Abstract
MicroRNAs (miRNAs) are emerging as a novel class of molecular targets and therapeutics to control gene expression for tissue repair and regeneration. However, a safe and effective transfection of miRNAs to cells has been a major barrier to their applications. In this work, a multifunctional polyplex micelle named PPP-RGI was developed as a non-viral gene vector for the efficient transfection of miR-218 (an osteogenic miRNA regulator) to bone marrow-derived mesenchymal stem cells (BMSCs) for accelerated osteogenic differentiation. PPP-RGI was designed and synthesized via conjugation of a multifunctional R9-G4-IKVAVW (RGI) peptide onto an amphiphilic poly(lactide-co-glycolide)-g-polyethylenimine-b-polyethylene glycol (PPP) copolymer. PPP-RGI self-assembled into polyplex micelles and strongly condensed miR-218 to prevent its RNase degradation. When the PPP-RGI/miR-218 complex was brought into contact with BMSCs, it exhibited high internalization efficiency and a fast escape from endo/lysosomes of the BMSCs. Subsequently, miR-218 released from the PPP-RGI/miR-218 complex regulated gene expressions and significantly enhanced the osteogenic differentiation of BMSCs. The multifunctional peptide conjugated nanocarrier serves as an effective miRNA delivery vector to promote osteogenesis.
1. Introduction
MicroRNAs (miRNAs) are emerging as a class of promising molecules for tissue repair and regeneration.1 These short non-coding RNAs (approximately 22 nucleotides in length) modulate numerous cellular activities by binding to the 3′-untranslated regions (3′-UTR) of their target mRNAs.2,3 The specific binding between an miRNA and the target mRNA leads to degradation of the targeted mRNA and subsequently the suppression of protein synthesis. Therefore, miRNAs can be utilized in tissue engineering to precisely regulate gene expression within cells at a post-transcriptional level.4 For example, miR-145 reduced the expression of osterix, thus repressing the osteogenic differentiation of C2C12 and MC3T3-E1 cells.5
Naked miRNAs are highly susceptible to degradation in the presence of nucleases and have a short half-life. Moreover, direct miRNA injection has low transfection efficiency because miRNAs have difficulty passing through the phospholipid bilayer cell membrane. Therefore, the development of effective miRNA delivery has become a prerequisite for miRNA application in bone tissue engineering. A number of vectors have been developed to deliver miRNAs.6 While viral vectors are the most efficient for miRNA delivery, low loading capacity, high toxicity, and strong immunogenicity limit their clinical applications. In comparison with viral vectors, non-viral vectors can be constructed to have low toxicity, high condensing capacity, and high biocompatibility. Hence, the development of non-viral vectors for safe and efficient gene delivery has attracted increasing attention in recent years.7
A number of polycations, such as polyethylenimine (PEI), poly(L-lysine), and chitosan, have been developed as non-viral gene delivery vectors.8–10 PEI is one of the most effective non-viral gene vectors among these polycation vectors due to its high charge density and intrinsic “proton sponge effect”. However, PEI exhibits ineluctable disadvantages—high cytotoxicity with increased transfection efficiency.11 PEI-based gene vectors are often PEGylated or modified by hydrophobic groups to decrease cytotoxicity.12,13 Nevertheless, these modification approaches inevitably decreased transfection efficiency.14–16 Therefore, when PEI-based non-viral gene vectors are used to regulate gene expression, they need to be further optimized to improve transfection efficiency and retain high biocompatibility.
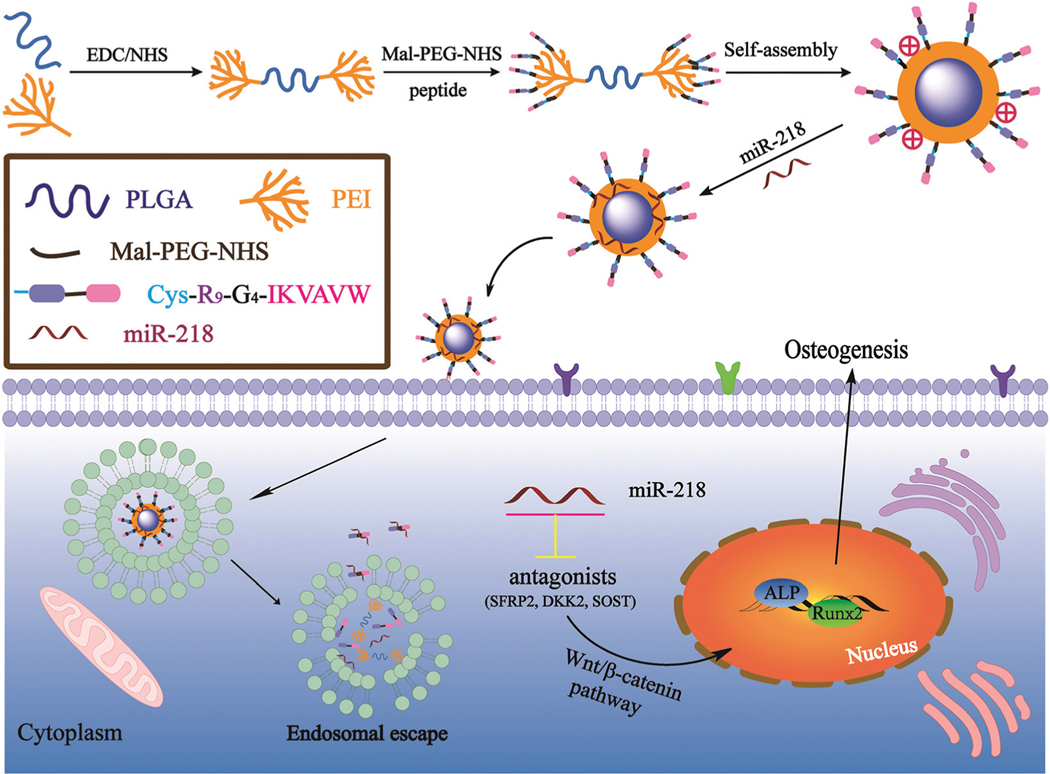
Herein, we designed and synthesized multifunctional, peptide-conjugated, polyplex micelles to efficiently deliver miR-218 for accelerated osteogenic differentiation of bone marrow-derived mesenchymal stem cells (BMSCs). The multi-functional polyplex micelles were synthesized via conjugation of a multifunctional R9-G4-IKVAVW (RGI) peptide onto an amphiphilic poly(lactide-co-glycolide)-g-polyethylenimine-b-polyethylene glycol (PPP) copolymer. We designed and synthesized a multifunctional RGI peptide in which R9 is a cell-penetrating peptide (CPP)—a powerful cargo holder to deliver plasmid DNA ( pDNA) and small interfering RNAs through the cell membrane to effectively enhance cellular uptake.17,18 In addition, the IKVAV peptide (Ile-Lys-Val-Ala-Val) is derived from the laminin A protein chain and was used to recognize and target BMSCs (binding α3β1 integrin) and promote proliferation19 by activating the phosphorylation of Akt and ERK1/Furthermore, G4 is a glycine residue and acted as a flexible spacer to support the function of R9 and IKVAV.20 Consequently, we hypothesized that the incorporation of RGI into the PEI-based vector would greatly enhance cellular transfection efficiency and promote BMSC growth. PPP-RGI conveniently self-assembled into stable polyplex micelles due to the incorporation of hydrophobic segment PLGA and hydrophilic segment PEG in the PPP copolymer. The formation of polyplex micelles and the gene delivery process are shown in Scheme 1. Meanwhile, miR-218 was recently identified to directly target secreted frizzled-related protein 2 (SFRP2), sclerostin (SOST), and dickkopf WNT signaling pathway inhibitor 2 (DKK2), which are three negative regulators of the Wnt/β-catenin signaling pathway.21–23 Therefore, miR-218 is a potential osteogenic regulator of BMSCs via regulation of Wnt/β-catenin signaling. To date, miR-218 has not been found to promote the osteogenesis of BMSCs in tissue regeneration. In this study, we first synthesized and characterized multifunctional PPP-RGI polyplex micelles. Next, we investigated the transfection of PPP-RGI/miR-218 to BMSCs for enhanced osteogenic differentiation of the BMSCs.
Scheme 1.
Illustration of the preparation of the PPP-RGI/miR-218 complex and the delivery of miR-218 from the complex to promote osteogenic differentiation.
2. Material and methods
2.1. Materials
Polyethylenimine (branched PEI, Mw = 10 000 g mol−1), succinic anhydride, stannous octoate (Sn(Oct)2), chloroform, 4-di-methylaminopyridinec (DMAP), N-hydroxysulfosuccinimide (NHS), dimethyl sulfoxide (DMSO), 1,4-dioxane, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT), nitroblue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3′-indolyl phosphate p-toluidine salt (BCIP), as well as HSA-MIR-218 (miR-218) were obtained from Sigma-Aldrich (St Louis, MO). N-Hydroxysuccinimide (NHS) and 1,8-octanediol were purchased from Acros Organics (New Jersey, NJ). 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) was purchased from Thermo Scientific (Rockford, IL). L-Lactide (LLA), glycolide (GA) and cetylpyridinium chloride were obtained from TCI America (Montgomeryville, PA). Maleimide-PEG-NHS (Mal-PEG-NHS) was purchased from Nanocs (Natick, MA). C-R9-G4-IKVAVW was bought from SciLight Biotechnology (Beijing, China). Hoechst 33342, Alexa Fluor647 labeled-miR-218 (AF647-miR-218) and miRNA Control (miRCon) were purchased from Thermo Scientific (Rockford, IL). LysoTracker® Green DND-26 (LysoTracker) was ordered from Cell Signaling Technology (Danvers, MA). Mir-X miRNA FirstStrand Synthesis and SYBR qRT-PCR kits were obtained from Takara Bio (Mountain View, CA). GoScript™ Reverse Transcription Mix was purchased from Promega (Madison, WI). PowerUp™ SYBR® Green Master Mix was purchased from Applied Biosystems (Foster City, CA). PD98059 was obtained from APExBIO (Houston, TX). Wortmannin was obtained from J&K Scientific (San Jose, CA).
2.2. Synthesis and characterization of PPP-RGI
2.2.1. Synthesis and characterization of PLGA-PEI.
Poly (lactic-co-glycolic acid) (PLGA) was synthesized by ring opening polymerization (ROP) of GA and LLA using 1,8-octanediol as the initiator and Sn(Oct)2 as the catalyst. Briefly, 0.056 g (0.38 mmol) of 1,8-octanediol, 0.62 g (5.34 mmol) of GA, and 4.38 g (30.42 mmol) of LLA were weighed in a Schlenk flask and subjected to vacuum/nitrogen cycles. The Schlenk flask was sealed under dry nitrogen and immersed in an oil bath at 100 °C for 24 h. The product was then purified via precipitation in n-hexane three times and air-dried. Next, the obtained PLGA (3.0 g, 0.242 mmol), succinic anhydride (0.49 g, 4.84 mmol), Et3N (200 μL), and DMAP (0.59 g, 4.84 mmol) were dissolved in 20 mL of dry 1,4-dioxane in a round-bottom flask. The reaction was performed under a dry nitrogen atmosphere at 25 °C for 24 h. The crude product was obtained by precipitation in cold n-hexane and was re-dissolved in chloroform. The chloroform solution was washed three times with a hydrochloric acid aqueous solution (10% in v/v), four times with a saturated sodium bicarbonate solution, and finally three times with a saturated sodium chloride solution. The organic phase was isolated and dried over anhydrous sodium sulphate and filtered. Afterwards, the solution was precipitated in cold n-hexane and dried under vacuum to obtain PLGA-COOH. To synthesize PLGA-PEI, PLGA-COOH (0.5 g, 0.038 mmol), EDC (0.1179 g, 0.615 mmol), and NHS (0.078 g, 0.677 mmol) were dissolved in 10 mL of DMSO and stirred at room temperature for 1 h, followed by dropwise addition into 2 mL of DMSO containing 1.15 g (0.115 mmol) of PEI. The reaction mixture was stirred for 24 h, then dialyzed and lyophilized to obtain the triblock copolymer PLGA-PEI. To characterize the synthetic polymer, the 1H NMR spectra of the copolymers were recorded with a JEOL Delta NMR spectrometer (JEOL Ltd, Akishima, Japan) operating at 500 MHz in DMSO-d6. Moreover, the FT-IR spectra were obtained using an FT-IR spectrometer (Thermo Scientific Nicolet iS10, USA).
2.2.2. Synthesis of PPP-RGI.
The RGI peptide was conjugated to PLGA-PEI through a Mal-PEG-NHS linker. Briefly, PLGA-PEI (8 mg, 0.24 μmol) and Mal-PEG-NHS (8.16 mg, 2.4 μmol) were dissolved in dimethyl sulfoxide (DMSO) and stirred at room temperature for 2 h. The RGI peptide (1.2 mg, 0.48 μmol) was dissolved in PBS ( pH = 7.4) and slowly dropped in the DMSO solution and stirred overnight. The reaction mixture was purified by dialysis to obtain PLGA-PEI-PEG-Cys-R9-G4-IKVAVW (abbreviated as PPP-RGI1). Similarly, PPP-RGI2 was synthesized from PLGA-PEI (8 mg, 0.24 μmol), Mal-PEG-NHS (8.16 mg, 2.4 μmol) and the RGI peptide (5.9 mg, 2.4 μmol) following the same method.
2.3. Preparation and characterization of PPP-RGI micelles and PPP-RGI/miR-218 complexes
2.3.1. Preparation of PPP-RGI micelles and PPP-RGI/miR-218 complexes.
PPP-RGI was dissolved in DMSO to prepare the solution, at a concentration of 5.0 mg mL−1. 1.0 mL of the solution was added dropwise to 5 mL of ultrapure water under vigorous stirring to form micelles. After 2 h, the micelles were dialyzed to remove DMSO. miR-218 was then diluted with DEPC-treated water to a concentration of 28.8 μg mL−1. The PLGA-PEI/miR-218, PPP/miR-218, PPP-RGI1/miR-218, and PPP-RGI2/miR-218 complexes were prepared by mixing the copolymer and miR-218 solutions with various weight ratios, respectively.
2.3.2. Characterization of the PPP-RGI micelles and PPP-RGI/miR-218 complexes.
The weight percentage of the immobilized RGI peptide in the PPP-RGI micelles was determined by using a TECAN infinite M200 fluorescence plate reader based on the emission wavelength of tryptophan at 348 nm upon excitement at a 280 nm wavelength. The size and zeta potential of the PPP-RGI micelles and PPP-RGI/miR-218 complexes were measured using NanoBrook 90Plus PALS at a constant angle of 90°. Agarose gel electrophoresis was used to evaluate the miR-218 condensation ability of the complexes. Briefly, different ratios of the complexes were first prepared as described above. Then, 2 μL of 6× loading buffer was mixed with 10 μL of the complex suspension and analyzed using 2% agarose gel containing 5 μL of GelRed® in 1× TAE buffer at 100 V for 30 min. The miR-218 retardation was performed using a UV illuminator. In addition, an agarose gel electrophoresis assay was performed to evaluate the ability of PPP-RGI to protect miR-218 against RNase degradation. Before running agarose gel electrophoresis, the PLGA-PEI/miR-218, PPP/miR-218, PPP-RGI1/miR-218, and PPP-RGI2/miR-218 complexes were incubated with RNase for 4 h for digestion. Naked miR-218 was used as a control group.
2.4. Culture of BMSCs
Mouse BMSCs were isolated from the bone marrow of 6–8 week-old C57BL/6 mice as described by Peister et al.24 Briefly, the femur and tibia of mice legs were removed, sterilized, and washed with DPBS. Each end of the tibia and femur was cut and rinsed with α-MEM to collect the marrow cells. The cell pellets were cultured in a red blood cell lysis buffer for 5 min, then centrifuged and re-suspended in α-MEM culture medium, followed by filtration through a 100 μm nylon mesh filter. The isolated BMSCs were then cultured in α-MEM completed medium with 10% fetal bovine serum and 1% penicillin/streptomycin (P/S) in a humidified atmosphere containing 5% CO2 at 37 °C. The BMSCs at passage 2 to passage 6 (P2-P6) were collected and stocked for further experiments.
2.5. In vitro cytotoxicity
The MTT assay was used to evaluate the in vitro cytotoxicity of the gene complexes. The BMSCs were seeded in a 96-well plate with a density of 5 × 103 cells per well and cultured in α-MEM containing 10% FBS for 24 h. Next, the medium was replaced with a serum-free α-MEM medium, and miR-218 (in concentrations ranging from 0.4 to 4.5 μg mL−1) was added into the medium. The cells were cultured in an incubator for 3 days. Afterwards, an MTT solution was added, followed by culturing for another 4 h. Next, 150 μL of DMSO was added to dissolve formazan crystals formed by viable cells. The optical density (OD) of the formazan solution was acquired using a SpectraMax 250 microplate spectrophotometer (Molecular Devices, USA).
2.6. Cellular uptake and intercellular distribution
The BMSCs were seeded in a 6-well plate with a density of 2 × 105 cells per well and cultured in α-MEM containing 10% FBS for 24 h. Before transfection, the medium was replaced with serum-free α-MEM; the PLGA-PEI/miR-218, PPP/miR-218, PPP-RGI1/miR-218, and PPP-RGI2/miR-218 complexes were added and cultured for 4 h. The transfected cells were washed with DPBS and trypsinized with 0.25% trypsin. After the cells were centrifuged and re-suspended in 300 μL DPBS ( pH = 7.4), their cellular uptake and mean fluorescence intensity (MFI) were evaluated using flow cytometry (BD FACS Canto II Flow Cytometry, BD Biosciences, USA).
To evaluate intercellular distribution, 75 nM per well of LysoTracker Green DND-26 was added into the transfected cell medium and incubated for 1 h. Next, 2 μg mL−1 Hoechst 33342 was added and incubated for 20 min. The cells were washed with DPBS (pH = 7.4), and directly imaged by confocal microscopy (Leica STP6000, Germany). The excitation wavelengths were 649 nm, 504 nm, and 350 nm for AF647 (red), LysoTracker Green (green), and Hoechst 33342 (blue), respectively. The co-localization rate (CLR) of endo/lysosomes was calculated using the Image-Pro Plus 6.0 software according to the following equation:
In the equation, the red pixels correspond to the AF647-oligonucleotide in the cytoplasm, and the yellow pixels correspond to the AF647-oligonucleotide in endo/lysosomes.
2.7. Cell proliferation assay
The colony formation assay was used to evaluate the cell proliferation of the transfected BMSCs. Briefly, the BMSCs were first transfected with the PLGA-PEI/miR-218, PPP-RGI1/miR-218, or PPP-RGI2/miR-218 complexes and cultured for 48 h. Next, 1 × 103 transfected cells were trypsinized and seeded in a 6-well plate, and then cultured for 2 weeks. The colonies were washed with PBS, fixed with 4% PFA for 15 min, dyed with crystal violet, and counted under an Olympus SZX16 stereomicroscope (Olympus, Tokyo, Japan). The number of regenerated colonies (>50 cells each) was scored to determine the efficiency of colony formation. In addition, to further evaluate the cell proliferation ability, the PLGA-PEI/miR-218, PPP-RGI1/miR-218 or PPP-RGI2/miR-218 complex transfected BMSCs were stained with the calcein AM dye and imaged under confocal microscopy.
2.8. Alkaline phosphatase (ALP) and Alizarin Red S (ARS) staining
BMSCs were seeded in a 24-well plate with a density of 5 × 104 cells per well. After being transfected with the PLGA-PEI/miR-218, PPP-RGI1/miR-218, or PPP-RGI2/miR-218 complexes, the cells were washed with DPBS twice and cultured with an osteogenic induction medium containing 1 mM dexamethasone, 1 M β-glycerophosphate, and 10 mM ascorbic acid. ALP and ARS staining was examined at day 6 and day 14. For the ALP staining assay, the cells were washed with DPBS and fixed with 4% paraformaldehyde for 20 min. Subsequently, the cells were equilibrated with ALP buffer (100 mM NaCl, 100 mM Tris-HCl pH 9.5, and 50 mM MgCl2) for 5 min and added to the ALP substrate solution (5 μl NBT and 3.75 μl BCIP into 1 mL ALP buffer). Each well was added with 0.3 mL of the ALP substrate solution and incubated for 20 min at 37 °C. The reaction was stopped by washing the cells with distilled water. For ARS staining, the transfected cells were washed with DPBS twice and fixed with 4% paraformaldehyde for 20 min. The cells were then stained with the ARS solution (pH 4.2) for 30 min at room temperature and washed three times. Both ALP and ARS staining were photographed with an Olympus SZX16 stereomicroscope. In addition, to quantify the amount of ARS bound to the mineralized nodules, the bound ARS was dissolved in 10% cetylpyridinium chloride (CPC), and OD values at a 570 nm wavelength were measured with a SpectraMax 250 microplate spectrophotometer.
2.9. qRT-PCR analysis
Total RNA was isolated from the transfected cells with the TRIzol reagent. For miRNA qRT-PCR, RNAs were reverse-transcribed into cDNA using Mir-X miRNA First Strand Synthesis and SYBR qRT-PCR Kits. All the reverse transcription and PCR amplification conditions were set according to the manufacturer’s protocol. Primers for miR-218 or U6 were mixed with the template as well as the Mir-X miRNA First Strand Synthesis and SYBR qRT-PCR Kits. Transcript levels were normalized to U6. For mRNA qRT-PCR, RNAs were reverse-transcribed into cDNA using GoScript™ Reverse Transcription Mix. The resulting cDNAs were used as templates for qRT-PCR using PowerUp™ SYBR® Green Master Mix which was performed using Bio-Red CFX96™ (BioRad, USA). GAPDH was used as an internal normalizing control for mRNA. Relative quantification of gene expression was analyzed using the 2−ΔΔCT method. The primer sequences are listed in Table S1.†
2.10. Western blotting
The transfected cells were washed three times with cold PBS (pH = 7.4) and the total protein was extracted with RIPA lysis buffer. After 30 min, the lysates were centrifuged at 12 000 rpm at 4 °C for 10 min. The total protein was quantified by using a BCA protein assay kit and denatured by adding 4× SDS. The proteins were separated via SDS-PAGE, transferred to PVDF membranes, and incubated with fat-free milk. The PVDF membranes were subjected to immunoblotting using the indicated antibodies. The proteins were then visualized using an enhanced chemiluminescence method and imaged by using a ChemiDoc™ Touch Imaging System (BioRad, USA). The following primary antibodies were used: ALP (sc-166261), RUNX2 (sc-101145), OCN (sc-365797), Col1 (ab-34710), SFRP2 (af1169), DKK2 (af2435), SOST (af1589), and β-actin (sc-47778).
2.11. Statistical analysis
The data were analyzed using the SPSS software (version 19.0). The results were expressed as mean ± standard deviation. Each experiment was repeated 3 times. The differences among the groups were compared by one-way ANOVA. To test the significance of the observed differences between two groups, an unpaired Student’s t-test was applied. The significance level was set at p < 0.05.
3. Results
3.1. Synthesis and characterization of PPP-RGI
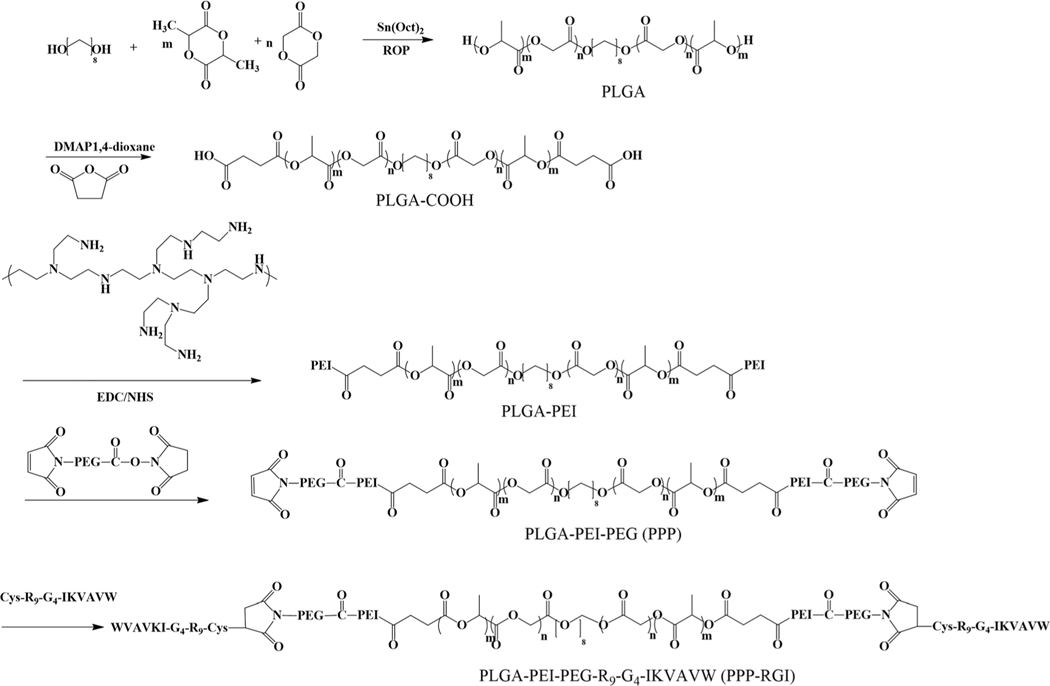
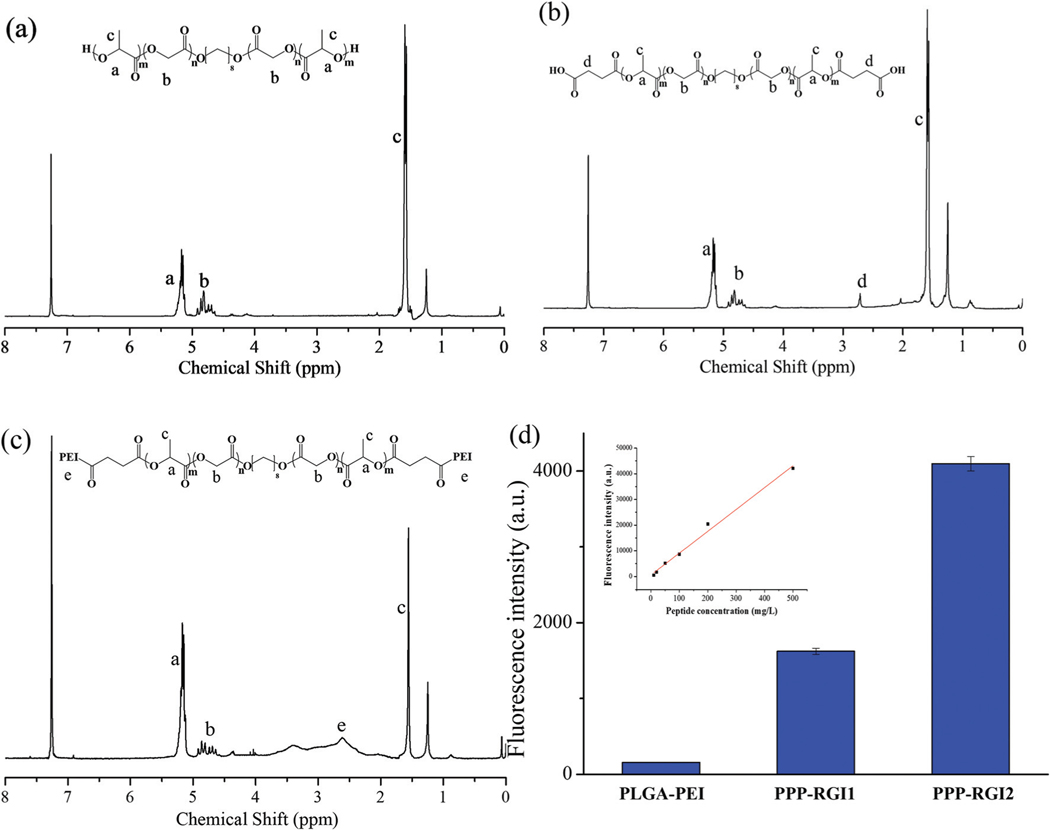
The synthetic route of PPP-RGI is shown in Fig. 1. First, the block copolymer PLGA-PEI was synthesized through a three-step process, including the formation of dihydroxyl PLGA via the ring opening polymerization (ROP) of GA and LLA, the transformation of PLGA-OH to PLGA-COOH, and the reaction of PLGA-COOH with PEI to form amphiphilic PLGA-PEI. Next, the PLGA-PEI was functionalized by Mal-PEG-NHS to obtain PPP, which was further reacted with RGI to form the final product PPP-RGI. The number average molecular weight of the PLGA was 12388 with a polydispersity index of 1.50. Fig. 2 shows the 1H NMR spectra of the synthesized PLGA, PLGA-COOH, and PLGA-PEI. The peak at 4.80 ppm (OOCCH2, 2H) represents the GA residue, while the peaks at 5.17 ppm (OOCCHCH3, 1H) and 1.56 ppm (OOCCHCH3, 3H) represent the LA residue in the copolymer. The peaks at 2.71 ppm (OCCH2CH2CO, 4H) and 2.2–3.0 ppm (NH-CH2-CH2, PEI) indicate the successful syntheses of PLGA-COOH and PLGA-PEI. The synthetic polymers were further characterized by FT-IR (Fig. S1†). The signal at 1750 cm−1 is assigned to the carboxylate group (–O–CO–) in the PLGA. After grafting the PEI, the symmetric stretching characteristic peaks of the primary amine (–NH2) of the PEI were observed at 3273 and 1644 cm−1, and the characteristic peak of the amine (–CO–NH–) of the PLGA-PEI was observed at 1533 cm−1.
Fig. 1.
Synthesis route of multifunctional PPP-RGI.
Fig. 2.
Characterization of PPP and PPP-RGI. (a–c) 1H NMR spectra of PLGA (a), PLGA-COOH (b), and PLGA-PEI (c). (d) The fluorescence intensity of PPP-RGI1 and PPP-RGI2.
The content of the RGI peptide in PPP-RGI was determined by fluorescence spectroscopy based on the chromophoric group (indole) of tryptophan in PPP-RGI (Fig. 2d). Low and high concentrations of the RGI peptide were conjugated separately onto the PPP copolymer. The obtained PPP-RGI1 and PPP-RGI2 contained 6.2 wt% and 20.7 wt% of RGI, respectively. Both PPP-RGI1 and PPP-RGI2 were tested in the following experiments.
PPP-RGI self-assembled into the micelles and formed PPP-RGI/miR-218 complexes when miR-218 was condensed in the PPP-RGI micelles. PPP-RGI completely condensed miR-218 at the w/w ratio of 5 (Fig. S2a†). Higher w/w ratios led to more positive charges on the surface of the micelles and increased the cytotoxicity. Therefore, the w/w ratio of PPP-RGI/miR-218 = 5/1 was selected in this study. With the addition of exogenous RNase, PPP-RGI/miR-218 suppressed the digestion of miR-218, whereas free miR-218 was gradually degraded (Fig. S2b†). These results indicated that miR-218 was well protected by the PPP-RGI micelles.
The sizes of the PLGA-PEI/miR-218, PPP/miR-218, and PPP-RGI/miR-218 complexes ranged from 145 to 235 nm, and did not vary significantly with the ratios of miR-218 in the complexes (Fig. S2c†). However, the zeta potential of the complexes increased with the ratios of miR-218 in the complexes (Fig. S2d†). At the same ratio, the PPP/miR-218 complex exhibited a lower surface charge density than the PLGA-PEI/miR-218 complex due to the shield effect of PEG in the PPP copolymer. Meanwhile, increasing the amount of the RGI peptide increased the zeta potential of the PPP-RGI/miR-218 complex.
3.2. Cellular uptake and intercellular distribution
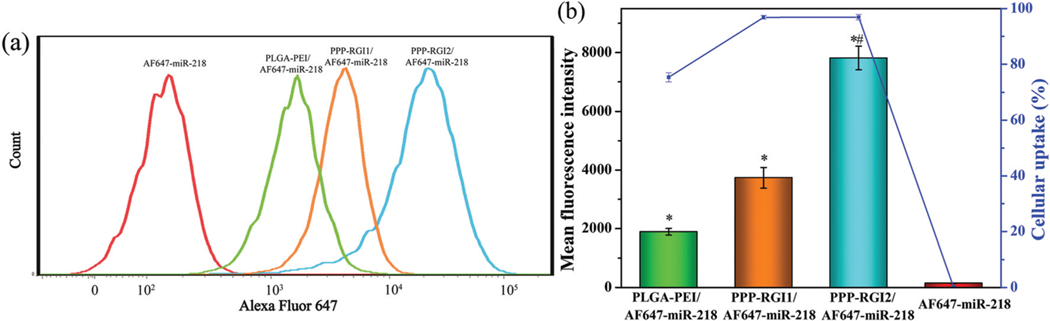
Flow cytometry was performed to quantitatively evaluate the cellular uptake of miR-218 using AF647-miR-218 as a reporter gene. As shown in Fig. 3, the cellular uptake of naked miR-218 was negligible, while miR-218 in the PPP-RGI/AF647-miR-218 group exhibited more than 75% cellular uptake. Meanwhile, the mean fluorescence intensity (MFI) values of PPP-RGI/AF647-miR-218 were 2–4 fold higher than that of PLGA-PEI/AF647-miR-218. In addition, the MFI value of PPP-RGI2/AF647-miR-218 was 2.1 fold higher than that of PPP-RGI1/AF647-miR-218, owing to the higher amount of the R9 membrane-penetration peptide in PPP-RGI2.
Fig. 3.
(a) Cellular uptake of micelle/AF647-miR-218 complexes 4 h after transfection. (b) Mean fluorescence intensity (MFI) measured by flow cytometry and percentages of cellular uptake measured by confocal microscopy. N = 3, *P < 0.05 vs. AF-647-miR-218 group, #P < 0.05 vs. PLGA-PEI/AF647-miR-218.
PLGA-PEI/AF647-miR-218 and PPP-RGI/AF647-miR-218 were incubated with the BMSCs for 4 h and 24 h to investigate the intracellular distribution of the complexes (Fig. 4). The co-localization ratio (CLR) value of the endo/lysosomes in PPP-RGI/AF647-miR-218 was higher than that of PLGA-PEI/AF647-miR-218 at 4 h. In addition, PPP-RGI2/AF647-miR-218 showed the highest endo/lysosome CLR of 79.6 ± 3.2% at 4 h. From 4 h to 24 h, the endo/lysosome CLR of PPP-RGI2/AF647-miR-218 reduced to 24.1 ± 3.6%, indicating that 69.7% of the complexes in the endo/lysosomes escaped from the endo/lyso-somes and released into the cytoplasm. In contrast, the escape efficiency in PPP-RGI1/AF647-miR-218 and PLGA-PEI/miR-218 was 47.7% and 37.9%, respectively. Furthermore, Fig. S3† showed that after being treated with PPP-RGI2/AF647-miR-218 for 24 h, the MFI exhibited no significant decrease compared to the group treated for 4 h. These results demonstrated that the RGI peptide accelerated the endo/lysosome escape of the complexes. In addition, the nucleus CLR was hardly detected, indicating that miR-218 did not accumulate in the nucleus.
Fig. 4.
(a) CLSM images of intracellular trafficking of micelle/AF647-miR-218 complexes at 4 h. (b) CLSM images of intracellular trafficking of micelle/AF647-miR-218 complexes at 24 h. (c) The endo/lysosome co-localization ratio (CLR) values at 4 and 24 h.
3.3. Cell viability and proliferation
The cytotoxicity of the micelle/miR-218 was evaluated using an MTT assay. As shown in Fig. S4,† all four complexes showed a higher cell viability when compared to PEI/miR-218. In addition, the BMSCs treated with PPP/miR-218 and PPP-RGI/miR-218 showed higher cell viabilities than PLGA-PEI/miR-218. At a low miR-218 concentration (0.4 μg mL−1), PPP-RGI1/miR-218 and PPP-RGI2/miR-218 showed a relative cell viability of 108.7% and 122.5%, respectively.
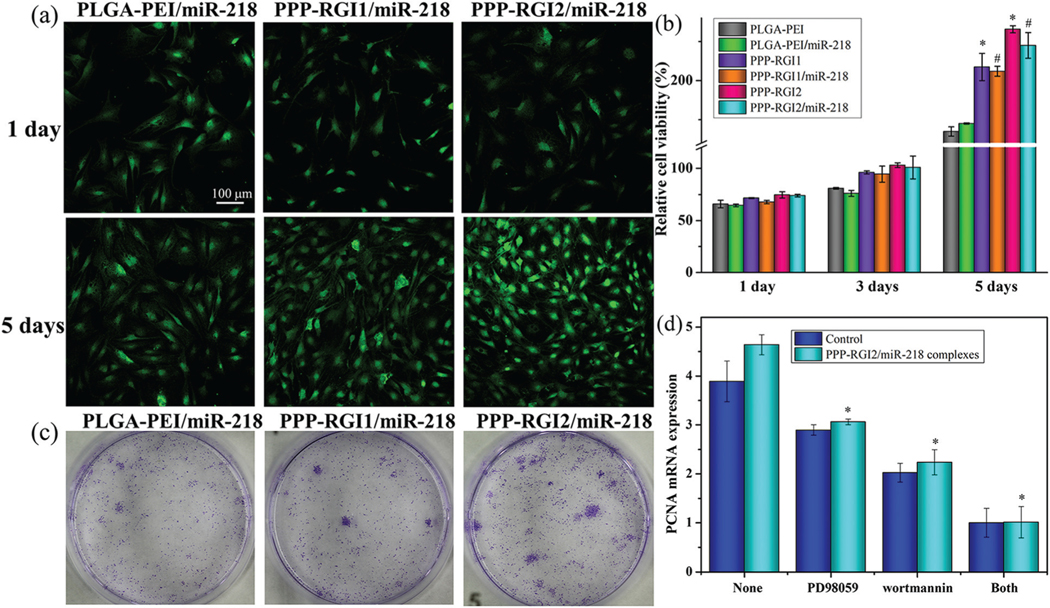
Next, the BMSCs were treated with the complexes for up to 5 days to evaluate cell proliferation. At day 1, the treated BMSCs showed no significant difference in cell proliferation among the transfected groups (Fig. 5a). The number of BMSCs increased with culture time in all groups. Meanwhile, the cell proliferation rates between the micelles and corresponding complexes showed no significant difference at the miR-218 concentration of 2.9 μg mL−1. However, the cell numbers in the PPP-RGI2 and PPP-RGI2/miR-218 groups were 1.39 and 1.29 fold higher than those of the PLGA-PEI and PLGA-PEI/ miR-218 groups, respectively, indicating that the introduction of the IKVAV peptide promoted BMSC proliferation.
Fig. 5.
(a and b) The relative cell viability of BMSCs treated with micelles and complexes at the miR-218 concentration of 2.9 μg mL−1 for up to 5 days. (c) Colony formation assay of the BMSCs treated with complexes. (d) The effects of PD98059 and wortmannin on the PCNA synthesis of BMSCs transfected with PPP-RGI2/miR-218 complexes.
The proliferation of the transfected BMSCs was further confirmed with a plate colony formation assay. Compared with PLGA-PEI/miR-218, the colony-forming ability of PPP-RGI/miR-218 was significantly increased (Fig. 5c). In addition, the PPP-RGI2/miR-218 group had higher colony-forming ability than that of the PPP-RGI1/miR-218 group.
The BMSCs were pretreated with PD98059 (a MAPK/ERK pathway inhibitor) and wortmannin (a PI3K/Akt pathway inhibitor) and then transfected with PPP-RGI2/miR-218 to determine whether the MAPK/Akt signaling pathways were activated by the PPP-RGI complexes. As shown in Fig. 5d, the proliferating cell nuclear antigen (PCNA) expression was reduced to 66.0% by PD98059, to 48.3% by wortmannin, and to 21.8% by both inhibitors. This result indicated that the MAPK/Akt signaling pathways were involved in the proliferation of the PPP-RGI2/miR-218-treated BMSCs.
3.4. miR-218 expression, RT-PCR, western blotting, and in vitro mineralization
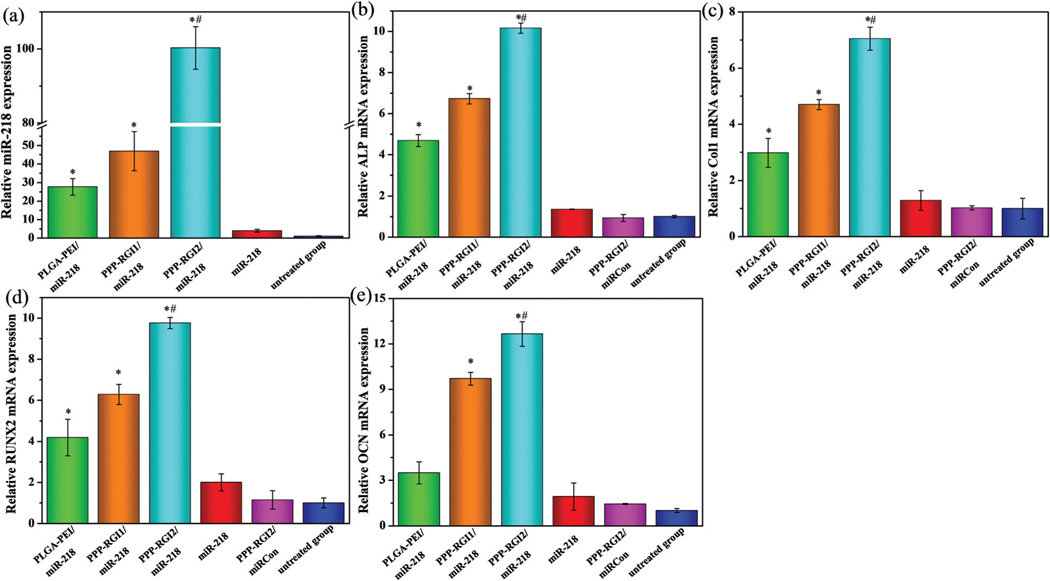
RT-PCR was conducted to detect the miR-218 expression in BMSCs after transfection with PPP-RGI/miR-218 (Fig. 6a). Compared to the control group, the miR-218 expression increased in the BMSCs transfected with PLGA-PEI/miR-218. In addition, the BMSCs transfected with PPP-RGI/miR-218 exhibited significantly higher miR-218 expression after the introduction of the RGI peptide. In the PPP-RGI2/miR-218 group, the miR-218 expression was 2.0 fold higher than that of the PPP-RGI1/miR-218 group and 3.6 fold higher than that of the PLGA-PEI/miR-218 group, separately. The results indicated that PPP-RGI/miR-218, especially PPP-RGI2/miR-218 with a higher content of RGI, was effective at up-regulating the intracellular miR-218 level.
Fig. 6.
Relative miR-218 and osteogenic marker expressions of different groups transfected with miR-218-containing complexes. (a) miR-218, (b) ALP, (c) Col1, (d) RUNX2, and (e) OCN. N = 3, *P < 0.05 vs. untreated group, #P < 0.05 vs. PLGA-PEI/miR-218.
The osteogenic markers (RUNX2, ALP, Col1, and OCN) were evaluated after the BMSCs transfected with PPP-RGI/miR-218 were cultured in an osteogenic induction medium for 14 days (Fig. 6). The expression levels of ALP, Col1, RUNX2, and OCN in the PPP-RGI2/miR-218 group were 2.2, 2.4, 2.3 and 3.6 fold higher than those in the PLGA-PEI/miR-218 group, and 1.5, 1.5, 1.6, and 1.3 fold higher than those in the PPP-RGI1/miR-218 group, respectively. Meanwhile, the PPP-RGI2/miRCon complex-treated group had similar mRNA levels to the control group, confirming the osteogenic capability of miR218 in the complexes.
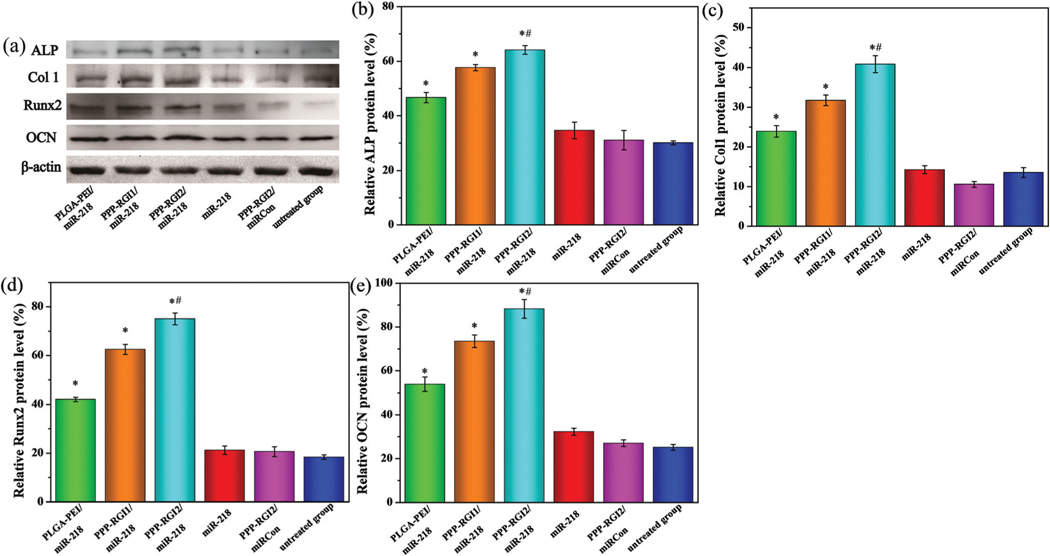
The expressions of osteogenic genes (ALP, Col1, RUNX2, and OCN) were measured at the protein level by western blotting analysis. The expressions of the osteogenic proteins in the transfected BMSCs were significantly higher than those of the control group 14 days after osteogenic induction (Fig. 7). In addition, the protein expression levels of ALP, Col1, RUNX2, and OCN in the PPP-RGI2/miR-218 group were 11.0%, 28.8%, 20.1%, and 20.2% higher than those in the PPP-RGI1/ miR-218 group, respectively.
Fig. 7.
(a) Western blotting analysis and the protein expression of (b) ALP, (c) Col1, (d) RUNX2, and (e) OCN 14 days after osteogenic induction. N = 3, *P < 0.05 vs. untreated group, #P < 0.05 vs. PLGA-PEI/miR-218.
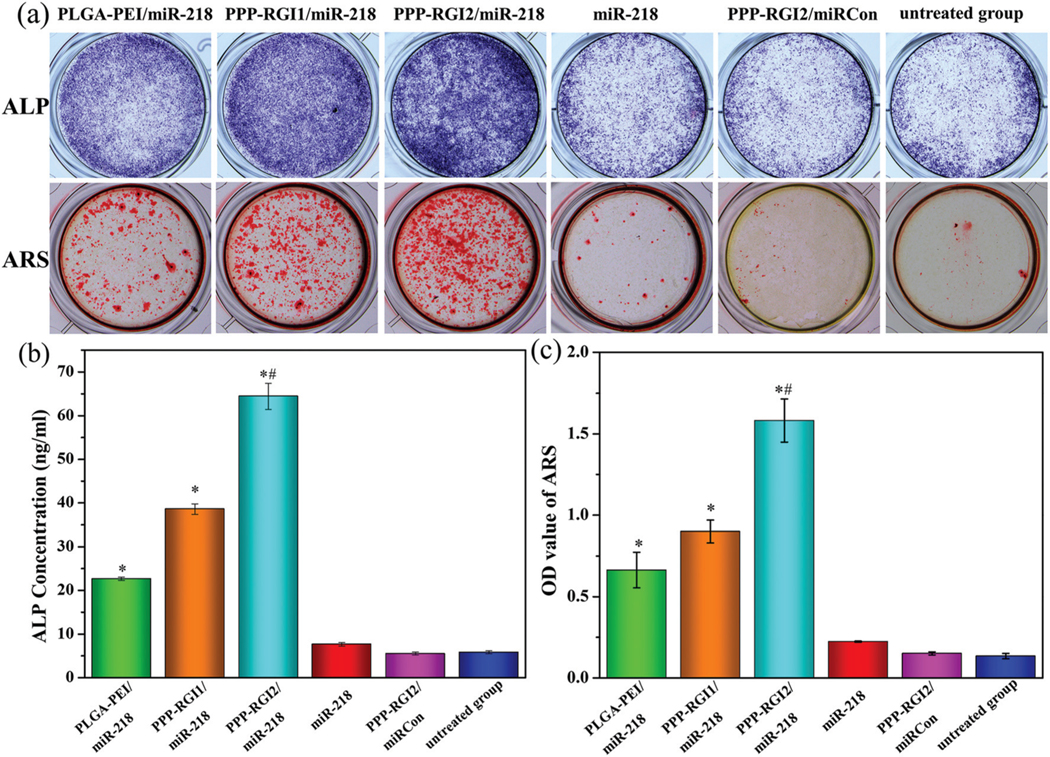
In vitro mineralization was performed by ALP staining at day 6 and ARS staining at day 14 (Fig. 8). The ALP concentration of the PLGA-PEI/miR-218 group (22.7 ng mL−1) was approximately 3 fold higher than that of the miR-218 group (Fig. 8b). Subsequently, the ALP concentration of the PPP-RGI2/miR-218 group was 1.7 fold and 2.8 fold higher than that of the PPP-RGI1/miR-218 group and the PLGA-PEI/miR-218 group, respectively.
Fig. 8.
(a) ALP and ARS staining after BMSCs were transfected with the micelle/miR-218 complexes for 6 days and 14 days, separately. (b) The ALP concentrations in different groups. (c) The OD values of ARS in different groups. N = 3, *P < 0.05 vs. untreated group, #P < 0.05 vs. PLGA-PEI/miR-218.
Furthermore, ARS staining at day 14 revealed a dramatic improvement in calcium deposition in the PPP-RGI2/miR-218 group compared with other groups. The relative calcium deposition of the PPP-RGI2/miR-218 group was 1.8 fold and 2.4 fold higher than those of the PPP-RGI1/miR-218 and PLGA-PEI/miR-218 groups, respectively (Fig. 8c). Collectively, these data suggested that PPP-RGI2/miR-218 enhanced the cellular uptake of miR-218 and played a positive role in regulating the osteogenic differentiation of BMSCs.
3.5. miR-218 regulates the osteogenic differentiation of BMSCs via the Wnt signaling pathway
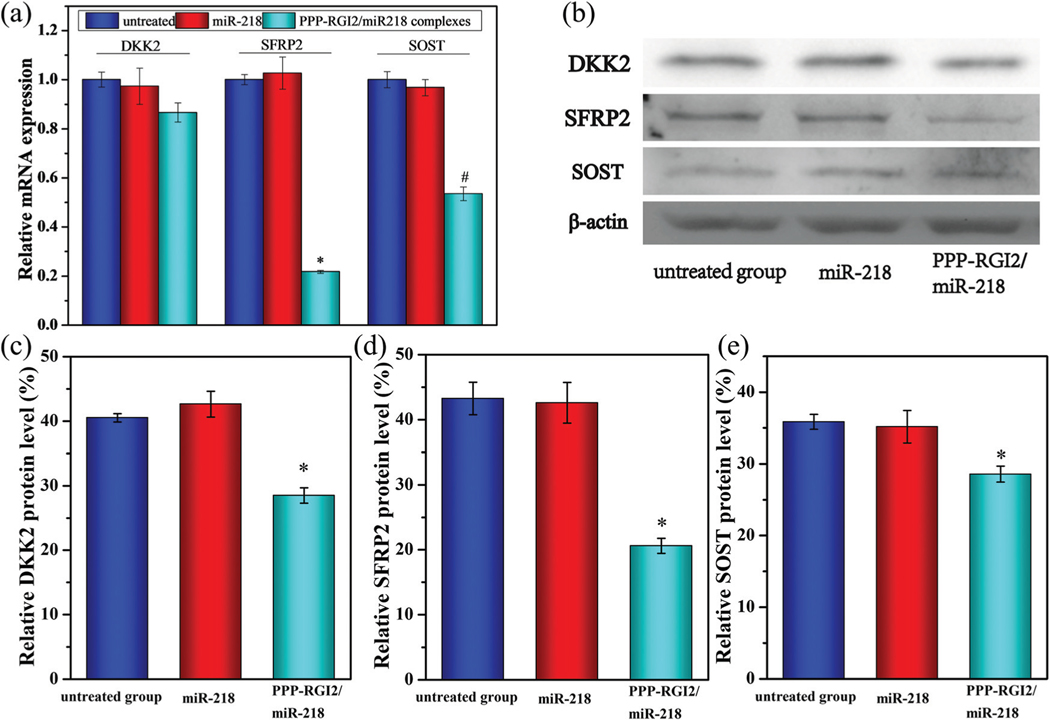
The expressions of DKK2, SFRP2, and SOST (three Wnt targeting molecules) between the untreated group and the nuke miR-218 treated group had no significant differences at the mRNA and protein levels. The PPP-RGI2/miR-218 treated group reduced the mRNA levels of DKK2 by 13.3%, SFRP2 by 74.0%, and SOST by 46.5%, separately (Fig. 9a). In addition, the western blotting result showed that after transfection with the PPP-RGI2/miR-218 complexes, the expressions of DKK2, SFRP2, and SOST also reduced by 33.3%, 51.6%, and 18.5%, respectively (Fig. 9c–e). These results indicate that miR-218 stimulated BMSC differentiation by targeting multiple Wnt signaling pathway inhibitors.
Fig. 9.
(a) The expressions of miR-218 in untreated, miR-218, and PPP-RGI2/miR-218 groups. (b–e) Western blotting analyses of the expressions of DKK, SFRP2, and SOST at the protein level. N = 3, *P < 0.05 vs. miR-218 treated group, #P < 0.05 vs. miR-218 treated group.
4. Discussion
A suitable miRNA vector needs to have several crucial features, including low toxicity, high cellular uptake, high endo/lysosome escape, and excellent biodegradability. Several miRNA delivery strategies were recently developed in an attempt to promote the osteogenic differentiation of BMSCs.25 For example, arginine-rich low molecular weight protamine (LMWP) was used as an miR-29b vector to regulate the osteoblastic differentiation of human BMSCs in vitro.26 However, the instability of the LMWP vector and the low endo/lysosome escape affected the delivery efficacy. In another study, PEI-capped gold nanoparticles were used as a delivery vector for miR-29b to regulate osteogenesis.27 Although inorganic gold nanoparticles exhibited relatively high gene transfection efficiency and low cytotoxicity in vitro, the non-biodegradation of the vector is a concern for in vivo applications. Herein, we designed and synthesized a multifunctional, peptide-conjugated, non-viral miRNA vector PPP-RGI to address the above challenges. We first synthesized a biodegradable PLGA-based copolymer as the building material of the miRNA vector. We then introduced PEG to reduce the cytotoxicity of the vector. Next, to enhance cellular uptake and endo/lysosome escape, we conjugated the multifunctional peptide on the biodegradable copolymer PLGA-PEI. The RGI contains four unique components: R9-G4-IKVAV-W, in which R9 acts as a cell-enetrating peptide to enhance cellular uptake, the IKVAV peptide is used to enhance BMSC adhesion and proliferation, G4 serves as a flexible spacer to regulate the activity of R9 and IKVAV, and the W residue provides a fluorescence spectrum at 348 nm to trace gene delivery and quantify the multifunctional peptide. Cell-penetrating peptides (CPPs), also known as protein transduction domains (PTDs), are a collection of different families of short peptides that enter cells by penetrating cell membranes.28 Some of the most commonly used peptides include Tat and polyarginine. These CPPs have successfully delivered proteins, nucleic acids, and nanocarriers into different cell types.29–31 Studies have shown that R9 was 20-fold more efficient than Tat 49–57 in terms of cellular uptake, as determined by the Michaelis–Menton kinetic analysis.32 Therefore, R9 was selected as the penetration peptide for intracellular delivery in this work.
Owing to the positive charge of PEI and cationic R9 on micelle surfaces, PPP-RGI effectively condensed miR-218 and formed positive charged micelle/miR-218 complexes. The micelle/miR-218 complexes displayed sizes ranging from 145 nm to 235 nm, which are suitable for endocytosis via the clathrin-mediated pathway.33 By the assistance of R9, the cellular uptake of the PPP-RGI2/AF647-miR-218 complex increased to 97% (Fig. 3). Meanwhile, strong intracellular trafficking capability to escape from endosomes after endocytosis is crucial for gene transfection. Owing to the “proton sponge effect”,34 PEI in the PPP complex ruptured the lysosomal membrane and the complex was subsequently released into the cytoplasm. In addition, the endo/lysosome escape of the micelle/miR-218 complex increased with the amount of the RGI peptide in the micelle. This response occurred probably because arginine-rich R9 destabilized anionic endosomal membranes.35 However, CPP endosomal escape is an inefficient process, and endosomal fusion to lysosomes leads to the degradation of peptides and proteins.36 The combination of R9 and PEI in PPP-RGI exhibited a synergistic effect on promoting the release of miR-218 into the cytoplasm (Fig. 4).
In the process of bone formation, BMSC proliferation is desirable prior to differentiation because it leads to more osteoblasts, differentiating into osteocytes and therefore accelerating bone tissue formation.37,38 As the active sequence of laminin, the IKVAV peptide was used to promote the adhesion, growth and proliferation of neural stem cells.39,40 The IKVAV peptide was also found to enhance BMSC proliferation through activating the Akt and ERK1/2 signaling pathways.19 Hence, we introduced the IKVAV peptide into the PPP copolymer. Compared with the PLGA-PEI and PLGA-PEI/miR-218 treated groups, the PPP-RGI2 and PPP-RGI2/miR-218 transfected BMSCs showed higher proliferation rates. Furthermore, the PPP-RGI2/miR-218 transfected BMSCs had higher colony-forming ability than PPP-RGI1/miR-218 due to the higher ratio of the IKVAV peptide in PPP-RGI2 (Fig. 5c). These results demonstrated that the incorporation of the IKVAV peptide in the PLGA-PEI micelles efficiently enhanced BMSC proliferation.
PPP-RGI/miR-218 promoted the osteogenic differentiation of BMSCs with enhanced expressions of osteogenic markers (ALP, Col1, OCN, and RUNX2) and calcium nodule formation. ALP is highly expressed at the early stage of osteogenic differentiation. ALP promotes mineralization by hydrolyzing pyrophosphate and generating inorganic phosphate, thus regulating bone formation.41 Col1 and OCN serve as early and late markers of osteogenic differentiation, respectively, whereas RUNX2 is a transcription factor that regulates the differentiation and maturation of osteogenic differentiation.42 The expression levels of ALP, Col1, RUNX2, and OCN in the PPP-RGI2/miR-218 group were all up-regulated compared to other groups. These results were further verified at the protein level and with the in vitro calcium deposition (Fig. 7 & 8).
MiRNAs are crucial post-transcriptional repressors in supporting osteoblast differentiation by compromising mRNA stability and/or by blocking protein translation. A number of miRNAs, including miR-218, have been used as regulators of osteoblasts.25 MiR-218 was significantly up-regulated during osteoblast differentiation, and was predicted to target multiple inhibitors of Wnt signaling.22,23 We examined three potential targets (DKK2, SFRP2, and SOST) of miR-218 to understand the mechanism by which miR-218 controls the osteogenic differentiation of BMSCs. Our results show that miR-218 directly down-regulated the expressions of the three targets at the transcriptional and translational levels, except that DKK2 had no significant difference at the transcriptional level. While DKK2 is an antagonist of canonical Wnt signaling, DKK2 was also found to be an agonist in terminal osteoblast differentiation into mineralized matrices.43 These results suggest that the precise role of DKK2 in the regulation of BMSCs needs further investigation. Together, our data show that miR-218 targeted multiple Wnt signal pathway inhibitors and can be used to promote osteoblast differentiation.
In the future work, we will first incorporate the PPP-RGI/miR-218 complex into a 3D biomimetic scaffold for controlled miR-218 delivery. After optimizing the delivery of miR-218, we will implant the PPP-RGI/miR-218/scaffold construct into a rat alveolar bone defect44 to accelerate bone regeneration in vivo. Our ultimate goal is to develop a PPP-RGI/miR-218/scaffold for the treatment of periodontitis-induced alveolar bone loss, which is beyond the scope of our current work and will be reported separately in the future.
5. Conclusion
A novel non-viral gene vector PPP-RGI was designed and synthesized by conjugating a multifunctional peptide RGI with an amphipathic, biodegradable, block copolymer PPP. The synthetic PPP-RGI exhibited high stability, high cellular uptake efficiency, and improved proliferation owing to the incorporation of functional units PEI, R9 and IKVAV in PPP-RGI. Furthermore, PPP-RGI/miR-218 significantly promoted the osteogenic differentiation of BMSCs. Multifunctional PPP-RGI is an efficient miR-218 delivery vector for accelerated osteogenesis.
Supplementary Material
Acknowledgements
This work was supported by NIH/NIDCR DE024979 and DE029860 and Texas A&M Triads for Transformation (T3). We acknowledge Dr Cancan Xu and Yi Hong (UT Arlington) for the assistance with 1H NMR analysis, and Dr Uday Kumar Chintapula and Kytai Truong Nguyen (UT Arlington) for their assistance with particle size and zeta potential analyses. We thank Dr Xianghong Luan for her assistance with signaling pathway analysis. We also thank Meghann Holt for her assistance with the editing of this article.
Footnotes
Conflicts of interest
The authors declare that they have no known competing financial interest.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1nr02638k
References
- 1.Gori M, Trombetta M, Santini D. and Rainer A, Expert Opin. Biol. Ther, 2015, 15, 1601–1622. [DOI] [PubMed] [Google Scholar]
- 2.Chen K. and Rajewsky N, Nat. Rev. Genet, 2007, 8, 93–103. [DOI] [PubMed] [Google Scholar]
- 3.Peng S, Gao D, Gao C, Wei P, Niu M. and Shuai C, Mol. Med. Rep, 2016, 14, 623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong E. and Reddi AH, Tissue Eng., Part B, 2012, 18, 445–453. [DOI] [PubMed] [Google Scholar]
- 5.Jia J, Tian Q, Ling S, Liu Y, Yang S. and Shao Z, FEBS Lett, 2013, 587, 3027–3031. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Wang Z. and Gemeinhart RA, J. Controlled Release, 2013, 172, 962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang W, Wang F, Feng L, Yan S. and Guo R, Curr. Gene Ther, 2018, 18, 21–28. [DOI] [PubMed] [Google Scholar]
- 8.Guo Y, Zhou L, Wang M, Li Y. and Lei B, Chem. Eng. J, 2021, 405, 127085. [Google Scholar]
- 9.Kim H, Kim D, Ku SH, Kim K, Kim SH and Kwon C, J. Biomater. Sci., Polym. Ed, 2017, 28, 1070–1085. [DOI] [PubMed] [Google Scholar]
- 10.Wu D, Zhang Y, Xu X, Guo T, Xie D, Zhu R, Chen S, Ramakrishna S. and He L, Acta Biomater, 2018, 72, 266–277. [DOI] [PubMed] [Google Scholar]
- 11.Nouri F, Sadeghpour H, Heidari R. and Dehshahri A, Int. J. Nanomed, 2017, 12, 5557–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mastorakos P, Zhang C, Berry S, Oh Y, Lee S, Eberhart CG, Woodworth GF, Suk JS and Hanes J, Adv. Healthcare Mater, 2015, 4, 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L, Shu S, Cheung GS and Wei X, BioMed Res. Int, 2016, 2016, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu L, Faig A, Abdelhamid D. and Uhrich K, Acc. Chem. Res, 2014, 47, 2867–2877. [DOI] [PubMed] [Google Scholar]
- 15.Kolte A, Patil S, Lesimple P, Hanrahan JW and Misra A, Int. J. Pharm, 2017, 524, 382–396. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Zhang X, Zhang J, Mu X, Duan Q, Wang T. and Tian H, J. Mater. Sci.: Mater. Med, 2018, 29, 1–10. [DOI] [PubMed] [Google Scholar]
- 17.Hu H, Wang J, Wang H, Tan T, Li J, Wang Z, Sun K, Li Y. and Zhang Z, Theranostics, 2018, 8, 3597–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banerjee P, Pal S, Kundu N, Mondal D. and Sarkar N, Chem. Commun, 2018, 54, 11451–11454. [DOI] [PubMed] [Google Scholar]
- 19.Li B, Qiu T, Zhang P, Wang X, Yin Y. and Li S, Cell Proliferation, 2014, 47, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wriggers W, Chakravarty S. and Jennings PA, Biopolymers, 2005, 80, 736–746. [DOI] [PubMed] [Google Scholar]
- 21.Fang S, Deng Y, Gu P. and Fan X, Int. J. Mol. Sci, 2015, 16, 8227–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassan MQ, Maeda Y, Taipaleenmaki H, Zhang W, Jafferji M, Gordon JA, Li Z, Croce CM, Van Wijnen AJ and Stein JL, J. Biol. Chem, 2012, 287, 42084–42092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, Qiao X, Zhang Z. and Li C, Biosci. Rep, 2020, 40, BSR20182121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peister A, Mellad JA, Larson BL, Hall BM, Gibson LF and Prockop DJ, Blood, 2004, 103(5), 1662–1668. [DOI] [PubMed] [Google Scholar]
- 25.Leng Q, Chen L. and Lv Y, Theranostics, 2020, 10, 3190–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suh JS, Lee JY, Choi YS, Chung CP and Park YJ, Biomaterials, 2013, 34, 4347–4359. [DOI] [PubMed] [Google Scholar]
- 27.Pan T, Song W, Gao H, Li T, Cao X, Zhong S. and Wang Y, ACS Appl. Mater. Interfaces, 2016, 8, 19217–19227. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Jin Y, Plummer MR, Pooyan S, Gunaseelan S. and Sinko PJ, Mol. Pharmaceutics, 2009, 6, 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng Z, Luan L, Kang Z, Feng S, Meng Q. and Liu K, J. Mater. Chem. B, 2017, 5, 74–84. [DOI] [PubMed] [Google Scholar]
- 30.Moku G, Layek B, Trautman L, Putnam S, Panyam J. and Prabha S, Cancers, 2019, 11, 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang S, Meng Z, Kang Z, Sun C, Wang T, Feng S, Meng Q. and Liu K, RSC Adv, 2018, 8, 28356–28366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L. and Rothbard JB, Proc. Natl. Acad. Sci. U. S. A, 2000, 97, 13003–13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rejman J, Oberle V, Zuhorn IS and Hoekstra D, Biochem. J, 2004, 377, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benjaminsen RV, Mattebjerg MA, Henriksen JR, Moghimi SM and Andresen TL, Mol. Ther, 2013, 21, 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tseng Y-C and Huang L, J. Biomed. Nanotechnol, 2009, 5, 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen JK, Brock DJ, Kondow-McConaghy HM and Pellois J-P, Biomolecules, 2018, 8, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Ju W, Shang P, Lei L. and Nie H, J. Mater. Chem. B, 2015, 3, 1907–1920. [DOI] [PubMed] [Google Scholar]
- 38.Yan M, Ni J, Shen H, Song D, Ding M. and Huang J, RSC Adv, 2017, 7, 19621–19629. [Google Scholar]
- 39.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA and Stupp SI, Science, 2004, 303, 1352–1355. [DOI] [PubMed] [Google Scholar]
- 40.Kubinová Š, Horák D, Kozubenko N, Vaněček V, Proks V, Price J, Cocks G. and Syková E, Biomaterials, 2010, 31, 5966–5975. [DOI] [PubMed] [Google Scholar]
- 41.Li N, Zhou L, Xie W, Zeng D, Cai D, Wang H, Zhou C, Wang J. and Li L, Chem. Eng. J, 2019, 371, 618–630. [Google Scholar]
- 42.Wang Q, Zhao G, Xing Z, Zhan J. and Ma J, Exp. Ther. Med, 2019, 17, 764–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, Liu P, Liu W, Maye P, Zhang J, Zhang Y, Hurley M, Guo C, Boskey A. and Sun L, Nat. Genet, 2005, 37, 945–952. [DOI] [PubMed] [Google Scholar]
- 44.Li X, Liu X, Ni S, Liu Y, Sun H. and Lin Q, J. Craniofac. Surg, 2019, 47, 1147–1154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.