Significance
Major immune response pathways control the expression of hundreds of genes that represent potential effectors of the immune response. The Drosophila Toll pathway is required in the host defenses against several Gram-positive bacterial infections as well as against fungal infections. The current paradigm is that peptides secreted in the hemolymph during the systemic immune response are either bona fide antimicrobial peptides or likely ones. The finding of a dual role for one Toll pathway effector in the resilience to both Enterococcus faecalis and Metarhizium robertsii infections underscores an original concept in insect innate immunity. Evolution can select effectors tailored to protect the host from the action of microbial toxins of prokaryotic or eukaryotic origin, independently of antibodies or detoxification pathways.
Keywords: Baramicin A, microbial toxins, Destruxin A, enterocin O16, resilience/disease tolerance
Abstract
The Drosophila systemic immune response against many Gram-positive bacteria and fungi is mediated by the Toll pathway. How Toll-regulated effectors actually fulfill this role remains poorly understood as the known Toll-regulated antimicrobial peptide (AMP) genes are active only against filamentous fungi and not against Gram-positive bacteria or yeasts. Besides AMPs, two families of peptides secreted in response to infectious stimuli that activate the Toll pathway have been identified, namely Bomanins and peptides derived from a polyprotein precursor known as Baramicin A (BaraA). Unexpectedly, the deletion of a cluster of 10 Bomanins phenocopies the Toll mutant phenotype of susceptibility to infections. Here, we demonstrate that BaraA is required specifically in the host defense against Enterococcus faecalis and against the entomopathogenic fungus Metarhizium robertsii, albeit the fungal burden is not altered in BaraA mutants. BaraA protects the fly from the action of distinct toxins secreted by these Gram-positive and fungal pathogens, respectively, Enterocin V and Destruxin A. The injection of Destruxin A leads to the rapid paralysis of flies, whether wild type (WT) or mutant. However, a larger fraction of wild-type than BaraA flies recovers from paralysis within 5 to 10 h. BaraAs' function in protecting the host from the deleterious action of Destruxin is required in glial cells, highlighting a resilience role for the Toll pathway in the nervous system against microbial virulence factors. Thus, in complement to the current paradigm, innate immunity can cope effectively with the effects of toxins secreted by pathogens through the secretion of dedicated peptides, independently of xenobiotics detoxification pathways.
The study of host defense against infections has essentially focused on the immune response and the mechanisms used by the organism to directly attack, kill, or neutralize invading pathogens. This dimension of host defense is known as resistance and in insects is mediated primarily by antimicrobial peptides (AMPs) and also by the cellular immune response and melanization (1–4). However, there is a second complementary dimension known as disease tolerance or resilience whereby the organism is able to withstand and, in some cases, repair damages inflicted by the virulence factors of pathogens or the host’s own immune response (5–7). Some instances of resilience have been reported in Drosophila, e.g., the removal of oxidized lipids by Malpighian tubules through the lipid-binding protein Materazzi, the requirement for CrebA in regulating secretion during the immune response or the enterocyte cytoplasmic purge against pore-forming toxins (8–10). One way to discriminate between resistance and resilience is to monitor the microbial burden of infected hosts. It will be increased during infection of immunodeficient as compared to immunocompetent hosts. In contrast, it will not change much in organisms with defective resilience, which will tend to succumb to a lower load of pathogens, as monitored by measuring the pathogen load upon death (PLUD) (11, 12).
In Drosophila, the Toll pathway is one of the two NF-κB (nuclear factor kappa light chain enhancer of activated B cells) pathways that regulate the systemic immune response to microbial infections and through the MyD88 adapter complex is required in the host defense against many Gram-positive and fungal infections. It regulates the expression of more than 250 genes (9, 13–16). A few AMPs active against filamentous fungi have been identified (Drosomycin, Metchnikowin, and Daisho) (17–19). However, effectors solely regulated by the Toll pathway able to attack pathogenic yeasts or Gram-positive bacteria in vitro have not been described so far. Mass spectrometry analysis performed on the hemolymph of single immune-challenged flies has led to the identification of more than 30 peaks corresponding to Drosophila immune-induced molecules (DIMs) (20, 21). Some of them correspond to known AMPs, whereas others belong to a family of 12 proteins that contain a domain known as the Bomanin domain (21, 22). Ten such Bomanin genes are located at the 55C locus, including DIMs 1 to 3, now referred to BomS1 to S3. The deletion of this locus strikingly phenocopies the Toll mutant phenotype, being sensitive to filamentous fungi, pathogenic yeasts, and Gram-positive bacteria such as Enterococcus faecalis (22). Some short Bomanins that essentially contain only the Bomanin domain may be active against Candida glabrata in vivo (23).
Several DIMs (5, 6, 8, 10, 12, 13, 22, 24) are actually derived from a polyprotein precursor known as IMPPP and until recently their function has not been understood (20, 21). A recent study renamed this protein as Baramicin A (BaraA) and proposed that some of the derived peptides function as antifungal AMPs (24). Here, we report our analysis of BaraA mutants. While we confirm a sensitivity to entomopathogenic fungi, our data clearly establish a susceptibility also to E. faecalis, but not to other pathogens we have tested. Interestingly, the fungal burden does not appear to be altered in the mutants, from the beginning to the end of the infections. Our data indicate that a major function of BaraA is in the resilience against distinct toxins, Destruxin A (DtxA), a pore-forming toxin, and Enterocin V (EntV), a bacteriocin, respectively, secreted by Metarhizium robertsii and E. faecalis. BaraA helps the host recover from DtxA-induced paralysis and appears to be required in glial cells but not in neurons.
Results
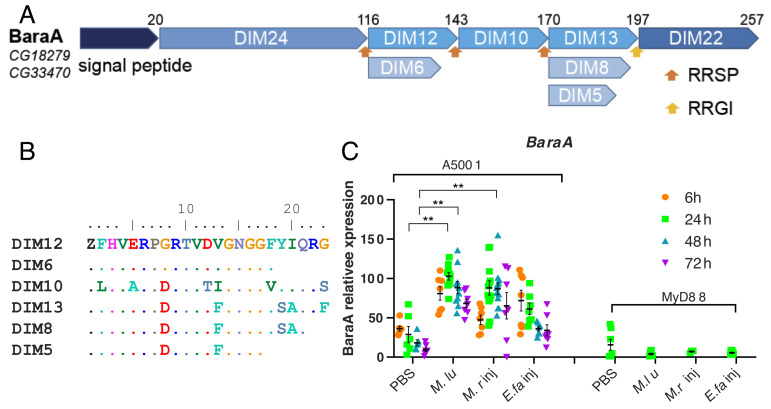
The BaraA locus encodes a polyprotein precursor that is likely processed by a furin-like enzymatic activity, which leads to the release in the hemolymph of multiple DIM peptides. These peptides share extensive sequence similarity, except for the N-terminal DIM24 protein that defines an evolutionarily conserved independent domain (25) (Fig. 1 A and B). For convenience, we shall refer to specific BaraA-derived peptides by their DIM names. Of note, BaraA lies next to the CG18278 gene and the two genes are found as a perfect duplication in some wild and laboratory lines (25) (SI Appendix, Fig. S1 A and B).
Fig. 1.
Structure of the BaraA precursor protein and induction of BaraA expression by an immune challenge. (A) Schematic structure of the BaraA polyprotein. The name of the peptides derived from the processing of the precursor upon furin cleavage is shown as Drosophila-induced immune molecules (DIM), their original name. The type of internal furin-like cleavage sites is indicated by orange and yellow arrows (RRSP, RRGI). (B) Alignment of the short DIM peptides derived from BaraA, referred to by their DIM numbers. (C) Expression of the BaraA gene monitored by qRT-PCR at various time points after the injection of the indicated microbes; M. luM. luteus; M. rM. robertsii; E. fa: E. faecalis. The measured expression of BaraA 24 h after a M. luteus challenge is taken as reference for all other data points and given a 100% value. The means ± SEM are shown in black. Pooled data from three independent experiments, **P < 0.01.
BaraA gene expression is induced by a challenge with the Gram-positive bacteria Micrococcus luteus (used as a reference) and E. faecalis and by injected M. robertsii spores in a MyD88-dependent manner (Fig. 1C), in keeping with previous data at the transcriptional and peptide levels (20, 24). In contrast, the CG18278 gene does not appear to be induced by any of these challenges (SI Appendix, Fig. S2A) and also not upon natural infection (SI Appendix, Fig. S2B).
BaraA Contributes to the Host Defense against E. faecalis.
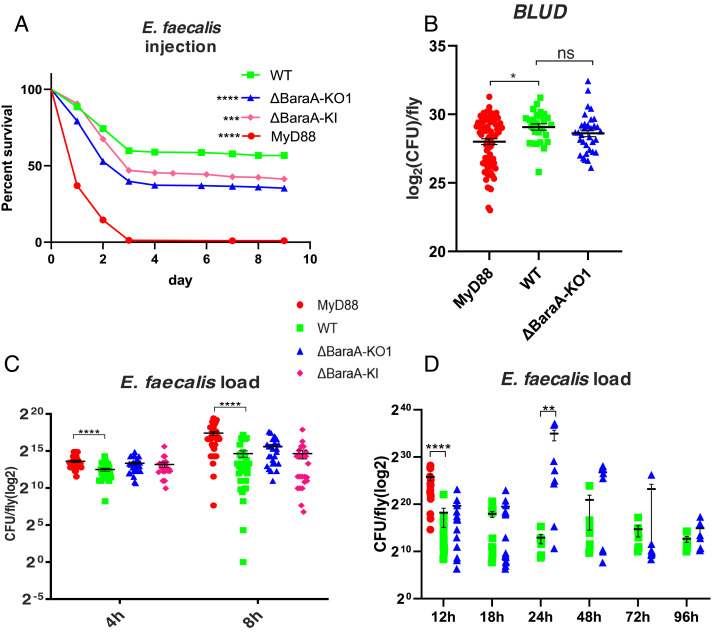
In this work, we have generated a CRISPR–Cas9-mediated knock-out (KO) line, a mCherry knock-in (KI) line, and also used an RNAi line for knockdown (KD) experiments (SI Appendix, Fig. S1 A, C and D). In these lines, the induction of BaraA expression by an immune challenge is hardly detected, both at the transcriptional (SI Appendix, Fig. S1E) and protein levels (SI Appendix, Fig. S3 and Table S1). We have also generated two CG18278 KO lines and tested them along a KD line with an E. faecalis challenge: No consistent susceptibility phenotype was detected (SI Appendix, Fig. S2 C–F). In contrast, we observed a significant susceptibility of BaraA mutant lines isogenized in the wild-type (WT) wA5001 background after the injection of this Gram-positive bacterial strain (Fig. 2A). Contrary to the MyD88 line, measurements of the bacterial burden did not reveal any difference between the KO/KI lines and the isogenic wA5001 control line except for the 24 h time point, also within 30 min after death (bacterial load upon death: BLUD (11); Fig. 2 B–D). Note that at the 24 h and 48 h time points, the distribution of bacterial loads is already bimodal in the BaraA mutant, reflecting that the fate of infected flies is likely settled earlier in the mutant than in the wild-type flies (11). These observations at 24 to 48 h opened the possibility that BaraA may be partially involved in resistance to E. faecalis. We therefore monitored the melanization arm of the immune response by visualizing the proteolytic cleavage that activates pro-phenol-oxidase (PPO) into a catalytically active enzyme after a microbial challenge; we observed a lessened maturation of PPO1 into PO1 in the BaraA KO mutant in three out of four experiments (SI Appendix, Fig. S4 A and A’). As regards the cellular immune response, we noted that the mCherry gene that replaces the BaraA–CG18278 locus was more strongly expressed in adult hemocytes after an E. faecalis challenge (SI Appendix, Fig. S4 B and B’ ). As expected, the BaraA KD line led to an enhanced sensitivity to E. faecalis and to M. robertsii when the short RNA hairpin was ubiquitously expressed (SI Appendix, Fig. S4 C and D). When we silenced BaraA by RNAi in hemocytes, we did observe that these flies were significantly more susceptible to injected E. faecalis bacteria (SI Appendix, Fig. S4E).
Fig. 2.
Susceptibility of BaraA mutant flies to E. faecalis infection. (A) Survival curves of the isogenic BaraA KO and KI flies infected with E. faecalis NCTC 775. The WT corresponds to a wild-type wA5001 line isogenized in parallel to the KO and KI lines, which behaves like the wA5001 line used for isogenization. Pooled data from six independent experiments, **P < 0.01, ***P < 0.001, ****P < 0.0001. (B) Bacterial load upon death (BLUD) of E. faecalis OG1RF in WT and BaraA KO and KI lines. Pooled data from three independent experiments. (C and D) Bacterial load of E. faecalis NCTC 775 (C) and OG1RF (D) in WT and BaraA KO and KI lines from early time points to 6 d after infection. No significant differences were detected between WT and BaraA mutants at each time point, except for the 24 h one, **P < 0.01. MyD88 was significantly different from WT, ****P < 0.0001. The caption applies to panels B–D. Pooled data from three independent experiments. Data are expressed as means ± SEM.
We conclude that the BaraA mutant lines display an intermediate sensitivity to E. faecalis infection. It is however not fully clear whether the altered bacterial burden measured at 24 h results from a lessened activation of melanization, or a hemocyte-mediated response, albeit an impaired antimicrobial activity can also not be excluded.
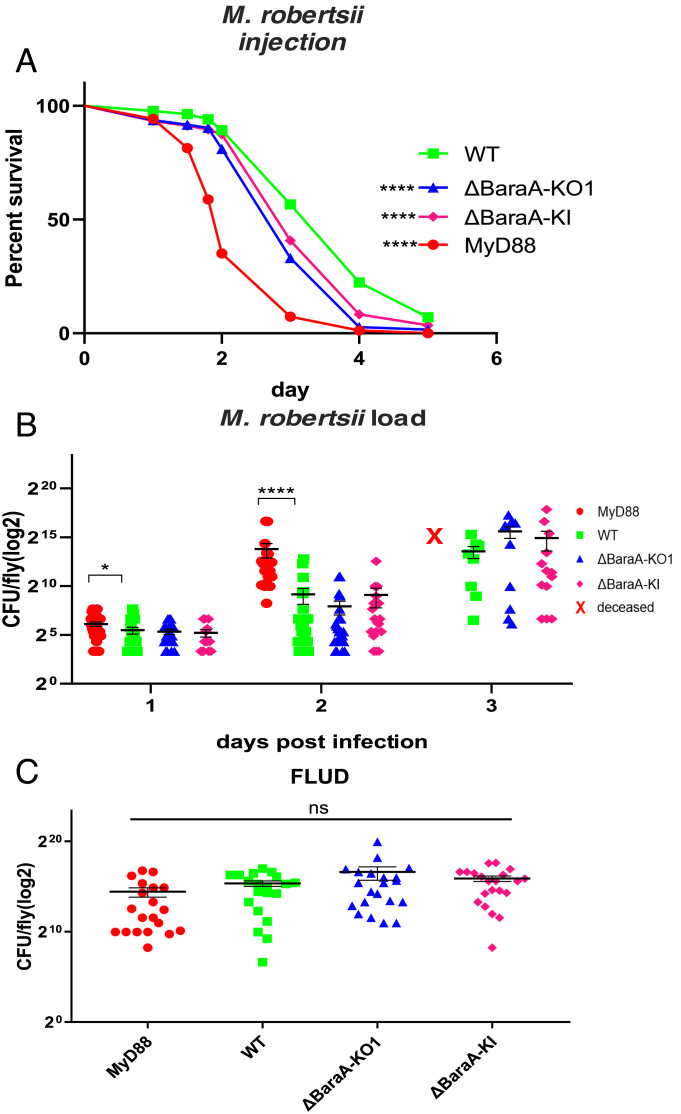
The BaraA Mutant Is Susceptible to M. robertsii Infection Only in the Septic Injury Model.
The BaraA KO and KI lines as well as the KD line consistently exhibited a moderately enhanced sensitivity to the injection of 50 M. robertsii conidia (Fig. 3A and SI Appendix, Fig. S4C). We did not detect an increased microbial titer in these mutants compared to wild-type controls during the infection, in contrast to MyD88; the fungal loads upon death (FLUD) were also similar (Fig. 3 B and C). Interestingly, no susceptibility to M. robertsii in the natural infection model was observed, even though the BaraA gene is induced by this challenge (SI Appendix, Fig. S5 A and A’ ). We have also tested a panel of other bacterial and fungal strains and did not observe any sensitivity to those infections (SI Appendix, Fig. S5 B–F).
Fig. 3.
Susceptibility of BaraA mutants to M. robertsii infection. (A) Survival of isogenic BaraA mutants injected with 50 M. robertsii conidia. Pooled data from 10 independent experiments. Statistical significance between WT and the KO or KI mutants: ****P < 0.0001. (B) Kinetics over 3 d of the fungal load of the BaraA KO and KI mutants injected with 50 M. robertsii conidia. No significant difference was detected between WT and BaraA mutants at each time point. (C) Fungal load upon death (FLUD) of single isogenic BaraA KO and KI flies injected with 50 M. robertsii conidia. No significant differences between WT and the isogenic mutant flies were detected. Three independent experiments have been performed and pooled (B and C). Data are expressed as means ± SEM.
In conclusion, we have found that BaraA appears to be required rather specifically in the host defense against a bacterial opportunistic pathogen, E. faecalis, and an entomopathogenic fungus, M. robertsii. Interestingly, the fungal burden did not appear to be altered in the BaraA mutants, which suggests that BaraA is not required in the resistance against M. robertsii.
The Transgenic Overexpression of BaraA Rescues the Sensitivity of MyD88 Flies to E. faecalis and to M. robertsii to a Limited Degree.
A complementary strategy to the loss-of-function analysis reported above consists in overexpressing the BaraA gene in a wild-type context, thus determining whether it might constitute a limiting factor in host defense against infections. The overexpression of BaraA at the adult stage using transgenic lines failed to enhance the protection of wild-type hosts against M. robertsii and to E. faecalis (SI Appendix, Fig. S6 A and B) yet rescued the BaraA sensitivity phenotype to these pathogens (SI Appendix, Fig. S6 C and D).
We next tested the overexpression of BaraA in a sensitized MyD88 background. The BaraA transgene partially rescued the sensitivity of MyD88 flies to M. robertsii and E. faecalis (SI Appendix, Fig. S6 A and B), suggesting that BaraA can function in the absence of Toll-induced Bomanins. We also checked by Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) spectrometry that the transgenic polyprotein was correctly processed, in the MyD88 background, so that the endogenous signal would not mask that of the transgene-derived protein. As shown in SI Appendix, Fig. S6E and Table S2, MyD88 is not required for the processing of the precursor into the DIM10, DIM12, DIM13, or DIM24 proteins by a putative furin. We also infer that the Toll pathway-dependent Bombardier activity needed to stabilize the expression of short Bomanins is not required for the stability of DIM10, DIM12, and DIM13 (26).
We conclude that the transgenic overexpression of BaraA is not sufficient to confer additional protection against E. faecalis or M. robertsii in the context of a wild type but can partially compensate for the Toll-deficient host defense against these two pathogens.
BaraA Does Not Modulate the Induction of the Toll Pathway.
Besides a potential role of effectors, proteins that are induced by immune signaling pathways may play a role in their feedback regulation. We therefore monitored Toll pathway activation using the steady-state mRNA levels of AMP genes known to be regulated by the Toll pathway such as Drosomycin, Metchnikowin, and IM1 (27–29). As shown in SI Appendix, Fig. S7, we did not observe any influence of the isogenized BaraA KO or KI null mutations over 48 h on their expressions.
BaraA Protects Drosophila from the Action of Secreted Microbial Toxins.
Our data thus far are not in keeping with a function for BaraA in resistance against M. robertsii and are compatible with this possibility as regards E. faecalis. The PLUD data are not indicative of a function in resilience.
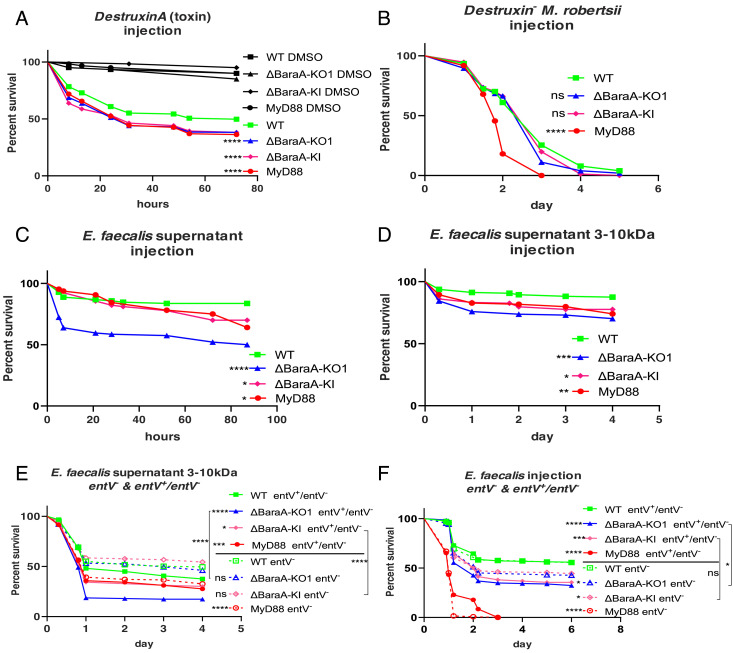
The concept of pathogen load and PLUD relies on the assumption that the virulence of the pathogen correlates with the microbial burden. We have recently established that the function of the Toll pathway in the host defense against Aspergillus fumigatus is not to directly fight off this pathogen, as immunodeficient flies are killed by a limited number of pathogens that are trapped at the injection site. Rather, we have discovered that Toll function in the host defense against A. fumigatus is to limit or counteract some of its secreted mycotoxins (30). As mycotoxins, namely Destruxins (Dtx), have been described as important virulence factors from generalist Metarhizium entomopathogenic fungi (31, 32), we therefore injected DtxA into wild-type and BaraA flies. Interestingly, BaraA KO and KI mutants as well as MyD88 flies reproducibly succumbed to a larger extent than wild-type flies to the injection of DtxA (Fig. 4A), a result confirmed in axenic flies (SI Appendix, Fig. S8A). We next determined that BaraA mutants are not more susceptible than wild-type flies to a challenge with a Dtx mutant M. robertsii strain (32) (Fig. 4B). Taken together, these results suggest that a major function of BaraA in the host defense against M. robertsii is to alleviate or counteract the effects of Destruxins secreted by the fungus in the septic injury model.
Fig. 4.
BaraA-dependent protection of Drosophila flies from the noxious effects of microbial toxins. (A) Mutant flies were injected with 4.6 nl, 8 mM Destruxin A toxin. 80% DMSO was injected as vehicle control. Pooled data from eight independent experiments. Statistically significant differences between wild-type and BaraA mutants, ****P < 0.0001. (B) BaraA mutants were injected with 50 spores of DestruxinS1− M. robertsii mutant strain in which the biosynthesis of Destruxins is blocked. No significant difference was observed between wild-type and BaraA flies; pooled data from five independent experiments. (C) Wild-type, MyD88, and BaraA KO1 and KI mutant flies were injected with the concentrated supernatant from overnight E. faecalis OG1RF cultures. About 50% of BaraA KO1 mutant but not wild-type flies succumbed to this challenge, while 30% of the KI and MyD88 flies succumbed to this challenge. Pooled data from four independent experiments, *P < 0.05, ****P < 0.0001. (D) Same as (C), except that the supernatant was size filtered to retain molecules ranging from 3 to 10 kDa. Pooled data from four independent experiments, *P < 0.05, **P < 0.01, ***P < 0.001. (E) The 3 to 10 kDa fraction supernatant from E. faecalis OG1RF was collected from entV− and entV+/entV− strains. The supernatant from the entV− strain killed BaraA mutants at the same rate as wild-type flies, whereas BaraA KO1 mutants were killed by the complemented entV+/entV− supernatant significantly faster than wild-type flies. Pooled data from eight independent experiments. (F) 0.5 OD, 4.6 nl of the mutant and rescued E. faecalis OG1RF strains were injected. Rescued strain entV+/entV− killed BaraA KO1 faster than wild-type flies. Significant differences between wild-type and BaraA mutant flies were detected upon entV− infection. BaraA KO1 infected by entV+/entV− strain was killed faster than BaraA KO1 infected by entV− strain, while no such significant difference was observed in the case of the BaraA KI line. Pooled data from four independent experiments. (E and F) For each condition (above or below the line in the caption), mutant flies are compared to wild-type flies submitted to the same challenge for statistical analysis, *P < 0.05; ***P < 0.001; ****P < 0.0001.
We then wondered whether BaraA might function in a similar manner in the host defense against E. faecalis. We therefore injected the E. faecalis culture supernatant into flies. Strikingly, whereas wild-type flies survived this challenge well, about 50% of BaraA KO flies and 20% of BaraA KI and MyD88 flies succumbed to the injected supernatant (Fig. 4C). Filtration experiments allowed us to determine that the toxic component of the supernatant can be recovered in a 3 to 10 kD fraction (Fig. 4D). Even though the noxious activity in the E. faecalis supernatant was heat resistant, it was nevertheless susceptible to proteinase K treatment, suggesting a protein component (SI Appendix, Fig. S8 B and C). Interestingly, it has been reported that the bacteriocin enterocin O16 is an E. faecalis virulence factor in Drosophila (33). Enterocin O16 is also known as EntV, which is heat resistant and able to kill some lactobacilli strain as well as to inhibit the hyphal growth, the virulence, and the biofilm formation of Candida albicans (34–36).
We therefore asked whether the toxic activity in the supernatant is still present when using a bacterial entV− strain. We observed that the supernatant from the complemented E. faecalis strain entV+/entV− behaved as that from the wild-type bacterial strain, that is, it killed MyD88 and BaraA KO and KI mutants more than wild-type flies. Strikingly, the supernatant from an entV− E. faecalis mutant strain killed wild-type and BaraA mutants at a similar rate, whereas MyD88 flies succumbed to the same extent to the mutant or complemented wild-type supernatants (Fig. 4E). Similarly, the direct infection-complemented E. faecalis strain entV+/entV killed the immunodeficient MyD88 and BaraA flies more than the wild-type control flies and thus behaved like the wild-type bacterial strain. In contrast, the entV− mutant E. faecalis strain did not kill BaraA KO flies to the same extent than the complemented bacterial strain (Fig. 4F). The EntV peptide derives from the open reading frame found in the ef1097/entV gene, which encodes a preproprotein. This precursor protein gets cleaved by the GelE protease into a 7.2 kDa active peptide (35, 37). As expected, a gelE E. faecalis mutant strain did not kill BaraA flies faster than wild-type flies (SI Appendix, Fig. S8D). We conclude that BaraA protects the flies from the action of the EntV bacteriocin.
Taken together, our data suggest that a major function of BaraA in Drosophila host defense is to protect the fly from specific secreted microbial toxins, whether of prokaryotic or eukaryotic origin.
BaraA Helps Drosophila Recover from DtxA-Induced Paralysis and Is Required in Glial Cells.
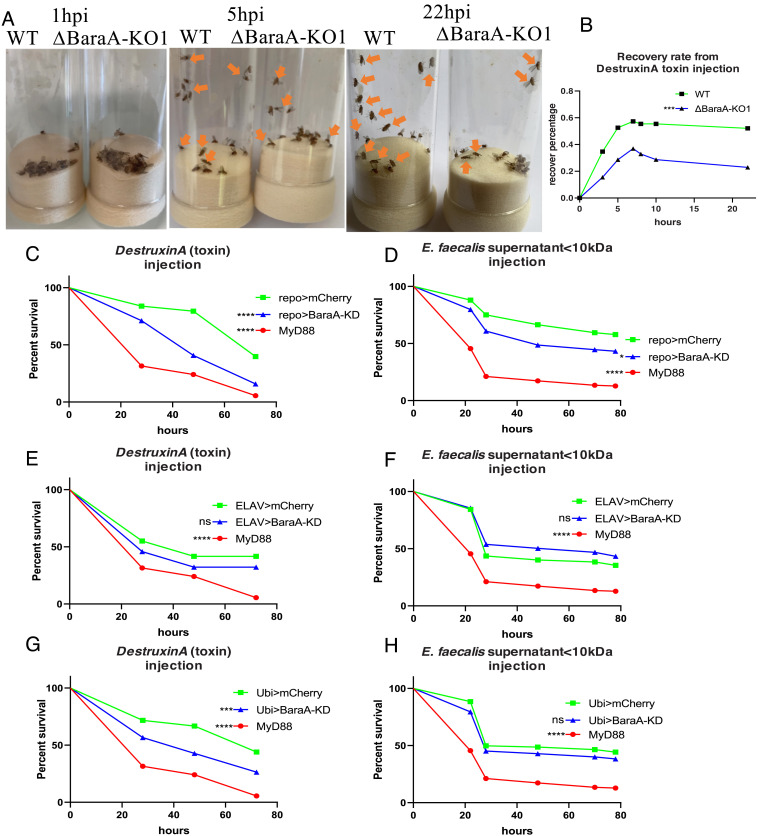
A striking phenotype observed upon the injection of DtxA is the immediate paralysis it induces as flies do not recover from anesthesia as untreated flies do. A careful scrutiny revealed that some 60% of wild-type flies progressively recovered their activity within 5 to 10 h of DtxA injection in contrast to less than 40% for BaraA KO flies (Fig. 5 A and B and Movies S1–S3). In contrast, the injection of the E. faecalis supernatant only temporarily slowed down the flies, a phenomenon difficult to quantify accurately.
Fig. 5.
BaraA counteracts the paralysis induced by exposure to Destruxin A and is required in glial cells. (A) Wild-type flies or BaraA-KO1 mutant flies were injected with 4.6 nl, 8 mM Destruxin A toxin. Pictures were taken at 1, 5, and 22 h postinfection (see also the corresponding Movies S1–S3). After 1 h postinfection, all flies were paralyzed. At 5 h postinfection, 11 wild-type flies woke up, whereas only six BaraA-KO1 mutant flies woke up from toxin injection. At 22 h postinfection, 12 wild-type flies woke up, whereas four BaraA-KO1 mutant flies woke up. (B) Quantification of (A). Pooled data from two independent experiments, ***P < 0.0001. (C and D) BaraA-KD flies were silenced in glial cells (repo-Gal4) and injected with 4.6 nl, 8 mM Destruxin A toxin (C) or 23 nl of E. faecalis supernatant <10 kDa (D). BaraA-KD flies displayed significant difference from wild-type control flies. Pooled data from four independent experiments, *P < 0.05, ****P < 0.0001. (E and F) BaraA-KD flies were silenced in neurons (elav-Gal4) and injected with 4.6 nl, 8 mM Destruxin A toxin (E) or 23 nl of E. faecalis supernatant <10 kDa (F). BaraA-KD flies showed no significant (ns) difference from wild-type control flies. Pooled data from four independent experiments. (G and H) BaraA-KD flies were silenced ubiquitously (ubi-Gal4) and were injected with 4.6 nl, 8 mM Destruxin A toxin (G) or 23 nl of E. faecalis supernatant <10 kDa (H). Ubi-Gal4>BaraA KD flies showed significant difference from wild-type control flies after Destruxin A injection (G). No significant difference between BaraA KD and wild-type control upon E. faecalis OG1RF supernatant injection. Pooled data from four independent experiments.
This set of data suggested that the toxins may somehow interfere with the nervous system. We therefore silenced BaraA gene expression either in neurons or in glia and monitored the survival of flies to injected DtxA or E. faecalis supernatant. Silencing BaraA expression in glial cells but not in neurons enhanced the sensitivity of flies to these challenges (Fig. 5 C–F). In the case of DtxA, the effect was as strong as that observed upon using a ubiquitous driver (Fig. 5 C and G). In contrast, the degree of enhanced susceptibility to the injection of the E. faecalis supernatant was modest and unexpectedly none was detected upon the ubiquitous silencing of BaraA (Fig. 5H).
We conclude that BaraA function is required in glial cells where it may mediate the protection against the effects of DtxA on the nervous system.
Discussion
Our analysis of the BaraA infection mutant phenotype revealed a sensitivity to specific pathogens and not to broad categories of microorganisms as is the case for Toll pathway mutants. Interestingly, we observed a susceptibility to E. faecalis and to M. robertsii, respectively, a Gram-positive bacterium and an entomopathogenic fungus. For both pathogens, specific secreted virulence factors killed a significant fraction of BaraA mutants, whereas the BaraA phenotype of enhanced sensitivity to infection was lost when the corresponding virulence factor genes were mutated in the pathogen. Taken together, these results indicate that a major function of BaraA in Drosophila host defense is to protect it from the action of specific secreted toxins. Indeed, whereas in a concurrent study we showed that Toll pathway mutant flies are sensitive to A. fumigatus restrictocin (30), BaraA mutants did not exhibit any enhanced sensitivity to restrictocin, nor to Beauvericin, a toxin made by Beauveria bassiana, another entomopathogenic fungus that has been reported to kill BaraA mutants faster than wild-type flies (SI Appendix, Fig. S9) (24).
A recently published study proposed that BaraA is involved in resistance to infection to entomopathogenic fungi as an AMP since, besides being sensitive to B. bassiana and Metarhizium rileyi, BaraA mutants exhibit an increased B. bassiana load 48 h after infection (24). In addition, BaraA-derived IM10-like peptides synergize with a membrane-active antifungal compound to kill C. albicans in vitro (24). The fact that BaraA is a polyprotein that produces multiple DIM10-like peptides and that the BaraA locus is found to be duplicated in about 14% of wild-type Drosophila strains caught at one location is in keeping with this possibility, as these strains would be expected to produce twice as much BaraA-derived peptides (25). Because BaraA encodes a polyprotein precursor, we cannot formally exclude such an AMP function for one or several of these BaraA-derived peptides, possibly acting locally to achieve an effective antimicrobial concentration, for instance in the brain. Indeed, the Bomanin family presents a similar situation: Whereas we have shown a function for some specific 55C Bomanins in the resilience to A. fumigatus mycotoxins (30), it is known that at least some Bomanin genes are required for resistance to E. faecalis (22), a finding we have directly confirmed for at least one 55C Bomanin gene (38). We note that if DIM10-like peptides indeed act as AMPs, they would need to act rather specifically against E. faecalis and M. robertsii since we did not detect an enhanced sensitivity to the other pathogens tested in this study. Very specific antibacterial functions for some Drosophila AMPs have been documented (39–41); however, we are not aware of AMPs having dual specificities against both particular bacterial and fungal species. We observed an increased E. faecalis burden at one specific time point, which supports a function in resistance that may be mediated through antimicrobial activity as discussed above, or through a function in mediating melanization or the cellular immune response (Fig. 2 and SI Appendix, Fig. S4). Nevertheless, the findings that bacterial and fungal toxin mutants kill BaraA mutant as efficiently as wild-type mutant flies and that ubiquitous BaraA overexpression does not enhance the protection against E. faecalis or M. robertsii support the concept that an important function of BaraA is to neutralize or counteract the action of specific secreted microbial toxins in the case of E. faecalis or M. robertsii infections.
Interestingly, several studies have shown that besides their direct antimicrobial functions, mammalian α-defensins have the remarkable property to neutralize some microbial pore-forming toxins or enzymes that need to cross the host cell plasma membrane to act on their intracellular targets (42 and references therein). The proposed mechanism of action of these AMPs relies on a common property of these microbial virulence factors: a relative thermodynamic instability that is required for the necessary flexibility to insert into or cross the plasma membrane. α-defensins are constituted by amphipatic α-helices that through hydrophobic interactions with the targeted enzymes are able to destabilize them (42). The unfolded proteins can then be degraded. We know little about the biochemistry of BaraA-derived peptides and no antimicrobial activity at physiological concentrations has been yet found in vitro in the absence of a cofactor. While a similar mechanism may be at play with the EntV protein, it is less likely to function with a hexadepsipeptide that is circular (31) and thus likely difficult to destabilize and degrade because of its circular conformation, even though it is rather hydrophobic.
A study on the evolution of BaraA as well as two related paralogs generated by independent duplication events suggests that the core domain of these three proteins is the N-terminal DIM24 domain, which is associated with only two DIM10-like domains in BaraB and none in BaraC (25). The expression of BaraB and BaraC has been reported not to be induced by an immune challenge (25), a finding we have independently confirmed for BaraB. We did not find a susceptibility of BaraB KO mutant to E. faecalis infection (43), in keeping with a reported lack of detectable immune function (25). BaraB function is essential in neurons, whereas BaraC appears to be expressed in glial cells (25). Interestingly, a BaraA expression fluorescent reporter is detected in brain tissues (24). Thus, it is likely that the DIM24 domain may have a function distinct from the DIM10-like peptides that are thought to act more like AMPs, although definitive evidence is presently lacking. These observations taken together with our finding of a requirement for BaraA expression in glial cells to protect the host against the noxious effects of DtxA therefore open the possibility that the DIM24 peptide might mediate this function, a proposition that requires experimental validation.
The exact mode of action of BaraA-derived peptides in the resilience to microbial toxins remains to be characterized, in as much as they act against distinct types of toxins. Destruxins have been isolated 60 y ago and they appear to act as ionophores that deplete cellular ions such as H+, Na+, and K+ through the formation of pores in the membranes in a reversible process (44), although many other functions have been proposed (31). ClassII bacteriocins also form pores on the membrane of targeted bacteria, but the specific molecular mechanism of action of EntV on eukaryotic cells remains unknown. It presents an activity against the formation of biofilms by the dimorphic yeast C. albicans or the monomorphic yeast C. glabrata. Furthermore, it prevents filamentation of the former in vitro and in vivo (34). Thus, it is unclear whether BaraA would act directly on both toxins, through the same or distinct BaraA-derived peptides, would counteract a common process triggered by Destruxins and EntV such as intracellular ion depletion, or would indirectly alter the physiology of cells exposed to the action of these toxins. It will be important to determine how the toxins act on the host and whether they target preferentially some tissues. For instance, it will be interesting to determine whether the function of BaraA in glial cells is linked to the blood–brain barrier. An emerging theme is that some of the Toll pathway effectors act in the brain and counteract the noxious effects of toxins that also act on the nervous system as exemplified here with the requirement for BaraA expression in glial cells. Interestingly, we have recently found that BomS6 overexpression in the brain protects the flies from the effects of the injected A. fumigatus toxin verruculogen (30).
A specificity of the Toll pathway is that it is required in the host defense against both prokaryotic and eukaryotic pathogens. As compared to the Immune deficiency (IMD) pathway, one interesting feature is that the Toll pathway can be activated by proteases secreted by invading pathogens (45–47). It is interesting to note here that the function of BaraA against two distinct secreted virulence factors, likely pore-forming toxins, provides another point of convergence for the dual role of the Toll pathway, this time at the effector level. It is thus an open possibility that one of the selective pressures that shaped the function of the Toll pathway would be the need to cope with pathogens secreting virulence factors in the extracellular compartment.
Taken together with a concurrent study (30), our work underscores that the Toll pathway mediates resilience against the action of multiple toxin types such as pore-forming toxins, ribotoxins, or tremorgenic toxins, which are mediated by specific Bomanins or BaraA-derived proteins. It is likely that other uncharacterized effectors are able to counteract other toxins to which Drosophila flies are exposed to in the wild. In contrast to the current paradigm according to which secreted peptides act as AMPs, our discoveries illustrate an original concept in insect innate immunity, the ability of the host to counteract secreted microbial virulence factors by dedicated effectors of the immune response.
Materials and Methods
Fly Strains.
Fly lines were raised on media at 25° with 65% humidity. For 25 L fly food medium, 1.2 kg cornmeal (Priméal), 1.2 kg glucose (Tereos Syral), 1.5 kg yeast (Bio Springer), and 90 g nipagin (VWR Chemicals) were diluted into 350 mL ethanol (Sigma-Aldrich), 120 g agar-agar (Sobigel), and water qsp.
wA5001(48) and occasionally yw flies were used as wild-type controls as needed. The positive controls for infection assays for Gram-positive/fungal infections and Gram-negative infections were, respectively, MyD88 and key in the wA5001background. Where stated, mutant flies were isogenized in the wA5001background. For RNAi experiments, virgin females carrying the Ubi-Gal4, ptub1-Gal80ts (Ubi-Gal4, Gal80ts), repo-Gal4, and elav-Gal4 transposon were crossed to Trip lines males carrying an UAS-RNAi (Upstream Activation Sequence-RNA interference) transgene (TRiP) from the Tsinghua RNAi Center: THU0393 (BaraA KD), THU02336.N (CG18278 KD). The control flies were the offspring of the cross of the driver to UAS-mCherry RNAi VALIUM20 (Bloomington Stock Center #BL35785). Crosses with the Ubi-Gal4, Gal80ts driver were performed at 25 °C for 3 d, then the progeny was left to develop at the nonpermissive 18 °C temperature. The hatched flies were kept at 29 °C for 5 d prior to the experiment to allow Gal4-mediated transcription. All crosses involving flies without RNAi expression were performed at 25 °C. Unless stated otherwise, female flies were 5 to 7 d old at the beginning of each experiment.
To generate axenic flies, standard fly media was autoclaved. Antibiotics were added (ampicillin 50 µg/mL, kanamycin 50 µg/mL, tetracyclin 50 µg/mL, erythromycin 15 µg/mL) when it cooled down to 50 to 60°. The embryos were bleached and then cultured on the sterilized media. The sterility of axenic flies (20 d old) was checked on Lysogeny Broth (LB), Bushnell-Haas medium (BHB), Yeast Extract–Peptone–Dextrose (YPD), and deMan, Rogosa, Sharpe (MRS) plates.
Pathogen Infections.
The bacterial strains used in this study include the Gram-negative bacterium Pectobacterium carotovorum carotovorum 15 (strain Ecc15, Optical Density (OD)600 = 50) and the Gram-positive strains E. faecalis National Collection of Type Cultures (NCTC) 775 (American Type Tissue Culture Collection (ATCC) 19433) or OG1RF (ATCC 47077) (OD = 0.1), Micrococcus luteus (OD = 200), and Staphylococcus albus (OD = 10), as well as gelE, entV−, and complemented entV+/entV− and, which are derivatives of the wild-type E. faecalis OG1RF strain (OD = 0.5) (kind gifts of Profs. Garsin and Lorenz, Houston, USA) (34). The fungal strains we used include filamentous fungi, A. fumigatus (5 × 107 spores/mL, 250 spores in 4.6 nL), M. robertsii (ARSEF2575, 1 × 107 spore/mL, natural infection 5 × 104/mL), and DestruxinS1 mutant strain (1 × 107 spore/mL), a kind gift from Prof. Wang, Shanghai, China (32). Besides, we used yeast as well, C. albicans (pricked) and C. glabrata (1 × 109 yeasts/mL). The following media were used to grow the strains: Yeast extract–Peptone–Glucose Broth Agar (YPDA, C. albicans and C. glabrata) or LB–all others at 29 °C (Ecc15, M. luteus, C. albicans, C. glabrata) or 37 °C, entV−, entV+/entV− E. faecalis, Brain Heart Infusion (BHI) medium, 37 °C overnight, rifampicin 100 µg/mL. Spores of M. robertsii and A. fumigatus were grown on Potato Dextrose Agar (PDA) plates at 25 °C or 29 °C (A. fumigatus) for approximately 1 wk or 3 wk (A. fumigatus) until sporulation. We injected 4.6 nL of the suspension into each fly thorax using a Nanoject III (Drummond). Natural infections were initiated by shaking anesthetized flies in 5 mL 0.01% tween-20 solution containing M. robertsii conidia at a concentration of 5 × 104/mL. Infected flies were subsequently maintained at 29 °C (C. albicans, C. glabrata, A. fumigatus, M. robertsii) or at 25 °C (for all other pathogens, except for experiments with RNAi KD flies performed at 29 °C). The flies were anesthetized with light CO2 for about 3 min during the injection procedure and were observed 3 h after injection to confirm recovery from manipulations. Survival experiments were usually performed on three batches of 20 flies tested in parallel and independent experiments pooled for statistical analysis using the log-rank test.
Quantification of the pathogen burden in infected flies.
To characterize the dynamics of within-host microbial loads or BLUDs or FLUDs, live flies were taken at each time point postinjection for pathogen load or flies were infected with E. faecalis or M. robertsii and vials were monitored every 30 min for newly dead flies (PLUD). These flies were then individually homogenized with a bead in 100 µL Phosphate Buffered Saline (PBS) with 0.01% tween-20 (PBST:Phosphate Buffered Saline tween) or PBS. Homogenates were diluted serially (10-fold dilutions, checked by fivefold dilutions for some BLUD experiments) in PBST (or PBS) and spread on LB (E. faecalis) or PDA (M. robertsii) plates for incubation at 37 °C (E. faecalis) or 25 °C (M. robertsii) overnight. Colonies were counted manually. Data were obtained from at least three independent experiments and pooled.
Western Blots.
For western blots, hemolymph samples were collected from 50 flies in a protease inhibitor solution (PBS+:phenylmethylsulfonyl fluoride (PMSF)). Protein concentration of the samples was determined by Bradford assay. 30 µg protein was separated on an 8% gel by SDS-PAGE and transferred to a PVDF membrane. After blocking in 5% bovine serum albumin in PBST for 1 h at room temperature, samples were incubated at 4 °C overnight with rabbit antibodies against Drosophila PPO1 at a 1:10,000 dilution (a kind gift from Prof. Erjun Ling). After washes, a goat anti-rabbit-horseradish peroxidase (HRP) secondary antibody at a 1:20,000 dilution was incubated for 1 h at room temperature. Enhanced chemiluminescence substrate (ECL, General Electric Healthcare) was used according to the manufacturer’s instructions to reveal the blot.
Gene Expression Quantitation.
We followed the protocol as described (49) using primer pairs displayed in SI Appendix, Table S4.
Survival Tests.
Survival tests were performed using 20 to 25 flies per vial in biological triplicates. Female adult flies used for survival tests were 5 to 7-d old. For survival tests using RNAi-silencing genes, flies were crossed at 25 °C for 3 d for laying eggs and then transferred to 18 °C; after hatching, flies were kept for at least 5 d at 29 °C prior to infections. Flies were counted every day. Each experiment shown is representative of at least two independent experiments.
Toxin Injection.
Destruxin A (MedChemExpress) was resuspended in high-quality-grade dimethyl sulfoxide (DMSO) and was diluted in PBS to a 8-mM concentration. 4.6 nL of the solution or of control DMSO diluted in PBS at the same concentration was injected into flies using the Nanoject III microinjector (Drummond). Restrictocin (Sigma) was resuspended in PBS to the concentration of 1 mg/mL, 4.6 nL was injected. Beauvericin (Sigma) was resuspended in high-quality-grade DMSO. 20 mM, 9.2 nL was injected.
Collection and Preparation of E. faecalis Supernatants.
Filter-sterilized supernatant phases were obtained from 10 mL overnight cultures grown in LB medium that were collected by centrifugation at 4,000 rpm for 10 min. The supernatants were sterilized by passing through a 0.2-µm-pore-size sterile syringe filter. The sterilized supernatants were centrifuged through a 15-mL Amicon Centricon filter (Millipore) to separately collect the molecules larger or lower than 10 kDa. 1.5 mL Eppendorf tubes were used to collect the supernatant lower than 10 kDa, which were vacuum freeze-dried for 24 h. The powder was resuspended with H2O and thus concentrated 10 to 20-fold. The solution was filtered on 3 kDa Amicon Centricon filter columns (Millipore) by centrifugation at 10,000 rpm for 30 min. The nonfiltered fraction was then injected into flies with a volume optimized according to the batch (16 to 69 nL) and the same volume of buffer was used for the controls. All experiments were performed at least three times.
Statistics.
Statistical analyses were performed using GraphPad software Prism 6 and R. Data were expressed as means ± SEM. RT-qPCR data were analyzed by ANOVA (one-way) with Dunnett’s multiple comparisons test, with a significance threshold of P = 0.05. Log-rank tests were used to determine whether survival curves of female flies were significantly different from each other. Experiments measuring microbial loads (log2 values) were analyzed using linear models (lm) or linear mixed-effect models (lmer, package lme4) (50) in order to include the different factors of the experiment, such as the fly line or the treatment, and to include random factors, such as the experimental replicates. Significance of interactions between factors was tested by comparing models fitting the data with and without the interactions, using ANOVA. Models were simplified when interactions were not significant. Pairwise comparisons of the estimates from fitted models were analyzed using lmerTest, lsmean, and multcomp packages. Details are included in the legend of each figure. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Supplementary Material
Appendix 01 (PDF)
Wild-type (left) and BaraA KO1 (right) flies were inspected one hour after the injection of Destruxin A.
Wild-type (top) and BaraA KO1 (bottom) flies were inspected five hours after the injection of Destruxin A.
Wild-type (left) and BaraA KO1 (right) flies were inspected 22 hours after the injection of Destruxin A.
Acknowledgments
We thank our colleagues at the Sino-French Hoffmann Institute for help and advice at various steps of this work. We are grateful to Prof. Erjun Ling for providing antibodies, to Prof. Chengshu Wang for the M. robertsii Destruxin− mutant strain, to Profs. Danielle Garsin and Michael Lorentz for the entV and gelE mutant strains (kindly sent by Melissa Cruz, Murray laboratory), to Dr. Lucas Waltzer for the PPO2-Gal4 stock, and the Bloomington and Tsinghua stock Centers for fly stocks. We are grateful to Dr. Inês Pais for expert help on statistical analysis. We would like to thank Sébastien Voisin from the BioPark for his help in mass spectrometry and Yongfang Zheng for expert technical help. We are indebted to the Guangzhou Drosophila Resource Center for the generation of CRISPR–Cas9 knock-out lines. We also thank Bruno Lemaitre and Mark Hanson for their cooperation and discussions on the BaraA gene. This work has been funded by the 111 Project (#D18010; China), the Incubation Project for Innovative Teams of the Guangzhou Medical University, the Open Project from State Key Laboratory of Respiratory Diseases, China, and the China High-end Foreign Talent Program to D.F. J.A.H. is supported by the University of Strasbourg Institute for Advanced Study and J.H. was partly supported by the Chinese Scholarship Council (fellowship #202008440686).
Author contributions
J.H., P.B., and D.F. designed research; J.H., Y.L., P.B., C.C., and K.M. performed research; J.L., P.B., and R.J. contributed new reagents/analytic tools; J.H., Y.L., P.B., S.L., Z.L., and D.F. analyzed data; S.L. helped supervised directly the work from students when in France and in China; Z.L. supervised work from students in China and managed the team as co-PI; and J.H., J.A.H., and D.F. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Raw data have been deposited in Figshare (51).
Supporting Information
References
- 1.Lemaitre B., Hoffmann J., The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Lazzaro B. P., Zasloff M., Rolff J., Antimicrobial peptides: Application informed by evolution. Science 368, eaau5480 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin S. J. H., Cohen L. B., Wasserman S. A., Effector specificity and function in Drosophila innate immunity: Getting AMPed and dropping Boms. PLoS Pathog. 16, e1008480 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanson M. A., Lemaitre B., New insights on Drosophila antimicrobial peptide function in host defense and beyond. Curr. Opin. Immunol. 62, 22–30 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Medzhitov R., Schneider D. S., Soares M. P., Disease tolerance as a defense strategy. Science 335, 936–941 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrandon D., The complementary facets of epithelial host defenses in the genetic model organism Drosophila melanogaster: From resistance to resilience. Curr. Opin. Immunol. 25, 59–70 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Soares M. P., Teixeira L., Moita L. F., Disease tolerance and immunity in host protection against infection. Nat. Rev. Immunol. 17, 83–96 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Li X., Rommelaere S., Kondo S., Lemaitre B., Renal purge of hemolymphatic lipids prevents the accumulation of ROS-induced inflammatory oxidized lipids and protects Drosophila from tissue damage. Immunity 52, 374–387.e376 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Troha K., Im J. H., Revah J., Lazzaro B. P., Buchon N., Comparative transcriptomics reveals CrebA as a novel regulator of infection tolerance in D. melanogaster. PLoS Pathog. 14, e1006847 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee K. Z., et al. , Enterocyte Purge and Rapid Recovery Is a Resilience Reaction of the Gut Epithelium to Pore-Forming Toxin Attack. Cell Host. Microbe. 20, 716–730 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Duneau D., et al. , Stochastic variation in the initial phase of bacterial infection predicts the probability of survival in D. melanogaster. eLife 6, e28298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lafont P., Lauzeral C., Duneau D., Ferdy J.-B., A within-host infection model to explore tolerance and resistance. bioRxiv (2021), 10.1101/2021.10.19.464998 (Accessed 20 October 2021). [DOI]
- 13.Irving P., et al. , A genome-wide analysis of immune responses in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 98, 15119–15124 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Gregorio E., Spellman P. T., Rubin G. M., Lemaitre B., Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. U.S.A. 98, 12590–12595 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boutros M., Agaisse H., Perrimon N., Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell 3, 711–722 (2002). [DOI] [PubMed] [Google Scholar]
- 16.De Gregorio E., Spellman P. T., Tzou P., Rubin G. M., Lemaitre B., The Toll and Imd pathways are the major regulators of the immune response in Drosophila. Embo J. 21, 2568–2579 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fehlbaum P., et al. , Septic injury of Drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. J. Biol. Chem. 269, 33159–33163 (1995). [PubMed] [Google Scholar]
- 18.Levashina E. A., et al. , Metchnikowin, a novel immune-inducible proline-rich peptide from Drosophila with antibacterial and antifungal properties. Eur. J. Biochem. 233, 694–700 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Cohen L. B., Lindsay S. A., Xu Y., Lin S. J. H., Wasserman S. A., The Daisho peptides mediate Drosophila defense against a subset of filamentous fungi. Front. Immunol. 11, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uttenweiler-Joseph S., et al. , Differential display of peptides induced during the immune response of Drosophila: A matrix-assisted laser desorption ionization time-of- flight mass spectrometry study. Proc. Natl. Acad. Sci. U.S.A. 95, 11342–11347 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy F., et al. , Peptidomic and proteomic analyses of the systemic immune response of Drosophila. Biochimie 86, 607–616 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Clemmons A. W., Lindsay S. A., Wasserman S. A., An effector peptide family required for Drosophila Toll-mediated immunity. PLoS Pathog. 11, e1004876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindsay S. A., Lin S. J. H., Wasserman S. A., Short-form bomanins mediate humoral immunity in Drosophila. J. Innate. Immun. 10, 306–314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanson M. A., et al. , The Drosophila Baramicin polypeptide gene protects against fungal infection. PLoS Pathog. 17, e1009846 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanson M. A., Bruno L., Repeated truncation of a modular antimicrobial peptide gene for neural context. PLoS Genet. 18, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin S. J. H., Fulzele A., Cohen L. B., Bennett E. J., Wasserman S. A., Bombardier enables delivery of short-form bomanins in the Drosophila Toll response. Front. Immunol. 10, 3040 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemaitre B., Nicolas E., Michaut L., Reichhart J. M., Hoffmann J. A., The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Ferrandon D., Imler J. L., Hetru C., Hoffmann J. A., The Drosophila systemic immune response: Sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 7, 862–874 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Busse M. S., Arnold C. P., Towb P., Katrivesis J., Wasserman S. A., A kappaB sequence code for pathway-specific innate immune responses. Embo J. 26, 3826–3835 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu R., et al. , The Toll pathway mediates Drosophila resilience to Aspergillus mycotoxins through specific Bomanins. EMBO Rep. e56036 (2022), 10.15252/embr.202256036. [DOI] [PMC free article] [PubMed]
- 31.Pedras M. S., Irina Zaharia L., Ward D. E., The destruxins: Synthesis, biosynthesis, biotransformation, and biological activity. Phytochemistry 59, 579–596 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Wang B., Kang Q., Lu Y., Bai L., Wang C., Unveiling the biosynthetic puzzle of destruxins in Metarhizium species. Proc. Natl. Acad. Sci. U.S.A. 109, 1287–1292 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teixeira N., et al. , Drosophila host model reveals new Enterococcus faecalis quorum-sensing associated virulence factors. PLoS One 8, e64740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graham C. E., Cruz M. R., Garsin D. A., Lorenz M. C., Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 114, 4507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dundar H., et al. , The fsr Quorum-sensing system and cognate gelatinase orchestrate the expression and processing of proprotein EF_1097 into the mature antimicrobial peptide Enterocin O16. J. Bacteriol. 197, 2112–2121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cruz M. R., et al. , Structural and functional analysis of EntV reveals a 12 amino acid fragment protective against fungal infections. Nat. Commun. 13, 6047 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown A. O., et al. , Antifungal activity of the Enterococcus faecalis peptide EntV requires protease cleavage and disulfide bond formation. mBio 10, e01334-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lou Y., Study of the Function of Bomanin Genes at the 55C Locus in Drosophila melanogaster Host Defense against Microbial Infections (Université de Strasbourg, 2021). [Google Scholar]
- 39.Unckless R. L., Howick V. M., Lazzaro B. P., Convergent balancing selection on an antimicrobial peptide in Drosophila. Curr. Biol. 26, 257–262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanson M. A., et al. , Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. eLife 8, e44341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanson M. A., Kondo S., Lemaitre B., Drosophila immunity: The Drosocin gene encodes two host defence peptides with pathogen-specific roles. Proc. Biol. Sci. 289, 20220773 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kudryashova E., et al. , Human defensins facilitate local unfolding of thermodynamically unstable regions of bacterial protein toxins. Immunity 41, 709–721 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang J., Study of Two Toll Pathway Effector Genes Involved in Resilience and Resistance to Microbial Infections in Drosophila (Université de Strasbourg, Strasbourg, 2022), p. 151. [Google Scholar]
- 44.Shanbhag S. R., Vazhappilly A. T., Sane A., D’Silva N. M., Tripathi S., Electrolyte transport pathways induced in the midgut epithelium of Drosophila melanogaster larvae by commensal gut microbiota and pathogens. J. Physiol. 595, 523–539 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gottar M., et al. , Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell 127, 1425–1437 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El Chamy L., Leclerc V., Caldelari I., Reichhart J. M., Sensing of "danger signals" and pathogen-associated molecular patterns defines binary signaling pathways "upstream" of Toll. Nat. Immunol. 9, 1165–1170 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Issa N., et al. , The circulating protease persephone is an immune sensor for microbial proteolytic activities upstream of the Drosophila Toll pathway. Mol. Cell 69, 539–550.e536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thibault S. T., et al. , A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36, 283–287 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Huang C., et al. , Differential requirements for mediator complex subunits in Drosophila melanogaster host defense against fungal and bacterial pathogens. Front. Immunol. 11, 478958 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bates D., Mächler M., Bolker B., Walker S., Fitting linear mixed-effects models using lme4. J. Stat. Software 67, 1–48 (2015). [Google Scholar]
- 51.Huang J., A toll pathway effector protects Drosophila specifically from distinct toxins secreted by a fungus or a bacterium. Figshare. 10.6084/m9.figshare.21967805.v3. Deposited 31 January 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Wild-type (left) and BaraA KO1 (right) flies were inspected one hour after the injection of Destruxin A.
Wild-type (top) and BaraA KO1 (bottom) flies were inspected five hours after the injection of Destruxin A.
Wild-type (left) and BaraA KO1 (right) flies were inspected 22 hours after the injection of Destruxin A.
Data Availability Statement
Raw data have been deposited in Figshare (51).