Abstract
The recent advent of human pluripotent stem cell (PSC) derived 3D brain organoids has opened a window into aspects of human brain development that were not accessible before, allowing tractable monitoring and assessment of early developmental processes. However, their broad and effective use for modeling later stages of human brain development and disease is hampered by the lack of a stereotypic anatomical organization, which limits maturation processes dependent upon formation of unique cellular interactions and short and long-range network connectivity. Emerging methods and technologies aimed at tighter regulatory control through bioengineering approaches, along with newer unbiased organoid analysis readouts should resolve several of the current limitations. Here, we review recent advances in brain organoid generation and characterization with a focus on highlighting future directions utilizing interdisciplinary strategies that will be important for improving the physiological relevance of this model system.
Keywords: Brain organoids, Neurodevelopmental Disorders, Bioengineering, Synthetic Biology, Extracellular Matrix, Regional Patterning, Microfluidics, Synthetic Scaffolding, Lineage Tracing, Neuronal Network Activity
1. Introduction
Can an in vitro system recapitulate the complex processes underlying human organogenesis? An astonishing level of self-organization can emerge from 3D aggregates of human pluripotent stem cells (hPSCs) which then continuously evolve through positive and negative feedback interactions to drive the differentiation and assembly of highly organized tissue patterns. The seminal work from the laboratory of Yoshiki Sasai showed how autonomous emergence of highly ordered neural structures can emerge from embryonic stem cells (ESCs) grown in 3D aggregates (Eiraku et al., 2008, Eiraku et al., 2011). In the last decade, building on this work a variety of protocols have been established for the generation of reductionist replicas of multiple brain regions as reviewed in (Quadrato et al., 2016, Di Lullo and Kriegstein, 2017, Quadrato and Arlotta, 2017, Paşca, 2019, Qian et al., 2019, Kelava and Lancaster, 2016). But how far can human brain development be recapitulated in vitro? The answer to this fundamental question is of enormous importance in determining which aspects of human brain development and disease can be currently modelled with high fidelity in organoids. Therefore, a systematic understanding of the principles guiding self-organization in these multicellular systems will enable us to harness their full potential and provide key information to overcome their limitations. Much work is still needed to address this point, but the general consensus in the field is that intrinsic self-organizing properties control cellular diversity and local tissue microarchitecture, while the number and localization of organ primordia is interdependent with external tissue cues and structural environment. For brain organoids, the current inability to precisely control both intrinsic and extrinsic cues results in lack of stereotypical anatomy and aberrant long-range connectivity that in turn affects levels of neuronal activity, cell maturation and overall physiological relevance of this model system. These limitations have affected data analysis and interpretations making classical analysis techniques underpowered (Atamian et al., 2020). Here we summarize, recent advances in brain organoid culturing and analysis techniques that have attempted to remedy some of these limitations. We discuss additional emerging technologies, spanning from cutting edge bioengineering innovations to tools for unbiased multimodal analysis that, combined with current methods, will allow for modeling and understanding aspects of human brain development and disease that are still inaccessible.
2. The Modern Art of Culturing Brain Organoids
2.1. Generating Self-Patterned and Patterned Organoids
Human brain development occurs in a tightly regulated series of events governed by long range morphogen driven gradients combined with local cell-cell and cell-extracellular matrix interactions that continuously evolve through positive and negative feedback regulation over the course of gestation. These complex and intricate regulatory networks allow for the formation of discrete structures and cellular stratification that contribute to the vast processing power of the human brain. Protocols for human brain organoid generation have taken inspiration from these in vivo patterning processes to recapitulate neurodevelopment in a dish. Current organoid protocols are divided into two major categories, patterned and self-patterned. Generally, in both approaches dissociated pluripotent stem cells are aggregated into 3D spheres or embryoid bodies to generate self-organizing 3D neuroectodermal structures. Self-patterned protocols utilize bFGF and retinoic acid during hPSC reaggregation to generate whole brain organoids. These protocols leverage the default bias of early ectoderm to generate dorsal forebrain fates (Espuny-Camacho et al., 2013). However, the efficiency and propensity of different human PSC lines to differentiate into ectoderm (Bock et al., 2011) and downstream cortical cell types varies widely (Quadrato et al., 2017). This leads to high variability among organoids generated within and across PSC lines. In addition, whole brain organoids contain a variety of neuronal and glial cell types from multiple brain regions and have been shown to contain a wide-breadth of non-ectodermal lineage cells as well (Lancaster et al., 2013, Lancaster et al., 2017, Quadrato et al., 2017, Ormel et al., 2018, Camp et al., 2015). However, in whole brain organoids this high degree of cellular diversity comes at the expense of the reproducible generation of distinct cell types causing substantial organoid-to-organoid variability (Quadrato et al., 2017). The stochastic nature of cellular diversity in self-patterned organoids makes this approach not well suited for disease modeling and drug screening purposes. Therefore, patterned protocols that improve regional accuracy and cellular reproducibility have emerged as a widely used alternative in the field (Figure 1).
Figure 1. Development-inspired strategies to recapitulate early human brain regionalization.
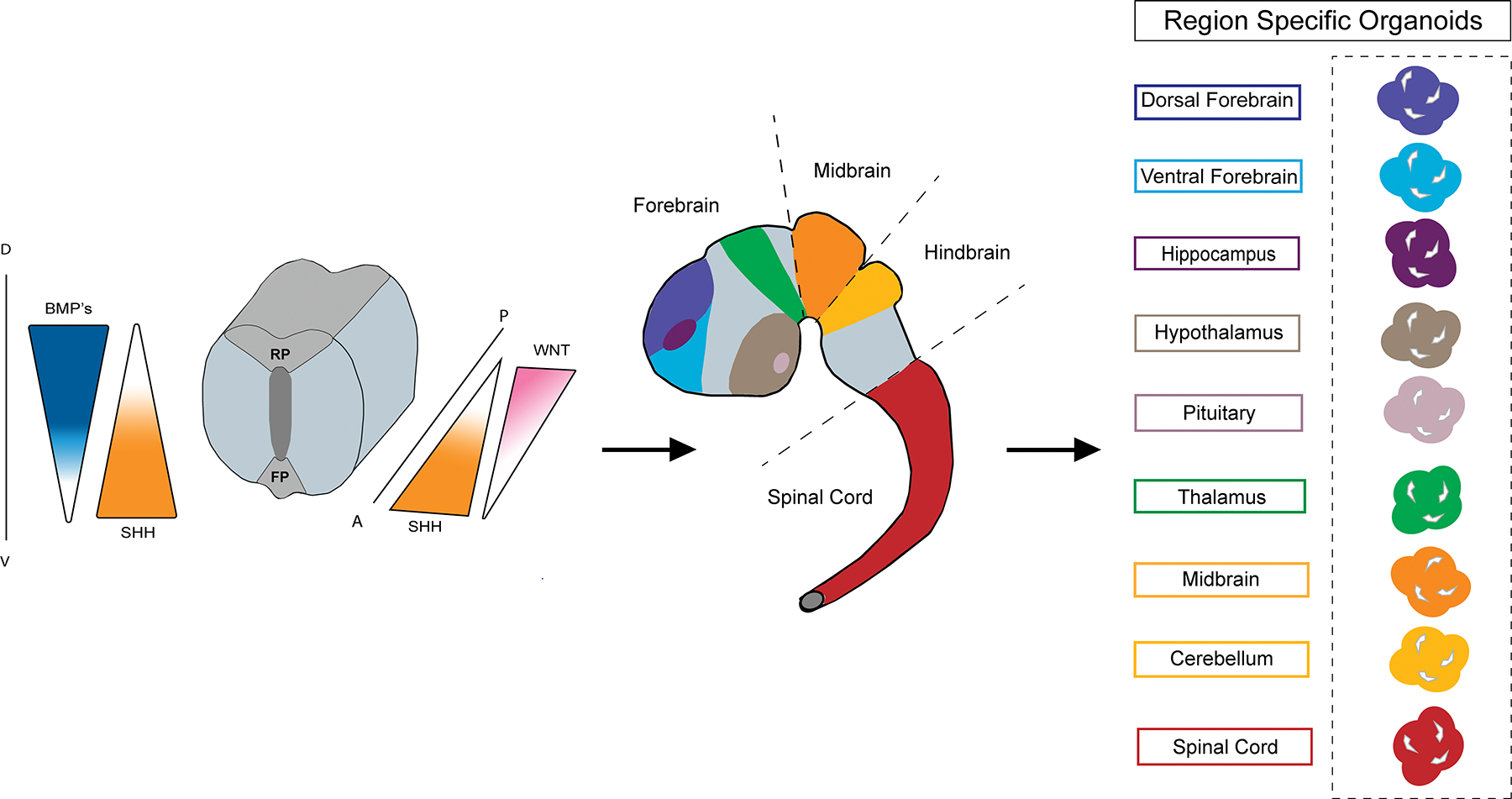
Morphogen gradients within the neural tube are critical in determining the rostro-caudal and dorsal-ventral axis of the developing brain and contribute to the formation of discrete regionalization. Several key signaling centers present in the neural tube including the Roof Plate (RP) and Floor Plate (FP) are a prominent source of BMP (bone morphogenic protein), WNT, SHH (sonic hedgehog), RA (retinoic acid) and FGFs (fibroblast growth factors) respectively. Patterning strategies have allowed for the establishment of protocols resembling distinct brain regions including the dorsal and ventral forebrain, hippocampus, hypothalamus, anterior pituitary, thalamus, midbrain, cerebellum, midbrain, spinal cord and choroid plexus. The bottom panel is a simplified description of the patterning strategies leveraged to create the region-specific brain organoids previously mentioned. The red asterisk indicates that a variation of SMAD inhibition (TGFb with or without BMP inhibition and or WNT inhibition) was employed to direct towards an initial neuroectodermal fate.
A unifying characteristic among most patterned protocols is an initial inhibition of the SMAD pathway (Chambers et al., 2009). The SMAD family of proteins act as signal transducers downstream of TGF-beta superfamily ligand binding to promote the generation of epidermis over neural ectoderm. Patterned protocols have generally employed the use of partial or dual SMAD inhibition through administration of small molecule inhibitors of the TGF-beta type I receptor, and/ or administration of inhibitors which target the activity of bone morphogenic proteins (BMPs). Additionally, WNT inhibition during early stages of organoid generation is often employed to repress the mesodermal lineage and in turn promote the production of anterior neuroectoderm (ten Berge et al., 2008). A number of variations of this basic patterning strategy are currently used for the generation of cortical organoids, leveraging the default bias of early ectoderm to generate dorsal forebrain fates (Kadoshima et al., 2013, Mariani et al., 2015, Paşca et al., 2015, Qian et al., 2016, Rigamonti et al., 2016, Bagley et al., 2017, Watanabe et al., 2017, Velasco et al., 2019). Despite patterning and culturing differences, cortical organoids across protocols can develop neural-tube-like neuroepithelial structures that follow a similar temporal developmental trajectory as in vivo. These structures give rise to a diverse range of cortical cell types as seen in the endogenous human forebrain including, inner and outer radial glia, intermediate progenitors, deep and upper layer cortical neurons, astroglia, and oligodendroglia precursor cells. The question of whether broad cell types in cortical organoids can be reproducibly generated has been addressed by Yoon et al. (2019), who were able to obtain consistent a cellular composition in cortical spheroids at different developmental stages. Further characterization of cellular diversity and reproducibility of cortical cell types generated in individual organoids, within and across lines, was thoroughly tested by Velasco et al. (2019), who demonstrated reproducible generation of all main classes of cortical progenitors and postmitotic cells of cortical organoids using a modified version of the Kadoshima et al. (2013) protocol. However, the molecular signature fidelity of cortical subtypes generated in organoids has been questioned in a recent paper that demonstrates increased metabolic stress due to in vitro culture conditions can alter subtype specification (Bhaduri et al., 2020). Cortical brain organoid research is rapidly evolving, and it will be exciting to follow how quickly other brain region organoid protocols improve.
Cortical organoids are widely used because of their relevance to human neurodevelopmental disorders, as defects in higher order cognitive abilities predominately stem from dysregulation within this brain region. However, in the last few years patterned protocols that overcome the default forebrain bias to differentiate into other brain regions have been developed. These include methods to generate human organoids of the thalamus (Xiang et al., 2019), hypothalamus (Qian et al., 2016), hippocampus (Sakaguchi et al., 2015), cerebellum (Muguruma et al., 2015), midbrain (Jo et al., 2016, Qian et al., 2016, Monzel et al., 2017), anterior pituitary (Ozone et al., 2016), and spinal cord (Ogura et al., 2018).
The thalamus emerges from the caudal region of the forebrain and its generation requires a delicate balance of morphogen gradients along the rostro-caudal axis. A recent thalamic organoid protocol attempted to mimic this through early insulin treatment to promote caudalization, which is later attenuated by MEK-ERK inhibition to block excessive caudalization towards a midbrain fate, and simultaneous BMP7 treatment to promote a thalamic specific fate (Xiang et al., 2019). This protocol has successfully generated distinct thalamic lineages including glutamatergic neurons and a small percentage of GABAergic and serotonergic neurons with undefined regional identity.
The hippocampus is derived from the dorsomedial telencephalon. Sakaguchi et al. (2015) recapitulated this specification through modulation of BMP4 and Wnt signaling (CHIR) to mimic the in vivo dorsal midline and cortical hem signaling center that is required for the generation of dorsomedial telencephalic tissue. The specific timing and duration of the morphogen treatment in this protocol allowed for the first time, generation of hippocampal granule neurons and CA3 pyramidal neurons in a 3D culture environment.
The midbrain is a centrally located ventral structure that represents the most superior portion of the brain stem. Jo et al. (2016) designed a patterning strategy for this region centered around promoting ventralization using SHH and determining the rostro-caudal positional identity through Fgf8 administration, ultimately resulting in a tissue type similar to the midbrain and capable of producing dopaminergic neurons. Qian et al. (2016) used a similar strategy with the addition of a WNT agonist to initially promote a floor plate-like identity before pushing for a midbrain specific identity to robustly generate dopaminergic neurons. An alternative approach in which neuroepithelial stem cells (NESCs) were used as the starting population for midbrain organoid generation was developed by Monzel et al. (2017). In this case the 3D NESC spheres generated were patterned with a similar approach as previous groups, using a SHH pathway agonist and a GSK3 inhibitor. These organoids contained post-mitotic dopaminergic neurons as in previous protocols, however astrocytes and functionally mature oligodendrocytes were found to be present as well.
The cerebellum is an elaborately organized hindbrain structure with several distinct progenitor zones that give rise to different neuronal populations, making the recapitulation of the structure in vitro complicated. Mugaruma et al. (2015), established a 3D cerebellar organoid protocol with the goal of generating Purkinje cells using FGF2, which has been shown to be a potent caudalizing factor at high concentrations. This caudalizing factor allowed for the generation of an isthmic organizer-like center, a region that expresses signaling molecules that promotes the generation of midbrain and hindbrain on either side of it. Differentiation occurs along this axis to result in a multilayered structure resembling the ventricular and rhombic lip germinal zones of early embryonic development. However, maturation of Purkinje cells to its recognizable “tree-like” structure could be achieved only following coculture with mouse cerebellar granule neurons.
The spinal cord is the most caudal portion of the central nervous system and functions to relay sensory information to and from the brain. The structural organization of the spinal cord is complex, involving the intermingling of more than twenty distinct neuronal subclasses. The generation of 3D spinal cord tissue has recently been accomplished by Ogura et al. (2018) who took inspiration from in vivo signaling centers pertinent to early stages of spinal cord development. This included the roof plate and floor plate which are largely responsible for producing BMP’s, Wnts and SHH, respectively. By utilizing these morphogens alongside retinoic acid to promote the caudalization of the organoid the authors successfully generated several subclasses of spinal cord neurons, including GABAergic, glutamatergic and cholinergic neurons within tissue reminiscent of the dorsal, intermediate and ventral spinal cord.
The Lancaster group has also recently explored the generation of choroid plexus-like tissue in brain organoids through the addition of the patterning factors CHIR and BMP4 (Pellegrini et al., 2020). In vivo, the choroid plexus functions to produce cerebrospinal fluid (CSF) which contains nutrients and signaling molecules important to brain development. In vitro, the Lancaster group was able to induce the formation of choroid plexus-like stroma and epithelial tissue capable of forming tight junctions and producing CSF. The induction of this functional structure in brain organoid cultures may contribute to the ability to mimic a physiologically relevant microenvironment in vitro.
While some studies addressing the fidelity and reproducibility of cell types generated in cortical organoids at different developmental stages have been carried out, the characterization of organoids of other brain regions is still in its infancy. Many outstanding questions related to the reproducibility of cell types generated within and across cell lines, as well as, the molecular fidelity of these cells to their endogenous counterparts need to be answered before they can be reliably used for disease modeling and drug screening.
2.2. “Integrated” Organoids
Patterning protocols have successfully generated distinct regions of the brain reminiscent of their in vivo counterparts by taking inspiration from morphogen gradients that guide human brain development (Figure 2). Currently, morphogen treatment of organoids in vitro is done via bath application, which does not wholly recapitulate the tightly regulated spatiotemporal gradients that are key to proper neurodevelopment. Although cells within organoids are capable of self-generated local gradients that mimic in vivo patterning, the reproducibility of these local cues and the extent to which they can influence the generation of discrete structures remains unclear. Generally, these directed differentiation approaches generate a single early brain region and do not recapitulate multiple brain regions or the long-range connectivity between them, which are necessary for refinement and specification of cells within a developing human brain. Additionally, when PSCs are directed towards a neuroectoderm fate using a bath of morphogens, non-ectodermal lineage cells are excluded. Importantly, these include yolk-sac derived microglia and other cells of mesodermal origin such as endothelial cells which form the vascular network. These non-ectodermal cell types play critical roles in brain development and their absence negatively impacts human brain organoid development. However, recent publications have explored alternative methods to incorporate non-ectodermal cells, improve cellular diversity, and enable co-development of multiple brain structures.
Figure 2. Comparison of patterned and self-patterning approaches in generating human brain organoids.
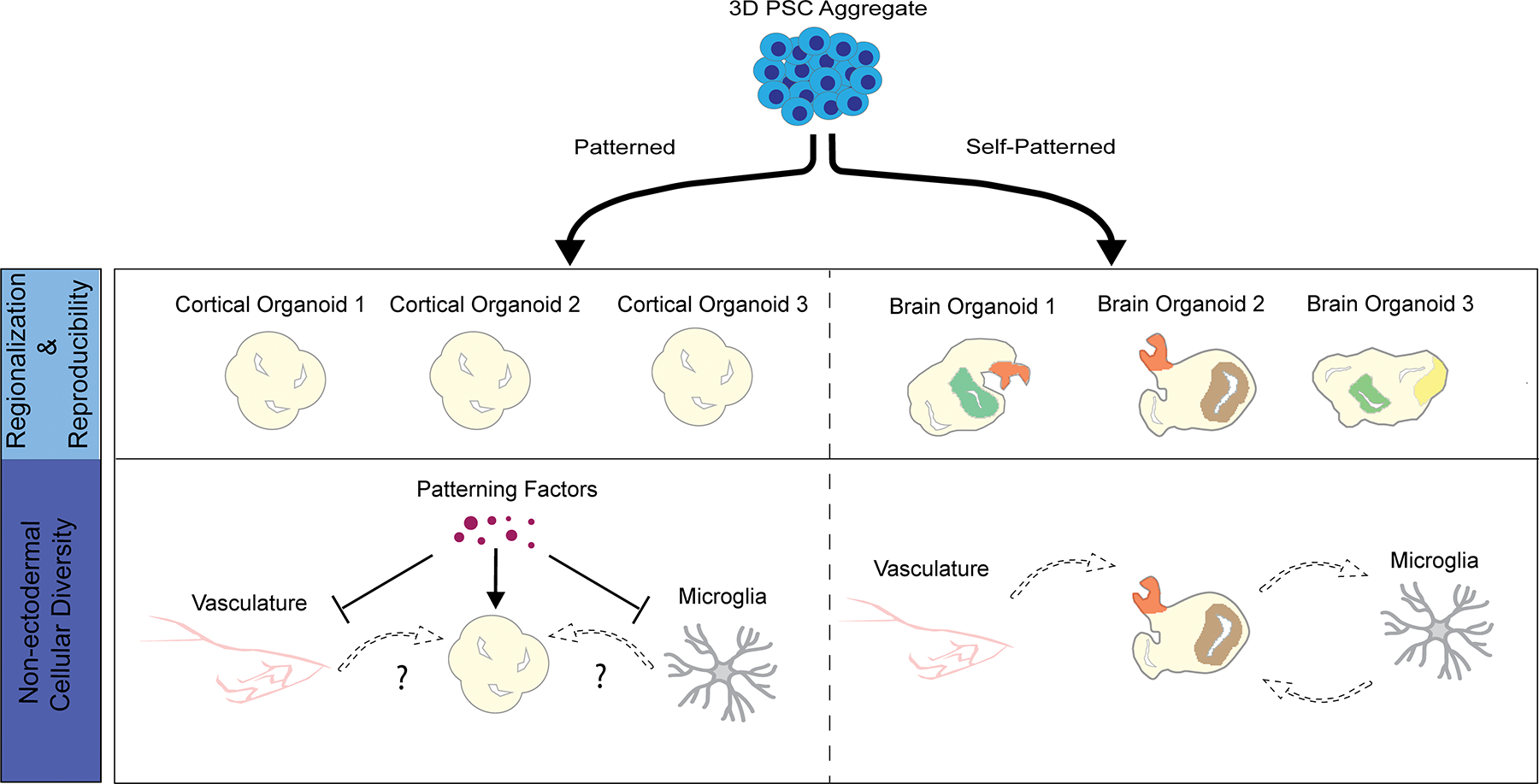
Protocols rely on either the use of morphogen driven patterning strategies to generate organoids reminiscent of a specific brain region, or the innate ability of aggregated pluripotent stem cells (PSC’s) to establish self-patterning morphogen gradients that drives neuroectodermal identity and produce organoids reminiscent of the whole brain. While organoids patterned towards the cortex have been shown to reproducibly generate cortical neurons in similar ratios, self-patterned whole brain organoids contain diverse but heterogeneous brain regions (top panels). For example, in patterned cortical organoids, all non-ectodermal cell types including microglia are omitted. In addition, the heavy patterning employed at early stages of organoid generation in these protocols may affect the integration of microglial and vascular cells into these cultures at this stage. Self-patterned brain organoids, on the other hand, allow for the spontaneous generation of microglial populations and due to the lack of exogenous morphogens, permit integration of non-ectodermal cell types (bottom panels).
2.2.1. Incorporation of diverse cell types
While neuronal populations have been a longstanding focus of brain research, glial cells have also emerged as key players in the human brain during development and throughout life (Zuchero and Barres, 2015). Oligodendrocytes and astrocytes are collectively termed macroglia and together constitute most of the cell types present in the brain. Gliogenesis follows neurogenesis, beginning mid gestation in humans and continuing beyond birth; encompassing developmental stages that are difficult to recapitulate in brain organoids. Currently it is possible to generate macroglia across several brain organoid generation protocols (Paşca et al., 2015, Quadrato et al., 2017, Bhaduri et al., 2020, Velasco et al., 2019, Sloan et al., 2017) but the level of maturity and functionality that they attain remains limited. In order to model later stages of development it is critical that macroglia be present, mature and functional, an endeavor pioneered by several groups that have modified culture conditions to promote this. The promotion of oligodendrocyte maturation has been accomplished in brain organoids through modulation of the culturing media to include factors known to promote oligodendrocyte differentiation and maturation while preserving a robust neuronal population. This has been accomplished by Madhavan et al. (2018) and Marton et al. (2019) who both recorded functionally mature oligodendrocytes after 28 and 30 weeks, respectively.
As previously mentioned, patterned brain organoids lack several non-ectodermal cell types. While oligodendrocytes and astrocytes are derived from neural progenitors within the neuroectoderm, microglia are generated separately from hematopoietic stem cells in the yolk sac during early embryogenesis and migrate into the brain parenchyma during early development. This process generally begins around the fifth week of gestation in humans, just following the onset of neurogenesis (Monier et al., 2007, Kierdorf et al., 2013). As microglia take residence in the brain, they receive and provide feedback to neuronal populations influencing migration, differentiation and synaptic connectivity as well as regulating immune responses (Cunningham et al., 2013, Squarzoni et al., 2014). These microglia dependent processes have been implicated in the pathogenesis of several neurodevelopmental disorders, creating a need to incorporate this glial population into brain organoid models (Tay et al., 2017, Lenz and Nelson, 2018). While protocols that rely on undirected neuronal differentiation (Lancaster et al., 2013) have observed that microglia can innately develop within brain organoids (Ormel et al., 2018), more reproducible directed protocols that employ SMAD inhibition to force a neuroectodermal fate, do not form microglia progenitors. This has led to the development of in vitro protocols to generate ESC and iPSC derived microglia (Muffat et al., 2016, Abud et al., 2017, Douvaras et al., 2017, Haenseler et al., 2017, McQuade et al., 2018), which can then be integrated into brain organoid cultures. This strategy has been used to study Alzheimer’s disease (Abud et al., 2017, Lin et al., 2018), basic neuro-immune interactions (Abreu et al., 2018, Muffat et al., 2018, Song et al., 2019) and to assess the transcriptomic changes upon microglia integration into brain organoids (Bejoy et al., 2019, Schmunk et al., 2020). While these studies have effectively demonstrated that exogenously derived microglia can integrate, generate an immune response, and participate in functional cross talk with neuronal populations within brain organoids, there is still much work to be done in optimizing this system. Currently, the time point for integration of microglia into organoids differs across protocols; however, the exact timing of integration may have drastically different effects on neuronal populations. Additionally, the number of successfully integrated microglia may impart differing effects on neuronal survival and immune response, and while attempts have been made to incorporate microglia in proportions similar to what is seen in vivo, there is yet no standardized protocol for the timing and amount of microglia to add. Lastly, the longevity of microglia in brain organoid cultures remains unclear, with the longest recorded time point being one month (Lin et al., 2018), and a majority of other protocols using a shorter time point of one week. Much of the appeal that surrounds brain organoids is the ability to maintain cultures over extended periods of time to allow for the emergence and functional maturation of distinct neuronal subtypes. Successful long-term culture of microglia integrated brain organoids will be crucial to understanding how neuroimmune interactions shape organoid cytoarchitecture and influence the functional maturity of neuronal populations during normal and aberrant development.
Brain organoids lack vasculature and therefore undergo chronic hypoxia in the innermost regions. In vivo, by six gestational weeks, a capillary plexus covers the entire cerebral cortex and by eight gestational weeks the intracerebral microvasculature in the brain begins to develop in a ventral to dorsal progression, providing oxygenation, and serving to deliver nutrients and eliminate waste (Marin-Padilla, 2012). As this neurovascular network expands in complexity and magnitude, it begins to innervate deeper regions of the brain and participates in dynamic crosstalk with neuronal progenitor populations to guide proliferation, migration and differentiation (Tata and Ruhrberg, 2018). Therefore, integrating a functional vascular network into 3D brain organoids is necessary to accurately model later stages of human brain development. Recently, Ham et al. (2020) treated brain organoids at early stages with vascular endothelial growth factor (VEGF) and Wnt7b to promote the formation of vascular endothelial cells and also pericyte-like supporting cells without compromising neuronal populations. This led to the formation of open-circle vascular structures that contained CD31 positive cells and modulation of gene networks associated with brain embryogenesis in neuronal populations, indicating crosstalk between neuronal and endothelial cell populations. Cakir et al. (2019) generated human cortical organoids that displayed vascular-like structures by co-culturing hPSCs engineered to overexpress ETV2, a transcription factor known to induce endothelial cell fate, with unedited hPSCs. This resulted in vascularized cortical brain organoids that displayed blood-brain barrier-like characteristics such as expression of tight junctions and nutrient transporters. Worsdorfer et al. (2019) developed a co-culture method in which mesodermal progenitor cells integrated into brain organoid cultures produced vascular networks with typical blood vessel ultrastructure after roughly eight weeks in vitro. Alternatively, Pham et al. (2018) co-cultured endothelial cells with brain organoids and supplemented the media with vascular endothelial growth factor (VEGF), resulting in the emergence of robust vascular networks after roughly four weeks in vitro. However, these in vitro based systems still lack a pump and therefore an active flow of media through the vascular structures.
Finally, Xenograft models have been developed to increase cellular diversity and functional neuronal connectivity. This method was pioneered by Mansour et al. (2018) where after several weeks of in vitro growth, organoids were intracerebrally grafted into a cleared cavity in the mouse cortex and allowed to grow for several more months. Upon analysis of the grafted organoids, it was revealed that they exhibited progressive neuronal differentiation and maturation, gliogenesis, integration of microglia and invasion of mouse vasculature, and outgrowth of axons that formed functional connections with the host brain. This model system will provide important insights for the development and refinement of completely in vitro-based approaches that achieve similar results, but in a higher throughput, human specific brain organoid model system.
2.2.2. The Fusion of Patterned Brain Organoids
To establish the formation of multiple regional identities, fusion protocols have been used, wherein organoids are individually directed toward a specific brain fate and subsequently fused together via co-culturing. This method has been utilized by several groups to combine dorsal and ventral forebrain organoids (Bagley et al., 2017, Birey et al., 2017, Xiang et al., 2017). In vivo, the ventral forebrain produces GABAergic inhibitory interneurons that migrate to dorsal regions of the forebrain and integrate themselves into cortical circuits during development. The groups were able to identify GABAergic inhibitory interneurons of several subtypes, including the medial ganglionic eminence derived somatostatin and parvalbumin positive interneuron subtypes and observed their migration into the dorsally patterned forebrain organoid, recapitulating what is seen in vivo. While it remains largely unknown in humans, during mouse brain development there are three waves of oligodendrocyte generation during embryonic development. The first two waves are transient and originate from the ventral regions of the brain while the latter is a permanent population that is generated from more dorsal brain regions. Kim et al. (2019) fused dorsal and ventral forebrain organoids to monitor this process of oligodendrocyte development. After fusion, fifteen weeks of culture, and five weeks of T3 (a thyroid hormone to promote oligodendrocyte maturation) treatment, the authors concluded that there are indeed similar waves of ventrally and dorsally derived oligodendrocytes. This fusion method has also been employed to interrogate the reciprocal neuronal projections between thalamic and cortical organoids (Xiang et al., 2019). These studies indicate that fused organoid models can form functionally relevant connections, improving functional maturation in some cases and pave the way for future studies to investigate long-range connectivity between multiple distinct brain regions.
3. Guiding self-organization in brain organoids
3.1. Engineering a Pseudo-Signaling Center
During human brain development, morphogen gradients are orchestrated by signaling centers. These gradients in turn, establish cell fate and polarity necessary for proper formation of the central nervous system. As current organoid generation protocols use a bath application of morphogens and lack a true signaling center to direct cell polarity, these organoids form multiple neural tube-like rosette structures that give rise to neural progenitors. Cederquist et al. (2019) probed the use of a pseudo Sonic Hedgehog (SHH) organizing center by creating an inducible SHH-expressing hPSC line (iSHH) that was embedded at one pole of a forebrain organoid during development. They observed that a SHH gradient established by this signaling center could induce the formation of forebrain subdivisions with in vivo-like topography. While SHH is a critical morphogen for the establishment of a dorsal-ventral and rostral-caudal axis during development, it acts in concert with an array of other factors to precisely pattern distinct regions. Having a model that uses a singular signaling center is a promising step forward in organoid generation but establishment of multiple synthetic signaling centers to generate interacting morphogen gradients would more closely recapitulate in vivo neurodevelopment.
Signaling centers can also be engineered via microfluidics. A microfluidic device contains a set of micro-channels designed to precisely control the delivery of media and/or gases to a culture chamber where nutrients can be exchanged, and wastes removed. Microfluidic devices have the potential to change patterning dynamics by facilitating orthogonal gradients of morphogens. This was demonstrated using gradients of retinoic acid, a major determinant of the rostral-caudal axis, and SAG, a ventrally secreted smoothened agonist, to mimic the combinatorial effect of these orthogonally distributed morphogens on specification of ventral lower layer motor neuron differentiation in the developing neural tube (Uzel et al., 2016). The Kirkeby lab has implemented an approach termed microfluidic-controlled stem cell regionalization (MiSTR), to model neural tube development by creating a WNT gradient during hPSC differentiation (Rifes et al., 2020). The gradient allowed cells to adopt a rostral-caudal organization and develop molecular characteristics of organizing centers, for example formation of an isthmic organizer. Collectively, these results indicate the possibility to modulate the self-organization of hPSC by extrinsically controlling the spatiotemporal formation of morphogen gradients via a microfluidic approach.
3.2. Engineering anatomical features
The lack of stereotypical anatomy found in cortical organoids hinders the ability to perform complex functional and tracing analysis. One reason hypothesized to contribute to this deficient layer separation is the limited diffusion of nutrients and oxygen at the cortical organoid core which may lead to organoids undergoing chronic hypoxia. To overcome this limitation, Qian et al. (2020) generated sliced neocortical organoids (SNOs) to bypass diffusion limitations, and showed reduced cell death and improved oxygenation in organoid cores. This method allowed organoids to maintain long-term neurogenesis, sustained progenitor expansion and separation of cortical layers. Similarly, the Lancaster group cultured sliced cortical organoids at the air-liquid-interface, termed ALI-COs (Giandomenico et al., 2019), and these organoids displayed improved neuronal survival compared to whole organoids, as well as mature morphological and functional features including formation of discrete long range axonal tracts that were able to make functional connections with mouse spinal cord explants. Discrete long-range axonal tracts have also been shown to form between cortical organoids in a static microculture device (Kirihara et al., 2019). Here, organoids were individually cultured in separate chambers, connected by a microchannel that was 9 mm in length in which axon fascicles were shown to form reciprocal connections. In this study, axonal tracts formed spontaneously, but made sporadic connections; however, a more robust system in which axons are guided will be required to establish reproducible and functional long-range connectivity between distinct brain structures. Additional engineering of hPSC using techniques such as the recently described optogenetic control of growth cone guidance in zebrafish motor neurons (Harris et al., 2020) or through the modulation of axonal guidance cues may facilitate future studies of long range connections between distinct organoids. The generation of distinct anatomical features in brain organoids remains challenging but emerging techniques such as sliced culturing protocols that improve neuronal survival, axonal growth, and connectivity provide direction to increase feasibility. In addition, axonal connectivity can be guided via microchannels or by optogenetic control, which opens the possibility of connecting brain organoids in reproducible architectures that will anatomically and functionally mimic in vivo brain circuits.
3.3. Engineering biomimetic cellular microenvironments
Extrinsic cues such as morphogens have been heavily used via bath application to control and direct the differentiation of hPSCs into brain organoids. This method, while effective, remains simplistic and alone does not encompass the complexity that is required to generate and maintain discrete brain regions in a reproducible manner. The inclusion of additional external cues will allow for further refinement of the model by providing additional layers of control. The extracellular matrix (ECM) is a 3D structure composed of insoluble components secreted from a diverse range of cell types. It plays a key role in maintaining structural integrity and participates in transducing cell-cell communication. Within the brain, the ECM is highly specialized and its composition varies based upon the region and developmental time point (Krishnaswamy et al., 2019). Dysregulation associated with the ECM is becoming increasingly implicated in neurodevelopmental and neuropsychiatric disorders, creating a need to accurately model this component of the brain in 3D brain organoid cultures (Beroun et al., 2019). However, several important cell types known to contribute to ECM formation and maintenance, such as microglia and vascular endothelial cells are not present in many current brain organoid cultures. While there have been several reports of synthetic hydrogels or decellularized human fetal ECM being used as an alternative to the intrinsically produced ECM in 2D culture conditions (Barry et al., 2017, Jin et al., 2018, Cho et al., 2019, Lam et al., 2019, Galarza et al., 2020, Reginensi et al., 2020), matrigel remains the preferred ECM-like component utilized in 3D brain organoid cultures. Although extensively used, the composition of matrigel is generic and largely uncharacterized, which can lead to variability and a lack of physiologically relevant biochemical cues during brain organoid generation. Sood et al. (2019) has recently employed the use of ECM derived from fetal porcine brains in combination with silk scaffolding to better recapitulate the in vivo ECM micro-environment. The authors observed that inclusion of the fetal porcine ECM improved the functional maturation of neuronal populations and supported their long-term survival (Sood et al., 2019). The use of PLGA, a synthetic microfilament fiber was used to provide a basement membrane-like scaffold for self-patterned brain organoids termed microfilament-engineered cerebral organoids (enCORs) (Lancaster et al., 2017). This additional scaffolding improved radial organization and cytoarchitecture within enCORs, demonstrating the benefits that structural cues have on brain organoid development. Finally, the field of bioprinting or microcontact printing has allowed for tightly regulated patterning of cells at specified locations within a culture device, providing spatial control during differentiation (Knight et al., 2018, Haremaki et al., 2019, Fedorchak et al., 2020). This approach can also be combined with microfluidic devices to add additional levels of patterning control and mechanical flow cues to organoids. Adopting more biologically relevant scaffolding for brain organoid cultures is another step of many that will need to be taken to improve organoid generation and the quality of data that can be extracted from them.
4. Emerging techniques for the Unbiased Analysis of Organoids
4.1. Cellular and Molecular Analysis at Single Cell Resolution
Over the past decade there has been incredible momentum within the field of brain organoids that has led to the establishment and improvement of a wide variety of protocols for their generation (Figure 3). As brain organoids become more complex, it is imperative that the technology used to analyze them is appropriately matched in complexity. Classical techniques that provide transcriptional information, such as bulk RNA-seq and qPCR, have been used extensively to characterize organoids, but do not provide enough resolution to resolve the complexity of human brain development. As technology has progressed, we have achieved single-cell resolution at the functional and molecular level which has been used to characterize brain organoids. Single-cell RNA-seq (scRNA-seq) in particular has been used extensively to characterize brain organoids across protocols with the use of both droplet-based and multi-well sorting methods (Birey et al., 2017, Quadrato et al., 2017, Xiang et al., 2017, Madhavan et al., 2018, Cakir et al., 2019, Giandomenico et al., 2019, Kanton et al., 2019, Klaus et al., 2019, Marton et al., 2019, Pollen et al., 2019, Velasco et al., 2019, Xiang et al., 2019). These studies have demonstrated the cellular diversity in patterned and self-patterned brain organoid protocols (as Reviewed in Atamian et al., 2020). An in silico analysis has been performed to compare the various single cell datasets of differentiation protocols to the developing fetal brain (Tanaka et al., 2020). These sets of analyses showed a similarity in cellular composition across protocols; however, individual protocols displayed distinct differentiation trajectories and unique developmental gene networks were identified when comparing oranoids to fetal brains.
Figure 3. Protocols that have impacted the human brain organoid field over the past decade.
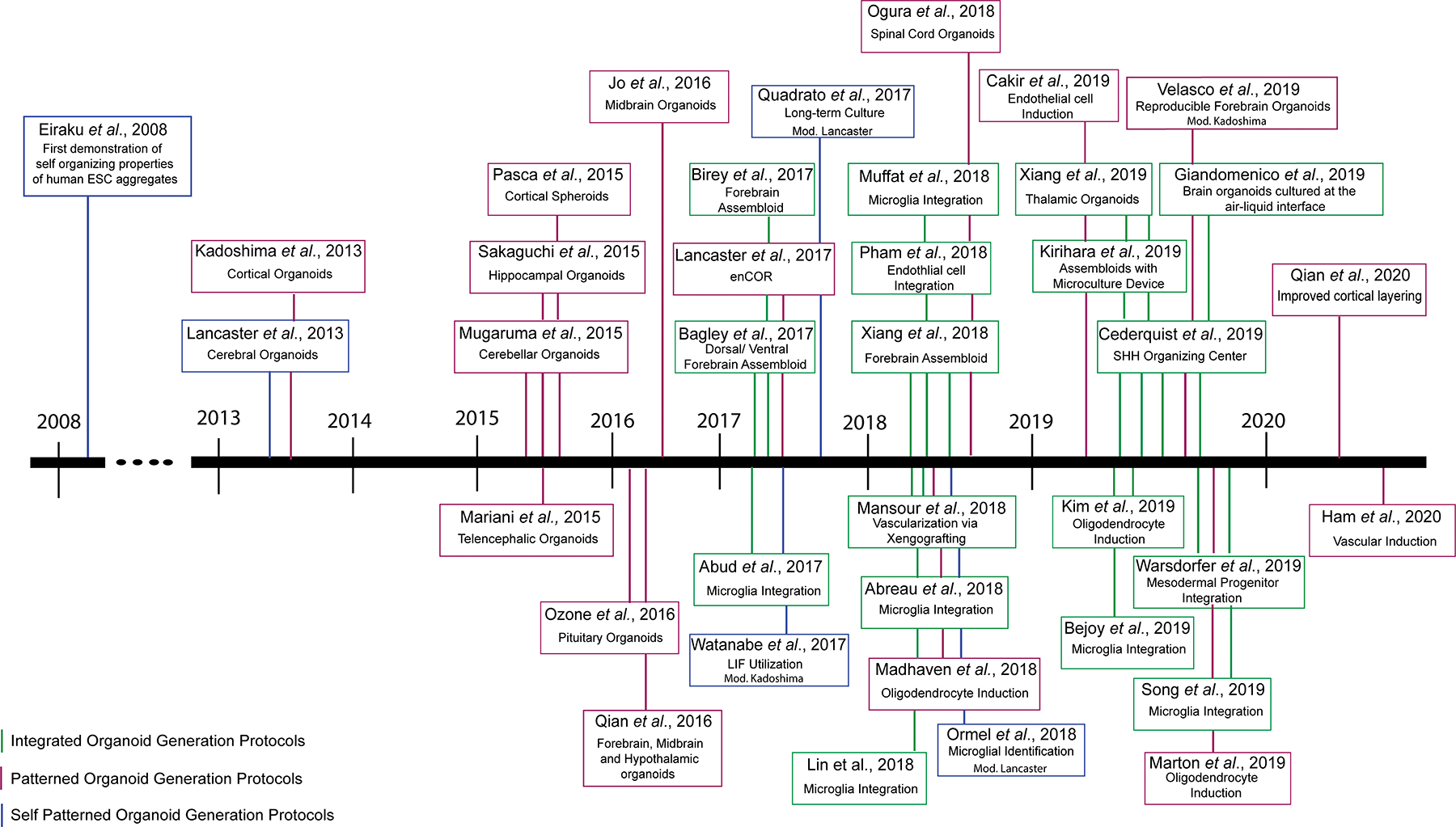
The use of brain organoids as a model system has gained incredible momentum over the past decade, leading to the establishment of diverse strategies to generate more complex organoids, and here we have classified as integrated (multiregional, multilineage, or including an artificial signaling center; green), patterned (magenta) or self-patterned (blue).
Several researchers have begun to probe the epigenetic landscape of cortical organoids using ATAC-seq (assay for transposase-accessible chromatin with high throughput sequencing). Trevino et al. (2020) found that after long term culture (20 months), organoids undergo dynamic chromatin state transitions that recapitulate primary brain tissue chromatin accessibility patterns suggesting organoids retain the intrinsic ability to progressively remodel their epigenetic landscape during protracted development. Single-cell ATAC-seq has also been used to identify human specific differences in chromatin accessibility between human and chimpanzee derived brain organoids (Kanton et al., 2019). This study found that a majority of the differentially accessible peaks between human and chimpanzee were specific to neural progenitor cells and neurons, suggesting a unique evolutionary divergence of these cells at the epigenetic level. Using single-nucleus ATAC-seq (snATAC-seq), comparisons of the cell type-specific chromatin landscape of cerebral organoids to primary developing cortex found organoids establish broad cell type-specific enhancer accessibility patterns similar to the developing cortex, but lack many putative regulatory elements identified in homologous primary cell types (Ziffra et al., 2019). Here, the authors utilize single-cell RNA-seq and snATAC-seq separately and perform a coembedding analysis to combine the datasets and make inferences on cell-type specific epigenetic status. These studies highlight the need for understanding the epigenetic similarities and limitations of brain organoids to facilitate improvement of culture conditions and future disease modelling.
Brain organoids give us for the first time the opportunity to investigate the emergence of cellular diversity and lineage relationships in human tissue. With scRNA-seq, several computational approaches have been developed to infer lineage relationships based on transcriptomic high-dimensional data. A detailed comparison of these methods and guidelines for users was recently provided by Saelens et al. (2019). In brain organoids, several papers have implemented different iterations of the Monocle package to construct pseudotime trajectories (Camp et al., 2015, Marton et al., 2019, Velasco et al., 2019, Ziffra et al., 2019, Sloan et al., 2017). As these computational approaches rely on making inferences, combining single cell sequencing with lineage tracing systems would improve the robustness of future cell lineage findings and allow for direct tracing of neuronal ontogeny within organoids. Broadly speaking, lineage tracing techniques can be categorized by their tagging system. Currently cell ontogeny can be traced based on somatic mutations, reporter systems, cell barcoding, and dynamic lineage tracing (Frieda et al., 2017, Yao et al., 2017, Alemany et al., 2018, Raj et al., 2018, Spanjaard et al., 2018, McKenna and Gagnon, 2019). A detailed description of the advantages and considerations of emerging lineage tracing approaches that can be combined with scRNA-seq are reviewed by Wagner and Klein (2020). Recently, scRNA-seq has been combined with an inducible CRISPR/Cas9-based barcoding system to reconstruct dynamic lineage trees within microdissected self-patterned organoids for the first time (He et al., 2020). This elegant approach, called iTracer, integrates the sleeping beauty transposon system (Ivics et al., 1997) with an inducible iCRISPR system (Gonzalez et al., 2014). Using this approach, progenitor-neuron lineage relationships were reconstructed revealing a clonal accumulation bias of iPSCs towards distinct brain regions. We envision that similar approaches will emerge and become commonplace for the analysis of patterned organoids in healthy and disease states.
Similarly, just as lineage tracing can be leveraged to determine neuronal ontogeny, spatial transcriptomics will allow for the probing of cellular spatial relationships during human brain organoid development. Several techniques that incorporate fluorescence in situ hybridization have emerged to identify and spatially resolve specific transcripts at the single cell level (Chen et al., 2015, Codeluppi et al., 2018, Salmen et al., 2018, Wang et al., 2018, Eng et al., 2019). Recently, the 10X Visium platform was used to perform spatial transcriptomics of iTracer (barcoded and scarred) cells within cerebral organoids (He et al., 2020). This analysis confirmed previously discussed findings of iPSC clonal bias in distinct brain organoid regions and marks the first use of spatial transcriptomics in the brain organoid field. The future incorporation of spatial registration combined with transcriptomic and epigenomic techniques will be critical in the analysis of brain organoids as they do not reproducibly recapitulate in vivo stereotypic anatomy.
To date large-scale, unbiased proteomic analysis has not been performed on brain organoids at single cell resolution. With the use of proteomics, researchers will no longer have to rely upon mRNA to determine expression of cell identifying marker genes. Strategies to profile the proteome of individual cells are starting to emerge (Doerr, 2019) and will provide additional cellular readouts in healthy and diseased organoids. The addition of emerging proteomics with current unbiased -omic modalities to brain organoid analysis will allow for the molecular, phenotypic and functional characterization of single cells. Though there are limitations to any single technique, the characterization of organoids will undoubtedly utilize complementary methods to compensate for their individual shortcomings (Figure 4). As sequencing technologies have advanced, several groups have been able to simultaneously perform multiplexing or multiple-omic assays at the single cell level. This represents a powerful new era in single cell technology wherein additional layers of information can be measured in a single experiment. This is of importance for brain organoids as the fidelity of in vitro cells types to endogenous tissue has been questioned at the transcriptomic, epigenetic, and spatial level. Thus, performing multiple measurements from a single cell will provide an accurate portrayal of molecular underpinnings in organoids.
Figure 4. Upgrading the Physiological Relevance of Human Brain Organoids.
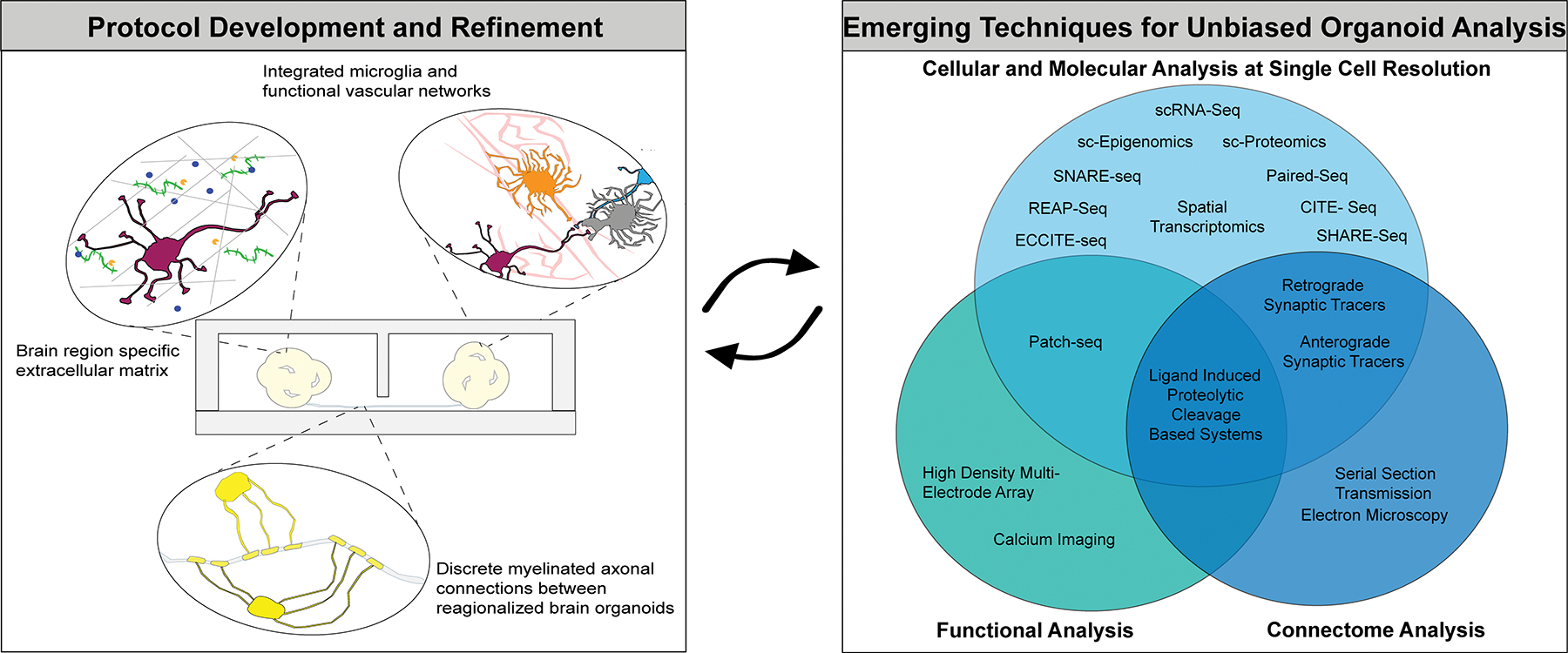
Multidisciplinary integration of emerging technologies to improve the generation (Panel 1) and characterization (Panel 2) of human brain organoids.
4.2. Functional Assessment of Neural Activity in Brain Organoids
Refinement of culture conditions has allowed for the protracted development of organoids and opened the door to understanding the functional physiological properties of neurons within organoids. Dendritic spines and structurally defined synaptic connections have been identified in both patterned and self-patterned protocols suggesting that 3D cultures favor the development of structural features characteristic of mature neurons. Given the presence of these mature neuronal populations, it is unsurprising that brain organoids can develop spontaneous neural network activity as evidenced by calcium fluxes as well as intracellular and extracellular recordings (Mariani et al., 2015, Muguruma et al., 2015, Paşca et al., 2015, Sakaguchi et al., 2015, Jo et al., 2016, Qian et al., 2016, Birey et al., 2017, Quadrato et al., 2017, Cakir et al., 2019, Marton et al., 2019, Song et al., 2019, Qian et al., 2020). Whole-cell patch-clamp, voltage clamp, and current clamp have been utilized extensively to characterize neurons in organoids, typically in sliced cultures or organoid edges where functional connectivity between neurons has been detected (Mariani et al., 2015, Muguruma et al., 2015, Paşca et al., 2015, Sakaguchi et al., 2015, Qian et al., 2016, Birey et al., 2017, Xiang et al., 2017, Marton et al., 2019, Smits et al., 2019, Song et al., 2019, Xiang et al., 2019, Qian et al., 2020). Though intracellular patch-clamp provides important information on the electrical properties and functional connectivity of neurons, it is not a high throughput approach, which is necessary to comprehensively characterize the complex neuronal network behaviour for the large number of neuronal subtypes present in organoids. The standard microelectrode arrays (MEAs) approach for extracellular recordings of in vitro assays uses planar electrodes arranged in grids. While these 2D MEAs are useful for detecting spikes in mono-layers and have been used extensively in the organoid field, they are limited to capturing the activity of neurons in the outer layers of the organoid (Giandomenico et al., 2019, Schmunk et al., 2020). High density silicon microelectrodes are an emerging technology that allows for insertion of probes into 3D tissue and can provide single cell resolution by leveraging the simultaneous recordings of each ‘spike’, across multiple spatially distributed channels, to improve the clustering of ‘spikes’ that originate from the same neuron. Simultaneously, it is also possible to record network burst activity indicated by recordings of neurons in multiple probes that fire coordinated bursts of action potentials in a temporal manner. In Quadrato et al (2017) this technology has been successfully used to characterize network behaviour in 8 months old self-patterning brain organoids. However, this technology is difficult to implement for high throughput experiments that require simultaneous recording of multiple organoids. Several studies have used calcium indicators to record transient calcium waves that arise during depolarization (Lancaster et al., 2013, Sakaguchi et al., 2015, Birey et al., 2017, Lancaster et al., 2017, Xiang et al., 2017, Park et al., 2018, Kim et al., 2019, Xiang et al., 2019, Samarasinghe et al., 2019). Calcium imaging is more amenable to large scale studies as compared to MEA technology and represents a powerful tool for the extraction of temporal information on neuronal firing patterns. However, currently there is no effective method to monitor the activity of a large number of neurons over long periods of time in organoids although this has been accomplished in vivo. Typically, imaging is performed at the organoid surface, yet with two-photon and light-sheet microscopy it is possible to acquire data from deeper organoid layers and eventually perform 4D imaging (Chen et al., 2014, Schoneberg et al., 2018, Lavagnino et al., 2016). Lineage tracking of early formed embryoid bodies (EBs) was recently performed using 4D light-sheet microscopy to visualize how single nuclei give rise to spatially restricted daughter cells, which only migrate short distances from parent cells (He et al., 2020). The development of miniscopes to perform long-term calcium imaging in freely moving rodents, suggest the exciting possibility that similar approaches could be adapted to perform much needed long-term recordings of neuronal activity in organoids (Gonzalez et al., 2019). The major drawback compared to other techniques result from the decaying dynamics of the calcium indicators and resolving signal over noise, which becomes increasingly difficult to distinguish at single-cell resolution as neuronal firing rates increase. A powerful yet low-throughput technique that enables correlation of transcriptomic and functional data at the single cell level to simultaneously combine functional and molecular profiling of neurons is Patch-seq (Cadwell et al., 2016, van den Hurk et al., 2018), which combines whole-cell patch-clamp recordings with scRNA-seq and morphological characterization. This recently developed method, in which a single neuron can be targeted by a microelectrode, recorded for its electrical function and thereafter aspired and prepared for sequencing, is critical for investigating how electrophysiological properties in neurons correlate to cell subtype specific transcriptomes. Application of this powerful technique to the analysis of brain organoids will provide the field with much needed understanding of the molecular determinants of human neuronal diversity and it will facilitate and improve the classification of neural cell types across different stages of neural development in a human cellular context. This knowledge will also be pivotal for the cell-type specific characterization of disease states.
4.3. Tracing Neural Connections in Brain Organoids
The wiring diagram of distinct or fused organoids has not been traced so far. Here we review established and emerging techniques that could be used to elucidate connectivity in this complex system. The traditional gold standard of electron microscopy provides a high-resolution view of cellular structural features including the connection points between neurons, synapses. Additionally, the use of scanning electron microscopy (SEM) on ultra-thin brain sections has allowed large volumes of brain tissue (several cubic millimeters) to be reconstructed (Witvliet et al., 2020, Hildebrand et al., 2017), and has been used for 3D reconstructions of synapses and dendritic spines in brain organoids (Quadrato et al., 2017). However, this technique is not well suited for high-throughput studies or to trace multiple long-range connections. Additionally, EM-based techniques such as SEM are performed on fixed tissue, limiting their use in a multimodal analysis with functional assays and genetic manipulations.
Viral-based retrograde (rabies, pseudorabies) and anterograde (HSV-1 strain H129) trans-synaptic tracers have been extensively used to elucidate connectivity in mammalian model systems (Dix et al., 1983, Strack et al., 1989, Zemanick et al., 1991, Ugolini, 1995, Aston-Jones and Card, 2000). Though initially developed using wild-type replication-competent viruses, these tracers have been genetically modified to improve their neurotropism, specificity, and safety. Though neurotropic viral tracers have been extensively used in neurobiology, recent evidence suggests that different retrograde viral strains have biased neurotropism and altered degrees of neurotoxicity (Sun et al., 2019). It is currently unclear how widespread this bias in tropism is among neurotropic viruses. As viral tracers result in neurotoxicity that increases with exposure time, the technique is difficult to implement with additional functional analyses. Currently efforts to reduce viral-mediated cytotoxicity are being made which can potentially allow the technique to be used in conjunction with other downstream analysis (Chatterjee et al., 2018).
Another set of tracing techniques being developed are the highly amenable ligand-induced intramembrane proteolytic cleavage-based systems (Huang et al., 2016, Morsut et al., 2016, Inagaki et al., 2012, Jagadish et al., 2014, Huang et al., 2017). These tracing approaches utilize ‘sender’ cells expressing an artificial ligand to bind and activate ‘receiver’ cells expressing an artificial receptor to perform trans-synaptic tracing. A detailed description of this mechanism and the various methods of implementation for synaptic tracing are reviewed in (Lee et al., 2017). To date, studies using this system have primarily been done in 2D cultures and model systems such as Drosophila. With additional engineering in hPSCs, these systems allow the capability of ligand-receptor induced trans-cellular expression of a transgene allowing for easily combining tracing with additional analyses. One potential caveat to using a ligand-induced intramembrane proteolytic cleavage-based system is the considerable amount of optimization required to ensure there is minimal leakiness of expression/activation of the receiver cells. The background signaling must be kept minimal to ensure the error of making a false-positive is kept low. Both viral-based and synthetic engineering-based tracing systems have the potential to be combined to assess circuit connectivity in organoids as well as in conjunction with spatial transcriptomics. This combination will allow for interrogation of dynamic cell-cell interactions with single cell resolution.
5. Concluding Remarks
Human brain organoids have revolutionized the field of developmental neurobiology by providing a tractable system to interrogate human specific aspects of neurodevelopmental processes in normal and diseased states. While the use of inhibitors of the WNT and SMAD pathways has improved cortical organoid-to-organoid reproducibility, their use might be problematic. Core components or modulators of WNT and SMAD signaling are causative of neurodevelopmental disorders (NDDs) (Kumar et al., 2019) and their inhibition by patterning molecules may affect disease phenotypes and drug screening. In addition, external patterning might negatively affect the survival and differentiation of other cell types, including microglial cells and endothelial cells that have recently been successfully integrated into brain organoids. Thus, alternative strategies to increase brain organoid reproducibility that do not interfere with key signaling pathways must be established for organoids to accurately model disease. The confluence of other disciplines including, tissue engineering and synthetic biology onto brain organoid generation will be required to achieve reproducibility in the absence of patterning, and surmount the obstacles surrounding the ability of organoids to achieve more mature cellular states. Tissue engineering approaches will be key to establish a controlled microenvironment and for engineering reproducible anatomical features. In addition, instructing organoid maturation through synthetic control of gene regulatory networks that govern cellular maturation across multiple lineages is an interesting concept for brain organoids and has already been successfully implemented in liver organoids (Velazquez et al., 2020). Improving brain organoids may also depend on biophysical mechanisms as kidney organoids showed improved vascularization and maturation when subjected to the mechanical shear stress forces within a microfluidics device that mimics fluid flow during in vivo development (Homan et al., 2019). Pursuing this logic, culturing ‘integrated’ organoids that contain vasculature-like structures within a fluidic device may aid in the maturation of the vascular network and neural populations. Recently, extracellular recordings of primary neuronal cultures have been performed as they differentiate and develop within a microfluidic device (Lam et al., 2019), thus demonstrating how the integration of tissue engineering approaches could be exploited to perform much needed functional longitudinal analyses of the organoids as they develop. As organoid generation protocols continue to be improved and refined, it is important that the technology used to analyze them is equipped to extract finely resolved data that encompasses the full potential and limitations of the system. Finally, we envision that the multidisciplinary integration of the emergent technologies highlighted in this review will inform the further improvement of organoid generation protocols in a continuous cycle of growth that will eventually result in “upgraded” brain organoids capable of more accurately modeling complex developmental processes.
Acknowledgments
We thank members of the Quadrato lab for insightful discussions and Alexander Atamian for careful reading and editing of the manuscript. This work was supported by a grant from the Baxter foundation (To G.Q.) and a Stem Cell T32 Training Fellowship (To J.P.U.). We apologize to colleagues whose work we could not cite due to space limitations.
References
- ABREU CM, GAMA L, KRASEMANN S, CHESNUT M, ODWIN-DACOSTA S, HOGBERG HT, HARTUNG T & PAMIES D 2018. Microglia Increase Inflammatory Responses in iPSC-Derived Human BrainSpheres. Front Microbiol, 9, 2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ABUD EM, RAMIREZ RN, MARTINEZ ES, HEALY LM, NGUYEN CHH, NEWMAN SA, YEROMIN AV, SCARFONE VM, MARSH SE, FIMBRES C, CARAWAY CA, FOTE GM, MADANY AM, AGRAWAL A, KAYED R, GYLYS KH, CAHALAN MD, CUMMINGS BJ, ANTEL JP, MORTAZAVI A, CARSON MJ, POON WW & BLURTON-JONES M 2017. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron, 94, 278–293 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALEMANY A, FLORESCU M, BARON CS, PETERSON-MADURO J & VAN OUDENAARDEN A 2018. Whole-organism clone tracing using single-cell sequencing. Nature, 556, 108–112. [DOI] [PubMed] [Google Scholar]
- ASTON-JONES G & CARD JP 2000. Use of pseudorabies virus to delineate multisynaptic circuits in brain: opportunities and limitations. J Neurosci Methods, 103, 51–61. [DOI] [PubMed] [Google Scholar]
- ATAMIAN A, CORDÓN-BARRIS L & QUADRATO G 2020. Taming human brain organoids one cell at a time. Seminars in Cell & Developmental Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAGLEY JA, REUMANN D, BIAN S, LEVI-STRAUSS J & KNOBLICH JA 2017. Fused cerebral organoids model interactions between brain regions. Nat Methods, 14, 743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARRY C, SCHMITZ MT, PROPSON NE, HOU Z, ZHANG J, NGUYEN BK, BOLIN JM, JIANG P, MCINTOSH BE, PROBASCO MD, SWANSON S, STEWART R, THOMSON JA, SCHWARTZ MP & MURPHY WL 2017. Uniform neural tissue models produced on synthetic hydrogels using standard culture techniques. Exp Biol Med (Maywood), 242, 1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEJOY J, YUAN X, SONG L, HUA T, JESKE R, SART S, SANG Q-XA & LI Y 2019. Genomics Analysis of Metabolic Pathways of Human Stem Cell-Derived Microglia-Like Cells and the Integrated Cortical Spheroids. Stem Cells International, 2019, 2382534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEROUN A, MITRA S, MICHALUK P, PIJET B, STEFANIUK M & KACZMAREK L 2019. MMPs in learning and memory and neuropsychiatric disorders. Cell Mol Life Sci, 76, 3207–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BHADURI A, ANDREWS MG, MANCIA LEON W, JUNG D, SHIN D, ALLEN D, JUNG D, SCHMUNK G, HAEUSSLER M, SALMA J, POLLEN AA, NOWAKOWSKI TJ & KRIEGSTEIN AR 2020. Cell stress in cortical organoids impairs molecular subtype specification. Nature, 578, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIREY F, ANDERSEN J, MAKINSON CD, ISLAM S, WEI W, HUBER N, FAN HC, METZLER KRC, PANAGIOTAKOS G, THOM N, O’ROURKE NA, STEINMETZ LM, BERNSTEIN JA, HALLMAYER J, HUGUENARD JR & PASCA SP 2017. Assembly of functionally integrated human forebrain spheroids. Nature, 545, 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOCK C, KISKINIS E, VERSTAPPEN G, GU H, BOULTING G, SMITH ZD, ZILLER M, CROFT GF, AMOROSO MW, OAKLEY DH, GNIRKE A, EGGAN K & MEISSNER A 2011. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell, 144, 439–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CADWELL CR, PALASANTZA A, JIANG X, BERENS P, DENG Q, YILMAZ M, REIMER J, SHEN S, BETHGE M, TOLIAS KF, SANDBERG R & TOLIAS AS 2016. Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat Biotechnol, 34, 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAKIR B, XIANG Y, TANAKA Y, KURAL MH, PARENT M, KANG YJ, CHAPETON K, PATTERSON B, YUAN Y, HE CS, RAREDON MSB, DENGELEGI J, KIM KY, SUN P, ZHONG M, LEE S, PATRA P, HYDER F, NIKLASON LE, LEE SH, YOON YS & PARK IH 2019. Engineering of human brain organoids with a functional vascular-like system. Nat Methods, 16, 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAMP JG, BADSHA F, FLORIO M, KANTON S, GERBER T, WILSCH-BRÄUNINGER M, LEWITUS E, SYKES A, HEVERS W, LANCASTER M, KNOBLICH JA, LACHMANN R, PÄÄBO S, HUTTNER WB & TREUTLEIN B 2015. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proceedings of the National Academy of Sciences, 112, 15672–15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CEDERQUIST GY, ASCIOLLA JJ, TCHIEU J, WALSH RM, CORNACCHIA D, RESH MD & STUDER L 2019. Specification of positional identity in forebrain organoids. Nat Biotechnol, 37, 436–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAMBERS SM, FASANO CA, PAPAPETROU EP, TOMISHIMA M, SADELAIN M & STUDER L 2009. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol, 27, 275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHATTERJEE S, SULLIVAN HA, MACLENNAN BJ, XU R, HOU Y, LAVIN TK, LEA NE, MICHALSKI JE, BABCOCK KR, DIETRICH S, MATTHEWS GA, BEYELER A, CALHOON GG, GLOBER G, WHITESELL JD, YAO S, CETIN A, HARRIS JA, ZENG H, TYE KM, REID RC & WICKERSHAM IR 2018. Nontoxic, double-deletion-mutant rabies viral vectors for retrograde targeting of projection neurons. Nat Neurosci, 21, 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN B-C, LEGANT WR, WANG K, SHAO L, MILKIE DE, DAVIDSON MW, JANETOPOULOS C, WU XS, HAMMER JA, LIU Z, ENGLISH BP, MIMORI-KIYOSUE Y, ROMERO DP, RITTER AT, LIPPINCOTT-SCHWARTZ J, FRITZ-LAYLIN L, MULLINS RD, MITCHELL DM, BEMBENEK JN, REYMANN A-C, BÖHME R, GRILL SW, WANG JT, SEYDOUX G, TULU US, KIEHART DP & BETZIG E 2014. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science, 346, 1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN KH, BOETTIGER AN, MOFFITT JR, WANG S & ZHUANG X 2015. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science, 348, aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHO AN, JIN Y, KIM S, KUMAR S, SHIN H, KANG HC & CHO SW 2019. Aligned Brain Extracellular Matrix Promotes Differentiation and Myelination of Human-Induced Pluripotent Stem Cell-Derived Oligodendrocytes. ACS Appl Mater Interfaces, 11, 15344–15353. [DOI] [PubMed] [Google Scholar]
- CODELUPPI S, BORM LE, ZEISEL A, LA MANNO G, VAN LUNTEREN JA, SVENSSON CI & LINNARSSON S 2018. Spatial organization of the somatosensory cortex revealed by osmFISH. Nat Methods, 15, 932–935. [DOI] [PubMed] [Google Scholar]
- CUNNINGHAM CL, MARTÍNEZ-CERDEÑO V & NOCTOR SC 2013. Microglia Regulate the Number of Neural Precursor Cells in the Developing Cerebral Cortex. The Journal of Neuroscience, 33, 4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DI LULLO E & KRIEGSTEIN AR 2017. The use of brain organoids to investigate neural development and disease. Nature Reviews Neuroscience, 18, 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIX RD, MCKENDALL RR & BARINGER JR 1983. Comparative neurovirulence of herpes simplex virus type 1 strains after peripheral or intracerebral inoculation of BALB/c mice. Infection and immunity, 40, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOERR A 2019. Single-cell proteomics. Nature Methods, 16, 20–20. [DOI] [PubMed] [Google Scholar]
- DOUVARAS P, SUN B, WANG M, KRUGLIKOV I, LALLOS G, ZIMMER M, TERRENOIRE C, ZHANG B, GANDY S, SCHADT E, FREYTES DO, NOGGLE S & FOSSATI V 2017. Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Reports, 8, 1516–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EIRAKU M, TAKATA N, ISHIBASHI H, KAWADA M, SAKAKURA E, OKUDA S, SEKIGUCHI K, ADACHI T & SASAI Y 2011. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature, 472, 51–6. [DOI] [PubMed] [Google Scholar]
- EIRAKU M, WATANABE K, MATSUO-TAKASAKI M, KAWADA M, YONEMURA S, MATSUMURA M, WATAYA T, NISHIYAMA A, MUGURUMA K & SASAI Y 2008. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell, 3, 519–32. [DOI] [PubMed] [Google Scholar]
- ENG CL, LAWSON M, ZHU Q, DRIES R, KOULENA N, TAKEI Y, YUN J, CRONIN C, KARP C, YUAN GC & CAI L 2019. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature, 568, 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESPUNY-CAMACHO I, MICHELSEN KIMMOA, GALL D, LINARO D, HASCHE A, BONNEFONT J, BALI C, ORDUZ D, BILHEU A, HERPOEL A, LAMBERT N, GASPARD N, PÉRON S, SCHIFFMANN SERGEN, GIUGLIANO M, GAILLARD A & VANDERHAEGHEN P 2013. Pyramidal Neurons Derived from Human Pluripotent Stem Cells Integrate Efficiently into Mouse Brain Circuits In Vivo. Neuron, 77, 440–456. [DOI] [PubMed] [Google Scholar]
- FEDORCHAK NJ, IYER N & ASHTON RS 2020. Bioengineering tissue morphogenesis and function in human neural organoids. Semin Cell Dev Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRIEDA KL, LINTON JM, HORMOZ S, CHOI J, CHOW KK, SINGER ZS, BUDDE MW, ELOWITZ MB & CAI L 2017. Synthetic recording and in situ readout of lineage information in single cells. Nature, 541, 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GALARZA S, CROSBY AJ, PAK C & PEYTON SR 2020. Control of Astrocyte Quiescence and Activation in a Synthetic Brain Hydrogel. Adv Healthc Mater, 9, e1901419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIANDOMENICO SL, MIERAU SB, GIBBONS GM, WENGER LMD, MASULLO L, SIT T, SUTCLIFFE M, BOULANGER J, TRIPODI M, DERIVERY E, PAULSEN O, LAKATOS A & LANCASTER MA 2019. Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat Neurosci, 22, 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GONZALEZ F, ZHU Z, SHI ZD, LELLI K, VERMA N, LI QV & HUANGFU D 2014. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell, 15, 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GONZALEZ WG, ZHANG H, HARUTYUNYAN A & LOIS C 2019. Persistence of neuronal representations through time and damage in the hippocampus. Science, 365, 821–825. [DOI] [PubMed] [Google Scholar]
- HAENSELER W, SANSOM SN, BUCHRIESER J, NEWEY SE, MOORE CS, NICHOLLS FJ, CHINTAWAR S, SCHNELL C, ANTEL JP, ALLEN ND, CADER MZ, WADE-MARTINS R, JAMES WS & COWLEY SA 2017. A Highly Efficient Human Pluripotent Stem Cell Microglia Model Displays a Neuronal-Co-culture-Specific Expression Profile and Inflammatory Response. Stem Cell Reports, 8, 1727–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAM O, JIN YB, KIM J & LEE MO 2020. Blood vessel formation in cerebral organoids formed from human embryonic stem cells. Biochem Biophys Res Commun, 521, 84–90. [DOI] [PubMed] [Google Scholar]
- HAREMAKI T, METZGER JJ, RITO T, OZAIR MZ, ETOC F & BRIVANLOU AH 2019. Self-organizing neuruloids model developmental aspects of Huntington’s disease in the ectodermal compartment. Nature Biotechnology, 37, 1198–1208. [DOI] [PubMed] [Google Scholar]
- HARRIS JM, WANG AY, BOULANGER-WEILL J, SANTORIELLO C, FOIANINI S, LICHTMAN JW, ZON LI & ARLOTTA P 2020. Long-Range Optogenetic Control of Axon Guidance Overcomes Developmental Boundaries and Defects. Dev Cell, 53, 577–588 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HE Z, GERBER T, MAYNARD A, JAIN A, PETRI R, SANTEL M, LY K, SIDOW L, CALLEJAL FS, RIESENBERG S, CAMP JG & TREUTLEIN B 2020. Lineage recording reveals dynamics of cerebral organoid regionalization. BioRxiv. [Google Scholar]
- HILDEBRAND DGC, CICCONET M, TORRES RM, CHOI W, QUAN TM, MOON J, WETZEL AW, SCOTT CHAMPION A, GRAHAM BJ, RANDLETT O, PLUMMER GS, PORTUGUES R, BIANCO IH, SAALFELD S, BADEN AD, LILLANEY K, BURNS R, VOGELSTEIN JT, SCHIER AF, LEE W-CA, JEONG W-K, LICHTMAN JW & ENGERT F 2017. Whole-brain serial-section electron microscopy in larval zebrafish. Nature, 545, 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOMAN KA, GUPTA N, KROLL KT, KOLESKY DB, SKYLAR-SCOTT M, MIYOSHI T, MAU D, VALERIUS MT, FERRANTE T, BONVENTRE JV, LEWIS JA & MORIZANE R 2019. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat Methods, 16, 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG T-H, VELHO T & LOIS C 2016. Monitoring cell-cell contacts <em>in vivo</em> in transgenic animals. Development, 143, 4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG TH, NIESMAN P, ARASU D, LEE D, DE LA CRUZ AL, CALLEJAS A, HONG EJ & LOIS C 2017. Tracing neuronal circuits in transgenic animals by transneuronal control of transcription (TRACT). Elife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INAGAKI HIDEHIKOK, BEN-TABOU DE-LEON S, WONG AM, JAGADISH S, ISHIMOTO H, BARNEA G, KITAMOTO T, AXEL R & ANDERSON DAVIDJ 2012. Visualizing Neuromodulation In Vivo: TANGO-Mapping of Dopamine Signaling Reveals Appetite Control of Sugar Sensing. Cell, 148, 1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IVICS Z, HACKETT PB, PLASTERK RH & IZSVÁK Z 1997. Molecular Reconstruction of Sleeping Beauty, a Tc1-like Transposon from Fish, and Its Transposition in Human Cells. Cell, 91, 501–510. [DOI] [PubMed] [Google Scholar]
- JAGADISH S, BARNEA G, CLANDININ TR & AXEL R 2014. Identifying functional connections of the inner photoreceptors in Drosophila using Tango-Trace. Neuron, 83, 630–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIN Y, LEE JS, KIM J, MIN S, WI S, YU JH, CHANG GE, CHO AN, CHOI Y, AHN DH, CHO SR, CHEONG E, KIM YG, KIM HP, KIM Y, KIM DS, KIM HW, QUAN Z, KANG HC & CHO SW 2018. Three-dimensional brain-like microenvironments facilitate the direct reprogramming of fibroblasts into therapeutic neurons. Nat Biomed Eng, 2, 522–539. [DOI] [PubMed] [Google Scholar]
- JO J, XIAO Y, SUN AX, CUKUROGLU E, TRAN HD, GOKE J, TAN ZY, SAW TY, TAN CP, LOKMAN H, LEE Y, KIM D, KO HS, KIM SO, PARK JH, CHO NJ, HYDE TM, KLEINMAN JE, SHIN JH, WEINBERGER DR, TAN EK, JE HS & NG HH 2016. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell, 19, 248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KADOSHIMA T, SAKAGUCHI H, NAKANO T, SOEN M, ANDO S, EIRAKU M & SASAI Y 2013. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell–derived neocortex. Proceedings of the National Academy of Sciences, 110, 20284–20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KANTON S, BOYLE MJ, HE Z, SANTEL M, WEIGERT A, SANCHIS-CALLEJA F, GUIJARRO P, SIDOW L, FLECK JS, HAN D, QIAN Z, HEIDE M, HUTTNER WB, KHAITOVICH P, PAABO S, TREUTLEIN B & CAMP JG 2019. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature, 574, 418–422. [DOI] [PubMed] [Google Scholar]
- KELAVA I & LANCASTER MA 2016. Dishing out mini-brains: Current progress and future prospects in brain organoid research. Dev Biol, 420, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIERDORF K, ERNY D, GOLDMANN T, SANDER V, SCHULZ C, PERDIGUERO EG, WIEGHOFER P, HEINRICH A, RIEMKE P, HOLSCHER C, MULLER DN, LUCKOW B, BROCKER T, DEBOWSKI K, FRITZ G, OPDENAKKER G, DIEFENBACH A, BIBER K, HEIKENWALDER M, GEISSMANN F, ROSENBAUER F & PRINZ M 2013. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci, 16, 273–80. [DOI] [PubMed] [Google Scholar]
- KIM H, XU R, PADMASHRI R, DUNAEVSKY A, LIU Y, DREYFUS CF & JIANG P 2019. Pluripotent Stem Cell-Derived Cerebral Organoids Reveal Human Oligodendrogenesis with Dorsal and Ventral Origins. Stem Cell Reports, 12, 890–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIRIHARA T, LUO Z, CHOW SYA, MISAWA R, KAWADA J, SHIBATA S, KHOYRATEE F, VOLLETTE CA, VOLZ V, LEVI T, FUJII T & IKEUCHI Y 2019. A Human Induced Pluripotent Stem Cell-Derived Tissue Model of a Cerebral Tract Connecting Two Cortical Regions. iScience, 14, 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLAUS J, KANTON S, KYROUSI C, AYO-MARTIN AC, DI GIAIMO R, RIESENBERG S, O’NEILL AC, CAMP JG, TOCCO C, SANTEL M, RUSHA E, DRUKKER M, SCHROEDER M, GOTZ M, ROBERTSON SP, TREUTLEIN B & CAPPELLO S 2019. Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia. Nat Med, 25, 561–568. [DOI] [PubMed] [Google Scholar]
- KNIGHT GT, LUNDIN BF, IYER N, ASHTON LMT, SETHARES WA, WILLETT RM & ASHTON RS 2018. Engineering induction of singular neural rosette emergence within hPSC-derived tissues. eLife, 7, e37549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KRISHNASWAMY VR, BENBENISHTY A, BLINDER P & SAGI I 2019. Demystifying the extracellular matrix and its proteolytic remodeling in the brain: structural and functional insights. Cell Mol Life Sci, 76, 3229–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUMAR S, REYNOLDS K, JI Y, GU R, RAI S & ZHOU CJ 2019. Impaired neurodevelopmental pathways in autism spectrum disorder: a review of signaling mechanisms and crosstalk. J Neurodev Disord, 11, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAM D, ENRIGHT HA, CADENA J, PETERS SKG, SALES AP, OSBURN JJ, SOSCIA DA, KULP KS, WHEELER EK & FISCHER NO 2019. Tissue-specific extracellular matrix accelerates the formation of neural networks and communities in a neuron-glia co-culture on a multi-electrode array. Sci Rep, 9, 4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANCASTER MA, CORSINI NS, WOLFINGER S, GUSTAFSON EH, PHILLIPS AW, BURKARD TR, OTANI T, LIVESEY FJ & KNOBLICH JA 2017. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol, 35, 659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANCASTER MA, RENNER M, MARTIN C-A, WENZEL D, BICKNELL LS, HURLES ME, HOMFRAY T, PENNINGER JM, JACKSON AP & KNOBLICH JA 2013. Cerebral organoids model human brain development and microcephaly. Nature, 501, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAVAGNINO Z, SANCATALDO G, D’AMORA M, FOLLERT P, DE PIETRI TONELLI D, DIASPRO A & CELLA ZANACCHI F 2016. 4D (x-y-z-t) imaging of thick biological samples by means of Two-Photon inverted Selective Plane Illumination Microscopy (2PE-iSPIM). Sci Rep, 6, 23923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE D, HUANG TH, DE LA CRUZ A, CALLEJAS A & LOIS C 2017. Methods to investigate the structure and connectivity of the nervous system. Fly (Austin), 11, 224–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENZ KM & NELSON LH 2018. Microglia and Beyond: Innate Immune Cells As Regulators of Brain Development and Behavioral Function. Front Immunol, 9, 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIN YT, SEO J, GAO F, FELDMAN HM, WEN HL, PENNEY J, CAM HP, GJONESKA E, RAJA WK, CHENG J, RUEDA R, KRITSKIY O, ABDURROB F, PENG Z, MILO B, YU CJ, ELMSAOURI S, DEY D, KO T, YANKNER BA & TSAI LH 2018. APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer’s Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron, 98, 1141–1154 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MADHAVAN M, NEVIN ZS, SHICK HE, GARRISON E, CLARKSON-PAREDES C, KARL M, CLAYTON BLL, FACTOR DC, ALLAN KC, BARBAR L, JAIN T, DOUVARAS P, FOSSATI V, MILLER RH & TESAR PJ 2018. Induction of myelinating oligodendrocytes in human cortical spheroids. Nat Methods, 15, 700–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANSOUR AA, GONCALVES JT, BLOYD CW, LI H, FERNANDES S, QUANG D, JOHNSTON S, PARYLAK SL, JIN X & GAGE FH 2018. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol, 36, 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARIANI J, COPPOLA G, ZHANG P, ABYZOV A, PROVINI L, TOMASINI L, AMENDUNI M, SZEKELY A, PALEJEV D, WILSON M, GERSTEIN M, GRIGORENKO EL, CHAWARSKA K, PELPHREY KA, HOWE JR & VACCARINO FM 2015. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell, 162, 375–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARIN-PADILLA M 2012. The human brain intracerebral microvascular system: development and structure. Front Neuroanat, 6, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTON RM, MIURA Y, SLOAN SA, LI Q, REVAH O, LEVY RJ, HUGUENARD JR & PASCA SP 2019. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat Neurosci, 22, 484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCKENNA A & GAGNON JA 2019. Recording development with single cell dynamic lineage tracing. Development, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCQUADE A, COBURN M, TU CH, HASSELMANN J, DAVTYAN H & BLURTON-JONES M 2018. Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol Neurodegener, 13, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MONIER A, ADLE-BIASSETTE H, DELEZOIDE A-L, EVRARD P, GRESSENS P & VERNEY C 2007. Entry and Distribution of Microglial Cells in Human Embryonic and Fetal Cerebral Cortex. Journal of Neuropathology and Experimental Neurology, 66, 372–382. [DOI] [PubMed] [Google Scholar]
- MONZEL AS, SMITS LM, HEMMER K, HACHI S, MORENO EL, VAN WUELLEN T, JARAZO J, WALTER J, BRUGGEMANN I, BOUSSAAD I, BERGER E, FLEMING RMT, BOLOGNIN S & SCHWAMBORN JC 2017. Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Reports, 8, 1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORSUT L, ROYBAL KT, XIONG X, GORDLEY RM, COYLE SM, THOMSON M & LIM WA 2016. Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell, 164, 780–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUFFAT J, LI Y, OMER A, DURBIN A, BOSCH I, BAKIASI G, RICHARDS E, MEYER A, GEHRKE L & JAENISCH R 2018. Human induced pluripotent stem cell-derived glial cells and neural progenitors display divergent responses to Zika and dengue infections. Proc Natl Acad Sci U S A, 115, 7117–7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUFFAT J, LI Y, YUAN B, MITALIPOVA M, OMER A, CORCORAN S, BAKIASI G, TSAI LH, AUBOURG P, RANSOHOFF RM & JAENISCH R 2016. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med, 22, 1358–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUGURUMA K, NISHIYAMA A, KAWAKAMI H, HASHIMOTO K & SASAI Y 2015. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep, 10, 537–50. [DOI] [PubMed] [Google Scholar]
- OGURA T, SAKAGUCHI H, MIYAMOTO S & TAKAHASHI J 2018. Three-dimensional induction of dorsal, intermediate and ventral spinal cord tissues from human pluripotent stem cells. Development, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ORMEL PR, VIEIRA DE SA R, VAN BODEGRAVEN EJ, KARST H, HARSCHNITZ O, SNEEBOER MAM, JOHANSEN LE, VAN DIJK RE, SCHEEFHALS N, BERDENIS VAN BERLEKOM A, RIBES MARTINEZ E, KLING S, MACGILLAVRY HD, VAN DEN BERG LH, KAHN RS, HOL EM, DE WITTE LD & PASTERKAMP RJ 2018. Microglia innately develop within cerebral organoids. Nat Commun, 9, 4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OZONE C, SUGA H, EIRAKU M, KADOSHIMA T, YONEMURA S, TAKATA N, OISO Y, TSUJI T & SASAI Y 2016. Functional anterior pituitary generated in self-organizing culture of human embryonic stem cells. Nat Commun, 7, 10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARK J, WETZEL I, MARRIOTT I, DREAU D, D’AVANZO C, KIM DY, TANZI RE & CHO H 2018. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat Neurosci, 21, 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAŞCA AM, SLOAN SA, CLARKE LE, TIAN Y, MAKINSON CD, HUBER N, KIM CH, PARK J-Y, O’ROURKE NA, NGUYEN KD, SMITH SJ, HUGUENARD JR, GESCHWIND DH, BARRES BA & PAŞCA SP 2015. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nature Methods, 12, 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAŞCA SP 2019. Assembling human brain organoids. Science, 363, 126–127. [DOI] [PubMed] [Google Scholar]
- PELLEGRINI L, BONFIO C, CHADWICK J, BEGUM F, SKEHEL M & LANCASTER MA 2020. Human CNS barrier-forming organoids with cerebrospinal fluid production. Science, eaaz5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHAM MT, POLLOCK KM, ROSE MD, CARY WA, STEWART HR, ZHOU P, NOLTA JA & WALDAU B 2018. Generation of human vascularized brain organoids. Neuroreport, 29, 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POLLEN AA, BHADURI A, ANDREWS MG, NOWAKOWSKI TJ, MEYERSON OS, MOSTAJO-RADJI MA, DI LULLO E, ALVARADO B, BEDOLLI M, DOUGHERTY ML, FIDDES IT, KRONENBERG ZN, SHUGA J, LEYRAT AA, WEST JA, BERSHTEYN M, LOWE CB, PAVLOVIC BJ, SALAMA SR, HAUSSLER D, EICHLER EE & KRIEGSTEIN AR 2019. Establishing Cerebral Organoids as Models of Human-Specific Brain Evolution. Cell, 176, 743–756 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QIAN X, NGUYEN HN, SONG MM, HADIONO C, OGDEN SC, HAMMACK C, YAO B, HAMERSKY GR, JACOB F, ZHONG C, YOON KJ, JEANG W, LIN L, LI Y, THAKOR J, BERG DA, ZHANG C, KANG E, CHICKERING M, NAUEN D, HO CY, WEN Z, CHRISTIAN KM, SHI PY, MAHER BJ, WU H, JIN P, TANG H, SONG H & MING GL 2016. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell, 165, 1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QIAN X, SONG H & MING G-L 2019. Brain organoids: advances, applications and challenges. Development, 146, dev166074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QIAN X, SU Y, ADAM CD, DEUTSCHMANN AU, PATHER SR, GOLDBERG EM, SU K, LI S, LU L, JACOB F, NGUYEN PTT, HUH S, HOKE A, SWINFORD-JACKSON SE, WEN Z, GU X, PIERCE RC, WU H, BRIAND LA, CHEN HI, WOLF JA, SONG H & MING G-L 2020. Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation. Cell Stem Cell, 26, 766–781.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QUADRATO G & ARLOTTA P 2017. Present and future of modeling human brain development in 3D organoids. Curr Opin Cell Biol, 49, 47–52. [DOI] [PubMed] [Google Scholar]
- QUADRATO G, BROWN J & ARLOTTA P 2016. The promises and challenges of human brain organoids as models of neuropsychiatric disease. Nat Med, 22, 1220–1228. [DOI] [PubMed] [Google Scholar]
- QUADRATO G, NGUYEN T, MACOSKO EZ, SHERWOOD JL, MIN YANG S, BERGER DR, MARIA N, SCHOLVIN J, GOLDMAN M, KINNEY JP, BOYDEN ES, LICHTMAN JW, WILLIAMS ZM, MCCARROLL SA & ARLOTTA P 2017. Cell diversity and network dynamics in photosensitive human brain organoids. Nature, 545, 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAJ B, WAGNER DE, MCKENNA A, PANDEY S, KLEIN AM, SHENDURE J, GAGNON JA & SCHIER AF 2018. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat Biotechnol, 36, 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REGINENSI D, ORTIZ D, PRAVIA A, BURILLO A, MORALES F, MORGAN C, JIMENEZ L, DAVE KR, PEREZ-PINZON MA & GITTENS RA 2020. Role of Region-Specific Brain Decellularized Extracellular Matrix on In Vitro Neuronal Maturation. Tissue Eng Part A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIFES P, ISAKSSON M, RATHORE GS, ALDRIN-KIRK P, MØLLER OK, BARZAGHI G, LEE J, EGEROD KL, RAUSCH DM, PARMAR M, PERS TH, LAURELL T & KIRKEBY A 2020. Modeling neural tube development by differentiation of human embryonic stem cells in a microfluidic WNT gradient. Nature Biotechnology. [DOI] [PubMed] [Google Scholar]
- RIGAMONTI A, REPETTI GG, SUN C, PRICE FD, RENY DC, RAPINO F, WEISINGER K, BENKLER C, PETERSON QP, DAVIDOW LS, HANSSON EM & RUBIN LL 2016. Large-Scale Production of Mature Neurons from Human Pluripotent Stem Cells in a Three-Dimensional Suspension Culture System. Stem Cell Reports, 6, 993–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAELENS W, CANNOODT R, TODOROV H & SAEYS Y 2019. A comparison of single-cell trajectory inference methods. Nat Biotechnol, 37, 547–554. [DOI] [PubMed] [Google Scholar]
- SAKAGUCHI H, KADOSHIMA T, SOEN M, NARII N, ISHIDA Y, OHGUSHI M, TAKAHASHI J, EIRAKU M & SASAI Y 2015. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat Commun, 6, 8896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SALMEN F, STAHL PL, MOLLBRINK A, NAVARRO JF, VICKOVIC S, FRISEN J & LUNDEBERG J 2018. Barcoded solid-phase RNA capture for Spatial Transcriptomics profiling in mammalian tissue sections. Nat Protoc, 13, 2501–2534. [DOI] [PubMed] [Google Scholar]
- SAMARASINGHE RA, MIRANDA OA, MITCHELL S, FERANDO I, WATANABE M, BUTH JE, KURDIAN A, GOLSHANI P, PLATH K, LOWRY WE, PARENT JM, MODY I & NOVITCH BG 2019. Identification of neural oscillations and epileptiform changes in human brain organoids. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHMUNK G, KIM CN, SOLIMAN SS, KEEFE MG, BOGDANOFF D, TEJERA D, ZIFFRA RS, SHIN D, ALLEN DE, CHHUN BB, MCGINNIS CS, WINKLER EA, ABLA AA, CHANG EF, GARTNER ZJ, MEHTA SB, PIAO X, HENGEN KB & NOWAKOWSKI TJ 2020. Human microglia upregulate cytokine signatures and accelerate maturation of neural networks. BioRxiv. [Google Scholar]
- SCHONEBERG J, DAMBOURNET D, LIU TL, FORSTER R, HOCKEMEYER D, BETZIG E & DRUBIN DG 2018. 4D cell biology: big data image analytics and lattice light-sheet imaging reveal dynamics of clathrin-mediated endocytosis in stem cell-derived intestinal organoids. Mol Biol Cell, 29, 2959–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SLOAN SA, DARMANIS S, HUBER N, KHAN TA, BIREY F, CANEDA C, REIMER R, QUAKE SR, BARRES BA & PASCA SP 2017. Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells. Neuron, 95, 779–790 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SMITS LM, REINHARDT L, REINHARDT P, GLATZA M, MONZEL AS, STANSLOWSKY N, ROSATO-SIRI MD, ZANON A, ANTONY PM, BELLMANN J, NICKLAS SM, HEMMER K, QING X, BERGER E, KALMBACH N, EHRLICH M, BOLOGNIN S, HICKS AA, WEGNER F, STERNECKERT JL & SCHWAMBORN JC 2019. Modeling Parkinson’s disease in midbrain-like organoids. NPJ Parkinsons Dis, 5, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SONG L, YUAN X, JONES Z, VIED C, MIAO Y, MARZANO M, HUA T, SANG QA, GUAN J, MA T, ZHOU Y & LI Y 2019. Functionalization of Brain Region-specific Spheroids with Isogenic Microglia-like Cells. Sci Rep, 9, 11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOOD D, TANG-SCHOMER M, POULI D, MIZZONI C, RAIA N, TAI A, ARKUN K, WU J, BLACK LD, SCHEFFLER B, GEORGAKOUDI I, STEINDLER DA & KAPLAN DL 2019. 3D extracellular matrix microenvironment in bioengineered tissue models of primary pediatric and adult brain tumors. Nature Communications, 10, 4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPANJAARD B, HU B, MITIC N, OLIVARES-CHAUVET P, JANJUHA S, NINOV N & JUNKER JP 2018. Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat Biotechnol, 36, 469–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SQUARZONI P, OLLER G, HOEFFEL G, PONT-LEZICA L, ROSTAING P, LOW D, BESSIS A, GINHOUX F & GAREL S 2014. Microglia modulate wiring of the embryonic forebrain. Cell Rep, 8, 1271–9. [DOI] [PubMed] [Google Scholar]
- STRACK AM, SAWYER WB, PLATT KB & LOEWY AD 1989. CNS cell groups regulating the sympathetic outflow to adrenal gland as revealed by transneuronal cell body labelling with pseudorabies virus. Brain Research, 491, 274–296. [DOI] [PubMed] [Google Scholar]
- SUN L, TANG Y, YAN K, YU J, ZOU Y, XU W, XIAO K, ZHANG Z, LI W, WU B, HU Z, CHEN K, FU ZF, DAI J & CAO G 2019. Differences in neurotropism and neurotoxicity among retrograde viral tracers. Mol Neurodegener, 14, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANAKA Y, CAKIR B, XIANG Y, SULLIVAN GJ & PARK IH 2020. Synthetic Analyses of Single-Cell Transcriptomes from Multiple Brain Organoids and Fetal Brain. Cell Rep, 30, 1682–1689 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TATA M & RUHRBERG C 2018. Cross-talk between blood vessels and neural progenitors in the developing brain. Neuronal Signaling, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAY TL, BECHADE C, D’ANDREA I, ST-PIERRE MK, HENRY MS, ROUMIER A & TREMBLAY ME 2017. Microglia Gone Rogue: Impacts on Psychiatric Disorders across the Lifespan. Front Mol Neurosci, 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TEN BERGE D, KOOLE W, FUERER C, FISH M, EROGLU E & NUSSE R 2008. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell, 3, 508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TREVINO AE, SINNOTT-ARMSTRONG N, ANDERSEN J, YOON S-J, HUBER N, PRITCHARD JK, CHANG HY, GREENLEAF WJ & PAȘCA SP 2020. Chromatin accessibility dynamics in a model of human forebrain development. Science, 367, eaay1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UGOLINI G 1995. Specificity of rabies virus as a transneuronal tracer of motor networks: transfer from hypoglossal motoneurons to connected second-order and higher order central nervous system cell groups. J Comp Neurol, 356, 457–80. [DOI] [PubMed] [Google Scholar]
- UZEL SG, AMADI OC, PEARL TM, LEE RT, SO PT & KAMM RD 2016. Simultaneous or Sequential Orthogonal Gradient Formation in a 3D Cell Culture Microfluidic Platform. Small, 12, 612–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN DEN HURK M, ERWIN JA, YEO GW, GAGE FH & BARDY C 2018. Patch-Seq Protocol to Analyze the Electrophysiology, Morphology and Transcriptome of Whole Single Neurons Derived From Human Pluripotent Stem Cells. Front Mol Neurosci, 11, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VELASCO S, KEDAIGLE AJ, SIMMONS SK, NASH A, ROCHA M, QUADRATO G, PAULSEN B, NGUYEN L, ADICONIS X, REGEV A, LEVIN JZ & ARLOTTA P 2019. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature, 570, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VELAZQUEZ JJ, LEGRAW R, MOGHADAM F, TAN Y, KILBOURNE J, HISLOP J, LIU S, CATS D, CHUVA DE SOUSA LOPES SM, PLAISIER C, CAHAN P, KIANI S & EBRAHIMKHANI MR 2020. Synthetic Maturation of Multilineage Human Liver Organoids via Genetically Guided Engineering. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAGNER DE & KLEIN AM 2020. Lineage tracing meets single-cell omics: opportunities and challenges. Nat Rev Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG X, ALLEN WE, WRIGHT MA, SYLWESTRAK EL, SAMUSIK N, VESUNA S, EVANS K, LIU C, RAMAKRISHNAN C, LIU J, NOLAN GP, BAVA FA & DEISSEROTH K 2018. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATANABE M, BUTH JE, VISHLAGHI N, DE LA TORRE-UBIETA L, TAXIDIS J, KHAKH BS, COPPOLA G, PEARSON CA, YAMAUCHI K, GONG D, DAI X, DAMOISEAUX R, ALIYARI R, LIEBSCHER S, SCHENKE-LAYLAND K, CANEDA C, HUANG EJ, ZHANG Y, CHENG G, GESCHWIND DH, GOLSHANI P, SUN R & NOVITCH BG 2017. Self-Organized Cerebral Organoids with Human-Specific Features Predict Effective Drugs to Combat Zika Virus Infection. Cell Rep, 21, 517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WITVLIET D, MULCAHY B, MITCHELL JK, MEIROVITCH Y, BERGER DR, WU Y, LIU Y, KOH WX, PARVATHALA R, HOLMYARD D, SCHALEK RL, SHAVIT N, CHISHOLM AD, LICHTMAN JW, SAMUEL ADT & ZHEN M 2020. Connectomes across development reveal principles of brain maturation in C. elegans. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WORSDORFER P, DALDA N, KERN A, KRUGER S, WAGNER N, KWOK CK, HENKE E & ERGUN S 2019. Generation of complex human organoid models including vascular networks by incorporation of mesodermal progenitor cells. Sci Rep, 9, 15663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XIANG Y, TANAKA Y, CAKIR B, PATTERSON B, KIM KY, SUN P, KANG YJ, ZHONG M, LIU X, PATRA P, LEE SH, WEISSMAN SM & PARK IH 2019. hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell, 24, 487–497 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XIANG Y, TANAKA Y, PATTERSON B, KANG YJ, GOVINDAIAH G, ROSELAAR N, CAKIR B, KIM KY, LOMBROSO AP, HWANG SM, ZHONG M, STANLEY EG, ELEFANTY AG, NAEGELE JR, LEE SH, WEISSMAN SM & PARK IH 2017. Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell, 21, 383–398 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAO Z, MICH JK, KU S, MENON V, KROSTAG AR, MARTINEZ RA, FURCHTGOTT L, MULHOLLAND H, BORT S, FUQUA MA, GREGOR BW, HODGE RD, JAYABALU A, MAY RC, MELTON S, NELSON AM, NGO NK, SHAPOVALOVA NV, SHEHATA SI, SMITH MW, TAIT LJ, THOMPSON CL, THOMSEN ER, YE C, GLASS IA, KAYKAS A, YAO S, PHILLIPS JW, GRIMLEY JS, LEVI BP, WANG Y & RAMANATHAN S 2017. A Single-Cell Roadmap of Lineage Bifurcation in Human ESC Models of Embryonic Brain Development. Cell Stem Cell, 20, 120–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOON S-J, ELAHI LS, PAȘCA AM, MARTON RM, GORDON A, REVAH O, MIURA Y, WALCZAK EM, HOLDGATE GM, FAN HC, HUGUENARD JR, GESCHWIND DH & PAȘCA SP 2019. Reliability of human cortical organoid generation. Nature Methods, 16, 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZEMANICK MC, STRICK PL & DIX RD 1991. Direction of transneuronal transport of herpes simplex virus 1 in the primate motor system is strain-dependent. Proc Natl Acad Sci U S A, 88, 8048–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZIFFRA RS, KIM CN, WILFERT A, HAEUSSLER M, CASELLA AM, PRZYTYCKI PF, KREIMER A, POLLARD KS, AMENT SA, EICHLER EE, AHITUV N & NOWAKOWSKI TJ 2019. Single cell epigenomic atlas of the developing human brain and organoids. BioRxiv. [Google Scholar]
- ZUCHERO JB & BARRES BA 2015. Glia in mammalian development and disease. Development (Cambridge, England), 142, 3805–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]


