Keywords: COVID-19, E2F1, hyaluronan, lung, proteomics, SARS-CoV-1, SARS-CoV-2
Abstract
The COVID-19 pandemic continues to impose a major impact on global health and economy since its identification in early 2020, causing significant morbidity and mortality worldwide. Caused by the SARS-CoV-2 virus, along with a growing number of variants, COVID-19 has led to 651,918,402 confirmed cases and 6,656,601 deaths worldwide (as of December 27, 2022; https://covid19.who.int/). Despite advances in our understanding of COVID-19 pathogenesis, the precise mechanism by which SARS-CoV2 causes epithelial injury is incompletely understood. In this current study, robust application of global-discovery proteomics identified highly significant induced changes by the Spike S1 protein of SARS-CoV-2 in the proteome of alveolar type II (ATII)-like rat L2 cells that lack ACE2 receptors. Systems biology analysis revealed that the S1-induced proteomics changes were associated with three significant network hubs: E2F1, CREB1/RelA, and ROCK2/RhoA. We also found that pretreatment of L2 cells with high molecular weight hyaluronan (HMW-HA) greatly attenuated the S1 effects on the proteome. Western blotting analysis and cell cycle measurements confirmed the S1 upregulation of E2F1 and ROCK2/RhoA in L2 cells and the protective effects of HMW-HA. Taken as a whole, our studies revealed profound and novel biological changes that contribute to our current understanding of both S1 and hyaluronan biology. These data show that the S1 protein may contribute to epithelial injury induced by SARS-CoV-2. In addition, our work supports the potential benefit of HMW-HA in ameliorating SARS CoV-2-induced cell injury.
INTRODUCTION
Coronavirus disease 2019 (COVID-19) is caused by a new β-coronavirus, SARS-CoV-2; its epicenter was in Wuhan, China. COVID-19 was identified as a pandemic by the World Health Organization (WHO) on March 11, 2020. As of December 27, 2022, there have been 651,918,402 cases worldwide resulting in 6,656,601 deaths (https://covid19.who.int/). This far exceeds the total deaths caused by both SARS CoV-1 and Middle Eastern respiratory syndrome (MERS). A large fraction of patients with COVID-19 develop acute respiratory distress syndrome (ARDS) within 24–48 h after onset of symptoms with more than a 50% mortality rate (1).
COVID-19 is caused by an enveloped, nonsegmented, positive-sense RNA new β-coronavirus, SARS-CoV-2; it belongs to the sarbecovirus, orthornaviridae subfamily, which is broadly distributed in humans and other mammals (2). It contains four main structural proteins including three glycoproteins [spike (S), envelope (E), and a membrane (M)], a nucleocapsid (N) protein, and several accessory proteins (3). The S glycoprotein is a transmembrane protein with a molecular weight of ∼150 kDa, found in the outer portion of the virus. It forms homotrimers protruding in the viral surface and facilitates binding of SARS CoV-2 to human ACE2 and CD147 receptors of host cells (4). The SARS CoV-2 S protein has significant homology with the SARS-CoV-1 (5), but binds with higher affinity to ACE2 receptors (6) due to a furin-like cleavage site (682RRAR/S686) inserted in the S1/S2 protease cleavage site (7). The S1 region of the Spike protein is responsible for binding to the host cell ACE2 receptor, whereas the S2 region is responsible for fusion of the viral RNA and cellular membrane. SARS-CoV-2 infects several different animal species like rats, ferrets, hamsters, nonhuman primates, minks, tree shrews, raccoon dogs, fruit bats, and rabbits (8).
Despite the introduction of several effective vaccines, the mutagenicity and transmissibility of SARS-CoV-2, as evidenced by the emergence of several variants, make the understanding of the pathophysiological mechanism involved and the identification of therapeutic agents an extremely high priority (9). COVID-19 is mainly a respiratory disease but it can lead to other systemic effects and eventually to a systemic disorder (10, 11). SARS-CoV-2 exhibits a very broad tissue tropism as it can infect and affect a variety of different organs and systems, including the musculoskeletal (12), nervous (13), renal (14), and cardiovascular systems (11).
The pathophysiology and immune response to SARS-CoV-2 infection are broad and involve signaling pathways from several cell and tissue types (15). In particular, although it is understood that SARS-CoV-2 used S-protein binding to ACE2 to enter cells, it is now increasingly reported that it binds to other receptors such as neurophilin-1, (16), CD147 (17), tyrosine-protein kinase receptor UFO (AXL) (18, 19), ASGR1, KREMEN1 (20), and others. Viral genetic material activates TLR7, and there have been several reports linking TLR7 deficiency with severe outcomes in COVID-19 (21–23). Less well understood is the role that S-protein-induced inflammation and cell injury may play in this disease. There is increasing evidence that the S-protein can activate innate immunity, including Toll-like receptors TLR2 and TLR4 (24–27). In addition, instillation of the SARS-CoV-2 protein in K18-hACE2 mice caused marked inflammatory response and pulmonary edema (28). Thus, it is important to understand the inflammatory effect of S1 on epithelial cells, independent of viral replication, if we are to grasp the full spectrum of SARS-CoV-2 effects on mammalian biology.
Hyaluronan or hyaluronic acid (HA) is a linear polymer formed by a repeating disaccharide structure of glucuronic acid and N-acetyl-glucosamine consisting of up to 25,000 disaccharide units (MW = 1–10 million Da). High molecular weight hyaluronan (HMW-HA) is a major structural component of the extracellular matrix; it promotes cell survival, has antiangiogenic properties, and anti-inflammatory effects on immune cells, some of these mediated via binding to its receptors CD44, TLR2, and TLR4 (29–32).
Dysregulation of HA expression is relevant in disease and injury (33). Reactive species fracture HMW-HA to lower fragments (LMW-HA ∼300 kDa) that act as endogenous innate immune ligand, and promote inflammatory responses, angiogenesis, and epithelial to mesenchymal transition (30, 31, 33–35). LMW-HA increases vascular permeability by activating RhoA and ROCK (its downstream kinase), inducing cytoskeletal reorganization and inhibiting cell-cell contacts (36, 37). LMW-HA is increased in severe COVID-19 (38–40). On the contrary, administration of HMW-HA protects from the development of lung injury in ozone exposure (41), bleomycin administration (42), smoke inhalation, and sepsis (43, 44) and has been used in the treatment of COVID-19 (clinicaltrials.gov, NCT04830020). A pharmaceutical preparation of HMW-HA (Yabro; ∼1,000 kDa) has been shown to prevent exercise-induced bronchoconstriction in patients with asthma (45). No protective effects were seen following administration of LMW-HA (46). Several studies indicate that the relevant factor for HMW-HA protection is their size and not the source from which HMW-HA is derived (31, 43, 44). Thus, HMW-HA can be a useful anti-inflammatory agent in SARS-CoV-2-induced cell injury and inflammation. To better understand both organ-specific and global effects of COVID-19, in addition to better drug targets, a growing number of bioinformatics studies include the mining of public data repositories containing –omics datasets derived from patients with COVID-19 and translational models (13, 14).
Herein, we used an unsupervised global proteomics analysis followed by systems biology analysis to identify changes in the proteome of an ATII-like rat lung epithelial cell line (L2) following incubation with SARS-CoV-2 Spike S1 recombinant protein, independent of the S-protein-ACE2 interaction, since rat ACE2 has very low affinity to S1. AT2 cells, even though less numerous than AT1 cells, play a crucial role in lung physiology, as surfactant producers, orchestrators of the immune response, and AT1 progenitors. AT2 cells also stabilize the alveolar-capillary barrier and are involved in lung fluid homeostasis and prevention of pulmonary edema through sodium transport (47). It is also believed that the inflammatory response in ATII cells plays an outsized role in COVID-19 and that ATII cells may be the main target of SARS-CoV-2 and a major hub for severe COVID-19 development (48). Therefore, we believe that our choice of L2 cells make sense in the biological context of severe COVID-19. Our findings showed the S1 treatment alone resulted in significant modification to the rat L2 cellular proteome. Network analysis indicated increased activation of E2F1, CREB1, and RhoA/ROCK2 networks, which were confirmed by Western blot studies. Conversely, preincubation of L2 cells with HMW-HA before addition of S1 prevented these changes. Previous protein-protein interaction studies along with analysis of differentially expressed genes identified within RNA sequencing datasets from patients with COVID-19 highlighted the transcription factor E2F1 as an important player (49).
MATERIALS AND METHODS
Materials
SARS-CoV-2 (2019-nCoV) Spike S1-His recombinant protein (Cat. No 40591-V08H) was purchased from Sino Biological Inc. Yabro (pure, pharmaceutical-grade HMWHA, MW, 800,000–1,200,000, at a concentration of 3 mg/mL) was provided by IBSA Farmaceutici Italia srl, Italy. The chemical reagents dithiothreitol (Cat. No. D9779) and iodoacetamide (Cat. No. I1149) were purchased from Sigma-Aldrich (St. Louis, MO). Acetonitrile (ACN) was purchased from Thermo Fisher Scientific (Cat. No. A996SK; St. Louis, MO). All other disposables are referenced within the manuscript.
Cell Culture and Exposure to S1 Protein
Rat lung type II-like epithelial cells (L2) were provided to us by Dr. Judith Creighton (deceased). Their origin was characterized and verified using the presence of human and rat genes using PCR, immunohistochemistry, and Western blots of appropriate markers (Ref. 50; Supplemental Fig. S1). Cells were cultured in DMEM media supplemented with 10% FBS and 1% antibiotics in a humidified incubator with 95% air and 5% CO2 at 37°C until they became confluent as determined by light microscopy examination. For the experiment, cells were seeded in six-well plates at 5 × 104 cells/cm2 and incubated at 37°C until they became confluent in a humidified atmosphere of 95% air and 5% CO2. On the day of the experiment, some wells were pretreated with HMW-HA (200 ng/mL) for 1 h, and after that, cells were treated with S1 protein (100 ng/0.5 mL) for 24 h, in the presence of HMW-HA, at which time they were used for various experiments.
Proteomics Analysis
Proteomics analysis was carried out as previously described (51). The protein fractions of L2 cells were quantified using an 8-point Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Cat. No. PI23225) and 20 µg of protein per sample diluted to 35 µL using NuPAGE LDS sample buffer (1x final conc., Invitrogen, Cat. No. NP0007). Proteins were then reduced with dithiothreitol (DTT) and denatured at 70°C for 10 min before loading onto Novex NuPAGE 10% Bis-Tris Protein gels (Invitrogen, Cat. No. NP0315BOX) and separated half way (15 min at 200 constant voltage). The gels were stained overnight with Novex Colloidal Blue Staining kit (Invitrogen, Cat. No. LC6025). Following destaining, each lane was partitioned into three separate molecular weight fractions and equilibrated in 100 mM ammonium bicarbonate (Millipore SIGMA; CAS No. 1066-33-7). Each gel plug was then digested overnight with Trypsin Gold, Mass Spectrometry Grade (Promega, Cat. No. V5280) following manufacturer’s instruction. Peptide extracts were reconstituted in 0.1% formic acid and double distilled H2O at 0.1 µg/µL.
nLC-ESI-MS2 Analysis and Database Searches
Peptide digests (8 µL each) were injected onto a 1260 Infinity nHPLC stack (Agilent Technologies) and separated using a 100 µm ID × 13.5 cm pulled tip C-18 column (Jupiter C-18 300 Å, 5 micron, Phenomenex, Torrance, CA). This system runs in-line with a Thermo Orbitrap Velos Pro hybrid mass spectrometer, equipped with a nanoelectrospray source (Thermo Fisher Scientific), and all data were collected in collision-induced dissociation (CID) mode. The nHPLC was configured with binary mobile phases that included solvent A [0.1% formic acid (FA) in double distilled H2O] and solvent B (0.1% FA in 15% double distilled H2O-85% acetonitrile), programmed as follows: 10 min @ 5% solvent B (2 µL/min, load), 90 min @ 5%–40% solvent B (linear: 0.5nL/min, analyze), 5 min @ 70% solvent B (2 µL/min, wash), and 10 min @ 0% solvent B (2 µL/min, equilibrate). Following each parent ion scan (300–1,200 m/z @ 60k resolution), fragmentation data (MS2) were collected on the top most intense 15 ions. For data-dependent scans, charge state screening and dynamic exclusion were enabled with a repeat count of two, repeat duration of 30 s, and exclusion duration of 90 s.
The XCalibur*.raw files were collected in profile mode, centroided, and converted to MzXML using ReAdW v. 3.5.1. The data were searched using SEQUEST, which was set for two maximum missed cleavages, a precursor mass window of 20 ppm, trypsin digestion and variable modifications of cysteine residues involved in disulfide bridges capped/alkylated by iodoacetamide treatment in in-gel digestion process, resulting in carbamidomethylation with 57.0293 Da shift and methionine residues oxidized with a mass shift of 15.9949 Da. Searches were performed separately with both human and rat species-specific subsets of the UniProtKB databases. Following this analysis, we used the high-confidence Rattus norvegicus to human tryptic peptide homolog data to avoid confusion for downstream Systems Analysis.
Peptide Filtering, Grouping, and Quantification
The list of peptide IDs generated based on SEQUEST (Thermo Fisher Scientific) search results were filtered using Scaffold (Protein Sciences, Portland, OR). Scaffold filters and groups all peptides to generate and retain only high-confidence IDs while also generating normalized spectral counts (N-SC’s) across all samples for the purpose of relative quantification. The filter cut-off values were set with minimum peptide length of >five amino acids (AAs), with no (m/z + 1) charge states, with peptide probabilities of >80% confidence intervals (CIs), and with the number of peptides per protein ≥2. The protein probabilities were then set to a >99.0% CI and a false discovery rate (FDR) <1.0. Scaffold incorporates the two most common methods for statistical validation of large proteome datasets, the false discovery rate (FDR), and protein probability (52–54). Relative quantification across experiments were then performed via spectral counting (55), and when relevant, spectral count abundances were then normalized between samples (56).
Statistical and Systems Biology Analysis
For the proteomic data generated, two separate nonparametric statistical analyses were performed between each pair-wise comparison. These nonparametric analyses include 1) the calculation of weight values by significance analysis of microarray (SAM; cutoff >|0.6|) combined with 2) t test (single tail, unequal variance, cutoff of P < 0.05), which then were sorted according to the highest statistical relevance in each comparison. For SAM (57), whereby the weight value (W) is a statistically derived function that approaches significance as the distance between the means (μ1 − μ2) for each group increases, and the SD (δ1 − δ2) decreases using the formula, W = (μ1 − μ2)/(δ1 − δ2). For protein abundance ratios determined with N-SC’s, we set a 1.5–2.0 fold change as the threshold for significance, determined empirically by analyzing the innerquartile data from the control experiment indicated above using ln-ln plots, where the Pierson’s correlation coefficient (R) was 0.98, and >99% of the normalized intensities fell between ±1.5 fold. In each case, any two of the three tests (SAM, t test, or fold change) had to be satisfied.
Gene ontology assignments and pathway analysis were carried out using MetaCore (GeneGO Inc., St. Joseph, MI). Interactions identified within MetaCore are manually correlated using full-text articles. Detailed algorithms have been described previously (58).
Measurement of RhoA Activity
The Rho activity was measured and quantified by using the RhoA Activation Assay Biochem Kit (Cat. No. BK036; Cytoskeleton) based on the Rhotekin pull-down assay as per the manufacturer’s instructions as described previously (36). In brief, L2 cells (n = 10) were cultured in six-well culture plates and few wells were pretreated with HMW-HA (200 ng/mL) for 1 h and then treated with or without spike S1 (100 ng) protein for 30 min at 37°C. The cell lysate was prepared in radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (Cat. No. 78425; Thermo Fisher Scientific) immediately after treatment and protein estimation was done, and equal amounts of protein were incubated with Rhotekin-RBD beads (Cat. No. RT02) for 1 h at 4°C. After the beads were washed with wash buffer, proteins were removed from the beads with Laemmli buffer and then subjected to Western blotting.
Measurement of E2F1 and ROCK2 Phosphorylation
In brief, L2 cells (n = 10) were cultured in six-well culture plates and few wells were pretreated with HMW-HA (200 ng/mL) for 1 h and then treated with or without spike S1 (200 ng/mL; 100 ng total) protein for 24 h at 37°C. The cell lysates were prepared in RIPA buffer containing protease inhibitors (Thermo Fisher Scientific, Rockford, IL) immediately after treatment. Protein estimation was carried out using the BCA assay. Equal amounts of protein in Laemmli buffer were loaded in 4%–20% gradient Tris·HCl Criterion precast gels, and proteins were transferred to PVDF membranes. Membranes were probed with p-E2F1 (1:1,000 dilution; Cat. No. MABE1782; MilliporeSigma) or E2F1 (1:1,000 dilution; Cat. No. 3742S; Cell Signaling Technology, Beverly, MA) and ROCK2 (1:1,000 dilution; Cat. No. 8236; Cell Signaling Technology, Beverly, MA) or pTyr722-ROCK2 antibody (Cat. No. SAB4301564; MilliporeSigma). Bands were detected by the chemiluminescent horseradish peroxidase substrate. Protein loading was normalized by reprobing the membranes with an antibody specific to β-actin.
Cell Cycle Analysis by Flow Cytometry
Cell cycle was analyzed using staining with DAPI nucleic acid stain (ThermoFisher, Waltham, MA) in cells following dissociation and fixation in single-cell suspension. DAPI fluorescence was detected using flow cytometry in the UAB Comprehensive Flow Cytometry Core facility using an LSR II Flow Cytometer (Beckton Dickinson, Franklin Lakes, NJ) and cell cycle was determined based on nucleic acid content/cell. Cells associated with the peak at the lower fluorescence intensity were deemed to be G0/G1 cells, cells in the peak at the higher fluorescence were deemed G2M phase, and cells with intermediate fluorescence were determined to be S phase.
Statistical Analysis for Nonproteomics Studies
Figures were generated and statistics were performed using GraphPad Prism version 8 for Windows (GraphPad Software, San Diego, CA). The mean ± SEM were calculated in all experiments, and statistical significance was determined by one-way ANOVA followed by the Tukey’s multiple comparisons test.
Pathway Associations with Human Cell Lines Exposed to SARS-CoV-2
To determine whether the associated functions observed in the S1-Tx proteomics L2 cells are correlated with the transcriptomic data, the RNA-seq transcriptomic data of five human lung cell lines that were exposed to SARS-CoV-2 were downloaded from public domain (Supplemental Table S2). The Gene Set Enrichment Analysis (GSEA v4.0.3; https://www.gsea-msigdb.org/gsea/index.jsp) was conducted against the Hallmark and C2 curated canonical pathways datasets in the Molecular Signatures Database (MSigDB v7.4) with 1,000 permutations (min size 15, max size 500). Gene sets with adjusted FDR q-value less than 0.25 were considered as significant.
RESULTS
Spike S1 Effects on the ATII-Like Rat L2 Proteome
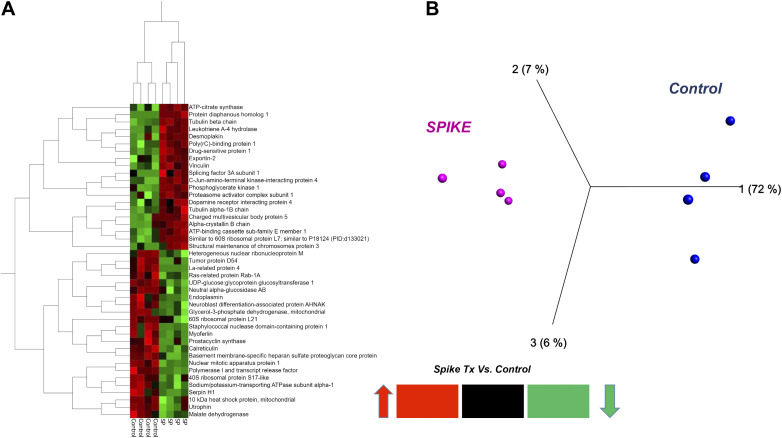
We tested whether treatment of ATII-like rat L2 cells with SARS-CoV-2 Spike S1-recombinant protein for 24 h causes significant changes to its proteome (Tx-S1 vs. vehicle control/C, n = 4). The proteomics analysis revealed 1,678 altered proteins, 730 of which had <1% false discovery rate (FDR) with at least 3 out of 4 replicates measuring nonzero values across all groups tested (Supplemental Table S1). Ninety-nine proteins were significantly changed in abundance by S1 treatment, 51 of which increased (Table 1) and 48 decreased (Table 2). This dataset was best visualized using a volcano plot (Fig. 1), in addition to a two-dimensional (2-D) hierarchical heat map (HCA-HM, Fig. 2A) followed by principal component analysis (PCA, Fig. 2B). All statistical analysis and visualizations were carried out using the data matrix found in Supplemental Table S1.
Table 1.
List of 51 proteins of rat L2 cells that were increased significantly 24 h post-S1
| Statistics |
Metacore Definitions |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| UniProtKB Name | UniProt ID | Entrez ID | Network ID | Sam | t-Test (P Value) | Fold Δ (S1/C) | Object Type | Fig. 1 | Fig. 5 |
| Protein diaphanous homolog 1 | O60610 | 1729 | DIA1 | 2.67 | 0.00548 | 3.4 | Generic binding protein | X | X |
| Structural maintenance of chromosomes protein 3 | Q9UQE7 | 9126 | SMC3 | 1.18 | 0.02235 | 3.2 | Generic binding protein | X | |
| Leukotriene A-4 hydrolase | P09960 | 4048 | LTA4H | 1.42 | 0.01370 | 3.2 | Generic enzyme | ||
| ATP-citrate synthase | P53396 | 47 | ACLY | 1.57 | 0.00275 | 3.0 | Generic enzyme | X | |
| C-Jun-amino-terminal kinase-interacting protein 4 | O60271 | 9043 | SPAG9 | 1.82 | 0.00251 | 2.7 | Generic binding protein | ||
| Splicing factor 3 A subunit 1 | Q15459 | 10291 | SF3A1 | 1.00 | 0.03406 | 2.7 | Generic binding protein | X | |
| Alcohol dehydrogenase [NADP(+)] | P14550 | 10327 | ALDX | 0.79 | 0.05316 | 2.7 | Generic enzyme | ||
| Poly(rC)-binding protein 1 | Q15365 | 5093 | PCBP-1 | 1.55 | 0.00584 | 2.5 | Generic binding protein | X | X |
| Vacuolar protein sorting-associated protein 26 A | O75436 | 9559 | Vps26A | 1.16 | 0.08734 | 2.5 | Generic binding protein | X | |
| α-Crystallin B chain | P02511 | 1410 | α Crystallin B | 1.32 | 0.00562 | 2.4 | Generic binding protein | X | |
| Dopamine receptor interacting protein 4 | Q4W4Y1 | 1.12 | 0.03228 | 2.3 | |||||
| Rab GDP dissociation inhibitor α | P31150 | 2664 | RABGDIA | 1.64 | 0.01578 | 2.3 | Regulators (GDI, GAP, GEF) | X | X |
| Protein DJ-1 | K7ELW0 | 11315 | DJ-1 | 0.98 | 0.01989 | 2.2 | Generic enzyme | X | |
| ATP-binding cassette subfamily E member 1 | P61221 | 6059 | RLI | 1.35 | 0.00696 | 2.2 | Generic binding protein | X | |
| Galectin-1 | P09382 | 3956 | Galectin-1 | 0.81 | 0.07362 | 2.2 | Receptor ligand | ||
| Exportin-2 | P55060 | 1434 | CSE1L | 1.18 | 0.00793 | 2.2 | Transporter | X | |
| Phosphoglycerate kinase 1 | P00558 | 5230 | PGK1 | 1.93 | 0.00097 | 2.2 | Generic kinase | X | X |
| Protein FAM49B | Q9NUQ9 | 51571 | FAM49B | 0.95 | 0.07184 | 2.1 | Protein | X | |
| S-phase kinase-associated protein 1 | P63208 | 6500 | SKP1 | 0.85 | 0.06385 | 2.1 | Generic binding protein | ||
| Adapter molecule crk | P46108 | 1398 | CRK | 1.19 | 0.00888 | 2.1 | Generic binding protein | ||
| Similar to 60S ribosomal protein L7 | O95036 | 2.11 | 0.00063 | 2.1 | X | ||||
| Vinculin | P18206 | 7414 | Vinculin | 1.33 | 0.00801 | 2.0 | Generic binding protein | X | |
| Heat shock protein β-1 | P04792 | 3315 | HSP27 | 0.79 | 0.07920 | 2.0 | Generic binding protein | X | |
| Rab GDP dissociation inhibitor β | P50395 | 2665 | GDI2 | 0.81 | 0.05170 | 2.0 | Generic binding protein | X | |
| Ephrin type-A receptor 2 | P29317 | 1969 | Ephrin-A receptor 2 | 0.84 | 0.04238 | 2.0 | Receptor with enzyme activity | X | |
| Transgelin-2 | P37802 | 8407 | TAGLN2 | 1.02 | 0.02116 | 2.0 | Protein | ||
| Proteasome subunit β type-5 | P28074 | 5693 | PSMB5 | 0.91 | 0.03810 | 2.0 | Generic protease | ||
| S-methyl-5′-thioadenosine phosphorylase | Q13126 | 4507 | MTAP | 0.77 | 0.04806 | 2.0 | Generic enzyme | ||
| Golgin subfamily A member 3 | Q08378 | 2802 | Golgin-160 | 1.11 | 0.05072 | 1.9 | Generic binding protein | X | |
| Regulator of nonsense transcripts 1 | Q92900 | 5976 | RENT1 | 0.79 | 0.04820 | 1.9 | Generic binding protein | X | |
| 26S proteasome non-ATPase regulatory subunit 12 | O00232 | 5718 | PSMD12 | 1.33 | 0.01118 | 1.9 | Generic binding protein | X | |
| Fructose-bisphosphate aldolase C | P09972 | 230 | ALDOC | 1.50 | 0.01419 | 1.8 | Generic enzyme | X | X |
| Pyruvate dehydrogenase E1 component subunit β | P11177 | 5162 | PDH | 0.82 | 0.03006 | 1.8 | Generic enzyme | X | |
| 60S ribosomal protein L6 | Q02878 | 6128 | RPL6 | 0.98 | 0.01818 | 1.8 | Generic binding protein | X | |
| Peroxiredoxin-6 | P30041 | 9588 | NSGPeroxidase | 0.74 | 0.04281 | 1.8 | Generic phospholipase | ||
| COP9 signalosome complex subunit 4 | Q9BT78 | 51138 | COPS4 | 0.81 | 0.04235 | 1.8 | Generic binding protein | X | |
| Heat shock 70 kDa protein 4 | P34932 | 3308 | HSP70 | 0.95 | 0.02048 | 1.7 | Generic binding protein | X | |
| Peptidyl-prolyl cis-trans isomerase D | Q08752 | 5481 | PPID | 0.83 | 0.05670 | 1.7 | Generic enzyme | ||
| 40S ribosomal protein S26 | P62854 | 6231 | RPS26 | 0.95 | 0.02206 | 1.7 | Generic binding protein | X | |
| Heat shock protein 105 kDa | Q92598 | 10808 | HSP105 | 0.90 | 0.06162 | 1.7 | Generic binding protein | X | |
| Drug-sensitive protein 1 | Q9NZ23 | 1.77 | 0.00557 | 1.7 | X | ||||
| Desmoplakin | P15924 | 1832 | Desmoplakin | 1.33 | 0.01192 | 1.7 | Generic binding protein | X | |
| Actin-related protein 2/3 complex subunit 4 | P59998 | 10093 | ARPC4 | 0.97 | 0.03172 | 1.7 | Generic binding protein | ||
| Transcription elongation factor B polypeptide 2 | Q15370 | 6923 | Elongin B | 0.84 | 0.08657 | 1.6 | Generic binding protein | X | |
| Proteasome subunit α type-4 | P25789 | 5685 | PSMA4 | 0.94 | 0.04583 | 1.6 | Generic protease | ||
| Elongation factor 1-α 1 | P68104 | 1915 | eEF1A | 0.86 | 0.02515 | 1.6 | Generic binding protein | X | |
| Plectin | Q15149 | 5339 | Plectin 1 | 0.94 | 0.01884 | 1.5 | Generic binding protein | X | |
| Tubulin β-2A chain | Q13885 | 7280 | Tubulin β | 1.86 | 0.00221 | 1.5 | Generic binding protein | X | |
| Kinesin-1 heavy chain | P33176 | 3799 | KIF5B | 0.78 | 0.03425 | 1.5 | Generic binding protein | X | |
| 60S ribosomal protein L5 | P46777 | 6125 | RPL5 | 1.07 | 0.02048 | 1.5 | Generic binding protein | X | X |
Table 2.
List of 48 proteins of rat L2 cells that were significantly decreased 24 h post-S1
| Statistics |
Metacore Definition |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| UniProtKB Name | UniProt ID | Entrez ID | Network ID | SAM | t Test (P Value) | Fold Δ (S1/C) | Object Type | Fig. 1 | Fig. 5 |
| Trifunctional enzyme subunit α, mitochondrial | P40939 | 3030 | HADHA | 1.18 | 0.06945 | −4.6 | Generic enzyme | X | |
| Basement membrane-specific heparan sulfate proteoglycan core protein | P98160 | 3339 | Perlecan | 1.54 | 0.00765 | −3.7 | Generic binding protein | X | |
| Nuclear mitotic apparatus protein 1 | Q14980 | 4926 | NUMA1 | 1.99 | 0.00737 | −3.2 | Generic binding protein | ||
| Signal recognition particle 54 kDa protein | P61011 | 6729 | SRP54 | 1.26 | 0.04139 | −3.0 | Generic binding protein | X | |
| Ras-related protein Rab-5B | P61020 | 5869 | RAB-5B | 0.97 | 0.02599 | −2.8 | RAS - superfamily | X | X |
| Clathrin light chain A | P09496 | 1211 | Clathrin light chain | 0.96 | 0.04513 | −2.6 | Generic binding protein | ||
| NAD-dependent malic enzyme, mitochondrial | P23368 | 4200 | ME2 | 0.86 | 0.03056 | −2.4 | Generic enzyme | ||
| Glycerol-3-phosphate dehydrogenase, mitochondrial | P43304 | 2820 | GPD2 | 1.33 | 0.01184 | −2.3 | Generic enzyme | ||
| Pyridoxal-dependent decarboxylase domain-containing protein 1 | Q6P996 | 23042 | LP8165 | 1.42 | 0.03052 | −2.3 | Generic binding protein | X | |
| Heterogeneous nuclear ribonucleoprotein M | P52272 | 4670 | hnRNP M | 1.36 | 0.00843 | −2.3 | Generic binding protein | X | |
| Prostacyclin synthase | Q16647 | 5740 | PTGIS | 0.88 | 0.04089 | −2.3 | Generic enzyme | X | |
| Tumor protein D54 | O43399 | 7165 | D54 | 1.16 | 0.01435 | −2.3 | Generic binding protein | X | X |
| Nucleobindin-1 | Q02818 | 4924 | Nucb1 | 0.98 | 0.03256 | −2.3 | Generic binding protein | ||
| Vacuolar protein sorting-associated protein 13 A | Q96RL7 | 23230 | VPS13A | 1.28 | 0.04082 | −2.3 | Transporter | X | |
| Cleavage and polyadenylation specificity factor subunit 5 | O43809 | 11051 | CPSF5 | 1.02 | 0.03985 | −2.2 | Generic enzyme | X | |
| Pre-mRNA-processing factor 19 | Q9UMS4 | 27339 | NMP200 | 1.24 | 0.00673 | −2.2 | Generic binding protein | X | X |
| La-related protein 4 | Q71RC2 | 113251 | LARP4 | 2.21 | 0.00918 | −2.2 | Generic binding protein | X | |
| Lanosterol 14-α demethylase | Q16850 | 1595 | CYP51A1 | 0.79 | 0.05002 | −2.1 | Generic enzyme | X | |
| RNA-binding protein FUS | P35637 | 2521 | FUS | 0.91 | 0.04299 | −2.1 | Generic binding protein | X | |
| 10 kDa heat shock protein, mitochondrial | P61604 | 3336 | HSP10 (mitochondrial) | 1.27 | 0.00704 | −2.0 | Generic binding protein | X | |
| UDP-glucose:glycoprotein glucosyltransferase 1 | Q9NYU2 | 56886 | UGCGL1 | 1.89 | 0.00092 | −2.0 | Generic enzyme | ||
| Hypoxia upregulated protein 1 | Q9Y4L1 | 10525 | HYOU1 | 0.91 | 0.02828 | −2.0 | Generic binding protein | X | |
| Staphylococcal nuclease domain-containing protein 1 | Q7KZF4 | 27044 | SND1 | 1.80 | 0.01223 | −2.0 | Generic binding protein | X | |
| ELAV-like protein 1 | Q15717 | 1994 | ELAVL1 (HuR) | 0.93 | 0.02877 | −2.0 | Generic binding protein | X | |
| B-cell receptor-associated protein 31 | P51572 | 10134 | BCAP31 | 1.02 | 0.02750 | −1.9 | Generic binding protein | ||
| 26S proteasome non-ATPase regulatory subunit 7 | P51665 | 5713 | PSMD7 | 0.79 | 0.04622 | −1.9 | Generic binding protein | X | |
| Elongation factor Tu, mitochondrial | P49411 | 7284 | TUFM | 0.85 | 0.02811 | −1.9 | Generic binding protein | ||
| Prolyl 4-hydroxylase subunit α-2 | O15460 | 8974 | P4HA | 0.82 | 0.03704 | −1.9 | Generic enzyme | X | |
| P37 AUF1 | Q12771 | 0.71 | 0.04582 | −1.8 | |||||
| Ornithine aminotransferase, mitochondrial | P04181 | 4942 | OAT | 0.81 | 0.05087 | −1.8 | Generic enzyme | ||
| Actin-related protein 2/3 complex subunit 1B | O15143 | 10095 | ARPC1 | 0.93 | 0.01943 | −1.7 | Generic binding protein | X | |
| Utrophin | P46939 | 7402 | Utrophin | 1.48 | 0.00458 | −1.7 | Generic binding protein | X | X |
| Vesicle-associated membrane protein-associated protein A | Q9P0L0 | 9218 | VAPA | 1.11 | 0.01762 | −1.7 | Generic binding protein | X | |
| Ras-related protein Rab-1A | P62820 | 5861 | Rab-1A | 1.08 | 0.01950 | −1.7 | RAS - superfamily | ||
| Calnexin | P27824 | 821 | Calnexin | 0.79 | 0.04528 | −1.7 | Generic binding protein | X | |
| α-Aminoadipic semialdehyde dehydrogenase | P49419 | 501 | AL7A1 | 1.82 | 0.03742 | −1.7 | Generic enzyme | X | |
| Ras-related protein Rab-1B | Q9H0U4 | 81876 | Rab-1B | 0.99 | 0.01658 | −1.7 | RAS - superfamily | ||
| Calreticulin | P27797 | 811 | Calreticulin | 1.55 | 0.00609 | −1.7 | Generic binding protein | X | |
| Polymerase I and transcript release factor | Q6NZI2 | 284119 | PTRF | 1.73 | 0.01208 | −1.7 | Generic binding protein | X | X |
| Neutral α-glucosidase AB | Q14697 | 23193 | GANAB | 1.30 | 0.00599 | −1.6 | Generic enzyme | ||
| Pyruvate dehydrogenase E1 component subunit α | P08559 | 5160 | PDH | 0.88 | 0.07592 | −1.6 | Generic enzyme | ||
| Coatomer subunit α | P53621 | 1314 | COPA | 1.06 | 0.01301 | −1.6 | Generic binding protein | X | X |
| Heterogeneous nuclear ribonucleoprotein F | P52597 | 3185 | hnRNP F | 0.75 | 0.04082 | −1.6 | Generic binding protein | ||
| Serpin H1 | P50454 | 871 | HSP47 | 1.25 | 0.02301 | −1.6 | Generic binding protein | X | |
| Nischarin | Q9Y2I1 | 11188 | IRAS | 1.03 | 0.02231 | −1.6 | Generic binding protein | ||
| Neuroblast differentiation-associated protein AHNAK | Q09666 | 79026 | AHNAK | 1.38 | 0.00585 | −1.6 | Generic binding protein | X | |
| Endoplasmin | P14625 | 7184 | Endoplasmin | 1.21 | 0.02263 | −1.5 | Generic binding protein | X | |
| 60 kDa heat shock protein, mitochondrial | P10809 | 3329 | HSP60 | 1.05 | 0.03181 | −1.5 | Generic binding protein | ||
| Malate dehydrogenase | Q6FHZ0 | 4191 | MDH2 | 1.15 | 0.01381 | −1.5 | Generic enzyme | X | |
X denotes whether a protein was included in the indicated analysis and whether its decrease by S1 was prevented by pretreatment of L2 cells with HMW-HA. HA, hyaluronan (hyaluronic acid); HMW, high molecular weight.
Figure 1.
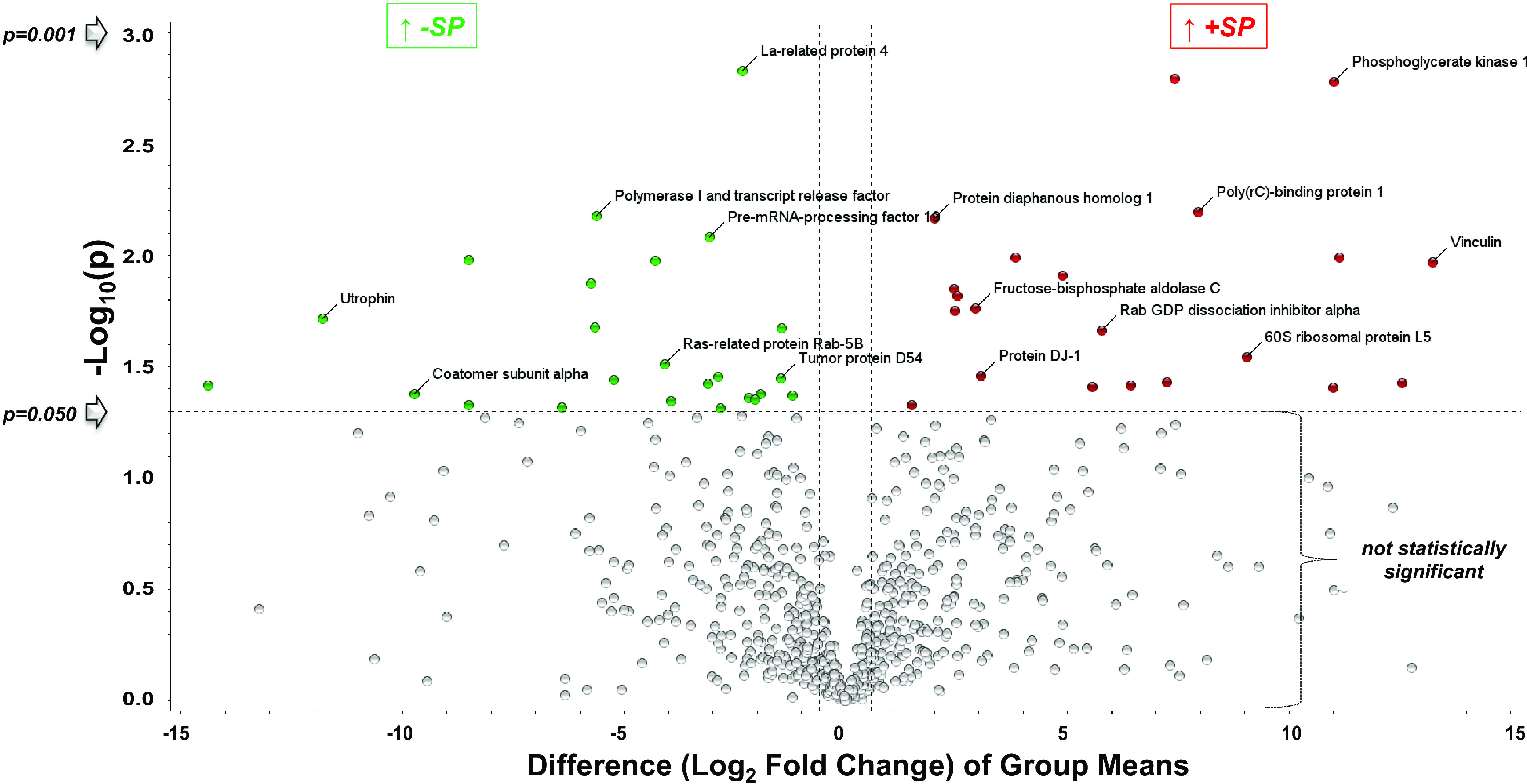
Volcano plot of proteins increased or decreased in abundance following addition of S1 vs. vehicle 24 h later. Ninety-nine proteins were significantly changed in abundance by S1 treatment, 51 of which increased (Table 1), and 48 decreased (Table 2). The y-axis is the negative log (base 10) of the P value. This results in highly significant points with low P values appearing toward the top of the plot. The log of the fold change between the two conditions is plotted on the x-axis in base 2 (ex: Log2 Fold 35 = 5). Points found toward the top of the plot that are either on the left- or right-hand sides represent values that display large-magnitude fold changes as well as high statistical significance. Values are means of four different experiments.
Figure 2.
Two-dimensional (2-D) HCA heat map (A) and PCA plot (B) of the 36 most significantly changed proteins from rat L2 cells 24 h following S1 treatment vs. control. Each column in the HCA heat map and each circle in the PCA plot represent different experiments. HCA, hierarchical heat map; PCA, principal component analysis.
Spike S1 Associated Biological Processes and Pathways via Systems Biology
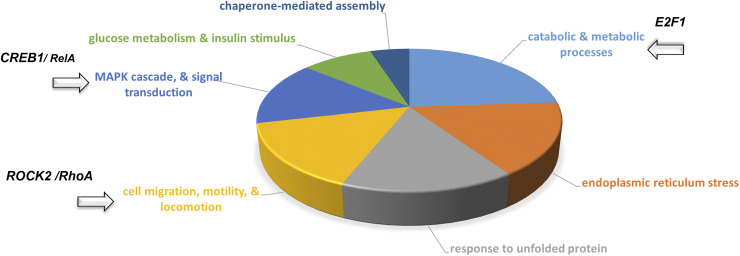
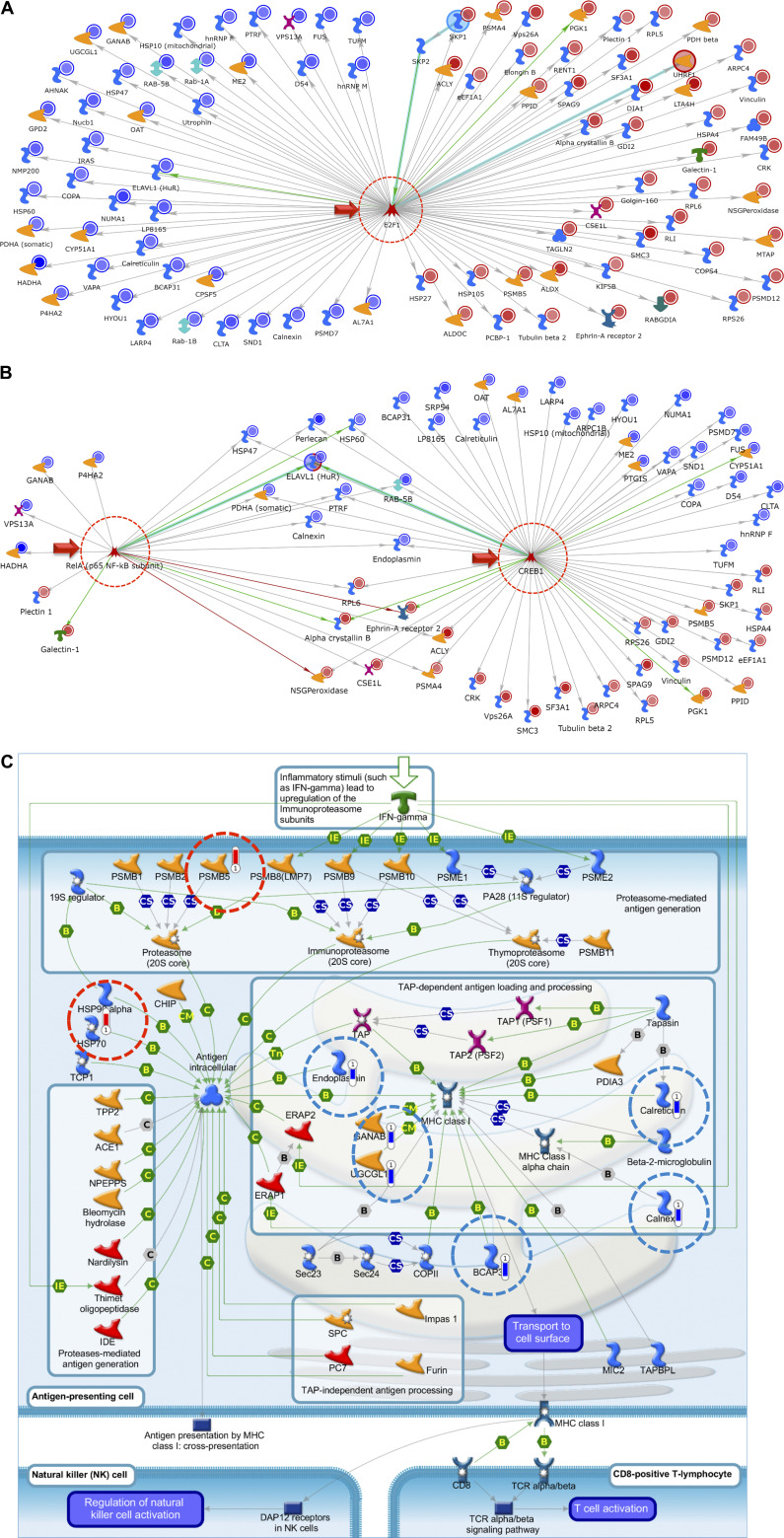
To identify key biological processes altered following treatment of L2 cells with S1 proteins, we performed in-depth systems biology analysis using the data listed in Tables 1 and 2. Based on the results of this analysis, we generated a pie chart to better visualize the more common Gene Ontology (GO)-based process networks that the most changed proteins are associated with (Fig. 3). This analysis suggests that treatment of L2 cells with the S1 protein alters numerous cellular functions, including catabolic and glucose metabolism, endoplasmic reticulum (ER) stress, cell migration, and signal transduction. We then performed unbiased network analysis using Metacore’s Systems Biology software (https://www.nihlibrary.nih.gov/about-us/news/metacore-enabling-systems-biology-research-through-pathway-analysis). This analysis identified three important network hubs that cross correlate with the E2F1 pathway (Fig. 4A), CREB/RelA (Fig. 4B), and ROCK2/RhoA (Table 3). To better understand the network and pathway figures, we have included a complete explanation of the various symbols in Supplemental Fig. S1 As illustrated in Fig. 4A, nearly the entire list of significantly changed proteins after S1 addition (listed as network IDs in Tables 1 and 2) is associated with E2F1 signaling. Similarly, as shown in Fig. 4B, both CREB1 and RelA transcription factors are highlighted as central network hubs. Further analysis illustrated in Table 3 highlights networks such as cell adhesion, cytoskeleton associated (actin filaments, rearrangement, spindle and cytoplasmic tubules), in addition to cell cycle mitosis and meiosis. Also of interest, there was an apparent pronounced effect on cellular MHC class I immune response as the top pathway (Fig. 4C).
Figure 3.
Top process networks pie chart from systems biology analysis carried out on the most significantly changed proteins (Tables 1 and 2), from rat L2 cells 24 h after treatment with SARS-CoV-2 S1 or vehicle. Highlighted segments contain proteins associated with ROCK2/RhoA (cell migration, motility, and locomotion), CREB1/RelA (MAPK cascade, signal transduction), and E2F1 (catabolic and metabolic processes).
Figure 4.
Top network map with the use of the 99 most significantly changed proteins at 24 h post-S1 identified in Tables 1 and 2, using Metacore’s Systems Biology software to carry out network analysis. E2F1 (A) and ReA/p65, NF-κB (B) were identified as a major centralized network hub. Nearly the entire list of proteins identified were shown to interact/associate in some way with E2F1 as a direct results of treatment of L2 cells with SARS-CoV-2 S1 protein for 24 h. Top network map for SPIKE S1 treatment; RelA and CREB1 presented as minor and major network hubs, respectively. Blue circles indicate a decrease, whereas red circles indicate an increase in protein abundance at 24 h postincubation with S1 as compared to vehicle. Detailed description of all symbols is shown in Supplemental Fig. S1. C: top pathway map for SPIKE S1 treatment: immune response-antigen presentation by major histocompatibility (MHC) class I, classical pathway. Blue bars indicate a decrease and red bars indicate an increase in protein abundance 24 h post-S1 incubation compared to vehicle control.
Table 3.
Enrichment by process networks (S1 treated vs. vehicle)
| # | Network | Total | P Value | FDR | In Data | Network Objects from Active Data |
|---|---|---|---|---|---|---|
| 1 | Protein folding_Response to unfolded proteins | 71 | 1.1E-11 | 8.8E-10 | 12 | HSP90, HSP27, HSP47, Endoplasmin, HSP70, HSP60, Calreticulin, HYOU1, HSP105, HSP10 (mitochondrial), HSPA4, Calnexin |
| 2 | Protein folding_Folding in normal condition | 119 | 5.5E-09 | 2.1E-07 | 12 | HSP90, HSP27, HSP47, α crystallin B, Endoplasmin, HSP70, HSP60, HYOU1, HSP105, HSP10 (mitochondrial), HSPA4, PPID |
| 3 | Protein folding_ER and cytoplasm | 44 | 4.4E-07 | 1.1E-05 | 7 | HSP90, HSP47, HSP70, Calreticulin, UGCGL1, HYOU1, Calnexin |
| 4 | Immune response_Phagosome in antigen presentation | 241 | 6.3E-05 | 1.2E-03 | 11 | HSP90, PSMA4, CRK, Endoplasmin, PSMD7, HSP70, Calreticulin, Vinculin, PSMB5, Calnexin, PSMD12 |
| 5 | Cytoskeleton_Regulation of cytoskeleton rearrangement (RhoA) | 183 | 1.8E-04 | 2.8E-03 | 9 | ARPC1B, CRK, ARPC1, Vinculin, ARPC4, Plectin 1, Tubulin β, DIA1, Tubulin β 2 |
| 6 | Cytoskeleton_Intermediate filaments (RhoA) | 81 | 2.6E-04 | 2.9E-03 | 6 | KIF5B, Kinesin heavy chain, Plectin 1, Tubulin β, Tubulin β 2, Desmoplakin |
| 7 | Immune response_Antigen presentation | 193 | 2.6E-04 | 2.9E-03 | 9 | HSP90, PSMA4, Endoplasmin, PSMD7, HSP70, Calreticulin, PSMB5, Calnexin, PSMD12 |
| 8 | Cytoskeleton_Actin filaments (RhoA) | 176 | 7.0E-04 | 6.8E-03 | 8 | Utrophin, ARPC1B, CRK, ARPC1, Vinculin, ARPC4, Plectin 1, DIA1 |
| 9 | Proteolysis_Ubiquitin-proteasomal proteolysis | 166 | 2.3E-03 | 2.0E-02 | 7 | HSP90, PSMA4, PSMD7, HSP70, PSMB5, PSMD12, Elongin B |
| 10 | Protein folding_Protein folding nucleus | 58 | 3.8E-03 | 3.0E-02 | 4 | HSP90, HSP70, HYOU1, HSPA4 |
| 11 | Transcription_mRNA processing | 160 | 8.5E-03 | 6.0E-02 | 6 | PCBP-1, SF3A1, hnRNP M, CPSF5, NMP200, hnRNP F |
| 12 | Cell adhesion_Integrin-mediated cell-matrix adhesion (RhoA) | 214 | 9.4E-03 | 6.1E-02 | 7 | Galectin-1, CRK, Vinculin, Tubulin β, DIA1, Tubulin β 2, IRAS |
| 13 | Signal transduction_Insulin signaling | 171 | 4.1E-02 | 2.4E-01 | 5 | PDH, PDH β, ACLY, PDH α, PDHA (somatic) |
| 14 | Translation_Elongation-Termination | 232 | 4.3E-02 | 2.4E-01 | 6 | eEF1A, RPS26, eEF1A1, SRP54, RPL6, RPL5 |
| 15 | Cell cycle_Mitosis | 179 | 4.8E-02 | 2.5E-01 | 5 | NUMA1, CRK, Tubulin β, SMC3, CSE1L |
| 16 | Cell cycle_Meiosis | 106 | 1.1E-01 | 5.3E-01 | 3 | HSP70, Tubulin β, SMC3 |
| 17 | Cytoskeleton_Spindle microtubules (RhoA) | 109 | 1.2E-01 | 5.3E-01 | 3 | NUMA1, Tubulin β, SMC3 |
| 18 | Translation_Translation initiation | 171 | 1.2E-01 | 5.3E-01 | 4 | RPS26, RPL6, RENT1, RPL5 |
| 19 | Cytoskeleton_Cytoplasmic microtubules (RhoA) | 115 | 1.4E-01 | 5.6E-01 | 3 | KIF5B, Kinesin heavy chain, Tubulin β |
| 20 | Signal transduction_Androgen receptor nuclear signaling | 126 | 1.6E-01 | 6.4E-01 | 3 | HSP90, Calreticulin, ELAVL1 (HuR) |
| 21 | Apoptosis_Endoplasmic reticulum stress pathway | 89 | 2.6E-01 | 9.3E-01 | 2 | Endoplasmin, BCAP31 |
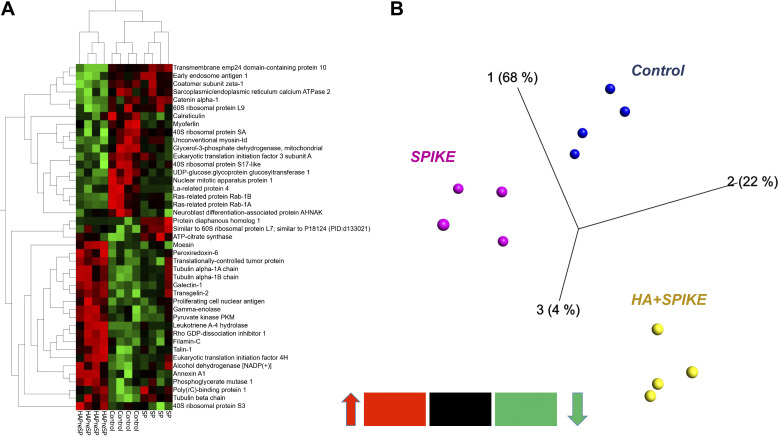
HMW-HA Pretreatment Attenuates Spike S1-Induced Changes to the Proteome of Rat L2 Cells
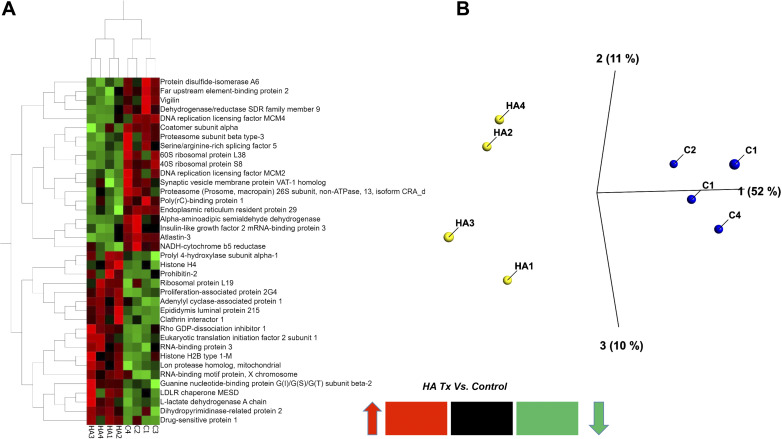
As shown in the heat map of Fig. 5A, incubation of L2 cells with HMW-HA for 1 h before the addition of S1 reversed most of the S1 changes to their proteome. PCA analysis of the data from L2 cells treated with vehicle, S1, and preincubated with HMW-HA showed tight clustering and clear separation among groups (Fig. 5B).
Figure 5.
Heat map (A) and three-dimensional (3-D) PCA plot of control (vehicle; B) vs. S1 (SPIKE), and HMW-HA pretreatment prior to S1 treatment of rat L2 cells, stemming from the most significant protein differences by ANOVA (analysis of variance) using the data matrix from Table 4. Each column represents a different experiment. Note the tight grouping of the various experiments in the 3-D PCA plot (each point represents a different experiment). HA, hyaluronan (hyaluronic acid); HMW, high molecular weight; PCA, principal component analysis.
HMW-HA Effects on the Rat L2 Proteome and Associated Pathways
In a separate follow-up proteomics experiment, we found that the addition of HMW-HA to rat L2 cells for 24 h (in the absence of the S1 protein) caused significant changes in 75 proteins (37 increased and 38 decreased, HMW-HA vs. C; Table 4). A 2 D-HCA heat map of the top most significantly changed 37 proteins (Fig. 6A) is illustrated along with a separate PCA plot (Fig. 6B).
Table 4.
Seventy-five most significantly changed proteins in HMW-HA v vehicle groups (37 increased and 38 decreased)
| Vehicle |
HMW-HA Treated |
Stats (HMW-HA vs. Vehicle) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UniProtKB Name | Acc# | C1 | C2 | C3 | C4 | HA1 | HA2 | HA3 | HA4 | SAM | t Test | Fold (HMW-HA vs. Contr.) |
| Epididymis luminal protein 215 | Q6PUJ7 | 2.0 | 1.1 | 1.1 | 3.8 | 5.3 | 10.7 | 5.6 | 8.2 | 1.44 | 0.007 | 3.7 |
| 116 kDa U5 small nuclear ribonucleoprotein component | Q15029 | 5.0 | 1.1 | 2.2 | 3.8 | 10.6 | 7.1 | 19.6 | 4.1 | 0.87 | 0.057 | 3.4 |
| Endothelin-1 | P05305 | 4.0 | 4.3 | 3.4 | 2.8 | 6.3 | 19.6 | 4.7 | 9.2 | 0.86 | 0.077 | 2.7 |
| Eukaryotic translation initiation factor 2 subunit 1 | P05198 | 3.0 | 1.1 | 4.5 | 1.9 | 5.3 | 4.5 | 9.3 | 9.2 | 1.10 | 0.015 | 2.7 |
| Coiled-coil domain-containing protein 47 | Q96A33 | 2.0 | 1.1 | 1.1 | 6.3 | 1.8 | 3.1 | 0.81 | 0.111 | 2.7 | ||
| RNA-binding protein 3 | P98179 | 1.0 | 2.1 | 1.9 | 3.6 | 4.7 | 5.1 | 1.99 | 0.005 | 2.7 | ||
| Clathrin interactor 1 | Q14677 | 1.0 | 3.2 | 1.1 | 1.9 | 3.2 | 7.1 | 3.7 | 5.1 | 1.07 | 0.017 | 2.7 |
| Drug-sensitive protein 1 | Q9NZ23 | 4.0 | 12.9 | 3.4 | 3.8 | 17.9 | 14.3 | 18.7 | 9.2 | 1.01 | 0.014 | 2.5 |
| Prohibitin-2 | Q99623 | 2.0 | 2.1 | 1.9 | 7.4 | 4.5 | 4.7 | 3.1 | 1.49 | 0.024 | 2.4 | |
| Lon protease homolog, mitochondrial | P36776 | 3.0 | 2.1 | 3.4 | 0.9 | 5.4 | 6.5 | 5.1 | 1.80 | 0.002 | 2.4 | |
| Rho GDP-dissociation inhibitor 1 | P52565 | 5.0 | 3.2 | 8.9 | 3.8 | 10.6 | 9.8 | 16.8 | 12.3 | 1.24 | 0.006 | 2.4 |
| Heparin-binding protein HBp15 | Q7Z4W8 | 2.0 | 2.1 | 2.2 | 0.9 | 1.1 | 6.2 | 4.7 | 5.1 | 0.86 | 0.057 | 2.3 |
| LDLR chaperone MESD | Q14696 | 3.2 | 2.2 | 1.9 | 6.3 | 5.4 | 7.5 | 3.1 | 1.21 | 0.018 | 2.3 | |
| Proliferation-associated protein 2G4 | Q9UQ80 | 4.0 | 2.1 | 4.5 | 1.9 | 6.3 | 8.0 | 5.6 | 8.2 | 1.52 | 0.002 | 2.2 |
| Cysteine and glycine-rich protein 1 | P21291 | 3.0 | 2.1 | 2.2 | 0.9 | 3.2 | 7.1 | 3.7 | 0.86 | 0.080 | 2.2 | |
| Glycylpeptide N-tetradecanoyltransferase 1 | P30419 | 4.0 | 1.1 | 2.2 | 7.4 | 3.7 | 5.1 | 0.89 | 0.049 | 2.2 | ||
| 60S ribosomal protein L7 | P18124 | 4.0 | 8.6 | 4.5 | 6.6 | 16.9 | 7.1 | 8.4 | 19.4 | 0.86 | 0.049 | 2.2 |
| RNA-binding motif protein, X chromosome | P38159 | 1.0 | 1.1 | 3.4 | 4.7 | 5.3 | 6.2 | 6.5 | 4.1 | 1.03 | 0.018 | 2.2 |
| Proteasome subunit β type-7 | Q99436 | 2.0 | 1.1 | 0.9 | 2.7 | 3.7 | 2.0 | 1.02 | 0.039 | 2.1 | ||
| Histone H2B type 1-M | Q99879 | 3.0 | 2.1 | 4.5 | 1.9 | 4.2 | 5.4 | 7.5 | 1.00 | 0.038 | 2.0 | |
| Calpain small subunit 1 | P04632 | 2.0 | 1.1 | 0.9 | 2.1 | 2.7 | 3.7 | 2.0 | 0.95 | 0.026 | 2.0 | |
| Histone H4 | P62805 | 5.0 | 3.2 | 2.2 | 3.8 | 7.4 | 8.9 | 4.7 | 1.03 | 0.045 | 2.0 | |
| B-cell receptor-associated protein 31 | P51572 | 3.2 | 2.2 | 1.9 | 2.1 | 5.4 | 6.5 | 5.1 | 0.91 | 0.042 | 2.0 | |
| Ubiquitin-conjugating enzyme E2 N | P61088 | 2.0 | 1.1 | 1.9 | 3.2 | 2.7 | 4.7 | 2.0 | 0.92 | 0.037 | 1.9 | |
| l-lactate dehydrogenase A chain | P00338 | 6.0 | 5.4 | 4.5 | 2.8 | 8.4 | 6.2 | 12.1 | 7.2 | 0.96 | 0.026 | 1.8 |
| C-Jun-amino-terminal kinase-interacting protein 4 | O60271 | 1.0 | 1.1 | 1.1 | 2.1 | 0.9 | 2.8 | 0.85 | 0.129 | 1.8 | ||
| Phospholipase D3 | Q8IV08 | 2.0 | 1.1 | 2.2 | 3.8 | 4.2 | 3.6 | 5.6 | 3.1 | 0.83 | 0.028 | 1.8 |
| Similar to 60S ribosomal protein L7 | O95036 | 6.0 | 13.9 | 7.8 | 5.7 | 16.9 | 11.6 | 11.2 | 20.4 | 0.81 | 0.032 | 1.8 |
| Adenylyl cyclase-associated protein 1 | Q01518 | 2.0 | 3.2 | 2.2 | 3.8 | 5.4 | 4.7 | 5.1 | 1.91 | 0.003 | 1.8 | |
| Src substrate cortactin | Q14247 | 4.0 | 4.3 | 2.8 | 7.1 | 4.7 | 8.2 | 1.15 | 0.044 | 1.8 | ||
| Dihydropyrimidinase-related protein 2 | Q16555 | 9.0 | 5.4 | 3.4 | 4.7 | 10.6 | 8.0 | 11.2 | 9.2 | 1.07 | 0.016 | 1.7 |
| 40S ribosomal protein S6 | P62753 | 8.0 | 8.6 | 7.8 | 9.4 | 7.4 | 13.4 | 15.0 | 20.4 | 0.92 | 0.064 | 1.7 |
| Destrin | P60981 | 2.0 | 2.1 | 3.4 | 3.8 | 6.3 | 4.5 | 4.7 | 3.1 | 0.82 | 0.035 | 1.6 |
| Prolyl 4-hydroxylase subunit α-1 | P13674 | 9.0 | 8.6 | 4.5 | 10.4 | 15.8 | 15.2 | 13.1 | 9.2 | 0.94 | 0.019 | 1.6 |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit β-2 | P62879 | 5.0 | 5.4 | 2.2 | 4.7 | 6.3 | 6.2 | 9.3 | 6.1 | 0.90 | 0.022 | 1.6 |
| Small nuclear ribonucleoprotein Sm D2 | P62316 | 2.0 | 1.1 | 1.9 | 3.2 | 2.7 | 2.0 | 0.92 | 0.045 | 1.6 | ||
| Ribosomal protein L19 | J3QR09 | 9.0 | 11.8 | 6.7 | 5.7 | 12.7 | 12.5 | 10.3 | 15.3 | 0.92 | 0.022 | 1.5 |
| Poly(rC)-binding protein 1 | Q15365 | 23.1 | 16.1 | 16.8 | 19.8 | 12.7 | 8.9 | 10.3 | 16.4 | 1.06 | 0.011 | −1.6 |
| Coatomer subunit α | P53621 | 23.1 | 21.4 | 20.1 | 25.5 | 20.1 | 14.3 | 9.3 | 13.3 | 1.23 | 0.012 | −1.6 |
| Insulin-like growth factor 2 mRNA-binding protein 3 | O00425 | 3.0 | 4.3 | 3.8 | 2.1 | 2.8 | 2.0 | 1.29 | 0.021 | −1.6 | ||
| 26S proteasome non-ATPase regulatory subunit 14 | O00487 | 14.1 | 9.6 | 8.9 | 8.5 | 8.4 | 7.1 | 6.5 | 3.1 | 0.82 | 0.030 | −1.6 |
| 60S ribosomal protein L23a | P62750 | 18.1 | 16.1 | 22.3 | 9.4 | 6.3 | 12.5 | 10.3 | 11.2 | 0.80 | 0.046 | −1.6 |
| SAP domain-containing ribonucleoprotein | P82979 | 2.0 | 3.2 | 2.8 | 1.8 | 0.9 | 2.0 | 0.92 | 0.044 | −1.7 | ||
| Ran-specific GTPase-activating protein | P43487 | 4.0 | 4.3 | 5.7 | 4.2 | 1.8 | 2.8 | 2.0 | 0.98 | 0.024 | −1.7 | |
| DNA replication licensing factor MCM4 | P33991 | 7.0 | 7.5 | 7.8 | 4.7 | 3.2 | 5.4 | 3.7 | 3.1 | 1.19 | 0.008 | −1.8 |
| Endoplasmic reticulum resident protein 29 | P30040 | 7.0 | 7.5 | 6.7 | 5.7 | 4.2 | 1.8 | 3.7 | 5.1 | 1.38 | 0.007 | −1.8 |
| Ras-related protein Rab-5C | P51148 | 13.1 | 9.6 | 6.7 | 11.3 | 8.4 | 7.1 | 4.7 | 2.0 | 0.83 | 0.028 | −1.8 |
| Small ubiquitin-related modifier 1 | P63165 | 2.0 | 3.2 | 2.2 | 1.9 | 1.1 | 1.8 | 0.9 | 1.02 | 0.021 | −1.9 | |
| Heme oxygenase 2 | P30519 | 1.0 | 1.1 | 2.2 | 2.8 | 1.1 | 0.9 | 0.9 | 0.84 | 0.081 | −1.9 | |
| E3 ubiquitin-protein ligase HUWE1 | Q7Z6Z7 | 14.1 | 7.5 | 6.7 | 14.2 | 6.3 | 5.4 | 3.7 | 7.2 | 0.90 | 0.043 | −1.9 |
| Synaptic vesicle membrane protein VAT-1 homolog | Q99536 | 12.1 | 10.7 | 8.9 | 16.0 | 2.1 | 5.4 | 7.5 | 10.2 | 0.88 | 0.024 | −1.9 |
| Serine/arginine-rich splicing factor 5 | Q13243 | 5.0 | 3.2 | 5.7 | 3.2 | 2.7 | 1.9 | 2.0 | 1.18 | 0.039 | −1.9 | |
| 40S ribosomal protein S23 | P62266 | 7.0 | 5.4 | 8.9 | 6.6 | 6.3 | 2.7 | 4.7 | 1.0 | 0.87 | 0.030 | −1.9 |
| U2 small nuclear RNA auxiliary factor 2 isoform b | B5BU25 | 2.0 | 3.2 | 5.6 | 3.8 | 1.1 | 2.7 | 1.9 | 2.0 | 0.80 | 0.048 | −1.9 |
| Vigilin | Q00341 | 19.1 | 9.6 | 14.5 | 11.3 | 4.2 | 9.8 | 8.4 | 6.1 | 0.98 | 0.022 | −1.9 |
| Protein disulfide-isomerase A6 | Q15084 | 33.2 | 19.3 | 26.8 | 25.5 | 17.9 | 12.5 | 15.0 | 9.2 | 1.33 | 0.006 | −1.9 |
| 40S ribosomal protein S8 | P62241 | 24.1 | 22.5 | 32.4 | 31.1 | 19.0 | 12.5 | 11.2 | 13.3 | 1.61 | 0.002 | −2.0 |
| Small glutamine-rich tetratricopeptide repeat-containing protein α | O43765 | 4.0 | 3.4 | 2.8 | 1.1 | 1.8 | 0.9 | 3.1 | 1.07 | 0.018 | −2.0 | |
| NADH-cytochrome b5 reductase | Q6ZVI6 | 8.0 | 13.9 | 8.9 | 9.4 | 3.2 | 1.8 | 8.4 | 6.1 | 0.93 | 0.019 | −2.1 |
| Proteasome subunit β type-3 | P49720 | 6.0 | 3.2 | 4.5 | 7.5 | 3.2 | 3.6 | 1.9 | 1.0 | 0.95 | 0.023 | −2.2 |
| Serine-protein kinase ATM | Q13315 | 2.0 | 1.1 | 3.4 | 0.9 | 0.9 | 1.0 | 0.99 | 0.106 | −2.3 | ||
| Endoplasmic reticulum resident protein 44 | Q9BS26 | 3.0 | 5.4 | 2.2 | 1.8 | 1.9 | 1.0 | 0.95 | 0.080 | −2.3 | ||
| 10 kDa heat shock protein, mitochondrial | P61604 | 20.1 | 22.5 | 7.8 | 11.3 | 6.3 | 8.9 | 4.7 | 7.2 | 0.99 | 0.042 | −2.3 |
| 60S ribosomal protein L38 | P63173 | 3.0 | 5.4 | 6.7 | 7.5 | 3.2 | 1.8 | 2.8 | 2.0 | 1.22 | 0.020 | −2.3 |
| Atlastin-3 | Q6DD88 | 8.0 | 10.7 | 7.8 | 8.5 | 3.2 | 6.2 | 1.9 | 3.1 | 1.62 | 0.002 | −2.4 |
| DNA replication licensing factor MCM2 | P49736 | 5.0 | 8.6 | 10.1 | 15.1 | 1.1 | 3.6 | 4.7 | 6.1 | 0.92 | 0.030 | −2.5 |
| ER membrane protein complex subunit 8 | O43402 | 3.2 | 1.1 | 2.8 | 0.9 | 0.9 | 1.0 | 1.22 | 0.077 | −2.5 | ||
| α-Aminoadipic semialdehyde dehydrogenase | P49419 | 2.0 | 4.3 | 2.2 | 3.8 | 1.1 | 1.8 | 0.9 | 1.0 | 1.24 | 0.019 | −2.6 |
| Far upstream element-binding protein 2 | Q92945 | 15.1 | 8.6 | 11.2 | 11.3 | 1.1 | 8.0 | 3.7 | 5.1 | 1.26 | 0.005 | −2.6 |
| Proteasome (Prosome, macropain) 26S subunit, non-ATPase, 13, isoform CRA_d | B4DJ66 | 3.0 | 4.3 | 2.2 | 4.7 | 2.1 | 0.9 | 0.9 | 1.23 | 0.011 | −2.7 | |
| Dehydrogenase/reductase SDR family member 9 | Q9BPW9 | 4.0 | 2.1 | 2.2 | 2.8 | 1.1 | 0.9 | 1.0 | 1.95 | 0.012 | −2.8 | |
| Talin-2 | Q9Y4G6 | 27.9 | 13.4 | 18.9 | 10.6 | 3.6 | 7.2 | 1.20 | 0.036 | −2.8 | ||
| Coatomer subunit gamma-1 | Q9Y678 | 5.0 | 4.3 | 12.3 | 9.4 | 2.1 | 1.8 | 0.9 | 6.1 | 0.82 | 0.036 | −2.8 |
| Spectrin-like protein of the nuclear envelope and Golgi | Q7RTM4 | 2.0 | 1.1 | 6.7 | 2.8 | 1.1 | 0.9 | 1.0 | 0.85 | 0.089 | −3.2 | |
| Serine/arginine repetitive matrix protein 2 | Q9UQ35 | 3.0 | 2.1 | 6.7 | 2.8 | 1.1 | 0.9 | 1.0 | 1.25 | 0.039 | −3.7 | |
HA, hyaluronan (hyaluronic acid); HMW, high molecular weight.
Figure 6.
Multivariate analysis carried out on the 37 top most significantly changed proteins between HMW-HA vs. vehicle (C) using the data from Table 4. Two-dimensional (2-D) HCA heat map (A) and three-dimensional (3-D) PCA plot (B) of control (C) vs. HMW-HA-treated (HA) rat L2 cells. Each column of the heat map and each point of the PCA represents a different experiment. HA, hyaluronan (hyaluronic acid); HMW, high molecular weight; PCA, principal component analysis.
Together these data indicate that HMW-HA induced a profound effect on the rat L2 proteome. The top 75 significantly differential protein list was then used for systems biology analysis. This analysis led to the identification of a number of significant Network Hubs and GO Biological Processes that include strong associations with E2F1, E2F5, and MCM-complex-associated cellular response to stress, DNA replication, and cell cycle regulation (Table 5). In addition, p53, NF-κB, and RAD54B-associated protein SUMOylation and DNA repair were also significantly affected by HMW-HA.
Table 5.
Top network list (HMW-HA vs. vehicle)
| # | Network | GO Processes | Total Nodes | Seed Nodes | P Value | z Score | G Score |
|---|---|---|---|---|---|---|---|
| 1 | E2F5, Histone H1, G0/G1switch 2, CDK3, Ebp1 | Cellular response to stress (38.4%; 1.909e-13), nucleic acid metabolic process (41.4%; 3.344e-13), establishment of localization (58.6%; 1.255e-12), cellular macromolecule catabolic process (25.3%; 4.243e-12), cellular localization (45.5%; 9.177e-12) | 100 | 81 | 6.1E-284 | 440.04 | 440.04 |
| 2 | MCM4, MCM2, E2F1, MCM10, Geminin | G1/S transition of mitotic cell cycle (20.2%; 2.689e-20), cell cycle G1/S phase transition (20.2%; 3.197e-20), DNA replication initiation (11.7%; 6.811e-18), DNA replication (19.1%; 1.542e-17), nucleic acid metabolic process (45.7%; 1.169e-15) | 95 | 77 | 2.9E-268 | 429.18 | 429.18 |
| 3 | Histone H2B, RBP-J kappa (CBF1), c-Jun, NF-κB, REV-ERBα | Negative regulation of gene expression (37.6%; 5.810e-10), cellular macromolecule catabolic process (23.5%; 2.256e-09), macromolecule catabolic process (23.5%; 4.291e-08), positive regulation of cellular process (61.2%; 4.466e-08), viral process (21.2%; 7.526e-08) | 85 | 65 | 1.0E-219 | 382.99 | 382.99 |
| 4 | HIP1, VAMP2, Synaptotagmin I, Caspase-3, Clathrin | Response to abiotic stimulus (45.0%; 3.968e-30), regulation of transport (50.7%; 9.340e-29), response to organonitrogen compound (42.9%; 2.997e-28), response to nitrogen compound (43.6%; 1.069e-27), positive regulation of transport (39.3%; 2.645e-27) | 154 | 63 | 4.3E-188 | 280.3 | 280.3 |
| 5 | MAP-1B, ERK1/2, WNT, L-Glutamic acid extracellular region, BDNF | Establishment of localization in cell (51.8%; 2.860e-17), establishment of localization (68.7%; 6.767e-17), transport (66.3%; 7.749e-16), localization (73.5%; 2.307e-15), cellular localization (54.2%; 2.654e-15) | 93 | 56 | 1.4E-178 | 318.87 | 318.87 |
| 6 | NF-κB1 (p50), DLC1 (Dynein LC8a), MTA1, ESR1 (nuclear), PR (nuclear) | Nucleic acid metabolic process (48.1%; 2.864e-10), cellular nitrogen compound metabolic process (57.7%; 5.046e-10), nucleobase-containing compound metabolic process (50.0%; 4.050e-09), organic cyclic compound metabolic process (53.8%; 4.856e-09), metabolic process (84.6%; 4.918e-09) | 53 | 44 | 7.9E-149 | 328.32 | 328.32 |
| 7 | NF-κB1 (p50), DLC1 (Dynein LC8a), REA, ESR1 (nuclear), PR (nuclear) | Nucleic acid metabolic process (48.1%; 2.864e-10), cellular nitrogen compound metabolic process (57.7%; 5.046e-10), nucleobase-containing compound metabolic process (50.0%; 4.050e-09), organic cyclic compound metabolic process (53.8%; 4.856e-09), metabolic process (84.6%; 4.918e-09) | 53 | 44 | 7.9E-149 | 328.32 | 328.32 |
| 8 | Histone H2, Testosterone intracellular, Activin A, Inhibin, NCOA4 (ARA70) | Positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway (21.2%; 1.831e-14), activin receptor signaling pathway (13.5%; 1.389e-12), response to organic substance (65.4%; 3.507e-12), regulation of transmembrane receptor protein serine/threonine kinase signaling pathway (23.1%; 7.720e-12), positive regulation of follicle-stimulating hormone secretion (9.6%; 3.467e-11) | 56 | 36 | 6.6E-115 | 266.1 | 266.1 |
| 9 | p53, NF-κB, MDM2, BRIP1, RAD54B | Protein sumoylation (17.2%; 2.807e-15), double-strand break repair (22.4%; 1.676e-14), cellular response to stress (48.3%; 3.211e-13), positive regulation of protein metabolic process (46.6%; 2.677e-12), positive regulation of cellular protein metabolic process (44.8%; 4.875e-12) | 58 | 34 | 1.8E-105 | 242.48 | 242.48 |
| 10 | MAT2A, Bcl-XS, IGF-2, Ethanol intracellular, IGF-1 | Phosphatidylinositol 3-kinase signaling (23.5%; 2.938e-15), phosphatidylinositol-mediated signaling (26.5%; 5.712e-14), response to peptide (47.1%; 6.707e-14), inositol lipid-mediated signaling (26.5%; 8.453e-14), response to peptide hormone (44.1%; 1.502e-13) | 50 | 19 | 1.4E-55 | 157.34 | 157.34 |
| 11 | ERP5, ATF-4, GRP78, 19S regulator, JIK | Response to endoplasmic reticulum stress (45.7%; 9.974e-21), cellular response to stress (74.3%; 6.606e-19), intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress (25.7%; 7.354e-17), response to stress (88.6%; 7.790e-17), regulation of endoplasmic reticulum unfolded protein response (22.9%; 1.856e-16) | 39 | 13 | 2.7E-36 | 113.01 | 113.01 |
GO, Gene Ontology; HA, hyaluronan (hyaluronic acid); HMW, high molecular weight.
S1 Increased E2F1, RhoA, and ROCK2 Phosphorylation, Which Were Prevented by Preincubation with HMW-HA
Western blot analysis carried out on L2 cells treated with S1 protein for 24 h resulted in a significant increase of E2F1 phosphorylation as compared with controls (Fig. 7A). Pretreatment with HMW-HA before S1 addition prevented E2F1 phosphorylation when normalized to either actin or total E2F1 (Fig. 7A). There were no changes in total E2F1 as compared with actin for any condition (data not shown). Similarly, and in agreement with the proteomics outcome, as illustrated in Fig. 7A, incubation of L2 cells with the S1 protein induced phosphorylation (activation) of both RhoA (Fig. 7B) and ROCK2 (Fig. 7C) within 30 min following incubation, whereas pretreatment with HMW-HA significantly inhibited these S1-associated effects.
Figure 7.
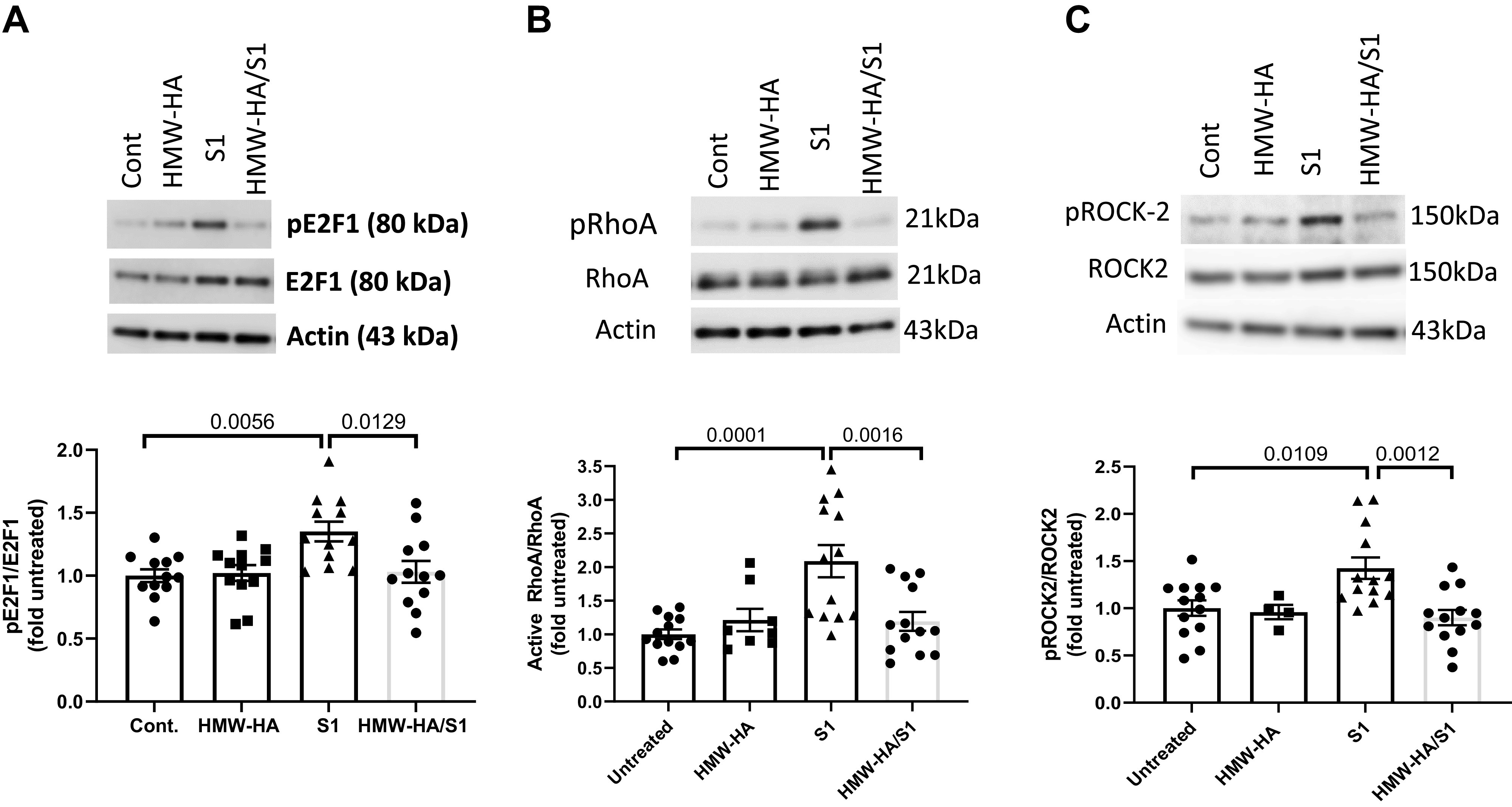
Western blots of confluent rat L2 cells, treated with PBS (Cont), HMW-HA (200 ng/mL) for 24 h, S1 (100 ng) for 24 h or HMW-HA for 1 h followed by S1 for 24 h. A: immunoblotting was performed using specific antibodies against E2F1, its phosphorylated form and actin (top). The intensities of the shown bands were quantified and the ratios of pE2F1 to E2F1 are shown at bottom. B and C: immunoblotting was performed using specific antibodies against phosphRhoA (active RhoA), total RhoA, phosphoROCK2, or total ROCK2 (top). The intensities of the various bands were quantified and the ratios of the phosphorylated to total forms are shown (bottom). Incubation of rat L2 cells with S1 upregulated the activities of the phosphorylated forms of E2F1, RhoA, and its downstream kinase, ROCK2. Preincubation with HMW-HA prevented these effects. Values are means with standard error of the means and individual points from three different batches of cells. One-way analysis of variance followed by the Tukey’s t test for multiple comparisons. HA, hyaluronan (hyaluronic acid); HMW, high molecular weight.
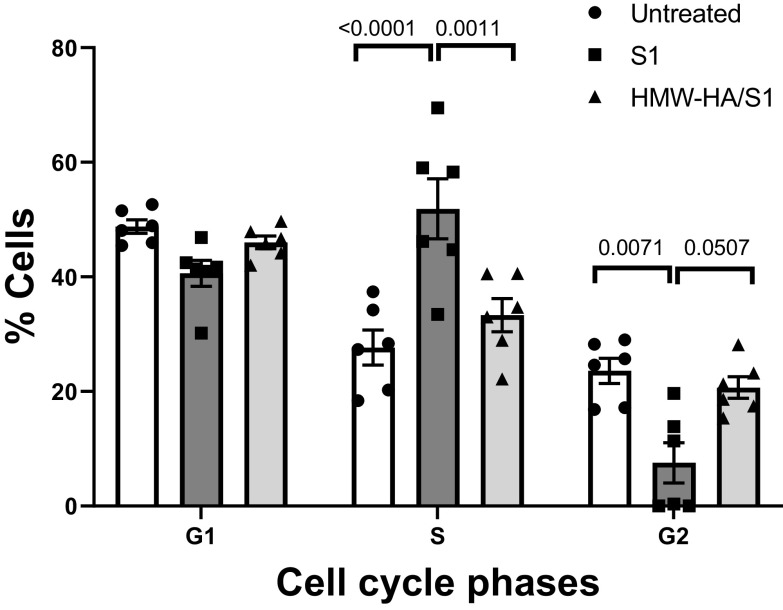
As E2F1 plays an important role in cell cycle regulation, we assessed changes in cell cycle by S1 and its reversal by HMW-HA. As shown in Fig. 8, treatment of L2 cells with the S1 protein for 24 h resulted in a significant increase in S phase accumulation with a concomitant decrease in the G2 phase as compared with the nontreated control, and both of these changes were either reversed or blocked with 1 h pretreatment with HMW-HA.
Figure 8.
Cell cycle analysis of confluent rat L2 cells incubated with S1 protein (100 ng) for 24 h or pretreated with HMW-HA (200 ng/mL) for 1 h prior to incubation with S1 protein. Cell cycle analysis was conducted as described in the materials and methods. Values are means with standard error of the means and individual values. One-way analysis of variance followed by the Tukey’s t test for multiple comparisons. Data from two different batches of cells. HA, hyaluronan (hyaluronic acid); HMW, high molecular weight.
Bioinformatic Networks Correlation of Spike S1-Exposed Rat L2 Proteome Data to SARS-CoV-2-Exposed Human Cells Transcriptome
The list of 99 proteins stemming from statistically significant changes induced by 24 h Tx-S1 in rat L2 cells at the beginning of these studies (Tables 1 and 2) was strongly correlated with a number of pathways, including E2F1, cell cycle, Rock2/RhoA, and CREB1. Following our validation, we were interested to see if there would be further correlation of these pathways at transcriptomic level using RNA-seq data from human lung cell lines exposed to live SARS-CoV-2. The Gene Set Enrichment Analysis (GSEA) results revealed that E2F signaling, E2F-mediated DNA metabolism, G2M checkpoint, and/or cell cycle were significantly correlated in three of the cell lines, and CREB was also correlated in two of these lung-derived human cell lines (Table 6).
Table 6.
Rat L2 proteomic S1-Tx cross correlated with pathway associations using transcriptomic data from five human cell lines exposed to SARS-CoV-2 virus
| Geneset | Pathway | #Targets | NES | P Value | q Value |
|---|---|---|---|---|---|
| 1. NHBE (Normal Human Bronchial Epithelial) | |||||
| HALLMARK | E2F | 195 | −1.4 | 0.0091 | 0.2044 |
| 2. A549 (adenocarcinoma human alveolar basal epithelial cells) | |||||
| HALLMARK | E2F | 197 | −3.1 | 0.0000 | 0.0000 |
| HALLMARK | G2M checkpoint | 194 | −2.7 | 0.0000 | 0.0000 |
| HALLMARK | Fatty acid metabolism | 150 | −1.5 | 0.0105 | 0.0320 |
| 3. A549-ACE2 | |||||
| HALLMARK | Fatty acid metabolism | 150 | −2.3 | 0.0000 | 0.0000 |
| HALLMARK | E2F | 196 | −1.8 | 0.0000 | 0.0002 |
| REACTOME | E2F-mediated DNA replication | 22 | −1.9 | 0.0056 | 0.0051 |
| BIOCARTA | CREB | 22 | −1.6 | 0.0295 | 0/0661 |
| 4. HAE (Human Airway Epithelial) | |||||
| HALLMARK | Fatty acid metabolism | 151 | −1.6 | 0.0000 | 0.0177 |
| BIOCARTA | CREB | 21 | −1.7 | 0.0037 | 0.1270 |
| 5. Calu (nonsmall-cell lung cancer) | |||||
| HALLMARK | E2F | 197 | −3 | 0.0000 | 0.0000 |
| HALLMARK | G2M checkpoint | 194 | −2.3 | 0.0000 | 0.0000 |
| HALLMARK | Fatty acid metabolism | 147 | −2.0 | 0.0000 | 0.0000 |
| REACTOME | E2F-mediated DNA replication | 22 | −1.9 | 0.0030 | 0.0119 |
| KEGG | Cell cycle | 124 | −1.6 | 0.0081 | 0.0510 |
DISCUSSION
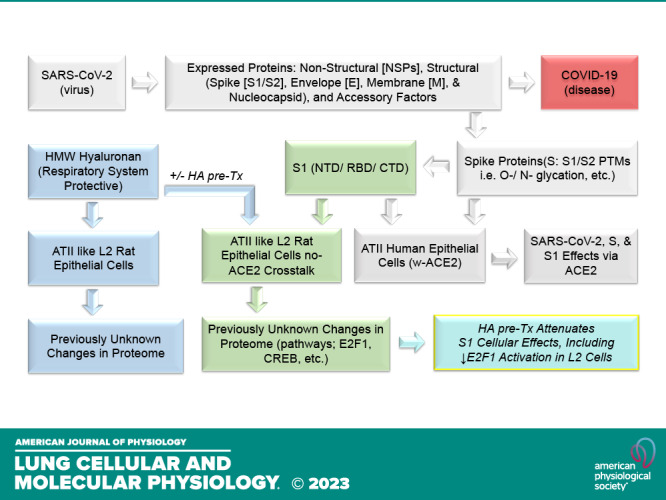
In this study, we addressed two key questions: 1) does human SARS-CoV-2 Spike S1 protein elicit a global proteomic effect on a rat lung-derived ATII-like L2 cell line that is expected to be primarily rat-ACE2 independent when exposed to S1 protein (59) and 2) does HWW-HA attenuate these effects? Our findings support a novel proposed model of S1-protein-induced cell injury (Fig. 9). Beyond the well-described interaction of the S-protein with host alveolar epithelia via the receptor human angiotensin-converting enzyme 2 (hACE2) (5, 60, 61), the S1-protein also exerts pathological effects (changes in proteome, E2F1, CREB, p65 activation) independent of ACE2. Furthermore, HMW-HA, which is naturally found in mammalian airways (29, 62), and can also be given pharmacologically (63), attenuates the deleterious effects of S1-protein (Fig. 9). Thus, our findings suggest a novel injury pathway and a novel treatment agent for COVID-19-associated lung injury.
Figure 9.
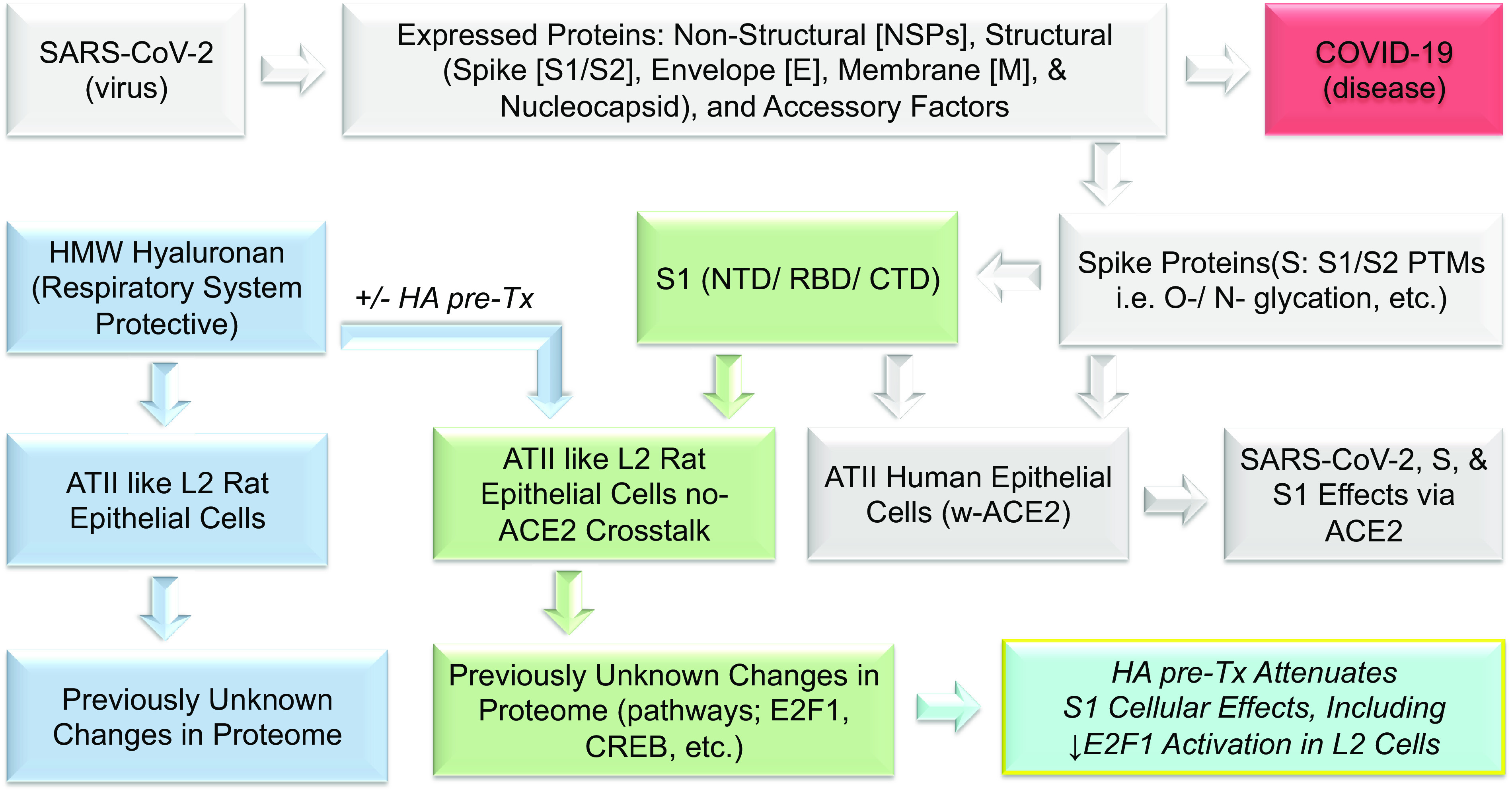
Experimental workflow. Similar to other coronaviruses, the makeup of SARS-CoV-2 is composed of four structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. S, E, and M proteins are glycosylated, and the N protein is phosphorylated. The S protein is involved in the interaction with the host receptor human angiotensin-converting enzyme 2 (hACE2), which is also heavily glycosylated. The S protein makes up two parts, the S1 and S2 spike proteins, which are heavily glycosylated, and S1 contains the receptor-binding domain (RBD). S1 is known to present with biological activities that are presumed largely to be due to interactions with hACEII receptors in human ATII cells (gray background). However, previously unreported, S1 also induces significant changes in ATII-like rat L2 cells that appear independent of hACEII as measured via proteomics analysis (green background). Similarly, and also previously unreported, HA induces significant changes in L2 cells (blue background) that greatly attenuate the deleterious effects of S1-Tx (turquois; bottom right). We have reported these observations here.
It is becoming increasingly evident that S-protein may impact cellular biology independent of interactions with the ACE2 receptor. Recently, there have been reports that the SARS-CoV-2 S protein interacts with other receptors such as neurophilin-1, (16), CD147 (17), tyrosine-protein kinase receptor UFO (AXL) (18, 19), ASGR1 and KREMEN1 (20), and others. In addition, the S-protein induces innate immune activation via Toll-like receptors TLR2 and TLR4 (24, 25, 27, 64) and in silico modeling suggests that it may direct interact with TLRs (65). TLR expression levels correlate with markers of organ injury in COVID-19 (66) while genetic polymorphisms in TLR2 and TLR4 are associated with COVID-19 outcomes (67, 68). Thus, the TLR axis may contribute significantly to the inflammation observed with SARS-CoV2 infection. Our results support this conclusion. Our unbiased analysis suggested p65, a central downstream mediator of TLR activation (69), as a major hub of our proteomic changes. Furthermore, other major hubs like CREB1 and E2F are modulators of TLR signaling (70, 71).
E2F is mostly described for its role in regulation of the cell cycle, with additional roles in metabolism and cell fate determination. However, there is an emerging pattern of E2F playing a role in inflammation as well (70, 72). In particular, recent papers suggest that the E2F activation may play a role in COVID-19 inflammatory cascade and cytokine storm (73). Our unbiased approach supports this conclusion and further suggests that this effect may be impacting the innate immune pathway, at least partly downstream of the S1-protein. It is also interesting that we identified ROCK/RhoA as a top enriched gene pathway. Interestingly, a recent report showed that the S1 protein caused a concentration-dependent decrease in transendothelial electrical resistance of human lung microvascular cells, an event that is associated with activation of the RhoA/ROCK pathway (74). In addition, recent papers highlight a role for these kinases in cellular injury and disruption of the cell barrier in COVID-19 (75, 76), thus supporting that our findings have biological relevance. Importantly, we have shown in several papers that RhoA/ROCK mediate noninfectious lung injury (36, 77–81), a process which can be inhibited by HMW-HA, a striking parallel to the finding in this work.
In aggregate, our proteomic results support that S-protein induces a robust innate immune activation cascade that can be a target of therapeutic interventions. HMW-HA is such a potential intervention. It is naturally present in mammalian airways (29, 62) and has been extensively studied for its ability to modulate TLR signaling (82). Importantly, therapeutic application of HMW-HA has been shown to have beneficial effects, including in infectious and inflammatory lung disease (63, 78, 79, 81). Given that it is a cheap, natural, biological compound with essentially no adverse effects, HMW-HA is therefore attractive as a first-line agent in emerging infectious diseases like COVID-19.
The addition of HMW-HA to L2 cells, even in the absence of S-protein, resulted in significant changes to the proteome. Systems biology analysis led to the identification of a number of significant Network Hubs and GO Biological Processes that include strong associations with E2F1, E2F5, DNA replication, cell cycle regulation, p53, NF-κB, and DNA repair. Given that a number of reports have presented beneficial effects of HMW-HA on pulmonary pathologies (36, 37, 41, 81), it is possible that some of the effects of HMW-HA in our model are mediated by a pro-homeostatic effect of HMW-HA that is not specific to S-protein. Indeed, HMW-HA is known to ameliorate inflammation downstream of LPS/TLR4 signaling (83), thus again supporting a global activity pattern as an anti-inflammatory mediator. In further support of this interpretation, instillation of S1 protein in K18-hACE2 mice that overexpress human ACE2 receptors resulted in robust activation of STAT3 and NF-κB inflammatory response, suggesting that modulation of the innate immune response may indeed significantly alter the inflammatory response in COVID-19 (74).
In conclusion, this work demonstrates that S-protein exposure in epithelial cells leads to a robust proteomic response, even in the absence of ACE2 engagement. We identify several novel signaling hubs, including E2F1, CREB, and ROCK/RhoA, and demonstrate that HMWHA, a natural component of airway lining, can ameliorate these changes and promote cellular homeostasis. Our findings shed new light on the pathogenesis of COVID-19 and suggest novel angles of treatment that will ameliorate cell injury and lung dysfunction in this devastating disease.
DATA AVAILABILITY
The mass spectrometry proteomics data will be made available upon request.
SUPPLEMENTAL DATA
Supplemental Tables S1 and S2 and Supplemental Fig. S1: https://doi.org/10.6084/m9.figshare.21785384.v1.
GRANTS
Funding was provided by the CounterACT Program, National Institutes of Health Office of the Director, the National Institute of Neurological Disorders and Stroke, and the National Institute of Environmental Health Sciences, Grants 5UO1 ES026458, 3UO1 ES026458 03S1, and 1R21 ES032956 to S. Matalon; UAB Comprehensive Cancer Center, agency: National Institutes of Health, institute: National Cancer Institute, Project no. P30CA013148 to J. A. Mobley; and a REINVENT grant from the Department of Anesthesiology and Perioperative Medicine to S. Matalon and ES102605, ES103342 to S. Garantziotis.
DISCLOSURES
Sadis Matalon receives an honorarium from American Physiological Society for his service as Editor of Physiological Reviews. None of the other authors has any conflicts of interest, financial or otherwise, to disclose.
AUTHOR CONTRIBUTIONS
J.A.M., T.J., J.-L.L., S.G., and S.M. conceived and designed research; J.A.M., A.M., K.K., I.A., and S.M. performed experiments; J.A.M., K.K., I.A., J.-L.L., and S.M. analyzed data; J.A.M., I.A., T.J., J.-L.L., S.G., and S.M. interpreted results of experiments; J.A.M., J.-L.L., and S.M. prepared figures; J.A.M., I.A., S.G., and S.M. drafted manuscript; J.A.M., A.M., K.K., T.J., J.-L.L., S.G., and S.M. edited and revised manuscript; J.A.M., A.M., K.K., T.J., J.-L.L., S.G., and S.M. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors are grateful to Zhihong Yu for technical assistance with these studies.
Preprint is available at https://doi.org/10.1101/2022.08.31.506023.
REFERENCES
- 1. Wu Z, McGoogan JM. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA 323: 1239, 2020. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 2. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395: 497–506, 2020. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiang S, Hillyer C, Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol 41: 355–359, 2020. [Erratum in Trends Immunol 41: 545, 2020]. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, Huan Y, Yang P, Zhang Y, Deng W, Bao L, Zhang B, Liu G, Wang Z, Chappell M, Liu Y, Zheng D, Leibbrandt A, Wada T, Slutsky AS, Liu D, Qin C, Jiang C, Penninger JM. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med 11: 875–879, 2005. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11: 1620, 2020. [Erratum in Nat Commun 12: 2144, 2021]. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367: 1260–1263, 2020. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 176: 104742, 2020. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Banerjee A, Mossman K, Baker ML. Zooanthroponotic potential of SARS-CoV-2 and implications of reintroduction into human populations. Cell Host Microbe 29: 160–164, 2021. doi: 10.1016/j.chom.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saag M. Wonder of wonders, miracle of miracles: the unprecedented speed of COVID-19 science. Physiol Rev 102: 1569–1577, 2022. doi: 10.1152/physrev.00010.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ahmad S, Matalon S, Kuebler WM. Understanding COVID-19 susceptibility and presentation based on its underlying physiology. Physiol Rev 102: 1579–1585, 2022. doi: 10.1152/physrev.00008.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ji HL, Zhao R, Matalon S, Matthay MA. Elevated plasmin(ogen) as a common risk factor for COVID-19 susceptibility. Physiol Rev 100: 1065–1075, 2020. doi: 10.1152/physrev.00013.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Manzano GS, Woods JK, Amato AA. Covid-19-associated myopathy caused by type I interferonopathy. N Engl J Med 383: 2389–2390, 2020. doi: 10.1056/NEJMc2031085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iadecola C, Anrather J, Kamel H. Effects of COVID-19 on the nervous system. Cell 183: 16–27.e1, 2020. doi: 10.1016/j.cell.2020.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nadim MK, Forni LG, Mehta RL, Connor MJ Jr, Liu KD, Ostermann M, Rimmelé T, Zarbock A, Bell S, Bihorac A, Cantaluppi V, Hoste E, Husain-Syed F, Germain MJ, Goldstein SL, Gupta S, Joannidis M, Kashani K, Koyner JL, Legrand M, Lumlertgul N, Mohan S, Pannu N, Peng Z, Perez-Fernandez XL, Pickkers P, Prowle J, Reis T, Srisawat N, Tolwani A, Vijayan A, Villa G, Yang L, Ronco C, Kellum JA. COVID-19-associated acute kidney injury: consensus report of the 25th Acute Disease Quality Initiative (ADQI) workgroup. Nat Rev Nephrol 16: 747–764, 2020. [Erratum in Nat Rev Nephrol 16: 765, 2020]. doi: 10.1038/s41581-020-00356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Romagnoli S, Peris A, De Gaudio AR, Geppetti P. SARS-CoV-2 and COVID-19: from the bench to the bedside. Physiol Rev 100: 1455–1466, 2020. doi: 10.1152/physrev.00020.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Daly JL, Simonetti B, Klein K, Chen KE, Williamson MK, Antón-Plágaro C, Shoemark DK, Simón-Gracia L, Bauer M, Hollandi R, Greber UF, Horvath P, Sessions RB, Helenius A, Hiscox JA, Teesalu T, Matthews DA, Davidson AD, Collins BM, Cullen PJ, Yamauchi Y. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370: 861–865, 2020. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang K, Chen W, Zhang Z, Deng Y, Lian JQ, Du P, et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther 5: 283, 2020. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ng PS, Foo K, Sim S, Wang G, Huang C, Tan LH, Poulsen A, Liu B, Tee DHY, Ahmad NHB, Wang S, Ke Z, Lee MA, Kwek ZP, Joy J, Anantharajan J, Baburajendran N, Pendharkar V, Manoharan V, Vuddagiri S, Sangthongpitag K, Hill J, Keller TH, Hung AW. Fragment-based lead discovery of indazole-based compounds as AXL kinase inhibitors. Bioorg Med Chem 49: 116437, 2021. doi: 10.1016/j.bmc.2021.116437. [DOI] [PubMed] [Google Scholar]
- 19. Wang S, Qiu Z, Hou Y, Deng X, Xu W, Zheng T, Wu P, Xie S, Bian W, Zhang C, Sun Z, Liu K, Shan C, Lin A, Jiang S, Xie Y, Zhou Q, Lu L, Huang J, Li X. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res 31: 126–140, 2021. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gu Y, Cao J, Zhang X, Gao H, Wang Y, Wang J, He J, Jiang X, Zhang J, Shen G, Yang J, Zheng X, Hu G, Zhu Y, Du S, Zhu Y, Zhang R, Xu J, Lan F, Qu D, Xu G, Zhao Y, Gao D, Xie Y, Luo M, Lu Z. Receptome profiling identifies KREMEN1 and ASGR1 as alternative functional receptors of SARS-CoV-2. Cell Res 32: 24–37, 2022. [Erratum in Cell Res 32: 600, 2022]. doi: 10.1038/s41422-022-00654-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. El-Hefnawy SM, Eid HA, Mostafa RG, Soliman SS, Omar TA, Azmy RM. COVID-19 susceptibility, severity, clinical outcome and Toll-like receptor (7) mRNA expression driven by TLR7 gene polymorphism (rs3853839) in middle-aged individuals without previous comorbidities. Gene Rep 27: 101612, 2022. doi: 10.1016/j.genrep.2022.101612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Q, Bastard P; COVID Human Genetic Effort, Cobat A, Casanova JL. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature 603: 587–598, 2022. doi: 10.1038/s41586-022-04447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Q, Matuozzo D, Le PJ, Lee D, Moens L, Asano T, et al. Recessive inborn errors of type I IFN immunity in children with COVID-19 pneumonia. J Exp Med 219: e20220131, 2022. doi: 10.1084/jem.20220131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aboudounya MM, Heads RJ. COVID-19 and toll-like receptor 4 (TLR4): SARS-CoV-2 may bind and activate TLR4 to increase ACE2 expression, facilitating entry and causing hyperinflammation. Mediators Inflamm 2021: 8874339, 2021. doi: 10.1155/2021/8874339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Khan S, Shafiei MS, Longoria C, Schoggins JW, Savani RC, Zaki H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-κB pathway. eLife 10: e68563, 2021. doi: 10.7554/eLife.68563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shirato K, Kizaki T. SARS-CoV-2 spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages. Heliyon 7: e06187, 2021. doi: 10.1016/j.heliyon.2021.e06187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zheng M, Karki R, Williams EP, Yang D, Fitzpatrick E, Vogel P, Jonsson CB, Kanneganti TD. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat Immunol 22: 829–838, 2021. doi: 10.1038/s41590-021-00937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xiong C, Yang G, Kumar S, Aggarwal S, Leustik M, Snead C, Hamacher J, Fischer B, Umapathy NS, Hossain H, Wendel A, Catravas JD, Verin AD, Fulton D, Black SM, Chakraborty T, Lucas R. The lectin-like domain of TNF protects from listeriolysin-induced hyperpermeability in human pulmonary microvascular endothelial cells - a crucial role for protein kinase C-alpha inhibition. Vascul Pharmacol 52: 207–213, 2010. doi: 10.1016/j.vph.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Forteza R, Lieb T, Aoki T, Savani RC, Conner GE, Salathe M. Hyaluronan serves a novel role in airway mucosal host defense. FASEB J 15: 2179–2186, 2001. doi: 10.1096/fj.01-0036com. [DOI] [PubMed] [Google Scholar]
- 30. Garantziotis S, Li Z, Potts EN, Lindsey JY, Stober VP, Polosukhin VV, Blackwell TS, Schwartz DA, Foster WM, Hollingsworth JW. TLR4 is necessary for hyaluronan-mediated airway hyperresponsiveness after ozone inhalation. Am J Respir Crit Care Med 181: 666–675, 2010. doi: 10.1164/rccm.200903-0381OC. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 31. Jiang D, Liang J, Noble PW. Regulation of non-infectious lung injury, inflammation, and repair by the extracellular matrix glycosaminoglycan hyaluronan. Anat Rec (Hoboken) 293: 982–985, 2010. doi: 10.1002/ar.21102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scheibner KA, Lutz MA, Boodoo S, Fenton MJ, Powell JD, Horton MR. Hyaluronan fragments act as an endogenous danger signal by engaging TLR2. J Immunol 177: 1272–1281, 2006. doi: 10.4049/jimmunol.177.2.1272. [DOI] [PubMed] [Google Scholar]
- 33. Jiang D, Liang J, Noble PW. Hyaluronan as an immune regulator in human diseases. Physiol Rev 91: 221–264, 2011. doi: 10.1152/physrev.00052.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lazrak A, Song W, Zhou T, Aggarwal S, Jilling T, Garantziotis S, Matalon S. Hyaluronan and halogen-induced airway hyperresponsiveness and lung injury. Ann N Y Acad Sci 1479: 29–43, 2020. doi: 10.1111/nyas.14415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li Y, Jiang D, Liang J, Meltzer EB, Gray A, Miura R, Wogensen L, Yamaguchi Y, Noble PW. Severe lung fibrosis requires an invasive fibroblast phenotype regulated by hyaluronan and CD44. J Exp Med 208: 1459–1471, 2011. doi: 10.1084/jem.20102510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lazrak A, Creighton J, Yu Z, Komarova S, Doran SF, Aggarwal S, Emala CW Sr, Stober VP, Trempus CS, Garantziotis S, Matalon S. Hyaluronan mediates airway hyperresponsiveness in oxidative lung injury. Am J Physiol Lung Cell Mol Physiol 308: L891–L903, 2015. doi: 10.1152/ajplung.00377.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lennon FE, Singleton PA. Hyaluronan regulation of vascular integrity. Am J Cardiovasc Dis 1: 200–213, 2011. [PMC free article] [PubMed] [Google Scholar]
- 38. Hellman U, Karlsson MG, Engström-Laurent A, Cajander S, Dorofte L, Ahlm C, Laurent C, Blomberg A. Presence of hyaluronan in lung alveoli in severe Covid-19: an opening for new treatment options? J Biol Chem 295: 15418–15422, 2020. doi: 10.1074/jbc.AC120.015967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kratochvil MJ, Kaber G, Demirdjian S, Cai PC, Burgener EB, Nagy N, Barlow GL, Popescu M, Nicolls MR, Ozawa MG, Regula DP, Pacheco-Navarro AE, Yang S, de Jesus Perez VA, Karmouty-Quintana H, Peters AM, Zhao B, Buja ML, Johnson PY, Vernon RB, Wight TN, Milla CE, Rogers AJ, Spakowitz AJ, Heilshorn SC, Bollyky PL. Biochemical, biophysical, and immunological characterization of respiratory secretions in severe SARS-CoV-2 infections. JCI Insight 7: e152629, 2022. doi: 10.1172/jci.insight.152629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Queisser KA, Mellema RA, Middleton EA, Portier I, Manne BK, Denorme F, Beswick EJ, Rondina MT, Campbell RA, Petrey AC. COVID-19 generates hyaluronan fragments that directly induce endothelial barrier dysfunction. JCI Insight 6: e147472, 2021. doi: 10.1172/jci.insight.147472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Garantziotis S, Li Z, Potts EN, Kimata K, Zhuo L, Morgan DL, Savani RC, Noble PW, Foster WM, Schwartz DA, Hollingsworth JW. Hyaluronan mediates ozone-induced airway hyperresponsiveness in mice. J Biol Chem 284: 11309–11317, 2009. [Erratum in J Biol Chem 291: 19257–19258, 2016]. doi: 10.1074/jbc.M802400200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 42. Jiang D, Liang J, Fan J, Yu S, Chen S, Luo Y, Prestwich GD, Mascarenhas MM, Garg HG, Quinn DA, Homer RJ, Goldstein DR, Bucala R, Lee PJ, Medzhitov R, Noble PW. Regulation of lung injury and repair by Toll-like receptors and hyaluronan. Nat Med 11: 1173–1179, 2005. doi: 10.1038/nm1315. [DOI] [PubMed] [Google Scholar]
- 43. Huang PM, Syrkina O, Yu L, Dedaj R, Zhao H, Shiedlin A, Liu YY, Garg H, Quinn DA, Hales CA. High MW hyaluronan inhibits smoke inhalation-induced lung injury and improves survival. Respirology 15: 1131–1139, 2010. doi: 10.1111/j.1440-1843.2010.01829.x. [DOI] [PubMed] [Google Scholar]
- 44. Liu YY, Lee CH, Dedaj R, Zhao H, Mrabat H, Sheidlin A, Syrkina O, Huang PM, Garg HG, Hales CA, Quinn DA. High-molecular-weight hyaluronan–a possible new treatment for sepsis-induced lung injury: a preclinical study in mechanically ventilated rats. Crit Care 12: R102, 2008. doi: 10.1186/cc6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Petrigni G, Allegra L. Aerosolised hyaluronic acid prevents exercise-induced bronchoconstriction, suggesting novel hypotheses on the correction of matrix defects in asthma. Pulm Pharmacol Ther 19: 166–171, 2006. doi: 10.1016/j.pupt.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 46. Telenga ED, Kerstjens HA. Effect of inhaled hyaluronic acid (HA) on exercised induced bronchoconstriction (EIB). Pulm Pharmacol Ther 21: 430, 2008. doi: 10.1016/j.pupt.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 47. Matalon S, O'Brodovich H. Sodium channels in alveolar epithelial cells: molecular characterization, biophysical properties, and physiological significance. Annu Rev Physiol 61: 627–661, 1999. doi: 10.1146/annurev.physiol.61.1.627. [DOI] [PubMed] [Google Scholar]
- 48. Carcaterra M, Caruso C. Alveolar epithelial cell type II as main target of SARS-CoV-2 virus and COVID-19 development via NF-Kb pathway deregulation: a physio-pathological theory. Med Hypotheses 146: 110412, 2021. doi: 10.1016/j.mehy.2020.110412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shahjaman M, Rezanur Rahman M, Rabiul Auwul M. A network-based systems biology approach for identification of shared Gene signatures between male and female in COVID-19 datasets. Inform Med Unlocked 25: 100702, 2021. doi: 10.1016/j.imu.2021.100702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ahmad I, Molyvdas A, Jian MY, Zhou T, Traylor AM, Cui H, Liu G, Song W, Agarwal A, Jilling T, Aggarwal S, Matalon S. AICAR decreases acute lung injury by phosphorylating AMPK and upregulating heme oxygenase-1. Eur Respir J 58: 2003694, 2021. doi: 10.1183/13993003.03694-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Addis DR, Aggarwal S, Doran SF, Jian MY, Ahmad I, Kojima K, Ford DA, Matalon S, Mobley JA. Vascular permeability disruption explored in the proteomes of mouse lungs and human microvascular cells following acute bromine exposure. Am J Physiol Lung Cell Mol Physiol 319: L337–L359, 2020. doi: 10.1152/ajplung.00196.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392, 2002. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 53. Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658, 2003. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 54. Weatherly DB, Atwood JA 3rd, Minning TA, Cavola C, Tarleton RL, Orlando R. A Heuristic method for assigning a false-discovery rate for protein identifications from Mascot database search results. Mol Cell Proteomics 4: 762–772, 2005. doi: 10.1074/mcp.M400215-MCP200. [DOI] [PubMed] [Google Scholar]
- 55. Liu H, Sadygov RG, Yates JR 3rd.. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76: 4193–4201, 2004. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 56. Beissbarth T, Hyde L, Smyth GK, Job C, Boon WM, Tan SS, Scott HS, Speed TP. Statistical modeling of sequencing errors in SAGE libraries. Bioinformatics 20: i31–i39, 2004. doi: 10.1093/bioinformatics/bth924. [DOI] [PubMed] [Google Scholar]
- 57. Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286: 531–537, 1999. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 58. Bhatia VN, Perlman DH, Costello CE, McComb ME. Software tool for researching annotations of proteins: open-source protein annotation software with data visualization. Anal Chem 81: 9819–9823, 2009. doi: 10.1021/ac901335x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Conceicao C, Thakur N, Human S, Kelly JT, Logan L, Bialy D, Bhat S, Stevenson-Leggett P, Zagrajek AK, Hollinghurst P, Varga M, Tsirigoti C, Tully M, Chiu C, Moffat K, Silesian AP, Hammond JA, Maier HJ, Bickerton E, Shelton H, Dietrich I, Graham SC, Bailey D. The SARS-CoV-2 spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biol 18: e3001016, 2020. doi: 10.1371/journal.pbio.3001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271–280.e8, 2020. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mason RJ. Pathogenesis of COVID-19 from a cell biology perspective. Eur Respir J 55: 2000607, 2020. doi: 10.1183/13993003.00607-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Abbadi A, Lauer M, Swaidani S, Wang A, Hascall V. Hyaluronan rafts on airway epithelial cells. J Biol Chem 291: 1448–1455, 2016. doi: 10.1074/jbc.M115.704288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Galdi F, Pedone C, McGee CA, George M, Rice AB, Hussain SS, Vijaykumar K, Boitet ER, Tearney GJ, McGrath JA, Brown AR, Rowe SM, Incalzi RA, Garantziotis S. Inhaled high molecular weight hyaluronan ameliorates respiratory failure in acute COPD exacerbation: a pilot study. Respir Res 22: 30, 2021. doi: 10.1186/s12931-020-01610-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhao Y, Kuang M, Li J, Zhu L, Jia Z, Guo X, Hu Y, Kong J, Yin H, Wang X, You F. SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res 31: 818–820, 2021. doi: 10.1038/s41422-021-00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Choudhury A, Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J Med Virol 92: 2105–2113, 2020. doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sultan RH, Elesawy BH, Ali TM, Abdallah M, Assal HH, Ahmed AE, Ahmed OM. Correlations between kidney and heart function bioindicators and the expressions of toll-like, ACE2, and NRP-1 receptors in COVID-19. Vaccines (Basel) 10: 1106, 2022. doi: 10.3390/vaccines10071106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Minashkin MM, Grigortsevich NY, Kamaeva AS, Barzanova VV, Traspov AA, Godkov MA, Ageev FA, Petrikov SS, Pozdnyakova NV. The role of genetic factors in the development of acute respiratory viral infection COVID-19: predicting severe course and outcomes. Biomedicines 10: 549, 2022. doi: 10.3390/biomedicines10030549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Taha SI, Shata AK, El-Sehsah EM, Mohamed MF, Moustafa NM, Youssef MK. Comparison of COVID-19 characteristics in Egyptian patients according to their Toll-Like Receptor-4 (Asp299Gly) polymorphism. Infez Med 30: 96–103, 2022. doi: 10.53854/liim-3001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Guha M, Mackman N. LPS induction of gene expression in human monocytes. Cell Signal 13: 85–94, 2001. doi: 10.1016/s0898-6568(00)00149-2. [DOI] [PubMed] [Google Scholar]
- 70. Lim CA, Yao F, Wong JJ, George J, Xu H, Chiu KP, Sung WK, Lipovich L, Vega VB, Chen J, Shahab A, Zhao XD, Hibberd M, Wei CL, Lim B, Ng HH, Ruan Y, Chin KC. Genome-wide mapping of RELA(p65) binding identifies E2F1 as a transcriptional activator recruited by NF-kappaB upon TLR4 activation. Mol Cell 27: 622–635, 2007. doi: 10.1016/j.molcel.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 71. Wen AY, Sakamoto KM, Miller LS. The role of the transcription factor CREB in immune function. J Immunol 185: 6413–6419, 2010. doi: 10.4049/jimmunol.1001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yang B, Fukuda N, van Hoek A, Matthay MA, Ma T, Verkman AS. Carbon dioxide permeability of aquaporin-1 measured in erythrocytes and lung of aquaporin-1 null mice and in reconstituted proteoliposomes. J Biol Chem 275: 2686–2692, 2000. doi: 10.1074/jbc.275.4.2686. [DOI] [PubMed] [Google Scholar]
- 73. Ali N, Prasad K, AlAsmari AF, Alharbi M, Rashid S, Kumar V. Genomics-guided targeting of stress granule proteins G3BP1/2 to inhibit SARS-CoV-2 propagation. Int J Biol Macromol 190: 636–648, 2021. doi: 10.1016/j.ijbiomac.2021.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Colunga Biancatelli RML, Solopov PA, Gregory B, Khodour Y, Catravas JD. HSP90 Inhibitors Modulate SARS-CoV-2 spike protein subunit 1-induced human pulmonary microvascular endothelial activation and barrier dysfunction. Front Physiol 13: 812199, 2022. doi: 10.3389/fphys.2022.812199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ashour L. Roles of the ACE/Ang II/AT1R pathway, cytokine release, and alteration of tight junctions in COVID-19 pathogenesis. Tissue Barriers. In press. doi: 10.1080/21688370.2022.2090792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Barman RK, Mukhopadhyay A, Maulik U, Das S. A network biology approach to identify crucial host targets for COVID-19. Methods 203: 108–115, 2022. doi: 10.1016/j.ymeth.2022.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Addis DR, Aggarwal S, Lazrak A, Jilling T, Matalon S. Halogen-induced chemical injury to the mammalian cardiopulmonary systems. Physiology (Bethesda) 36: 272–291, 2021. doi: 10.1152/physiol.00004.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Garantziotis S, Matalon S. Sugarcoating lung injury: a novel role for high-molecular-weight hyaluronan in pneumonia. Am J Respir Crit Care Med 200: 1197–1198, 2019. doi: 10.1164/rccm.201908-1554ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Johnson CG, Stober VP, Cyphert-Daly JM, Trempus CS, Flake GP, Cali V, Ahmad I, Midura RJ, Aronica MA, Matalon S, Garantziotis S. High molecular weight hyaluronan ameliorates allergic inflammation and airway hyperresponsiveness in the mouse. Am J Physiol Lung Cell Mol Physiol 315: L787–L798, 2018. doi: 10.1152/ajplung.00009.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lazrak A, Yu Z, Doran S, Jian MY, Creighton J, Laube M, Garantziotis S, Prakash YS, Matalon S. Upregulation of airway smooth muscle calcium-sensing receptor by low-molecular-weight hyaluronan. Am J Physiol Lung Cell Mol Physiol 318: L459–L471, 2020. doi: 10.1152/ajplung.00429.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhou T, Yu Z, Jian MY, Ahmad I, Trempus C, Wagener BM, Pittet JF, Aggarwal S, Garantziotis S, Song W, Matalon S. Instillation of hyaluronan reverses acid instillation injury to the mammalian blood gas barrier. Am J Physiol Lung Cell Mol Physiol 314: L808–L821, 2018. doi: 10.1152/ajplung.00510.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tighe RM, Garantziotis S. Hyaluronan interactions with innate immunity in lung biology. Matrix Biol 78–79: 84–99, 2019. doi: 10.1016/j.matbio.2018.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Campo GM, Avenoso A, Campo S, D'Ascola A, Nastasi G, Calatroni A. Molecular size hyaluronan differently modulates toll-like receptor-4 in LPS-induced inflammation in mouse chondrocytes. Biochimie 92: 204–215, 2010. doi: 10.1016/j.biochi.2009.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Tables S1 and S2 and Supplemental Fig. S1: https://doi.org/10.6084/m9.figshare.21785384.v1.
Data Availability Statement
The mass spectrometry proteomics data will be made available upon request.