Abstract
The chromosome axis plays a crucial role in meiotic recombination. Here, we study the function of ASY1, the Arabidopsis homolog of the yeast chromosome axis-associated component Hop1. Specifically, we characterized cross-over (CO) distribution in female and male meiosis by deep sequencing of the progeny of an allelic series of asy1 mutants. Combining data from nearly 1,000 individual plants, we find that reduced ASY1 functionality leads to genomic instability and sometimes drastic genomic rearrangements. We further observed that COs are less frequent and appear in more distal chromosomal regions in plants with no or reduced ASY1 functionality, consistent with previous analyses. However, our sequencing approach revealed that the reduction in CO number is not as dramatic as suggested by cytological analyses. Analysis of double mutants of asy1 with mutants with three other CO factors, MUS81, MSH4, and MSH5, as well as the determination of foci number of the CO regulator MLH1 demonstrates that the majority of the COs in asy1, similar to the situation in the wildtype (WT), largely belong to the class I, which are subject to interference. However, these COs are redistributed in asy1 mutants and typically appear much closer than in the WT. Hence, ASY1 plays a key role in CO interference that spaces COs along a chromosome. Conversely, since a large proportion of chromosomes do not receive any CO, we conclude that CO assurance, the process that ensures the obligatory assignment of one CO per chromosome, is also affected in asy1 mutants.
Significance statement.
The regulation of the number and placement of cross-overs (COs) during meiosis is critical to ensure meiotic fidelity and promote new genetic combinations. Here, we investigated the function of ASY1, which resides on the chromosome axis and plays a key role in CO formation. Our results show that COs in asy1 mutants are positioned closer to each other than in the wildtype and that, despite a roughly similar number of COs, not every chromosome receives a CO. With this, our results shed light on the mechanisms regulating two important but still poorly understood aspects of meiosis: CO assurance, which safeguards at least one CO per chromosome pair, and CO interference, which prevents two COs from occurring close to each other.
Introduction
Meiotic cross-overs (CO) are critical to the generation of genetic diversity. Through COs, segments of homologous chromosomes (homologs) are exchanged during the first meiotic division, creating new and unique combinations of alleles. Furthermore, COs are crucial for the equal segregation of chromosomes at the end of meiosis I since they physically link homologs and by that position them so that each cell pole will receive one homolog.
To generate COs, DNA double strand breaks (DSBs) are deliberately introduced into chromosomes in early prophase I. A fraction of these DSBs is then repaired by homologous recombination (HR) between the homologs, resulting in COs (1). HR relies on the resection of DSBs by the MRN complex generating 3’-overhanging single-stranded DNA, the search for sequence homology between the homologs, and subsequent strand invasion mediated by the RecA-related recombinases radiation sensitive51 (RAD51) and disrupted meiotic cDNA (DMC1). Following DNA synthesis of the invading strand, and the capture of the second end of the initial DSB, a tetrahedral cross-strand intermediate is formed that is known as double Holliday junction (dHJ), which, when resolved, can result in a CO or a nonCO (NCO) (2, 3). NCOs can also be generated without the formation of dHJs (4).
COs are typically grouped into two classes, which can be distinguished by the molecular machinery that catalyzes the resolution of the dHJ. Type I COs rely on the ZMM group of proteins, including the DNA mismatch repair proteins mutl homologue1/3 (MLH1/3) (1). A hallmark of type I COs is that they experience positive interference, i.e., COs are more distantly spaced from each other than expected by chance (5). This results in a gamma-shaped distribution (with a shape distribution parameter larger than one) of COs across the genome. In contrast, type II COs are formed by a different pathway, which involves the endonuclease MMS4 AND UV SENSITIVE81 (MUS81) and are not subjected to interference (1).
Of key importance for meiotic recombination is the formation of the chromosome axis, by which a proteinaceous scaffold is formed along the DNA, and from which chromatin loops are thought to extrude (6). The chromosome axis plays a key role in DSB induction in many species and early steps of CO formation (7). Later in meiosis I, this axis becomes the lateral element in the synaptonemal complex (SC) that zips the two homologs together and promotes the maturation of COs in many species.
An important protein, which is associated with the chromosome axis, is Hop1 and its orthologs, such as ASYNAPTIC1 (ASY1) in Arabidopsis (8, 9). Earlier work has implicated ASY1/Hop1 in many central aspects of meiotic recombination, such as promoting interhomolog-biased DNA repair, chromosome pairing, SC formation, and CO production (8–14). In addition to these conserved functions, there are also species-specific functions of Hop1-type proteins for meiotic recombination, e.g., Hop1 is strictly required for DSB formation in yeast while DSBs are still formed in asy1 mutants in Arabidopsis (10, 15).
The number and pattern of CO formation varies between species, and can be modulated by environmental conditions (16). The number of COs is also different between male and female meiosis. In Arabidopsis, there are usually 10 to 12 COs formed during male meiosis, but typically only 6 to 7 during female meiosis (17). The majority of COs, i.e., approximately 85%, are type I COs, and the 2 to 3 COs per chromosome are typically evenly distributed along the chromosome in male meiocytes (18). In contrast, a single CO is typically observed per chromosome in female meiocytes, and it is more often located in the pericentromeric regions (17). Importantly, all chromosomes typically undergo at least one CO in both sexes. This is insured through a mechanism referred to as CO assurance, which has been detected in many species (19–21).
How CO interference and assurance are brought about is currently not understood. In yeast, several mutants have been identified that affect CO interference, including topoisomerase II (TOPO II) (20). Whether these components function in a similar manner in other organisms remains unclear. One attractive model of how interference could be established is the “beam-film model” (22, 23). This model predicts that COs are formed at sites of high mechanical stress between the engaged homologs. Once a CO is formed, it locally releases this stress, and this relaxation spreads along the chromosomes laterally, preventing the formation of a CO nearby (22, 23). However, how this tension stress and its relaxation are transmitted is not clear. Recent studies suggest an alternative model proposing the diffusion-mediated coarsening of the proCO factor HEI10 (ZHP-3/4 in Caenorhabditis elegans), a RING finger-containing E3 ubiquitin ligase, that accounts for the CO positioning and interference (24, 25).
CO site selection appears to involve both epigenetic information, such as DNA methylation (26–29), and genetic factors such as the activity of the cell cycle kinase CYCLIN DEPENDENT KINASE A;1 (CDKA;1) (30, 31), as for instance shown in Arabidopsis. Interestingly, the number and position of COs have often been found to be modulated in polyploid plants, possibly to adapt to the challenge of faithfully assorting multiple chromosome sets during meiosis (32). Typically, polyploid species show a reduced CO number, and the remaining COs are often located at distal positions, a pattern which is also often found in diploid crop species such as maize (33). How this change in pattern is accomplished is not clear. It is also not clear whether allelic variants of the core meiotic machinery genes that appear to be associated with polyploidization, including alleles for ASY1, are functionally associated with this change in CO number and distribution (34).
Here, we have assessed the role of ASY1 in CO formation and distribution, using a genomics approach. To this end, we generated recombination maps of female and male meiosis for nearly 700 asy1 mutant plants, and characterized their CO patterns in comparison to the WT. First, we observed many instances of full or partial chromosome aneuploidy in the progenies of the asy1 mutants, confirming the importance of ASY1 for meiotic fidelity. Our work revealed that the overall number of COs is reduced in asy1 mutants but this trend is less pronounced in the surviving progeny than previously estimated by cytological studies. We show that COs in asy1 largely belong to the class I of COs and tend to closely localize at the telomeric and subtelomeric regions, highlighting a role for ASY1 in CO interference. Furthermore, the distribution of COs per chromosome is altered in plants with reduced ASY1 functionality and a large percentage of chromosomes being devoid of any CO, suggesting that ASY1 is a crucial factor for CO assurance as well.
Methods
Plant materials and growing conditions
The A. thaliana accessions Columbia (Col‐0) and Landsberg erecta (Ler-1) were used as the WT references in this study. The asy1 T-DNA insertion line (SALK_046,272) in Col-0 background (35) and mlh1-3 (SK25975) was obtained from the T‐DNA mutant collection at the Salk Institute Genomics Analysis Laboratory (SIGnAL, http://signal.salk.edu/cgi-bin/tdnaexpress) via NASC (http://arabidopsis.info/). The hypomorphic non-phosphorylatable mutant asy1T142V in Col-0 background was generated previously in Yang et al. (36, 37). The asy1 mutant in Ler background was generated in this study by CRISPR/Cas9 approach (Fig. S1). All plants were grown in growth chambers with a 16 h light/21°C and 8 h dark/18°C cycle at 60% humidity. The generation of genome sequencing populations was described in the main text. For this procedure, homozygous asy1 mutants (asy1Col-0, asy1T142V,and asy1ler-1) were used.
Genomic sequencing and raw read processing
For Arabidopsisgenomic DNA extraction, about 2 cm2 of leaf was harvested and used for DNA extraction using the SPRI beads-based method (38). Subsequently, genomic DNAs were quantified using a plate fluorometer with SYBR Green I, and normalized. After normalization, the samples were checked on agarose for uniformity. The generation of the sequencing libraries was performed following the Rowan protocol (39). Briefly, samples were fragmented, adapters were added, the fragments were amplified with indexed P5 and P7 primers, and visualization on agarose before being pooled (see Supplementary Material 1 for index and pooling information). Pools then underwent size selection with magnetic beads before measuring DNA concentration using a Qubit (Invitrogen) and verifying size distribution with the Agilent Bioanalyzer. Pools were sequenced on an Illumina HiSeq 4000 sequencer to obtain PE150 reads. Raw reads were checked for sequence quality, trimmed, and demultiplexed using custom Python scripts, as described previously, using allprep-15.py (40). After demultiplexing, each individual retained reads corresponding to 1.7x coverage of the haploid A. thaliana genome. Next, they were aligned to the TAIR10 genomic reference (https://www.arabidopsis.org) using BWA and default parameters (41), producing sam files (bwa-doall.py version 0.7.17) appropriate for dosage analysis (see below). For the purpose of genotyping and the creation of genetic maps, the data was also concatenated into a single mpileup.txt file using samtools (42). Finally, this mpileup file was further processed to obtain the percentages of each base call at each position and each sample using the script called mpileup-parser.py (beta-mpileup-parser_vMEM-O.py, https://github.com/Comai-Lab/mpileup-tools).
Dosage analysis and genotyping
Aneuploidy and dosage variation detection were performed as previously described (43). Briefly, the genome was divided into consecutive nonoverlapping 100 kb bins and reads that mapped to each bin were counted for each sample using a custom python script (bin-by-sam-v7.py, https://github.com/Comai-Lab/bin-by-sam). For each bin, read counts were normalized to account for variation in total read counts, as well as compared to the mean values for the whole population, in order to detect variation in dosage relative to the expected value of two in a diploid background. Data were plotted (Fig. 1 for an example), and aneuploidy was detected visually, as a variation in copy number that is consistent across at least three consecutive bins.
Fig. 1.
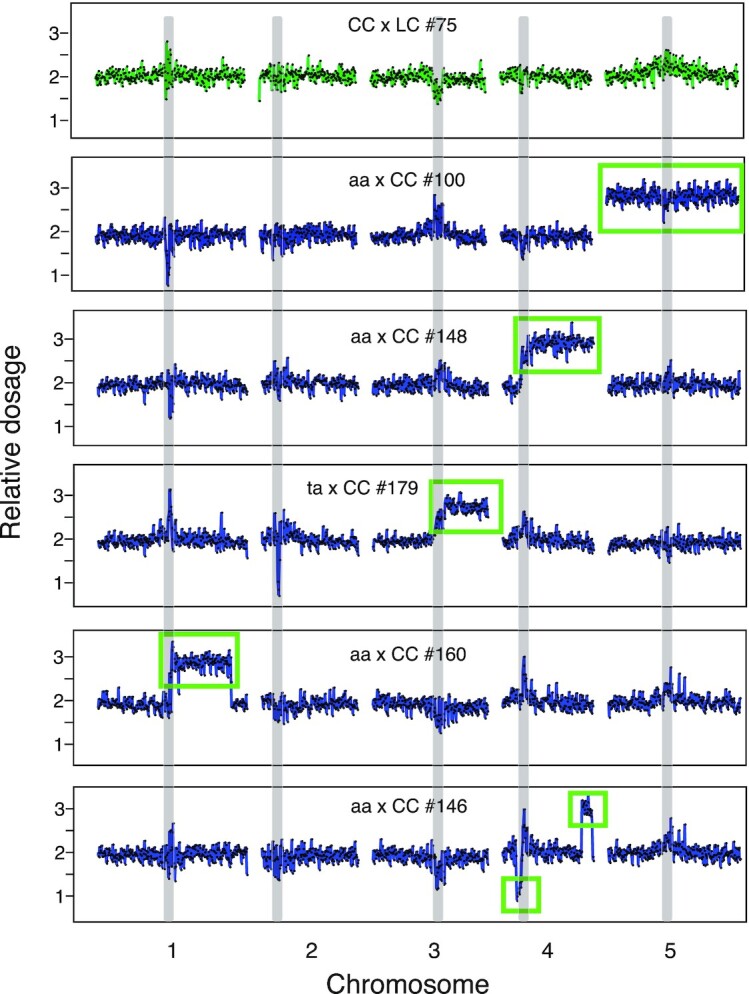
Aneuploidy in the progeny of the asy1 mutants. Dosage variation in six representative individuals from different crosses (CC x LC #75, aa x LC #100, aa x CC #148, ta x CC #179, aa x CC #160, and aa x CC #146). Each panel represents one individual and, for each individual, the five chromosomes are shown. For each individual, relative dosage, e.g., ∼ 2 for normal diploids, is depicted based on short-read sequence coverage variation after being normalized to the whole population. Dots represent consecutive nonoverlapping 100 kb bins. None of the progeny of wild-type control crosses (CC x LC and LC x CC) exhibited aneuploidy (top panel, shown in green). Crosses involving asy1 or asy1T142V mutants produce many aneuploid progeny (panel 2 to 6, colored in blue), including full trisomics (panel 2, aa x LC #100), partial trisomics (3rd panel, aa x CC #148), or more complex aneuploids (panels 5 and 6, aa x CC #160 and aa x CC #146). Many of the progeny of the crosses involving the hypomorphic asy1T142V mutant carried an extra copy of the bottom arm of chromosome 3 (Chr. 3B) (Table S2), suggesting that it originated from the parental line (panel 4, ta x CC #179). The regions that are present in additional or missing copies are indicated by the green rectangles. Centromeric and pericentromeric regions (gray vertical bands) consistently exhibited variation that mimicked dosage variation, but more likely originated from allelic variation between Col-0 and Ler-1 in these repeat-rich regions. These variations were consistently observed in all populations and were not recorded as aneuploidy.
Determination of CO locations and data analysis
A list of Col-0/Ler-1 SNPs was derived from a previously published list of ∼870 K SNP positions, (http://mtweb.cs.ucl.ac.uk/mus/www/19genomes/variants.SDI/ler_0.v7c.sdi (44)). We first filtered this list by selecting positions that had been detected by both the IMR and DENOM software, that did not include any ambiguous bases (only A, T, G, and C were allowed), with phred scores >100, and for which the reported consensus base SNP and the high mapping quality consensus base SNP were in agreement. This reduced the SNP set to ∼418 K positions. Next, we further filtered this list by only retaining positions that were covered at least once in at least 500/1108 of our samples and for which two both expected alleles were detected in our population (at least 5% of each allele type). The resulting ∼125 K positions were used for genotyping. For each sample, the allele calls at each of these SNP positions were recorded. Because we sequenced each sample at low coverage, allele calls were pooled in order to obtain robust and complete genotype calls for each sample. Specifically, allele calls were pooled into a total of 239 consecutive nonoverlapping 500 kb bins, and a per-bin genotype was derived for each individual by calculating the mean percentage of Col-0 allele in each bin, as previously described (45). The resulting genotype information was input into R/qtl (version 1.42 to 8) for the creation of genetic maps for each cross (est.map function), and the detection of COs (countXO function) and the determination of their position (locateXO function) (46).
CO interference
The testcross design described above was used to assess interference. A Python script was used to divide the genome into nonoverlapping intervals of arbitrary length. Recorded recombination breakpoints were counted, inferring COs per interval. The recombination frequency of any interval was equal to the (number of COs)/(number of scored chromatids). The predicted double COs number was determined by taking the square of the recombination rate. Double COs instances were scored based on COs pairs. Ten instances of triple or higher number CO's were observed in the 4.5 Mb bin analysis, and scored as individual instances. For example, an interval with three COs would be scored as having two double COs, one between CO1 and CO2, the second between CO2 and CO3. These data are summarized in Supplementary Material 2.
Cytologenetic analyses
Meiotic chromosome spreads and fluorescence in situ hybridization (FiSH) were performed according to procedures previously described (47). The following DNA probes were used: pTa71 [45S ribosomal DNA (rDNA), pTa71 of Triticum aestivum; (48)] and pCT4.2 [5S 133 rDNA; (49)].
Immunolocalization procedures were conducted following the method detailed in Armstrong et al. (50), with modifications detailed in Varas and Pradillo (51). The primary antibodies used were kindly provided by Professor Chris Franklin (University of Birmingham): anti-AtASY1 (rat; 1/1000), anti-AtZYP1 (rat; 1/500), anti-MLH1 (rabbit, 1/500), anti-RAD51 (rabbit, 1/300). Secondary antibodies were anti-rat and anti-rabbit IgG FITC conjugated (Agrisera) and anti-rabbit and anti-rat IgG Alexa Fluor 555 conjugated (Molecular Probes).
Slides were examined on an Olympus BX61 epifluorescence microscope and imaged with a CCD Olympus DP71 camera and computer using analySIS software (Soft Imaging System). For foci scoring, blind-coded digital images were used. Adobe Photoshop (CC 2018 v.19.0) and ImageJ (version 1.51 s) were applied for image processing.
Results
Creation of mapping populations to study ASY1 function in meiotic recombination
To study the effect of ASY1 on meiotic recombination, we sought to dissect CO distribution through a backcross design of the two different Arabidopsis thaliana accessions Columbia-0 and Landsberg erecta (Ler-1). Therefore, we first generated an asy1 null mutant using a CRISPR/Cas9 approach in the background of the accession Landsberg erecta (Ler-1) (52). Cas9 was targeted to the ASY1 coding region at the first exon-intron junction by a single guide RNA harboring 20 bases (Fig. S1A). A mutant allele (asy1Ler-1) harboring a guanine (G) insertion at the beginning of the second exon of ASY1 was subsequently identified from the T2 generation that did not harbor the CRISPR/Cas9 T-DNA anymore. This G insertion results in a frameshift and premature stop codon after the translation of the first 13 amino acids. Homozygous asy1Ler-1 mutants showed high levels of pollen mortality, and a high percentage of aborted seeds, matching the mutant phenotype of the previously identified asy1 null mutant in the Columbia-0 accession (asy1Col-0) (Fig. S1B to E) (12, 53). That this allele represents a loss-of-function mutant was confirmed by the absence of ASY1 from male meiocytes by immuno-detection (Fig. S1F). A complementation test using the previously generated functional ASY1 reporter (PROASY1: ASY1: GFP) (36) showed that the fertility defects of asy1Ler-1 mutants were fully rescued by this reporter, corroborating that the meiotic defects of this CRISPR/Cas9-induced allele are due to the loss of ASY1 function (Fig. S1B to E).
Next, the asy1Ler-1 mutant was combined with either an asy1Col-0 null mutant, or a previously generated hypomorphic asy1 mutant, called asy1T142V (both in the Col-0 genetic background), in which the threonine residue of the presumptive CDKA;1 phosphorylation site at position 142 is replaced by the nonphosphorylatable amino acid valine (36). In vitro, the nonphosphorylatable mutant asy1T142V has reduced binding affinity to ASY3, a key component of the chromosome axis, and shows decreased chromosome association in vivo (36, 54). These combinations then resulted in the following Col-Ler F1 hybrids: the first carries two null alleles of ASY1 (referred to as aa from now on), and the second carries a null allele in Ler-1 and the hypomorphic allele in Col-0 (asy1T142V-Col-0 asy1Ler-1 referred to as ta from now on).
Finally, we produced six mapping populations by backcrossing these hybrids to WT Col-0 plants (Table S1): specifically, the hybrids with reduced ASY1 functionality were crossed as female parents to the Col-0 WT (referred to as CC from now on) to assess the importance of ASY1 in female meiosis. These crosses are designated in the following as aa x CC (n = 207) for crosses with the asy1 null mutant and ta x CC (n = 213) for crosses with the hypomorphic mutant asy1T142V. Reciprocal crosses were performed to monitor male meiosis, designated as CC x aa (n = 216) for crosses with the asy1 null mutant and CC x ta (n = 214) for crosses with the hypomorphic mutant asy1T142V. As a control, we also generated F1 hybrids between the WT parental lines Col-0 and Ler-1, and crossed them reciprocally with a Col-0 tester line giving rise to two additional populations, named LC x CC (n = 117) and CC x LC (n = 119). To characterize recombination in asy1 mutants, the progeny of these six populations were subjected to whole-genome sequencing.
Reduction of ASY1 functionality results in aneuploid offspring and genome instability
Low recombination frequencies often cause the formation of univalents and hence unequal chromosome distributions and aneuploidy in the resulting spores (11). Previous cytological data (12, 10, 53, 54) suggested that CO numbers might be reduced in asy1 mutants since not all homologs are able to form bivalents. Consistent with these earlier studies, we also found univalents in meiotic chromosome spread analyses of male meiocytes produced by ta plants (Fig. S2A). To investigate the effect of these univalents on the karyotype of the resulting progenies, we screened each of these populations for the presence of aneuploidy. Looking for variation in sequencing read coverage across the whole genome, we found aneuploid plants in the progenies of all four crosses involving asy1 mutants, while none were found in the control WT crosses (Table S2 and Figs. 1 and 2).
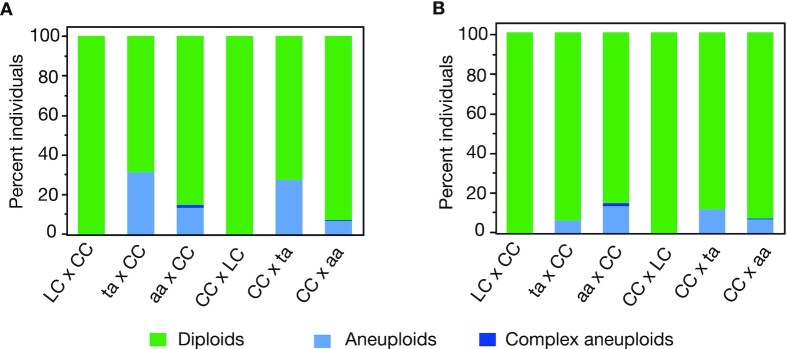
Fig. 2.
Percentage of diploid and aneuploid individuals in the various crosses. For each cross-type, the percentage of diploid (green), simple aneuploids (light blue), and complex aneuploids (dark blue) are represented. Complex aneuploids represent individuals with partial chromosomes, suggesting the presence of at least two breaks in a single chromosome. The overall percentage of aneuploid individuals is highest in the crosses involving the ta mutant (panel A). In those crosses, extra copies of the bottom arm of chromosome 3 (Chr. 3B) possibly originated from the parental line. Those individuals are discarded in panel B.
Most of the aneuploids recovered were trisomics, carrying one additional copy of a single chromosome (Fig. 1, second panel). Trisomies of all chromosome types were recovered, except for chromosome 1 (Table S2), perhaps because of more severe selection against gametes carrying additional copies of this particular chromosome, as observed previously (55, 56), or because of the more severe phenotypic consequences associated with additional copies of chromosome 1 in A. thaliana (57). Conversely, we did not detect any instance of a missing chromosome, probably because it is lethal or severely detrimental at the gametic stage, consistent with previous work (58–60).
Interestingly, we also found examples of segmental aneuploids, carrying only a part of an additional chromosome (Fig. 1, panels 3 to 6 and Table S2). The karyotypes of some of segmental aneuploids could have originated from a single miss-repaired DNA DSB (Fig. 1, panels 3, 4), in which case only a terminal segment remains. In addition, we find individuals carrying signs of two or more breaks (Fig. 1, panels 5, 6), resulting in more drastically rearranged genomes. Comparing the approximate location of these breaks with the expected position of the centromeres (the Arabidopsis genome initiative), suggested that more than half (9/17) of these miss-repaired breaks occurred at or around the centromeres (Table S2, second column). While this pattern possibly suggests a direct or indirect role of ASY1 in the formation and/or repair of DSB near centromeres, breaks at centromeres are also often enriched after gamma radiation (59), in the progeny of aneuploids (56), and after genome elimination (58), suggesting that centromeric regions are in particular sensitive to DNA stresses.
Most of the individuals exhibiting segmental aneuploidy originated from crosses involving the asy1 mutants as a female (Table S2). This overrepresentation in females is consistent with the idea that segmental aneuploids are less fit than euploids and hence are counter-selected during pollen tube growth. Moreover, segmental aneuploids appeared much more frequently in crosses involving asy1 null mutants, as we only found one case originating from a cross with the hypomorphic mutant (Table S2), suggesting that genomic instability scales with the level of ASY1 reduction.
Strikingly, one individual (#102) from the aa x CC cross exhibited signs of high levels of genomic instability (Fig. S3). Plant #102 carried an extra copy of the top arm of chromosome 1 but coverage information for that region of the genome was highly variable compared to the diploid individual from the same cross (Fig. S3, panel 1, 3). Comparison with other segmental aneuploids of chromosome 1 confirmed that this level of variability did not originate from methodological artifacts associated with the presence of a third copy of the corresponding sequences (Fig. S3, panel 2). Such a catastrophic genome restructuring event is reminiscent of chromothripsis, when laggard chromosomes are incorporated into micronuclei, and undergo massive DSBs, resulting in restructuring and copy number variation clustered on a single chromosome (61). Chromothripsis is often associated with cancer in humans (61), but has also been observed in A. thaliana following CENH3-mediated genome elimination (58). To confirm the variability observed in our low coverage data, we sequenced this individual deeper and confirmed the observed dosage variation (62). Further analysis of the structure of this chromosome confirms the presence of hundreds of novel DNA junctions clustered on the top arm of chromosome 1, consistent with the consequences of chromothripsis (62). It is likely that other plants carrying severely rearranged chromosomes were strongly counter-selected and are not found in our experiments. Thus, this sample provides a unique opportunity to witness a snapshot of the extent of meiotic instability of asy1 mutants.
Unexpectedly, both of the crosses involving the hypomorphic mutant produced a population of plants enriched in segmental trisomy for chromosome 3B (Fig. 1, individual ta x CC #179). The fact that this particular karyotype was clearly overrepresented in those two crosses (56/213 for ta x CC and 36/214 for CC x ta), but never found in the other four crosses (Table S2), suggests that the hybrid plant carrying the hypomorphic asy1 mutation itself carried this segmental aneuploidy, and transmitted it to a subset of its progeny. Possibly, this additional copy of chromosome 3B originated from defective meiosis in the parental asy1T142V or asy1ler-1mutants that were used to produce the F1 hypomorphic ta mutant (see the "Methods" section). Since the additional copy of a chromosomal segment would strongly interfere with the identification of COs on this particular chromatid using the method applied in this study, we discarded all aneuploid progenies from further analysis. The final number of diploid individuals remaining for each population ranged between 117 and 200 (Table S2).
We cannot completely exclude that the likely aneuploidy in the original ta line could have affected meiosis beyond producing an over-representation of chromosome 3 aneuploids and hence, the results from this line have to be taken cautiously. However, the comparison between WT and aa remains unchanged and notably, the results from the ta line are in accordance with the results obtained for aa (see below). Thus, we believe that the analysis of the ta plants brings additional confidence to our conclusions.
Number and position of COs are affected by reduced functionality of ASY1
To characterize the number and position of COs in the WT and asy1 mutant lines, each remaining diploid individual was genotyped at each of 239 nonoverlapping 500 kb marker bins covering the entire A. thaliana genome. The data were input into r-qtl (see the "Methods" section), and separate genetic maps were created for the six populations (Fig. S4). In all cases, the marker order was consistent with the physical order of the bins. We next identified the position of all COs detectable in these samples (Supplementary Material 2).
The mean number of COs per chromosome varied depending on whether the asy1 mutant was the male or the female, the chromosome type, and the level of ASY1 functionality (Fig. 3). Specifically, the number of COs per chromosome was not significantly different for the control or mutant crosses for the subtelocentric chromosomes (chromosomes 2 and 4). On the other hand, there was a significant decrease in CO number for chromosomes 1, 3, and 5 on the male side and chromosomes 1 and 3 on the female side (t-test P values <0.05 or less, Fig. 3). In most of the cases for female meiosis, the hypomorphic allele exhibited CO numbers that were intermediate between the control crosses and the crosses involving the asy1 null allele. A similar finding was also obtained for male meiosis in the asy1 hypomorphs, but only when all chromosomes were combined (Fig. 3). However, it is important to note that, in comparison to the cytological method, a sequencing approach is biased towards viable progenies, especially in mutants where the meiotic process is severely defective. This bias might underestimate the extent of meiotic defects actually present.
Fig. 3.
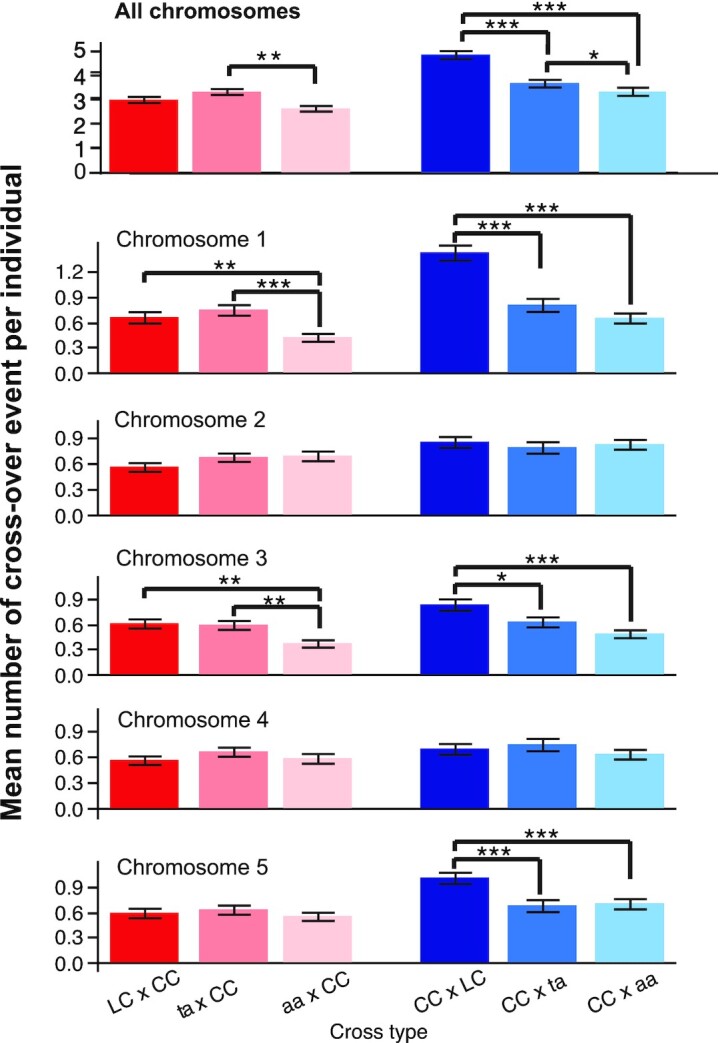
The mean number of CO per individual. For each cross-type, the mean number of CO events was calculated for each individual (top panel) and for each chromosome and each individual (all other panels). CO events were only detectable for the hybrid (Ler-Col) parent, not the Col-0 parent, and therefore represent either events that occurred on the paternal side (blue bars) or the maternal side (red/pink bars) only. Since each meiosis produces four gametic cells and each CO event can only be detected from two of the four gametes, the average number of CO per chromosome per individual indicates only half of the CO events formed in each meiosis. For each cross type and chromosome, the mean and SE are represented. Within a cross type and a cross direction, significant differences between the means were tested on a pair-wise basis using Student's t-test. Significant comparisons are indicated as follows: ***: P value <0.0001, **: P value <0.01, *: P value <0.05. Lack of significance level between two specific means indicates that the comparison was not significant.
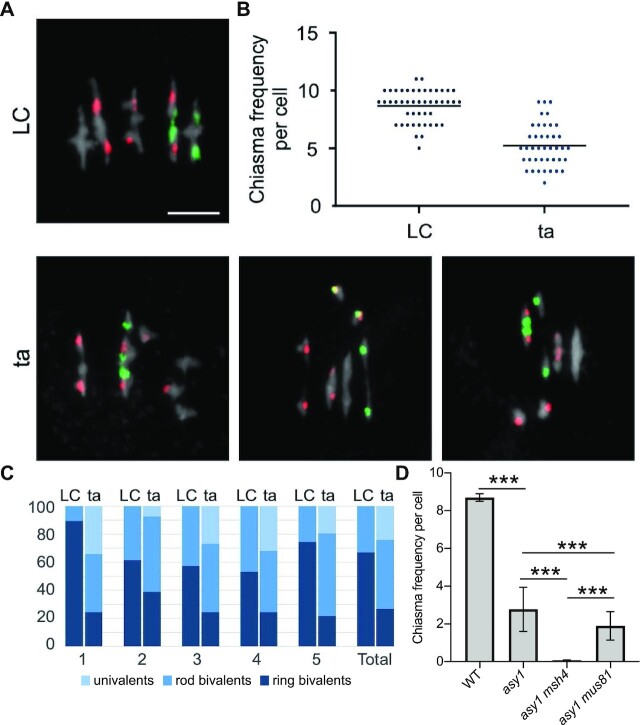
Next, we analyzed the chiasma formation in asy1 mutants by chromosome spreads of male meiocytes at metaphase I. This analysis revealed a significant decrease in the chiasma frequency of asy1 mutants (asy1Ler-1: 2.62 ± 1.39, n = 47; asy1Col-0: 2.78 ± 1.17, n = 65; asy1T142V: 5.2 ± 0.3, n = 41 vs. 8.7 ± 0.2 in the WT, n = 47, Fig. 4). To have a resolution at chromosomal level, FISH experiments were performed to distinguish different chromosomes using the 5S and 45S rDNA probes in ta mutants in which the occurrence of aneuploids is not so severe as that in asy1 null mutants. We found that all chromosomes, except for chromosome 2, showed a reduction in CO formation (Fig. 5 and Table S3), and this reduction was higher for the large chromosomes 1 and 5, consistent with earlier work analyzing asy1 null mutants (12, 53) (Fig. 5C).
Fig. 4.
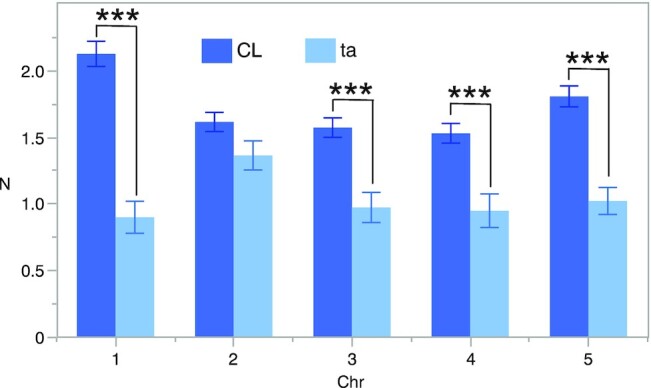
Mean chiasma frequencies per chromosome. The percentages represent the chromosome contribution to the total chiasma frequency. Means and SE values are shown. The mean values were significantly different (t-test P value <0.001) for all chromosomes, except for chromosome 2. The decrease in chiasma frequency was highest for the large chromosomes 1 and 5.
Fig. 5.
Determination of the chiasmata number in asy1 mutants using FISH. (A) Representative metaphase I nuclei of LC (up) and ta (down) meiocytes using FISH to identify the different chromosome types. Red signals (5S rDNA loci) are present in chromosomes 3, 4, and 5, whereas green signals (45S rDNA loci, NORs) appear in the acrocentric chromosomes 2 and 4. DAPI staining appears in gray. Examples of ta cells include univalents in different chromosomes: 1 and 3 (left); 4 (middle); 5 (right). Bars represent 5 µm. (B) Quantification of chiasmata in metaphase I cells. Each dot represents an individual cell and bars indicate the mean. (C) Percentage of pairs of univalents, rod bivalents (with chiasmata only in one chromosome arm), and ring bivalents (with at least one chiasma on both chromosome arms) in the WT LC hybrids (left) and the ta hybrids (right). (D) Quantification of chiasmata in metaphase I cells in WT, asy1, asy1 msh4, and asy1 mus81 mutant plants. *** P < 0.0001.
To examine whether the reduction in bivalent formation in asy1 mutants is due to a reduction in the formation of DSBs, we analyzed RAD51 foci in leptotene/zygotene. We found that RAD51 foci numbers in male meiosis of aa (136.8 ± 10.9; n = 14) and ta (134.1 ± 17.6, n = 15) were not significantly different from those of the WT LC hybrids (134.6 ± 14.2, n = 12; aa vs. LC P = 0.89; ta vs LC P = 0.99) (Fig. S5). Therefore, the observed reduction in bivalent number in the asy1 hybrids does most likely not originate from reduced DSB formation, consistent with previous analysis of asy1 null mutants (10).
The discrepancy between the observed chiasmata and the COs identified by sequencing, especially for chromosome 4, for which we did not find any significant reduction in CO number in our genetic maps, prompted us to analyze CO distribution within each chromosome (Fig. 6 and Fig. S6). For ease of visualization and comparison, we expressed CO position as the distance to the telomeres, expressed in 0.5 Mb units, and are presenting data from the left and right arms separately to distinguish the effect of the mutations from those of the short arm of the subtelocentric chromosomes. The distribution of COs was markedly different for the ta and aa crosses compared to the control crosses in both cross directions (Fig. 6A). Specifically, COs were inclined to localize near the telomeres in the asy1 mutant crosses, while they were more evenly distributed along the length of the chromosomes in the WT control crosses. The hypomorphic mutant exhibited an intermediate behavior. This trend was not only visible when data for all chromosomes were pooled (Fig. 6A), but also for each individual chromosome type (Fig. 6B), with the exception of the left arms of chromosomes 2 and 4, which are too short to detect changes in distribution. This telomere-proximal accumulation of COs is consistent with previous and our cytological analyses of spread chromosomes, which revealed long-stretched bivalents due to the chiasmata in chromosomal arm ends (60% in asy1, 30 out of 50 bivalents) (Fig. 5) (12, 53). These findings also resolved the apparent difference to the cytological work that does not have the resolution to reveal closely spaced COs.
Fig. 6.
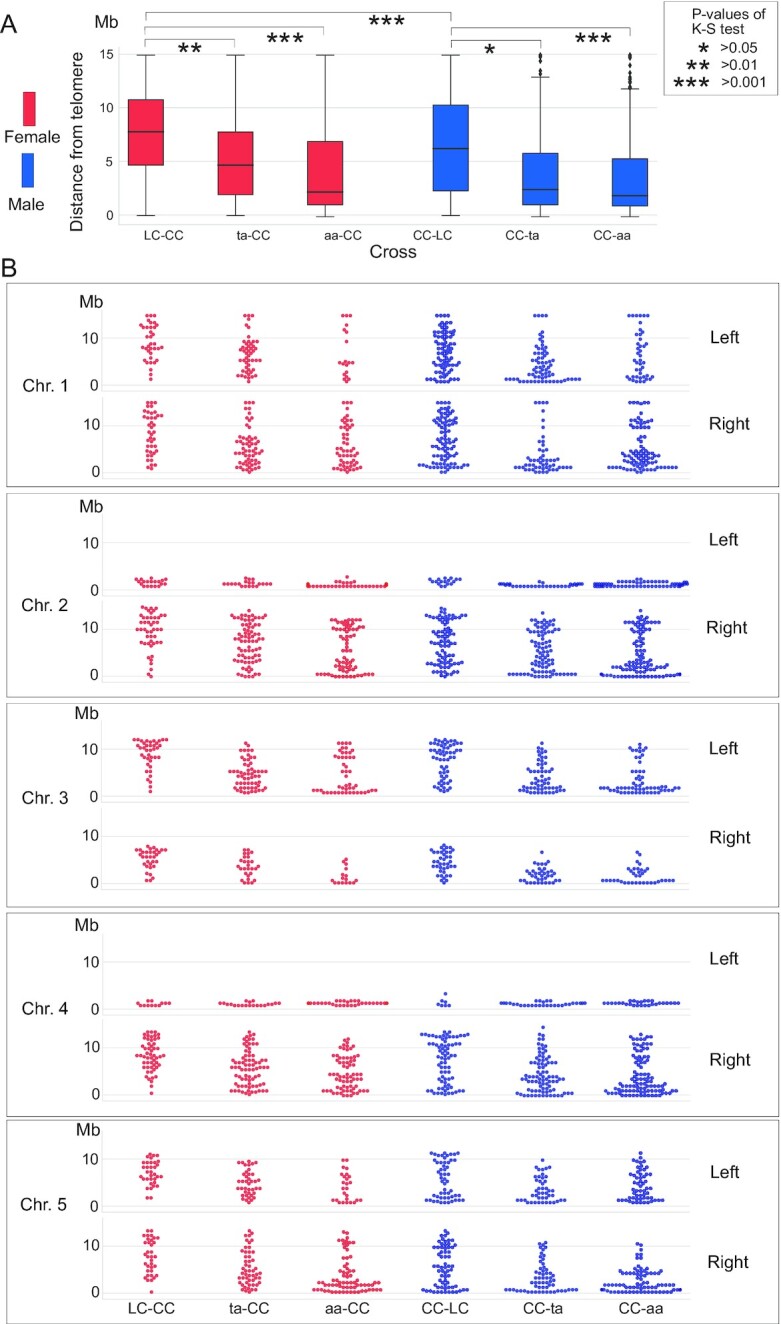
Distribution of CO position in the different crosses. (A) Box plot of the cumulative distributions of CO-telomere distance. The significance of comparisons was tested by the Kolmogorov–Smirnov test (K-S). (B) Swarm plots of CO-telomere distance per chromosome. The distance from the CO to the telomeres is shown and the maximum distance for each arm is at the centromere.
CO interference is reduced in asy1 mutants
The observed overaccumulation of COs in distal chromosome positions could be due to two different mechanisms. On the one hand, the number of interference-sensitive type I COs could be reduced at the benefit of additional type II COs, which are not subject to interference. On the other hand, type I COs, which are sensitive to interference, might lose their sensitivity in meiocytes with reduced or no ASY1 functionality.
To discriminate between these two possibilities, we generated double mutants of asy1Col-0 with mutants in MSH4 (also in Col-0 background), one key component of type I CO formation, and double mutants of asy1 with mus81 (in Col-0 as well) in which type II COs are reduced. Depleting MSH4 in asy1 caused a drastic reduction of chiasma numbers, i.e., in 25 meiocytes of the asy1 msh4 double mutants a chiasma frequency of only 0.07 CO per meiocyte was observed, while this number, as shown above, is 2.78 ± 1.17 (n = 65) in asy1 single mutants.
Thus, the majority of the remaining COs in asy1 belong to the type I class of COs. Consistently, the asy1 mus81 double mutants did not show a strong reduction of chiasmata i.e., from 57 meiocytes, a chiasmata frequency of 1.9 ± 0.75 per meiocyte was obtained, suggesting that type II COs are present only at a small frequency in asy1 mutants. Thus, the loss of ASY1 does apparently not grossly alter the relative proportion of type I to type II COs found in the WT.
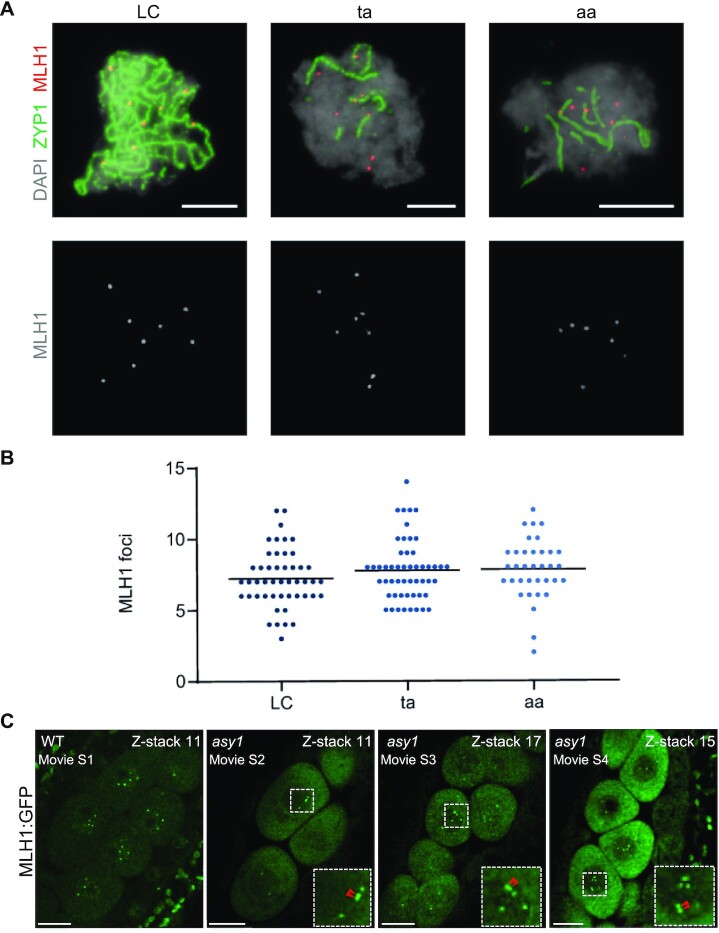
To complement the genetic analysis, we performed immunolocalization analysis of MLH1, which marks type I COs, in male meiocytes (Fig.7A and B). In WT-LC hybrids, we observed an average of 7.2 ± 0.3 MLH1 foci per meiocyte (n = 48), matching previous analyses. Strikingly, we did not find significant changes in the frequency of MLH1 foci in the aa plants (7.8 ± 0.3, n = 35; P = 0.406). A similar number was also found in the ta plants (7.8 ± 0.3, n = 52; P = 0.405). The discrepancy between the determination of COs in asy1 mutants by sequencing with the number of MLH1 foci possibly indicates that not all MLH1 foci maturate into COs. Notably, MLH1 foci were found on both synapsed and nonsynapsed chromosome regions. However, although the MLH1 antibody and scoring of MLH1 foci have been often used in the past, we could fully exclude technical difficulties and for instance sometimes chromosomes are not clearly stained making it difficult to evaluate the results of the immuno-detection.
Fig. 7.
Analysis of MLH1 foci. (A) Representative images showing pachytene (LC) and pachytene-like (ta, aa) meiocytes after immunolabeling for detecting the class I CO marker MLH1 (red). The central element protein of the SC ZYP1 is detected in green and DAPI staining appears in gray. Bars represent 5 µm. (B) Comparison of the number of MLH1 foci. Each dot represents an individual cell and bars indicate the mean. (C) Presence of MLH1 foci in WT and asy1 mutant plants. One of the z-stacks was shown from Supplementary Movies 1 to 4. Red arrows indicate the closely located MLH1 foci. Bars represent 10 µm.
Therefore, we sought to complement the MLH1 localization by a previously established live cell imaging setup of meiocytes. To achieve this, we first generated a genomic reporter of MLH1 in which the GFP fluorescent tag was inserted immediately before the stop codon (PROMLH1: MLH1: GFP, called MLH1: GFP), and then transformed this construct into mlh1 mutants. In contrast to the severe fertility reduction of mlh1 mutant plants, mutants harboring the MLH1: GFP construct were almost fully rescued, as shown by the fully developed silique and good pollen viability, suggesting that MLH1: GFP is functional (Fig. S7A to C).
Next, the expression and localization of MLH1: GFP at different stages of male meiosis were imaged by laser scanning confocal microscopy. We found that at premeiosis, i.e., S-phase, MLH1-GFP was expressed in entire meiocytes with many bright foci in the nuclei presumably highlighting DNA mismatch repair sites, which arise during replication (Fig. S7D). Subsequently, from early to mid prophase I, MLH1: GFP was only diffusely present in both the cytosol and nuclei of male meiocytes, and no particular pattern was observed (Fig. S7D). However, when meiocytes reached late prophase I, e.g., late pachytene or diplotene stages, clear foci appeared in the nuclei, presumably marking type I CO intermediates (Fig. S7D).
Next, to check CO distribution, MLH1: GFP reporter was introgressed into asy1 mutant plants and compared to wild-type plants bearing this construct. To cover all MLH1 foci in a meicocyte, z-stacks with 0.8 μm spacing intervals were acquired (Supplementary Movies 1 to 4), and the foci were counted using the image analysis software Fiji. Consistent with our immunodetection (Fig. 7B), we found that there was a similar number of MLH1 foci in asy1 mutant (9.13 ± 1.52, n = 37) when compared to the WT (9.42 ± 1.36, n = 34), corroborating not only the presence of type I COs in asy1 mutants but also that apparently not all MLH1 foci in asy1 mutants maturate into COs. Notably, compared to the WT, closely located MLH1 foci were frequently observed in asy1 mutants (Fig. 7C, Supplementary Movies1 to 4).
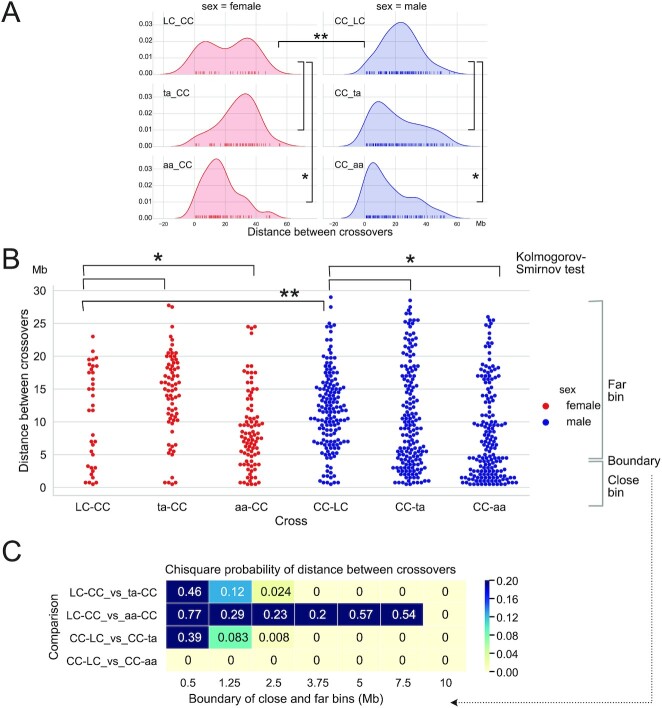
The preference of telomere-proximal formation of CO in asy1 mutant, together with the observation that these COs are of type I with largely the same number of MLH1-positive foci suggest that ASY1 might play a role in CO interference. To further investigate this possibility, we compared the distribution of distances between adjacent COs in the six different crosses (Fig. 8). Inter-CO distance followed a broad distribution that appeared skewed toward short values for the ta and aa plants. Both female and male distributions in the aa mutant were significantly different from that of the corresponding WT crosses (Fig. 8A and B, Kolmogorov–Smirnov adjusted for multiple testing, P = 0.03, 0.02, respectively). To further probe this skewness, we divided inter-COs segments into two classes, close and far, arbitrarily changing the dividing line between these two classes and observing the effect on the results (Fig. 8B and C). The female control cross only yielded few instances of double COs (Fig. 8B, LC-CC cross), restricting our ability to observe significant differences between cross types. The ta-CC cross was significantly enriched in distant double COs (Fig. 8A and B). Nevertheless, significant enrichment for close events in female meiosis was demonstrated in the aa-CC cross when close and far bins were divided at 10 Mb (Fig. 8C, 10 Mb units). The higher recombination frequency in male meiosis provided more cases of syntenic COs than in female meiosis. Thus, the male CC-aa cross proved significant for any size bin and the male ta mutant displayed strong and significant differences above 1.255 Mb (Fig. 8C).
Fig. 8.
The Frequency of COs and the distance between COs. CO to CO distance distribution displayed as (A) Kernel Density Estimation and (B) swarm plots, using in 1 Mb units. Knock out of ASY1 is associated with reduced distance between COs. C. Chi-square probability for comparison of close vs. far binned CO–CO distances. The respective upper limit of the close bin varies and is shown below each column of P values. Yellow denotes significant difference by the Chi square test. Note that significance for the ta-CC cross was caused by an increase of the distant CO class, as opposed to an increase in the number of close COs, as observed in the aa crosses. Significance thresholds: * <0.05, ** <0.01.
A direct measurement of interference, such as those obtained by the classical rate of observed vs. expected double COs, was not applicable because the number of individuals used for each cross limited the number of COs and statistical power. Nonetheless, measuring interference on a sliding, nonoverlapping window of 4.5 Mb, suggested reduction of interference at multiple genomic intervals for both mutants, the aa null mutation displaying stronger reduction than the ta mutation (Fig. S8). The overall distribution of interference values for WT control crosses and asy1 mutant crosses was highly significantly different (Fig. S8, Kolmogorov–Smirnov test adjusted for multiple testing, P < 0.001 for all comparisons).
CO assurance is affected in the ASY1 mutants
The above data indicated that ASY1 is required for the proper number and distribution of COs along chromosomes. However, a telomeric or subtelomeric CO is sufficient to keep bivalents together, preventing a random distribution of homologs in meiosis I, and subsequent aneuploidy, as seen for instance in many crop species in which COs are usually distally located (33, 63, 64). Likewise, a low number of COs does not necessarily lead to the formation of univalents as exemplified by female meiosis in Arabidopsis, during which only about 6 to 7 COs are formed but every chromosome receives at least one CO (17). Thus, according to our sequencing data and chiasma counting (Fig. 5 and Table S3), the 5 to 6 COs formed in female meiosis and especially the 6 to 7 COs (reflecting the situation in female WT meiosis) formed in the male meiosis of asy1 null and hypomorphic mutants could be roughly sufficient to equip every chromosome with a CO obeying CO assurance. However, asy1 mutants show a high degree of univalent formation (Fig. 5C, Tables S4 and S5), as also reported previously (12, 53). Consistently, we also found significantly more nonrecombined chromosomes in our asy1 backcross populations compared to the WT control cross (Fig. 9 and Table S6). This suggested impaired CO assurance as well.
Fig. 9.
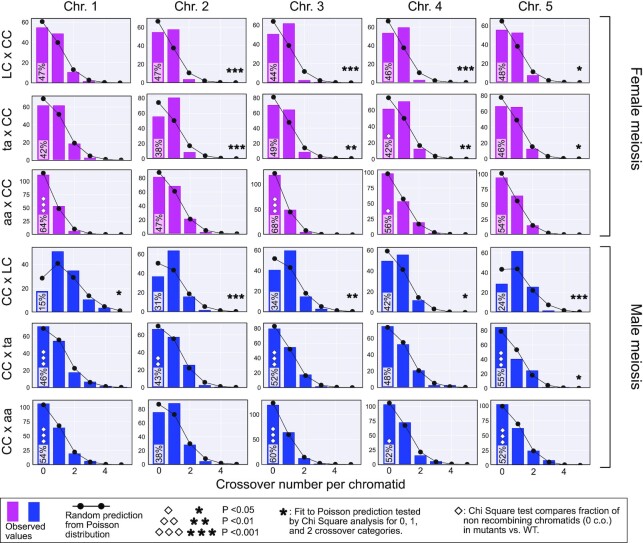
Comparison of CO numbers per chromatid in WT and asy1 mutants. The bar plots illustrate the count of chromatid classes for given genotypes and chromosomes. Pink and blue bars are for female and male meiosis, respectively. The random expectation according to the Poisson distribution is displayed by the black line with round black circles. The star markers display significance of the random fit (low P corresponds to significant deviation). Note that almost all chromosomes for both test crosses significantly deviate from a random distribution while all asy1 null mutants and most of the asy1 hypomorphic mutant show a pattern that matches a poisson distribution. The % of the 0 CO chromatid class is displayed inside the 0 CO bar. For this fraction of 0 CO chromatids, the P of the Chi Square comparison with WT values is marked with diamonds (low P corresponds to significant deviation). The exact number of instances in each category can be found in Table S6.
The hallmark of CO assurance is the presence of one obligatory CO that causes a nonrandom distribution of COs over the genome, resulting in a deviation from a poisson distribution of COs and an overrepresentation of chromatids exhibiting a single CO (Fig. 9, LC x CC cross). In contrast, in the ta x CC progenies and to an even larger extent in the aa x CC progenies, we consistently observed distributions of COs that were closer to the expected random distribution (asterisks, significance test in Fig. 9). If the CO distribution deviates in the asy1 crosses from a random distribution, it is usually due to an underrepresentation of the 1-CO class, e.g., chromosome 1 in male meiosis (Fig. 9, lower three panels), combined with an overrepresentation of both nonrecombined chromatids (diamonds, significance test in Fig. 9), and chromatids with more than one CO. Thus, we conclude that another key feature of COs, i.e., CO assurance, is also affected by the loss of ASY1 function, namely, ASY1 is required for the acquisition of one obligatory CO for each homolog pair.
Discussion
ASY1 and its homolog Hop1 are important for several aspects of meiotic recombination (8–12). Here, we have focused on the role of ASY1 in CO placement in female and male meiosis. Mapping COs in hybrids between two different Arabidopsis accessions that are mutant for ASY1 has allowed us to reveal several important and previously not recognized aspects of the function of ASY1, foremost that ASY1 mediates CO interference and plays an important role for CO assurance. Using two asy1 alleles with different strengths shows that these two aspects are sensitive to the level of ASY1 functionality. Our findings are consistent with another report by Lambing et al. that appeared while our paper was in revision (65). Lambing et al. demonstrate that ASY1 is involved in CO interference and counteracts telomere-led CO placements in male meiosis.
Interestingly, CO interference was also reduced in yeast cells mutant for the AAA-ATPase PACHYTENE CHECKPOINT2 (PCH2) (66–70). PCH2 has been previously found to be important for the removal of ASY1 and Hop1 to promote assembly of the SC (69, 71, 72). This raises the question of why loss of ASY1 and a failure to remove it can cause similar mutant phenotypes. However, recent work indicated that PCH2, at least in Arabidopsis, also plays a role in delivering ASY1 into the nucleus, and in the absence of PCH2 or its cofactor COMET, ASY1 strongly accumulated in the cytoplasm while ASY1 is a solely nuclear localized protein in the WT (36, 73). Thus, it is possible that the timely and correct association of ASY1 with the axis, especially during early phases of axis formation, could be compromised in pch2 reconciling the pch2 and asy1 mutant effects with respect to CO interference. Conversely, we postulate that interference should also be affected in comet mutants.
Pairing and synapsis usually start in the distal regions of homologs in Arabidopsis (74–76). Notably, despite the general failure of chromosome alignment and synapsis in the asy1 mutant, telomere clustering and the pairing of telomeres of homologs has been found to occur during meiotic interphase and early prophase I (50). Thus, the placement of COs in the distal regions of homologs in asy1, as observed here and in previous studies, is consistent with this observation (12, 53). With that respect, it is interesting to note that synapsis occurs in only short chromosomal sections in Sordaria mer2 mutants and COs tend to cluster in these sections, thus also modulating the strength of interference (77).
We also observed that not all MLH1 foci in asy1 maturated into COs and hence, ASY1 directly or indirectly is involved in CO maturation. Since several MLH1 foci were found outside of synapsed regions, it is an intriguing question to test in the future whether only MLH1 foci on synapsed regions will mature into COs. However, recent studies have shown that loss of ZYP1 in Arabidopsis does not abolish COs and hence, synapsis per se is not a prerequisite for CO formation (78–80). Instead, the maturation of MLH1 foci has been previously found to depend on the pairing regulator Ph1 in rye wheat hybrids (81). However, how Ph1 charges MLH1 foci is not clear and whether there is a direct link to ASY1 action at the chromosome axis needs to be resolved in the future. Remarkably, even the hypomorphic asy1 mutant showed a strong distalization of COs. Therefore, it is plausible that even a slight modulation of the axis and/or the speed by which the axis is formed could restrict COs to distal positions while maintaining a high level of fertility, as seen in the hypomorphic asy1T142 mutant, which is semifertile (36).
In this context, it is interesting to note that many polyploid species show a distalization of COs based on cytological analyses (32, 82). Moreover, mutations in ASY1 and of the axis component ASY3 have been found to be specifically associated with tetraploidy in the Arabidopsis arenosa species, hinting at an adaptive advantage of these mutations in polyploids (34, 83). While the benefit of distalized COs in polyploids is obscure, CO numbers have often been found to be reduced in polyploids, and this effect would be very beneficial for the faithful segregation of species with more than two homologs, as it would reduce the level of chromosomes that are interconnected by COs and hence prone to missegregation (32). Reduced CO number in polyploids has been speculated to be due to increased interference (32). Our findings offer an alternative explanation, i.e., reduction/slowing down of pairing with concomitant reduction of interference. Moreover, our findings would also explain why COs are distalized in polyploids. However, whether an asy1-dependent decrease in interference represses missegregation in polyploids can only be a valid explanation if the remaining COs that form in the distal region of chromosomes are more likely to all concern the same pair of homologs. Such an exclusive CO position could possibly be forced by sterical constraints between different homologs. The determination of CO patterns by sequencing in polyploid species will help resolve this question. In addition, live cell imaging of meiosis, as recently established for Arabidopsis (84), and tracking of chromosomes in polyploid species could be a very powerful tool to address this point.
Why interference is reduced in asy1 mutants remains unclear and can, as far as we can tell, not be explained by the so far known functions of ASY1 in meiosis, e.g., interhomolog versus intersister repair bias. Applying the beam-film model of interference to our data (22), leads to the speculation that a reduction in functional ASY1 protein levels would prevent the tension relaxation brought about by a CO to spread along the chromosome, possibly because the chromosome axis is not connected by ASY1. Consistently, the ASY1 homolog Hop1 has been proposed, based on in vitro data, to build long head-to-tail polymers (85, 86). Such a polymer could possibly transmit a relaxation force. However, at least in such a model, it seems unlikely that ASY1 itself contributes to the tension since, in that case, no COs would be expected in asy1 mutants in the first place. Hence, more likely under the assumption of the beam-film model, an ASY1 polymer, if built in vivo, could serve as a platform for other proteins that help relaxing the mechanical stress, such as topo II (87). Interestingly, TOPO II has been co-precipitated with ASY1 from Brassica oleracea meiocytes (88). However, it is currently not clear whether this interaction is direct or whether TOPO II in Brassica is also involved in interference control.
In the light of the recently proposed model of CO interference by diffusing mediated coarsening of the proCO factor HEI10 (24), one could speculate that loss of ASY1 limits HEI10 to telomeric/subtelomeric regions and by that promotes the formation of type I COs there. However, why this should be the case is not clear as yet and needs further exploration.
Our work showed that ASY1 regulates not only interference but also CO assurance. The beam-film model predicts that CO assurance is not a function of interference, although the initial obligatory CO drives interference subsequently by providing the starting gun for mechanical stress. While some mutants that affect interference in yeast, were found to still undergo an obligatory CO, such as mutants in TOPO II, CO assurance and interference were concomitantly compromised in others mutants, e.g., in mutants of MutS HOMOLOG4 (MSH4) (20, 89). We currently do not know whether the effect of ASY1 on interference and assurance have the same biochemical foundation, or reflect two different functions of ASY1.
Moreover, the observation that the class with one CO is often underrepresented in asy1 mutant populations in combination with closely spaced COs in the telomeric and subtelomeric regions suggests that asy1 mutants could represent a rare case of negative interference, i.e., the attraction of COs after a first CO is formed (5). Notably, we found that the extent of interference and assurance reduction in asy1 mutants appears to depend on the individual chromosome for yet unknown reasons. Thus, the exploration of the dynamics and function of the chromosome axis in CO interference and CO assurance, ideally with chromosome-specific resolution, remains an exciting question.
Acknowledgement
We thank Beth Rowan for assistance with the preparation of the DNA sequencing libraries and Meric Lieberman for bioinformatics assistance. We are grateful to Kirsten Bomblies (ETH Zurich), Nancy Hollingsworth (Stony Brook University, New York), and Erik Wijnker (University of Wageningen) and for critical reading and helpful comments on the manuscript. The ASY1 and MLH1 primary antibodies were kindly provided by Chris Franklin (University of Birmingham, United Kingdom). This manuscript was posted as a preprint: https://doi.org/10.1101/2022.03.17.484635.
Contributor Information
Gaetan Pochon, Department of Developmental Biology, University of Hamburg, 22609 Hamburg, Germany.
Isabelle M Henry, Genome Center and Department of Plant Biology, UC Davis, Davis, CA 95618, USA.
Chao Yang, Department of Developmental Biology, University of Hamburg, 22609 Hamburg, Germany; National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China.
Niels Lory, Department of Developmental Biology, University of Hamburg, 22609 Hamburg, Germany.
Nadia Fernández-Jiménez, Department of Genetics, Physiology and Microbiology, University Complutense of Madrid, 28040 Madrid, Spain.
Franziska Böwer, Department of Developmental Biology, University of Hamburg, 22609 Hamburg, Germany.
Bingyan Hu, Department of Developmental Biology, University of Hamburg, 22609 Hamburg, Germany.
Lena Carstens, Department of Developmental Biology, University of Hamburg, 22609 Hamburg, Germany.
Helen T Tsai, Genome Center and Department of Plant Biology, UC Davis, Davis, CA 95618, USA.
Monica Pradillo, Department of Genetics, Physiology and Microbiology, University Complutense of Madrid, 28040 Madrid, Spain.
Luca Comai, Genome Center and Department of Plant Biology, UC Davis, Davis, CA 95618, USA.
Arp Schnittger, Department of Developmental Biology, University of Hamburg, 22609 Hamburg, Germany.
Supplementary Material
Supplementary Material is available at PNAS Nexus online.
Funding
N.F.J. is a PhD fellow funded by the FPU programme of the Spanish Ministry of Education (FPU16/02772). This research was supported by the European Union (Marie Curie ITN, COMREC 606956, and MEICOM 765212) to M.P. and A.S. M.P. and A.S. are members of the COST Action n°CA 16212 “In Depth” http://www.cost.eu/COST_Actions/ca/CA16212. M.P. acknowledges the support of the Ministry of Economy and Competitiveness of Spain (AGL2015-67349-P). M.P. is part of the International Plant Nucleus Consortium (IPNC, https://radar.brookes.ac.uk). L.C. and I.M.H. are supported by the Innovative Genomics Institute. C.Y. is supported by funds from the National Natural Science Foundation of China (32170354 to C.Y.) and Huazhong Agricultural University (101–11042110006 to C.Y.). A.S. kindly acknowledges the core funding of the University of Hamburg.
Authors' Contributions
A.S., I.H., and Lu.C. designed the research; G.P., I.M., C.Y., N.F.J., N.L., L.C., H.T.T., and M.P., performed the experiments. G.P., I.M., C.Y., M.P., Lu.C., and A.S. analyzed the data. I.H., C.Y., M.P., Lu.C., and A.S. wrote the manuscript.
Data Availability
All material generated in this study is freely available from A.S. Sequence data from this article can be found and is freely accessible in the NCBI SRA database under BioProject number PRAJNA 723,952.
References
- 1. Hunter N. 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol. 7:a016618–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Holliday R. 1964. A mechanism for gene conversion in fungi. Genet Res. 5:282–304. [DOI] [PubMed] [Google Scholar]
- 3. Gaillard P-HL, Noguchi E, Shanahan P, Russell P. 2003. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol Cell. 12:747–759. [DOI] [PubMed] [Google Scholar]
- 4. Allers T, Lichten M. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 106:47–57. [DOI] [PubMed] [Google Scholar]
- 5. Berchowitz L, Copenhaver G. 2010. Genetic interference: dont stand so close to me. Curr Genomics. 11:91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zickler D, Kleckner N. 2015. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol. 7:a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zickler D, Kleckner N. 1999. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 33:603–754. [DOI] [PubMed] [Google Scholar]
- 8. Caryl AP, Armstrong SJ, Jones GH, Franklin FC. 2000. A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma. 109:62–71. [DOI] [PubMed] [Google Scholar]
- 9. Hollingsworth NM, Goetsch L, Byers B. 1990. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell. 61:73–84. [DOI] [PubMed] [Google Scholar]
- 10. Sanchez-Moran E, Santos JL, Jones GH, Franklin FCH. 2007. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 21:2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hollingsworth NM, Byers B. 1989. HOP1: a yeast meiotic pairing gene. Genetics. 121:445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ross KJ, et al. 1997. Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA-transformed lines. Chromosome Res. 5:551–559. [DOI] [PubMed] [Google Scholar]
- 13. Smith AV, Roeder GS. 1997. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol. 136:957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loidl J, Klein F, Scherthan H. 1994. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J Cell Biol. 125:1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woltering D, et al. 2000. Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol Cell Biol. 20:6646–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zelkowski M, Olson MA, Wang M, Pawlowski W. 2019. Diversity and determinants of meiotic recombination landscapes. Trends Genet. 35:359–370. [DOI] [PubMed] [Google Scholar]
- 17. Giraut L, et al. 2011. Genome-Wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-Specific patterns along chromosomes. PLos Genet. 7:e1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M. 2015. The molecular biology of meiosis in plants. Annu Rev Plant Biol. 66:297–327. [DOI] [PubMed] [Google Scholar]
- 19. Jones GH, Franklin FCH. 2006. Meiotic crossing-over: obligation and interference. Cell. 126:246–248. [DOI] [PubMed] [Google Scholar]
- 20. Wang S, Zickler D, Kleckner N, Zhang L. 2015. Meiotic crossover patterns: obligatory crossover, interference and homeostasis in a single process. Cell Cycle. 14:305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shinohara M, Oh SD, Hunter N, Shinohara A. 2008. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat Genet. 40:299–309. [DOI] [PubMed] [Google Scholar]
- 22. Zhang L, Liang Z, Hutchinson J, Kleckner N. 2014. Crossover patterning by the beam-film model: analysis and implications. PLos Genet. 10:e1004042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kleckner N, et al. 2004. A mechanical basis for chromosome function. Proc Natl Acad Sci U S A. 101:12592–12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morgan C, et al. 2021. Diffusion-mediated HEI10 coarsening can explain meiotic crossover positioning in Arabidopsis. Nat Commun. 12:4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang L, Köhler S, Rillo-Bohn R, Dernburg AF. 2018. A compartmentalized signaling network mediates crossover control in meiosis. Elife. 7.e30789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Melamed-Bessudo C, Levy AA. 2012. Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc Natl Acad Sci U S A. 109:E981–E988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mirouze M, et al. 2012. Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc Natl Acad Sci U S A. 109:5880–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Underwood CJ, et al. 2018. Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation. Genome Res. 28:519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Colomé-Tatché M, et al. 2012. Features of the Arabidopsis recombination landscape resultingfrom the combined loss of sequence variation and DNA methylation. Proc Natl Acad Sci U S A. 109:16240–16245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wijnker E, et al. 2019. The Cdk1/Cdk2 homolog CDKA;1 controls the recombination landscape in Arabidopsis. Proc Natl Acad Sci U S A. 116:12534–12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nowack MK, et al. 2012. Genetic framework of cyclin-dependent kinase function inArabidopsis. Dev Cell. 22:1030–1040. [DOI] [PubMed] [Google Scholar]
- 32. Bomblies K, Jones G, Franklin C, Zickler D, Kleckner N. 2016. The challenge of evolving stable polyploidy: could an increase in “crossover interference distance” play a central role?. Chromosoma. 125:287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Luo C, Li X, Zhang Q, Yan J. 2019. Single gametophyte sequencing reveals that crossover events differ between sexes in maize. Nat Commun. 10:785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hollister JD, et al. 2012. Genetic adaptation associated with genome-Doubling in autotetraploid arabidopsis arenosa. PLos Genet. 8:e1003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Crismani W, et al. 2013. MCM8 Is required for a pathway of meiotic double-Strand break repair independent of DMC1 in Arabidopsis thaliana. PLos Genet. 9:e1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang C, et al. 2019. The arabidopsis Cdk1/Cdk2 homolog CDKA;1 controls chromosome axis assembly during plant meiosis. EMBO J. 39:e101625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dissmeyer N, Schnittger A. 2011. Use of phospho-site substitutions to analyze the biological relevance of phosphorylation events in regulatory networks. Methods Mol Biol. 779:93–138. [DOI] [PubMed] [Google Scholar]
- 38. Rowan BA, Seymour DK, Chae E, Lundberg DS, Weigel D. 2017. Methods for genotyping-by-Sequencing. Methods Mol Biol. 1492:221–242. [DOI] [PubMed] [Google Scholar]
- 39. Rowan BA, et al. 2019. An ultra high-Density arabidopsis thaliana crossover map that refines the influences of structural variation and epigenetic features. Genetics. 213:771–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Henry IM, Zinkgraf MS, Groover AT, Comai L. 2015. A system for dosage-Based functional genomics in Poplar. Plant Cell. 27:2370–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li H, et al. 2009. The sequence alignment/map format and samtools. Bioinformatics. 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tan E, Comai L, Henry I. 2016. Chromosome dosage analysis in plants using whole genome sequencing. BIO-PROTOCOL. 6(13):e1854. [Google Scholar]
- 44. Gan X, et al. 2011. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 477:419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Henry IM, et al. 2014. The BOY NAMED SUE quantitative trait locus confers increased meiotic stability to an adapted natural allopolyploid of Arabidopsis. Plant Cell. 26:181–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Broman KW, Wu H, Sen S, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 19:889–890. [DOI] [PubMed] [Google Scholar]
- 47. Martinez-Garcia M, Pradillo M. 2017. Functional analysis of arabidopsis argonautes in meiosis and DNA repair. Methods Mol Biol. 1640:145–158. [DOI] [PubMed] [Google Scholar]
- 48. Gerlach WL, Bedbrook JR. 1979. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 7:1869–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Campell BR, Song Y, Posch TE, Cullis CA, Town CD. 1992. Gene. 112:225–228. [DOI] [PubMed] [Google Scholar]
- 50. Armstrong SJ, Caryl AP, Jones GH, Franklin FCH. 2002. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci. 115:3645–3655. [DOI] [PubMed] [Google Scholar]
- 51. Varas J, Pradillo M. 2018. Immunolabeling protocols for studying meiosis in plant mutants defective for nuclear envelope components. Methods Mol Biol. 1840:237–247. [DOI] [PubMed] [Google Scholar]
- 52. Schiml S, Fauser F, Puchta H. 2014. The CRISPR/Cas system can be used as nuclease for in plantagene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 80:1139–1150. [DOI] [PubMed] [Google Scholar]
- 53. Sanchez Moran E, Armstrong SJ, Santos JL, Franklin FC, Jones GH. 2001. Chiasma formation in arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Res. 9:121–128. [DOI] [PubMed] [Google Scholar]
- 54. Ferdous M, et al. 2012. Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLos Genet. 8:e1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Henry IM, Dilkes BP, Tyagi AP, Lin H-Y, Comai L. 2009. Dosage and parent-of-origin effects shaping aneuploid swarms in A. thaliana. Heredity. 103:458–468. [DOI] [PubMed] [Google Scholar]
- 56. Koornneef M, Van der Veen JH. 1983.Trisomics in Arabidopsis thaliana and the location of linkage groups. Genetica. 61:41–46. [Google Scholar]
- 57. Henry IM, Dilkes BP, Miller ES, Burkart-Waco D, Comai L. 2010. Phenotypic consequences of aneuploidy in Arabidopsis thaliana. Genetics. 186:1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tan EH, et al. 2015. Catastrophic chromosomal restructuring during genome elimination in plants. Elife. 4:e06516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vizir IY, Mulligan BJ. 1999. Genetics of gamma-irradiation-induced mutations in Arabidopsis thaliana: large chromosomal deletions can be rescued through the fertilization of diploid eggs. J Hered. 90:412–417. [DOI] [PubMed] [Google Scholar]
- 60. Siegel JJ, Amon A. 2012. New insights into the troubles of aneuploidy. Annu Rev Cell Dev Biol. 28:189–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stephens PJ, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 144:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Guo W, Comai L, Henry IM. 2022. Chromoanagenesis in the asy1 meiotic mutant of Arabidopsis. G3. 10.1093/g3journal/jkac185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zickler D, Kleckner N. 1998. The leptotene-zygotene transition of meiosis. Annu Rev Genet. 32:619–697. [DOI] [PubMed] [Google Scholar]
- 64. Osman K, Higgins JD, Sanchez-Moran E, Armstrong SJ, Franklin FCH. 2011. Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol. 190:523–544. [DOI] [PubMed] [Google Scholar]
- 65. Lambing C, Kuo PC, Tock AJ, Topp SD, Henderson IR. 2020. ASY1 acts as a dosage-dependent antagonist of telomere-led recombination and mediates crossover interference in Arabidopsis. Proc Natl Acad Sci U S A. 117:13647–13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zanders S, Alani E. 2009. The pch2Δ mutation in Baker's yeast alters meiotic crossover levels and confers a defect in crossover interference. PLos Genet. 5:e1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. San-Segundo PA, Roeder GS. 1999. Pch2 links chromatin silencing to meiotic checkpoint control. Cell. 97:313–324. [DOI] [PubMed] [Google Scholar]
- 68. Joshi N, Barot A, Jamison C, Börner GV. 2009. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLos Genet. 5:e1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen C, Jomaa A, Ortega J, Alani EE. 2014. Pch2 is a hexameric ring atpase that remodels the chromosome axis protein Hop1. Proc Natl Acad Sci. 111:E44–E53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. da Silva RC, da Silva RC, Vader G. 2021. Getting there: understanding the chromosomal recruitment of the AAA atpase Pch2/TRIP13 during meiosis. Curr Genet. 67:553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Börner GV, Barot A, Kleckner N. 2008. Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proc Natl Acad Sci U S A. 105:3327–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lambing C, et al. 2015. Arabidopsis PCH2 mediates meiotic chromosome remodeling and maturation of crossovers. PLos Genet. 11:e1005372–E9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Balboni M, Yang C, Komaki S, Brun J, Schnittger A. 2020. COMET functions as a PCH2 cofactor in regulating the HORMA domain protein ASY1. Curr Biol. 30:4113–4127.e6. [DOI] [PubMed] [Google Scholar]
- 74. López E, Pradillo M, Romero C, Santos JL, Cuñado N. 2008. Pairing and synapsis in wild type Arabidopsis thaliana. Chromosome Res. 16:701–708. [DOI] [PubMed] [Google Scholar]
- 75. Armstrong SJ, Franklin FC, Jones GH. 2001. Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J Cell Sci. 114:4207–4217. [DOI] [PubMed] [Google Scholar]
- 76. Hurel A, et al. 2018. A cytological approach to studying meiotic recombination and chromosome dynamics in Arabidopsis thalianamale meiocytes in three dimensions. Plant J. 95:385–396. [DOI] [PubMed] [Google Scholar]
- 77. Tessé S, et al. 2017. Asy2/Mer2: an evolutionarily conserved mediator of meiotic recombination, pairing, and global chromosome compaction. Genes Dev. 31:1880–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. France MG, et al. 2021. ZYP1 is required for obligate cross-over formation and cross-over interference in Arabidopsis. Proc Natl Acad Sci. 118(14):e2021671118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Capilla-Pérez L, et al. 2021. The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis. Proc Natl Acad Sci U S A. 118(12):e2023613118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yang C, et al. , Bipartite recruitment of PCH2 and COMET to the synaptonemal complex drives chromosome axis reconstruction leading to crossover restriction. 10.1101/2022.03.23.485444. [DOI] [PMC free article] [PubMed]
- 81. Martín AC, Shaw P, Phillips D, Reader S, Moore G. 2014. Licensing MLH1 sites for crossover during meiosis. Nat Commun. 5:4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sybenga J. 1975. Meiotic configurations. Monographs on Theoretical and Applied Genetics. Berlin, Heidelberg: Springer. 10.1007/978-3-642-80960-6. [DOI] [Google Scholar]
- 83. Yant L, et al. 2013. Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr Biol. 23:2151–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Prusicki MA, et al. 2019. Live cell imaging of meiosis in Arabidopsis thaliana. Elife. 8.e42834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. West AMV, Komives EA, Corbett KD. 2018. Conformational dynamics of the Hop1 HORMA domain reveal a common mechanism with the spindle checkpoint protein Mad2. Nucleic Acids Res. 46:279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Rosenberg SC, Corbett KD. 2015. The multifaceted roles of the HORMA domain in cellular signaling. J Cell Biol. 211:745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang L, et al. 2014. Topoisomerase II mediates meiotic crossover interference. Nature. 511:551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Osman K, et al. 2018. Affinity proteomics reveals extensive phosphorylation of the Brassica chromosome axis protein ASY1 and a network of associated proteins at prophase I of meiosis. Plant J. 93:17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Krishnaprasad GN, et al. 2015. Variation in crossover frequencies perturb crossover assurance without affecting meiotic chromosome segregation in Saccharomyces cerevisiae. Genetics. 199:399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All material generated in this study is freely available from A.S. Sequence data from this article can be found and is freely accessible in the NCBI SRA database under BioProject number PRAJNA 723,952.