Abstract
A major opportunity in nuclear cardiology is the many significant artificial intelligence (AI) applications that have recently been reported. These developments include using deep learning (DL) for reducing the needed injected dose and acquisition time in perfusion acquisitions also due to DL improvements in image reconstruction and filtering, SPECT attenuation correction using DL without need for transmission images, DL and machine learning (ML) use for feature extraction to define myocardial left ventricular (LV) borders for functional measurements and improved detection of the LV valve plane and AI, ML, and DL implementations for MPI diagnosis, prognosis, and structured reporting. Although some have, most of these applications have yet to make it to widespread commercial distribution due to the recency of their developments, most reported in 2020. We must be prepared both technically and socio-economically to fully benefit from these and a tsunami of other AI applications that are coming.
Keywords: Nuclear cardiology, Absolute myocardial blood flow, Myocardial flow reserve, Artificial intelligence, Machine learning, Deep learning
Introduction
Over the past 50 years, the nuclear cardiology imaging field has evolved and developed enormously, becoming an integral part of current clinical practice and playing crucial roles in detection, risk stratification, and treatment selection for patients with known or suspected cardiovascular disease [1, 2]. Single-photon emission computed tomography (SPECT) myocardial perfusion imaging (MPI) is the most widely performed noninvasive technique to detect coronary artery disease (CAD) — the single most common cause of death in the developed world — with several millions procedures performed worldwide annually [3, 4]. More recently, the introduction of cardiac positron emission tomography (PET) to clinical environments with its ability to quantify absolute myocardial blood flow has exhibited improved image quality, reduction in radiation dose, and greater diagnostic and prognostic value [5, 6]. While nuclear imaging modalities are powerful tools to study cardiovascular pathophysiology, a key aspect for their widespread clinical usage over other modalities has been the continuous development of algorithms and techniques for automated processing, quantification, and display [7–9].
In recent years, artificial intelligence (AI)-based techniques and methods have crossed over from applications such as search engines, natural language processing, and self-driving cars to healthcare and medicine. AI developers have found in nuclear cardiology imaging a particularly rich and adapt field, already inherently quantitative due to a wealth of parameters and indexes with essential diagnostic and prognostic value routinely extracted [7–9]. The field of nuclear cardiology turned early on to computer-aided diagnosis methods and expert systems, which can be described as early expressions of AI-based techniques. Of note, an expert system developed at Emory used AI to automatically generate a structured natural language report with a diagnostic performance comparable to those of experts [10]. This is one of few AI-driven diagnostic systems that have been cleared by the Food and Drug Administration (FDA) and commercially available. But the field is bound to further benefit from the introduction of highly efficient AI tools such as machine learning (ML), deep learning (DL), and particularly convolutional neural networks (CNNs), which have been shown to be very effective in many imaging applications. These techniques have already been applied to a variety of tasks including enhancing image quality, image segmentation, diagnosis, and especially risk stratification and outcome prediction. The ability of these techniques to utilize large amounts of heterogeneous data, directly learning from it, and to extrapolate patterns often invisible to the human eye moves clinicians and scientists closer to a tailored clinical and therapeutical approach to each patient based on his/her characteristics. Although most ML- and DL-based published methods have not entered clinical routine yet, and important limitations need to be addressed and emphasized, in the last few years, a number of investigations have demonstrated not only the feasibility of AI techniques in nuclear cardiology, but also its usefulness.
Recent AI Applications in Nuclear Cardiology
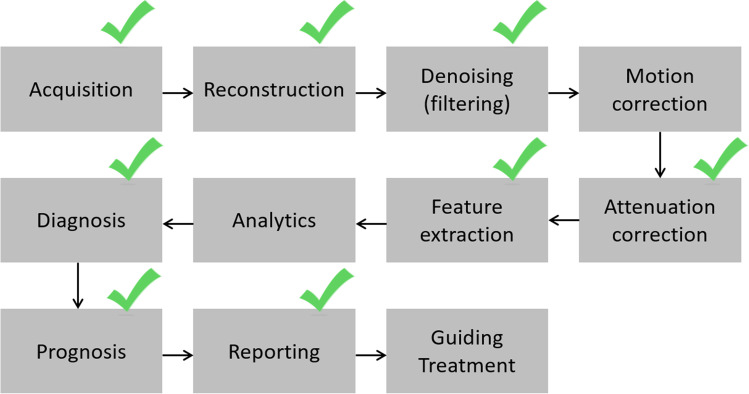
As AI-based techniques have entered cardiac medical imaging, they have been primarily used in image processing, diagnosis, and risk stratification. But other equally important fields of nuclear imaging, such as dose reduction and image reconstruction techniques, are expected to experience new developments [11]. Figure 1 illustrates the chain of major applications commonly applied to a nuclear cardiology tomographic study.
Fig. 1.
Chain of individual applications required for a clinical myocardial perfusion tomographic study. Green checkmarks signify the specific applications that have been improved with machine learning and deep learning algorithms covered in this article
Tomographic Acquisition, Reconstruction, and Dose Reduction
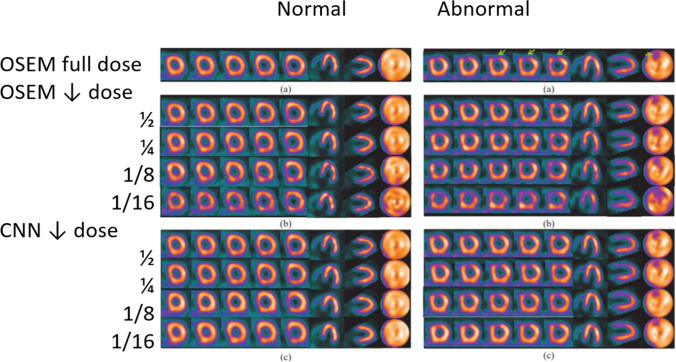
Shiri et al. [12] recently proposed a CNN method aimed at the reduction of acquisition time, and consequently of radiation dose, for SPECT MPI by either reducing the acquisition time per projection by 50% or by halving the number of angular projections. For each patient in a cohort of 363, four sets of projections were generated from the acquisition list mode data: full time (FT) with 20 s per projection, half time (HT) with 10 s per projection, full projection (FP) with 32 projections and half projection (HP) with 16. The convolution network performed an image-to-image transformation to predict FT from HT and FP from HP. Their results, though preliminary, showed that the predicted images obtained with HT projection achieved a better image quality than the ones predicted from HP. Other feasibility studies investigated the applicability of low-dose imaging [13, 14] by developing DL-based techniques for image denoising that outperformed traditional image processing techniques. Particularly, Ramon et al. [13] used a supervised DL approach to remove the high image noise typical of low-dose acquisitions by using pairs of full-dose and low-dose images. The study included 1,052 SPECT MPI datasets and tested two common reconstruction techniques, filtered back projection (FBP), and ordered-subsets expectation–maximization (OSEM). The results indicate that DL methods can significantly improve noise reduction while enhancing diagnostic accuracy. Figure 2 depicts the results of the technique in the presence of a perfusion defect with progressive decreases of injected dose.
Fig. 2.
Results from DL-based denoising technique for low-dose SPECT MPI [13]. Images for a normal male subject (left panels) and a CAD male subject (right panels) with: a OSEM from full dose data, b OSEM from reduced dose data, and c DL processing. A normal perfusion distribution is shown in the left panels (normal (a)). A perfusion defect in the LAD territory is indicated with green arrow in the right panel (abnormal (a)). In b and c, the rows from top to bottom correspond to 1/2, 1/4, 1/8, and 1/16 dose, respectively. As compared to OSEM, the normal regions of the LV myocardial wall are more uniform after DL processing in both left and right panels. In the DL processing, the extent of the defect is better preserved than in OSEM processing even at much lowered dose levels. CAD, coronary artery disease, LAD, left anterior descending artery, DL, deep learning, OSEM, ordered-subsets expectation–maximization. Figure
adapted from slide courtesy of MA King and AJ Ramon
For decades, stress-only protocols have been proposed as a mean to reduce effective radiation exposure by up to 60% and cancel unnecessary tests. Yet, these protocols remain severely underutilized. A ML-based algorithm was recently proposed [15] for risk prediction based on only stress MPI and clinical data to select the low-risk patients that could benefit from rest test cancelation. The study included 20,414 subjects with 98 features trained and tested to provide risk of major adverse cardiovascular events (MACE) as a continuous probability of future events indicated as ML score. Three different thresholds of ML score were derived by matching the cancellation rates achieved by a physician interpretation and two clinical guidelines, and the annual MACE rates for patients selected for rest test cancellation were compared between the different approaches. Patients selected for rest cancellation with ML-guided cancellation recommendations exhibited a lower MACE rates than those chosen based on physician’s interpretation or clinical guidelines.
Attenuation Correction
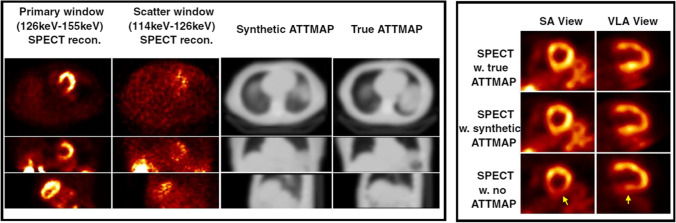
Another important area is that of attenuation correction (AC) with low-dose CT. Decades of investigations on SPECT MPI have shown that AC reduces ambiguity and increases diagnostic accuracy. But since SPECT/CT devices are expensive and not always available, non-AC SPECT images are commonly used for diagnosis. DL methods have recently presented scientists with a possible solution in the form of generative adversarial networks (GANs) and their specific application in creating images of one modality from those of another. A feasibility study for generating AC maps from nuclear SPECT images was recently published. Shi et al. [16] developed a 3D model based on GANs that was trained to estimate attenuation maps solely from SPECT images. The study included 65 patients (normal and abnormal) with SPECT and AC CT. The synthetic attenuation maps were qualitatively and quantitatively consistent with the true maps with a mean absolute error of 3.6%. Analogously, the two sets of AC-corrected SPECT images (true and with synthetic AC) were highly similar with a mean absolute error of 0.26%. The trained DL model was able to generate a synthetic AC map from SPECT images in 1 s. Figure 3 shows for two patients the true vs synthetic AC maps, as well as the final perfusion polar maps obtained with true and synthetic AC vs without AC.
Fig. 3.
Results from DL-based generation of attenuation correction maps from SPECT MPI images [16]. Left panel shows images of the primary, scatter window SPECT reconstructions, the synthetic attenuation maps, and CT-based attenuation (true) maps in the axial, coronal, and sagittal views. Right panel shows SPECT reconstructed images corrected using CT-based (true) attenuation maps, synthetic attenuation maps generated by the Generative Adversarial Network (GAN) and without attenuation correction in short axis (SA) and vertical long axis (VLA) views. Note in the right panel the agreement between the true corrected and corrected with synthetic DL-generated ATTMAP short axis (SA) views and vertical long axis (VLA) views. Note that the uncorrected bottom slices show inferior wall attenuation artifacts absent in the corrected ones. ATTMAP, attenuation map, w, with, w.no, without. Reprint with permission from [16], Shi et al., Eur J Nucl Med Mol Imag, 2020
Image Segmentation
Accurate left ventricular (LV) segmentation is crucial for a reliable assessment of myocardial perfusion and function. Many LV segmentation algorithms have been previously proposed using conventional analysis for this task [17]. Yet, recent ML and DL technological advancements have further increased the LV segmentation robustness. A quality control algorithm was developed to evaluate the automatically obtained contours and detects segmentation failure [18]. Two error flags were implemented to check the LV shape and the location of the mitral valve. The software was validated against expert readers in the identification of segmentation errors and significantly reduced human supervision. Further, ML techniques were recently used in the implementation of an algorithm for the automated identification of the valve plane (VP) from SPECT images [19]. The quantitative assessment of ischemia greatly relies on correct rest and stress relative perfusion maps, which in turn depend on the proper VP identification during segmentation. The authors created high-level image features — 22 features related to VP location, shape and intensity as well as from gated acquisitions — as input into an algorithm based on support vector machine (SVM) techniques that efficiently include expert knowledge and the anatomical variations of VP location. Coronary CT angiography was used to validate the VP location, and total perfusion deficit (TPD) was employed to assess diagnostic accuracy of the model. This fully automated method exhibited a diagnostic performance equivalent to that obtained after correction by experienced readers. Another team leveraged the potential of DL techniques to develop a model for the delineation of LV epicardial and endocardial profiles from gated SPECT images [20]. The method was implemented as an end-to-end fully CNN trained with LV contours delineated by clinicians. Preliminary results on 56 subjects demonstrated excellent precision in assessing LV myocardium volumes with an error of 1.09 ± 3.66%.
Image Interpretation and Outcome Prediction
Image-based diagnosis and outcome prediction is possibly the sector of nuclear cardiology that has experienced the largest progress since the latest technological and methodological developments brought about by ML and DL techniques. In early approaches to CAD investigations with AI methods [21–27], artificial neural networks (ANNs) have been employed to determine if ischemia was present, to what extent, and which coronary was likely to have a stenotic lesion by using sample of myocardial perfusion data as input and corresponding invasive coronary angiography (ICA) results as output. Recently, using a fully ML-based SVM technique [28], Arsanjani et al. [29] combined several quantitative variables to detect CAD from a cohort of 957 SPECT studies. The algorithm integrated perfusion and functional indexes, namely TPD, ischemic changes, and ejection fraction (EF) changes between rest and stress, and regional LV wall motion and thickening scores. The diagnostic accuracy was compared to visual interpretation by two expert readers and validated with ICA results. Each individual quantitative measure had been extensively used to assess CAD, but their integration has remained challenging to perform. The study showed an improved diagnostic accuracy for the SVM technique with a receiver operating characteristic (ROC) area under the curve (AUC) of 0.92 vs 0.88 and 0.87 for the two readers. In a follow-up investigation [29] which included 1181 rest/stress SPECT studies, the authors demonstrated additional improvements achieved with ML when quantitative perfusion measures and clinical variables were combined. The ML results were once again tested against an expert reader with an AUC of 0.94 vs 0.89 (p < 0.001). As integration of numerous heterogeneous variables can prove challenging and given that MPI interpretation largely depends on readers’ experience, the inclusion of such automated assessment could provide fundamental assistance to diagnosticians in the future, particularly in less experienced centers. The search for improved diagnostic accuracy has as its ultimate goal outcome prediction, and particularly, in the case of CAD, the identification of subjects that would benefit from earlier intervention. In Arsanjani et al. [30], the authors aimed at investigating if early revascularization could be effectively predicted by combining clinical data with quantitative SPECT MPI-derived features. A database of 713 rest/stress studies with a total of 372 revascularization events was analyzed, and an automated feature selection algorithm was used to extract the relevant parameters from a collection of 33 indexes and variables. The performance of the ML model was compared to that of two readers. The results were mixed: in predicting revascularization, the ML algorithm exhibited sensitivity comparable to that of one experienced reader, higher than the second one, but similar to standalone perfusion measures; its specificity was better than both readers’ but similar to TPD. In a multicenter study, Hu et al. [31] also used a ML algorithm to predict per-vessel early revascularization within 90 days from SPECT MPI. The model used 18 clinical variables, 9 from stress testing plus additional 28 imaging features from a database of 1980 patients for a total of 5590 vessel observations. The AUC of the ML method was compared to expert analysis and quantitative assessment; it outperformed current quantitative methods per-patient and per-vessel and also expert interpretation. The prediction of the need for revascularization is an important clinical need in CAD assessment, as a consistent number of patients that are scheduled for invasive angiography ultimately do not have obstructive disease.
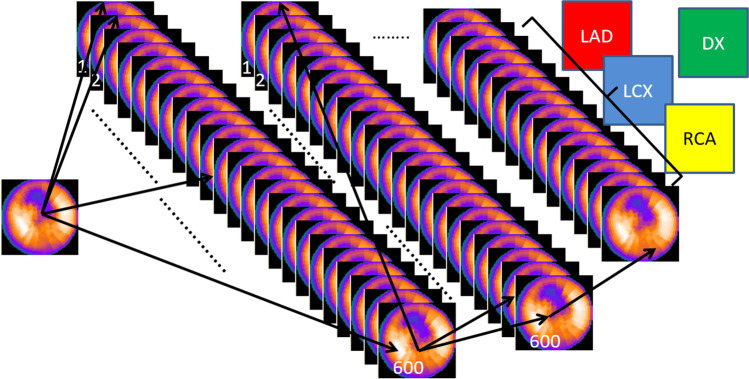
The ML-based ANN approaches have had reasonable diagnostic performance when using limited inputs like the 17 LV segments perfusion scores (SSS, SDS). Figure 4 illustrates how when the level of diagnostic complexity needed raises to that where all the LV voxels are fully connected to each other, the simple ANN approach becomes inefficient and where DL CNN approach has major benefits. DL-based techniques have also been extensively used in CAD diagnosis and prognosis studies, with the algorithms identifying patterns independently. Its first use is described in Betancur et al. [32] with a deep CNN modeled to directly interpret perfusion polar maps and identify obstructive disease. Figure 5 describes the simplicity of the CNN approach in these complex feature detection challenges [33]. In Betancur’s approach, a database of 1638 patients was used to implement the model and was compared to quantitative assessment by TPD: per-vessel sensitivity improved from 64.4% (TPD) to 69.8% (CNN) by only using as input imaging data. The technique was further developed to include upright and supine data [34], an acquisition approach routinely used to mitigate attenuation artifacts. The DL algorithm was trained on polar maps, hypoperfusion defects, and hypoperfusion severity and similarly evaluated to detect obstructive CAD. Compared to combined quantitative assessment, the DL model showed improved AUC in predicting disease per-patient and vessel, as well as enhanced sensitivity and specificity. The same team also developed a ML model for risk stratification and prediction of MACE by combining clinical data and imaging features from 2619 SPECT [35]. When ML was compared to expert visual interpretation, automated stress TPD and automated ischemic TPD, the AUCs were respectively 0.81 vs 0.65 vs 0.73 vs 0.71 respectively (p < 0.01).
Fig. 4.
Limitations of the artificial neural network (ANN) architecture. ANN approaches have found some success when using limited input variables and three-layer feed-forward architecture where the ANN is fed with perfusion scores from the 17 myocardial segments in order to reach a conclusion as to whether the left ventricular myocardium demonstrates ischemia. When more image detail is required such as shown in this illustration, and each of the 600 voxels in a polar map has to be connected to every other voxel, the first ANN layer alone would require 600 × 600 connections, huge databases and extensive computer time to train. Thus, the success of the deep learning approaches showed in Figs. 5 and 6. LAD, left anterior descending vascular territory, LCX, left circumflex vascular territory, RCA, right coronary vascular territory, DX, diagnostic conclusion
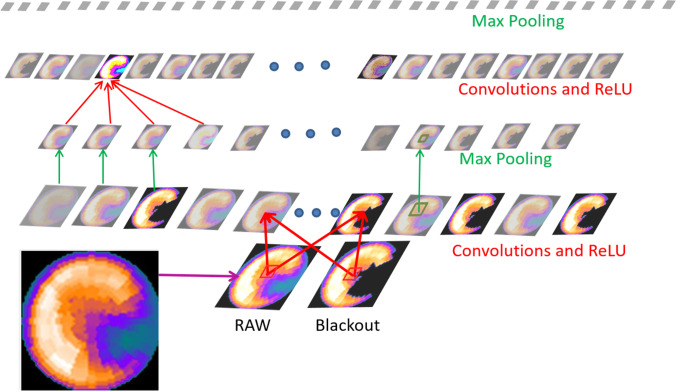
Fig. 5.
Example of inner workings of a convolutional neural network. The outputs of each layer of a typical convolutional network architecture applied to the images of a raw myocardial perfusion polar map and a blackout polar map after comparison to normal limits emulated from Betancur et al. [32]. Simple matrix operators are convolved with each image to extract desired features. Shown here are two operators: rectified linear unit (ReLU) which applies a threshold from the input to the next output layer, and Max Pooling, which applies a filter to a 2 × 2 image patch reducing image dimensions. Each rectangular image is a feature map corresponding to the output for one of the learned features, detected at each of the image positions. Information flows bottom up, and a score is computed for each image class in output. Illustration
adapted from LeCun et al. [33]
Efforts to reduce complexity in input data and features selection are also taking place. Juarez-Orozco et al. [36] developed an ML algorithm from easily accessible clinical and functional variables for the detection of myocardial ischemia as determined through quantitative PET-derived myocardial flow reserve (MFR) < 2.0. A database of 1234 patients was used for the model implementation and testing, and its performance was evaluated against a simple logistic regression model with the same variables. ML achieved an AUC of 0.72 compared to 0.67 obtained with traditional regression techniques. Similarly, Alonso et al. [37] estimated patient’s risk of cardiac death based on SPECT MPI and clinical data with multiple ML models by selecting subsets of variables from a group of 122 potential features. Models were trained and tested on 8321 patients and compared to standard regression techniques. The authors identified a model with 6 features and an AUC of 0.77 that outperformed more complex regression models that used up to 14 features.
Key Socio-economic Questions
The remainder of this article suggests how the various nuclear cardiology stakeholders should prepare for the AI revolution to exploit the opportunities that it brings and avoid potential threats. Similar perspectives have been offered for all of healthcare [38].
Institutional Issues
Perhaps the most important role in preparing for the AI revolution lies with the clinical institutions that generate data, including both actual images and clinical variables. This is because the ML and DL algorithms are mostly driven by data rather than by the sophistication or cleverness of the scientific code. In fact, the majority of the code needed for this AI may be obtained for free as open-source code, and it is the data that trains the algorithms and where the value lies. Thus the saying “Data is the new oil.” Following this analogy, it may be argued that most healthcare systems are sitting on top of major oil deposits and do not even know it. Figure 6 illustrates the increase in performance when using large databases (Big Data) as compared to conventional algorithms.
Fig. 6.
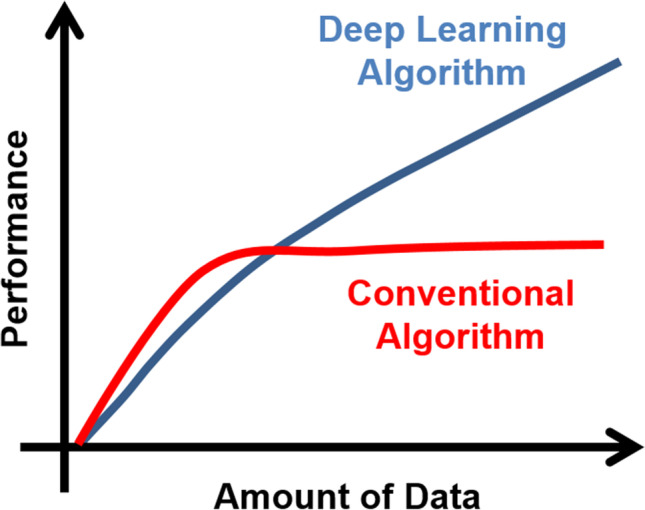
Why deep learning. Contrast of deep learning (DL) vs. conventional algorithm performance as a function of the amount of data used for training. Note that when data is limited conventional algorithms exhibit better performance, but this advantage is quickly reversed as more training data is used. ML and DL algorithms are mostly driven by data rather than by the sophistication or cleverness of the scientific code and thus the benefit of a much faster implementation compared to conventional approaches
Some institutions do know the newfound value of clinical data and are exploiting it. Sloan-Kettering Cancer Center is providing data to Paige. AI in the form of 25 million image sets of patient tissue slides and pathologists findings to develop AI algorithms to assist in cancer diagnosis [39, 40]. Allegedly, the center provides exclusive use of these data for a 9% equity stake in the company. In another deal, St. Louis, Missouri–based Ascension, a Catholic chain of 2600 hospitals, agreed to share data with Google in a project with the code name “Project Nightingale” [41]. The reported shared data [41] include radiology scans, laboratory tests, hospitalization records, medical conditions, and patient’s personal information. These deals and many others follow the footsteps of IBM which in 2016 purchased Truven Health Analytics for $2.6 billion for their healthcare data on tens of millions of medical records and years of health insurance data [41].
Legal and ethical issues aside, which will be discussed below, these major financial transactions form part of the foundation for the fast-approaching AI-tsunami. Just like oil needs to be extracted and refined to be useful, institutions would benefit from having personnel to curate their clinical data and to have systems that can transparently access it. The purpose to this ready access to carefully curated, annotated clinical data is twofold. Access is both a source of valuable data for AI training but also more importantly to be able to benefit from the algorithms developed by others including academic investigators and industry.
Professional Societies’ Role
Professional societies have a role, perhaps a responsibility, in becoming a catalyst to expedite the deployment of AI in our field. Just as is commonly done by computer societies, healthcare societies can provide a major service by being one source of the needed big databases from contributions by many institutions for training algorithms. Although in the past this has created legal and financial concerns, the time is now to resolve it. Creative business models may be developed to reward all concerned and move the field forward. These databases, by their annotated nature, would also provide a source of validation, a valuable asset for AI algorithm development and regulatory clearance. Finally, by being an integral part of the development and validation process, societies could become more relevant by fast tracking the new AI algorithms into their imaging guidelines, vastly reducing the many years that it now takes from innovation to recommendation.
Regulatory Issues
Initially, there was concern as to how to regulate AI algorithms that came in the form of a black box, i.e., with no description as to how the algorithm reached conclusions or made decisions as these algorithms are mostly a set of training weights. The FDA, realizing that the coming AI-tsunami has the potential to transform healthcare, has adapted its vision as described in their recently published action plan [42] “…with appropriately tailored total product lifecycle-based regulatory oversight, AI/ML-based Software as a Medical Device (SaMD) will deliver safe and effective software functionality that improves the quality of care that patients receive”. This follows the FDA’s publication [43] on a proposed regulatory framework describing their foundation for a potential approach to premarket review for AI and ML-driven software modifications. This action plan recognizes and embraces the iterative improvement power of AI/ML/DL allowing a post-market mechanism for continuously improving the algorithms as new data is available. The process would be constrained to specific procedures and meeting certain target accuracy thresholds. These FDA actions are major encouragements for the medical device industry to fully utilize all the benefits of AI.
A similar AI vision and concessions are also required from the radiopharmaceutical division of the USA FDA. Devices and imaging radiopharmaceuticals have a symbiotic relationship in that the radiopharmaceuticals’ FDA clinical trials follow a rigorous imaging protocol that then becomes part of the product’s package insert. To change the package insert usually requires another expensive clinical trial. Deviation from the package insert may be done under the practice of medicine but may not be promoted by either the pharmaceutical or the device company. A simple example is when devices are improved, such as DL-assisted reconstruction to reduce the needed injected dose, and the new required dose falls below the lower limit listed in the radiopharmaceutical package insert. Even though this could result in a reduction of radiation to the patient and a potential financial saving to the healthcare industry, there is no present regulatory framework for its implementation. Coalitions of radiopharmaceutical companies should prioritize discussions with the FDA to implement these frameworks. Consideration should be given to both retrospective and prospective trials. In this context, retrospective trials are FDA trials that are completed resulting in FDA approval of the radiopharmaceutical imaging agent, and now, there is data to modify the package insert based on algorithm improvements applied to the same original data. For prospective trials that companies are designing now, strong consideration should be given to permanently storing the original imaging data in its most elementary acquired raw form so it may be reused each time new algorithms improve efficiency but affect the original imaging protocols. In this scenario, FDA should be able to accept these results as those coming from a “new” clinical trial that allows the change in package insert.
Legal, Ethical Issues
As usual, legal and ethical norms woefully drag behind technological developments. This is even truer for the AI-tsunami because the legal and ethical issues are intertwined. There are two main legal implications: who owns the patients’ data, for example their MPI studies, and who is liable for using the results of an AI algorithm. Legally in the US, clinical data are considered the property of the provider organization and can be sold by the organization as long as patient access and privacy protection requirements are met [39]. “The Immortal Life of Henrietta Lacks” is a book and film about a true story of the case that helped provide the legal precedence to the ownership court decision [44].
The question of liability for clinical malpractice in the US is always dependent on the judgment of jurors and thus if sued, for whatever reason, one needs a defense. Perhaps unexpectedly, a recent study [45] sampling a representative set of 2000 adults presented with four scenarios in which the AI system provided treatment recommendations to a physician indicated that physicians who receive advice from an AI system to provide standard care can reduce their risk of liability by accepting, rather than rejecting, that advice, all else being equal. However, when an AI system recommends nonstandard care, there is no similar shielding effect of rejecting that advice and so providing standard care. These investigators concluded that tort law system is unlikely to undermine the use of AI precision medicine tools and may even encourage the use of these tools [45].
Ethicists have not reached consensus as to ethical norms for using and sharing clinical imaging data for AI. It has been proposed, and generally accepted, that all who participate in the health care system, including patients, have a moral obligation to contribute to improving that system [46] and thus to allow the use of their imaging studies and clinical data for the development of AI algorithms. What is contested is whether it is ethical for this valuable clinical data to be sold or should it become public to benefit future patients [40]. Since it is legal to sell the data as a significant revenue producing component of a healthcare system, history dictates that the data will not be given away for free. A possible exception is a scenario where multiple institutions participate voluntarily, and to a limited degree, to create a big database for the benefit of the field.
Conclusion
AI-based methodologies have the potential to revolutionize the way we approach patient’s care and medical imaging, including nuclear cardiology. The research reviewed in this article illustrates the vast applicability of these techniques. Nuclear cardiology has already benefited from a wealth of techniques for reliable image acquisition, processing, diagnosis, prognosis, and report generation. AI’s progress to date and the fast approaching tsunami of new AI applications provide a novel collection of tools that should be embraced to enhance the future of our field. Continued progress resides in our commitment as scientists to thoughtfully and critically invest in new technologies with patients’ care in mind.
Declarations
Conflict of Interest
Dr. Ernest Garcia receives royalties from the sale of the Emory Cardiac Toolbox and has equity positions with Syntermed, Inc. The terms of these arrangements have been reviewed and approved by Emory University in accordance with its conflict of interest policies. Marina Piccinelli reports no conflict of interest.
Ethics Approval
For this review, ethical approval is not required.
Informed Consent
For this review, consent is not required.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Salerno M, Beller GA. Noninvasive assessment of myocardial perfusion. Circ Cardiovasc Imaging. 2009;2:412–424. doi: 10.1161/CIRCIMAGING.109.854893. [DOI] [PubMed] [Google Scholar]
- 2.Einstein AJ, Pascual TNB, Mercuri M, Karthikeyan G, Vitola JV, Mahmarian JJ, et al. Current worldwide nuclear cardiology practices and radiation exposure: results from the 65 country IAEA Nuclear Cardiology Protocols Cross-Sectional Study (INCAPS) Eur Heart J. 2015;36:1689–1696. doi: 10.1093/eurheartj/ehv117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slomka P, Xu Y, Berman D, Germano G. Quantitative analysis of perfusion studies: strengths and pitfalls. J Nucl Cardiol. 2012;19:338–346. doi: 10.1007/s12350-011-9509-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nuclear Medicine Market Outlook Report 2018. Accessed on 12/30/2020. Available from: https://imvinfo.com/product/nuclear-medicine-market-outlook-report-2018/
- 5.Schindler T, Schelbert HR, Quercioli A, Dilsizian V. Cardiac PET for the detection and monitoring of coronary artery disease and microvascular health. JACC Cardiovasc Imag. 2010;3:623–640. doi: 10.1016/j.jcmg.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Al Badarin FJ, Malhotra S. Diagnosis and prognosis of coronary artery disease with SPECT and PET. Curr Cardiol Rep. 2019;21:57. doi: 10.1007/s11886-019-1146-4. [DOI] [PubMed] [Google Scholar]
- 7.Garcia E, Slomka P, Moody JB, Germano G, Ficaro EP. Quantitative clinical nuclear cardiology, Part 1: established applications. J Nucl Med. 2019;60:1507–1516. doi: 10.2967/jnumed.119.229799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slomka PJ, Moody JB, Miller RJ, et al. Quantitative clinical nuclear cardiology, Part 2: evolving/emerging applications. J Nucl Med. 2020. 10.2967/jnumed.120.242537. Online ahead of print.
- 9.Motwani M, Berman DS, Germano G, Slomka PJ. Automated quantitative nuclear cardiology methods. Cardiol Clin. 2016;34:47–57. doi: 10.1016/j.ccl.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia EV, Klein JL, Moncayo V, Cooke CD, Del'Aune C, Folks R, et al. Diagnostic performance of an artificial intelligence-driven cardiac-structured reporting system for myocardial perfusion SPECT imaging. J Nucl Cardiol. 2020;27:1652–1664. doi: 10.1007/s12350-018-1432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia EV: Deep learning, another important tool for improving acquisition efficiency in SPECT MPI Imaging. (Editorial) J Nucl Cardiol. 2020. 10.1007/s12350-020-02188-z [DOI] [PubMed]
- 12.Shiri I, Sabet KA, Arabi H, Pourkeshavarz M, Teimourian B, Ay MR, et al. Standard SPECT myocardial perfusion estimation from half-time acquisitions using deep convolutional residual neural networks. J Nucl Cardiol. 2020. 10.1007/s12350-020-02119-y. Online ahead of print. [DOI] [PubMed]
- 13.Ramon AJ, Yang Y, Pretorius PH, Johnson KL, King MA, Wernick MN. Improving diagnostic accuracy in low-dose SPECT myocardial perfusion imaging with convolution denoising networks. IEEE Trans Med Imaging. 2020;39:2893–2903. doi: 10.1109/TMI.2020.2979940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song C, Yang Y, Wernick MN, Pretorius PH, King MA. Low-dose cardiac-gated SPECT studies using a residual convolutional neural network. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), 2019, 653–656.
- 15.Hu LH, Miller RJH, Sharir T, Commandeur F, Rios R, Einstein AJ, et al. Prognostically safe stress-only single-photon emission computed tomography myocardial perfusion imaging guided by machine learning: report from REFINE SPECT. Eur Heart J Cardiovasc Imaging. 2020; 00–1–10. [DOI] [PubMed]
- 16.Shi L, Onofrey JA, Liu H, Liu YH, Liu C. Deep learning-based attenuation map generation for myocardial perfusion SPECT. Eur J Nucl Med Mol Imag. 2020;47:2383–2395. doi: 10.1007/s00259-020-04746-6. [DOI] [PubMed] [Google Scholar]
- 17.Gomez J, Doukky R, Germano G, Slomka P. New trends in quantitative nuclear cardiology methods. Curr Cardiovasc Imaging Rep. 2018;11:1. doi: 10.1007/s12410-018-9443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Kavanaugh PB, Fish MB, Gerlach J, Ramesh A, Lemley M, et al. Automated quality control for segmentation of myocardial perfusion. J Nucl Med. 2009;50:1418–1426. doi: 10.2967/jnumed.108.061333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betancur J, Rubeaux M, Fuchs TA, Otaki Y, Arnson Y, Slipczuk L, et al. Automatic valve plane localization in myocardial perfusion SPECT/CT by machine learning: anatomic and clinical validations. J Nucl Med. 2017;58:961–967. doi: 10.2967/jnumed.116.179911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T, Lei Y, Tang H, He Z, Castillo R, Wang C, et al. A learning-based automatic segmentation and quantification method of left ventricle in gated myocardial perfusion SPECT imaging: a feasibility study. J Nucl Cardiol. 2020;27:976–987. doi: 10.1007/s12350-019-01594-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujita H, Katafuchi T, Uehara T, Nishimura T. Application of neural network to computer-aided diagnosis of coronary artery disease in myocardial SPECT bull’s eye images. J Nucl Med. 1992;33:272–276. [PubMed] [Google Scholar]
- 22.Porenta G, Dorffner G, Kundrat S, Petta P, Duit-Schedlmayer J, Sochor H. Automated interpretation of planar thallium-201-dipyridamole stress-redistribution scintigrams using artificial neural networks. J Nucl Med. 1994;35:2041–2047. [PubMed] [Google Scholar]
- 23.Hamilton D, Riley PJ, Miola UJ, Amro AA. A feed-forward neural network for classification of bull’s eye myocardial perfusion images. Eur J Nucl Med. 1995;22:108–115. doi: 10.1007/BF00838939. [DOI] [PubMed] [Google Scholar]
- 24.Lindhal D, Palmer J, Ohlsson M, Peterson C, Lundlin A, Edenbrandt Automated interpretation of myocardial SPECT perfusion images using artificial neural networks. J Nucl Med. 1997;38:1870–1875. [PubMed] [Google Scholar]
- 25.Lindhal D, Palmer J, Edenbrandt L. Myocardial SPECT: artificial neural networks describe extent and severity of perfusion defects. Clin Physiol. 1999;19:497–503. doi: 10.1046/j.1365-2281.1999.00203.x. [DOI] [PubMed] [Google Scholar]
- 26.Lindhal D, Toft J, Hesse B, Palmer J, Ali S, Lundin A, et al. Scandinavian test of artificial neural network for classification of myocardial perfusion images. Clin Physiol. 2000;20:253–261. doi: 10.1046/j.1365-2281.2000.00255.x. [DOI] [PubMed] [Google Scholar]
- 27.Allison JS, Heo J, Iskandrian AE. Artificial neural network modeling of stress single-photon emission computed tomographic imaging for detecting extensive coronary artery disease. Am J Cardiol. 2005;95:178–181. doi: 10.1016/j.amjcard.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Arsanjani R, Xu Y, Dey D, Fish M, Dorbala S, Hayes S, et al. Improved accuracy of myocardial perfusion SPECT for the detection of coronary artery disease using a support vector machine algorithm. J Nucl Med. 2013;54:549–555. doi: 10.2967/jnumed.112.111542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arsanjani R, Xu Y, Dey D, Vahistha V, Shalev A, Nakanishi R, et al. Improved accuracy of myocardial perfusion SPECT for detection of coronary artery disease by machine learning in a large population. J Nucl Cardiol. 2013;20:553–562. doi: 10.1007/s12350-013-9706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arsanjani R, Dey D, Khachatryan T, Shalev A, Hayes SW, Fish M, et al. Prediction of revascularization after myocardial perfusion SPECT by machine learning in a large population. J Nucl Cardiol. 2015;22:877–884. doi: 10.1007/s12350-014-0027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu LH, Betancur J, Sharir T, Einstein AJ, Bokhari S, Fish MB, et al. Machine learning predicts per-vessel early coronary revascularization after fast myocardial perfusion SPECT: results from multicenter REFINE SPECT registry. Eur Heart J Cardiovasc Imaging. 2020;12:549–559. doi: 10.1093/ehjci/jez177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betancur J, Commandeur F, Motlagh M, Sharir T, Einstein AJ, Bokhari S, et al. Deep learning for prediction of obstructive disease from fast myocardial perfusion SPECT: a multicenter study. JACC Cardiovasc Imaging. 2018;11:1654–1663. doi: 10.1016/j.jcmg.2018.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LeCun Y, Bengio Y, Hinton G. Deep Learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 34.Betancur J, Hu LH, Commandeur F, Sharir T, Einstein AJ, Fish MB, et al. Deep learning analysis of upright-supine high-efficiency SPECT myocardial perfusion imaging for prediction of coronary artery disease: a multicenter study. J Nucl Med. 2019;60:664–670. doi: 10.2967/jnumed.118.213538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Betancur J, Otaki Y, Motwani M, Fish MB, Lemley M, Dey D, et al. Prognostic value of combined clinical and myocardial perfusion imaging data using machine learning. JACC Cardiovasc Imaging. 2018;11:1000–1009. doi: 10.1016/j.jcmg.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juarez-Orozco LE, Knol RJJ, Sanches-Catasus CA, Martinez-Manzanera O, van der Zant FM, Knuuti J. Machine learning in the integration of simple variables for identifying patients with myocardial ischemia. J Nucl Cardiol. 2020;27:147–155. doi: 10.1007/s12350-018-1304-x. [DOI] [PubMed] [Google Scholar]
- 37.Alonso DH, Wernick MN, Yang Y, Germano G, Berman DS, Slomka P. Prediction of cardiac death after adenosine myocardial perfusion SPECT based on machine learning. J Nucl Cardiol. 2019;26:1746–1754. doi: 10.1007/s12350-018-1250-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sung J, Stewart CL, Freedman B. Artificial intelligence in health care: preparing for the fifth Industrial Revolution. MJA 213(6), 2020. 10.5694/mja2.50755 [DOI] [PubMed]
- 39.Ornstein C, Thomas K. Sloan Kettering’s cozy deal with start-up ignites a new uproar. New York Times. https://www.nytimes.com/2018/09/20/ health/memorial-sloan-kettering-cancer-paige-ai.html. Published September 20, 2018. Accessed May 5, 2021.
- 40.Larson DB, Magnus DC, Lungren MP, Shah NH, Langlotz CP. Ethics of using and sharing clinical imaging data for artificial intelligence: a proposed framework. Radiology. 2020;295:675–682. doi: 10.1148/radiol.2020192536. [DOI] [PubMed] [Google Scholar]
- 41.Copeland R. Google’s ‘Project Nightingale’ gathers personal health data on millions of Americans. Wall Street Journal. https://www.wsj.com/articles/ google-s-secret-project-nightingale-gathers-personal-health-data-on-millionsof-americans-11573496790?mod=article_inline. Published November 11, 2019. Accessed May 5, 2021.
- 42.Artificial Intelligence/Machine Learning (AI/ML)-Based software as a medical device (SaMD) Action Plan (2021) https://www.fda.gov/media/145022/download Accessed May 5, 2021
- 43.Proposed regulatory framework for modifications to artificial intelligence/machine learning (AI/ML)-based software as a medical device (SaMD) - discussion paper and request for feedback. (2019) https://www.fda.gov/media/122535/download Accessed May 5, 2021
- 44.Skloot R. The Immortal Life of Henrietta Lacks. New York: Penguin Random House; 2010. [Google Scholar]
- 45.Tobia K, Nielsen A, Stremitzer S. When does physician use of AI increase liability? J Nucl Med. 2021;62:17–21. doi: 10.2967/jnumed.120.256032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faden RR, Kass NE, Goodman SN, Pronovost P, Tunis S, Beauchamp TL. An ethics framework for a learning health care system: a departure from traditional research ethics and clinical ethics. Hastings Cent Rep. 2013;43(Spec No):S16–S27. doi: 10.1002/hast.134. [DOI] [PubMed] [Google Scholar]