Abstract
Class 2 CRISPR effectors Cas9 and Cas12 may have evolved from nucleases in IS200/IS605 transposons. IscB is about 2/5 the size of Cas9 but shares similar domain organization. The associated ωRNA plays the combined role of crRNA and tracrRNA to guide dsDNA cleavage. Here we report a 2.78 Å cryo-EM structure of IscB-ωRNA bound to dsDNA target, revealing the architectural and mechanistic similarities between IscB and Cas9 RNPs. Target-adjacent motif recognition, R-loop formation, and DNA cleavage mechanisms are explained at high resolution. ωRNA plays the equivalent function of REC domains in Cas9, and contacts the RNA/DNA heteroduplex. The IscB-specific PLMP domain is dispensable for RNA-guided DNA cleavage. The transition from ancestral IscB to Cas9 involved dwarfing the ωRNA and introducing protein domain replacements.
One Sentence Summary:
Structure defines architectural and mechanistic similarities between IscB-ωRNA and Cas9-crRNA-tracrRNA, revealing possible evolutionary connection.
Increasing evidence points to the possibility that the core components of the CRISPR-Cas adaptive immune systems evolved from genes in mobile genetic elements. The class 2 CRISPR effectors Cas9 (Type II) and Cas12 (Type V) are believed to have independently evolved from an ancestral TnpB-like nuclease, which is still commonly found in insertion sequence (IS) elements today (1–3). Cas9 appears to have emerged from a distinct branch of IS elements within the IS200/605 superfamily harboring IscB (2). IscB and Cas9 share a common domain architecture at the sequence level (Fig. 1A–B). Both contain an arginine-rich bridge helix and an HNH endonuclease domain inserted into a RuvC endonuclease domain (1, 2). The bridge helix in Cas9 plays a crucial role in mediating complex ribonucleoprotein (RNP) formation with two non-coding RNAs, crRNA and tracrRNA (4–8). HNH and RuvC endonucleases are used by Cas9 to cleave the target and nontarget DNA strands, respectively (4). During CRISPR interference, the DNA substrate is validated through R-loop formation, which involves DNA unwinding and RNA/DNA heteroduplex formation (7, 9–14). IscB was found to assemble with a single large (>200 nt) noncoding RNA encoded by the transposon, ωRNA (OMEGA: obligate mobile element guided activity) (2). Together IscB-ωRNA mediates RNA-guided DNA cleavage similar to the Cas9-crRNA-tracrRNA RNP (2). To avoid self-targeting and to reduce search space, Cas9 further specifies a protospacer adjacent motif (PAM) adjacent to the target site (15, 16). This mechanism is conserved in IscB-ωRNA, and the equivalent target-adjacent motif (TAM) is recognized by a TAM interaction domain (TID) in IscB (2). All IscBs further encode a PLMP-motif containing domain at the N-terminus (2). This domain is not found in Cas9 and has an undefined function (Fig. 1B).
Fig.1. Cryo-EM reconstruction and structure of IscB RNP bound to target DNA.
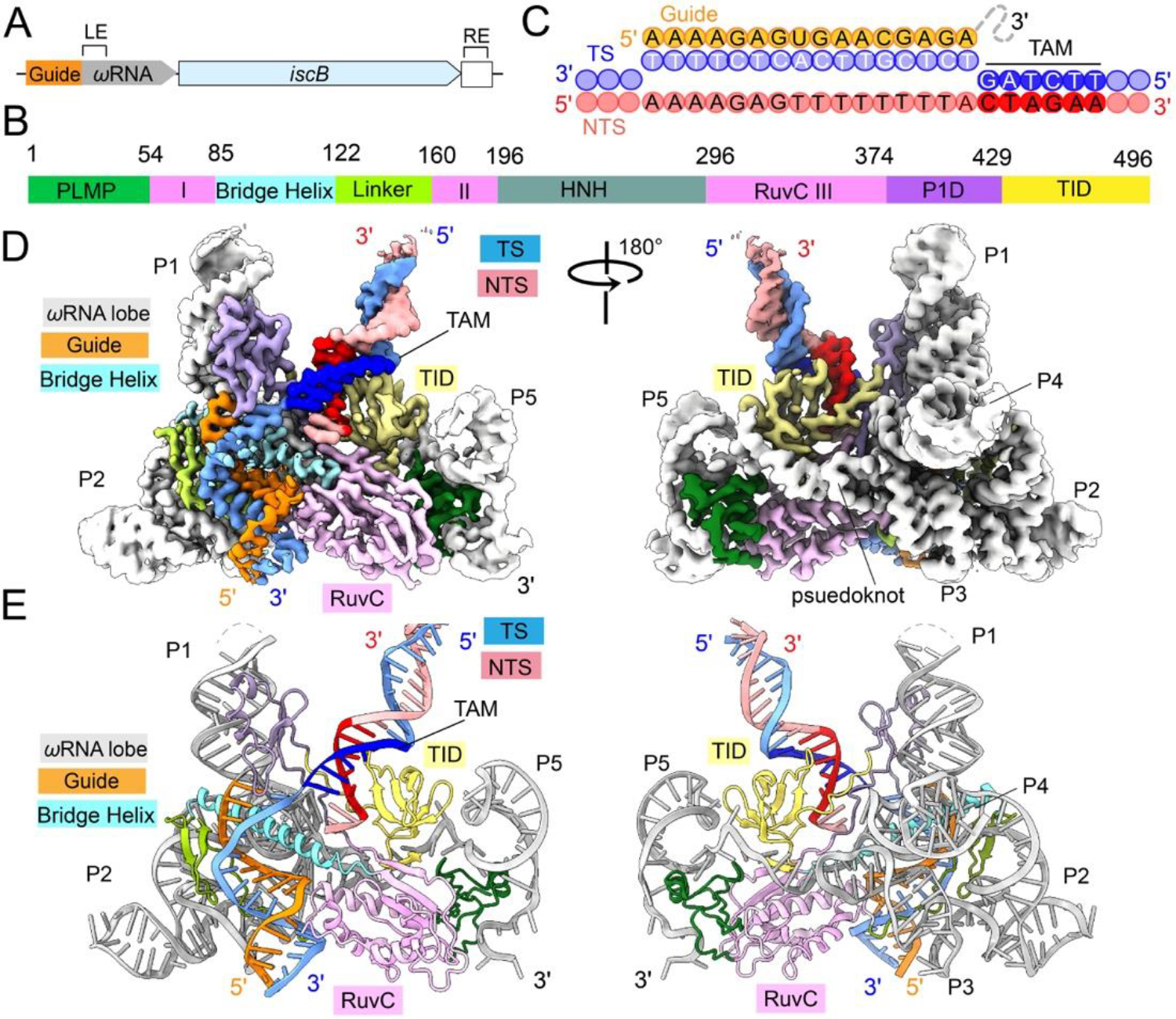
(A) Arrangement of the OgeuIscB and ωRNA in its native IS element defined by the left (LE) and right (RE) ends of the transposon. (B) Domain organization of IscB. P1D, P1 interaction domain; TID, TAM-interaction domain. RuvC domain is separated into three segments: RuvC I, II, and III. Color scheme is conserved throughout Fig. 1. (C) Diagram of R-loop formed between guide RNA and target DNA. TAM sequence is read 5’-CTAGAA-3’ on the non-target strand. (D, E) cryo-EM reconstruction at 2.78 Å and cartoon representations of the IscB-ωRNA/target DNA complex.
To understand the RNA-guided DNA cleavage mechanism by the compact IscB-ωRNA RNP and its relationship with Cas9-crRNA-tracrRNA, we determined a 2.78 Å structure of the gut microbiome derived OgeuIscB-ωRNA RNP complex (2) bound to target DNA using cryo-electron microscopy (cryo-EM) (Figs. 1, S1–3; Movie S1). Whereas the majority of the 496-aa IscB and 222-nt ωRNA could be unambiguously resolved, only a portion of the 60-bp DNA target could be reliably modeled. These include 13-bp of the TAM-proximal double-stranded (ds)DNA, the entire 16-nt target strand (TS) single-stranded (ss) DNA and 2-nt non-target strand (NTS) ssDNA in the R-loop region (Fig. 2A). The TAM-distal DNA is missing from the EM density due to molecular motion rather than cleavage and dissociation, because phosphorothioate modifications have been introduced into the DNA backbone at the HNH and RuvC cleavage sites (Fig. S1H) (2).
Fig. 2. Structural organization of the ωRNA and comparison to Cas9 crRNA-tracrRNA.
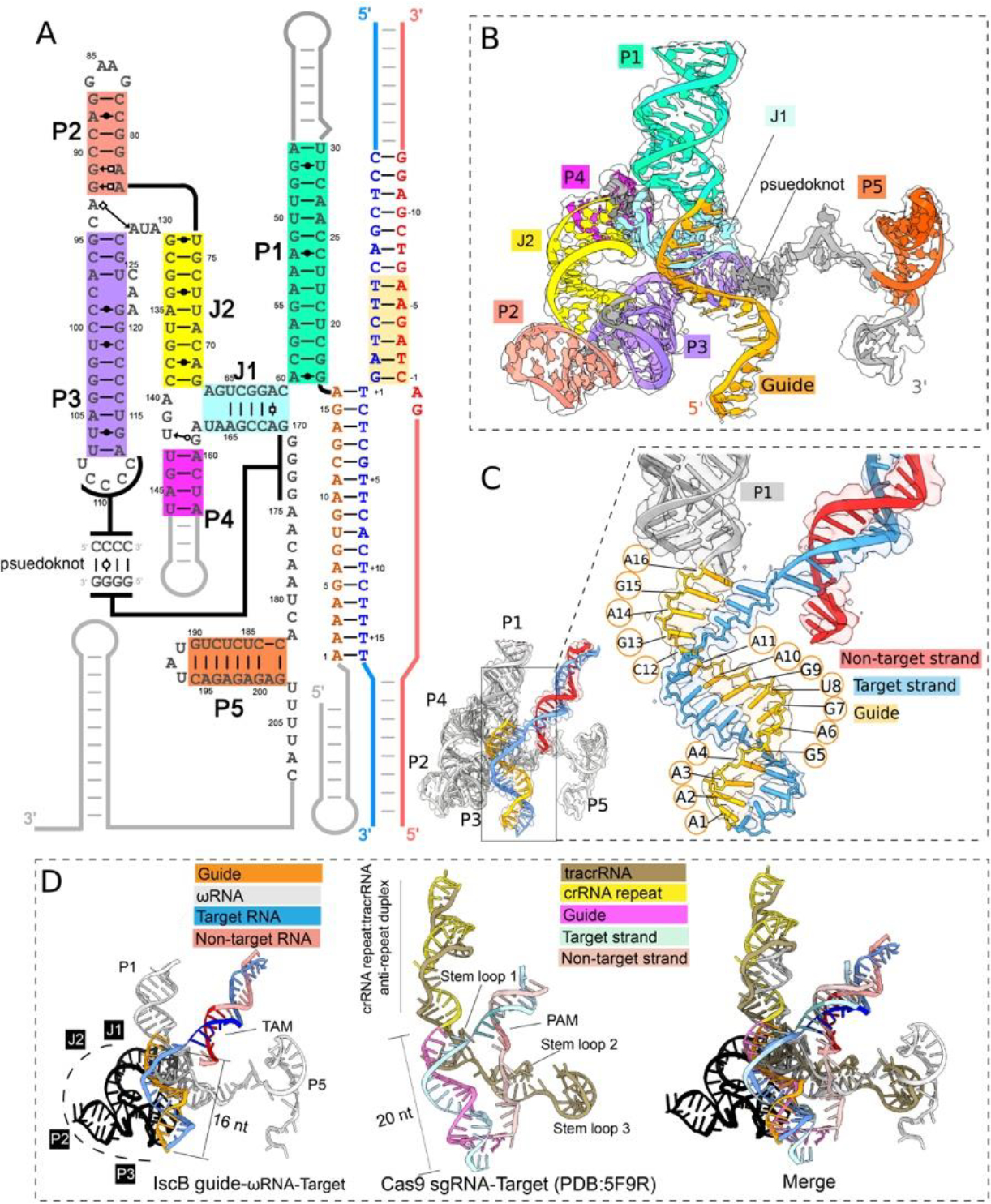
(A) Schematic of ωRNA depicting secondary and tertiary interactions. Non-target strand, red; target strand, blue; guide RNA, orange. (B) Atomic model of ωRNA. (C) Close-up view depicting R-loop base pairing between guide RNA and target strand DNA. (D) Structural alignment of ωRNA and tracrRNA-crRNA in SpCas9 RNP showing conserved RNA structures in guide RNAs, P1 with SpCas9 tracrRNA-crRNA helix, J1 with SpCas9 tracrRNA stem loop 1, P3 pseudoknot with SpCas9 tracrRNA stem loop 2, and P5 with SpCas9 tracrRNA stem loop 3. Colored in black is the region of the ωRNA replaced by the REC lobe in Cas9.
We found that the architectural organization, domain functionality, and nucleic acid binding mode are similar between IscB-ωRNA and Cas9 RNP. IscB-ωRNA adopts a similar two-lobed architecture, although its overall shape is much flatter, because several surface domains in Cas9 are missing in IscB (Fig. S4; Movie S2). Structural alignments revealed that the P1 stem loop of ωRNA is the functional equivalent of the crRNA repeat-tracrRNA anti-repeat duplex in the Cas9 RNP. It occupies the same location in the RNP and assists R-loop formation in a similar manner, by stabilizing the guide-RNA/TS-DNA heteroduplex through continuous base stacking (Fig. 1C–E; Movie S3). The TAM-containing dsDNA and the guide-RNA/TS-DNA heteroduplex in the R-loop region are accommodated by IscB-ωRNA at similar locations as in Cas9s, through conceptually similar mechanisms (Figs. 1D–E; S4). The TS-DNA base-pairs with the 16-nt guide RNA. The first 12-bp of the DNA/RNA heteroduplex adopts a distorted A-form due to IscB contacts, with a widened major groove and base-stacking almost perpendicular to the helical axis. The last 4-bp of the heteroduplex adopts a canonical A-form geometry (Fig. 1D–E).
Architecturally, the biggest structural difference between IscB and Cas9 is its lack of a polypeptide-based recognition (REC) lobe (Fig. S4; Movie S2). The functional replacement is the ωRNA lobe (from J1 to the pseudoknot), which folds into a sophisticated tertiary RNA structure (Fig. 2A). The structured portion of ωRNA was previously identified as HEARO RNA (HNH Endonuclease-Associated RNA and ORF) (17). This RNA and its associated HNH-containing ORF together was speculated to constitute a mobile genetic element (17). Here the 3D structure supports the secondary structure models from previous studies (2, 17). The central portion of ωRNA is a tail-to-tail stacked P2-P3 superhelix. J2 helix extrudes from the P2-P3 junction, then bifurcates into P4 and J1 at its end. While P4 projects away, J1 projects towards the apex of P3. The following residues zip up with the apical loop of P3 through a 4-bp G/C-rich pseudoknot (Fig. 2A–B). Following the pseudoknot, ωRNA extends horizontally along the backside of the IscB as a conserved ss-linker and a terminator-like element (P5, followed by four consecutive Us) (Fig. 2B). A conserved and highly structured RNA typically mediates either catalysis, ligand binding, or RNP formation (17). Our structure does not support a direct involvement of ωRNA in RNA-guided DNA cleavage because the bulk of ωRNA is insulated from the guide-RNA/TS-DNA heteroduplex by a layer of protein elements from IscB (Figs. 1C–D, 2C; Movie S1). Our structure further suggests the evolutionary trend from ancestral IscB to Cas9 involves replacing the structural roles of ωRNA with protein domains. However, the crRNA-tracrRNA of SpCas9 and NmeCas9 RNPs still contain structural elements reminiscent of P1, J1, pseudoknot, and terminator in ωRNA (Fig. 2D, S5) presumably because these elements are indispensable for RNP assembly.
Opposite from the ωRNA lobe, the equivalent of the Cas9 nuclease (NUC) lobe contains the RuvC nuclease as its platform. RuvC is woven together from three split polypeptide elements (Figs. 1B, S6A; Movie S1). It projects structural domains to various regions of the RNP. These elements are rich in positive surface charges, making favorable contacts with nucleic acids in different regions (Fig. S6A). The N-terminal PLMP motif-containing domain is packed at the edge of the NUC lobe to capture the terminator-like structure in ωRNA (Fig. S6B). The Arg-rich bridge helix is regarded as one of the most conserved structural elements in Cas9 (7, 8). It plays an equally important function in IscB-ωRNA RNP. Projected from RuvC, the bridge helix travels underneath the guide RNA, along the pseudoknot and J1, and at the base of P1, making multiple electrostatic contacts to the sugar-phosphate backbones. A line of consecutive arginine and lysine residues along one phase of the bridge helix make consecutive phosphate contacts to seven residues in the RNA guide (U8-A14), immobilizing the seed region of the guide in place for TS-DNA base-pairing (Fig. S6C). A β-hairpin followed by a flexible linker connects the bridge helix back to RuvC. Although very degenerate in size and structural complexity, this flexible structural elements “glues” ωRNA and middle portion of the guide RNA together with its positive Arg/Lys residues (Fig. S6D). The HNH nuclease domain is projected internally from RuvC. Like in many Cas9s, this domain is not well resolved in the averaged EM density map due to conformational flexibility. RuvC sends P1D domain to recognize the P1 helix of ωRNA; its functional equivalence is the WED domain in Cas9 (Figs. S6E, S4) (7, 10, 16). Finally, P1D connects with the TAM-interaction domain (TID) situated above RuvC through flexible linkers.
The OgeuIscB-ωRNA/R-loop structure explains the RNA-guided target recognition mechanism in high resolution (Fig. 3A; Movie S4). TAM (5’-NWRRNA-3’ (2); actual sequence: CTAGAA) in the dsDNA target is captured from the major groove side by the TID domain of IscB and from the minor groove side by the P1D linker (Figs. 3B, 4C, 4F). No contact was found at −1 TAM position. The −2 TAM position is recognized from the minor groove side by His397 and K380 in P1D linker to O2 of TNTS-2 and N3 of ATS-2, respectively. G-C pairs may be rejected in either combination due to the steric clash caused by the N2 protrusion from guanosine into the minor groove. The −3 and −4 of TAM appear to be probed indirectly for shape complementarity. It is likely that only purines in the NTS support the Van der Waals contacts to the backbone of Glu459 and Gly460 in TID; pyrimidines are too recessed. The −6 TAM position is recognized through hydrophobic contacts to the methyl groups of TTS-6 in the major grove, by Tyr468 and Trp478 in TID, respectively. Many IscB homologs encode smaller TID domains and specify less stringent TAM codes (2). Domain swapping attempts, structure-guided design, and directed evolution could lead to more versatile IscB-ωRNA tools with expanded TAM codes.
Fig. 3. Structural basis for TAM recognition and R-loop formation by IscB-ωRNA.
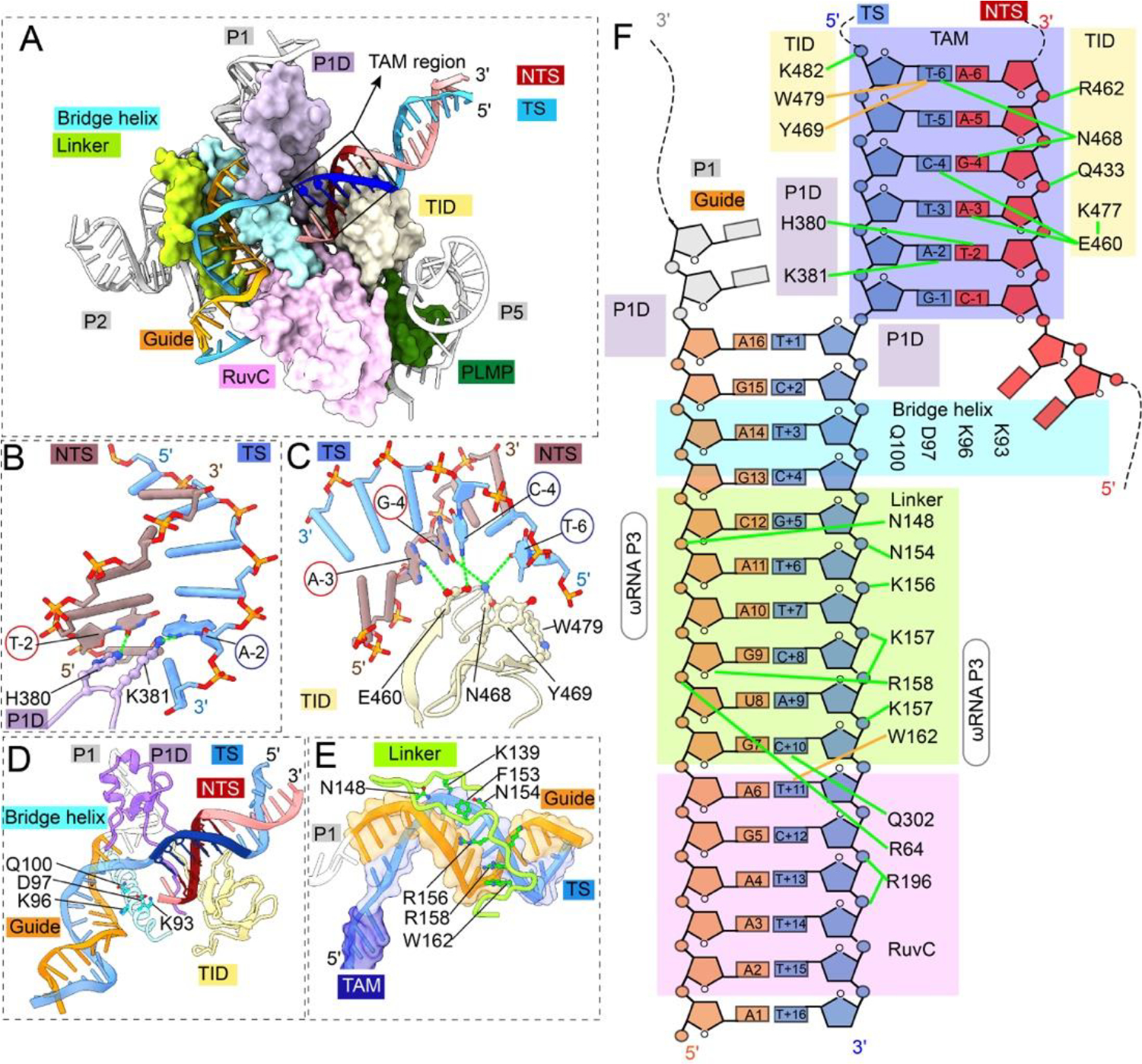
(A) TAM recognition and R-loop specification by domains of IscB. Color scheme is consistent with Fig. 1. (B) Close-up view of P1 interaction domain (P1D) linker residues recognizing TAM-2 basepair (target adjacent motif) from the DNA minor groove side. (C) Close-up view of the IscB TAM interaction domain (TID) making base-specific contacts from the DNA major groove side. (D) Close-up view of the bridge helix and P1D making contacts with the beginning portion of the DNA/RNA heteroduplex in the R-loop region. (E) Close-up view of the β-hairpin+linker domain specifying meandering the minor groove of the middle portion of the DNA/RNA heteroduplex. (F) Diagram of IscB contacts to TAM and DNA/RNA heteroduplex in the R-loop. Positioning of bridge helix domain separating the R-loop from the core of ωRNA in light blue. Green lines denote electrostatic contacts and brown lines denote hydrophobic contacts. TAM highlighted with purple box (ideal TAM sequence: 5’-NWRRNA-3’). guide RNA (orange), target strand DNA (blue), non-target strand DNA (red).
Figure 4. Mechanistic dissection of RNA-guided DNA cleavage by IscB.
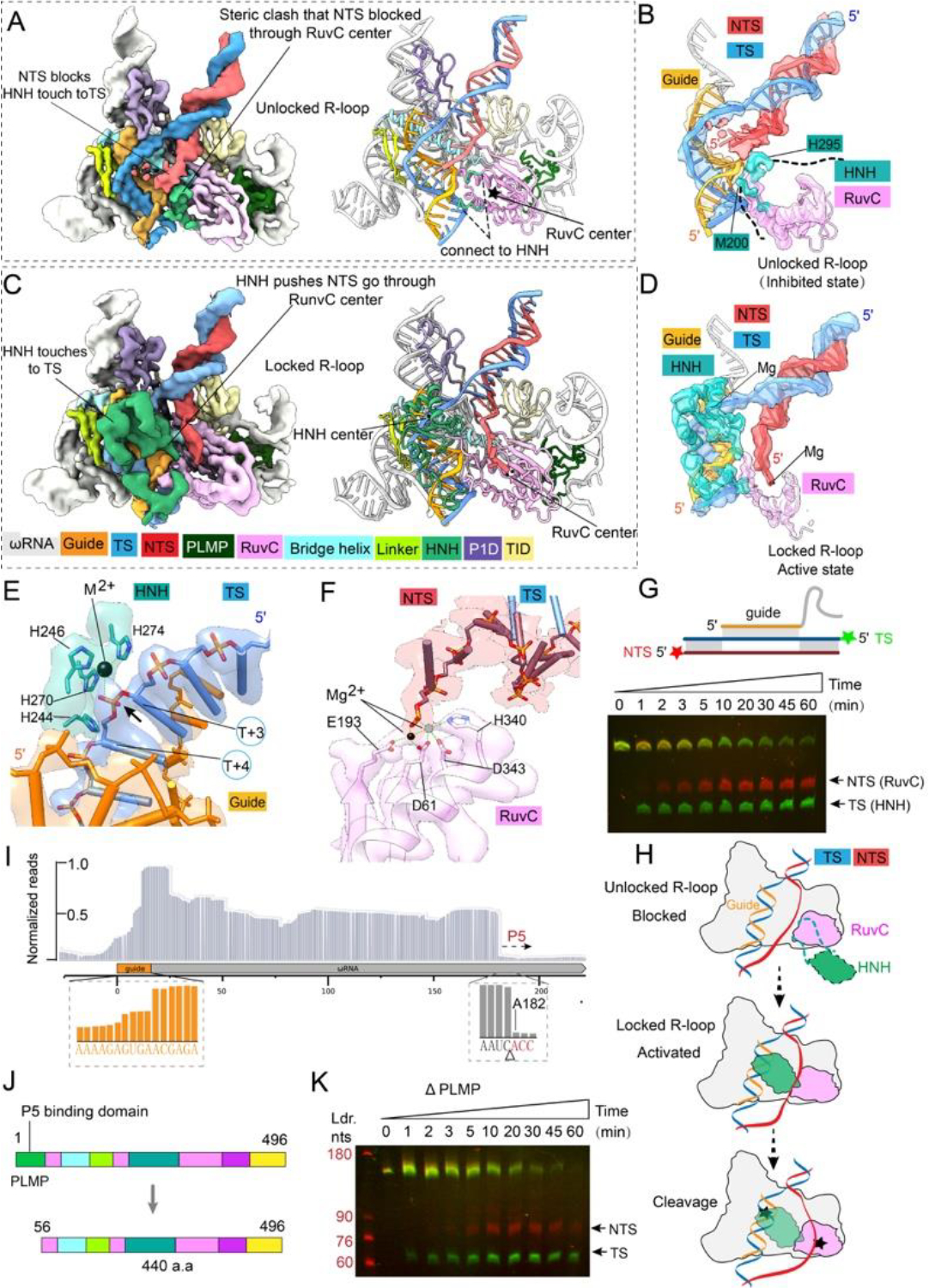
(A) 3.7 Å EM map and atomic model depicting the unlocked R-loop state. Color scheme is consistent with that in Fig. 1. (B) Focused view of DNA, guide RNA, and nuclease densities seen in the unlocked R-loop state. Note that NTS is blocked from entering the RuvC cleavage site by the anchor of HNH to RuvC. (C) 3.8 Å EM map and atomic model of the locked R-loop state. Alphafold predicted HNH domain structure (in green) is docked unambiguously into the EM density. Linker between HNH and RuvC domains can be seen interacting with the TAM-distal portion of the R-loop. (D) Focused view of HNH densities in the locked (active) state. Note that the NTS density is now allowed into the RuvC active site. (E) Close-up view of the HNH active site in the locked state. Catalytic metal ion (black) is seen coordinated to the TS substrate. A second metal ion is required for cleavage (ball with dash line). It is repelled from the active site by the phosphothioate modification in DNA. (F) Close-up view of the RuvC active site in the locked R-loop state. The coordinated catalytic metal ion (black) is seen contacting the backbone of the incoming NTS DNA. (G) Urea-PAGE showing time-resolved DNA cleavage. TS is cleaved by HNH prior to NTS cleavage by RuvC, supporting the unlocked/locked R-loop cleavage model. (H) Proposed mechanistic model explaining ordered strand cleavage by IscB. (I) Small RNA-seq of purified IscB-RNP, showing partial degradation of the guide RNA and a predictable cleavage site preceding stemloop P5. (J) Domain organization of wild-type and ΔPLMP IscB. (K) Urea-PAGE showing time-resolved DNA cleavage by IscB ΔPLMP.
A recent Cas9 study showed that off-targeting is inversely correlated with the extent of protein contacts to the guide-RNA/TS-DNA heteroduplex; the more local interactions to specify an A-form geometry, the less mismatch tolerance therein (14). In this regard, our structural analysis identified extensive R-loop contacts (Fig. 3A, 4D–F), which implies that IscB-ωRNA can specify a DNA target stringently despite its miniature size and shorter R-loop specification. A P1D loop (aa 396–408) specifies the first two base-pairs of the guide-RNA/TS-DNA heteroduplex from the minor groove side. The bridge helix and the following β-hairpin and linker specifies the middle portion of the heteroduplex (bp 2–9) from major and minor sides, respectively. ωRNA provides the platform support for these contacts, and a portion of the ωRNA backbone (P2, nt 114–116) directly contacts the backbone of guide RNA (bp 10–11). The RuvC domain then contacts the minor groove of bp 9–13. Basepairs 14–16 are not contacted and have weaker density. As shown later, this region is recognized when HNH docks onto the DNA/RNA heteroduplex.
To gain insight into the DNA cleavage mechanism, we analyzed the conformational dynamics in the IscB-ωRNA/R-loop EM reconstruction. Finer conformational sampling revealed two predominant conformational states. In the unlocked R-loop state (Figs. 4A–B, S7), the 3.1 Å map shows the NTS-DNA traveling near the RNA-bound TS-DNA. NTS-DNA is blocked from accessing the RuvC active site due to a steric clash with the anchor connecting HNH to RuvC (Fig. 4A). Although unresolved in EM density, HNH is likely part of the blocking mechanism as well. Its approximate location can be inferred by comparing it to the NmeCas9 apo structure (12). In contrast, the 3.2 Å locked R-loop state (Fig. 4C–D) shows HNH docking onto the RNA/TS-DNA heteroduplex and caging it with the rest of the IscB elements mentioned previously (Fig. 3A, 4C; Movie S5). The entry and exiting linkers from RuvC to HNH probe for shape complementarity with the bottom and middle portions of the DNA/RNA heteroduplex, respectively. The body of HNH sinks into the major groove of the DNA/RNA heteroduplex (Fig. 4C). These close contacts are expected to further reduce mismatch tolerance. An Alphafold (18) predicted HNH structure was docked into EM map (Fig. 4C, 4D, S8). While the HNH core structure agreed with the density very well, manual adjustments were needed to fit the predicted linker structures into density (Fig. S8). The HNH nuclease “bites” onto the sugar-phosphate backbone of TS-DNA in the heteroduplex. The His-rich active site coordinates a catalytic metal ion towards the phosphate of the 4th residue in TS-DNA (Fig. 4E, S9), which would leave 3-nt at the TS-DNA side after cleavage, consistent with the biochemistry (2). Topologically, the observed docking movement is only possible if HNH passes underneath NTS-DNA, in a game of limbo with NTS-DNA, which in turn clears the roadblock that previously denied NTS access to RuvC (Movie S6). A continuous corridor of density reveals TAM-proximal NTS-DNA entering the RuvC active site, coordinated by a metal ion therein (Fig. 4F, S10–11). The order of events explains the biochemical observation that TS-DNA cleavage precedes the NTS cleavage (Fig. 4G–H). Previously, RuvC in SpCas9 was found to be allosterically controlled by HNH conformational changes (19), and its cleavage rate trails behind HNH (20). Our structural analysis defines the structural basis for the allosteric control in IscB (Fig. 4H). The same mechanism is likely present in Cas9 RNP.
Given the robust RNA-guided DNase activity in vitro, it is puzzling to observe only weak genome editing activity from OgeuIscB-ωRNA in human cells (2). We noticed the presence of multiple RNA species in the purified OgeuIscB-ωRNA RNP and subjected the sample for RNA deep-sequencing. The sequence coverage dropped immediately before the terminator-like P5 element of ωRNA (Fig. 4I). This is rather surprising because P5 density is clearly present in the RNP structure. We speculated that the cryo-EM particle picking and 3D reconstruction process might have inadvertently biased towards P5-containing single particles (Fig. S2A). Given the high DNA cleavage activity in our OgeuIscB-ωRNA RNP, we probed into the possibility that the PLMP-P5 interaction may be dispensable for RNA-guided DNA cleavage. Indeed, OgeuIscB-ωRNA with a structure-guided PLMP domain truncation (Δaa1–55) was only slightly slower than the wild-type RNP in target DNA cleavage (Fig. 4J–K, S12). This result argues that the PLMP domain is not ubiquitously essential for RNA-guided DNA cleavage among IscB homologs (2). We speculate that the PLMP-P5 interaction may instead be important for the biogenesis of IscB-ωRNA, by controlling the readthrough and termination ratio at ωRNA P5 in order to achieve copy number balance between IscB and ωRNA. Alternatively, these domains may be important for the transposition of IS200/IS605. The sequencing result further revealed a stepwise decrease in coverage for the guide (after the 6th and 10th nucleotide; Fig. 4I). This pattern is consistent with the observed guide accessibility in the IscB-ωRNA structure (Fig. 1). Whether the guide RNA stability may have been the cause of low genome editing activity in IscB is worth investigation in the future. In fact, naturally occurring tracrRNA variants containing a 11-nt-long guide were shown to convert SpCas9 from a nuclease to an RNA-guided transcriptional repressor (21). Chemical modification efforts also revealed that the guide RNA integrity could influence the in vivo activity of Cas9 significantly (22).
In summary, our structural analysis provides a high-resolution explanation for the relationship between IscB-ωRNA and Cas9-crRNA-tracrRNA. ωRNA was speculated to transpose by itself based on informatic searches (17), therefore it may play more active roles than what the structures revealed. Such functions might no longer be needed, or even be detrimental, when IscB-ωRNA established co-option with the CRISPR system. This may have led to the observed adaptation in Cas9-crRNA-tracrRNA, where the body of ωRNA was entirely replaced by protein domains. The remaining portion only serves two essential functions: guiding RNP assembly and guiding target searching. On the application side, there has been a strong interest to miniaturize Cas9 for expanded usage. For example, it would be desirable to package the next-generation Cas9-based genome editors (23–27) into mature delivery tools, such as the adenovirus-associated virus (AAV) vectors. Neither structure-guided approach nor directed evolution was particularly successful at miniaturizing the RNA-guided nucleases. By peeking into nature’s winning solutions, we gain a fresh starting point to develop a new generation of powerful genome editing tools, packageable into AAV. Fifty-five amino acids have already been removed from IscB without abolishing its activity (Fig. 4I). Further structure-guided efforts will likely lead to smaller, more robust, and more active versions of genome editors.
Materials and Methods
Cloning, expression, and purification
An IscB from a human gut metagenome (Genbank: OGEU01000025.1, CDS: 120729-122219) was codon optimized and synthesized (GeneUniversal) with an N-terminal 6x His, thrombin, Twin-Strep-tag, HRV 3C protease site, sumo protease site, SV40NLS and C-terminal nucleoplasm NLS. This IscB construct was cloned into pCDFDuet™-1 (Novagen) vector between the NcoI and BamHI sites. The IscBΔPLMP expression vector was constructed using PCR mutagenesis using F_remove_PLMP CTGGTTCTGGGTATTGATCCG and R_remove_PLMP AGATCCCACCTTCCGTTTC. The ωRNA (Genbank: OGEU01000025.1, 120523-120728) sequence as defined previously (Altae-Tran et al. 2021) was synthesized (GeneUniversal) and cloned into pUC57-Kan between the HindIII and EcoRI sites. Upstream of the ωRNA was a T7 promoter, csy4 stem loop, and 16nt guide. A T7 terminator was placed downstream of the ωRNA.
IscB and ωRNA plasmids were co-transformed into E. coli T7 Express cells (New England Biolabs). The cell culture was grown in LB medium supplemented with 0.75g L-cysteine/L at 37°C until the optical density at 600nm reached 0.8. Expression was induced by adding isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.5mM at 16°C overnight. Cells were collected by centrifugation and lysed by sonication in buffer A (175mM NaCl, 50mM HEPES pH7.25, 2mM TCEP, 5% glycerol, 2.5mM MgCl2) with 1mM phenylmethylsulfonyl fluoride (PMSF). The lysate was centrifuged at 12,000 r.p.m. for 60 minutes at 4°C, and the supernatant was applied onto a pre-equilibrated strep-tactin resin (iba lifesciences). Resin was then washed with 15mL of buffer A, 25mL of buffer A with 0.1mM CaCl2 and 2μg DNaseI (Gold Biotechnology), 20mL buffer B (1M NaCl, 50mM HEPES pH7.25, 2mM TCEP, 2.5mM MgCl2), and 40mL buffer A. Resin was resuspended in buffer A and incubated with 3C protease at 4°C overnight. The flow through buffer containing the 3C cleaved IscB was then concentrated and further purified by anion chromatography (MonoQ 5/50GL; Cytiva) with a gradient elution beginning with buffer A and increasing the percent of buffer B. Peak fractions were tested for cleavage activity and pooled. Pooled fractions were concentrated and further purified by size-exclusion chromatography (Superdex 200 Increase 10/300 GL; Cytiva) equilibrated with buffer C (175mM NaCl, 50mM HEPES pH7.25, 2mM TCEP, 2.5mM MgCl2). The first peak was collected, concentrated, and flash frozen with liquid nitrogen.
DNA substrate preparation
DNA oligonucleotides for cryo-EM were synthesized (Integrated DNA Technologies). NT_Fam_O3_target_PS /56-FAM/CGCCCCACGAGGGTACGGCAAAAGA*G*T*T*T*T*T*TTTACTAGAAGTCGAGGTCAGCCCGTGGC, T_O3_target_PS GCCACGGGCTGACCTCGACTTCTAGT*C*T*C*G*T*T*CACTCTTTTGCCGTACCCTCGTGGGGCG (*phosphorothioate bond). Oligonucleotides were annealed in duplex buffer (30 mM HEPES pH 7.5, 100 mM potassium acetate) by heating to 95°C for 5 min and slowly cooling. Annealed Oligonucleotides were purified in a 10% native PAGE gel. The template strand for DNA cleavage assays was synthesized by (Integrated DNA Technologies) Template_cleavage_target CCCACGAAGGGTTACGGCAAAGCATCATCAAAAAGAGTGAACGAGACTAGAAGTCTGAAAAGGTCATTTTTTAAAGCC. DNA substrate for cleavage assays was produced using PCR using F_cleavage_target /Cy3/CCGCAAGAGGATGATTCGGGTGCGGCAACGGAAGGGGAGGGCCCCACGAAGGGTTACGG R_cleavage_target /Cy5/GCTGATCTGATGCAGTTAAGTGCCTGCTGGGCTTTAAAAAATGACCTTTTCAGAC. PCR products were agarose gel purified using GeneJet gel extraction kit (Thermo Scientific).
Cleavage assays
The cleavage assays were performed as follows. 10μL reactions were prepared where 20nM target DNA was incubated with 1μM IscB in cleavage buffer (50mM NaCl, 50 mM HEPES pH7.25, 2mM βME, 5mM MgCl2) and incubated at 37°C for 1 hour. For time course experiment reactions were quenched with the addition of EDTA to 150mM (final concentration) and an equal volume of 100% formamide. 2mM MnCl2 was added to the cleavage buffer for phosphorothioate bond cleavage rescue experiment. Samples were heated to 95°C for 10 minutes and run on 12% urea-PAGE. Fluorescent signals were imaged using ChemiDoc (BioRad) and quantified using Image Lab.
RNA extraction, urea gel running, and RNA sequencing
20 μL of IscB sample and 20 μL phenol-chloroform solution was mixed together and vortexed vigorously at room temperature. The aqueous and organic phases were separated by 13,000 rpm centrifuge for 2 minutes at room temperature. 10 μL sample was taken from the aqueous phase (top layer), mixed with 10 μL of formamide loading dye, heat-denatured at 95 °C for 10 min, and immediately loaded to a 12% urea-polyacrylamide (PAGE) gel. After 50 minutes of electrophoresis at 25 watts, the gel was stained with EtBr to for 10 min, destained in water for 10 minutes, and scanned with the ChemiDoc imaging system (Bio-Rad) at appropriate wavelength.
Small RNA sequencing
Phenol-choloroform extracted RNA was ethanol precipitated with 9x volumes of chilled 100% ethanol and 1μL of GlycoBlue (Invitrogen) and stored at −80°C. Precipitated RNA was centrifuged at 13,000 rpm for 30 minutes at 4°C. Ethanol was removed, the RNA pellet was dried, and resuspended in nuclease free water. RNA was sent for the Cornell TREx facility for NEBNext small RNA library prep and Illumina sequencing. Library was sequenced to a depth of 10 million reads with a read length of 75nt. The Cornell TREx facilities processed the raw single-end reads with trim-galore package to trim low quality bases and adapter sequences. Trimmed reads were aligned to the T7 Express E. coli genome (Genbank: CP014268.2) and IscB expression plasmids using STAR v2.7. BAM files were visualized in Integrated Genome Browser (IGV).
Cryo-EM sample preparation, data acquisition, and processing
IscB was incubated for 15 minutes at 37°C with the target DNA in cleavage buffer. DNA was supplied at a 3 fold molar excess to IscB (0.5mg/mL final concentration). 3.5μL of were applied to a Quantifoil holey carbon grid (1.2/1.3, 200 mesh) which had been glow-discharged with 20mA at 0.39 mBar for 30 seconds (PELCO easiGlow). Grids were blotted with Vitrobot blotting paper (Electron Microscopy Sciences) for 6.5 s at 4 °C, 100% humidity, and plunge-frozen in liquid ethane using a Mark IV FEI/Thermo Fisher Vitrobot. Data were collected on a Krios G3i Cryo Transmission Electron Microscope (Thermo Scientific) with a Ceta 16M CMOS camera 300kV, Gatan K3 direct electron detector. The total exposure time of each movie stack led to a total accumulated dose of 50 electrons per Å2 which fractionated into 50 frames. Dose-fractionated super-resolution movie stacks collected from the Gatan K3 direct electron detector were binned to a pixel size of 1.1 Å. The defocus value was set between −1.0 μm to −2.5 μm.
Motion correction, CTF-estimation, blob particle picking, 2D classification, 3D classification and non-uniform 3D refinement were performed in cryoSPARC v.2 (28). Refinements followed the standard procedure, a series of 2D and 3D classifications with C1 symmetry were performed as shown in Fig. S2 to generate the final maps. A solvent mask was generated and was used for all subsequent local refinement steps. CTF post refinement was conducted to refine the beam-induced motion of the particle set, resulting in the final maps. The detailed data processing and refinement statistics for cryo-EM structures are summarized in Fig. S2, S7 and Table S1.
Supplementary Material
Acknowledgements
We thank F. Ding for carrying in vitro RNA transcriptions to test IscB RNP activities, Q. Sun for providing initial IscB Alphafold predictions, and A. Pyle for advice on cryo-EM grid preparation.
Funding
National Institutes of Health grant GM118174 (AK)
Department of Defense through the National Defense Science & Engineering Graduate Fellowship Program (GS)
National Science Foundation Materials Research Science and Engineering Centers program grant DMR-1719875 (Cornell Center for Materials Research Shared DOE Office of Biological and Environmental Research grant KP1607011 (Laboratory for BioMolecular Structure (LBMS))
Footnotes
Competing Interests
The authors declare competing financial interests on IscB-ωRNA structure-inspired genome editing applications.
Data and materials availability
The IscB-ωRNA/R-loop coordinates and cryo-EM density map have been deposited in the Protein Data Bank (PDB:7UTN) and the Electron Microscopy Data Bank (EMD-26782).
The RNA-sequencing data has been deposited in the Sequence Read Archive (PRJNA838366).
Plasmids used in this study are available upon request.
References and Notes
- 1.Kapitonov VV, Makarova KS, Koonin EV, ISC, a Novel Group of Bacterial and Archaeal DNA Transposons That Encode Cas9 Homologs. J Bacteriol 198, 797–807 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altae-Tran H et al. , The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science 374, 57–65 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karvelis T et al. , Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature 599, 692–696 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M et al. , A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L et al. , Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasiunas G, Barrangou R, Horvath P, Siksnys V, Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A 109, E2579–2586 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishimasu H et al. , Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shmakov S et al. , Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol Cell 60, 385–397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang F et al. , Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–871 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jinek M et al. , Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343, 1247997 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishimasu H et al. , Crystal Structure of Staphylococcus aureus Cas9. Cell 162, 1113–1126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun W et al. , Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States. Mol Cell 76, 938–952 e935 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das A et al. , The molecular basis for recognition of 5’-NNNCC-3’ PAM and its methylation state by Acidothermus cellulolyticus Cas9. Nat Commun 11, 6346 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bravo JPK et al. , Structural basis for mismatch surveillance by CRISPR-Cas9. Nature 603, 343–347 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mojica FJM, Diez-Villasenor C, Garcia-Martinez J, Almendros C, Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology (Reading) 155, 733–740 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Anders C, Niewoehner O, Duerst A, Jinek M, Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinberg Z, Perreault J, Meyer MM, Breaker RR, Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature 462, 656–659 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jumper J et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sternberg SH, LaFrance B, Kaplan M, Doudna JA, Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 527, 110–113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong S, Yu HH, Johnson KA, Taylor DW, DNA Unwinding Is the Primary Determinant of CRISPR-Cas9 Activity. Cell reports 22, 359–371 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Workman RE et al. , A natural single-guide RNA repurposes Cas9 to autoregulate CRISPR-Cas expression. Cell 184, 675–688 e619 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Mir A et al. , Heavily and fully modified RNAs guide efficient SpyCas9-mediated genome editing. Nat Commun 9, 2641 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaudelli NM et al. , Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anzalone AV et al. , Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR, Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anzalone AV et al. , Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat Biotechnol, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ioannidi EI et al. , Drag-and-drop genome insertion without DNA cleavage with CRISPR-directed integrases. bioRxiv, 2021.2011.2001.466786 (2021). [Google Scholar]
- 28.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


