Abstract
Background & Aims
T helper 1 (Th1) effector cells are implicated in inflammatory bowel disease. The stimulator of interferon genes (STING), an intracellular DNA sensor, has been shown to regulate infection and various cancers. However, whether and how intrinsic STING signaling in Th1 cells regulates colitis is still unknown.
Methods
Dextran sodium sulfate–induced colitis and wild-type/STING-deficient CD4+T cell adoptive transfer models were used to analyze the role of STING in regulating colitis. The effect of STING on Th1 cells was determined by flow cytometry, RNA sequencing, metabolic assays, and mitochondrial functions. 16S ribosomal RNA sequencing and germ-free mice were used to investigate whether the microbiota were involved. The in vivo effect of STING agonist in murine colitis was determined. The expression and role of STING in human T cells were also determined.
Results
Activation of STING transformed proinflammatory IFNγ+Th1 cells into IL-10+IFNγ+Th1 cells, which were dramatically less pathogenic in inducing colitis. STING promoted Th1 interleukin (IL)-10 production by inducing STAT3 translocation into nuclear and mitochondria, which promoted Blimp1 expression and mitochondrial oxidation, respectively. Blockade of glucose or glutamine-derived oxidation, but not lipid-derived oxidation, suppressed STING induction of IL-10. Gut microbiota were changed in STING–/– mice, but the altered microbiota did not mediate STING effects on intestinal CD4+T cell production of IL-10. Translationally, STING agonists suppressed both acute and chronic colitis. Intestinal STING+ CD4+T cells were increased in inflammatory bowel disease patients, and STING agonists upregulated IL-10 production in human CD4+T cells.
Conclusions
These findings establish a crucial role of T cell–intrinsic STING in switching off the pathogenic programs of Th1 cells in intestinal inflammation.
Keywords: STING, Th1 Cells, Colitis, Mitochondrial Oxidation
Graphical abstract
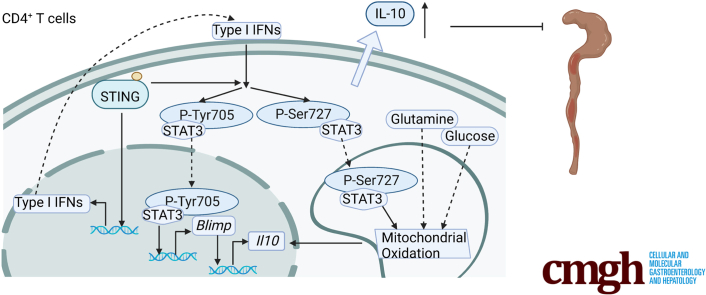
Summary.
STING decreases the pathogenecity of effector Th1 cells in inducing colitis by upregulating the anti-inflammatory cytokine IL-10 through STAT3 translocation into nuclear and mitochondria, which promotes Blimp1 expression and mitochondrial oxidation, respectively.
The role of CD4+ T cells in host protective and homeostatic responses has been well established.1, 2, 3 Different subsets of T cells have been shown to play various roles in mediating autoimmune inflammatory diseases.4 T helper (Th) 1 effector cells mediate the pathogenesis of diabetes, rheumatoid arthritis, and inflammatory bowel disease (IBD), mainly through the secretion of proinflammatory cytokines.5,6 It is well established that several factors contribute to the inflammatory program of Th1 effector cells, including transcriptional factors, microbiota, tissue inflammatory milieu, and cellular metabolism.7, 8, 9 There are also multiple mechanisms for restricting excessive effector T cell responses, including the critical anti-inflammatory cytokine interleukin (IL)-10 produced by many immune cells.10 T effector cell production of IL-10 has been considered a self-limiting mechanism to prevent an exaggerated T cell response, which otherwise would be detrimental.11 However, the intrinsic factors that switch off the inflammatory programs in Th1 cells are still not completely understood.
The detection of pathogen-derived nucleic acid is the key feature of innate immunity. cGAS, 1 of 3 major DNA sensors, protects the host from pathogens by inducing type I interferon production and interferon-stimulated genes, mediated by stimulator of interferon genes (STING).12 Accumulating data have also indicated the role of STING in adaptive immune responses in tumors and autoimmunity as well as in intestinal inflammation and colorectal tumorigenesis.13, 14, 15, 16, 17 However, the role of STING in intestinal homeostasis is still largely unclear. STING signaling has been reported to promote apoptosis in CD4+ T cells18,19 and regulate CD4+ cell differentiation and function.20,21 However, the role of intrinsic STING signaling in regulating Th1 cell function to maintain intestinal homeostasis remains unclear.
In this study, we demonstrate that STING switches off the pathogenic program of Th1 cells by transforming proinflammatory Th1 cells into IL-10–producing Th1 cells. STING-deficient T cells induce more severe colitis, and STING agonist-pretreated Th1 cells induce less severe colitis mediated by IL-10. Both transcriptional regulation and metabolic modulation mediate STING upregulation of the Th1 cell IL-10 program.
Results
STING-Deficient CD4+ T Cells Are More Pathogenic in Inducing Intestinal Inflammation
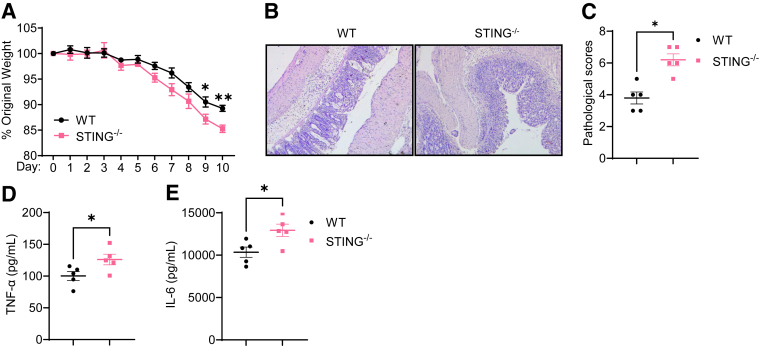
To investigate the role of STING signaling in regulating intestinal homeostasis, we first induced colitis in wild-type (WT) and STING–/– mice using dextran sulfate sodium (DSS). Mouse weights were monitored daily. We found that STING–/– mice showed more weight loss than WT mice (Figure 1A). When the mice were sacrificed, the severity of intestinal inflammation was determined. STING–/– mice demonstrated more cell infiltration in the mucosa and submucosal, as well as more severe ulceration in the intestine compared with WT mice (Figure 1B and C). In addition, proinflammatory cytokines, including tumor necrosis factor α (TNF-α) and IL-6, were higher in the colon of STING–/– mice (Figure 1D and E). These data suggested that deficiency of STING aggravates colitis.
Figure 1.
STING–/–mice are more susceptible to DSS-induced acute colitis. WT mice and STING–/– mice were administered 2% DSS (w/v) in drinking water for 7 days and then normal drinking for additional 3 days. Mice were sacrificed on day 10. (A) Weight change. (B) Representative colonic histological images. (C) Pathological scores. (D) Intestinal TNF-α production. (E) Intestinal IL-6 production. Data were shown as mean ± SEM. One representative of 2 independent experiments. (A, D, E) Unpaired Student’s t test; (C) Mann-Whitney U test. ∗P < .01, ∗∗P < .01.
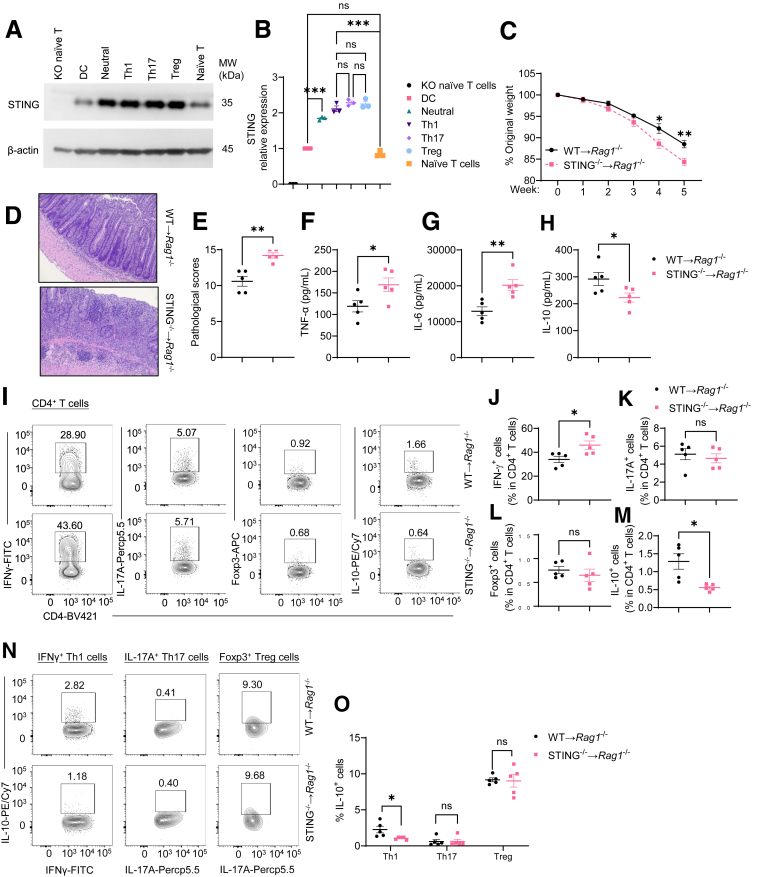
Given that intestinal immune responses are critical in the pathogenesis of colitis and that both innate and adaptive immune cells express STING, we then compared the STING expression in dendritic cells and different subsets of CD4+ T cells. As shown in Figure 2A and B, naïve CD4+ T cells expressed STING at similar levels with dendritic cells. STING expression was increased once the CD4+ T cells were activated. Furthermore, there was no significant difference in STING expression among Th1, Th17, and regulatory T (Treg) cells (Figure 2A and B). CD4+ T cells have been shown crucial in mediating colitis development. Considering that CD4+ T cells, especially activated T cells, highly expressed STING, we determined whether STING altered CD4+ T cells’ pathogenicity to induce colitis. WT and STING-deficient CD4+ CD45Rbhi T cells were transferred into Rag1–/– mice, and the recipient mice were sacrificed 5 weeks post-cell transfer. We found that STING-deficient CD4+ T cells induced more severe colitis, as shown by more weight loss (Figure 2C), more cell infiltrates and higher pathological scores (Figure 2D and E), and increased levels of TNF-α and IL-6 in the intestines (Figure 2F and G). Interestingly, intestinal production of IL-10 was decreased in the recipients of STING-deficient CD4+ T cells (Figure 2H). To define the role of STING in regulating CD4+ T cell responses under inflammatory conditions, we determined the phenotypes of intestinal CD4+ T cells by flow cytometry. We found that the STING-deficient CD4+ T cells differentiated into more IFNγ-producing Th1 cells, but not IL-17–producing Th17 cells and Foxp3+ Treg cells, in the colon of Rag1–/– mice (Figure 2I–L). In addition, IL-10 production was decreased in intestinal CD4+ T cells of Rag1–/– recipients of STING-deficient CD4+ T cells (Figure 2I and M). Specifically, STING-deficient Th1 cells, but not Th17 and Treg cells, produced a decreased level of IL-10 (Figure 2N and O). Overall, these data indicated that STING protects against intestinal inflammation by modulating CD4+ T cell responses.
Figure 2.
STING-deficient CD4+T cells induce more severe colitis in Rag1–/–mice. (A, B) STING and β-actin levels were determined in WT dendritic cells (DCs), naïve WT T cells, naïve STING–/– (knockout [KO]) T cells, and WT T cells cultured on neutral, Th1, Th17, and Treg conditions by Western blot (n = 3/group). (A) Western blot bands. (B) STING relative expression. (C–O) WT or STING-deficient CD4+ CD45Rbhi T cells were intravenously transferred into Rag1–/– mice (n = 5/group). The recipient Rag1–/– mice were sacrificed 5 weeks later. (C) The weight changes of the recipient Rag1–/– mice. (D) Representative intestinal histological images. (E) Pathological scores. (F) Intestinal TNF-α production. (G) Intestinal IL-6 production. (H) Intestinal IL-10 production. (I) Representative flow cytometry plots of intestinal IFNγ+, IL-17A+, Foxp3+, and IL-10+ CD4+ T cells. (J–M) Dot plots of IFNγ+, IL-17A+, Foxp3+, and IL-10+ CD4+ T cells. (N) Representative flow cytometry plots of IL-10+ Th1, IL-10+ Th17, and IL-10+ Treg cells. (O) Dot plots of IL-10+ Th1, IL-10+ Th17, and IL-10+ Treg cells. Data were shown as mean ± SEM. (A, B) Data were pooled from 3 independent experiments. (C–O) One representative of 3 independent experiments. (A) 1-way analysis of variance with Tukey’s multiple comparisons test; (C, F, G, I, J, K, L, M, N) unpaired Student’s t test; (E) Mann-Whitney U test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. ns, not significant.
STING Limits the Pathogenecity of Th1 Effector Cells via IL-10 Production
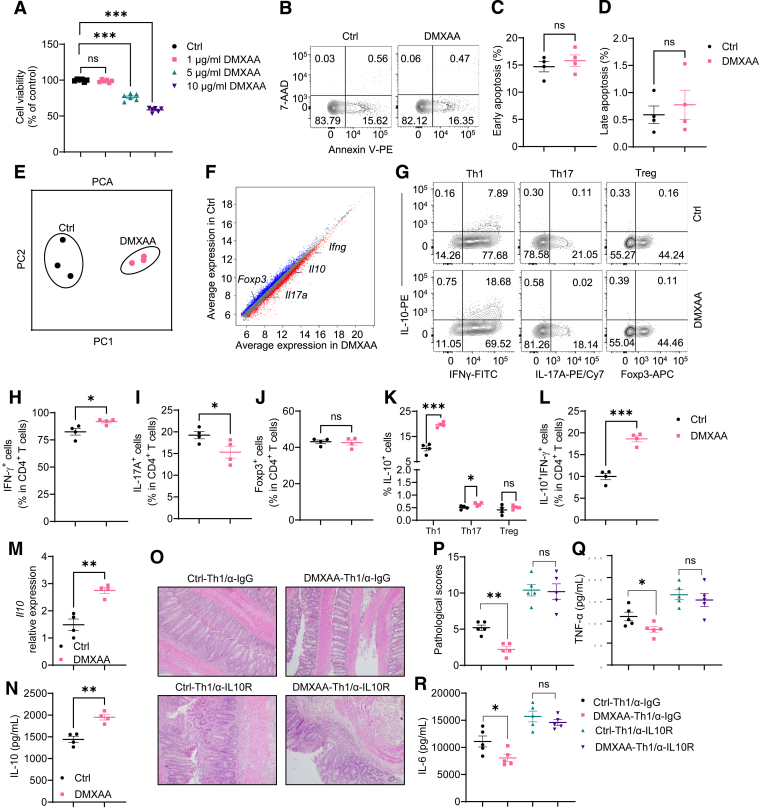
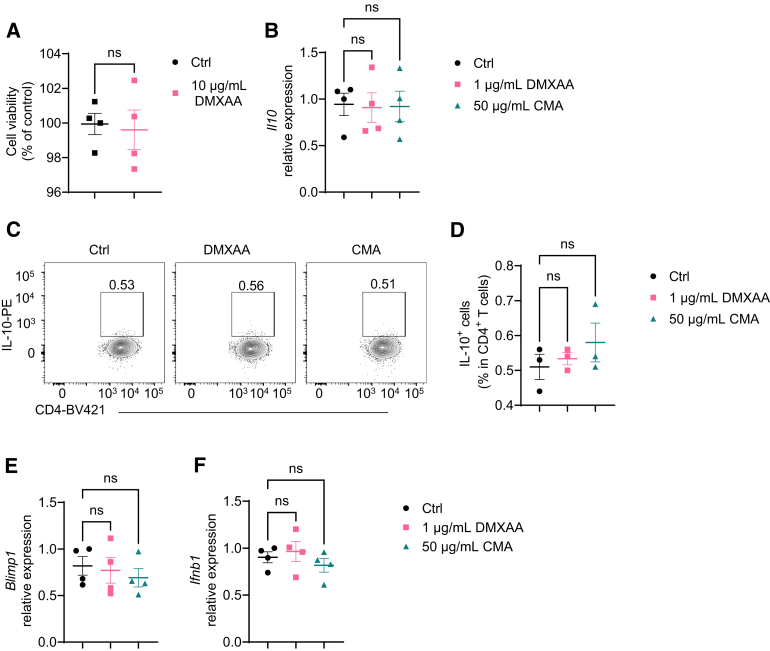
To explore the role of STING in CD4+ T cell responses, we first determined whether STING affects T cell viability. Consistent with the previous studies,18,19 10 μg/mL of DMXAA, a STING agonist, reduced WT, but not STING-deficient, T cell viability (Figures 3A and 4A). In addition, DMXAA reduction of viability was dose-dependent in WT T cells. 1 μg/mL of DMXAA did not affect WT T cell viability (Figure 3A). Furthermore, we found that 1 μg/mL of DMXAA did not affect WT CD4+ T cell apoptosis (Figure 3B–D). Therefore, we chose the DMXAA at the concentration of 1 μg/mL in all the following experiments in this study.
Figure 3.
STING agonist suppresses Th1 pathogenicity in inducing colitis through upregulation of IL-10. (A) Mouse splenic CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA at the concentration of 1 μg/mL, 5 μg/mL, or 10 μg/mL. The cell viability was determined by the Resazurin viability assay 2 days later (n = 6/group). (B–D) Mouse splenic CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (1 μg/mL) for 2 days (n = 4/group). (B) Representative flow cytometry plots of apoptosis in T cells. (C, D) Dot plots of early and late apoptosis levels. (E, F) Mouse splenic CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (1 μg/mL) for 2 days. Cells were collected for bulk RNA sequencing (n = 3/group). (E) Principal component analysis (PCA) of gene profile. (F) Scatter plot of gene expression. (G–L) Mouse splenic CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (1 μg/mL) under Th1, Th17, or Treg conditions, for 5 days (n = 4/group). (G) Representative flow cytometry plots of IFNγ+, IL-17A+, Foxp3+, and IL-10+ CD4+ T cells. (H–L) Dot plots of IFNγ+, IL-17A+, Foxp3+, and IL-10+ CD4+ T cells, and IL-10+ IFNγ+ CD4+ T cells. (M, N) Mouse splenic CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (1 μg/mL) under Th1 conditions for 2 days (n = 4/group). (M) Il10 relative expression. (N) IL-10 production in cell culture supernatants. (O–R) Control or DMXAA-pretreated Th1 cells were intravenously transferred to Rag1–/– mice. The recipient Rag1–/– mice were peritoneally injected with anti-IL-10R antibody (25 mg/kg) or anti-IgG as control every other day (n = 5/group). The recipient Rag1–/– mice were sacrificed after 6 weeks. (O) Representative intestinal histological images. (P) Pathological scores. (Q) Intestinal TNF-α production. (R) Intestinal IL-6 production. Data were shown as mean ± SEM. One representative of (O–R) 2 or (A–D, G–N) 3 independent experiments. (A) One-way analysis of variance with Dunnett's multiple comparisons test; (C, D, H, I, J, K, L, M, N, Q, R) unpaired Student’s t test; (P) Mann-Whitney U test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
Figure 4.
STING agonists do not act on STING-deficient T cells. (A) STING-deficient CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (10 μg/mL). The cell viability was determined by the Resazurin viability assay 2 days later (n = 4/group). (B–F) STING-deficient CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (1 μg/mL) or CMA (50 μg/mL) under Th1 conditions. (B) Il10 relative expression on day 2. (C) Representative flow cytometry plots of IL-10+ CD4+ T cells on day 5. (D) Dot plots of IL-10+ CD4+ T cells on day 5. (E) Blimp1 relative expression on day 2. (F) Ifnb relative expression on day 2. Data were shown as mean ± SEM. One representative of 2 independent experiments. (A) Unpaired Student’s t test; (B, D, E, F) 1-way analysis of variance with Dunnett's multiple comparisons test.
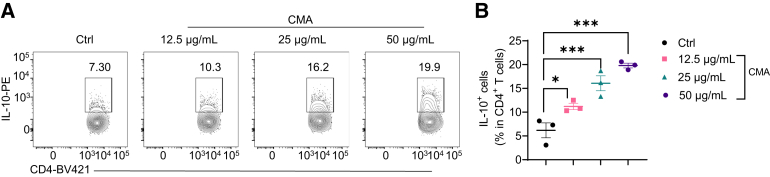
To determine how STING regulates T cell gene expression, we performed a transcriptomics analysis. We activated splenic CD4+ T cells with anti-CD3 and anti-CD28 antibodies in the presence or absence of DMXAA under neutral conditions for 2 days and determined gene expressions by RNA sequencing. As shown in Figure 3E, DMXAA altered the gene expression profile of CD4+ T cells. DMXAA increased the expression of Ifng and decreased the expression of Foxp3 but had no effect on Il17a (Figure 3F). Interestingly, DMXAA promoted T cell expression of Il10 (Figure 3F), which is critical for restraining the T cell inflammatory program to inhibit inflammation and maintain homeostasis.11,22 To further investigate whether DMXAA affects T cell differentiation, we cultured the CD4+ T cells in the presence or absence of DMXAA under Th1, Th17, or Treg conditions. DMXAA slightly promoted Th1 polarization and decreased Th17 differentiation (Figure 3G–I). There was no difference in Foxp3 expression in T cells treated with or without DMXAA under Treg conditions (Figure 3G and J), suggesting that STING does not affect Treg differentiation. Although IL-10 production was increased in T cells under Th1 and Th17 conditions, the STING induction of IL-10 was much more striking in Th1 cells than in Th17 cells (Figure 3G and K). Interestingly, STING agonist switched the Th1 cell pattern from IFNγ+ T cells to IFNγ+ IL-10+ T cells (Figure 3G and L), suggesting that STING may transform proinflammatory Th1 cells into suppressive IL-10–producing Th1 cells. In line with these data, DMXAA increased Il10 messenger RNA (mRNA) levels in T cells under Th1 conditions (Figure 3M), and IL-10 production was increased in culture supernatants in T cells treated with DMXAA under Th1 conditions (Figure 3N). In addition, CMA, another STING agonist, also promoted T cell IL-10 production under Th1 conditions (Figure 5A and B). DMXAA and CMA did not affect IL-10 mRNA expression and production in STING–/– T cells (Figure 4B–D), indicating that these 2 agonists act on T cells specifically through activation of STING pathway.
Figure 5.
CMA promotes IL-10 production in Th1 cells. Mouse splenic CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of CMA at indicated concentrations under Th1 conditions for 5 days (n = 4/group). (G) Representative flow cytometry plots of IL-10+ CD4+ T cells. (H–L) Dot plots of IL-10+ IFNγ+ CD4+ T cells. Data were shown as mean ± SEM. One representative of 3 independent experiments. (B) One-way analysis of variance with Dunnett's multiple comparisons test. ∗P < .05, ∗∗∗P < .001.
To determine whether treatment with STING agonist affects the pathogenicity of Th1 cells in inducing colitis, we treated T cells with or without DMXAA under Th1 conditions for 5 days and then transferred these T cells into Rag1–/– mice. To define the role of STING agonist–induced T cell IL-10 production in the regulation of colitis development, we treated groups of Rag1–/– recipients with an anti-IL-10 receptor antibody or anti-IgG antibody as controls. Five weeks post–cell transfer, mice were sacrificed for analysis of intestinal inflammation. DMXAA-pretreated Th1 cells induced less severe colitis than control Th1 cells in Rag1–/– recipients treated with anti-IgG antibody, as demonstrated by lower pathological scores and decreased intestinal proinflammatory cytokines (Figure 3O–R). Treatment of anti-IL10R antibody aggravated intestinal inflammation in Rag1–/– recipients of control Th1 cells or DMXAA-pretreated Th1 cells. There were no differences in the pathological score and intestinal proinflammatory cytokines between these 2 groups of mice (Figure 3O–R). These data suggest that STING limits the pathogenecity of effector Th1 cells via IL-10 production.
Blimp1 Mediates the STING Regulation of Th1 Cell IL-10 Production
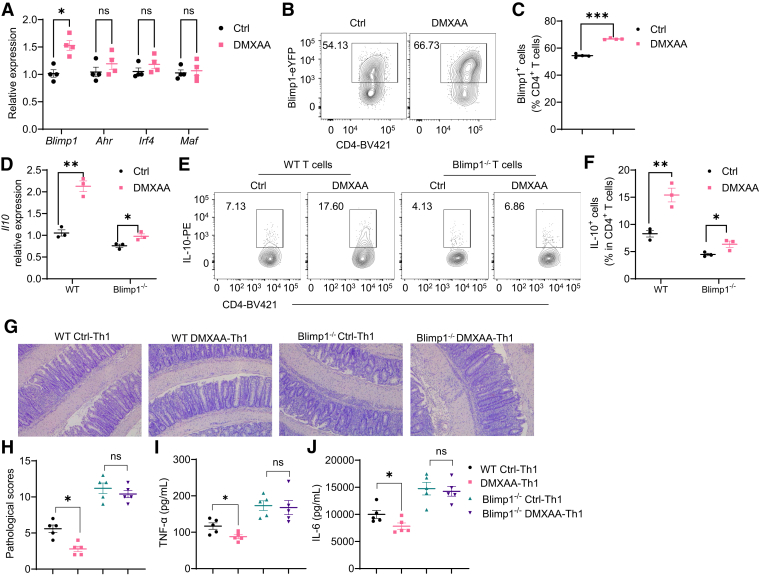
To investigate how STING promotes Th1 production of IL-10, we determined the expression of key transcription factors associated with IL-10, including Blimp1, AhR, IRF4, and c-Maf,23 in T cells treated with DMXAA under Th1 conditions. We found that DMXAA promoted T cell expression of Blimp1 but not AhR, Irf4, and Maf (Figure 6A). Consistently, DMXAA increased Blimp1 protein expression in T cells (Figure 6B and C). DMXAA and CMA did not affect Blimp1 expression in STING–/– T cells (Figure 4E).To investigate whether Blimp1 mediates the STING induction of IL-10 in T cells, we treated WT and Blimp1-deficient CD4+ T cells with or without DMXAA and found that the STING induction of IL-10 was compromised in Blimp1-deficient CD4+ T cells compared with WT T cells at both mRNA and protein levels (Figure 6D–F), indicating that Blimp1 mediates IL-10 production induced by STING signaling.
Figure 6.
Blimp1 mediates STING regulation of Th1 cell pathogenicity. (A, B) Mouse splenic CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (1 μg/mL) under Th1 conditions for 2 days (n = 4/group), and Blimp1, Ahr, Irf4, and Maf were determined by quantitative reverse-transcriptase polymerase chain reaction. (B, C) Splenic CD4+ T cells were isolated from Blimp1-eYFP mice and activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (1 μg/mL) under Th1 conditions for 3 days (n = 4/group). (B) Representative flow cytometry plots of Blimp1+ CD4+ T cells. (C) Dot plots of Blimp1+ CD4+ T cells. (D-F) WT and Blimp1-deficient CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (1 μg/mL) under Th1 conditions for (D) 2 or (E, F) 5 days (n = 3/group). (D) Il10 relative expression was determined. (E) Representative flow cytometry plots of IL-10+ CD4+ T cells. (F) Dot plots of IL-10+ CD4+ T cells. (G–J) Control or DMXAA-pretreated WT or Blimp1-deficient Th1 cells were intravenously transferred to Rag1–/– mice. The recipient Rag1–/– mice were sacrificed after 6 weeks (n = 5/group). (G) Representative intestinal histological images. (H) Pathological scores. (I) Intestinal TNF-α production. (J) Intestinal IL-6 production. Data were shown as mean ± SEM. One representative of (G–J) 2 or (A–F) 3 independent experiments. (A, C, D, F, I, J) Unpaired Student’s t test; (H) Mann-Whitney U test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
To investigate whether Blimp1 mediates STING limitation of effector Th1 cell pathogenecity in inducing colitis, we transferred DMXAA-pretreated or control WT and Blimp1-deficient Th1 cells into Rag1–/– mice. Intestinal inflammation was determined 5 weeks later. Consistent with the data in Figure 3, Rag1–/– recipients of DMXAA-pretreated WT Th1 cells developed less severe colitis and produced lower TNF-α and IL-6 than Rag1–/– recipients of control WT Th1 cells (Figure 6G–J). However, DMXAA-pretreated Blimp1-deficient Th1 cells induced similar levels of colitis and produced similar levels of TNF-α and IL-6 compared with control Blimp1-deficient T cells (Figure 6G–J). Overall, these data suggest that Blimp1 is indispensable in STING protection against colitis induced by T cells.
Type I IFN/STAT3 Pathway Partially Regulates STING-Induced IL-10 in Th1 Cells
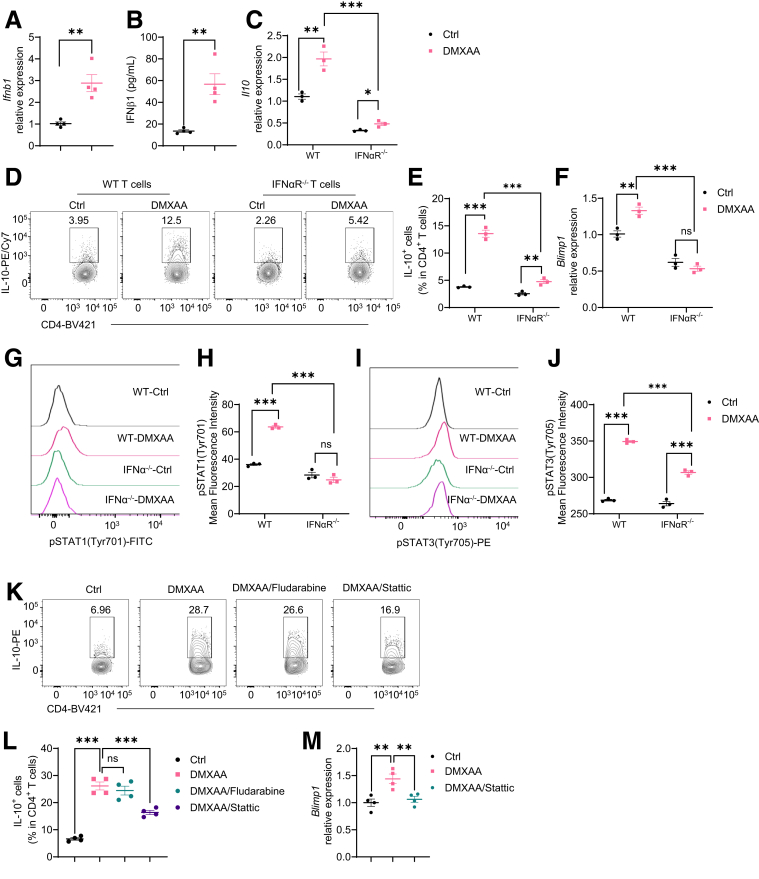
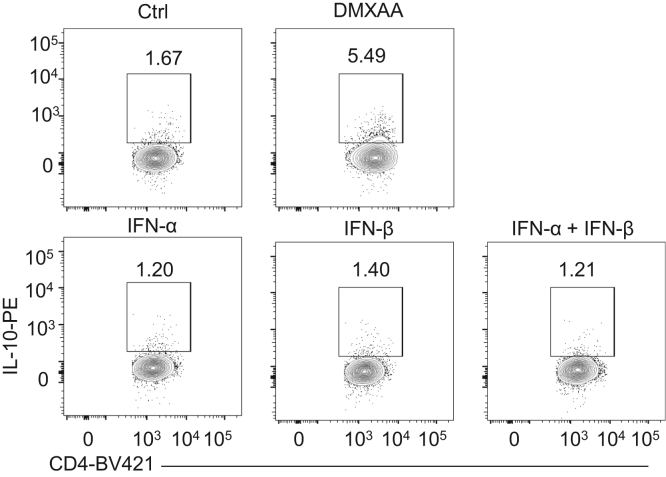
As STING activates type I IFN signaling,24 we investigated whether type I IFNs mediate STING induction of IL-10 in T cells. We found that treatment with DMXAA and CMA significantly increased IFNβ, one of the major type I IFNs, at mRNA and protein levels in WT T cells under Th1 conditions (Figure 7A and B), while DMXAA and CMA did not affect IFNβ expression in STING–/– T cells (Figure 4F). We then treated type I IFN receptor IFNα receptor–deficient and WT T cells with DMXAA under Th1 conditions and found that although DMXAA induced IL-10 production in IFNα receptor–deficient T cells, the induction level was compromised (Figure 7C–E). However, IFNα and IFNβ failed to increase IL-10 production in WT T cells (Figure 8). These data indicated that type I IFNs together with other pathways mediate STING induction of IL-10 production in T cells. Interestingly, the STING agonist failed to increase Blimp1 expression in IFNαR-deficient T cells (Figure 7F). Collectively, these data suggest that type I IFNs mediate the STING induction of IL-10 through the Blimp1 pathway.
Figure 7.
Type I IFN/STAT3 pathway is involved in STING induction of IL-10 production in Th1 cells. (A, B) Mouse splenic CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (1 μg/mL) under Th1 conditions for 2 days (n = 4/group). (A) Ifnb1 relative expression. (B) IFNβ1 production in cell culture supernatants. (C-F) WT and IFNαR-deficient CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (1 μg/mL) under Th1 conditions for (C, F) 2 or (D, E) 5 days (n = 3/group). (C) Il10 relative expression was determined. (D) Representative flow cytometry plots of IL-10+ CD4+ T cells. (E) Dot plots of IL-10+ CD4+ T cells. (F) Blimp1 relative expression. (G–J) WT and IFNα receptor (IFNαR)–deficient CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (1 μg/mL) under Th1 conditions for 48 hours. (G) Representative flow cytometry plots of phosphorylated STAT1 (pSTAT1) site Tyr701. (H) Dot plots of pSTAT1 Tyr701 mean fluorescence intensity. (I) Representative flow cytometry plots of phosphorylated STAT3 (pSTAT3) site Tyr705. (J) Dot plots of pSTAT3 Tyr705 mean fluorescence intensity. (K–M) Mouse splenic CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (1 μg/mL) with or without fludarabine (2.5 μM)/static (2.5 μM) under Th1 conditions for (M) 2 or (K, L) 5 days. (K) Representative flow cytometry plots of IL-10+ CD4+ T cells. (L) Dot plots of IL-10+ CD4+ T cells. (M) Blimp1 relative expression. Data were shown as mean ± SEM. One representative of 3 independent experiments. (A, B, C, E, F, H, J) Unpaired Student’s t test; (L, M) 1-way analysis of variance with Dunnett's multiple comparisons test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
Figure 8.
Type I IFN alone does not induce IL-10 production in T cells. Mouse splenic CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (1 μg/mL), IFNα (5 ng/mL), IFNβ (5 ng/mL), or IFNα (5 ng/mL) and IFNβ (5 ng/mL) for 5 days. Representative flow cytometry plots of IL-10+ CD4+ T cells. Data were shown as mean ± SEM. One representative of 2 independent experiments.
Next, we defined how type I IFN regulates Blimp1 expression induced by STING. As STAT1 and STAT3 can be activated in response to STING/type I IFN signaling,25,26 we first determined if STING activates STAT1 and STAT3 activation in CD4+ T cells. We found that DMXAA promoted the phosphorylation of STAT1 tyrosine (Tyr)701 and STAT3 site Tyr705 in WT T cells (Figure 7G–J). However, DMXAA failed to increase the phosphorylation of STAT1 Tyr701, and DMXAA induction of STAT3 Tyr705 was compromised in IFNα receptor–deficient T cells (Figure 7G–J). These data indicate that type I IFNs are indispensable in STING activation of STAT1, and STING activates STAT3 is partially dependent on the type I IFN pathway. To define whether STAT1 and/or STAT3 mediate STING induction of IL-10 production, we treated T cells with DMXAA under Th1 conditions in the presence of STAT1 inhibitor, Fludarabine, or STAT3 inhibitor, Stattic. Blockade of the STAT3 suppressed IL-10 production induced by DMXAA (Figure 7K and L). However, inhibiting STAT1 by Fludarabine did not reduce the effect of DMXAA on IL-10 production (Figure 7K and L), which indicates that STAT3 might be more important in the STING induction of IL-10 in T cells. In addition, the STAT3 inhibitor decreased Blimp1 expression (Figure 7M). These data indicate that STAT3 mediates the STING induction of IL-10 in Th1 cells.
STING Regulates T Cell Metabolism to Promote IL-10 Expression in Th1 Cells
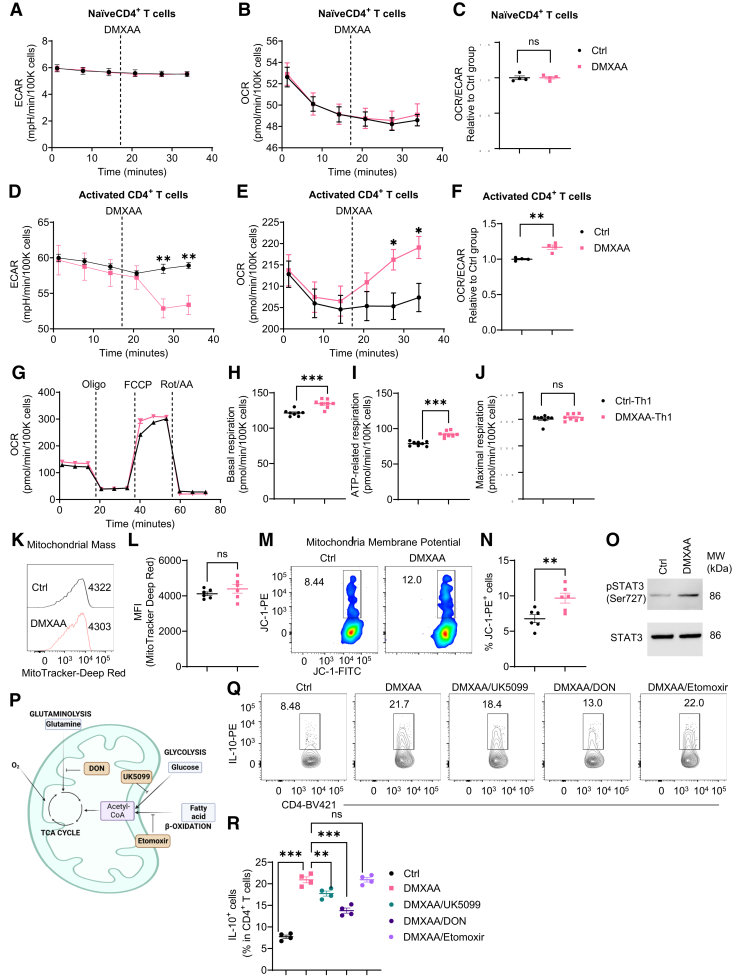
Cell metabolism has been reported to control T cell differentiation and fate.8,27 Therefore, we investigated whether STING affects T cell IL-10 production by regulating cell metabolic pathways. First, we measured the effect of STING agonist on glycolysis and mitochondrial oxidation in naïve and activated CD4+ T cells in real time using an extracellular flux Seahorse analyzer. We found that DMXAA did not affect the extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) in naïve T cells, which primarily represent glycolysis and mitochondrial oxidation individually (Figure 9A–C). However, DMXAA downregulated ECAR but upregulated OCR, leading to an increased OCR/ECAR ratio in activated T cells (Figure 9D–F), suggesting that STING agonist increases mitochondrial activity in activated T cells but not in naïve T cells. Then, we analyzed the key parameters of mitochondrial oxidation in control and DMXXAA-treated Th1 cells using the Mito stress test kit. DMXAA-treated Th1 cells showed higher levels of basal and adenosine triphosphate–related respiration and a similar level of maximum respiration compared with control Th1 cells (Figure 9G–J). Furthermore, DMXAA-treated Th1 cells displayed an increase in mitochondrial membrane potential but not mitochondrial mass (Figure 9K–N). Additionally, DMXAA promoted the phosphorylation of mitochondrial STAT3 site serine (Ser)727 (mitoSTAT3) (Figure 9O), which has been found to localize mitochondria and promote mitochondrial respiration.28, 29, 30 Overall, these data suggest that STING increases mitochondrial oxidation in Th1 cells through activation of mitoSTAT3.
Figure 9.
STING agonist modulates T cell metabolism to upregulate IL-10 production in Th1 cells. (A–C) The real-time levels of (A) ECAR and (B) OCR in splenic naïve CD4+ T cells were measured before and after treatment of DMXAA (1 μg/mL) using an extracellular flux Seahorse analyzer (n = 4/group). (C) The ratio of OCR/ECAR at the last time point. (D–F) The real-time levels of (D) ECAR and(E) OCR in splenic activated CD4+ T cells were measured before and after treatment of DMXAA (1 μg/mL) using an extracellular flux Seahorse analyzer (n = 4/ group). (F) The ratio of OCR/ECAR at the last time point. (G-J) The parameters of mitochondrial respiration in control Th1 and DMXAA-pretreated Th1 cells were measured by a Mito Stress Test Kit (n = 8/group). (G) The OCR profile. (H) Basal respiration. (I) Adenosine triphosphate (ATP)–related respiration. (J) Maximal respiration. (K, L) Representative flow cytometry plots and dot plots of mitochondrial mass in control Th1 and DMXAA-pretreated Th1 cells. (M, N) Representative flow cytometry plots and dot plots of mitochondrial membrane potential in control Th1 and DMXAA-pretreated Th1 cells. (O) Mouse splenic CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (1 μg/mL) under Th1 conditions for 6 hours, and the phosphorylated STAT3 (pSTAT3) site Ser727 and total STAT3 were determined by Western blot. (P) A diagram of different metabolic pathways contributing to mitochondrial oxidation. (Q–R) Mouse splenic CD4+ T cells were activated with anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) antibodies in the presence or absence of DMXAA (1 μg/mL) with or without UK5099 (30 μM)/DON (1 μM)/etomoxir (50 μM) under Th1 conditions for 5 days. (Q) Representative flow cytometry plots of IL-10+ CD4+ T cells. (R) Dot plots of IL-10+ CD4+ T cells. Data were shown as mean ± SEM. One representative of 2 (A-J) or 3 (K-O and Q-R) independent experiments. (C, F, H, I, J, L, N) Unpaired Student’s t test; (R) 1-way analysis of variance with Dunnett's multiple comparisons test. ∗∗P < .01, ∗∗∗P < .001.
As glucose, glutamine, and fatty acid are the major fuel for mitochondrial oxidation,8 and each pathway can be inhibited by specific inhibitors (Figure 9P), we next determined which pathways are involved in the STING-induced IL-10 production. Inhibition of glucose oxidation or glutamine oxidation by UK5099 and DON suppressed IL-10 production induced by STING agonist, while blockade of fatty acid oxidation did not affect IL-10 production (Figure 9Q and R). These data suggest that glucose and glutamine metabolisms are critical in STING induction of IL-10 in T cells.
STING Regulation of Microbiota Does Not Affect Intestinal CD4+ T Cell Production of IL-10
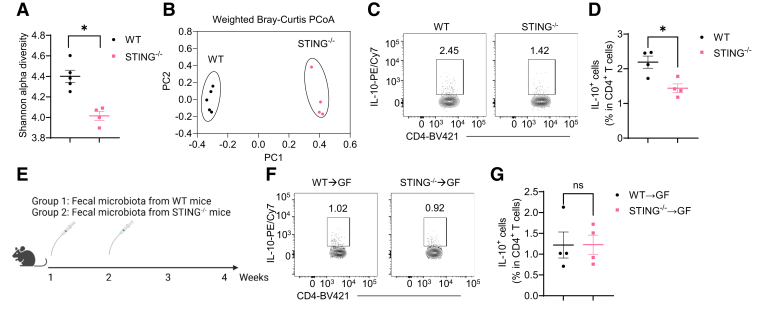
STING has been reported to regulate gut microbiota.14 We then collected the feces from WT and STING–/– mice and compared the microbiota diversity and composition using 16S ribosomal RNA sequencing analysis. We found that STING–/– mice showed a decreased Shannon alpha diversity (Figure 10A), which represents microbiota richness and evenness. In addition, principal coordinate analysis plot showed that the gut microbiota composition was different between WT and STING–/– mice (Figure 10B).
Figure 10.
STING altered gut microbiota do not mediate STING induction of T cell production of IL-10. (A, B) The Shannon alpha diversity and the principal coordinate analysis (PCoA) of gut microbiota from WT and STING–/– mice (n = 4/group). (C, D) Representative flow cytometry plots and dot plots of intestinal IL-10+ CD4+ T cells in WT and STING–/– mice (n = 4/group). (E–G) Fecal microbiota from WT or STING–/– mice were orally transferred to GF mice twice in the first 2 weeks, and bacteria-recipient mice were sacrificed 4 weeks post–first transfer (n = 4/group). (E) The diagram of the experiment design. (F) Representative flow cytometry plots of intestinal IL-10+ CD4+ T cells. (G) The Dot plots of intestinal IL-10+ CD4+ T cells. Data were shown as mean ± SEM. (A) Mann-Whitney U test; (D and G) unpaired Student’s t test. ∗P < .05.
STING–/– mice showed a decreased population of IL-10+ CD4+ T cells in the intestinal compared with WT mice (Figure 10C and D). To investigate whether the altered microbiota in STING–/– mice affect CD4+ T cell production of IL-10, we transferred fecal microbiota from WT and STING–/– mice to WT germ-free (GF) mice by gavage (Figure 10E). Four weeks post–first gavage, mice were sacrificed, and intestinal CD4+ T cell expression of IL-10 was determined by flow cytometry. We found no difference in CD4+ T cell production of IL-10 between GF recipients of WT and STING–/– fecal microbiota (Figure 10F and G). These data suggest that STING affects intestinal microbiota, but such altered microbiota do not affect intestinal CD4+ T cell production of IL-10.
STING Agonist 2,3-cGAMP Suppresses Intestinal Inflammation
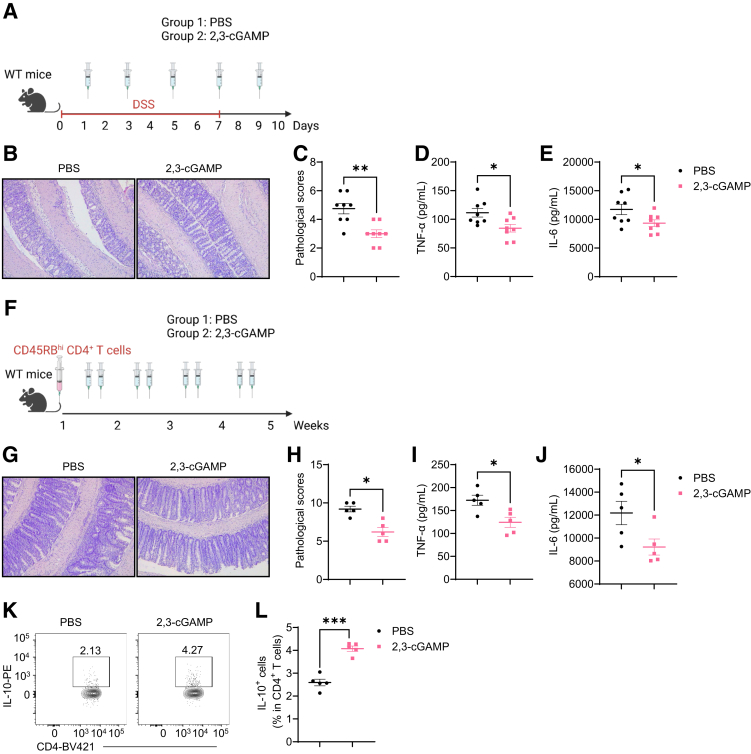
To investigate whether STING agonists could treat intestinal inflammation, we utilized 2,3-cGAMP, a STING agonist working in both mice and humans,31 to treat mice under inflammatory conditions. First, we induced acute colitis in WT mice using DSS, and these mice were administered 2,3-cGAMP or carrier alone daily (Figure 11A). We found that treatment of 2,3-cGAMP inhibited DSS-induced colitis development, as evidenced by lower pathological scores and decreased intestinal TNF-α and IL-6 (Figure 11B–E).
Figure 11.
2,3-cGAMP suppresses intestinal inflammation in DSS-induced acute colitis and T cell-induced chronic colitis models. (A–F) WT mice were administered with 2% DSS (w/v) in drinking water for 7 days and then normal drinking for the last 3 days. One group of mice was intraperitoneally injected with 2,3-cGAMP (0.5 mg/kg) and another group was given phosphate-buffered saline (PBS) as control animals (n = 8/group). Mice were sacrificed on day 10. (A) The diagram of the experiment design. (B) Representative intestinal histological images. (C) Pathological scores. (D) Intestinal TNF-α production levels. (E) Intestinal IL-6 production levels. (F–L) WT CD4+ CD45Rbhi T cells were intravenously transferred into Rag1–/– mice (n = 5/group). The recipient Rag1–/– mice were sacrificed after 5 weeks. (F) The diagram of the experiment design. (G) Representative intestinal histological images. (H) Pathological scores. (I) Intestinal TNF-α production levels. (J) Intestinal IL-6 production levels. (K) Representative flow cytometry plots of intestinal IL-10+ CD4+ T cells. (L) The Dot plots of intestinal IL-10+ CD4+ T cells. Data were shown as mean ± SEM. One representative of 2 independent experiments. (D, E, I, J, L) Unpaired Student’s t test; (C, H) Mann-Whitney U test. ∗P < .01; ∗∗P < .01, ∗∗∗P < .001.
Next, we induced chronic colitis using the CD4+ CD45RBhi T cell transfer model and treated these mice with 2,3-cGAMP or carrier alone every other day (Figure 11F). Similar to DSS-induced colitis, Rag1–/– recipient mice developed less severe colitis and produced less intestinal TNF-α and IL-6 when treated with 2,3-cGAMP (Figure 11G–J). Furthermore, 2,3-cGAMP induced more IL-10-producing intestinal CD4+ T cells (Figure 11K–L).
STING Promotes Human CD4+ T Cell Expression of IL-10
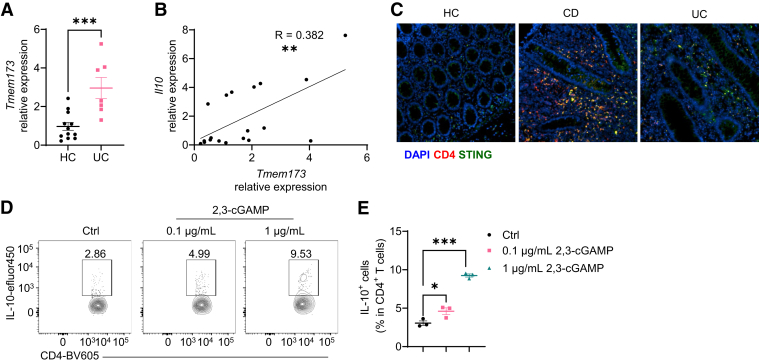
Considering that the STING agonist inhibited colitis in mice (Figure 11), we investigated whether STING functions similarly in IBD patients. We analyzed the STING expression in intestinal biopsies from healthy controls and patients with ulcerative colitis (UC). We found that intestinal STING expression was increased in patients with UC compared with healthy controls (Figure 12A). Interestingly, intestinal STING expression was positively correlated with the level of IL-10 (Figure 12B). To further locate the STING expression and CD4+ T cells in the human intestine, we stained the colon tissue array by immunofluorescence staining. We found that STING+ CD4+ T cells were increased in UC patients and patients with Crohn’s disease (Figure 12C). Then, we investigated whether STING promotes IL-10 production in human CD4+ T cells and found that treatment with 2,3-cGAMP significantly increased IL-10 production in human CD4+ T cells in a dose-dependent manner (Figure 12D and E).
Figure 12.
2,3-cGAMP upregulates IL-10 production in human CD4+T cells. (A, B) Colonic biopsies were obtained from healthy control (HC) subjects (n = 12) and patients with UC (n = 7). (A) Temem173 (Sting) expression in colonic biopsies from HC subjects and UC patients. (B) The correlation between Tmem173 and Il10 in intestinal biopsies. (C) Representative immunofluorescence staining of CD4 and STING in colonic tissues microarray. (D, E) Human peripheral blood mononuclear cells were activated with anti-CD3 and anti-CD28 antibodies and then nucleofected with 2,3-cGAMP at the dose of 0.1 μg/mL and 1 μg/mL or phosphate-buffered saline as control condition. Cells were then cultured for 5 days. (D) Representative flow cytometry plots of IL-10+ CD4+ T cells. (E) Dot plots of IL-10+ CD4+ T cells. Data were shown as mean ± SEM. (D, E) One representative of 3 independent experiments. (A) Unpaired Student’s t test; (B) Person correlation analysis; (E) 1-way analysis of variance with Dunnett's multiple comparisons test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
Discussion
Th1 effector cells play a critical role in the development of intestinal inflammation, and several factors have been found to contribute to the polarization and pathogenicity of Th1 cells.32 However, the intrinsic molecular mechanisms by which switch off the inflammatory programs of Th1 cells are poorly understood. Our current study demonstrated that STING, a cytosolic DNA sensor, negatively regulates the proinflammatory program of Th1 cells through the upregulation of anti-inflammatory cytokine IL-10, thereby limiting the pathogenicity of Th1 cells to maintain intestinal homeostasis.
Recognized as the sensor of exogenous and endogenous DNA, the role of cGAS-STING signaling in innate immune responses and host defense has been well established.24 More recently, the effect of STING on tumors and autoimmunity has also been explored.17 However, the role of STING signaling in intestinal homeostasis is controversial.13, 14, 15, 16 In this study, we found that STING–/– mice were more susceptible to DSS-induced acute colitis and STING-deficient T cells induced more exaggerated chronic colitis, which establishes the importance of STING in protecting against both acute and chronic intestinal inflammation and limiting the pathogenicity of T cells in inducing colitis. While a previous study demonstrated the role of STING signaling in promoting pro- and anti-inflammatory cytokines in innate cells,25 we revealed the effect of STING activation on anti-inflammatory cytokine IL-10 in Th1 cells. STING agonists have been found to induce T cell apoptosis.18,19 Although innate immune cells, but not CD4+ T cells, play the role of recognizing viruses, some viruses (e.g., HIV) could directly attack CD4+ T cells and replicate within them.33 Therefore, it is plausible that sensing viruses is also sufficient to induce T cell functions through Sting pathway. Consistent with previous studies,18, 19 we found that DMXAA at doses higher than 5 μg/mL decreased viability and enhanced apoptosis in T cells. Interestingly, 1 μg/mL of DMXAA promoted Th1 production of IL-10 without affecting cell viability and apoptosis in vitro. These data suggest that different doses of STING agonists diversely control the fate of CD4+ T cells in vitro. However, based on these data, we cannot make a definite conclusion about whether the different effects on T cells induced by different doses of STING agonist reflect physiologically different responses to real situations. In addition, we confirmed that STING activation suppresses IL-17 but promotes IL-10 in Th17 cells in vitro, which was reported in a recent study.21 Among T cell cytokines, IL-10 was decreased in STING-deficient intestinal T cells, specifically Th1 cells but not Th17 or Treg cells, in Rag1–/– recipients. As effector T cell expression of IL-10 is essential for restricting excessive T cell responses in a self-limiting manner,11 the STING pathway appears to be critical in switching off the proinflammatory program of Th1 effector cells in the intestine.
As the downstream product of the STING pathway, type I IFNs regulate a series of immune responses.24 Our current study demonstrated that the STING induction of IL-10 in Th1 cells partially depended on the type I IFN pathway. It has been reported that IFN regulatory factor 3 and nuclear factor-κB p53, which are activated by TBK1 but not type I IFN pathway, are contributing to STING-triggered T cell death,19 suggesting that different doses of STING agonists drive T cell death and affect T cell production of IL-10 through different pathways. In addition, deficiency in the receptor for type I IFNs resulted in decreased Th1 expression of Blimp1, which was induced by STING agonists in WT Th1 cells. However, STING activation did not affect the expression of AhR, which has been reported to mediate STING induction of IL-10 production in Th17 cells.21 This difference might be attributed to the lower AhR expression but higher Blimp1 expression in Th1 cells compared with Th17 cells.34,35
Accumulating evidence highlights the importance of cellular metabolism in regulating T cell functions and autoimmunity.8 STING has also been reported to affect lipid metabolism in Drosophila and mice36,37 and glycolysis in intestinal epithelial cells.16 Here, we demonstrated that STING activation altered the OCR/ECAR ratio in activated T cells, indicating that STING regulates the metabolic program in T cells. More specifically, STING agonist-pretreated Th1 cells showed increased mitochondrial oxidation, which was similar to the metabolic profile in anti-inflammatory Treg cells.38,39 In addition, we also demonstrated that the mitochondrial oxidation of glucose and glutamine, but not lipids, mediated the STING induction of IL-10 in Th1 cells.
As a transcription factor, the role of STAT3 has been widely reported to mediate a variety of autoimmune diseases by regulating gene expression.40 In this study, we found that activation of STING induced phosphorylation of STAT3 at the site of Tyr705, leading to STAT3 homodimerization, nuclear translocation, and DNA binding.41,42 Blimp1 is one of the downstream target genes of nuclear STAT3.43 In addition to Tyr705, STING signaling also induced STAT3 Ser727 phosphorylation. Besides translocation to nuclear, STAT3 is also present in mitochondria, where it regulates mitochondrial respiration. Although both Tyr705 and Ser727 phosphorylation of STAT3 are present in mitochondria, Ser727 is critical for mitochondrial activity.29,30
Emerging evidence indicates the importance of gut microbiota in immune responses and intestinal homeostasis.9,44 Large amounts of bacteria-derived nucleic acids exist in the intestines, which activates DNA sensors to regulate the intestinal mucosal immune system.45 A recent study has shown that commensal bacteria influence STING signaling.25 Our study indicated that STING–/– mice displayed an altered gut microbiota, indicating the role of STING in regulating the intestinal commensal bacteria community. However, the transfer of gut microbiota from STING–/– mice induced similar levels of IL-10–producing T cells in GF mice compared with the microbiota from WT mice, which exclude the microbiota in the process of STING induction of IL-10 production in T cells. However, our recent study has reported that STING affect gut microbiota to regulate intestinal IgA reponses.46 Threfore, altered microbiota might be involved in other cytokine production and immune responses, which needs investigation in the future.
As DMXAA and CMA only work on murine cells,47,48 it is important to test the effect of 2,3-cGAMP, a STING agonist working in vivo in both humans and mice,31 in vitro and in vivo. Here, we found that 2,3-cGAMP promoted human T cell expression of IL-10 and administered with 2,3-cGAMP inhibits acute and chronic colitis, which provides a foundation for developing STING agonists as therapeutics for colitis. Overall, this study demonstrates the role of STING signaling in switching off the proinflammatory pattern of Th1 effector cells in the intestine.
Materials and Methods
Mouse Strains
C57BL/6J WT mice, B6(Cg)-Sting1tm1.2Camb/J (STING–/–) mice, B6.129S7-Rag1tm1Mom/J (Rag1–/–) mice, B6.Cg-Tg(Cd4-cre)1Cwi/BfluJ (Cd4cre) mice, B6.129-Prdm1tm1clme/J (Prdm1fl/fl) mice, B6.Cg-Tg(Prdm1-EYFP)1Mnz/J (Blimp1-eYFP) mice, and B6.129S2-Ifnar1tm1Agt/Mmjax (IFNAR–/–) mice were purchased from the Jackson Laboratory (Bar Harbor, ME). CD4creBlimp1fl/fl mice were generated by crossing Cd4cre mice with Prdm1fl/fl mice. All the above mice were maintained in the specific pathogen-free animal facility, and GF C57BL/6 mice were bred and maintained in the GF mouse facility of the University of Texas Medical Branch. All the mouse experiments in this study were reviewed and approved by the Institutional Animal Care and Use Committee.
Human Samples
Human colonic biopsies
All the participants, including healthy control subjects and patients with ulcerative colitis (UC), were recruited at the Department of Gastroenterology, Affiliated Hospital of Jining Medical University. The diagnosis of UC follows the standard criteria, including clinical features, laboratory tests, and endoscopic and histological findings. The colonic biopsies for determining the mRNA expression of STING were collected during endoscopy. The experiments were approved by the Institutional Review Board for Clinical Research of Affiliated Hospital of Jining Medical University.
Human colon tissue array
For the immunofluorescence staining, the human paraffin-embedded colon tissue arrays (CO245a and CO246) were ordered from US Biomax (Derwood, MD).
Human peripheral blood mononuclear cells
The human peripheral blood mononuclear cells were generously provided by Dr Haitao Hu at the University of Texas Medical Branch.
Murine CD4+ T cell isolation, activation, and polarization
After grinding and red blood cell lysis, splenic cells were incubated with anti-mouse CD4 magnetic particles to purify CD4+ T cells. CD4+ T cells were cultured on the anti-CD3 antibody (5 μg/mL)–coated plates in RPMI media supplemented with fetal bovine serum (10%, v/v), sodium pyruvate (1 mM), L-glutamine (2.05 mM), 2-mercaptoethanol (50 μM), HEPES (10 μM), penicillin (100 units/mL), and streptomycin (100 μg/mL) with anti-CD28 antibody (2 μg/mL) under neutral, Th1 (IL-12, 10 ng/mL), Th17 (transforming growth factor β, 2 ng/mL; IL-6, 50 ng/mL; anti-IFNγ antibody, 10 μg/mL; anti-IL-4, 5 μg/mL), and Treg (transforming growth factor β, 5 ng/mL; anti-IFNγ antibody, 5 μg/mL) conditions.
Murine Colitis Models and Treatment
All the mice used for colitis models were sex and age matched.
DSS-induced acute colitis
Mice were treated with 2% (w/v) of DSS in drinking water for 7 days. DSS was changed to regular drinking water for another 3 days. Mice were then sacrificed to analyze the severity of intestinal inflammation.
CD45Rbhi CD4+ T cell transfer–induced chronic colitis
CD45Rbihi CD4+ T cells (200,000 cells/mouse) were intravenously injected into Rag1–/– mice. The recipient Rag1–/– mice were then sacrificed 5 weeks later.
Th1 cell transfer–induced chronic colitis
CD4+ T cells were activated and cultured under Th1 conditions for 5 days. Th1 cells were collected, and dead cells were removed using the MojoSort Mouse Dead Cell Removal Kit (Biolegend). Th1 cells (1 million/mouse) were then intravenously injected into Rag1–/– mice. The recipient Rag1–/– mice were then sacrificed 6 weeks later.
Treatment
In vivo treatment of 2,3-cGAMP: mice were peritoneally injected 2,3-cGAMP (0.5 mg/kg) every day (DSS-induced acute colitis model) or every other day (CD45Rbhi CD4+ T cell transfer induced chronic colitis model).
Fecal Microbiota Transplantation
Feces were freshly collected from WT and STING–/– mice, resuspended in the 1× phosphate-buffered saline supplemented with L-cysteine hydrochloride (0.05%, w/v). After homologation, large particles were removed from fecal suspension through centrifugation and strainers. Fecal bacteria suspension was then orally transferred to GF mice via gavage twice in the first 2 weeks. GF recipients of fecal bacteria were sacrificed 4 weeks post–first gavage for analyzing IL-10-producing CD4+ T cells.
Hematoxylin and Eosin Staining and Pathological Scoring
Colonic tissues were rolled from the distal to the proximal end, fixed in the 10% neutral buffered formalin, dehydrated with a series of alcohol, and embedded in paraffin. After cutting, colonic samples were stained with hematoxylin and eosin. The severity of intestinal inflammation was assessed following the criteria below. DSS-induced chronic colitis model: the pathological score was determined by cell infiltration and architecture change; CD45Rbhi CD4+ T cell transfer–induced chronic colitis and Th1 cell transfer–induced chronic colitis models: the pathological score was calculated by summing the scores of 6 parameters, including the levels of lamina propria inflammation, goblet cell boss, abnormal crypt, the presence or absence of crypt abscesses, and the severity of mucosal erosion and ulceration.
Isolation of Intestinal Lamina Propria Cells
After removing the intestinal epithelial cell using 0.5 mM of EDTA, colonic tissues were digested with 0.5 mg/mL of collagenase IV. Intestinal lamina propria cells were purified by cell strainer and 40%/75% Percoll (Sigma-Aldrich, St Louis, MO).
Western Blot
Cellular protein was extracted using RIPA lysis buffer supplemented with PMSF and protease and phosphatase inhibitor cocktails. A total of 10 μg of total protein was loaded into 4%–15% gels, separated by electrophoresis, and then transferred from the gels to the PVDF membranes. After blocking with 1% casein blocker (w/v) for 1 hour at room temperature, the membrane was incubated with primary antibodies (β-actin, 1:1000; STING, 1:1000; pSTAT1, 1:1000; STAT1, 1:1000; pSTAT3 Tyr705, 1:1000; pSTAT3 Ser727, 1:1000) overnight at 4°C. After incubation with a secondary antibody (1:2000), membranes were merged into chemiluminescent substrates, and the chemiluminescent signals were captured using an imaging system.
Quantitative Polymerase Chain Reaction
Cellular RNA was extracted and purified by TRIzol, chloroform, and isopropanol. Complementary DNA was generated from RNA by reverse transcription using the Quantabio qScript cDNA Synthesis Kit (Quantabio, Beverly, MA). Quantitative polymerase chain reaction was performed to measure the mRNA levels of Gapdh, Il10, Blimp1, Ahr, Irf4, Maf, and Ifnb1. The primers used in this study were listed in Table 1.
Table 1.
Primers for Quantitative Reverse-Transcriptase Polymerase Chain Reaction in This Study
| Genes | Species | Forward Primer | Reverse Primer |
|---|---|---|---|
| Gapdh | Mouse | GGTTGTCTCCTGCGACTTCA | TGGTCCAGGGTTTCTTACTCC |
| Il10 | Mouse | AGCCGGGAAGACAATAACTG | GGAGTCGGTTAGCAGTATGTTG |
| Blimp1 | Mouse | CTCAACACTCTCATGTAAGAGGC | AGCATGACCTGACATTGACACC |
| Ahr | Mouse | TTGGTTGTGATGCCAAAGGGC | CATGCGGATGTGGGATTCTGC |
| Irf4 | Mouse | TCCGACAGTGGTTGATCGAC | CCTCACGATTGTAGTCCTGCTT |
| Maf | Mouse | GGAGACCGACCGCATCATC | TCATCCAGTAGTAGTCTTCCAGG |
| Ifnb1 | Mouse | GCCTTTGCCATCCAAGAGATGC | ACACTGTCTGCTGGTGGAGTTC |
| Gapdh | Human | CAGGAGGCATTGCTGATGAT | GAAGGCTGGGGCTCATTT |
| Il10 | Human | TCTCCGAGATGCCTTCAGCAGA | TCAGACAAGGCTTGGCAACCCA |
| Tmem173 | Human | CCTGAGTCTCAGAACAACTGCC | GGTCTTCAAGCTGCCCACAGTA |
RNA Sequencing
Splenic CD4+ T cells were activated with anti-CD3 antibody (5 μg/mL) and anti-CD28 antibody (2 μg/mL) in the presence or absence DMXAA (1 μg/mL) under neutral conditions for 2 days. Total RNA was extracted by TRIzol, chloroform, and isopropanol. RNA was quantified and qualified by NanoDrop (Thermo Fisher Scientific, Waltham, MA), agarose gel electrophoresis, and Agilent 2100 (Agilent, Santa Clara, CA). The library was constructed using NEBNext Ultra RNA Library Prep Kit for Illumina at Novogene (Sacramento, CA). Qualified libraries were sequenced using a paired-end 150 run on an Illumina NovaSeq Platform (Illumina, San Diego, CA). Data were analyzed by Novogene and visualized by iDEP.49
Enzyme-Linked Immunosorbent Assay
The supernatants of cell culture or colonic tissue culture were collected by centrifugation to remove cell or tissue debris. The 96-well plates were precoated with capture antibodies (1:200) overnight at 4°C. After blocking with 1% bovine serum albumin for 1 hour at room temperature, supernatants were added to the plates, followed by incubation with detection antibodies (1:200) and horseradish peroxidase for 2 hours. Subsequently, plates were incubated with the substrate, and the reaction was quenched with 1 M H2SO4. The concentration of the cytokines in the supernatants was determined by measuring the absorbance at 450 nm using a multimode reader.
Flow Cytometry Staining
Cytokine and transcription factor staining
Cells were treated with 20 ng/mL of Phorbol-12-myristate 13-acetate and 750 ng/mL of ionomycin for 5 hours and 5 μg/mL of Brefeldin A for 3 hours. Then, cells were Fc blocked with anti-CD16/32 antibody and stained with live dye using LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit (Thermo Fisher Scientific). After staining with surface marker CD4, cells were fixed and permeabilized using the Foxp3/Transcription Factor Fixation/Permeabilization set (Thermo Fisher Scientific) and then incubated with antibodies (IFNγ, 1:100; IL-17A, 1:200; IL-10, 1:200; Foxp3, 1:200).
Phosphoflow
Cells were stained with live dye using LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit, and then fixed by IC Fixation Buffer and permeabilized using 100% methanol. Cells were than Fc blocked with anti-CD16/32 antibody and stained with antibodies (pSTAT1 Tyr701, 1:50; pSTAT3 Tyr 705, 1:50).
Apoptosis assay
After washing, cells were resuspended in binding buffer and stained with Annexin V and 7-AAD for 15 minutes at room temperature.
Mitochondrial mass and mitochondrial membrane potential
After staining with live dye and anti-CD4 antibody, cells were stained with MitoTracker Deep Red (50 μM) (Thermo Fisher Scientific) and JC-1 (5 μM) for 15 minutes at 37°C.
Cells were collected on a BD LSRFortessa or a BD FACSymphony A5 (BD Biosciences, Franklin Lakes, NJ) and analyzed by FlowJo version 10 (FlowJo, Ashland, OR).
Resazurin Viability Assay
Resazurin was added to the cell culture media at a concentration of 44 μM. After 2 days, cell viability was determined by subtracting the absorbance at 595 nm from the absorbance at 570 nm.
16S Ribosomal RNA Sequencing
Fecal pellets were freshly collected from WT and STING–/– mice, followed by bacterial DNA extraction and library construction by universal 16S ribosomal RNA V3-V4 region primers.50 Then, the samples were barcoded for multiplexing and sequenced on an Illumina MiSeq instrument. The raw reads were trimmed to 300 bases and analyzed using CLC Genomics Workbench 9.5 Microbial Genomics Module (Qiagen, Hilden, Germany). Reference-based OTU picking was performed using the SILVA SSU v132 97% database.51 Sequences that were present in more than 1 copy but were not clustered in the database were then placed into de novo OTUs (97% similarity) and aligned against the database with an 80% similarity threshold. Alpha diversity was determined using the Shannon diversity index at the genus level, and the beta diversity was analyzed by weighted Bray-Curtis dissimilarity matrices.
Seahorse Metabolic Assays
Sensor cartridges were hydrated with sterile water overnight and prewarmed XF Calibrant (Agilent) for 1 hour in a 37°C non-CO2 incubator. Naïve CD4+ T cells (200,000 cells/well), activated CD4+ T cells (100,000 cells/well), and Th1 cells (100,000 cells/well) were seeded onto the Seahorse XF Culture 96-well plates (Agilent). DMXAA or dimethyl sulfoxide was loaded into the cartridge to analyze the real-time effect of DMXAA on ECAR and OCR in naïve and activated T cells. The Mito Stress Test kit (Agilent) was used to determine the key parameters of mitochondrial respiration. ECAR and OCR levels were normalized to the values of 100,000 cells.
Immunofluorescence Staining
Colon tissue arrays (CO245a and CO246) were heated at 60°C for 1 hour in a horizontal position and dehydrated in a series of ethanol. After antigen retrieval, tissues were permeabilized with triton, blocked with 10% goat serum, and incubated with antibodies (CD4, 1:1000; STING, 1:2000) in a humid chamber at 4°C overnight. After washing, tissues were stained with secondary antibody (1:1000) at room temperature for 2 hours. Images were captured using a Cytation 5 imager (BioTek, Winooski, VT).
Analysis
All the data in this study were presented as mean ± SEM, and statistical analyses were performed using GraphPad Prism 9.0 (GraphPad Software, San Diego, CA). According to the type of quantitative data and the number of groups, we analyzed the data using an unpaired 2-tailed Student t test, Mann-Whitney U test, or 1-way analysis of variance, which were indicated in each figure. P < .05 was considered statistically significant.
Acknowledgments
The graphic abstract and Figures 9P, 10E, 11A, and 11F were created with BioRender.com. Flow cytometry analysis in this paper was done on instruments in the University of Texas Medical Branch Flow Cytometry and Cell Sorting Core Lab. RNA sequencing and 16S ribosomal RNA sequencing data were deposited in the National Center for Biotechnology Information sequencing read archive under BioProject PRJNA885518 and PRJNA885485, respectively. Any additional information or materials required to reanalyze the data reported in this article is available from the corresponding author upon request with a material transfer agreement (MTA).
CRediT Authorship Contributions
Wenjing Yang, MD, PhD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Project administration: Lead; Validation: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Tianming Yu, MD, PhD (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Validation: Equal; Visualization: Equal; Writing – original draft: Equal; Writing – review & editing: Equal)
Guangxi Zhou, MD, PhD (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Resources: Supporting; Writing – review & editing: Supporting)
Suxia Yao, MD (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Maki Wakamiya, PhD (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Haitao Hu, PhD (Resources: Supporting; Writing – review & editing: Supporting)
Slobodan Paessler, PhD (Resources: Supporting; Writing – review & editing: Supporting)
Jiaren Sun, MD, PhD (Resources: Supporting; Writing – review & editing: Supporting)
Yingzi Cong, PhD (Conceptualization: Lead; Data curation: Lead; Funding acquisition: Lead; Investigation: Lead; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflict of interest The authors disclose no conflicts.
Funding This work was supported by National Institutes of Health grants DK105585, DK124132, AI150201, DK112436, and DK125011.
References
- 1.Swain S.L., McKinstry K.K., Strutt T.M. Expanding roles for CD4⁺ T cells in immunity to viruses. Nat Rev Immunol. 2012;12:136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skapenko A., Leipe J., Lipsky P.E., et al. The role of the T cell in autoimmune inflammation. Arthritis Res Ther. 2005;7:S4–S14. doi: 10.1186/ar1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tay R.E., Richardson E.K., Toh H.C. Revisiting the role of CD4(+) T cells in cancer immunotherapy-new insights into old paradigms. Cancer Gene Ther. 2021;28:5–17. doi: 10.1038/s41417-020-0183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jäger A., Kuchroo V.K. Effector and regulatory T cell subsets in autoimmunity and tissue inflammation. Scand J Immunol. 2010;72:173–184. doi: 10.1111/j.1365-3083.2010.02432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lafaille J.J. The role of helper T cell subsets in autoimmune diseases. Cytokine Growth Factor Rev. 1998;9:139–151. doi: 10.1016/s1359-6101(98)00009-4. [DOI] [PubMed] [Google Scholar]
- 6.Imam T., Park S., Kaplan M.H., et al. Effector T helper cell subsets in inflammatory bowel diseases. Front Immunol. 2018;9:1212. doi: 10.3389/fimmu.2018.01212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu J., Jankovic D., Oler A.J., et al. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 2012;37:660–673. doi: 10.1016/j.immuni.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang W., Yu T., Cong Y. CD4(+) T cell metabolism, gut microbiota, and autoimmune diseases: implication in precision medicine of autoimmune diseases. Precis Clin Med. 2022;5:pbac018. doi: 10.1093/pcmedi/pbac018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang W., Cong Y. Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell Mol Immunol. 2021;18:866–877. doi: 10.1038/s41423-021-00661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saraiva M., O'Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 11.Jankovic D., Kugler D.G., Sher A. IL-10 production by CD4+ effector T cells: a mechanism for self-regulation. Mucosal Immunol. 2010;3:239–246. doi: 10.1038/mi.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motwani M., Pesiridis S., Fitzgerald K.A. DNA sensing by the cGAS-STING pathway in health and disease. Nat Rev Genet. 2019;20:657–674. doi: 10.1038/s41576-019-0151-1. [DOI] [PubMed] [Google Scholar]
- 13.Zhu Q., Man S.M., Gurung P., et al. Cutting edge: STING mediates protection against colorectal tumorigenesis by governing the magnitude of intestinal inflammation. J Immunol. 2014;193:4779–4782. doi: 10.4049/jimmunol.1402051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canesso M.C.C., Lemos L., Neves T.C., et al. The cytosolic sensor STING is required for intestinal homeostasis and control of inflammation. Mucosal Immunol. 2018;11:820–834. doi: 10.1038/mi.2017.88. [DOI] [PubMed] [Google Scholar]
- 15.Martin G.R., Blomquist C.M., Henare K.L., et al. Stimulator of interferon genes (STING) activation exacerbates experimental colitis in mice. Sci Rep. 2019;9 doi: 10.1038/s41598-019-50656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Y., Yang W., Bilotta A.J., et al. STING controls intestinal homeostasis through promoting antimicrobial peptide expression in epithelial cells. Faseb j. 2020;34:15417–15430. doi: 10.1096/fj.202001524R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barber G.N. STING: infection, inflammation and cancer. Nat Rev Immunol. 2015;15:760–770. doi: 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larkin B., Ilyukha V., Sorokin M., et al. Cutting edge: activation of STING in T cells induces type I IFN responses and cell death. J Immunol. 2017;199:397–402. doi: 10.4049/jimmunol.1601999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulen M.F., Koch U., Haag S.M., et al. Signalling strength determines proapoptotic functions of STING. Nat Commun. 2017;8:427. doi: 10.1038/s41467-017-00573-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benoit-Lizon I., Jacquin E., Rivera Vargas T., et al. CD4 T cell-intrinsic STING signaling controls the differentiation and effector functions of T(H)1 and T(H)9 cells. J Immunother Cancer. 2022;10 doi: 10.1136/jitc-2021-003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damasceno L.E.A., Cebinelli G.C.M., Fernandes M.F., et al. STING is an intrinsic checkpoint inhibitor that restrains the T(H)17 cell pathogenic program. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyer S.S., Cheng G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit Rev Immunol. 2012;32:23–63. doi: 10.1615/critrevimmunol.v32.i1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubo M., Motomura Y. Transcriptional regulation of the anti-inflammatory cytokine IL-10 in acquired immune cells. Front Immunol. 2012;3:275. doi: 10.3389/fimmu.2012.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishikawa H., Ma Z., Barber G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahn J., Son S., Oliveira S.C., et al. STING-dependent signaling underlies IL-10 controlled inflammatory colitis. Cell Rep. 2017;21:3873–3884. doi: 10.1016/j.celrep.2017.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.González-Navajas J.M., Lee J., David M., et al. Immunomodulatory functions of type I interferons. Nat Rev Immunol. 2012;12:125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Neill L.A., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tammineni P., Anugula C., Mohammed F., et al. The import of the transcription factor STAT3 into mitochondria depends on GRIM-19, a component of the electron transport chain. J Biol Chem. 2013;288:4723–4732. doi: 10.1074/jbc.M112.378984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wegrzyn J., Potla R., Chwae Y.J., et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gough D.J., Corlett A., Schlessinger K., et al. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324:1713–1716. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuriakose T., Kanneganti T.D. cGAMP: A tale of two signals. J Exp Med. 2017;214:3471–3473. doi: 10.1084/jem.20171925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shale M., Schiering C., Powrie F. CD4(+) T-cell subsets in intestinal inflammation. Immunol Rev. 2013;252:164–182. doi: 10.1111/imr.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orzalli M.H., Knipe D.M. Cellular sensing of viral DNA and viral evasion mechanisms. Annu Rev Microbiol. 2014;68:477–492. doi: 10.1146/annurev-micro-091313-103409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veldhoen M., Hirota K., Westendorf A.M., et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 35.Salehi S., Bankoti R., Benevides L., et al. B lymphocyte-induced maturation protein-1 contributes to intestinal mucosa homeostasis by limiting the number of IL-17-producing CD4+ T cells. J Immunol. 2012;189:5682–5693. doi: 10.4049/jimmunol.1201966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akhmetova K., Balasov M., Chesnokov I. Drosophila STING protein has a role in lipid metabolism. Elife. 2021;10 doi: 10.7554/eLife.67358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vila I.K., Chamma H., Steer A., et al. STING orchestrates the crosstalk between polyunsaturated fatty acid metabolism and inflammatory responses. Cell Metab. 2022;34:125–139.e8. doi: 10.1016/j.cmet.2021.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinberg S.E., Singer B.D., Steinert E.M., et al. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature. 2019;565:495–499. doi: 10.1038/s41586-018-0846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beier U.H., Angelin A., Akimova T., et al. Essential role of mitochondrial energy metabolism in Foxp3⁺ T-regulatory cell function and allograft survival. Faseb j. 2015;29:2315–2326. doi: 10.1096/fj.14-268409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gharibi T., Babaloo Z., Hosseini A., et al. Targeting STAT3 in cancer and autoimmune diseases. Eur J Pharmacol. 2020;878 doi: 10.1016/j.ejphar.2020.173107. [DOI] [PubMed] [Google Scholar]
- 41.Zhong Z., Wen Z., Darnell J.E., Jr. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 42.Durant L., Watford W.T., Ramos H.L., et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diehl S.A., Schmidlin H., Nagasawa M., et al. STAT3-mediated up-regulation of BLIMP1 Is coordinated with BCL6 down-regulation to control human plasma cell differentiation. J Immunol. 2008;180:4805–4815. doi: 10.4049/jimmunol.180.7.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng D., Liwinski T., Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okude H., Ori D., Kawai T. Signaling through nucleic acid sensors and their roles in inflammatory diseases. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.625833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu T., Yang W., Yao S., et al. STING Promotes Intestinal IgA Production by Regulating Acetate-producing Bacteria to Maintain Host-microbiota Mutualism. Inflamm Bowel Dis. 2023 Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 47.Conlon J., Burdette D.L., Sharma S., et al. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J Immunol. 2013;190:5216–5225. doi: 10.4049/jimmunol.1300097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cavlar T., Deimling T., Ablasser A., et al. Species-specific detection of the antiviral small-molecule compound CMA by STING. Embo j. 2013;32:1440–1450. doi: 10.1038/emboj.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ge S.X., Son E.W., Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics. 2018;19:534. doi: 10.1186/s12859-018-2486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klindworth A., Pruesse E., Schweer T., et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yilmaz P., Parfrey L.W., Yarza P., et al. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014;42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]