Abstract
As single-cell RNA sequencing (scRNA-seq) has emerged as a great tool for studying cellular heterogeneity within the past decade, the number of available scRNA-seq datasets also rapidly increased. However, reuse of such data is often problematic due to a small cohort size, limited cell types, and insufficient information on cell type classification. Here, we present a large integrated scRNA-seq dataset containing 224,611 cells from human primary non-small cell lung cancer (NSCLC) tumors. Using publicly available resources, we pre-processed and integrated seven independent scRNA-seq datasets using an anchor-based approach, with five datasets utilized as reference and the remaining two, as validation. We created two levels of annotation based on cell type-specific markers conserved across the datasets. To demonstrate usability of the integrated dataset, we created annotation predictions for the two validation datasets using our integrated reference. Additionally, we conducted a trajectory analysis on subsets of T cells and lung cancer cells. This integrated data may serve as a resource for studying NSCLC transcriptome at the single cell level.
Subject terms: High-throughput screening, Data integration
Introduction
The technology of whole-transcriptome single-cell RNA sequencing (scRNA-seq) was first introduced in 20091. Since then, this technique has rapidly emerged as a powerful tool for studying cellular heterogeneity in various fields, including Oncology2,3. The number of publicly available scRNA-seq datasets containing samples from various tissues and species greatly increased within the past decade, with the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO)4,5 being one of the most popular platforms dedicated to deposition of such data. However, small cohort size, inclusion of limited cell types, and insufficient annotation of cell populations are common obstacles to efficient reuse of the data, often slowing down the analysis. Therefore, several strategies have been developed for integration of the scRNA-seq data and correction of technical differences between the samples, also termed as batch effect6.
Among these strategies, Harmony7 and Seurat8 are commonly recommended9,10. Seurat identifies pairs of cells in a similar biological state across the datasets, termed anchors, and uses them to organize the data into a single integrated, corrected expression matrix. In this approach, cell subpopulations shared between different datasets are identified using canonical correlation analysis (CCA) and mutual nearest neighbours (MNNs)11,12. Seurat also enables data transfer between scRNA-seq datasets. In data transfer, principal component (PC) structure of a reference dataset is projected onto the query based on transfer anchors, and annotation predictions are generated for query cells11. In contrast to Seurat, Harmony integration operates on the PCs values, which represent a low-dimensional embedding of the original expression matrix and projects cells from different batches into a new shared embedding. Rather than using CCA, Harmony clusters cells in a way to obtain a balanced ratio of cells from different batches in each cluster, via k-means clustering and cluster centroid correction10,13. In our analysis, we decided to perform the integration and batch correction using Seurat.
Lung and bronchus cancer is the leading cause of cancer mortalities worldwide, with non-small cell lung cancer (NSCLC) accounting for the majority of new lung cancer cases14,15. Histologically, NSCLC is commonly classified as one of the two most common subtypes, including lung adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC)16. LUAD has been confirmed to originate mostly from type two alveolar epithelial cells of the lung, whereas LUSC can arise either from basal cells of the bronchial epithelium, club cells, or alveolar cells16. Growing evidence suggests a prognostic and predictive value of diverse cell types in NSCLC, including fibroblasts, immune, and endothelial cells17–19. A detailed single-cell atlas exploring a variety of cell populations would thus provide insight into the tumor microenvironment and help unveil novel markers for improvement of NSCLC therapy.
Until now, published integrated lung datasets have been established for healthy tissue or single cell types20,21. However, a large-scale integrated data set of NSCLC, comprising data from several studies, and a variety of cell populations is still missing, up until very recently, there is a high-resolution single-cell atlas of the tumor microenvironment in NSCLC specifically22. Here, we present an integrated single-cell transcriptomic dataset for human NSCLC, containing 224,611 cells, with a thorough characterization of present cell types on two levels of annotation (Fig. 1). Our integrated transcriptome data may serve as a vast resource for studying gene expression patterns between cell types, reconstructing cellular trajectories and identification of potential novel biomarkers in NSCLC.
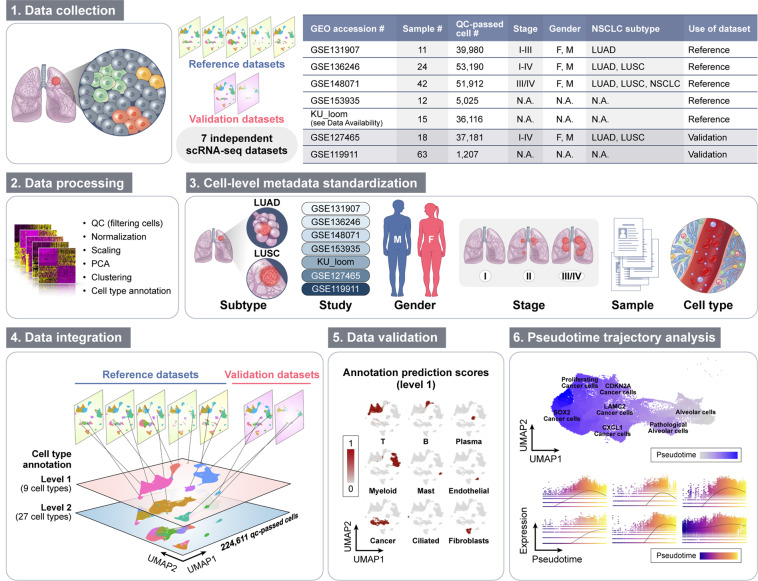
Fig. 1.
Study design. Seven independent datasets were collected, pre-processed, and clustered using the Seurat package. Cell-level metadata on cell type classification and sample clinical information was standardized for all datasets. To obtain a large reference dataset, five datasets were integrated in an anchor-based manner. The cells of the integrated reference were subjected to a standard workflow for clustering and cell type annotation. The integrated reference was used for annotation of the validation dataset and the two datasets were then merged into the final dataset. Additionally, pseudotime trajectory analysis of selected clusters was conducted.
Results
Generation of an integrated reference dataset of NSCLC tumors
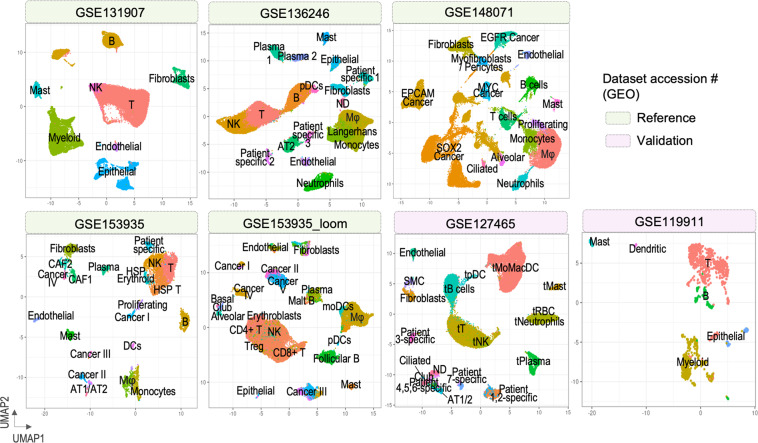
For generation of the large-scale integrated dataset, we collected seven publicly available scRNA-seq datasets comprising of 185 NSCLC human tumor samples in total. Among the seven datasets, five were used to construct an integrated reference and the remaining two served as validation. Details on samples included in the analysis are summarized in Tables 1,2. Using the R Seurat package (v 4.1.0)8 we followed a standard workflow for quality control and clustering of cells (Table 3). Each dataset was processed individually, including only human tumor samples. We identified diverse cell populations which were clearly separated on Uniform Manifold Approximation and Projection (UMAP) embeddings (Fig. 2).
Table 1.
Naming and basic information on the datasets used in the study.
| GEO accession # | Sample # | QC-passed cell # | Stage | Gender | NSCLC subtype | Use of dataset |
|---|---|---|---|---|---|---|
| GSE131907 | 11 | 39,980 | I-III | F, M | LUAD | Reference |
| GSE136246 | 24 | 53,190 | I-IV | F, M | LUAD, LUSC | Reference |
| GSE148071 | 42 | 51,912 | III/IV | F, M | LUAD, LUSC, NSCLC | Reference |
| GSE153935 | 12 | 5,025 | N.A. | N.A. | N.A. | Reference |
| Loom files (see Data Availability) | 15 | 36,116 | N.A. | N.A. | N.A. | Reference |
| GSE127465 | 18 | 37,181 | I-IV | F, M | LUAD, LUSC | Validation |
| GSE119911 | 63 | 1,207 | N.A. | N.A. | N.A. | Validation |
Table 2.
Accession numbers of the samples from each study used in the reanalysis.
Table 3.
Analysis of the seven scRNA-seq datasets in R Seurat- workflow.
| Dataset | GSE131907 | GSE136246 | GSE148071 | GSE153935 | KU_loom | GSE127465 | GSE119911 |
|---|---|---|---|---|---|---|---|
| Step 1: QC | nFeature_RNA > 200 & < 3000; Percent_mt <20 | ||||||
| Step 2: Normalization | |||||||
| Step 3: Identification of variable features | |||||||
| Step 4: Scaling the data | |||||||
| Step 5: PCA dimensional reduction | |||||||
| Step 6: Determine no. of PCs | 20 | 20 | 20 | 20 | 20 | 20 | 20 |
| Step 7: Cell clustering | |||||||
| Step 8: UMAP plotting | |||||||
| Step 9: Cell type annotation | Annotation provided | Annotation provided | Own annotation | Own annotation | Own annotation | Annotation provided | Own annotation |
Fig. 2.
UMAP plots of the seven datasets after individual clustering analysis.
Subsequently, we integrated the five reference datasets using identified integration anchors and performed the downstream analysis. The reference dataset comprised of 186,223 cells, distributed among 27 clusters (Supplementary Fig. 1a–e). By examining expression patterns of canonical marker genes (see details in the “Methods” section), we performed a two-level classification of clusters, in which 9 and 27 cell types were identified for level 1 and 2 annotation, respectively (Fig. 3a, Supplementary Fig. 1f, g). The main cell types include immune (T, B, plasma, mast, and myeloid cells), epithelial (cancer and ciliated cells), and stromal cells (fibroblasts and endothelial cells), all of which were further divided into subtypes in level 2 annotation.
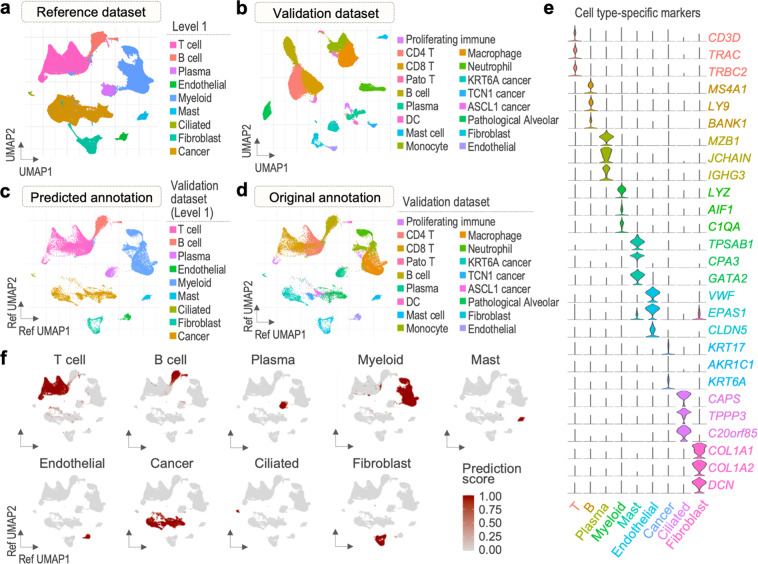
Fig. 3.
Transfer of reference cell type labels to cells of the validation dataset. (a) Cell types of the reference dataset (level 1). (b) Cell types of the validation dataset (original annotation). (c) Cells of the validation dataset projected in a UMAP structure of the reference with predicted annotation (level 1). (d) Cells of the validation dataset projected in a UMAP structure of the reference with original annotation. (e) Violin plots of reference cell type-specific markers expression in the validation cells (level 1). (f) Validation cell type prediction scores (level 1).
Use of the reference dataset for annotation of query datasets
We integrated the two validation datasets via the anchor-based approach to obtain one validation dataset comprised of 39,511 cells. We clustered the cells of the validation dataset into 17 clusters, in which we initially classified independently of the reference dataset using canonical marker genes (Fig. 3b, Supplementary Fig. 2a–f). To assess the validity of the reference dataset, we conducted a cell type label transfer from the reference onto the validation dataset (Fig. 3c, Supplementary Fig. 2g). As a result, we obtained two levels of predicted annotations for the validation dataset. Cells of the validation dataset were well distributed in UMAP structure of the reference dataset, and all cell types defined in the reference were identified in the validation. We observed a satisfactory match between the original and predicted validation annotation in terms of main cell types, supporting the technical quality of our integrated data as an annotation reference atlas (Fig. 3d).
Next, we assessed the accuracy of the annotation predictions obtained in the mapping process. The cells of the validation dataset showed cell type-specific expression of marker genes (Fig. 3e) and high prediction score computed by the Seurat for all major cell types (Fig. 3f, Supplementary Fig. 3), supporting the credibility of the predicted annotations. To avoid inclusion of faultily classified validation cells in the final dataset, only the cells with high prediction score (>0.5) were merged into the final dataset and were selected as default identities of the validation dataset for further analyses.
Cell type classification of the final dataset
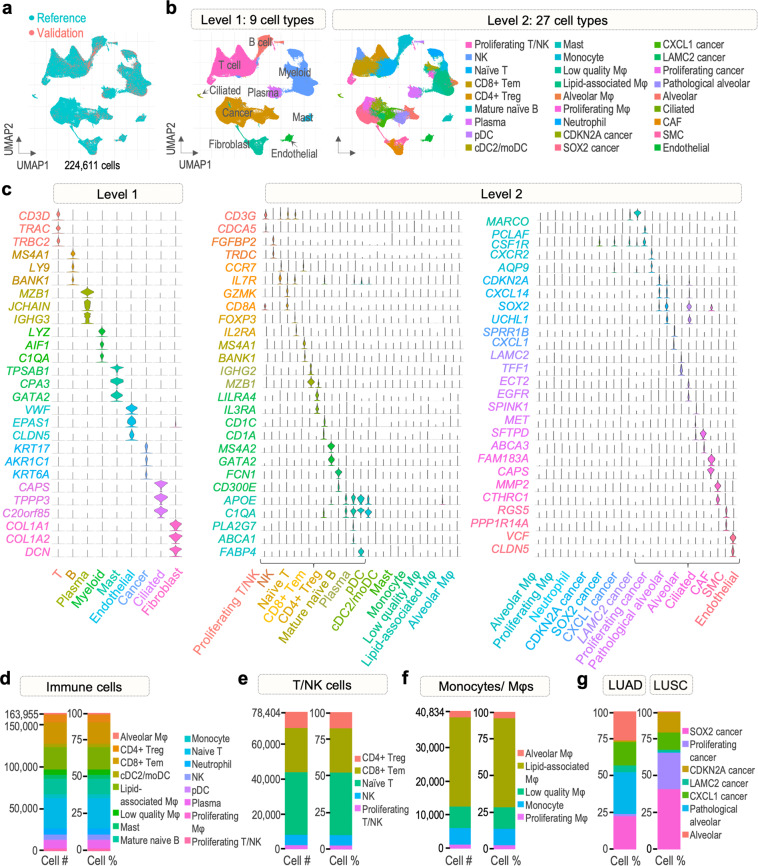
We merged the reference and validation datasets into a final dataset comprised of 224,611 cells. The UMAP plot in Fig. 4a shows a clear overlap of cells from the validation dataset with the reference in a single UMAP embedding, demonstrating a successful incorporation of the two datasets. We defined the previously generated two levelled annotation as final cell type classification of the final dataset (Fig. 4b,c).
Fig. 4.
Cell types of the final dataset. (a) Distribution of cells derived from the reference and validation datasets in the final dataset. (b) Two levels of cluster annotation. (c) Violin plots of cell type-specific gene markers used for level 1 and 2 annotations. (d) Number of cells from immune cell population and their proportion. (e) Number of cells from population of T and NK cells and their proportion. (f) Number of cells from population of monocytes and macrophages and their proportion. (g) Proportion of cells from cancer cell population in LUAD and LUSC samples.
We aimed at thoroughly characterizing the immune infiltrate and expression patterns of immune cells that reside in the tumor microenvironment (TME), including diverse subpopulations of T cells and myeloid cells (Fig. 4d–f, Supplementary Fig. 4). Subtyping of the T cell cluster revealed that naïve T cells accounted for majority of all T cells (45.41%), followed by CD8+ effector memory T cells (Tem), CD4+ regulatory T cells (Treg), NK, and proliferating T cells (32.29, 11.76, 7.68, and 2.88%, respectively). We found lipid-associated macrophages to be the most abundant subtype of the monocyte/macrophage group (64.70%). The remaining subtypes included low-quality macrophages, monocytes, alveolar, and proliferating macrophages (15.56, 12.01, 4.75, and 2.98%, respectively). Among other immune cells, we found a considerable amount of mature naïve B cells (11.24% of all immune cells), plasma cells (5.66%), and neutrophils (4.45%). Moreover, a detectable level of mast cells (2.85%) and dendritic cells (conventional/monocyte-derived 2.48%, plasmacytoid 0.59%) was identified. These results highlight the diversity of the immune cell population in the TME of NSCLC and provide a field of action for future studies.
We next identified seven subclusters in the cancer cluster, including alveolar cells, pathological alveolar cells and five cancer cell subtypes. We classified the cancer cells into the five cancer subtypes as CDKN2A, SOX2, CXCL1, LAMC2, and proliferating cancer based on the top markers that are highly expressed in each cluster. Interestingly, we found substantial differences in proportions of cancer cell subtypes between LUAD and LUSC samples (Fig. 4g). In LUAD, the proportion of alveolar (21.05% vs 0.5%), pathological alveolar (30.07% vs 0.34%) was much higher comparing to LUSC, in line with the previously reported LUAD developing from alveolar cells16. LUAD samples were also characterized by a higher percentage of LAMC2 (4.97% vs 1.79%) and CXCL1 cells (16.95% vs 12.5%). As CXCL1 and LAMC2 are associated with recruitment of neutrophils and macrophages into tumor tissue23,24, these results demonstrate the significant role of immune cell population in LUAD growth. In contrast to LUAD, LUSC samples were more abundant in CDKN2A (14.65% vs 1.05%), proliferating (26.33% vs 1.73%), and SOX2 cancer cells (43.89% vs 24.18%). Tumor suppressor CDKN2A regulates the cell cycle and is frequently altered in LUSC25. Similarly, SOX2 controls cell proliferation and is commonly amplified in LUSC, promoting its growth by maintaining stem cell-like phenotype of cancer cells26. Together, these three cell subtypes account for over 80% of all cancer cells derived from LUSC samples, indicating the highly malignant nature of this tumor subtype.
Assessment of the validity of the final integrated dataset
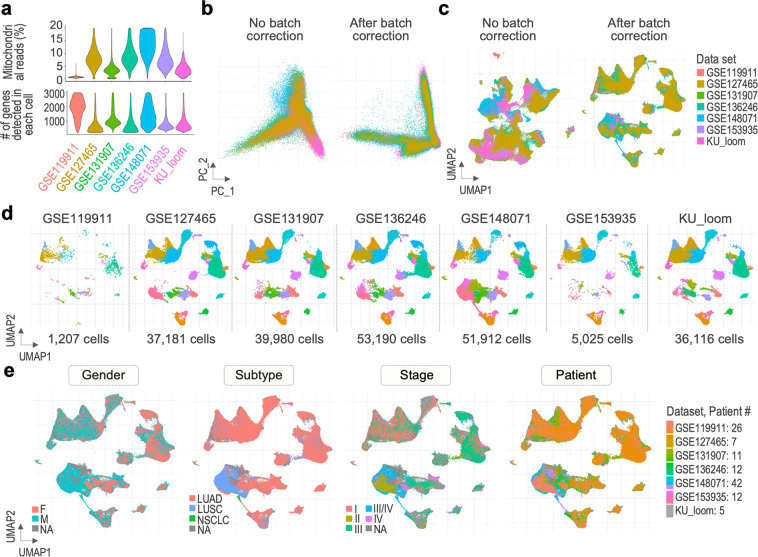
For quality control of our final dataset, we applied commonly used quality metrics such as percentage of counts from mitochondrial genes and number of features (Fig. 5a). Cells that have more than 20% of mitochondria-related read counts or unique feature counts over 3,000 and less than 200 were filtered out. To visualize the efficiency of the integration process, we generated PC and dimensional reduction plots comparing our final dataset and a dataset comprised of the same datasets, merged without batch correction. The resulting plots in Fig. 5b show a major disconnection between the merged data when colored by dataset in the first two PCs. In contrast, the final data clearly overlays between the source datasets, suggesting that the effect of non-biological variances have been corrected. Cells of the batch-uncorrected dataset are separated by study of origin, rather than cell type, whereas those of the batch-corrected final dataset are distributed more evenly according to study in every cluster (Fig. 5c), suggesting cells are grouped by cell type that account for the most variance in the data. The distribution of cells in the UMAP plot visualized in Fig. 5d once again shows that cells from each study can be found in each cluster, suggesting that the differences in contribution to formation of the clusters arise from the count of cells in the initial data sets, rather than differences in cell type composition. Altogether, these results indicate that the process of integration and data transfer with Seurat was completed successfully, minimizing the effect of technical batches on cell clustering. An additional value of our dataset is the collected metadata containing clinical information on patients included in the study, such as gender, histological subtype, and stage of the tumor (Fig. 5e).
Fig. 5.
Quality of the final dataset. (a) Violin plots showing QC metrics of cells included in the final dataset, including percentage of counts from mitochondrial genes (Percent_mt) and number of genes (nFeature_RNA), split by study. (b) Removal of the batch effect presented by PCA and dimensional reduction plots (c) for the final dataset, in a comparison to a merged dataset without batch correction. (d) Distribution of cells from the seven analyzed datasets in the clusters of the final dataset. (e) UMAP plots of the final dataset cells grouped by clinical metadata, including gender, subtype, stage, and patient.
Pseudotime trajectory analysis
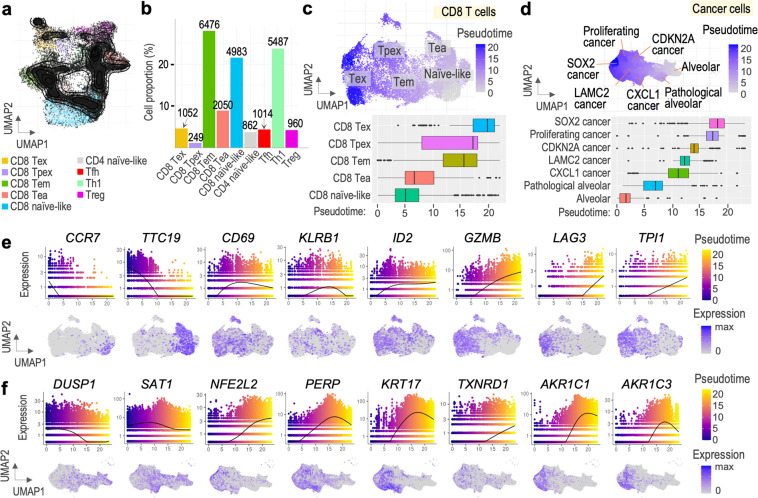
T cells are the main target of immunotherapy in NSCLC27,28. According to current understanding of CD8+ T cell differentiation, upon activation naïve T cells differentiate into different effector and memory T cells. In tumors, chronic T cell stimulation leads to disturbance in their differentiation toward dysfunction and exhaustion characterized by loss of effector function and expression of inhibitory receptors29. To depict the different states of CD8+ T cells, we conducted a pseudotime trajectory analysis using the R Monocle3 package7. Specifically, we extracted T cells from our final dataset and reanalyzed their cell states using R ProjecTILs package30. We projected our query cells on the reference map provided by ProjecTILs and calculated the number of cells in each state (Fig. 6a,b, Supplementary Fig. 5). In total, 14,810 cells were classified as ‘CD8_NaiveLike’, ‘CD8_EarlyActiv’, ‘CD8_EffectorMemory’, ‘CD8_Tpex’, or ‘CD8_Tex’ cells for subsequent analyses. The extracted cells were re-clustered using Seurat and subjected to trajectory analysis via Monocle3. As T cells differentiate from naïve to effector to memory and exhausted states, we specified the trajectory to start from CD8_NaiveLike cells. The UMAP plot in Fig. 6c shows the population of CD8+ T cells colored by pseudotime, suggesting a continuous progression of cells from naïve-like to exhausted state. Ordering the five cell states by median pseudotime revealed a transition from naïve-like cells to early activated, followed by effector memory, precursor exhausted, and exhausted cells (Fig. 6c, bottom). Importantly, although the median pseudotime of Tpex cluster is higher than that of Tem, it exhibits a wider spectrum of pseudotime values, suggesting that initiation of T cell exhaustion may start upon activation. We further verified these results by analysing genes which showed significant expression changes in pseudotime. We observed clear differences in expression of naïve (CCR7, TTC19), memory (CD69, ID2), cytotoxicity (KLRB1, GZMB), and exhaustion-related genes (LAG3, TPI1) in pseudotime, supporting a consistent shift of T cells towards differentiation and exhaustion (Fig. 6f).
Fig. 6.
Pseudotime trajectory analysis. (a) Distribution of query T cells on ProjecTILs reference map. (b) Percentage and number of T cells from each functional state. (c) UMAP plot of CD8+ T cell subset colored by pseudotime (top) and boxplot showing median pseudotime of each cell type (bottom). (d) UMAP plot of cancer cell subset colored by pseudotime (top) and boxplot showing median pseudotime of each cell type (bottom). (e) Chosen genes of the CD8+ T cell subset showing changing expression in pseudotime. (f) Chosen genes of the cancer cell subset showing changing expression in pseudotime.
Lastly, we performed a joint trajectory analysis of all 46,450 cells of the cancer cluster. Starting from alveolar cells, the cells transformed into pathological alveolar cells, CXCL1, LAMC2, CDKN2A, proliferating, and SOX2 cancer cells as they progressed in pseudotime (Fig. 6d). We identified distinct changes in expression of reactive oxygen species (ROS) genes in pseudotime (Fig. 6f). Expression of DUSP1 was the highest at the beginning of pseudotime, as opposed to TXNRD1 which was mainly expressed in late pseudotime. It has been suggested that high expression of DUSP1 is correlated with better prognosis, whereas TXNRD1, with poor patient prognosis in lung cancer31. These results demonstrate progression of cancer cells in the trajectory towards more resistant phenotype. Moreover, few genes have been reported to be implicated in p53 signalling (SAT1, PERP, KRT17)32–34 or ferroptosis (SAT1, NFE2L2, AKR1C1, AKR1C3)32,35,36. Together, the presented dataset reveals complete cancer cell landscape of NSCLC tumor progression, associated with ROS metabolism and p53 activity.
Discussion
In this study, we generated a large-scale scRNA-seq dataset of human primary NSCLC tumors containing both LUAD and LUSC samples, from early to advanced stages, of both genders. While each dataset used to generate the presented data contains a limited number of tumor cells, our integrated dataset may provide a more comprehensive cell landscape of NSCLC specifically. We thoroughly annotated the presented scRNA-seq data to facilitate the re-use of our data for novel cell type discovery and extensive characterization of diverse cell subpopulations, including immune cells residing in tumor microenvironment. In addition, inclusion of patients from different studies with standardized cell-level metadata may enable the study of NSCLC transcriptome on a wider spectrum of samples than the analysis of single study or dataset having limited number of QC-passed cells.
Since in this analysis we reused data from published studies, we observed substantial batch effects arising from the technical differences in library preparation and data processing. According to several benchmarking studies evaluating performance of available batch effect correction methods, Harmony and Seurat are described as tools suitable for scRNA-seq analysis. As Harmony utilizes PCA subspace as input for further transformations, it is often noted to be faster and require less memory. However, it limits its usability in gene-based analyses in which expression matrix is the input, such as pseudotime or identification of differentially expressed genes9,10. Integration with Seurat usually requires more memory and a longer runtime. Nevertheless, it can precisely merge batches while producing a corrected gene expression matrix, useful for downstream analysis9,10. Seurat also enables data transfer between scRNA-seq datasets. In data transfer, PCA structure of a reference dataset is projected onto the query based on transfer anchors, and annotation predictions are generated for query cells. This workflow does not require CCA, which substantially reduces the runtime11. Taken together, although Harmony may be faster in the process of integration itself, we employed functions of the Seurat, which allows a wider range of downstream analyses, to integrate the datasets and correct for batch effects. In addition, we used the same pre-processing and clustering workflow on each dataset prior to integration, to minimize potential differences between them. We used PCA and UMAP to visualize batch effect correction, which showed good batch mixing results in our final dataset in comparison to a dataset obtained using basic merging function.
Through extensive analysis of marker genes’ expression, we classified the 224,611 cells into nine main cell types, which were further divided into twenty-seven subtypes primarily consisting of immune cell populations. Apart from cell types commonly described in NSCLC microenvironment, we identified a subtype of low-quality macrophages characterized by elevated expression of mitochondrial genes and genes encoding for ribosomal proteins, suggesting damage or stress of the cells. We found that the most abundant subtype of macrophages show a lipid-related signature, with expression of PLA2G7, ABCA1, FOLR2, APOE, CTSB/D, and C1QA/B/C, which is associated with phagocytosis and immunosuppression37. Sub-clustering of T cells further revealed the presence of naïve, helper and cytotoxic cells, as well as NK and proliferating T cells. Comparing cell type abundances between our dataset and the recently published NSCLC atlas (Salcher et al.)22, we observed several differences in fractions of cell types. Interestingly, neutrophils, which are short-lived cells, often underrepresented in scRNA-seq studies, in our dataset account for 3.25% of all cell populations, while in the Salcher et al. dataset22, only 1.5%. In addition, fractions of epithelial cells and B cells are higher in our dataset (20.99% vs ~15% and 8.64% vs ~5.5%, respectively). In contrast, abundance of macrophages/monocytes is lower in our dataset than in the Salcher et al. dataset (18.18% vs 28,5%). Nevertheless, we identified several subtypes of myeloid cells showing distinct signatures, as noted above.
We conducted an additional functional state analysis of the T cells using ProjecTILs30 and subjected a subset of CD8+ cells to pseudotime trajectory analysis via Monocle37. The mouse-derived reference map provide by ProjecTILs may attribute to a large number of our query cells that were filtered out during QC process. Species-specific differences in gene expression may have contributed to failure in detecting the query cells as “pure” T cells. However, we believe that the remaining QC-passed 14,810 cells which were successfully assigned to reference functional states were sufficient to perform a trajectory analysis. Our analyses revealed a dynamic functional spectrum of CD8+ T cells from naïve to exhausted state in NSCLC, showing effective data reuse.
Finally, we identified seven cancer subclusters and analyzed possible dynamics between them in pseudotime. The seven subclusters included alveolar cells, pathological alveolar cells expressing both normal respiratory cell markers (SFTPB, AGR3) and genes related to cancer progression (SPINK1, MET), as well as five cancer subsets. We observed considerable differences in abundance of cells from each of the seven subtypes between LUAD and LUSC samples, implicating stem cell-like phenotype of LUSC cells and immune infiltration promotion by LUAD. Pseudotime trajectory analysis revealed a dynamic path in which normal epithelial cells went under a transformation to cancer cells. This process was accompanied by changes in expression of genes related to p53 signaling and ROS metabolism, showing further differences in progression of the two tumor subtypes. Interestingly, several genes (PERP, KRT17, AKR1C1) have been recently reported as potential NSCLC biomarkers36,38. As we previously noted, LUAD and LUSC showed distinct differences in cancer cell subtype content. The cell types more abundant in LUSC (SOX2, CDKN2A, proliferating cancer) were placed later in pseudotime than the LUAD-specific cell types (alveolar, pathological alveolar, CXCL1 cancer). Altogether, these results suggest that LUSC cells show more aggressive and resistant characteristics. In conclusion, these results demonstrate the usefulness and technical validity of our integrated scRNA-seq dataset. Reuse of this large-scale dataset may contribute to further understanding of NSCLC.
Methods
Data collection and pre-processing
Seven publicly available scRNA-seq datasets were collected, comprising of 185 NSCLC human primary tumor samples in total. Datasets GSE13190728,39, GSE13624640,41, GSE14807142,43, GSE15393544,45, and KU_loom (https://gbiomed.kuleuven.be/scRNAseq-NSCLC)46,47 were used to create a large reference dataset, whereas datasets GSE12746527,48 and GSE11991149,50 served as validation. Details on samples included in the analysis are summarized in Tables 1, 2. Using Seurat package (v 4.1.0)8 in R (v 4.1.1), a standard workflow for data pre-processing and the clustering of cells was followed. Briefly, the seven scRNA-seq datasets were analyzed individually, including quality control (QC), normalization, feature selection, data scaling, dimensional reduction by principal component analysis (PCA), clustering, Uniform Manifold Approximation and Projection (UMAP) reduction, and visualization of clusters. From each dataset, human tumor samples were extracted and loaded into respective Seurat objects. QC of the gene-cell matrix consisted of filtering the cells such that cell with counts from mitochondrial genes below 20 percent and number of features more than 200 and less than 3000 were included. Detailed information on quality control and subsequent steps of the single data sets analysis are described in Table 3. Gene expression normalization was applied to each dataset using LogNormalize method. The number of principal components (PCs) to include in further analysis was determined based on JackStraw plots and Elbow plots generated for each dataset. Cell clustering was conducted using FindNeighbors and FindClusters functions, and non-linear dimensional reduction was managed by RunUMAP function. For datasets GSE13190728,39, KU_loom (https://gbiomed.kuleuven.be/scRNAseq-NSCLC)46,47, and GSE12746527,48, metadata on cell type annotation of the single cells was provided by the authors. Clusters from the remaining datasets were assigned to specific cell types considering positive (avglog2FC > 0) cell type-specific markers found via FindAllMarkers function. For visualization, UMAP plots showing obtained annotated clusters were generated (Fig. 2).
Integration of reference datasets
To establish a single reference dataset, five datasets (GSE13190728,39, GSE13624640,41, GSE14807142,43, GSE15393544,45, and KU_loom (https://gbiomed.kuleuven.be/scRNAseq-NSCLC)46,47) were integrated and analyzed using functions of the Seurat package, following the workflow proposed by Satija Lab11,12 (https://satijalab.org/seurat/articles/integration-introduction.html). A list consisting of five pre-processed datasets previously specified as reference was created and features repeatedly shared within the objects were identified using Seurat’s SelectIntegrationFeatures function. Subsequently, FindIntegrationAnchors function enabled selection of a set of 219,432 cell pairs in a similar biological state (anchors), which were then utilized in the integration process via IntegrateData function. Once the integration process was executed successfully, the integrated assay was specified as default for downstream analysis.
Reference dataset analysis
The integrated dataset comprised of 186223 cells. Standard steps leading to clustering of the cells were conducted, including identification of highly variable features, scaling of the data, PCA, UMAP (no. of dims = 30), and finding neighbours (Supplementary Fig. 1). Identification of clusters was performed at resolutions 0.02 and 0.5 respectively, to obtain two versions of dimension reduction plots containing different number of clusters (level1 and level2). The clusters were classified using two types of gene markers: positive biomarkers detected using FindAllMarkers function, and markers conserved across the datasets detected via FindConservedMarkers function (grouping.var = Study, DefaultAssay = RNA). The cell type identities were firstly assigned to clusters based on the conserved markers, while the general biomarkers were a secondary source of information for both levels of annotation. Since identification of conserved markers is based on differential expression testing, the RNA assay was used in this analysis instead of the integrated assay, to include more potential markers. Features conserved among the data sets were identified using study of origin as the grouping variable. Cell type specificity of the markers was further confirmed using several recent publications28,37,51–78. As a result, 9 and 27 cell types were found for level 1 and 2 of annotation, respectively (Supplementary Fig. 1, Table 4).
Table 4.
Additional marker genes used for cell type classification of clusters (level 2).
| Level2 annotation – additional markers | |
|---|---|
| Proliferating T/NK | TOP2A, MKI67, NUSAP1 |
| NK | KLRF1, KLRD1, KLRB1, GNLY, NKG7 |
| Naïve T | PTPRC |
| CD8+ Tem | GZMA, GZMM, CD8 |
| CD4+ Treg | CTLA4, CD4 |
| Mature naïve B | CD22, CD53, CD79A |
| Plasma | IGHA2, IGHM, TNFRSF17 |
| pDCs | IRF7, IRF8 |
| cDC2/moDCs | CLEC10A |
| Mast | KIT, CPA3, CD63 |
| Monocytes | CD14, CSF3R |
| Low quality Mφ | LYZ, FTL, high number of MT- and RPL/S genes |
| Lipid-associated Mφ | MS4A7, IL1B, IL4I1, FOLR2, APOE, C1QA/B/C, CTSB/D |
| Alveolar Mφ | MCEMP1, PPARG, MRC1 |
| Proliferating Mφ | CDCA8, MKI67, CENPF, CD14, TOP2A |
| Neutrophils | FCGR3B, CSF3R, S100A12, S100A8 |
| CDKN2A Cancer | CDK4, PUM3, NTS, EPCAM |
| SOX2 Cancer | KRT17, S100A2, SFN, PTHLH, PERP |
| CXCL1 Cancer | SPRR3, AGR2, CEACAM6 |
| LAMC2 Cancer | FGB, FGA, FGG, PAEP, TESC |
| Proliferating Cancer | MKI67, TOP2A, CENPF, CDC20 |
| Pathological Alveolar | SFTPB, WFDC2, AGR2, AGR3, MUC1 |
| Alveolar | SFTPC, AQP4, SCGB3A1 |
| Ciliated | FOXJ1, CDHR3 |
| CAF | SPARC, FAP, PDGFRB |
| SMC | ACTA2, CALD1, TAGLN |
| Endothelial | FLT1, PECAM1 |
Validation dataset analysis
Datasets GSE12746527,48 and GSE11991149,50 acquired from NCBI GEO were processed individually with the previously described workflow for clustering analysis (Table 3). For dataset GSE12746527,48 identified clusters were annotated based on metadata provided by the authors, whereas clusters of dataset GSE11991149,50 were annotated manually based on canonical markers (Fig. 2). Due to a small number of QC-passed cells from dataset GSE11991149,50 (1359 cells), Seurat anchor-based integration of cells from the two validation datasets was conducted to form a single validation dataset (see “Integration of reference datasets”). The integrated validation dataset included 39511 cells and was subjected to clustering analysis (no. of dims = 30). At resolution 0.5, 17 clusters were obtained and initially classified according to expression of conserved gene markers (see “Reference dataset analysis”, Supplementary Fig. 2a–f).
Cell type label transfer from reference to validation dataset
Following Seurat anchor-based methodology for data transfer11 (https://satijalab.org/seurat/articles/multimodal-reference-mapping.html), cell type classifications of the integrated dataset were transferred onto the validation dataset. Cells of our reference dataset were utilized as reference and validation dataset, as query. Transfer anchors were identified using FindTransferAnchors function, with LogNormalize as normalization method, PCA as reduction, and number of dimensions 30. The anchorset was applied in the label transfer using MapQuery function, leading to creation of two levels of predicted annotations (Fig. 3, Supplementary Fig. 2g). To assess efficiency of the new query annotations, prediction scores were generated for each of the query cells. Cells with high prediction score (predicted.celltype.score >0.5) were included in further analysis. The integrated and validation datasets were merged into a final dataset comprising 224611 cells and visualized in UMAP embedding of the reference. Additionally, feature plots showing strength of the cell type predictions were generated (Fig. 3f, Supplementary Fig. 3). The two levelled annotation was used as final classification of cells of the final dataset. Expression of marker genes and proportions of cell types were investigated (Fig. 4, Supplementary Fig. 4).
Visualization of batch effect correction and final dataset quality
Violin plots showing QC metrics applied during pre-processing of the seven datasets were generated, including percentage of mitochondrial reads and number of genes detected in each cell (Fig. 5a). To assess the efficiency of the integration process, several visualization methods were used to compare our final dataset with a simply merged dataset without batch effect correction. A list of the seven pre-processed datasets was created and all respective Seurat objects were merged using Merge_Seurat_List function. The merged dataset was subjected to clustering analysis in a way corresponding to clustering of the reference and validation datasets (identification of highly variable features, scaling of data, PCA, UMAP (30 dims), finding neighbours, identification of clusters at resolution 0.5). PCA and dimensional reduction plots were visualized for both the final and merged datasets (Fig. 5b,c). UMAP plot of the cells of the final dataset split by study of origin was made to observe the placement of cells from each dataset (Fig. 5d). In addition, plots of the final UMAP structure colored by collected metadata were generated, including gender, histological subtype, stage of the tumor, and patient id (Fig. 5e).
Pseudotime trajectory analysis
CD8+ T cells
Cells of the T cell cluster according to level 1 of annotation were extracted from the final dataset into a new Seurat object. Cell states of the T cells were re-evaluated using ProjecTILs R package (v 3.0)30. Reference atlas of tumor-infiltrating T lymphocytes was loaded from ProjecTILs Git repository. Our query T cells were filtered and projected on the reference map (Fig. 6a). Cell states predictions were generated according to gene expression signatures pre-determined by the package for specific T cell subtypes (Fig. 6b, Supplementary Fig. 5). Cells predicted as belonging to CD8+ T cell functional clusters were selected for further analysis, including CD8_NaiveLike, CD8_EarlyActiv, CD8_EffectorMemory, CD8_Tpex, and CD8_Tex. The newly obtained subset of cells was pre-processed using Seurat functions (FindVariableFeatures, ScaleData, RunPCA, FindNeighbors (dims = 1:20), FindClusters (resolution = 0.5), RunUMAP) and visualized using the annotations predicted by ProjecTILs. Pre-processed Seurat object was converted to an object of cell dataset class using as.cell_data_set function and data size factors were calculated using estimate_size_factors for trajectory analysis in Monocle37 (v 1.0.0). Cell and gene-level metadata, counts and cluster information, as well as previously obtained UMAP embedding were retrieved from the Seurat object to the cell dataset object. All cells were assigned to a single partition and the trajectory graph was learned using learn_graph function. To place the cells in pseudotime, cells which belong to CD8_NaiveLike cluster were assigned as “roots” of the trajectory. Obtained cell pseudotime information was stored in the T cell Seurat object’s metadata for visualization purposes (Fig. 6c). Differential expression analysis was performed to identify genes of which expression changes in pseudotime (Fig. 6e). The top genes were found by arranging the results by q_value and status (status == “OK”).
Cancer cells
Cells belonging to clusters “Alveolar”, “CDKN2A Cancer”, “CXCL1 Cancer”, “LAMC2 Cancer”, “Pathological Alveolar”, “Proliferating Cancer”, and “SOX2 Cancer” in level 2 of annotation were extracted from the final dataset into a new Seurat object. The Seurat object containing cancer cells was converted to an object of cell dataset class. Size factors for each cell were estimated using estimate_size_factors function. Necessary metadata etc. was retrieved from the Seurat object as described above for the T cell analysis. The trajectory graph was learned using learn_graph function and cells belonging to the Alveolar cluster were assigned as “roots” of the trajectory for pseudotime analysis. Obtained cell level pseudotime information was stored in the cancer cell Seurat object’s metadata (Fig. 6d). Accordingly, differential expression analysis was performed to identify genes with changing expression in pseudotime, and the top genes were found by arranging the results by q_value and status (status == “OK”) (Fig. 6f).
Supplementary information
Acknowledgements
This work was conceived and carried out at the Ajou Precision Medicine Laboratory at the Department of Biochemistry and Molecular Biology, Ajou University School of Medicine. We acknowledge support provided by the National Research Foundation (NRF) of Korea (2020R1A6A1A03043539, 2020M3A9D8037604, and 2022R1C1C1004756). S.B.L. is supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HR22C1734).
Author contributions
S.B.L. conceptualized and designed the study. S.B.L. and K.P. developed the R pipeline to establish the integrated data set. K.P. analyzed and interpreted the data. Both authors reviewed and contributed to the manuscript.
Data availability
Among input data processed in the reanalysis, six datasets were acquired from NCBI GEO (GSE13190728,39 (2020), GSE13624640,41 (2021), GSE14807142,43 (2021), GSE15393544,45 (2020), GSE12746527,48 (2019), GSE11991149,50 (2022)). Dataset referred to as KU_loom was downloaded from resources of the Ku Leuven Laboratory for Functional Epigenetics as “all cells” loom file (https://gbiomed.kuleuven.be/scRNAseq-NSCLC (2018)46,47). Set of samples used in this study is summarized Table 2. Seurat object of our final scRNA-seq dataset with UMAP embeddings can be found at figshare (10.6084/m9.figshare.c.6222221.v3)79. Associated data, including matrix of raw and normalized counts, and metadata (two levels of cell type annotation, validation dataset prediction scores, QC metrics, patient id, gender, study of origin, tumor subtype and stage) are available under the same figshare project as “RNA_rawcounts_matrix”, “Integrated_normalized_counts”, and “Metadata” files, respectively.
Code availability
The main computational tools used in this study are R language based. Seurat8 was used for data pre-processing, integration, and label transfer between reference and validation datasets. ProjecTILs30 was used for interpretation of T cell states, and Monocle37 was used for pseudotime trajectory analysis. The R codes used for pre-processing of the used datasets, reference and validation datasets analysis, and pseudotime trajectory analysis can be found at figshare as “NSCLC_data_reanalysis_codes” file (10.6084/m9.figshare.c.6222221.v3)79.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-023-02074-6.
References
- 1.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, et al. Single-cell RNA sequencing in cancer research. J Exp Clin Cancer Res. 2021;40:81. doi: 10.1186/s13046-021-01874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seow JJW, Wong RMM, Pai R, Sharma A. Single-Cell RNA Sequencing for Precision Oncology: Current State-of-Art. J Indian Inst Sci. 2020;100:579–588. doi: 10.1007/s41745-020-00178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett T, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luecken MD, et al. Benchmarking atlas-level data integration in single-cell genomics. Nat Methods. 2022;19:41–50. doi: 10.1038/s41592-021-01336-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao J, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tran HTN, et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020;21:12. doi: 10.1186/s13059-019-1850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chazarra-Gil R, van Dongen S, Kiselev VY, Hemberg M. Flexible comparison of batch correction methods for single-cell RNA-seq using BatchBench. Nucleic Acids Res. 2021;49:e42. doi: 10.1093/nar/gkab004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stuart T, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902 e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korsunsky I, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 15.Qu J, et al. The progress and challenge of anti-PD-1/PD-L1 immunotherapy in treating non-small cell lung cancer. Ther Adv Med Oncol. 2021;13:1758835921992968. doi: 10.1177/1758835921992968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sainz de Aja J, Dost AFM, Kim CF. Alveolar progenitor cells and the origin of lung cancer. J Intern Med. 2021;289:629–635. doi: 10.1111/joim.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catacchio I, Scattone A, Silvestris N, Mangia A. Immune Prophets of Lung Cancer: The Prognostic and Predictive Landscape of Cellular and Molecular Immune Markers. Transl Oncol. 2018;11:825–835. doi: 10.1016/j.tranon.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu H, et al. Three subtypes of lung cancer fibroblasts define distinct therapeutic paradigms. Cancer Cell. 2021;39:1531–1547 e1510. doi: 10.1016/j.ccell.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goveia J, et al. An Integrated Gene Expression Landscape Profiling Approach to Identify Lung Tumor Endothelial Cell Heterogeneity and Angiogenic Candidates. Cancer Cell. 2020;37:21–36 e13. doi: 10.1016/j.ccell.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Schupp JC, et al. Integrated Single-Cell Atlas of Endothelial Cells of the Human Lung. Circulation. 2021;144:286–302. doi: 10.1161/CIRCULATIONAHA.120.052318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Travaglini KJ, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature. 2020;587:619–625. doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salcher S, et al. High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell. 2022;40:1503–1520 e1508. doi: 10.1016/j.ccell.2022.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan M, et al. Tumor-Derived CXCL1 Promotes Lung Cancer Growth via Recruitment of Tumor-Associated Neutrophils. J Immunol Res. 2016;2016:6530410. doi: 10.1155/2016/6530410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu M, et al. LAMC2 promotes the proliferation of cancer cells and induce infiltration of macrophages in non-small cell lung cancer. Ann Transl Med. 2021;9:1392. doi: 10.21037/atm-21-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang P, et al. TP53 and CDKN2A mutations in patients with early-stage lung squamous cell carcinoma: an analysis of the correlations and prognostic outcomes. Ann Transl Med. 2021;9:1330. doi: 10.21037/atm-21-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samulin Erdem J, et al. Mutations in TP53 increase the risk of SOX2 copy number alterations and silencing of TP53 reduces SOX2 expression in non-small cell lung cancer. BMC Cancer. 2016;16:28. doi: 10.1186/s12885-016-2061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zilionis R, et al. Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity. 2019;50:1317–1334 e1310. doi: 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim N, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun. 2020;11:2285. doi: 10.1038/s41467-020-16164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang W, et al. Exhausted CD8+T Cells in the Tumor Immune Microenvironment: New Pathways to Therapy. Front Immunol. 2020;11:622509. doi: 10.3389/fimmu.2020.622509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andreatta M, et al. Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat Commun. 2021;12:2965. doi: 10.1038/s41467-021-23324-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leone A, Roca MS, Ciardiello C, Costantini S, Budillon A. Oxidative Stress Gene Expression Profile Correlates with Cancer Patient Poor Prognosis: Identification of Crucial Pathways Might Select Novel Therapeutic Approaches. Oxid Med Cell Longev. 2017;2017:2597581. doi: 10.1155/2017/2597581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ou Y, Wang SJ, Li D, Chu B, Gu W. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc Natl Acad Sci USA. 2016;113:E6806–E6812. doi: 10.1073/pnas.1607152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen K, Luo Z, Li Z, Liu Y, Zhao Q. PERP gene therapy attenuates lung cancer xenograft via inducing apoptosis and suppressing VEGF. Cancer Biol Ther. 2011;12:1114–1119. doi: 10.4161/cbt.12.12.18435. [DOI] [PubMed] [Google Scholar]
- 34.Baraks G, et al. Dissecting the Oncogenic Roles of Keratin 17 in the Hallmarks of Cancer. Cancer Res. 2022;82:1159–1166. doi: 10.1158/0008-5472.CAN-21-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu S, et al. The role of ferroptosis in lung cancer. Biomark Res. 2021;9:82. doi: 10.1186/s40364-021-00338-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang F, Zheng Y, Li X, Luo H, Luo L. Ferroptosis-related gene AKR1C1 predicts the prognosis of non-small cell lung cancer. Cancer Cell Int. 2021;21:567. doi: 10.1186/s12935-021-02267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma RY, Black A, Qian BZ. Macrophage diversity in cancer revisited in the era of single-cell omics. Trends Immunol. 2022;43:546–563. doi: 10.1016/j.it.2022.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Chen JW, Dhahbi J. Lung adenocarcinoma and lung squamous cell carcinoma cancer classification, biomarker identification, and gene expression analysis using overlapping feature selection methods. Sci Rep. 2021;11:13323. doi: 10.1038/s41598-021-92725-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim N, 2020. GEO. //identifiers.org/geo/GSE131907
- 40.Maroni G, et al. Identification of a targetable KRAS-mutant epithelial population in non-small cell lung cancer. Commun Biol. 2021;4:370. doi: 10.1038/s42003-021-01897-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maroni G, 2021. GEO. //identifiers.org/geo/GSE136246
- 42.Wu F, et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat Commun. 2021;12:2540. doi: 10.1038/s41467-021-22801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu F, 2021. GEO. //identifiers.org/geo/GSE148071
- 44.Hanley CJ, et al. Single-cell analysis reveals prognostic fibroblast subpopulations linked to molecular and immunological subtypes of lung cancer. Nat Commun. 2023;14:387. doi: 10.1038/s41467-023-35832-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanley CJ, Waise S. 2020. GEO. //identifiers.org/geo/GSE153935
- 46.Lambrechts D, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24:1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 47.Lambrechts, D. et al. Phenotype molding of stromal cells in the lung tumor microenvironment.https://gbiomed.kuleuven.be/scRNAseq-NSCLC (2018). [DOI] [PubMed]
- 48.Zilionis R, 2019. GEO. //identifiers.org/geo/GSE127465
- 49.Li Q, et al. Molecular profiling of human non-small cell lung cancer by single-cell RNA-seq. Genome Med. 2022;14:87. doi: 10.1186/s13073-022-01089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li, Q. & Wang, R. GEO.https://identifiers.org/geo/GSE119911 (2022).
- 51.Chen C, et al. Analysis of the Expression of Cell Division Cycle-Associated Genes and Its Prognostic Significance in Human Lung Carcinoma: A Review of the Literature Databases. Biomed Res Int. 2020;2020:6412593. doi: 10.1155/2020/6412593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J, et al. Single-cell transcriptome and antigen-immunoglobin analysis reveals the diversity of B cells in non-small cell lung cancer. Genome Biol. 2020;21:152. doi: 10.1186/s13059-020-02064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colpitts SL, Dalton NM, Scott P. IL-7 receptor expression provides the potential for long-term survival of both CD62Lhigh central memory T cells and Th1 effector cells during Leishmania major infection. J Immunol. 2009;182:5702–5711. doi: 10.4049/jimmunol.0803450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crinier A, et al. High-Dimensional Single-Cell Analysis Identifies Organ-Specific Signatures and Conserved NK Cell Subsets in Humans and Mice. Immunity. 2018;49:971–986 e975. doi: 10.1016/j.immuni.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daniel JM, et al. Regulator of G-Protein Signaling 5 Prevents Smooth Muscle Cell Proliferation and Attenuates Neointima Formation. Arterioscler Thromb Vasc Biol. 2016;36:317–327. doi: 10.1161/ATVBAHA.115.305974. [DOI] [PubMed] [Google Scholar]
- 56.Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat Immunol. 2013;14:986–995. doi: 10.1038/ni.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gutiontov SI, et al. CDKN2A loss-of-function predicts immunotherapy resistance in non-small cell lung cancer. Sci Rep. 2021;11:20059. doi: 10.1038/s41598-021-99524-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karachaliou N, Rosell R, Viteri S. The role of SOX2 in small cell lung cancer, lung adenocarcinoma and squamous cell carcinoma of the lung. Transl Lung Cancer Res. 2013;2:172–179. doi: 10.3978/j.issn.2218-6751.2013.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim EY, et al. Early lung carcinogenesis and tumor microenvironment observed by single-cell transcriptome analysis. Transl Oncol. 2022;15:101277. doi: 10.1016/j.tranon.2021.101277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kosibaty Z, Murata Y, Minami Y, Noguchi M, Sakamoto N. ECT2 promotes lung adenocarcinoma progression through extracellular matrix dynamics and focal adhesion signaling. Cancer Sci. 2021;112:703–714. doi: 10.1111/cas.14743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Liu W, Zhang X, Wang Y. Cancer-associated fibroblast-secreted collagen triple helix repeat containing-1 promotes breast cancer cell migration, invasiveness and epithelial-mesenchymal transition by activating the Wnt/beta-catenin pathway. Oncol Lett. 2021;22:814. doi: 10.3892/ol.2021.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell. 2019;176:775–789 e718. doi: 10.1016/j.cell.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu T, et al. Cancer-associated fibroblasts: an emerging target of anti-cancer immunotherapy. J Hematol Oncol. 2019;12:86. doi: 10.1186/s13045-019-0770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol. 2012;12:191–200. doi: 10.1038/nri3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Minegishi K, et al. TFF-1 Functions to Suppress Multiple Phenotypes Associated with Lung Cancer Progression. Onco Targets Ther. 2021;14:4761–4777. doi: 10.2147/OTT.S322697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moon YW, et al. LAMC2 enhances the metastatic potential of lung adenocarcinoma. Cell Death Differ. 2015;22:1341–1352. doi: 10.1038/cdd.2014.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morgan D, Tergaonkar V. Unraveling B cell trajectories at single cell resolution. Trends Immunol. 2022;43:210–229. doi: 10.1016/j.it.2022.01.003. [DOI] [PubMed] [Google Scholar]
- 68.Mould, K. J., Jackson, N. D., Henson, P. M., Seibold, M. & Janssen, W. J. Single cell RNA sequencing identifies unique inflammatory airspace macrophage subsets. JCI Insight4, 10.1172/jci.insight.126556 (2019). [DOI] [PMC free article] [PubMed]
- 69.Rindler, T. N. et al. Alveolar injury and regeneration following deletion of ABCA3. JCI Insight2, 10.1172/jci.insight.97381 (2017). [DOI] [PMC free article] [PubMed]
- 70.Shaykhiev R, et al. Smoking-induced CXCL14 expression in the human airway epithelium links chronic obstructive pulmonary disease to lung cancer. Am J Respir Cell Mol Biol. 2013;49:418–425. doi: 10.1165/rcmb.2012-0396OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su W, et al. Smooth muscle-selective CPI-17 expression increases vascular smooth muscle contraction and blood pressure. Am J Physiol Heart Circ Physiol. 2013;305:H104–113. doi: 10.1152/ajpheart.00597.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szabo PA, et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat Commun. 2019;10:4706. doi: 10.1038/s41467-019-12464-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang C, Liu Y, Kessler PS, Vaughan AM, Oram JF. The macrophage cholesterol exporter ABCA1 functions as an anti-inflammatory receptor. J Biol Chem. 2009;284:32336–32343. doi: 10.1074/jbc.M109.047472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van der Leun AM, Thommen DS, Schumacher TN. CD8(+) T cell states in human cancer: insights from single-cell analysis. Nat Rev Cancer. 2020;20:218–232. doi: 10.1038/s41568-019-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yao J, et al. UCHL1 acts as a potential oncogene and affects sensitivity of common anti-tumor drugs in lung adenocarcinoma. World J Surg Oncol. 2022;20:153. doi: 10.1186/s12957-022-02620-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu S, et al. CXCL1 as an Unfavorable Prognosis Factor Negatively Regulated by DACH1 in Non-small Cell Lung Cancer. Front Oncol. 2019;9:1515. doi: 10.3389/fonc.2019.01515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Z, et al. Identification of small proline-rich protein 1B (SPRR1B) as a prognostically predictive biomarker for lung adenocarcinoma by integrative bioinformatic analysis. Thorac Cancer. 2021;12:796–806. doi: 10.1111/1759-7714.13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhuang Q, et al. Single-Cell Transcriptomic Analysis of Peripheral Blood Reveals a Novel B-Cell Subset in Renal Allograft Recipients With Accommodation. Front Pharmacol. 2021;12:706580. doi: 10.3389/fphar.2021.706580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prazanowska K, Lim SB. 2022. An integrated single-cell transcriptomic dataset for non-small cell lung cancer. figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Kim N, 2020. GEO. //identifiers.org/geo/GSE131907
- Maroni G, 2021. GEO. //identifiers.org/geo/GSE136246
- Wu F, 2021. GEO. //identifiers.org/geo/GSE148071
- Hanley CJ, Waise S. 2020. GEO. //identifiers.org/geo/GSE153935
- Zilionis R, 2019. GEO. //identifiers.org/geo/GSE127465
- Prazanowska K, Lim SB. 2022. An integrated single-cell transcriptomic dataset for non-small cell lung cancer. figshare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Among input data processed in the reanalysis, six datasets were acquired from NCBI GEO (GSE13190728,39 (2020), GSE13624640,41 (2021), GSE14807142,43 (2021), GSE15393544,45 (2020), GSE12746527,48 (2019), GSE11991149,50 (2022)). Dataset referred to as KU_loom was downloaded from resources of the Ku Leuven Laboratory for Functional Epigenetics as “all cells” loom file (https://gbiomed.kuleuven.be/scRNAseq-NSCLC (2018)46,47). Set of samples used in this study is summarized Table 2. Seurat object of our final scRNA-seq dataset with UMAP embeddings can be found at figshare (10.6084/m9.figshare.c.6222221.v3)79. Associated data, including matrix of raw and normalized counts, and metadata (two levels of cell type annotation, validation dataset prediction scores, QC metrics, patient id, gender, study of origin, tumor subtype and stage) are available under the same figshare project as “RNA_rawcounts_matrix”, “Integrated_normalized_counts”, and “Metadata” files, respectively.
The main computational tools used in this study are R language based. Seurat8 was used for data pre-processing, integration, and label transfer between reference and validation datasets. ProjecTILs30 was used for interpretation of T cell states, and Monocle37 was used for pseudotime trajectory analysis. The R codes used for pre-processing of the used datasets, reference and validation datasets analysis, and pseudotime trajectory analysis can be found at figshare as “NSCLC_data_reanalysis_codes” file (10.6084/m9.figshare.c.6222221.v3)79.