Abstract
Addictions are heritable and unfold dynamically across the lifespan. One prominent neurobiological theory proposes that substance-induced changes in neural circuitry promote the progression of addiction. Genome-wide association studies have begun to characterize the polygenic architecture undergirding addiction liability and revealed that genetic loci associated with risk can be divided into those associated with a general broad spectrum liability to addiction and those associated with drug-specific addiction risk. In this Perspective, we integrate these genomic findings with our current understanding of the neurobiology of addiction to propose a new genetically informed neurobiology of addiction (GINA) model.
Introduction
The loss of life and socioeconomic costs associated with addictive substances burden the world1. There are vast individual differences in patterns of substance use, which range from casual or occasional to excessive and disordered. Addiction emerges when, following chronic regular use, the presence of a substance helps maintain homeostasis. Behaviorally and psychologically addiction typically results in the attenuation of reward elicited by initial substance use and the development of compulsive use [G] to ameliorate negative affect as well as psychological and physiological stress states and withdrawal symptoms that arise when a substance is absent. Increasing quantities, dosage, and potencies of substances are often pursued in addiction in an attempt to obtain the increasingly fleeting ‘highs’ experienced during initial use. Box 1 provides an outline of the correspondence between this definition of addiction with the definitions of substance use and substance use disorders (SUDs).
Box 1|. Clinical and genetic distinctions between substance use and addiction.
Not all substance use reflects or results in addiction (nor is addiction only relevant to psychotropic substances3,232–234, although here we restrict our discussion to substance addiction). In this article, we broadly define addiction as the stage at which the pleasurable aspects of substance use are attenuated and compulsive use emerges to ameliorate the negative affect and stress states that arise in the absence of the substance.
Various diagnostic schema define substance use disorders (SUDs) in a substance-specific manner as the syndromes that arise from excessive drug involvement accompanied by loss of control over use, use despite deleterious consequences, and impairment. For instance, according to the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5), a specific substance use disorder (such as opioid use disorder) can be diagnosed when an individual qualifies for 2 or more of 11 criteria that assess physiological, psychological and interpersonal problems235. The 11th edition of the International Classification of Diseases (ICD-11), on the other hand, includes separate entries for substance dependence, harmful patterns of use, and hazardous patterns of use236. Harmful or problematic substance use is also evaluated in healthcare settings using short screeners 237. Therefore, the definition of addiction that we use here approximates the DSM-5 diagnosis of a moderate or severe SUD or the ICD-11 diagnosis of harmful use and dependence.
Both propensity for substance use and risk of developing SUDs are heritable and genome-wide association studies (GWASs) provide insight into the extent and nature of the genetic influence on the use of and addiction to various substances50,54,46,47,238,50,239–241,81,242 (see the Table).
| Substance | Use or Use Disorder | Sample Size of Current Largest GWASs | SNP-heritability | Number of independent variants/genes identified | Genetic correlation between Use and Use Disorder | Reference |
|---|---|---|---|---|---|---|
| Alcohol | Alcohol Use Disorder/Problem Alcohol Use | 435,563 | 0.07 | 29 / 66 genes | 0.77 | 49,53,54 |
| Drinks/week | 941,280 | 0.04 | 99 / 362 genes | 47 | ||
| Tobacco/Nicotine | Nicotine Dependence | 58,000 | 0.09 | 5 / 16 genes | 0.4 – 0.5 | 238 |
| Ever smoked | 1,232,091 | 0.08 | 378 / 833 genes | 47 | ||
| Cannabis | Cannabis Use Disorder | 384, 925 | 0.12 | 2 / 3 genes | 0.50 | 46 |
| Ever used cannabis | 184,765 | 0.11 | 8 / 35 genes | 51 | ||
| Opioids | Opioid use disorder | 639, 709a | 0.13 | 10 / 4 genes | N/A | 48,53,239–241 |
| Cocaine | Cocaine use disorder | 6,546b | 0.30 | 1 / 5 genes | N/A | 81,242 |
Note: this GWAS of European and African ancestries has a case N = 20,858, but another recent GWAS reports a larger case N = 31,473. This second GWAS reports a smaller N overall (N = 425,944) but a slightly more diverse sample (including European Americans, African Americans, and Hispanic Americans)240,241
GWASs also suggest distinctions between substance use and SUDs. There is a moderate to high genetic correlation between substance use (encompassing ever using a substance and use in daily life) and problematic or disordered use46,50,53,54. However, substance use and SUDs differ in their genetic associations with other psychosocial factors and psychiatric and medical comorbidities. For instance, while problem drinking is genetically correlated with negative health outcomes, many studies document that alcohol consumption is genetically correlated with higher educational attainment and a lower risk for cardiometabolic disease and is not significantly related to genetic risk for other psychopathologies (reviewed in61). How often someone drinks is confounded with higher socio-economic advantage, thus biasing genetic correlations identified by GWASs that focus on measures of drinking in daily life60,62. Likewise, cannabis use and cannabis use disorder exhibit opposing correlations with educational attainment, body mass index, and intracranial volume, but both are genetically related to more serious psychiatric outcomes, such as schizophrenia and depression46,51. This difference is not as evident for nicotine, where both smoking initiation and nicotine dependence appear to be linked to a greater genetic risk for psychosocial disadvantage, and psychiatric and somatic illness47.
Translational neuroscience research has transformed our understanding of addiction and led to its re-characterization as a neurobiological state rather than a controllable moral failing2. A largely independent line of research has shown that the moderate-to-large heritability [G] of SUDs is undergirded by a polygenic [G] architecture that is associated with broad spectrum liability to addiction as well as distinct genetic architectures [G] associated with substance-specific risks3–5. This genetic risk is compounded by environmental factors that are substance-specific (such as policy-related access to specific substances) and associated with factors related to general psychiatric liability (such as socioeconomic status).
In this Perspective, we incorporate knowledge from human genetic studies of addiction into brain disease models. First, we provide an overview of the three-stage neurobiological model of addiction, which postulates that substance-induced neural changes are the predominant contributor to the etiology of SUDs. We then showcase how contemporary genetic work has begun to identify the polygenic architecture underlying addiction risk. Here, we highlight the utility of genetically-informed study designs to probe the plausibility of proposed models of addiction. Last, we integrate genetic research into an expanded version of the stage-based neurobiological model of addiction by proposing a new model: the genetically informed neurobiology of addiction (GINA) model. In this model, the interplay between genetic liability [G] and substance-induced changes in the neural substrates of positive reinforcement [G] (thought to drive binging [G], intoxication and escalating use), negative urgency [G] (thought to take place during withdrawal [G] and negative affect) and executive function [G] and/or regulatory capacity (important for the preoccupation with and/or withdrawal from substance use), as well as neural and peripheral substance-specific pathways, contribute to SUD development.
Brain-based models of addiction
According to the most prominent neurobiological model of addiction6–10, substance and/or experience-dependent alterations in cortiostriatal11 and corticolimbic12 circuitry (Fig. 1) drive three recurring and non-mutually exclusive stages of addiction: binge-intoxication, withdrawal-negative affect and preoccupation-anticipation.
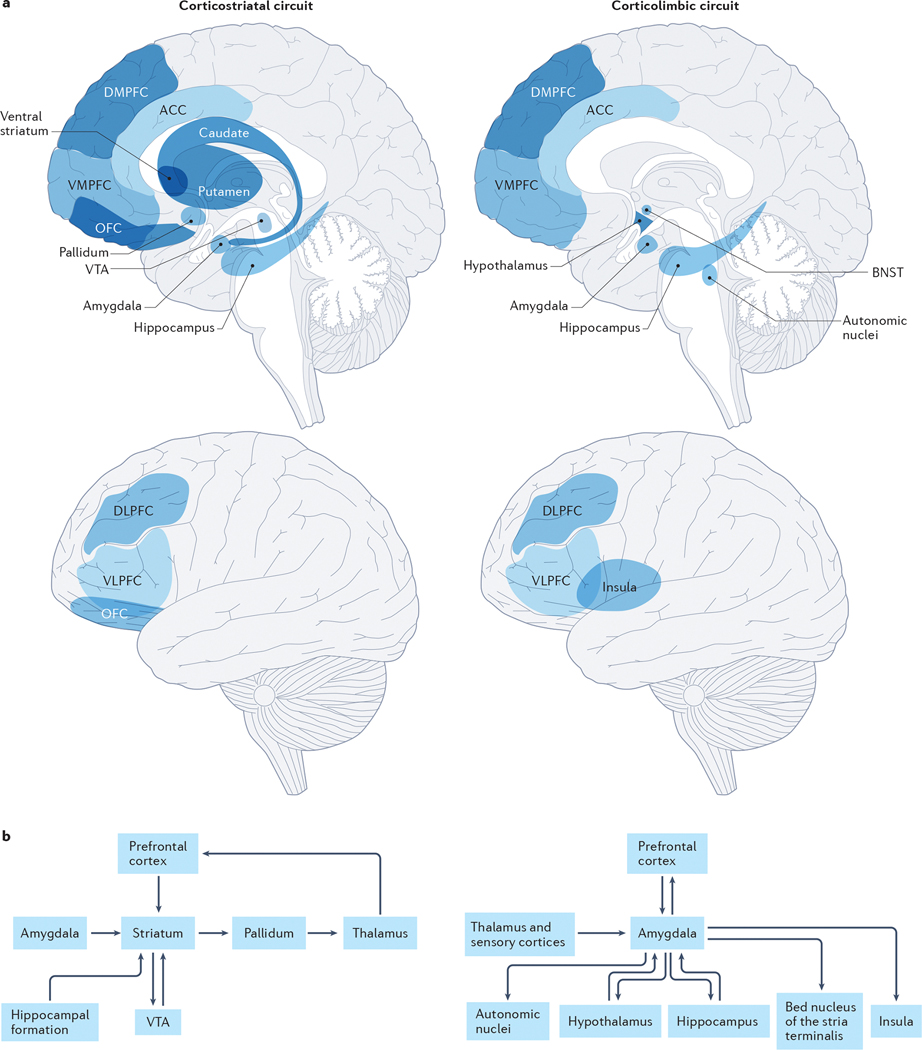
Fig 1: Corticostriatal and corticolimbic circuits underlying addiction.
Anatomical locations of (a) and connections between (b) the primary nodes within the corticostriatal and corticolimbic circuits that support reward, emotion, and their regulation and are proposed to influence the binge-intoxication, withdrawal-negative affect, and preoccupation-anticipation stages of addiction. The corticostriatal circuit is critical for reward processing and largely contributes to the binge-intoxication stage of addiction. The striatum (comprised of the putamen, caudate and ventral striatum) is the primary node of this network. Through its connections with other nodes, the striatum supports learning reward contingencies, hedonic responsiveness, generating motivation to pursue rewards and goals, forming and implementing plans to obtain reward, adjusting behavior and plans according to changing contingencies, and coordinating motor movements in the service of obtaining reward11. More specifically, dopaminergic projections from the ventral tegmental area to the nucleus accumbens within the ventral striatum support reward prediction and learning in combination with multimodal sensory information received from the basolateral amygdala and contextual information from the hippocampal formation. Projections from the striatum to the pallidum support hedonic responsiveness through endogenous opioid stimulation and provide motivational signals to the VMPFC (supporting integration of contextual and interoceptive information, bottom-up drive and top-down regulation) and the DLPFC (supporting goal-directed planning) through thalamic relays. Afferents from the PFC to the ventral striatum further serve to facilitate the implementation of plans to obtain reward (DLPFC) as well as flexible behavioral adjustment when expected actions do not obtain predicted outcomes (ACC) and can facilitate or inhibit the motivational significance of reward predictive cues in the environment. The corticolimbic circuit is critical for affective processing and behavioral vigilance; it largely contributes to the withdrawal-negative affect stage of addiction. The amygdala (inclusive of the amygdala and the extended amygdala) is the primary node of this network; through its connections with other nodes it supports responses to environmental challenges, including threat and stress, by generating and regulating emotional responses12. Low and high resolution sensory information arrives in the basolateral complex of the amygdala from the thalamus and sensory cortices, respectively. Efferent projections from the centromedial and extended amygdala, including the bed nucleus of the stria terminalis (BNST), to autonomic nuclei (such as the parabrachial nucleus), the hypothalamus and the hippocampus drive emotional responses, including fear conditioning and the generation of stress-related physiological changes. Direct and indirect connections between the amygdala and insula facilitate interoception (awareness and importance of our physiological states). Projections from the nucleus basalis of Meynert in the extended amygdala facilitate amygdala-driven arousal and sensitivity of the cortex. Projections from the amygdala to the VMPFC promote subjective awareness and evaluation of emotion and the integration of affective information (such as motivational information conveyed by the ventral striatum projections shown in the upper panel). Projections from the DLPFC and VLPFC to the amygdala through the DMPFC and VMPFC promote the regulation of affective responses and physiological arousal. Both the corticostriatal and corticolimbic circuits support executive function and the regulation of behavior to influence the preoccupation-anticipation stage of addiction by contributing to incentive salience (such as the ventral striatum projections to the VMPFC and OFC within the corticostriatal circuit), interoceptive signals associated with withdrawal physiology and affect (such as the insula within the corticolimbic circuit), as well as the regulation of behavior (through the DLPFC, VLPFC and ACC in both circuits). While there are many additional connections within and between these circuits, we present a heuristic model focusing on those most well linked to addiction. These circuits are explained in greater detail in prior publications11,12. Note: Unlike prior depictions of the stage-based neurobiological model which show 3 circuits corresponding to each stage, we present the corticostriatal and corticolimbic circuits, which are hypothesized to predominantly drive the binge-intoxication and withdrawal-negative affect stages, respectively. The preoccupation-anticipation stage is undergirded by prefrontal connections within and across these circuits in this model.
During the binge-intoxication stage of the neurobiological model of addiction, substance-induced stimulation of neural reward circuitry provides positive reinforcement. In support of this idea, all addictive substances have been shown to directly or indirectly elicit fast and large increases in dopamine (DA) release in the nucleus accumbens (NAc) 13,14 that resemble predictive reward signals [G] 15,16. Extrinsic cues (such as with whom, where and when one uses a substance) and intrinsic cues (such as mood and physiology) are quickly and strongly paired to drug-reward and become conditioned predictors that motivate substance use17,18. This stage exerts its largest influence on addiction during the initial escalating and episodic heavy use of a substance (particularly during adolescence and young adulthood), the re-emergent escalating use that follows abstinence, and the use of substance types with increasing psychoactive impact (higher doses or potency) following addiction progression.9,19,20
With continued heavy substance use and progression towards severe SUD, it is proposed that the reinforcing properties of substances shift to negative reinforcement [G] 8,9,21,22. This transition to the withdrawal-negative affect stage of addiction is marked by distress and anhedonia [G] 23,24,25, as well as by the aversive physiological states (such as blackouts, nausea and insomnia) and psychological states (such as anxiety, depression and heightened stress) that arise in the absence of a drug’s effects.9 During this stage, substance use is compulsive and functions to provide relief from these aversive states by returning the body to a state of homeostasis. Such homeostasis can now only be achieved when the drug is present10, because a series of neuroadaptations (such as fewer DA receptors and attenuated reward-related DA release)13 have taken place as natural adjustments to repeated drug exposure. These adaptations also promote anhedonia, through which other non-drug rewards (such as social interactions, achievement, food and sex) lose their reinforcing properties. Thus, increasing quantities of the addictive substance are required to reach homeostasis, with even greater amounts being required to ‘chase the highs’ that were associated with the binge-intoxication stage17. The chronic use of addictive substances also promotes stress and negative affect (through, for example, elevations of corticotropin-releasing hormone in the extended amygdala)26. During substance abstinence, withdrawal and related negative affect leads to heightened interoceptive salience, through which the physiological arousal associated with withdrawal and negative emotionality potentiates negative reinforcement-related craving [G] 10,27.
In the third stage of this model, the repeated pairing of drug use with reward and relief results in a cognitive preoccupation-anticipation of the drug in expectation of these effects. This is often characterized by the subjective experience of drug craving. Building on dual systems models that postulate that self-control arises when deliberative executive function counteracts more automatic emotionally driven behavior28,29, this stage of addiction is proposed to be defined by a loss of this regulatory capacity as a result of substance-induced impairment of top-down executive function (such as reduced prefrontal control over striatal and other limbic circuits)8,30,31. Thus, it is proposed that heavy and sustained use causes the prefrontal cortex to become less efficient at minimizing the direct incentive salience [G] evoked by substance cues and emergent negative emotionality, leaving individuals with less intrinsic ability to combat the throes of addiction, even if there are deep subjective desires to stop.
This model emphasizes how substance-induced neural changes contribute to the development and maintenance of moderate to severe SUDs. Given that drugs of abuse impact neurotransmitters and neuromodulators at a magnitude that does not intrinsically occur17,32, it might be assumed that resulting brain changes would be so penetrant that addiction would be a foregone conclusion in anyone with escalating levels of use. In reality, only some individuals exposed to addictive substances develop addiction33 and a significant minority of individuals with SUDs recover34. The neurobiological stage-based model of addiction therefore acknowledges that factors that influence an individual’s predisposition to addiction must be considered as major contributors to the disorder5,9.
The neurobiological stage-based model provides a framework of addiction susceptibility that is not substance-specific. While the concept of an addiction risk that is shared across substances is supported by evidence that SUDs are frequently comorbid with one another35 and share similar neural36, genetic37,38 and environmental39 correlates, emerging evidence reported in a recent preprint highlights substance-specific risk factors that take the form of variants in genes within substance-specific metabolic and signaling pathways37. Thus, we can hypothesize that predispositional liability to the addictive properties of a specific substance (or a non-substance-related behavior) may set the pace for transitions between the neurobiological stages of addiction, and intensify the subjective experience of each stage.
Developmental vulnerability [G] and trait-like vulnerability [G] have been highlighted by other neurobiological theories of addiction. For instance, arising from developmental psychology40, the neurodevelopmental model of addiction41 has theorized that adolescence and young adulthood confer broad spectrum addiction risk owing to typical patterns of brain maturation that initially prioritize emotional and social processing over cognitive control and regulation. This promotes risk-taking behavior42 as well as increased impulsive attempts to cope with negative emotion, placing adolescents and young adults at risk for both the positive and negative reinforcing aspects of SUD risk (particularly in the context of underdeveloped physiological tolerance and still-developing regulatory capacity). In this model, the typical earlier maturation of reward-related and stress and negative affect-related neural circuitry and relatively delayed prefrontal development are seen as addiction risk factors. It is speculated that substance use may more profoundly shape neural circuitry, and especially prefrontal development, during these periods of extensive neural maturation.
The neurodevelopmental and stage-based neurobiological models of addiction have unique origins and differentially weight predispositional liability [G] and substance-induced alterations. The neurobiological model arose primarily from data on functional differences in brain activity and receptor densities and emphasizes the role of substance-induced neural plasticity in the etiology of addiction. On the other hand, the neurodevelopmental model relies predominantly on emergent structural changes during adolescence and emphasizes predispositional developmental liability. However, both models highlight the contributions of impulsivity, negative affect, and executive function (and their neural substrates) in addiction vulnerability.
Over the past five years, we have witnessed immense progress in human genetics (reviewed below) that can shed further light on the mechanisms of addiction. With this in mind, it is now important to begin to measure and integrate predispositional genetic risk into brain disease models of addiction.
Genetics of addiction
Genetics of substance use and SUDs
Historically, the search for genetic variation underlying SUDs has focused on the genes encoding substance-specific neurotransmitters or metabolic enzymes (such as opioid or nicotinic receptors, alcohol dehydrogenase and cytochrome p450) and genes encoding proteins involved in canonical systems that have a widespread effect on psychiatrically relevant behaviors (such as dopaminergic and serotonergic receptors and transporters)43. As in studies of other complex phenotypes, studies were conducted on relatively modestly sized samples and single gene variants [G], genes, or haplotypes were examined, with exonic variation prioritized. However, the introduction of genome-wide association studies (GWASs) [G] to the field led to a transition from candidate gene [G] validation to genetic exploration, with some unexpected consequences.
For many psychiatric disorders (such as schizophrenia or major depression), the candidate genes that had been hypothesized to be involved in disease liability were found by GWASs to be no more likely to be associated with disease risk than those selected at random and, with a few exceptions, novel loci were associated with these disorders44,45. However, in the case of substance use and addiction, some of the strongest significant signals (in addition to novel loci) in GWASs were in candidate genes known to regulate metabolism (such as ADH1B for alcohol and CYP2A6 for nicotine), encode receptor binding sites (such as CHRNA5 for nicotine and OPRM1 for opioids)46–54 and implicated in addiction models (such as corticotropin-releasing factor receptor 1 (CRHR1))34,50,55. Recent comprehensive reviews of these GWAS findings are available5,43 (Box 1).
More generally, GWASs of complex traits (including substance use and SUDs) have revealed that, with a few exceptions (such as the effects of the single nucleotide polymorphism (SNP) [G] rs1229984 in ADH1B on alcohol use49), single genetic variants have small effect sizes and that these traits are highly polygenic49. Notably, while twin studies [G] suggest a heritability of ~50% for a range of addictions, GWASs and whole exome analyses56,57 can explain, at best, only a quarter of this heritability58, although the inclusion of less common variants does improve the heritability of some traits57. This discrepancy is typical of most complex traits; it is possible that some of this ‘missing heritability’ resides in rare variants that will be identified using sequencing technologies59. However, the high polygenicity of addictions also suggest that larger sample sizes may be required to identify additional novel common variants than is the case for other complex traits5.
Three other insights unique to SUDs are notable. First, while genetic correlations between liability to substance use (for example likelihood of ever using or frequency of use) and problematic or disordered use are moderate to high46,50, the genetics of disordered use faithfully reproduce a pattern of correlated medical comorbidities — both psychiatric and somatic — and potential indicators of negative life outcomes (such as lower education attainment) whereas the genetics of substance use has been related to adaptive psychosocial correlates and inconclusively linked with psychopathology (Box 1). Accordingly, the risk of substance use may represent a mixture of risk for future problems and resilience to them 46,49,60–62.
A second insight recapitulates prior evidence from twin studies that genetic liability for SUDs is largely shared across substances, but that there is also important substance-specific liability (Fig. 2)37,63. A recent preprint reports that the polygenic architecture underlying the general liability to SUDs includes loci that regulate dopaminergic signaling37, including signals linked to the D2 dopamine receptor (DRD2) and cAMP-specific 3’,5’-cyclic phosphodiesterase 4B (PDE4B) 37,50,64. As PDE4B is instrumental in neuroplasticity within prefrontal dopaminergic pathways and is associated with stress and negative affect in animal models65–67 it represents an intriguing locus for addiction that is consistent with the stage-based neurobiological model of addiction. Another recent GWAS identified genes contributing to shared liability to substance use, addictions and other related behaviors (such as a large number of sexual partners) 64. Genes identified as being associated with traits with externalizing features included cell adhesion molecule 2 (CADM2), which has also been linked to general liability to SUDs37 as well as to substance use, risky sexual behavior, self-control and obesity68,69. It is thus possible that CADM2 impacts addiction liability by influencing early risk-taking and self-control more broadly70 (as opposed to influencing addiction progression). Intriguingly, despite the identification of overlapping loci and a high genetic correlation between the externalizing GWAS64 and a factor identified in the recently-reported GWAS representing common addiction liability37, many novel loci are associated with the latter addiction factor, implying that addiction pathology is partially genetically distinct from general liability to externalizing behaviors37.
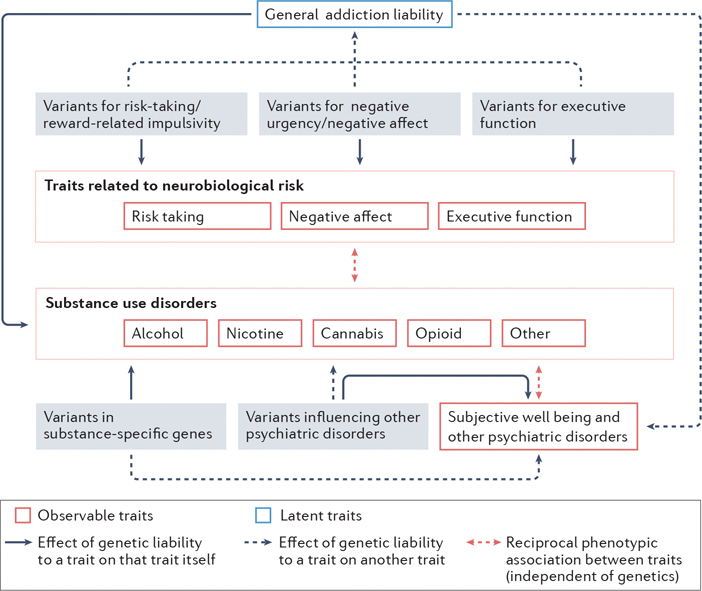
Fig. 2: The genomic architecture of substance use disorders.
The genetic contribution to individual substance use disorders (SUDs) is attributable to variants that influence general addiction liability and substance-specific variants63. General addiction liability is driven by variants influencing traits that correspond to the 3 stages of the neurobiological model of addiction: reward and risk-taking, negative affect and urgency, and executive functioning. In contrast, variants in receptors that respond to the psychoactive components of individual substances or those in genes metabolizing individual drugs directly influence each substance use disorder in a substance-specific manner. Furthermore, genetic variants that influence other psychiatric disorders may also independently influence SUDs (solid arrows indicate the effects of variants on a specific trait/phenotype, whereas the dashed lines indicate cross-trait effects). Reciprocally, the genetics underlying general addiction liability may impact other psychiatric disorder risk (gray dashed arrownot shown). Small effects of substance-specific genetic variants on other psychiatric disordersare also predicted (gray dashed arrow). In addition to these genetic pathways, prolonged substance use and SUDs may phenotypically influence risk-taking, negative affect, executive functioning, as well as psychiatric health and well-being (double headed dashed red arrows depict phenotypic associations). Note that alcohol, nicotine, cannabis, cocaine and opioid use disorders are shown, as there are current large genome-wide association studies of these SUDs; however, the genetics of many other SUDs could be similarly classified.
Third and finally, these GWASs have shown that, after the common genetic liability to addiction is taken into account, residual and substance-specific variation is often conferred by variants in genes encoding metabolic factors and substance-specific receptors37. This substance-specific variation is also polygenic; however the effect sizes of some individual variants are an order of magnitude larger than those of the variants that are common across addictions37,64
Correspondence between GWAS and molecular genetics
That addictions originate and induce perturbations in brain-based genetic pathways is supported by sources of genetic data in addition to GWASs. Many loci linked to addiction through GWASs have been shown to be expression quantitative trait loci (eQTL) [G] across developmental stages in tissue obtained from brain regions implicated in the three-stage neurobiological model of addiction46,71–75. For instance, variants linked to substance use were associated with gene expression and co-expression network modules in the NAc, anterior cingulate cortex (ACC) (Fig. 1), cerebellum, dorsolateral prefrontal cortex and other brain regions71–73. Gene co-expression patterns were preserved across these brain regions, supporting a generalized overall enrichment of these variants in the brain rather than in specific brain regions71. Beyond eQTL effects, heritability enrichment analyses of GWASs have revealed that the genetics of substance use and addiction liability is enriched for tissue- and cell-specific regulatory elements [G] specifically related to chromatin architecture76 (that is, DNA folding) in primary brain regions specified in the 3-stage neurobiological model of addiction. Genes linked to alcohol and tobacco use and problematic use also show higher expression in excitatory neurons in cortical and midbrain regions, as well as the hippocampus, thalamus and amygdala76. Thus, addiction-relevant GWAS have begun to reveal evidence that is convergent with the 3-stage neurobiological model of addiction and highlights the role of predisposition within this framework.
Drug exposure-induced transcriptomic changes have also been observed. In one study, human alcohol dependence GWAS-associated genes showed networks of co-expression in the prefrontal cortex, NAc and ventral tegmental area of ethanol-exposed mouse brains77. Another found cross-species conservation in gene expression changes associated with cocaine exposure within the hippocampus and ventral tegmental area that mapped to well-established addiction pathways (such as dopaminergic networks)78. These studies suggest that, just as the brain responds dynamically to repeated drug exposure, so does the transcriptome and epigenome. However, this genomic and neural plasticity may not be a consequence of drug exposure alone: novel studies that integrate GWAS signals with multi-omics data have suggested that a subset of the genes that are differentially expressed also show enriched genetic associations with substance use 79. In one study, genes that were differentially expressed in the dorsolateral prefrontal cortex (DLPFC) of individuals that were alcohol-dependent coalesced into two co-expression modules that included genes that were enriched in alcohol use and addiction GWASs80. In other instances, genes identified as having significant association with substance use in GWASs were not differentially expressed, but genes within their co-expression networks were81. Therefore, GWAS data in combination with multi-omics and cross-species findings can provide insights into the neural gene networks vulnerable to substance-induced modulation.
Brain imaging genetics
The conceptualization of brain structure and function as intermediate phenotypes, or endophenotypes82(which are hypothesized to lie between genes and/or environmental experiences and disease processes) generated enthusiasm that genetic research on neural phenotypes would help characterize genetic architecture underlying complex behavior, including addiction. For example, the high heritability of brain structure83 alongside its objective quantification, reliability and proximity to gene function were the basis for presuming that GWASs of brain structure would yield loci with large effects82. Meta-analyses of the GWASs of structural brain phenotypes found, instead, that brain imaging phenotypes are highly polygenic and complex and that their genetic correlation with behavioral outcomes is far more modest than hypothesized84–87. For example, initial genetic correlations (rG) estimated across GWASs of substance involvement (including use, problematic use and SUDs) and brain structure have revealed that, much like the genetic correlations between behavioral phenotypes and substance use phenotypes, the genetic correlations between substance use and/ or SUDs and brain structure are modest in magnitude.88,89,90. These data suggest that, while there is gene–brain–behavior genetic overlap, it will be difficult to characterize without large samples. Notably, these genetic correlations may be constrained by small effects of brain–behavior associations91.
Unraveling the genetic architecture of functional neuroimaging phenotypes (such as task-related activity or resting-state functional connectivity [G]) has been even more challenging. GWASs of task-related functional magnetic resonance imaging (t-fMRI) and resting state functional connectivity phenotypes in the UK Biobank have revealed low heritability and identified few loci86,87,92. This may be partially attributable to the low reliability of traditional t-fMRI93 and the need for large amounts of resting state functional connectivity data to facilitate its reliable measurement94. Thus, despite the prominence of functional neuroimaging studies of addiction95, their vanishingly low heritability, psychometric challenges and practical difficulties in harmonizing the findings of different studies have led to relatively few well-powered genetically-informed investigations
Predispositional and/or Causal?
As reviewed above, GWASs and transcriptomic studies of addiction-related phenotypes highlight the role of predisposition within the 3-stage neurobiological model as well as potential substance-induced changes in epigenetic structure and the transcriptomic landscape. By contrast, the vast majority of brain-imaging addiction-related science has interpreted cross-sectional associations between substance involvement and brain phenotypes to putatively reflect causative substance-induced brain alterations. Below we review evidence from longitudinal and genetically-informed designs that can be used to infer whether substance-related variability in brain structure may plausibly reflect predispositional risk and/or sequela of substance involvement. Much like GWAS, these data highlight the need to incorporate genetic predisposition into neurobiological models of addiction.
Longitudinal studies.
Longitudinal studies of substance involvement have revealed that changes in brain structure and function are associated with escalating substance use. For example, the National Consortium on Alcohol and Neurodevelopment in Adolescence (NCANDA)96 has found that heavy alcohol use in adolescence is associated with, and precedes, accelerated cortical gray matter decline, particularly in the medial and dorsal prefrontal cortices97, as well as a decline in white matter integrity 98. Similarly, in a cohort examined as part of the IMAGEN study 99, the initiation of cannabis use was associated with cortical thinning in the superior and anterior medial prefrontal cortex. This change occurred over a 5 year period among participants who were cannabis naïve at baseline (age 14) and in a dose dependent manner that was also associated with impulsivity100. Other longitudinal studies, however, have found evidence supportive of the predispositional model. For instance, reduced DLPFC volume in substance-naïve children was associated with an earlier age of drinking initiation when those children reached adolescence and an attenuation of the typical reduction in drinking that occurs in young adulthood 73. Similarly, two studies found that lower orbitofrontal cortex (OFC) volume in early adolescence preceded and was associated with a subsequent onset of cannabis use101,102 (but see 103).
Two considerations are noteworthy when interpreting longitudinal studies. First, adolescence is characterized by dynamic changes in brain development. As such, individual differences in the trajectories of brain development in youth who initiate substance use may reflect substance-induced changes and/or gradations of predispositional influences on neurodevelopment. While experimental evidence in non-human animals shows that heavy substance use can reduce markers of neurogenesis and brain growth104–107, other evidence suggests that neural development is significantly genetic in origin108–111. Therefore, disentangling the aspects of brain development that are attributable to genetic predisposition from those that are sequelae of substance use is challenging, especially when the genes contributing to brain development may exert pleiotropic effects [G] on substance involvement.
Second, while a dose-response relationship (in which the more one uses a substance, the stronger the association with brain metrics) may reflect causal effects112, it is also possible that those with preexisting neurobiological liabilities that manifest in altered neurodevelopmental trajectories in adolescence may be more vulnerable to escalating and disordered substance use112,113. Indeed, GWASs show that increasing severity of drug use is associated with a polygenic signal that is partially distinct from the genes influencing lighter or milder use and is more likely to exert pleiotropic effects on brain development (even when we make simplified assumptions that the effects are linear)114.
Genetic causal modeling.
Genomic data can be used to examine the plausibility of hypotheses of phenotypic causality. Because modest but significant genetic correlations have been demonstrated between psychiatric phenotypes and brain phenotypes46,49,90, the associations between variants at addiction-relevant loci and neural phenotypes have been partially attributed to the pleiotropic effects of these variants. However, if addiction is conceptualized as the escalation of drug exposure and the brain phenotype as the target of such prolonged exposure, then any effect of variants in drug-related loci on the brain may represent genetic causality (and this could be tested via Mendelian Randomization115; Fig. 3). Modern genetic causality approaches that account for the polygenic nature of SUDs can estimate the proportion of shared genetic liability or identify genetic variants that might be causal116. One such analysis found no support for a genetically causal effect of problematic alcohol use on brain structure phenotypes89. By contrast, and consistent with a predispositional model, it did provide support for the idea that differences in brain structure (including a lower volume of the basal forebrain and a greater volume of the pars opercularis) may plausibly contribute to problematic alcohol use.
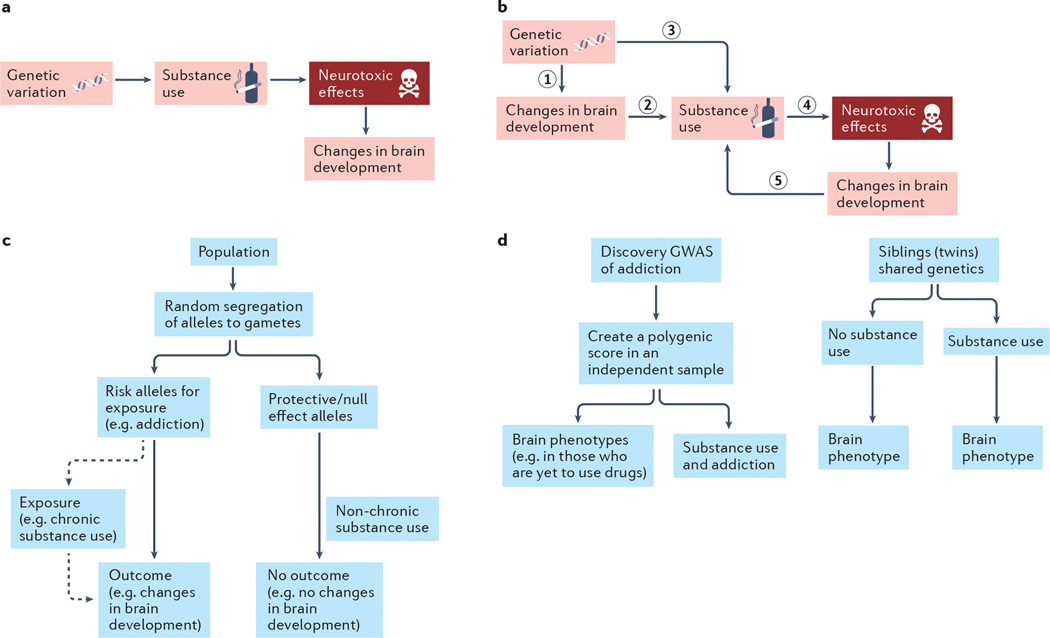
Fig. 3: Using genomics to validate hypotheses of addiction.
a| According to neurobiological models of addiction, genetic variation influences substance use, which may, in turn, exert neurotoxic effects that alter brain development. b| According to predispositional models of addiction, genetic risk for substance use disorders impacts brain development (1) prior to or concurrent with the onset of and escalating substance use and sets the neurobiological stage for substance use and future addiction (2). Consequent substance involvement (3) (also influenced by genetic risk that is not associated with neural phenotypes) may then causally influence the brain, via neurotoxic mechanisms, to further potentiate problematic substance use (4). Cyclically, these brain-related changes may further enhance risk for addiction progression (5). c| Mendelian randomization115 and other genetic causal methods can be used to evaluate these models. These approaches are based on the fact that parental genotypes conferring risk of exposure (i.e., chronic substance use) are equally as likely to be inherited by the offspring as genotypes that are protective or of no effect. Individuals inheriting risk alleles or polygenic risk of substance use will subsequently be more likely to use drugs; we can then test whether this chronic use causally alters brain development. In this method, the individual risk alleles or the polygenic risk of drug exposure is the genetic instrument and an independent association between this genetic instrument and the outcome (changes in brain development), as shown in the flow chart, is possible evidence for causal effects of substance exposure on the brain. The genetic instrument is assumed to influence the outcome (changes in brain development) solely via its influence on chronic substance use (dashed line). For a greater discussion of Mendelian Randomization approaches as well as their limitations see115. d| Testing the association between polygenic risk140 for addiction and brain imaging phenotypes, including trajectories, in drug-naïve individuals (left flow chart) is an ideal approach to assess whether pre-existing brain-related differences precede addiction. Here, the effects of genetic variants are taken from a discovery GWAS of addiction and applied to a sample, ideally of individuals without a history of substance use (e.g., children), which has brain data. A polygenic score is created in this new independent sample. It is expected that this polygenic score will eventually be associated with substance use and addiction in this sample. However, if it is also associated with brain phenotypes prior to use of substances, then we can infer that genetic risk that precedes onset of substance use contributes to brain development (part b, step 1) and later substance use (part b, step.3), rather than a causal effect of substance use on the brain alone (part b, step.4). Alternatively, examining twins (or similarly aged non-twin siblings) that are discordant117 for substance involvement can provide information on whether substance-related neural phenotypes arise from predispositional influences and/or are induced through substance involvement (right flow chart). If the brains of genetically similar individuals differ as a function of their substance use, then non-genetic mechanisms, including substance-induced changes, might be implicated. However, if they brain phenotypes are similar among those discordant for substance use, this would suggest that predispositional effects including shared genetic variation and environmental exposures are responsible for their associations with substance involvement.
Discordant Designs.
Monozygotic (identical) twins that are discordant for drug exposure serve as a natural quasi-experiment for the study of causal effects of drugs in humans (Fig. 3)117. If the correlation between drug use and brain structure is entirely genetic, the brain structure in monozygotic pairs discordant for substance use (or SUDs) would not be different. If, on the other hand, there is a difference in brain structure, then contributors beyond shared genetics and prenatal and familial environment are implicated. These contributors may be exposure-specific (that is, causal effects) or due to a person-specific third variables (such as early trauma that motivates substance use and, independently, modifies brain structure in that twin). While it is ideal to study identical twins, dizygotic twins and even non-twin siblings (close in age) can also be used to further parse non-genetic sources of similarity (for example, dizygotic twins are more closely matched for prenatal exposures than non-twin siblings).
Several studies have examined structural brain differences in twin pairs who vary in their drug exposure. In data from the Human Connectome Project118, it was shown that an association between cannabis use and reduced volume of subcortical structures were no longer apparent when cannabis-using individuals were compared with their co-twins or age-approximate siblings, consistent with the robust estimates of genetic correlation between cannabis use and subcortical brain volume119. Similarly, it was discovered that the correlations between alcohol consumption and insula and DLPFC volumes were primarily attributable to predispositional factors73. Instead of relying on discordancy for alcohol use alone, this study also contrasted brain structure in twin pairs in which both individuals were heavy drinkers as well as in pairs in which both individuals were light drinkers. Heavy and light drinking twins from discordant pairs did not differ in their gray matter volume and also did not differ from twins in pairs where both were heavy drinkers, suggesting that the reduced gray matter volume that is associated with alcohol use does not arise as a consequence of use. A recent series of studies of 436 24-year old twins from the Minnesota Twin Family Study120 document support for both predispositional and causal contributors to associations between substance involvement and cortical thickness. For instance, a thinner medial OFC among those with alcohol, tobacco and/or cannabis use disorders was attributable to predispositional risk, with some evidence that alcohol or cannabis use disorder may also contribute to these reductions121. In the same sample, an index of alcohol (but not cannabis) use was associated with a thinner cortex overall, with evidence that this reflects both predispositional risk and a potential consequence of alcohol exposure 122. Collectively, these analyses reveal that brain structure correlates of substance use and SUDs may reflect a predispositional liability to substance involvement, as well as a potential consequence of exposure.
Genetic predisposition.
Evidence for correlations between genetic variants identified in GWASs of brain imaging and those identified in GWASs of substance use and SUDs 89,90 suggest that associations between brain structure and addiction-related behavior that occur after the onset of exposure are confounded by preexisting pleiotropic liability. Thus, nearly any evidence for causation would also support predisposition. Ideal support for predispositional effects arises from studies of brain development in individuals before their drug exposure. Family studies provide persuasive support for such pre-existing brain differences in those genetically enriched for addiction liability123,124. Youth with a family history of alcohol use disorder have thinner frontal and parietal cortices111 and smaller frontal gray matter volume125, as well as larger gray matter volumes of the cerebellar lobes126, the NAc127 and the amygdala110,128, and task-related response variability in brain regions related to reward response and decision-making129–133 than those without such a family history. These studies provide compelling support for the association between family history of addiction and brain structure and function, which (in some instances) was investigated prior to substance use onset. They further support the neurodevelopmental hypothesis because family history of alcohol use disorder was also associated with early behavioral undercontrol134.
However, family history is an amalgamation of inherited genetic risk and genetic nurture[G] 135 and can be biased by lack of adequate measurement of familial density of risk136. Twin studies disentangle these familial effects to some degree by either explicitly modeling genetic nurture or separating genetic and family environmental factors within the offspring 137,138. Well-powered neuroimaging studies that also assess family history and future substance use, particularly during the developmental period prior to onset of substance use, are rare; however, the Adolescent Brain and Cognitive Development (ABCD study) provides an opportunity to evaluate the interrelationships between brain and substance use development in individuals from ages 9–10 years into early adulthood139. In the baseline data from this sample, total brain and regional cortical and subcortical volumes, cortical thickness and surface area, fractional anisotropy [G] and mean diffusivity indices were examined for their association with polygenic liability to alcohol consumption and problem drinking114 (Fig. 3). The polygenic risk score (PRS, the aggregated effects of risk alleles associated with a trait)140 for problematic alcohol use was associated with lower volume of the left frontal pole and greater cortical thickness of the right supramarginal gyrus, although nominally significant associations for both typical and problematic alcohol use PRS and insula metrics were evident114. In another analysis, the PRS for cannabis use disorder, but not cannabis use, was associated with lower white matter volume46. In each of these analyses, data were excluded from the small subset of youths who report substance use 141. However, an even earlier epoch of substance exposure — prenatal exposure — also merits consideration. In the ABCD sample, prenatal exposure to cannabis, particularly beyond the first trimester, is correlated with psychopathology (but not global brain structure) outcomes142 and persists as children enter their teens143. Therefore, the study of predisposition that is exclusively related to genotype requires consideration of family history, genetic nurture and other third variable confounders, as well as prenatal exposure to substance use. However, even studies of prenatal exposure are confounded by intergenerational transmission of genetic predisposition144–151.
The GINA Model
Brain imaging studies of addiction have tended to invoke drug-induced mechanisms of effect, whereas genomic studies have mostly relied on predispositional aspects of vulnerability. Each domain implicitly considers the relevance of the other to some degree. For instance, the neurodevelopmental model references common latent genetic influences on behavioral undercontrol and substance use152–156. Similarly, genomic studies are identifying drug-induced epigenetic alterations in relevant brain regions157.
Based on these foundational discoveries we outline an integrated framework for the development of addiction, the Genetically Informed Neurobiology of Addiction (GINA) model (Fig. 4). At the core of the GINA model is polygenic liability. The neural circuitry underlying reward, negative affect and executive function, as well as drug-specific pathways (such as those featuring drug receptors or enzymes involved in drug metabolism) serve as the substrates within which polygenic liability, risk and sequela of addiction unfold. Environmental factors serve as the filter through which gene–brain associations influence addiction-related behavior. While the GINA model is presented as a framework for integrating imaging and genetics studies of addiction in both clinical and population cohorts, it can also be extended to evaluate onset of use, occasional use and milder forms of SUD.
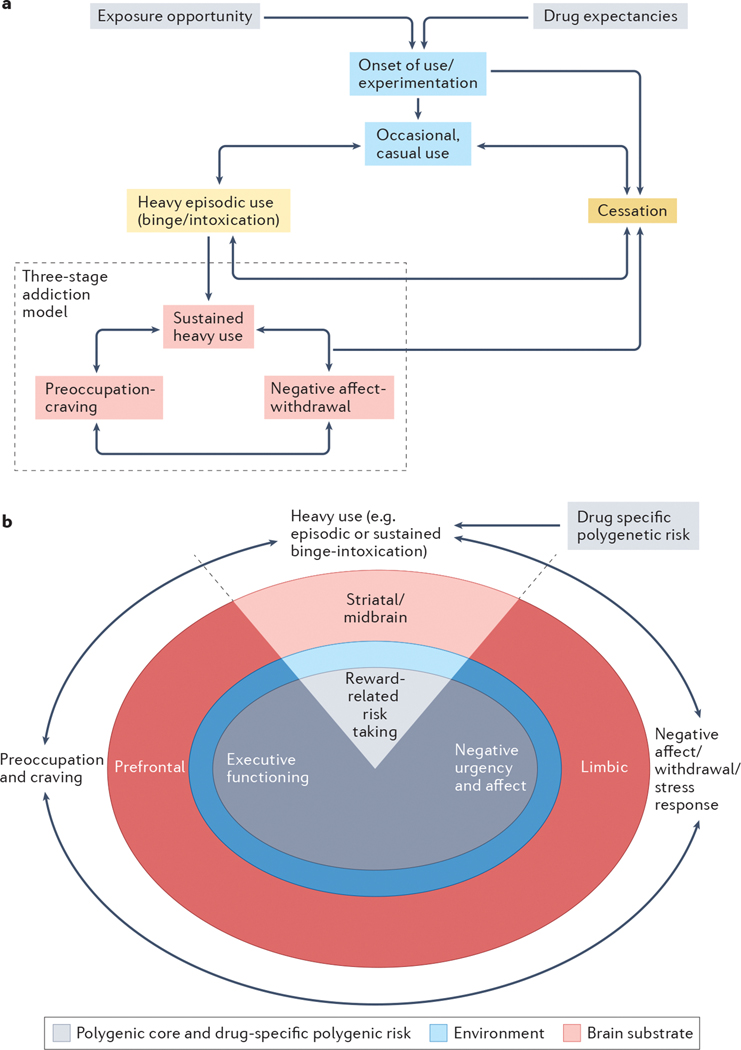
Fig. 4: A genetically informed neurobiology of addiction (GINA) model.
Addiction may be conceptualized as a developmental process or as a syndrome comprised of stages of escalating problem use. While the GINA model described here outlines a testable gene–brain–behavior mechanism underpinning the stages of addiction, it is scalable and can be extended to advance our understanding of the process of addiction. a| An illustration of the process of addiction, and those that lead into addiction, serves as a framework for understanding the GINA model. Exposure opportunity, availability193,194 and initial expectations surrounding the anticipated subjective effects of substance use serve as early contributors to drug-seeking behaviors and increase the likelihood of substance use195–198. Onset of substance use occurs in a subset of individuals, with some further entering a phase of casual but repeated substance use. Depending on the addictive potential of the substance, progression through periods of heavy episodic use and cessation may then occur (intervening aspects of these processes are not depicted). For some substances, periods of primarily reward-related occasional or casual use, heavy episodic use and cessation may occur (e.g., heavy drinking limited to college), during which time individuals may even meet criteria for milder forms of SUD199–201. Not shown are the numerous genetic and environmental influences that promote or deter progression through these substance-interfacing behaviors. For a further subset of individuals, heavy episodic use advances into a phase of sustained heavy use, wherein the pleasurable aspects of substance use are attenuated and compulsive use emerges to ameliorate negative affect, psychological and/or physiological stress states, and physiological withdrawal symptoms164. Withdrawal, and related negative mood, following substance abstinence leads to potentiated interoceptive salience through which physiological arousal associated with withdrawal and negative emotionality are potentiated21. We propose that this phase reflects moderate to severe forms of SUDs. b| The neurobiological model of addiction in the GINA framework. The GINA model places the 3 stages of addiction (shown around the outside of the image) within the context of a polygenic core (grey), environmental filter (blue) and brain substrate (pink; width of circles does not correspond to any relative magnitude of effect, i.e., genes and environment may be equally important). Each stage aligns most closely with genetic predispositions that act via specific posited brain mechanisms (these are shown “stacked” below that stage of the addiction cycle). All of the addiction stages are influenced by a polygenic core, which broadly corresponds to trait representations of substance-induced stages of sustained heavy use (binge-intoxication), negative affect (withdrawal-negative affect), and preoccupation/craving (preoccupation-anticipation) and by additional drug-specific polygenic risk that influences addiction, partly via the brain (for example, variants in genes encoding neurotransmitters) as well as via non-brain mechanisms (such as metabolic variants). Polygenic liability to reward-related risk-taking contributes to initial phases of binge-intoxication and may promote later escalating use (shown as heavy - episodic and sustained - use), which plays a role in promoting the reward related neural response to pleasurable aspects of substance use (e.g., striatal brain regions6). On the other hand, chronic substance use induces brain-related alterations that culminate in heightened stress states and negative affect (for instance, those with polygenic liability to negative urgency may be more vulnerable to this pathway) via a modified limbic response6. Furthermore, polygenic liability to executive function is likely to be instrumental in drug craving via changes in prefrontal brain function that results in increasing difficulties regulating the emotional salience of substance-related stimuli, despite the potential of subjective and cognitive desires to stop. Despite the appearance in this schematic of a one-to-one correspondence between polygenic liability, brain region and addiction stage, the gene–brain–behavior map is likely to be more interconnected. For instance, sustained heavy use in the context of negative affect may be influenced by polygenic risk to negative urgency and affect, via limbic pathways as well as substance-induced alterations in striatal circuits. The environment provides a filter for genetic liability (i.e., the magnitude and nature of genetic effects may be different in differing environmental contexts) and also directly underpins addictions. The brain is depicted as the outer substrate from which psychological aspects of addiction emerge. While distinct brain systems are illustrated, it is likely that networks of brain regions correspond to the three stages of addiction. While not noted here, aspects of addiction arise from and impact other bodily systems as well as the brain.
Polygenic core
As a simplifying principle, we posit that four key domains of genetic risk form the polygenic core of addictions. Three are common to all addictions: genes affecting reward and risk-taking (notably in the context of positive urgency [G]), genes affecting negative affect and/or susceptibility to negative urgency and genes affecting executive functioning and/or regulation. Genetic measurement of these domains, especially as they pertain to addiction liability, remains incomplete and underspecified. For instance, negative urgency is a hallmark characteristic of SUDs and some comorbid mood disorders26,158. However, current GWASs of negative affect rely on heterogeneous constructs (such as depression or neuroticism)159,160. The genetic disarticulation of the sub-facets of these composites161 (such as negative urgency)70,161,162 as well as well-powered GWASs of addiction-relevant indices of negative affect (such as using substances to cope or stress-responsivity163,164), using approaches such as genomic structural equation modeling [G] 165 will be required to fine-tune this polygenic core from an index of generalized risk for psychopathology to an addiction-specific liability factor.
The fourth source of genetic variability arises from genes encoding drug-specific metabolic factors and receptors; while polygenic in architecture, some of the drug-specific single loci may exert relatively large effects. For instance, the effect sizes of variants in alcohol metabolizing enzymes166 can approach those seen for the apolipoprotein e4 variant and Alzheimer disease risk167. Studying such pronounced (but scarce) genetic effects alongside polygenic patterns of common and drug-specific genetic risk requires novel statistical approaches that can handle mixtures of distributions of genetic effect sizes and conditional analyses. Drug-specific loci that encode the neurotransmitter targets for a drug are, however, rarely so specific. For instance, the rs16969968 variant in CHRNA5, encoding a nicotinic receptor, was shown to be highly significant for tobacco use phenotypes and also associated with schizophrenia and educational attainment168,169. Neuroimaging studies of this variant (rs16969968) have linked the risk allele to greater hippocampal activation in response to smoking cues170 and with reduced resting-state connectivity between the dorsal ACC and the ventral-striatopallidal circuit171 but with null effects on brain differences in light smoking adolescents172. Drug-specific loci also capture some variability in responses to existing treatments for addiction but findings are mixed173 and specific GWASs of pharmacogenomics response are needed174. Notably, drug-specific loci are evident in GWASs of both substance use and addiction (for example, the rs1229984 variant in ADH1B is significant for typical, maximum habitual and problem and/or disordered drinking)47,50,175, suggesting that their influence on addiction may be routed via their regulation of substance consumption and the subjective, and possibly interoceptive, effects associated with use176–179. Therefore, in the GINA model, we place drug-specific polygenic liability in the context of exposure, and most notably, heavy episodic and heavy sustained use (Fig. 4), where it regulates subjective response and sets the pace for entry into, and progression within, the three-stage addiction model.
Brain substrate
The GINA model provides a framework for gene–brain–addiction mechanisms from which we can develop testable hypotheses. For example, polygenic liability to executive function deficits might modify prefrontal regulatory capacity and, in turn, potentiate preoccupation with drugs. It would, however, be reductive to assume a one-to-one correspondence between polygenic liability, brain region and behavioral manifestation. For instance, it is highly likely that striatal circuitry is sensitive to the stages of addiction that evoke positive urgency and that polygenic liability to risk-taking as well as executive function (that is, undercontrol) undergird positive urgency. Polygenic liability to risk-taking is also likely to contribute to other stages of substance use and addiction (such as relapse)180 and to affect other brain regions beyond striatal regions (Fig. 4). Multi-method studies that integrate polygenic risk with whole brain and multivariate behavioral phenotypes will be necessary to broaden the scope of gene–brain–addiction connections. For example, machine learning [G] based approaches coupled with large-scale data181 could be used to perform a systematic, data driven study of the complexity underlying the GINA model.
Environmental filter.
While not detailed in this Perspective, the GINA model incorporates environment as the filter through which addiction emerges. Similar to genetics, some environmental factors (such as life stress) will generalize across substances and other psychopathology while others (such as policy, taxation and distance to alcohol outlet) are likely to be more substance-specific (although policies do have cross-cutting effects182). A proportion of the environmental impact on addiction also involves neurobiological mechanisms. For instance, trauma (especially when occurring during early life) is associated with brain development and exacerbates addiction risk183–185. However, the environment and genetic susceptibility may be related. Some traumatic experiences are correlates of genetic risk (such as passive exposure to early adverse environments that are a product of parental genotype), while others are modifiers of polygenic liability (such as trauma that moderates polygenic liability to addiction) and gene expression (such as trauma-induced epigenetic changes)186–189.
Characterizing addiction in the GINA model.
The GINA model characterizes addiction as a transition from episodic to sustained heavy use in the context of emerging negative affect and preoccupation (Fig. 4). From a diagnostic perspective, this coincides with moderate and severe SUD, as defined in DSM-5190. Recently, individuals endorsing between 2 to 5 DSM-5 criteria (mild or moderate SUD) were classified as being in a high-risk, sub-threshold state of preaddiction191 (similar to pre-diabetes) where interventions may be maximally beneficial. While the GINA model is more closely aligned with addiction per se, those with the higher preaddiction scores, depending on their individual symptomatology, may well be described by the GINA model. While reminiscent of the three-stage neurobiological model, the GINA model separates the broader binge-intoxication stage into heavy use that is either episodic or sustained. The former represents intermittent reward-motivated accelerations in use (such as alcohol consumption in college students) and may also capture mild SUDs as defined in DSM-5 192 and thus, is somewhat distinct from addiction. The latter represents the form of escalating chronic use that aligns with current conceptualizations of heavy use in the context of addiction. While behaviorally distinct, these aspects of binge-intoxication may share genetic and neurobiological contributors.
Characterizing substance use in the GINA model.
It could be argued that, rather than being classified as disorders in their own right, addictions would be better represented as a process or as a series of interactions with psychoactive substances that – in some instances – becomes disordered. From this perspective, the addiction process (Fig. 4) begins with exposure opportunity and expectations regarding the drug use experience. These early stages are strongly motivated by environmental factors, although gene–environment correlations influence drug availability and exposure opportunity193,194. Upon onset, initial experiences (which may be heritable) and subsequent experiences (which may be subjective) with individual drugs motivate or deter further use195–198. Continued use represents a mixture of pathways – heavy episodic use may become entrenched and transition to addiction or, for substances with lower addiction potential, settle into patterns of socially accepted or intermittent use. Many individuals attempt to quit using drugs in their 20s and 30s and the GINA model features both this early199–201 and later cessation. The same genetic core that contributes to the stage-based neurobiological model is also likely to contribute to these aspects of the addiction process. For instance, genetic propensity to risk-taking may motivate early drug-seeking behaviors64 and some neuroimaging studies suggest pre-existing brain differences in youth at risk for substance use onset73,101,102. Likewise, initial and typical subjective responses to individual substances may be influenced by variation in drug-specific loci176,196.
Relationship with other heuristics
The GINA model represents our conceptualization of the vast complexities that underlie addictions and is inspired by the extensive output of psychology, psychiatry, neuroscience, genetics, and translational research generated by international teams of scientists, particularly those who forecasted a need to bridge brain and genome research120,123,202–206. It is certainly not unique in adopting a multi-factorial view. Instead, it represents a conceptual increment that has resulted from novel study designs, genetic discoveries and rapid increases in availability of genetically informed neuroimaging data. For instance, the Addictions Neuroclinical Assessment (ANA)207 was developed to guide researchers in designing studies that might test the three-stage neurobiological model of addiction. However, the GINA model provides a framework from which a series of hypotheses can be tested using the ANA-derived data. A previous study also integrated evidence from behavior genetic and neuroimaging studies to provide a framework for a common liability to addictions208 and tested it using multiple sources of data209. These studies were prescient in anticipating the role of dopaminergic pathways on common addiction risk, although the GINA model has the advantage of leveraging contemporary insights from GWAS to advance a polygenic framework. Other schema, such as the Hierarchical Taxonomy of Psychopathology (HiTOP)210, aim to outline the common genetic and neurobiological underpinnings of a broad range of psychopathologies, including addictions. While the GINA model includes cross-disorder components (Fig. 2), it is clear that addictions are not merely the product of generalized genetic liability to broad-spectrum psychopathology and chronic substance use63. In spirit, the GINA model is aligned with the newly hypothesized Etiologic, Theory-Based, Ontogenetic Hierarchical Framework of Alcohol Use Disorder (ETOH)211 which highlights the importance of distinguishing between common and drug-specific mechanisms of risk and considering premorbid and drug-acquired pathways of influence. However, the ETOH model is finely tailored towards advancing our understanding of alcohol use disorder, whereas the GINA model focuses on addictions more broadly in the context of common and drug-specific genetic liability.
Predisposition in the context of drug-induced change.
The central aim of the GINA model is to establish the role of genetic predisposition as a source of individual differences in substance-related neural phenotypes. Most neuroimaging studies speculate neurobiological associations arise as sequelae of drug use, while widespread evidence of pleiotropy212 motivates geneticists to be partial to correlational mechanisms. The GINA model presents one possible reconciliation of these disparate perspectives. Simply put, while the consequence of chronic drug use may be evident in the brain, genetic factors that predate onset of substance use may influence whether or not an individual will initiate drug use, escalate in use, and progress to addiction. Furthermore, trait correlates of substance-induced states serve as additional sources of individual differences (Fig. 4). For example, we theorize that limbic adaptation to chronic drug use is likely to induce a negative affect state in most individuals, but that those with a genetic predisposition to negative affect may be more susceptible to this transition. Thus, individual differences in the developmental tempo and severity of the individual stages are likely to be due to polygenic liability – either via direct effects of genomic variation or via early influences on brain variability – similar to environmental moderators of risk.
Conclusions and perspectives
Future directions
Heuristic models, such as the GINA model, are intended to be dynamic and riddled with chasms that anticipated advances in genetics, neuroscience, and psychiatry can bridge. Recent criticism of genetic approaches surrounds the questionable prognostic utility of addiction PRSs213. Neuroimaging data are also associated with small effects91,214,215 and researchers in this field have grappled with their own methodological challenges, such as the reliability and heritability of task-based functional MRI studies86,93. The following emerge as priority areas for improvement.
First, there is a need to incorporate evidence from studies of the trajectories of brain development and studies of brain function. Most addiction neuroscience work has focused on brain function95. However, typical measurements of t-fMRI and resting-state functional connectivity are characterized by low heritability87, which may arise (at least in part) from reliability challenges93,94. Going forward, outlining the genetic architecture of brain function requires better phenotyping. In addition to improving the reliability of univariate metrics (such as dense-sampling94), multivariate approaches may have great utility. Indeed, polyneural phenotypes (that is, those with differentially weighted regional associations, much like polygenic risk scores) may be required to meaningfully predict individual differences in complex behavior. While such ‘lumping’ approaches may disappoint those hoping to link complex behavior and genetic variation to readily interpretable brain regions according to their known role in behavior, they may be replicable and reliable, similar to polygenic approaches216. Characterizing the longitudinal trajectories of brain development that contribute to periods of addiction vulnerability is equally important. Efforts targeting brain development217, especially large longitudinal samples such as ABCD of middle childhood-young adulthood 139 and the emerging HEALthy Brain and Cognitive Development (HBCD) study, which begins during the prenatal period, will be critical for this process.
Second, as genomic and imaging data become available in larger, population-representative settings that span developmental periods, the GINA model will require retooling to account for lighter and moderate levels of substance use and milder forms of addiction. Distinctions between levels of addiction severity may impact the magnitude of neurogenetic associations or may map onto qualitatively different polygenic signals and brain regions. From a genomics perspective, larger scale discovery GWAS will profoundly impact the GINA model. Currently, far too few GWAS are well-powered to identify genetic signals in individuals of ancestries other than Europeans, which limits the equitable application of PRS218. In addition, while smaller studies of copy number variants exist219,220, large-scale meta-analyses of structural variants221 are needed. Transcriptomic analyses provided some of earliest validation of GWAS discoveries; however, most have been conducted in bulk tissue and greater specificity is needed. Results for other psychiatric disorders have highlighted the importance of studying the enrichment of genetic signals in single cells or single nuclei, thus partitioning heritability (even within a single brain region) into specific cell types 222,223. Beyond eQTL, other cell-type specific annotations, such as enhancer effects224 could also be incorporated into PRS development. Such data are essential to providing a neuro-cellular perspective to the GINA model; however, at this time, larger GWASs are likely to be the most influential source of data225. Nonetheless, multi-omics data also provide valuable data matrices for emerging methods for developing functional PRS226–228 which appear to be more portable across ancestral populations than traditional effect size based PRS229.
A Consideration
Brain neuroimaging and genetics have substantially advanced our understanding of the biological bases of addiction. Both approaches have opponents who have argued that the conceptualization of addiction as a ‘brain disease’ discounts the role of socio-political factors, which may be more modifiable than one’s biology230,231. Yet, influential environmental provocations may in fact not be predictable or malleable. There are also deeper ethical issues surrounding the use of either genetics or neurobiology – or for that matter, environmental factors – to prospectively categorize an individual by their future addiction diagnosis. It is therefore worth considering the tradeoff between using genetics and neuroscience to prognosticate addiction risk as a source of stigma versus the potential for predispositional mechanisms to unburden individuals and societies of ill-construed notions regarding the moral valence of persons using substances.
Summary
Weaving genetics and neuroscience together, especially in the context of environmental considerations, can provide appreciable insights into the origins of addiction. It may even provide clues for preventing the critical transition from non-problematic to maladaptive use and illuminate efficacious therapeutic pathways. By proposing the integrative GINA model, we encourage the adoption of a multifactorial perspective of addiction – a process that represents a dynamic cascade of genetic predisposition and environmental risk and resilience that is enacted via developmentally relevant brain maturation and substance-induced brain alterations.
Acknowledgments.
The authors acknowledge the following funding from the United States National Institutes of Health: R.B. (R01DA54750; R21AA27827, U01DA055367), A.S.H. (T32DA007261, K01AA030083), E.C.J. (K01DA51759), A.A. (K02DA32573, R01DA54750). Funders were not involved in the preparation of this manuscript in any way.
Glossary
- Compulsive use
Drug consumption that is not under control and typically functions to achieve drug-present homeostasis and alleviation of negative affect/withdrawal as opposed to drug-induced euphoric reward.
- Heritability
The proportion of total variation in a phenotype that is due to genetic factors.
- Polygenic
The genetic characteristic of traits that is due to the aggregated small effects of many genetic variants.
- Genetic architectures
Distinct genetic factors that influence one or more traits.
- Genetic liability
The contribution of genetic factors to the likelihood of observing a phenotype.
- Positive reinforcement
Reward obtained after a stimulus and/or behavior.
- Binging
Consuming a large amount of a substance (typically alcohol) in a short period of time.
- Negative urgency
A personality facet related to impulsive behavior in the context of negative mood or experiences.
- Withdrawal
Physical (e.g., headaches and insomnia) and psychological (e.g., depressed mood) aversive experiences that occur when use of substance is discontinued.
- Executive function
Complex mental processes and cognition (e.g, working memory) that control skills (e.g., organizing, solving) and regulate emotion and behavior.
- Predictive reward signals
Neural signals that demarcate the expected delivery of reward following extrinsic and/or intrinsic cues.
- Negative reinforcement
The removal of something unpleasant or uncomfortable by a stimulus and/or behavior.
- Anhedonia
The loss of pleasure or lack of reactivity to pleasurable stimuli.
- Craving
A persistent desire to use a substance.
- Incentive salience
A cognitive process that motivates behavior toward reward.
- Developmental vulnerability
Vulnerability to a given outcome that arises in the context of typical development.
- Trait-like vulnerability
Vulnerability to a given trait.
- Predispositional liability
The aspect of an outcome that is attributable to predipositional (i.e., genetic variation, prior experiences) factors.
- Gene variants
Sections of DNA sequence that differ across groups of individuals.
- Genome-wide association studies (GWASs)
A hypothesis-free analysis of the association between common genetic variation across the genome and a phenotype.
- Candidate gene
A gene posited to be associated with a phenotype based on prior knowledge.
- Single nucleotide polymorphism (SNP)
A single base pair in the genome that varies across individuals.
- Twin studies
Comparisons of phenotype correlations in identical and fraternal twins to parse the role of genetic and environmental effects on a given phenotype or set of phenotypes.
- Expression quantitative trait loci (eQTL)
Genetic variants that modify the expression of a gene by acting upon the regulatory elements of the gene.
- Regulatory elements
Components of a gene, such as the promoter and introns, that regulate its expression.
- Resting-state functional connectivity
Correlated signal between brain regions in the absence of any stimulus or task.
- Pleiotropic effects
The influences of a variant, gene, or groups of variants on multiple phenotypes.
- Genetic nurture
The effect of genetically-influenced parent behavior on the offspring’s behavior.
- Fractional anisotropy
A measure of the degree of anisotropy of a diffusion process ranging from 0–1. In the context of diffusion tensor imaging it reflects the uniform directionality of white-matter fibers in the brain and is often conceptualized as an index of white matter integrity and structural connectivity.
- Positive urgency
A facet of personality related to impulsive behavior in the context of anticipated reward.
- Genomic structural equation modeling
A statistical genetics method for identifying genetic variants that influence multiple phenotypes as well as each individual phenotype.
- Machine learning
A data-driven approach that iteratively examines a training dataset for patterns across large numbers and diverse types of variables associated with an outcome, and, upon ‘learning’ these data patterns, can be used to test whether these patterns accurately predict the outcome in independent datasets.
Footnotes
Competing interests
The authors declare no competing interests
REFERENCES
- 1.Peacock A et al. Global statistics on alcohol, tobacco and illicit drug use: 2017 status report. Addiction 113, 1905–1926 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Volkow ND, Gordon JA & Koob GF Choosing appropriate language to reduce the stigma around mental illness and substance use disorders. Neuropsychopharmacology 46, 2230–2232 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agrawal A et al. The genetics of addiction-a translational perspective. Transl Psychiatry 2, e140 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kendler KS et al. Recent advances in the genetic epidemiology and molecular genetics of substance use disorders. Nat Neurosci 15, 181–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gelernter J & Polimanti R Genetics of substance use disorders in the era of big data. Nat Rev Genet 22, 712–729 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koob GF & Volkow ND Neurocircuitry of addiction. Neuropsychopharmacology 35, 217–38 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volkow ND, Koob GF & McLellan AT Neurobiologic Advances from the Brain Disease Model of Addiction. N Engl J Med 374, 363–71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koob GF & Volkow ND Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry 3, 760–773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volkow ND & Boyle M Neuroscience of Addiction: Relevance to Prevention and Treatment. Am J Psychiatry 175, 729–740 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Koob GF Drug Addiction: Hyperkatifeia/Negative Reinforcement as a Framework for Medications Development. Pharmacol Rev 73, 163–201 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haber SN Corticostriatal circuitry. Dialogues Clin Neurosci 18, 7–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janak PH & Tye KM From circuits to behaviour in the amygdala. Nature 517, 284–92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volkow ND, Fowler JS, Wang GJ, Baler R & Telang F Imaging dopamine’s role in drug abuse and addiction. Neuropharmacology 56 Suppl 1, 3–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volkow ND, Michaelides M & Baler R The Neuroscience of Drug Reward and Addiction. Physiol Rev 99, 2115–2140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schultz W Dopamine reward prediction error coding. Dialogues Clin Neurosci 18, 23–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keiflin R & Janak PH Dopamine Prediction Errors in Reward Learning and Addiction: From Theory to Neural Circuitry. Neuron 88, 247–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volkow ND & Morales M The Brain on Drugs: From Reward to Addiction. Cell 162, 712–25 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Filbey FM et al. fMRI study of neural sensitization to hedonic stimuli in long-term, daily cannabis users. Hum Brain Mapp 37, 3431–43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perry JL & Carroll ME The role of impulsive behavior in drug abuse. Psychopharmacology (Berl) 200, 1–26 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Willuhn I, Burgeno LM, Groblewski PA & Phillips PE Excessive cocaine use results from decreased phasic dopamine signaling in the striatum. Nat Neurosci 17, 704–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koob GF Negative reinforcement in drug addiction: the darkness within. Curr Opin Neurobiol 23, 559–63 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Koob GF & Le Moal M Plasticity of reward neurocircuitry and the ‘dark side’ of drug addiction. Nat Neurosci 8, 1442–4 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Ahmed SH & Koob GF Transition from moderate to excessive drug intake: change in hedonic set point. Science 282, 298–300 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Martz ME et al. Association of Marijuana Use With Blunted Nucleus Accumbens Response to Reward Anticipation. JAMA Psychiatry 73, 838–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balodis IM & Potenza MN Anticipatory reward processing in addicted populations: a focus on the monetary incentive delay task. Biol Psychiatry 77, 434–44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zorrilla EP, Logrip ML & Koob GF Corticotropin releasing factor: a key role in the neurobiology of addiction. Front Neuroendocrinol 35, 234–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naqvi NH, Gaznick N, Tranel D & Bechara A The insula: a critical neural substrate for craving and drug seeking under conflict and risk. Ann N Y Acad Sci 1316, 53–70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClure SM & Bickel WK A dual-systems perspective on addiction: contributions from neuroimaging and cognitive training. Ann N Y Acad Sci 1327, 62–78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heather N The concept of akrasia as the foundation for a dual systems theory of addiction. Behav Brain Res 390, 112666 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Cadet JL & Bisagno V Neuropsychological Consequences of Chronic Drug Use: Relevance to Treatment Approaches. Front Psychiatry 6, 189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brockett AT, Pribut HJ, Vazquez D & Roesch MR The impact of drugs of abuse on executive function: characterizing long-term changes in neural correlates following chronic drug exposure and withdrawal in rats. Learn Mem 25, 461–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerdeman GL, Partridge JG, Lupica CR & Lovinger DM It could be habit forming: drugs of abuse and striatal synaptic plasticity. Trends Neurosci 26, 184–92 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Grant BF et al. Epidemiology of DSM-5 Drug Use Disorder: Results From the National Epidemiologic Survey on Alcohol and Related Conditions-III. JAMA Psychiatry 73, 39–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha R New findings on biological factors predicting addiction relapse vulnerability. Curr Psychiatry Rep 13, 398–405 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stinson FS et al. Comorbidity between DSM-IV alcohol and specific drug use disorders in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Drug Alcohol Depend 80, 105–16 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Mackey S et al. Mega-Analysis of Gray Matter Volume in Substance Dependence: General and Substance-Specific Regional Effects. Am J Psychiatry 176, 119–128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatoum AS et al. Multivariate genome-wide association meta-analysis of over 1 million subjects identifies loci underlying multiple substance use disorders. Preprint at https://www.medrxiv.org/content/10.1101/2022.01.06.22268753v1 (2022). [DOI] [PMC free article] [PubMed]
- 38.Schoeler T et al. Novel biological insights into the common heritable liability to substance involvement: a multivariate genome-wide association study. Biological Psychiatry 10.1016/j.biopsych.2022.07.027 (2022). [DOI] [Google Scholar]
- 39.Zucker RA et al. Assessment of culture and environment in the Adolescent Brain and Cognitive Development Study: Rationale, description of measures, and early data. Dev Cogn Neurosci 32, 107–120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chassin L, Sher KJ, Hussong A & Curran P The developmental psychopathology of alcohol use and alcohol disorders: research achievements and future directions. Dev Psychopathol 25, 1567–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casey BJ & Jones RM Neurobiology of the adolescent brain and behavior: implications for substance use disorders. J Am Acad Child Adolesc Psychiatry 49, 1189–201 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rapuano KM et al. Behavioral and brain signatures of substance use vulnerability in childhood. Dev Cogn Neurosci 46, 100878 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deak JD & Johnson EC Genetics of substance use disorders: a review. Psychol Med 51, 2189–2200 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson EC et al. No Evidence That Schizophrenia Candidate Genes Are More Associated With Schizophrenia Than Noncandidate Genes. Biol Psychiatry 82, 702–708 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Border R et al. No Support for Historical Candidate Gene or Candidate Gene-by-Interaction Hypotheses for Major Depression Across Multiple Large Samples. Am J Psychiatry 176, 376–387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson EC et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry 7, 1032–1045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu M et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 51, 237–244 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polimanti R et al. Leveraging genome-wide data to investigate differences between opioid use vs. opioid dependence in 41,176 individuals from the Psychiatric Genomics Consortium. Mol Psychiatry 25, 1673–1687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walters RK et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci 21, 1656–1669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou H et al. Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat Neurosci 23, 809–818 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pasman JA et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci 21, 1161–1170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu K et al. Genome-wide association study of smoking trajectory and meta-analysis of smoking status in 842,000 individuals. Nat Commun 11, 5302 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou H et al. Association of OPRM1 Functional Coding Variant With Opioid Use Disorder: A Genome-Wide Association Study. JAMA Psychiatry 77, 1072–1080 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kranzler HR et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun 10, 1499 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gelernter J et al. Genome-wide Association Study of Maximum Habitual Alcohol Intake in >140,000 U.S. European and African American Veterans Yields Novel Risk Loci. Biol Psychiatry 86, 365–376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brazel DM et al. Exome Chip Meta-analysis Fine Maps Causal Variants and Elucidates the Genetic Architecture of Rare Coding Variants in Smoking and Alcohol Use. Biol Psychiatry 85, 946–955 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jang SK et al. Rare genetic variants explain missing heritability in smoking. Nat Hum Behav 10.1038/s41562-022-01408-5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sullivan PF & Geschwind DH Defining the Genetic, Genomic, Cellular, and Diagnostic Architectures of Psychiatric Disorders. Cell 177, 162–183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet 47, 1114–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mallard TT et al. Item-Level Genome-Wide Association Study of the Alcohol Use Disorders Identification Test in Three Population-Based Cohorts. Am J Psychiatry, 179, 58–70(2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanchez-Roige S, Palmer AA & Clarke TK Recent Efforts to Dissect the Genetic Basis of Alcohol Use and Abuse. Biol Psychiatry 87, 609–618 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marees AT et al. Genetic correlates of socio-economic status influence the pattern of shared heritability across mental health traits. Nat Hum Behav 5, 1065–1073 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hatoum AS et al. The addiction risk factor: A unitary genetic vulnerability characterizes substance use disorders and their associations with common correlates. Neuropsychopharmacology 47, 1739–1745 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karlsson Linner R et al. Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction. Nat Neurosci 24, 1367–1376 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu X, Zhong P, Vickstrom C, Li Y & Liu QS PDE4 Inhibition Restores the Balance Between Excitation and Inhibition in VTA Dopamine Neurons Disrupted by Repeated In Vivo Cocaine Exposure. Neuropsychopharmacology 42, 1991–1999 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tibbo AJ & Baillie GS Phosphodiesterase 4B: Master Regulator of Brain Signaling. Cells 9, 1254 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuroiwa M et al. Phosphodiesterase 4 inhibition enhances the dopamine D1 receptor/PKA/DARPP-32 signaling cascade in frontal cortex. Psychopharmacology (Berl) 219, 1065–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arends RM et al. Associations between the CADM2 gene, substance use, risky sexual behavior, and self-control: A phenome-wide association study. Addict Biol, 26, e13015 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morris J et al. Genetic variation in CADM2 as a link between psychological traits and obesity. Sci Rep 9, 7339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanchez-Roige S et al. CADM2 is implicated in impulsive personality traits by genome- and phenome-wide association studies in humans, with further support from studies of Cadm2 mutant mice. Preprint at https://www.medrxiv.org/content/10.1101/2022.01.29.22270095v3 (2022). [DOI] [PMC free article] [PubMed]
- 71.Gerring ZF, Vargas AM, Gamazon ER & Derks EM An integrative systems-based analysis of substance use: eQTL-informed gene-based tests, gene networks, and biological mechanisms. Am J Med Genet B Neuropsychiatr Genet 186, 162–172 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kapoor M et al. Multi-omics integration analysis identifies novel genes for alcoholism with potential overlap with neurodegenerative diseases. Nat Commun 12, 5071 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baranger DAA et al. Convergent Evidence for Predispositional Effects of Brain Gray Matter Volume on Alcohol Consumption. Biol Psychiatry 87, 645–655 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marees AT et al. Post-GWAS analysis of six substance use traits improves the identification and functional interpretation of genetic risk loci. Drug Alcohol Depend 206, 107703 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin H et al. Prefrontal cortex eQTLs/mQTLs enriched in genetic variants associated with alcohol use disorder and other diseases. Epigenomics 12, 789–800 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sey NYA et al. Chromatin architecture in addiction circuitry identifies risk genes and potential biological mechanisms underlying cigarette smoking and alcohol use traits. Mol Psychiatry 27, 3085–3094 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mignogna KM, Bacanu SA, Riley BP, Wolen AR & Miles MF Cross-species alcohol dependence-associated gene networks: Co-analysis of mouse brain gene expression and human genome-wide association data. PLoS One 14, e0202063 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huggett SB, Bubier JA, Chesler EJ & Palmer RHC Do gene expression findings from mouse models of cocaine use recapitulate human cocaine use disorder in reward circuitry? Genes Brain Behav 20, e12689 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Palmer RHC et al. Multi-omic and multi-species meta-analyses of nicotine consumption. Transl Psychiatry 11, 98 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kapoor M et al. Analysis of whole genome-transcriptomic organization in brain to identify genes associated with alcoholism. Transl Psychiatry 9, 89 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huggett SB & Stallings MC Genetic Architecture and Molecular Neuropathology of Human Cocaine Addiction. J Neurosci 40, 5300–5313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gottesman II & Gould TD The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry 160, 636–45 (2003). [DOI] [PubMed] [Google Scholar]
- 83.Jansen AG, Mous SE, White T, Posthuma D & Polderman TJ What twin studies tell us about the heritability of brain development, morphology, and function: a review. Neuropsychol Rev 25, 27–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grasby KL et al. The genetic architecture of the human cerebral cortex. Science 367, eaay6690 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hofer E et al. Genetic correlations and genome-wide associations of cortical structure in general population samples of 22,824 adults. Nat Commun 11, 4796 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Elliott LT et al. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature 562, 210–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smith SM et al. An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat Neurosci 24, 737–745 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bulik-Sullivan B et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 47, 1236–41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hatoum AS, Johnson EC, Agrawal A & Bogdan R Brain structure and problematic alcohol use: a test of plausible causation using latent causal variable analysis. Brain Imaging Behav 15, 2741–2745 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rabinowitz JA et al. Shared Genetic Etiology between Cortical Brain Morphology and Tobacco, Alcohol, and Cannabis Use. Cereb Cortex 32, 796–807 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marek S et al. Reproducible brain-wide association studies require thousands of individuals. Nature 603, 654–660 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guimaraes JPOFT, Sprooten E, Beckmann CF, Franke B & Bralten J Shared genetic influences on resting-state functional networks of the brain. Hum Brain Mapp 43, 1787–1803 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Elliott ML et al. What Is the Test-Retest Reliability of Common Task-Functional MRI Measures? New Empirical Evidence and a Meta-Analysis. Psychol Sci 31, 792–806 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gordon EM et al. Precision Functional Mapping of Individual Human Brains. Neuron 95, 791–807 e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hayes A, Herlinger K, Paterson L & Lingford-Hughes A The neurobiology of substance use and addiction: evidence from neuroimaging and relevance to treatment. BJPsych Advances 26, 367–378 (2020). [Google Scholar]
- 96.Brown SA et al. The National Consortium on Alcohol and NeuroDevelopment in Adolescence (NCANDA): A Multisite Study of Adolescent Development and Substance Use. J Stud Alcohol Drugs 76, 895–908 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pfefferbaum A et al. Altered Brain Developmental Trajectories in Adolescents After Initiating Drinking. Am J Psychiatry 175, 370–380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao Q et al. Association of Heavy Drinking With Deviant Fiber Tract Development in Frontal Brain Systems in Adolescents. JAMA Psychiatry 78, 407–415 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mascarell Maricic L et al. The IMAGEN study: a decade of imaging genetics in adolescents. Mol Psychiatry 25, 2648–2671 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Albaugh MD et al. Association of Cannabis Use During Adolescence With Neurodevelopment. JAMA Psychiatry 78, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Luby JL et al. Developmental Trajectories of the Orbitofrontal Cortex and Anhedonia in Middle Childhood and Risk for Substance Use in Adolescence in a Longitudinal Sample of Depressed and Healthy Preschoolers. Am J Psychiatry 175, 1010–1021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheetham A et al. Orbitofrontal Cortex Volume and Effortful Control as Prospective Risk Factors for Substance Use Disorder in Adolescence. Eur Addict Res 23, 37–44 (2017). [DOI] [PubMed] [Google Scholar]
- 103.Wade NE et al. Orbitofrontal cortex volume prospectively predicts cannabis and other substance use onset in adolescents. J Psychopharmacol 33, 1124–1131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Taffe MA et al. Long-lasting reduction in hippocampal neurogenesis by alcohol consumption in adolescent nonhuman primates. Proc Natl Acad Sci U S A 107, 11104–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shnitko TA, Liu Z, Wang X, Grant KA & Kroenke CD Chronic Alcohol Drinking Slows Brain Development in Adolescent and Young Adult Nonhuman Primates. eNeuro 6, ENEURO.0044–19.2019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kibaly C, Xu C, Cahill CM, Evans CJ & Law PY Non-nociceptive roles of opioids in the CNS: opioids’ effects on neurogenesis, learning, memory and affect. Nat Rev Neurosci 20, 5–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Coleman LG Jr., He J, Lee J, Styner M & Crews FT Adolescent binge drinking alters adult brain neurotransmitter gene expression, behavior, brain regional volumes, and neurochemistry in mice. Alcohol Clin Exp Res 35, 671–88 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Douet V, Chang L, Cloak C & Ernst T Genetic influences on brain developmental trajectories on neuroimaging studies: from infancy to young adulthood. Brain Imaging Behav 8, 234–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brouwer RM et al. The Speed of Development of Adolescent Brain Age Depends on Sex and Is Genetically Determined. Cereb Cortex 31, 1296–1306 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dager AD et al. Shared genetic factors influence amygdala volumes and risk for alcoholism. Neuropsychopharmacology 40, 412–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Henderson KE et al. Cortical Thickness in Adolescents with a Family History of Alcohol Use Disorder. Alcohol Clin Exp Res 42, 89–99 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Baranger DAA & Bogdan R Editorial: Causal, Predispositional, or Correlate? Group Differences in Cognitive Control-Related Brain Function in Cannabis-Using Youth Raise New Questions. J Am Acad Child Adolesc Psychiatry 58, 665–667 (2019). [DOI] [PubMed] [Google Scholar]
- 113.Gage SH, Munafo MR, MacLeod J, Hickman M & Smith GD Cannabis and psychosis. Lancet Psychiatry 2, 380 (2015). [DOI] [PubMed] [Google Scholar]
- 114.Hatoum AS et al. Polygenic risk scores for alcohol involvement relate to brain structure in substance-naive children: Results from the ABCD study. Genes Brain Behav, 20, e12756 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Munafo MR, Higgins JPT & Smith GD Triangulating Evidence through the Inclusion of Genetically Informed Designs. Cold Spring Harb Perspect Med 11, a040659 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.O’Connor LJ & Price AL Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat Genet 50, 1728–1734 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gillespie NA & Kendler KS Use of Genetically Informed Methods to Clarify the Nature of the Association Between Cannabis Use and Risk for Schizophrenia. JAMA Psychiatry 78, 467–468 (2021). [DOI] [PubMed] [Google Scholar]
- 118.Elam JS et al. The Human Connectome Project: A retrospective. Neuroimage 244, 118543 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pagliaccio D et al. Shared Predisposition in the Association Between Cannabis Use and Subcortical Brain Structure. JAMA Psychiatry 72, 994–1001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Iacono WG & McGue M Minnesota Twin Family Study. Twin Res 5, 482–7 (2002). [DOI] [PubMed] [Google Scholar]
- 121.Harper J et al. Orbitofrontal cortex thickness and substance use disorders in emerging adulthood: causal inferences from a co-twin control/discordant twin study. Addiction 116, 2548–2558 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Harper J et al. The Effects of Alcohol and Cannabis Use on the Cortical Thickness of Cognitive Control and Salience Brain Networks in Emerging Adulthood: A Co-twin Control Study. Biol Psychiatry 89, 1012–1022 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hill SY Trajectories of alcohol use and electrophysiological and morphological indices of brain development: distinguishing causes from consequences. Ann N Y Acad Sci 1021, 245–59 (2004). [DOI] [PubMed] [Google Scholar]
- 124.Rangaswamy M & Porjesz B Uncovering genes for cognitive (dys)function and predisposition for alcoholism spectrum disorders: a review of human brain oscillations as effective endophenotypes. Brain Res 1235, 153–71 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Weiland BJ et al. Substance abuse risk in emerging adults associated with smaller frontal gray matter volumes and higher externalizing behaviors. Drug Alcohol Depend 137, 68–75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hill SY, Lichenstein SD, Wang S & O’Brien J Volumetric Differences in Cerebellar Lobes in Individuals from Multiplex Alcohol Dependence Families and Controls: Their Relationship to Externalizing and Internalizing Disorders and Working Memory. Cerebellum 15, 744–754 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cservenka A, Gillespie AJ, Michael PG & Nagel BJ Family history density of alcoholism relates to left nucleus accumbens volume in adolescent girls. J Stud Alcohol Drugs 76, 47–56 (2015). [PMC free article] [PubMed] [Google Scholar]
- 128.Hill SY et al. Right amygdala volume in adolescent and young adult offspring from families at high risk for developing alcoholism. Biol Psychiatry 49, 894–905 (2001). [DOI] [PubMed] [Google Scholar]
- 129.Acheson A et al. Increased forebrain activations in youths with family histories of alcohol and other substance use disorders performing a Go/NoGo task. Alcohol Clin Exp Res 38, 2944–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cservenka A, Herting MM & Nagel BJ Atypical frontal lobe activity during verbal working memory in youth with a family history of alcoholism. Drug Alcohol Depend 123, 98–104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cservenka A & Nagel BJ Risky decision-making: an FMRI study of youth at high risk for alcoholism. Alcohol Clin Exp Res 36, 604–15 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yau WY et al. Nucleus accumbens response to incentive stimuli anticipation in children of alcoholics: relationships with precursive behavioral risk and lifetime alcohol use. J Neurosci 32, 2544–51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Heitzeg MM, Nigg JT, Yau WY, Zucker RA & Zubieta JK Striatal dysfunction marks preexisting risk and medial prefrontal dysfunction is related to problem drinking in children of alcoholics. Biol Psychiatry 68, 287–95 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zucker RA, Heitzeg MM & Nigg JT Parsing the Undercontrol/Disinhibition Pathway to Substance Use Disorders: A Multilevel Developmental Problem. Child Dev Perspect 5, 248–255 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kong A et al. The nature of nurture: Effects of parental genotypes. Science 359, 424–428 (2018). [DOI] [PubMed] [Google Scholar]
- 136.Molina BS, Donovan JE & Belendiuk KA Familial loading for alcoholism and offspring behavior: mediating and moderating influences. Alcohol Clin Exp Res 34, 1972–84 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Balbona JV, Kim Y & Keller MC Estimation of Parental Effects Using Polygenic Scores. Behav Genet 51, 264–278 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Maes HH et al. Cross-Cultural Comparison of Genetic and Cultural Transmission of Smoking Initiation Using an Extended Twin Kinship Model. Twin Res Hum Genet 21, 179–190 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Volkow ND et al. The conception of the ABCD study: From substance use to a broad NIH collaboration. Dev Cogn Neurosci 32, 4–7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wray NR et al. From Basic Science to Clinical Application of Polygenic Risk Scores: A Primer. JAMA Psychiatry 78, 101–109 (2021). [DOI] [PubMed] [Google Scholar]
- 141.Lisdahl KM et al. Substance use patterns in 9–10 year olds: Baseline findings from the adolescent brain cognitive development (ABCD) study. Drug Alcohol Depend 227, 108946 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Paul SE et al. Associations Between Prenatal Cannabis Exposure and Childhood Outcomes: Results From the ABCD Study. JAMA Psychiatry 78, 64–76 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Baranger DAA et al. Association of mental health burden with prenatal cannabis exposure from childhood to early adolescence: longitudinal findings from the Adolescent Brain Cognitive Development (ABCD) study. JAMA Pediatrics 10.1001/jamapediatrics.2022.3191 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Roffman JL et al. Association of adverse prenatal exposure burden with child psychopathology in the Adolescent Brain Cognitive Development (ABCD) Study. PLoS One 16, e0250235 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.El Marroun H et al. Prenatal tobacco exposure and brain morphology: a prospective study in young children. Neuropsychopharmacology 39, 792–800 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ross EJ, Graham DL, Money KM & Stanwood GD Developmental consequences of fetal exposure to drugs: what we know and what we still must learn. Neuropsychopharmacology 40, 61–87 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.D’Onofrio BM, Sjolander A, Lahey BB, Lichtenstein P & Oberg AS Accounting for Confounding in Observational Studies. Annu Rev Clin Psychol 16, 25–48 (2020). [DOI] [PubMed] [Google Scholar]
- 148.Quinn PD et al. Association Between Maternal Smoking During Pregnancy and Severe Mental Illness in Offspring. JAMA Psychiatry 74, 589–596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Micalizzi L et al. A sibling-comparison study of smoking during pregnancy and risk for reading-related problems. Neurotoxicol Teratol 84, 106961 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Donald KA et al. Neuroimaging effects of prenatal alcohol exposure on the developing human brain: a magnetic resonance imaging review. Acta Neuropsychiatr 27, 251–69 (2015). [DOI] [PubMed] [Google Scholar]
- 151.Easey KE et al. Characterization of alcohol polygenic risk scores in the context of mental health outcomes: Within-individual and intergenerational analyses in the Avon Longitudinal Study of Parents and Children. Drug Alcohol Depend 221, 108654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vrieze SI, McGue M, Miller MB, Hicks BM & Iacono WG Three mutually informative ways to understand the genetic relationships among behavioral disinhibition, alcohol use, drug use, nicotine use/dependence, and their co-occurrence: twin biometry, GCTA, and genome-wide scoring. Behav Genet 43, 97–107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dick DM, Adkins AE & Kuo SI Genetic influences on adolescent behavior. Neurosci Biobehav Rev 70, 198–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kendler KS, Prescott CA, Myers J & Neale MC The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Arch Gen Psychiatry 60, 929–37 (2003). [DOI] [PubMed] [Google Scholar]
- 155.Iacono WG, Malone SM & McGue M Behavioral disinhibition and the development of early-onset addiction: common and specific influences. Annu Rev Clin Psychol 4, 325–48 (2008). [DOI] [PubMed] [Google Scholar]
- 156.McGue M, Irons D & Iacono WG The adolescent origins of substance use disorders: a behavioral genetic perspective. Nebr Symp Motiv 61, 31–50 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hamilton PJ & Nestler EJ Epigenetics and addiction. Curr Opin Neurobiol 59, 128–136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pang RD, Farrahi L, Glazier S, Sussman S & Leventhal AM Depressive symptoms, negative urgency and substance use initiation in adolescents. Drug Alcohol Depend 144, 225–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Levey DF et al. Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat Neurosci 24, 954–963 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nagel M et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet 50, 920–927 (2018). [DOI] [PubMed] [Google Scholar]
- 161.Nagel M, Watanabe K, Stringer S, Posthuma D & van der Sluis S Item-level analyses reveal genetic heterogeneity in neuroticism. Nat Commun 9, 905 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sanchez-Roige S et al. Genome-Wide Association Studies of Impulsive Personality Traits (BIS-11 and UPPS-P) and Drug Experimentation in up to 22,861 Adult Research Participants Identify Loci in the CACNA1I and CADM2 genes. J Neurosci 39, 2562–2572 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zorrilla EP & Koob GF Impulsivity Derived From the Dark Side: Neurocircuits That Contribute to Negative Urgency. Front Behav Neurosci 13, 136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Koob GF Anhedonia, Hyperkatifeia, and Negative Reinforcement in Substance Use Disorders. Curr Top Behav Neurosci 58, 147–165 (2022). [DOI] [PubMed] [Google Scholar]
- 165.Grotzinger AD et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav 3, 513–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Edenberg HJ & McClintick JN Alcohol Dehydrogenases, Aldehyde Dehydrogenases, and Alcohol Use Disorders: A Critical Review. Alcohol Clin Exp Res 42, 2281–2297 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Andrews SJ, Fulton-Howard B & Goate A Interpretation of risk loci from genome-wide association studies of Alzheimer’s disease. Lancet Neurol 19, 326–335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Okbay A et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Janes AC et al. Association between CHRNA5 genetic variation at rs16969968 and brain reactivity to smoking images in nicotine dependent women. Drug Alcohol Depend 120, 7–13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hong LE et al. A genetically modulated, intrinsic cingulate circuit supports human nicotine addiction. Proc Natl Acad Sci U S A 107, 13509–14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chaarani B et al. Low Smoking Exposure, the Adolescent Brain, and the Modulating Role of CHRNA5 Polymorphisms. Biol Psychiatry Cogn Neurosci Neuroimaging 4, 672–679 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hartwell EE et al. Systematic review and meta-analysis of the moderating effect of rs1799971 in OPRM1, the mu-opioid receptor gene, on response to naltrexone treatment of alcohol use disorder. Addiction 115, 1426–1437 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Biernacka JM et al. Genetic contributions to alcohol use disorder treatment outcomes: a genome-wide pharmacogenomics study. Neuropsychopharmacology 46, 2132–2139 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Deak JDL, D.; Wendt FR; Zhou H; Galimberti M; Kranzler HR; Gaziano JM; Stein MB; Polimanti R; The Million Veteran Program; Gelernter J;. Genome-wide investigation of maximum habitual alcohol intake (MaxAlc) in 247,755 European and African Ancestry U.S. Veterans informs the relationship between habitual alcohol consumption and alcohol use disorder Preprint at 10.1101/2022.05.02.22274580 (2022). [DOI]
- 176.Sherva R et al. Association of a single nucleotide polymorphism in neuronal acetylcholine receptor subunit alpha 5 (CHRNA5) with smoking status and with ‘pleasurable buzz’ during early experimentation with smoking. Addiction 103, 1544–52 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.McCarthy DM, Pedersen SL, Lobos EA, Todd RD & Wall TL ADH1B*3 and response to alcohol in African-Americans. Alcohol Clin Exp Res 34, 1274–81 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Duranceaux NC et al. Associations of variations in alcohol dehydrogenase genes with the level of response to alcohol in non-Asians. Alcohol Clin Exp Res 30, 1470–8 (2006). [DOI] [PubMed] [Google Scholar]
- 179.Jensen KP et al. A CHRNA5 Smoking Risk Variant Decreases the Aversive Effects of Nicotine in Humans. Neuropsychopharmacology 40, 2813–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kreek MJ, Nielsen DA, Butelman ER & LaForge KS Genetic influences on impulsivity, risk taking, stress responsivity and vulnerability to drug abuse and addiction. Nat Neurosci 8, 1450–7 (2005). [DOI] [PubMed] [Google Scholar]
- 181.Wright SN & Little AR NIDA vision for big data science to understand the biological underpinnings of substance use disorders. Neuropsychopharmacology 46, 262 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Veligati S et al. Changes in alcohol and cigarette consumption in response to medical and recreational cannabis legalization: Evidence from U.S. state tax receipt data. Int J Drug Policy 75, 102585 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kendler KS et al. Childhood sexual abuse and adult psychiatric and substance use disorders in women: an epidemiological and cotwin control analysis. Arch Gen Psychiatry 57, 953–9 (2000). [DOI] [PubMed] [Google Scholar]
- 184.Sartor CE, Agrawal A, McCutcheon VV, Duncan AE & Lynskey MT Disentangling the complex association between childhood sexual abuse and alcohol-related problems: a review of methodological issues and approaches. J Stud Alcohol Drugs 69, 718–27 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tomasi D & Volkow ND Associations of family income with cognition and brain structure in USA children: prevention implications. Mol Psychiatry 26, 6619–6629 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yehuda R & Lehrner A Intergenerational transmission of trauma effects: putative role of epigenetic mechanisms. World Psychiatry 17, 243–257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Meyers J et al. Early Sexual Trauma Exposure and Neural Response Inhibition in Adolescence and Young Adults: Trajectories of Frontal Theta Oscillations During a Go/No-Go Task. J Am Acad Child Adolesc Psychiatry 58, 242–255 e2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Meyers JL et al. Psychosocial moderation of polygenic risk for cannabis involvement: the role of trauma exposure and frequency of religious service attendance. Transl Psychiatry 9, 269 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Szutorisz H & Hurd YL Feeding the Developing Brain: The Persistent Epigenetic Effects of Early Life Malnutrition. Biol Psychiatry 80, 730–732 (2016). [DOI] [PubMed] [Google Scholar]
- 190.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, (American Psychiatric Association, Washington, DC, 2013). [Google Scholar]
- 191.McLellan AT, Koob GF & Volkow ND Preaddiction-A Missing Concept for Treating Substance Use Disorders. JAMA Psychiatry 79, 749–751 (2022). [DOI] [PubMed] [Google Scholar]
- 192.Lane SP, Steinley D & Sher KJ Meta-analysis of DSM alcohol use disorder criteria severities: structural consistency is only ‘skin deep’. Psychol Med 46, 1769–84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hines LA et al. Overlap of heritable influences between cannabis use disorder, frequency of use and opportunity to use cannabis: trivariate twin modelling and implications for genetic design. Psychol Med 48, 2786–2793 (2018). [DOI] [PubMed] [Google Scholar]
- 194.Gillespie NA et al. Longitudinal modeling of genetic and environmental influences on self-reported availability of psychoactive substances: alcohol, cigarettes, marijuana, cocaine and stimulants. Psychol Med 37, 947–59 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Agrawal A, Madden PA, Bucholz KK, Heath AC & Lynskey MT Initial reactions to tobacco and cannabis smoking: a twin study. Addiction 109, 663–71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Edwards AC et al. Meta-Analysis of Genetic Influences on Initial Alcohol Sensitivity. Alcohol Clin Exp Res 42, 2349–2359 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Grant JD et al. Subjective reactions to cocaine and marijuana are associated with abuse and dependence. Addict Behav 30, 1574–86 (2005). [DOI] [PubMed] [Google Scholar]
- 198.Lyons MJ et al. How do genes influence marijuana use? The role of subjective effects. Addiction 92, 409–17 (1997). [PubMed] [Google Scholar]
- 199.Windle M A Multilevel Developmental Contextual Approach To Substance Use and Addiction. Biosocieties 5, 124–136 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lee MR & Sher KJ “Maturing Out” of Binge and Problem Drinking. Alcohol Res 39, 31–42 (2018). [PMC free article] [PubMed] [Google Scholar]
- 201.Verges A et al. Refining the notion of maturing out: results from the national epidemiologic survey on alcohol and related conditions. Am J Public Health 103, e67–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Iacono WG et al. The utility of twins in developmental cognitive neuroscience research: How twins strengthen the ABCD research design. Dev Cogn Neurosci 32, 30–42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Porjesz B, Jones K & Begleiter H The genetics of oscillations in the human brain. Suppl Clin Neurophysiol 57, 441–9 (2004). [DOI] [PubMed] [Google Scholar]
- 204.Anokhin AP Genetic psychophysiology: advances, problems, and future directions. Int J Psychophysiol 93, 173–97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mackey S et al. Genetic imaging consortium for addiction medicine: From neuroimaging to genes. Prog Brain Res 224, 203–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Van Essen DC et al. The WU-Minn Human Connectome Project: an overview. Neuroimage 80, 62–79 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kwako LE, Momenan R, Litten RZ, Koob GF & Goldman D Addictions Neuroclinical Assessment: A Neuroscience-Based Framework for Addictive Disorders. Biol Psychiatry 80, 179–89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Vanyukov MM et al. Liability to substance use disorders: 1. Common mechanisms and manifestations. Neurosci Biobehav Rev 27, 507–15 (2003). [DOI] [PubMed] [Google Scholar]
- 209.Vanyukov MM et al. Liability to substance use disorders: 2. A measurement approach. Neurosci Biobehav Rev 27, 517–26 (2003). [DOI] [PubMed] [Google Scholar]
- 210.Latzman RD, DeYoung CG & Hitop Neurobiological Foundations W Using empirically-derived dimensional phenotypes to accelerate clinical neuroscience: the Hierarchical Taxonomy of Psychopathology (HiTOP) framework. Neuropsychopharmacology 45, 1083–1085 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Boness CL, Watts AL, Moeller KN & Sher KJ The Etiologic, Theory-Based, Ontogenetic Hierarchical Framework of Alcohol Use Disorder: A Translational Systematic Review of Reviews. Psychol Bull 147, 1075–1123 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Watanabe K et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet 51, 1339–1348 (2019). [DOI] [PubMed] [Google Scholar]
- 213.Barr PB et al. Using polygenic scores for identifying individuals at increased risk of substance use disorders in clinical and population samples. Transl Psychiatry 10, 196 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Dick AS et al. Meaningful associations in the adolescent brain cognitive development study. Neuroimage 239, 118262 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Baranger DAA et al. Borderline Personality Traits Are Not Correlated With Brain Structure in Two Large Samples. Biol Psychiatry Cogn Neurosci Neuroimaging 5, 669–677 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bogdan R, Baranger DAA & Agrawal A Polygenic Risk Scores in Clinical Psychology: Bridging Genomic Risk to Individual Differences. Annu Rev Clin Psychol 14, 119–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Frangou S et al. Cortical thickness across the lifespan: Data from 17,075 healthy individuals aged 3–90 years. Hum Brain Mapp 43, 431–451 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Martin AR et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet 51, 584–591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Li D et al. Genome-wide association study of copy number variations (CNVs) with opioid dependence. Neuropsychopharmacology 40, 1016–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sulovari A, Liu Z, Zhu Z & Li D Genome-wide meta-analysis of copy number variations with alcohol dependence. Pharmacogenomics J 18, 398–405 (2018). [DOI] [PubMed] [Google Scholar]
- 221.Mukamel RE et al. Protein-coding repeat polymorphisms strongly shape diverse human phenotypes. Science 373, 1499–1505 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Skene NG et al. Genetic identification of brain cell types underlying schizophrenia. Nat Genet 50, 825–833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ruzicka WB et al. Single-cell dissection of schizophrenia reveals neurodevelopmental-synaptic axis and transcriptional resilience. Preprint at https://www.medrxiv.org/content/10.1101/2020.11.06.20225342v1 (2020).
- 224.Nasser J et al. Genome-wide enhancer maps link risk variants to disease genes. Nature 593, 238–243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Baranger DAA et al. Multi-omics analyses cannot identify true-positive novel associations from underpowered genome-wide association studies of four brain-related traits. Preprint at https://www.biorxiv.org/content/10.1101/2022.04.13.487655v1 (2022).
- 226.Liang Y et al. Polygenic transcriptome risk scores (PTRS) can improve portability of polygenic risk scores across ancestries. Genome Biol 23, 23 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Marquez-Luna C et al. Incorporating functional priors improves polygenic prediction accuracy in UK Biobank and 23andMe data sets. Nat Commun 12, 6052 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hu Y et al. Leveraging functional annotations in genetic risk prediction for human complex diseases. PLoS Comput Biol 13, e1005589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Amariuta T et al. Improving the trans-ancestry portability of polygenic risk scores by prioritizing variants in predicted cell-type-specific regulatory elements. Nat Genet 52, 1346–1354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Hall W, Carter A & Forlini C Brain disease model of addiction: misplaced priorities? Lancet Psychiatry 2, 867 (2015). [DOI] [PubMed] [Google Scholar]
- 231.Risch N & Merikangas K The future of genetic studies of complex human diseases. Science 273, 1516–7 (1996). [DOI] [PubMed] [Google Scholar]
- 232.Potenza MN et al. Gambling disorder. Nat Rev Dis Primers 5, 51 (2019). [DOI] [PubMed] [Google Scholar]
- 233.Slutske WS et al. Common genetic vulnerability for pathological gambling and alcohol dependence in men. Arch Gen Psychiatry 57, 666–73 (2000). [DOI] [PubMed] [Google Scholar]
- 234.Kaye WH et al. Does a shared neurobiology for foods and drugs of abuse contribute to extremes of food ingestion in anorexia and bulimia nervosa? Biol Psychiatry 73, 836–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hasin DS et al. DSM-5 criteria for substance use disorders: recommendations and rationale. Am J Psychiatry 170, 834–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Saunders JB, Degenhardt L, Reed GM & Poznyak V Alcohol Use Disorders in ICD-11: Past, Present, and Future. Alcohol Clin Exp Res 43, 1617–1631 (2019). [DOI] [PubMed] [Google Scholar]
- 237.Saunders JB, Aasland OG, Babor TF, de la Fuente JR & Grant M Development of the Alcohol Use Disorders Identification Test (AUDIT): WHO Collaborative Project on Early Detection of Persons with Harmful Alcohol Consumption--II. Addiction 88, 791–804 (1993). [DOI] [PubMed] [Google Scholar]
- 238.Quach BC et al. Expanding the genetic architecture of nicotine dependence and its shared genetics with multiple traits. Nat Commun 11, 5562 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Gaddis N et al. Multi-trait genome-wide association study of opioid addiction: OPRM1 and Beyond. Preprint at https://www.medrxiv.org/content/10.1101/2021.09.13.21263503v1 (2021). [DOI] [PMC free article] [PubMed]
- 240.Kember RL et al. Cross-ancestry meta-analysis of opioid use disorder uncovers novel loci with predominant effects in brain regions associated with addiction. Nat. Neurosci 10.1038/s41593-022-01160-z (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Deak JD et al. Genome-wide association study in individuals of European and African ancestry and multi-trait analysis of opioid use disorder identifies 19 independent genome-wide significant risk loci. Molecular Psychiatry 10.1038/s41380-022-01709-1 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Cabana-Dominguez J, Shivalikanjli A, Fernandez-Castillo N & Cormand B Genome-wide association meta-analysis of cocaine dependence: Shared genetics with comorbid conditions. Prog Neuropsychopharmacol Biol Psychiatry 94, 109667 (2019). [DOI] [PubMed] [Google Scholar]