosa represents a novel and unique genetic source for improving oilseed rape resistance against SSR.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00122-023-04335-9.
Introduction
The necrotrophic fungus Sclerotinia sclerotiorum (Lib.) de Bary, a soil-borne fungal pathogen, causes the disease Sclerotinia stem rot (SSR) in rapeseed oil. The fungus embraces a broad spectrum of host plants with more than 400 species including many economically important crops (Boland and Hall 1994; Bolton et al. 2006). S. sclerotiorum overwinters as sclerotia in the soil which are the main inoculum for SSR epidemics in oilseed rape. In spring, apothecia growing on sclerotia carpogenically germinate producing ascospores that are released into air currents and deposited to aboveground parts of the host plant such as the stem axils. When conditions are favorable the fungus starts to grow and infect healthy stem tissue (Derbyshire and Denton-Giles 2016). The pronounced virulence of the fungus is among others attributed to a broad repertoire to produce cell wall-degrading enzymes, phytotoxins, and secreted effector proteins (Amselem et al. 2011). Most common symptoms are bleached lesions traversed by white mycelium in the stem or branch and the formation of black sclerotia inside the infected tissue (Bolton et al. 2006). SSR is one of the most destructive diseases in many oilseed rape cultivation areas worldwide. The tremendous increase in cultivation area of oilseed rape in combination with shorter crop rotation cycles favored SSR dispersal over the past decades. Stems of infected plants tend to burst and shatter. The weakened stem stability and resulting lodging of the plants can cause severe yield and quality losses in oilseed rape cultivation (Derbyshire and Denton-Giles 2016). Resistance to Sclerotinia is mainly measured via leaf-, petiole-, or stem-inoculations on the basis of Zhao et al. (2004) and Zhao and Meng (2003). Studies attempting to assess correlations between the different resistance traits reported contradictory results (Mei et al. 2011, 2013; Taylor et al. 2018; Uloth et al. 2013; You et al. 2016) and more effort is needed to determine their genetic link. Though SSR can be effectively controlled by application of fungicides, the increasing restriction of fungicide use due to its potential environmental and health hazards and the emergence of resistant isolates ask for alternative control strategies worldwide (Derbyshire and Denton-Giles 2016; Wang et al. 2014; Zhou et al. 2014).
Breeding for resistant varieties is an important method in plant disease management. So far, no effective genetic resistance against SSR is available in the B. napus gene pool (Derbyshire and Denton-Giles 2016). Few B. napus genotypes that feature partial Sclerotinia-resistance were reported (Li et al. 2009b; Wang et al. 2002; Zhang et al. 2002). QTLs for Sclerotinia-resistance were also identified in various B. napus mapping populations (Behla et al. 2017; Wei et al. 2014; Wu et al. 2013; Yin et al. 2010; Zhao et al. 2006; Zhao and Meng 2003) and genome-wide association studies (GWAS) identified single-nucleotide polymorphisms (SNPs) associated with Sclerotinia-resistance in numerous B. napus accessions (Gyawali et al. 2016; Wei et al. 2016; Wu et al. 2016).
Therefore, efforts have been made to transfer high SSR-resistance from interspecific crosses to the primary gene pool of B. napus (Chen et al. 2007; Garg et al. 2010). The Brassica oleracea complex, including B. incana, B. rupestris, B. insularis and B. villosa, was identified as valuable pool of high Sclerotinia-resistance (Mei et al. 2011; Taylor et al. 2018). Mei et al. (2013) identified QTLs for SSR-resistance in a mapping population from an interspecific cross between the wild B. incana (resistant) and the cultivated B. oleracea var. alboglabra (susceptible). The resistance was partially transferred into the B. napus gene pool (Mei et al. 2015, 2020). The transcriptome analysis of B. incana linked the resistance to an increased oxidative burst at the infection site balanced by Ca2+ signaling and a suppressed degradation of the plant cell wall by S. sclerotiorum (Ding et al. 2019). These studies highlight the B. oleracea gene pool as important source for introgression of improved resistance to Sclerotinia into the primary gene pool of B. napus.
In this study, we report genetic mapping and QTL analysis of Sclerotinia-resistance in the wild cabbage species Brassica villosa and present for the first time the QTLs for Sclerotinia-resistance in this genome. The comparison with previously identified QTLs in the wild B. incana (Mei et al. 2013) allows for evaluating the resistance mechanisms existing in different Brassica species. In addition, transcriptome analysis was performed to provide molecular insights into the early defense response in B. villosa. We identified 58 defense-related genes to be specifically upregulated in the resistant B. villosa in response to Sclerotinia-inoculation and a cluster of five putative RLKs and two PR proteins which are co-localized within one QTL on the chromosome C07 of the B. oleracea reference genome. Furthermore, the transcriptome analysis suggests that the distinct activation of signaling pathways mediated by ET may play a pivotal role in the Sclerotinia-resistance that is associated with a strong immune response, a negative regulation of cell death, and an elevated phytoalexin biosynthesis in B. villosa.
Materials and methods
Plant material and population structure
Seeds of wild Brassica species (BRA1909, BRA2886, BRA3044, BRA2856, BRA2945, BRA1166, BRA2923, BRA1262, BRA2918, BRA1896) were obtained from the Institute of Plant Genetics and Crop Plant Research (Gatersleben, Germany) and screened for Sclerotinia-resistance with the detached leaf- and petiole-assays (Mei et al. 2011; Zhao et al. 2004) under greenhouse conditions. From this set, the highly resistant wild species B. villosa (BRA1896) was crossed with the susceptible B. oleracea (BRA1909) (Supplementary Data S1). Two mapping populations with 252 and 258 F2 individuals (referred to as Population A and B) were cultivated under greenhouse conditions in two separate years, of which 234 and 258 F2 individuals were screened for Sclerotinia-resistance. For each population, two 15-k Brassica SNP-chip assays (TraitGenetics, Gatersleben, Germany) were used with each chip carrying a total of 96 samples including two NTCs. Therefore, we selected 187 and 184 F2 individuals for genotyping from Population A and B, respectively. The remaining slots on the chips were reserved for genotyping of the parental species. The 184 F2 genotyped individuals from Population B were re-evaluated twice for Sclerotinia-resistance.
Resistance screening and population comparison
Resistance evaluation was performed with the leaf- and petiole-inoculation (Mei et al. 2011; Zhao et al. 2004). Though the stem-inoculation is the most comparable assay to the natural Sclerotinia infection, early and late flowering phenotypes which started to shoot when other plants were already in senescence as well as diverse morphology made an evaluation for stem-resistance difficult and was therefore not performed in our mapping populations. We used a S. sclerotiorum strain originally isolated from an oilseed rape field in Chongqing, China (Mei et al. 2011). The fungus was cultured on potato dextrose agar (PDA; 20 g/l PDB, 15 g/l Bacto agar) plates with a pH of 5.6 at 21 °C and transferred to a new PDA plate two days before inoculation with a cork corer (Ø 8 mm). Leaf resistance was evaluated with at least three Sclerotinia-plugs on detached leaves with the mycelia-site facing the leaf surface. The third and fourth leaves (counted from the apical meristem) and their detached petioles were used for inoculation. Petiole resistance was evaluated with Sclerotinia-plugs stuck on the open cut of the petioles with 1-ml pipette tips. Detached leaves and petioles were placed in a tray with wetted paper towel placed around open cut surfaces and sealed with foil. Leaf-lesion area and petiole-lesion length were measured at 2 days post-infection (dpi). The leaf-lesion area was calculated with the following equation:
‘a’ equals the semi-major axis and ‘b’ indicates the semi-minor axis of a lesion ellipse. The mean leaf- and petiole-lesion value was calculated for each individual plant. Lesion values of the parental plants were used to characterize F2 individuals for their resistance in the whole population screenings. Plants with lesion values smaller than that of the resistant parent (B. villosa) were classified as resistant while plants with lesion values larger than that of the susceptible parent (B. oleracea) were classified as susceptible. Plants with lesion values between those of B. villosa and B. oleracea were defined as intermediate. All statistical analyses were performed via the R software (R Core Team 2021). Data handling in R was mainly performed with the dplyr package (Wickham et al. 2021). Parental lesions in each population were compared via a linear model. Analysis of variance (ANOVA) was followed by multiple contrast tests with the multcomp package (Hothorn et al. 2008). Pearson’s correlation analysis was performed between leaf- and petiole-lesions in each inoculation-assay. Figures were created via the ggplot2 package (Wickham 2016). The phenotype data is available in the Supplementary Data S2–S4.
Trypan blue staining
Detached leaves of B. villosa and B. oleracea were placed in petri dishes, inoculated with PDA-plugs of actively growing Sclerotinia and sealed with Parafilm. After 2 dpi, leave tissue of the junction between necrotic and non-necrotic material was hand-dissected in small rectangles, placed into petri dishes and stained with Trypan blue staining solution according to Fernández-Bautista et al. (2016). Samples were visualized and taken with a SteREO Discovery.V20 microscope (Carl Zeiss AG, Oberkochen, Germany), an AxioCam MRc microscope-cam (Carl Zeiss AG, Oberkochen, Germany), and the AxioVision software (Carl Zeiss AG, Oberkochen, Germany).
Genotyping and genetic map construction
DNA was isolated from leaves following the cetyltrimethylammonium bromide (CTAB) method (Rogers and Bendich 1985). DNA concentration was adjusted to 20 ng/µl with 1% agarose gel with Lambda-DNA (Thermo Fisher Scientific, Massachusetts, USA) and the Gel Doc™ Image Lab Software (Bio-Rad Laboratories, California, USA). Plants were genotyped by TraitGenetics (Gatersleben, Germany) with the 15-k Brassica SNP-chip assay (TraitGenetics, unpublished). The chip carries a total of 13,714 SNP markers. We evaluated the publicly available Brassica reference genomes by RNAseq read-based alignment analysis and found that the genome of B. oleracea (cv. TO1000) represents the best reference genome for this study. The SNP marker sequences were searched against the B. oleracea TO1000 reference genome (Parkin et al. 2014) via the BLAST + software (Altschul et al. 1990; Camacho et al. 2009) with the following options: -evalue 1e−5; -max_target_seqs 2; -max_hsps 1; -outfmt 6. SNPs were transformed into ABH alleles via custom-written python code. Genotypes of both mapping populations were merged for construction of a common genetic map to enable a better comparison of QTLs between both mapping populations which had no influence on the QTL analysis itself. The genetic map was constructed with the R/qtl package (Broman et al. 2003). The linkage groups were assigned to the chromosomes of the B. oleracea TO1000 reference genome via the best hits from the BLAST + search. The genetic and physical positions of all SNP marker are available in Supplementary Data S5.
QTL mapping
The QTL analysis was performed with the R/qtl package according to the workflow described in Broman and Sen (2009) separately for each mapping population. A single-QTL model scan (‘scanone’ function) was performed with the Haley–Knott regression (Haley and Knott 1992) followed by a scan with a nonparametric model which considers the rank-based phenotypes (model = ‘np’) when the first scan detected no QTLs. Peak markers of identified QTLs were used as covariates in the single-QTL model to scan for additive and interactive effects of these markers to other loci followed by a two-dimensional QTL scan considering epistatic effects. A multiple-QTL model was set up according to the identified loci from the scans and screened for additional (‘addqtl’ function) and interacting (‘addint’ function) QTLs. The model was adjusted and finally fitted with the forward/backward model selection algorithm with the Haley–Knott method via the ‘stepwiseqtl’ function. The effect and amount of explainable phenotypical variance by each QTL was estimated with an ANOVA of the final multiple-QTL model. QTL intervals were estimated with the 95% Bayes credible interval method in R/qtl. QTLs with overlapping intervals were classified as common QTL. The significance thresholds were determined via genome-scan-adjusted P values based on permutation tests (10,000 permutations for the single-QTL scans; 2000 permutations for the two-dimensional scans). The mapping data is available in Supplementary Data S6.
Library preparation and RNA sequencing
RNA was isolated with the innuPREP Plant RNA Kit (Analytik Jena AG, Jena, Germany) of Sclerotinia-inoculated and mock-treated petioles from B. villosa and B. oleracea according to the manufacturer’s recommendation. For this, three independent biological replications of each sample were harvested at 8 h post-inoculation (hpi) and immediately frozen in liquid nitrogen. Samples were kept at − 80 °C until further processing. One biological replication consisted of pooled material of eight petioles (1 cm in length from the inoculation site) from four plants. For the transcriptome analysis, we chose 8 hpi to study the early transcriptome response. As we observed that at this time point, the petioles of the resistant plants showed no clear symptoms, while the petioles of the susceptible plants already exhibited profound necrotic lesions at the inoculation sites. This is in accordance with Rietz et al. (2012) and Ding et al. (2019). RNA quality and concentration were determined on 1.3% agarose gel as well as with the NanoVue Plus Spectrophotometer (GE Healthcare, Illinois, USA). Samples were sent to Novogene (Beijing, China) for library preparation and sequencing on the Illumina HiSeq 4000 system. Adapters and reads containing unknown nucleotides (> 10%) and low-quality bases (Q-score ≤ 5) with more than 50% of the total bases were removed by Novogene. Raw sequencing data is available at the NCBI Sequence Read Archive (PRJNA706136).
RNAseq analysis
Data analysis was performed with reference-based and de novo-based RNAseq software tools to bypass limitations of reference-based transcriptome analysis and to provide a more detailed insight into the transcriptome profiles of the wild Brassica species. Briefly, raw reads were processed by removing reads with an average quality less than Q30 (AVGQUAL: 30) via the Trimmomatic software (Bolger et al. 2014). Clean reads were aligned to the B. oleracea TO1000 reference genome and assembled to a reference transcriptome via the ‘new Tuxedo’ protocol including HISAT2 (Kim et al. 2019) and StringTie (Pertea et al. 2015) according to Pertea et al. (2016). SAM files were sorted and converted to BAM files via SAMtools (Li et al. 2009a). The GFF utilities gffread and gffcompare (Pertea and Pertea 2020) were used to extract reference transcript sequences and to retrieve transcriptome assembly statistics. The reference gene count matrix was extracted with the enclosed python script in the StringTie software package. Unmapped reads from the reference transcriptome assembly were then extracted from the BAM files via SAMtools (‘samtools view’ command) with the following parameters: − f 12; − F 256. Unmapped BAM files were converted to fastq format via the ‘bamtofastq’ utility from BEDtools (Quinlan and Hall 2010) and re-aligned to the S. sclerotiorum 1980 reference genome (Amselem et al. 2011). Unmapped reads that neither aligned to B. oleracea nor to S. sclerotiorum were then re-converted to fastq format and assembled de novo via Trinity (Grabherr et al. 2011). Counts of de novo transcripts were estimated via RSEM (Li and Dewey 2011) and the count matrix was filtered for transcripts with at least 10 counts in each of the three biological replications of each sample. Left-over transcripts were loosely defined as genes and merged with the gene count matrix from the reference transcriptome assembly. Differentially expressed gene (DEG) analysis was performed with the merged raw count matrix via the DESeq2 software (Love et al. 2014). The model included the comparison of Sclerotinia-inoculated samples to mock-treated samples of each genotype as well as the interaction term. Genes were considered to be statistically differentially expressed with an adjusted P value ≤ 0.05. Regularized (rlog)-transformed samples were checked with the sample-to-sample distance matrix and the principal component analysis (PCA) in DESeq2. Graphical illustrations and data handling were mainly performed in R via the ggplot2 and the dplyr packages. The RNAseq gene table is available in Supplementary Data S7.
Gene annotation and Gene Ontology (GO) enrichment analysis
The major isoform, measured by the highest averaged FPKM value of all isoforms for a gene across all samples, was used for in silico gene annotation and GO enrichment analysis. The TransDecoder software (https://github.com/TransDecoder/TransDecoder/wiki) was used to convert transcript sequences into protein sequences and to identify functional protein domains. The ‘TransDecoder.LongOrfs’ tool was used to predict longest open reading frames (ORFs) which were then used for a homology-based coding region identification in Pfam (El-Gebali et al. 2019) via the HMMER software (http://hmmer.org/) and in a protein sequence database of Arabidopsis (organism: 3702) downloaded from Uniprot (The UniProt Consortium 2019) via BLASTp + and the following options: -evalue 1e5; -max_target_seqs 1; -max_hsps 1; -outfmt 6. The Pfam and BLAST + results were integrated into the final coding prediction with the ‘TransDecoder.Predict’ tool. The closest homologs in A. thaliana were used for GO annotations of the Brassica genes via the KOBAS database from Xie et al. (2011). Additionally, all genes were searched via BLAST + against the B. napus Refseq database (taxid: 3708) downloaded from the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/). GO enrichment analysis was performed with the goseq package (Young et al. 2010) taking the gene length bias of RNAseq into account. The P values were adjusted via the FDR method by Benjamini and Hochberg (1995) and GO terms were considered to be significant with a FDR ≤ 0.05. The comparative GO analysis was performed and output tables were created with custom-written R-scripts. The GO enrichment results for biological processes is available in Supplementary Data S8.
Real-time quantitative PCR (RT-qPCR) analysis
For RT-qPCR, RNA was isolated at 8 hpi and 16 hpi as described before from three independent biological replications of Sclerotinia- and mock-treated petioles from B. villosa and B. oleracea. Experimental design was identical to the RNAseq experiment. Synthesis of cDNA was performed with 500 µg of RNA treated with RNAse-free DNase I (Thermo Fisher Scientific, Massachusetts, USA) in a volume of 20 µl with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Massachusetts, USA) according to the manufacturer’s instructions. Primer targets were checked by PRIMER-BLAST (Ye et al. 2012). Two microliters (1:5 diluted) were mixed with 18 µl Master Mix as described in the manual of the qPCRBIO SyGreen Mix (PCR Biosystems Inc., Pennsylvania, USA). RT-qPCR was performed on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, California, USA). Conditions for the reactions were as follows: 3 min at 95 °C; 40 cycles of 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 20 s. Gene expression was normalized to BolACT7 (Bo3g005290) and relative quantification between Sclerotinia- and mock-treated samples was calculated according to Pfaffl (2001). The stable expression of BolACT7 was checked by RNAseq. Analysis of primer efficacy was determined by a standard curve of pooled cDNA from all samples for each gene. Information about primer is provided in the Supplementary Table S1. Statistical analysis was performed via a linear model using generalized least squares with the nlme package (Pinheiro et al. 2021) which included the factors genotype, treatment and hpi, as well as their interaction terms. The residuals were assumed to be normally distributed and to be heteroscedastic based on a graphical residual analysis. ANOVA was conducted and followed by multiple contrast tests with the multcomp package (Hothorn et al. 2008). Sanger sequencing of selected templates at Eurofins Scientific SE (Luxembourg, Luxembourg) in Hamburg, Germany, validated targets in the wild Brassica species.
Results
Identification of B. villosa as source of high Sclerotinia-resistance
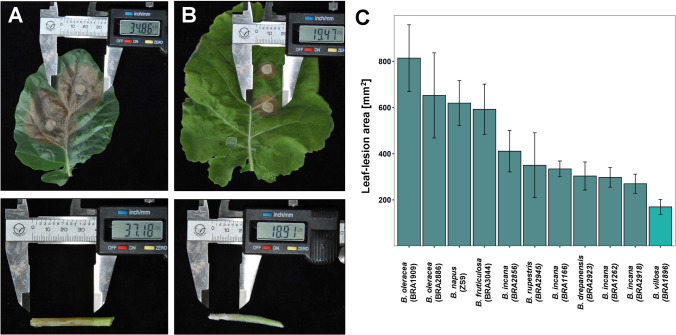
To evaluate the potential of wild Brassica species for Sclerotinia-resistance, we screened a set of wild Brassica species for their resistance with the widely established leaf- and petiole-inoculation assays (Mei et al. 2011; Zhao et al. 2004) under greenhouse conditions. Because the wild Brassica species used in this study exhibited a high degree of diversity in stem development, as described by Taylor et al. (2018), the stem-inoculation method was not feasible for this study. As a result, B. villosa (BRA1896) was identified to be highly resistant to Sclerotinia infection as compared with B. oleracea (BRA1909; Fig. 1A, B) and four B. incana species (BRA1166, BRA1262, BRA2856, BRA2918; Fig. 1C). In support for this, we observed noticeable differences in the fungal spread on infected leaves between the resistant BRA1896 and the susceptible BRA1909 plants by Trypan blue staining assays. A dense and compact structured growth mainly within the necrotic tissue with a sharply delimited junction between healthy and infected tissue was characteristic for the susceptible B. oleracea (Supplementary Figure S1). In the resistant B. villosa, the fungal expansion was less structured, mainly centered on the leaf surface with no sharply delimited changeover between healthy and infected tissue and strongly pronounced infection cushions. For genetic analysis of Sclerotinia-resistance in B. villosa (BRA1896), two segregating F2 populations (Population A, Population B) were generated from an interspecific cross between the resistant B. villosa (BRA1896) and a wild susceptible B. oleracea (BRA1909) in two years.
Fig. 1.
Detached leaf- and petiole-assay of wild Brassica species. A Leaf- (top) and petiole- (bottom) lesions of the susceptible B. oleracea (BRA1909) and B leaf- (top) and petiole- (bottom) lesions of the resistant B. villosa (BRA1896). Lesion sizes were measured 2 days post-inoculation (dpi). C Leaf-lesion sizes of a collection of wild Brassica species from the gene bank of the Institute of Plant Genetics and Crop Plant Research in Gatersleben, Germany. Lesion sizes were measured at 2 dpi. Error bars represent standard errors from three biological replicates
Evaluation of F2 populations for Sclerotinia-resistance
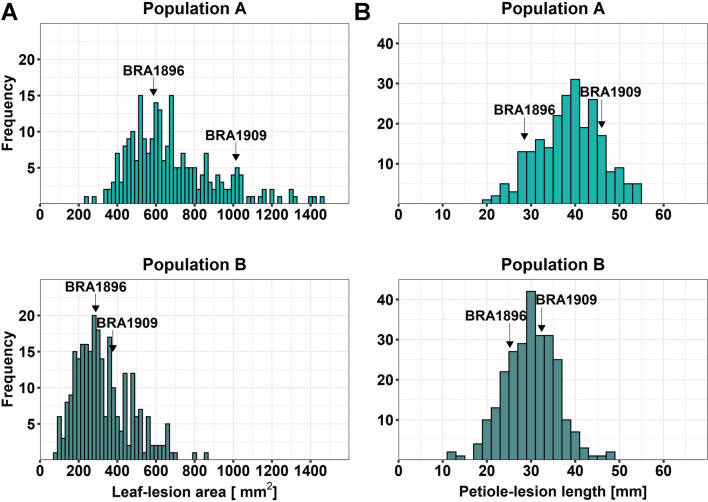
In total, 234 F2 plants in Population A and 258 F2 plants in Population B were separately evaluated for Sclerotinia-resistance with the detached leaf- and petiole-assay, respectively. Leaf-lesion values ranged from 241 to 1452 mm2 in Population A and from 78 to 867 mm2 in Population B, reflecting a slower disease development in the leaf assay in the second year (Fig. 2A.) The petiole-assays produced similar lesion size distributions in the two populations, ranging from 19 to 55 mm in Population A and from 13 to 48 mm in Population B (Fig. 2B). In each population, we compared the leaf- and petiole-lesion sizes of the F2 plants with the parental plants and divided individual F2 plants into three categories (resistant, intermediate, susceptible) and compared the numbers of plants showing a similar response in both assays. Thereby, we identified 15 F2 individuals from Population A and 32 F2 individuals from Population B, which showed a higher resistance level than B. villosa in both assays (Supplementary Figure S2). In total, we identified 207 F2 plants from both populations which were classified into the same category of resistant, intermediate, and susceptible in both assays.
Fig. 2.
Lesion size distributions in two F2 mapping populations from a cross between B. villosa (BRA1896) and B. oleracea (BRA1909). A Leaf-lesion size and B petiole-lesion size distribution in Population A (top, light green) and Population B (bottom, dark green). Positions of B. villosa (BRA1896) and B. oleracea (BRA1909) are indicated by arrows
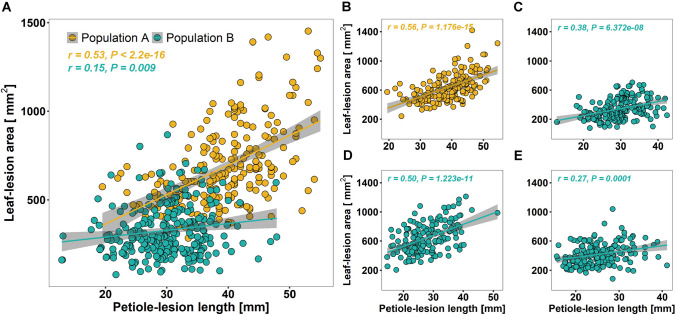
Next, we determined the correlation between leaf- and petiole-lesions in both populations and identified significant correlations explaining about 28% of variance (r = 0.53) in Population A and about 2% of variance (r = 0.15) in Population B (Fig. 3A). The lower correlation in Population B was obviously caused by the slower lesion development. However, re-evaluation of Sclerotinia-resistance with a subset of Population B confirmed the positive correlation with an explainable variance of 25% (r = 0.50) and 7% (r = 0.27) in the two replications, respectively (Fig. 3D, E). Consequently, we selected the 207 F2 plants that were classified by both leaf and petiole tests in conformance with each other and 164 F2 individuals that showed moderate variation from the three categories from the both populations for genotyping with the 15-k Brassica SNP-chip assay (TraitGenetics, unpublished). The correlation between leaf- and petiole-lesions with the selected F2 plants from the both populations increased to 31% in Population A (r = 0.56) and to 14% (r = 0.38) in Population B (Fig. 3B, C). Lesion size distributions of all genotyped F2 individuals are shown in Supplementary Figure S3.
Fig. 3.
Scatterplots of leaf- and petiole-lesions in the two Brassica populations. A Lesion values of all F2 individuals in Population A (yellow) and Population B (cyan). B Lesion values of all genotyped F2 individuals in Population A. C–E Lesion values of all genotyped F2 individuals in Population B in all three resistance screenings. C First assay, D second assay, and E third assay. Correlations were calculated with Pearson’s correlation analysis. r coefficient of correlation, P value
Construction of a high-density genetic map
Overall, 9333 (68%) and 9156 (66%) SNPs were successfully called in B. oleracea and B. villosa. Of these, 392 and 215 SNPs were unique to B. oleracea and B. villosa, respectively. Filtering for polymorphic SNPs between both Brassica species reduced the set of markers to 2806. We combined the genotypes of all F2 individuals from both populations for genetic map construction. After quality filtering and removing duplicate markers, a genetic map was constructed from 1118 SNP marker with data from 361 F2 individuals (Supplementary Table S2 and Data S5). The core markers were ordered into 10 linkage groups with a total length of 784.9 cM and an average distance of 0.7 cM between adjacent markers. Linkage groups were assigned to chromosomes of the B. oleracea TO1000 reference genome (Parkin et al. 2014) according to a BLAST + search (Altschul et al. 1990; Camacho et al. 2009) of the SNP marker sequences. Low coverage of markers corresponding to the B. oleracea reference chromosome C04 resulted in two separate linkage groups (C04a and C04b). Overall, genetic positions of SNP markers were concordantly with their assumed physical positions in the B. oleracea TO1000 reference genome (Supplementary Data S5).
QTL mapping for Sclerotinia-resistance in the wild Brassica populations
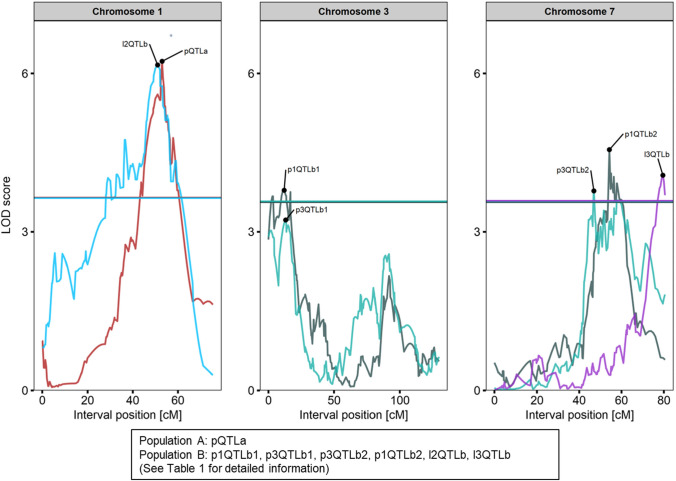
In total, seven QTLs were identified in the two mapping populations (Fig. 4, Table 1). We identified one QTL (pQTLa) for petiole-resistance on linkage group C01 in Population A explaining 15.8% of the phenotypical variance (Table 1). No additional or interacting QTLs were detected in Population A. The QTL peak was detected at the marker Bn-scaff_19564_1-p17934 which was mapped to Scaffold01187 (~ 23 kb) of the B. oleracea TO1000 genome. Flanking markers were Bn-scaff_15749_1-p118178 (26,828,052 bp) and Bn-scaff_16929_1-p495739 (29,084,454 bp). The alleles of B. villosa in pQTLa reduced the petiole-lesions on average by 18% in comparison to B. oleracea. Six QTLs were additionally identified in Population B of which two QTLs (l2QTLb, l3QTLb) accounted for leaf resistance and four QTLs (p1QTLb1, p1QTLb2, p3QTLb1, p3QTLb2) for petiole-resistance (Table 1). In total, the QTLs for leaf- and petiole-resistance in Population B explained 26.9 and 26.6% of phenotypic variance, respectively. Two QTLs (p1QTLb1, p3QTLb2) on chromosome C03 and two QTLs (p1QTLb2, p3QTLb2) on chromosome C07 were repeatedly identified in Population B. The alleles of B. villosa reduced leaf-lesions by 7% and 35%, while petiole-lesions were reduced on average by 15%. Alleles from the susceptible B. oleracea were dominant in four of the six identified QTLs. All QTLs showed additive effects and no epistatic interactions were identified. The QTL for leaf resistance on chromosome C01 (l2QTLb2) explained 16.5% of phenotypic variance and overlapped with the QTL (pQTLa) for petiole-resistance from Population A. The peak was detected at Bn-scaff_22790_1-p152675 which corresponds to position 16,593,775 bp in the B. oleracea reference genome between the flanking markers Bn-scaff_15747_1-p105633 (14,270,425 bp) and Bn-scaff_22790_1-p1026422 (17,467,522 bp). The QTL for leaf resistance on chromosome C07 (l3QTLb) explained 10.4% of variance. The QTL peak was detected between the markers Bn-scaff_16110_1-p975852 (47,352,014 bp) and Bn-scaff_16110_1-p426547 (47,901,219 bp) corresponding to a 550 kbp region in the B. oleracea reference genome. The QTLs for petiole-resistance on chromosome C03 (p1QTLb1, p3QTLb1) explained 3.8% to 8.1% of variance. Both QTL peaks were detected between the markers Bn-scaff_16614_1-p734250 (2,054,448 bp) and Bn-scaff_16614_1-p174856 (3,106,932 bp). The QTLs for petiole-resistance on chromosome C07 (p1QTLb2, p3QTLb2) explained 4.8% to 9.9% of phenotypic variance. The QTL peaks were detected at Bn-scaff_16069_1-p2611780 (42,321,768 bp) and Bn-scaff_16069_1-p4306874 (44,016,862 bp) corresponding to a 1.7 mbp region in the B. oleracea reference genome. Logarithm of odds (LOD) profiles for all linkage groups are available in Supplementary Figure S4.
Fig. 4.
Logarithm of the odds (LOD) profiles of the identified QTL on chromosomes C01, C03, and C07 in the two F2 mapping populations. QTLs were labeled by trait (p = petiole, l = leaf) with numbers representing the replication and by mapping population (a = Population A, b = Population B) followed by a second number to distinguish multiple QTLs from one assay. LOD profiles from different assays are highlighted in different colors. Vertical lines indicate significance thresholds. Detailed information about each QTL are given in Table 1
Table 1.
Identified QTLs for Sclerotinia-resistance in the wild Brassica populations
| Trait | QTL | LOD | LG | Position [cM] | Peak-markera | P value | Var [%] | Addb | Domc |
|---|---|---|---|---|---|---|---|---|---|
| Petiole | pQTLa | 6.2 | C01 | 53 | Bn-scaff_19564_1-p17934 | < 0.001 | 15.8 | 3.8 | − 0.6 |
| Leaf | l2QTLb | 6.2 | C01 | 51 | Bn-scaff_22790_1-p152675 | < 0.001 | 16.5 | 124 | 88.2 |
| Leaf | l3QTLb | 4.1 | C07 | 79.6 | Bn-scaff_16110_1-p976517 | 0.018 | 10.4 | 15.9 | − 90.3 |
| Petiole | p1QTLb1 | 3.8 | C03 | 12 | Bn-scaff_18936_1-p269153 | 0.032 | 8.1 | 2.0 | 1.3 |
| Petiole | p1QTLb2 | 4.6 | C07 | 54.3 | Bn-scaff_16069_1-p4306874 | < 0.01 | 9.9 | 2.4 | 1.1 |
| Petiole | p3QTLb1 | 3.2 | C03 | 13.1 | Bn-scaff_18936_1-p269153 | 0.09 | 3.8 | 1.6 | − 0.6 |
| Petiole | p3QTLb2 | 3.8 | C07 | 46.9 | Bn-scaff_16069_1-p2611780 | 0.03 | 4.8 | 1.3 | 1.6 |
QTLs were labeled by trait (p = petiole, l = leaf) with numbers representing the replication and by mapping population (a = Population A, b = Population B) followed by a second number to distinguish multiple QTLs from one assay
LOD Logarithm of the odds, LG Linkage group, cM centiMorgan
aMarker at peak or nearest to the peak
bAdditive effect. Positive value indicates that alleles from the susceptible parent (BRA1909) increase lesion values
cDominant effect. Positive value indicates alleles from the susceptible parent (BRA1909) are dominant
Comparative transcriptome analysis
To analyze plant transcriptional response to Sclerotinia infection, we applied RNAseq for a comparative transcriptome analysis on both B. villosa and B. oleracea at 8 hpi. Since the B. villosa genome is not yet available, we combined reference- and de novo-based RNAseq analyses to include genes that are not present in the B. oleracea TO1000 reference genome (presence/absence variants; PAVs). The alignment rate of RNAseq samples of mock-treated B. oleracea and B. villosa were on average 93.64% and 87.99%, respectively (Supplementary Table S3). In Sclerotinia-inoculated samples, the alignment rate of the sequences decreased to about 77.70% in B. oleracea and 74.59% in B. villosa, respectively. Hierarchical clustering and PCA showed that RNAseq samples grouped accordingly to species and treatments (Supplementary Figure S5). Almost all variance (97%) between the samples was explained by the factors species (58%) and treatment (39%). Overall, 63,995 expressed genes were identified in the wild Brassica species of which 15,251 transcripts (putative PAVs) could not be aligned to the B. oleracea reference genome. In total, 8,459 DEGs were identified in the resistant and 10,775 DEGs in the susceptible species, respectively (Fig. 5A), from which 5095 up- and 751 downregulated DEGs were common in both species (Fig. 5B).
Fig. 5.
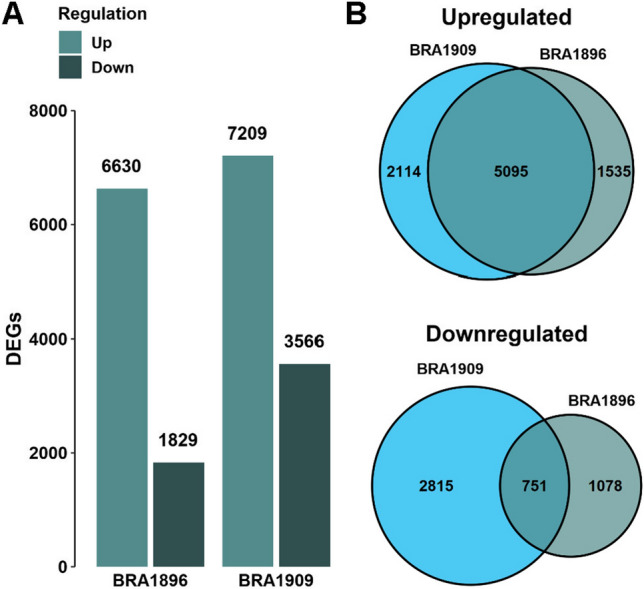
Differentially expressed gene (DEG) analysis between Sclerotinia- and mock-inoculated petioles of B. villosa (BRA1896) and B. oleracea (BRA1909). A Total amount of up- and downregulated DEGs in B. villosa and B. oleracea. B Comparison of up- and downregulated DEGs between the wild Brassica species. Genes were considered as differentially expressed in comparison to the mock-inoculated control based on an adjusted P value ≤ 0.05
Transcriptomic data suggested that the Sclerotinia infection induced an early transcriptional response in both species, which were associated with genes involved in ‘defense response,’ ‘response to chitin,’ ‘immune response,’ and ‘response to decreased oxygen levels,' as indicated by GO enrichment analysis. In total, 111 biological processes were commonly enriched in the resistant and the susceptible species, of which the ‘response to chitin’ (BRA1896: P adj. = 2.31e−60; BRA1909: P adj. = 3.59e−38) represented the most enriched process in both species. Further on, the comparison of the DEGs associated with this biological process identified 130 DEGs common to both species and 39 DEGs specific for the resistant B. villosa. The resistance-specific DEGs included homologs of WRKY, NAC, and zinc finger transcription factors (TFs). For instance, a homolog (Unigene.4449) of the zinc finger protein ZAT12 showed a 5.3-fold (log2-fold: 2.41) induction in B. villosa but no significant induction (0.38-fold; P adj. = 0.73) in B. oleracea. ZAT12 is a key component in oxidative stress response signaling in A. thaliana (Rizhsky et al. 2004). In addition, significant responses of ET-, abscisic acid (ABA)-, salicylic acid (SA)-, and jasmonic acid (JA)-activated pathways were observed in both species. While phytoalexin-related processes were highly enriched in B. villosa, the secondary metabolic processes identified in B. oleracea were mainly associated with glucosinolate metabolism.
Pathways and genes specifically enriched in B. villosa
Diverse biological processes were identified to be specifically activated in B. villosa. The processes were among others enriched with DEGs that are associated with ‘positive regulation of defense response,’ ‘negative regulation of cell death,’ and ‘response to reactive oxygen species.’ From them, 25 genes were specifically induced in the resistant B. villosa, including several homologs of ET responsive factors, receptor-like proteins (RLPs), and RLKs associated with the perception of necrosis-inducing proteins (Supplementary Table S4). Of these, seven de novo genes (genes with a ‘Trinity_’ prefix) were not found in the B. oleracea TO1000 reference genome and not expressed (zero read count) in samples of the susceptible B. oleracea. Interestingly, one homolog (Unigene.13474) of the multifunctional RLK BOTRYTIS INDUCED KINASE 1 (BIK1) was specifically induced in B. villosa with 1.9-fold (log2-fold: 0.91) in response to the Sclerotinia-inoculation. We also identified one homolog (Trinity_DN2466_c0_g1_i7) of the RECEPTOR-LIKE PROTEIN 30 (RLP30) to be specifically expressed 2.63-fold (log2-fold: 1.4) in the resistant B. villosa. RLP30 was shown to mediate enhanced resistance to necrotrophic pathogens such as S. sclerotiorum and B. cinerea in A. thaliana (Zhang et al. 2013). In total, we identified 413 de novo DEGs significantly induced in B. villosa. Of these 58 are functionally associated with the plant defense response of which 34 were not detectable (zero read count) in B. oleracea (Table 2). As revealed by in silico sequence analysis, these genes encode among others for putative RLPs, chitinases, disease resistance proteins, zinc finger family proteins, ET response factors (ERFs), and WRKY TFs.
Table 2.
De novo assembled genes specifically expressed in B. villosa (BRA1896) and associated with plant defense response
| Gene_ID | Log2-fold change | Homolog in A. thaliana | Putative annotation | |
|---|---|---|---|---|
| BRA1896 | BRA1909 | |||
| Trinity_DN1159_c1_g1_i10 | 1.68 | 0.00 | AT1G11310 | Transmembrane domain protein |
| Trinity_DN2585_c0_g1_i2 | 2.68 | 0.00 | AT2G33580 | Receptor-like kinase |
| Trinity_DN1605_c0_g2_i5 | 3.24 | 0.00 | AT2G38470 | WRKY-transcription factor |
| Trinity_DN2365_c0_g1_i5 | 1.22 | 0.29 | AT5G28040 | Leucine zipper transcription factor |
| Trinity_DN46357_c0_g1_i1 | 2.20 | 2.35 | AT1G70130 | Protein kinase |
| Trinity_DN628_c1_g1_i3 | 1.34 | 0.00 | AT5G18370 | Leucine-rich repeat domain receptor |
| Trinity_DN1154_c0_g1_i3 | 1.73 | 0.00 | AT1G61390 | Protein kinase |
| Trinity_DN557_c0_g1_i10 | 2.22 | 3.81 | AT1G15520 | ABC-transporter |
| Trinity_DN5013_c0_g1_i25 | 2.52 | 0.00 | AT3G09830 | Receptor-like kinase |
| Trinity_DN7883_c0_g1_i17 | 1.68 | 0.00 | AT1G74360 | Serine/threonine kinase |
| Trinity_DN5417_c0_g1_i33 | 2.81 | 0.00 | AT5G27420 | Ubiquitin ligase |
| Trinity_DN4012_c0_g1_i4 | 4.94 | 0.00 | AT4G23310 | Cysteine-rich receptor-like protein kinase |
| Trinity_DN21071_c0_g2_i2 | 5.05 | 2.23 | AT2G02220 | Receptor-like protein |
| Trinity_DN1154_c0_g1_i46 | 3.11 | 0.00 | AT1G61390 | Protein kinase |
| Trinity_DN2466_c0_g1_i7 | 1.40 | 0.00 | AT3G05360 | Receptor-like protein |
| Trinity_DN3122_c1_g3_i4 | 1.43 | 0.00 | AT2G43590 | Chitinase protein |
| Trinity_DN242_c0_g1_i19 | 1.30 | 0.00 | AT1G10170 | Transcription repressor |
| Trinity_DN4251_c0_g1_i18 | 5.13 | 0.50 | AT1G02305 | Capase |
| Trinity_DN7129_c0_g1_i1 | 1.89 | 0.00 | AT4G02420 | Protein kinase |
| Trinity_DN651_c3_g2_i2 | 2.48 | 0.00 | AT4G17490 | Ethylene response factor |
| Trinity_DN1154_c0_g1_i11 | 3.06 | 0.00 | AT1G61380 | Receptor-like kinase |
| Trinity_DN12674_c0_g2_i4 | 7.19 | 0.00 | AT2G40140 | Zinc finger family protein |
| Trinity_DN1095_c0_g1_i27 | 1.60 | − 0.47 | AT1G19180 | Nuclear-localized protein |
| Trinity_DN1554_c0_g1_i16 | 2.55 | 1.87 | AT2G37940 | Inositol phosphorylceramide synthase |
| Trinity_DN3122_c1_g3_i1 | 1.81 | 0.49 | AT2G43590 | Chitinase protein |
| Trinity_DN6286_c0_g1_i18 | 3.44 | 4.00 | AT3G04720 | Chitin-binding protein |
| Trinity_DN8160_c0_g1_i1 | 1.02 | − 0.54 | AT2G37040 | Phenylalanine ammonia-lyase |
| Trinity_DN1605_c0_g2_i6 | 3.01 | − 2.37 | AT2G38470 | WRKY-transcription factor |
| Trinity_DN770_c0_g1_i6 | 2.73 | 0.00 | AT2G03760 | Sulfotransferase |
| Trinity_DN4012_c0_g1_i2 | 1.94 | − 3.39 | AT4G23310 | Cysteine-rich receptor-like protein kinase |
| Trinity_DN624_c0_g1_i2 | 1.24 | 0.00 | AT5G03320 | Protein kinase |
| Trinity_DN1627_c0_g2_i4 | 2.34 | 0.00 | AT2G32240 | PAMP-induced protein |
| Trinity_DN787_c0_g3_i1 | 4.25 | 0.00 | AT2G32680 | Receptor-like protein |
| Trinity_DN882_c0_g2_i3 | 1.46 | 0.00 | AT2G37040 | Phenylalanine ammonia-lyase |
| Trinity_DN10838_c0_g1_i8 | 1.62 | 1.45 | AT4G34131 | UDP-glucosyl transferase |
| Trinity_DN159_c0_g3_i18 | 3.00 | − 0.89 | AT2G21660 | Glycine-rich RNA binding protein |
| Trinity_DN590_c0_g1_i10 | 2.36 | 0.00 | AT3G05200 | Putative ring-h2 zinc finger protein |
| Trinity_DN2969_c0_g1_i1 | 3.51 | 0.00 | AT1G80840 | WRKY-transcription factor |
| Trinity_DN5502_c0_g1_i19 | 1.13 | − 0.47 | AT4G25030 | Plastid localized protein |
| Trinity_DN871_c0_g1_i3 | 1.79 | 0.00 | AT5G64120 | Peroxidase |
| Trinity_DN3122_c1_g3_i2 | 1.57 | 1.45 | AT2G43590 | Chitinase protein |
| Trinity_DN1154_c0_g1_i24 | 2.59 | 0.00 | AT1G61390 | Protein kinase |
| Trinity_DN4012_c0_g3_i1 | 2.34 | 0.43 | AT4G23180 | Receptor-like protein |
| Trinity_DN839_c0_g1_i16 | 2.61 | 0.00 | AT5G06320 | Disease resistance gene |
| Trinity_DN1154_c0_g1_i51 | 1.92 | 0.00 | AT1G61380 | Receptor-like kinase |
| Trinity_DN1607_c1_g4_i1 | 1.41 | 0.00 | AT5G47220 | Ethylene response factor |
| Trinity_DN787_c0_g3_i2 | 2.24 | 0.00 | AT2G32680 | Receptor-like protein |
| Trinity_DN1900_c2_g1_i4 | 1.22 | 0.66 | AT5G48380 | Receptor-like kinase |
| Trinity_DN1607_c1_g6_i1 | 3.30 | − 2.70 | AT2G44840 | Ethylene response factor |
| Trinity_DN1095_c0_g1_i8 | 1.02 | 0.00 | AT1G19180 | Nuclear-localized protein |
| Trinity_DN686_c0_g1_i20 | 1.64 | 0.88 | AT1G80820 | Cinnamoyl CoA-reductase |
| Trinity_DN7883_c0_g1_i10 | 2.06 | 0.00 | AT1G74360 | Serine/threonine kinase |
| Trinity_DN9783_c0_g1_i3 | 1.62 | − 0.08 | AT1G58602 | Disease resistance protein |
| Trinity_DN1605_c0_g3_i6 | 3.52 | − 0.47 | AT2G38470 | WRKY-transcription factor |
| Trinity_DN21071_c0_g2_i1 | 1.84 | 0.45 | AT2G02220 | Receptor-like protein |
| Trinity_DN1154_c0_g1_i45 | 1.79 | 0.00 | AT1G61390 | Protein kinase |
| Trinity_DN1795_c0_g2_i9 | 1.61 | 0.00 | AT3G09980 | Microtubules-associated protein |
| Trinity_DN1605_c0_g3_i1 | 5.41 | − 0.47 | AT2G38470 | WRKY-transcription factor |
Significant log2-fold change is marked in bold (P adj. ≤ 0.05). BRA1909 = B. oleracea
Enhanced ET-mediated signaling in B. villosa
Genes with significant differences in their expression patterns between the both species were denoted as interaction DEGs (iDEGs) for further analysis. From 854 iDEGs, 542 were significantly induced in B. villosa, while 312 were stronger induced in B. oleracea. Strikingly, the GO enrichment analysis with the iDEGs showed that most of these genes were among others associated with ‘response to chitin,’ ‘defense response,’ ‘hormone-mediated signaling pathway,’ ‘immune system process,’ and ‘ET-activated signaling pathway.’ In particular, 23 ET–related iDEGs noticeably differed between the both species (Supplementary Table S5). Most of these iDEGs encode putative ERFs and were stronger induced in B. villosa. For example, a homolog of ETHYLENE RESPONSE FACTOR 1 (ERF1; Unigene.2457) was found to be significantly induced in B. villosa (11.7-fold) compared with B. oleracea (1.3-fold). By contrast, ABA-related genes were overall stronger induced in B. oleracea. For example, a homolog (Unigene.6344) of the E3 ubiquitin–protein ligase RHA2B was 5.1-fold (log2-fold: 2.35, P adj. = 0) induced in B. oleracea, while no significant induction (log2-fold: 0.45, P adj. = 0.56) was observed in B. villosa (Data S7). RHA2B is involved in the positive regulation of ABA-mediated signaling (Li et al. 2011).
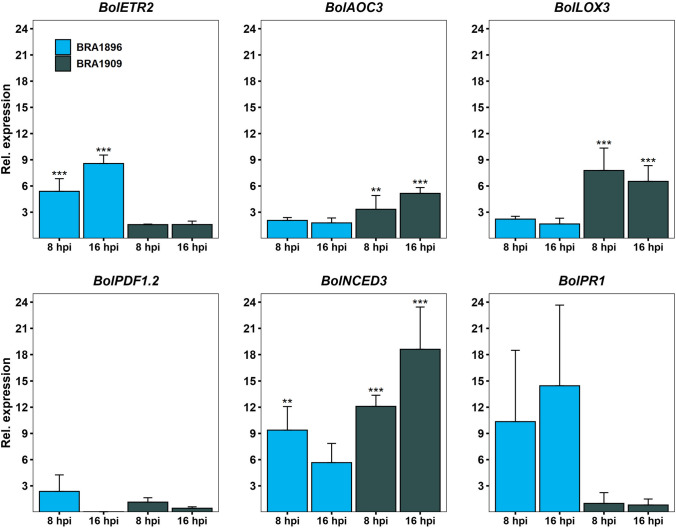
To monitor changes in the hormone-mediated pathways, transcriptional profiling of known marker genes was conducted by RT-qPCR for both species at 8 hpi and 16 hpi. They include ALLENE OXIDASE CYCLASE 3 (AOC3; Bo9g075870) and LIPOXYGENASE 3 (LOX3; Bo8g067210) for JA; ETR2 (Unigene.2465) for ET; PLANT DEFENSIN 1.2 (PDF1.2; Bo2g086460) for the JA/ET branch; NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 3 (NCED3; Bo5g130280) for ABA and PATHOGENESIS-RELATED GENE 1 (PR1; Bo3g088360) for SA (Fig. 6). As expected, RT-qPCR data at 8 hpi were overall in accordance with the RNAseq data (Supplementary Figure S6). Thereby the expression of BolETR2 (Unigene.2465) increased from 5.39-fold at 8 hpi to 8.57-fold at 16 hpi in the resistant B. villosa, while only about 1.5-fold induction occurred in the susceptible B. oleracea at both time points. In contrast, the JA marker genes BolAOC3 (Bo9g075870) and BolLOX3 (Bo8g067210) were induced up to 7.8-fold in the susceptible B. oleracea at 8 hpi and 16 hpi, whereas their expression in B. villosa increased about 2-fold on average. Unexpectedly, no significant change in BolPDF1.2 (Bo2g086460) was observed in both Brassica species. The expression level of BolNCED3 (Bo5g130280) was higher in B. oleracea than in B. villosa and increased from 12.1-fold to 18.6-fold in B. oleracea and decreased from 9.38-fold to 5.66-fold in B. villosa, respectively. Also, BolPR1 (Bo3g088360) expression patterns varied between B. villosa (10.4-fold at 8 hpi and 14.5-fold at 16 hpi) and B. oleracea (0.99-fold at 8 hpi to 0.81-fold at 16 hpi). Taken together, RT-qPCR data support the differences in the ET-and ABA-mediated signaling pathways between B. villosa and B. oleracea as observed from the RNAseq data.
Fig. 6.
Marker gene expression analysis by reverse-transcribed quantitative PCR (RT-qPCR) in petioles of B. villosa (BRA1896, blue) and B. oleracea (BRA1909, gray) at 8 hpi to 16 hpi. Asterisks indicate a significant induction compared to the control group calculated by a linear model using generalized least squares and multiple contrast tests. **P adj. < 0.01, ***P adj. < 0.001. Error bars represent standard deviation of at least two biological replications. Primer used for RT-qPCR are listed in Supplementary Table S1 (colour figure online)
Identification of a RLK gene cluster in the QTL on chromosome C07
To identify genes that may contribute to Sclerotinia-resistance in B. villosa, we surveyed the genomic regions of the QTLs identified in the B. oleracea TO1000 reference genome with our RNAseq data. As a result, 36 genes were found to reside in the QTLs which showed a stronger expression in the resistant B. villosa and are functionally related to plant defense response. They include putative disease resistance proteins (e.g., Unigene.2315, Unigene.27237), MYB TFs (Unigene.7579), ET receptors (Unigene.2258), and RLKs (Unigene.27214; Table 3). Moreover, we identified a cluster of five putative cysteine-rich repeat RLKs on chromosome C07, which were significantly and/or more strongly induced in the resistant B. villosa in responsive to the Sclerotinia infection. Adjacent to the five RLKs, a homolog (Unigene.27237) of a toll-interleukin-resistance (TIR) domain protein and a homolog (Bo7g109370) of a thaumatin-like protein were identified as well, which showed a 4.79- and 4.92-fold increased expression in resistant petioles, respectively.
Table 3.
Pathogenesis-related (PR) genes in the identified QTLs in the B. oleracea TO1000 reference genome
| Chr | Position [bp] | Gene_ID | Log2-fold change | Homolog in A. thaliana | Putative annotation | |
|---|---|---|---|---|---|---|
| BRA1896 | BRA1909 | |||||
| pQTLa | ||||||
| C1 | 27,090,180–27,094,365 | Unigene.2258 | 0.56 | − 0.27 | AT1G60060 | ET receptor |
| C1 | 27,961,418–27,964,840 | Unigene.2305 | 0.28 | − 0.05 | AT5G09890 | Protein kinase |
| C1 | 28,166,345–28,169,898 | Unigene.2315 | 1.23 | − 0.14 | AT1G61100 | Disease resistance protein |
| C1 | 28,665,765–28,668,996 | Unigene.2328 | 1.37 | 0.28 | AT1G61380 | Receptor kinase |
| C1 | 28,684,815–28,689,499 | Unigene.2332 | 0.82 | 0.20 | AT1G61490 | Protein kinase |
| C1 | 28,775,666–28,778,647 | Bo1g098240 | 1.56 | − 0.30 | AT1G61590 | Protein kinase |
| l2QTLb | ||||||
| C1 | 14,675,631–14,677,152 | Unigene.1621 | 1.52 | − 0.68 | AT4G32480 | Sugar phosphate exchanger |
| C1 | 15,950,476–15,952,442 | Unigene.1707 | 2.41 | 0.64 | AT4G16260 | Beta-1,3-endoglucanase |
| C1 | 16,945,305–16,951,076 | Bo1g057070 | 3.77 | 0.15 | AT5G38340 | Disease resistance protein |
| C1 | 17,209,332–17,213,228 | Bo1g057280 | 1.25 | 0.02 | AT4G14940 | Copper amine oxidase |
| l3QTLb | ||||||
| C7 | 47,411,341–47,413,075 | Unigene.27801 | 0.80 | 0.03 | AT4G37150 | Methyl salicylate esterase |
| C7 | 47,471,969–47,474,259 | Unigene.27814 | 1.13 | 0.79 | AT4G36900 | ERF/AP2 transcription factor |
| C7 | 47,507,341–47,508,305 | Unigene.27815 | 1.61 | 2.67 | AT4G37290 | PAMP-induced peptide |
| C7 | 47,591,970–47,593,598 | Unigene.27828 | 0.42 | − 0.07 | AT4G37610 | TAZ-domain protein |
| C7 | 47,645,847–47,647,163 | Bo7g118680 | 2.05 | − 1.10 | AT4G37690 | Glycosyltransferase |
| p1QTLb1/p3QTLb1 | ||||||
| C3 | 2,464,575–2,468,509 | Unigene.7573 | 0.86 | − 0.92 | AT5G11250 | TIR-NBS-LRR protein |
| C3 | 2,505,710–2,510,061 | Unigene.7579 | 0.33 | − 1.22 | AT5G11510 | MYB-transcription factor |
| C3 | 2,590,387–2,591,964 | Unigene.7588 | 2.17 | 0.76 | AT1G05890 | RING/U-box superfamily protein |
| C3 | 2,678,919–2,679,799 | Unigene.7604 | 2.12 | 0.21 | NA | NA |
| C3 | 2,925,738–2,929,237 | Bo3g008890 | 2.86 | − 2.40 | AT5G12920 | WD40 repeat-like superfamily protein |
| p2QTLb2/p3QTLb2 | ||||||
| C7 | 42,428,839–42,430,131 | Bo7g108060 | 1.22 | − 0.33 | AT4G22680 | MYB-domain protein |
| C7 | 42,449,575–42,451,152 | Unigene.27187 | 0.63 | 0.00 | AT4G22690 | Cytochrome P450 protein |
| C7 | 42,731,387–42,734,266 | Unigene.27214 | 1.16 | − 0.57 | AT4G23130 | Cysteine-rich receptor-like kinase |
| C7 | 42,737,429–42,739,520 | Unigene.27215 | 3.29 | 0.94 | AT4G23130 | Cysteine-rich receptor-like kinase |
| C7 | 42,744,618–42,745,142 | Unigene.27216 | 4.72 | 2.52 | AT4G23130 | Cysteine-rich receptor-like kinase |
| C7 | 42,793,701–42,796,988 | Unigene.27221 | 1.18 | 0.49 | AT4G23230 | Cysteine-rich receptor-like kinase |
| C7 | 42,800,478–42,803,336 | Unigene.27222 | 2.39 | 0.42 | AT4G23240 | Cysteine-rich receptor-like kinase |
| C7 | 42,926,260–42,931,088 | Unigene.27237 | 2.26 | 0.51 | AT4G23515 | TIR-domain protein |
| C7 | 42,967,394–42,969,656 | Unigene.27241 | 0.99 | − 0.01 | AT4G23570 | Protein SGT1 homolog A |
| C7 | 43,082,687–43,083,634 | Unigene.27253 | 3.96 | 0.79 | AT4G23720 | Transmembrane protein (DUF119) |
| C7 | 43,297,480–43,298,989 | Bo7g109370 | 2.30 | − 1.88 | AT4G24180 | Thaumatin-like protein |
| C7 | 43,317,954–43,319,839 | Unigene.27284 | 0.82 | 0.06 | AT4G24240 | Calmodulin-binding protein |
| C7 | 43,411,597–43,412,228 | Unigene.27297 | 2.74 | − 0.03 | AT4G30400 | RING/U-box superfamily protein |
| C7 | 43,515,228–43,516,110 | Unigene.27316 | 2.50 | 1.12 | AT4G24960 | HVA22-like protein |
| C7 | 43,568,299–43,569,252 | Unigene.27323 | 2.08 | − 1.18 | AT4G25130 | Methionine sulfoxide reductase |
| C7 | 43,865,115–43,870,847 | Bo7g110600 | 2.22 | − 1.36 | AT4G25960 | P-glycoprotein |
Discussion
B. villosa is a promising source of genetic resistance against Sclerotinia
Mei et al. (2011) and Taylor et al. (2018) reported wild B. incana to be highly resistant to Sclerotinia. Here, we demonstrate for the first time that a wild B. villosa accession (BRA1896) represents a novel and more promising genetic source for breeding Sclerotinia-resistance in oilseed rape. The partial transfer of the Sclerotinia-resistance of the wild B. incana into the B. napus has been demonstrated by Mei et al. (2020) leading to improvement of Sclerotinia-resistance to an average of 35% as compared with the partially resistant Zhongshuang 9. The QTLs from B. incana explained an average effect of ca. 11% of phenotypical variance (Mei et al. 2020). Although the QTLs identified from B. villosa showed a similar effect magnitude but with a higher level of quantitative resistance, it is reasonable to believe that the introgression of them into the B. napus will be worth the effort for breeding Sclerotinia-resistant oilseed rape.
The petiole-assay is an efficient and reliable method to assess Sclerotinia-resistance in wild Brassica species
Leaf- and petiole-inoculation assays were both applied for this study. We observed that a fast drying of PDA plugs, which were used as medium for Sclerotinia, on the leaf-surfaces and irregularities of the leaf-surfaces severely impeded the inoculation process of the leaf assay in some cases. In the petiole-assay, PDA-plugs were well protected from dehydration by the surrounding pipette tip and the fungus could easily infiltrate the petiole via the open cut of the petiole tissue. This may explain the discrepancy of our phenotypic data obtained from two populations: A decelerated symptom development in the leaf assay and smaller leaf-lesion sizes in Population B than in Population A. However, the petiole-assays showed lesion sizes in a similar range for both populations, thus be able to discriminate the resistant and susceptible parent species with high significance (P values < 0.01, Supplementary Figure S7). This may explain why the detached petiole-assays yielded more stable QTLs than the leaf assay in this study. Though the inoculation of leaves is a well-established Sclerotinia-inoculation technique (Joshi et al. 2016; Mei et al. 2011; Zhao and Meng 2003), our data suggest that the detached petiole-assay is a more efficient and reliable screening method, especially when the stem-inoculation is not possible as in our case.
There are contradictory reports in regarding the correlation between leaf-, stem-, and petiole-resistance. Taylor et al. (2018) found no significant correlation between leaf- and petiole-resistance results by analyzing a set of wild Brassica species, whereas it was given between petiole- and stem-resistance in a collection of B. napus accessions. Uloth et al. (2013) and You et al. (2016) found no association between leaf- and stem-resistance under field conditions after investigation on diverse Brassica species. But, a correlation between leaf- and stem-resistance was reported by Mei et al. (2011, 2013) even under field as well as controlled environments by artificial inoculation. In this study, we demonstrate a significant correlation (P values > 0.01) between leaf- and petiole-resistance across all assays. A rather lower explainable variance, ranging from 2 to 31%, might be mainly attributed to the technical sensitivity of the leaf assay, which masked the correlation analysis. It should be noted that the results of these two assays may reflect different resistance mechanisms: The leaf assay includes the additional effect from preformed resistance, a physical barrier to the fungus in general, while the petiole-assay mainly reflects the plant defense response induced by the fungal infection. Thus, the combination of leaf- and petiole-assays as did in this study may facilitate the screening and identifying of effective Sclerotinia-resistance. Further studies, e.g., by the stem-inoculation under natural conditions, are needed to substantiate the functional significance of the QTLs identified in this study.
Diverse Sclerotinia-resistance mechanisms existing in different wild Brassica species
Seven QTLs for Sclerotinia-resistance in B. villosa were identified, from which two were from leaf- and five from the petiole-assays. Strikingly, the major QTL identified from the petiole-assay (pQTLa) in Population A is overlapping with the major QTL for leaf resistance (l2QTLb) in Population B, with both accounting for approx. 16% of phenotypic variance, respectively. This finding suggests that the underlying resistance mechanism may be relying on common genetic resistance factors. We also detected two partially overlapping QTLs from the petiole-assay (p1QTLb2, p3QTLb2) and one QTL from the leaf assay (l3QTLb) on chromosome C07, but due to their genetic and physical distance, these QTLs do not appear to be directly connected to each other. Mei et al. (2013) analyzed one F2 mapping population from a cross between wild B. incana (resistant) and the cultivated B. oleracea var. alboglabra (susceptible). They detected a major QTL for Sclerotinia-resistance on chromosome C09 explaining up to 28.4% of phenotypical variance as well as minor QTLs on chromosomes C01, C03, C04, and C07. But, the use of different markers from various references as well as a different B. oleracea reference genome (JZS; B. oleracea sp. capitata) makes a direct comparison with our results not possible. Nevertheless, we compared our results with those reported by Li et al. (2015) who had physically integrated QTLs for Sclerotinia-resistance from several studies including those reported by Mei et al. (2013) to the B. napus genome. Conspicuously the QTLs on chromosome C01 from our study are physically overlapped with those reported by Mei et al. (2013), unfortunately there is no overlapping QTL on chromosome C07, where most B. villosa QTLs reside. Recently, SNPs significantly associated with SSR-resistance were detected on chromosome C03 in B. napus (Shahoveisi et al. 2021; Roy et al. 2021). Comparing the physical positions reported in the B. napus genome with those of the reference genome revealed that one SNP at position 7.893,201 bp in B. napus (Roy et al. 2021) is corresponding to 3.334.677 bp in the B. oleracea genome which is close to the QTL marker Bn-scaff_16614_1-p174856 (3,106,932 bp) in this study.
It is to note that the QTLs on chromosome C01 in our mapping populations contributed to phenotypic variance of about 16%, while only 8.4% of phenotypic variance was reported by Mei et al. (2013). These data suggest that there exist distinct Sclerotinia-resistance mechanisms in different wild Brassica species, but some genetic factors being involved in resistance seem to be conserved, thus supporting that the B. oleracea complex represents a valuable genetic source for breeding Sclerotinia-resistance in oilseed rape.
Sclerotinia-resistance in B. villosa is linked to the ET-activated signaling pathway
The enriched biological processes and pathways identified in this study differ from those in B. incana (Ding et al. 2019). The resistance in B. incana is mainly associated with an enhanced cell wall integrity and an accelerated Ca2+ signaling which, as the authors suggested, regulates the production of an early respiratory burst via the accumulation of reactive oxygen species (ROS; Ding et al. 2019). Furthermore, they demonstrated that S. sclerotiorum takes over control of the copper ion homeostasis in susceptible host genotypes to scavenge and detoxify plant ROS to repress the oxidative burst in the early stage of infection, while resistant host genotypes such as B. incana were less disturbed in their copper ion homeostasis (Ding et al. 2020). However, the reported mechanisms are not linked to the previously identified QTLs in B. incana. Thus, the main genetic factors and mechanisms governing the resistance in this species remain unsolved. Yet, we did not find any evidence for the involvement of the copper ion homeostasis in B. villosa, but identified processes related to the perception of ROS, the regulation of cell death, as well as an enhanced immune and defense response. Our data, however, underpin the role of initiation of an early respiratory burst by the fungal infection in plant resistance to S. sclerotiorum. Several related genes showed contrasting expression patterns in resistant and susceptible species, including, e.g., Unigene.4449, a homolog of the key gene ZAT12 in oxidative stress response signaling in A. thaliana (Rizhsky et al. 2004) that was 5.3-fold induced in B. villosa but not significantly induced in B. oleracea.
The early respiratory burst was apparently differently regulated in the resistant and susceptible species. In B. villosa, it was found to be associated with an enhanced expression of genes related to the ET-activated signaling pathway, whereas the JA-mediated signaling pathway was less activated. This may be a result of the interference of JA- and SA-pathways as, in support of this, a highly elevated transcript abundance of PR1 was concurrently observed in B. villosa as compared to B. oleracea. As confirmed by RT-qPCR, the expression level of ETR2, an ET-marker gene, significantly increased from 8 to 16 hpi only in B. villosa, but not in the susceptible B. oleracea, whereas two JA-marker genes AOC3 and LOX3 were in contrast induced only in B. oleracea but not in B. villosa at both time points, respectively.
The ET- and JA-mediated signaling pathways are key components in regulating plant defense to necrotrophic pathogens by synergizing the ERF branch via ERF1/ORA59 (Broekgaarden et al. 2015; Pré et al. 2008). A key marker gene of the ERF branch is PDF1.2 that is regulated by ORA59, an essential integrator of the ET- and JA-signal transduction pathway (Pré et al. 2008). Interestingly, our RNAseq data revealed a significant elevation of the expression level of one ORA59-like gene (Bo8g114710) in B. villosa but without induction of PDF1.2 in both Brassica species. Thus, we speculate that the ET- and JA-signaling pathways do not act through the common ERF branch but trigger different immune responses in the two Brassica species. An early oxidative burst in B. villosa could be a result of the ET-activated signaling. To clarify this, more experiments are needed.
Yang et al. (2017) reported that infection of resistant rice cultivars with the blast fungus Magnaporthe oryzae also induced the ET-mediated signaling pathway that increased ROS accumulation and the production of phytoalexins. An significant enrichment of DEGs associated with the phytoalexin metabolism was also identified in B. villosa, thus suggesting a signaling network similar to that described in rice (Yang et al. 2017). In the susceptible B. oleracea, we found that the glucosinolate and sulfur compound metabolic processes were enhanced after Sclerotinia infection, in line with the observation in B. napus as a result of an enhanced JA-mediated signaling (Wei et al. 2016). In addition, we noticed the activation of 19 iDEGs involved in the ABA-signaling pathway in B. oleracea. Interestingly, the orthologue Bo2g159220 of the MYC branch marker gene VSP2 was neither significantly induced in B. oleracea nor in B. villosa. The JA-mediated MYC branch, antagonistic to the ERF branch, proved to be responsible for defense against herbivores and co-regulated by ABA (Broekgaarden et al. 2015; Vos et al. 2015). These data suggest a complex interplay of hormone-mediated signalings occurs during resistance to Sclerotinia in B. villosa.
Possible candidate genes for Sclerotinia-resistance
A large set of candidate genes were identified in the resistant B. villosa, which are functionally associated with the immune response or PR genes and linked to our QTLs. Notably, a small cluster of RLKs together with two putative disease resistance proteins reside in the QTL on chromosome C07. Most of the RLK genes were induced specifically in the resistant B. villosa. Two putative disease resistance genes were also found in the resistant B. villosa in the QTLs on chromosome C01. Further analyses are needed to clarify whether these genes are involved in Sclerotinia-resistance. Since the reference-based analysis relies on the B. oleracea reference genome, we integrated the de novo transcriptome assembler Trinity (Grabherr et al. 2011) into our RNAseq analysis to identify putative PAVs specific for B. villosa but not present in the susceptible B. oleracea. This approach let to identification of additional 34 putative PAVs associated with the plant defense response in B. villosa. Because alignment with the reference genome failed, no genetic linkage could be established with the identified QTLs. Thus, re-sequencing of the B. villosa genome will shed more light on the role of these genes in Sclerotinia-resistance and facilitate the identification of candidate genes for breeding Sclerotinia-resistant oilseed rape in the future.
Conclusion
By QTL mapping and transcriptome analysis, we demonstrate for the first time that the wild accession B. villosa is a novel and valuable genetic source of quantitative resistance against the fungal pathogen S. sclerotiorum. The ET-activated signaling may represent a key signaling pathway to the activation of plant S. sclerotiorum defense response, associated with an early ROS production and an increased production of phytoalexins in the resistant B. villosa. Moreover, the genes and the QTLs identified in this study are promising candidates for investigation on molecular plant-S. sclerotiorum interactions as well as for breeding of resistant oilseed rape varieties against S. sclerotiorum infection.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We gratefully thank Dr. Plieske from TraitGenetics for the helpful support, Prof. Dr. Broman for the helpful advice with R/qtl, Annalena Hartmann and Martina Wittke for the excellent technical assistance. The authors thank the Stiftung Schleswig-Holsteinische Landschaft for providing scholarship to Thomas Bergmann (2018/06), as well as DAAD and BLE for travel grants (Grant no. 57317839, 13/14-15-CHN).
Author contribution statement
TB performed the main phenotyping and genotyping, constructed the genetic map, designed and conducted the RNAseq and RT-qPCR experiment, performed statistical, genetic as well bioinformatical analyses and drafted the manuscript. JM and WY were involved in phenotyping and genotyping experiments. MS supported computational analysis. MH supported the statistical analysis. SR and GL provided materials and supported the project. DG conceived and directed the project and finalized the manuscript. All authors read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was financially supported by the Bundesministerium für Bildung und Forschung (BMBF, Grant No. 0315637 B), the Bundesministerium für Ernährung und Landwirtschaft (BMEL) and the Fachagentur für Nachwachsende Rohstoffe (Grant No. 22410312, FKZ22006516).
Availability of data and material
The main data is provided in the electronic supplementary material. Additional data and pre-calculated permutations for the QTL analysis are available on request. Raw sequencing data is available at the NCBI Sequence Read Archive (PRJNA706136).
Code availability
The main code is available at https://zenodo.org/record/5524469.
Declarations
Conflicts of interest
The authors declare no competing interests.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, Fournier E, Gout L, Hahn M, Kohn L, Lapalu N, Plummer KM, Pradier J-M, Quévillon E, Sharon A, Simon A, Have A ten, Tudzynski B, Tudzynski P, Wincker P, Andrew M, Anthouard V, Beever RE, Beffa R, Benoit I, Bouzid O, Brault B, Chen Z, Choquer M, Collémare J, Cotton P, Danchin EG, Da Silva C, Gautier A, Giraud C, Giraud T, Gonzalez C, Grossetete S, Güldener U, Henrissat B, Howlett BJ, Kodira C, Kretschmer M, Lappartient A, Leroch M, Levis C, Mauceli E, Neuvéglise C, Oeser B, Pearson M, Poulain J, Poussereau N, Quesneville H, Rascle C, Schumacher J, Ségurens B, Sexton A, Silva E, Sirven C, Soanes DM, Talbot NJ, Templeton M, Yandava C, Yarden O, Zeng Q, Rollins JA, Lebrun M-H, Dickman M (2011) Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet 7:e1002230. 10.1371/journal.pgen.1002230 [DOI] [PMC free article] [PubMed]
- Behla R, Hirani AH, Zelmer CD, Yu F, Fernando WGD, McVetty P, Li G. Identification of common QTL for resistance to Sclerotinia sclerotiorum in three doubled haploid populations of Brassica napus (L.) Euphytica. 2017;213:88. doi: 10.1007/s10681-017-2047-5. [DOI] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (methodol) 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Boland GJ, Hall R. Index of plant hosts of Sclerotinia sclerotiorum. Can J Plant Path. 1994;16:93–108. doi: 10.1080/07060669409500766. [DOI] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton MD, Thomma BPHJ, Nelson BD. Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol Plant Pathol. 2006;7:1–16. doi: 10.1111/J.1364-3703.2005.00316.X. [DOI] [PubMed] [Google Scholar]
- Broekgaarden C, Caarls L, Vos IA, Pieterse CMJ, van Wees SCM. Ethylene: traffic controller on hormonal crossroads to defense. Plant Physiol. 2015;169:2371–2379. doi: 10.1104/pp.15.01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Sen S. A guide to QTL mapping with R/qtl. New York NY: Springer; 2009. [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-F, Wang H, Li Z-Y. Production and genetic analysis of partial hybrids in intertribal crosses between Brassica species (B. rapa, B. napus) and Capsella bursa-pastoris. Plant Cell Rep. 2007;26:1791–1800. doi: 10.1007/s00299-007-0392-x. [DOI] [PubMed] [Google Scholar]
- Derbyshire MC, Denton-Giles M. The control of sclerotinia stem rot on oilseed rape (Brassica napus): current practices and future opportunities. Plant Pathol. 2016;65:859–877. doi: 10.1111/ppa.12517. [DOI] [Google Scholar]
- Ding Y, Mei J, Chai Y, Yu Y, Shao C, Wu Q, Disi JO, Li Y, Wan H, Qian W. Simultaneous transcriptome analysis of host and pathogen highlights the interaction between Brassica oleracea and Sclerotinia sclerotiorum. Phytopathology. 2019;109:542–550. doi: 10.1094/PHYTO-06-18-0204-R. [DOI] [PubMed] [Google Scholar]
- Ding Y, Mei J, Chai Y, Yang W, Mao Y, Yan B, Yu Y, Disi JO, Rana K, Li J, Qian W. Sclerotinia sclerotiorum utilizes host-derived copper for ROS detoxification and infection. PLoS Pathog. 2020;16:e1008919. doi: 10.1371/journal.ppat.1008919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Bautista N, Domínguez-Núñez J, Moreno MM, Berrocal-Lobo M. Plant tissue trypan blue staining during phytopathogen infection. Bio-Protoc. 2016;6:e2078. doi: 10.21769/BioProtoc.2078. [DOI] [Google Scholar]
- Garg H, Atri C, Sandhu PS, Kaur B, Renton M, Banga SK, Singh H, Singh C, Barbetti MJ, Banga SS. High level of resistance to Sclerotinia sclerotiorum in introgression lines derived from hybridization between wild crucifers and the crop Brassica species B. napus and B. juncea. Field Crops Res. 2010;117:51–58. doi: 10.1016/j.fcr.2010.01.013. [DOI] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, Di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyawali S, Harrington M, Durkin J, Horner K, Parkin IAP, Hegedus DD, Bekkaoui D, Buchwaldt L. Microsatellite markers used for genome-wide association mapping of partial resistance to Sclerotinia sclerotiorum in a world collection of Brassica napus. Mol Breed. 2016;36:72. doi: 10.1007/s11032-016-0496-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity (edinb) 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- Joshi RK, Megha S, Rahman MH, Basu U, Kav NNV. A global study of transcriptome dynamics in canola (Brassica napus L.) responsive to Sclerotinia sclerotiorum infection using RNA-Seq. Gene. 2016;590:57–67. doi: 10.1016/j.gene.2016.06.003. [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CX, Liu SY, Sivasithamparam K, Barbetti MJ. New sources of resistance to Sclerotinia stem rot caused by Sclerotinia sclerotiorum in Chinese and Australian Brassica napus and B. juncea germplasm screened under Western Australian conditions. Australas Plant Pathol. 2009;38:149. doi: 10.1071/AP08087. [DOI] [Google Scholar]
- Li H, Jiang H, Bu Q, Zhao Q, Sun J, Xie Q, Li C. The Arabidopsis ring finger E3 ligase RHA2b acts additively with RHA2a in regulating abscisic acid signaling and drought response. Plant Physiol. 2011;156:550–563. doi: 10.1104/pp.111.176214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhao Z, Hayward A, Cheng H, Fu D. Integration analysis of quantitative trait loci for resistance to Sclerotinia sclerotiorum in Brassica napus. Euphytica. 2015;205:483–489. doi: 10.1007/s10681-015-1417-0. [DOI] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei J, Qian L, Disi JO, Yang X, Li Q, Li J, Frauen M, Cai D, Qian W. Identification of resistant sources against Sclerotinia sclerotiorum in Brassica species with emphasis on B. oleracea. Euphytica. 2011;177:393–399. doi: 10.1007/s10681-010-0274-0. [DOI] [Google Scholar]
- Mei J, Ding Y, Lu K, Wei D, Liu Y, Disi JO, Li J, Liu L, Liu S, McKay J, Qian W. Identification of genomic regions involved in resistance against Sclerotinia sclerotiorum from wild Brassica oleracea. Theor Appl Genet. 2013;126:549–556. doi: 10.1007/s00122-012-2000-x. [DOI] [PubMed] [Google Scholar]
- Mei J, Liu Y, Wei D, Wittkop B, Ding Y, Li Q, Li J, Wan H, Li Z, Ge X, Frauen M, Snowdon RJ, Qian W, Friedt W. Transfer of sclerotinia resistance from wild relative of Brassica oleracea into Brassica napus using a hexaploidy step. Theor Appl Genet. 2015;128:639–644. doi: 10.1007/s00122-015-2459-3. [DOI] [PubMed] [Google Scholar]
- Mei J, Shao C, Yang R, Feng Y, Gao Y, Ding Y, Li J, Qian W. Introgression and pyramiding of genetic loci from wild Brassica oleracea into B. napus for improving Sclerotinia resistance of rapeseed. Theor Appl Genet. 2020;133:1313–1319. doi: 10.1007/s00122-020-03552-w. [DOI] [PubMed] [Google Scholar]
- Parkin IAP, Koh C, Tang H, Robinson SJ, Kagale S, Clarke WE, Town CD, Nixon J, Krishnakumar V, Bidwell SL, Denoeud F, Belcram H, Links MG, Just J, Clarke C, Bender T, Huebert T, Mason AS, Pires JC, Barker G, Moore J, Walley PG, Manoli S, Batley J, Edwards D, Nelson MN, Wang X, Paterson AH, King G, Bancroft I, Chalhoub B, Sharpe AG. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 2014;15:R77. doi: 10.1186/gb-2014-15-6-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea G, Pertea M. GFF Utilities: GffRead and GffCompare. F1000Res. 2020;9:304. doi: 10.12688/f1000research.23297.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D (2021). Nlme: linear and nonlinear mixed effects models: R package version 3.1–144
- Pré M, Atallah M, Champion A, de Vos M, Pieterse CMJ, Memelink J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008;147:1347–1357. doi: 10.1104/pp.108.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2021) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
- Rietz S, Bernsdorff FEM, Cai D. Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum. J Exp Bot. 2012;63:5507–5519. doi: 10.1093/jxb/ers203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem. 2004;279:11736–11743. doi: 10.1074/jbc.m313350200. [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol. 1985;5:69–76. doi: 10.1007/bf00020088. [DOI] [PubMed] [Google Scholar]
- Roy J, Shaikh TM, del Rio ML, Hosain S, Chapara V, Rahman M. Genome-wide association mapping and genomic prediction for adult stage sclerotinia stem rot resistance in Brassica napus (L.) under field environments. Sci Rep. 2021;11:21773. doi: 10.1038/s41598-021-01272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahoveisi F, Oladzad A, del Rio ML, Hosseinirad S, Ruud S, Rissato B. Assessing the effect of phenotyping scoring systems and SNP calling and filtering parameters on detection of QTL associated with reaction of Brassica napus to Sclerotinia sclerotiorum. PhytoFront. 2021;1:135–148. doi: 10.1094/PHYTOFR-10-20-0029-R. [DOI] [Google Scholar]
- Taylor A, Rana K, Handy C, Clarkson JP. Resistance to Sclerotinia sclerotiorum in wild Brassica species and the importance of Sclerotinia subarctica as a Brassica pathogen. Plant Pathol. 2018;67:433–444. doi: 10.1111/ppa.12745. [DOI] [Google Scholar]
- The UniProt Consortium UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uloth MB, You MP, Finnegan PM, Banga SS, Banga SK, Sandhu PS, Yi H, Salisbury PA, Barbetti MJ. New sources of resistance to Sclerotinia sclerotiorum for crucifer crops. Field Crop Res. 2013;154:40–52. doi: 10.1016/j.fcr.2013.07.013. [DOI] [Google Scholar]
- Vos IA, Moritz L, Pieterse CMJ, van Wees SCM. Impact of hormonal crosstalk on plant resistance and fitness under multi-attacker conditions. Front Plant Sci. 2015;6:639. doi: 10.3389/fpls.2015.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HZ, Liu GH, Zheng YB, Wang XF, Yang Q. Breeding of Brassica napus cultivar Zhongshuang No. 9 with resistance to Sclerotinia sclerotiorum. Chin J Oil Crop Sci. 2002;24:71–73. [Google Scholar]
- Wang Y, Hou Y-P, Chen C-J, Zhou M-G. Detection of resistance in Sclerotinia sclerotiorum to carbendazim and dimethachlon in Jiangsu Province of China. Australas Plant Pathol. 2014;43:307–312. doi: 10.1007/s13313-014-0271-1. [DOI] [Google Scholar]
- Wei D, Mei J, Fu Y, Disi JO, Li J, Qian W. Quantitative trait loci analyses for resistance to Sclerotinia sclerotiorum and flowering time in Brassica napus. Mol Breed. 2014;34:1797–1804. doi: 10.1007/s11032-014-0139-7. [DOI] [Google Scholar]
- Wei L, Jian H, Lu K, Filardo F, Yin N, Liu L, Qu C, Li W, Du H, Li J. Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol J. 2016;14:1368–1380. doi: 10.1111/pbi.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H, Romain F, Lionel H, Müller K (2021). dplyr: a grammar of data manipulation: R package version 1.0.5. https://CRAN.R-project.org/package=dplyr
- Wickham H (2016). ggplot2: Elegant graphics for data analysis. Use R! Springer, Cham
- Wu J, Zhao Q, Liu S, Shahid M, Lan L, Cai G, Zhang C, Fan C, Wang Y, Zhou Y. Genome-wide association study identifies new loci for resistance to sclerotinia stem rot in Brassica napus. Front Plant Sci. 2016;7:1418. doi: 10.3389/fpls.2016.01418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Cai G, Tu J, Li L, Liu S, Luo X, Zhou L, Fan C, Zhou Y (2013) Identification of QTLs for resistance to sclerotinia stem rot and BnaC.IGMT5.a as a candidate gene of the major resistant QTL SRC6 in Brassica napus. PLoS One 8:e67740. 10.1371/journal.pone.0067740 [DOI] [PMC free article] [PubMed]
- Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li C-Y, Wei L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Li W, Cao J, Meng F, Yu Y, Huang J, Jiang L, Liu M, Zhang Z, Chen X, Miyamoto K, Yamane H, Zhang J, Chen S, Liu J. Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J. 2017;89:338–353. doi: 10.1111/tpj.13388. [DOI] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Yi B, Chen W, Zhang W, Tu J, Fernando WGD, Fu T. Mapping of QTLs detected in a Brassica napus DH population for resistance to Sclerotinia sclerotiorum in multiple environments. Euphytica. 2010;173:25–35. doi: 10.1007/s10681-009-0095-1. [DOI] [Google Scholar]
- You MP, Uloth MB, Li XX, Banga SS, Banga SK, Barbetti MJ. Valuable new resistances ensure improved management of sclerotinia stem rot (Sclerotinia sclerotiorum) in horticultural and oilseed brassica species. J Phytopathol. 2016;164:291–299. doi: 10.1111/jph.12456. [DOI] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JF, Fu S, Qi C, Pu H, Chen Y, Gu B, Chen X, Gao J. Breeding and utilization of Ning RS-1 resistance to sclerotinia stem rot in rapeseed (B. napus L.) Chin J Oil Crop Sci. 2002;24:6–9. [Google Scholar]
- Zhang W, Fraiture M, Kolb D, Löffelhardt B, Desaki Y, Boutrot FFG, Tör M, Zipfel C, Gust AA, Brunner F. Arabidopsis receptor-like protein 30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell. 2013;25:4227–4241. doi: 10.1105/tpc.113.117010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Meng J. Genetic analysis of loci associated with partial resistance to Sclerotinia sclerotiorum in rapeseed (Brassica napus L.) Theor Appl Genet. 2003;106:759–764. doi: 10.1007/s00122-002-1171-2. [DOI] [PubMed] [Google Scholar]
- Zhao J, Peltier AJ, Meng J, Osborn TC, Grau CR. Evaluation of sclerotinia stem rot resistance in oilseed Brassica napus using a petiole inoculation technique under greenhouse conditions. Plant Dis. 2004;88:1033–1039. doi: 10.1094/PDIS.2004.88.9.1033. [DOI] [PubMed] [Google Scholar]
- Zhao J, Udall JA, Quijada PA, Grau CR, Meng J, Osborn TC. Quantitative trait loci for resistance to Sclerotinia sclerotiorum and its association with a homeologous non-reciprocal transposition in Brassica napus L. Theor Appl Genet. 2006;112:509–516. doi: 10.1007/s00122-005-0154-5. [DOI] [PubMed] [Google Scholar]
- Zhou F, Zhang X-L, Li J-L, Zhu F-X. Dimethachlon Resistance in Sclerotinia sclerotiorum in China. Plant Dis. 2014;98:1221–1226. doi: 10.1094/PDIS-10-13-1072-RE. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data is provided in the electronic supplementary material. Additional data and pre-calculated permutations for the QTL analysis are available on request. Raw sequencing data is available at the NCBI Sequence Read Archive (PRJNA706136).
The main code is available at https://zenodo.org/record/5524469.