Abstract
Background
To assess the radiosensitivity of liver tumors harboring different genetic mutations, mouse liver tumors were generated in vivo through the hydrodynamic injection of clustered regularly interspaced short palindromic repeat/caspase 9 (CRISPR/Cas9) constructs encoding single-guide RNAs (sgRNAs) targeting Tp53, Pten, Nf1, Nf2, Tsc2, Cdkn2a, or Rb1.
Methods
The plasmid vectors were delivered to the liver of adult C57BL/6 mice via hydrodynamic tail vein injection. The vectors were injected into 10 mice in each group. Organoids were generated from mouse liver tumors. The radiation response of the organoids was assessed using an ATP cell viability assay.
Results
The mean survival period of mice injected with vectors targeting Nf2 (4.8 months) was lower than that of other mice. Hematoxylin and eosin staining, immunohistochemical (IHC) staining, and target sequencing analyses revealed that mouse liver tumors harbored the expected mutations. Tumor organoids were established from mouse liver tumors. Histological evaluation revealed marked morphological similarities between the mouse liver tumors and the generated tumor organoids. Moreover, IHC staining indicated that the parental tumor protein expression pattern was maintained in the organoids. The results of the ATP cell viability assay revealed that the tumor organoids with mutated Nf2 were more resistant to high-dose radiation than those with other gene mutations.
Conclusions
This study developed a radiation response assessment system for mouse tumors with mutant target genes using CRISPR/Cas9 and organoids. The Tp53 and Pten double mutation in combination with the Nf2 mutation increased the radiation resistance of tumors. The system used in this study can aid in elucidating the mechanism underlying differential intrinsic radiation sensitivity of individual tumors.
Keywords: CRISPR-Cas systems, liver, organoids, radiation tolerance, tumor
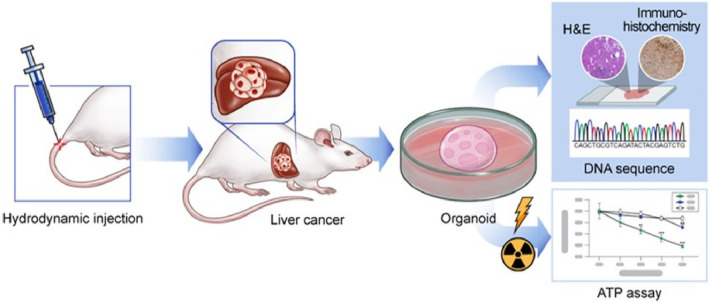
Graphical Abstract.
This is a visual representation of the abstract.
Introduction
More than 900,000 cases of liver cancer were reported in 2020. Liver cancer is the sixth most common cancer worldwide and the second leading cause of cancer-related mortality.1
Various local treatments (including surgery, trans-arterial chemoembolization (TACE), and radiation therapy) and systemic therapies have been developed and clinically applied for patients with liver cancer. However, limited studies have used tumor models targeting specific gene mutations.
Clustered regularly interspaced short palindromic repeat-caspase 9 (CRISPR-Cas9) is a useful gene editing and genetic screening tool.2–5 CRISPR library screening has enabled the identification of various oncogenes and tumor suppressors in different types of cancer.5–7 However, the role of tumor suppressor genes in accelerating the formation of liver cancer in different genetic backgrounds is not clear. Depending on the genetic background, mutations in tumor suppressor genes may differentially affect the progression of liver tumors. TP53 is the most frequently mutated gene in human cancers. The liver-specific knockout of Pten promotes lipid accumulation and induces late-onset liver cancer in mice.8,9 Mutations in TP53 and PTEN have been frequently reported in several types of human cancers, including human cholangiocarcinoma.10 Additionally, the double mutation of Tp53 and Pten generated via hydrodynamic injection of the CRISPR-Cas9 system can cause liver tumors in the mouse model.11 In this study, mouse models were generated to determine the effect of mutations in tumor suppressor genes on the acceleration of liver tumor formation in Tp53−/−; Pten−/− genetic background.
Organoid technology, a state-of-the-art cell culture tool, has revolutionized research on development, regeneration, and disease. The term “organoid” was previously used to refer to various three-dimensional culture systems similar to organs modeled to varying degrees.12,13 Huch et al.14 developed a culture system of leucine-rich repeat-containing G protein-coupled receptor 5+ (Lgr5+) murine bile duct stem cell-derived organoids that can grow indefinitely in vitro. The authors generated clonal hepatocytes from human epithelial cell adhesion molecules + (EpCAM+) cells around the adult bile duct.15 The aim of organoid research is to describe the developmental process and the organization of pathological organs in vitro to develop and test treatment options. The potential utilization of organoids in various clinical applications, such as disease modeling, regenerative medicine, drug discovery, and personalized medicine has piqued the interest of the scientific community. Several studies have demonstrated the application of organoids in anticancer drug sensitivity tests or radiation sensitivity tests.16,17
A method to analyze the genotype of clinical tumor samples and establish a radiotherapy strategy can be a new direction for personalized cancer treatment. Recently, personalized treatment based on the individual characteristics of carcinoma has become a basic consideration in cancer treatment. The identification of the type and rate of mutated genes in the cancer tissues is a shortcut to personalized medicine.
This study identified specific gene mutations that cause an aggressive liver cancer phenotype using the CRISPR/Cas9 system and evaluated their effects on radiation sensitivity using organoids derived from the generated mouse liver tumor.
Materials and Methods
Experimental Mice
Male C57BL/6 (aged 8-9 weeks) were purchased from Central Laboratory Animal Incorporated (Seoul, Korea). All animal protocols were approved by the Institutional Review Board (AEC-2017-009, AEC 2018-012). Reporting of this study conforms to ARRIVE 2.0 guidelines.18 The animal care protocols were based on the Guide for the Care and Use of Laboratory Animals, eighth edition.19
Construction of Plasmids
To generate the CRISPR/Cas9 constructs, the px458 (Addgene #48138) plasmid that transiently expresses single-guide RNAs (sgRNAs) and wild-type SpCas9 was used. The sgTp53 sequence was inserted into the px458 plasmid and linearized using BbsI. The U6 promoter and the sgPten sequence were polymerase chain reaction (PCR)-amplified and inserted into the px458 plasmid encoding Tp53 sgRNA. Next, the U6 promoter and sgRNAs targeting Nf1, Nf2, Tsc2, Cdkn2a, or Rb1 were sequentially PCR-amplified and inserted into px458 encoding sgRNAs targeting Tp53 and Pten. All primers used for cloning are listed in Supplementary Table 1.
Generation of Mouse Liver Tumor Using CRISPR/Cas9
The CRISPR/Cas9 construct was delivered to the liver of adult C57BL/6 mice using hydrodynamic tail vein injection, which can deliver DNA to hepatocytes for transient expression.20 Plasmid constructs were extracted using the NucleoBond Xtra Maxi-EF kit (MN). The plasmids (60 μg) were prepared and diluted in 2 mL saline. The solution was hydrodynamically injected through the lateral tail vein using a 31-gauge syringe in 5–7 s.21 Plasmids with Tp53 + Pten, Tp53 + Pten + Nf1, Tp53 + Pten + Nf2, Tp53 + Pten + Tsc2, Tp53 + Pten + Cdkn2a, or Tp53 + Pten + Rb1 sgRNAs were injected into 9–10 mice in each group. Mice were raised in a clean room to observe the development of liver tumors. The number of mice required in each experimental group was determined in a small-scale preliminary study and was based on the number of animals that died during the experiment and the minimum number of groups required to obtain the research data. Alfaxalone (60 mg/kg bodyweight) and xylazine (10 mg/kg bodyweight) were used as anesthetics for pain relief during euthanization. After the intraperitoneal injection, euthanasia was performed via asphyxiation using carbon dioxide gas.22
Survival Analysis for the Cancer Genome Atlas (TCGA) Data
Survival analysis and graph preparations were performed using the R (version 4.2.1) package “survival.”23,24 Mice with Tp53 + Pten, Tp53 + Pten + Nf1, Tp53 + Pten + Nf2, Tp53 + Pten + Tsc2, Tp53 + Pten + Cdkn2a, or Tp53 + Pten + Rb1 mutations were subjected to survival analysis. Clinical information on all tumor types was collected from TCGA. The retrieved information was filtered according to TP53 and PTEN double mutation. The data on the expression levels of NF1, NF2, TSC2, CDKN2A, and RB1 were collected from the cBioportal database. Survival analysis was performed according to each target gene by filtering only clinical information under low expression conditions.
Histological Evaluation and Immunohistochemistry
To perform histological analysis, all the tumor tissues were fixed with 4% phosphate-buffered formalin, embedded into paraffin, and sectioned into 4-μm thick sections. The sections were stained with hematoxylin and eosin and imaged using a light microscope (Nikon Eclipse 80i; Nikon Corporation; Tokyo, Japan). The images were analyzed using J program software (NIH, Bethesda, MD, USA).
To perform immunohistochemical analyses, the tumor tissues were embedded in paraffin and sectioned into 4-μm thick sections. The tissue sections were blocked with goat serum for 1 h and incubated overnight with primary monoclonal antibodies against Tp53 (1:100, Santa Cruz Biotechnology, CA, #SC-126), Pten (1:200, Santa Cruz Biotechnology, #SC-7974), Nf2 (1:150; Santa Cruz Biotechnology, #SC-331), or Yap1 (1:50; Abcam, Cambridge, MA, #ab52771). Next, the sections were incubated with horseradish peroxidase-conjugated secondary antibodies using a diaminobenzidine substrate kit (Dako, Glostrup, Denmark). All sections were counterstained with Mayer's hematoxylin. The histological characteristics of the tumor tissues were examined using an optical microscope (Nikon Eclipse 80i; Nikon Corporation, Tokyo, Japan).
Organoid Culture
For each liver sample, biopsies were obtained from non-tumorous or tumor tissues. The non-tumorous and tumor tissues were washed with Hank's balanced salt solution (HBSS; STEMCELL Technologies) and minced using sterile scalpel blades. The minced tissues were transferred into 50-mL conical tubes containing 30 mL of HBSS, inverted 10 times, and centrifuged at 350 g for 5 min (this step was repeated twice for washing). The tissues were resuspended in 8 mL of digestion buffer (7.2 mL HBSS, 800 µL collagenase P, and 80 µL DNAse I) at 230 rpm and 37 °C for 40 min. The digestion was stopped with the addition of 10 mL HBSS. The samples were then centrifuged at 350 g for 5 min. The dissociated tissues were resuspended with 10 mL of HBSS and passed through a 70-µm cell strainer (Corning). Dissociated cell clusters (small pieces of tissues) were centrifuged. The pellet was resuspended in 100% Matrigel (Corning) and plated in a 120 μL drop in the middle of one well of a pre-coated 12-well plate (Corning). To allow solidification, the drop was incubated at 37 °C and 5% CO2 for 2 min. Next, 1 mL of the organoid culture isolation medium was added to the well for the initial 3–4 days, and the medium was replaced with the organoid culture expansion medium. During culture, the medium was replaced once every 3 days. Organoids were passaged at a split ratio of 1:3 every 7–10 days. The organoid isolation medium comprises AdDMEM/F12 (Thermo Scientific, with 10 mM HEPES, 1× GlutaMax, and 1× penicillin-streptomycin) plus 2 nM Wnt surrogate–Fc Fusion protein (ImmunoPrecise), 25 ng/mL hNoggin (Peprotech), 5% RSPO1 conditioned medium (home-made), 1× B27, 50 ng/mL epidermal growth factor (Peprotech), 1 mM N-acetylcysteine (Sigma), 10 nM gastrin (Sigma), 50 ng/mL hepatocyte growth factor (Peprotech), 100 ng/mL FGF10 (Peprotech), 10 mM nicotinamide (Sigma), and 10 mM Rho inhibitor g-27632 (Calbiochem). After expansion, organoid isolation medium without 2 nM Wnt surrogate–Fc fusion protein (ImmunoPrecise) and 25 ng/mL hNoggin (Peprotech) was used. For tumoroid culture, the organoid expansion medium with 2 µM Nutlin-3 (Sigma) was used.
Targeted Deep Sequencing
Targeted deep sequencing was performed as previously described.25 Briefly, genomic DNA from mouse liver organoids was isolated using the Wizard® genomic DNA purification kit (Promega). Genomic DNA was PCR-amplified using Phusion polymerase (New England Biolabs) with primers spanning the target sequence from approximately 75 bp upstream to 75 bp downstream of the cleavage site of CRISPR-Cas9. For the first round of PCR amplification, 100 ng of genomic DNA from each sample was used as a template. Meanwhile, for the second round of PCR amplification, 20 ng of purified PCR products from the first round of PCR amplification were annealed with both Illumina adapter and barcode sequences. The primers used for PCR are listed in Supplementary Table S1. The resulting products were isolated, purified, mixed, and subjected to 150 paired-end sequencing using the HiSeq system (Illumina). Deep sequencing data were sorted and analyzed with a reference wild-type sequence (no indel) using Cas-Analyzer26 (http://www.rgenome.net/cas-analyzer) with the comparison range (R) parameter, minimum frequency (n), and wild-type (WT) marker range (r) set to 50, 1, and 5, respectively. To increase accuracy, the extracted data were filtered based on the read number. The read numbers of WT sequences (same target sequence but in different contexts because of sequencing errors) were combined as a single WT sequence.
Cell Viability Assay After Irradiation
The dome organoid was embedded and dislodged by pipetting with 1 mL organoid harvest solution (R&D Systems). The contents of the wells were transferred to a 15-mL tube and incubated on ice for 30 min. The samples were incubated with the same volume of phosphate-buffered saline and centrifuged at 1200 rpm for 5 min. The supernatant was aspirated and resuspended in pre-warmed TrypLE for 5 min with gentle pipetting to obtain a single-cell suspension. The single-cell suspension was transferred to microtubes (five groups for irradiation with 0, 1, 2, 4, and 8 Gy). Irradiation was performed using an electron beam from a 6 MeV C-band linear accelerator.27 The irradiated cells were centrifuged, and the supernatant was aspirated. The pellets were mixed with Matrigel and plated in a 120 μL drop in the middle of one well of a pre-coated 12-well plate (Corning) with organoid expansion media with or without Nutlin-3 for 7 days. Cell viability was assayed using CellTiter-Glo 3D (Promega), following the manufacturer's instructions, after 7 days of irradiation. Data analyses were performed using the GraphPad Prism 8 software.
Statistical Analyses
Data are presented as mean ± standard error. Cell viability assay data were analyzed using multiple t-tests to examine the effect of each radiation dose without assuming a consistent standard deviation. Statistical analyses were performed using GraphPad Prism 8 software. Differences were considered significant at P < 0.05.
Results
Generation of Mouse Liver Tumor Model
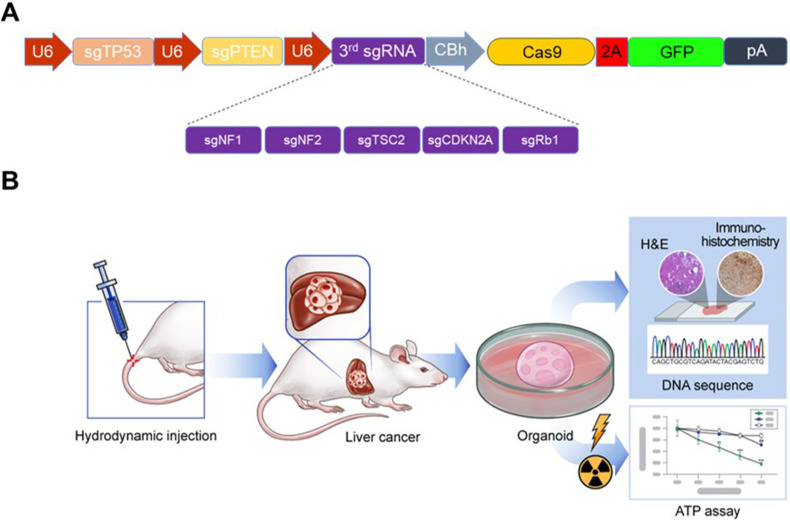
To investigate the effects of gene mutations, additional gene mutations were introduced in the Tp53 and Pten double mutation background. Tp53 and Pten double mutation is reported to induce liver tumors in approximately 4 months.7 In this study, five known tumor suppressor genes (Nf1, Nf2, Tsc2, Cdkn2a, and Rb1) were selected.5,6,28 Nf1, Nf2, and Tsc2 were selected as they have been identified as liver tumor suppressor genes through genome-wide ex vivo CRISPR screening in the Tp53−/− and Myc overexpression genetic background.6 Cdkn2a and Rb1, which are frequently mutated in human cancer, were selected as CDKN2A and RB1 mutations co-occur with TP53 mutation in human hepatocellular carcinoma (HCC) and/or bile duct cancer.28 To simultaneously introduce mutations in three genes, one vector system expressing three sgRNAs (sgTp53, sgPten, and sgRNA against target gene) was generated (Fig. 1A). The CRISPR-Cas9 construct was delivered into mice via hydrodynamic injection. After the generation of the liver tumor mouse model, liver tumor organoids were cultured and used for irradiation experiments to evaluate radiation sensitivity (Fig. 1B).
Figure 1.
DNA constructs and overview of the experimental setup. (A) Schematic map of the plasmid expressing three single-guide RNAs (sgRNAs) simultaneously. The sgRNA is expressed under the control of the U6 promoter, while the expression of spCas9 and enhanced green fluorescent protein is driven by the CBh (chicken beta-actin hybrid) promoter. A third sgRNA was introduced to target Nf1, Nf2, Tsc2, Cdkn2a, or Rb1 along with the first and second sgRNAs to target Tp53 and Pten, respectively. P2A, 2A self-cleaving peptide; pA; poly A signal. (B) Schematic representation of the experimental setup. After the establishment of the mouse tumor model through hydrodynamic tail vein injection of the clustered regularly interspaced short palindromic repeat-caspase 9 (CRISPR-Cas9) constructs, organoids were prepared from the generated mouse liver tumor to analyze their sensitivity to radiation.
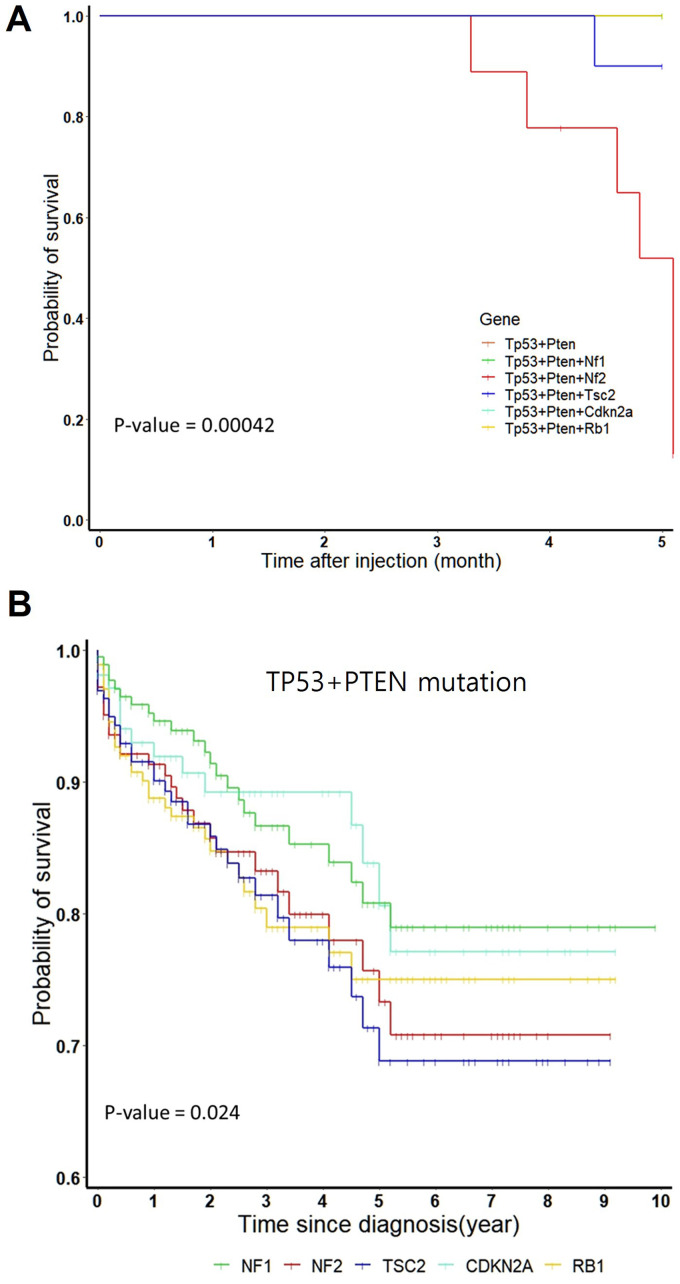
Survival Analysis for Mouse Model and Human Clinical Data
The mouse survival period varied depending on the mutated genes in the Tp53 and Pten double mutation background. Most mice with Tp53 and Pten double mutation survived for 6 months post-hydrodynamic injection, whereas mice with Tp53, Pten, and Nf2 triple mutation began to die 3 months post-hydrodynamic injection. Meanwhile, mice with Tp53, Pten, and Tsc2 triple mutation began to die 4 months post-hydrodynamic injection (Fig. 2A).
Figure 2.
Survival rates of mouse models and human cohort from the cancer genome atlas (TCGA) datasets. (A) Survival plots of mouse models with Nf1, Nf2, Tsc2, Cdkn2a, or Rb1 mutations in the Tp53 and Pten double mutation background. Significant differences in the survival rates between the six groups were examined using the log-rank test (P = 0.00042). Survival rate was significantly low in the Tp53 + Pten + Nf2 (n = 10) and Tp53 + Pten + Tsc2 (n = 10) groups. The Tp53 + Pten (n = 9), Tp53 + Pten + Nf1 (n = 10), Tp53 + Pten + Cdkn2a (n = 10), and Tp53 + Pten + Rb1 (n = 10) groups exhibited the same survival rate with overlapping survival curves. (B) Survival plots for human clinical data with low expression levels of five genes (NF1, NF2, TSC2, CDKN2A, or RB1) in the cohort with TP53 and PTEN double mutation background retrieved from TCGA database. Significance differences in the survival rates between the six groups were assessed using the log-rank test (P = 0.024, PTEN + TP53 group not shown).
Human Data Correlation
To determine if the effects of mutations in five target genes (Nf1, Nf2, Tsc2, Cdkn2a, and Rb1) on tumor formation in mice are similar to those in humans, human clinical data were analyzed. In total, 10967 human clinical datasets with TP53 and PTEN mutations were retrieved from TCGA database. In the TP53 and PTEN mutation genetic background, the number of samples with mutations in five genes (NF1, NF2, TSC2, CDKN2A, and RB1) was not sufficient for statistical analysis. Hence, the transcriptional data of the five genes were analyzed. The number of samples with downregulated expression of NF1, NF2, TSC2, CDKN2A, and RB1 were 203, 174, 203, 123, and 206, respectively. As shown in Fig. 2B, the survival rate in the groups with low NF2 or TSC2 expression was lower than that in other groups. These findings indicate a concordance in survival rates between mouse models and human clinical data.
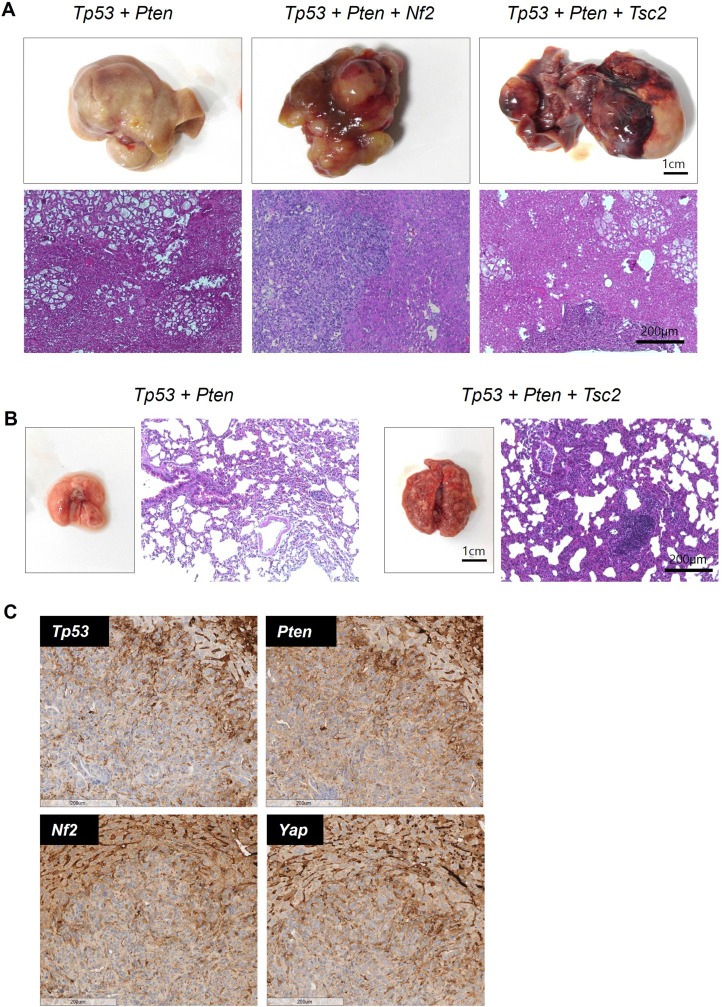
Tumor Phenotype and Immunohistochemical Staining According to Liver and Lung Metastases
Mice, including those with lung metastases in the group in which liver tumors were established, were subjected to gross morphology and tissue analyses. Tumor formation was observed in all groups. This is consistent with the results of previous studies, which reported that Tp53 and Pten double mutation induces liver tumors in approximately 4 months.20 Nf2 or Tsc2 mutation accelerated tumor formation in the Tp53 and Pten double mutation background (Fig. 2A). Therefore, liver tumor formation was further investigated in these two groups. The phenotypes of mouse liver and lung metastatic tumors (scale; 1 cm) according to each gene mutation (Tp53 + Pten, Tp53 + Pten + Nf2, and Tp53 + Pten + Tsc2) are shown in Fig. 3A–B. The differential histological characteristics between the tumor and the surrounding non-tumor tissues were evaluated by subjecting the tissue sections (scale; 200 μm) to hematoxylin and eosin staining. The tumor tissue exhibited irregular infiltration in an undifferentiated state. Nf2 can regulate the Hippo pathway and modulate the expression level of YAP1. Hence, this study examined Nf2 mutation using immunohistochemical staining. Immunohistochemical staining revealed that the tumor exhibited an Nf2-negative phenotype when compared with the surrounding healthy liver stromal tissue (Fig. 3C).
Figure 3.
Representative images of tumors with different gene mutations. (A) Gross morphology, nodule phenotypes, and hematoxylin and eosin staining of tumors with Tp53 + Pten, Tp53 + Pten + Nf2, and Tp53 + Pten + Tsc2 mutations. (B) Gross morphology, nodule phenotypes, and hematoxylin and eosin staining of metastatic tumors with Tp53 + Pten and Tp53 + Pten + Tsc2 mutations. (C) Immunohistochemical analyses of Tp53, Pten, Nf2, and Yap1. Staining of other stromal cells in the liver served as a positive control.
Establishment of Clonal Tp53 + Pten and Tp53 + Pten + Nf2 Knockout Mouse Liver Organoid Lines
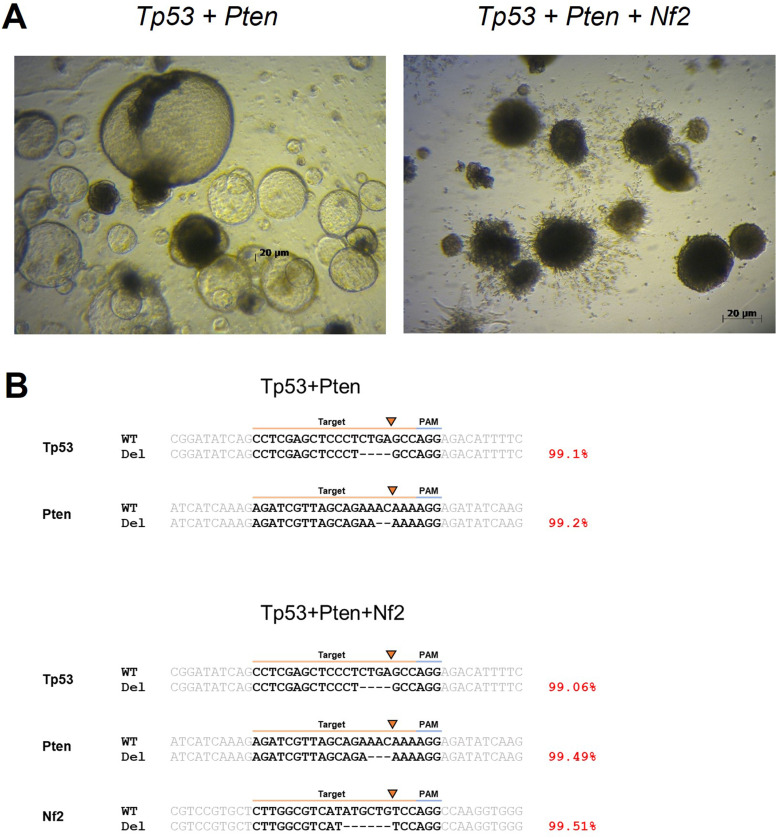
Tissue samples were obtained from surgically resected and histologically diagnosed non-tumorous and tumorous liver tissues. The culture was initiated using the same organoid medium for 4 days. After several days, the cultures were exposed to Nutlin-3 to select organoids with the mutant Tp53 + Pten and Tp53 + Pten + Nf2 phenotypes. Figure 4A shows the representative images of non-tumorous and Tp53 + Pten and Tp53 + Pten + Nf2 mutant organoids. These organoid lines exhibited differential morphological phenotypes. Non-tumorous and Pten + Tp53 mutant organoids exhibited a cystic structure, whereas Tp53 + Pten + Nf2 mutant organoids exhibited a compact morphology with no lumen. Targeted deep sequencing was performed to analyze the indel frequencies of candidate genes using CRISPR-Cas9 in single-cell-mediated mouse liver organoids. The CRISPR-Cas9 construct was highly mutated near the cut site of the spacer (average 99.27% mutation). Consistently, DNA sequencing confirmed that organoids with Nf2 mutation were well established, whereas organoids with Tsc2 mutation were not well established.
Figure 4.
Representative images of organoids. (A) Mouse liver organoids with Tp53 + Pten and Tp53 + Pten + Nf2 mutations after plating. Scale bar: 20 µm (B) Sequence analysis of representative indel patterns induced by clustered regularly interspaced short palindromic repeat-caspase 9 (CRISPR-Cas9) constructs. For each gene, the wild-type sequence is shown at the top containing the context sequence (indicated with gray text) and target sites (indicated with black text and highlighted in orange line). The PAM sequence is highlighted with a blue line. Deletions are shown as black dashes. The arrow indicates the cut site of the CRISPR-Cas9 construct. The mutation frequencies are shown on the right.
Organoids with Mutation Exhibited Differential Sensitivity to Radiation
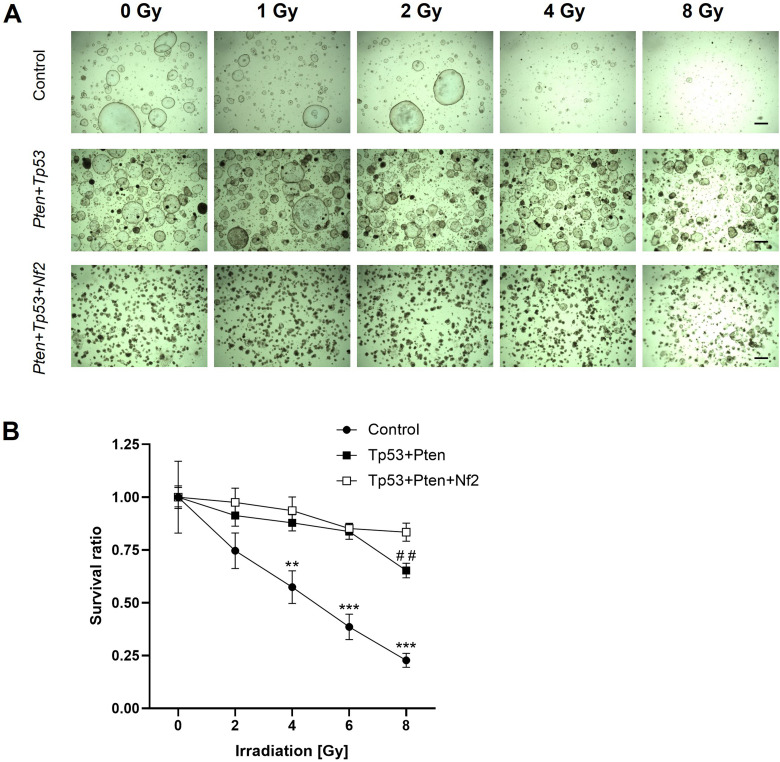
To evaluate the response of the organoids to radiation, the organoids were irradiated with the indicated doses for 7 days. The cell survival rate was examined using CellTiter-Glo reagent. As shown in the bright-field images and cell survival rate results (Fig. 5A–B), radiation dose-dependently exerted cytotoxic effects on non-tumorous organoids. In contrast, radiation did not exert cytotoxic effects on Tp53 + Pten Tp53 + Pten + Nf2 mutant organoids at all tested doses, except at 8 Gy. These data indicated that patients with Tp53/Pten/Nf2 mutation exhibit resistance to radiation therapy. In the non-irradiated group, organoid formation rate and size in the Tp53 + Pten + Nf2 group were lower than those in the Tp53 + Pten group.
Figure 5.
Organoids exhibit differential sensitivity to radiation. (A) Representative bright-field images of the organoids exposed to radiation at the indicated doses for 7 days are shown. Scale bar: 20 µm (B) Cell survival rate was recorded at day 7 post-irradiation. This experiment was repeated two times in triplicate. The results are expressed as mean ± standard error. **P < 0.01, ***P < 0.001 (control vs Tp53 + Pten or Tp53 + Pten + Nf2) and ##P < 0.01 (Tp53 + Pten vs Tp53 + Pten + Nf2). This experiment was repeated two times in triplicate.
Discussion
Tumor organoids are an important model for optimizing treatment strategies and examining the therapeutic mechanisms against cancer resistance. This is because mutated models can be generated using tumor organoids by focusing on a subset of the genetic characteristics of cancer.29 In this study, mouse liver cancer and its organoids were generated by introducing driver mutations in specific genes using the CRISPR/Cas9 gene editing system. Additionally, the effect of these mutations on radiation resistance was investigated.30 Cell viability assay revealed that the tumor organoid model with Nf2 mutation in the Tp53 and Pten double knockout background exhibited enhanced radiation resistance. This may be because the morphology of the Nf2−/− cancer organoid was more compact than that of other organoids. The compact shape may be one of the factors contributing to the differential response to radiation.
Pten and Tp53 suppress stress factors associated with the tumorigenesis of liver cancer through distinct mechanisms. The simultaneous loss of both nuclear PTEN and TP53 promotes oxidative stress in hepatocytes, leading to the development of HCC and intrahepatic cholangiocarcinoma.31 Additionally, the deletion of Nf2 in mouse HCC is known to modulate EGFR signaling and Hippo-dependent pathways and is functionally associated with the development of HCC.32
As HCC exhibits extremely diverse molecular characteristics, models with specific gene mutations can serve as research models that can consistently predict treatment responses. In particular, the comparative analysis of radiation resistance of cancer organoid models with and without Nf2 mutation can aid in predicting radiation resistance by introducing additional mutations in a specific gene, as well as in determining optimal treatment. Identifying genetic mutations through tissue and genomic analysis of HCC can aid in establishing various personalized treatment strategies, including radiation therapy.33
The effects of mutated tumor suppressor genes may vary depending on the genetic background. Tp53−/−; Myc-overexpressing cells form tumors slowly but the inactivation of additional tumor suppressors accelerates tumor formation.6 In the Tp53−/−; Myc overexpression genetic background, CRISPR-Cas9 screening revealed that Nf1, Nf2, Tsc2, and other genes accelerate tumor formation. Additionally, the findings of experiments with the Tp53−/−; Myc overexpression; Nf1−/− mouse model, which was established via hydrodynamic injection, confirmed significant acceleration of liver tumor formation in mice.6 However, in this study, liver tumor formation was accelerated in mice with Nf2 and Tsc2 mutations but not in those with Nf1 mutation in the Tp53−/−; Pten−/− genetic background. This may be attributed to the epistatic network of interacting tumor suppressor genes. Zhao et al reported that TP53 cooperated with NF2 and/or PTEN synergistically in breast cancer.7 TCGA clinical dataset analysis indicated that NF2 mutation in the TP53−/−; PTEN−/− genetic background accelerated tumor formation and affected overall survival. A combination of mouse model and human clinical data analysis is useful for studying the effects of tumor suppressor genes on tumor formation in different genetic backgrounds.
A recent study treated a rare type of primary HCC using patient-derived tumor organoids. The case was classified as HCC-neuroendocrine carcinoma and the best anticancer treatment option was suggested based on the establishment of organoids from the patient-derived tissues within 3 weeks after performing tissue and genetic analysis of HCC.34 Additionally, one study examined hepatitis B virus (HBV)-induced HCC using a liver organoid model. Customized and precise treatment was possible through analysis of genetic variations associated with the occurrence of liver cancer in HBV-infected patients.35
The establishment of non-tumorous liver organoids and tumoroids from biopsies is advantageous because the models can be established at diagnosis, during treatment, and at the time of recurrence from minute amounts of biopsy material. The in vitro sensitivity of tumors to radiation was assayed in this study (Figs. 4 and 5). These assays could provide a rationale for testing new therapies that are demonstrated to be effective in vitro and in vivo using the tumor tissue of the patient as an embedded source of information. Further studies are needed to establish the usefulness of this approach, including a prospective evaluation of the concordance between patient and tumor responses to standard therapies, including chemotherapy and radiation therapy. Additionally, after confirmation with a large sample size, an ex vivo radiation platform could be used to assess patient responses before initiating radiation treatment, which will potentially avoid the toxic side effects of radiation in patients with radiation-resistant tumors.36
Accumulation of these data will contribute to devising various drug treatments, including immunotherapy targeting specific genes, through the elucidation of signaling pathways involved in the pathogenesis of HCC. The concept used in this study will aid in the elucidation of factors related to the differential intrinsic radiation sensitivity of individual tumors.
This study is associated with some limitations. First, this study involved a mouse tumor model and a small sample size. Hence, the applicability of the findings of mouse studies to human cancer models is limited. Additionally, the tumor organoids generated in this study were confirmed using DNA sequencing. The NF2 mutant organoids exhibited good growth. However, the TSC2 mutant organoid model could not be established and hence its sensitivity to irradiation could not be determined.
Conclusions
In this study, mouse liver tumors were generated using the CRISPR/Cas9 gene editing system. Additionally, the effects of individual tumor suppressor gene mutations on the radiation sensitivity of tumors were examined. The results of this study will aid in determining patient-specific optimal treatment by examining the response to radiation, as well as in identifying the optimal drug treatment, in the future.
Supplemental Material
Supplemental material, sj-xlsx-1-tct-10.1177_15330338231165125 for Evaluation of Radiation Sensitivity Differences in Mouse Liver Tumor Organoids Using CRISPR/Cas9-Mediated Gene Mutation by Wan Jeon, Se Yeon Jung, Chae Young Lee, Won-Tae Kim, Hyun Kim, Kyoung Won Jang, Heuijin Lim, Manwoo Lee, Dong Hyeok Jeong, Sung Dae Kim, In Ah Kim, Si Ho Choi, Tae Gen Son and Kyung Su Kim in Technology in Cancer Research & Treatment
Acknowledgements
We would like to thank Editage (www.editage.co.kr) for English language editing.
Abbreviations
- ATP
adenosine triphosphate
- CDKN2A
cyclin dependent kinase inhibitor 2A
- EpCAM
epithelial cell adhesion molecules
- IHC
immunohistochemistry
- Lgr5
leucine-rich and repetitively-containing G protein-coupled receptor 5
- NF1
neurofibromin1
- NF2
neurofibromin2
- PCR
polymerase chain reaction
- PTEN
phosphatase and tensin homolog
- Rb1
retinoblastoma transcriptional corepressor 1
- sgRNA
single guide RNA
- TCGA
the cancer genomic atlas
- Tp53
tumor protein P53
- TSC2
tuberous Sclerosis Complex 2
- YAP
yes-associated protein.
- WT
wild type
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics: All experiment protocols were approved by Dongnam Institute of Radiological and Medical Sciences (DIRAMS AEC-2017-009, DIRAMS AEC-2018-012).
Date of Approval: 2017.03.15–2018.12.31
Funding: This work was supported by the Dongnam Institute of Radiological and Medical Sciences (DIRAMS) grant funded by the Korea government (MSIT) (50592-2022, 50591-2023, 50495-2022, 50493-2023). This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No.2021R1A2C1095736, No. 2022R1A2C1092928).
ORCID iD: Wan Jeon https://orcid.org/0000-0002-0849-8312
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. May 2021;71(3):209-249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. Feb 15 2013;339(6121):823-826. doi: 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. Nov 2013;8(11):2281-2308. doi: 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konermann S, Brigham MD, Trevino AE, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. Jan 29 2015;517(7536):583-588. doi: 10.1038/nature14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shalem O, Sanjana NE, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. Jan 3 2014;343(6166):84-87. doi: 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song CQ, Li Y, Mou H, et al. Genome-wide CRISPR screen identifies regulators of mitogen-activated protein kinase as suppressors of liver tumors in mice. Gastroenterology. 2017;152(5):1161-1173.e1. doi: 10.1053/j.gastro.2016.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao X, Li J, Liu Z, Powers S. Combinatorial CRISPR/Cas9 screening reveals epistatic networks of interacting tumor suppressor genes and therapeutic targets in human breast cancer. Cancer Res. Dec 15 2021;81(24):6090-6105. doi: 10.1158/0008-5472.CAN-21-2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldser DM, Kostova KK, Winslow MM, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. Nov 25 2010;468(7323):572-575. doi: 10.1038/nature09535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horie Y, Suzuki A, Kataoka E, et al. Hepatocyte-specific pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest. 2004;113(12):1774-1783. doi: 10.1172/JCI20513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ong CK, Subimerb C, Pairojkul C, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet. May 6 2012;44(6):690-693. doi: 10.1038/ng.2273 [DOI] [PubMed] [Google Scholar]
- 11.Xue Z, Wu M, Wen K, et al. CRISPR/Cas9 mediates efficient conditional mutagenesis in Drosophila. G3 (Bethesda). Sep 5 2014;4(11):2167-2173. doi: 10.1534/g3.114.014159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reza HA, Okabe R, Takebe T. Organoid transplant approaches for the liver. Transpl Int. Nov 2021;34(11):2031-2045. doi: 10.1111/tri.14128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prior N, Inacio P, Huch M. Liver organoids: From basic research to therapeutic applications. Gut. Dec 2019;68(12):2228-2237. doi: 10.1136/gutjnl-2019-319256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huch M, Dorrell C, Boj SF, et al. In vitro expansion of single Lgr5+liver stem cells induced by Wnt-driven regeneration. Nature. Feb 14 2013;494(7436):247-250. doi: 10.1038/nature11826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huch M, Gehart H, van Boxtel R, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. Jan 15 2015;160(1-2):299-312. doi: 10.1016/j.cell.2014.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vlachogiannis G, Hedayat S, Vatsiou A, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. Feb 23 2018;359(6378):920-926. doi: 10.1126/science.aao2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park M, Kwon J, Kong J, et al. A patient-derived organoid-based radiosensitivity model for the prediction of radiation responses in patients with rectal cancer. Cancers (Basel). Jul 27 2021;13(15). doi: 10.3390/cancers13153760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Percie du Sert N, Hurst V, Ahluwalia A, et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. Br J Pharmacol. Aug 2020;177(16):3617-3624. doi: 10.1111/bph.15193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Research Council Committee for the Update of the Guide for the C, Use of Laboratory A. The National Academies Collection: Reports funded by National Institutes of Health. Guide for the Care and Use of Laboratory Animals. National Academies Press (US) Copyright © 2011, National Academy of Sciences; 2011.
- 20.Xue W, Chen S, Yin H, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. Oct 16 2014;514(7522):380-384. doi: 10.1038/nature13589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An SB, Yang K, Kim CW, et al. Longitudinal imaging of liver cancer using MicroCT and nanoparticle contrast agents in CRISPR/Cas9-induced liver cancer mouse model. Technol Cancer Res Treat. Jan-Dec 2021;20:15330338211016466. doi: 10.1177/15330338211016466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navarro KL, Huss M, Smith JC, Sharp P, Marx JO, Pacharinsak C. Mouse anesthesia: The art and science. ILAR J. 2021;62(1-2):238-273. doi: 10.1093/ilar/ilab016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore DF. SpringerLink (Online service). Applied Survival Analysis Using R.XIV, 226 p. 66 illus., 26 illus. in color. 10.1007/978-3-319-31245-3. [DOI]
- 24.Therneau TM, Grambsch PM. Modeling survival data: Extending the Cox model. Statistics for biology and health. Springer; 2000:xiii, 350 p.
- 25.Jung SB, Lee CY, Lee KH, Heo K, Choi SH. A cleavage-based surrogate reporter for the evaluation of CRISPR-Cas9 cleavage efficiency. Nucleic Acids Res. Sep 7 2021;49(15):e85. doi: 10.1093/nar/gkab467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J, Lim K, Kim JS, Bae S. Cas-analyzer: An online tool for assessing genome editing results using NGS data. Bioinformatics. Jan 15 2017;33(2):286-288. doi: 10.1093/bioinformatics/btw561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim H, Jeong DH, Jang KW, et al. Implementation of ultra-high dose-rate electron beam from 6-MeV C-band linear accelerator for preclinical study. J Instrum. 2020;15(09):P09013. doi: 10.1088/1748-0221/15/09/P09013 [DOI] [Google Scholar]
- 28.Cancer Genome Atlas Research Network. Electronic address wbe, cancer genome atlas research N. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. Jun 15 2017;169(7):1327-1341.e23. doi: 10.1016/j.cell.2017.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu H, Jiao D, Liu A, Wu K. Tumor organoids: Applications in cancer modeling and potentials in precision medicine. J Hematol Oncol. May 12 2022;15(1):58. doi: 10.1186/s13045-022-01278-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramakrishna G, Babu PE, Singh R, Trehanpati N. Application of CRISPR-Cas9 based gene editing to study the pathogenesis of colon and liver cancer using organoids. Hepatol Int. Dec 2021;15(6):1309-1317. doi: 10.1007/s12072-021-10237-z [DOI] [PubMed] [Google Scholar]
- 31.Kato T, Murata D, Anders RA, Sesaki H, Iijima M. Nuclear PTEN and p53 suppress stress-induced liver cancer through distinct mechanisms. Biochem Biophys Res Commun. Apr 16 2021;549:83-90. doi: 10.1016/j.bbrc.2021.02.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urtasun R, Latasa MU, Demartis MI, et al. Connective tissue growth factor autocriny in human hepatocellular carcinoma: Oncogenic role and regulation by epidermal growth factor receptor/yes-associated protein-mediated activation. Hepatology. Dec 2011;54(6):2149-2158. doi: 10.1002/hep.24587 [DOI] [PubMed] [Google Scholar]
- 33.Nuciforo S, Fofana I, Matter MS, et al. Organoid models of human liver cancers derived from tumor needle biopsies. Cell Rep. Jul 31 2018;24(5):1363-1376. doi: 10.1016/j.celrep.2018.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meier MA, Nuciforo S, Coto-Llerena M, et al. Patient-derived tumor organoids for personalized medicine in a patient with rare hepatocellular carcinoma with neuroendocrine differentiation: A case report. Commun Med (Lond). 2022;2:80. doi: 10.1038/s43856-022-00150-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Crignis E, Hossain T, Romal S, et al. Application of human liver organoids as a patient-derived primary model for HBV infection and related hepatocellular carcinoma. Elife. Jul 30 2021;10. doi: 10.7554/eLife.60747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schrag D, Weiser MR, Goodman KA, et al. Neoadjuvant chemotherapy without routine use of radiation therapy for patients with locally advanced rectal cancer: A pilot trial. J Clin Oncol. 2014;32(6):513-518. doi: 10.1200/JCO.2013.51.7904 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xlsx-1-tct-10.1177_15330338231165125 for Evaluation of Radiation Sensitivity Differences in Mouse Liver Tumor Organoids Using CRISPR/Cas9-Mediated Gene Mutation by Wan Jeon, Se Yeon Jung, Chae Young Lee, Won-Tae Kim, Hyun Kim, Kyoung Won Jang, Heuijin Lim, Manwoo Lee, Dong Hyeok Jeong, Sung Dae Kim, In Ah Kim, Si Ho Choi, Tae Gen Son and Kyung Su Kim in Technology in Cancer Research & Treatment