Abstract
Periodontal ligament (PDL) is assembled from highly organized collagen fiber bundles (PDL principal fibers) that are crucial in supporting teeth and buffering mechanical force. Therefore, regeneration of PDL needs to reconstruct these well-ordered fiber bundles to restore PDL functions. However, the formation of PDL principal fibers has long been a challenge due to the absence of an effective three-dimensional (3D) matrix to guide the growth of periodontal ligament stem cells (PDLSCs) and to inhibit the osteogenic differentiation of PDLSCs during the PDL principal fibers deposition. In this work, we designed and fabricated a bio-inspired tubular 3D matrix to guide the migration and growth of human PDLSCs and form well-aligned PDL principal fibers. As a biomimetic 3D template, the tubular matrix controlled PDLSCs migration inside the tubules and aligned the cells to the designated direction. Inside the tubular matrix, the PDLSCs expressed PDL markers and formed oriented fiber bundles with the same size and density as those of natural PDL principal fibers. Furthermore, the tubular matrix downregulated the osteogenic differentiation of PDLSCs. A mechanism study revealed that the Yap1/Twist1 signaling pathway was involved in the inhibition of PDLSCs osteogenesis within the tubular matrix. This work provides an effective approach to induce PDLSCs to form principal fibers and gives insight into the underlying mechanism of inhibiting the osteogenic differentiation of PDLSCs in biomimetic tubular matrices.
Keywords: principal fibers, periodontal ligament, periodontal regeneration, tubular matrix, osteogenic differentiation
Graphical Abstract
1. INTRODUCTION
Periodontitis is a common chronic disease that degrades periodontal tissues, including the periodontal ligament (PDL). In the United States, 47% of adults are affected by periodontitis.1,2 Current clinical treatments for periodontitis are local inflammation control and plaque removal, including gross debridement, scaling and root planing, and surgery. Although these therapies relieve symptoms and prevent progression of the disease, they cannot rebuild the lost periodontal tissues or restore the original attachment of teeth to the alveolar bone. Tissue-engineering-based strategies have been extensively explored to regenerate periodontal tissues and restore their biological functions in recent years.3–8
As a highly organized fibrous tissue, PDL mainly consists of principal fibers with one side inserted into cementum and the other side embedded in alveolar bone at various insertion angles. Our recent work further indicates that the sizes of the terminal ends for the principal fibers (Sharpey’s fibers) are in the range of 5−10 μm with a density of 5000−10000 fibers/mm2.9 These PDL principal fiber bundles form a continuous anastomosing network between teeth and alveolar bone and play the primary role in supporting teeth and absorbing masticatory force. Therefore, it is crucial to reconstruct the well-organized PDL principal fiber bundles during periodontal tissue regeneration.
To date, there are no effective biomimetic scaffolds to guide periodontal ligament stem cells (PDLSCs) and form well-organized principal fiber bundles. For example, trilayered and biphasic scaffolds only regenerated disordered fibrous tissue on the surface of a tooth root.10,11 Likewise, a micropatterning approach was used to arrange PDL cells with actin filaments parallel to the strips.12 While this work demonstrated the effectiveness of geometrical cues to regulate cell alignment, the micropatterned substrate was two-dimensional (2D) and could not be used to guide PDL principal fiber formation. Another work reported the use of a three-dimensional (3D) printing microgroove pattern to guide cell and collagen fiber orientation.13 An in vivo study showed that fibers only inside the microgrooves followed the intended orientation, while there were randomized fibrous tissues between the microgrooves. Furthermore, there was a lack of connection between the oriented fibers and the alveolar bone/cementum within the scaffold.13 At present, most of the approaches cannot mimic the nanofibrous architecture of natural extracellular matrix (ECM) to guide principal fiber formation.12–14
Meanwhile, PDL is a soft tissue situated between two mineralized tissues (tooth root and alveolar bone), and this soft tissue is regenerated from PDLSCs that have the osteogenic differentiation capacity to form new bone and cellular cementum.15,16 Therefore, it is crucial to have strict spatial control of biomineralization during periodontal tissue regeneration.17 Specifically, it is crucial to prevent osteogenic differentiation of PDLSCs inside the PDL matrix. While a number of scaffolds have been used for PDL regeneration, the mechanisms of the interactions between the PDL scaffolds and PDLSCs are largely unknown.5,18,19
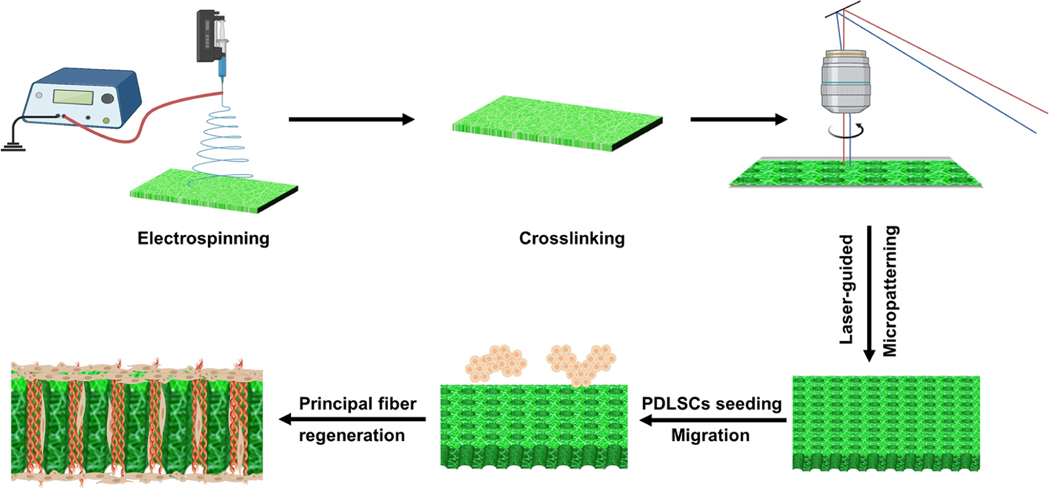
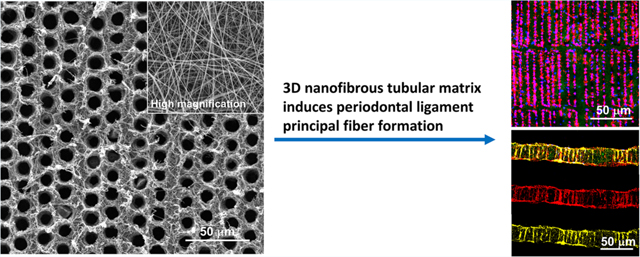
Herein, we report the development of a 3D nanofibrous tubular matrix to guide PDLSCs alignment, form well-ordered PDL principal fiber bundles, and inhibit osteogenic differentiation of PDLSCs. The nanofibrous tubular matrix was prepared via a combination of electrospinning and laser-guided micropatterning processes (Scheme 1). Electrospinning was adopted to mimic the nanofibrous structure of natural ECM. Gelatin was selected to mimic the chemical composition of collagen that is the main component of the ECM in PDL. More importantly, a laser-guided micropatterning process was used to create well-organized tubules inside the matrix to control principal fiber formation. To control the sizes and distributions of regenerated principal fibers, the tubules were fabricated with the same size and density of natural principal fiber bundles (diameters of 5−10 μm and density of 5000−10000 fibers/mm2). In vitro cell culture experiments demonstrated that this biomimetic matrix provided excellent biophysical cues to guide PDLSCs migration and form PDL principal fiber bundles. In addition, the effect of the 3D nanofibrous tubular architecture on the osteogenic differentiation of PDLSCs was examined. A mechanism study further revealed that the Yap1/Twist1 signaling pathway was involved in the inhibition of PDLSCs osteogenesis in the biomimetic tubular matrix.
Scheme 1.
Illustration of the Fabrication of the 3D Nanofibrous Tubular Matrix and Use of the 3D Tubular Matrix for PDL Principal Fiber Regeneration
2. MATERIALS AND METHODS
2.1. Materials.
Gelatin, alizarin red, dimethyl sulfoxide, penicillin−streptomycin, ROCK inhibitor Y27632, scramble-esiRNA, and Twist1-esiRNA were purchased from Sigma-Aldrich (St. Louis, MO). Triton X-100, ethyl acetate, and paraformaldehyde were obtained from VWR (Radnor, USA). Alpha-minimum essential medium (α-MEM), fetal bovine serum (FBS), and goat serum were purchased from Gibco (Carlsbad, CA). Hoechst 33342 was purchased from Pierce Biotechnology (Waltham, MA). The RNeasy Mini Kit was purchased from Qiagen (Hilden, Germany). The SoAdvancedTM Universal SYBR Green Supermix and iScriptTM gDNA Clear cDNA Synthesis Kit were purchased from BioRad (Hercules, CA). Anti-Collagen I (ab34710), Anti-Periostin (ab14041), Anti-Collagen III (ab7778), Anti-Yap1 (ab39361), and Anti-Twist1 antibodies (ab50581) were purchased from Abcam (Cambridge, UK).
2.2. Preparation of Nanofibrous Tubular 3D Gelatin Matrices.
Nanofibrous gelatin matrices were fabricated by using electrospinning as previously described.20 Briefly, gelatin21 was dissolved in an acetic acid/ethyl acetate/distilled water/hexafluor-oisopropanol (2.5/1.5/1/5) solvent mixture. Nanofibrous gelatin matrices were prepared at a voltage of 12 kV and a feeding rate of 0.5 mL/h. The nanofibrous gelatin matrices were cross-linked as described in our previous study.22 The matrices were soaked in a glycine aqueous solution (8 g/L) to block the reaction, followed by water washing and vacuum-dried. Later, the matrices were fixed on membrane glass slides for laser microdissection using a Leica LMD 7000 system as described in our previous work.23 The parameters that used to produce tubules in Figure 1 were as follows. Large: laser power = 60, laser aperture = 20, laser speed = 47, and laser pulse frequency = 105 Hz. Medium: laser power = 60, laser aperture = 10, laser speed = 47, and laser pulse frequency = 105 Hz. Small: laser power = 50, laser aperture = 5, laser speed = 47, and laser pulse frequency = 105 Hz. The matrices were observed under a scanning electronic microscope (SEM) (JSM6010, JEOL, Japan). The diameters and densities of nanofibers were analyzed by Image Pro Plus 6.0 software. At least 50 tubules in each group were measured, and 10 areas of interest were used to measure the density in each group. The 3D tubular gelatin matrices had an average tensile strength of 7.67 MPa under dry conditions and 2.79 MPa under wet conditions.23 In addition, the degradation of the tubular gelatin matrices was precisely controlled by the cross-linking density.23
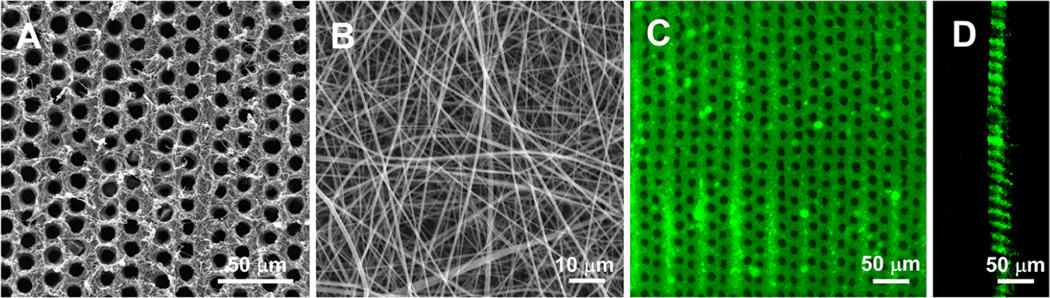
Figure 1.
Characterizations of 3D tubular matrices. (A) An SEM image of a nanofibrous tubular matrix. (B) High magnification of the tubular matrix showing that the matrix was composed of collagen-like nanofibers. (C) Top view of a tubular matrix under a confocal microscope. (D) Cross-section views of the tubular matrix showing that the tubules were open at both ends.
2.3. Cell Seeding on Gelatin Matrices.
Gelatin matrices were fixed onto CellCrownTM24NX Scaffdex devices. After sterilization, the assembled matrices were placed into 24-well culture plates. Human periodontal ligament stem cells were seeded onto gelatin matrices with/without tubules (flat/tubular matrices) and cultured at 37 °C and 5% CO2 (the cell culture condition was the same in the following experiments). To characterize the morphology of the PDLSCs, PDLSCs were seeded on a Petri dish and cultured for 3 and 7 days. The photos were taken under an integrated microscope (SZX16, Olympus) (Figure S1). Cell proliferation was determined by MTT assays according to the manufacturer’s instruction.
2.4. Adhesion and Migration Assay.
PDLSCs were seeded under the condition described in Section 2.3 and allowed to attach to the matrices and migrate inside tubules for 2, 6, 12, and 24 h. Afterward, the matrices were gently washed with PBS and fixed with paraformaldehyde. The PDLSCs adhered to the nanofibrous matrices were pretreated with 0.3% Triton X-100 PBS. After being blocked with 5% goat serum for 30 min, the samples were stained with CF633 Phalloidine at 37 °C for 60 min. Next, the samples were stained with Hoechst 33342 and observed under a TSC SP5 microscope (Leica, Germany) for top view images. For a cross-section view, the matrices were dehydrated in 30% sucrose and embedded in OCT for frozen sections. The frozen sections were performed in the interval of 25 μm and observed under an SP5 microscope. Migration distances were measured from the top of the matrix to the end of migrating cells, and 30 cells in each group were measured by using Image Pro Plus 6.0 software. Cells that stayed on the surface of the matrix, migrated inside tubules, and migrated to the other side of the matrix were included in the measurement. Migration ratios were calculated by number of migrated cells/total number of cells. Six fields that were randomly selected from each sample were used for the analysis.
2.5. Immunofluorescence Staining.
To assess the in vitro PDL-like tissue formation inside nanofibrous tubular matrices, PDLSCs were seeded on the matrices as described in Section 2.3 and cultured in a collagen-inducing medium containing 50 mM ascorbic acid. After 7 and 14 days, samples were collected, fixed, and pretreated as described in Section 2.3. After being blocked with goat serum, the matrices were stained with the Anti-Collagen I antibody (1:500), Anti-Collagen III antibody (1:200), or Anti-Periostin antibody (1:200) overnight together with CF633 Phalloidine (10 U/mL) at 4 °C. The samples were stained with the Alexa Fluor Plus 555 sary antibody (1:200) for 2 h, followed by 1 μg/mL Hoechst 33342 for 20 min. The samples were dehydrated, embedded, and frozen as described in the Section 2.3. Images were taken under an SP5 confocal microscope, and the expressions of PDL markers were analyzed by using Image Pro Plus 6.0.
To assess Yap1 and Twist1 expressions in PDLSCs, PDLSCs were seeded on matrices as described in Section 2.2. After 3 and 7 days, samples were collected and treated as described above for immunofluorescence staining of Yap1 (1:200) and Twist1 (1:200). For Yap1 images, total Yap1 and Yap1 expressions in the nuclei were measured by using Image Pro Plus 6.0.
2.6. Sirius Red Staining and Alizarin Red Staining.
Three weeks after being cultured in collagen-inducing medium, the samples were fixed by paraformaldehyde for 30 min, dehydrated in ethanol, and embedded in paraffin. The paraffin sections were cut to a thickness of 25 μm and mounted on slides. A Sirius red staining process was prepared according to our previous study.9 Alizarin red staining was conducted to measure calcified tissue formation on the matrices as described previously.24
2.7. Yap1 and Twist1 Inhibition Study.
PDLSCs were seeded on flat and tubular matrices for 24 h. Next, 2 μM ROCK inhibitor Y-27632 was added into the complete culture medium. On day 7, samples were collected for a real-time PCR analysis. To inhibit the Twist1 expression, the endoribonuclease-prepared small interfering (esi) RNA of the Twist1 gene was transfected into PDLSCs. The PDLSCs were seeded on the flat and tubular matrices as described above and then transfected with a Lipofectamine iMAX reagent using 0.2 μg of esiRNA according to protocols of the manufacturer. On day 7, a RT-PCR was performed to evaluate the osteogenesis of PDLSCs. There were six treatment groups: flat group, flat + scramble-esiRNA group (control esiRNA), flat + Twist1-esiRNA group, tubular group, tubular + scramble-esiRNA group, tubular + Twist1-esiRNA group, and tubular group.
2.8. Real-Time PCR.
Total RNA was extracted from the PDLSCs with an RNeasy Mini Kit after they were cultured for 3 and 7 days on the flat and tubular matrices and reverse transcribed into cDNA using the iScriptTM gDNA Clear cDNA Synthesis Kit. For Yap1 and Twist1 inhibition study, total RNA was extracted 7 days after PDLSCs were treated with ROCK inhibitor or Twist1-esiRNA. The resultant cDNA was prepared for a real-time PCR together with SYBR Green Supermix and the gene specific primers: osteocalcin (Ocn) (F: 5-TCACACTCCTCGCCCTATTG-3; R: 5-GGGTCTCTTCACT-ACCTCGC-3), runt-related transcription factor 2 (Runx2) (F: 5-GTGAAGACGGTTATGGTCAAGG-3; R: 5-CAGATGGG-ACTGTGGTTACTGT-3), Osterix (Sp7) (F: 5-GAGTGG-AACAGGAGTGGAGC-3; R: 5-CAGGCAGATGGAGAGAGCTG-3), yes1 associated transcriptional regulator (Yap1) (F: 5-CCCTCGTTTTGCCATGAACC-3; R: 5-CATCTGTTGCTGCT-GGTTGG-3), Twist family BHLH transcription factor 1 (Twist1) (F: 5-AGCCACTGAAAGGAAAGGCA-3; R: 5-CAGGCCAGTTT-GATCCCAGT-3), and glyceraldehyde-3-phosphate dehydrogenase (Gapdh) (F: 5-GAATGGGCAGCCGTTAGGAA-3; R: 5-AGGAGAAATCGGGCCAGCTA-3). A CFX96TMreal-time system (BioRad, USA) was used to perform the reactions. The relative mRNA expressions of target genes were normalized by Gapdh.
2.9. RNA-Seq and Bioinformatics Analysis.
Total RNA was isolated from PDLSCs cultured for 3 days on the flat and tubular scaffolds. RNA integrity number (RIN) values greater than 7.8 were used for library preparation and sequencing. After the cDNA library was prepared, the sequencing was performed on the NextSeq 550 platform (Illumina, USA). The differential expression was evaluated by the edgeR package.25 The flat group was used as a control, and the data were analyzed on WebGestalt for gene ontology (GO) enrichment.26
2.10. Statistical Analysis.
All experiments were repeated twice. The normality of data was tested by skewness, kurtosis, and histograms. For all the quantitative analyses (except cell migration distances), results were normally distributed and presented as mean ± standard error. The results of the cell migration distances were not normally distributed and presented as median and interquartile range. Data were analyzed by using SPSS 22.0 software. An unpaired Student’s t test was conducted to compare the differences between each group for PDL marker expression and gene expression results. A one-way ANOVA test followed by an unpaired Student’s t test of a post hoc test was conducted to compare the differences among each group for microtunnel characterizations and cell migration ratios. A Kruskal−Wallis H test followed by a Mann−Whitney U test of a post hoc test was conducted to compare the differences among each group in cell migration distances. P < 0.05 was considered as significantly different.
3. RESULTS
3.1. Nanofibrous Tubular Matrices Guided PDLSC Migration.
Highly ordered nanofibrous tubular matrices were fabricated by a combination of electrospinning and laser-guided micropatterning (Figure 1). The diameter and density of tubules in the matrix were 10 μm and 8000 tubules/mm2, respectively, which mimicked the size and density of the principal fibers in PDL.9 The gelatin matrix was composed of nanofibers with an average diameter of ∼200 nm (Figure 1B). The confocal image showed that all the tubules were open on both sides of the 3D matrix (Figure 1C,D), which is crucial for cell migration and principal fiber formation. The thickness of the matrix was ∼40 μm and can be readily tailored via electrospinning time. In addition, the diameters of the tubules were precisely controlled from several to tens of micrometers by adjusting the fabrication parameters (Figure S2). Similarly, low and high densities of tubules in the 3D matrix were fabricated under different micropatterning conditions (Figure S2). The biocompatibility of the matrices was confirmed by MTT assays (Figure S3).
When PDLSCs were cultured on a tubular matrix for 24 h, the cells exhibited spindle shapes and aligned with the micropatterned matrix (Figure 2A). Cross-section images indicated that many PDLSCs migrated inside of the tubules, and some of the cells even reached the other side of the matrix (Figure 2C). A quantitative analysis showed that 74 ± 11% of the PDLSCs migrated into the tubules. In contrast, PDLSCs randomly spread on the flat (nontubular) matrix surface, and no cells were capable of migrating into the matrix (Figure 2B,D).
Figure 2.
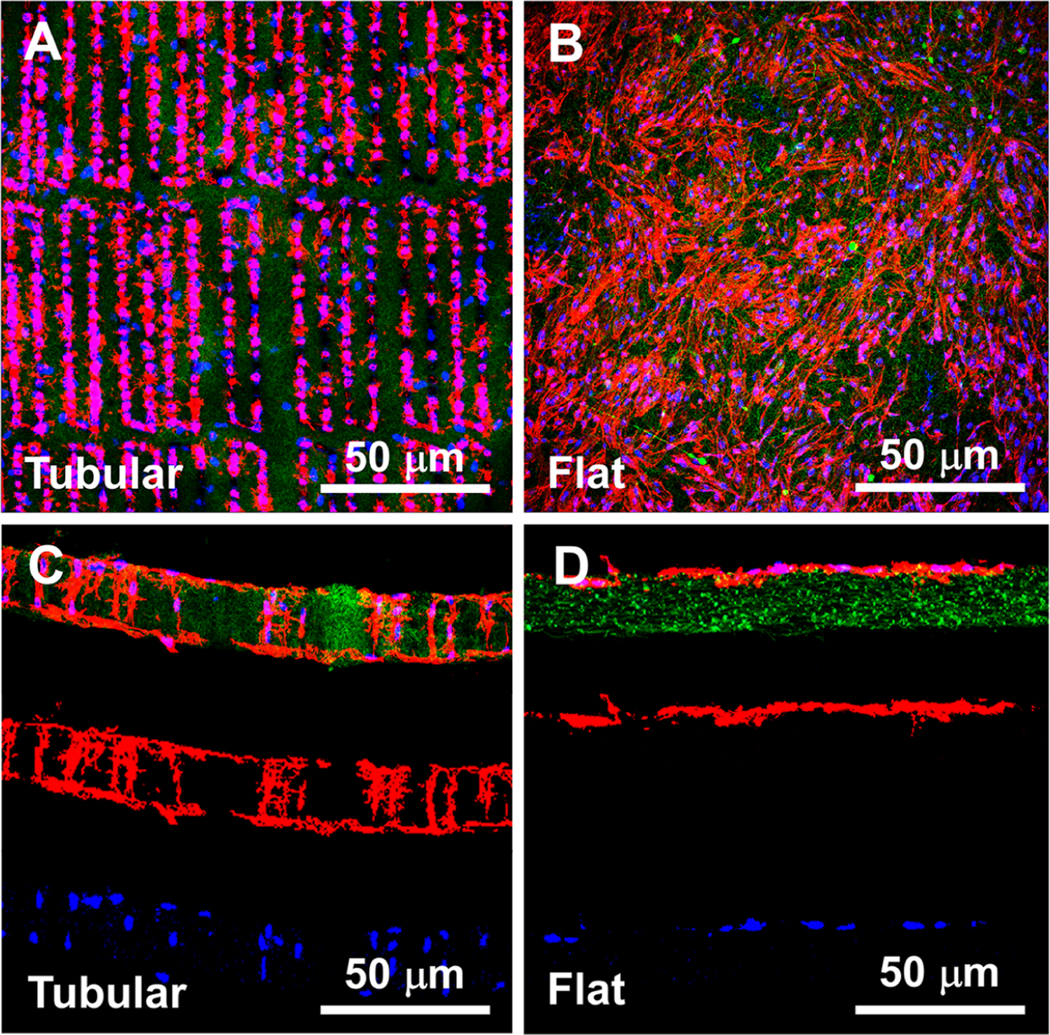
Migration of PDLSCs in the tubular matrices (A, C) and on the flat matrices (B, D). (A) and (B) are top view images, (C) and (D) are cross-section view images. Green: gelatin matrices. Red: actin. Blue: DAPI.
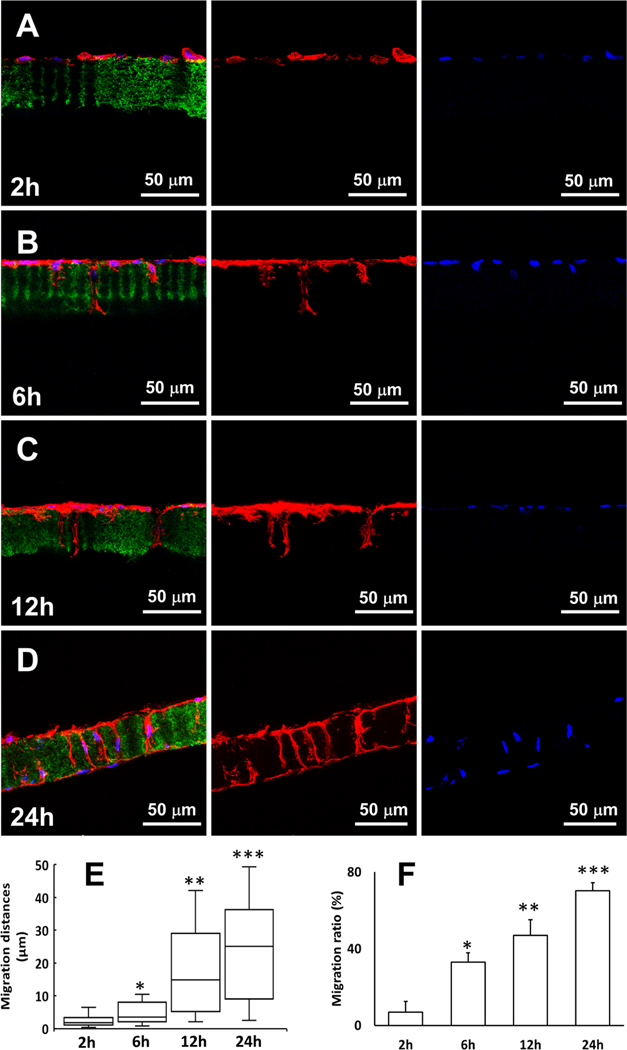
Confocal microcopy examination revealed that the migration of PDLSCs into the tubules was a dynamic process (Figure 3). Two hours after cell seeding, the PDLSCs attached to the surface of the tubular matrix, and no cell migration was observed (Figure 3A). A small number of PDLSCs migrated into the tubules at 6 h (33 ± 5%, n = 30) (Figure 3B). The cell number in the tubules increased with a median migration distance of 15 μm (IQR: 5.3 μm, 28.2 μm, n = 30) at 12 h (Figure 3C). After 24 h, the other side of the matrix was also occupied by the PDLSCs, and cell distribution in the tubules reached a balance (Figure 3D). Overall, the median migration distance of the PDLSCs increased from approximately 2 μm (IQR: 1.1 μm, 3.5 μm, n = 30) to 25 μm (IQR: 9.8 μm, 36.0 μm, n = 30) from 2 to 24 h (Figure 3E).
Figure 3.
(A−D) Dynamic migration process of PDLSCs in the tubular matrix from 2 to 24 h. Green: gelatin matrix. Red: actin. Blue: DAPI. (E) Migration distances of PDLSCs from 2 to 24 h. (F) Migration ratios of PDLSCs from 2 to 24 h. (∗) Compared to the 2 h group; (∗∗) compared to the 6 h group, and (∗∗∗) compared to the 12 h group.
3.2. Nanofibrous Tubular Matrices Regulated Principal Fiber Bundles Formation and Promoted PDL Markers Expressions.
Collagen I, Collagen III, and Periostin (PDL markers) were stained to show PDL-like tissue formation. The expressions of Collagen I, Collagen III, and Periostin from day 7 to day 14 are shown in Figure 4. On day 14, most of the tubules inside the matrix were deposited with the newly secreted Collagen I, III, and Periostin which were secreted from the PDLSCs. When stained with Sirius red and observed under polarized light, the secreted collagen displayed well-ordered fiber bundles in the tubules, indicating that the nanofibrous tubular matrix guided PDL-like fiber formation (Figure 4C).
Figure 4.
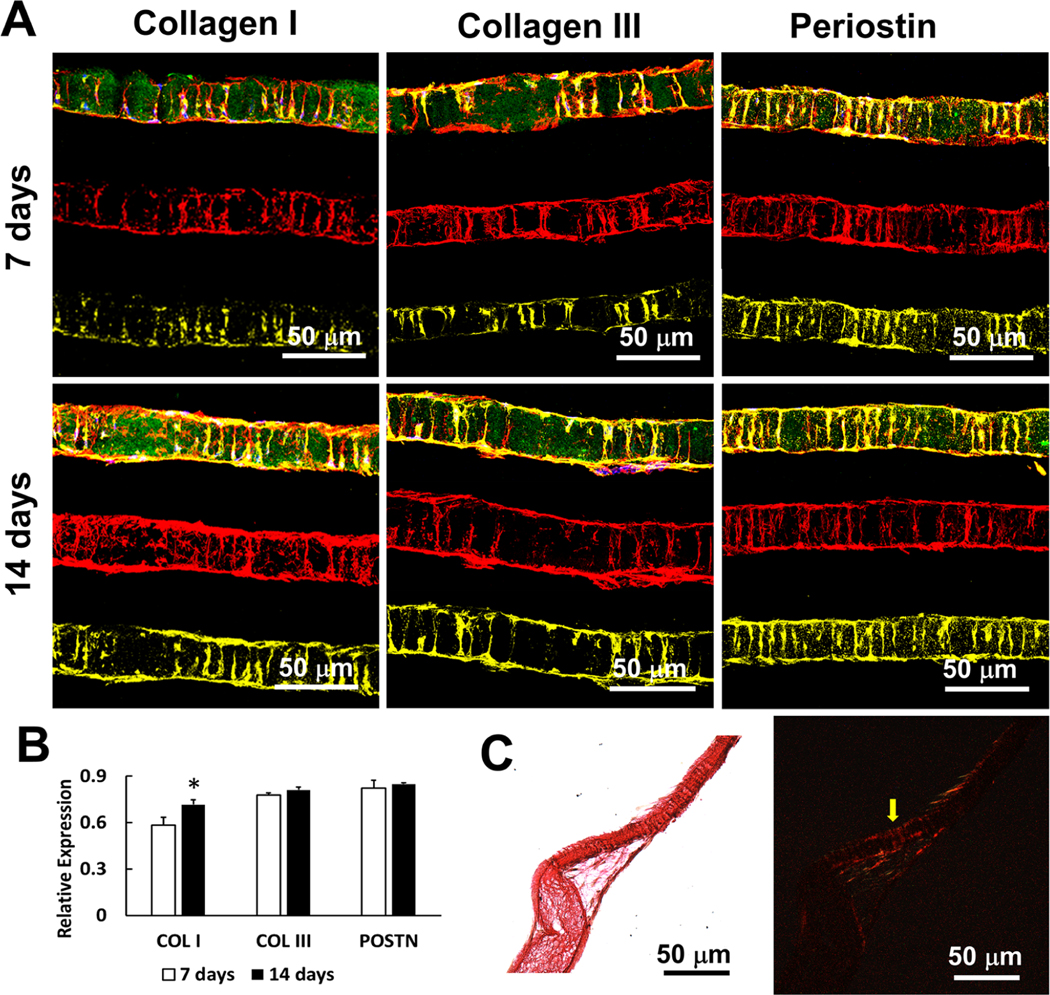
PDL-like tissue formation in tubular matrices. (A) The expressions of PDL markers (Collagen I, Collagen III, and Periostin) in the tubular matrices. Green: gelatin matrices. Red: actin. Blue: DAPI. Yellow: PDL markers. (B) Quantitative analyses of the expressions from Collagen I, Collagen III and Periostin. (C) Sirius red staining of the tubular construct after 21 days. Collagen fiber bundles formed inside of the tubules (yellow arrow). Images were taken under bright light (left) and polarized light (right) conditions, separately.
3.3. Nanofibrous Tubular Matrices Inhibited Osteogenesis of PDLSCs through Yap1/Twist1 Signaling Pathway.
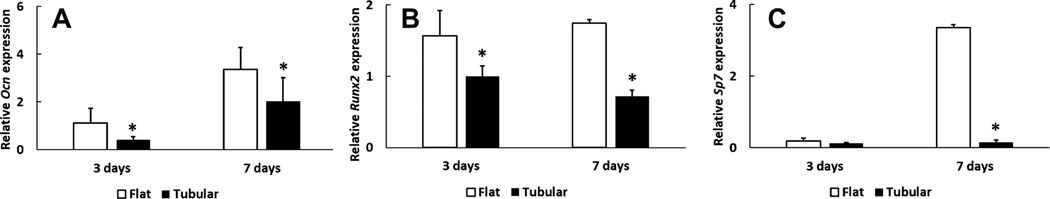
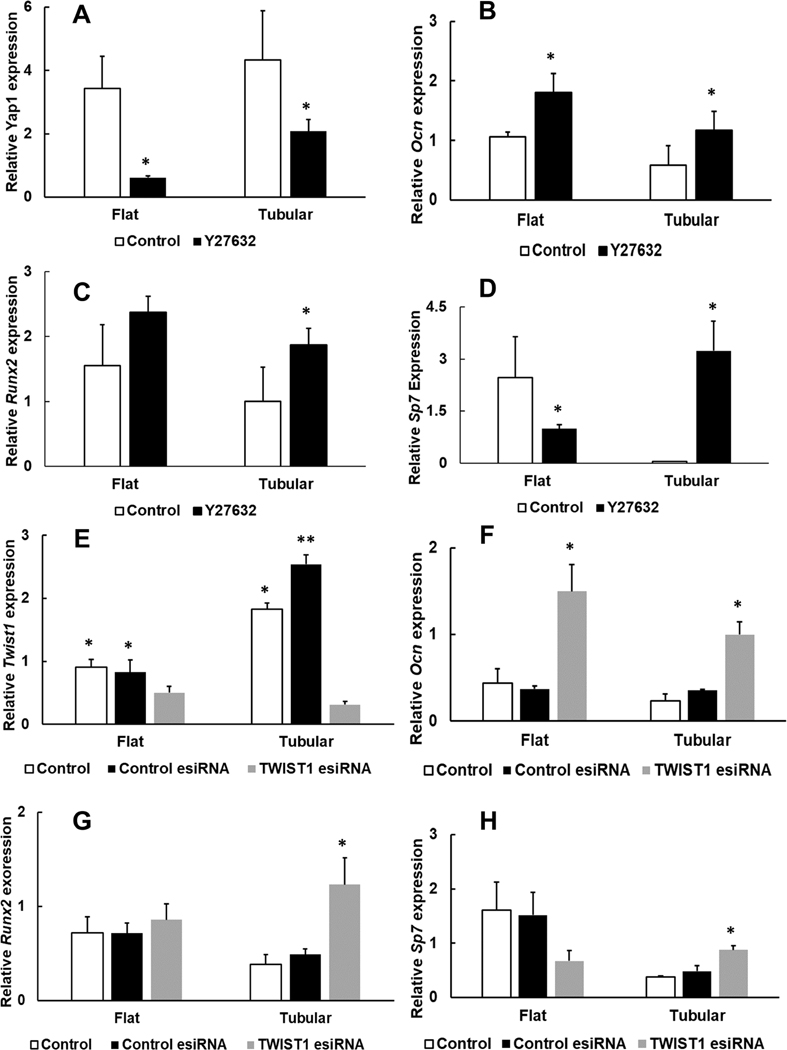
The expressions of Ocn, Runx2, and Sp7 (osteogenic markers) in the tubular matrix were lower than those in the flat matrix on day 3 and day 7 (Figure 5). Specifically, the Ocn expression in the tubular group was approximately 30% (0.41 ± 0.14 to 1.11 ± 0.61, n = 3) and 60% (2.03 ± 0.98, 3.35 ± 0.90, n = 3) to that of the flat group on day 3 and day 7, respectively. On day 3, the Runx2 level in the tubular matrix was 40% (1.00 ± 0.14 to 1.57 ± 0.34, n = 3) lower than that in the control group. From day 3 to day 7, the expression of Runx2 in the tubular group downregulated to approximately 30% (from 1.00 ± 0.14 to 0.72 ± 0.09, n = 3), while it remained at a similar level in the flat group. The expression of Sp7 in the tubular matrix was about 1/30 to that of the flat matrix on day 7 (0.15 ± 0.16 to 3.36 ± 0.08, n = 3). There was no mineralized tissues formation on both the flat and tubular matrix (Figure S4).
Figure 5.
Expressions of osteogenic genes on the tubular and flat matrices on day 3 and day 7. The three osteogenic genes are (A) Ocn, (B) Runx2, and (C) Sp7.
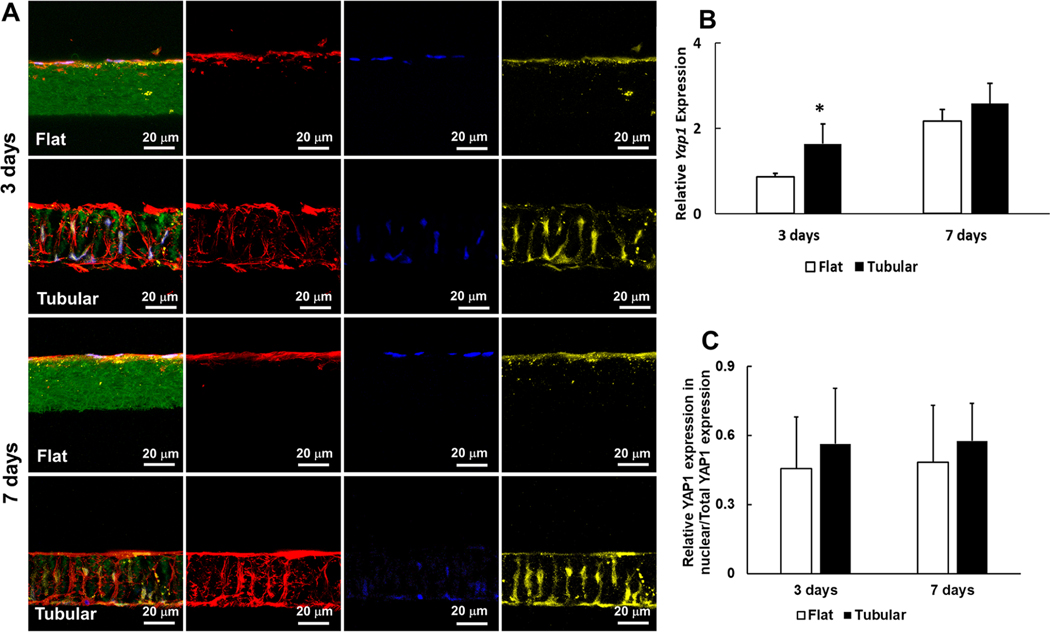
PDLSCs expressed Yes associated protein 1 (Yap1) on flat and tubular matrices on day 3 and day 7 (Figure 6). The level of Yap1 in the tubular group was significantly higher than that in the flat group on day 3 (1.64 ± 0.07 to 0.86 ± 0.08, n = 3) (Figure 6B). Meanwhile, the expression of Twist family bHLH transcription factor 1 (Twist1) in the tubular matrix was 3-fold higher than that in the flat matrix on day 3 (1.64 ± 0.17 to 0.45 ± 0.08, n = 3) (Figure S5).
Figure 6.
Yap1 expressions on the tubular and flat matrices on day 3 and day 7. (A) Immunofluorescence of PDLSCs on the tubular and flat matrices. Green: gelatin matrices. Red: actin. Blue: DAPI. Yellow: Yap1. (B) Relative Yap1 gene expressions on the tubular and flat matrices on day 3 and day 7. (C) Ratios of Yap1expression in nuclei/total Yap1 expression on the tubular and flat matrices, separately.
The addition of Y27632 (a ROCK inhibitor) inhibited the expression of Yap1 (Figure 7A). Consequently, the expressions of Ocn and Runx2 had 2-fold upregulation in the tubular group (Figure 7B,C). More strikingly, the inhibition of Yap1 led to a 65-fold increase of the Sp7 expression in the tubular group (3.24 ± 0.85 to 0.04 ± 0.01, n = 3), whereas the expression of Sp7 reduced to ∼50% in the flat group (2.46 ± 1.17 to 1 ± 0.11, n = 3) (Figure 7D). Similarly, the addition of Twist-esiRNA inhibited the expression of Twist1 and increased the expressions of Ocn and Runx2 in both the tubular and control groups (Figure 7F,G). In addition, inhibition of Twist1 significantly increased the expression of Sp7 in the tubular group (Figure 7H), suggesting that the Twist1 signaling pathway was involved in the inhibition of PDLSCs osteogenesis in the bio-inspired tubular matrix.
Figure 7.
(A−D) Yap1 and osteogenic gene expressions on the tubular and flat matrices after the treatment of ROCK inhibitor for 7 days. (E−H) Twist1 and osteogenic gene expressions on the tubular and flat matrices after the treatment of Twist1 esiRNA for 7 days. P < 0.05 was considered statistically significant. (∗) Compared to the control group.
3.4. Nanofibrous Tubular Architecture Downregulated Pathways Related to Cell–ECM Interaction, ECM Metabolism, and Osteogenesis.
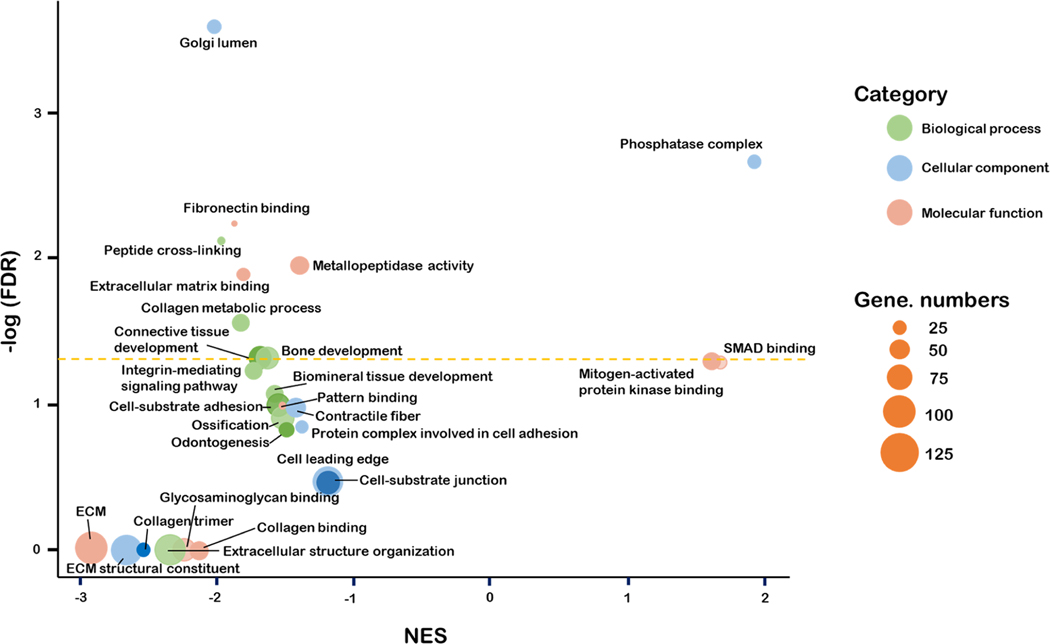
RNA sequencing results showed that the differentially expressed genes were enriched in gene sets that are related to cell−ECM interaction, ECM production and metabolism, and osteogenic differentiation (Figure 8). Among them, the gene sets related to cell−ECM interaction are “cell−substrate adhesion” (extracellular matrix protein 2, collagen 3A1, discoidin domain receptor tyrosine kinase 2, etc.), “cell−substrate binding” (collagen 16A1, collagen 8A1, extracellular matrix protein 2, etc.), “extracellular matrix binding” (decorin, biglycan, collagen 11A1, etc.), and “protein complex involved in cell adhesion” (ankyrin 3, extracellular matrix protein 2, C-X-C motif chemokine ligand 12, etc.). Their normalized enrichment scores (NES) were mostly negative, indicating that the tubular architecture in the matrix was more likely to decrease the adhesion of PDLSCs to the matrix. The gene sets related to the osteogenic differentiation of PDLSCs included “biomineral tissue development” (inorganic pyrophosphate transport regulator, bone morphogenic protein 2, bone morphogenic protein 4, etc.), “odontogenesis” (alkaline phosphatase, collagen 1A1, asporin, bone morphogenic protein 2, etc.), “bone development” (alkaline phosphatase, bone morphogenic protein 4, collagen 12 A1, etc.), and “ossification” (alkaline phosphatase, bone morphogenic protein 4, collagen 1A1, etc.) and had negative NES values, confirming that the 3D tubular matrix inhibited the osteogenic differentiation of PDLSCs.
Figure 8.
Gene ontology enrichment analysis of the gene expressions of PDLSCs in the tubular matrix using PDLSCs on the flat matrix as a control.
4. DISCUSSION
Restoration of functional PDL requires the regeneration of well-organized PDL with aligned principal fibers to resist the occlusal force and mechanical stresses. Previous work on PDL restoration only obtained randomized amorphous and non-functional fibrous structures. In addition, the interactions of the matrix and PDLSCs as well as the underlying mechanism were seldom explored. In this study, we developed a bio-inspired nanofibrous tubular matrix to guide PDLSCs migration and form well-aligned PDL principal fibers. Furthermore, we investigated the mechanism of the interactions between the tubular matrix and the PDLSCs.
Electrospinning has been widely used to fabricate nanofibrous matrices. The nanofibrous architecture simulates the fibrous ECM of PDL and promotes periodontal tissue regeneration.27
Previous studies reported that aligned nanofibrous matrices could guide cell dispersion and fiber orientation.28,29 However, cells proliferated, migrated, and secreted ECM only on the surfaces of the matrices in those studies. In other words, those matrices served as 2D substrates but not 3D matrices. To create 3D channels to guide cell migration inside matrices, a 3D printing microchannel technique was developed.13 This 3D matrix induced cell migration in the tubules and secreted collagen fibers aligned inside of the microchannels. However, the microchannels were not open on the other side of the matrix; therefore, the regenerated collagen fibers terminated at the end of the matrices and could not integrate with alveolar bone or cementum. Meanwhile, none of the above synthetic matrices took the microstructure of PDL into account and hence could not regenerate the principal fibers of PDL.
In this work, we have developed a laser-guided micro-patterning process to introduce tubules (∼10 μm in diameter) in nanofibrous matrices, which emulates the microstructure of PDL and provides 3D microchannels for PDLSC migration and principal fiber formation. Our laser-guided micropatterning process has the advantage of precisely controlling tubular size and density, which are modulated by many parameters, including laser pulse frequency, laser aperture, speed, laser power, and repeat time. In this study, we controlled the laser power and aperture to fabricate tubules with various diameters and densities in the nanofibrous matrix. The medium-sized tubules have similar diameter (about 8−10 μm) and density (5000−10000 fibers/mm2) to Sharpey’s fibers (the ends of PDL principal fibers)9 and were used in this study. The side-view confocal image showed that the tubules connected two sides of the nanofibrous matrix, providing microchannels for cell migration from one side to the other side (Figure 1). This characteristic enables the regenerated PDL principal fibers to further insert into the alveolar bone and cementum to form Sharpey’s fibers, resulting in the formation of an alveolar bone−PDL−cementum complex to restore the functions of the periodontium.
Guiding PDLSCs alignment along with the tubules is a prerequest for oriented PDL principal fiber formation. Therefore, the synthetic nanofibrous tubular matrix was first examined to guide PDLSCs migration and alignment. At 24 h, PDLSCs migrated through the tubules of the matrix. During this process, the long axis of the PDLSCs aligned with the tubules and formed long spindle shapes within the tubules. This result indicated that the 3D matrix guided PDLSCs alignment along with the direction of the tubules within 24 h.
Next, we extended the culture time and examined collagen fiber deposition. Our results showed that the PDLSCs secreted ECM that accumulated inside the tubules to form PDL-like fibers. The expression of Col I, Col III, and POSTN gradually increased and accumulated inside of the tubules (Figure 4). The majority of PDL principal fibers are composed of Col I and Col III;30 thus, the expressions and accumulation of Col I and Col III indicated the regeneration of PDL fibers. POSTN is a key ECM protein and is involved in cell migration and guiding PDL collagen fiber bundle formation.31–33 POSTN has been used as a periodontal regeneration marker,34,35 indicating the aligned and mature PDL fibers that bear mechanical strength.36 Therefore, increasing POSTN expression over time is a symbol of oriented PDL principal fiber regeneration (Figure 4). The induction of PDL principal fiber regeneration was further demonstrated by Sirius red staining. Sirius red molecules enhance the birefringence of parallel collagen bundles; hence, the stained collagen remained bright while other tissues remained dark under polarized light.9 After 21 days, the regenerated fibers inside the tubules were stained by Sirius red and were detected under polarized light, indicating the regeneration of oriented and mature collagen fiber bundles (Figure 4C). Therefore, the synthetic tubular matrix successfully induced PDLSCs to differentiate and form PDL principal fibers.
Inhibition of the osteogenic differentiation of PDLSCs is critical for PDL regeneration. The osteogenic markers of PDLSCs in the tubular matrix were lower than that in the control. Yap1 is critical in mediating cell−matrix interaction, and surface topographies of biomaterials influence Yap1 expression.12,22 Twist1 contributes to cell migration in the epithelial mesenchymal transition.37 It was reported that Yap1 and downstream Twist1 promoted fibroblast proliferation, migration, and collagen production.38,39 Besides, Yap1 and Twist1 negatively regulated the osteogenic differentiation of human Adipose-derived stem cells.40 Our experiment showed that the expressions of Yap1 and Twist1 genes in the tubular matrix were upregulated at a higher level than that in the flat group, while the Runx2 expression in the tubular group was significantly lower compared to the flat group. It was documented that the increase of nuclear Yap1 and Twist1 suppressed the activity of Runx2 by forming Runx2/Yap1 and Runx2/Twist1 complexes.41–44 The high amount of Runx2/Yap1 and Runx2/Twist1 complexes reduced free Runx2 in nuclei. Consequently, the low activity of Runx2 as a transcriptional factor decreased the expressions of other osteogenic markers and inhibited osteogenic differentiation.22,43,45 The addition of Y-27632 and Twist1-esiRNA inhibited the formation of Runx2/Yap1 and Runx2/Twist1 complexes; therefore, it increased free Runx2 in nuclei and other osteogenic marker expressions. Runx2 directly regulates Ocn and Sp7 by interacting with their promoters in osteoblasts.46–48 However, the regulation of osteogenesis is a complicated process, and it is possible that other pathways and transcription factors are also involved in regulating Sp7 expression.48 This explains why the Sp7 expression on the flat matrix was suppressed after the addition of Yap1 and Twist1. Moreover, the GO enrichment analysis further shows that the gene sets related to bone development, ossification, and odontogenesis decreased in the tubular matrix, which was consistent with the cell culture experiments and the RT-PCR result. For example, genes related to cell−ECM adhesion were downregulated which may facilitate cell migration through the tubular structure. Also, genes related to osteogenesis and tissue mineralization were also decreased, consistent with the result of the osteogenic differentiation. Besides, The GO enrichment analysis also provides valuable information such as the gene expression of cell leading edge, the change of Golgi lumen, and other signaling pathways. This information is not directly related to this study and not discussed in this paper but may provide clues for studies about cell organs, polarization, and so on. Overall, the study of the matrix−PDLSCs interaction indicated that the tubular matrix downregulated PDLSC osteogenesis and the Yap1/Twist1 pathway was involved in this process. In our next step, we will integrate the tubular matrix into a hierarchical periodontal scaffold that contains two other components for cementum and alveolar bone regeneration. Through this hierarchical scaffold, we will examine functional PDL principal fibers and Sharpey’s fiber regeneration using a periodontitis rat model, which is beyond the scope of current study and will be reported separately in the future.
5. CONCLUSION
A bio-inspired tubular matrix was designed and fabricated via the combination of electrospinning and laser-guided micro-patterning processes. The tubular matrix mimicked the physical architecture (nanofibers, principal fiber bundle size, and density) and chemical compositions of the ECM of PDL and was an excellent template to guide PDLSCs migration and form well-ordered collagen fibers inside the tubules. In addition, the tubular matrix inhibited the osteogenesis of PDLSCs by reducing the expressions of Ocn, Runx2, and Sp7. The Yap1/Twist1 signaling pathway was involved in the inhibition of PDLSC osteogenesis in the bioinspired tubular matrix.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Bei Chang, Qian Li, Chi Ma, Zhiai Hu, and Yuejia Deng for their technical support. The authors also thank Meghann Holt for her assistance with the editing of this article.
Funding
This study was supported by NIH/NIDCR DE029808 and Texas A&M University Presidential Impact Fellow (X.L.).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.2c09420.
Characterization of tubular matrix; Twist1 expressions on the tubular and flat matrices (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsami.2c09420
The authors declare no competing financial interest.
Contributor Information
Yongxi Liang, Department of Biomedical Sciences, Texas A&M University College of Dentistry, Dallas, Texas 75246, United States.
Ajay Shakya, Department of Biomedical Sciences, Texas A&M University College of Dentistry, Dallas, Texas 75246, United States.
Xiaohua Liu, Department of Biomedical Sciences, Texas A&M University College of Dentistry, Dallas, Texas 75246, United States.
REFERENCES
- (1).Papapanou PN; Susin C. Periodontitis Epidemiology: Is Periodontitis Under-Recognized, Over-Diagnosed, or Both? Periodontol. 2000 2017, 75, 45–51. [DOI] [PubMed] [Google Scholar]
- (2).Slots J. Periodontitis: Facts, Fallacies and the Future. Periodontol. 2000 2017, 75, 7–23. [DOI] [PubMed] [Google Scholar]
- (3).Tsumanuma Y; Iwata T; Washio K; Yoshida T; Yamada A; Takagi R; Ohno T; Lin K; Yamato M; Ishikawa I; Okano T; Izumi Y. Comparison of Different Tissue-Derived Stem Cell Sheets for Periodontal Regeneration in a Canine 1-Wall Defect Model. Biomaterials 2011, 32, 5819–5825. [DOI] [PubMed] [Google Scholar]
- (4).Liu Y; Zheng Y; Ding G; Fang DJ; Zhang CM; Bartold PM; Gronthos S; Shi ST; Wang SL Periodontal Ligament Stem Cell-Mediated Treatment for Periodontitis in Miniature Swine. Stem Cells 2008, 26, 1065–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Chen FM; Liu XH Advancing Biomaterials of Human Origin for Tissue Engineering. Prog. Polym. Sci. 2016, 53, 86–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hu Z; Ma C; Rong X; Zou S; Liu X. Immunomodulatory ECM-like Microspheres for Accelerated Bone Regeneration in Diabetes Mellitus. ACS Appl. Mater. Interfaces 2018, 10, 2377–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Li XW; Liu XH; Ni SL; Liu YN; Sun HC; Lin Q. Enhanced Osteogenic Healing Process of Rat Tooth Sockets Using a Novel Simvastatin-Loaded Injectable Microsphere-Hydrogel System. J. Craniomaxillofac Surg. 2019, 47, 1147–1154. [DOI] [PubMed] [Google Scholar]
- (8).Sculean A; Nikolidakis D; Nikou G; Ivanovic A; Chapple ILC; Stavropoulos A. Biomaterials for Promoting Periodontal Regeneration in Human Intrabony Defects: A Systematic Review. Periodontol. 2000 2015, 68, 182–216. [DOI] [PubMed] [Google Scholar]
- (9).Liang Y; Hu Z; Chang B; Liu X. Quantitative Characterizations of the Sharpey’s Fibers of Rat Molars. J. Periodontal. Res.2020, 55, 307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sowmya S; Mony U; Jayachandran P; Reshma S; Kumar RA; Arzate H; Nair SV; Jayakumar R. Tri-Layered Nano-composite Hydrogel Scaffold for the Concurrent Regeneration of Cementum, Periodontal Ligament, and Alveolar Bone. Adv. Healthc. Mater. 2017, 6, 1601251. [DOI] [PubMed] [Google Scholar]
- (11).Vaquette C; Saifzadeh S; Farag A; Hutmacher DW; Ivanovski S. Periodontal Tissue Engineering with a Multiphasic Construct and Cell Sheets. J. Dent. Res. 2019, 98, 673–681. [DOI] [PubMed] [Google Scholar]
- (12).Zheng L; Jiang J; Gui J; Zhang L; Liu X; Sun Y; Fan Y. Influence of Micropatterning on Human Periodontal Ligament Cells’ Behavior. Biophys. J. 2018, 114, 1988–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Pilipchuk SP; Monje A; Jiao Y; Hao J; Kruger L; Flanagan CL; Hollister SJ; Giannobile WV Integration of 3D Printed and Micropatterned Polycaprolactone Scaffolds for Guidance of Oriented Collagenous Tissue Formation In Vivo. Adv. Healthc. Mater. 2016, 5, 676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Park CH; Kim KH; Lee YM; Giannobile WV; Seol YJ 3D Printed, Microgroove Pattern-Driven Generation of Oriented Ligamentous Architectures. Int. J. Mol. Sci. 2017, 18, 1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Seo BM; Miura M; Gronthos S; Bartold PM; Batouli S; Brahim J; Young M; Robey PG; Wang CY; Shi ST Investigation of Multipotent Postnatal Stem Cells from Human Periodontal Ligament. Lancet 2004, 364, 149–155. [DOI] [PubMed] [Google Scholar]
- (16).Gay IC; Chen S; MacDougall M. Isolation and Characterization of Multipotent Human Periodontal Ligament Stem Cells. Orthod. Craniofac. Res. 2007, 10, 149–60. [DOI] [PubMed] [Google Scholar]
- (17).Liang Y; Luan X; Liu X. Recent Advances in Periodontal Regeneration: A biomaterial Perspective. Bioact. Mater. 2020, 5, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Reis ECC; Borges APB; Araujo MVF; Mendes VC; Guan LM; Davies JE Periodontal Regeneration Using a Bilayered PLGA/Calcium Phosphate Construct. Biomaterials 2011, 32, 9244–9253. [DOI] [PubMed] [Google Scholar]
- (19).Costa PF; Vaquette C; Zhang QY; Reis RL; Ivanovski S; Hutmacher DW Advanced Tissue Engineering Scaffold Design for Regeneration of the Complex Hierarchical Periodontal Structure. J. Clin. Periodontol. 2014, 41, 283–294. [DOI] [PubMed] [Google Scholar]
- (20).Ma C; Chang B; Jing Y; Kim H; Liu X. Bio-Inspired Micropatterned Platforms Recapitulate 3D Physiological Morphologies of Bone and Dentinal Cells. Adv. Sci. 2018, 5, 1801037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Sachar A; Strom TA; San Miguel S; Serrano MJ; Svoboda KK; Liu X. Cell-Matrix and Cell-Cell Interactions of Human Gingival Fibroblasts on Three-Dimensional Nanofibrous Gelatin Scaffolds. J. Tissue Eng. Regen. Med. 2014, 8, 862–873. [DOI] [PubMed] [Google Scholar]
- (22).Chang B; Ma C; Liu X. Nanofibers Regulate Single Bone Marrow Stem Cell Osteogenesis via FAK/RhoA/YAP1 Pathway. ACS Appl. Mater. Interfaces 2018, 10, 33022–33031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Ma C; Qu T; Chang B; Jing Y; Feng JQ; Liu X. 3D Maskless Micropatterning for Regeneration of Highly Organized Tubular Tissues. Adv. Healthc. Mater. 2018, 7, 1700738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Liang Y; Hu Z; Li Q; Liu X. Pyrophosphate Inhibits Periodontal Ligament Stem Cell Differentiation and Mineralization Through MAPK Signaling Pathways. J. Periodontal Res. 2021, 56, 982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Robinson MD; Smyth GK Small-Sample Estimation of Negative Binomial Dispersion with Applications to SAGE Data. Biostatistics 2007, 9, 321–32. [DOI] [PubMed] [Google Scholar]
- (26).Wang J; Vasaikar S; Shi Z; Greer M; Zhang B. WebGestalt 2017: A More Comprehensive, Powerful, Flexible and Interactive Gene Set Enrichment Analysis Toolkit. Nucleic Acids Res. 2017, 45, w130–w137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Rnjak-Kovacina J; Weiss AS Increasing the Pore Size of Electrospun Scaffolds. Tissue Eng. Part B Rev. 2011, 17, 365–372. [DOI] [PubMed] [Google Scholar]
- (28).Ren S; Yao Y; Zhang H; Fan R; Yu Y; Yang J; Zhang R; Liu C; Sun W; Miao L. Aligned Fibers Fabricated by Near-Field Electrospinning Influence the Orientation and Differentiation of hPDLSCs for Periodontal Regeneration. J. Biomed. Nanotechnol. 2017, 13, 1725–1734. [DOI] [PubMed] [Google Scholar]
- (29).Shang S; Yang F; Cheng X; Walboomers XF; Jansen JA The Effect of Electrospun Fibre Alignment on the Behaviour of Rat Periodontal Ligament Cells. Eur. Cells Mater. 2010, 19, 180–192. [DOI] [PubMed] [Google Scholar]
- (30).Nanci A. Ten Cate’s Oral Histology Development, Structure, and Function, 8/e; Elsevier India: 2008. [Google Scholar]
- (31).Yamada S; Tauchi T; Awata T; Maeda K; Kajikawa T; Yanagita M; Murakami S. Characterization of a Novel Periodontal Ligament-Specific Periostin Isoform. J. Dent. Res. 2014, 93, 891–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Tabata C; Hongo H; Sasaki M; Hasegawa T; de Freitas PH; Yamada T; Yamamoto T; Suzuki R; Yamamoto T; Oda K; Li M; Kudo A; Iida J; Amizuka N. Altered Distribution of Extracellular Matrix Proteins in the Periodontal Ligament of Periostin-Deficient Mice. Histol. Histopathol. 2014, 29, 731–742. [DOI] [PubMed] [Google Scholar]
- (33).Wu Z; Dai W; Wang P; Zhang X; Tang Y; Liu L; Wang Q; Li M; Tang C. Periostin Promotes Migration, Proliferation, and Differentiation of Human Periodontal Ligament Mesenchymal Stem Cells. Connect. Tissue Res. 2018, 59, 108–119. [DOI] [PubMed] [Google Scholar]
- (34).Kudo A. Periostin in Fibrillogenesis for Tissue Regeneration: Periostin Actions Inside and Outside the Cell. Cell. Mol. Life Sci. 2011, 68, 3201–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Park CH; Rios HF; Jin Q; Sugai JV; Padial-Molina M; Taut AD; Flanagan CL; Hollister SJ; Giannobile WV Tissue Engineering Bone-Ligament Complexes Using Fiber-Guiding Scaffolds. Biomaterials 2012, 33, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Du J; Li M. Functions of Periostin in Dental Tissues and Its Role in Periodontal Tissues’ Regeneration. Cell. Mol. Life Sci. 2017, 74, 4279–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Duan Y; He Q; Yue K; Si H; Wang J; Zhou X; Wang X. Hypoxia Induced Bcl-2/Twist1 Complex Promotes Tumor Cell Invasion in Oral Squamous Cell Carcinoma. Oncotarget 2017, 8, 7729–7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Chen Y; Zhao X; Sun J; Su W; Zhang L; Li Y; Liu Y; Zhang L; Lu Y; Shan H; Liang H. YAP1/Twist Promotes Fibroblast Activation and Lung Fibrosis That Conferred by MiR-15a Loss in IPF. Cell Death Differ. 2019, 26, 1832–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Wang Y; Li J; Qiu Y; Hu B; Chen J; Fu T; Zhou P; Song J. Low-Intensity Pulsed Ultrasound Promotes Periodontal Ligament Stem Cell Migration Through TWIST1-Mediated SDF-1 Expression. Int. J. Mol. Med. 2018, 42, 322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Quarto N; Senarath-Yapa K; Renda A; Longaker MT TWIST1 Silencing Enhances In Vitro and In Vivo Osteogenic Differentiation of Human Adipose-Derived Stem Cells by Triggering Activation of BMP-ERK/FGF Signaling and TAZ Upregulation. Stem Cells 2015, 33, 833–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Bialek P; Kern B; Yang X; Schrock M; Sosic D; Hong N; Wu H; Yu K; Ornitz DM; Olson EN; Justice MJ; Karsenty G. A Twist Code Determines the Onset of Osteoblast Differentiation. Dev. Cell 2004, 6, 423–435. [DOI] [PubMed] [Google Scholar]
- (42).Zhang XW; Zhang BY; Wang SW; Gong DJ; Han L; Xu ZY; Liu XH Twist-Related Protein 1 Negatively Regulated Osteoblastic Transdifferentiation of Human Aortic Valve Interstitial Cells by Directly Inhibiting Runt-Related Transcription Factor 2. J. Thorac. Cardiovasc. Surg. 2014, 148, 1700–1708. [DOI] [PubMed] [Google Scholar]
- (43).Lee MS; Lowe GN; Strong DD; Wergedal JE; Glackin CA TWIST, A Basic Helix-Loop-Helix Transcription Factor, Can Regulate the Human Osteogenic Lineage. J. Cell Biochem. 1999, 75, 566–577. [DOI] [PubMed] [Google Scholar]
- (44).Zhou M; Gao S; Zhang X; Zhang T; Zhang T; Tian T; Li S; Lin Y; Cai X. The Protective Effect of Tetrahedral Framework Nucleic Acids on Periodontium Under Inflammatory Conditions. Bioact. Mater. 2021, 6, 1676–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Zhang H; Cooper LF; Zhang X; Zhang Y; Deng F; Song J; Yang S. Titanium Nanotubes Induce Osteogenic Differentiation Through the FAK/RhoA/YAP Cascade. RSC Adv. 2016, 6, 44062–44069. [Google Scholar]
- (46).Ducy P; Zhang R; Geoffroy V; Ridall AL; Karsenty G. Osf2/Cbfa1: A Transcriptional Activator of Osteoblast Differentiation. Cell 1997, 89, 747–754. [DOI] [PubMed] [Google Scholar]
- (47).Franceschi RT; Xiao G; Jiang D; Gopalakrishnan R; Yang S; Reith E. Multiple Signaling Pathways Converge on the Cbfa1/ Runx2 Transcription Factor to Regulate Osteoblast Differentiation. Connect. Tissue Res. 2003, 44, 109–116. [PMC free article] [PubMed] [Google Scholar]
- (48).Yang D; Okamura H; Qiu L. Upregulated Osterix Expression Elicited by Runx2 and Dlx5 Is Required for the Accelerated Osteoblast Differentiation in PP2A Calpha-Knockdown Cells. Cell Biol. Int. 2018, 42, 403–410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.