Significance
Staphylococcus aureus is a versatile opportunistic pathogen responsible for a significant proportion of hospital- and community-acquired infections. S. aureus cell wall-anchored (CWA) surface proteins enhance virulence, promoting host colonization and infection. There are important questions remaining about the mechanisms supporting the surface display of CWA proteins during cell wall biogenesis and division. We show that cell wall hydrolases, including the major S. aureus autolysin, Atl, are important for the display of CWA proteins. We also demonstrate that the type V glycopeptide antibiotic complestatin is a potent antiadhesive compound with promising antivirulence properties. Our findings can be used to guide therapeutic interventions taking aim at virulence as an orthogonal approach to reduce the occurrence and/or severity of S. aureus infections.
Keywords: Staphylococcus aureus, host adhesion, antibiotic resistance, virulence, autolysins
Abstract
Peptidoglycan hydrolases, or autolysins, play a critical role in cell wall remodeling and degradation, facilitating bacterial growth, cell division, and cell separation. In Staphylococcus aureus, the so-called “major” autolysin, Atl, has long been associated with host adhesion; however, the molecular basis underlying this phenomenon remains understudied. To investigate, we used the type V glycopeptide antibiotic complestatin, which binds to peptidoglycan and blocks the activity of autolysins, as a chemical probe of autolysin function. We also generated a chromosomally encoded, catalytically inactive variant of the Atl enzyme. Autolysin-mediated peptidoglycan hydrolysis, in particular Atl-mediated daughter cell separation, was shown to be critical for maintaining optimal surface levels of S. aureus cell wall-anchored proteins, including the fibronectin-binding proteins (FnBPs) and protein A (Spa). As such, disrupting autolysin function reduced the affinity of S. aureus for host cell ligands, and negatively impacted early stages of bacterial colonization in a systemic model of S. aureus infection. Phenotypic studies revealed that Spa was sequestered at the septum of complestatin-treated cells, highlighting that autolysins are required to liberate Spa during cell division. In summary, we reveal the hydrolytic activities of autolysins are associated with the surface display of S. aureus cell wall-anchored proteins. We demonstrate that by blocking autolysin function, type V glycopeptide antibiotics are promising antivirulence agents for the development of strategies to control S. aureus infections.
Staphylococcus aureus is a colonizing opportunistic pathogen causing infections when the host’s immune defenses are breached (1). The cell wall-anchored surface (CWA) proteins of S. aureus are important virulence-associated factors that support processes such as immune evasion, iron acquisition, biofilm formation, and host adhesion (1–3). CWA proteins adhere to different molecules and surfaces within the body, and consequently contribute to the ability of S. aureus to cause a multitude of different diseases (1). There are critical questions remaining about the mechanisms facilitating the surface display of CWA proteins during cell wall biogenesis and cell division. While identifying genetic loci impacting the ability of S. aureus to adhere to different host ligands, we demonstrated a central requirement for the S. aureus so-called “major” murein hydrolase autolysin (Atl) in this process (4). Indeed, there is an extensive body of literature supporting an important role for Atl in surface adhesion. Atl and the functionally interchangeable Staphylococcus epidermidis AtlE enzyme (5) have been associated with adhesion of staphylococci to abiotic surfaces such as polystyrene (6), and host-derived proteins, including fibronectin (6, 7), fibrinogen (6), vitronectin (6, 8), heparin (7), gelatin (7), heat shock cognate 71-kDa protein (Hsc70) (9), and thrombospondin (8). Our genome-wide adhesion screen supported these observations and identified keratin as a host ligand for which binding by S. aureus was affected by loss of Atl (4).
Here, we sought to examine the molecular basis underlying the role of Atl in S. aureus host adhesion, hypothesizing that autolysin function could be associated with the surface display of CWA proteins with affinity for host ligands such as fibronectin, keratin, and fibrinogen. To investigate, we utilized a type V glycopeptide antibiotic, complestatin, as a chemical probe of autolysin function; this antibiotic was recently shown to bind peptidoglycan and block the hydrolytic activities of multiple autolysins essential for remodeling of the cell wall during growth (10). At concentrations that were subinhibitory for growth, we demonstrate that complestatin is a potent antiadhesive inhibitor, reducing the affinity of S. aureus for various clinically important host ligands, including immunoglobulins. Complestatin was subsequently shown to reduce the surface levels of the CWA fibronectin-binding proteins (FnBPs) and protein A (Spa). We also generated a chromosomally encoded, catalytically inactive variant of the Atl enzyme, which was produced to levels equivalent to those in the isogenic wild-type strain. Similar to complestatin-treated cells, Atl loss-of-function reduced the surface levels of CWA proteins, and consequently the affinity of S. aureus for host ligands. Cellular localization studies revealed the FnBPs and Spa were differentially affected by autolysin loss-of-function, and phenotypic studies showed that Spa was sequestered at the septum of complestatin-treated cells. Using a mouse model of infection, we also revealed that complestatin exhibited antivirulence properties. In summary, this study provides a more complete model of the mechanisms supporting the surface display of S. aureus CWA adhesins, highlighting an important virulence-associated role for peptidoglycan hydrolases.
Results
Type V Glycopeptide Antibiotics Inhibit S. aureus Adhesion to Host Ligands.
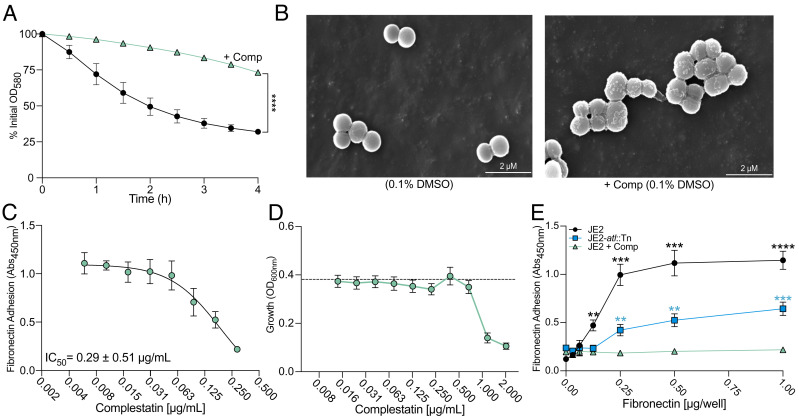
Through binding to peptidoglycan and blocking the activities of autolysins, the type V glycopeptide antibiotics complestatin and corbomycin can be uniquely utilized as chemical probes of peptidoglycan hydrolases (10). Since inactivation of the gene encoding the major S. aureus autolysin, atl, significantly reduced the affinity of S. aureus for host ligands (4), we hypothesized complestatin and corbomycin could attenuate S. aureus adhesion to host ligands, by blocking the function of Atl. We first confirmed that complestatin-treated cells exhibited impaired autolysis and cell division defects in methicillin-resistant S. aureus (MRSA) USA300 JE2 clone Los Angeles County (LAC) jail using detergent-induced autolysis assays (Fig. 1A) and scanning electron microscopy (SEM) (Fig. 1B). We then performed dose-dependent fibronectin adhesion assays in the presence of varying concentrations of complestatin to determine the half-maximal adhesion inhibitory concentration (Fig. 1C). In these assays, the host ligand was immobilized to the surface of microtiter plates and S. aureus adhesion to fibronectin was detected using our optimized enzyme-linked immunosorbent assay (ELISA)-based approach (4). Complestatin reduced the affinity of S. aureus for fibronectin (Fig. 1C) at concentrations that were subinhibitory for growth (Fig. 1D). Indeed, adhesion was reduced to a level greater than an isogenic atl transposon mutant (atl::Tn) (Fig. 1E). We additionally investigated the antiadhesive properties of the type V antibiotic corbomycin, which is also documented to bind peptidoglycan and block the activity of autolysins (10). At concentrations that were subinhibitory for growth (SI Appendix, Fig. S1A), corbomycin similarly inhibited fibronectin adhesion (SI Appendix, Fig. S1B); however, the compound was not as potent as complestatin. Finally, we demonstrated these compounds were broad-spectrum antiadhesives that additionally attenuated S. aureus adhesion to the host ligands keratin (SI Appendix, Fig. S1C) and fibrinogen (SI Appendix, Fig. S1D). In summary, we reveal that complestatin and corbomycin are potent antiadhesive agents with activity against MRSA. Since complestatin was the more potent of the two, all subsequent studies were performed with this compound.
Fig. 1.
Complestatin inhibits S. aureus adhesion to fibronectin at concentrations subinhibitory for growth. (A) Detergent-induced whole-cell autolysis assay of MRSA USA300 JE2 clone LAC (JE2) (black) and complestatin-treated cells (green). (B) Scanning electron microscopy (SEM) analysis of JE2 and complestatin-treated cells (0.5 µg/mL). (C) Measurement of the complestatin half-maximal inhibitory concentration (IC50) using ELISA-based fibronectin (1.0 µg/well) adhesion assays (Abs450nm). (D) End-point readings to assess the growth (OD600nm) of JE2 in the presence of complestatin. Each data point represents the average of three biological replicates ± the SD, and each biological replicate represents the mean of eight technical replicates. The dashed black line is the mean of 24 replicates of JE2 grown in the presence of 1% DMSO as a no drug solvent control. Adhesion assays were performed at complestatin concentrations that were subinhibitory (≤0.5 µg/mL) for growth, as determined by statistical analysis and comparison to the solvent control. (E) Fibronectin adhesion assay of JE2 (black), complestatin-treated cells (0.5 µg/mL) (green), and an atl inactivated mutant (JE2-atl::Tn) (blue). All data points represent the average of three biological replicates ± the SD. P-values were calculated using the two-tailed unpaired Student’s t test. Statistical analyses of complestatin-treated cells compared with JE2 (1% DMSO) are represented by black asterisks. Complestatin compared with JE2-atl::Tn (1% DMSO) is represented by blue asterisks. Statistically significant decreases are denoted as P ≤ 0.01**; P ≤ 0.001***, P ≤ 0.0001****, related to SI Appendix, Fig. S1.
Atl Catalytic Function Is Required for S. aureus Adhesion to Host Ligands.
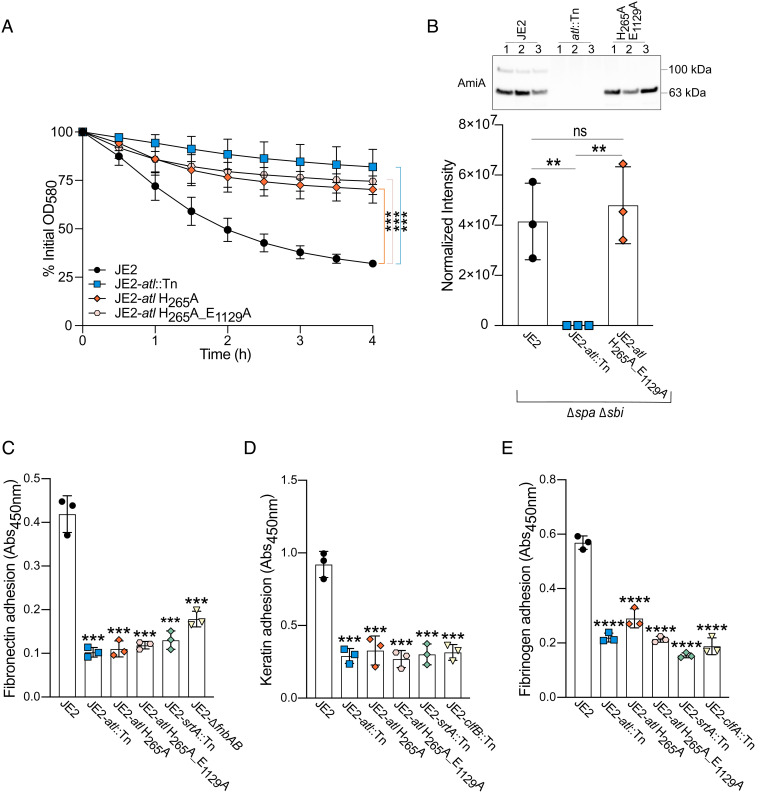
Since the inactivation of atl significantly reduced the affinity of S. aureus for host ligands (4), we investigated whether this phenotype was associated with catalytic function. Following cell division, Atl hydrolyzes peptidoglycan covalently tethering daughter cells (5, 11, 12). The Atl proform is posttranslationally processed to generate two distinct enzymes: an N-acetylmuramyl-L-alanine amidase (AmiA) and a β-N-acetylglucosaminidase (NAGase) (SI Appendix, Fig. S2A) (12). In AmiA, H265 forms part of a Zn2+ binding pocket and H265A mutagenesis inhibits peptidoglycan cleavage while the substrate is still able to bind the enzyme (13, 14). For NAGase, mutagenesis of E1129A inhibits peptidoglycan degradation (15). Using CRISPR/Cas9-mediated counterselection (16), we generated in-frame point mutations of these two key active site residues within the genome of USA300 JE2 (17) (SI Appendix, Fig. S2B). Cell separation defects were confirmed using SEM (SI Appendix, Fig. S3) and detergent-induced autolysis assays (Fig. 2A). We also confirmed the mutants did not exhibit a fitness cost when cultured in microtiter plates with linear shaking (SI Appendix, Fig. S4). Finally, immunoblotting demonstrated that the catalytically inactive AmiA enzyme was comparably abundant to the wild-type enzyme in culture supernatants (Fig. 2B and SI Appendix, Fig. S2C).
Fig. 2.
Autolysin (Atl) catalytic function is required for S. aureus adhesion to host ligands. (A) Whole-cell detergent-induced autolysis assays confirmed that the Atl catalytic mutant strains were inactive. Each data point represents the average of three biological replicates ± the SD. (B) Immunoblots of culture supernatants from JE2 (Δspa Δsbi) (black) and the catalytically inactive atl variant (JE2-atl H265A_E1129A Δspa Δsbi) (orange). JE2-atl::Tn (blue) was included as a negative control. Each data point represents the average of three technical replicates for a single biological replicate ± the SD. Representative immunoblots of one technical replicate of each biological replicate (labeled 1 to 3) are shown. AmiA intensity was normalized to total protein (SI Appendix, Fig. S2C) using stain-free imaging. ELISA-based adhesion assays (Abs450nm) of (C) fibronectin (0.5 μg/well), (D) keratin (0.125 μg/well), and (E) fibrinogen (0.25 μg/well). Isogenic adhesion defective control strains were also included [atl transposon mutant (JE2-atl::Tn), sortase A transposon mutant (JE2-srtA::Tn), and mutants of the host ligand’s respective cell wall-anchored adhesins (fibronectin-fnbAB; keratin-clfB; fibrinogen-clfA)]. Each data point represents a single biological replicate ± the SD. P-values were calculated using the two-tailed unpaired Student’s t test comparing each mutant with the wild-type strain (JE2). Statistically significant decreases are denoted as P ≤ 0.01**, P ≤ 0.001***, and P ≤ 0.0001****. NS = not significant, related to SI Appendix, Fig. S5.
Adhesion to fibronectin, keratin, and fibrinogen was then assessed using three different detection methods: an ELISA-based adhesion assay (Fig. 2 C–E), crystal violet staining of adherent cells (SI Appendix, Fig S5 A–C), and the manual enumeration of preadhesion and postadhesion colony-forming units (CFUs) (SI Appendix, Fig. S5 D–F). Overall, our findings demonstrated that the AmiA mutant (JE2-atl H265A) exhibited significantly impaired adhesion to all three host ligands, comparable to that of the atl:Tn mutant (Fig. 2 and SI Appendix, Fig. S5). However, some discrepancies were observed when assessing fibrinogen adhesion using crystal violet and CFU enumeration as detection methods, which we posit was due to extensive clumping later in the growth cycle. Taken together, these data indicate that Atl catalytic function, particularly AmiA-mediated cleavage of peptidoglycan cross-links, is associated with S. aureus adhesion to host ligands.
The Overexpression of Cell Wall-Anchored Adhesins Did Not Restore Adhesion in Autolysin-Defective Cells.
Since we demonstrated that Atl-mediated peptidoglycan hydrolysis was important for S. aureus adhesion, we were curious to ascertain whether CWA adhesins could enhance adhesion in autolysin-defective cells. CWA adhesins play an important role in mediating the affinity of S. aureus for host ligands; for example, loss of sortase A (SrtA) (18), the housekeeping transpeptidase responsible for anchoring CWA proteins to the peptidoglycan, renders S. aureus devoid of most CWA adhesins (18). As such, adhesion to host ligands, including keratin, fibronectin, and fibrinogen (SI Appendix, Fig. S1), is significantly impaired (4). We subsequently sought to explore the consequence of overexpressing a CWA fibronectin adhesin, fibronectin-binding protein A (FnBPA, encoded by fnbA), in an autolysin-defective cell. The overexpression of fnbA (pALC2073:fnbA) significantly increased adhesion of the wild-type strain to fibronectin, and this phenotype was inhibited by the presence of complestatin (SI Appendix, Fig. S6A). Similar to JE2 treatment with complestatin, the overexpression of fnbA did not restore adhesion in an atl inactivated mutant (SI Appendix, Fig. S6B); however, there was a modest increase in adhesion that was lost in the presence of complestatin. We also investigated this phenomenon using a purR mutant (purR::Tn). Mutations in the transcriptional repressor of purine biosynthesis, purR, increase epithelial cell invasion and enhance the virulence of USA300 (19, 20). This hypervirulent phenotype is correlated with the increased production of FnBPs, which are ordinarily tightly regulated (19, 20). The purR-associated overproduction of the FnBPs did not restore adhesion of complestatin-treated cells (SI Appendix, Fig. S6C). Thus, these results demonstrate that autolysin function is important for FnBP-mediated adherence of S. aureus to fibronectin.
Autolysin-Mediated Peptidoglycan Hydrolysis Is Associated with the Surface Display of Cell Wall-Anchored Proteins.
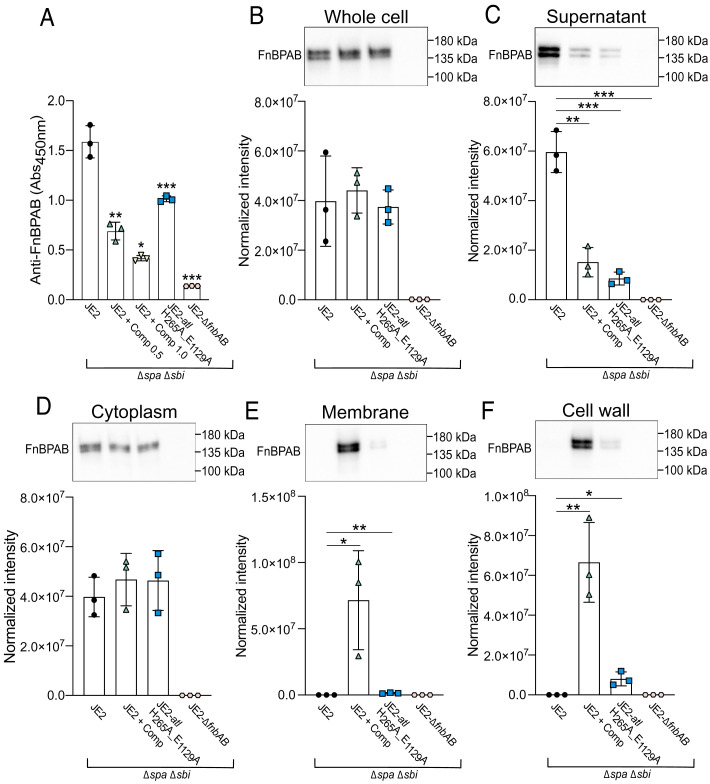
We next sought to investigate the fate of CWA proteins in autolysin-defective cells. To assess whether the FnBP surface levels were affected, we took an ELISA-based approach using an anti-FnBP antibody with affinity for the D1-D3 domains of these proteins, and whole cells immobilized to the surface of microtiter plates. These assays were performed in a USA300 strain devoid of the immunoglobulin binding protein Spa and the second immunoglobulin-binding protein (Sbi). While the bacterial cells equally adhered to the microtiter plate surface (SI Appendix, Fig. S7A), the proportion of the FnBPs accessible to the antibody at the cell surface was significantly reduced in both complestatin-treated cells and the catalytically inactive Atl mutant (Fig. 3A). Immunoblotting of whole-cell lysates revealed the FnBPs were produced at equivalent levels in the complestatin-treated cells and in the catalytically inactive Atl mutant (Fig. 3B and SI Appendix, Fig. S7B), yet there was a significant reduction of these proteins in the culture supernatant (Fig. 3C and SI Appendix, Fig. S7B). Cellular fractionation revealed equivalent cytoplasmic levels of FnBP-A/B (Fig. 3D and SI Appendix, Fig. S7B), with a significant increase in FnBP-A/B in the membrane (Fig. 3E and SI Appendix, Fig. S7B) and cell wall fractions (Fig. 3F and SI Appendix, Fig. S7B) of autolysin-defective cells. In particular, the abundance of the FnBPs in the membrane and cell wall fractions was significantly higher in the complestatin-treated cells compared with the catalytically inactive Atl mutant. We did not detect the FnBPs in the untreated wild-type samples when they were assessed alongside the complestatin-treated samples, suggesting the high abundance of the FnBPs in the complestatin-treated cells was obscuring the wild-type signal. The wild-type fractions were subsequently assessed in the absence of the complestatin-treated samples, enabling measurement of FnBP abundance and cellular localization (SI Appendix, Fig. S8). The increased FnBP abundance in the membrane of complestatin-treated cells could reflect defects in cell wall anchoring, since srtA inactivation induces CWA protein mislocalization (18). However, whole-cell immunoblotting of a srtA inactivated mutant (JE2-srtA::Tn) revealed the presence of a higher molecular weight variant, which likely corresponds to unprocessed CWA proteins, and was absent in the autolysin-defective samples (SI Appendix, Fig. S9).
Fig. 3.
Autolysin function is important for maintaining optimal surface levels of the S. aureus fibronectin-binding proteins (FnBPs). (A) ELISA-based (Abs450nm) detection of surface-exposed FnBPs on JE2 (Δspa Δsbi), complestatin-treated (0.5 and 1.0 µg/mL) cells (Δspa Δsbi) and JE2-atl H265A_E1129A (Δspa Δsbi). A mutant lacking the FnBPs (JE2-∆fnbAB Δspa Δsbi) was included as a negative control. (B) Immunoblot analysis of FnBP abundance in lysed whole cells, (C) culture supernatants, (D) cytoplasmic fractions, (E) cell wall fractions, and (F) membrane fractions. For all immunoblots, the data points represent the cumulative normalized intensity of FnBPA and FnBPB from the average of three technical replicates of a single biological replicate ± the SD. Representative immunoblots from one biological replicate are shown. P-values were calculated using the two-tailed unpaired Student’s t test comparing each mutant with the wild-type strain (JE2). Statistically significant decreases in adhesion and normalized intensity compared with JE2 were denoted as P ≤ 0.05*, P ≤ 0.01**, P ≤ 0.001***, and P ≤ 0.0001****. Protein abundance was normalized to total protein using a stain-free approach, related to SI Appendix, Figs. S7 and S8.
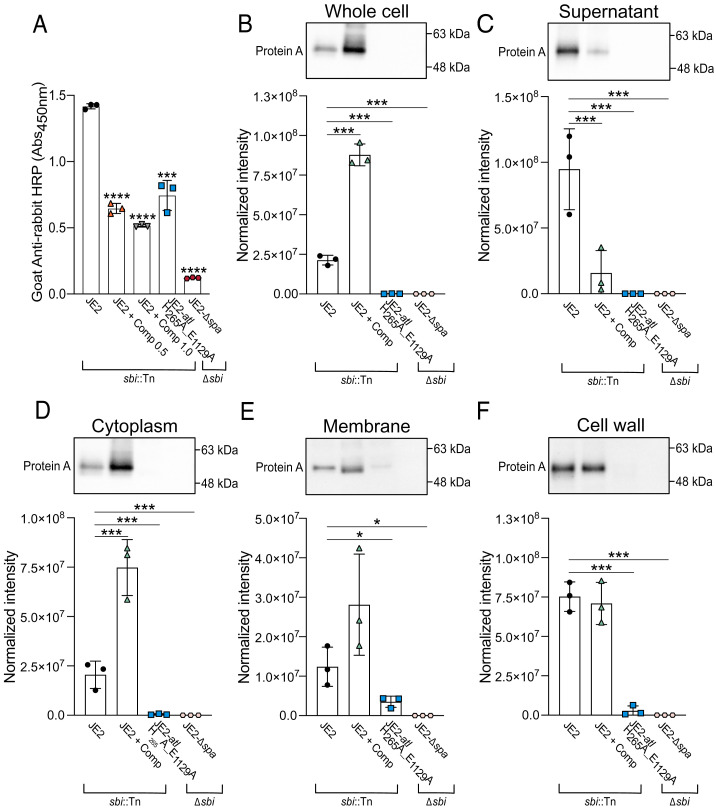
We also assessed the surface levels of Spa, a CWA protein that nonspecifically adheres to immunoglobulins (21–23), to determine if autolysins were associated with the surface presentation of other CWA proteins. To measure Spa IgG affinity, a mutant devoid of Sbi (JE2-sbi::Tn) and the same ELISA-based approach was used. Overall, the complestatin-treated cells and the catalytically inactive Atl variant exhibited significantly decreased affinity for IgG (Fig. 4A), while the strains adhered comparably to the microtiter plates (SI Appendix, Fig. S10A). Immunoblotting revealed the overall cellular abundance and release of Spa into the culture supernatants was significantly reduced in the catalytically inactive Atl mutant (Fig. 4 B and C and SI Appendix, Fig. S10B, respectively), consistent with previous studies revealing transcriptional repression (24). However, the cellular and cytoplasmic abundance of Spa was significantly higher in complestatin-treated cells, yet there was a significant reduction of Spa in the culture supernatants (Fig. 4 B–D and SI Appendix, Fig. S10B, respectively). The abundance of Spa in the membrane and cell wall of complestatin-treated cells was comparable to that of the untreated cells (Fig. 4 E and F and SI Appendix, Fig. S10B). Overall, these findings suggest that autolysin function is important for maintaining optimal surface levels of CWA proteins.
Fig. 4.
Autolysin function is important for maintaining optimal surface levels of S. aureus protein A (Spa). (A) ELISA-based (Abs450nm) detection of IgG-antibody adhesion to Spa exposed on the surface of immobilized whole cells of JE2 (sbi::Tn), complestatin-treated (0.5 and 1.0 µg/mL) JE2-sbi::Tn and JE2-atl H265A_E1129A cells (∆spa). A mutant lacking Spa (JE2-∆spa ∆sbi) was included as a negative control. (B) Immunoblot analysis of Spa abundance in lysed whole cells, (C) culture supernatants, (D) cytoplasm fractions, (E) cell wall fractions, and (F) membrane fractions. For each immunoblot, data points represent the average of three technical replicates for a single biological replicate ± the SD. Representative immunoblots from one biological replicate are shown. P-values were calculated using the two-tailed unpaired Student’s t test comparing each mutant with the wild-type strain (JE2). Statistically significant decreases in adhesion and normalized intensity compared with JE2 were denoted as P ≤ 0.05*, P ≤ 0.01**, P ≤ 0.001***, and P ≤ 0.0001****. Protein abundance was normalized to total protein using a stain-free approach, related to SI Appendix, Fig. S10.
Sequestration of Spa at the Septum of Complestatin-Treated Cells.
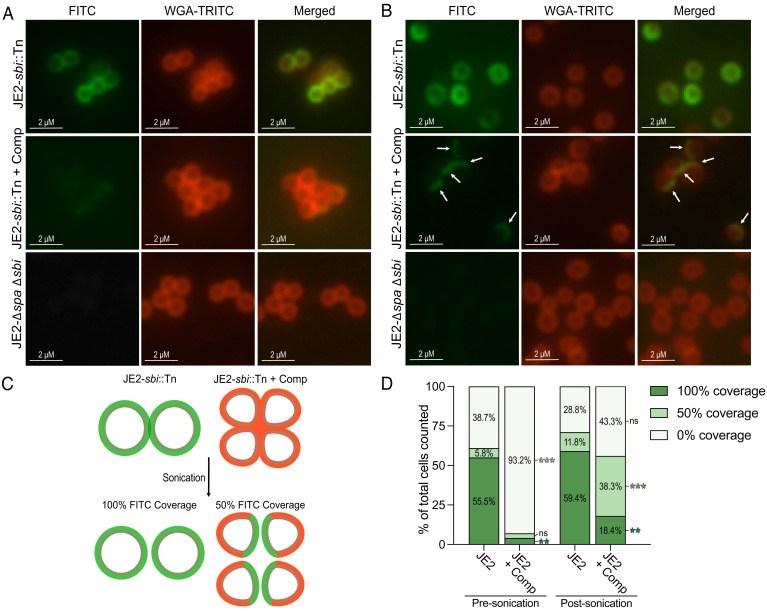
We next sought to further explore the requirement for autolysin activity in the surface display of Spa. Using immunofluorescence microscopy, we investigated the cellular localization of Spa in complestatin-treated cells. Consistent with the ELISA-based approach (Fig. 4A), complestatin reduced the surface levels of Spa, and the fluorescence signal was poorly detectable (Fig. 5A and SI Appendix, Fig. S11). We then mechanically separated complestatin-induced cell clusters using gentle sonication, revealing that Spa was significantly concentrated on one face of the separated cells (Fig. 5 B–D and SI Appendix, Fig. S11). Spa retained a concentric distribution on the surface of the wild-type untreated cells following gentle sonication (Fig. 5 B–D).
Fig. 5.
Spa is masked at the septum of complestatin-treated S. aureus cells. (A) Representative immunofluorescence microscopy images assessing Spa localization [IgG goat anti-mouse FITC-conjugated antibody (green)] on the surface of JE2 (JE2-sbi::Tn) cells ± complestatin (0.5 µg/mL). WGA-TRITC (red) was used to stain the cell wall. JE2-∆spa ∆sbi was included as a negative control. (B) Representative immunofluorescence microscopy images assessing Spa localization following mechanical separation using gentle sonication. The arrows indicate Spa concentrated on one face of the separated cells. (C) Schematic representation of the immunofluorescence microscopy results. Green represents visible FITC and red represents visible WGA-TRITC in the absence of FITC on complestatin-treated cells. Mechanical disruption of complestatin-induced cell clusters liberated Spa from the division septum. (D) Measuring FITC coverage in complestatin-treated cells compared with the untreated cells, prior to and following mechanical separation of cell clusters using gentle sonication. 100% coverage = concentric Spa distribution; 50% coverage = Spa concentration on one face of the cell; 0% = minimal to no detectable signal. An average of 280 cells were counted per sample for a total of three biological replicates per strain both presonication and postsonication. For each strain, the average percentage of total cells counted for three biological replicates is shown. Statistically significant differences in Spa localization of complestatin-treated cells compared with JE2 are denoted as P ≤ 0.01**, P ≤ 0.001***. P-values were calculated using the two-tailed unpaired Student’s t test. (Scale bars, left, 2 µM.) FITC: Fluorescein isothiocyanate; WGA-TRITC: wheat germ albumin-tetramethylrhodamine, related to SI Appendix, Fig. S11.
The Mechanical and Enzymatic Separation of Autolysin-Associated Cell Clumping Enhanced S. aureus Adhesion to Host Ligands.
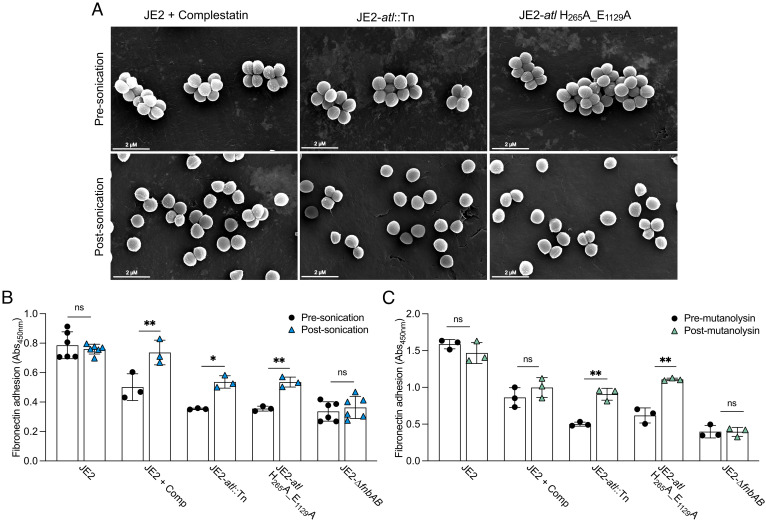
We next investigated whether the separation of cell clusters induced by autolysin loss-of-function impacted the affinity of S. aureus for host ligands. Here, cell clusters were mechanically separated using gentle sonication (Fig. 6A). Additionally, we used mutanolysin, a muramidase produced by Streptomyces globisporus, to enzymatically separate cell clusters. This enzyme has been shown to separate cell clusters in USA300 atl inactivated mutants (25, 26). CFUs were enumerated prior to and following cell separation, demonstrating the cells were viable after separation using both methods (SI Appendix, Fig. S12). Overall, fibronectin adhesion was significantly enhanced in Atl-deficient cells following the dispersal of cell clusters using sonication and mutanolysin (Fig. 6 B and C). Adhesion was also enhanced in complestatin-treated cells following sonication; however, adhesion was unaffected by mutanolysin since complestatin blocks peptidoglycan hydrolysis by this murein hydrolase (10) (Fig. 6C). Thus, the separation of cell clusters induced by autolysin loss-of-function both increases the accessible surface abundance of Spa (Fig. 5A) and increases the affinity of S. aureus for fibronectin (Fig. 6), which we believe is due to the liberation of CWA adhesins from the division septum.
Fig. 6.
Mechanical and enzymatic separation of autolysin-associated cell clusters enhanced S. aureus adhesion to fibronectin. (A) SEM analysis of autolysin-deficient strains presonication and postsonication. (B) Fibronectin ELISA-based whole-cell adhesion assays (Abs450nm) presonication (black) and postsonication (blue). (C) Fibronectin ELISA-based whole-cell adhesion assays premutanolysin (black) and postmutanolysin treatment (green) of Atl-inactivated strains. A mutant lacking the FnBPs (JE2-∆fnbAB) was included as an adhesion defective control. Each data point represents a single biological replicate ± the SD. P-values were calculated using the two-tailed unpaired Student’s t test comparing each strain with the presonication samples. Statistically significant differences were denoted as P ≤ 0.05*, P ≤ 0.01**, related to SI Appendix, Fig. S12.
Assessing the Antivirulence Potential of Complestatin.
Previous studies demonstrated that complestatin reduced the bacterial burden in an established S. aureus superficial skin infection in mice (10). We hypothesized complestatin could also exhibit antivirulence properties, by reducing the surface levels of the FnBPs, which could impact the immediate colonization of major organs. Thus, we evaluated the in vivo efficacy and antivirulence potential of complestatin during bacteremia using a murine infection model. The wild-type JE2 strain was propagated to the mid-exponential phase of growth (~OD600nm 1.5) in the presence of subinhibitory concentrations of complestatin (0.5 μg/mL). For each test group (± complestatin), 12 mice were infected via the tail vein with 1 × 107 CFU. Overall, we observed that even the transient presence of complestatin during bacterial growth was sufficient to significantly reduce the bacterial burden in all major organs at 24 h post infection (hpi) (SI Appendix, Fig. S13).
Discussion
Although Atl has long been associated with surface adhesion (5–8), the molecular basis underlying this phenomenon remains understudied in S. aureus. While Atl exhibits affinity for host ligands (5, 6, 8, 9), through the generation of a catalytically inactive Atl variant, we highlight an important role for the peptidoglycan hydrolytic function of Atl in the maintenance of CWA protein surface levels. Particularly, we associate the daughter cell separating properties of Atl with this process. Atl-mediated peptidoglycan hydrolysis plays an important role in the final step of cell division; the two Atl catalytic subunits, AmiA and NAGase, hydrolyze the peptidoglycan that covalently tethers recently divided daughter cells (5, 14, 27, 28). Since S. aureus divides perpendicularly to previous division planes, Atl loss-of-function causes tetrameric cell structures to initially form, followed by large cell clusters later in the cell cycle (SI Appendix, Fig. S3) (28). Our findings suggest this cell-separating function underlies the importance of Atl in adhesion.
There are different mechanistic possibilities underlying this phenomenon; for example, the cell clumping phenotype induced by Atl loss-of-function reduces the accessible surface area of S. aureus, which could in turn reduce the proportion of surface-exposed CWA proteins that can interact with host ligands and antibodies. Indeed, the mechanical disruption of these cell clumps enhanced S. aureus adhesion to fibronectin (Fig. 6). However, adhesion was only restored to ~60 to 75% of the wild-type levels (Fig. 6). If the phenotype was solely reflective of reduced surface area due to cell clumping, then one would anticipate the disruption of cell clusters would completely restore adhesion. Based on our findings, we instead hypothesize autolysins are important for liberating CWA proteins from the peptidoglycan synthesized at the division septum. CWA proteins containing a YSIRK/R motif in their signal peptides—e.g., the FnBPs (fibronectin adhesins), ClfB (keratin adhesin), ClfA (fibrinogen adhesin), and Spa—are secreted at the septal region of dividing cells (29). SrtA covalently anchors these proteins to the peptidoglycan at the septum, followed by incorporation into the growing peptidoglycan by the cell wall synthesis machinery (29) (SI Appendix, Fig. S14). The final step of cell division involves Atl-mediated daughter cell separation, a process that likely liberates YSIRK-containing CWA proteins from the division septum (SI Appendix, Fig. S2) (5, 14, 27, 28). These proteins can then be detected across the cell surface in a ring-like/hemispherical distribution (29). Atl loss-of-function induces morphological changes in the peptidoglycan composition, including apparent cell wall thickening, with the daughter cells remaining tethered by thread-like structures of peptidoglycan (28). These morphological changes could induce sequestration and occlusion of CWA proteins in the peptidoglycan at the septal region, impacting the surface display of CWA adhesins, and consequently adhesion of S. aureus to host ligands. Consistent with this notion, the cellular and cytoplasmic abundance of the FnBPs was unaffected in an Atl-deficient cell, yet there was a significant increase in their abundance in the membrane and cell wall. However, the abundance of the FnBPs on the cell surface and in the culture supernatant was significantly reduced. Our preliminary analyses suggest the FnBPs are anchored to the cell wall in autolysin-defective conditions, since we do not detect unprocessed proteins that are present in a srtA-inactivated mutant (SI Appendix, Fig. S9), and mechanically disrupted cells exhibit increased adhesion to host ligands (Fig. 6). Future studies should investigate peptidoglycan composition in autolysin-defective conditions, including the proportion of CWA proteins covalently attached to the pentaglycine cross bridges in the cell envelope.
Through binding to the peptidoglycan, the type V glycopeptide antibiotic complestatin blocks the activity of autolysins and can therefore be used as a chemical probe to study multiple autolysins simultaneously (10). Since atl inactivation significantly reduces the affinity of S. aureus for host ligands (Fig. 1 and SI Appendix, Fig. S1) (4), our findings suggest that complestatin and corbomycin likely block the hydrolytic activity of Atl. Previous studies demonstrated these compounds inhibited peptidoglycan digestion by lysozyme (from hen egg white), mutanolysin from S. globisporus, and the Bacillus subtilis endopeptidases CwlK (YcdD) and CwlO (YcvE) (10). We have added to these findings by observing that complestatin reduced adhesion of the wild-type strain to a level greater than an isogenic atl transposon mutant (atl::Tn) (Fig. 1E and SI Appendix, Fig. S1). There were also significantly lower levels of CWA proteins on the surface of complestatin-treated cells compared with the catalytically inactive Atl mutant strain (Figs. 3 and 4). Thus, we propose other autolysins may also be associated with the surface presentation of CWA proteins, since complestatin simultaneously inhibits multiple peptidoglycan hydrolases (10).
Complestatin also allowed us to study the role of autolysins in the surface presentation of Spa; as described, atl inactivation induces a significant reduction in the cellular levels of Spa (Fig. 4B) (24). Since Spa is the most abundant S. aureus CWA protein (30–34), we hypothesize S. aureus down-regulates the protein when atl is inactivated, to reduce the potentially detrimental accumulation of large proportions of Spa at the division septum. In contrast, the cellular and cytoplasmic abundance of Spa was increased in complestatin-treated cells, yet the protein was poorly detected on the cell surface and in the culture supernatants (Fig. 4). This observation suggests Spa is not displayed on the surface of the cell and is instead accumulating in the cytoplasm. The abundance of Spa in the cell wall and membrane fractions was not significantly different to that of the untreated cells, which could be due to saturation of the secretory apparatus and peptidoglycan at the division septum, due to the ordinarily high abundance of Spa in the wild-type untreated cells (30–34). Indeed, we observed a different phenotype when studying the cellular localization of the FnBPs, which are not as abundant. The cellular and cytoplasmic levels were unchanged, yet there was a significant increase in the proportions of the proteins in the membrane and cell wall fractions of complestatin-treated cells. Thus, we posited the increased abundance of the FnBPs in the complestatin-treated samples was saturating the secretory apparatus and the peptidoglycan at the division septum. Taken together, these data suggest the CWA proteins are secreted and incorporated into the peptidoglycan synthesized at the division septum, yet they are not displayed on the cell surface. Indeed, this finding supports our hypothesis that morphological changes in the peptidoglycan composition, including cell wall thickening and defects in cell separation, cause sequestration and occlusion of CWA proteins at the septal region. Consistent with this hypothesis, the mechanical disruption of complestatin-treated cells liberated Spa from the division septum (Fig. 5), and increased adhesion of S. aureus to host ligands (Fig. 6). This observation also implies the association of autolysins with adhesion is not primarily associated with reduced surface area due to cell clumping; immunofluorescence revealed that Spa was not detected on the surface of complestatin-treated cells, including the periphery of the cell clumps, yet was visibly concentrated on one side of the bacterial cell when the cell clumps were dispersed (Fig. 5).
We propose a working model to describe the role of autolysins in the surface presentation of CWA proteins such as Spa and the FnBPs, which involves the liberation of these proteins from the peptidoglycan at the septal region, facilitating surface display. In the absence of autolysin function, cell wall thickening and defects in cell separation cause occlusion of CWA proteins, rendering them inaccessible to ligands and antibodies (SI Appendix, Fig. S14). Autolysins are therefore important for CWA protein-mediated virulence, and we show that at concentrations that are subinhibitory for growth, complestatin is a potent antiadhesive agent reducing the bacterial burden in a sepsis model in all organs assessed (SI Appendix, Fig. S13). The systemic administration of this compound could further improve efficacy during bacteremia since it is possible that complestatin became depleted and/or dissociated from the cell surface since the bacteria were preincubated with the compound prior to inoculation.
In summary, autolysins have previously been associated with peptidoglycan remodeling to enable heme acquisition (35), the assembly of protein complexes such as pili (36), and for the release of cytoplasmic proteins (24) and virulence factors (37). Here, we associate autolysin function with the surface display of S. aureus CWA proteins and thus the virulence of this pathogen. Such information could be exploited for the generation of therapies to treat and control S. aureus infections, and complestatin and corbomycin are particularly promising examples of such an approach.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
MRSA LAC JE2 clone USA300 (CMRSA-10 in Canada) (17), which is the most clinically prevalent MRSA clone in North America (38, 39), was used for all purposes of this study. Strains of S. aureus were routinely propagated in tryptic soy broth (TSB; Wisent Inc.) and Escherichia coli strains in Lysogeny broth (LB; BioShop Canada). Unless otherwise stated, all strains were propagated at 37 °C with aeration at 220 rpm (25-mm throw), except for microtiter plates, which were aerated with 900 rpm using an incubator with a 3-mm throw (INFORS HT Multitron). When studying adhesion, strains were routinely grown to an OD600nm of ~0.4 for fibronectin adhesion, ~0.6 for keratin, and ~1.0 for fibrinogen (4). Antibiotics used in this study were erythromycin (5 μg/mL), chloramphenicol (10 μg/mL), and ampicillin (100 μg/mL). Complestatin and corbomycin were kindly provided by Dr. Gerard Wright (McMaster University); complestatin was used at a concentration of 0.5 µg/mL unless stated otherwise. pCas9counter and pCN-EC2132tet plasmids (16) were obtained from Addgene (Plasmid # 107192; RRID:Addgene_107192 and Plasmid # 107191; RRID:Addgene_107191). E. coli IM08B (40) (BEI resources) was used as a routine cloning host prior to transformation of plasmids into S. aureus. All strains and plasmids used in this study are listed in SI Appendix, Tables S1 and S2.
ELISA-Based Adhesion Assays.
The ELISA-based adhesion assay was performed as previously described (4). Briefly, NUNC Maxisorp microtiter plates (Thermo Scientific) were coated with 100 µL host ligands and incubated at 4 °C for 18 h. Keratin (Millipore Sigma) was diluted in a carbonate buffer (15 mM Na2HCO3, 35 mM NaHCO3 [pH 9.6]). Fibronectin and fibrinogen (Millipore Sigma) were diluted in phosphate-buffered saline (PBS) [pH 7.4]. The plates were blocked with 300 μL 2% (w/v) bovine serum albumin (BSA; HyClone™) in PBS for 1 h at room temperature. 100 μL bacterial cultures were then applied to the coated plates, which were sealed and agitated for 5 min (900 rpm), followed by incubation for 1 h at room temperature with gentle shaking (100 rpm). During the final 20 min of incubation, the microtiter plate lids were removed, and the cultures irradiated under ultraviolet (UV) light for 20 min. 100 μL PBS was then applied before irradiating under UV light for a further 20 min. The PBS was removed and 100 μL IgM mouse anti-S. aureus monoclonal primary antibody (2 μg/mL in 1% (w/v) BSA in PBS) (Invitrogen), raised against UV-inactivated S. aureus, was applied and incubated for 1 h at room temperature. For detection of adhered cells, 100 μL goat anti-mouse IgM (heavy chain) HRP-conjugated secondary antibody (Invitrogen) (125 ng/mL in 1% BSA (w/v) in PBS) was applied and incubated for 1 h. Finally, following extensive washing of the wells with PBS, 100 μL room temperature equilibrated 1-step™ Ultra 3,3′,5,5′-tetramethylbenzidine (TMB; Thermo Scientific) colorimetric substrate was applied and incubated for 20 min. The reaction was quenched with 100 μL 2M H2SO4, and the Abs450nm was measured using a BioTek Synergy H1 microplate reader. Wash steps involved four washes with 400 μL PBS using a BioTek 405 microplate washer, except for host ligand aspiration, where the plates were only washed once with 400 μL PBS. When assessing fibrinogen adhesion, the PBS was supplemented with 1.7 mM of CaCl2.
To assess the antiadhesive potential of complestatin and corbomycin, the compounds were reconstituted and titrated twofold in 100% dimethyl sulfoxide (DMSO) starting at a high dose of 2.0 µg/mL. Overnight cultures were prepared for three biological replicates of each strain of interest, followed by standardization to an OD600nm of 0.1 in 0.85% (w/v) NaCl, which was then diluted 1:200 in TSB. 100 μL of the cell inoculum were applied to microtiter round-bottom plates in the presence of each compound dilution [1% (v/v) DMSO]. The plates were incubated at 37 °C until an appropriate OD600nm (4) was reached. Growth (OD600nm) was measured using a BioTek Synergy H1 microplate reader to assess the minimum inhibitory concentration of each compound. To assess fibronectin adhesion, 50 μL cells and 50 μL PBS were applied to fibronectin-coated wells, and the ELISA was performed as described. The antiadhesive potential of complestatin on JE2-purR::Tn was also assessed using this method.
To assess S. aureus dose-dependent adhesion to immobilized polymers, three biological replicates of each strain were propagated in 3 mL cultures for 18 h, diluted 1:100 in 5 mL TSB and grown to the desired OD600nm. MaxiSorp™ microtiter plates were coated with increasing concentrations of host ligands, and the ELISA was performed as described. Where appropriate for comparison, atl mutant strains, JE2-srtA::Tn, and/or the isogenic mutants of the host ligand cognate adhesins were used as controls. Once the compound potency was determined for fibronectin adhesion using the checkerboard assay described above, all experiments using complestatin were performed at a concentration of 0.5 µg/mL unless otherwise stated. When utilizing the pALC2073 vector (41), chloramphenicol was added to a final concentration of 10 μg/mL.
Detergent-Induced Autolysis Assays.
Autolysis activity was assessed using whole-cell assays as previously described (15). Each strain was assessed using three biological replicates. A single colony was inoculated into 3 mL TSB, and the strains were grown overnight at 37 °C. The overnight cultures were diluted to an OD600nm of 0.1 in TSB supplemented with 1 M NaCl in an Erlenmeyer flask, followed by incubation at 37 °C for 3 h. Complestatin was added to a final concentration of 1.25 µg/mL. The bacterial cells were harvested by centrifugation and washed twice with 12.5 mL 4 °C dH2O. The bacterial cells were resuspended in 50 mM Tris-HCl [pH 7.2] with 0.05% (v/v) Triton X-100 to a final OD600nm <1.0. 100 µL of each cell suspension was arrayed into a 96-well round bottom microtiter plate and incubated at 30 °C with linear shaking (250 rpm) in a BioTek Synergy H1 microplate reader. The OD580nm was measured with 30-min intervals for 4 h.
Scanning Electron Microscopy.
Strains of interest were propagated in TSB to an OD600nm of 0.6 and 1.0. Complestatin-treated cells (0.5 µg/mL) were propagated to an OD600nm of 0.3. The cells were harvested by centrifugation (4,000 × g, 4 °C) and washed three times in 0.85% (w/v) NaCl. The samples were adhered to carbon planchets, fixed with 2.5% (v/v) glutaraldehyde (1 h) and 1% osmium tetroxide (30 min), dried under ambient conditions, and coated with 15 nm of gold using a sputter coater (Denton Desk V TSC). The samples imaged presonication and postsonication were subjected to an ethanol dehydration series and fully dried using a Denton DCP-1 critical point dryer. Images were acquired using an FEI Quanta FEG 250 scanning electron microscope (Thermo Fisher Scientific) operated at 5.0 kV under high vacuum.
Growth Profiling of Autolysin Inactivated Mutant Strains.
For growth profiling, single colonies were used to inoculate TSB, and the cultures were propagated overnight at 37 °C. Cell inoculum was prepared by standardizing all strains to an OD600nm of 0.1 in 0.85% (w/v) NaCl followed by a 1:200 dilution into TBS. The OD600nm was measured every 15 min for 24 h using a BioTek Synergy H1 microplate reader.
CRISPR/Cas9-Mediated Counterselection to Generate Atl Active Site Mutants.
Precision genome engineering using CRISPR/Cas9-mediated counterselection was performed as previously described (16) with minor modifications. sgRNAs for pCas9counter and recombineering oligonucleotides (Integrated DNA Technologies, Inc.) can be found in SI Appendix, Table S2. To increase recombineering efficiency, the cells were incubated at 30 °C for an additional 18 h following electroporation. To confirm successful active site mutations, the entire atl gene was amplified and assessed using Sanger sequencing with the following primers: AmiA_F_check, AmiA_R_check, NAGase_F_check, NAGase_R_check, Atl_FullFwd_Check, Atl_FullRvs_Check and the Atl_Check primers 1-11 (SI Appendix, Table S2).
Generating Markerless Knockout Mutants Using Allelic Exchange.
Allelic exchange was used as previously described (42). Briefly, the upstream and downstream flanking regions of the sbi and spa genes were amplified using the following: primers PrimerA_Spa, PrimerB_Spa, PrimerC_Spa, and PrimerD_Spa, and PrimerA_Sbi, PrimerB_Sbi, PrimerC_Sbi and PrimerD_Sbi (SI Appendix, Table S2). The amplicons were recombined into pJB38 and passaged through E. coli IM08B prior to electroporation into strains of interest. Strains containing the plasmid of interest were streaked from a glycerol stock and incubated at 43 °C overnight on TSA with chloramphenicol. Large colonies, which had undergone a single recombination event, were isolated and propagated on TSA with chloramphenicol at 43 °C overnight. A single colony was propagated in 3 mL TSB at 30 °C overnight to induce plasmid loss and further recombination. The cultures were diluted 1:500 into TSB for four consecutive days and plated onto TSA with 2 µg/mL, 1 µg/mL, and 0 µg/mL anhydrotetracycline. Colonies were patched onto TSA and TSA with chloramphenicol. Any colonies that had lost chloramphenicol resistance were screened using PCR with PrimerA_Spa and PrimerD_Spa, and PrimerA_Sbi and PrimerD_Sbi (SI Appendix, Table S2) to confirm gene deletions.
Detection of Atl in Culture Supernatants.
Immunoblots were performed with concentrated culture supernatants using an anti-AmiA antibody. The anti-AmiA antibody was generated by Pacific Immunology using their custom monospecific antibody production service. The antibodies were purified from the serum of rabbits (13-wk protocol) immunized with a synthetic peptide (AmiA–DYIRKNNLKAPKIEEDYTS-Cys). The peptide was selected based on predictions of regions that were unique, immunogenic, and surface exposed, using Pacific Immunology’s proprietary combination of algorithms. To ensure the antibody was specific to AmiA, the detected bands were excised, and mass spectrometry was used to confirm that the product was AmiA. Additionally, loss of immunoreactivity was confirmed by the lack of signal in a mutant devoid of Atl (JE2-atl::Tn). A JE2-∆spa∆sbi background was utilized for these studies. Three biological replicates were grown at 37 °C for 18 h. The cultures were diluted 1:100 in 50 mL TSB in an Erlenmeyer flask, followed by incubation at 37 °C with aeration for 3 h until the cells reached stationary phase. The cells were harvested by centrifugation and the supernatants were concentrated using Amicon Ultra-15 3,000 Da MWCO centrifugal filter units (Millipore Corporation) to ~250 to 500 µL. Protein concentrations were determined using a Bradford assay (Biorad) following the manufacturer’s suggested guidelines.
The concentrated supernatants were boiled with 4× Laemmli loading buffer [0.25 M Tris [pH 6.8], 8% (v/v) SDS, 40% (v/v) glycerol, 20% (v/v) 2-mercapto-ethanol, 0.04% (w/v) bromophenol blue], and 12 µg total protein was separated using SDS-PAGE with 4 to 15% Mini-PROTEAN TGX Precast stain-free acrylamide gels (BioRad). Prior to transfer, the stain-free gel was irradiated using the ChemiDoc XRS+ UV light for 5 min to activate the trihalo compound in the stain-free gels. The proteins were then transferred overnight onto low-fluorescence polyvinylidene difluoride (LF-PVDF) membranes (BioRad) using the wet tank transfer system. The membrane was washed three times with TBS (10 mM Tris-HCl, 150 mM NaCl) and blocked for 1 h with 5% (w/v) skim milk in TBS + 0.1% (v/v) Tween-20. The blot was washed twice with Tris Buffered Saline Tween20 Triton X-100 (TBSTT) [20 mM Tris-HCl, 500 mM NaCl, 0.05% (v/v) Tween-20 and 0.2% (v/v) Triton-X 100] and once with TBS. The blot was then incubated overnight at 4 °C with the anti-AmiA primary antibody [1:100 in 5% (w/v) skim milk in TBS + 0.1% (v/v) Tween-20]. The blot was washed twice with TBSTT and once with TBS, followed by incubation with a goat anti-Rabbit IgG (H+L) HRP-conjugated secondary antibody (Invitrogen) [1:5,000 in 5% (w/v) skim milk in TBS+ 0.1% (v/v) Tween-20] for 1 h at room temperature. The blot was washed four times with TBSTT and once with TBS. To measure total protein, the membrane was imaged using the stain-free blot protocol (Biorad) prior to development using the Immobilon Crescendo Western chemiluminescent HRP substrate (Millipore Sigma). The blots were visualized using a Biorad ChemiDoc XRS+ system. Protein intensity was normalized to the total protein using ImageLab and stain-free technology (BioRad).
Crystal Violet Adhesion Assays.
Crystal violet adhesion assays were performed as described for the ELISA-based adhesion assays, with crystal violet being used to detect surface adhesion. Following UV irradiation, the PBS was removed, and 100 µL 0.5% (w/v) crystal violet [20% (v/v) methanol] was applied to each well followed by a 4-min incubation at room temperature. The crystal violet was removed, and the wells washed four times by hand with 400 µL PBS. The crystal violet was solubilized using 7% (v/v) acetic acid, agitated for 5 min at 600 rpm, and the Abs595nm was measured using a BioTek Synergy H1 microplate reader.
Colony-Forming Unit (CFU) Adhesion Assays.
Each strain was propagated in 3 mL TSB for 18 h followed by a 1:100 dilution in TSB. The diluted cultures were grown in Erlenmeyer flasks to the corresponding OD600nm for each polymer. Once the appropriate OD600nm was reached, 60 μL of the bacterial cells and 60 μL PBS were applied to MaxiSorp™ microtiter plates coated with 0.25 μg/well (fibronectin), 0.125 μg/well (keratin), and 0.0625 µg/well (fibrinogen). Three technical replicates were performed for each biological replicate. The MaxiSorp™ plates were washed and blocked as described. To enumerate applied CFUs, 20 μL of each culture was immediately removed and diluted 1:100 in sterile 0.85% (w/v) NaCl to a final dilution of 10−5 (fibronectin and fibrinogen) and 10−6 (keratin). The remaining 100 μL applied bacterial cultures were agitated for 5 min (900 rpm) and incubated for 1 h at room temperature with gentle shaking (100 rpm). The plates were washed as described above, and adherent cells were detached with 100 μL prewarmed trypsin-EDTA [0.25% (w/v) trypsin (Sigma), 0.5 mM EDTA in PBS lacking Ca2+ or Mg2+], followed by incubation for 4 min at 37 °C. To quench the reaction, 200 μL sterile 0.85% (w/v) NaCl was applied and gently mixed to detach any remaining cells. Adhered CFUs were manually enumerated: 20 μL was removed and diluted 1:100 in sterile 0.85% (w/v) NaCl to a final dilution of 10−3 (fibronectin) and 10−4 (keratin and fibrinogen). Then, 100 μL of the final two dilutions for both the applied and adhered cells were manually enumerated following incubation on TSA at 37 °C for 18 h.
Measuring the Abundance of CWA Proteins on the Surface of S. aureus.
CWA protein surface abundance was measured using an ELISA-based adhesion assay. These assays were performed in backgrounds devoid of the Spa and Sbi (JE2-∆sbi and JE2-ΔspaΔsbi). Three biological replicates of each strain were assessed, and each biological replicate was assessed using three technical replicates; a single colony for each replicate was used to inoculate 3 mL TSB, followed by growth at 37 °C for 18 h. Saturated overnight cultures were diluted 1:100 in 5 mL TSB. Complestatin (100% v/v DMSO) was added to a final concentration of 0.5 µg/mL and 1.0 µg/mL. Equivalent volumes of 100% v/v DMSO were included as a solvent control. The cultures were incubated at 37 °C until they reached an OD600nm of 0.4 (FnBPs) and 1.0 (Spa), harvested by centrifugation at 4,000 × g for 10 min at 4 °C, washed in PBS, and then standardized to an OD600nm of 2.0. The standardized bacterial cultures (200 µL) were applied to Nunc Maxisorp microtiter plates (Thermo Scientific) and serially diluted 1:2 in PBS. The plates were sealed and agitated for 5 min (900 rpm), followed by incubation for 1 h at room temperature with gentle shaking (100 rpm). During the final 20 min of incubation, the microtiter plate lids were removed, and the cultures irradiated under ultraviolet (UV) light for 20 min. Then, 100 µL PBS was applied before irradiating under UV light for a further 20 min. The PBS was aspirated and 300 µL 2% (w/v) BSA (in PBS) was applied to each well followed by static incubation for 1 h at room temperature. To ensure the different strains equally adhered to the polystyrene, two identical Maxisorp plates were prepared for all the strains: One plate was used to detect CWA protein surface levels (described below), and the other plate was used to assess adherence of the bacteria to the plate using crystal violet, as described above. To detect Spa, 100 µL of an IgG goat anti-rabbit (H+L) HRP-conjugated secondary antibody (Invitrogen) (1:15,000 in 1% BSA) was applied to the immobilized cells, followed by agitation for 5 min at 600 rpm, and incubation for 1 h at room temperature with gentle shaking (100 rpm). To measure the abundance of surface exposed FnBPs, a mouse anti-FnBPAB antibody was used. To generate this antibody, the D1 motif of fnbA was cloned and synthesized by ProSci, and the antibodies were raised against this motif in mice. Loss of reactivity was confirmed by the complete lack of signal in an isogenic mutant devoid of FnbpA and FnbpB (JE2-∆fnbAB ∆spa∆sbi). Then, 100 µL of the mouse anti-FnBPAB primary antibody (1:10,000 dilution in 1% (w/v) BSA) was applied, agitated for 5 min at 600 rpm, and incubated with gentle shaking (100 rpm) for 1 h at room temperature. The wells were washed with PBS, and 100 µL peroxidase AffiniPure F(ab′)2 fragment goat anti-mouse IgG (H+L) HRP-conjugated secondary antibody (1:10,000 in 1% (w/v) BSA; Jackson ImmunoResearch) was applied to each well. The plate was agitated for 5 min at 600 rpm followed by a 1-h incubation at room temperature with gentle shaking (100 rpm). Finally, following extensive washing of the wells with PBS, for detection of both the FnBPs and Spa, 100 μL -room temperature equilibrated 1-step™ Ultra TMB colorimetric substrate (Thermo Scientific) was applied and incubated for 20 min. The reaction was quenched with 100 μL 2M H2SO4, and the Abs450nm was measured using a BioTek Synergy H1 microplate reader. Wash steps involved four washes with 400 μL PBS using a BioTek 405 microplate washer, except for bacterial culture aspiration, where the plates were only washed once with 400 μL PBS.
Cellular Fractionation.
Cell fractionation preparation was performed as previously described (43), with minor modifications. Strains of interest were assessed in biological triplicate and grown overnight in 3 mL TSB for 18 h at 37 °C. A JE2-∆spa∆sbi background was utilized for detection of FnBPs and a JE2-∆sbi background for detection of Spa. Overnight cultures were diluted 1:100 into 50 mL TSB and propagated to the appropriate OD600nm (0.4 for the detection of FnBPs, and 1.0 for Spa), harvested by centrifugation at 4,000 × g for 10 min at 4 °C, and washed once with PBS. The pellet was suspended in 750 µL Tris-buffered saline with EDTA-free Pierce™ protease inhibitor minitablets (TBSI, 50 mM Tris-HCl [pH 7.5], 100 mM NaCl). Cell suspensions were lysed with 0.1 µM ceramic beads (OMNI International) using a Bead Mill 4 (Fisher Brand) on setting 5, for 10 cycles of 40 s. The samples were incubated on ice for 2 min between cycles. Following lysis, the beads were removed by centrifugation at 2,000 × g, and the supernatant was transferred to a microcentrifuge tube followed by centrifugation at 5,000 × g to remove intact cells. Then, 50 µL was reserved for the whole-cell fraction. The supernatants were then centrifuged at 18,000 × g for 10 min at 4 °C to isolate the cell wall. The supernatants containing the cytoplasmic and membrane fractions were collected, and the cell wall fraction was washed in TBSI and resuspended in 30 µL TBSI. The membrane fraction was collected by ultracentrifugation at ~130,000 × g. Following centrifugation, the supernatant was collected as the cytoplasmic fraction and the pellet containing the membrane fraction was washed once in TBSI before being resuspended in 100 µL TBSI.
Immunoblotting of Cellular Fractions.
The fractionated samples were quantified using a BCA assay (Pierce), and 10 µg protein from each sample was boiled with 4 × Laemmli loading buffer. 20 µL of each sample was separated by SDS–PAGE using 4 to 15% Mini-PROTEAN TGX Precast stain-free acrylamide gels (BioRad). Prior to transfer, the gels were imaged using the stain-free Gel protocol described earlier. The proteins were transferred overnight onto LF-PVDF membranes (BioRad) using the wet tank transfer system. The membrane was washed three times with TBS and blocked for 1 h with 5% (w/v) skim milk in TBS, followed by a further three washes in TBS + 0.01% (v/v) Tween-20 (TBST). The membrane was incubated for 1 h with a rabbit anti-mouse IgG (H+L) HRP-conjugated secondary antibody in 5% (w/v) skim milk in TBST (1:20,000 for whole cell and supernatant, and 1:10,000 for cytoplasm, cell wall and membrane), followed by three washes with TBST. To quantify the abundance of FnBPs, the mouse anti-FnBPAB antibody [1:2000 in 5% (w/v) skim milk in TBST] was incubated with the membrane for 1 h, washed three times with TBST, and then incubated for 1 h with a peroxidase AffiniPure F(ab′)2 fragment goat anti-mouse IgG (H+L) HRP-conjugated secondary antibody (1:3,000 in TBST, 5% (w/v) skim milk; Jackson ImmunoResearch). The membranes were then washed three times with TBST. Total protein was measured using the stain-free blot protocol (Biorad) as described above, prior to development using the Immobilon Crescendo Western chemiluminescent HRP substrate (Millipore Sigma). The ChemiDoc XRS+ system (Biorad) was used to measure the chemiluminescence, which was normalized to the total protein using ImageLab. Three biological replicates were assessed using three independent technical replicates.
Adhesion Assays Assessing Mechanically and Enzymatically Disrupted Cell Clusters.
Three biological replicates of each strain were propagated for 18 h at 37 °C. The overnight cultures were diluted 1:100 into 8 mL TSB in test tubes and grown to the desired OD600nm before being harvested by centrifugation at 4,000 × g for 10 min at 4 °C. Two subcultures were generated for each strain: one was reserved as a “presonication” control, and the second was gently sonicated (“postsonication”). The bacterial cells were washed once with PBS. The presonication samples were standardized to an OD600nm of ~0.5, and the postsonication samples were suspended in 2 mL PBS, followed by measurement of the OD600nm. To disrupt the cell clusters, the postsonication samples were sonicated on ice using a sonicator micro tip for 8 min (20 s on, 20 s off) at an amplitude of 10%.
For digestion with mutanolysin, the strains were treated as above. Two subcultures were generated: “premutanolysin” and “postmutanolysin”. Both subcultures were pelleted and resuspended in 3 mL PBS and the mutanolysin-treated samples were supplemented with 10 units/mL mutanolysin (Sigma). Premutanolysin and postmutanolysin subcultures were incubated at 37 °C for 30 min.
Following sonication and mutanolysin-treatment, the cultures were standardized to a final OD600nm of 0.5. Then, 100 µL of the standardized cells was then applied to fibronectin-coated Maxisorp plates and dose-dependent adhesion was measured using the ELISA-based adhesion assay described above.
To assess viability, the samples were manually enumerated to determine the CFU/mL pretreatment and posttreatment. The standardized cultures were serially diluted 1:100 in sterile 0.85% (w/v) NaCl to a final dilution of 10−6. Then, 100 µL of the final two dilutions (10−5 and 10−6) for presonication and postsonication and mutanolysin samples was plated onto TSA and incubated at 37 °C for 18 h before manually enumerating. Each sample was assessed in biological triplicate.
Assessing the Cellular Localization of Spa.
Immunofluorescence microscopy was used to visualize the cellular localization of Spa as described previously, with minor modifications (44, 45). Three biological replicates of each strain were propagated at 37 °C for 18 h, diluted 1:100 into 8 mL TSB, and incubated at 37 °C until an OD600nm of 1.0 was reached. The cells were harvested by centrifugation at 4,000 × g for 10 min at 4 °C and washed once in 2 mL PBS. The cells were resuspended in 2 mL PBS and separated for treatment with and without sonication. The presonication cells were harvested by centrifugation and resuspended in 1 mL PBS. The postsonication samples were resuspended in 2 mL PBS and sonicated as described above. Following sonication, the cells were harvested by centrifugation and resuspended in 1 mL PBS. Following the wash steps described above, 250 µL of the cell suspension for both presonication and postsonication samples were incubated with 250 µL fixing solution [2.4% (v/v) paraformaldehyde, 0.01% (v/v) glutaraldehyde in PBS] at room temperature for 20 min. The cells were washed twice in 1 mL PBS and harvested by centrifugation at 18,000 × g, with vortexing between two washes. The samples were resuspended in 100 µL PBS and 50 µL was applied to poly-L-lysine coated 10-well glass slides (MP Biomedicals). Following a 5-min incubation, the wells were washed twice with PBS and blocked with 3% (w/v) BSA in PBS for 30 min at room temperature. For detection of Spa, 50 µL goat anti-mouse IgG fluorescein isothiocyanate (FITC)-conjugated antibody [1:100 in 3% (w/v) BSA in PBS; ThermoScientific] was applied and incubated for 1 h at room temperature in the dark. The wells were then washed 10 times with PBS. To stain the cell wall, wheat germ albumin conjugated to tetramethylrhodamine (WGA-TRITC; ThermoScientific) was diluted 1:500 in PBS to a final concentration of 2 ng/mL, and 50 µL was applied to each well. The slides were incubated for 10 min in the dark, followed by three washes and the addition of SlowFade™ Diamond Antifade Mountant (Invitrogen) and a coverslip. The slides were sealed and stored at 4 °C prior to imaging.
The images were acquired sequentially using a Leica DM5500 upright epifluorescence microscope (Leica, Mannheim, Germany) equipped with a 63× (NA = 1.3) glycerol immersion objective. Images for each biological replicate were acquired with identical settings and all images were obtained under the same magnification. To perform cell count analysis to determine the relative proportion of cells with 0%, 50%, and 100% coverage, a region of interest of consistent size was selected and ~200 to 350 cells were counted within the region of interest. Of note, 0% coverage was designated to cells with minimal to no visible FITC signal, 50% coverage was designated to cells with FITC signal concentrated on one face of the cells, and 100% coverage was designated to cells with concentric FITC signal surrounding the entire cell. Three biological replicates were counted for each sample presonication and postsonication, and the data were normalized by the percentage of total cells.
Murine Models of Infection.
All animal experiments were performed in compliance with guidelines set out by the Canadian Council on Animal Care. All animal protocols (protocol #2017-028) were reviewed and approved by the University of Western Ontario Animal Use Subcommittee, a subcommittee of the University Council on Animal Care. For bloodstream infection, the wild-type strain (JE2) was grown in TSB ± 0.5 µg/mL complestatin to an OD600nm of 1.5, washed twice with PBS, and resuspended in PBS. In the untreated cells, equal concentrations of DMSO were included as a solvent control. Female 8 to 10-wk-old BALB/c mice (n = 6 per group) (Charles River Laboratories) were infected via tail vein with 1 to 2 × 107 CFU. At 24 hours post infection (hpi) the animals were sacrificed, and the organs were harvested in 3 mL PBS + 0.1% (v/v) TritonX-100, followed by homogenization in a Bullet Blender homogenizer (Speed 6, 2 × 5 min runs). Viable counts preinfection and postinfection were performed on TSA following sonication.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Dr. Elyse Roach and the late Dr. Bob Harris (University of Guelph) for their assistance with SEM sample preparation, and Nicole Garnier, Alyssa Banaag, and Dr. Michaela Struder–Kypke for guidance and assistance with immunofluorescence microscopy. We also thank Mackenzie Verhoef (University of Guelph) for technical assistance with aspects of the adhesion assays. Special thanks to Dr. Gerard Wright, Dr. Min Xu, and Dr. Beth Culp (McMaster University) for their kind gift of complestatin and corbomycin. Additionally, we would like to thank Dr. Simon Foster and Dr. Mariana Tinajero-Trejo (University of Sheffield) for guidance with cellular fractionation. This study was supported by funding through a New Frontiers in Research Fund-Exploration Grant awarded to G.C. (NFRFE-2018-01058), the Canada Foundation for Innovation (JELF 37730) to G.C., by Canadian Institutes of Health Research grants awarded to D.E.H. (PJT-168842) and G.C. (PJT-436506), and a Doctoral Research Award awarded to A.C.L. through the Canada Graduate Scholarships program.
Author contributions
D.E.H. and G.C. designed research; A.C.L., M.I.G., S.E.G., H.S., L.E.P., and L.K.T. performed research; M.I.G., C.M.K., and D.E.H. contributed new reagents/analytic tools; A.C.L., M.I.G., S.E.G., H.S., L.E.P., and C.M.K. analyzed data; and A.C.L., M.I.G., D.E.H., and G.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Berry K. A., Verhoef M. T. A., Leonard A. C., Cox G., Staphylococcus aureus adhesion to the host. Ann. N. Y. Acad. Sci. 1515, 75–96 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Foster T. J., Geoghegan J. A., Ganesh V. K., Höök M., Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leonard A. C., Petrie L. E., Cox G., Bacterial anti-adhesives: Inhibition of Staphylococcus aureus nasal colonization. ACS Infect. Dis. 5, 1668–1681 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Petrie L. E., Leonard A. C., Murphy J., Cox G., Development and validation of a high-throughput whole-cell assay to investigate Staphylococcus aureus adhesion to host ligands. J. Biol. Chem. 295, 16700–16712 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas R., et al. , Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. 259, 260–268 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Heilmann C., Hussain M., Peters G., Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 5, 1013–1024 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Porayath C., et al. , Autolysin mediated adherence of Staphylococcus aureus with Fibronectin, Gelatin and Heparin. Int. J. Biol. Macromol. 110, 179–184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohler T. P., et al. , Repeating structures of the major staphylococcal autolysin are essential for the interaction with human thrombospondin 1 and vitronectin. J. Biol. Chem. 289, 4070–4082 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirschhausen N., et al. , A novel staphylococcal internalization mechanism involves the major autolysin Atl and heat shock cognate protein Hsc70 as host cell receptor. Cell. Microbiol. 12, 1746–1764 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Culp E. J., et al. , Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling. Nature 578, 582–587 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Sugai M., et al. , Identification of endo-beta-N-acetylglucosaminidase and N-acetylmuramyl-L-alanine amidase as cluster-dispersing enzymes in Staphylococcus aureus. J. Bacteriol. 177, 1491–1496 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oshida T., et al. , A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: Cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. U.S.A. 92, 285–289 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoi K.-J., et al. , Molecular properties of the putative autolysin Atl(WM) encoded by Staphylococcus warneri M: Mutational and biochemical analyses of the amidase and glucosaminidase domains. Gene 416, 66–76 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Zoll S., et al. , Structural basis of cell wall cleavage by a staphylococcal autolysin. PLoS Pathog. 6, e1000807 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bose J. L., Lehman M. K., Fey P. D., Bayles K. W., Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS One 7, e42244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penewit K., et al. , Efficient and scalable precision genome editing in Staphylococcus aureus through conditional recombineering and CRISPR/Cas9-mediated counterselection. mBio 9, e00067-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fey P. D., et al. , A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4, e00537-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazmanian S. K., Liu G., Jensen E. R., Lenoy E., Schneewind O., Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. U.S.A. 97, 5510–5515 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goncheva M. I., et al. , Stress-induced inactivation of the Staphylococcus aureus purine biosynthesis repressor leads to hypervirulence. Nat. Commun. 10, 775 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goncheva M. I., Flannagan R. S., Heinrichs D. E., De novo purine biosynthesis is required for intracellular growth of Staphylococcus aureus and for the hypervirulence phenotype of a purR mutant. Infect. Immun. 88, e00104-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forsgren A., Sjöquist J., “Protein a” from S. aureus: I. Pseudo-immune reaction with human γ-globulin. J. Immunol. 97, 822–827 (1966). [PubMed] [Google Scholar]
- 22.Inganas M., Comparison of mechanisms of interaction between protein A from Staphylococcus aureus and human monoclonal IgG, IgA and IgM in relation to the classical Fcgamma and the alternative F(ab’)2gamma protein A interactions. Scand. J. Immunol. 13, 343–352 (1981). [DOI] [PubMed] [Google Scholar]
- 23.Peterson P. K., Verhoef J., Sabath L. D., Quie P. G., Effect of protein A on staphylococcal opsonization. Infect. Immun. 15, 760–764 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasztor L., et al. , Staphylococcal major autolysin (Atl) is involved in excretion of cytoplasmic proteins. J. Biol. Chem. 285, 36794–36803 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi J., et al. , Molecular characterization of an atl null mutant of Staphylococcus aureus. Microbiol. Immunol. 46, 601–612 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Zheng X., Ma S. X., St John A., Torres V. J., The major autolysin Atl regulates the virulence of Staphylococcus aureus by controlling the Sorting of LukAB. Infect. Immun. 90, e0005622 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamada S., et al. , An autolysin ring associated with cell separation of Staphylococcus aureus. J. Bacteriol. 178, 1565–1571 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nega M., Tribelli P. M., Hipp K., Stahl M., Götz F., New insights in the coordinated amidase and glucosaminidase activity of the major autolysin (Atl) in Staphylococcus aureus. Commun. Biol. 3, 695 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeDent A., Bae T., Missiakas D. M., Schneewind O., Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. EMBO J. 27, 2656–2668 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nandakumar R., Nandakumar M. P., Marten M. R., Ross J. M., Proteome analysis of membrane and cell wall associated proteins from Staphylococcus aureus. J. Proteome Res. 4, 250–257 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Gatlin C. L., et al. , Proteomic profiling of cell envelope-associated proteins from Staphylococcus aureus. Proteomics 6, 1530–1549 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Taverna F., et al. , Characterization of cell wall associated proteins of a Staphylococcus aureus isolated from bovine mastitis case by a proteomic approach. Vet. Microbiol. 119, 240–247 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Jones R. C., Deck J., Edmondson R. D., Hart M. E., Relative quantitative comparisons of the extracellular protein profiles of Staphylococcus aureus UAMS-1 and its sarA, agr, and sarA agr regulatory mutants using one-dimensional polyacrylamide gel electrophoresis and nanocapillary liquid chromatography coupled with tandem mass spectrometry. J. Bacteriol. 190, 5265–5278 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ventura C. L., et al. , Identification of a novel Staphylococcus aureus two-component leukotoxin using cell surface proteomics. PLoS One 5, e11634 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farrand A. J., et al. , An iron-regulated autolysin remodels the cell wall to facilitate heme acquisition in Staphylococcus lugdunensis. Infect. Immun. 83, 3578–3589 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vollmer W., Joris B., Charlier P., Foster S., Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32, 259–286 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Berry A. M., Lock R. A., Hansman D., Paton J. C., Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect. Immun. 57, 2324–2330 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tenover F. C., Goering R. V., Methicillin-resistant Staphylococcus aureus strain USA300: Origin and epidemiology. J. Antimicrob. Chemother. 64, 441–446 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Diekema D. J., et al. , Continued emergence of USA300 methicillin-resistant Staphylococcus aureus in the United States: Results from a nationwide surveillance study. Infect. Control Hosp. Epidemiol. 35, 285–292 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Monk I. R., Tree J. J., Howden B. P., Stinear T. P., Foster T. J., Complete bypass of restriction systems for major Staphylococcus aureus lineages. mBio 6, e00308-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corrigan R. M., Foster T. J., An improved tetracycline-inducible expression vector for Staphylococcus aureus. Plasmid 61, 126–129 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Bose J. L., Fey P. D., Bayles K. W., Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl. Environ. Microbiol. 79, 2218–2224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tinajero-Trejo M., et al. , The Staphylococcus aureus cell division protein, DivIC, interacts with the cell wall and controls its biosynthesis. Commun. Biol. 5, 1228 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu W., Missiakas D., Schneewind O., Septal secretion of protein A in Staphylococcus aureus requires SecA and lipoteichoic acid synthesis. Elife 7, e34092 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scaffidi S. J., Shebes M. A., Yu W., Tracking the subcellular localization of surface proteins in Staphylococcus aureus by immunofluorescence microscopy. Bio. Protoc. 11, e4038 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.