Abstract
Lipid transfer proteins (LTPs) are widely distributed in plants and play an important role in the response to stress. Potato ( Solanum tuberosum L.) is sensitive to a lack of water, and drought stress is one of the limiting factors for its yield. Therefore, mining candidate functional genes for drought stress and creating new types of potato germplasm for drought resistance is an effective way to solve this problem. There are few reports on the LTP family in potato. In this study, 39 members of the potato LTP family were identified. They were located on seven chromosomes, and the amino acid sequences encoded ranged from 101 to 345 aa. All 39 family members contained introns and had exons that ranged from one to four. Conserved motif analysis of potato LTP transcription factors showed that 34 transcription factors contained Motif 2 and Motif 4, suggesting that they were conserved motifs of potato LTP. Compared with the LTP genes of homologous crops, the potato and tomato ( Solanum lycopersicum L.) LTPs were the mostly closely related. The StLTP1 and StLTP7 genes were screened by quantitative reverse transcription PCR combined with potato transcriptome data to study their expression in tissues and the characteristics of their responses to drought stress. The results showed that StLTP1 and StLTP7 were upregulated in the roots, stems, and leaves after PEG 6000 stress. Taken together, our study provides comprehensive information on the potato LTP family that will help to develop a framework for further functional studies.
Keywords: expression analysis, genome‐wide identification, LTP, potato
1. INTRODUCTION
Potato (Solanum tuberosum L.) is one of the most important food crops and economic crops in China, and the amounts of its area and yield rank first in the world. Inner Mongolia is one of the primary bases for the production of potato seed and commercial potatoes in China. The annual rainfall is approximately 200–350 mm, and the region is arid and semiarid (du et al., 2013; Zhang, Li, et al., 2022). Drought stress is by far the most complex abiotic stress that affects plant development, survival, and crop productivity on a global scale (Ahuja et al., 2010). Thus, studies on drought resistance in potato have always been major issues of concern. Currently, researchers have conducted extensive research on the physiological, biochemical, and morphological indicators related to potato drought resistance, but less research has been conducted on the molecular mechanisms used by potato to respond to drought stress and the mining of drought resistance genes (Chen et al., 2022; Wang & Qin, 2021). When potato plants are subjected to drought stress, they produce a series of complex signal pathway activation responses, which induce the expression of drought resistance‐related genes, and then cause changes in the levels of proteins and metabolites to resist the deleterious effects of disease and drought. The continuous improvement in potato transcriptome data and the rapid development of bioinformatics provide effective means and new research ideas to analyze the mechanism of potato drought resistance.
Lipid transfer proteins (LTPs) are basic small proteins that are 7 to 10 kDa. They are composed of a folded peptide chain that is formed by an α‐helix that is connected by four disulfide bonds, and they are members of the class of small molecular weight antimicrobial peptides (Salminen et al., 2016). LTPs were first observed in spinach (Spinacea oleracea L.) leaves, maize (Zea mays L.) coleoptiles, and barley (Hordeum vulgare L.) aleurone (Wei & Zhong, 2014). It was reported that the LTP genes were highly adaptable to stress in maize (Noonan et al., 2017), rice (Oryza sativa L.) (Lin et al., 2017), cotton (Gossypium hirsutum L.) (Zhang, Zhao, et al., 2022), wheat (Triticum aestivum L.) (Fang et al., 2020), potato (Shang et al., 2022), Arabidopsis (Arabidopsis thaliana [L.] Henyh.) (Zou et al., 2013), and other crops. LTP genes can induce the expression of lipid transporter genes when the plants adapt to or resist stress environments, such as drought, cold, high salt stress, and pathogen infection, and participate in plant growth, seed germination, and pollen development (Duo et al., 2021; Vangelisti et al., 2022). Highland barley and maize can induce the production of multiple LTP genes under drought stress (Fang et al., 2023; Wei & Zhong, 2014). LTP3 in wheat has antifungal functions (Kirubakaran et al., 2008), and the expression of VrLTPs in mung bean (Vigna radiata [L.] Wilezek) increases under water stress (Liang et al., 2020). Various tertiary structures of lipid transporters that form lipid complexes are the primary substances that regulate the resistance of plants to drought (Edstam et al., 2011; Salminen et al., 2016). Studies have shown that the primary protein in cabbage (Brassica oleracea L. var. capitata) epidermal wax is a lipid transporter, and wax can decrease the loss of water on the epidermal surface. This affects the ability of plants to show resistance to drought stress (Ji et al., 2018). Gao et al. (2008) introduced the LTP gene into potato plants and found that the gene functions in the resistance to salt and drought. Moraes et al. (2015) studied the tolerance and disease resistance genotypes of 11 genetic isoforms of rice LTP1 proteins, and Wang et al. (2014) found that LTP3 could be overexpressed in transgenic Arabidopsis under thermotolerance. It was reported that the cis‐regulatory elements of the NtLTP genes in Nicotiana sylvestris promoters were involved in abiotic stress and phytohormone responsiveness (Yang et al., 2022). The study found that abscisic acid (ABA) and ethylene significantly upregulated the ZmLTP63 gene, whereas gibberellin downregulated the ZmLTP63 gene. The ZmLTP63 gene has various stress response elements, such as drought and low temperature (Sun et al., 2014).
Rainfall is infrequent in northern China, and the growth and development of potatoes are often hindered, and the yield reduced, owing to a lack of water during the critical period of potato growth. LTP is not only involved in the transport of glycolipids, fatty acids, and phospholipids between biofilms but also effectively resists abiotic stresses, such as drought (Missaoui et al., 2022). However, few studies have reported on the function of the potato LTP gene family. Therefore, mining genes related to potato drought resistance to improve this parameter is valuable for research and application prospects. The gene sequence of maize ZmLTPL63 was compared with that of the potato LTP gene, and the potato gene was found to be 98% homologous with the maize gene. In this study, the method of bioinformatics combined with quantitative reverse transcription PCR (RT‐qPCR) was used to identify the potato LTP gene family. These methods were combined with transcriptome data to screen the highly expressed LTP drought resistance genes to explore their level of expression after induction by PEG 6000 stress. Mechanistic studies provide reliable information to advance the process of creating transgenic potatoes.
2. MATERIALS AND METHODS
2.1. Test materials
Tissue culture seedlings of potato variety “Kexin No.1” were used as experimental materials and provided by the Potato Genetics and Breeding Research Group of the School of Life Science and Technology, Jining Normal University (Jining, China).
2.2. Identification of LTP members in potato
The potato LTP sequence was searched in the Phytozome database using the keyword PF00234, and the protein sequence with the LTP conserved domain in the potato genome was searched in the HMMER software. Pfam (http://pfam.xfam.org/search) was used to analyze whether it was conserved. The LTP domain was deleted, and the LTP sequences whose conserved domains were less than 50% were deleted. Finally, the potato LTP family members were obtained by screening. The corresponding nucleic acid sequences were downloaded, and the genes were annotated on the identification result. The amino acid composition, isoelectric point, molecular weight, and hydropathic index of the potato LTPs were analyzed using the ProtParam tool (https://web.expasy.org/protparam/). PSORT Prediction (http://psort1.hgc.jp/form.html) was used to predict their subcellular localization.
2.3. Potato LTP gene structure and phylogenetic analysis
The amino acid sequences of the potato LTP family members were aligned by Muscle in MEGA 6.0 software (parameter default), and a developmental tree was constructed by the neighbor‐joining (NJ) method in phylogeny. The bootstrap parameter was set to 1000, and the gaps processing method was used. The pairs were deleted. The potato LTP genome sequence and coding sequence (CDS) were downloaded from the JGI official website. ClustalX was used to align the 39 LTP sequences, and the alignment results were saved in the NWK format. The LTP gene structure map was drawn using the sequence (FASTA) format of GSDS 2.0. The conservation of 39 potato LTP transcription factors (TFs) was analyzed using WebLogo (http://weblogo.berkeley.edu/logo.cgi), which revealed the characteristic sequences of the LTP domain. InterPro database (http://www.ebi.ac.uk/interpro/) was used to predict the protein domains of StLTP1 and StLTP7.
2.4. Phylogenetic analysis of the LTP members among different crops
A phylogenetic analysis was constructed to further explore the evolutionary relationships among the four plant species, including potato, soybean (Glycine max L.), tomato, and A. thaliana. The LTP member protein sequences of soybean, tomato, and A. thaliana were downloaded from the Phytozome database. The LTP sequences of the four species were completely aligned by Muscle in MEGA 6.0 software to construct a phylogenetic tree.
2.5. Gene mapping
The potato genome data was downloaded from the NCBI (https://www.ncbi.nlm.nih.gov/) database, and the location information of the potato LTP family members and their chromosomal location were determined from the JGI (Setaria italica v2.2) database. The distribution of LTP genes on 10 potato chromosomes was drawn using Map Gene 2 Chromosome.
2.6. Promoter analysis
The promoters of the potato LTP family members were analyzed using Plantscare. The whole potato genome sequence and the GFF3 file were downloaded in Phytozome, and the LTP sequence was then extracted by the FASTA Extractor of TBtools software. The promoter that was located 2000 bp upstream of the start codon of the LTP member coding region was analyzed.
2.7. Stress treatment analysis
The tissue culture seedlings were placed in a light incubator for 15–20 days under 16 h of light and 8 h of dark conditions. When the seedlings had grown to 10–12 cm, a solution of 20% PEG 6000 was used to stress the tissue culture seedlings at 0, 4, 8, 12, 18, and 24 h. The roots, stems, and leaves of the tissue culture seedlings were quickly frozen in liquid nitrogen, stored at −80 C, and the RNA was extracted in the next step.
2.8. RNA extraction and reverse transcription
The roots, stems, and leaves treated with different times of stress were fully ground, and the RNA was extracted using an OminiPlant RNA Kit (DNase I) (CWBIO, Inc., Beijing, China). The integrity of RNA was detected by 1.5% agarose gel electrophoresis. The total amount of complete RNA was uniformly set at 1000 ng, and the total RNA was reverse transcribed using a HiScript Q RT SuperMix for qPCR (+gDNA wiper) kit (Vazyme, Nanjing, China). Three biological replicates and three technical replicates were performed.
2.9. RT‐qPCR analysis
According to the potato transcriptome data, two LTP genes that were closely related to drought resistance were screened and numbered StLTP1 and StLTP7. Premier5.0 software was used to design fluorescent quantitative primers. Actin was used as the internal reference gene, and a LineGene 9600 plus type fluorescence quantitative PCR instrument (Hangzhou Bioer Technology, Co., Ltd., Hangzhou, China) was used for the PCR analysis. ChamQ SYBR COLOR qPCR Master Mix 2X reagent (Vazyme) was used for the RT‐qPCR analysis of StLTP1 and StLTP7 in the roots, stems, and leaves after stress treatments. The reaction program was as follows: 95°C for 5 min (95°C for 30 s, 56°C for 30 s, 72°C for 40 s) × 40 cycles; and 72°C for 40 s. The reaction system is shown in Table S1, and the primer sequences are shown Table S2.
3. RESULTS AND ANALYSIS
3.1. Phylogenetic analysis of the LTP families in different crops
To explore the phylogenetic relationship of the LTP families in different crops, the NJ method of MEGA 6.0 was used to analyze the LTP structures of soybean (23), tomato (19), Arabidopsis (23), and potato (39). The amino acid sequences of the domains were aligned, and a phylogenetic tree was constructed (Figure 1). The results showed that the 104 LTP genes were divided into approximately eight branches, and the 39 LTP genes of potato were distributed in six branches. Among them, there were 20 LTPs genes in branch II and seven LTPs genes in branch III. In general, the LTP genes of potato are closely related to those of tomato.
FIGURE 1.
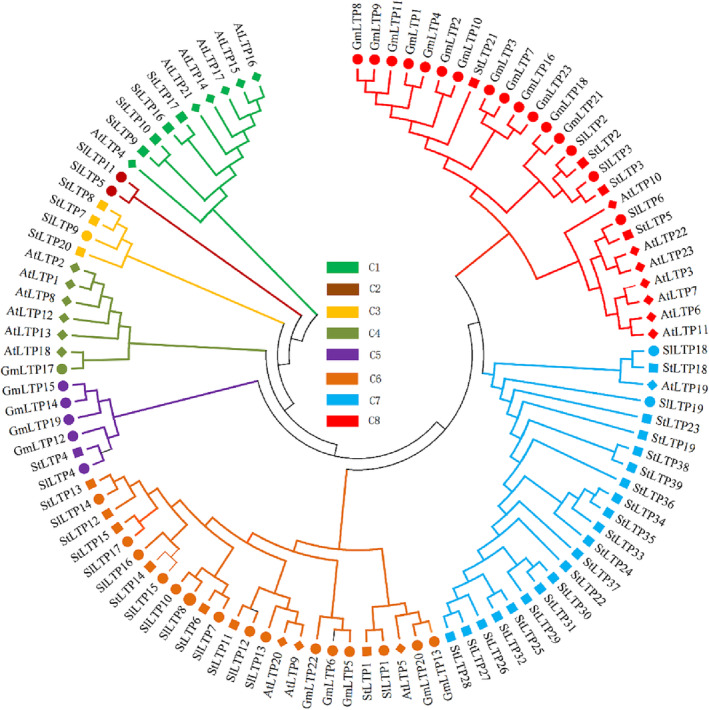
Phylogenetic tree of the lipid transfer proteins (LTPs) from soybean (23), tomato (19), Arabidopsis (23), and potato (39): The complete amino acid sequences of LTPs were aligned by MUSCLE, and the phylogenetic tree was constructed using the maximum‐likelihood method in MEGA 6.0. The boot‐strap value was 1000 replications. St: S. tuberosum L., At: A. thaliana , Gm: G. max (Linn.) Merr., SI: Solanum lycopersicum L.. The 104 LTP genes are classified into eight distinct branches, and these branches are shown in different colors in the phylogenetic tree.
3.2. Identification of the LTPs gene family members
The potato LTP sequence was searched by the keyword PF00234 in the Phytozome database, and the protein sequence with the LTP conserved domain in the potato genome was searched in the HMMER software. Pfam was used to remove the redundancy. Finally, 39 LTP family members (StLTP1–StLTP39) were identified. The structural domains of the potato LTP family were analyzed by Conserved Domain Database (CDD) (Table S3). The results showed that there were between 52 and 103 amino acid residues in the conserved domains of the members of the StLTP family, and the total number of amino acids was between 101 and 345. The isoelectric points were between 4.23 and 9.53, and the molecular weights were between 10.71 and 26.19 kDa. The protein hydrophilicity index indicated that the StLTP4, StLTP9, StLTP10, StLTP11, StLTP15, and StLTP16 family members were hydrophilic proteins, and the proteins encoded by StLTP15 were the most hydrophilic. The StLTP9 protein was predicted to be located in the mitochondrial matrix, and the StLTP12 protein was located in the chloroplast thylakoid space and endoplasmic reticulum membrane. The StLTP13 and StLTP39 proteins were located in the cytoplasm, and the StLTP21 protein was located in the endoplasmic reticulum. The rest were detected extracellularly, and some were located in the vacuole.
3.3. Analysis of the phylogeny, gene structure, and protein domain of the StLTPs
A phylogenetic tree was constructed to explore the phylogenetic relationship of the 39 LTP family members in potato using the NJ method with 1000 bootstrap values. The results showed that the 39 LTP family members can be divided into seven branches (Figure 2). The first branch includes 21 family members. The second subfamily includes five family members, and the third includes three. The fourth subfamily includes five family members. The fifth subfamily includes only StLTP4, and the sixth subfamily and the seventh subfamily each include two family members. A structural diagram of the potato LTP gene family (Figure 2) indicates that StLTP10 and StLTP11 contain three introns. StLTP9, StLTP16, and StLTP17 contain one intron, and the rest of the genes have no introns. A total of 16 LTP family members only contained CDS sequences, whereas 15 LTP family members contained upstream, downstream, and CDS sequences. StLTP39 contained downstream and CDS sequences; StLTP30 and StLTP31 contained upstream and CDS sequences; and StLTP17 contained 5′ UTR, 3′ UTR, intron, and CDS sequences.
FIGURE 2.
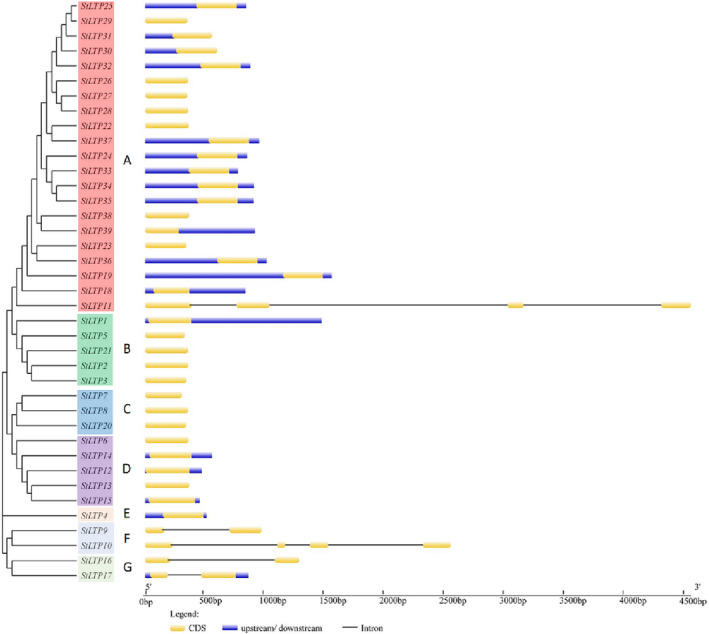
Phylogenetic relationships and gene structure of the potato LTPs genes: The phylogenetic tree was generated using a MEGA 6.0 program with the full‐length amino acid sequences of the potato LTPs proteins obtained by a neighbor‐joining (NJ) method, including 1000 boot‐strap replications. Exon/intro organization of the LTPs genes is also shown as follows: The introns and exons are represented by black lines and yellow boxes, respectively, and the upstream and downstream regions are represented by blue boxes. The lengths of introns and exons of each gene can be estimated using the scale detailed at the bottom. Subfamilies of LTPs genes (A–G) are highlighted with different colored backgrounds.
A conserved motif analysis of the potato LTP TFs revealed 16 motifs (Figure 3), and the amino acid sequences of 39 LTP TFs showed conserved motifs. Among the motif‐containing sequences, 35 LTP TFs contained Motif 4; 34 LTP TFs contained Motif 2; 19 LTP TFs contained Motif 1; 19 LTP TFs contained Motif 3; 15 LTP TFs contained Motif 5; 13 LTP TFs contained Motif 6; and the remaining motifs were contained in the LTP TFs. There were few motifs in the LTP TFs, and they ranged from two to six. A total of 34 TFs contained Motif 2 and Motif 4, which appeared the most frequently. They constitute the domain characteristic sequence of LTP and are usually in a series. Thus, it was hypothesized that Motif 2 and Motif 4 are conserved motifs of the potato LTP (Figure 4). The conservation of 39 potato LTP TFs was analyzed using WebLogo, which revealed the characteristic sequences of the LTP domain (Figure 5).
FIGURE 3.
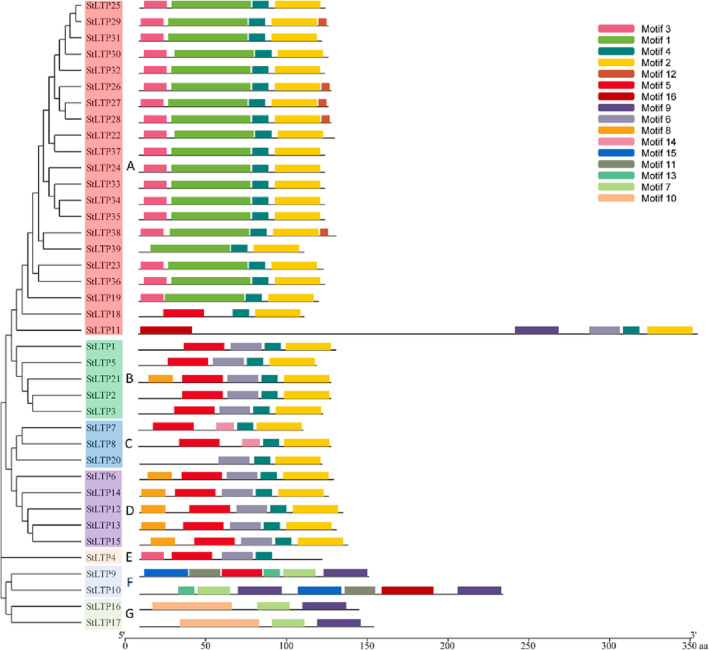
Conserved motifs in the lipid transfer proteins (LTPs): Each colored box represents a motif in each of the LTPs, with the motif's name indicated in the box on the right. The length of each protein can be estimated using the scale at the bottom.
FIGURE 4.

The conserved Motif 2 and Motif 4 of lipid transfer protein (LTP) transcription factors in potato. The height of amino acid letters at different positions represents the level of conservation. The conserved Motif 2 and Motif 4 was analyzed using WebLogo (http://weblogo.berkeley.edu/logo.cgi).
FIGURE 5.
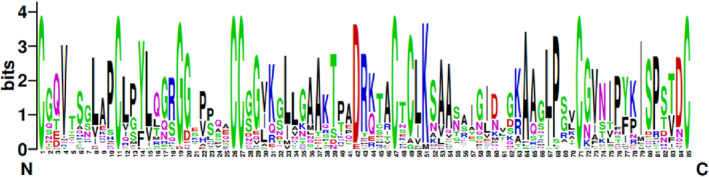
The domains of 39 lipid transfer protein (LTP) transcription factors (TFs) family in potato. The height of amino acid letters at different positions represents the level of conservation. The conservation of 39 potato LTP TFs was analyzed using WebLogo (http://weblogo.berkeley.edu/logo.cgi).
We used the InterPro database to predict the protein domains of StLTP1 and StLTP7. The results showed that StLTP1 encodes a protein of 121 amino acid residues, and the conserved domain is 85 amino acids. The starting position of the protein domain is 33–117 (Figure 6a). Its protein structure represents a structural domain that consists of four helices with a folded leaf topology, which forms a right‐handed superhelix. The prediction of StLTP1 protein structure revealed a Signal Peptide N Region, SignalP‐TM, TMhelix, Signal Peptide H Region, Non Cytoplasmic Domain, Signal Peptide, SignalP‐noTM, and Signal Peptide N Region. StLTP7 encodes a protein of 101 amino acid residues with a conserved domain of 86 amino acids. The starting position of the protein domain is 14–99 (Figure 6b). The protein structure of StLTP7 is the same as that of StLTP1.
FIGURE 6.
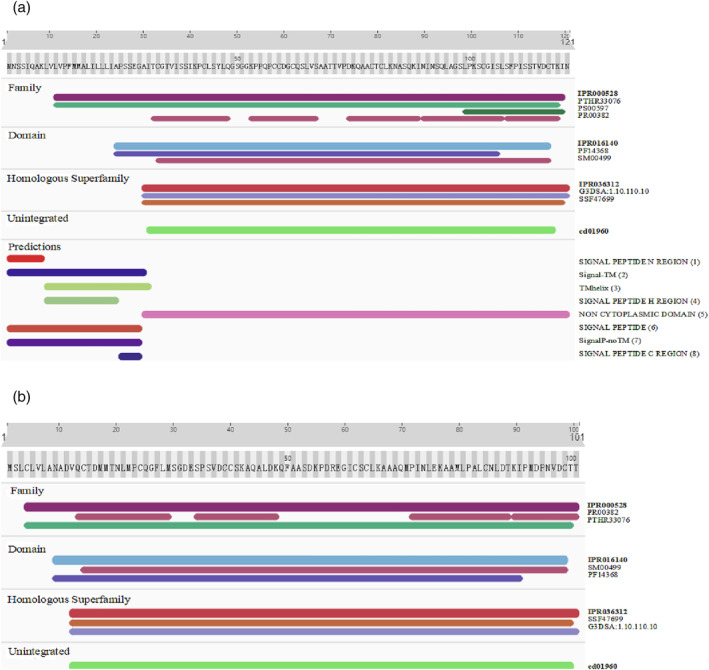
Prediction of the StLTP1 and StLTP7 protein domains. The protein domains of StLTP1 and StLTP7 were predicted by InterPro database (http://www.ebi.ac.uk/interpro/). (a) is the domain prediction of StLTP1 protein, and (b) is the domain prediction of StLTP7 protein. Different domains are represented by different colors.
3.4. Potato LTP gene family promoter analysis
An analysis of the cis‐acting elements of 39 StLTP family members showed that the cis‐acting elements involved in the promoter region of potato LTP family included low temperature, drought, dehydration, salt stress, defense and wound response elements, light response elements, and plant hormone (auxin, ABA, and gibberellin) response elements (Table S4). The response elements included in the promoter regions of the 39 StLTP genes were the TATA‐box and CAAT‐box.
Chromosomal location analysis of the potato LTP gene family.
The Map Gene 2 Chromosome tool was used to draw the distribution map of 39 LTP genes on the potato chromosome (Figure 7). The results showed that all 39 LTP members were located on the potato chromosome, and they were uneven. Twenty were distributed on Chromosome 10. The LTP genes were concentrated in the proximal end of chromosomes, and only one LTP gene was distributed on Chromosome 2, which is located at the end of chromosomes. The rest were scattered on Chromosomes 1, 6, 7, 8, and 9. StLTP9, StLTP10, StLTP16, and StLTP17 were located at the end, and the rest were concentrated at the proximal end.
FIGURE 7.
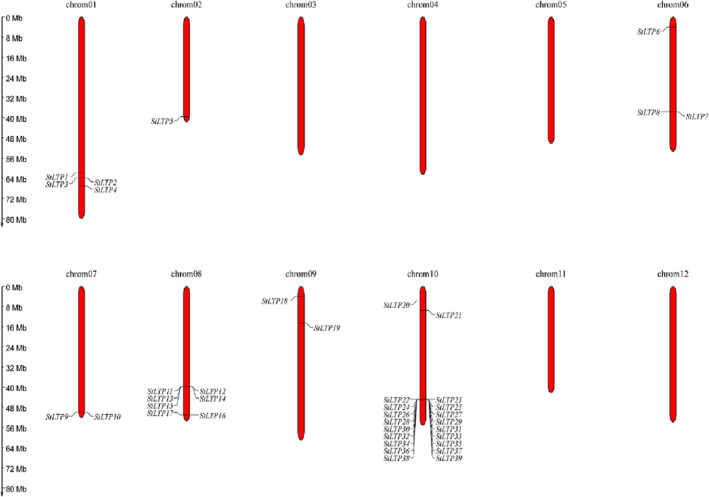
Chromosomal distribution of 39 lipid transfer protein (LTP) genes in potato. The scale bar on the left indicates the size of chromosomes. The 39 LTP genes were located on the seven potato chromosomes, and the concrete positions of 39 LTP genes were labeled on chromosomes using linea nigra and gene name. The size of each LTP gene can be estimated using the scale at the left.
3.5. Analysis of the expression of potato LTP gene family
StLTP1 (Soltu.DM.01G028720.1) and StLTP7 (Soltu.DM.06G016150.1) have strong anti‐stress functions in the transcriptome data published by the Phytozome potato genome. This study examined the drought resistance functions of the StLTP1 and StLTP7 genes that were identified following stress induction by treatment with PEG 6000. The results showed that the expression of StLTP1 in potato roots (Figure 8a) and stems (Figure 8b) showed a trend of upregulation first and then downregulation. The leaves (Figure 8c) showed a trend of upregulation and then downregulation followed by upregulation. StLTP7 was expressed the most highly in the potato roots at 12 h (Figure 8d), which was nearly 180‐fold greater than that of the unstressed roots. The overall expression was upregulated first. The trend of downregulation and then upregulation resulted in the highest level of expression in the stems at 18 h (Figure 8e), which was nearly 50‐fold that of the unstressed time. The overall trend included upregulation and then downregulation. The leaf genes were expressed the most at 24 h (Figure 8f), which was nearly 110‐fold that of the non‐stressed state and trended upward. In conclusion, the levels of expression of these two StLTP genes were upregulated to varying degrees under drought stress, indicating that they were regulated by StLTP1 and StLTP7 under abiotic stress.
FIGURE 8.
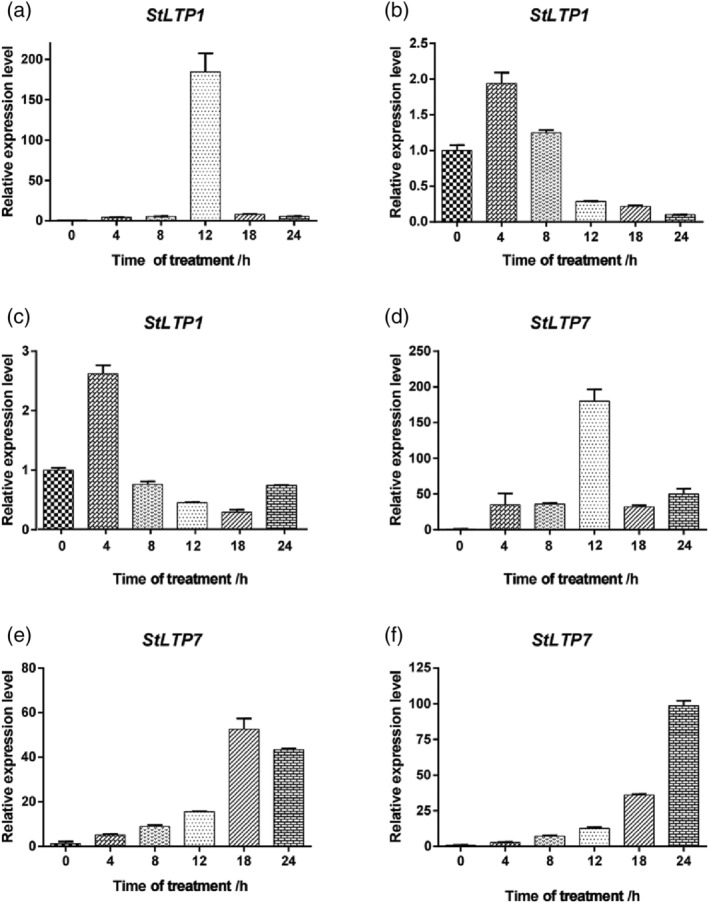
Relative level of expression of the StLTP genes following treatment with PEG 6000. (a–f) are the levels of expression of StLTP1 and StLTP7 under the process of PEG 6000 drought in the roots, stems, and leaves for 0, 4, 8, 12, 18 and 24 h. (a) expression of StLTP1 in the root volume. (b) expression of StLTP1 in the stems. (c) expression of StLTP1 in the leaves. (d) expression of StLTP7 in the root volume. (e) expression of StLTP7 in the stems. (f) expression of StLTP7 in the leaves. The x‐axes show the duration of treatment, and the y‐axes depict the relative level of expression. Error bars indicate the SD. The data are shown as means of three experiments and three biological repeats ±SD. SD, standard deviation.
4. DISCUSSION
When plants are under stress, they will re‐mobilize the defense system in their tissues, which prompts a series of physiological and biochemical reactions (Jacq et al., 2017). Some TFs related to stress resistance begin to be synthesized and expressed, so that some functional genes are regulated. These functional genes are used to complete various physiological functions, such as protein synthesis, activation, inhibition, degradation, and physiological metabolism. These defensive proteins in plants play a very important role in adapting to and resisting external stresses (Wang et al., 2009). Previous studies have found that the tertiary structure of lipid transporters can form various lipid complexes, which are the primary substances that regulate plant drought resistance. They participate in the formation of cuticle and constitute epidermal wax, whereas lipid transporters exist between the cuticle and cell wall interface. The hydrophobic waxy moiety can be transported into the epidermal cells through the hydrophilic cell wall matrix, and studies suggest that LTPs are involved in this process, thereby enabling the plants to exhibit drought resistance (Jin et al., 2015). Studies have shown that lipid transporters can induce multiple genes in plants, including mung bean and wheat, under drought conditions (Boutrot et al., 2007; Li et al., 2006).
LTP is a TF that is closely related to stress, which can combine with various cis‐acting elements in the promoter region to regulate gene expression. The number of LTP genes also varies among different species. For example, Arabidopsis has 15 LTPs; pepper (Capsicum annuum L.) has six LTPs, and rice has 53 LTPs, which could be related to genome‐wide duplication (Maximiano & Franco, 2021). There are substantial differences in the number of amino acids, isoelectric points, and molecular weights contained in the LTPs, which could be owing to the different functions of each family member in the growth and development of different species. The molecular weight and total number of amino acid residues indicate that LTP can be divided into two categories, which include LTP I and LTP II. The molecular weight of LTP I is approximately 10 kDa, and it is composed of approximately 90 amino acid residues, which are primarily distributed in the aerial parts of plants. The molecular weight of LTP II is approximately 7 kDa, and it has approximately 70 amino acid residues that are primarily distributed in the root (Li et al., 2006; Maximiano & Franco, 2021). In this study, the physicochemical properties of the potato LTPs were analyzed, and the results showed that there were between 52 and 103 amino acid residues in the 39 LTP members of potato; there were between 101 and 345 amino acids in total; the isoelectric point was between 4.23 and 9.53, and the molecular weight was between 10.71 and 26.19 kDa. It is hypothesized that StLTP11 and StLTP12 could be members of LTP I, and the rest are members of LTP II. Related studies have shown that there is an independent hydrophobic cavity in the tertiary structure of LTP, which can complete transmembrane transport. The hydrophilicity prediction of PdbLTP indicated that the LTPs of Chinese pistache (Pistacia chinensis) are all hydrophobic proteins and have certain functions in transmembrane transport and hydrophobic interactions as described by Sun et al. (2022). Except for StLTP4, StLTP9, StLTP10, StLTP11, StLTP15, and StLTP16, the 39 potato LTP members in this study are all hydrophobic proteins and can participate in transmembrane transport.
It was reported that nsLTPs are associated with stress resistance, including increased cold tolerance and drought resistance and salt tolerance (Hincha, 2002). A total of 19 tomato LTP genes were downloaded in this study. The phylogenetic tree of 104 LTP genes from different crops showed the following: 39 LTP genes of potato were distributed in six branches, and branch II had 20 StLTP genes and two SILTP tomato genes. There were seven StLTP genes and nine SILTP tomato genes in the third branch. Compared with A. thaliana and soybean, these two have the closest relationship. Research by Sun et al. (2014) on maize found that some hormones can significantly upregulate the level of expression of the ZmLTP63 gene, and the ZmLTP63 gene promoter contains a variety of abiotic stress response and hormone response elements.
The cis‐regulatory elements in the promoter sequence can regulate the level of transcription of genes. This study found that the potato LTP gene promoter has stress response elements, such as drought and low temperature, and hormone response elements, such as methyl jasmonate (MeJA). Among them, MYB‐binding sites (MBS) and dehydration‐responsive element (DRE) are cis‐acting elements responsible for drought response, and the TC‐rich repeats are cis‐acting elements responsible for defense and stress responses. They were detected in StLTP1 and StLTP7, indicating that the potato StLTP1 and StLTP7 are key genes related to stress. We analyzed the cis‐acting elements in the LTP promoter sequence to better understand how the LTP gene performs biological functions under environmental stress. Plant hormones are secondary metabolites that play an important role in chemical signal transduction and the response to biotic or abiotic stresses. Different plants can induce the expression of a large number of defense‐related genes after the application of MeJA. Different organs of plants synthesize ABA to initiate defense mechanisms, which results in the expression of a large number of defense‐related genes, thereby helping the plants to resist stress and survive periods that are not conducive to growth. Most LTP genes have more than one cis‐acting element in response to MeJA and ABA, indicating that the LTP genes may be among the important genes related to secondary metabolism that plays an important role in response to various stress conditions. The published transcriptome data of potato showed that StLTP1 and StLTP7 were significantly expressed under drought stress. To further analyze the function of StLTP1 and StLTP7, we also studied the expression of StLTP1 and StLTP7 in different tissues. In this study, the levels of expression of StLTP1 and StLTP7 in the roots, stems, and leaves of potato differed to varying degrees after stress was induced by treatment with 20% PEG 6000. The level of expression of StLTP1 was high in potato roots but very low in the stems and leaves. StLTP7 was more highly expressed in the potato roots and leaves, indicating that StLTP1 is specifically expressed in particular tissues, and StLTP7 is expressed in one or more tissues. This indicates that the LTPs may be involved in the growth and development process. It also shows that StLTP1 and StLTP7 may be closely related to plant hormone signaling pathways and are key genes for the response to drought stress in potato. The study lays a foundation to reveal the function of the potato LTP family and discover genes related to potato stress resistance, but it has not verified the important functions of the remaining members. Further research is merited to provide the basis for potato stress resistance breeding and new drought resistant varieties. Other studies provide a theoretical basis.
5. CONCLUSIONS
A bioinformatics analysis of the potato LTP genes and their level of expression following stress identified 39 members of the potato LTP family, which were distributed on six chromosomes. They are hydrophobic proteins. The response elements of the promoter regions of these 39 LTP family members include response elements, such as dehydration, low temperature, salt stress, and drought, and hormone response elements, such as MeJA. Most LTPs can be detected in the extracellular milieu and in vacuoles. Signal and phylogenetic tree results indicated that the 39 proteins were distributed on six points. After induction by 20% PEG 6000 stress, the levels of expression of the potato StLTP1 and StLTP7 genes were upregulated to varying degrees in the roots, stems, and leaves, indicating that the LTP genes play an important role in regulating the response to stress. The results of this study provide a basis to reveal the function of potato LTPs and discover the resistance genes of potato, and simultaneously, it can provide excellent engineering materials for the subsequent development of new potato varieties.
AUTHOR CONTRIBUTIONS
All authors contributed to the study conception and design. Dan Wang and Jian Song equally contributed to the article regarding original draft preparation. Zhicheng Zhang helped in review process. Tuanrong Lin and Yuhe Yin provided technical assistance, and Junxiang Mu and Yaqin Wang provided help in the process of experiment. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supporting information
Table S1. RT‐qPCR reaction system.
Table S2. RT‐qPCR primer sequences.
Table S3. StLTPs gene family information.
Table S4. Cis‐acting regulatory elements in the promoter of StLTPs gene family.
ACKNOWLEDGMENTS
We would like to thank the College of Life Sciences and Technology for providing the experiment platform and Ulanqab Academy of Agricultural and Forestry Research Sciences for providing the Virus‐Free Potato.
Wang, D. , Song, J. , Lin, T. , Yin, Y. , Mu, J. , Liu, S. , Wang, Y. , Kong, D. , & Zhang, Z. (2023). Identification of potato Lipid transfer protein gene family and expression verification of drought genes StLTP1 and StLTP7 . Plant Direct, 7(3), e491. 10.1002/pld3.491
Dan Wang and Jian Song contributed equally to this work.
Funding information This study was supported by Inner Mongolia Autonomous Region potato seed industry technology innovation center project (grant number: HX202101), the Jining Normal University Ph.D. Fund, Jining, China (grant number: jsbsjj1707), and the Natural Science Foundation of Inner Mongolia (grant number: 2021BS03010).
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- Ahuja, I. , de Vos, R. C. H. , Bones, A. M. , & Hall, R. D. (2010). Plant molecular stress responses face climate change. Trends in Plant Science, 15, 664–674. 10.1016/j.tplants.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Boutrot, F. , Meynard, D. , Guiderdoni, E. , Joudrier, P. , & Gautier, M. F. (2007). The Triticum aestivum non‐specific lipid transfer protein (TaLtp) gene family: Comparative promoter activity of six TaLtp genes in transgenic rice. Planta, 225, 843–862. 10.1007/s00425-006-0397-7 [DOI] [PubMed] [Google Scholar]
- Chen, Z. Y. , Yang, X. Q. , Tang, M. H. , Wang, Y. J. , Zhang, Q. , Li, H. Y. , Zhou, Y. , Sun, F. J. , & Cui, X. Y. (2022). Molecular characterization and drought resistance of GmNAC3 transcription factor in Glycine max (L.) Merr. International Journal of Molecular Sciences, 23(20), 12378. 10.3390/ijms232012378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- du, J. , Tian, Z. D. , Liu, J. , Vleeshouwers, V. G. A. A. , Shi, X. L. , & Xie, C. H. (2013). Functional analysis of potato genes involved in quantitative resistance to Phytophthora infestans . Molecular Biology Reports, 40(2), 957–967. 10.1007/s11033-012-2137-3 [DOI] [PubMed] [Google Scholar]
- Duo, J. , Xiong, H. , Wu, X. , Li, Y. , Si, J. , Zhang, C. , & Duan, R. (2021). Genome‐wide identification and expression profile under abiotic stress of the barley non‐specific lipid transfer protein gene family and its Qingke orthologues. BMC Genomics, 22(1), 674. 10.1186/s12864-021-07958-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edstam, M. M. , Viitanen, L. , Salminen, T. A. , & Edqvist, J. (2011). Evolutionary history of the non‐specific lipid transfer proteins. Molecular Plant, 4, 947–964. 10.1093/mp/ssr019 [DOI] [PubMed] [Google Scholar]
- Fang, C. , Wu, S. , Li, Z. , Pan, S. , Wu, Y. , An, X. , Long, Y. , Wei, X. , & Wan, X. (2023). A systematic investigation of lipid transfer proteins involved in male fertility and other biological processes in maize. International Journal of Molecular Sciences, 24(2), 1660. 10.3390/ijms24021660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Z. W. , He, Y. Q. , Liu, Y. K. , Jiang, W. Q. , Song, J. H. , Wang, S. P. , Ma, D. F. , & Yin, J. L. (2020). Bioinformatic identification and analyses of the non‐specific lipid transfer proteins in wheat. Journal of Integrative Agriculture, 19(5), 1170–1185. 10.1016/S2095-3119(19)62776-0 [DOI] [Google Scholar]
- Gao, G. , Ren, C. H. , Jin, L. P. , Xie, K. Y. , & Qu, D. Y. (2008). Cloning, expression and characterization of a non‐specific lipid transfer protein gene from potato. Acta Agronomica Sinica, 34, 1510–1517. 10.3724/SP.J.1006.2008.01510 [DOI] [Google Scholar]
- Hincha, D. K. (2002). Cryoprotectin: A plant lipid‐transfer protein homologue that stabilizes membranes during freezing. Philosophical Transactions of the Royal Society of London, 357(1423), 909–916. 10.1098/rstb.2002.1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacq, A. , Pernot, C. , Martinez, Y. , Domergue, F. , Payré, B. , Jamet, E. , Burlat, V. , & Pacquit, V. B. (2017). The Arabidopsis lipid transfer protein 2 (AtLTP2) is involved in cuticle‐cell wall interface integrity and in etiolated hypocotyl permeability. Front. Plant Science, 8, 263. 10.3389/fpls.2017.00263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, J. L. , Lv, H. H. , Yang, L. M. , Fang, Z. Y. , Zhuang, M. , Zhang, Y. Y. , Liu, Y. M. , & Li, Z. S. (2018). Genome‐wide identification and characterization of non‐specific lipid transfer proteins in cabbage. Peer J, 6, e5379. 10.7717/peerj.5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, D. , Yang, J. , Li, F. , & Lin, F. (2015). Cloning and expression analysis of non‐specific lipid transfer protein gene from Nicotiana tabacum . Tobacco Science and Technology, 48, 12–20. 10.16135/j.issn1002-0861.20150103 [DOI] [Google Scholar]
- Kirubakaran, S. I. , Begum, S. M. , Ulaganathan, K. , & Sakthivel, N. (2008). Characterization of a new antifungal lipid transfer protein from wheat. Plant Physiology and Biochemistry, 46, 918–927. 10.1016/j.plaphy.2008.05.007 [DOI] [PubMed] [Google Scholar]
- Li, B. B. , Shi, Q. S. , Shu, X. L. , Ou, Y. Y. S. , & Chen, Y. B. (2006). Plants antimicrobial proteins nsLTPs. Plant Physiology Communications, 42, 539–544. CNKI:SUN:ZWSL.0.2006‐03‐047. [Google Scholar]
- Liang, Z. J. , Zhu, P. X. , Ji, Z. L. , Chen, X. R. , & Zhu, F. (2020). Cloning of LTP1 in Nicotiana benthamiana and construction of VIGS silencing and over expression vectors. Molecular Plant Breeding, 18, 924–930. 10.13271/j.mpb.018.000924 [DOI] [Google Scholar]
- Lin, K. C. , Wu, T. M. , Chandrika, N. N. P. , Chou, S. J. , & Hong, C. Y. (2017). Molecular characterization and subcellular localization of salt‐inducible lipid transfer proteins in rice. Biologia Plantarum, 61, 501–510. 10.1007/s10535-016-0671-x [DOI] [Google Scholar]
- Maximiano, M. R. , & Franco, O. L. (2021). Biotechnological applications of versatile plant lipid transfer proteins (LTPs). Peptides, 140, 170531. 10.1016/j.peptides.2021.170531 [DOI] [PubMed] [Google Scholar]
- Missaoui, K. , Gonzalez‐Klein, Z. , Jemli, S. , Garrido‐Arandia, M. , Diaz‐Perales, A. , Tome‐Amat, J. , & Brini, F. (2022). Identification and molecular characterization of a novel non‐specific lipid transfer protein (TdLTP2) from durum wheat. PLoS ONE, 17(4), e0266971. 10.1371/journal.pone.0266971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes, G. P. , Benitez, L. C. , do Amaral, M. N. , Vighi, I. L. , Auler, P. A. , da Maia, L. C. , Bianchi, V. J. , & Braga, E. J. B. (2015). Expression of LTP genes in response to saline stress in rice seedlings. Genetics and Molecular Research, 14, 8294–8305. 10.4238/2015.July.27.18 [DOI] [PubMed] [Google Scholar]
- Noonan, J. , Williams, W. P. , & Shan, X. (2017). Investigation of antimicrobial peptide genes associated with fungus and insect resistance in maize. International Journal of Molecular Sciences, 18(9), 1938. 10.3390/ijms18091938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen, T. A. , Blomqvist, K. , & Edqvist, J. (2016). Lipid transfer proteins: Classification, nomenclature, structure, and function. Planta, 244, 971–997. 10.1007/s00425-016-2585-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, K. , Xu, Y. , Cao, W. L. , Xie, X. Y. , Zhang, Y. R. , Zhang, J. F. , Liu, H. M. , Zhou, S. M. , Zhu, X. P. , & Zhu, C. X. (2022). Potato (Solanum tuberosum L.) non‐specific lipid transfer protein StLTP6 promotes viral infection by inhibiting virus‐induced RNA silencing. Planta, 256, 54. 10.1007/s00425-022-03948-6 [DOI] [PubMed] [Google Scholar]
- Sun, Q. , Wu, Y. H. , & Zhang, Y. X. (2022). Bioinformatic analysis and expression pattern of LTP family genes in Populus davidiana x P. albs var. pyramidalis . Bulletin of Botanical Research, 42, 211–223. 10.7525/j.issn.1673-5102.2022.02.006 [DOI] [Google Scholar]
- Sun, X. Y. , Zhu, Y. , Zhao, M. M. , Li, Z. X. , & Zhou, H. W. (2014). Cloning and characterization of a lipid transfer protein gene, ZmLTP3, from maize. Journal of Maize Sciences, 22(1), 62–66. 10.13597/j.cnki.maize.science.2014.01.013 [DOI] [Google Scholar]
- Vangelisti, A. , Simoni, S. , Usai, G. , Mascagni, F. , Ventimiglia, M. , Natali, L. , Cavallini, A. , & Giordani, T. (2022). In silico genome‐wide characterisation of the lipid transfer protein multigenic family in sunflower (H. annuus L.). Plants (Basel), 11(5), 664. 10.3390/plants11050664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , Zang, X. S. , Kabir, M. R. , Liu, K. L. , Liu, Z. S. , Ni, Z. F. , Yao, Y. Y. , Hu, Z. R. , Sun, Q. X. , & Peng, H. R. (2014). A wheat lipid transfer protein 3 could enhance the basal thermotolerance and oxidative stress resistance of Arabidopsis. Gene, 550(1), 18–26. 10.1016/j.gene.2014.08.007 [DOI] [PubMed] [Google Scholar]
- Wang, G. B. , & Qin, L. (2021). Transcriptome sequencing and drought resistance gene annotation in Quercus liaotungensis leaves. Acta Physiologiae Plantarum, 43, 120. 10.1007/s11738-021-03294-2 [DOI] [Google Scholar]
- Wang, X. F. , Wang, H. , Li, Y. L. , Cao, K. M. , & Ge, X. C. (2009). A rice lipid transfer protein binds to plasma membrane proteinaceous sites. Molecular Biology Reports, 36, 745–750. 10.1007/s11033-008-9238-z [DOI] [PubMed] [Google Scholar]
- Wei, K. , & Zhong, X. (2014). Non‐specific lipid transfer proteins in maize. BMC Plant Biology, 14, 281. 10.1186/s12870-014-0281-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. X. , Li, P. , Liu, C. , Wang, P. , Cao, P. J. , Ye, X. F. , & Li, Q. C. (2022). Systematic analysis of the non‐specific lipid transfer protein gene family in Nicotiana tabacum reveal its potential roles in stress responses. Plant Physiology and Biochemistry, 172, 33–47. 10.1016/j.plaphy.2022.01.002 [DOI] [PubMed] [Google Scholar]
- Zhang, D. Y. , Li, J. N. , Li, M. M. , Cheng, Z. M. , Xu, Q. A. , Song, X. , Shang, X. , & Guo, W. (2022). Overexpression of a cotton nonspecific lipid transfer protein gene, GhLTP4, enhances drought tolerance by remodeling lipid profiles, regulating abscisic acid homeostasis and improving tricarboxylic acid cycle in cotton. Environmental and Experimental Botany, 201, 104991. 10.1016/j.envexpbot.2022.104991 [DOI] [Google Scholar]
- Zhang, H. L. , Zhao, Y. N. , Zhao, X. J. , Zhang, Z. H. , Liu, J. , Shi, M. H. , & Song, B. T. (2022). Methylation level of potato gene OMT30376 regulates tuber anthocyanin transformations. Frontiers in Plant Science, 13, 1021617. 10.3389/fpls.2022.1021617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, H. W. , Tian, X. H. , Ma, G. H. , & Li, Z. X. (2013). Isolation and functional analysis of ZmLTP3, a homologue to Arabidopsis LTP3. International Journal of Molecular Sciences, 14, 5025–5035. 10.3390/ijms14035025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. RT‐qPCR reaction system.
Table S2. RT‐qPCR primer sequences.
Table S3. StLTPs gene family information.
Table S4. Cis‐acting regulatory elements in the promoter of StLTPs gene family.
Data Availability Statement
Not applicable.


