Abstract
Objectives:
Increased numbers of tumor infiltrating lymphocytes (TIL) in endometrial cancer (EC) are associated with improved survival, but it is unclear how this prognostic significance relates to the underlying EC molecular subtype. In this explorative hypothesis-generating study, we sought to define the immune signatures associated with the molecular subtypes of EC (i.e., POLE-mutated, microsatellite unstable (MSI-high), copy number (CN)-low, and CN-high) and to determine their correlation with patient outcomes.
Methods:
RNA-sequencing and molecular subtype data of 232 primary ECs were obtained from The Cancer Genome Atlas. Deconvolution of bulk gene expression data was performed using single sample Gene Set Enrichment Analysis (ssGSEA) and Cell type Identification By Estimating Relative Subsets Of known RNA Transcripts (CIBERSORT). The association of the resultant immune signatures with overall survival was determined across molecular subtypes.
Results:
Statistically significant differences in enrichment were identified in 16/30 and 6/23 immune gene sets by ssGSEA and CIBERSORT, respectively. Signature of CD8+ cells in ECs of CN-high molecular subtype was associated with improved overall survival by ssGSEA (p=0.0108), while CD8 signatures did not appear to be prognostic in MSI-high (p=0.74) or CN-low EC molecular subtypes (p=0.793). Of all molecular subtypes, CN-high ECs exhibited the lowest levels of CD8+ T cell infiltration. Consistent with antigen-induced T cell activation and exhaustion, enrichment for immunomodulatory receptors was predominantly observed in ECs of MSI-high and POLE-mutated molecular subtypes.
Conclusions:
Deconvolution of bulk gene expression data can be used to identify populations of immune infiltrated endometrial cancers with improved survival. These data support the existence of unique mechanisms of immune resistance within molecular subgroups of the disease.
Keywords: endometrial cancer, molecular subtype, deconvolution, RNA-sequencing, immune infiltration
1. INTRODUCTION
Despite the significant initial promise of immune checkpoint blockade (ICB) in some cancer types, the majority of endometrial cancers (ECs), with the exception of mismatch repair (MMR)-deficient tumors, are resistant to single agent ICB [1-4]. Combination of the multi-targeted tyrosine kinase inhibitor (TKI) lenvatinib with pembrolizumab (anti-PD1) in advanced EC demonstrated response rates of close to 40%, irrespective of MMR status or histology [5]. Aside from MMR status, however, the relationship between immunotherapy response and the underlying EC subtype remains poorly understood.
Comprehensive molecular profiling of ECs by The Cancer Genome Atlas (TCGA) revealed four molecular subtypes of the disease [6]. ECs of copy number (CN)-high (serous-like) subtype are characterized by high levels of CN alterations, highly recurrent TP53 mutations, a relatively low tumor mutational burden (TMB), and poor outcomes. The CN-low (endometrioid) group consists of predominantly low-grade, microsatellite stable ECs with few CN alterations, and CTNNB1 mutations in about half of the cases. The microsatellite unstable (MSI-high)/ hypermutated ECs are characterized by a high TMB and few CN alterations. Finally, ECs of POLE ultramutated subtype have the highest TMB due to mutations in the proofreading domain of the DNA polymerase epsilon (POLE) and lack CN alterations; this group also has the best prognosis [6]. These integrated molecular subtypes provide information beyond the routine histologic classification or traditional clinico-pathologic binary classification of ECs [6-8]. Within the context of each molecular subtype of EC, however, a variation of clinical outcomes exists [9, 10] and additional factors driving these prognostic differences remain largely unknown.
In particular, the relationship of the EC molecular subtypes with the immune system has not been adequately characterized. The degree and composition of tumor infiltrating lymphocytes (TILs) in the tumor microenvironment (TME) has been shown to be associated with outcomes and response to therapy in many solid tumors, including EC [11, 12]. While presence of TILs appears to be associated with improved outcomes in EC in general, at present, the relationship between TILs and prognosis within the context of each molecular subtype of EC remains poorly characterized. Furthermore, evaluation of the TME for prognostic and potentially predictive biomarkers relies on reproducible identification and quantification of not only TILs in general, but additional leukocyte subsets and immunoregulatory markers.
Traditionally TILs have been identified through immunohistochemical (IHC) or immunofluorescence analyses. Further characterization and identification of specific immune cell population can be achieved through flow cytometry, however these analyses are only possible in freshly-resected specimens and may be subject to bias introduced by tissue dissociation and processing [13]. Newer technologies have allowed for the inference of immune cell infiltration within solid tumors by using bulk transcriptomic data derived from high throughput RNA-sequencing [14]. These deconvolution approaches are able to use either gene expression signatures or cell-specific marker genes to infer relative fractions of immune cell types of interest. Such methods do not depend on surface marker presentation and are not subject to the same artifacts of cellular dissociation seen in flow cytometry, and they allow for more uniform sample processing and collection, which may improve the reproducibility of TIL identification, while still providing data in a quantitative fashion [14]. When utilizing the quantitative or semi-quantitative assessments of the TME as prognostic indicators or potential treatment biomarkers, uniformity of processing, and reproducibility of results is imperative to the correlation of this data to clinical outcomes. This may prove to be particularly true in EC where efforts to correlate the TME to outcomes have provided inconsistent correlations [15]. Given the wide variation in clinical outcomes within each of the TCGA molecular subtypes of EC [9, 10], we performed an exploratory hypothesis-generating subgroup re-analysis of TCGA data to determine whether quantification of the immune cell fractions and other TME parameters from bulk tumor transcriptomic data could provide prognostic stratification of patients within the context of each EC molecular subtype.
2. MATERIAL AND METHODS
2.1. Patients and data source
Publicly available RNA-sequencing data and corresponding clinical data from the 232 treatment-naïve primary ECs with molecular subtype classification from TCGA [6] were retrieved from the Genomic Data Commons (Downloaded from https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp). Of the 232 ECs, 90 were of CN-low molecular subtype, 60 were CN-high, 65 were MSI-high and 17 were POLE as defined by TCGA [6]. Adjuvant therapy data were available for 43% of patients and none of the patients received immunotherapy in the adjuvant setting. No information about receipt of immunotherapy in subsequent settings was available.
2.2. Estimation of tumor-infiltrating immune cell population
Sequence read pairs for each case were aligned to the reference genome GRCh37 using STAR [16]. Aligned read counts were calculated using HTSeq, converted to normalized Reads Per Kilobase Million (RPKM) counts per gene, and two computational approaches for deconvolution of bulk gene transcriptional data were employed, as previously described [17]. First, Cell type Identification By Estimating Relative Subsets Of known RNA Transcripts (CIBERSORT) was utilized, a deconvolution algorithm based on normalized gene expression profiles described by Newman et al. [18]. CIBERSORT predicts immune cell fractions, thus the output estimates for each individual sample were normalized to 1. Second, immune gene cell signatures were defined using Single Sample Gene Set Enrichment Analysis (ssGSEA)[19], which identifies genes specific for individual leukocyte classes that are differentially enriched in a single sample by comparison to sorted validated cell populations. We utilized the immune cell signatures as described by Bindea et al. based on the expression of 570 genes [19].
2.3. Immune profiles and survival outcomes
The immune enrichment data were generated for 26 leukocyte cell types and 4 immunomodulatory proteins by ssGSEA and 23 leukocyte cell types were obtained by CIBERSORT. The distributions for the immune profiles in ECs stratified according to their TCGA molecular subtypes [6] were assessed using Wilcoxon rank-sum test using STATA IC (v10 StataCorp). Statistically significant differences amongst the four TCGA molecular subtypes were defined as a p<0.05.
The median enrichment for CD8+ cells was identified for each TCGA subset subdividing the population into an enriched “CD8-high” population above the median, as well as “CD8-low” population below the median. The clinicopathologic characteristics for the CD8-high and CD8-low populations were compared using Fisher’s exact for categorical variables and Wilcoxon rank-sum test for continuous variables. Overall survival days were extracted from TCGA, using the diagnosis date as the time zero. Median survival and survival rate at certain year were calculated using Kaplan Meier method. Log-rank test or Log-rank with permutation test (if very few event number in certain level) [20] was applied to obtain p-value for comparisons. Survival curves were generated using the Kaplan-Meier method. Significance was defined as a p<0.05; all reported p-values are two-sided. Analyses were performed using R 3.6.0 (https://www.R-project.org/).
3. RESULTS
3.1. Enrichment of immune signatures in EC molecular subtypes
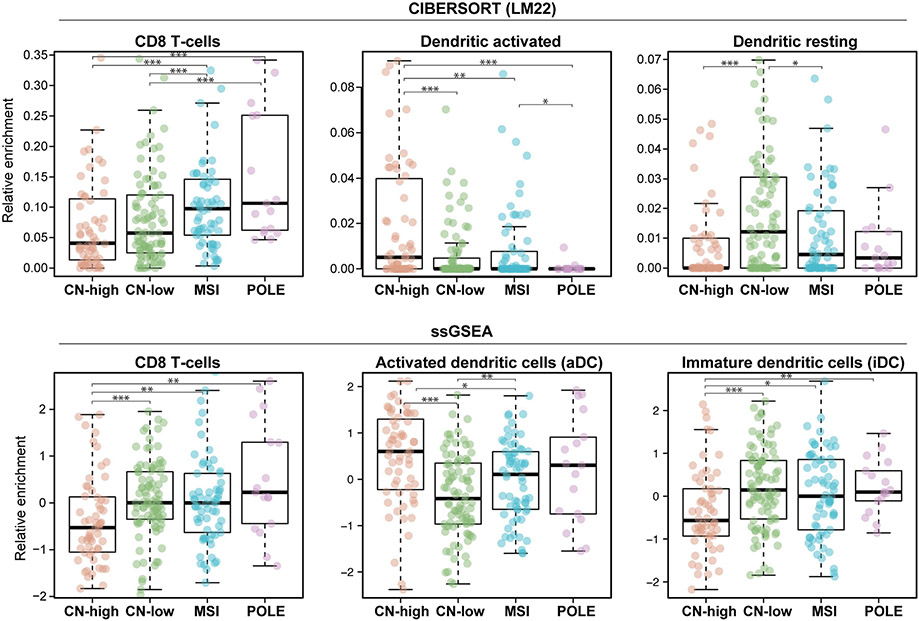
We first defined the immune landscape of primary untreated endometrioid and serous ECs classified into the four molecular subtypes by TCGA based on their bulk transcriptomic profiles using CIBERSORT and ssGSEA. Statistically significant differences as obtained by ANOVA comparisons in gene expression amongst the four molecular subtypes were identified in 16/26 leukocyte subset gene sets by ssGSEA, in 4/4 immunomodulatory marker gene sets by ssGSEA, and in 6/23 leukocyte subset gene sets by CIBERSORT (Supplementary Table 1). When comparing the results obtained by ssGSEA and CIBERSORT, we observed significant enrichment differences in at least one EC TCGA subtype for CD8+ cells, activated dendritic cells, and inactive/resting dendritic cells by both deconvolution methods (Figure 1). In particular, CN-high ECs had higher enrichments for activated dendritic cells when compared to CN-low (ssGSEA p<0.001, CIBERSORT p<0.001) and MSI-high ECs (ssGSEA p<0.01, CIBERSORT p<0.01). Statistically significant differences in enrichment for inactive/resting dendritic cell gene signatures between the different molecular subtypes were also noted. CN-high ECs had lower enrichment for inactivated/resting dendritic cell when compared to CN-low (ssGSEA p<0.001, CIBERSORT p<0.001) and MSI-high ECs (ssGSEA p<0.01, CIBERSORT p<0.01) (Figure 1).
Figure 1: Immune cell enrichment in endometrial carcinomas stratified by molecular subtype using CIBERSORT and ssGSEA.
Box plots displaying the relative enrichment for leukocyte subsets in the four molecular subtypes of endometrial cancer using CIBERSORT (top) or ssGSEA (bottom). Subgroups were compared using Wilcoxon rank-sum. Statistical significance was defined as a p<0.05 and annotated as ***P<0.001, **P< 0.01, *P<0.05. Comparisons not annotated were not statistically significant. CN-high, copy number-high; CN-low, copy number-low; MSI, microsatellite unstable/ hypermutated; POLE, polymerase epsilon-mutant/ ultramutated.
Consistent with the high levels of TILs observed by pathology review in DNA mismatch repair-deficient and POLE ECs [21-25], deconvolution analysis revealed that ECs of POLE (ultramutated) and MSI-high (hypermutated) molecular subtypes were characterized by the highest median enrichment for CD8+ cells (Figure 1). In contrast, CN-high ECs had the lowest enrichment for CD8+ T-cells (p<0.01) by both CIBERSORT and ssGSEA compared to ECs of all of the molecular subtypes analyzed.
3.2. Association of immune signatures and outcomes in EC molecular subtypes
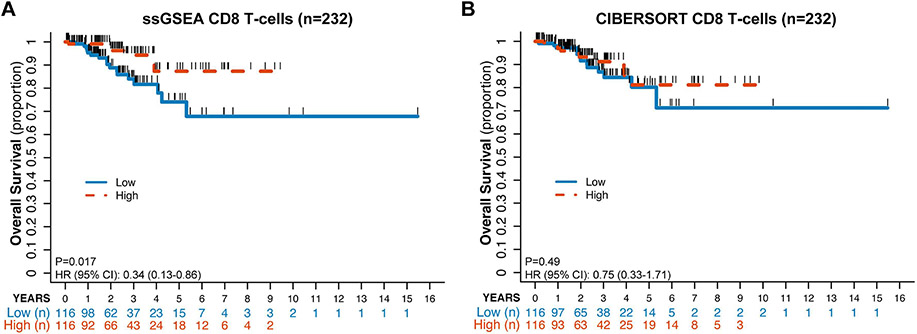
Evaluation of the entire cohort of ECs dichotomized by ssGSEA CD8+ T-cell enrichment around the median revealed statistically significant differences in overall survival (OS) by Kaplan-Meyer estimates (p=0.017), with ECs patients with high CD8+ T-cell enrichment having a better outcome (Figure 2A); however, this did not reach statistical significance using CIBERSORT (p=0.49; Figure 2B). It should be noted that 10.3% (12/116) of the CD8-high ECs by ssGSEA and 9.5% (11/116) of the CD8-high ECs by CIBERSORT were of POLE molecular subtype. On the other hand, approximately one third of the ECs classified as CD8-low (35.3 %, 41/116, ssGSEA; 32.8%, 38/116; CIBERSORT) were of CN-low molecular subtype. Interestingly, the difference in progression-free survival (PFS) was not statistically significant by either method (ssGSEA: p=0.084, CIBERSORT: p=0.318), however for 16/232 (8/46 with progression) ECs PFS days are not available in TCGA.
Figure 2: Kaplan-Meyer estimate of overall survival for patients with endometrial carcinoma according to CD8+ infiltration defined by ssGSEA and CIBERSORT.
Endometrial cancers from TCGA of all molecular subtypes (n=232) were classified into CD8+ high (blue solid line) or CD8+ low (orange dashed line) defined by their distribution around the median CD8+ infiltration for the entire cohort using ssGSEA (A) or CIBERSORT (B). Kaplan-Meyer estimates are shown.
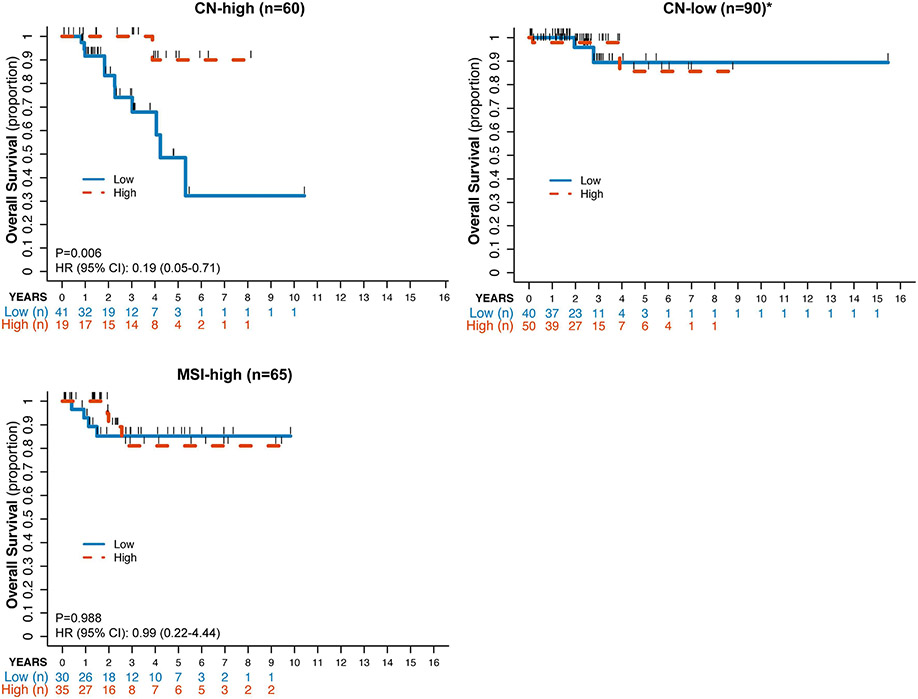
When each individual molecular subtype group was stratified by CD8+ T cell enrichment (high vs low relative to the median), statistically significant differences in OS were seen in the CN-high EC subtype but not in the other molecular subtypes by ssGSEA (p=0.0108; Figure 3). The statistical significance was not maintained when performing the same analysis by CIBERSORT, however (p=0.641; Supplementary Figure 1). Baseline patient characteristics between the CN-high ECs classified as CD8-low or CD8-high by either method are summarized in Table 1. There were no statistically significant differences in age, FIGO 2009 stage and histologic grade in the ssGSEA analysis. In the CIBERSORT analysis, the CD8-high patients were younger (66 vs 77, p=0.015) and more commonly had ECs of endometrioid histology (10% vs 43.3%) though 76% cases had grade 3(G3)/serous/mixed histology. Similar to the analysis of the entire cohort, no statistically significant differences were seen in PFS (ssGSEA, p=0.084; CIBERSORT, p=0.318) for the whole cohort analyses, possibly due to missing PFS data for a subset of the patients. Median overall survival was not reached in the CD8+ high CN-high ECs as assessed by either deconvolution method. There were no statistically significant differences in OS between the CD8-high and CD8-low groups for the MSI-high and CN-low EC molecular subtypes (Figure 3).
Figure 3: Kaplan-Meyer estimate of overall survival for ECs by molecular subtype according to CD8+ infiltration defined by ssGSEA.
ECs of CN-high, CN-low, and MSI-high molecular subtypes were stratified into CD8+ high (blue solid line) or CD8+ low (orange dashed line) defined by their distribution around the median CD8+ infiltration for the entire cohort (n=232) using ssGSEA. *Due to only 4 events in OS, no HR and p-value were reported for the CN-low cohort.
Table 1:
Clinicopathologic characteristics of endometrial cancers of copy number-high (serous-like) molecular subtype by CD8+ infiltration.
| Total Cohort n (%) |
ssGSEA | CIBERSORT | |||||
|---|---|---|---|---|---|---|---|
| CD8+ low | CD8+ high |
p- value |
CD8+ low | CD8+ high |
p- value |
||
| Age (years) | |||||||
| Median (Mean) | 68 (69.05) | 67 (68.87) | 69 (69.23) | 0.584 | 71 (71.73) | 66 (66.37) | 0.015 |
| Range | 44-90 | 58-90 | 44-87 | 58-87 | 44-90 | ||
| Stage | |||||||
| I/II | 33 (55%) | 17 (56.7%) | 16 (53.3%) | 1 | 14 (46.7%) | 19 (63.3%) | 0.503 |
| III | 19 (31.7%) | 9 (30%) | 10 (33.3%) | 11 (36.7%) | 8 (26.7%) | ||
| IV | 8 (13.3%) | 4 (13.3%) | 4(13.3%) | 5 (16.7%) | 3 (10%) | ||
| Histology | |||||||
| Endometrioid | 16 (26.7%) | 6 (20%) | 10 (33.3%) | 0.382 | 3 (10%) | 13 (43.3%) | 0.007 |
| Serous/Mixed | 44 (73.3%) | 24 (80%) | 20 (66.7%) | 27 (90%) | 17 (56.7%) | ||
| Tumor grade (endometrioid) | |||||||
| G1 | 1 (1.7%) | 0 (0%) | 1 (3.3%) | ** | 0 (0%) | 1 (3.3%) | ** |
| G2 | 6 (10%) | 2 (6.7%) | 4 (13.3%) | 0 (0%) | 6 (20%) | ||
| G3 and serous/ mixed | 53 (88.3%) | 28(93.3%) | 25 (83.3%) | 30 (100%) | 23 (76.7%) | ||
| BMI kg/m2 (categorical; missing, n=3) | |||||||
| Normal (BMI<25) | 16 (28.1%) | 7 (24.1%) | 9 (32.1%) | 0.724 | 9 (31%) | 7 (25%) | 0.827 |
| Overweight (BMI:25-30) | 11 (19.3%) | 5 (17.2%) | 6 (21.4%) | 6 (20.7%) | 5 (17.9%) | ||
| Obese (BMI≥30) | 30 (52.6%) | 17 (58.6%) | 13 (46.4%) | 14 (48.3%) | 16 (57.1%) | ||
Clinicopathologic characteristics of copy number-high (serous-like) endometrial cancers by CD8+ infiltration high vs low as defined by ssGSEA and CIBERSORT using median for cutoff. All p-values reported here are unadjusted, using Fisher’s exact test for categorical variables and Wilcoxon-rank sum test for continuous variables.
p-value not provided if certain level has counts <5. BMI, body mass index.
3.3. Signatures of immune dysfunction and angiogenesis in EC molecular subtypes
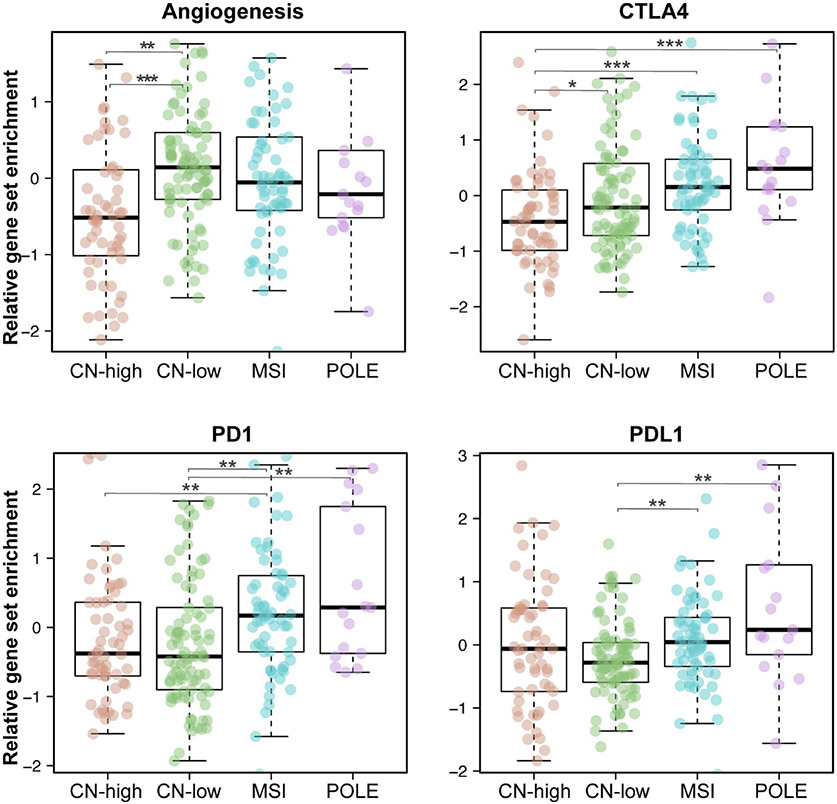
Given the known differential responsiveness of EC molecular subtypes to immunotherapy, we sought to determine whether markers of immune dysfunction, which are known to be associated with tumor antigen reactivity, are differentially expressed among the distinct EC molecular subtypes using the ssGSEA analysis (Figure 4). Both ECs of POLE (ultramutated) and MSI-high (hypermutated) subtypes were found to have high enrichment for gene signatures associated with T cell activation/dysfunction and inflammatory response including enrichment for CTLA-4 (p<0.001), PD-1 (p <0.001) and PD-L1 (p=0.004). Despite their enrichment in the POLE and MSI-high molecular subtypes, it should be noted that in ECs of the CN-high and CN-low subtypes there were also subsets of patients with high enrichment for these markers. CN-low ECs were found to have higher enrichment for angiogenesis signatures in comparison to the other subtypes, with CN-high ECs having the lowest. Within ECs of CN-high and CN-low subtypes, there was a subset of ECs with high enrichment for the ssGSEA angiogenesis signature and PD-L1 expression (CN-high, 7/60, 11.6%; CN-low, 8/90, 8.9%; Figure 4).
Figure 4: Relative gene set enrichment of selected immunoregulatory markers in ECs stratified by molecular subtype.
Comparison of selected immunoregulatory marker enrichment using ssGSEA including angiogenesis, CTLA4, PD1 and PD-L1. Subgroups were compared using Wilcoxon rank-sum. Statistical significance was defined as a p<0.05 and annotated in the figure with ***P<0.001, **P<0.01, *P<0.05. Comparisons not annotated were not statistically significant.
4. DISCUSSION
In this study we utilized two methods of deconvolution of publicly available RNA-sequencing data to analyze the TME of ECs and their relation to the four TCGA molecular subtypes. The use of deconvolution of bulk RNA-sequencing data has been used to define the relationship between the TME and other solid tumor types, which led to the identification of prognostic associations between TME components and outcomes in breast, colon and lung cancers [14, 18, 26-28]. These data have been used to help delineate the genes, pathways, and leukocyte subtypes that drive the association between immune infiltration and survival [28]. These methods potentially have advantage of uniformity and reproducibility in comparison to IHC or flow cytometry, in particular when it comes to the identification of specific leukocyte subsets [29]. In breast cancer, for example, Nederlof et al showed that interpathologist concordance for leukocyte subtype specific quantification is lower than the quantification of the immune infiltration in general when compared to deconvolution methods [30]. In addition, identification of lymphocyte infiltration in the TME does not provide information about their activation status. For example, measurement of exhausted tumor antigen-specific T cell subsets has been suggested to be better measured by the cell’s transcriptional activity rather than visual identification using the complex interplay of cell surface immunomodulatory proteins [14, 31].
Prior investigation of the EC TME using enrichment scores from ssGSEA estimates resulted in clustering of ECs into 3 unique separate immunophenotypes based on relative immune cell abundance [32]. Cluster I, the cluster with highest immune cell infiltration, was found to be associated with higher TMB, upregulation of co-stimulating and immune checkpoint proteins and improved survival [32]. Our data expand these analyses by defining the TME phenotypes within the context each TCGA subtype, highlighting that while specific immune subsets are preferentially enriched within each TCGA subtype, a heterogeneity of immune phenotypes nevertheless does exist within a given molecular subtype group. The largest influence of immune cell infiltration on prognosis was observed in the ECs of CN-high subtype. Understanding the differences in immune response in the MSI-high/POLE ECs may require further immunophenotyping to determine the mechanisms of immune resistance in these immunogenic tumors.
Given their central role in the anti-tumor immune response, CD8+ T cells have long been a front runner as a prognostic biomarker in solid tumors [33, 34]. Our analysis confirmed that the ECs of hypermutated and ultramutated TCGA molecular subtypes are associated with marked leukocyte infiltration, particularly CD8+ T cells [35-37]. Consistent with prior data, analysis of the entire TCGA cohort by enrichment for CD8+ T cells demonstrates survival difference, however, this type of analysis is confounded by the predominance of MSI-high/ POLE ECs in the CD8-high group as well as CN-high ECs in the CD8-low group. Consistent with this hypothesis, Talhouk et al assessed TIL infiltration via IHC in ECs grouped similarly to TCGA genotypes, demonstrating that outcomes are more driven by molecular subtype rather than markers of immune infiltration [38].
Our exploratory analysis takes it a step further and attempts to assess within each TCGA subclass to further stratify outcomes based on TME. We find that the poor prognosis associated with lack of CD8+ T cell infiltration is indeed largely driven by the CN-high cancers, a subset of which is depleted for CD8+ T cell-associated expression signature. Notably, within the CN-high molecular subtype there exists a population of ECs with high levels of CD8+ T cell infiltration as defined by ssGSEA, which is associated with superior OS. This population may represent a lower-risk cohort of CN-high tumors and may have therapeutic implications for treatment decision-making, especially within the context of adjuvant therapy where biomarkers to identify high-risk CN-high ECs are currently inadequate. This contrasts to the CN=L and MSI-H TCGA subtypes, which did not display this relationship. These findings are supported by a recent IHC analysis of PORTEC-1 and PORTEC-2 data by Horeweg and colleagues, demonstrating the association between intraepithelial CD8+ T cell infiltration and survival outcomes, whereby the association was greatest in patients with TP53-mutant tumors (known to be predominantly present in the CN-high subgroup)[39].
We found that ssGSEA was able to identify signature-specific prognostic differences, which in comparison to CIBERSORT, quantifies each cell subset individually using gene lists, rather than relative to the entire population-which may offer an advantage over CIBERSORT. We used the LM22 MATRIX for CIBERSORT [18], which allows RNA-sequencing data to be used for deconvolution rather than microarray-derived transcriptomic data for which CIBERSORT was originally developed. It has been reported that samples analyzed by the LM22 matrix may have a higher frequency of p-values>0.05 [40]. This is largely due to the differing dynamic range of RNA-Sequencing and microarray gene expression data and may not accurately reflect the quality of the deconvolution results [40].
Differences in quantification of the leukocyte subtypes by CIBERSORT and ssGSEA is to be expected given their distinctive methods, assumptions, and leukocyte specific gene sets [28]. Signatures attributed to gene expression are seldom specific to only one cell type, bulk data do not distinguish the location of T cells (i.e., tumor vs stroma) and thus analyses can be influenced by tumor purity of the samples. When comparing these methods head-to-head no one method outperforms others consistently across benchmarking datasets [41]. Differences in deconvolution estimates can be both leukocyte subtype-specific and tumor-specific, suggesting that these methods may need to be further tailored to specific cancer types taking into consideration the optimal leukocyte-specific signatures for each tumor type [36, 42]. The advantage of ssGSEA over CIBERSORT is the assessment of phenotypic differences in cell types allowing for insight into pathways and biologic processes, including for example angiogenesis. This may have greater clinical utility in EC when applying these pathways to more contemporary populations treated with anti-angiogenic and immune modulating therapies.
This study has several limitations, including that it is based on a retrospective subgroup analysis of a singular patient cohort. Due to the exploratory nature of our study, adjustment for multiple testing was not performed to increase the discovery of potential new biological associations and moderate inflation of type error II. We acknowledge the limitation of such approach and reiterate the importance of an independent validation of our findings using independent patient cohorts/ transcriptomic data with treatment and outcome information, particularly in the setting of immune checkpoint blockade are warranted. In addition, the TCGA data set lacks patient-level clinical-pathologic data on some well-described prognostic factors in EC, including lymphovascular space invasion. In addition, these patients were all treated in the pre-ICI era for MMR-deficient/MSI tumors, which likely impacts the survival outcomes of these patients. Finally, we focused on findings consistently identified using both deconvolution methods given that differences were observed between them. The use of orthogonal approaches, including IHC or immunofluorescence, could provide direct information on which of the two deconvolution methods is most representative for the analysis of the TME in ECs.
Validation of our findings using independent patient cohorts/ transcriptomic data with treatment and outcome information, particularly in the setting of immune checkpoint blockade are warranted. In addition, we focused on relationships consistently identified using both deconvolution methods given that differences were observed between both methods. The use of orthogonal methods, including IHC or immunofluorescence, could provide direct information on which of the two methods is most appropriate for analysis of the TME in ECs.
In conclusion, deconvolution of bulk gene expression data using ssGSEA and CIBERSORT allowed for the description of the EC TME according to molecular subtype. Our analysis revealed the existence of a CD8-high subset of CN-high ECs, which may confer significantly longer OS than those with low CD8 expression, while CD8 signatures appeared not to have prognostic significance in other molecular subtypes of EC. These findings highlight that mechanisms of immune escape may be different among the CN-high and CN-low tumors, which may have implications for the development of biomarkers and combination immunotherapies for patients with EC.
Supplementary Material
Supplementary Table 1: CIBERSORT and ssGSEA comparison for leukocyte subsets in endometrial carcinomas obtained from The Cancer Genome Atlas.
Supplementary Figure 1: Kaplan-Meyer estimate of overall survival for ECs by molecular subtype according to CD8+ infiltration defined by CIBERSORT.
Highlights.
Distinct molecular subtypes of endometrial cancer (EC) exhibit divergent immune infiltration signatures
CN-high ECs exhibit the lowest levels of CD8+ T cell infiltration of all molecular subtypes
CD8 T cell infiltration transcriptional signatures are prognostic in CN-high ECs
ACKNOWLEDGEMENTS
Research reported in this publication was supported in part by a Cancer Center Support Grant of the NIH/NCI (Grant No. P30CA008748). D.Z. is supported by the Ovarian Cancer Research Foundation Liz Tilberis Award, and the Department of Defense Ovarian Cancer Research Academy (OC150111). B.W. is supported in part by Cycle for Survival grants. J.S. Reis-Filho and B. Weigelt are funded in part by NIH/NCI P50 CA247749 01 and Breast Cancer Research Foundation grants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICTS OF INTEREST
N.R. Abu-Rustum reports Stryker/ Novadaq and GRAIL grants paid to the institution, outside the current study. J.S. Reis-Filho reports consulting fees from Goldman Sachs, REPARE Therapeutics, Paige.AI and Bain Capital, (ad hoc) membership of the scientific advisory boards of VolitionRx, REPARE Therapeutics, Paige.AI, Roche Tissue Diagnostics, Personalis, Daiichi Sankyo and Merck, and membership of the Board of Directors of Grupo Oncoclinicas, outside the scope of this study. J.S. Reis-Filho has stocks/ stock options with Oncoclinicas and Paige.AI. D. Zamarin reports institutional research support from AstraZeneca, Roche, Plexxikon and Synthekine; consulting fees from Agenus, Hookipa Biotech, Synthekine, Tessa Therapeutics, Xencor, Memgen, Takeda, Celldex, Astellas, AstraZeneca, Cown Biosciences, Roche, GSK, ImmunOS, Kalivir, Synlogic Therapeutics and Targovax; royalties from Merck; and stock options from Accurius Therapeutics, ImmunOS Therapeutics and Calidi Biotherapeutics; and a patent for use of Newcastle Disease Virus for cancer therapy, outside the submitted work. B. Weigelt reports a research grant from REPARE Therapeutics paid to the institution, outside the submitted work. N. Riaz reports research support by Pfizer, REPARE Therapeutics and Invitae, and consulting fees from Illumina and REPARE Therapeutics, outside the scope of this study. The remaining authors have no conflicts of interest to declare.
References
- [1].Ott PA, Bang YJ, Berton-Rigaud D, Elez E, Pishvaian MJ, Rugo HS, et al. Safety and Antitumor Activity of Pembrolizumab in Advanced Programmed Death Ligand 1-Positive Endometrial Cancer: Results From the KEYNOTE-028 Study. J Clin Oncol. 2017;35:2535–41. [DOI] [PubMed] [Google Scholar]
- [2].Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science (New York, NY). 2017;357:409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].O'Malley DM, Bariani GM, Cassier PA, Marabelle A, Hansen AR, De Jesus Acosta A, et al. Pembrolizumab in Patients With Microsatellite Instability-High Advanced Endometrial Cancer: Results From the KEYNOTE-158 Study. J Clin Oncol. 2022;40:752–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Oaknin A, Gilbert L, Tinker AV, Brown J, Mathews C, Press J, et al. Safety and antitumor activity of dostarlimab in patients with advanced or recurrent DNA mismatch repair deficient/microsatellite instability-high (dMMR/MSI-H) or proficient/stable (MMRp/MSS) endometrial cancer: interim results from GARNET-a phase I, single-arm study. J Immunother Cancer. 2022;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Makker V, Taylor MH, Aghajanian C, Oaknin A, Mier J, Cohn AL, et al. Lenvatinib Plus Pembrolizumab in Patients With Advanced Endometrial Cancer. J Clin Oncol. 2020;38:2981–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].N. Cancer Genome Atlas Research, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15:10–7. [DOI] [PubMed] [Google Scholar]
- [8].Setiawan VW, Yang HP, Pike MC, McCann SE, Yu H, Xiang YB, et al. Type I and II endometrial cancers: have they different risk factors? J Clin Oncol. 2013;31:2607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Momeni-Boroujeni A, Dahoud W, Vanderbilt CM, Chiang S, Murali R, Rios-Doria EV, et al. Clinicopathologic and Genomic Analysis of TP53-Mutated Endometrial Carcinomas. Clin Cancer Res. 2021;27:2613–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Momeni-Boroujeni A, Nguyen B, Vanderbilt CM, Ladanyi M, Abu-Rustum NR, Aghajanian C, et al. Genomic landscape of endometrial carcinomas of no specific molecular profile. Mod Pathol. 2022;35:1269–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gooden MJ, de Bock GH, Leffers N, Daemen T, Nijman HW. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. Br J Cancer. 2011;105:93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Swartz MA, Iida N, Roberts EW, Sangaletti S, Wong MH, Yull FE, et al. Tumor microenvironment complexity: emerging roles in cancer therapy. Cancer Res. 2012;72:2473–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chang Q, Hedley D. Emerging applications of flow cytometry in solid tumor biology. Methods. 2012;57:359–67. [DOI] [PubMed] [Google Scholar]
- [14].Finotello F, Trajanoski Z. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol Immunother. 2018;67:1031–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Guo F, Dong Y, Tan Q, Kong J, Yu B. Tissue Infiltrating Immune Cells as Prognostic Biomarkers in Endometrial Cancer: A Meta-Analysis. Dis Markers. 2020;2020:1805764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell. 2017;171:934–49 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–95. [DOI] [PubMed] [Google Scholar]
- [20].Heller G, Venkatraman ES. Resampling Procedures to Compare Two Survival Distributions in the Presence of Right-Censored Data Biometrics. 1996;52:1204–13. [Google Scholar]
- [21].Shia J, Black D, Hummer AJ, Boyd J, Soslow RA. Routinely assessed morphological features correlate with microsatellite instability status in endometrial cancer. Hum Pathol. 2008;39:116–25. [DOI] [PubMed] [Google Scholar]
- [22].Suemori T, Susumu N, Iwata T, Banno K, Yamagami W, Hirasawa A, et al. Intratumoral CD8+ Lymphocyte Infiltration as a Prognostic Factor and Its Relationship With Cyclooxygenase 2 Expression and Microsatellite Instability in Endometrial Cancer. Int J Gynecol Cancer. 2015;25:1165–72. [DOI] [PubMed] [Google Scholar]
- [23].Imboden S, Nastic D, Ghaderi M, Rydberg F, Rau TT, Mueller MD, et al. Phenotype of POLE-mutated endometrial cancer. PLoS One. 2019;14:e0214318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hussein YR, Weigelt B, Levine DA, Schoolmeester JK, Dao LN, Balzer BL, et al. Clinicopathological analysis of endometrial carcinomas harboring somatic POLE exonuclease domain mutations. Mod Pathol. 2015;28:505–14. [DOI] [PubMed] [Google Scholar]
- [25].Raffone A, Travaglino A, Raimondo D, Boccellino MP, Maletta M, Borghese G, et al. Tumor-infiltrating lymphocytes and POLE mutation in endometrial carcinoma. Gynecol Oncol. 2021;161:621–8. [DOI] [PubMed] [Google Scholar]
- [26].Petitprez F, Vano YA, Becht E, Giraldo NA, de Reynies A, Sautes-Fridman C, et al. Transcriptomic analysis of the tumor microenvironment to guide prognosis and immunotherapies. Cancer Immunol Immunother. 2018;67:981–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bense RD, Sotiriou C, Piccart-Gebhart MJ, Haanen J, van Vugt M, de Vries EGE, et al. Relevance of Tumor-Infiltrating Immune Cell Composition and Functionality for Disease Outcome in Breast Cancer. J Natl Cancer Inst. 2017;109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hendry S, Salgado R, Gevaert T, Russell PA, John T, Thapa B, et al. Assessing Tumor-Infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method from the International Immuno-Oncology Biomarkers Working Group: Part 2: TILs in Melanoma, Gastrointestinal Tract Carcinomas, Non-Small Cell Lung Carcinoma and Mesothelioma, Endometrial and Ovarian Carcinomas, Squamous Cell Carcinoma of the Head and Neck, Genitourinary Carcinomas, and Primary Brain Tumors. Adv Anat Pathol. 2017;24:311–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nederlof I, De Bortoli D, Bareche Y, Nguyen B, de Maaker M, Hooijer GKJ, et al. Comprehensive evaluation of methods to assess overall and cell-specific immune infiltrates in breast cancer. Breast Cancer Res. 2019;21:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pauken KE, Wherry EJ. Overcoming T cell exhaustion in infection and cancer. Trends Immunol. 2015;36:265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cai Y, Chang Y, Liu Y. Multi-omics profiling reveals distinct microenvironment characterization of endometrial cancer. Biomed Pharmacother. 2019;118:109244. [DOI] [PubMed] [Google Scholar]
- [33].Barnes TA, Amir E. HYPE or HOPE: the prognostic value of infiltrating immune cells in cancer. Br J Cancer. 2017;117:451–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. [DOI] [PubMed] [Google Scholar]
- [35].Mehnert JM, Panda A, Zhong H, Hirshfield K, Damare S, Lane K, et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Invest. 2016;126:2334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].van Gool IC, Eggink FA, Freeman-Mills L, Stelloo E, Marchi E, de Bruyn M, et al. POLE Proofreading Mutations Elicit an Antitumor Immune Response in Endometrial Cancer. Clin Cancer Res. 2015;21:3347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Victoor J, Borght SV, Spans L, Lehnert S, Brems H, Laenen A, et al. Comprehensive immunomolecular profiling of endometrial carcinoma: A tertiary retrospective study. Gynecol Oncol. 2021;162:694–701. [DOI] [PubMed] [Google Scholar]
- [38].Talhouk A, Derocher H, Schmidt P, Leung S, Milne K, Gilks CB, et al. Molecular Subtype Not Immune Response Drives Outcomes in Endometrial Carcinoma. Clin Cancer Res. 2019;25:2537–48. [DOI] [PubMed] [Google Scholar]
- [39].Horeweg N, de Bruyn M, Nout RA, Stelloo E, Kedziersza K, Leon-Castillo A, et al. Prognostic Integrated Image-Based Immune and Molecular Profiling in Early-Stage Endometrial Cancer. Cancer Immunol Res. 2020;8:1508–19. [DOI] [PubMed] [Google Scholar]
- [40].Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jimenez-Sanchez A, Cast O, Miller ML. Comprehensive Benchmarking and Integration of Tumor Microenvironment Cell Estimation Methods. Cancer Res. 2019;79:6238–46. [DOI] [PubMed] [Google Scholar]
- [42].Racle J, de Jonge K, Baumgaertner P, Speiser DE, Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: CIBERSORT and ssGSEA comparison for leukocyte subsets in endometrial carcinomas obtained from The Cancer Genome Atlas.
Supplementary Figure 1: Kaplan-Meyer estimate of overall survival for ECs by molecular subtype according to CD8+ infiltration defined by CIBERSORT.