Abstract
The critical requirements in developing clinical-grade human-induced pluripotent stem cells–derived neural precursors (hiPSCs-NPCs) are defined by expandability, genetic stability, predictable in vivo post-grafting differentiation, and acceptable safety profile. Here, we report on the use of manual-selection protocol for generating expandable and stable human NPCs from induced pluripotent stem cells. The hiPSCs were generated by the reprogramming of peripheral blood mononuclear cells with Sendai-virus (SeV) vector encoding Yamanaka factors. After induction of neural rosettes, morphologically defined NPC colonies were manually harvested, re-plated, and expanded for up to 20 passages. Established NPCs showed normal karyotype, expression of typical NPCs markers at the proliferative stage, and ability to generate functional, calcium oscillating GABAergic or glutamatergic neurons after in vitro differentiation. Grafted NPCs into the striatum or spinal cord of immunodeficient rats showed progressive maturation and expression of early and late human-specific neuronal and glial markers at 2 or 6 months post-grafting. No tumor formation was seen in NPCs-grafted brain or spinal cord samples. These data demonstrate the effective use of in vitro manual-selection protocol to generate safe and expandable NPCs from hiPSCs cells. This protocol has the potential to be used to generate GMP (Good Manufacturing Practice)-grade NPCs from hiPSCs for future clinical use.
Keywords: human-induced pluripotent stem cells (hiPSCs), neural precursor cells (NPCs), manual selection, spinal cord grafting, brain grafting, immunodeficient rat
Introduction
The discovery of human somatic cell reprogramming-induced pluripotent stem cells (hiPSCs) technology1,2 provided remarkable progress in the potential development of patient-specific cell-replacement-based therapies. Besides, the generation of specific cell types from hiPSCs derived from patients with a defined genetic mutation proved to be a valuable platform for drug target discovery or creating human disease modeling both in vitro and in vivo3–6.
Previous experimental and ongoing clinical studies have established a therapeutic potential of human fetal tissue–derived, embryonic stem cells (ESs)–derived, or iPSC-derived neural precursor cells (hiPSC-NPCs) in the treatment of a variety of neurodegenerative disorders, including spinal cord/brain traumatic injury7,8, stroke9–11, Alzheimer’s and Parkinson’s disease12–14, and ALS15. The underlying rationale for using cell replacement therapies is to (1) repopulate the areas affected by neuronal or glial degeneration, (2) provide mechanical support of previously injured tissue, or (3) provide regional neuroprotection by releasing a variety of neurotrophic factors16–20.
Several factors are being considered in the process of hiPSC generation particularly if generated cell lines or their lineage-committed derivatives are planned to be used in prospective human clinical trials. First is the source of somatic cells to be used in reprogramming to generate hiPSCs. The most frequently used cell types are skin fibroblasts harvested by biopsies21. The key, but relative, limitation in using fibroblasts for reprogramming is the invasive nature to obtain the biopsy material and the need for the initial in vitro expansion of collected fibroblasts before the reprogramming process can be initiated22. The use of peripheral blood mononuclear cells (PBMCs) has been recently used for reprogramming23,24. The primary advantage in using PBMCs is a relative simplicity in obtaining the blood samples and the ability to collect a sufficient number of cells from a single blood draw needed for reprogramming (ie, no need for cell expansion). Also, the collection can be repeated if additional samples are needed. Second, is the use of reprogramming vector(s). The majority of initial studies have employed integrating viral vectors, including retrovirus for reprogramming1,25. The major limitation in using integrating vectors for reprogramming is the risk of mutagenesis and resulting tumorigenicity after in vivo grafting26–28. More recently, the use of several non-integrating vectors to generate hiPSCs from adult somatic cells was reported29. One of the non-integrating reprogramming vectors, the Sendai-virus (SeV; a single-stranded RNA virus), has been demonstrated to reprogram somatic cells with high efficiency, low cytotoxicity, and minimal risk of being integrated into the host genome30. Using SeV (encoding Yamanaka factors: SOX2, c-MYC, Klf4, and OCT3/4), several research groups generated the hiPSCs by reprogramming human PBMCs23,24,31,32. SeV-reprogrammed hiPSCs were further successfully used for the generation of lineage-committed cell line derivatives, including photoreceptor-like cells33, and cardiomyocytes34.
Here, we report on a successful generation of in vitro expandable SeV-hiPSCs-NPCs. Established NPCs show a long-term in vitro expandability while maintaining a stable karyotype, expression of neural precursor markers but with a lack of pluripotent markers. Upon transplantation into the striatum or spinal cord of immunodeficient rat grafted NPCs acquire a neuronal and glial phenotype with no detectable tumor formation for up to 6 months post-grafting. These data show that the NPCs manual-selection derivation protocol by using SeV-generated hiPSCs can successfully be used and considered for the future generation of clinical GMP-grade NPCs to be used in prospective clinical trials in the treatment of several neurodegenerative disorders, including spinal ischemic or traumatic injury, ALS, stroke, or brain trauma.
Materials and Methods
Development of hiPSC-NPCs
This study was approved by the University of California, San Diego (UCSD) Internal Review Board (IRB) (approval ID 101323).
Human-induced pluripotent stem cells
Human peripheral mononuclear cells were isolated from blood samples (33-year-old woman; Lonza, Basel, Switzerland) and reprogrammed by SeV-encoding Yamanaka factors1,25. Reprogramming was performed under feeder-free (FF)/xeno-free (XF) condition35.
Human iPSCs expansion
After reprogramming, cells were plated on Laminin (Laminin 511 E8, iMatrix-511; Nippi, Incorporated. Tokyo; Product code: 892001/892002)-coated six-well plate (cat. no. 3471, CORNING, Corning, NY, USA) in 1.4 × 104 cells/well in iPSC media (AK03N, Ajinomoto, Chuo City, Tokyo, Japan) containing 10 µM ROCK inhibitor (Y37632, Stem Cell Technologies, Cambridge, MA, USA). After 7 days, the hiPSC colonies were dissociated with trypsin solution (2 ml of TripLE solution, Life technologies, Carlsbad, CA, USA; 2 ml of DPBS (Dulbecco’s phosphate-buffered saline); 2 µl of 0.5M EDTA (Ethylenediaminetetraacetic acid)) and passaged. Cells were passaged at least three times before the embryoid bodies (EBs) formation was initiated. A subpopulation of established hiPSCs colonies was used to prepare a single-cell suspension and cells stored in LN vapor for potential future use (AMSBIO, Cambridge, MA, USA).
Quantitative polymerase chain reaction assessment of residual SeV
RNAs were prepared from (1) non-infected PBMCs (negative control), (2) SeV-infected PBMCs (3 days post-SeV infection; positive control), and (3) established iPSC line (LPF11; passage 5). cDNAs were synthesized using 1,000 ng of RNAs with Qiagen Quantitect RT Kit (in 20 μl of reaction volume). Quantitative polymerase chain reaction (Qpcr) was performed with TaqMan probes against SeV (custom-made probe targeting the SeV genome located between the coding region of NP and P protein) and β-actin. cDNAs were synthesized from 6.25, 6.25, and 1.56 ng of RNA prepared from negative-control PBMCs, positive-control PBMCs (3 days post-SeV infection), and established iPSC line (LPF11; passage 5), respectively. cDNA of positive control was diluted 160-fold. Conditions of PCR reaction are as follows: 50°C for 2 min, 95°C for 10 min and 40 cycles of (95°C for 15 s and 60°C for 1 min).
EB generation
hiPSC colonies were dissociated with trypsin solution (0.05%) and transferred into low attachment six-well plate (CORNING) containing neural induction medium (Stem cell technologies, Cambridge, MA, USA).
Rosettes formation
Four to six days in vitro–cultured EBs were transferred to poly-L-ornithine/laminin (PLO/L)-coated dishes. Cells were cultured in DMEM/F12-based NPCs media containing 0.5% N2 supplement (Life Technologies), 1% B27 supplement (Life Technologies), 1% glutamate (Life Technologies), and 1% penicillin-streptomycin (Life Technologies) supplemented with 10 ng/ml recombinant human basic fibroblast growth factor (bFGF) (Thermo Fisher Scientific, Waltham, MA, USA) as a solo mitogen.
NPCs isolation
Once rosettes-like structures were observed (typically 3–4 days after EBs plating), approximately 100 to 200 morphologically defined NPCs that appeared outside of each rosette were manually picked up by pipette tips and transferred in PLO/L-coating 48-well plates. Isolated NPCs were cultured in the same media as described above for the whole duration of cell expansion. Cells were repeatedly passaged over 10 times. To examine the genetic stability of generated NPCs, karyotype was examined at passage 12 (Cell Line Genetics, Madison, WI, USA).
Doubling time
The NPCs were cultured to passage 20, seeded at 2 × 105 cells per well in PLO/L-coated six-well plates. Three wells per day were then separately dissociated with Trypsin and collected in separate tubes to be counted using the Countess II automated cell counter (Thermo Fisher Scientific). The doubling time was calculated using the following equation:
Differentiation From the Developed NPCs to Neuron and Glial Cells In Vitro
NPC differentiation
The NPCs were plated onto chamber slides or coverslips and allowed to reach confluence. The differentiation was initiated with differentiation-induction media (DMEM/F12) containing 0.5% N2 supplement (Life Technologies), 1% B27 supplement (Life Technologies), 1% glutamate (Life Technologies), and 1% penicillin-streptomycin (Life Technologies) supplemented with 10 µg/ml BDNF (brain-derived neurotrophic factor) (PeproTech, Rocky Hill, NJ, USA), 10 µg/ml GDNF (glial cell line-derived neurotrophic factor) (PeproTech) with/without 1% fetal bovine serum (FBS). The medium was changed every other day until the cells were fixed with 4% paraformaldehyde (PFA) or used for calcium oscillation assay (~1–4 months after induction).
Calcium Oscillation in hiPSC-NPCs Derived Differentiated Neurons In Vitro
Time-lapse calcium oscillation was analyzed to detect functional neurons in induced hiPSC-NPCs. Four month–induced NPCs cultured on glass coverslips were loaded with Fluo-4AM (reconstituted in DMSO) in Hank’s Balanced Salt Solution (HBSS) for 40 min at room temperature in accordance with manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). Cells were rinsed with HBSS two times after Fluo-4AM loading and were then incubated at room temperature in the dark for 30 min until data acquisition. Fluorescence imaging of intracellular Ca2+ dynamics for periods of 5 min 17 s was performed with a Olympus BX51W1 fixed-stage upright fluorescence microscope (Olympus Corp., Center Valley, PA, USA) equipped with a SPOT-Xplorer™—XS 1.4 MP monochrome camera (Diagnostic Instruments, Inc., Sterling Heights, MI, USA). The imaging system was controlled by SPOT 5.2 ADVANCED Software (Diagnostic Instruments, Inc.). Neurons were imaged in time lapse with a water immersion 40× magnification, 0.8 numerical aperture (NA) LUMPlanFL N objective (Olympus). Images were captured every 1.035 s under low-light level conditions. Images were collected at a slow transfer rate, which reduces background noise, and binned (2 × 2). Digital image processing was performed by using SPOT 5.2 ADVANCED Software (Diagnostic Instruments, Inc.), image processing program ImageJ (NIH, Bethesda, MD, USA) and graphics and data analysis software SigmaPlot 12.5 (Systat Software, Inc., San Jose, CA, USA).
Animal Studies
All animal studies were approved by the UCSD Institutional Animal care and Use Committee (Protocol No. S01193).
NPCs Grafting and Differentiation In Vivo
Intra-striatal cell transplantation
Adult immunodeficient rats (Crl:NIH-Foxn1rnu; Charles River; n = 8) (Charles River, Wilmington, MA, USA) were anesthetized with isoflurane (5% induction, 1.5%–2% maintenance) and placed in a stereotaxic apparatus to keep the head in a fixed position. After the scalp was shaved, a sagittal midline skin incision was performed to expose the skull. To permit an intra-parenchymal brain injection, a small borrow hole was drilled into the skull using a dental drill. The NPCs (30,000 cells/µl/injection) were injected into the striatum (stereotaxic X-Y-Z coordinates: bregma 0.5 mm, lateral to ± 3.0 mm, and 3.8, 5.0, and 6.2 mm depth: three injections delivered at each depth bilaterally) using a 34G needle interconnected with a digital microinjector (Tritech Research, San Diego, CA, USA).
Spinal cord parenchyma cell transplantation
After laminectomy at the T13-L1 level, rats (Crl: NIH-Foxn1rnu; Charles River; n = 8) received five bilateral spinal cord gray matter injections of NPCs (30,000 cells/µl/injection; X-Y-Z coordinates: lateral to midline ± 0.8 mm and ~ 1.0 mm depth). Cells were injected by using a Hamilton Syringe and homemade glass capillary (inner diameter: 80 µm, outer diameter: 100 µm) interconnected with a digital microinjector (Tritech Research).
Same animals received both intra-spinal and intra-cerebral NPCs injections.
Animal Perfusion-Fixation and Immunofluorescence Staining
At 2 months (n = 3) or 6 months (n = 5) after in vivo NPCs grafting rats were terminally anesthetized with pentobarbital (100 mg/kg; i.p.) and transcardially perfused with heparinized saline (100 ml) followed by 4% PFA in PBS. The brain and spinal cord tissues were then dissected, post-fixed with 4% PFA overnight at 4°C, and cryoprotected in 30% sucrose for a minimum of 5 days. Coronal brain or transverse spinal cord sections were then cut on a cryostat (20–30 µm thick) and stained by using a standard immunofluorescence (IF) protocol. Sections were first washed with 0.3 % Triton-100-containing PBS (TX-PBS) 3 × 10 min and incubated with a blocking solution containing 5% normal donkey serum in 0.3% TX-PBS for 60 min. This was followed by incubation in primary antibodies (Table 1) in blocking solution overnight at 4°C. Sections were then washed 3 × 10 min in PBS and incubated with fluorophore-conjugated secondary antibodies diluted in 0.3% TX-PBS for 1 h. After 3× wash with PBS, nuclear staining was performed using DAPI solution (0.1 µg/ml) followed by three times rinses in PBS. Sections were then mounted on slides and covered with anti-fade mounting medium (ProLong; Thermo Fisher Scientific).
Table 1.
Antibodies Used for Flow Cytometry and Immunofluorescence Staining.
| Catalog no. | Name | Company |
|---|---|---|
| Antibodies used for flow cytometry | ||
| 560341 | Nestin—Alexa Fluor 647 | BD Biosciences |
| 555427 | CD24—FITC | BD Biosciences |
| 561664 | PAX6—Alexa Fluor 488 | BD Biosciences |
| 561610 | Sox2—V450 | BD Biosciences |
| 560791 | Nanog—Alexa Fluor 488 | BD Biosciences |
| Primary antibodies used for immunofluorescence staining | ||
| AF1759 | OCT-3/4 | R&D systems |
| MAB1435 | SSEA4 | R&D systems |
| AB5603 | SOX2 | Millipore Sigma |
| AF1997 | Nanog | R&D systems |
| MAB6326 | Nestin | Millipore Sigma |
| 610920 | N-cadherin | BD Biosciences |
| 339100 | ZO1 | Invitrogen |
| ab104854 | Plzf | abcam |
| AB5934 | SOX1 | Millipore Sigma |
| AF1979 | Otx2 | R&D Systems |
| AF8150 | Pax6 | R&D Systems |
| 134014 | NeuN | Abcam |
| MAB377 | NeuN | Millipore Sigma |
| ABN78 | NeuN | Millipore Sigma |
| S4403 | MAP2 | Sigma |
| Sc-8066 | DCX | Santa Cruz |
| G3893 | GFAP | Sigma |
| AB5541 | GFAP | Chemicon |
| TA302094 | Human GFAP | Origene |
| 50-6525-80 | Synaptophysin | eBioSciences |
| VP-S285 | Synaptophysin | Vector laboratories |
| Sc-73650 | GAD65 | Santa Cruz biotechnology |
| AB5905 | VGLUT1 | Millipore Sigma |
| AB2251-1 | VGLUT2 | Millipore Sigma |
| AB5421-1 | VGLUT3 | Millipore Sigma |
| Ab9610 | Olig2 | Millipore Sigma |
| AB5733 | Vimentin | EMD Millipore |
| Ab16667 | Ki67 | abcam |
| Ab36999 | Human NuMA | abcam |
| AB951 | Human NSE | Chemicon |
| A-2052 | GABA | Sigma |
| 147011 | Gephyrin | Synaptic Systems |
| ab13970 | GFP | abcam |
| AB144P | ChAT | Millipore Sigma |
| 131002 | VGAT | Synaptic Systems |
For in vitro cultured cells, NPCs plated on glass coverslips were fixed in 4% PFA for 20 min and washed 3 × 5 min in PBS. After blocking with 5% normal donkey serum for 30 min, cells were incubated with primary antibodies (Table 1) in blocking solution overnight at 4°C. After 3 × 5 min rinses in PBS, the cells were then incubated with fluorophore-conjugated secondary antibodies in 0.2% TX-PBS for 1 h followed by 3 × 5 min rinses in PBS. Nuclei were stained with DAPI solution (0.1µg/ml) for 3 min followed by 3 × 5min wash in PBS. Coverslips were then mounted on slides with anti-fade mounting medium (ProLong; Thermo Fisher Scientific). The images were acquired using fluorescence (Zeiss Axio Imager M2 Microscope(Zeiss, Oberkochen, Germany) with Stereo Investigator software (MBF Bioscience, Williston, VT, USA), and using a confocal microscope (Fluoview FV1000, Olympus) with Olympus FV10-ASW Viewer software.
Flow Cytometry
Flow cytometry analysis was performed by following the company’s instruction with modification (BD Biosciences, San Jose, CA, USA). At first, the established NPCs were cultured in DMEM/F12 media supplemented with 10 ng/ml bFGF as described before. Once cells reached confluence, cells were dissociated with Accutase (cat. no. AT-104, Innovative Cell Technologies co., San Diego, CA, USA) and a single-cell suspension (1 × 106 cells per 1 ml sample) prepared and incubated with Aqua dye (no. L34965, Thermo Fisher Scientific) to detect live/dead cells for 30 min. After washing the cells with FACS buffer consisting of 97% DPBS, 2% FBS, and 1% (5 mM) EDTA, cells were fixed with 4% PFA in PBS. Then, permeabilization was done with a buffer (BP Phosflow Perm. Buffer III, cat. no. 558050, BD Biosciences) for 15 min. Cells were washed with FACS (fluorescence-activated cell sorting) buffer 2 ×, blocked with 5% normal donkey serum in FACS buffer, and then incubated with each antibody (Table 1) for 30 min in the dark. Expression of extracellular and intracellular protein markers was detected using flow cytometry using BD LSRFortessa (BD Biosciences). The data were analyzed by Flowjo analysis software (FLOWJO, Ashland, OR, USA).
Statistical Analysis
All data are reported as the mean ± SEM. An unpaired two-tailed Student’s t test was used for single comparisons of grafted cells at the 2-month and 6-month post-grafting. In each case, *P < 0.05 and **P < 0.01 were considered to be statistically significant. The GraphPad Prism software (version 6.0c; GraphPad Software Inc., San Diego, CA, USA) was used for all analyses.
Results
Lack of Residual SeV in Established Pluripotent LPF11 Cells
Using qPCR, we first probe for the residual presence of SeV (targeting the SeV genome located between the coding region of NP and P protein) in established pluripotent LPF11 cells. Compared with positive control (PBMCs 3 days post-SeV infection), no residual SeV was detected (Supplementary Fig. 1).
Generation of NPCs from hiPSCs
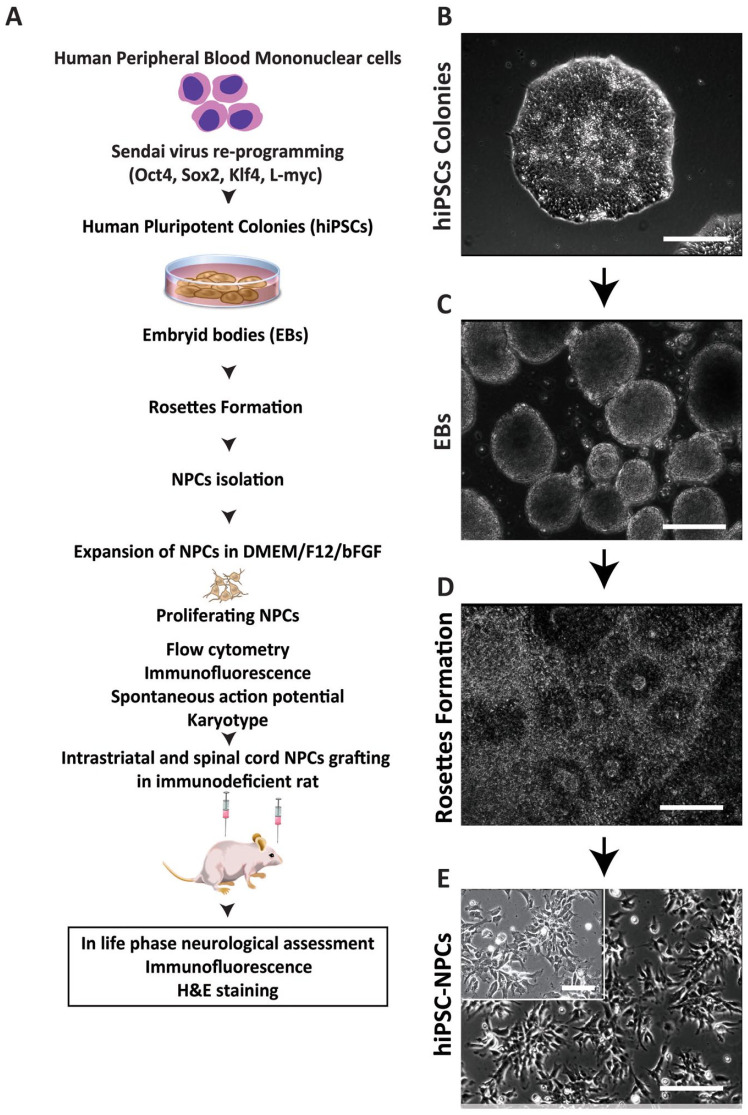
To generate the hiPSC-NPCs, a previously developed protocol established to isolate the NPCs from human pluripotent ESs and porcine iPSCs was employed36,37. First, the peripheral mononuclear blood cells were isolated from human blood samples and reprogrammed by SeV vectors encoding octamer-binding transcription factor 4 (OCT4), (sex-determining region Y)-box2 (SOX2), Kruppel-like factor 4 (KLF4), and myelocytomatosis viral oncogene (L-MYC). After reprogramming, cells were cultured on matrix-coating dishes as a monolayer (Fig. 1A, B).
Figure 1.
Strategy for development and in vitro and in vivo characterization of hiPSC-NPCs. (A) Schematic diagram depicting the experimental design of in vitro hiPSC-NPCs generation and in vitro and in vivo cell characterization. (B) Morphology of established hiPSCs colony. (C) Morphology of free-floating EBs. (D) A colony of neural rosettes cultured on P/L coated dish. (E) Representative image of established hiPSCs-NPCs. Scale bars: 100 µm (B, C, D, E). hiPSCs-NPC: human-induced pluripotent stem cells–derived neural precursors; EB: embryonic body, DMEM (Dulbecco’s Modified Eagle Medium).
Established pluripotent colonies (Fig. 1B) were manually dissociated and transferred into low attachment dishes to induced EBs (Fig. 1C). After 4 to 6 days, established EBs were transferred onto PLO/L pre-coated dishes and induced using NPCs induction media (see “Material and Methods” for details). Attached EBs gradually flattened in shape and the appearance of neural rosettes (NRs) was seen in about 4 to 7 days after EBs plating/induction (Fig. 1D).
Several cell clusters originating from NRs but localized at the edges of NRs were then manually picked and re-plated to P/L coating dishes. These cell populations were passaged several times until morphologically defined NPCs-like cell groups were identified. Colonies of NPCs-like cells were then manually harvested, expanded on P/L coated dishes (Fig. 1E), and used in all subsequent in vitro and in vivo grafting experiments (Fig. 1A).
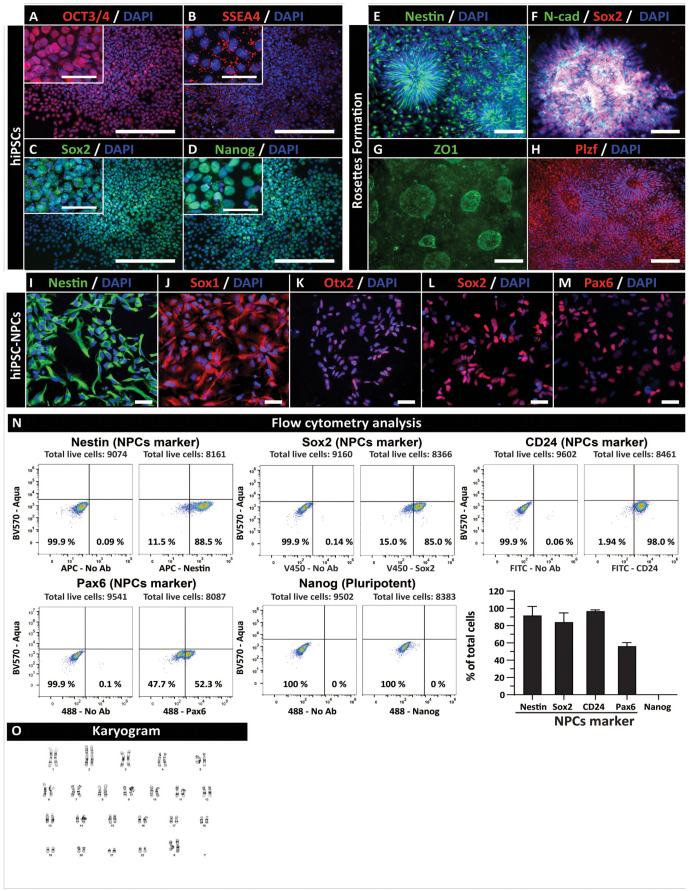
The expression of pluripotent markers, NRs-associated markers, and proliferating NPCs markers was analyzed periodically during the process of NPCs generation by IF and flow cytometry. In established pluripotent colonies, iPSC marker(s) expression, including OCT3/4, SSEA4, SOX2, and Nanog, was consistently seen (Fig. 2A–D). Staining of NRs showed a high level of expression for Nestin, N-cadherin, Sox2, ZO1, and Plzf (Fig. 2E–H). Tight junction protein ZO1 was detected asymmetrically in the central part of rosettes formations (Fig. 2G). Analysis of NPCs markers, including nestin, SOX1, OTX2, SOX2, and PAX6 showed consistent presence of all markers in established (passage 19) NPCs (Fig. 2I–M). Quantitative flow cytometry analysis of NPC markers showed 52.20% Pax6, 84.86% SOX2, 97.94% CD24, and 88.41% nestin-positive cells. No expression of the pluripotency marker (Nanog) was seen (Fig. 2N). Established NPCs (passage 12) showed normal karyotype (Fig. 2O).
Figure 2.
In vitro characterization of pluripotent and hiPSC-derived NPCs. Immunofluorescent staining of pluripotent hiPSCs colonies with (A) OCT4, (B) SSEA4, (C) SOX2, and (D) Nanog. Established neural rosette(s) expressing (E) Nestin, (F) (N-cadherin; N-cad), Sox2 (G) ZO1, and (H) Plzf. Proliferating NPCs at passage 19 stained with (I) Nestin, (J) Sox1, (K) Otx2, (L) Sox2, and (M) PAX6 antibodies. (N) Flow cytometry analysis of proliferating NPCs at passage 20 shows the consistent presence of NPC markers (Pax6, SOX2, CD24, and Nestin) but lack of pluripotent marker Nanog. (O) Normal karyogram in established NPCs (passage 12). Scale bars: 50 µm (A–D) and 20 µm (E–M, A–D-inserts). hiPSC: human-induced pluripotent stem cell; NPC: neural precursors.
Established hiPSCs-NPCs Differentiate Into Functional Neurons and Glial Cells In Vitro
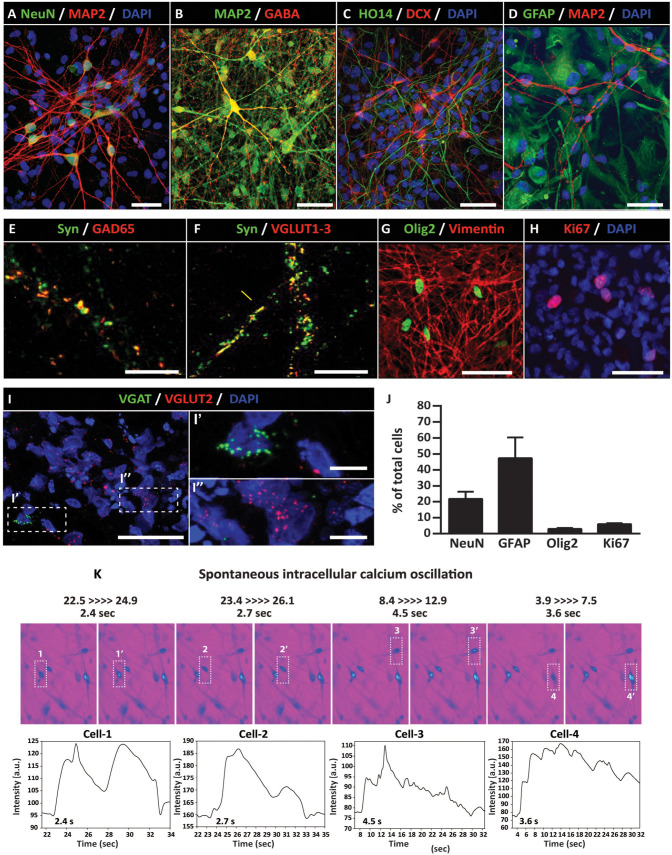
A defining characteristic of multipotent NPCs is their ability to generate functional neurons and glial cells (astrocytes and oligodendrocytes) after differentiation. To probe for the multi-lineage potency of established NPCs (passages 11–15), cells were treated with differentiation media (containing 10 ng/ml BDNF and 10 ng/ml GDNF) for 2 months. Induced cells were analyzed by (1) IF using neuronal and glial markers, (2) in situ fluorescence hybridization to identify glutamatergic or GABAergic mRNA transcripts, and, (3) by calcium oscillation imaging to confirm the presence of action potential-generating neurons.
At 2 months after differentiation in neuron-inducing media, a subpopulation of cells formed radiated neuron-like morphology and showed expression of immature- (DCX—doublecortin) and mature- (MAP2—microtubule-associated protein 2), (NeuN—DNA-binding neuron-specific protein) and (HO14-human-specific axonal neurofilament) neuronal markers (Fig. 3A–D). Staining with neurotransmitter phenotypic markers shows co-localization of synaptophysin+ puncta with GAD65 (GABAergic marker) and VGLUT1-3 (glutamatergic marker) (Fig. 3E, F). Staining with astrocyte marker [glial fibrillary acidic protein (GFAP)], oligodendrocyte marker (Olig2), and early glial marker (vimentin) shows a consistent presence of all three markers (Fig. 3D, G). Ki67 (mitotic marker) immunoreactivity was seen in a small population of cells scattered through the analyzed cell population (Fig. 3H). Staining with GABA antibodies showed well-established GABAergic neurons (Fig. 3B). FISH analysis using VGLUT2 (glutamate transporter 2) and VGAT (vesicular GABA transporter) probes confirmed the presence of both excitatory glutamatergic and GABAergic neurons (Fig. 3I). Quantitative analysis of relative numbers of NeuN+, GFAP+, OLIG2+, and Ki67+ cells is presented in Fig. 3J.
Figure 3.
Induced hiPSC-NPCs generate functional neurons in vitro. Immunofluorescence staining of 2 month–induced NPCs show expression of neuronal markers including (A) NeuN/MAP2, (B) MAP2/GABA, and, (C) HO14/DCX. (D) Expression of astrocyte marker (GFAP). (E, F) Co-localization of Syn/GAD65 and Syn/VGLUT1-3 in synaptic terminals. (G) Expression of Olig2 and Vimentin in immature glial cells. (H) Ki67 (mitotic marker) is seen in a small subpopulation of cells. (I) FISH shows the presence of VGLUT2 and VGAT transcripts in a separate population of induced neurons. (J) Quantification of NeuN, GFAP, Olig2, and mitotically active Ki67+ cells. (K) Presence of spontaneous intracellular calcium oscillation in induced neurons at 4 months after differentiation. Scale bars: 50 µm (I), 10 µm (A, B, C, D, E, F, I′, I″), 5 µm (G, H). DCX: doublecortin; GFAP: glial fibrillary acidic protein; hiPSCs-NPC: human-induced pluripotent stem cells–derived neural precursors; NeuN: DNA-binding neuron-specific protein; VGAT: vesicular GABA transporter.
To study the ability of induced neurons to generate action, potential cells were loaded with Fluo-4 AM and spontaneous calcium oscillation measured using high-resolution fluorescence microscopy. Numerous neurons with calcium oscillation patterns consistent with spiking neurons (ie, periodic, quasi-periodic, or chaotic spiking’s, as well as bursting’s activities) were seen (Fig. 3K).
Taken together, these data demonstrate that established NPCs have multi-lineage potential and can generate mature-functional excitatory inhibitory neurons and glial cells (astrocytes and oligodendrocytes).
In Vivo Grafted NPCs Are Non-Tumorogenic and Differentiates Into Neurons and Glial Cells in the Striatum and Spinal Cord of the Immunodeficient Rat
To characterize the safety and differentiation potential of established NPCs, cells were grafted into striata or lumbar spinal cord in the adult immunodeficient rat. Animals were sacrificed at 2 or 6 months post-grafting and the presence of grafted cells analyzed by H&E staining or IF using human-specific and non-specific antibodies (Fig. 1A).
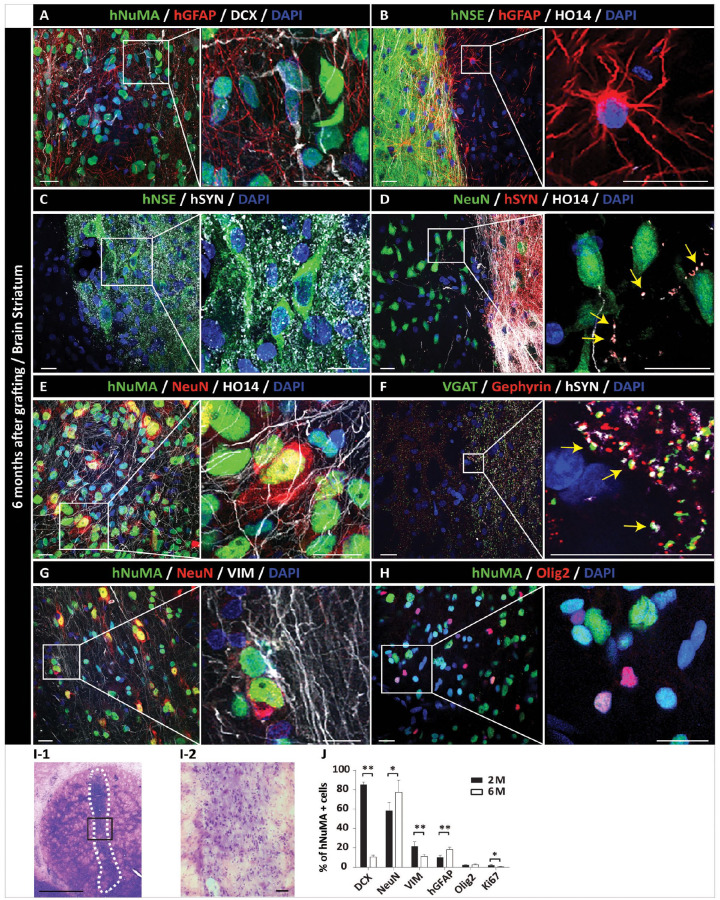
IF analysis of NPCs-grafted striata and spinal cords at 2 months postgrafting showed a consistent presence of grafted human cells stained with a human-specific nuclear marker (hNUMA), (Supplementary Figs. 2A and 3A). Staining with early (DCX) and late (NeuN, hNSE, and HO14) neuronal markers showed the presence of all neuronal markers in individual grafts. In general, the most intense expression was seen for early neuronal marker DCX and relatively lower expression was seen for late neuronal markers (NeuN, hNSE) (Supplementary Figs. 2A–E and 3A–C). Moderate intensity of human-specific synaptophysin staining was seen throughout the grafts with some hSYN+ puncta associated with human axons (HO14) (Supplementary Figs. 2C, D, and 3B). Staining with VGAT (pre-synaptic inhibitory marker) and gephyrin (post-synaptic glycine receptor-associated protein) showed the presence of putative synaptic inhibitory contacts between grafted neurons and neurons of the host (Supplementary Figs. 2F and 3F). Analysis of glial markers showed a relatively low expression of GFAP and Olig2 but with a high level of expression for vimentin (early glial marker) (Supplementary Figs. 2A, B, G, H, and 3A, C, D, E).
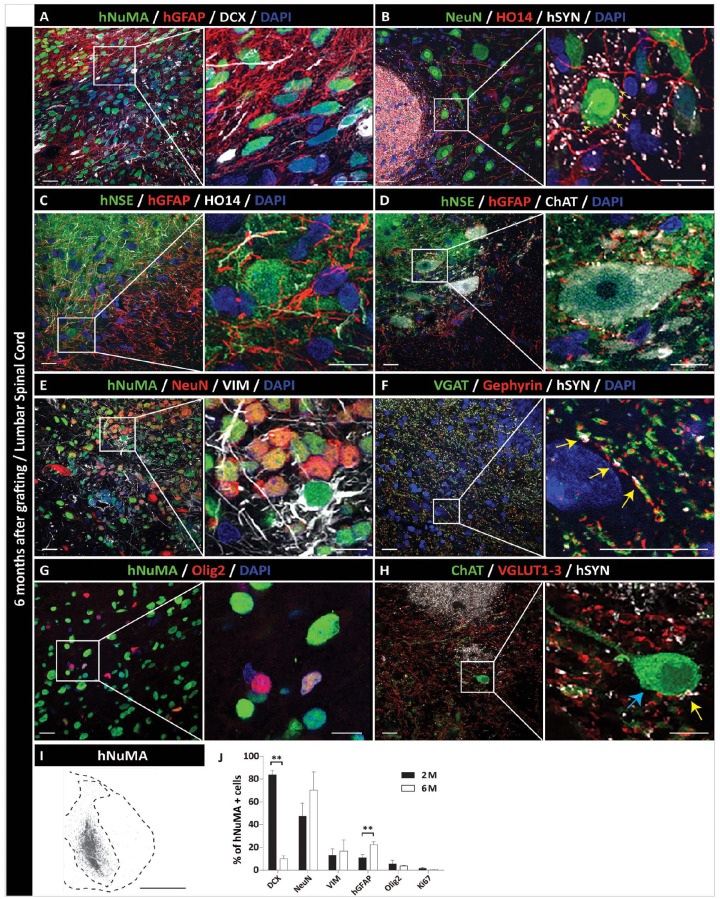
IF analysis at 6 months post-grafting showed a more advanced stage of grafted NPCs maturation in both striatum and spinal cord. Staining with mature neuronal markers (hNSE and NeuN) showed a high number of intensely hNSE-stained and NeuN-stained neurons throughout the grafts (Figs. 4B, C, E, G and 5C, D, E). An intense hSYN in the graft and often associated with peripherally projecting HO14+ human axons was also seen (Figs. 4C, D, and 5B). Co-staining with hSYN and VGAT or VGLUT1-3 showed the presence of both inhibitory (VGAT+) and excitatory (VGLUT1-3) puncta co-localizing with grafted neurons-derived hSYN+ terminals (Figs. 4F and 5F, H). Compared with 2 months post-grafting, the density of GFAP+ astrocytes was clearly increased in the graft as well as at the border of the graft with the host tissue (Figs. 4A, B, and 5A, C). In the same regions, a continuing presence of early glial marker vimentin was seen, suggesting an ongoing glial proliferation (Figs. 4G and 5E). Oligodendrocyte staining with Olig2 antibody (co-staining with hNUMA) showed the majority of double-stained grafted cells within the grafts (Figs. 4H and 5G). Hematoxylin and eosin (H&E) staining of striatal sections showed normally appearing mature neural grafts with no sign of hyper-cellularity (which would be indicative of tumor formation) (Fig. 4I-1, 2). Quantitative analysis of DCX-, NeuN-, vimentin-, hGFAP-, Olig2-, and Ki67-positive cells at 2 and 6 months is provided in Figs. 4J and 5J.
Figure 4.
Long-term (6 months) engraftment and neuronal and glial differentiation of hiPSC-NPCs after grafting into striata of the immunodeficient rat. (A) IF analysis of the coronal brain section showed a consistent presence of hNUMA+ grafts in the targeted striatal region. Staining with human-specific GFAP shows the homogenous distribution of grafted NPC-derived astrocytes throughout the graft. (B) Intense human-specific NSE expression (green area) with well morphologically defined human astrocytes (red signal). (C) Areas occupied with mature grafted human neurons (hNSE) show intense human-specific synaptophysin (hSYN) expression. (D) hSYN expression is primarily present in areas with a high density of grafted neuron-derived human axons (HO14+), (yellow arrows). (E) Expression of mature neuronal marker NeuN in hNUMA+ grafted neurons. (F) A high density of VGAT punctate-like expression in areas of hSYN immunoreactivity. hSYN+ terminals show co-localization with VGAT immunoreactivity and are opposed to post-synaptic gephyrin immunoreactivity (yellow arrows). (G) Presence of vimentin+ glial progenitors at the periphery of hNUMA+ graft. (H) Expression of Olig2 (oligodendrocyte marker) immunoreactivity in hNUMA+ grafted cells. (I) Hematoxylin-eosin-stained section with normally appearing graft. No hyper-cellularity (indicative of tumor formation) can be identified (I-1, I-2). (J) Quantitative analysis of neuronal and glial marker(s) expression. Scale bars: 20 µm (A–H), 10 µm (A–H; inserts). *P < 0.05; **P < 0.01, all data are presented as mean ± SEM, n = 3, two-tailed t test. DCX: doublecortin; GFAP: glial fibrillary acidic protein; hiPSCs-NPC: human induced pluripotent stem cells–derived neural precursors; hNUMA: human-specific nuclear marker; IF: immunofluorescence; NeuN: DNA-binding neuron-specific protein; SEM: standard error of the mean; VGAT: vesicular GABA transporter.
Figure 5.
Long-term (6 months) engraftment and neuronal and glial differentiation of hiPSC-NPCs after grafting into the lumbar spinal cord of the immunodeficient rat. (A) IF staining of transverse lumbar spinal cord section with hNUMA antibody shows the homogeneous distribution of grafted cells. In the same areas, a high density of human astrocytes (hGFAP+ cells) and residual DCX immunoreactivity is seen. (B) Intense human-specific synaptophysin (hSYN) immunoreactivity within the graft. A dense axonal network (HO14) derived from grafted cells and expressing hSYN can also be seen. (C, D) Expression of mature neuronal marker (hNSE), glial marker (hGFAP), and human axonal neurofilament (HO14). (E) NeuN expression in hNUMA+ grafted neurons in the core of the graft. Vimentin immunoreactivity in the same region can also be seen. (F) Co-localization of hSYN puncta with VGAT immunoreactivity in the proximity of post-synaptic gephyrin+ puncta (yellow arrows). (G) Expression of Olig2 (mature oligodendrocyte marker) in grafted hNUMA+ cells. (H) Expression of VGLUT1-3 immunoreactivity in grafted neuron-derived hSYN puncta (yellow arrows) and residing on the CHAT+ α-motoneuron of the host (blue arrows). (I) hNUMA positive graft (black color–stained cells), representing a single injection site localized between the center of the dorsal horn and ventral horn. (J) Quantitative analysis of neuronal and glial marker(s) expression. Scale bars: 20 µm (A–H), 10 µm (A–H; inserts). **P < 0.01, all data are presented as mean ± SEM, n = 3, two-tailed t test. DCX: doublecortin; GFAP: glial fibrillary acidic protein; hiPSCs-NPC: human induced pluripotent stem cells–derived neural precursors; hNUMA: human-specific nuclear marker; IF: immunofluorescence; NeuN: DNA-binding neuron-specific protein; SEM: standard error of the mean; VGAT: vesicular GABA transporter.
Discussion
Using SeV-reprogrammed hiPSCs, we demonstrate an effective generation of NPCs by using a manual-selection protocol. Established NPCs line can be expanded for over 20 passages while maintaining a stable karyotype, expression of neural precursor markers, and with no signs of spontaneous differentiation. No expression of pluripotent markers was seen in established NPCs. In vitro–induced NPCs show differentiation toward glial phenotype (oligodendrocytes and astrocytes) and excitatory and inhibitory neurons capable of generating spontaneous action potentials. Established NPCs grafted into striata or spinal cord of immunodeficient rats show robust engraftment and neuronal and glial differentiation at 2 or 6 months post-grafting. No tumor formation was seen in NPCs-grafted regions.
Strategies for Purification/Enrichment and Expansion of NPCs From Pluripotent Stem Cells
Several previous studies have demonstrated the successful use of fluorescence-activated cell sorting, by using several combinations of neural and/or neuronal cell surface markers to isolate/enrich the population of expandable neural/neuronal precursors38–43. While effective in generating well-defined NPC populations, these protocols require the use of GMP-grade antibodies and cell-sorting equipment(s). As such, routine use of the FACS protocol for clinical use is prohibitively expensive. In our previous study, we have developed and characterized a manual-selection protocol that permits a reliable morphologically defined selection of NPCs from human ES cell lines37. Our current study, which employed the same protocol, demonstrates that the NPCs can similarly be isolated from established hiPSCs. Comparably, as previously shown for ES-derived NPCs, a stable karyotype and lack of pluripotent markers were seen after long-term passaging of hiPSCs-NPCs. Mature neurons derived from established NPCs showed action potential-generating properties and expression of both excitatory and inhibitory neurotransmitter phenotype. These data are similar to other studies that show the appearance of functional neurons after induction of porcine iPSCs-NPCs in vitro36 or long-term maintained hiPSC-derived brain organoids44,45.
Selecting the Optimal In Vitro NPCs Culture Protocol to Generate Transplantable NPCs Cell Bank
One of the key determinants defining a successful and effective translation of cell-replacement-based therapies into clinical practice is the format on how the established NPCs cell lines are cultured, stored, and prepared for in vivo grafting. In general, two basic protocols are being employed to culture and expand NPCs whether derived from fetal central nervous system (CNS) tissue, ESs, or iPSC lines. First, the NPCs are cultured in the form of neurospheres46. For passage, the individual neurospheres are enzymatically or mechanically dissociated and then subcultured. While the in vitro culturing of established neurospheres is relatively straightforward and does not require any coating of tissue culture wells, there are three key limitations from the perspective of effective use of neurospheres in a clinical setting. (1) The freezing of established neurospheres is typically associated with a poor post-freezing cell/neurospheres recovery thus limiting the use of previously frozen neurospheres for routine clinical use. (2) Because the size of individual neurospheres varies significantly the required cell dosing to be delivered is difficult to control unless the filtering of prepared neurospheres culture is performed before grafting. (3) Finally, a relatively large size of neurospheres and tendency of aggregation prohibit the use of small diameter needles (~30G) for intra-parenchymal delivery. This can be of particular concern when the spinal delivery of cells (which requires smaller needles relative to brain injection cannulas) is desired47–49. Second, the NPCs can be cultured as a cell monolayer on previously coated tissue culture wells. For coating, a combination of PLO/L is typically used50. This culturing protocol has successfully been used to expand and bank clinical-grade NPCs derived from fetal tissue or ESs lines49,51. Our current protocol used a comparable approach with NPCs expanded as a monolayer on a previously P/L-coated surface. While relatively more laborious, this protocol has several specific advantages (compared with neurosphere technology) once considered for potential clinical application: (1) a consistent neural precursor cell morphology can be seen in established-proliferating NPCs, (2) minimal or no spontaneous differentiation is typically seen even with extensive passaging, (3) over 90% cell viability is usually seen in previously frozen cells after thawing, and (4) a relatively smaller injection needle (~30g) can be used without difficulties to deliver single-cell suspension into the brain or spinal cord in a large animal model(s) (such as pig or NHP) and in human36,47–49,52,53. As such, the use of monolayer NPCs culturing and expansion is a preferable method in producing NPC cell bank(s) intended for clinical use.
Manually Selected and In Vitro–Expanded hiPSCs-NPCs Show Long-Term In Vivo Engraftment in the Absence of Tumor Formation
In our current study, in vivo grafted NPCs into striata and spinal cord of immunodeficient rats showed a robust neural differentiation with the presence of mature neurons and glial cells (oligodendrocytes and astrocytes). No tumor formation was seen at 2 or 6 months post-grafting. Mature grafted neurons showed extensive axodendritic sprouting and development of putative inhibitory and excitatory synaptic contacts with the host interneurons and CHAT+ α-motoneurons. Qualitatively, these data are very similar to the differentiation profile of human fetal spinal cord-, ES-, or iPSCs-derived NPCs grafted spinally in naïve immunosuppressed adult pig or immunodeficient rat53,54, spinally injured rat or non-human primates52,55–58, or in rat with previous spinal ischemic injury59,60. Similarly as shown in previous studies, no excessive proliferation of grafted cells was seen and the only occasional presence of grafted Ki67+ cell was detected. Jointly these data demonstrate a high degree of phenotypic/functional equivalency in established iPSCs-NPCs to fetal tissue-derived or ES-derived NPCs after long-term in vivo grafting. More recently, using SeV reprogrammed adult pig skin fibroblasts-derived iPSC-NPCs, we have demonstrated a comparable long-term (2–8 months) engraftment and functionality of NPCs after intra-striatal grafting in immunodeficient rat or after spinal grafting in syngeneic naive pig or allogeneic transiently immunosuppressed pig with previous spinal cord traumatic injury36.
In summary, we demonstrate a successful generation of expandable SeV-reprogrammed hiPSCs-NPCs line. Long-term expanded NPCs showed a stable karyogram and expression of typical neural precursor markers. In vitro–induced NPCs differentiate toward functional neurons, astrocytes, and oligodendrocytes. Long-term (6 months) in vivo–grafted iPSC-NPCs show a comparable engraftment/differentiation profile and acceptable safety as previously reported for fetal tissue–derived or ES-derived clinical-grade NPCs. Accordingly, this hiPSCs-NPCs cell line and/or NPCs-selection protocol may represent an effective technology for generating clinical-grade NPCs to be used in the perspective human clinical trial(s) in the treatment of several spinal neurodegenerative disorders such as spinal ischemic/traumatic injury, ALS, or multiple sclerosis.
Supplemental Material
Supplemental material, sj-jpg-1-cll-10.1177_09636897231163232 for Derivation of Sendai-Virus-Reprogrammed Human iPSCs-Neuronal Precursors: In Vitro and In Vivo Post-grafting Safety Characterization by Michiko Shigyo, Yoshiomi Kobayashi, Oleksandr Platoshyn, Silvia Marsala, Tomohisa Kato Jr, Naoki Takamura, Kenji Yoshida, Akiyoshi Kishino, Mariana Bravo-Hernandez, Stefan Juhas, Jana Juhasova, Hana Studenovska, Vladimir Proks, Joseph D. Ciacci and Martin Marsala in Cell Transplantation
Supplemental material, sj-jpg-2-cll-10.1177_09636897231163232 for Derivation of Sendai-Virus-Reprogrammed Human iPSCs-Neuronal Precursors: In Vitro and In Vivo Post-grafting Safety Characterization by Michiko Shigyo, Yoshiomi Kobayashi, Oleksandr Platoshyn, Silvia Marsala, Tomohisa Kato Jr, Naoki Takamura, Kenji Yoshida, Akiyoshi Kishino, Mariana Bravo-Hernandez, Stefan Juhas, Jana Juhasova, Hana Studenovska, Vladimir Proks, Joseph D. Ciacci and Martin Marsala in Cell Transplantation
Supplemental material, sj-jpg-3-cll-10.1177_09636897231163232 for Derivation of Sendai-Virus-Reprogrammed Human iPSCs-Neuronal Precursors: In Vitro and In Vivo Post-grafting Safety Characterization by Michiko Shigyo, Yoshiomi Kobayashi, Oleksandr Platoshyn, Silvia Marsala, Tomohisa Kato Jr, Naoki Takamura, Kenji Yoshida, Akiyoshi Kishino, Mariana Bravo-Hernandez, Stefan Juhas, Jana Juhasova, Hana Studenovska, Vladimir Proks, Joseph D. Ciacci and Martin Marsala in Cell Transplantation
Supplemental material, sj-jpg-4-cll-10.1177_09636897231163232 for Derivation of Sendai-Virus-Reprogrammed Human iPSCs-Neuronal Precursors: In Vitro and In Vivo Post-grafting Safety Characterization by Michiko Shigyo, Yoshiomi Kobayashi, Oleksandr Platoshyn, Silvia Marsala, Tomohisa Kato Jr, Naoki Takamura, Kenji Yoshida, Akiyoshi Kishino, Mariana Bravo-Hernandez, Stefan Juhas, Jana Juhasova, Hana Studenovska, Vladimir Proks, Joseph D. Ciacci and Martin Marsala in Cell Transplantation
Footnotes
Author Contributions: M.S., T.K., N.T., K.Y., and A.K. performed in vitro iPSC-NPCs generation, expansion, and characterization. O.P. performed calcium imaging. Y.K. performed in vivo cell grating, tissue immunofluorescence staining, and image analysis. S.M. and M.B.-H. were the project managers and supervised immunofluorescence in vitro and animal tissue staining. S.J., J.J., H.S., V.P., J.D.C., and M.M. designed the study and prepared the manuscript. All authors contributed to the final editing and approval of the manuscript.
Availability of Data and Materials: All requests for raw and analyzed data and materials are promptly reviewed by the University of California, San Diego (Material Transfer office), to verify whether the request is subject to any intellectual property or confidentiality obligations. Any data and materials that can be shared will be released via a Material Transfer Agreement.
Ethical Approval: This study was approved by our institutional review board.
Statement of Human and Animal Rights: This article does not contain any studies with human subjects.
Research Ethics and Statement of Informed Consent: All animal studies were approved by the University of California, San Diego, Institutional Animal Care and Use Committee (Protocol Number: S01193). There are no human subjects in this article and informed consent is not applicable.
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: M. Marsala is the scientific founder of Neurgain Technologies, Inc., and has an equity interest in the company. In addition, he serves as a consultant to Neurgain Technologies, Inc., and receives compensation for these services. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its policies on conflict of interests. N. Takamura, K. Yoshida, and A. Kishino are employed by Sumitomo Dainippon Pharma Co., Ltd., Kobe, Japan. All other authors declare that no competing interests exist.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by Sanford Porcine Center, UCSD.
ORCID iD: Martin Marsala  https://orcid.org/0000-0001-5048-6422
https://orcid.org/0000-0001-5048-6422
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72. [DOI] [PubMed] [Google Scholar]
- 2. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–20. [DOI] [PubMed] [Google Scholar]
- 3. Doss MX, Sachinidis A. Current challenges of iPSC-based disease modeling and therapeutic implications. Cells. 2019;8(5):403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rowe RG, Daley GQ. Induced pluripotent stem cells in disease modelling and drug discovery. Nat Rev Genet. 2019;20(7):377–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elitt MS, Barbar L, Tesar PJ. Drug screening for human genetic diseases using iPSC models. Hum Mol Genet. 2018;27(R2):R89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, Hefferan MP, Van Gorp S, Nazor KL, Boscolo FS, Carson CT, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482(7384):216–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okubo T, Nagoshi N, Kohyama J, Tsuji O, Shinozaki M, Shibata S, Kase Y, Matsumoto M, Nakamura M, Okano H. Treatment with a gamma-secretase inhibitor promotes functional recovery in human iPSC-derived transplants for chronic spinal cord injury. Stem Cell Reports. 2018;11(6):1416–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin GQ, He XF, Liang FY, Guo Y, Sunnassee G, Chen J, Cao XM, Chen YY, Pan GJ, Pei Z, Tan S. Transplanted human neural precursor cells integrate into the host neural circuit and ameliorate neurological deficits in a mouse model of traumatic brain injury. Neurosci Lett. 2018;674:11–17. [DOI] [PubMed] [Google Scholar]
- 9. Hicks AU, Lappalainen RS, Narkilahti S, Suuronen R, Corbett D, Sivenius J, Hovatta O, Jolkkonen J. Transplantation of human embryonic stem cell-derived neural precursor cells and enriched environment after cortical stroke in rats: cell survival and functional recovery. Eur J Neurosci. 2009;29(3):562–74. [DOI] [PubMed] [Google Scholar]
- 10. Bühnemann C, Scholz A, Bernreuther C, Malik CY, Braun H, Schachner M, Reymann KG, Dihné M. Neuronal differentiation of transplanted embryonic stem cell-derived precursors in stroke lesions of adult rats. Brain. 2006;129(Pt 12):3238–48. [DOI] [PubMed] [Google Scholar]
- 11. Daadi MM, Lee SH, Arac A, Grueter BA, Bhatnagar R, Maag AL, Schaar B, Malenka RC, Palmer TD, Steinberg GK. Functional engraftment of the medial ganglionic eminence cells in experimental stroke model. Cell Transplant. 2009;18(7):815–26. [DOI] [PubMed] [Google Scholar]
- 12. Ben-Menachem-Zidon O, Ben-Menahem Y, Ben-Hur T, Yirmiya R. Intra-hippocampal transplantation of neural precursor cells with transgenic over-expression of IL-1 receptor antagonist rescues memory and neurogenesis impairments in an Alzheimer’s disease model. Neuropsychopharmacology. 2014;39(2):401–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blurton-Jones M, Kitazawa M, Martinez-Coria H, Castello NA, Muller FJ, Loring JF, Yamasaki TR, Poon WW, Green KN, LaFerla FM. Neural stem cells improve cognition via BDNF in a transgenic model of Alzheimer disease. Proc Natl Acad Sci U S A. 2009;106(32):13594–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li X, Zhu H, Sun X, Zuo F, Lei J, Wang Z, Bao X, Wang R. Human neural stem cell transplantation rescues cognitive defects in APP/PS1 model of Alzheimer’s disease by enhancing neuronal connectivity and metabolic activity. Front Aging Neurosci. 2016;8:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hefferan MP, Galik J, Kakinohana O, Sekerkova G, Santucci C, Marsala S, Navarro R, Hruska-Plochan M, Johe K, Feldman E, Cleveland DW, et al. Human neural stem cell replacement therapy for amyotrophic lateral sclerosis by spinal transplantation. PLoS One. 2012;7(8):e42614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Feo D, Merlini A, Laterza C, Martino G. Neural stem cell transplantation in central nervous system disorders: from cell replacement to neuroprotection. Curr Opin Neurol. 2012;25(3):322–33. [DOI] [PubMed] [Google Scholar]
- 17. Åkesson E, Sundström E. Human neural progenitor cells in central nervous system lesions. Best Pract Res Clin Obstet Gynaecol. 2016;31:69–81. [DOI] [PubMed] [Google Scholar]
- 18. Cossetti C, Alfaro-Cervello C, Donegà M, Tyzack G, Pluchino S. New perspectives of tissue remodelling with neural stem and progenitor cell-based therapies. Cell Tissue Res. 2012;349(1):321–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trueman RC, Klein A, Lindgren HS, Lelos MJ, Dunnett SB. Repair of the CNS using endogenous and transplanted neural stem cells. Curr Top Behav Neurosci. 2013;15:357–98. [DOI] [PubMed] [Google Scholar]
- 20. Chohan MO, Moore H. Interneuron progenitor transplantation to treat CNS dysfunction. Front Neural Circuits. 2016;10:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karumbayaram S, Lee P, Azghadi SF, Cooper AR, Patterson M, Kohn DB, Pyle A, Clark A, Byrne J, Zack JA, Plath K, et al. From skin biopsy to neurons through a pluripotent intermediate under good manufacturing practice protocols. Stem Cells Transl Med. 2012;1(1):36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Y, Nguyen HV, Tsang SH. Skin biopsy and patient-specific stem cell lines. Methods Mol Biol. 2016;1353:77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Agu CA, Soares FA, Alderton A, Patel M, Ansari R, Patel S, Forrest S, Yang F, Lineham J, Vallier L, Kirton CM. Successful generation of human induced pluripotent stem cell lines from blood samples held at room temperature for up to 48 hr. Stem Cell Reports. 2015;5(4):660–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ohmine S, Dietz AB, Deeds MC, Hartjes KA, Miller DR, Thatava T, Sakuma T, Kudva YC, Ikeda Y. Induced pluripotent stem cells from GMP-grade hematopoietic progenitor cells and mononuclear myeloid cells. Stem Cell Res Ther. 2011;2(6):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. [DOI] [PubMed] [Google Scholar]
- 26. Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–17. [DOI] [PubMed] [Google Scholar]
- 27. Giorgetti A, Montserrat N, Aasen T, Gonzalez F, Rodriguez-Piza I, Vassena R, Raya A, Boue S, Barrero MJ, Corbella BA, Torrabadella M, et al. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell. 2009;5(4):353–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ye Z, Zhan H, Mali P, Dowey S, Williams DM, Jang YY, Dang CV, Spivak JL, Moliterno AR, Cheng L. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009;114(27):5473–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haridhasapavalan KK, Borgohain MP, Dey C, Saha B, Narayan G, Kumar S, Thummer RP. An insight into non-integrative gene delivery approaches to generate transgene-free induced pluripotent stem cells. Gene. 2019;686:146–59. [DOI] [PubMed] [Google Scholar]
- 30. Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85(8):348–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Churko JM, Burridge PW, Wu JC. Generation of human iPSCs from human peripheral blood mononuclear cells using non-integrative Sendai virus in chemically defined conditions. Methods Mol Biol. 2013;1036:81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Soares FA, Pedersen RA, Vallier L. Generation of human induced pluripotent stem cells from peripheral blood mononuclear cells using sendai virus. Methods Mol Biol. 2016;1357:23–31. [DOI] [PubMed] [Google Scholar]
- 33. Komuta Y, Ishii T, Kaneda M, Ueda Y, Miyamoto K, Toyoda M, Umezawa A, Seko Y. In vitro transdifferentiation of human peripheral blood mononuclear cells to photoreceptor-like cells. Biol Open. 2016;5(6):709–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Riedel M, Jou CJ, Lai S, Lux RL, Moreno AP, Spitzer KW, Christians E, Tristani-Firouzi M, Benjamin IJ. Functional and pharmacological analysis of cardiomyocytes differentiated from human peripheral blood mononuclear-derived pluripotent stem cells. Stem Cell Reports. 2014;3(1):131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nakagawa M, Taniguchi Y, Senda S, Takizawa N, Ichisaka T, Asano K, Morizane A, Doi D, Takahashi J, Nishizawa M, Yoshida Y, et al. A novel efficient feeder-free culture system for the derivation of human induced pluripotent stem cells. Sci Rep. 2014;4:3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Strnadel J, Carromeu C, Bardy C, Navarro M, Platoshyn O, Glud AN, Marsala S, Kafka J, Miyanohara A, Kato T, Jr, Tadokoro T, et al. Survival of syngeneic and allogeneic iPSC-derived neural precursors after spinal grafting in minipigs. Sci Transl Med. 2018;10(440):eaam6651. [DOI] [PubMed] [Google Scholar]
- 37. Bohaciakova D, Hruska-Plochan M, Tsunemoto R, Gifford WD, Driscoll SP, Glenn TD, Wu S, Marsala S, Navarro M, Tadokoro T, Juhas S, et al. A scalable solution for isolating human multipotent clinical-grade neural stem cells from ES precursors. Stem Cell Res Ther. 2019;10(1):83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pruszak J, Sonntag KC, Aung MH, Sanchez-Pernaute R, Isacson O. Markers and methods for cell sorting of human embryonic stem cell-derived neural cell populations. Stem Cells. 2007;25(9):2257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pruszak J, Ludwig W, Blak A, Alavian K, Isacson O. CD15, CD24, and CD29 define a surface biomarker code for neural lineage differentiation of stem cells. Stem Cells. 2009;27(12):2928–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sundberg M, Jansson L, Ketolainen J, Pihlajamäki H, Suuronen R, Skottman H, Inzunza J, Hovatta O, Narkilahti S. CD marker expression profiles of human embryonic stem cells and their neural derivatives, determined using flow-cytometric analysis, reveal a novel CD marker for exclusion of pluripotent stem cells. Stem Cell Res. 2009;2(2):113–24. [DOI] [PubMed] [Google Scholar]
- 41. Peh GS, Lang RJ, Pera MF, Hawes SM. CD133 expression by neural progenitors derived from human embryonic stem cells and its use for their prospective isolation. Stem Cells Dev. 2009;18(2):269–82. [DOI] [PubMed] [Google Scholar]
- 42. Golebiewska A, Atkinson SP, Lako M, Armstrong L. Epigenetic landscaping during hESC differentiation to neural cells. Stem Cells. 2009;27(6):1298–308. [DOI] [PubMed] [Google Scholar]
- 43. Yuan SH, Martin J, Elia J, Flippin J, Paramban RI, Hefferan MP, Vidal JG, Mu Y, Killian RL, Israel MA, Emre N, et al. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS One. 2011;6(3):e17540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trujillo CA, Gao R, Negraes PD, Gu J, Buchanan J, Preissl S, Wang A, Wu W, Haddad GG, Chaim IA, et al. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell. 2019;25(4):558–569.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eura N, Matsui TK, Luginbühl J, Matsubayashi M, Nanaura H, Shiota T, Kinugawa K, Iguchi N, Kiriyama T, Zheng C, Kouno T, et al. Brainstem organoids from human pluripotent stem cells. Front Neurosci. 2020;14:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Svendsen CN, ter Borg MG, Armstrong RJ, Rosser AE, Chandran S, Ostenfeld T, Caldwell MA. A new method for the rapid and long term growth of human neural precursor cells. J Neurosci Methods. 1998;85(2):141–52. [DOI] [PubMed] [Google Scholar]
- 47. Mazzini L, Gelati M, Profico DC, Sgaravizzi G, Projetti Pensi M, Muzi G, Ricciolini C, Rota Nodari L, Carletti S, Giorgi C, Spera C, et al. Human neural stem cell transplantation in ALS: initial results from a phase I trial. J Transl Med. 2015;13:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Riley J, Federici T, Polak M, Kelly C, Glass J, Raore B, Taub J, Kesner V, Feldman EL, Boulis NM. Intraspinal stem cell transplantation in amyotrophic lateral sclerosis: a phase I safety trial, technical note, and lumbar safety outcomes. Neurosurgery. 2012;71(2):405–416; discussion 416. [DOI] [PubMed] [Google Scholar]
- 49. Curtis E, Martin JR, Gabel B, Sidhu N, Rzesiewicz TK, Mandeville R, Van Gorp S, Leerink M, Tadokoro T, Marsala S, Jamieson C, et al. A first-in-human, phase I study of neural stem cell transplantation for chronic spinal cord injury. Cell Stem Cell. 2018;22(6):941–950.e6. [DOI] [PubMed] [Google Scholar]
- 50. Johe KK, Hazel TG, Muller T, Dugich-Djordjevic MM, McKay RD. Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes Dev. 1996;10(24):3129–40. [DOI] [PubMed] [Google Scholar]
- 51. Keirstead HS, Nistor G, Bernal G, Totoiu M, Cloutier F, Sharp K, Steward O. Human embryonic stem cell-derived oligodendrocyte progenitor cell transplants remyelinate and restore locomotion after spinal cord injury. J Neurosci. 2005;25(19):4694–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kumamaru H, Lu P, Rosenzweig ES, Kadoya K, Tuszynski MH. Regenerating corticospinal axons innervate phenotypically appropriate neurons within neural stem cell grafts. Cell Rep. 2019;26(9):2329–2339.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Usvald D, Vodicka P, Hlucilova J, Prochazka R, Motlik J, Kuchorova K, Johe K, Marsala S, Scadeng M, Kakinohana O, Navarro R, et al. Analysis of dosing regimen and reproducibility of intraspinal grafting of human spinal stem cells in immunosuppressed minipigs. Cell Transplant. 2010;19(9):1103–22. [DOI] [PubMed] [Google Scholar]
- 54. Kakinohana O, Juhasova J, Juhas S, Motlik J, Platoshyn O, Galik J, Hefferan M, Yuan SH, Vidal JG, Carson CT, van Gorp S, et al. Survival and differentiation of human embryonic stem cell-derived neural precursors grafted spinally in spinal ischemia-injured rats or in naive immunosuppressed minipigs: a qualitative and quantitative study. Cell Transplant. 2012;21(12):2603–19. [DOI] [PubMed] [Google Scholar]
- 55. van Gorp S, Leerink M, Kakinohana O, Platoshyn O, Santucci C, Galik J, Joosten EA, Hruska-Plochan M, Goldberg D, Marsala S, Johe K, et al. Amelioration of motor/sensory dysfunction and spasticity in a rat model of acute lumbar spinal cord injury by human neural stem cell transplantation. Stem Cell Res Ther. 2013;4(3):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sevc J, Goldberg D, van Gorp S, Leerink M, Juhas S, Juhasova J, Marsala S, Hruska-Plochan M, Hefferan MP, Motlik J, Rypacek F, et al. Effective long-term immunosuppression in rats by subcutaneously implanted sustained-release tacrolimus pellet: effect on spinally grafted human neural precursor survival. Exp Neurol. 2013;248:85–99. [DOI] [PubMed] [Google Scholar]
- 57. Kobayashi Y, Okada Y, Itakura G, Iwai H, Nishimura S, Yasuda A, Nori S, Hikishima K, Konomi T, Fujiyoshi K, Tsuji O, et al. Pre-evaluated safe human iPSC-derived neural stem cells promote functional recovery after spinal cord injury in common marmoset without tumorigenicity. PLoS One. 2012;7(12):e52787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nori S, Okada Y, Yasuda A, Tsuji O, Takahashi Y, Kobayashi Y, Fujiyoshi K, Koike M, Uchiyama Y, Ikeda E, Toyama Y, et al. Grafted human-induced pluripotent stem-cell-derived neurospheres promote motor functional recovery after spinal cord injury in mice. Proc Natl Acad Sci U S A. 2011;108(40):16825–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Marsala M, Kakinohana O, Yaksh TL, Tomori Z, Marsala S, Cizkova D. Spinal implantation of hNT neurons and neuronal precursors: graft survival and functional effects in rats with ischemic spastic paraplegia. Eur J Neurosci. 2004;20(9):2401–14. [DOI] [PubMed] [Google Scholar]
- 60. Cizkova D, Kakinohana O, Kucharova K, Marsala S, Johe K, Hazel T, Hefferan MP, Marsala M. Functional recovery in rats with ischemic paraplegia after spinal grafting of human spinal stem cells. Neuroscience. 2007;147(2):546–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-jpg-1-cll-10.1177_09636897231163232 for Derivation of Sendai-Virus-Reprogrammed Human iPSCs-Neuronal Precursors: In Vitro and In Vivo Post-grafting Safety Characterization by Michiko Shigyo, Yoshiomi Kobayashi, Oleksandr Platoshyn, Silvia Marsala, Tomohisa Kato Jr, Naoki Takamura, Kenji Yoshida, Akiyoshi Kishino, Mariana Bravo-Hernandez, Stefan Juhas, Jana Juhasova, Hana Studenovska, Vladimir Proks, Joseph D. Ciacci and Martin Marsala in Cell Transplantation
Supplemental material, sj-jpg-2-cll-10.1177_09636897231163232 for Derivation of Sendai-Virus-Reprogrammed Human iPSCs-Neuronal Precursors: In Vitro and In Vivo Post-grafting Safety Characterization by Michiko Shigyo, Yoshiomi Kobayashi, Oleksandr Platoshyn, Silvia Marsala, Tomohisa Kato Jr, Naoki Takamura, Kenji Yoshida, Akiyoshi Kishino, Mariana Bravo-Hernandez, Stefan Juhas, Jana Juhasova, Hana Studenovska, Vladimir Proks, Joseph D. Ciacci and Martin Marsala in Cell Transplantation
Supplemental material, sj-jpg-3-cll-10.1177_09636897231163232 for Derivation of Sendai-Virus-Reprogrammed Human iPSCs-Neuronal Precursors: In Vitro and In Vivo Post-grafting Safety Characterization by Michiko Shigyo, Yoshiomi Kobayashi, Oleksandr Platoshyn, Silvia Marsala, Tomohisa Kato Jr, Naoki Takamura, Kenji Yoshida, Akiyoshi Kishino, Mariana Bravo-Hernandez, Stefan Juhas, Jana Juhasova, Hana Studenovska, Vladimir Proks, Joseph D. Ciacci and Martin Marsala in Cell Transplantation
Supplemental material, sj-jpg-4-cll-10.1177_09636897231163232 for Derivation of Sendai-Virus-Reprogrammed Human iPSCs-Neuronal Precursors: In Vitro and In Vivo Post-grafting Safety Characterization by Michiko Shigyo, Yoshiomi Kobayashi, Oleksandr Platoshyn, Silvia Marsala, Tomohisa Kato Jr, Naoki Takamura, Kenji Yoshida, Akiyoshi Kishino, Mariana Bravo-Hernandez, Stefan Juhas, Jana Juhasova, Hana Studenovska, Vladimir Proks, Joseph D. Ciacci and Martin Marsala in Cell Transplantation