Abstract
Hemorrhagic transformation (HT) is one of the most serious complications after endovascular thrombectomy (EVT) in acute ischemic stroke (AIS) patients. The purpose of this study is to develop and validate deep‐learning (DL) models based on multiparametric magnetic resonance imaging (MRI) to automatically predict HT in AIS patients. Multiparametric MRI and clinical data of AIS patients with EVT from two centers (data set 1 for training and testing: n = 338; data set 2 for validating: n = 54) were used in the DL models. The acute infarction area of diffusion‐weighted imaging (DWI) and hypoperfusion of perfusion‐weighted imaging (PWI) was labeled manually. Two forms of data sets (volume of interest [VOI] data sets and slice data sets) were analyzed, respectively. The models based on single parameter and multiparameter models were developed and validated to predict HT in AIS patients after EVT. Performance was evaluated by area under the receiver‐operating characteristic curve (AUC), accuracy (ACC), sensitivity, specificity, negative predictive value, and positive predictive value. The results showed that the performance of single parameter model based on MTT (VOI data set: AUC = 0.933, ACC = 0.843; slice data set: AUC = 0.945, ACC = 0.833) and TTP (VOI data set: AUC = 0.916, ACC = 0.873; slice data set: AUC = 0.889, ACC = 0.818) were better than the other single parameter model. The multiparameter model based on DWI & MTT & TTP & Clinical (DMTC) had the best performance for predicting HT (VOI data set: AUC = 0.948, ACC = 0.892; slice data set: AUC = 0.932, ACC = 0.873). The DMTC model in the external validation set achieved similar performance with the testing set (VOI data set: AUC = 0.939, ACC = 0.884; slice data set: AUC = 0.927, ACC = 0.871) (p > 0.05). The proposed clinical, DWI, and PWI multiparameter DL model has great potential for assisting the periprocedural management in the early prediction HT of the AIS patients with EVT.
Keywords: deep learning, endovascular thrombectomy, hemorrhagic transformation, magnetic resonance imaging, stroke
The proposed DWI, PWI, and clinical multiparameter DL model may provide a potential tool for predicting information before therapy to assist the periprocedural management in AIS patients with EVT.
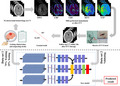
1. INTRODUCTION
Acute ischemic stroke (AIS), which has high mortality and disability rates, is seriously harmful to human life [1]. Endovascular thrombectomy (EVT) has been proven to benefit AIS patients with large vessel occlusion (LVO) [2, 3, 4]. However, the serious complication of hemorrhagic transformation (HT) often occurs after EVT [5]. Recent studies have shown that the incidence of HT after EVT can be as high as 31.9% [6] and increased morbidity and mortality [7]. Identifying this serious complication could help to adjust periprocedural management, especially by predicting needs for intensive care among patients at high risk of HT.
Neuroimaging, especially magnetic resonance imaging (MRI), has shown to be effective in identifying tissue at risk of infarction [8]. Additionally, diffusion‐weighted imaging (DWI) and perfusion‐weighted imaging (PWI) can detect tissue ischemia earlier than other conventional neuroimaging modalities in experimental and clinical settings, which may inform on the risk of HT [9]. Previous studies have reported that the large initial lesion volume on DWI, a large area of perfusion loss, and regions with very low cerebral blood volume (CBV) were important for predicting HT [6, 10, 11, 12]. However, as a single measure, none of these MRI‐based indices has been shown to be able to reliably identify tissue at risk of HT prior to EVT. In addition, automated MRI PWI–DWI mismatch estimation may be significantly different in individual patients when using different software packages [13]. Thus, because of the various risk factors, postprocessing software, settings, and chosen parameters, it is still difficult to predict the HT after EVT early in AIS patients in the clinical workflow.
Deep learning (DL) is a form of representation learning—in which a machine is fed with raw data and develops its own representations needed for pattern recognition—that is composed of multiple layers of representations [14]. Although DL algorithms require a large amount of data to function, they have exceeded the capabilities of classical statistical machine‐learning (ML) techniques on specific imaging tasks like multiclass classification [14, 15]. Convolutional neural networks (CNNs), a type of DL algorithm, have grown to be central in this field. CNN methods take image data as input and iteratively warp it through a series of convolutional and nonlinear operations until the original raw data matrix is transformed into a probability distribution over potential image classes [16]. CNNs have shown strong performances in the prediction of tissue outcome [17] and detection of penumbral tissue in AIS [18]. It also has the advantage of including simultaneously both multiple‐input biomarkers and spatial information and being capable of modeling complex interplays between the input images. Overall, the predictive results from CNNs yield an infarction probability, providing a much‐needed certainty level, and CNN may be a suitable candidate for predicting HT in AIS patients receiving EVT, which is not yet reported.
In this study, we developed and validated DL models to automatically predict HT in AIS patients receiving EVT by using multiparameter on DWI and PWI images. We hypothesized that the CNN model can be used to provide predictive information before therapy for assisting the periprocedural management in AIS patients with EVT.
2. MATERIALS AND METHODS
2.1. Patient selection and clinical data
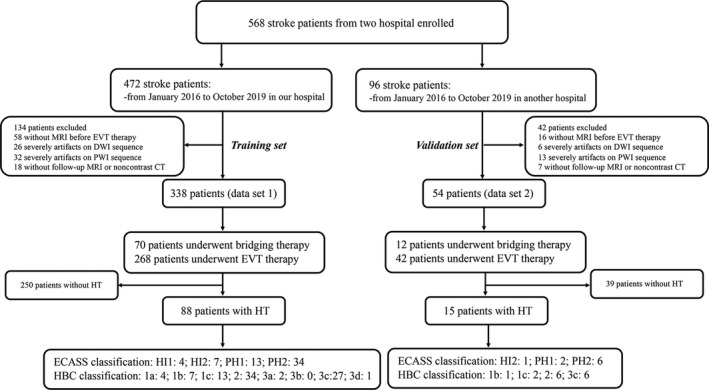
From January 2016 to October 2019, data from Nanjing First Hospital and the Affiliated Jiangning Hospital of Nanjing Medical University were collected. Patients with AIS were included in this study if (1) they are first‐ever AIS within 24 h from the onset, (2) DWI and PWI examinations were performed before EVT therapy, (3) receiving EVT therapy or bridging therapy (both intravenous thrombolysis [IVT] and EVT) according to the guidelines for managing AIS, and (4) follow‐up MRI or noncontrast CT within 24 h after EVT therapy. Patients with previous intracranial hemorrhage, brain surgery, large territorial lesion, or subarachnoid hemorrhage after EVT therapy were excluded. All patients in this study provided written informed consent before examination and treatment. The study was approved by the local ethics committee of the Nanjing Medical University. Finally, a total of 338 patients from Nanjing First Hospital (data set 1) and 54 patients from the Affiliated Jiangning Hospital of Nanjing Medical University (data set 2) were included. Data set 1 was used to train and test the models and data set 2 was preserved as an independent external validation set. The flowchart of this study is shown in Figures 1 and 2.
FIGURE 1.
The demographic data of the cohorts
FIGURE 2.
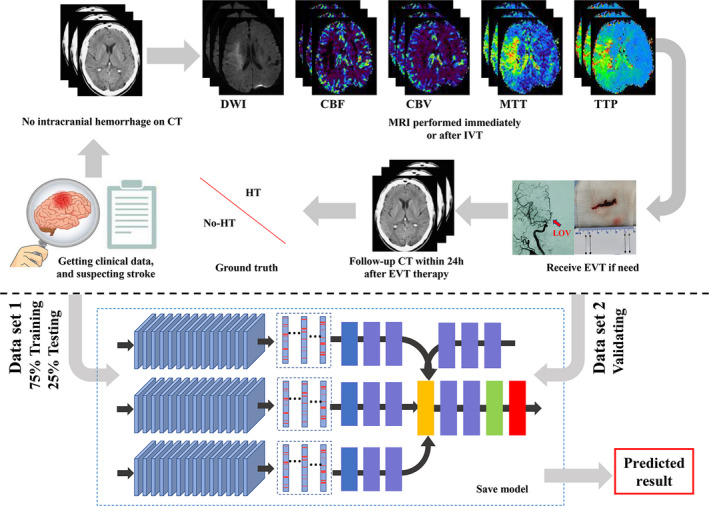
The flowchart of the study. (1) Data acquisition: all patients suspected of acute stroke underwent head CT examination to exclude hemorrhage. Clinical data were collected and normalized by min–max normalization method. Patients who met the criteria for intravenous thrombolysis (IVT) therapy received IVT after CT scanning, and MRI examination was followed scanned immediately. Then patients who met the criteria for endovascular thrombectomy (EVT) therapy performed EVT immediately. All patients underwent CT after EVT within 24 h. According to the follow‐up CT, the patients were divided into hemorrhagic transformation (HT) group and the no HT group. (2) Model training: data set 1 was split into training (75%) and testing (25%) subsets by stratified random sampling, and data set 2 was used for validating. The labels of two forms (VOI data sets and slice data sets) were trained and validated. Then the final result was evaluated [Colour figure can be viewed at wileyonlinelibrary.com]
HT was independently reviewed on follow‐up noncontrast CT or cranial MRI within 24 h after EVT therapy by two neuroradiology staff (Y‐CC, attending doctor with 4 years of experience in neuroradiology, and XY, director with 10 years of experience in neuroradiology). In case of discrepant assessment results between the two readers, a consensus was established. HT was categorized according to the Heidelberg Bleeding classification (HBC) [19] and European Collaborative Acute Stroke Study (ECASS) classification [20] (see Table 1). HT and contrast agent extravasation were distinguished by comparing the previous CT or MRI scans, and the result was confirmed on CT or MRI scans at 2–7 days after the therapy.
TABLE 1.
Overview of bleeding events, categorized with HBC and the ECASS classification, according to anatomical, descriptive, and clinical features
| HBC | ECASS | Patients with HT (data set 1; n = 88) | Patients with HT (data set 2; n = 15) | Description |
|---|---|---|---|---|
| 1a | HI1 | 4 (4.55%) | 0 | Scattered small petechia, no mass effect |
| 1b | HI2 | 7 (7.95%) | 1 (6.67%) | Confluent petechia, no mass effect |
| 1c | PH1 | 13 (14.77%) | 2 (13.33%) | Hematoma within infarcted tissue, occupying <30%, no substansive mass effect |
| 2 | PH2 | 34 (38.64%) | 6 (40.00%) | Hematoma occupying >30% or more of the infarcted tissue, with obvious mass effect |
| 3a | – | 2 (2.27%) | 0 | Parenchymal hematoma remote from infarcted brain tissue |
| 3b | – | 0 | 0 | Intraventricular hemorrhage |
| 3c | – | 27 (30.68%) | 6 (40.00%) | Subarachnoid hemorrhage |
| 3d | – | 1 (1.14%) | 0 | Subdural hemorrhage |
Abbreviations: ECASS, European Collaborative Acute Stroke Study; HBC, Heidelberg Bleeding classification; HI, hemorrhagic infarction; HT, hemorrhagic transformation; PH, parenchymatous hematoma.
2.2. MRI protocols and analysis
MRI on admission and follow‐up MRI were performed on 3.0 Tesla MRI scanner (Ingenia, Philips Medical Systems) with an eight‐channel receiver array head coil. The DWI images were acquired using a spin‐echo (SE) sequence with the following parameters: repetition time (TR), 2501 ms; echo time (TE), 98 ms; acquisition matrix, 152 × 122; three directions; field of view (FOV), 230 × 230 mm; flip angle (FA), 90°; slices, 18; slice thickness, 6 mm; intersection gap, 1.3 mm; and b values, 0 and 1000 s/mm2. DSC‐PWI images were acquired using a T2*‐weighted gradient recalled echo (T2*GRE) sequence with the following parameters: TR, 2000 ms; TE, 30 ms; acquisition matrix, 96 × 93; FOV, 224 × 224 mm; FA, 90°; slice thickness, 4 mm; and scan time, 88 s. Fifty phases and 20 images were obtained from each phase. During dynamic acquisition, a dose of 0.1 mmol/kg of contrast agent (Magnevist, Bayer Schering Pharma, Germany) was injected at a rate of 4 mL/s.
The PWI data were analyzed by using a Philips advanced workstation. The arterial input function (AIF) was selected by manually identifying the M2 segment of the MCA ipsilateral to the acute infarction. The cerebral blood flow (CBF), CBV, time to peak (TTP), and mean transit time (MTT) maps were generated from circular singular value decomposition of the concentration–time curve.
2.3. Preprocessing
As DICOMs with different modalities might have different pixel intensity ranges, we converted the pixel intensity of DWI into [−2200, 2800] and PWI into [0, 4095] to keep the intensity range consistent. In order to achieve the same contrast and brightness and easy to display lesions, we found that the lesions display of all patients was the best when the window width and level were 0–255. Therefore, we linearly compressed the pixel intensity range into [0, 255] of all DICOMs. Then we used the OpenCV library to histogram equalize the image to enhance contrast. Images were then saved as PNG files.
Furthermore, the clinical data (age, gender, NIHSS score on admission, time from MRI to onset, time from MRI to EVT therapy, and history of hypertension, diabetes mellitus, hyperlipidemia, homocysteine levels, and atrial fibrillation) were collected (Table 2). The patient's clinical data were entered into the CNN model in text form. Age was encoded by means of normalization. Sex, smoking, alcohol drinking, diabetes mellitus, hypertension, atrial fibrillation, hyperlipidemia, and homocysteine were encoded using a one‐hot method. The NIHSS was encoded into a one‐dimensional vector. Finally, a total of the 10‐dimensional vector was extracted.
TABLE 2.
Comparison of no HT and HT group in acute stroke patients after EVT
| Data set 1 | Data set 2 | |||||
|---|---|---|---|---|---|---|
| No HT (n = 250) | HT (n = 88) | p value | No HT (n = 39) | HT (n = 15) | p value | |
| Gender (male), n (%) | 165 (66.00%) | 48 (55.55%) | 0.072 | 23 (58.97%) | 8 (53.33%) | 0.765 |
| Age (years), mean ± SD | 65.78 ± 9.68 | 70.93 ± 11.29 | <0.001 | 64.22 ± 8.57 | 71.38 ± 10.19 | <0.001 |
| Time from MRI to onset (min), mean ± SD | 202.72 ± 77.36 | 218.17 ± 114.19 | 0.159 | 217.19 ± 89.39 | 220.03 ± 102.41 | 0.211 |
| Time from EVT to onset (min), mean ± SD | 285.25 ± 137.27 | 314.47 ± 104.56 | 0.070 | 301.36 ± 124.71 | 319.07 ± 128.11 | 0.127 |
| Smoking, n (%) | 57 (22.80%) | 16 (18.18%) | 0.452 | 7 (17.95%) | 2 (13.33%) | 1.000 |
| Alcohol drinking, n (%) | 25 (10.00%) | 15 (17.05%) | 0.086 | 4 (10.26%) | 2 (13.33%) | 1.000 |
| Diabetes mellitus, n (%) | 60 (24.00%) | 25 (28.41%) | 0.475 | 8 (20.51%) | 4 (26.67%) | 0.719 |
| Hypertension, n (%) | 197 (78.80%) | 72 (81.82%) | 0.645 | 31 (79.49%) | 12 (80.00%) | 1.000 |
| Atrial fibrillation, n (%) | 102 (40.80%) | 46 (52.27%) | 0.080 | 14 (35.90%) | 7 (46.67%) | 0.541 |
| Hyperlipidemia, n (%) | 22 (8.80%) | 9 (10.23%) | 0.672 | 3 (7.69%) | 2 (13.33%) | 0.610 |
| Homocysteine, n (%) | 18 (7.20%) | 7(7.95%) | 0.815 | 3 (7.69%) | 2 (13.33%) | 0.610 |
| NIHSS on admission, n (%) | 10.91 ± 3.87 | 13.90 ± 2.46 | <0.001 | 11.26 ± 4.41 | 14.79 ± 6.65 | <0.001 |
| Reperfusion therapy, n (%) | 0.362 | 1.000 | ||||
| EVT | 195 (78.00%) | 73 (82.95%) | 30 (76.92%) | 12 (80.00%) | ||
| IVT + EVT | 55 (22.00%) | 15 (17.05%) | 9 (23.08%) | 3 (20.00%) | ||
Abbreviations: EVT, endovascular thrombectomy; HT, hemorrhagic transformation; IVT, intravenous thrombolysis; NIHSS, National Institutes of Health Stroke Scale.
All DWI and PWI images of AIS patients were derived from DICOM format and then converted all DICOM format into NII format by using MRIcron software (https://www.nitrc.org/projects/mricron). The high‐intensity signal infarction area on DWI and abnormal perfusion area on CBF, CBV, MTT, and TTP were drawn as volumes of interest (VOIs) manually using ITK‐SNAP software (http://www.itksnap.org/pmwiki/pmwiki.php). All VOIs were performed in consensus by two board‐certified neuroradiologists (LJ, 9 years of experience in neuroradiology, H‐YC, 8 years of experience in neuroradiology) who were blinded to the clinical data. We developed and evaluated the HT prediction model in two forms. First, the VOIs of DWI, CBF, CBV, MTT, and TTP were analyzed as labels individually or together (VOI data set). Then, the axial images of VOIs (DWI, CBF, CBV, MTT, and TTP) were analyzed as labels individually or together (slice data set).
2.4. Deep neural network architecture and experiments
2.4.1. Single parameter model
For detecting the clinical features in predicting HT in AIS patients after EVT, the clinical model was built, which involved three fully connected layers and one sigmoid layer. The network used in our study of radiomics model was the Inception V3 [21], which offers compact end‐to‐end CNN structures that maintain high‐resolution multiscale features. The schematic diagram of the basic CNN model was shown in Figure 3. The feature vectors (1024 × 1 × N) extracted from all the images (N means the number of images of one patient) of a given single parameter were merged into a one‐dimensional vector (1024 × 1) to form the input tensor of the patient‐level model with a Max pooling layer. This layer was used to uniform the different image sequence lengths. At the end of the model, two FC layers (the first layer had 1024 nodes, and the second layer had two nodes for HT and no HT) were added, and the prediction of the HT risk used the Softmax function.
FIGURE 3.
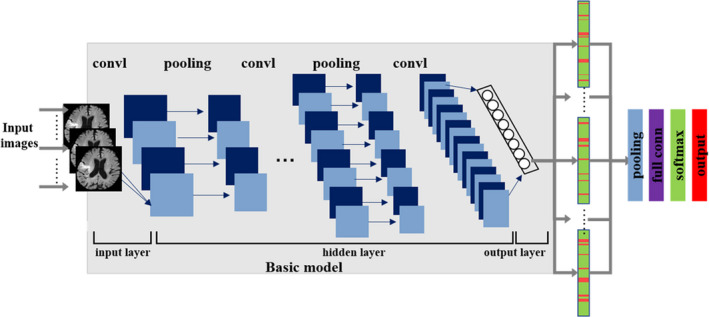
The schematic diagram of the basic CNN model [Colour figure can be viewed at wileyonlinelibrary.com]
2.4.2. Multiparameters model
Based on the single parameter models (DWI, CBF, CBV, MTT, and TTP), the multiparameter model was further established by considering the patient's multiparameter image features synthetically (Figure 4A). The multiparameter models were an ensemble of single parameter models and included four kinds of multiparameters: multiparameter PWI model (combining two PWI parameters of the best prediction efficiency); clinical + PWI parameters model (combining clinical data and the above two PWI parameters); DWI + PWI parameters model (combining DWI parameter and the above two PWI parameters); and clinical + MRI parameters model (combining clinical data, DWI parameter, and the above two PWI parameters). The methods are as follows: first, these separated single‐sequence models were trained and optimized, respectively. All models were CNN trained with the combined set of training images. We used Inception V3 as the convolutional neural network architecture that was pretrained on ImageNet. Then the features extracted from each pretrained single parameter model were concatenated to a tensor. Later, two layers of fully connected layer and Softmax layer after were added following the concatenation layer to classify HT or no HT. Furthermore, the compounded model was developed and validated by incorporating the multiparameter radiomic and clinical features (Figure 4B). The clinical feature tensor was got through three fully connected layers. The weights of each pretrained single parameter model and clinical model were frozen as the weight of the corresponding channel, and only the following fully connected layers were refined during the training process. Of note, “MT” represents the multiparameter model of MTT and TTP; “MTC” represents the multiparameter model of MTT and TTP and clinical; “DMT” represents the multiparameter model of DWI and MTT and TTP; and “DMTC” represents the multiparameter model of DWI and MTT and TTP and clinical.
FIGURE 4.
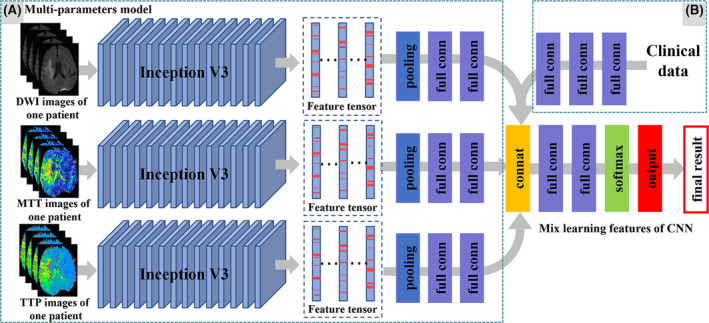
An overview of the patient‐level multiparameter model. (A) Patient‐level single parameter module: first, learning intraimage features (1024 × 1) from one patient's N image based on image‐level model (Inception V3) and concatenating features into two‐dimensional vectors (1024 × N). And then merge the two‐dimensional vectors into a one‐dimensional vector by the max pooling layer. Finally, the interimage features are learned through two FC layers. Based on the patient‐level single parameter module, the patient‐level multiparameter module was developed and validated by considering the patient's multiparameter radiomic features. (B) Furthermore, using the concatenation layer to concatenate the multiparameter radiomic features and clinical characteristics, then two layers of fully connected layer and Softmax layer were added [Colour figure can be viewed at wileyonlinelibrary.com]
2.4.3. Training details
As a result of the disequilibrium of our data set, during the training process, the HT data set was dynamically oversampled on the patient level to get a balance between HT and no HT data set. The training data set was augmented by adding rotation (rotation range = [−20°, 20°]), two shifts (width shift range = [0, 0.2]), height shift range = [0, 0.2]), and zoom (zoom ratio = 0.2) variants for each image. Volumes were randomly extracted from preprocessed input and label images for training. With sufficient data augmentation, the network could be prevented from overfitting.
The stochastic gradient descent (SGD) optimizer was used as the optimization method for model training. The learning rate was 0.01, the momentum was 0.9, and the decay rate was 1.0 × 10−6. The mini‐batch size was set to 16. The models were trained with the iteration stopping criteria, when the validation loss drops <0.03% within 10 epochs or the iteration reaches 200 epochs, the training is stopped. All models were implemented using the KERAS framework and all experiments were performed on a workstation equipped with an Intel(R) Xeon(R) E5‐2650 v4 CPUs @ 2.20 GHz (two CPUs, 24 cores, two threads/core, 128 GB of memory) and an NVIDIA Tesla M4.
2.4.4. Performance assessment
Data set 1 were split into training (75%) (HT: 187 patients; no HT: 66 patients) and testing (25%) (HT: 63 patients; no HT: 22 patients) subsets by stratified random sampling, and we ensured that images from the same patient remained in the same split to avoid training and testing on the same patient. Then parameters were trained and tested with two forms of labels (VOI and slice image), respectively. Model parameter exploration was performed by five‐fold cross‐validation on the training data set. During the validation phase, all the metrics were calculated based on the average five‐fold cross‐validation results. Then, a better model structure was chosen, the model was retrained with all the training data, and the model parameters were saved. To eliminate contingencies in the test results and evaluate the performance of the HT classification model, the results were compared with neuroradiologists’ conclusions and evaluated by several metrics, including accuracy (ACC), SEN, SPC, negative predictive value (NPV), positive predictive value (PPV), receiver‐operating characteristic (ROC) curves, and AUC. After the aforementioned process, a final model with the best performance was chosen and validated in the external validation set (data set 2).
2.5. Statistical analysis
All statistical analyses for clinical data were conducted using commercially available software (SPSS for Windows, version 19.0). Continuous data are shown as the mean ± SD and were analyzed by using an independent samples t‐test or Fisher's exact test, whereas categorical variables are presented as absolute and relative frequencies and were analyzed by using the χ 2 test. p < 0.05 was considered to be statistically significant. Kappa values were used to determine interrater agreement. The ROC differences of the testing set and validating set were evaluated according to Delong et al. [22].
3. RESULTS
3.1. Clinical and demographic information
Of 338 patients enrolled from data set 1, 88 patients (26.04%) had HT after EVT therapy. Of 54 patients enrolled from data set 2, 15 patients (27.78%) had HT after EVT therapy. The interobserver agreement for HT was k = 0.96 (95% CI, 0.93–0.99). Bleeding classification specifics are shown in Table 1. The clinic and demographic information are shown in Table 2. In both data set 1 and data set 2, the age in HT group was older (data set 1: 70.93 ± 11.29 vs. 65.78 ± 9.68, p < 0.001; data set 2: 71.38 ± 10.19 vs. 64.22 ± 8.57, p < 0.001) than that of no HT group, and the NIHSS on admission in HT group was higher (data set 1: 13.90 ± 2.46 vs. 10.91 ± 3.87, p < 0.001; data set 2: 14.79 ± 6.65 vs. 11.26 ± 4.41, p < 0.001) than that of no HT group. There were no significant differences in gender, smoking, alcohol drinking, diabetes mellitus, hypertension, atrial fibrillation, hyperlipidemia, and homocysteine between the two groups of both data set 1 and data set 2 (p > 0.05).
3.2. Evaluation of single parameter model
The results are listed in Tables 3 and 4. For the single parameter model, the result of the single clinical model (AUC = 0.680) to predict HT after EVT was the lowest, with the ACC was 0.659. The results of the model based on image features were significantly better than those based on clinical features. The VOI data set test results of AUCs from MTT (AUC = 0.933) and TTP (AUC = 0.916) were larger than other parameters (Figure 5A), with the ACC was 0.843 and 0.873, respectively. The slice data set test results of AUCs from MTT (AUC = 0.945) and TTP (AUC = 0.889) to predict HT after EVT in AIS patients were also larger than other parameters (Figure 5B), with the ACC was 0.833 and 0.818, respectively. Other metrics such as SEN, SPC, PPV, and NPV in Tables 3 and 4 presented similar trends as the AUCs. For the single parameter model, the results of the slice data set were worse than the VOI data set.
TABLE 3.
Performance comparison of the VOI CNN methods by using different MRI parameters
| Model | Parameters | ACC | SEN | SPC | PPV | NPV | AUC |
|---|---|---|---|---|---|---|---|
| The single parameter model | Clinical | 0.659 | 0.778 | 0.613 | 0.420 | 0.885 | 0.680 |
| DWI | 0.815 | 0.700 | 0.847 | 0.560 | 0.910 | 0.830 | |
| CBF | 0.773 | 0.9375 | 0.720 | 0.517 | 0.973 | 0.835 | |
| CBV | 0.823 | 0.815 | 0.827 | 0.629 | 0.925 | 0.878 | |
| MTT | 0.843 | 0.926 | 0.813 | 0.641 | 0.968 | 0.933 | |
| TTP | 0.873 | 0.889 | 0.867 | 0.706 | 0.956 | 0.916 | |
| The multiparameter model | MT | 0.863 | 0.926 | 0.840 | 0.676 | 0.969 | 0.924 |
| MTC | 0.882 | 0.895 | 0.880 | 0.630 | 0.973 | 0.924 | |
| DMT | 0.873 | 0.852 | 0.867 | 0.700 | 0.941 | 0.933 | |
| DMTC | 0.892 | 0.864 | 0.900 | 0.704 | 0.960 | 0.948 | |
| DMTC* | 0.884 | 0.859 | 0.890 | 0.703 | 0.955 | 0.939 |
Abbreviations: ACC, accuracy; AUC, area under the receiver‐operating characteristic curve; CBF, cerebral blood flow; CBV, cerebral blood volume; CNN, convolutional neural network; DMT, DWI & MTT &TTP; DMTC*, DWI & MTT & TTP & clinical of external validation set; DMTC, DWI & MTT & TTP & Clinical; DWI, diffusion‐weighted imaging; MT, MTT&TTP; MTC, MTT&TTP& Clinical; MTT, mean transit time; NPV, negative predictive value; PPV, positive predictive value; SEN, sensitivity; SPC, specificity; TTP, time to peak; VOI, volume of interest.
TABLE 4.
Performance comparison of the Slice CNN methods by using different MRI parameters
| Model | Parameters | ACC | SEN | SPC | PPV | NPV | AUC |
|---|---|---|---|---|---|---|---|
| The single parameter model | Clinical | 0.659 | 0.778 | 0.613 | 0.420 | 0.885 | 0.680 |
| DWI | 0.667 | 0.579 | 0.690 | 0.333 | 0.860 | 0.609 | |
| CBF | 0.742 | 0.625 | 0.78 | 0.476 | 0.867 | 0.689 | |
| CBV | 0.667 | 0.929 | 0.58 | 0.417 | 0.967 | 0.702 | |
| MTT | 0.833 | 1.000 | 0.78 | 0.592 | 1.000 | 0.945 | |
| TTP | 0.818 | 0.875 | 0.80 | 0.583 | 0.952 | 0.889 | |
| The multi parameter model | MT | 0.853 | 0.925 | 0.827 | 0.658 | 0.969 | 0.896 |
| MTC | 0.863 | 0.882 | 0.859 | 0.556 | 0.973 | 0.913 | |
| DMT | 0.871 | 0.818 | 0.919 | 0.786 | 0.932 | 0.921 | |
| DMTC | 0.873 | 0.850 | 0.878 | 0.630 | 0.960 | 0.932 | |
| DMTC* | 0.871 | 0.820 | 0.865 | 0.611 | 0.943 | 0.927 |
Abbreviations: ACC, accuracy; AUC, area under the receiver‐operating characteristic curve; CBF, cerebral blood flow; CBV, cerebral blood volume; CNN, convolutional neural network; DMT, DWI & MTT &TTP; DMTC*, DWI & MTT & TTP & clinical of external validation set; DMTC, DWI & MTT & TTP & clinical; DWI, diffusion‐weighted imaging; MT, MTT&TTP; MTC, MTT&TTP& Clinical; MTT, mean transit time; NPV, negative predictive value; PPV, positive predictive value; SEN, sensitivity; SPC, specificity; TTP, time to peak.
FIGURE 5.
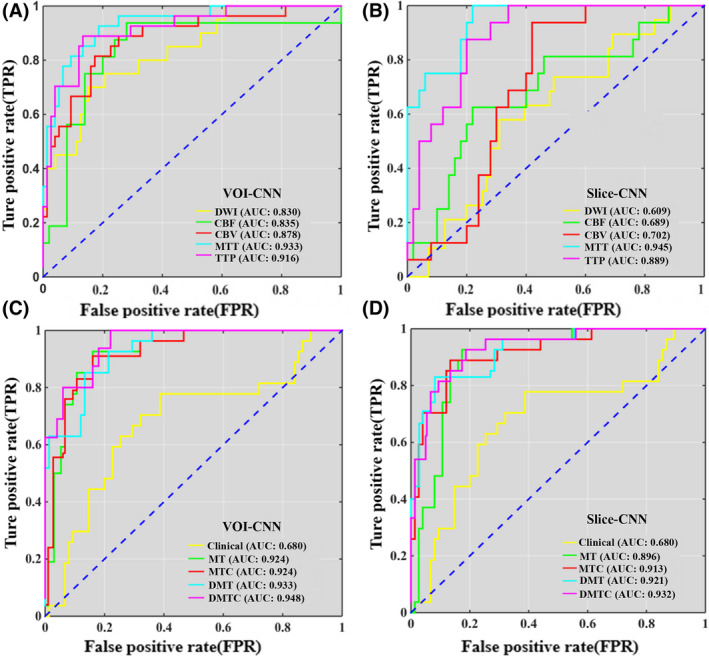
The ROC results are based on the VOI and Slice data set. (A) The ROCs of VOI data set trained with single MRI parameters. (B) The ROCs of the slice data set trained with single MRI parameters. (C) The ROCs of the VOI data set were trained with clinical and multi‐MRI parameters. (D) The ROCs of Slice data set were trained with clinical and multi‐MRI parameters. Note: MT: MTT & TTP; MTC: MTT & TTP & Clinical; DMT: DWI & MTT & TTP; DMTC: DWI & MTT & TTP & Clinical [Colour figure can be viewed at wileyonlinelibrary.com]
3.3. Evaluation of multiparameter model
MTT and TTP with high prediction accuracy in the PWI parameters were selected for the multiparameter model, and the DWI was also selected for the multiparameter model because of the necessary sequence of AIS diagnosis. The evaluation of test results is depicted in Tables 3 and 4. On the VOI data set, compared with the average AUC of all single parameter models (AUC = 0.845), the AUCs of the MT model (AUC = 0.924), and MTC model (AUC = 0.924) for predicting the HT after EVT in AIS were increased by 7.90%, with the ACCs were 0.863 and 0.882, respectively; DMT model (AUC = 0.933) was increased by 8.80% with the ACC was 0.873; DMTC model (AUC = 0.948) was increased by 10.30% with the ACC was 0.892, which was the highest of all models (Figure 5C). On the slice data set, compared with the average AUC of all single parameter models (AUC = 0.752), the AUC of MT model (AUC = 0.896) for predicting the HT after EVT in AIS were increased by 13.80% with the ACC was 0.853; MTC (AUC = 0.913) was increased by 16.10% with the ACC was 0.863; DMT model (AUC = 0.921) was increased by 16.90% with the ACC was 0.871; DMTC model (AUC = 0.932) was increased by 18.00% with the ACC was 0.873 (Figure 6), which was also the highest of all models (Figure 5D). Other metrics such as SEN, SPC, PPV, and NPV in Tables 3 and 4 also presented similar trends as the AUCs.
FIGURE 6.
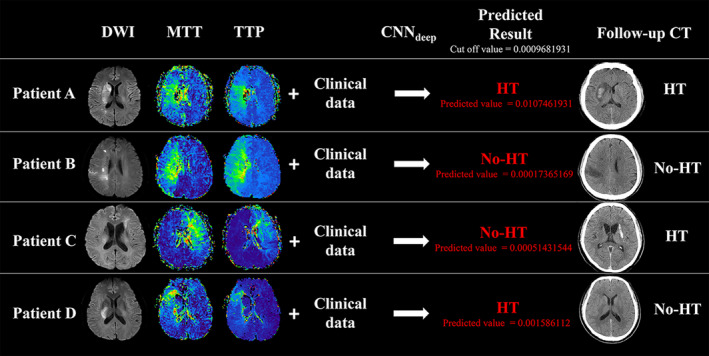
Results from the predictive models for patients from the test set. Four biomarkers were used for predictions (diffusion‐weighted imaging [DWI], mean transit time [MTT], time to peak [TTP], and Clinical data). After deep convolutional neural network (CNNdeep), the cutoff value of the DMTC model in predicting the hemorrhagic transformation (HT) is 0.0009681931. (A) Patient A is a 61‐year‐old man (NIHSS = 15), scanned 3.5 h after symptom onset. The predicted value is 0.0107461931, which means the patient having HT after therapy. Follow‐up CT at 24 h showed HT, which is consistent with the model prediction. (B) Patient B is a 58‐year‐old man (NIHSS = 18), scanned 3 h after symptom onset. The predicted value is 0.00017365169, which means the patient having no HT after therapy. Follow‐up CT at 24 h showed no HT, which is consistent with the model prediction. (C) Patient C is a 60‐year‐old man (NIHSS = 13), scanned 3.5 h after symptom onset. The predicted value is 0.00051431544, which means the patient having no HT after therapy while follow‐up CT at 24 h showed HT. (D) Patient D is a 65‐year‐old man (NIHSS = 16), scanned 4 h after symptom onset. The predicted value is 0.001586112, which means the patient having HT after therapy while follow‐up CT at 24 h showed no HT. Patients C and D showed that the prediction model may give erroneous results [Colour figure can be viewed at wileyonlinelibrary.com]
3.4. Independent external validation
The final model with the best performance (DMTC model) was validated in an independent external validation set. On the VOI data set, the AUC of the validation set was 0.939, and the accuracy, sensitivity, and specificity were 0.884, 0.859, and 0.890, respectively. On the slice dataset, the AUC of the validation set was 0.927, and the accuracy, sensitivity, and specificity were 0.871, 0.820, and 0.865, respectively. The results are depicted in Tables 3 and 4. There were no significant differences between the AUC of the testing set and the validation set (VOI data set: z = 1.479, p = 0.147; slice data set: z = 0.346, p = 0.729).
4. DISCUSSION
Our study showed that DL was able to use information from clinical variables and MRI images to transform them into accurate predictions of HT in AIS patients after EVT. The DMTC model in the VOI data set and slice data set had a good performance and good generalization ability for predicting HT of AIS patients. Thus, DL model using multiparameter MRI could be decisive in identifying patients who present with higher risks of HT and should not be treated with EVT therapy.
The exact pathophysiology of HT is still unclear. One of the possible mechanisms of HT is blood–brain barrier (BBB) disruption [23]. Slevin et al. [24] found that Monomeric C‐reactive protein (mCRP) could increase vascular monolayer permeability and gap junctions and produced hemorrhagic angiogenesis in mouse matrigel implants. Multiple MRI techniques, especially PWI, are based on this concept of disruption of the BBB. In connection with this, the value of DWI for evaluating the degree of ischemia is well known, and previous studies have demonstrated that the low ADC values were associated with HT [25]. In this context, the fact that DL models can predict HT from DWI and PWI is particularly encouraging. For conventional MRI, T1WI and FLAIR could also be an indication of HT [25, 26], however, conventional MRI showed lower sensitivity than advanced MRI. In addition, the onset time of AIS patients in our study is short (mean onset time: 306 min), and about 38% of patients did not show high signal on FLAIR in the acute infarction area, leading to difficulties of accurately delineating the VOI of acute infarction on FLAIR. Therefore, advanced MRI techniques (DWI and PWI) in our study were used to predict HT in AIS patients.
Recently, several studies have used ML to predict HT in AIS patients [26, 27, 28, 29]. However, the diagnostic performance of multiparameter MRI using DL for the prediction of HT in AIS has not yet been systematically evaluated. In this study, we first trained the DL model based on a single parameter, and the results showed that the performance of DL models based on MTT and based on TTP was better than those of the other MRI parameters. Clinical features can reflect a few focus information at the pathologic level and the MRI images could obtain more factors affecting HT in AIS patients. Also, the possible reason for PWI over DWI may be that the PWI could reflect the blood vessel integrity damaged which usually causes HT, whereas the DWI change is a downstream effect of ischemia on neurons [11]. The MTT is usually used to predict vulnerable brain tissue that may evolve from the infarction, and the TTP describes the time to reach the CBF’s highest value at the target tissue location [30].
We also trained the multiparameter model to investigate their accuracy in predicting HT after EVT. The results showed that the multiparameter model was better than the single parameter model, and the AUC of the DMTC model was the highest (VOI data set: 0.948; slice data set: 0.932), which was also higher than that of Suh et al. (AUC: 0.85) [10]. Considering the heterogeneity in stroke pathophysiology, the CNN model can retain and process complex information, so as to find a more accurate connection between the input and the output. This enables CNN to predict the HT after EVT more accurately in an automatic and user‐independent manner and also may be the reason that the multiple sequence model was better than the single sequence model or conventional statistical methods. The sensitivity of our study (VOI data set: 0.864; slice data set: 0.850) was slightly lower than that of Suh et al. (AUC: 0.920) [10], probably because of the unbalanced data in our study, and the inclusion of more HT patients may improve the sensitivity of DL models.
In our study, we validated the DMTC model in an independent external validation set, and comparing it with the training set, both the VOI data set and the slice data set had good performance in predicting HT, which showed good generalization ability. It is worth mentioning that the DMTC model based on the VOI data set and slice data set for predicting HT had similar performance. VOI data set is drawn by doctors manually, which needs a heavy workload and is easy to make boundary information loss because of different experiences. The slice data set was composed of axial MRI images selected based on the lesion of VOI, which has the advantages of easily choosing, high repeatability, and inputting full‐layer image information. Therefore, we found that the slice dataset may use for model training to replace the VOI data set.
Some limitations must be acknowledged. First, the sample size is relatively small. Although it is unclear how many patients are needed for the models to reach a plateau in accuracy, a larger data set would likely provide an increase in accuracy. Second, the data used in this study were retrospectively acquired. To mimic a prospective study, we grouped the data according to the admission time, in which 75% of patients in the early time were trained and 25% of patients in later time were tested. Third, other MRI sequences (T1WI, T2WI, or FLAIR) were not included in this study. Finally, as a result of the sample size, patients receiving bridging therapy were also enrolled, Health Quality Ontario [31] demonstrated that EVT did not show an increased incidence of clinically relevant HT in comparison with IVT. The therapy methods also showed no significant differences between the HT group and the no HT group in our study.
5. CONCLUSION
The multiparameter model of DMTC had high accuracy and good generalization ability in predicting HT after EVT in AIS patients. The proposed DWI, PWI, and clinical multiparameter DL model may provide a potential tool for predicting information before therapy to assist the periprocedural management in AIS patients with EVT.
CONFLICT OF INTEREST
The authors declare that there is no potential conflict of interest regarding the publication of this paper.
AUTHOR CONTRIBUTIONS
Yu‐Chen Chen and Xindao Yin designed the study and supervised the conduct of the study. Liang Jiang and Leilei Zhou contributed to the data collection. Wei Yong, Jinluan Cui, and Wen Geng provided methodologic advice. Huiyou Chen, Jianjun Zou, and Yang Chen performed the data analysis and results interpretation. Liang Jiang and Leilei Zhou drafted the manuscript which is reviewed and approved for publication by all authors.
ACKNOWLEDGMENTS
We thank Prof Guan Gui from Nanjing University of Posts and Telecommunications for his excellent technical assistance and thank Prof Hong Zhang from the Affiliated Jiangning Hospital of Nanjing Medical University for providing data for external validation. This work was funded by the Jiangsu Provincial Special Program of Medical Science (No. BE2021604), Natural Science Foundation of Jiangsu Province (No. BK20201118), and 333 High‐level Talents Training Project of Jiangsu Province (No. BRA2019122).
Jiang L, Zhou L, Yong W, Cui J, Geng W, Chen H, et al. A deep learning‐based model for prediction of hemorrhagic transformation after stroke. Brain Pathol. 2023;33:e13023. 10.1111/bpa.13023
Liang Jiang and Leilei Zhou are contributed equally to this work.
Contributor Information
Xindao Yin, Email: y.163yy@163.com.
Yu‐Chen Chen, Email: chenyuchen1989@126.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author.
REFERENCES
- 1. Pandian JD, Gall SL, Kate MP, Silva GS, Akinyemi RO, Ovbiagele BI, et al. Prevention of stroke: a global perspective. Lancet. 2018;392(10154):1269–78. [DOI] [PubMed] [Google Scholar]
- 2. Berkhemer OA, Fransen PS, Beumer D, van den Berg LA, Lingsma HF, Yoo AJ, et al. A randomized trial of intraarterial treatment for acute ischemic stroke. N Engl J Med. 2015;372(1):11–20. [DOI] [PubMed] [Google Scholar]
- 3. Goyal M, Menon BK, van Zwam WH, Dippel DW, Mitchell PJ, Demchuk AM, et al. Endovascular thrombectomy after large‐vessel ischaemic stroke: a meta‐analysis of individual patient data from five randomised trials. Lancet. 2016;387(10029):1723–31. [DOI] [PubMed] [Google Scholar]
- 4. Panni P, Gory B, Xie Y, Consoli A, Desilles JP, Mazighi M, et al. Acute stroke with large ischemic core treated by thrombectomy. Stroke. 2019;50(5):1164–71. [DOI] [PubMed] [Google Scholar]
- 5. Sporns PB, Kemmling A, Schwake M, Minnerup J, Nawabi J, Broocks G, et al. Triage of 5 noncontrast computed tomography markers and spot sign for outcome prediction after intracerebral hemorrhage. Stroke. 2018;49(10):2317–22. [DOI] [PubMed] [Google Scholar]
- 6. Neuberger U, Kickingereder P, Schönenberger S, Schieber S, Ringleb PA, Bendszus M, et al. Risk factors of intracranial hemorrhage after mechanical thrombectomy of anterior circulation ischemic stroke. Neuroradiology. 2019;61(4):461–9. [DOI] [PubMed] [Google Scholar]
- 7. Paciaroni M, Agnelli G, Corea F, Ageno W, Alberti A, Lanari A, et al. Early hemorrhagic transformation of brain infarction: rate, predictive factors, and influence on clinical outcome: results of a prospective multicenter study. Stroke. 2008;39(8):2249–56. [DOI] [PubMed] [Google Scholar]
- 8. Wu O, Dijkhuizen RM, Sorensen AG. Multiparametric magnetic resonance imaging of brain disorders. Top Magn Reson Imaging. 2010;21(2):129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elsaid N, Mustafa W, Saied A. Radiological predictors of hemorrhagic transformation after acute ischemic stroke: An evidence‐based analysis. Neuroradiol J. 2020;33(2):118–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suh CH, Jung SC, Cho SJ, Woo DC, Oh WY, Lee JG, et al. MRI for prediction of hemorrhagic transformation in acute ischemic stroke: a systematic review and meta‐analysis. Acta Radiol. 2020;61(7):964–72. [DOI] [PubMed] [Google Scholar]
- 11. Campbell BC, Christensen S, Parsons MW, Churilov L, Desmond PM, Barber PA, et al. Advanced imaging improves prediction of hemorrhage after stroke thrombolysis. Ann Neurol. 2013;73(4):510–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. El Nawar R, Yeung J, Labreuche J, Chadenat ML, Duong DL, De Malherbe M, et al. MRI‐based predictors of hemorrhagic transformation in patients with stroke treated by intravenous thrombolysis. Front Neurol. 2019;10:897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deutschmann H, Hinteregger N, Wiesspeiner U, Kneihsl M, Fandler‐Hofler S, Michenthaler M, et al. Automated MRI perfusion‐diffusion mismatch estimation may be significantly different in individual patients when using different software packages. Eur Radiol. 2021;31(2):658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, et al. A guide to deep learning in healthcare. Nat Med. 2019;25(1):24–9. [DOI] [PubMed] [Google Scholar]
- 15. Borstelmann SM. Machine learning principles for radiology investigators. Acad Radiol. 2020;27(1):13–25. [DOI] [PubMed] [Google Scholar]
- 16. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. [DOI] [PubMed] [Google Scholar]
- 17. Nielsen A, Hansen MB, Tietze A, Mouridsen K. Prediction of tissue outcome and assessment of treatment effect in acute ischemic stroke using deep learning. Stroke. 2018;49(6):1394–401. [DOI] [PubMed] [Google Scholar]
- 18. Wang K, Shou Q, Ma SJ, Liebeskind D, Qiao XJ, Saver J, et al. Deep learning detection of penumbral tissue on arterial spin labeling in stroke. Stroke. 2020;51(2):489–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. von Kummer R, Broderick JP, Campbell BC, Demchuk A, Goyal M, Hill MD, et al. The Heidelberg Bleeding Classification: classification of bleeding events after ischemic stroke and reperfusion therapy. Stroke. 2015;46(10):2981–6. [DOI] [PubMed] [Google Scholar]
- 20. Hacke W, Kaste M, Fieschi C, von Kummer R, Davalos A, Meier D, et al. Randomised double‐blind placebo‐controlled trial of thrombolytic therapy with intravenous alteplase in acute ischaemic stroke (ECASS II). Second European‐Australasian Acute Stroke Study Investigators. Lancet. 1998;352(9136):1245–51. [DOI] [PubMed] [Google Scholar]
- 21. Mukherjee L, Singh V, Dyer CR. Half‐integrality based algorithms for cosegmentation of images. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2009;2028–35. [PMC free article] [PubMed] [Google Scholar]
- 22. DeLong ER, DeLong DM, Clarke‐Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–45. [PubMed] [Google Scholar]
- 23. Yaghi S, Willey JZ, Cucchiara B, Goldstein JN, Gonzales NR, Khatri P, et al. Treatment and outcome of hemorrhagic transformation after intravenous alteplase in acute ischemic stroke: a scientific statement for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 2017;48(12):e343–61. [DOI] [PubMed] [Google Scholar]
- 24. Slevin M, Matou S, Zeinolabediny Y, Corpas R, Weston R, Liu D, et al. Monomeric C‐reactive protein–a key molecule driving development of Alzheimer's disease associated with brain ischaemia? Sci Rep. 2015;5:13281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shinoda N, Hori S, Mikami K, Bando T, Shimo D, Kuroyama T, et al. Prediction of hemorrhagic transformation after acute thrombolysis following major artery occlusion using relative ADC ratio: A retrospective study. J Neuroradiol. 2017;44(6):361–6. [DOI] [PubMed] [Google Scholar]
- 26. Jha R, Battey TW, Pham L, Lorenzano S, Furie KL, Sheth KN, et al. Fluid‐attenuated inversion recovery hyperintensity correlates with matrix metalloproteinase‐9 level and hemorrhagic transformation in acute ischemic stroke. Stroke. 2014;45(4):1040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kassner A, Liu F, Thornhill RE, Tomlinson G, Mikulis DJ. Prediction of hemorrhagic transformation in acute ischemic stroke using texture analysis of postcontrast T1‐weighted MR images. J Magn Reson Imaging. 2009;30(5):933–41. [DOI] [PubMed] [Google Scholar]
- 28. Deo RC. Machine learning in medicine. Circulation. 2015;132(20):1920–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bouts MJ, Tiebosch IA, Rudrapatna US, van der Toorn A, Wu O, Dijkhuizen RM. Prediction of hemorrhagic transformation after experimental ischemic stroke using MRI‐based algorithms. J Cereb Blood Flow Metab. 2017;37(8):3065–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Copen WA, Schaefer PW, Wu O. MR perfusion imaging in acute ischemic stroke. Neuroimaging Clin N Am. 2011;21(2):259–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Health QO. Mechanical thrombectomy in patients with acute ischemic stroke: a health technology assessment. Ont Health Technol Assess Ser. 2016;16(4):1–79. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.


