Significance
The recent publication of predicted structures of the entire human proteome was a landmark achievement. Combining automatic pipelines and manual curation, we identified 47,576 globular domains from predicted structures of the human proteome and determined their evolutionary relationships to known domains. A quarter of these domains lacked structural data before the release of these predictions. Investigation into these domains and the thousands of disease-causing single amino acid variations within them is expected to reveal insights into protein function and disease mechanisms, as illustrated by examples discussed here. Finally, comparison with automatically classified domains from multiple model organisms revealed differences associated with the unique physiology of humans, such as expansion in cytokines and depletion of odor-sensing domains.
Keywords: protein structure, domain classification, structural prediction, bioinformatics
Abstract
Recent advances in protein structure prediction have generated accurate structures of previously uncharacterized human proteins. Identifying domains in these predicted structures and classifying them into an evolutionary hierarchy can reveal biological insights. Here, we describe the detection and classification of domains from the human proteome. Our classification indicates that only 62% of residues are located in globular domains. We further classify these globular domains and observe that the majority (65%) can be classified among known folds by sequence, with a smaller fraction (33%) requiring structural data to refine the domain boundaries and/or to support their homology. A relatively small number (966 domains) cannot be confidently assigned using our automatic pipelines, thus demanding manual inspection. We classify 47,576 domains, of which only 23% have been included in experimental structures. A portion (6.3%) of these classified globular domains lack sequence-based annotation in InterPro. A quarter (23%) have not been structurally modeled by homology, and they contain 2,540 known disease-causing single amino acid variations whose pathogenesis can now be inferred using AF models. A comparison of classified domains from a series of model organisms revealed expansions of several immune response-related domains in humans and a depletion of olfactory receptors. Finally, we use this classification to expand well-known protein families of biological significance. These classifications are presented on the ECOD website (http://prodata.swmed.edu/ecod/index_human.php).
Protein function stems from structure, which is determined by sequence. We use sequence, structure, and functional similarity to detect homology between proteins, identify their domains, and infer function (1). Domains represent the structural, functional, and evolutionary units of proteins. Their definitions are collected in both structure-based domain classifications, such as in Evolutionary Classification of protein Domains (ECOD) (2–4), SCOP (5), and CATH (6), and sequence-based domain classifications, such as Pfam (7–9) and CDD (10). In the past, structure-based domain classifications have been limited to a small fraction of the protein universe with experimentally determined structures. However, studies suggested that nearly all possible folds have been represented by domains in the experimental structures (11–13).
Our classification, the Evolutionary Classification of protein Domains (ECOD), differs principally from other structure-based classifications in its hierarchy, which favors homology over topology (2), and in the degree of automation, which facilitates high-throughput classification. We have demonstrated that ECOD’s approach is suitable for the large-scale classification of experimental structures (SI Appendix, Fig. S1) (14). ECOD’s top level is architecture, a broad category based on the content and arrangement of secondary structures. Beneath it lies the level of possible homology (X-groups), approximately analogous to the fold level of SCOP (5). Domains sharing homology deduced from sequence and profile similarity or revealing structural similarity coupled with functional evidence are grouped into H-groups (homology groups). Domains in the same H-group are partitioned into topology groups (T-groups) if conformational switching or fold change between them has led to substantial structural differences. Proteins with confident evolutionary relationships (i.e., common ancestry) will be grouped in the same H-group, while significant structural changes between homologous proteins are permitted (2, 15). The finest classification level in ECOD is the family, or F-group, where closely related domains that are expected to have similar functions are grouped.
The recent Critical Assessment of techniques in Structure Prediction (CASP14) showcased a breakthrough in structure prediction (16, 17). AlphaFold (AF), developed by DeepMind, demonstrated its ability to predict three-dimensional structures of proteins from their sequences with accuracy approaching that of experimental methods (16, 18, 19). After the publication of AF, a flood of predicted structures was released to the public in the AlphaFold protein structure Database (AFDB) (20), including a complete set of predictions for the human proteome. These predictions significantly increased the structurally characterized fraction of the human proteome (21). However, these AF models are heterogeneous in compactness and prediction confidence (22), illustrating the need to identify domains from the confident regions of these models.
Here, we present our classification of domains from human proteins in the AFDB, using ECOD as the reference. We integrate our established pipeline for classifying domains from experimental structures with additional tools we developed to specifically address the nonglobular nature of AF models rich in flexible or helical interdomain linkers. These automatic pipelines are expected to detect ~99% of domains from AF models, and they can confidently assign 98% of these domains to the existing ECOD hierarchy, leaving 2%, which require manual curation. Consequently, we identified 47,576 domains from 20,296 human protein models, including 2,994 domains not previously annotated in the InterPro database (23, 24) and 10,994 domains whose 3D structures cannot be modeled by homology. The classification of these domains allowed us to populate ECOD homologous groups of crucial biological relevance (such as G-protein coupled receptors) and to view disease-causing single amino acid variations (SAVs) in previously unrecognized domains. Finally, a crosscomparison of domains classified from multiple eukaryotic model organisms reveals the expansion and depletion of protein families, enabling the specific adaptation of humans during evolution(25).
Results and Discussion
Domain Classification of the Human Proteome by Sequence and Structure.
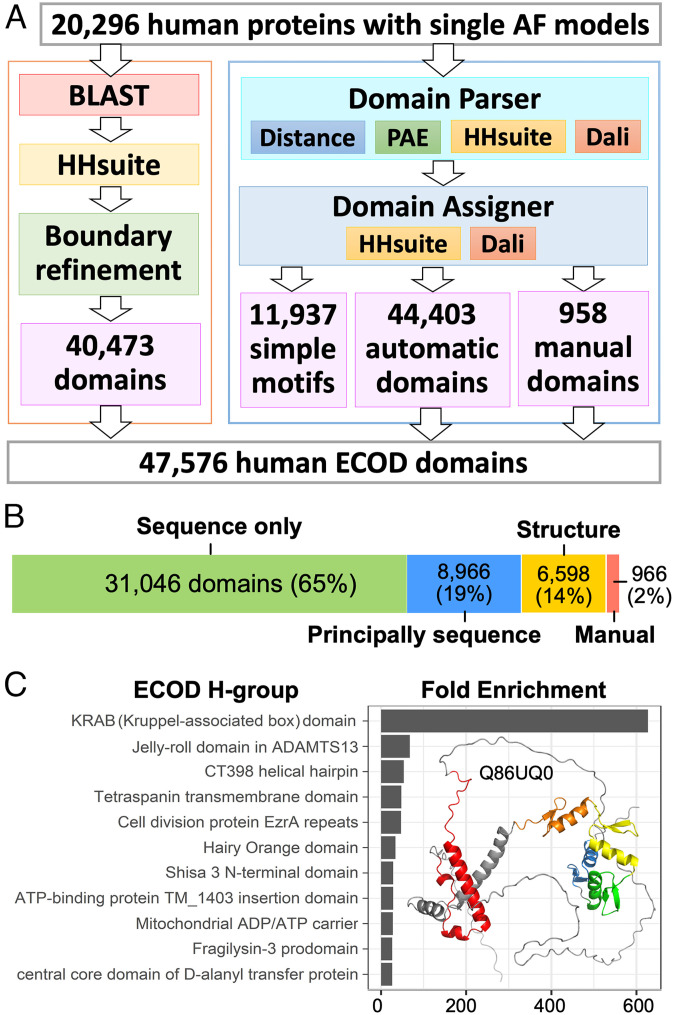
We classified domains in the 20,296 AF models of the human proteome based on their homology to ECOD domains using two pipelines (Fig. 1A). The first is our established pipeline to classify domains from experimental structures. This pipeline is primarily based on sequence similarities detected by BLAST (26) and HHsuite (27) against ECOD domains and previously classified PDB chains (2, 28). This pipeline identified at least one putative domain for 16,868 of the 20,296 AF models; it predicted a total of 40,473 domains, which we call “sequence domains.” 28,393 of these sequence domains were assigned by BLAST, and 12,080 were assigned by HHsuite. 10,968 domains showed ≥99% sequence identity to ECOD domains, suggesting that their structures had been determined experimentally and classified into ECOD.
-
•
These sequence domains only consist of 5.4 million (M) residues, around 51% of the 10.5M residues in the input models. AF models can be less globular and contain more repetitive regions and flexible loops than experimentally determined structures (22). Although manual inspection of residues outside our sequence domains revealed many disordered regions and simple helical structures (e.g., linkers and coiled coils) (SI Appendix, Fig. S2), it was unclear how many domains were missed due to the lack of confident similarity in sequences or sequence profiles to ECOD domains. We developed a dedicated tool, Domain Parser for AlphaFold Models (DPAM), which is described in a separate paper (29). The DPAM recognizes globular domains from AF models based on a) interresidue distances, b) predicted aligned errors (PAE) between residues, and candidate homologous ECOD domains found by c) HHsuite and d) Dali (30).
Fig. 1.
Overview of our domain classification pipeline and results. (A) Our pipeline to classify domains in AF models of human proteins. (B) The number of domains classified using different types of evidence. (C) The most enriched ECOD H-groups among human proteome compared to representative ECOD domains (filtered by 99% identity) from experimental structures. The most enriched H-group, KRAB domains (red), come from KRAB-C2H2 zinc fingers (orange, yellow, green, and blue) that are abundant in humans, and one such example is shown on the right.
Based on our previous benchmark, DPAM was able to distinguish globular domains from domain linkers and recognize 98.8% of domains from AF models. The DPAM pipeline identified 57,298 domains. 11,937 of these domains were excluded because they appear to be simple structure motifs like single helices or helical hairpins (Methods). The remaining 45,361 domains are referred to as “DPAM domains.” To assign DPAM domains to the ECOD hierarchy, we developed an automatic domain assigner to evaluate the probability for a reference ECOD domain to be from the same T-group as a query domain based on sequence and structure similarities (see Methods). This domain assigner can confidently classify most domains (44,403 out of 45,361) to ECOD based on the top-ranking ECOD reference by DPAM probabilities. The remaining 966 domains (2%) require manual curation.
The domains annotated by both pipelines were integrated to generate a final set of 47,576 domains. 31,046 of these domains were consistently predicted by both pipelines or assigned only by the sequence-based pipeline, i.e., they could have been correctly delineated and assigned merely by sequences. Another set of 8,966 domains (principally sequence) could have been correctly detected by sequences, but structures were needed to refine the boundaries of these domains. The remainder (6,598 domains) required both sequence and structural evidence to define domain boundaries and/or to assign them to the ECOD hierarchy (Fig. 1B). We previously observed that DPAM domains frequently have more accurate boundaries than those of homology-based domain predictions (29), presumably because structural data were directly used in their definition. Additionally, the DPAM probability demonstrated superior performance (by receiver operating characteristic curves) in assigning domains to the ECOD hierarchy than HHsuite probability (SI Appendix, Fig. S9). Therefore, in cases where sequence domains overlap with DPAM domains but show significantly different boundaries, we chose to use the DPAM assignments in the final dataset.
The inclusion of AF models significantly expands the available structural data in ECOD. We compared the human domains against previously classified domains from experimental structures in ECOD. The distribution of these domains among ECOD H-groups was compared against the ECOD representative domains filtered at 99% identity. The top 10 most enriched H-groups are shown in Fig. 1C, and the most prominent case is the Kruppel-associated box (KRAB) domains (ECOD H-group id:556.1). KRAB domains are found in KRAB-C2H2 zinc-fingers that underwent an expansion in tetrapods (31). Only a single KRAB domain (PDB: 1V65) has been structurally characterized by NMR (32), and it is not well ordered without the presence of binding partners. Due to the lack of rigid structure on its own, KRAB domains were largely not detected by the DPAM pipeline but they can be confidently recognized by the sequence-based pipeline. The AF human models increase the number of KRAB domains classified in ECOD to 240. Similarly, the “Jelly-roll domain in ADAMTS13” H-group (ECOD id:10.21) was previously represented by a single ECOD domain (PDB: 3GHN). Subsequently, 26 human homologs of this domain were classified. The “fragilysin-3 prodomain” H-group (ECOD id: 3338.1) is represented by a single bacterial protein possibly transferred from eukaryotes to prokaryotes (33). Twenty human domains expanded this H-group.
Classification of AF models into ECOD allowed us to gain functional insights about proteins using structural and evolutionary data, as we illustrate below. Additionally, comparative analysis of AF models and experimental structures classified into the same homologous group will facilitate future critical analysis of AF models. Despite the overall high quality of AF models, we did observe potential errors in AF models through our manual analysis and comparison to homologous experimental structures. Several such examples are included in the supplemental materials (SI Appendix, Figs. S3 and S4).
AF Models Reveal Previously Unclassified ECOD Domains in Human Proteins and Provide Structural Context for Disease-Causing SAVs.
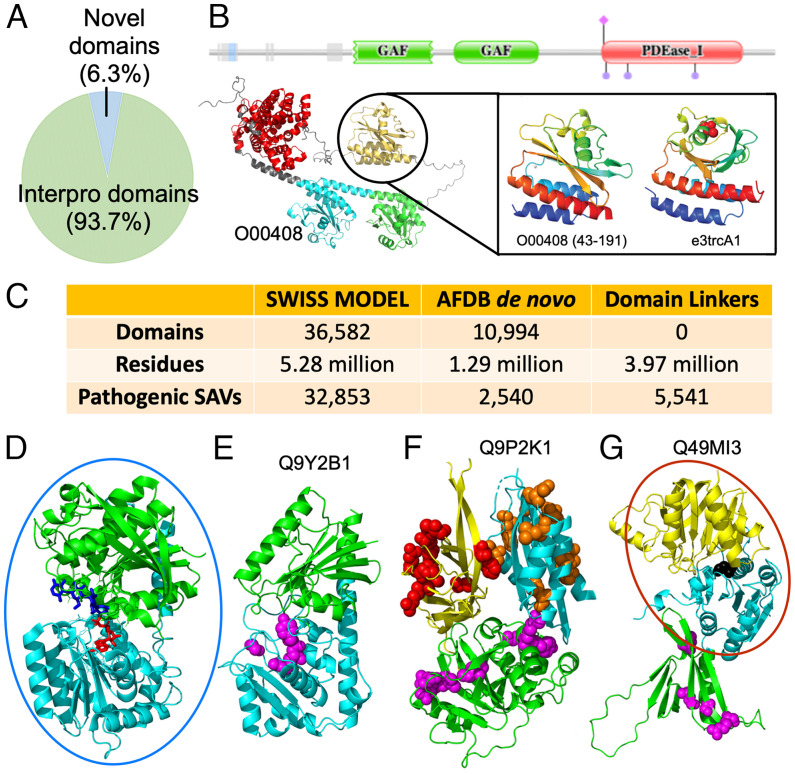
To seek biological insights revealed by AF models, we analyzed the domains we detected in humans against several databases. First, we utilized the InterPro database, a collection of sequence domains annotated from different resources, such as Pfam. A majority (93.7%) of our defined domains belong to known sequence domains, but a small fraction (6.3%, Dataset S1) was not previously annotated by sequence (Fig. 2A). Focusing on these domains in future studies might reveal unique insights about protein function. For example, phosphodiesterase 2 (PDE2A2), a protein that responds to the second messengers cAMP and cGMP, is known to contain two GAF domains and a catalytic PDEaseI domain (Fig. 2B). The first GAF acts as a dimerization domain, whereas the second binds cAMP and cGMP. The model of PDE2A2 reveals another domain at the N terminus that has not been structurally characterized and is bordered by disordered regions. This domain was recognized by DPAM and assigned as another GAF domain. The function of this GAF-like domain remains to be explored experimentally.
Fig. 2.
Insights about human proteins revealed by AF models and ECOD classification. (A) Fraction of unique domains outside annotated domains in the InterPro database. (B) Protein PDeaseI (UNP: O00408) contains three domains according to Pfam (Upper), but the AF model (Bottom Left) reveals 4 ECOD domains: 1 PDease (red) and 3 GAF domains (cyan, green, and orange). The extra domain is homologous to a known GAF domain (Bottom Right). (C) The number of domains that cannot be modeled by homology based on the SWISS-MODEL repository and the number of residues and known pathogenic SAVs in these domains. (D) Representative UDP-glycosyltransferase (PDB: 6GNE) has duplicated domains (green and cyan) that bind ATP (red stick) and oligosaccharide (blue stick) at the interface. (E) Pathogenic SAVs (magenta sphere) in RXYLT1 N-terminal domain (cyan) line the active site of the assigned UDP-glycosyltransferase fold. (F) CC2D2A model places a known N-terminal C2 domain (yellow) with disease-causing SAVs (red spheres) in between a TGL-like fold (green) with disease-causing SAVs (Magenta spheres) and a C-terminal DLT-like fold with pathogenic SAVs (orange spheres). (G) CERKL active site (black spheres) at the interface of the known catalytic class I glutamine amidotransferase-like fold (cyan) and NAD kinase beta-sandwich domain-like fold (yellow). An N-terminal PH domain (green) includes disease-causing SAVs (magenta spheres), with one on the interface with the catalytic domain. In (D–G), experimentally determined domains and domains that can be modeled by homology were placed in blue and red circles, respectively; other domains were structures predicted by AF with no experimentally determined structure in the PDB nor homology models in SWISS-MODEL.
Second, we analyzed homology-based models of human proteins in the SWISS-MODEL repository (updated in 2022). Almost a quarter of human domains we classified (10,994) were not modeled by homology, and we refer to them as de novo domains. These de novo domains contain 1.29M residues and are linked to 2,540 known disease-causing SAVs, according to UniProt (Fig. 2C). These pathogenic SAVs are concentrated in 786 (7%) de novo domains (Dataset S2), and the single-domain protein, glucose-6-phosphatase catalytic subunit 1 (UNP: P35575), contains the largest number of disease-causing SAVs, which are associated with the glycogen storage disease 1A (34). We further computed the number of associated SAVs mapping to de novo domains for each genetic disorder (Dataset S3). The de novo domains from AF models remarkably expanded the available structural information to offer possible molecular mechanisms for a number of diseases, such as nephrotic syndrome 1, Leber congenital amaurosis 1, and macular corneal dystrophy.
We manually studied human ECOD domains that are not annotated by InterPro, not modeled by SWISS-MODEL, but with pathogenic SAVs, and several interesting examples are discussed below. The positions of these SAVs in the AF models may suggest their roles in disease, especially in the context of the functions of evolutionarily related domains. SAVs in the core of a domain might affect structural stability or disturb enzymic sites; SAVs in the interfaces between domains may affect crucial interdomain interactions; surface-exposed SAVs may disrupt a protein’s interaction with other molecules, affecting its subcellular localization and function.
Ribitol-5-phosphate xylosyltransferase 1 (RXYLT1) functions in phosphorylated O-mannosyl trisaccharide biosynthesis. Pathogenic SAVs in RXYLT1 are implicated in severe cobblestone lissencephaly (35) and muscular dystrophies (36). While RXYLT1 does not have a homology-based model in SWISS-MODEL repository, the AF model confidently predicts two domains following the N-terminal TMH and disordered region. DPAM assigns both RXYLT1 domains as UDP-glycosyltransferase homologs based on their structural similarity. Related UDP-glycosyltransferases include duplicated domains, with the N-terminal domain binding ATP and the C-terminal domain coordinating oligosaccharide substrate at the domain interface (Fig. 2D). The RXYLT1 fold has diverged significantly from the closest structure, but all the three pathogenic SAVs map to the N-terminal domain near the putative active site (Fig. 2E), supporting the hypothesis that loss of phosphorylated O-mannosyl trisaccharide activity leads to disease.
The coiled-coil and C2 domain-containing protein 2A (CC2D2A) plays a critical role in cilia formation. The CC2D2A AF model includes two domains with known pathogenic SAVs in addition to the known C-terminal C2 domain: a transglutaminase-like (TGL) cysteine proteinase domain (37) and a dynein light chain (DLC) domain. The evolutionary roots of the TGLs suggest a relationship with dynein ATPases (37), and the coexistence of TGL and DCL domains in CC2D2A provides additional support for this relationship. C2, TGL, and DLC domains confidently interact with each other according to the PAEs of the model. Pathogenic SAVs in CC2D2A that cause Meckel syndrome type 6 or Joubert syndrome type 9 map to all the three domains (Fig. 2F). Several of these SAVs are at the domain interface, while others cluster at the surface. The C2 domain is thought to localize the protein to membranes (37). Thus, the surface cluster of disease-causing SAVs might contribute to membrane localization. The TGL domain includes pathogenic SAVs surrounding an active site lacking catalytic residues, suggesting that the fold has evolved to bind substrates without modifying them. Alteration of this binding activity by SAVs probably leads to disease.
The ceramide kinase-like (CERKL) protein is associated with an autosomal recessive form of retinitis pigmentosa. As a ceramide kinase homolog, CERKL does not modify ceramide, and its substrate is currently unknown. While homology models exist in SWISS-MODEL for the C-terminal catalytic domain, the CERKL structure model by AF confidently positions (according to PAE) an N-terminal PH domain to interact with the catalytic CERKL domain with a class I glutamine amidotransferase-like fold (Fig. 2G). One disease-causing SAV is at the interface between the PH and catalytic domains, suggesting that this domain interaction is vital for function. Many PH domains bind phosphatidylinositol, including the N-terminal domain in the related enzymes sphingosine kinase II and ceramide kinase (38), suggesting that the PH domain localizes CERKL to the membrane where it modifies substrate. The positions of pathogenic SAVs suggest that altered domain positioning or membrane localization leads to disease.
Comparison of Domains Classified in Different Model Organisms.
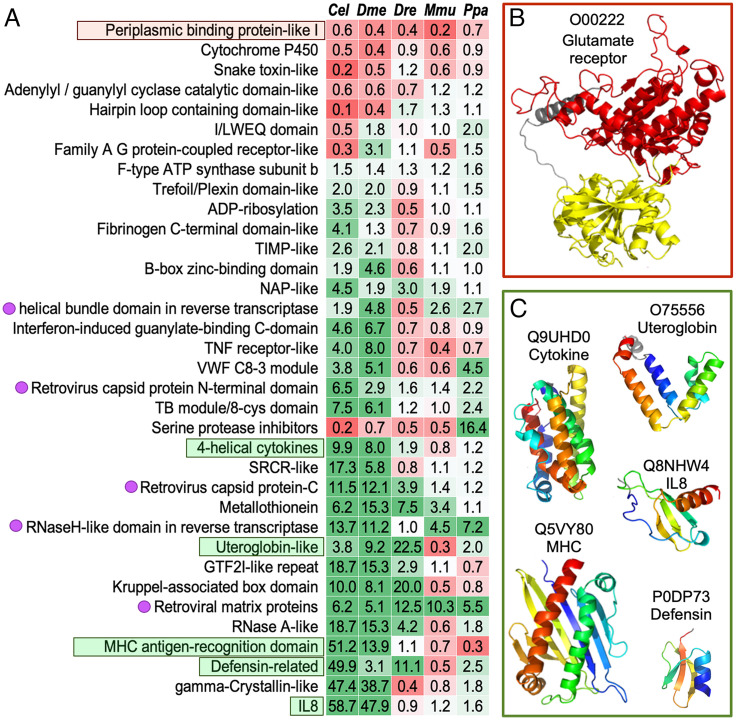
We classified domains in a series of model organisms, including Caenorhabditis elegans (worms, Cel), Drosophila melanogaster (flies, Dme), Danio rerio (fishes, Dre), Mus musculus (mice, Mmu), and Pan paniscus (chimpanzees, Ppa) using the DPAM pipeline. We compared the DPAM domains from human proteins against these model organisms and identified ECOD T-groups where human domains are significantly overrepresented or underrepresented. T-groups where human domains show significant changes relative to at least three other model organisms are shown in Fig. 3A. These domains tend to function in the communication between an organism and its environment, and a number of these changes might be associated with the unique features of humans and mammals. Most of these overrepresented and underrepresented T-groups in human could be revealed without AF models, because majority of these domains can be classified through sequence-based approaches.
Fig. 3.
Significant changes in the number of domains in ECOD T-groups among different model organisms. (A) Overrepresentation (green cells) and underrepresentation (red cells) of ECOD T-groups in human proteins compared to other species. The names of the T-groups are on the left, and the numbers in the table are , where is the fraction of human domains in this T-group and is the fraction for another species as labeled on the top. When a species lacks domains of a T-group, we used a pseudo count of 0.5, i.e., , and is the total number of domains in a species. T-groups illustrated in panels (B) and (C) are in red and green boxes, respectively. Magenta dots on the left label domains originated from retroviruses. (B) A representative structure for the periplasmic binding domains from glutamate receptors. (C) Representative structures for domains involved in immune responses.
Humans are depleted of periplasmic binding protein-like I, a group of domains found in glutamate receptors (Fig. 3B) (39). These receptors are responsible for gustatory and olfactory sensing, and humans were known to have experienced loss in odor and taste sensing during evolution (40), especially compared to rodents. In contrast, compared to lower Eukaryotes, mammals experienced an expansion in multiple ECOD T-groups involved in immune responses (Fig. 3C), including chemokine (IL8), cytokines, major histocompatibility complex (MHC) proteins, defensins, and uteroglobins. Genes involved in host defenses are frequently subject to rapid evolution (41). The expansion of genes in these homologous groups and subsequent functional divergence could be a mechanism for gaining immunity against pathogens.
The UniProt reference human proteome contains multiple retroviral domains (labeled by magenta dots in Fig. 3A) that are missing in other species. Many of these domains belong to the endogenous retrovirus group K member gag polyproteins (HERVK), which are known to display human-specific integrations and amplifications (42, 43). Several of these retroviral genes have been shown to be able to produce viral particles and infect humans today (44, 45). More details about these retroviral proteins encoded in the human genomes are in supplemental material. Other eukaryotic genomes in our comparison also contain many genes or pseudogenes likely originated from retroviruses (46), but they are frequently removed from the annotated protein set due to the lack of evidence for their transcription and translation.
Identification of Distant GPCR-Like Folds in the Human Proteome.
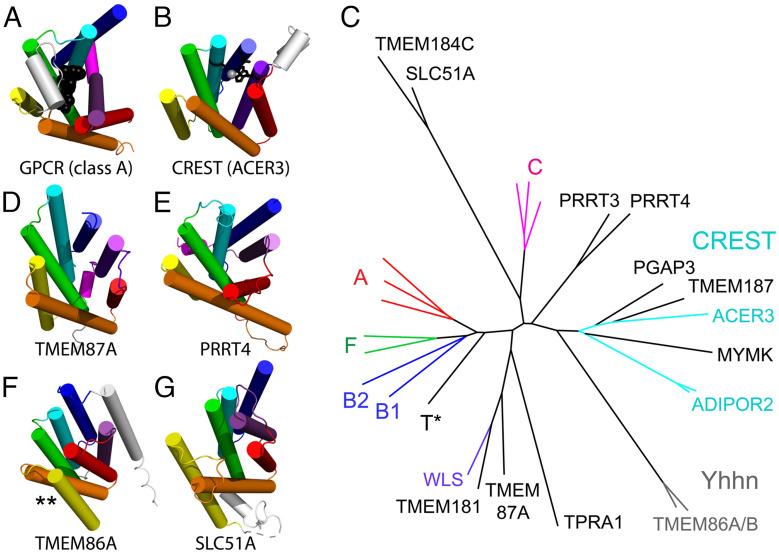
G-protein coupled receptors (GPCRs) are one of the most populated superfamilies in the human genome, with their members being targeted by over one-third of marketed drugs (47, 48). Human GPCRs are distributed among six main classes (A, B1, B2, C, F, and T) based on their phylogenetic and functional characteristics (49). The GPCR fold adopts a seven-transmembrane helix bundle that meanders with an up-and-down topology (Fig. 4A). GPCRs bind an extracellular ligand in the center of the bundle, which causes a conformational change that transmits a signal to the cytoplasm. A similar seven-transmembrane meandering topology is exhibited by members of a large and diverse superfamily of putative membrane-bound hydrolases called CREST (Fig. 4B) (50). The CREST superfamily binds substrates in a similar mode as the GPCRs. Based on this common active site and similar topology, ECOD classifies the GPCRs and CREST hydrolases as homologs.
Fig. 4.
Classification of GPCR-like folds. All folds are colored in a rainbow from TMH1 (blue) to TMH7 (purple) and labeled below. (A) Representative classic GPCR fold of Beta1 adrenergic receptor (class A, PDB: 7BU7) shown with a bound agonist (black spheres) that depicts the ligand binding site and a cytoplasmic helix that interacts with the heterotrimeric G-protein complex (magenta). (B) Representative CREST family enzyme alkaline ceramidase (ACER3, PDB: 6YXH) is shown with a catalytic zinc (gray sphere) and active site residues (black stick). (C) The structure tree for GPCR classification is colored and labeled by experimental structures for GPCR classes (A–C and F), CREST (cyan), and WLS (purple). Models are black, with a GPCR Taste receptor model indicated (T*), or gray (Yhhn family). (D) TMEM87A and (E) PRRT4 have a C-terminal helix (magenta). (F) The TMH4 (yellow) and TMH5 (orange) are flipped in TMEM86A (marked by **), and (G) SLC51A has a flattened meander of helices.
In our classification, we found 893 human proteins with GPCR-like domains (Dataset S4): Most of them could be detected by sequence, but 35 required support from structural data. A subset of these GPCR-like domains, especially those lacking substantial sequence similarity to classic GPCRs, were placed in a structure-based tree of different GPCR classes and CREST enzymes (Fig. 4C). The tree highlights the relationship between the main classes of classic GPCRs [24]. The class C GPCR structures, which include an extracellular β-hairpin insertion to the core 7-TMH fold, represent the most divergent structure class among the classic GPCRs. The class C glutamate receptor-like family is thought to be among the phylogenetically oldest of the classic GPCRs. The glutamate-binding activity expanded greatly to other functions (pheromone-, taste-, and calcium-sensing) during vertebrate evolution (51).
The CREST superfamily (50) structures (Fig. 4C, cyan) are distinct from the classic GPCRs in the structure tree. Several AF models of human proteins are grouped with CREST-like hydrolases. Among these, TMEM187 lacks the typical zinc-coordinating residues found in CREST hydrolases. However, its sequence and structure similarity to a known CREST family member, alkaline ceramidase 3 (ACER3), suggests that it might bind a similar lipid substrate but does not hydrolyze it using zinc. The human protein Myomaker (MYMK) is not known to be an enzyme. However, it controls mammalian myoblast fusion by mediating cell membrane lipid mixing (52, 53) and was previously identified as a CREST superfamily member (50). Interestingly, the MYMK-predicted structure includes several zinc-coordinating residues near the active site and might mediate the hydrolysis of membrane lipids.
Other GPCR-like AF models are distributed throughout the tree. Some of these include a short helix following the GPCR-like fold that might mediate interaction with cytoplasmic G-protein subunits. For example, the TMEM87A (Fig. 4D) and the PPRT (Fig. 4E) folds include a poorly modeled C-terminal helix. Additionally, the PPRT proteins possess long disordered segments in their termini. Several AF models are in long branches that question their classification as GPCR homologs. The long branch Yhhn structures have a flipped helix (Fig. 4F, marked by *) and require expert curation for their classification as questionable homologs (X-group) or as homologs with an alternate topology (T-group). The longest branch represents SLC51A (Fig. 4G) and TMEM184C, which adopt similar structures. These models have longer helices and a flattened helical meander with respect to the GPCR-like folds, suggesting that they should be classified with questionable homology (X-group). SLC51A is a component of the Ost-alpha/Ost-beta complex that transports bile from intestinal enterocytes into blood (54). The similarity between TMEM184C and SLC51A might suggest a similar transport function for TMEM184C.
Previously Unclassified ATP-Grasp/Protein Kinase Domains in AF Models.
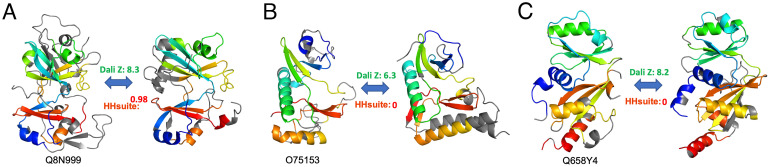
Our classification revealed previously undiscovered ATP-Grasp/protein kinase domains in three human proteins: C12orf29 (pfam: DUF5565), CLUH, and FAM91A1. Both C12orf29 and FAM91A1 adopt an ATP-grasp topology, whereas CLUH adopts a protein kinase-like topology. The closest homolog of C12orf29 with experimentally solved structures is an RNA ligase from Naegleria gruberi (PDB: 6VTB) (55). The ATP-Grasp/kinase domain in C12orf29 was supported by both structure similarity (Dali Z-score: 8.2) and sequence similarity (HHsuite probability: 0.97, Fig. 5A). C12orf29 possesses key catalytic residues found in RNA ligases and thus could be an active enzyme. The substrate of C12orf29 remains to be elucidated.
Fig. 5.
ATP-Grasp/protein kinase domains. (A) C12orf29 (Left) and its close structural homolog RNA ligase (right, PDB: 6VTB). (B) CLUH (Left) and its close structural homolog HopBF1 kinase (Right, PDB: 6PWD). (C) FAM91A1 (Left) and its close structural homolog RimK (Right, PDB: 4IWX). Dali Z-score and HHpred probability scores are shown for the pairs.
Previously unclassified ATP-Grasp/kinase domains in CLUH and FAM91A were only found by structural similarity (Dali) and not by sequence-based searches (HHsuite). CLUH is a multidomain protein functioning in mitochondrial fusion (56) and is widely distributed in eukaryotes. The ATP-Grasp/kinase domain (residues: 354 to 716, Fig. 5B) is located in the middle of CLUH, sandwiched by the N-terminal CLU_N domain and GSKIP domain and the C-terminal eIF3_p135 domain and ARM repeats. The closest known structure to the CLUH ATP-Grasp/kinase domain is a bacterial HopBF1 kinase (PDB: 6PWD) (57). However, the key catalytic residues in the CLUH ATP-Grasp/kinase domain are changed, suggesting the loss of kinase activity. FAM91A1 forms a complex with WDR11 and C17orf75 that functions in vesicle transport (58). The previously unclassified ATP-Grasp/kinase domain is at the C terminus of FAM91A1. Its N-terminal region has a couple of HTH domains and a minimal Rossman-fold domain. The closest known structure of the FAM91A1 ATP-Grasp/kinase domain (Fig. 5C) is Escherichia coli RimK (PDB: 4IWX) (59), an enzyme catalyzing the posttranslational addition of multiple glutamate residues to ribosomal protein S6. Compared to active enzymes in the same family (60), the FAM91A1 ATP-Grasp/kinase domain does not possess critical catalytic residues, suggesting the loss of enzymatic activity. The high sequence divergence in CLUH and FAM91A1 ATP-Grasp/kinase domains possibly explains why sequence-based methods did not detect them.
Altered catalytic residues in protein kinases, as observed in CLUH, often lead to a presumption of the family being pseudokinases. However, such pseudokinases may accommodate different ATP-binding orientations, exhibit catalytic site migration, and thus function as atypical protein kinases. Identification of atypical kinase families and elucidation of their activities has revealed diverse catalytic activities, such as RNA capping in SARS-CoV-2 (61), AMPylation by SelO (62), and glutamylation by the Legionella effector SidJ (63). Similar functional diversity may be revealed after identification of new families.
Conclusion
We developed tools to classify AF models into the ECOD hierarchy. Using these tools and manual curation, we detected and classified domains from AF models of the entire human proteome. On the one hand, this work serves as a prototype to expand ECOD to include the avalanche of predicted structures and to cover the entire protein universe eventually. On the other hand, our classification, presented on the ECOD website, will help the scientific community to utilize valuable structural data and gain functional insights about proteins. Comparison of classified domains from human proteins against other model organisms revealed the expansion and depletion of protein families during evolution. Additionally, our classification revealed domains not found by previous annotation efforts and structures that might explain the mechanisms of various diseases. Finally, our classification identified additional members from protein families of biological significance, such as GPCRs and protein kinases. Experimental characterization of these proteins might lead to the discovery of unique functions.
Materials and methods
Domain Classification Using Our Established Sequence-Based Pipeline.
We downloaded 23,391 models of human proteins from AFDB on July 1, 2021 (http://alphafold.ebi.ac.uk). Since long proteins were modeled in multiple overlapping segments, these models cover 20,504 human proteins, and we focused on the 20,296 proteins represented by single models. We sequentially used BLAST+ (e-value < 0.00005, hit coverage > 70%) against ECOD domains (v283), HHsuite (v3, probability > 90%, hit coverage > 70%) against ECOD F70 (from by HHsuite developers), and HHsuite against PDB70 (from https://wwwuser.gwdg.de/~compbiol/data/hhsuite/) to search homologs for each human protein. Homologous PDB entries (probability > 90%) were split into domains according to our previous ECOD classification. A domain was considered a valid homolog if the alignment covered > 70% of its residues. These homologs were used as a reference to split and assign domains in a query protein to the ECOD hierarchy. Where available, domain boundaries were optimized using structural domains identified by PDP by three principles. First, a PDP domain that significantly overlaps (> 70% bidirectional overlap between a PDP and sequence domain) with a sequence domain can shift the sequence domain’s boundaries. Second, short linkers between domains will be divided between these domains by assigning each residue to the domain with which it forms the most sidechain contacts. Third, we prefer to place domain boundaries between secondary structure elements. This pipeline is similar in form to the sequence-based classifier for experimental structures previously used in ECOD (1).
Domain Classification Using Our Pipeline Developed for AF Models.
Using a set of 18,759 AF models whose close homologs (sequence identity ≥ 95%) had been classified into ECOD as a benchmark, we have developed a Domain Parser for AF Models (DPAM) described in detail elsewhere (29). Briefly, DPAM first identified and excluded disordered regions or domain linkers, showing high PAE relative to other regions. DPAM then computed several interresidue measurements, including interresidue distance, PAE, and whether the two residues appear in the same HHsuite and Dali hit. These measures were converted to probabilities for two residues to be in the same domain based on regression analyses on the benchmark set. Finally, these probabilities were used to cluster 5-residue segments in an AF model into domains.
We developed a neural network (NN) to assign DPAM domains to the ECOD hierarchy using the DPAM benchmark set. This NN evaluates whether a candidate reference ECOD domain found by HHsuite or Dali belongs to the same ECOD T-group as the query domain. For each hit ECOD domain and a query DPAM domain, we computed the following parameters to evaluate their sequence similarity:
(1) Hprob: the HHsuite probability;
(2) Hcov: the coverage of the HHsuite alignment over the hit ECOD domain;
(3) Hrank: the rank of this ECOD hit’s H-group among all the H-groups detected by HHsuite.
To evaluate structural similarity, we first performed all-against-all Dali comparisons between ECOD domains in the same H-group, namely, internal comparisons. The following scores were used:
(4) Dzscore: Dali Z-score;
(5) Dsum: a summary score of aligned positions in the hit by Dali, where each position’s score was calculated as the fraction of internal comparisons where this position was aligned;
(6) Drank: the rank of this ECOD hit’s H-group among all the H-groups detected by Dali;
(7) Dztile: the quantile of Dzscore among internal comparisons for this hit.
(8) Dstile: the quantile of Dsum among internal comparisons for this hit.
Finally, we evaluated the consistency between HHsuite and Dali using two parameters:
(9) Cshift: the average shift in the index of aligned residue in the hit for the same query residue between HHsuite and Dali;
(10) Ccov: the fraction of residues in a hit domain that both HHsuite and Dali aligned.
In addition, we added the length of the domain, the number of alpha helices, and the number of beta strands in the domain as extra features. Each feature was rescaled to be between 0 and 1 using min-max normalization, and missing features were represented as “−1.” An NN (SI Appendix, Figs. S5 and S6) combined the above 13 features with three dense layers of 64 (activation: ReLU), 16 (activation: ReLU), and 2 (activation: softmax) neurons, respectively. The NN has 1,970 parameters in total. The output of the last dense layer (a vector with two values) represents 1) the probability for a query domain to be in the same T-group as a reference ECOD domain and 2) the probability for a query to be in a different T-group from reference. We trained this NN using the DPAM benchmark with 6,242,581 query-hit pairs for 60 epochs by minimizing the categorical cross entropy. We added regularization of the parameters with L2 norm (coefficient: 0.0002) and used a learning rate of 0.00001 and Adam optimizer.
Fourfold crossvalidation (SI Appendix, Fig. S7) was performed to train and test the model, and the loss for the training and testing set was very similar, suggesting the lack of overtraining. The NN outputs the probability for a hit domain to be in the same ECOD T-group as the query domain, which we dub the “DPAM probability” (SI Appendix, Fig. S8). DPAM probability was used to rank the ECOD hits for a query domain from high to low. The domain with the highest probability was used as a reference to assign the query domain to the ECOD hierarchy. We evaluated the application of DPAM probability to assign a query domain and found that it outperforms HHsuite probability or Dali Z-score (SI Appendix, Fig. S9).
Application of Domain Classification Pipelines and Filtering of the Results.
The DPAM pipeline was used to classify the proteome of humans and the five other model organisms in Fig. 3. For the automatic classification of these proteomes, we used a DPAM probability cutoff of >0.9 to assign domains. The number of proteins and domains is summarized in Dataset S5; these domains are collected in the supplementary dataset associated with this publication, in a Zenodo repository (25), and will be monitored and updated on the ECOD website. Both our sequence-based pipeline and DPAM pipeline were used to classify human proteins. Looser cutoffs were used to assign a human protein by an ECOD reference: 1) DPAM probability above 0.8 or 2) DPAM probability above 0.5, but the query protein contains domains from the same H-group with a probability above 0.8. Cases assigned using the lower cutoffs were inspected manually. Domains assigned by both pipelines were combined. The sequence-based pipeline revealed 2,215 domains that were missed by the DPAM pipelines: These domains tend to be small and lack secondary structure elements, such as zinc fingers and KRAB domains. We added them to the final set of human domains.
Manual curation revealed two problems with the DPAM pipeline. First, domains that are not globular, i.e., elongated domains or those with secondary structure elements that loosely interact with the rest of the domains, may be split into several domains (SI Appendix, Fig. S10). Second, the pipeline may consider some simple structural motifs, like single bend helices, helical hairpins, or beta hairpins, as domains. To alleviate these problems, we merged neighboring partial domains assigned to the same T-group, removed single helices from the confidently assigned (DPAM probability > 0.8) domains, and discarded domains with less than four secondary structure elements if its DPAM probabilities to all ECOD domains were lower than 0.8. The last criterion was used because these domains with simple topologies, without confident sequence homology, cannot be assigned confidently even with careful manual curation.
Analysis of Classified Domains.
Information from InterPro, SWISS-MODEL, and UniProt was used to analyze the classified human domains. InterPro annotated every human protein based on an extensive collection of domain and protein family databases, and we used these annotations to identify unique human domains. The SWISS-MODEL repository provided homology-based models for every human protein. We mapped the human protein sequences to these models (sequence identity > 95%) and identified domains that did not have structures from SWISS-MODEL. Genetic variants collected in the UniProt database were downloaded and filtered to include only the missense SAVs annotated as “Pathogenic” or “Likely pathogenic” and linked to specific diseases. The distribution of these SAVs among human domains was analyzed.
We selected experimentally determined structure representatives from ECOD for each class of GPCRs (A: 7BU7, 6FKC, 7EO4; B1: 6WPW; B2: 7D77; C: 7EB2, 7M3G, 7MTS; F:6OT0, 7EVW), choosing those defined as in the active state by the GPCRdb (49). GPCR class T did not have an experimentally determined structure; thus, the AF models for the human taste receptors were used. CREST superfamily enzymes (6YXH and 3WXV) and WLS (7KC4) were selected from ECOD. All-against-all structure comparisons between these known structures and the identified models (Q86UW1, Q9NVA4, Q5FWE3, C9JH25, Q96FM1, Q14656, A6NI61, Q8N661, Q8N2M4, Q86W33, Q8NBN3, Q9P2C4, AND P59544) were performed using DaliLite (64). Pairwise (ZAB) and self (ZAA, ZBB) Z-scores were transformed into distances using the following equation: -ln[ZAB/(minimum of ZAA, ZBB)]. The structure-based tree was produced using the FITCH program (with global optimization) of the Phylip package (65). Branch lengths generated by these Dali distance measures should be interpreted with care. The distance between domains represents structural similarity rather than evolutionary distance.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Acknowledgments
Q.C. is a Southwestern Medical Foundation-endowed scholar. J.Z. is supported by a training grant RP210041 from the Cancer Prevention and Research Institute of Texas. This research is funded by NSF 2224128 Division of Biological Infrastructure (DBI) and NIH GM127390 to N.V.G. This research is also supported by grant I-2095-20220331 to Q.C. and I-1505 to N.V.G. from the Welch Foundation.
Author contributions
R.D.S., J.Z., and Q.C. designed research; R.D.S., J.Z., L.N.K., J.P., and Q.C. performed research; J.Z. and L.N.K. contributed new reagents/analytic tools; R.D.S., J.Z., L.N.K., and J.P. analyzed data; Q.C. and N.V.G. provided supervision and funding; and R.D.S., J.Z., L.N.K., J.P., Q.C., and N.V.G. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. N.P. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
Domain classification data have been deposited in Zenodo (10.5281/zenodo.6998803) (66). The DPAM pipeline is open-source and its repository is maintained at GitHub (https://github.com/CongLabCode/DPAM) (67).
Supporting Information
References
- 1.Cheng H., Liao Y., Schaeffer R. D., Grishin N. V., Manual classification strategies in the ECOD database. Proteins 83, 1238–1251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng H., et al. , ECOD: An evolutionary classification of protein domains. PLoS Comput. Biol. 10, e1003926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medvedev K. E., Kinch L. N., Schaeffer R. D., Grishin N. V., Functional analysis of Rossmann-like domains reveals convergent evolution of topology and reaction pathways. PLoS Comput. Biol. 15, e1007569 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaeffer R. D., Liao Y., Cheng H., Grishin N. V., ECOD: New developments in the evolutionary classification of domains. Nucleic Acids Res. 45, D296–D302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreeva A., Kulesha E., Gough J., Murzin A. G., The SCOP database in 2020: Expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res. 48, D376–D382 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sillitoe I., et al. , CATH: increased structural coverage of functional space. Nucleic Acids Res. 49, D266–D273 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buljan M., Bateman A., The evolution of protein domain families. Biochem. Soc. Trans. 37, 751–755 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Finn R. D., et al. , Pfam: Clans, web tools and services. Nucleic Acids Res. 34, D247–251 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baek M., et al. , Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu S., et al. , CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nepomnyachiy S., Ben-Tal N., Kolodny R., Global view of the protein universe. Proc. Natl. Acad. Sci. U.S.A. 111, 11691–11696 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Hubner I. A., Arakaki A. K., Shakhnovich E., Skolnick J., On the origin and highly likely completeness of single-domain protein structures. Proc. Natl. Acad. Sci. U.S.A. 103, 2605–2610 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadreyev R. I., Kim B. H., Grishin N. V., Discrete-continuous duality of protein structure space. Curr. Opin. Struct. Biol. 19, 321–328 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaeffer R. D., Kinch L. N., Pei J., Medvedev K. E., Grishin N. V., Completeness and consistency in structural domain classifications. ACS Omega 6, 15698–15707 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grishin N. V., Fold change in evolution of protein structures. J. Struct. Biol. 134, 167–185 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Jumper J., et al. , Applying and improving AlphaFold at CASP14. Proteins 89, 1711–1721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinch L. N., Pei J., Kryshtafovych A., Schaeffer R. D., Grishin N. V., Topology evaluation of models for difficult targets in the 14th round of the critical assessment of protein structure prediction (CASP14). Proteins 89, 1673–1686 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tunyasuvunakool K., et al. , Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.UniProt C., UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porta-Pardo E., Ruiz-Serra V., Valentini S., Valencia A., The structural coverage of the human proteome before and after AlphaFold. PLoS Comput. Biol. 18, e1009818 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thornton J. M., Laskowski R. A., Borkakoti N., AlphaFold heralds a data-driven revolution in biology and medicine. Nat. Med. 27, 1666–1669 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Apweiler R., et al. , The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29, 37–40 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blum M., et al. , The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344–D354 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaeffer R. D., Zhang J., Kinch L., Cong Q., Grishin N. V., DPAM Domain Classification of Human Proteins against ECOD Reference. Zenodo. 10.5281/zenodo.6998803. Deposited 11-28-2022. [DOI]
- 26.Camacho C., et al. , BLAST+: Architecture and applications. BMC Bioinform. 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinegger M., et al. , HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinform. 20, 473 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexandrov N., Shindyalov I., PDP: Protein domain parser. Bioinformatics 19, 429–430 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Zhang J., Schaeffer R. D., Durham J., Cong Q., Grishin N. V., DPAM: A domain parser for alphafold models. Protein Sci. 32, e4548 (2022), 10.1002/pro.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J., Schaeffer R. D., Durham J., Cong Q., Grishin N. V., DPAM: A domain parser for alphafold models (2022), 10.1101/2022.09.22.509116. accessed 23 September. [DOI] [PMC free article] [PubMed]
- 31.Urrutia R., KRAB-containing zinc-finger repressor proteins. Genome Biol. 4, 231 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huntley S., et al. , A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 16, 669–677 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goulas T., Arolas J. L., Gomis-Ruth F. X., Structure, function and latency regulation of a bacterial enterotoxin potentially derived from a mammalian adamalysin/ADAM xenolog. Proc. Natl. Acad. Sci. U.S.A. 108, 1856–1861 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beyzaei Z., Geramizadeh B., Molecular diagnosis of glycogen storage disease type I: A review. EXCLI J. 18, 30–46 (2019). [PMC free article] [PubMed] [Google Scholar]
- 35.Vuillaumier-Barrot S., et al. , Identification of mutations in TMEM5 and ISPD as a cause of severe cobblestone lissencephaly. Am. J. Hum. Genet. 91, 1135–1143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manya H., et al. , The muscular dystrophy gene TMEM5 encodes a ribitol beta1,4-xylosyltransferase required for the functional glycosylation of dystroglycan. J. Biol. Chem. 291, 24618–24627 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang D., Aravind L., Novel transglutaminase-like peptidase and C2 domains elucidate the structure, biogenesis and evolution of the ciliary compartment. Cell Cycle 11, 3861–3875 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Don A. S., Rosen H., A lipid binding domain in sphingosine kinase 2. Biochem. Biophys. Res. Commun. 380, 87–92 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiu J., DeSalle R., Lam H. M., Meisel L., Coruzzi G., Molecular evolution of glutamate receptors: A primitive signaling mechanism that existed before plants and animals diverged. Mol. Biol. Evol. 16, 826–838 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Niimura Y., Nei M., Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J. Hum. Genet. 51, 505–517 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nomiyama H., Osada N., Yoshie O., Systematic classification of vertebrate chemokines based on conserved synteny and evolutionary history. Genes. Cells 18, 1–16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medstrand P., Mager D. L., Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol. 72, 9782–9787 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barbulescu M., et al. , Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9, 861–868 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Turner G., et al. , Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11, 1531–1535 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Boller K., et al. , Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J. Gen. Virol. 89, 567–572 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Temin H. M., Reverse transcription in the eukaryotic genome: Retroviruses, pararetroviruses, retrotransposons, and retrotranscripts. Mol. Biol. Evol. 2, 455–468 (1985). [DOI] [PubMed] [Google Scholar]
- 47.Gloriam D. E., Fredriksson R., Schioth H. B., The G protein-coupled receptor subset of the rat genome. BMC Genomics 8, 338 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hauser A. S., Attwood M. M., Rask-Andersen M., Schioth H. B., Gloriam D. E., Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug. Discov. 16, 829–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kooistra A. J., et al. , GPCRdb in 2021: Integrating GPCR sequence, structure and function. Nucleic Acids Res. 49, D335–D343 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pei J., Millay D. P., Olson E. N., Grishin N. V., CREST–a large and diverse superfamily of putative transmembrane hydrolases. Biol. Direct 6, 37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strotmann R., et al. , Evolution of GPCR: Change and continuity. Mol. Cell Endocrinol. 331, 170–178 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Millay D. P., et al. , Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 499, 301–305 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leikina E., et al. , Myomaker and myomerger work independently to control distinct steps of membrane remodeling during myoblast fusion. Dev. Cell 46, 767–780.e767 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ballatori N., et al. , OSTalpha-OSTbeta: A major basolateral bile acid and steroid transporter in human intestinal, renal, and biliary epithelia. Hepatology 42, 1270–1279 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Unciuleac M. C., Goldgur Y., Shuman S., Caveat mutator: Alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation. Nucleic Acids Res. 48, 5603–5615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang H., et al. , Clueless/CLUH regulates mitochondrial fission by promoting recruitment of Drp1 to mitochondria. Nat. Commun. 13, 1582 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lopez V. A., et al. , A bacterial effector mimics a host HSP90 client to undermine immunity. Cell 179, 205–218.e221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Navarro Negredo P., Edgar J. R., Manna P. T., Antrobus R., Robinson M. S., The WDR11 complex facilitates the tethering of AP-1-derived vesicles. Nat. Commun. 9, 596 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao G., et al. , Structure and function of Escherichia coli RimK, an ATP-grasp fold, L-glutamyl ligase enzyme. Proteins 81, 1847–1854 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Galperin M. Y., Koonin E. V., A diverse superfamily of enzymes with ATP-dependent carboxylate-amine/thiol ligase activity. Protein Sci. 6, 2639–2643 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park G. J., et al. , The mechanism of RNA capping by SARS-CoV-2. Nature 609, 793–800 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sreelatha A., et al. , Protein AMPylation by an evolutionarily conserved pseudokinase. Cell 175, 809–821.e819 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Osinski A., et al. , Structural and mechanistic basis for protein glutamylation by the kinase fold. Mol. Cell 81, 4527–4539.e4528 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holm L., Kääriäinen S., Rosenström P., Schenkel A., Searching protein structure databases with DaliLite vol 3. Bioinformatics 24, 2780–2781 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Felsenstein J., An alternating least squares approach to inferring phylogenies from pairwise distances. Syst Biol. 46, 101–111 (1997). [DOI] [PubMed] [Google Scholar]
- 66.Schaeffer R. D., et al. , DPAM domain classification of human proteins against ECOD reference. Zenodo. https://zenodo.org/record/6998803#.Y_1KGz1By5c. Deposited 28 November 2022.
- 67.Zhang J., DPAM: A domain parser for alphafold models. Github. https://github.com/CongLabCode/DPAM. Deposited 15 December 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Data Availability Statement
Domain classification data have been deposited in Zenodo (10.5281/zenodo.6998803) (66). The DPAM pipeline is open-source and its repository is maintained at GitHub (https://github.com/CongLabCode/DPAM) (67).