Summary
Analysis of the human and murine transcriptomes has identified long noncoding RNAs (lncRNAs) as major functional components in both species. Transcriptional profiling of the murine limb led to our discovery of lncRNA-HIT, which our previous in vitro analyses suggested a potential role for this lncRNA in the development of limb, craniofacial, and genitourinary tissues (Carlson et al., 2015). To test this hypothesis, we developed a conditional lncRNA-HIT loss of function allele which uses Cre recombinase to activate an shRNA specific for lncRNA-HIT. Activation of the lncRNA-HIT shRNA allele resulted in a robust knock-down of lncRNA-HIT as well as co-activation of a mCherry reporter, confirming the efficacy of the shRNA allele to reduce endogenous lncRNA levels in a tissue- and cell-type specific manner. Developmental analyses of embryos expressing the activated shRNA and mCherry co-reporter revealed multiple malformations corresponding to the sites of shRNA activation, affecting craniofacial, limb, and genitourinary tissue development. These results confirm the efficacy of lncRNA-HIT shRNA allele to knock-down endogenous transcripts in tissue- and cell type specific manner and indicate a requirement for lncRNA-HIT in the development of these tissues.
Keywords: genetics, mammal, organism, process, tissue: craniofacial, tissue: limb, tissue: mesoderm, tissue: reproductive, tissue: skeletal
1 ∣. INTRODUCTION
Long noncoding RNAs (lncRNAs) are transcribed sequences ranging from 200 to 20,000 nucleotides in length that do not encode proteins. Current analyses of the human and murine transcriptomes have identified >96,000 loci encoding lncRNAs in humans with >87,000 being identified in mice (NONCODE-V5, noncode.org, Fang et al., 2018). A primary feature of lncRNAs is their lack of sequence conservation between species, making the identification of orthologous transcripts highly dependent on computational analyses that discern conserved structural motifs or through their association with cis-regulatory regions conserved among multiple species (Hezroni et al., 2015; Quinn et al., 2016). To date, roughly 1,000 lncRNA have been identified to exhibit evolutionary constraint among vertebrate species, many of which are expressed in precise temporally and spatially restricted domains during development (Hezroni et al., 2015; Perry & Ulitsky, 2016). More importantly, loss of function studies provide a growing body of evidence that lncRNAs are functional, controlling diverse cellular processes involved in development and/or disease including: chromosome replication timing, gene dosage, mesenchyme condensation, skeletal element length, digit number, apoptosis, cell proliferation, hypertrophy, tissue homeostasis, and metastasis (Carlson et al., 2015; Hou et al., 2017; Klattenhoff et al., 2013; Munschauer et al., 2018; Platt, Smith, & Thayer, 2018; Quinn et al., 2016; Wang et al., 2011; Zhang & Guo, 2019).
To accomplish these diverse roles, lncRNAs recruit proteins in the cytoplasm and/or nucleus to facilitate gene regulation through a myriad of molecular processes affecting mRNA stability, splicing, and translation or by controlling chromatin accessibly by recruiting factors essential for histone methylation and/or acetylation (Butler, Johnston, Kaur, & Lubin, 2019; Carlson et al., 2015; Kherdjemil et al., 2016; Wang et al., 2011; Wang et al., 2019). Finally, lncRNAs can also control gene expression by mimicking hormone receptor DNA binding sites, competing with receptor-mediated transcriptional regulation of target loci (Kino, Hurt, Ichijo, Nader, & Chrousos, 2010).
In the developing murine limb, we previously identified lncRNA-HIT as a novel nuclear lncRNA that plays an essential role mediating H3K27ac to control in vitro mesenchymal cell condensation and chondrogenesis (Carlson et al., 2015). To ascertain whether the in vitro functions of lncRNA-HIT are conserved in vivo during limb development, we generated a conditionally activated lncRNA-HIT shRNA loss of function allele. In this report, we describe the production of this allele and characterize its efficacy to reduce lncRNA-HIT mRNA levels in discrete embryonic regions. Broad activation of the conditional shRNA allele using CMV-Cre results in severe malformations affecting the head, CNS, limbs, and developing vasculature, suggesting lncRNA function is required for the development of multiple tissues. In the developing limb, activation of the shRNA allele using Hoxa13-Cre resulted in tissue-specific activation of the mCherry co-reporter and robust knock-down of lncRNA-HIT in the developing autopod, causing ectopic chondrogenic differentiation of the forelimb interdigital tissues. Activation of the shRNA allele using Col2-Cre also stimulated the knock-down of lncRNA-HIT in the autopod chondrocytes. Confocal analysis of mCherry-labeled chondrocytes in lncRNA-HIT deficient autopods revealed diffuse cellular contribution at several sites in the developing autopod skeletal anlagen, suggesting a requirement for lncRNA-HIT in the contribution of chondrocytes to the autopod skeletal tissues. Finally, activation of the shRNA allele in the developing genital tubercle (GT) using Shh-Cre also resulted in the robust knockdown of the lncRNA. Confocal analysis of the mCherry co-reporter in the GT revealed a delay in the closure of the urethral tube at the site of activation of the shRNA allele in the urethral plate epithelium, suggesting that lncRNA-HIT is required for closure of the GT. Taken together these results indicate efficacy of the Rosa26 lncRNA-HIT shRNA allele to reduce endogenous lncRNA-HIT levels in a tissue- and cell type-specific manner, and confirm an essential role for this lncRNA in embryonic development.
2 ∣. RESULTS AND DISCUSSION
2.1 ∣. shRNA selection
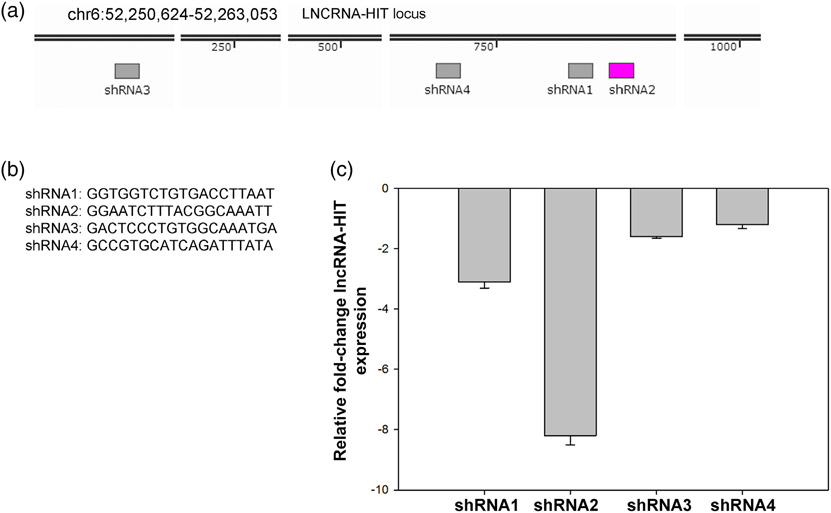
Four candidate lncRNA-HIT shRNA sequences were identified by the software pSico Oligomaker (version 1.5, Ventura et al., 2004) and synthesized (IDT Coralville, IA) with compatible cohesive ends for HpaI and XhoI restriction endonucleases. The shRNA DNA sequences were ligated into the HpaI-XhoI sites of pSico which contains a Cre-inducible shRNA system as described (Ventura et al., 2004). The pSico-shRNA plasmids were electroporated in combination with a Cre recombinase expression plasmid, pCMV-CRE; into an immortalized murine limb mesenchyme cell line previously described by Knosp, Scott, Bachinger, and Stadler (2004). Electroporations were accomplished using a Nucleofector II electroporation system and the Amaxa Nucleofector Kit (Lonza, Hayward, CA). Efficacy of the expressed shRNA sequences to knock-down endogenous levels of lncRNA-HIT was determined by qRTPCR using a Bio-Rad CFX96 Real Time PCR instrument as described (Carlson et al., 2015). Four independent electroporation and qRTPCR analyses were performed for each candidate shRNA (Figure 1). From this analysis, the second shRNA sequence (shRNA2) was identified as the most effective at stimulating the knock-down of lncRNA-HIT, producing a greater than eightfold reduction endogenous lncRNA-HIT levels (Figure 1). Based on these results, the shRNA2 sequence was selected for inclusion in the Rosa26 lncRNA-HIT shRNA targeting vector (Figure 2).
FIGURE 1.
Candidate lncRNA-HIT shRNA evaluation. (a) Location of candidate shRNA sequences within the lncRNA-HIT locus. (b) Candidate lncRNA-HIT shRNA sequences selected by the shRNA selection software pSicoOligmaker version 1.5. (c) qRTPCR analysis of lncRNA-HIT levels in an immortalized limb mesenchyme cell line following electroporation of the Cre-inducible pSico shRNA expression vector containing the candidate shRNA sequences and a pCMV-Cre expression plasmid. Bars represent the mean and standard error for four independent replicates
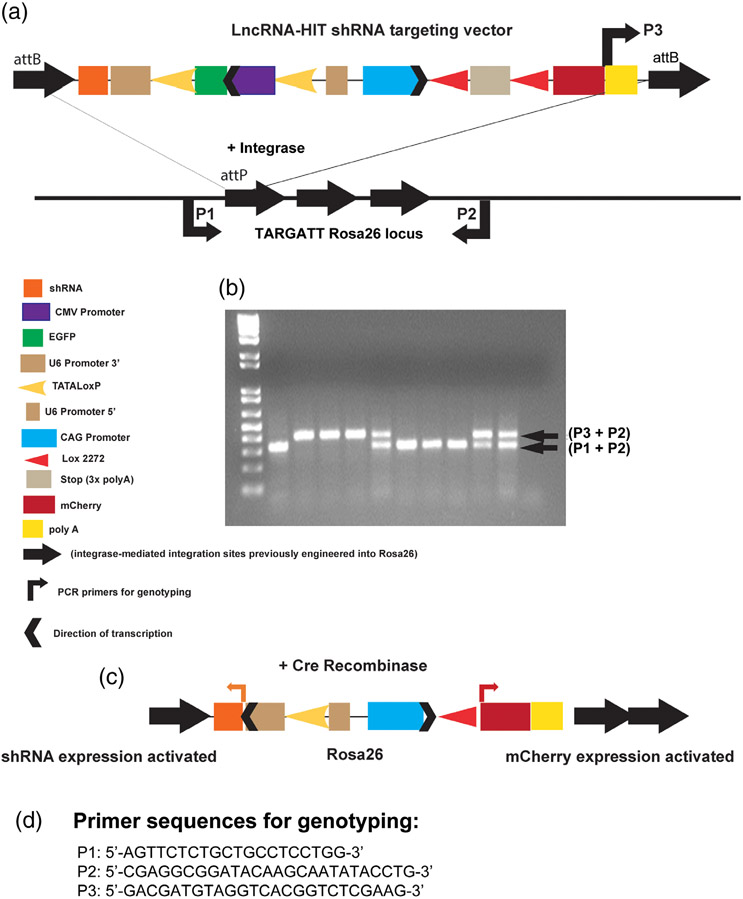
FIGURE 2.
Development of the LncRNA-HIT Rosa26 shRNA conditional mouse allele. (a) Functional components of the LncRNA-HIT Rosa26 shRNA conditional allele. Targeting to the Rosa26 locus is facilitated by integrase-mediated recombination of attB sites flanking the target vector with attP targeting vector attP sites with attB sites previously introduced at the Rosa26 locus as described (Tasic et al., 2011). (b) PCR genotyping of mouse tail DNA isolated from offspring derived from intercrosses of shRNAsilent/+mice. shRNAsilent allele is detected using primer combination P3 + P2. Wild type allele is detected using primer combination P1 + P2. (c) Schematic representation of the shRNAactive allele following Cre-mediated removal of loxP flanked EGFP cassette used to inactivate the U6 promoter necessary for shRNA expression. Activation of mCherry expression is also facilitated by Cre-mediated removal of the stop cassette which is flanked by heterologous Lox2272 sites. (d) Nucleotide sequences for primers P1, P2, and P3 used in genotyping
2.2 ∣. Production of the Cre-inducible lncRNA-HIT mouse allele
The location of lncRNA-HIT in the HoxA gene cluster places it within 1.4 kb of Hoxa13 (Carlson et al., 2015). The proximity of lncRNA-HIT to Hoxa13 raised the possibility that targeted disruption of the lncRNA-HIT locus might also affect cis-regulatory elements controlling Hoxa13 or other loci located in the gene-dense HoxA gene cluster, resulting in phenotypes reflecting both the loss and of lncRNA-HIT and decreased expression of nearby HoxA genes. To circumvent this possibility, we generated a Cre-inducible lncRNA-HIT shRNA allele which is expressed from the Rosa26 locus, allowing for RNA interference to knock-down endogenous levels of lncRNA-HIT, without disrupting any potential Hox cis-regulatory elements (Figure 2). The use of a trans-acting shRNA was also recommended in a recent series of guidelines focused on studying the developmental functions of lncRNAs located in gene-dense chromosomal regions (Bassett et al., 2014).
The Cre-inducible pSico plasmid containing the shRNA2 sequence s (Figure 1) was provided to Applied Stem Cell (Milpitas, CA) who subcloned it into their TARGATT system vector designed to facilitate integrase mediated recombination at the Rosa26 locus (https://www.appliedstemcell.com/products/targatt-genome-editing/transgenicmice; Tasic et al., 2011). A Cre-inducible mCherry co-reporter was included in final targeting vector to provide a mechanism to visualize tissues expressing the Cre-activated shRNA allele (Figure 2). Germline transmission of the Rosa26 LncRNA-HIT shRNA allele was verified by Applied Stem Cell and the heterozygous F1 Rosa26 LncRNA-HIT shRNA mice were shipped to the OHSU animal facility where they were intercrossed to produce homozygous pups bearing the Rosa26 LncRNA-HIT shRNA allele which is designated as shRNAinactive for this report (Figure 2). PCR genotyping of mouse tail DNA derived from the heterozygous intercrosses of shRNAinactive adults produced normal Mendelian ratios of homozygous shRNAinactive mice which exhibited no phenotypes. Verification of the correct targeting of lncRNA-HIT shRNA allele to the Rosa26 locus was determined by PCR amplification and sequencing of the targeted Rosa 26 locus from shRNAinactive pup DNA using primers specific for the Rosa26 locus as well as a reverse primer specific for the LncRNA-HIT shRNA targeting vector (Figure 2) as described (Tasic et al., 2011). DNA sequencing data for the shRNAinactive allele at the Rosa26 locus is provided in Supplemental Data File 1.
2.3 ∣. In vivo evaluation of the lncRNA-HIT shRNA allele
2.3.1 ∣. Whole embryo activation of the shRNA allele
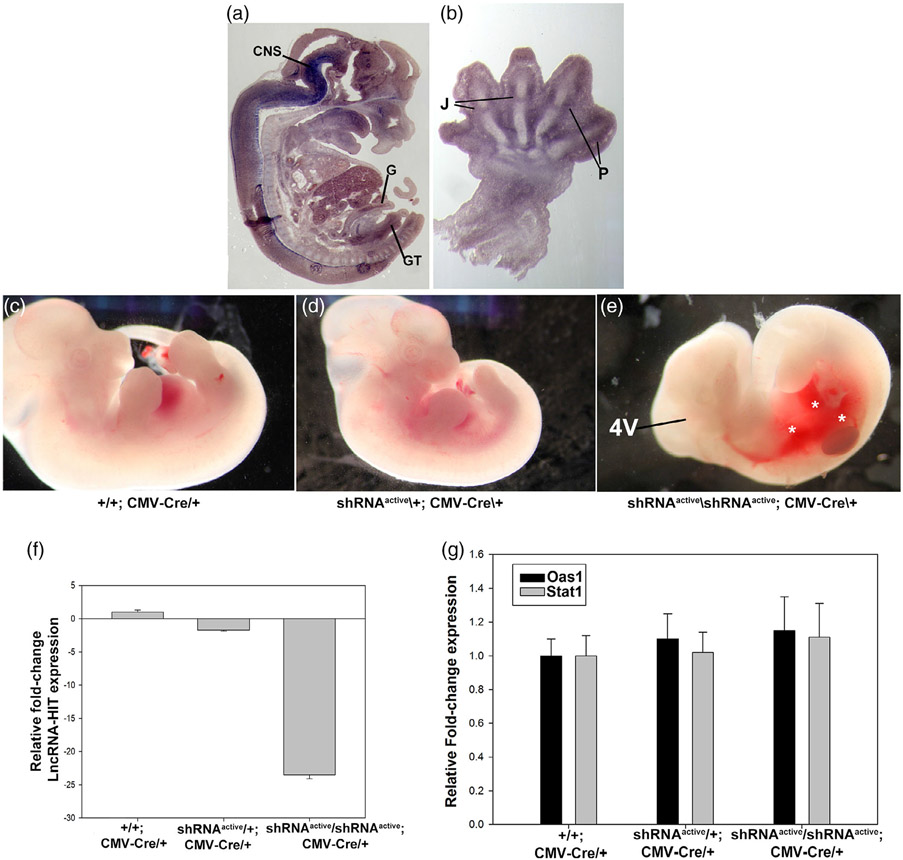
LncRNA-HIT is highly expressed in multiple tissues during murine development including the distal limb, gut, genital tubercle, central nervous system, and bladder (Figure 3) (Carlson et al., 2015). To determine the efficacy of the shRNA allele to knock-down endogenous lncRNA-HIT throughout the embryo, a CMV-Cre allele was used to broadly activate shRNA expression from the Rosa26 locus (Figure 2). For this report, the activated Rosa26 LncRNA-HIT allele is designated: shRNAactive/+; Cre/+. Compound heterozygous shRNAactive/+; CMV-Cre/+ mice were viable and fertile allowing for intercrosses to produce homozygous shRNAactive/shRNAactive; CMV-Cre/+ embryos. Analysis of E13.5 shRNAactive/shRNAactive; CMV-Cre/+ embryos revealed severe hemorrhaging of the homozygous shRNA active embryos as well as reductions in the fourth ventricle region of the developing central nervous system (CNS) (Figure 3). QRTPCR analysis of RNA isolated from E13.5 control and shRNAactive/shRNAactive; CMV-Cre/+ embryos revealed >24-fold reduction in embryonic lncRNA-HIT, confirming the efficacy of the shRNA allele to knock-down endogenous levels of lncRNA-HIT throughout the embryo (Figure 3). Next, because expressed shRNAs can mimic RNA virus infections and cause phenotypes due to their induction of a generalized interferon response (GIR), we examined whether two targets of GIR, Oas1, and Stat1, were affected in shRNAactive/shRNAactive; CMV-Cre/+ embryos (Bridge, Pebernard, Ducraux, Nicoulaz, & Iggo, 2003; Li et al., 2013). QRTPCR analysis of mRNA derived from control and homozygous shRNAactive/; CMV-Cre/+ embryos revealed no difference in Oas1 or Stat1 expression, suggesting that the phenotypes caused by the broad activation of the Rosa26 LncRNA-HIT shRNA allele by CMV-Cre are a consequence of the reduced levels of lncRNA-HIT (Figure 3).
FIGURE 3.
Whole embryo activation of the LncRNA-HIT shRNA allele using CMV-Cre. (a) LncRNA-HIT is expressed in multiple tissues during development. In situ hybridization using a whole embryo sagittal section at E13.5 reveals strong expression in the developing embryo. CNS = central nervous system. G = gut epithelium. B = bladder epithelium. GT = genital tubercle. (b) Section In situ hybridization of a E13.5 forelimb limb bud reveals strong expression in the developing joint fields (J) and perichondrial (P) tissues of the digits. (c–e) E13.5 heterozygous and homozygous shRNAactive; CMV-Cre/+ embryos exhibit severe hemorrhaging (asterisks) and reductions in the fourth ventricle region of the CNS compared to either heterozygous or wild type littermate controls. (f) qRTPCR analyses of E13.5 embryos reveal >24-fold reduction in lncRNA-HIT mRNA levels between control and homozygous shRNAactive/CMV-Cre/+ embryos. (h) qRTPCR analysis of Stat1 and Oas1 expression which function as downstream indicators of a generalized interferon response reveals no differences in expression between control and shRNAactive/shRNAactive; CMV-CRE embryos. Bars in panels (f) and (g) represent the mean and standard deviation for four independent replicates
2.3.2 ∣. Analysis of tissue specific restriction of the activated of shRNA allele
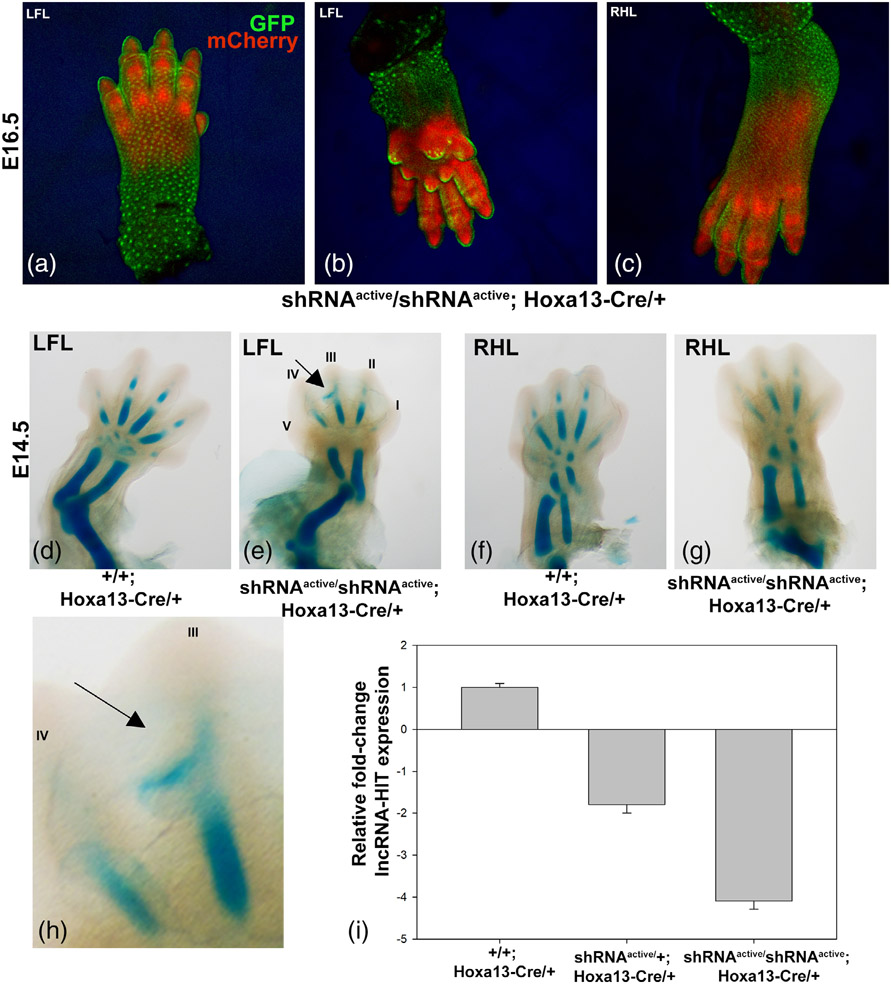
Next, to assess whether the activated shRNA allele recapitulates spatiotemporally restricted patterns of Cre expression/function, a Hoxa13-Cre allele was used which expresses Cre recombinase starting at E9.5 in the distal limb mesenchyme but not the overlying ectoderm (Scotti, Kherdjemil, Roux, & Kmita, 2015). Characterization of mCherry expression in shRNAactive/shRNAactive; Hoxa13-Cre/+ embryos at E16.5 revealed restriction of the activated shRNA allele to the underlying autopod mesenchyme with no expression of the mCherry co-reporter being detected in the overlying ectoderm (Figure 4). qRTPCR analysis of lncRNA-HIT levels in E16.5 shRNAactive/shRNAactive; Hoxa13-Cre/+ autopods revealed a >4-fold knock-down of lncRNA-HIT) confirming efficacy of the shRNA allele to knock-down lncRNA levels in the Hoxa13-specific autopod domain (Figure 4). Alcian blue staining of E14.5 limbs revealed a disruption of the digit III phalangeal element formation, resulting in a protrusion of alcian blue-positive tissue into the interdigital tissue of homozygous shRNAactive; Hoxa13-Cre/+ embryos (Figure 4).
FIGURE 4.
Tissue specific activation of the lncRNA-HIT shRNA allele by Hoxa13-Cre. (a–c) Confocal analysis of Hoxa13-Cre mediated activation of the lncRNA-HIT shRNA allele in E16.5 limbs. Note localization of the EGFP reporter to the overlying autopod ectoderm which indicates no activation of the shRNA allele whereas the subectodermal autopod mesoderm strongly expresses the mCherry co-reporter indicating activation of the shRNA allele. (d, e) Analysis of Hoxa13-Cre mediated activation of the lncRNA-HIT shRNA allele in the E14.5 littermate forelimbs revealed reduced skeletal element length and displacement of the digit III phalangeal cartilage into the interdigital tissue (arrow) in shRNAactive/shRNAactive; Hoxa13-Cre/+ embryos as detected by alcian blue staining. (f, g) Analysis Hoxa13-Cre mediated activation of the lncRNA-HIT shRNA allele in the E14.5 littermate hindlimbs revealed reduced skeletal element length and maturation as detected by alcian blue staining. (h) Higher magnification image of panel (e) indicating the displacement of the digit III phalangeal cartilage (arrow) into the interdigital tissue. (i) qRTPCR analyses of lncRNA-HIT expression in shRNAactive/shRNAactive; Hoxa13-Cre/+ limbs confirm robust knock-down (~4-fold) of the lncRNA by the activated shRNA allele. Bars represent the mean and standard error for four independent replicates
2.3.3 ∣. Analysis of cell-type-specific restriction of the shRNA allele
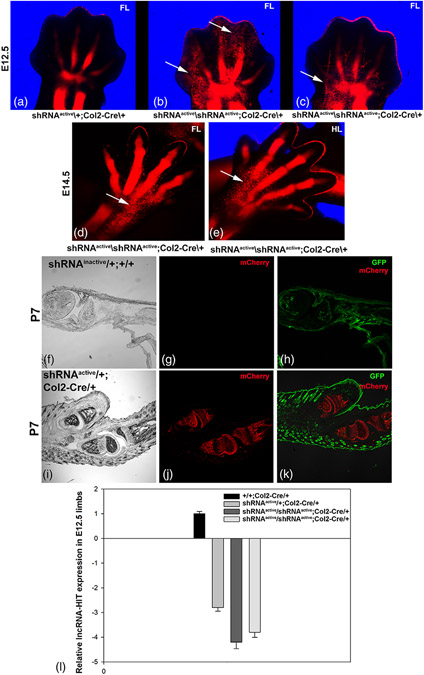
Next, to assess the efficacy of the Rosa26 LncRNA-HIT shRNA allele to reduce lncRNA levels in a specific cell type, a Col2-Cre allele was used to activate shRNA expression in chondrocytes which we previously demonstrated highly express lncRNA-HIT (Carlson et al., 2015). Characterization of the shRNAactive/shRNAactive; Col2-Cre/+ embryos revealed strong activation of the mCherry reporter in the E12.5 and E14.5 skeletal anlagen (Figure 5). Analysis of shRNAactive/+; Col2-Cre/+ and shRNAactive/shRNAactive; Col2-Cre/+ limbs revealed diffuse patches of chondrocytes expressing the mCherry co-reporter that were not incorporated into the developing autopod skeletal anlagen in homozygous shRNAactive limbs at both E12.5 and E14.5 (Figure 5). QRTPCR analysis of total RNA isolated from autopod limb tissues of shRNAactive/shRNAactive; Col2-Cre/+ embryos at E 12.5 revealed a 3.5–4-fold reduction in lncRNA-HIT confirming the efficacy of the shRNA allele to knock-down lncRNA levels in chondrocytes. Characterization of sectioned postnatal day (P) 7 limbs confirmed expression of the mCherry reporter is restricted to chondrocyte-derived skeletal tissues. Finally, confocal analysis of P7 sections of shRNAinactive/+; +/+ autopods revealed no mCherry fluorescence in limbs lacking Col2-Cre, confirming the efficacy of the CAG-Lox2272 stop Lox2272-mCherry cassette to conditionally regulate expression of the co-reporter (Figures 2 and 5).
FIGURE 5.
Chondrocyte-specific activation of the lncRNA-HIT shRNA allele by Col2-Cre. (a–c) Confocal analysis of E12.5 littermate forelimbs expressing the activated lncRNA-HIT shRNA allele reveal strong activation of the mCherry co-reporter in chondrocytes contributing to the developing limb skeletal anlagen. Arrows denote regions of poor mCherry-labeled chondrocyte contribution to the developing autopod skeletal anlagen in shRNAactive/shRNAactive; Col2-Cre/+limbs compared to shRNAactive/+; Col2-Cre/+ littermates. (d, e) Poor contribution of mCherry-expressing chondrocytes to the autopod skeletal anlagen was also detected at E14.5 in shRNAactive/shRNAactive; Col2-Cre/+ forelimbs and hindlimbs (arrows). (f–h) Confocal analysis of sagittal P7 forelimb sections of shRNAinactive/+; +/+ forelimbs reveal only GFP expression and no expression of mCherry co-reporter and confirming the absence of leakiness of the CAG-lox stop lox-mCherry cassette present in the LncRNA-HIT Rosa26 shRNA allele. (i–k) Confocal analysis of sagittal sections of P7 forelimbs in shRNAactive/+; Col2-Cre/+ mice confirm chondrocyte-specific activation of the mCherry co-reporter by Col2-Cre. (l) qRTPCR analyses of E12.5 limbs reveal robust knock-down of lncRNA-HIT by Col2-Cre activation in chondrocytes resulting in a 4–4.5-fold reduction in lncRNA levels in homozygous shRNAactive limbs compared to controls. Bars represent the mean and standard error for four independent replicates
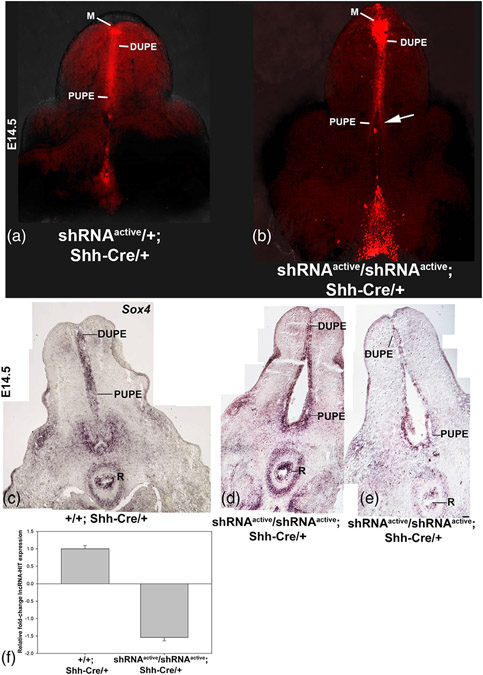
Finally, to assess the role of lncRNA-HIT in the developing genital tubercle (GT), we activated the shRNA allele in the GT urethral plate epithelium (UPE) using Shh-Cre, a structure that strongly expresses Shh to mediate urethral tube development (Harfe et al., 2004; Perriton, Powles, Chiang, Maconochie, & Cohn, 2002; Seifert, Bouldin, Choi, Harfe, & Cohn, 2009; Seifert, Zheng, Ormerod, & Cohn, 2010). Analysis of homozygous shRNAactive; Shh-Cre/+ embryos revealed an opening in the proximal urethral tube, which when examined as sections, revealed a malformation of the proximal urethral plate epithelium (PUPE) (Figure 6a-e). Using these same tissue sections, we examined whether the malformation of the PUPE reflected a loss of epithelia necessary for the urethral tube development. Characterization the GT epithelial marker, Sox4 (Dy et al., 2008), revealed strong expression in the UPE of homozygous shRNAactive embryos, suggesting that the malformation of the PUPE is not the result of epithelial cell loss (Figure 6c-e). The malformation of the PUPE in shRNAactive/shRNAactive; Shh-Cre/+ mice was surprising as our previous analysis of lncRNA-HIT indicates that the lncRNA can regulate Hoxa13 expression (Carlson et al., 2015). Based on this analysis, we predicted that lncRNA-HIT deficient GTs would exhibit a subset of the Hoxa13 null phenotype, which we previously reported affects distal GT structures including the meatus and distal urethral plate epithelium (DUPE) (Morgan, Nguyen, Scott, & Stadler, 2003). Instead, the knock-down of lncRNA-HIT in the GT preferentially affects proximal urethral tube development. These findings suggest that lncRNA-HIT may also function independently of Hoxa13 to facilitate urethral tube development. Finally, qRTPCR analysis of homozygous shRNAactive UPEs revealed a 1.6-fold knockdown of lncRNA-HIT. This result confirms a loss of the lncRNA at the site of the urethral tube malformation and suggests that even modest losses of the lncRNA (~1.6-fold) are sufficient to affect urethral tube development (Figure 6f).
FIGURE 6.
Activation of the lncRNA-HIT shRNA allele in the developing genital tubercle using Shh-Cre. (a, b) Confocal analyses of the developing genital tubercle in E14.5 shRNAactive; Shh-Cre/+ mice confirm activation of the mCherry co-reporter in the developing meatus (M), distal urethral plate epithelium (DUPE), and proximal urethral plate epithelium (PUPE). A comparison of heterozygous and homozygous shRNAactive; Shh-Cre/+ male genital tubercles revealed a delay in the urethral tube formation in shRNAactive embryos (arrow). (c–e) Section in situ hybridization analysis of homozygous shRNAactive; Shh-Cre/+ male embryos at E14.5 identifies the proximal UPE (PUPE) as the site of urethral tube malformation. Expression analysis of the genitourinary epithelial marker, Sox4, reveals no change in expression, suggesting that the malformation of the PUPE is not a consequence of epithelial tissue loss. R = rectum, DUPE = Distal urethral plate epithelium, PUPE = proximal urethral plate epithelium. Analysis of Sox4 expression, a general marker of genitourinary epithelia, confirms Sox4 expressing epithelia are present in the malformed PUPE, suggesting the malformation is not a consequence of epithelial tissue loss. (f) qRTPCR analyses of lncRNA-HIT expression in revealed a >1.6-fold knockdown of lncRNA-HIT in homozygous shRNAactive/Shh-Cre/+ embryos
3 ∣. METHODS
Following publication of this report, mice bearing the Rosa26 LncRNA-HIT shRNA allele will be made available for distribution to the research community upon request.
3.1 ∣. Cre mice used
Activation of the shRNA allele was accomplished by intercrosses of shRNAinactive mice with mice heterozygous for the following Cre expression alleles: CMV-Cre (Schwenk, Baron, & Rajewsky, 1995); Col2-Cre (Ovchinnikov, Deng, Ogunrinu, & Behringer, 2000), Hoxa13-Cre (Scotti et al., 2015), and Shh-Cre (Harfe et al., 2004). Genotyping for the presence of Cre recombinase in shRNAactive/+; Cre/+ mice was accomplished by PCR using genomic DNA and the following primers specific for Cre recombinase: CREF-5′GCGGTCTGGCAGTAAAAACTATC3′ and CRER-5′GTGAAACAGCATTGCTGTCACTT3′.
3.2 ∣. RNA isolation and qRTPCR analysis
Total RNA was isolated using TRIzol (Life Technologies, Grand Island, NY) as instructed. LncRNA-HIT expression was quantitated using a real-time quantitative reverse transcriptase polymerase chain reaction method (qRTPCR). First-strand cDNA was synthesized using an ImProm-II™ Reverse Transcription System (Promega, Madison, WI). A minimum of three independent samples were used for qRTPCR using a SYBR Green PCR Super Mix and a IQ5 thermal cycler according to the manufacturer's instructions (BioRad Hercules, CA). Fold change expression levels were determined after normalization of the amplification products to Gapdh expression using the BioRad IQ5 software suite as described (Carlson et al., 2015). Data was plotted using Sigmaplot 13.0 (Systat, San Jose, CA). Primers used to assess lncRNA-HIT expression were: LncRNA-HITF-5′GTTCCCAGACTCCCTGTGGC3′ and LncRNA-HITR-5′TCGTCAAGGTCAAGGTTTAAGGCC3′. Primers used to assess GAPDH expression were: GAPDHF-5′CCGGTGCTGAGTATGTCGTG3′ and GAPDHR-5′GGCGGAGATGATGACCCTTT3′. Primer sequences used to assess the expression of Oas1 were: OAS1F-5′CGTGCTG CCAGCCTATGATTT-3′ and OAS1R-5′TTGGTTGGGCGACAGTTCAG-3′. Primer sequences used to assess Stat1 expression were: STAT1F 5′-GCTGGGCGTCTATCCTGTGGT-3′ and STAT1R 5′-GCTCAGCTGGTCTGCGTTCA-3′.
3.3 ∣. Confocal analysis of the activated Rosa26 LncRNA-HIT shRNA allele
To follow sites of Cre-mediated activation of the Rosa26 LncRNA-HIT shRNA allele, a Cre-inducible mCherry fluorescent reporter (CAG promoter LoxP stop LoxP mCherry-poly A) was incorporated into the Rosa26 LncRNA-HIT shRNA targeting vector (Figure 2). In vivo activation of the mCherry reporter by Cre recombinase was verified by confocal microscopy using a Zeiss LSM 700 confocal microscope and the Zeiss Zen software (version 6.0). Excitation and emission wavelengths for EGFP were 480 nm and 530 nm respectively. Excitation and emission wavelengths for mCherry were 570 nm and 601 nm respectively.
3.4 ∣. Alcian blue staining of embryonic limbs
E14.5 embryos were collected in 1× phosphate buffered saline and fixed overnight in glass scintillation vials filled with 4% paraformaldehyde at 4°C. Following fixing, the embryos were washed 3 × 5 min using 70% ETOH. After the third wash, the scintillation vials were filled with 70% ethanol and the embryos were rocked for 60 min at room temperature. After the 60-min ethanol wash, the embryos were washed 3 × 5 min in water. After the final water wash, the embryos were placed in a filtered alcian blue staining solution consisting of 70 mL ETOH, 30 mL glacial acetic acid, and 20 mg alcian blue 8GX (Canemco Inc, Quebec, Canada) for 24 hr. After staining, the embryos were washed 3 × 10 min in 100% ETOH and cleared in Benzyl-Benzoate overnight. Cleared embryos were photographed using a Leica MZFLIII stereoscope fitted with a Canon EOS 40D digital camera.
3.5 ∣. Section in situ hybridization
A Sox4-specific antisense riboprobe template was produced by PCR amplification of a 551 bp region present in the 3′ UTR of murine Sox4 using genomic DNA and the following primers: Sox4 Forward 5′-TGGAGAGTAGAAGGAGAAAGGG-3′ and Sox4 Rev:5′-GTGATGCGTTTGGCATTTGT-3′. The amplified Sox4 template was cloned into a t-tailed vector, pXCMI, that contains T3 and T7 RNA polymerase promoters for riboprobe synthesis as described (Perez, Weller, Shou, & Stadler, 2010). Ten micron coronal sections containing the developing E14.5 male GT from shRNAsilent/; +/+ and shRNAactive/shRNAactive; Shh-Cre/+ embryos were processed for in situ hybridization using the Sox4 antisense riboprobe as described (Perez et al., 2010). For all analyses, control and homozygous shRNAactive; Shh-Cre/+ GT sections were treated identically using the same preparation and concentration of the Sox4 antisense riboprobe, hybridization buffer, temperatures, wash-, and colorimetric detection times. After stopping the colorimetric reactions, the sectioned GTs were covered with 10% glycerol/PBS, cover-slipped, and photographed using a Leica DMLB2 microscope equipped with DIC and a Canon EOS40D Camera. Individual images were assembled into photomontages using Adobe Photoshop Creative Cloud 2019.
ACKNOWLEDGMENTS
This work was funded by grants from the Shriners Hospitals for Children (85140 to HSS) and the National Institutes of Health (R56AR074931 to HSS).
Funding information
National Institute of Arthritis and Musculoskeletal and Skin Diseases, Grant/Award Number: R56AR074931 to HSS; Shriners Hospitals for Children, Grant/Award Number: Basic Science Grant 85140 to HSS
REFERENCES
- Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, & Ponting CP (2014). Considerations when investigating lncRNA function in vivo. eLife, 3, e03058. 10.7554/eLife.03058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, & Iggo R (2003). Induction of an interferon response by RNAi vectors in mammalian cells. Nature Genetics, 34(3), 263–264. 10.1038/ng1173 [DOI] [PubMed] [Google Scholar]
- Butler AA, Johnston DR, Kaur S, & Lubin FD (2019). Long noncoding RNA NEAT1 mediates neuronal histone methylation and agerelated memory impairment. Science Signaling, 12(588), eaaw9277. 10.1126/scisignal.aaw9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson HL, Quinn JJ, Yang YW, Thornburg CK, Chang HY, & Stadler HS (2015). LncRNA-HIT functions as an epigenetic regulator of chondrogenesis through its recruitment of p100/CBP complexes. PLoS Genetics, 11(12), e1005680. 10.1371/journal.pgen.1005680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dy P, Penzo-Mendez A, Wang H, Pedraza CE, Macklin WB, & Lefebvre V (2008). The three SoxC proteins–Sox4, Sox11 and Sox12–exhibit overlapping expression patterns and molecular properties. Nucleic Acids Research, 36(9), 3101–3117. 10.1093/nar/gkn162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Zhang L, Guo J, Niu Y, Wu Y, Li H, … Zhao Y (2018). NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Research, 46(D1), D308–D314. 10.1093/nar/gkx1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, & Tabin CJ (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell, 118(4), 517–528. 10.1016/j.cell.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, & Ulitsky I (2015). Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Reports, 11(7), 1110–1122. 10.1016/j.celrep.2015.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Long H, Zhou C, Zheng S, Wu H, Guo T, … Wang T (2017). Long noncoding RNA Braveheart promotes cardiogenic differentiation of mesenchymal stem cells in vitro. Stem Cell Research & Therapy, 8(1), 4. 10.1186/s13287-016-0454-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kherdjemil Y, Lalonde RL, Sheth R, Dumouchel A, de Martino G, Pineault KM, … Kmita M (2016). Evolution of Hoxa11 regulation in vertebrates is linked to the pentadactyl state. Nature, 539(7627), 89–92. 10.1038/nature19813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, & Chrousos GP (2010). Non-coding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Science Signaling, 3(107), ra8. 10.1126/scisignal.2000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, … Boyer LA (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell, 152(3), 570–583. 10.1016/j.cell.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knosp WM, Scott V, Bachinger HP, & Stadler HS (2004). HOXA13 regulates the expression of bone morphogenetic proteins 2 and 7 to control distal limb morphogenesis. Development, 131(18), 4581–4592. 10.1242/dev.01327 [DOI] [PubMed] [Google Scholar]
- Li J, Ding SC, Cho H, Chung BC, Gale M Jr., Chanda SK, & Diamond MS (2013). A short hairpin RNA screen of interferonstimulated genes identifies a novel negative regulator of the cellular antiviral response. MBio, 4(3), e00385–e00313. 10.1128/mBio.00385-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan EA, Nguyen SB, Scott V, & Stadler HS (2003). Loss of Bmp7 and Fgf8 signaling in Hoxa13-mutant mice causes hypospadia. Development, 130(14), 3095–3109. 10.1242/dev.00530 [DOI] [PubMed] [Google Scholar]
- Munschauer M, Nguyen CT, Sirokman K, Hartigan CR, Hogstrom L, Engreitz JM, … Lander ES (2018). The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature, 561 (7721), 132–136. 10.1038/s41586-018-0453-z [DOI] [PubMed] [Google Scholar]
- Ovchinnikov DA, Deng JM, Ogunrinu G, & Behringer RR (2000). Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis, 26(2), 145–146. [PubMed] [Google Scholar]
- Perez WD, Weller CR, Shou S, & Stadler HS (2010). Survival of Hoxa13 homozygous mutants reveals a novel role in digit patterning and appendicular skeletal development. Developmental Dynamics, 239, 446–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriton CL, Powles N, Chiang C, Maconochie MK, & Cohn MJ (2002). Sonic hedgehog signaling from the urethral epithelium controls external genital development. Developmental Biology, 247(1), 26–46. 10.1006/dbio.2002.0668 [DOI] [PubMed] [Google Scholar]
- Perry RB, & Ulitsky I (2016). The functions of long noncoding RNAs in development and stem cells. Development, 143(21), 3882–3894. 10.1242/dev.140962 [DOI] [PubMed] [Google Scholar]
- Platt EJ, Smith L, & Thayer MJ (2018). L1 retrotransposon antisense RNA within ASAR lncRNAs controls chromosome-wide replication timing. The Journal of Cell Biology, 217(2), 541–553. 10.1083/jcb.201707082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JJ, Zhang QC, Georgiev P, Ilik IA, Akhtar A, & Chang HY (2016). Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes & Development, 30(2), 191–207. 10.1101/gad.272187.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F, Baron U, & Rajewsky K (1995). A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Research, 23(24), 5080–5081. 10.1093/nar/23.24.5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti M, Kherdjemil Y, Roux M, & Kmita M (2015). A Hoxa13:Cre mouse strain for conditional gene manipulation in developing limb, hindgut, and urogenital system. Genesis, 53(6), 366–376. 10.1002/dvg.22859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert AW, Bouldin CM, Choi KS, Harfe BD, & Cohn MJ (2009). Multiphasic and tissue-specific roles of sonic hedgehog in cloacal septation and external genitalia development. Development, 136, 3949–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert AW, Zheng Z, Ormerod BK, & Cohn MJ (2010). Sonic hedgehog controls growth of external genitalia by regulating cell cycle kinetics. Nature Communications, 1, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Hippenmeyer S, Wang C, Gamboa M, Zong H, Chen-Tsai Y, & Luo L (2011). Site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proceedings of the National Academy of Sciences of the United States of America, 108(19), 7902–7907. 10.1073/pnas.1019507108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, … Jacks T (2004). Cre-lox-regulated conditional RNA interference from transgenes. Proceedings of the National Academy of Sciences of the United States of America, 101(28), 10380–10385. 10.1073/pnas.0403954101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, … Chang HY (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature, 472 (7341), 120–124. 10.1038/nature09819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen W, Lian J, Zhang H, Yu B, Zhang M, … Tang J (2019). The lncRNA PVT1 regulates nasopharyngeal carcinoma cell proliferation via activating the KAT2A acetyltransferase and stabilizing HIF-1alpha. Cell Death and Differentiation. 10.1038/s41418-019-0381-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, & Guo H (2019). Long non-coding RNA NORAD induces cell proliferation and migration in prostate cancer. The Journal of International Medical Research, 47(8), 3898–3904. 10.1177/0300060519862076 [DOI] [PMC free article] [PubMed] [Google Scholar]