Abstract
Purpose
To analyze the presence of potentially pathogenic variants of 29 candidate genes known to cause spermatogenic failure (SPGF) in patients with non-obstructive azoospermia (NOA) who underwent testicular histology.
Materials and Methods
Forty-eight patients with unexplained NOA referred to the Department of Transfusion Medicine and Transplantation Biology, University Hospital Center Zagreb, Zagreb, Croatia for testicular biopsy. They were divided into three groups: those who had cryptorchidism (n=9), those with varicocele (n=14), and those with idiopathic NOA (n=25). All included patients underwent blood withdrawal for next-generation sequencing (NGS) analysis and gene sequencing.
Results
We found a possible genetic cause in 4 patients with idiopathic NOA (16%) and in 2 with cryptorchidism (22%). No pathogenic or possibly pathogenic mutations were identified in patients with varicocele. Variants of undetermined significance (VUS) were found in 11 patients with idiopathic NOA (44%), 3 with cryptorchidism (33%), and 8 patients with varicocele (57%). VUSs of the USP9Y gene were the most frequently as they were found in 14 out of 48 patients (29%). In particular, the VUS USP9Y c.7434+14del was found in 11 patients. They showed varied histological pictures, including Sertoli cell-only syndrome, mixed atrophy, and hypospermatogenesis, regardless of cryptorchidism or varicocele. No direct correlation was found between the gene mutation/variant and the testicular histological picture.
Conclusions
Different mutations of the same gene cause various testicular histological pictures. These results suggest that it is not the gene itself but the type of mutation/variation that determines the testicular histology picture. Based on the data presented above, it remains challenging to design a genetic panel with prognostic value for the outcome of testicular sperm extraction in patients with NOA.
Keywords: Azoospermia, Male infertility, Next-generation sequencing, Spermatogenesis, Testicular histology
INTRODUCTION
Spermatogenesis is a very complex process, which takes about 74 days. It occurs in the seminiferous tubules where diploid spermatogonial stem cells differentiate into haploid spermatozoa. During spermatogenesis, numerous, finely regulated molecular mechanisms take place, most of which are only partially known. As a consequence, the etiology of spermatogenic failure (SPGF) is identified only in a minority of infertile patients. Indeed, an epidemiologic study reporting the presence of oligozoospermia in 20% of 8,518 male partners of infertile couples was able to identify its etiology in only 25% of cases [1]. Similarly, another study of 26,091 male partners of infertile couples reported a prevalence of oligozoospermia and non-obstructive azoospermia (NOA) of 49% and 11%, respectively. Noteworthy, the diagnosis could be reached in these couples in only 28%. Thus, as many as 72% of these couples remained of unknown etiology [2]. This finding highlights the need for studies that further clarify the etiology of SPGF. This need is becoming even more pressing as the prevalence of SPGF is set to increase further. Indeed, meta-regression data indicate a global trend toward decreasing sperm concentration and count [3].
Genetic screening of male infertility is of relevant importance for the diagnosis, prognosis, and decision-making clinical management of these patients [4]. In recent years, a deeper understanding of the molecular factors involved in mouse spermatogenesis has led to the search for homologous factors in humans and the design of suitable gene panels [5]. Blood testing for these gene panels in patients with SPGF has yielded encouraging results. For example, analysis of TEX11, NR5A1, and DMRT1 genes in 80 patients with NOA, normal karyotype, and without Yq microdeletions identified probable pathogenic mutations in 4 patients (5% of the whole cohort), thus increasing the diagnosis rate up to 25% [2]. Furthermore, the evaluation of a panel of 15 genes (USP9Y, NR5A1, KLHL10, ZMYND15, PLK4, TEX15, TEX11, MEIOB, SOHLH1, HSF2, SYCP3, TAF4B, NANOS1, SYCE1, and RHOXF2) in the blood of 25 patients with apparently idiopathic oligozoospermia or NOA led to the identification of pathogenic variants of the NR5A1 and TEX11 genes in 3 patients (12.0%) [6]. Similarly, the evaluation of 37 genes in 16 patients with NOA identified potential pathogenic gene variants in 6 of them [7]. These studies used a methodological approach based on next-generation sequencing (NGS) that allows for rapid and cost-effective analysis of a large number of genes.
It is known that more than 2,000 genes are involved in spermatogenesis [5], and without a careful genotype-phenotype analysis, the risk of distorted conclusions or false interpretations is high. Furthermore, only a few studies have explored the association between testicular histology and pathogenic variations of SPGF genes [8,9]. Further studies, therefore, still need to be performed to have firm conclusions. Therefore, this study aimed to evaluate the presence of potentially pathogenic variants of 29 SPGF-associated genes in a cohort of patients with NOA undergoing testicular histology.
MATERIALS AND METHODS
1. Patient selection
Sixty patients with NOA referred to the Department of Transfusion Medicine and Transplantation Biology, University Hospital Center Zagreb, Zagreb, Croatia for testicular biopsy were consecutively evaluated for eligibility. Each patient underwent testicular biopsy (after a complete diagnostic workup with their andrologist); their clinical data were carefully assessed. Twelve patients who had Klinefelter syndrome or other karyo-type abnormalities, Yq microdeletions, testicular trauma, torsion, cystic fibrosis transmembrane conductance regulator (CFTR) gene mutations, germ cell neoplasia in situ (GCNIS), or who had undergone chemotherapy were excluded from the study. Therefore, 48 patients were considered eligible and included in this study. They were divided into three groups: those who had cryptorchidism (n=9), those with varicocele (n=14), and those with idiopathic NOA (n=25) (Fig. 1). All included patients underwent blood draw for NGS analysis and candidate gene sequencing (Table 1).
Fig. 1. Flowchart of the patients included in the study. Out of the 60 patients assessed for eligibility, 48 were finally included. These were divided into 3 groups, those with cryptorchidism, varicocele, and idiopathic NOA. NOA: non-obstructive azoospermia, GCNIS: germ cell neoplasia in situ, CFTR: cystic fibrosis transmembrane conductance regulator.
Table 1. Spermatogenic failure genes sequenced in this study.
| Gene | Inheritance | OMIM number | OMIM phenotype | Mutation detection frequency | HGNC gene number |
|---|---|---|---|---|---|
| AURKC (STK13) | AR | 603495 | SPGF5 | 82.8% (72/87) | Aurora kinase C |
| BRDT | AR | 602144 | SPGF21 | 1 consanguineous family | Bromodomain testis associated |
| CATSPER1 | AR | 606339 | SPGF7 | Two 4-generation consanguineous families | Cation channel, sperm associated 1 |
| CFAP43 | AR | 617558 | SPGF19 | 13% (4/30) | Cilia and flagella associated protein 43 |
| CFAP44 | AR | 617559 | SPGF20 | - | Cilia and flagella associated protein 43 |
| DNAH1 | AR | 603332 | SPGF18 | 28%–44% | Dynein axonemal heavy chain 1 |
| DPY19L2 | AR | 613893 | SPGF9 | 66.7% (36/54) | Dpy-19-like 2 (C. elegans) |
| HSF2 | AD | 140581 | - | <1% (1/766) | Heat shock transcription factor 2 |
| KLHL10 | AD | 608778 | SPGF11 | 1.3% (7/556) | Kelch-like family member 10 |
| MEIOB | AR | 617670 | SPGF22 | 1 consanguineous family | Meiosis specific with OB domains |
| NANOS1 | AD | 608226 | SPGF12 | 2.6% (5/195) | Nanos homolog 1 (Drosophila) |
| NR5A1 | AR | 184757 | SPGF8 | 2% (7/315) | Nuclear receptor subfamily 5, group A, member 1 |
| PICK1 | - | 605926 | - | - | Protein interacting with PRKCA1 |
| PLCZ1 | AR | 608075 | SPGF17 | 2 family | Phospholipase C zeta 1 |
| PLK4 | AD | 605031 | - | 1.2% (1/81) | Polo like kinase 4 |
| RHOXF2 | - | 300447 | - | <1% (1/250) | Rhox homeobox family member 2 |
| SEPT12 | AD | 611562 | SPGF10 | 1.3% (2/160) | Septin 12 |
| SLC26A8 | AD | 608480 | SPGF3 | 2% (3/145) | Solute carrier family 26 (anion exchanger) member 8 |
| SOHLH1 | - | 610224 | - | 2% (2/100) | Spermatogenesis and oogenesis specific basic helix-loop-helix 1 |
| SPATA16 | AR | 609856 | SPGF6 | 1 consanguineous family | Spermatogenesis associated 16 |
| SYCE1 | AR | 611486 | SPGF15 | 1 consanguineous family | Synaptonemal complex central element protein 1 |
| SYCP3 | AD | 604759 | SPGF4 | 10.5% (2/19) | Synaptonemal complex protein 3 |
| SUN5 | AR | 613942 | SPGF16 | 47% (8/17) | Sad1 and UNC84 domain containing 5 |
| TAF4B | AR | 601689 | SPGF13 | 1 consanguineous family | TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| TEX11 | XLR | 300311 | SPGF, X-linked, 2 | 1-2.4% (7/289) azoospermia; 15% azoospermia with meiotic arrest | Testis expressed 11 |
| TEX15 | AR | 605795 | - | 2 family; 1 proband | Testis expressed 15, meiosis and synapsis associated |
| USP9Y | YL | 400005 | SPGF, Y-linked, 2 | 3 proband (4-db DEL; DEL incl. entire gene) | Ubiquitin specific peptidase 9, Y-linked |
| ZMYND15 | AR | 614312 | SPGF14 | 1 consanguineous family | Zinc finger, MYND-type containing 15 |
| ZPBP | - | 608498 | - | 2% (7/381) | Zona pellucida-binding protein |
OMIM: Online Mendelian Inheritance in Man, HCNC: HUGO Gene Nomenclature Committee, AR: autosomal recessive, AD: autosomal dominant, SPGF: spermatogenic failure, -: not available.
2. Ethic statement
The protocol of the study has been approved by the Ethical Committee of the University Hospital Zagreb has approved the study (reference: 8.1.-21/175-1; no.: 02/21 JG). Informed written consent was obtained from each participant after a full explanation of the purpose and nature of all procedures used. The study has been conducted according to the principles expressed in the Declaration of Helsinki.
3. Testicular histology
The samples were obtained following the open testicular biopsy procedure described by Holstein et al [10]. They were fixed in Bouin’s fluid immediately after excision inside the operational theatre. Afterwards, the tissue was washed several times in lithium carbonate solution, dehydrated, incubated in intermedium (methyl-benzoate), benzoyl and benzoyl-paraffin and then fixed in paraffin. The blocks were cut extensively by a rotary microtome Leitz 1512 (Leitz, Vienna, Austria) set at a section thickness of 4 µm. The obtained serial sections were then stained in hematoxylin and eosin (H&E stain) and prepared for histological analysis (the evaluation of preservation of seminiferous epithelium). The microscopic analysis was performed using Eclipse (E600) microscope (Nikon, Japan). The digital camera Axiocam has been employed for photo documentation (Carl Zeiss, Jena, Germany). The spermatogenesis status included the following histological pictures/classification: full spermatogenesis; hypospermatogenesis; maturation arrest; Sertoli cell-only syndrome (SCOS); tubular fibrosis and mixed atrophy.
4. Bioinformatics and genetic analysis
Genetic analysis was performed using the NGS approach and a custom-made gene panel designed to include the main genes involved in SPGF (Table 1). A MiSeq personal sequencer (Illumina, San Diego, CA, USA) was used for NGS. Sanger sequencing was performed when target region coverage was less than 10 reads. All laboratory methods have been described elsewhere [11]. We searched international databases dbSNP (https://www.ncbi.nlm.nih.gov/SNP/) and Human Gene Mutation Database professional (HGMD; http://www. biobase-international.com/product/hgmd) for all nucleotide changes. In silico evaluation of the pathogenicity of nucleotide changes in exons was performed using the Variant Effect Predictor tool (https://www.ensembl.org/Tools/VEP) and MutationTaster (https://www.mutationtaster.org). Minor allele frequencies (MAFs) were checked in the Genome Aggregation Database (gnomAD) (https://gnomad.broadinstitute.org/). All variants were evaluated according to American College of Medical Genetics and Genomics guidelines [12].
RESULTS
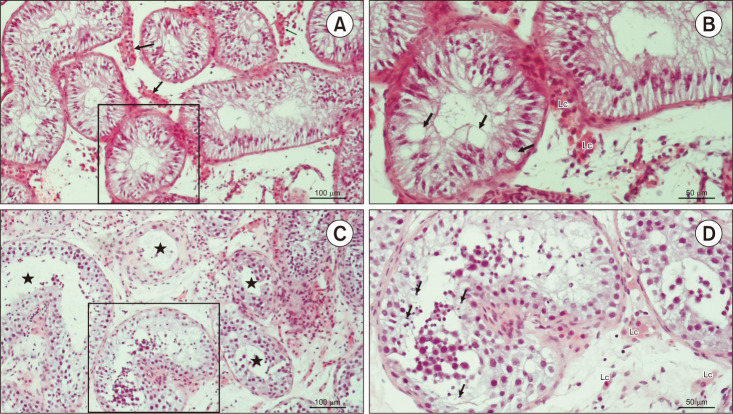
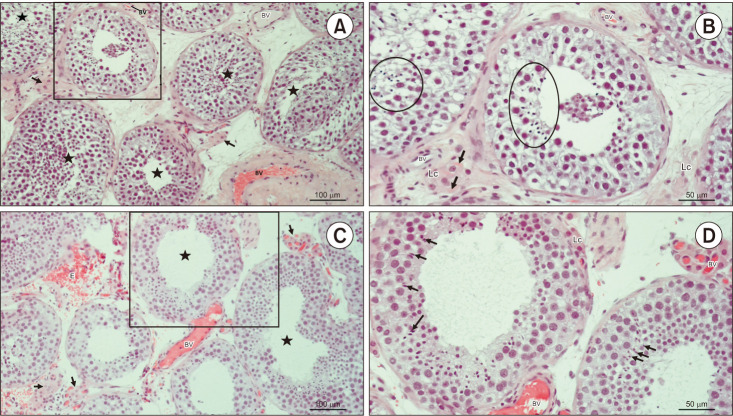
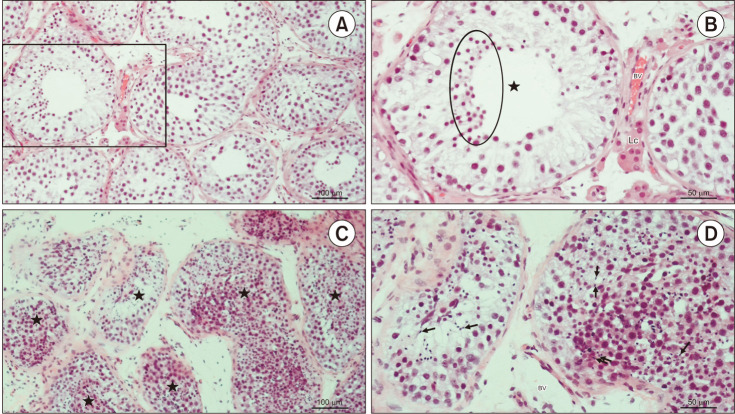
We found a possible genetic cause in 4 patients with idiopathic NOA (16%) and 2 with cryptorchidism (22%) since the mutations of these genes were predicted to be pathogenic or likely pathogenic. No pathogenic or likely pathogenic mutations were identified in patients with varicocele. Molecular and clinical data of these patients are summarized in Table 2. Their testicular histology is provided in Fig. 2, 3, 4.
Table 2. Pathogenic or likely pathogenic variants in genes identified in patients with non-obstructive azoospermia and their clinical characteristics.
| Gene symbol (OMIM number) | Patient code | Notes | FSH (IU/l) | Testis histology | Left/right testicular volume (mL) | Nucleotide change | Amino acid change | rs ID | Minor allele frequency (%) | In silico prediction |
|---|---|---|---|---|---|---|---|---|---|---|
| DNAH1 (603332) | 160585017 | Cryptorchidism | 12.9 | SCOS | 8.74/9 | NM_015512.5:c.6058G>T | NP_056327.4: pGlu2020* | - | - | Pathogenic |
| NANOS1 (608226) | 141879319 | Cryptorchidism | 11.3 | Mixed atrophy | Smaller than normal | NM_199461.4:c.262del | NP_955631.1: p.His88Thrfs*64 | rs1564744521 | - | Likely pathogenic |
| PLCZ1 (608075) | 265952375 | Idiopathic | N | Hypospermatogenesis | N | NM_033123.4:c.20del | NP_149114.2: p.Leu7Cysfs*15 | - | - | Pathogenic |
| ZPBP (608498) | 119526696 | Idiopathic | 11.48 | Hypospermatogenesis | NA | NM_007009.3:c.127+1G>A | NA | - | - | Pathogenic |
| 122836561 | Idiopathic | 10.1 | Mixed atrophy (left) Maturation arrest (right) |
N | NM_007009.3:c.4del | NP_008940.2: p.Glu2ArgfsTer54 | - | - | Pathogenic | |
| ZMYND15 (614312) | 163680875 | Idiopathic | N | Hypospermatogenesis | NA | NM_001136046.3: c.1260_1261del | NP_001129518.1: p.Gly422GlnfsTer79 | rs748488242 | 0.01 | Pathogenic |
| NM_001136046.3:c.1263del | NP_001129518.1: p. Gly422Alafs*25 | rs758076451 | 0.08 | Likely pathogenic |
OMIM: Online Mendelian Inheritance in Man, FSH: follicle stimulating hormone, SCOS: Sertoli cell-only syndrome, N: normal, NA: not assessed, -: not available.
Fig. 2. Testicular histology of patients code 160585017 and 141879319. (A) Testicular biopsy from an azoospermic patient. Seminiferous tubules display narrow lumen and are lined with Sertoli cells exclusively. Lc clusters are seen between the seminiferous tubules (→) (H&E, ×200, scale=100 µm). Patient 160585017. (B) Detail from panel A. Due to the depletion of spermatogenic cells, Sertoli cells display giant vacuoles in their cytoplasm (→) (H&E, ×200, scale=50 µm). Patient 160585017. (C) An azoospermic patient with mixed atrophy of seminiferous tubules. Seminiferous epithelium displays a variable status of spermatogenesis (★) (H&E, ×200, scale=100 µm). Patient 141879319. (D) Detail from panel C. In one of the seminiferous tubules, late spermatids (→) are clearly visible, despite the disorganisation of the seminiferous epithelium (H&E, ×200, scale bar=50 µm). Patient 141879319. Lc: Leydig cells.
Fig. 3. Testicular histology of patients code 265952375 and 119526696. (A) Testicular biopsy from a patient with hypospermatogenesis. In most tubules, (★) seminiferous epithelium is slightly disorganised but bears late spermatids. Within the loose connective tissue, there are clusters of Lc of moderate size (→) (H&E, ×200, scale bar=100 µm). Patient 265952375. (B) Detail from panel A. Despite changed stratification of spermatogenic cells, seminiferous tubules preserved the capacity to produce late spermatids (circled areas). The interstitial compartment bears small BV and Lc. Some Lc are rich in Reinke’s crystals (→) (H&E, ×200, scale bar=50 µm). Patient 265952375. (C) Testicular parenchyma sample from an azoospermic man. Seminiferous tubules display heterogeneous morphology of spermatogenesis (hypospermatogenesis, maturation arrest), including some tubules rich in late spermatids (★). Clusters of Lc (→) have an abundant network of accompanying capillaries. As a consequence of surgery, extravasated erythrocytes (E) are frequently encountered in the bioptic material (H&E, ×200, scale bar=100 µm). Patient 119526696. (D) Detail of panel C. These two seminiferous tubules have maintained a regular diameter and morphology of spermatogenesis. Some late spermatids (→) are in the final stage of spermiogenesis and will be released as spermatozoa in the lumen. Peritubular Lc are found adjacent to the lamina propria of the tubule. The interstitium is rich in BV (H&E, ×200, scale=50 µm). Patient 119526696. Lc: Leydig cells, BV: blood vessels.
Fig. 4. Testicular histology of patients code 122836561 and 16368087. (A) Testicular biopsy from an azoospermic patient with maturation arrest. The tubules are lined with Sertoli cells and all types of spermatogenic cells, apart from late spermatids (H&E, ×200, scale bar=100 µm). Patient 122836561. (B) Detail of panel A. Seminiferous tubule (★) with the “spermatid stop” (maturation arrest). Spermatogenesis progresses only to the stage of round (early) spermatids (circled area) (H&E, ×200, scale=50 µm). Patient 122836561. (C) A testicular tissue sample from an azoospermic patient with hypospermatogenesis. The stratification of seminiferous epithelium (★) is not maintained. However, all spermatogenic cells, including late spermatids, are visible (H&E, ×200, scale bar=100 µm). Patient 163680875. (D) Detail of panel C. Despite the disorganised seminiferous epithelium, many late spermatids (→) can be identified (H&E, ×200, scale=50 µm). Patient 163680875. Lc: Leydig cells, BV: blood vessels.
Variants of undetermined significance (VUS) were found in 11 patients with idiopathic NOA (44%), 3 with cryptorchidism (33%), and 8 patients with varicocele (57%). The molecular and clinical data of these patients are summarized in Table 3. VUS of the USP9Y gene was the most frequently found, being identified in 14 out of 48 patients (29%). In particular, the USP9Y c.7434+14del VUS was found in 11 patients. Five of these latter patients had testicular histology of mixed atrophy, 1 maturation arrest, 2 hypospermatogenesis, 1 SCOS in the right testis and mixed atrophy in the left one; 1 patient had hypospermatogenesis in the right testis and mixed atrophy in the left one.
Table 3. Variants with uncertain significance in genes identified in patients with non-obstructive azoospermia and their clinical characteristics.
| Gene symbol (OMIM number) | Patient code | Notes | FSH (IU/l) | Testis histology | Left/right testicular volume (mL) | Nucleotide change | Amino acid change | rs ID | Minor allele frequency (%) |
|---|---|---|---|---|---|---|---|---|---|
| CATSPER1 (606389) | 143808128 | Varicocele | NA | Mixed atrophy | NA | NM_053054.4:c.1270T>C | NP_444282.3:p.Trp424Arg | rs139761671 | 0.006 |
| CFAP44 (617559) | 115748838 | Idiopathic | N | Hypospermatogenesis | NA | NM_018338.3:c.2202_2204del | NP_060808.2:p.Glu737del | rs749873759 | - |
| DNAH1 (603332) | 111008936 | Idiopathic | 7.9 | Hypospermatogenesis | NA | NM_015512.5:c.4739C>G | NP_056327.4:p.Ala1580Gly | rs754588837 | 0.0008 |
| DPY19L2 (613893) | 125687856 | Idiopathic | N | Hypospermatogenesis | NA | NM_173812.5:c.1025A>C | NP_776173.3:p.Gln342Pro | - | - |
| NANOS1 (608226) | 105816704 | Varicocele | 8.7 | Hypospermatogenesis | NA | NM_199461.4:c.830_833del | NP_955631.1:p.Pro277Argfs*73 | rs746095721 | 0.002 |
| 131919035 | Varicocele | 26.05 | Mixed atrophy | NA | NM_199461.4:c.830_833del | NP_955631.1:p.Pro277Argfs*73 | rs746095721 | 0.002 | |
| PLK4 (605031) | 100132259 | Varicocele | 6.3 | Hypospermatogenesis | NA | NM_014264.5:c.17G>A | NP_055079.3:p.Gly6Glu | rs149003893 | 0.1 |
| 113090176 | Varicocele | 18 | Mixed atrophy | 3.5/3.5 | NM_014264.5:c.17G>A | NP_055079.3:p.Gly6Glu | rs149003893 | 0.1 | |
| SYCE1 (611486) | 163680875 | Idiopathic | N | Hypospermatogenesis | NA | NM_130784.3:c.52G>C | NP_570140.1:p.Glu18Gln | rs756386589 | 0.003 |
| SYCP3 (604759) | 130059718 | Varicocele | 27.3 | SCOS | 7.5/10 | NM_001177948.1:c.643del | NP_001171419.1:p.Ile215Leufs*2 | rs761136347 | 0.004 |
| SUN5 (613942) | 111008936 | Idiopathic | 7.9 | Hypospermatogenesis | NA | NM_080675.4:c.476G>A | NP_542406.2:p.Arg159Gln | rs144823079 | 0.09 |
| TEX11 (300311) | 125687856 | Idiopathic | N | Hypospermatogenesis | NA | NM_001003811.2:c.2186-6_2186-3del | - | - | - |
| TEX15 (605795) | 148743930 | Idiopathic | 23 | SCOS | NA | NM_001350162.2:c.541-4_541-3del | - | rs35279485 | 0.003 |
| 100132259 | Varicocele | 6.3 | Hypospermatogenesis | NA | NM_001350162.2:c.541-4_541-3del | - | rs35279485 | 0.003 | |
| USP9Y (400005) | 122354337 | Idiopathic | NA | Maturation arrest | NA | NM_004654.4:c.7434+14del | - | rs760325957 | 0.004 |
| 125687856 | Idiopathic | N | Hypospermatogenesis | NA | NM_004654.4:c.7434+14del | - | rs760325957 | 0.004 | |
| 128766983 | Idiopathic | N | Mixed atrophy | NA | NM_004654.4:c.6574del | NP_004645.2:p.Ala2192Glnfs*7 | - | - | |
| 128766983 | Idiopathic | N | Mixed atrophy | NA | NM_004654.4:c.7434+14del | - | rs760325957 | 0.004 | |
| 130424034 | Idiopathic | 28.9; 37 | Mixed atrophy | 12.6/7 | NM_004654.4:c.7434+14del | - | rs760325957 | 0.004 | |
| 148743930 | Idiopathic | 23 | SCOS | NA | NM_004654.4:c.7434+14del | - | rs760325957 | 0.004 | |
| 151184989 | Idiopathic | 28.9 | SCOS/mixed atrophy | N | NM_004654.4:c.7434+14del | - | rs760325957 | 0.004 | |
| 156833670 | Idiopathic | NA | Hypospermatogenesis/ mixed atrophy | N | NM_004654.4:c.7434+14del | - | rs760325957 | 0.004 | |
| 125938310 | Cryptorchidism | NA | Hypospermatogenesis | NA | NM_004654.4:c.449T>C | NP_004645.2:Ile150Thr | - | - | |
| 125938310 | Cryptorchidism | NA | Hypospermatogenesis | NA | NM_004654.4:c.7434+14del | - | rs760325957 | 0.004 | |
| 141879319 | Cryptorchidism | 11.3 | Mixed atrophy | Smaller than normal | NM_004654.4:c.7434+14del | - | rs760325957 | 0.004 | |
| USP9Y (400005) | 142201659 | Cryptorchidism | 26 | SCOS/Mixed atrophy | N/ Smaller than normal | NM_004654.4:c.7434+13_7434+14del | - | - | - |
| 143296311 | Cryptorchidism | NA | Hypospermatogenesis | NA | NM_004654.4:c.7434+14dup | - | rs760325957 | 0.08 | |
| 114516827 | Varicocele | N | Mixed atrophy | NA | NM_004654.4:c.7434+14del | - | rs760325957 | 0.004 | |
| 114709776 | Varicocele | N | Mixed atrophy | NA | NM_004654.4:c.7434+14del | - | rs760325957 | 0.004 | |
| 143808128 | Varicocele | NA | Mixed atrophy | NA | NM_004654.4:c.7434+14dup | - | rs760325957 | 0.08 | |
| ZMYND15 (614312) | 101797172 | Idiopathic | 6.39 | SCOS | NA | NM_001136046.3:c.1262G>A | NP_001129518.1:p.Arg421Gln | rs748907962 | 0.005 |
| 163680875 | Idiopathic | N | Hypospermatogenesis | NA | NM_001136046.3:c.1260_1262del | NP_001129518.1:p.Arg421del | rs866053815 | 0.0004 | |
| ZPBP (608498) | 120151485 | Idiopathic | 20 | SCOS | Smaller than normal | NM_007009.3:c.128-6del | - | rs202222027 | 0.02 |
OMIM: Online Mendelian Inheritance in Man, FSH: follicle stimulating hormone, NA: not assessed, N: normal, SCOS: Sertoli cell only syndrome, -: not available.
1. Genetic abnormalities in SCOS
Of the 48 included patients, SCOS was found in both testes of 5 patients with idiopathic NOA, 2 with cryptorchidism, and 1 with varicocele. The pathogenic c.6058G>T stopgain mutation in the DNAH1 gene was found in 1 patient with varicocele (Table 2, Fig. 2A, 2B). No other pathogenic mutations were found in the remaining patients. However, the ZPBP c.128-6del splice region and intron VUS and the SYCP3 c.643del frame-shift VUS were found in one patient with idiopathic NOA and one patient with varicocele, respectively (Table 3).
2. Genetic abnormalities in maturation arrest
Maturation arrest in both testes was found in only 1 patient with idiopathic NOA who carried the USP9Y c.7434+14del splice region and intron VUS (Table 3).
3. Genetic abnormalities in mixed atrophy
Overall, 4 patients with idiopathic NOA, 4 with cryptorchidism, and 10 with varicocele had mixed atrophy in both testes. The likely pathogenetic NANOS1 c.262del frameshift mutation was identified in 1 patient with cryptorchidism (Table 2, Fig. 2C, 2D). No other pathogenetic mutations were found in the remaining patients. The USP9Y c.7434+14del frameshift VUS was found in 1 patient, and the USP9Y c.7434+14del splice region and intron VUS was found in 2 patients with idiopathic NOA. In patients with varicocele and mixed atrophy, the PLK4 c.17G>A missense, the NANOS1 c.830_833del frameshift, the USP9Y c.7434+14del and c.7434+14del splice region, and intron VUS were identified (Table 3).
4. Genetic abnormalities in hypospermatogenesis
Hypospermatogenesis was found in both testes of 5 patients with idiopathic NOA, 2 patients with cryptorchidism, and 3 patients with varicocele. The pathogenetic ZPBP c.127+1G>A splice donor, the PLCZ1 c.20del frameshift, and the ZMYND15 c.1260_1261del and c.1263del frameshift mutations were identified in patients with idiopathic NOA (Table 2, Fig. 3, 4A, 4B). No other pathogenetic mutations were found. VUS were found in 3 patients with idiopathic NOA (CFAP44 c.2202_2204del inframe deletion, DNAH1 c.4739C>G missense, SUN5 c.476G>A missense, DPY19L2 c.1025A>C missense, TEX11 c.2186-6_2186-3del splice region and intron, USP9Y c.7434+14del splice region and intron, SYCE1 c.52G>C missense, and ZMYND15 c.1260_1262del inframe deletion variants), in 2 with cryptorchidism (USP9Y c.449T>C missense, USP9Y c.7434+14del splice region and intron, and USP9Y c.7434+14dup splice region and intron variant), and in 2 patients with varicocele (TEX15 c.541-4_541-3del splice region and intron, PLK4 c.17G>A missense, and NANOS1 c.830_833del frameshift variants) (Table 3).
DISCUSSION
Spermatogenesis is a complex mechanism, and various conditions can compromise it. More than 2000 genes are involved in this multifaceted process. Therefore, a genetic diagnosis can remain elusive in many cases [5]. Nowadays, NGS, a widely used method, allows the evaluation of a large number of genes simultaneously, rapidly, and inexpensively.
Few studies have evaluated mutations in genes known to cause SPGF in NOA patients with known testicular histology. A recent study by Krausz et al [8] assessed a panel of genes involved in meiosis in 147 patients with NOA and maturation arrest. At least 12 genes (ADAD2, TERB1, SHOC1, MSH4, RAD21L1, TEX11, TEX14, STAG3, MEIOB, DMRT1, MEI1, and SYCE) have been found associated with maturation arrest [8]. Therefore, the authors suggested that these genes may represent a gene panel to be used before sperm retrieval by testicular sperm extraction (TESE) in NOA patients. However, mutations/variations of the same gene seem capable of causing different testicular histological pictures. For example, partial deletion of the TEX11 gene resulted in maturation arrest in the study by Krausz et al [8], but other mutations (e.g. splice region and intron or missense) of the same gene are compatible with different testicular histological findings such as hypospermatogenesis (case ID 125687856 of the present study) or with oligozoospermia [6].
The present study was designed to understand whether variations of a homemade panel of genes involved in spermatogenesis can be associated with specific testicular histology in patients with NOA. Only SPGF-associated penetrating genes were selected. More specifically, the panel was developed based on our previous review study, in which its use led to the identification of pathogenic mutations in 12% (3/25) and likely pathogenic mutations in 44% (11/25) of patients with infertility [5]. These genes encode proteins with a role in spermatogonia proliferation, meiosis, crossing over, and spermiohistogenesis. Their variants have been reported in patients with infertility (Table 1). Furthermore, only those with more significant negative consequences (splicing, frameshift, missense, start loss, deletions) were chosen to reduce the possibility of benign variants. In contrast, those with a frequency greater than 0.05 in the general population were excluded. We found pathogenic variants of the following five genes in six patients (12.5%) using this panel.
DNAH1 encodes for a protein that belongs to the dynein family of proteins. These are microtubule-associated motor protein complexes. Specifically, the DNAH1 protein, an inner arm heavy chain dynein, has been identified in the full length of the sperm flagellum, and the gene mutations have been classically associated with multiple morphological abnormalities of sperm flagella [13]. However, DNAH1 mutations have been recently suggested deserving investigation not only in patients with asthenozoospermia but also in those with azoospermia [14]. Indeed, an observational study has analyzed DNAH1 gene mutations in a cohort of 200 patients with NOA, reporting the presence of pathogenic variants in 3 of them (1.5%) [14]. Accordingly, mutation of the DNAH5 gene, encoding for another component of the dynein family of proteins, has already been reported in a patient with NOA [15,16]. On this basis, the possible role of the DNAH1 gene in the pathogenesis of NOA needs to be further elucidated. We herein report a pathogenic c.6058G>T stopgain mutation of the DNAH1 gene in a patient with SCOS, borderline serum FSH levels, low testicular volume, and cryptorchidism. These data support the role of this gene in NOA.
NANOS1 encodes for a transcript involved in the modulation of germ cell proliferation and, therefore, acts in the very early phase of spermatogenesis [5]. Few reports have already suggested the link between mutations of this gene and SPGF. The heterozygous deletion of two single amino acids (p.Ser78del or p.Ala173del) has been reported in Polish patients with NOA, low testicular volume (6–10 mL), and elevated serum FSH levels (15.4–18.2 IU/L). The testicular histology was available only in one patient and showed SCOS [17]. In addition, the pArg246His and Arg276Tyr NANOS1 missense mutations have been described in patients with severe oligozoospermia [17]. Here we show a pathogenic frameshift NANOS1 mutation in a patient with cryptorchidism, low testicular volume, borderline FSH values, and mixed atrophy. Our findings expand the knowledge on the testicular histological features that can associate with NANOS1 mutations.
PLCZ1 encodes a protein of 608 amino acids expressed in spermatozoa and, specifically, in the acrosomal and post-acrosomal regions, in the intermediate tract, and the main part of the tail [18]. This protein represents the molecular trigger for the oocyte activation during fertilization, as human PLCZ1 has been observed to stimulate the activation of the mouse oocyte and the embryo's development up to the blastocyst stage [19]. Furthermore, PLCZ1 has been identified as a marker of human sperm health, and this protein’s total expression levels have been correlated with sperm parameters [20]. We found the c.20del frameshift pathogenic mutation of the PLCZ1 gene in a patient with apparently idiopathic NOA. He has normal testicular volume and serum FSH levels and hypospermatogenesis. There are no other studies that relate PLCZ1 to patients with NOA, most probably because the mutation of this gene is not usually searched in these patients. Further studies are needed to clarify better whether mutations of this gene are associated with hypospermatogenesis.
ZPBP encodes for a protein of 351 amino acids involved in acrosome compaction. The absence of this protein leads, in mice, to the fragmentation of the acrosome and the disruption of the Sertoli-spermatid junctions. These structural changes result in dysmorphic spermatozoa with a decreased ability to penetrate the zona pellucida [21]. However, disruption of the Sertolispermatid junction can also lead to abnormal spermatogenesis, resulting in azoospermia [22]. Despite this, no studies have described ZPBP gene mutations in patients with NOA. For the first time, we report ZPBP gene mutations in two patients with NOA, one with hypospermatogenesis and one with maturation arrest/mixed atrophy. This finding suggests that ZPBP gene mutations may play a pathogenic role in patients with NOA.
ZMYND15 encodes for a protein expressed in the testis, containing a zinc finger MYND motif and a nuclear localization signal. It is predicted to be a transcriptional repressor controlling normal temporal expression of haploid cell genes during spermiogenesis [23]. Mutations of this gene have already been associated with azoospermia due to maturation arrest [24] and in patients with severe oligozoospermia [25]. Herein, we show the presence of a pathogenic mutation of this gene in a patient with hypospermatogenesis. Interestingly, the expression of ZMYND15 mRNA in the seminal plasma may predict successful sperm retrieval with high sensitivity and specificity. Indeed, its expression was significantly decreased in patients with NOA and no sperm retrieval compared with NOA and successful sperm retrieval [26].
We found several VUS of the SPGF genes evaluated in patients with NOA. The majority of these VUSs are compatible with their testicular histological features but cannot be described as pathogenic because of the lack of functional studies. At least some of them will likely be considered pathogenic, thus further raising the diagnostic rate found in the present study. Among the genes we tested, we would like to underline the high prevalence of VUSs of the USP9Y gene and, in particular, the c.7434+14del variant. Furthermore, very interestingly, the USP9Y VUSs were found associated with different testicular histological features. These include SCOS, mixed atrophy, and hypospermatogenesis regardless of the presence of cryptorchidism or varicocele since they were also found in patients with idiopathic NOA. These findings lead to at least two major considerations: 1) the USP9Y gene may play a role in the pathogenesis of NOA; 2) there is not a direct correlation between a gene and specific testicular histology. More likely, the mutation/variation of a specific gene involved in spermatogenesis may cause different testicular histological features. This concept challenges the construction of custom-made gene panels for TESE prognostic purposes.
The USP9Y gene encodes for the ubiquitin-specific protease 9 and maps within the AZFa region of the Y chromosome. The hypothesis suggesting the role of the USP9Y gene in spermatogenesis failed when a case report documented normal spermatogenesis in a man with the complete deletion of this gene [27]. However, this is in sharp contrast with the mild testicular phenotype reported in two cases with complete USP9Y deletion [28]. Hence, still today, the role of this gene in spermatogenesis is a matter of debate. The data of the present study may suggest an association between VUSs of this gene and NOA.
The results of the present study must be interpreted with caution because of the absence of a control group consisting of fertile men and its relatively low sample size. Markedly, it was not possible to obtain testicular samples from fertile men as well the NGS analysis of their blood. This represents a general limitation of this kind of study as fertile men do not seek andrological counseling and have no reason to undergo testicular biopsy or genetic testing. On the other hand, the strict inclusion criteria, the exclusion of all possible acquired causes of NOA, the clear and detailed clinical caracterization, and the availability of testicular histology for each patient included in this study represent its main strengths.
CONCLUSIONS
In conclusion, the results of the present study showed that a relevant percentage of patients with apparently idiopathic NOA have mutations of SPGF genes. Furthermore, the data, taken together with those of other studies [6,8,9,17], suggest that not the gene itself but the type of mutation is associated with testicular histology. In fact, different mutations of the gene at stake cause various testicular histological features. This challenges the design of a gene panel with predictive value for sperm retrieval by TESE in patients with NOA.
Footnotes
Conflict of Interest: The authors have nothing to disclose.
Funding: This study was partially supported by the Scientific Center of Excellence for Reproductive and Regenerative Medicine, Republic of Croatia, and by the European Union through the European Regional Development Fund, under grant agreement No. KK.01.1.1.01.0008, project: “Reproductive and Regenerative Medicine - Exploring New Platforms and Potentials”.
- Conceptualization: RC, AEC.
- Data curation: DJ, MV, MB, RC.
- Formal analysis: DJ, MV, MB, RC.
- Funding acquisition: DJ.
- Investigation: AEC, RC, DJ.
- Methodology: MB, DJ, MV.
- Project administration: AEC, RC, DJ.
- Visualization: RAC, SLV.
- Writing – original draft: RC.
- Writing – review & editing: AEC, DJ, RC.
Data Sharing Statement
The data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.
References
- 1.Punab M, Poolamets O, Paju P, Vihljajev V, Pomm K, Ladva R, et al. Causes of male infertility: a 9-year prospective monocentre study on 1737 patients with reduced total sperm counts. Hum Reprod. 2017;32:18–31. doi: 10.1093/humrep/dew284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tüttelmann F, Ruckert C, Röpke A. Disorders of spermatogenesis: perspectives for novel genetic diagnostics after 20 years of unchanged routine. Med Genet. 2018;30:12–20. doi: 10.1007/s11825-018-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine H, Jørgensen N, Martino-Andrade A, Mendiola J, Weksler-Derri D, Mindlis I, et al. Temporal trends in sperm count: a systematic review and meta-regression analysis. Hum Reprod Update. 2017;23:646–659. doi: 10.1093/humupd/dmx022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asero P, Calogero AE, Condorelli RA, Mongioi' L, Vicari E, Lanzafame F, et al. Relevance of genetic investigation in male infertility. J Endocrinol Invest. 2014;37:415–427. doi: 10.1007/s40618-014-0053-1. [DOI] [PubMed] [Google Scholar]
- 5.Cannarella R, Condorelli RA, Duca Y, La Vignera S, Calogero AE. New insights into the genetics of spermatogenic failure: a review of the literature. Hum Genet. 2019;138:125–140. doi: 10.1007/s00439-019-01974-1. [DOI] [PubMed] [Google Scholar]
- 6.Cannarella R, Condorelli RA, Paolacci S, Barbagallo F, Guerri G, Bertelli M, et al. Next-generation sequencing: toward an increase in the diagnostic yield in patients with apparently idiopathic spermatogenic failure. Asian J Androl. 2021;23:24–29. doi: 10.4103/aja.aja_25_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Araujo TF, Friedrich C, Grangeiro CHP, Martelli LR, Grzesiuk JD, Emich J, et al. Sequence analysis of 37 candidate genes for male infertility: challenges in variant assessment and validating genes. Andrology. 2020;8:434–441. doi: 10.1111/andr.12704. [DOI] [PubMed] [Google Scholar]
- 8.Krausz C, Riera-Escamilla A, Moreno-Mendoza D, Holleman K, Cioppi F, Algaba F, et al. Genetic dissection of spermatogenic arrest through exome analysis: clinical implications for the management of azoospermic men. Genet Med. 2020;22:1956–1966. doi: 10.1038/s41436-020-0907-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salas-Huetos A, Tüttelmann F, Wyrwoll MJ, Kliesch S, Lopes AM, Goncalves J, et al. Disruption of human meiotic telomere complex genes TERB1, TERB2 and MAJIN in men with non-obstructive azoospermia. Hum Genet. 2021;140:217–227. doi: 10.1007/s00439-020-02236-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holstein AF, Schulze W, Breucker H. In: Male infertility. Hargreave TB, editor. London: Springer; 1994. Histopathology of human testicular and epididymal tissue; pp. 105–148. [Google Scholar]
- 11.Cannarella R, Precone V, Guerri G, Busetto GM, Di Renzo GC, Gerli S, et al. Clinical evaluation of a custom gene panel as a tool for precision male infertility diagnosis by next-generation sequencing. Life (Basel) 2020;10:242. doi: 10.3390/life10100242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben Khelifa M, Coutton C, Zouari R, Karaouzène T, Rendu J, Bidart M, et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X, Zhu D, Zhang H, Jiang Y, Hu X, Geng D, et al. Associations between DNAH1 gene polymorphisms and male infertility: a retrospective study. Medicine (Baltimore) 2018;97:e13493. doi: 10.1097/MD.0000000000013493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fliegauf M, Olbrich H, Horvath J, Wildhaber JH, Zariwala MA, Kennedy M, et al. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am J Respir Crit Care Med. 2005;171:1343–1349. doi: 10.1164/rccm.200411-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zariwala MA, Knowles MR, Omran H. Genetic defects in ciliary structure and function. Annu Rev Physiol. 2007;69:423–450. doi: 10.1146/annurev.physiol.69.040705.141301. [DOI] [PubMed] [Google Scholar]
- 17.Kusz-Zamelczyk K, Sajek M, Spik A, Glazar R, Jędrzejczak P, Latos-Bieleńska A, et al. Mutations of NANOS1, a human homologue of the Drosophila morphogen, are associated with a lack of germ cells in testes or severe oligo-astheno-teratozoospermia. J Med Genet. 2013;50:187–193. doi: 10.1136/jmedgenet-2012-101230. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Castro RA, Amoroso-Sanches F, Stokes JE, Graham JK, Carnevale EM. Localisation of phospholipase Cζ1 (PLCZ1) and postacrosomal WW-binding protein (WBP2 N-terminal like) on equine spermatozoa and flow cytometry quantification of PLCZ1 and association with cleavage in vitro. Reprod Fertil Dev. 2019;31:1778–1792. doi: 10.1071/RD19217. [DOI] [PubMed] [Google Scholar]
- 19.Cox LJ, Larman MG, Saunders CM, Hashimoto K, Swann K, Lai FA. Sperm phospholipase Czeta from humans and cynomolgus monkeys triggers Ca2+ oscillations, activation and development of mouse oocytes. Reproduction. 2002;124:611–623. doi: 10.1530/rep.0.1240611. [DOI] [PubMed] [Google Scholar]
- 20.Kashir J, Mistry BV, BuSaleh L, Abu-Dawas R, Nomikos M, Ajlan A, et al. Phospholipase C zeta profiles are indicative of optimal sperm parameters and fertilisation success in patients undergoing fertility treatment. Andrology. 2020;8:1143–1159. doi: 10.1111/andr.12796. [DOI] [PubMed] [Google Scholar]
- 21.Lin YN, Roy A, Yan W, Burns KH, Matzuk MM. Loss of zona pellucida binding proteins in the acrosomal matrix disrupts acrosome biogenesis and sperm morphogenesis. Mol Cell Biol. 2007;27:6794–6805. doi: 10.1128/MCB.01029-07. Erratum in: Mol Cell Biol 2008;28:2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berruti G, Paiardi C. The dynamic of the apical ectoplasmic specialization between spermatids and Sertoli cells: the case of the small GTPase Rap1. Biomed Res Int. 2014;2014:635979. doi: 10.1155/2014/635979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan W, Si Y, Slaymaker S, Li J, Zheng H, Young DL, et al. Zmynd15 encodes a histone deacetylase-dependent transcriptional repressor essential for spermiogenesis and male fertility. J Biol Chem. 2010;285:31418–31426. doi: 10.1074/jbc.M110.116418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayhan Ö, Balkan M, Guven A, Hazan R, Atar M, Tok A, et al. Truncating mutations in TAF4B and ZMYND15 causing recessive azoospermia. J Med Genet. 2014;51:239–244. doi: 10.1136/jmedgenet-2013-102102. [DOI] [PubMed] [Google Scholar]
- 25.Hu TY, Zhang H, Meng LL, Yuan SM, Tu CF, Du J, et al. Novel homozygous truncating variants in ZMYND15 causing severe oligozoospermia and their implications for male infertility. Hum Mutat. 2021;42:31–36. doi: 10.1002/humu.24138. [DOI] [PubMed] [Google Scholar]
- 26.Hashemi MS, Mozdarani H, Ghaedi K, Nasr-Esfahani MH. Could analysis of testis-specific genes, as biomarkers in seminal plasma, predict presence of focal spermatogenesis in non-obstructive azoospermia? Andrologia. 2020;52:e13483. doi: 10.1111/and.13483. [DOI] [PubMed] [Google Scholar]
- 27.Luddi A, Margollicci M, Gambera L, Serafini F, Cioni M, De Leo V, et al. Spermatogenesis in a man with complete deletion of USP9Y. N Engl J Med. 2009;360:881–885. doi: 10.1056/NEJMoa0806218. [DOI] [PubMed] [Google Scholar]
- 28.Krausz C, Degl'Innocenti S, Nuti F, Morelli A, Felici F, Sansone M, et al. Natural transmission of USP9Y gene mutations: a new perspective on the role of AZFa genes in male fertility. Hum Mol Genet. 2006;15:2673–2681. doi: 10.1093/hmg/ddl198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.