Abstract
Keratan sulfate (KS) is a glycosaminoglycan that is enriched in vertebrate cornea, cartilage, and brain. During embryonic development, highly sulfated KS (HSKS) is first detected in the developing notochord and then in otic vesicles; therefore, HSKS has been used as a molecular marker of the notochord. However, its biosynthetic pathways and functional roles in organogenesis are little known. Here, I surveyed developmental expression patterns of genes related to HSKS biosynthesis in Xenopus embryos. Of these genes, the KS chain-synthesizing glycosyltransferase genes, beta-1,3-N-acetylglucosaminyltransferase (b3gnt7) and beta-1,4-galactosyltransferase (b4galt4), are strongly expressed in the notochord and otic vesicles, but also in other tissues. In addition, their notochord expression is gradually restricted to the posterior end at the tailbud stage. In contrast, carbohydrate sulfotransferase (Chst) genes, chst2, chst3, and chst5.1, are expressed in both notochord and otic vesicles, whereas chst1, chst4/5-like, and chst7 are confined to otic vesicles. Because the substrate for Chst1 and Chst3 is galactose, while that for others is N-acetylglucosamine, combinatorial, tissue-specific expression patterns of Chst genes should be responsible for tissue-specific HSKS enrichment in embryos. As expected, loss of function of chst1 led to loss of HSKS in otic vesicles and reduction of their size. Loss of chst3 and chst5.1 resulted in HSKS loss in the notochord. These results reveal that Chst genes are critical for HSKS biosynthesis during organogenesis. Being hygroscopic, HSKS forms “water bags” in embryos to physically maintain organ structures. In terms of evolution, in ascidian embryos, b4galt and chst-like genes are also expressed in the notochord and regulate notochord morphogenesis. Furthermore, I found that a chst-like gene is also strongly expressed in the notochord of amphioxus embryos. These conserved expression patterns of Chst genes in the notochord of chordate embryos suggest that Chst is an ancestral component of the chordate notochord.
Keywords: proteoglycan, chordate bodyplan, gene duplication, evolution, morphogenesis, subfunctionalization
Introduction
Glycosaminoglycans (GAGs) are unbranched polysaccharides comprising the extracellular matrix (ECM) in animal tissues. GAGs consist of repeated disaccharide units of acidic sugars and N-acetylated amino sugars bearing sulfate groups, resulting in a high negative charge. On the basis of disaccharide composition, GAGs are categorized as heparan sulfate/heparin [D-glucuronic acid (GlcA) or L-iduronic acid (IdoA), and N-acetyl-glucosamine (GlcNAc)], chondroitin sulfate [GlcA and N-acetyl-galactosamine (GalNAc)], dermatan sulfate [IdoA and GalNAc], keratan sulfate [D-galactose (Gal) and GlcNAc], and hyaluronic acid [GlcA and GlcNAc] (Prydz and Dalen, 2000). Except for hyaluronic acid, GAGs are initiated on serine or threonine residues of core proteins, forming proteoglycans. A variety of core proteins, GAG types, and modification of GAGs, further increase the complexity of proteoglycans (Iozzo and Schaefer, 2015).
Molecular functions of proteoglycans in embryonic development, tissue differentiation, and disease have been thoroughly analyzed. Heparan sulfate proteoglycans function as scaffolds for cell-cell signal transduction by trapping morphogen ligands such as Bmp, Fgf, Hh, and Wnt (De Pasquale and Pavone, 2019; Mii, 2020). Chondroitin sulfate and dermatan sulfate proteoglycans are enriched in cartilage and brain, forming hydrogels for tissue homeostasis (Roughley and Mort, 2014; Hayes and Melrose, 2021). Keratan sulfate is also enriched in many tissues, such as cornea, central and peripheral nervous systems, and bone and cartilage, functioning in tissue hydration and cell-cell communication (Pomin, 2015; Caterson and Melrose, 2018). Cell type-specific distribution of proteoglycans has been revealed directly with biochemical studies, and indirectly via gene expression analysis of core proteins and modification enzymes. Here I focus on biosynthesis of keratan sulfate chains, because little is known about the role of keratan sulfate in early animal development.
Keratan sulfate chains are classified as KS-I, II, or III, based on differences in their linkages to core proteins (Caterson and Melrose, 2018). KS-I is attached to asparagine residues of core proteins via high mannose N-linked glycosylation, whereas KS-II and III are attached to serine/threonine residues of core proteins via O-linked glycosylation with O-Gal (KS-II) or O-mannose (KS-III). Abundance of these types vary between tissues: KS-I, II, and III are enriched in cornea, skeletal tissues, and nervous system, respectively. After initiation processes, keratan sulfate chains are synthesized and elongated by sequential reactions of β-1,3-N-acetyl-glucosaminyltransferase (β3GnT), N-acetylglucosaminyl-6-sulfotransferase (GlcNAc6ST), and β1,4-galactosyl transferase (β4GalT) (Caterson and Melrose, 2018). Then, Gal is sulfated by keratan sulfate galactosyl-6-sulfotransferase (KSGal6ST), activity of which depends heavily on sulfation of GlcNAc (Fukuta et al., 1997; Torii et al., 2000) (Figure 1A). Among seven β3GnT genes and seven β4GalT genes in humans, B3GNT7 and B4GALT4 encode the most specific enzymes catalyzing keratan sulfate biosynthesis (Seko et al., 2003; Seko and Yamashita, 2004) (Figure 1A). In humans, five GlcNAc6ST genes (CHST2, CHST4, CHST5, CHST6, CHST7) and two KSGal6ST genes (CHST1 and CHST3) have been identified (Uchimura and Rosen, 2006) (Figure 1A). CHST3 and CHST7 also exhibit chondroitin 6-O sulfation activity (Kitagawa et al., 2000; Yusa et al., 2006).
FIGURE 1.
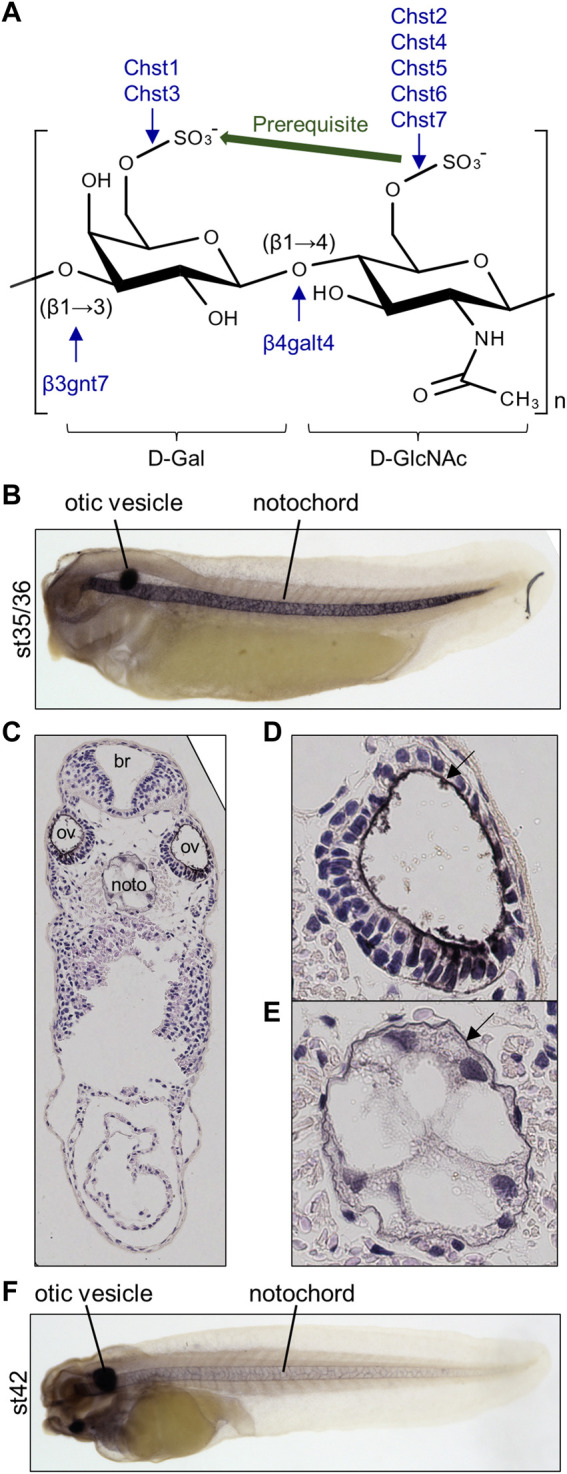
Highly sulfated keratan sulfate (HSKS) is enriched in the notochord and otic vesicles of Xenopus embryos. (A) Molecular structure of HSKS and enzymes catalyzing its biosynthesis are schematically represented. Sulfation of D-GlcNAc is required for sulfation of D-Gal. (B–F) Immunostaining using 5D4 monoclonal antibody demonstrates that HSKS is specifically enriched in the notochord and otic vesicles in Xenopus tropicalis tadpoles. In late tadpole stage (F), notochord staining becomes weaker, possibly due to vacuole growth and cell death, which makes the extracellular space smaller. Tadpoles cleared in BB/BA solution (benzyl benzoate: benzyl alcohol = 2:1) are shown in lateral view. (C–E) A cross-section of a tadpole (st. 35/36) immunostained with 5D4 antibody revealed that HSKS is enriched inside otic vesicles and outside notochord, as designated by arrows in magnified images of an otic vesicle (D) and notochord (E). Br, brain; ov, otic vesicle; noto, notochord.
In Xenopus and other vertebrate model systems, highly sulfated keratan sulfate (HSKS) is detected in the developing notochord and otic vesicles of early embryos by specific antibodies such as MZ-15 and 5D4 mouse monoclonal antibodies (Smith and Watt, 1985; Sugimoto et al., 2005) (Figures 1B–F). Therefore, HSKS has been used as a molecular marker of the notochord in developmental biology studies, e.g., detection of axis bifurcation. However, synthetic pathways, molecular functions, and evolutionary origins of HSKS in these tissues have never been investigated. In this study, I assessed these issues using Xenopus and amphioxus embryos.
Materials and methods
Animal experiments
Adult male and female Xenopus tropicalis (Nigerian BH strain) were provided by Hiroshima University Amphibian Research Center through the National BioResource Project (NBRP) of MEXT. All experiments with X. tropicalis were approved by the Animal Care and Use Committees at the RIKEN Yokohama Campus and Okinawa Institute of Science and Technology Graduate University.
Microinjection of Xenopus embryos
Xenopus tropicalis fertilized eggs were de-jellied and injected with antisense morpholino oligos (MOs). 1 nL of 0.5 mM MOs was injected into the animal pole region of both blastomeres at the two-cell stage (1 pmol per embryo). MOs were purchased from Gene Tools. MO sequences are as follows (antisense start codons are underlined for translation blocking MOs and antisense intron sequences are in small letters for splicing blocking MOs): standard control MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′; chst1 MO1, 5′-GCCTTCCAAGAACATTGCATGGCTG-3′; chst1 MO2, 5′-TCTGCTGTTGACTCTGTACCATAAG-3′; chst3 MO1, 5′-tcgctgacatttcttacTTTGAGAT-3′; chst3 MO2, 5′-TGGCAAAGGGAAAATTCCAATGACT-3′; chst3 MO3, 5′-GAGAACTTCCTGTCCCCCTTCATGA-3′; chst5.1 MO1, 5′-TTAGAGCCCGGAATCTGACCATGAC-3′; chst5.1 MO2, 5′-TCCAGCTTTCTGTTTTCTCAGCCTC-3′. The chst1 MO2, chst3 MO3, and chst5.1 MO2 were designed as second non-overlapping MOs targeting the 5′ untranslated region (5′UTR). To validate translational or splicing block by MOs, in vitro translation or RT-PCR was performed. For RT-PCR, embryos were collected at st. 28. Morphogenetic phenotypes were observed under a stereomicroscope (Zeiss Stemi 305), and otic vesicle sizes were calculated with Labscope v3.1.1 (Zeiss).
Genome editing of Xenopus embryos
To obtain genome-edited embryos, 1 ng of Cas9 protein (IDT, Alt-R® S.p. HiFi Cas9 Nuclease V3) and 200 ng of sgRNA were co-injected into the animal pole region of X. tropicalis embryos at the 1-cell stage. sgRNA was synthesized from PCR-assembled template DNA by in vitro transcription using a MEGAshortscript™ T7 Transcription Kit (Thermo Fisher Scientific, AM1354) as described (Nakayama et al., 2013; Sakane et al., 2017; Blitz and Nakayama, 2022). For PCR assembly of the template DNA, the 5′ oligonucleotide (5′-TAATACGACTCACTATAGG(N)18GTTTTAGAGCTAGAAATAGCAAG-3′, (N)18 corresponding to the target sequence in each gene of interest) and 3′ oligonucleotide (5′-AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3′) were used. Genome editing efficiency of each embryo was examined at the early tadpole stage using a DNeasy Blood & Tissue Kit (QIAGEN) for DNA extraction and an Alt-R® Genome Editing Detection Kit (IDT) for T7 endonuclease I (T7E1) assays. sgRNA for the tyrosinase gene [(N)18 is AACTGGCCCCTGCAAACA] was used as a control. sgRNA sequences were designed using CRISPRdirect (Naito et al., 2015), checking their specificity in the X. tropicalis genome v10 (see Supplementary Tables S1–S3). In vitro cleavage of target DNA with the Cas9-sgRNA complex was performed as described in the IDT protocol with a small modification, in which CutSmart buffer (NEB) was used to prepare the ribonucleoprotein complex and to digest DNA.
Whole mount in situ hybridization
Coding sequences of X. tropicalis genes and Branchiostoma floridae genes were PCR-amplified from cDNA pools of embryos and cloned into pCSf107 mT vectors (Mii and Taira, 2009) using an In-Fusion HD Cloning kit (Takara). Whole-mount in situ hybridization of Xenopus and amphioxus embryos was performed as previously described (Harland, 1991; Yu and Holland, 2009; Yasuoka et al., 2014), using digoxigenin-labelled anti-sense probes, which were transcribed from linearized plasmids. For Xenopus embryos, automated hybridization experiments were performed with InsituPro VSi (Intavis). Stained Xenopus embryos were bleached and observed under a stereomicroscope (Leica M205 FA). Stained amphioxus embryos were observed under a fluorescence microscope (Keyence BZ-X810).
Whole mount immunostaining
Whole-mount immunostaining was performed as described (Suga et al., 2006; Yasuoka et al., 2009; Yasuoka et al., 2014) with modifications for fluorescent imaging. Briefly, embryos were bleached before staining. 5D4 antibody (mouse monoclonal IgG) was used as the primary antibody (Cosmo Bio, PRPG-BC-M01, 1/100 diluted). HRP-conjugated anti mouse IgG (Promega, 1/500 diluted) or AlexaFluor488-conjugated anti-mouse IgG (Thermo Fisher Scientific, A-11001, 1/200 diluted) was used as the secondary antibody. For HRP staining, a Peroxidase Stain DAB Kit and Metal Enhancer for DAB Stain (Nacalai Tesque) were used. Automated immunolabelling experiments were performed with InsituPro VSi (Intavis). Some DAB-stained embryos were subjected to cross-sections stained with hematoxylin. Stained embryos were observed under a fluorescence microscope (Leica M205 FA or Keyence BZ-X810).
Phylogenetic analysis
To identify putative deuterostome Chst genes, protein-coding DNA sequences of X. tropicalis Chst1, Chst2, and Chst3 were submitted as queries to ORTHOSCOPE (v1.5.2), a species tree-based ortholog identification tool (Inoue and Satoh, 2019), with the following settings: analysis group, Deuterostomia; E-value threshold for reported sequences, 1e−5; number of hits to report per genome, 20; aligned site rate threshold within unambiguously aligned sites, 0.55; data set, DNA (Exclude third); rearrangement BS (bootstrap) value threshold, 60%. Using amino acid sequences reported by ORTHOSCOPE, an ML tree was constructed as described (Yasuoka et al., 2020).
Transcriptomic analysis
To examine expression profiles of chst16 during Xenopus development, I re-analyzed public time-course RNA-seq data (Owens et al., 2016). Fastq data of SRR1795535-1795624 (Clutch A polyA RNA-seq, 0-66 hpf) were mapped to the X. tropicalis genome v10.0 with NCBI gene models using STAR v2.7.1a (Dobin et al., 2013), and transcripts per embryo were calculated using RSEM v1.2.28 (Li and Dewey, 2011).
Results
HSKS is enriched inside otic vesicles and outside the notochord
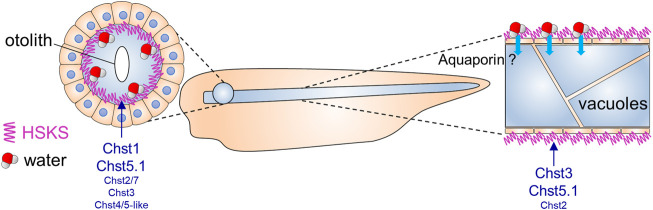
To determine the suborgan distribution of HSKS, I observed cross-sections of X. tropicalis embryos immunostained with 5D4 antibody at the early tadpole stage (Figures 1C–E). The result shows that HSKS is enriched in the ECM layer lining the lumen of otic vesicles (Figure 1D). On the other hand, HSKS is enriched in the ECM layer outside the notochord, which is called the notochordal sheath, but not in the vacuoles (Figure 1E). At later stages, HSKS is still abundant in otic vesicles, but decreases in the notochord, in which vacuoles grow larger (Figure 1F). These observations indicate that otic vesicles and notochord utilize HSKS for tissue hydration in different ways. Therefore, the lumen of otic vesicles must have evolved independently of the notochordal vacuole in vertebrates. Genetic mechanisms underlying these differences are further analyzed below.
HSKS synthetic genes are temporally syn-expressed during Xenopus development
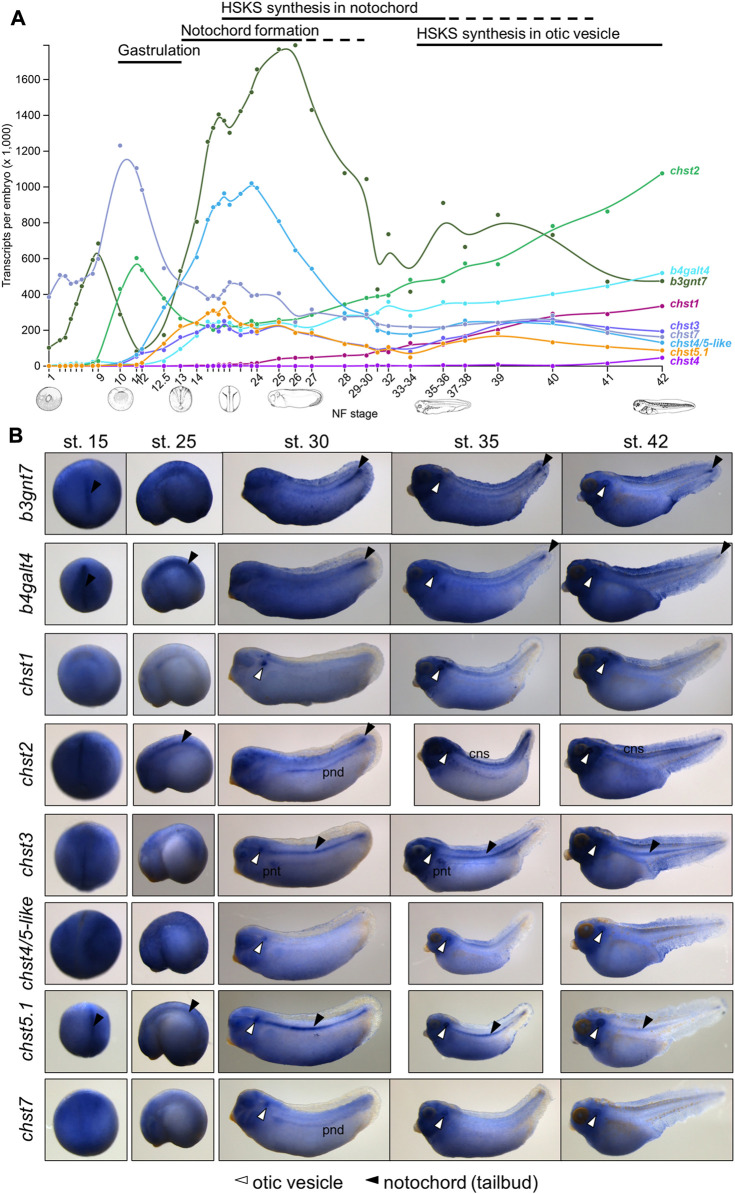
To examine fundamental roles of HSKS in early vertebrate embryos, I focused on expression profiles of the synthetic pathway genes, b3gnt7, b4galt4, chst1, chst2, chst3, chst4/5-like, chst5.1, and chst7 in X. tropicalis embryos. Names of carbohydrate sulfotransferase (chst) genes are in accordance with a recent comprehensive phylogenetic study of the carbohydrate 6-O sulfotransferase gene family (Daza and Haitina, 2020), in which chst4, chst5, and chst6 are renamed on the basis of their phylogenetic relationships (see Table 1; Supplementary Figures S1–S4 for details). Notably, chst4 and chst6 are lineage-specific paralogs of the chst4/5/6 gene in tetrapods and primates, possibly produced by local gene duplication. Another chst4/5-like gene is present in amphibians and some lepidosaurs, but its origin is uncertain. In frogs, chst5 is further duplicated into chst5.1 and chst5.2. Daza and Haitina (2020) reported that chst4 was lost and that chst5.1 and chst5.2 are present in the X. tropicalis genome, but the current genome assembly of X. tropicalis (v10) revealed the presence of chst4 and chst5.1 and the absence of chst5.2, with conserved microsyntenies around these genes (Supplementary Figures S1–S3). Evidently, the previous study overlooked X. tropicalis chst4, which is present in the genome assembly used (v9.1) (see Supplementary Figure S1). The presence of chst4 is plausible, since all other tetrapod genomes retain the gene, suggesting that it serves an indispensable role. The absence of chst5.2 in the current genome assembly is enigmatic, but this gene may be dying or may be undergoing neofunctionalization, given its accelerated evolutionary rate compared to chst5.1 (Daza and Haitina, 2020).
TABLE 1.
Modified gene names from the database (Xenbase and NCBI Entrez Gene).
| Names in this study | X. tropicalis | X. laevis L | X. laevis S |
|---|---|---|---|
| chst4 | chst5 | chst5.L | chst5.S |
| chst4/5-like | chst4 | a | chst4.S |
| chst5.1 | chst6 | chst6.L | chst6.S |
| chst5.2 | a | LOC108714029 | LOC108715076 |
| chst16 | LOC100485856 | LOC108710801 | a |
, gene losses.
Using publicly available time-course transcriptomic data during early embryogenesis of X. tropicalis (Owens et al., 2016), I first examined temporal expression patterns of HSKS synthetic pathway genes in Xenopus development (Figure 2A). The data showed that b3gnt7 and chst7 are maternally expressed, whereas others are zygotically expressed. Among zygotically expressed genes, chst2 expression initiates at the early gastrula stage (stage 10), earlier than others, whereas chst1 expression gradually appears from the pharyngula stage (stage 25). chst4 is almost silent during early embryogenesis, but is weakly expressed at the tadpole stage (stage 42). Remarkably, HSKS synthetic pathway genes, except chst1 and chst4, exhibit temporal synexpression patterns, corresponding to HSKS synthesis in the notochord and otic vesicles (Figures 1B–F). Because it was reported that temporal synexpression can be used to predict common gene functions in embryonic tissues (Owens et al., 2016), temporal synexpression of HSKS synthetic pathway genes suggests their coordinated functions during development.
FIGURE 2.
Spatio-temporal expression patterns of HSKS catalytic genes are linked to HSKS synthesis during Xenopus development (A) Expression levels of HSKS biosynthetic genes along the developmental time-course of Xenopus embryos are visualized in Xenbase (https://www.xenbase.org/) using an available RNA-seq dataset (Owens et al., 2016). Most genes showed elevation of expression levels corresponding to enrichment of HSKS in the notochord and otic vesicles. (B) Spatial expression patterns of Xenopus HSKS biosynthetic genes are represented with whole mount in situ hybridization from neurula to tadpole stages (st. 15–42). To detect their expression in the notochord, some embryos were overstained, resulting in higher background, especially in the head region of tadpoles. Therefore, it is difficult to distinguish precise expression domains of genes strongly expressed in the brain such as chst1, chst2, and chst3. Arrowhead, notochord; open arrowhead, otic vesicles; cns, central nervous system; pnd, pronephric duct; pnt, pronephric tubules.
Chst genes are expressed in the notochord and otic vesicles
Next, I examined spatial expression patterns of those enzymes by whole-mount in situ hybridization (Figure 2B). These data showed that each gene has distinct tissue-specific expression patterns. Among them, glycosyltransferases (b3gnt7 and b4galt4) are relatively ubiquitously expressed with strong expression in the notochord from neurula (st. 15) to pharyngula (st. 25) stages and in the tailbud region and otic vesicles at tadpole stages (st. 30–42).
Chst genes showed more restricted expression patterns (Figure 2B). chst1 manifests highly specific expression in otic vesicles and a small region of hindbrain. Similarly, chst4/5-like and chst7 are specifically expressed in otic vesicles. On the other hand, chst3 and chst5.1 are specifically expressed in the notochord and otic vesicles. chst2 presents a dynamic, complicated pattern. Its expression occurs in the notochord from neurula to pharyngula, but is then restricted to the tailbud region at early tadpole stage (st. 30). Finally, it is detected in otic vesicles and the central nervous system (st. 35–42). Notably, chst2 and chst7 are also expressed in the pronephric duct and chst3 is present in pronephric tubules. Similar expression patterns of chst2 and chst7 in tadpole stages (st. 30–42) may reflect their evolutionary relationship as “ohnologs”, paralogs generated by whole-genome duplication in vertebrates (Daza and Haitina, 2020). Consistent with the decreasing amount of HSKS in the notochord at late tadpole stage (Figures 1B–F), expression levels of HSKS synthetic genes are reduced.
These expression patterns of Chst genes suggest that tissue-specific expression of Chst genes is responsible for HSKS biosynthesis in the notochord and otic vesicles. The absence of HSKS in the pronephros is plausible because GlcNAc6ST (chst2/7) and KSGal6ST (chst3) are not co-expressed there, suggesting that Chst2/7 and Chst3 catalyze sulfation of different molecules in the pronephric system. In addition, chst1, chst2, and chst3 are also strongly expressed in the central nervous system, mainly brain, but HSKS is hardly detected in brains of tadpoles (Figures 1B–F), suggesting that those enzymes work in different parts of the brain and/or catalyze different substrates. Here I focused on the notochord and otic vesicles in early-stage embryos, but more detailed expression analysis of these enzymes in the pronephros and brain will reveal the biosynthetic pathway of HSKS in those tissues.
A recent comprehensive phylogenetic analysis of the carbohydrate 6-O sulfotransferase gene family revealed that frogs retain the chst16 gene, an ohnolog of chst1, which was lost in amniotes (Daza and Haitina, 2020). In the current X. tropicalis genome assembly (v10), chst16 is located on chromosome 3 and annotated as LOC100485856 (Tale 1 and Supplementary Figure S5A). In Xenopus laevis, chst16.S is possibly lost and chst16.L is annotated as LOC108710801 (Table 1). Otic vesicle expression of neighboring genes, tmem263.L and cry1.L, implies that chst16 is also expressed in otic vesicles under co-regulation in the same topologically associated domain (TAD) with tmem263 and cry1 (Supplementary Figure S5). However, in contrast to its ohnologs (chst1 and chst3), mRNA expression of chst16 is hardly detected in the time-course transcriptome data of X. tropicalis embryos (Supplementary Figure S6), suggesting that chst16 scarcely contribute to HSKS biosynthesis in early embryos.
Chst1, Chst3, and Chst5.1 have indispensable roles in HSKS biosynthesis during Xenopus development
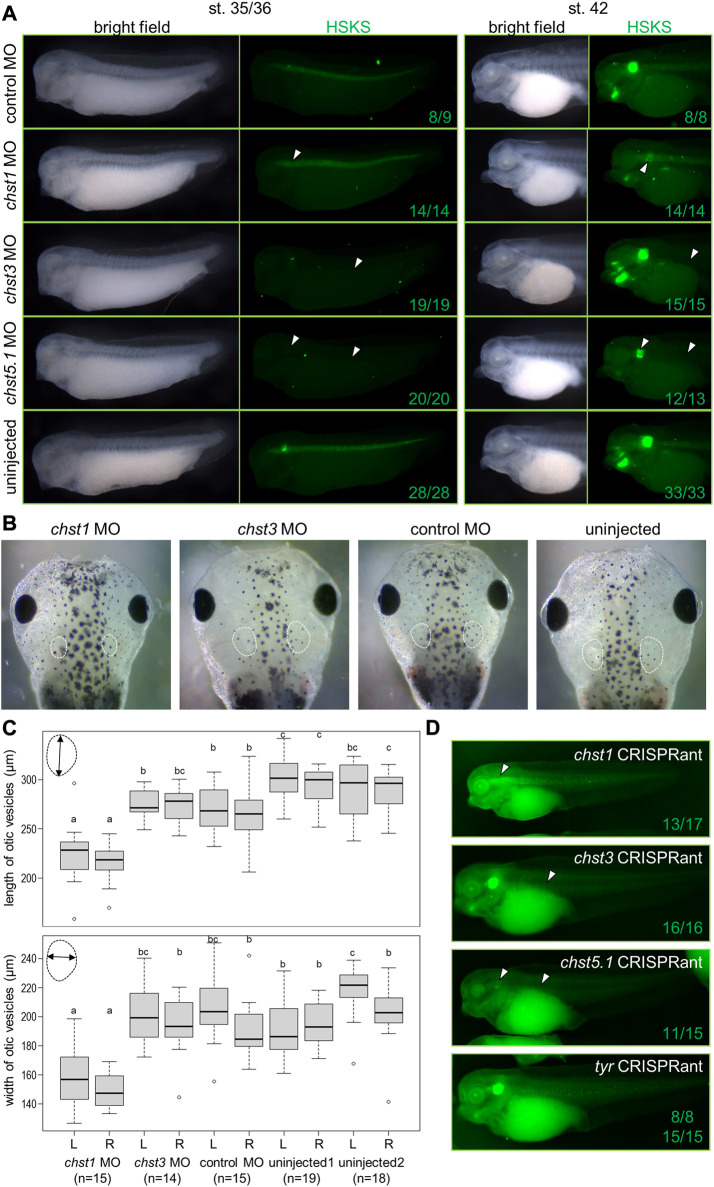
To examine Chst roles in HSKS formation, I performed loss of function experiments by microinjection of antisense morpholino oligos using X. tropicalis embryos (Figure 3A, see Supplementary Figures S7, S8 for validation of knock-down efficiency and specificity of morpholinos). Consistent with their expression domains, chst1 morphants exhibit loss of HSKS in otic vesicles, whereas chst3 and chst5.1 morphants do not produce HSKS in the notochord. The presence of HSKS in otic vesicles of chst3 and chst5.1 morphants indicates that other Chst genes compensate for their loss to synthesize HSKS in otic vesicles. Compared with chst3 morphants, chst5.1 morphants exhibit a more severe phenotype with reduced HSKS in otic vesicles. These results suggest that chst5.1 is the main contributor of GlcNAc6ST activity in both notochord and otic vesicles, and that chst1 and chst3 contribute to KSGal6ST activity in otic vesicles and notochord, respectively (Figure 4).
FIGURE 3.
Loss of function analysis revealed indispensable functions of Chst1, Chst3, and Chst5.1 for HSKS synthesis in Xenopus embryos. (A) Fluorescent immunostaining of HSKS of morphants at st. 35/36 and 42. Open arrowheads indicate lost or reduced enrichment of HSKS. See the text for a detailed explanation of phenotypes. (B) Smaller otic vesicles were observed in chst1 morphants at st. 45. White dashed lines indicate outlines of otic vesicles (C) Quantification of otic vesicle phenotypes. Box plots indicate length and width of otic vesicles on the left (L) or right (R) side in each sample. Each value (length of left vesicle, length of right vesicle, width of left vesicle, and width of right vesicle) was statistically analyzed with one-way ANOVA (p < 6.2 E−13, 9.9 E−11, 6.4 E−16, and 9.6 E−13, respectively), followed by Tukey’s honestly significant difference test with 95% confidence level (indicated with a, b, and c). The result demonstrated that otic vesicles of chst1 morphants are significantly smaller than those of chst3 morphants, control morphants, and uninjected controls (D) Genome editing experiments using the CRISPR-Cas9 system further demonstrated that chst1, chst3, and chst5.1 serve indispensable functions in HSKS in otic vesicles, notochord, and both, respectively (see Supplementary Figures S9–11 for more details). Numbers of embryos with observed phenotypes are indicated.
FIGURE 4.
Putative roles of HSKS in early Xenopus development.The contribution of HSKS to tissue hydration in Xenopus tadpoles is schematically represented. Accumulated HSKS in extra cellular matrix retains water, which may help otic vesicles and notochord vacuoles to swell. HSKS is enriched inside otic vesicles (Figure 1D), but outside of the notochord (Figure 1E), implying that aquaporin transports water from the extracellular space to intracellular vacuoles. Our results demonstrate that tissue-specifically expressed chst genes drive HSKS biosynthesis for normal development.
To validate specificities of morpholinos upon each gene function, second non-overlapping morpholinos were injected to X. tropicalis embryos. Because coding sequences of chst1 and chst5.1 are in single exons, splicing blocking morpholinos are unavailable for them. Therefore, chst1 MO2 and chst5.1 MO2 were designed to bind the 5′UTR to block translation (Supplementary Figure S7). Embryos injected with chst1 MO2 showed loss of HSKS in otic vesicles, further ensuring the HSKS synthetic function of Chst1 in otic vesicles. On the other hand, embryos injected with chst5.1 MO2 died during gastrulation, possibly due to its cytotoxicity. For chst3, two translation blocking morpholinos were examined (Supplementary Figure S8). Although both morpholinos were cytotoxic to some extent, chst3 MO3 is relatively safer than chst3 MO2, and embryos injected with chst3 MO3 exhibited reduction of HSKS in the notochord, supporting the hypothesis that Chst3 synthesizes HSKS in the notochord.
To investigate the impact of reduced HSKS on morphogenesis, otic vesicle sizes were compared between morphants at later tadpole stage (Figure 3B). Quantitative data regarding otic vesicle size indicate that otic vesicles of chst1 morphants are significantly smaller than other morphants and uninjected controls (Figure 3C). Together with the reduction of HSKS levels specifically in otic vesicles by chst1 knockdown (Figure 3A), HSKS effects on otic vesicle formation are assumed to retain water for lumen growth of otic vesicles. Despite the smaller size of otic vesicles, chst1 morphants formed otoliths normally, suggesting that HSKS does not affect the composition of the liquid inside otic vesicles.
Furthermore, I have also discovered a bent axis phenotype in chst5.1 morphants (Supplementary Figure S7F). This phenotype became evident gradually from early to late tadpole stages. Loss of HSKS in notochord of chst5.1 morphants may have impaired water retention and durability of the notochord, resulting in a bent axis. On the other hand, chst3 morphants did not show similar phenotypes, possibly due to substrate specificity. Monosulfated forms of keratan sulfate should remain in chst3 morphants and may be sufficient to permit the notochord to support the embryonic axis. In fact, keratan sulfates in human cornea comprise ∼4% unsulfated, 42% monosulfated, and 54% disulfated disaccharides (Plaas et al., 2001), suggesting that the monosulfated form is functional to some extent. More detailed comparison of notochord morphology, e.g., vacuole shape and size, will reveal molecular functions of chst3, chst5.1 and others in notochord development.
To further validate the functions of chst1, chst3, and chst5.1 in X. tropicalis embryos, I also performed genome editing experiments using the CRISPR-Cas9 system (Supplementary Figures S9–11). I designed two to four sgRNAs for each gene and their activity was confirmed by in vitro cleavage of target DNA. Then, preincubated Cas9-sgRNA complex was microinjected into fertilized eggs. Although some sgRNAs hardly exerted genome editing activity in embryos, I found several reliable sgRNAs for each gene. Consistent with morphant phenotypes, most of chst1, chst3, and chst5.1 CRISPRants exhibited loss of HSKS in the otic vesicle, notochord, and both, respectively (Figure 3D). Furthermore, otic vesicles of chst1 CRISPRants were smaller than those of control embryos, recapitulating the chst1 morphant phenotype (Figure 3C; Supplementary Figure S9G). Although bent axis phenotypes were not observed in F0 embryos of chst5.1 CRISPRants, the chst5.1 function in axial morphogenesis could be confirmed by examining their F1 or F2 embryos.
A chst gene is strongly expressed in amphioxus notochord
In tunicate (Ciona intestinalis) embryos, a glycosyltransferase gene (Ci-b4galt) and two carbohydrate 6-O sulfotransferase genes (Ci-C6ST-like1 and Ci-C6ST-like7) also exhibit restricted expression in the notochord (Katikala et al., 2013; Nakamura et al., 2014), suggesting that HSKS biosynthetic genes have participated in notochord formation since chordates arose. In fact, abnormal morphogenesis of the notochord was caused by knockdown of Ci-C6ST-like1 and Ci-C6ST-like7 (Nakamura et al., 2014). Because tunicate chst genes are quite distant from vertebrate genes (Daza and Haitina, 2020), substrates of Ci-C6ST-like1 and Ci-C6ST-like7 have not yet been determined. Enzymatic activity of Chst, such as GlcNAc6ST and KSGal6ST, may have evolved independently in each lineage.
To address the evolutionary origin of Chst gene expression in the notochord more deeply, I investigated expression patterns of Chst genes in amphioxus embryos (B. floridae). First I searched amphioxus Chst genes using ORTHOSCOPE (Inoue and Satoh, 2019). ORTHOSCOPE extracted 15 putative Chst genes from B. floridae gene models (assembly annotation: Bfl_VNyyK) (Supplementary Figure S12). Among them, six genes were annotated as “carbohydrate sulfotransferase 1-like,” seven were “carbohydrate sulfotransferase 3-like,” one was “carbohydrate sulfotransferase 5-like,” and one was “dermatan-sulfate epimerase-like”, but these gene names do not represent orthologous relationships to corresponding vertebrate genes, as shown by the phylogenetic analysis (Supplementary Figures S12, 13). To validate their expression patterns, four genes were successfully cloned by RT-PCR using a cDNA pool of B. floridae embryos (mid-neurula to mid-larva). Finally, I found that a Chst gene, LOC118425790, is strongly expressed in developing notochord of amphioxus neurulae to larvae (Figure 5A). This result suggests that notochord-specific expression of Chst genes is an ancestral feature of chordates (Figure 5B).
FIGURE 5.
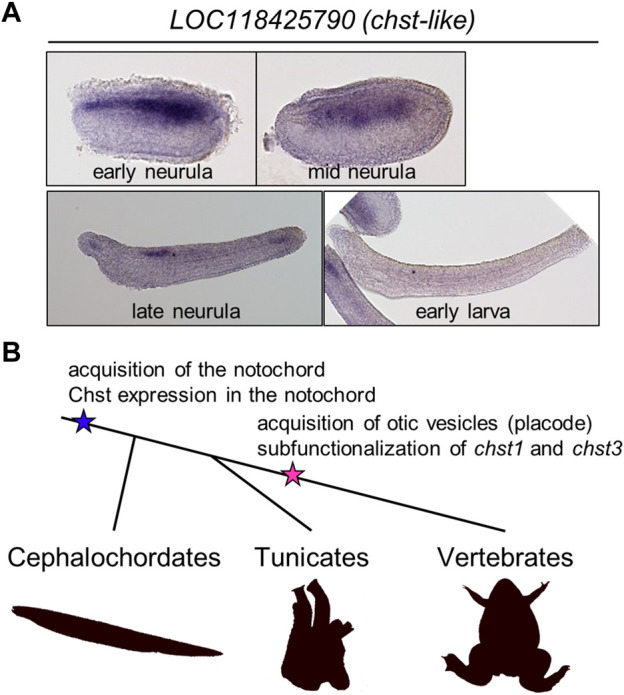
A chst gene is expressed in amphioxus notochord, suggesting evolutionary conservation of HSKS functions in chordates. (A) Whole-mount in situ hybridization using amphioxus embryos showed that expression of a putative Chst gene LOC118425790 is restricted to the notochord and a part of the neural tube. (B) An evolutionary scenario related to Chst genes in chordates. In this scenario, Chst expression is an ancestral feature of the notochord, and in association with the acquisition of otic vesicles, chst1 and chst3 (and others, possibly) are subfunctionalized to catalyze biosynthesis of HSKS in various tissues.
Discussion
In this study, I have demonstrated that HSKS biosynthetic genes are temporally and spatially syn-expressed for HSKS formation in the notochord and otic vesicles of Xenopus embryos (Figures 1, 2). Loss of function analysis revealed indispensable roles of chst1, chst3, and chst5.1 in HSKS synthesis, in accordance with their expression domains (Figures 2B, 3A). The small otic vesicle phenotype of chst1 morphants and CRISPRants demonstrated that HSKS is important for tissue morphogenesis (Figure 3C; Supplementary Figures S9G). These functions remain to be examined in more detail using other genetic tools such as genome editing. Although HSKS abundance has long been recognized in the notochord and otic vesicles, this is the first molecular demonstration of tissue-specific expression and functions of HSKS biosynthetic enzymes in vertebrate embryos. Various genetic programs underlying HSKS biosynthesis in the notochord and otic vesicles are feasible by virtue of their different developmental and evolutionary origins. The notochord develops from dorsal midline mesoderm and originates from a chordate ancestor, but otic vesicles develop from otic placode and originate from a vertebrate ancestor (Figure 5B). Furthermore, I have also shown that a Chst gene is strongly expressed in the amphioxus notochord (Figure 5A), suggesting that HSKS in the notochord is an ancestral feature of chordates and that the biosynthetic program was coopted to otic vesicles in vertebrates (Figure 5B).
Chst genes may have been subfunctionalized in vertebrates and other deuterostomes
Co-option of the HSKS biosynthetic pathway to otic vesicles may have resulted from subfunctionalization of Chst ohnologs. For example, chst1 and chst3 share KSGal6ST activity, but their expression domains are very different (Figures 1A, 2B). Similarly, chst2, chst4/5-like, chst5.1, and chst7 appear to be subfunctionalized with the same catalytic activity (GlcNAc6ST) (Figures 1A, 2B). In zebrafish, chst1 is specifically expressed in otic vesicles at the 14–19 somite stage (Thisse and Thisse, 2004). chst3a shows relatively ubiquitous expression with strong expression in somites and notochord at the five- and 15-somite stages, whereas chst3b displays only very weak expression (Habicher et al., 2015). chst5 (annotated as chst6 in NCBI and ZFIN databases) appears to be expressed in notochord and otic vesicles at somitogenesis stages. chst7 is strongly expressed in the notochord and tailbud at the five- and 15- somite stages and also in otic vesicles at the pharyngula stage (24 hpf) (Habicher et al., 2015). Taken together with my Xenopus data (Figure 2B), regulatory networks to control chst1 and chst5 expression may have been conserved in vertebrates, whereas those for chst3 and chst7 vary among amphibians and teleosts. Therefore, subfunctionalization of chst genes may have occurred multiple times independently, as long as HSKS are normally synthesized.
Although a previous study proposed that the vertebrate ancestor possessed an ancestral gene set comprising chst1/16, chst3, chst2/7, and chst4/5 before two rounds of whole-genome duplication (2R-WGD) (Daza and Haitina, 2020), I assume a more simplified ancestral gene set with one KSGal6ST gene (chst1/3/16) and one GlcNAc6ST gene (chst2/4/5/7). In this scenario, after the first WGD, chst1/16 and chst3 may have been subfunctionalized for development of otic vesicles and notochord (Figures 4, 5B). Similarly, subfunctionalization of chst2/7 and chst4/5 may have occurred in an early stage of vertebrate evolution. Subtle differences of amino acid sequences surrounding adenosine 3′-phosphate 5′-phosphosulfate binding motifs (Ong et al., 1999) in Chst proteins may have been associated with differentiation of substrates and their subfunctions (Supplementary Figure S14). More comprehensive studies on genomic synteny, expression profiles and substrate specificity should provide answers to these scenarios.
Because there are no clear orthologs of chst1/3/16 and chst2/4/5/7 in invertebrate deuterostome genomes (Supplementary Figures S12, 13), all chst genes for HSKS synthesis in vertebrates are probably derived from lineage-specific gene/genome duplication events. Notably, deuterostome chst genes tend to be duplicated in each lineage, as exemplified by 24, 11, 15, and 7 putative chst genes in acorn worms (Saccoglossus kowalevskii), starfish (Acanthaster planci), amphioxus (B. floridae), and ascidians (C. intestinalis), respectively (Supplementary Figures S12, 13). This character may have allowed GlcNAc6ST, KSGal6ST, and other Chst genes to evolve independently, and to increase complexity of biosynthetic reactions of GAG chains.
HSKS roles in development remain to be discovered
Here I address the biosynthetic pathway of HSKS, but core proteins for HSKS in notochord and otic vesicles are still unknown. It is possible that different core proteins contribute scaffolds of HSKS in the notochord and otic vesicles, as demonstrated for Chst genes in this study. In addition, not only highly sulfated forms of keratan sulfate, but also less sulfated forms should have some functions in embryos. Axis malformation in chst5.1 morphants, but not in chst3 morphants, suggests significant roles of less sulfated keratan sulfate (Figure 3D). Expression of chst2 and chst7 in the pronephric duct (Figure 2B) suggest enrichment of keratan sulfate chains (without sulfation of galactose) in that tissue. Together with chst3 expression in pronephric tubules, molecular functions of keratan sulfate and other sulfated GAGs in the pronephric system remain to be discovered. In terms of water metabolism, there may be similarities between the function of HSKS in the pronephric system and that in otic vesicles and notochord.
The function of HSKS in tissue morphology remains largely unknown, although some of it is revealed by this study. In zebrafish embryos, HSKS is not enriched in vacuoles of notochord cells, but in extracellular spaces (Ellis et al., 2013), which is consistent with our result in Xenopus (Figure 1E). Therefore, HSKS does not contribute to vacuolation of the notochord by serving as an osmolyte. Instead, I surmise that HSKS functions in tissue hydration to maintain turgor pressure of the notochord to support its rod-like structure (Figure 4). It has been shown that the highly sulfated form of keratan sulfate is enriched in cornea and cartilage and functions in tissue hydration (Roughley and Mort, 2014; Caterson and Melrose, 2018; Puri et al., 2020). Therefore, HSKS may also serve to keep water in the notochord and otic vesicles to maintain their morphology. That is, the notochord is a vacuolated support presenting a rod-like structure, and otic vesicles are spherical hollow organs forming otoliths inside. They are like water bags in embryos. In the case of notochord, aquaporin may be involved in transport of water absorbed by HSKS, although such functions of aquaporin genes in early notochord development have never been examined. As I proposed previously (Yasuoka, 2020), it is important to balance turgor pressure and sheath strength during notochord morphogenesis. HSKS may contribute to both by tissue hydration and extracellular matrix formation. In addition, HSKS may also have crucial roles in establishment of the biomineralization environment in the otic vesicle.
In Ciona, chst genes (Ci-C6ST-like1 and Ci-C6ST-like7) are required for notochord morphogenesis (Nakamura et al., 2014), but localization of HSKS has not been examined. Since Ciona notochord forms multicellular hollow tubes instead of becoming vacuolated (Dong et al., 2009), it would be valuable to investigate whether HSKS accumulates in the lumen, which is extracellular in Ciona notochord. Similarly, localization of HSKS in amphioxus embryos has not been analyzed. Together with functional assays of a chst gene expressed in the notochord (Figure 5A), further analysis of amphioxus embryos is needed to understand fundamental roles of HSKS in chordates.
Another role of HSKS could be signal transduction, as with other GAGs. Interaction between HSKS and signaling molecules such as Shh and Fgf2 has been reported (Weyers et al., 2013), and Chst2 KO mice were impaired in neural tube patterning by Shh signaling (Hashimoto et al., 2016). Compared to chondroitin sulfate and hyaluronic acid, keratan sulfate interacts with a larger number of neuroregulatory proteins such as Slit, Ephrin, and Semaphorin (Conrad et al., 2010; Melrose, 2019). Therefore, HSKS may modulate signal transduction by binding to ligands and receptors for inductive signals from notochord.
CHST3 is a susceptibility gene for lumbar disc degeneration
A genome-wide association study revealed that lumbar disc degeneration is associated with a variant (rs4148941) in the 3′UTR of CHST3 (Song et al., 2013). This disease-susceptibility allele enhances binding of a microRNA (miR-513a-5p) to the 3′UTR and reduces mRNA expression levels of CHST3 in annulus fibrosus, cartilage end-plate, and nucleus pulposus. Because the nucleus pulpo sus of intervertebral discs is a notochord remnant tissue, my finding of chst3 functions for HSKS synthesis in the notochord may be relevant to human diseases. That is, evolutionary and developmental remnant cells may be responsible for human back pain.
Conclusion
HSKS proteoglycans are synthesized through sequential reactions catalyzed by β3GnT, GlcNAc6ST, β4GalT, and KSGal6ST and are enriched in the notochord and otic vesicles of early vertebrate embryos. Some carbohydrate sulfotransferases exert GlcNAc6ST or KSGal6ST activity and their expression is restricted to those tissues. Remarkably, two KSGal6ST genes, chst1 and chst3, are differentially expressed and required for HSKS synthesis in otic vesicles and notochord in Xenopus embryos, suggesting subfunctionalization after gene/genome duplication. Notochordal expression of an amphioxus Chst-like gene further suggests that Chst contributed to chordate notochord development. Further studies on molecular functions of HSKS in development, and evolutionary comparisons between vertebrates and invertebrates should provide great insight into the origin of the notochord and contributions of glycobiology to human diseases.
Acknowledgments
I thank Noriyuki Satoh for his support of my research at OIST Graduate University, Hiroki Takahashi for kindly providing fixed embryos and total RNA of amphioxus, and Steven D. Aird for technical editing of the manuscript. I also thank all members of Marine Genomics Unit for discussion and Animal Resources Section at OIST Graduate University for animal care. I also thank all members of Laboratory for Comprehensive Genomic Analysis, especially Yuki Yasuoka, for technical support and animal care, Tomoko Hirata, Hiroko Kinoshita, Chitose Takahashi, Shohei Noma, and Tsugumi Kawashima for animal care, Kokoro Ozaki for discussion of CRISPR-Cas9 assays, and Yasushi Okazaki for his support of my research at RIKEN. I also thank Piero Carninci and his research team at RIKEN for permitting me to use their fluorescent microscope (Keyence BZ-X810). I also thank Mio Miyazawa of the Laboratory of Morphology and Image Analysis, Research Support Center, Juntendo University Graduate School of Medicine for preparing cross sections stained with hematoxylin.
Funding Statement
This work was supported in part by a Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) (Grant Nos. 17KT0114, 20H04875, and 22K06348).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by Animal Care and Use Committees at RIKEN Yokohama Campus and Okinawa Institute of Science and Technology Graduate University.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MO declared a shared parent affiliation with the author, YY, to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2023.957805/full#supplementary-material
References
- Blitz I. L., Nakayama T. (2022). CRISPR–Cas9 mutagenesis in Xenopus tropicalis for phenotypic analyses in the F0 generation and beyond. Cold Spring Harb. Protoc. 2022, pdb.prot106971–12. 10.1101/pdb.prot106971 [DOI] [PubMed] [Google Scholar]
- Caterson B., Melrose J. (2018). Keratan sulfate, a complex glycosaminoglycan with unique functional capability. Glycobiology 28, 182–206. 10.1093/glycob/cwy003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad A. H., Zhang Y., Tasheva E. S., Conrad G. W. (2010). Proteomic analysis of potential keratan sulfate, chondroitin sulfate A, and hyaluronic acid molecular interactions. Investig. Ophthalmol. Vis. Sci. 51, 4500–4515. 10.1167/iovs.09-4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daza D. O., Haitina T. (2020). Reconstruction of the carbohydrate 6-O sulfotransferase gene family evolution in vertebrates reveals novel member, CHST16, lost in amniotes. Genome Biol. Evol. 12, 993–1012. 10.1093/GBE/EVZ274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pasquale V., Pavone L. M. (2019). Heparan sulfate proteoglycans: The sweet side of development turns sour in mucopolysaccharidoses. Biochim. Biophys. Acta - Mol. Basis Dis. 1865, 165539. 10.1016/j.bbadis.2019.165539 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). Star: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B., Horie T., Denker E., Kusakabe T., Tsuda M., Smith W. C., et al. (2009). Tube formation by complex cellular processes in Ciona intestinalis notochord. Dev. Biol. 330, 237–249. 10.1016/j.ydbio.2009.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis K., Bagwell J., Bagnat M. (2013). Notochord vacuoles are lysosome-related organelles that function in axis and spine morphogenesis. J. Cell. Biol. 200, 667–679. 10.1083/jcb.201212095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuta M., Inazawa J., Torii T., Tsuzuki K., Shimada E., Habuchi O. (1997). Molecular cloning and characterization of human keratan sulfate Gal-6- sulfotransferase. J. Biol. Chem. 272, 32321–32328. 10.1074/jbc.272.51.32321 [DOI] [PubMed] [Google Scholar]
- Habicher J., Haitina T., Eriksson I., Holmborn K., Dierker T., Ahlberg P. E., et al. (2015). Chondroitin/dermatan sulfate modification enzymes in zebrafish development. PLoS One 10, 01219577–e122018. 10.1371/journal.pone.0121957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland R. M. (1991). Appendix G: In situ hybridization: An improved Whole-Mount method for Xenopus embryos. Methods Cell. Biol. 36, 685–695. 10.1016/S0091-679X(08)60307-6 [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Ishino Y., Jiang W., Yoshimura T., Takeda-Uchimura Y., Uchimura K., et al. (2016). Keratan sulfate regulates the switch from motor neuron to oligodendrocyte generation during development of the mouse spinal cord. Neurochem. Res. 41, 450–462. 10.1007/s11064-016-1861-9 [DOI] [PubMed] [Google Scholar]
- Hayes A. J., Melrose J. (2021). Neural tissue homeostasis and repair is regulated via CS and DS proteoglycan motifs. Front. Cell. Dev. Biol. 9, 696640. 10.3389/fcell.2021.696640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue J., Satoh N. (2019). Orthoscope: An automatic web tool for phylogenetically inferring bilaterian orthogroups with user-selected taxa. Mol. Biol. Evol. 36, 621–631. 10.1093/molbev/msy226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iozzo R. V., Schaefer L. (2015). Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 42, 11–55. 10.1016/j.matbio.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katikala L., Aihara H., Passamaneck Y. J., Gazdoiu S., José-Edwards D. S., Kugler J. E., et al. (2013). Functional brachyury binding sites establish a temporal read-out of gene expression in the Ciona notochord. PLoS Biol. 11. e1001697. 10.1371/journal.pbio.1001697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa H., Fujita M., Ito N., Sugahara K. (2000). Molecular cloning and expression of a novel chondroitin 6-O- sulfotransferase. J. Biol. Chem. 275, 21075–21080. 10.1074/jbc.M002101200 [DOI] [PubMed] [Google Scholar]
- Li B., Dewey C. N. (2011). Rsem: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinforma. 12, 323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melrose J. (2019). Keratan sulfate (KS)-proteoglycans and neuronal regulation in health and disease: The importance of KS-glycodynamics and interactive capability with neuroregulatory ligands. J. Neurochem. 149, 170–194. 10.1111/jnc.14652 [DOI] [PubMed] [Google Scholar]
- Mii Y. (2020). Heparan sulfate clusters regulate distribution and signaling of wnt morphogens. Trends Glycosci. Glycotechnol. 32, E205–E211. 10.4052/tigg.2006.7E [DOI] [Google Scholar]
- Mii Y., Taira M. (2009). Secreted Frizzled-related proteins enhance the diffusion of Wnt ligands and expand their signalling range. Development 136, 4083–4088. 10.1242/dev.032524 [DOI] [PubMed] [Google Scholar]
- Naito Y., Hino K., Bono H., Ui-Tei K. (2015). CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123. 10.1093/bioinformatics/btu743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura J., Yoshida K., Sasakura Y., Fujiwara S. (2014). Chondroitin 6-O-sulfotransferases are required for morphogenesis of the notochord in the ascidian embryo. Dev. Dyn. 243, 1637–1645. 10.1002/dvdy.24213 [DOI] [PubMed] [Google Scholar]
- Nakayama T., Fish M. B., Fisher M., Oomen-Hajagos J., Thomsen G. H., Grainger R. M. (2013). Simple and efficient CRISPR/Cas9-mediated targeted mutagenesis in Xenopus tropicalis. genesis 51, 835–843. 10.1002/dvg.22720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong E., Yeh J. C., Ding Y., Hindsgaul O., Pedersen L. C., Negishi M., et al. (1999). Structure and function of HNK-1 sulfotransferase. Identification of donor and acceptor binding sites by site-directed mutagenesis. J. Biol. Chem. 274, 25608–25612. 10.1074/jbc.274.36.25608 [DOI] [PubMed] [Google Scholar]
- Owens N. D. L., Blitz I. L., Lane M. A., Patrushev I., Overton J. D., Gilchrist M. J., et al. (2016). Measuring absolute RNA copy numbers at high temporal resolution reveals transcriptome kinetics in development. Cell. Rep. 14, 632–647. 10.1016/j.celrep.2015.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaas A. H., West L. A., Thonar E. J. A., Karcioglu Z. A., Smith C. J., Klintworth G. K., et al. (2001). Altered fine structures of corneal and skeletal keratan sulfate and chondroitin/dermatan sulfate in macular corneal dystrophy. J. Biol. Chem. 276, 39788–39796. 10.1074/jbc.M103227200 [DOI] [PubMed] [Google Scholar]
- Pomin V. H. (2015). Keratan sulfate: An up-to-date review. Int. J. Biol. Macromol. 72, 282–289. 10.1016/j.ijbiomac.2014.08.029 [DOI] [PubMed] [Google Scholar]
- Prydz K., Dalen K. T. (2000). Synthesis and sorting of proteoglycans. J. Cell. Sci. 113, 193–205. 10.1242/jcs.113.2.193 [DOI] [PubMed] [Google Scholar]
- Puri S., Coulson-Thomas Y. M., Gesteira T. F., Coulson-Thomas V. J. (2020). Distribution and function of glycosaminoglycans and proteoglycans in the development, homeostasis and pathology of the ocular surface. Front. Cell. Dev. Biol. 8, 731. 10.3389/fcell.2020.00731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughley P. J., Mort J. S. (2014). The role of aggrecan in normal and osteoarthritic cartilage. J. Exp. Orthop. 1, 8–11. 10.1186/s40634-014-0008-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakane Y., Suzuki K. T., Yamamoto T. (2017). A simple protocol for loss-of-function analysis in Xenopus tropicalis founders using the CRISPR-cas system. Methods Mol. Biol. 1630, 189–203. 10.1007/978-1-4939-7128-2_16 [DOI] [PubMed] [Google Scholar]
- Seko A., Dohmae N., Takio K., Yamashita K. (2003). β1,4-galactosyltransferase (β4GalT)-IV is specific for GlcNAc 6-O-sulfate: β4GalT-IV acts on keratan sulfate-related glycans and a precursor glycan of 6-sulfosialyl-Lewis X. J. Biol. Chem. 278, 9150–9158. 10.1074/jbc.M211480200 [DOI] [PubMed] [Google Scholar]
- Seko A., Yamashita K. (2004). beta1,3-N-Acetylglucosaminyltransferase-7 (beta3Gn-T7) acts efficiently on keratan sulfate-related glycans. FEBS Lett. 556, 216–220. 10.1016/S0014-5793(03)01440-6 [DOI] [PubMed] [Google Scholar]
- Smith J. C., Watt F. M. (1985). Biochemical specificity of Xenopus notochord. Differentiation 29, 109–115. 10.1111/j.1432-0436.1985.tb00302.x [DOI] [PubMed] [Google Scholar]
- Song Y. Q., Karasugi T., Cheung K. M. C., Chiba K., Ho D. W. H., Miyake A., et al. (2013). Lumbar disc degeneration is linked to a carbohydrate sulfotransferase 3 variant. J. Clin. Invest. 123, 4909–4917. 10.1172/JCI69277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga A., Hikasa H., Taira M. (2006). Xenopus ADAMTS1 negatively modulates FGF signaling independent of its metalloprotease activity. Dev. Biol. 295, 26–39. 10.1016/j.ydbio.2006.02.041 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Hayata T., Asashima M. (2005). XBtg2 is required for notochord differentiation during early Xenopus development. Dev. Growth Differ. 47, 435–443. 10.1111/j.1440-169X.2005.00819.x [DOI] [PubMed] [Google Scholar]
- Thisse B., Thisse C. (2004). Fast release clones: A high throughput expression analysis. ZFIN Direct Data Submission. Avaialable at: http://zfin.org . [Google Scholar]
- Torii T., Fukuta M., Habuchi O. (2000). Sulfation of sialyl N-acetyllactosamine oligosaccharides and fetuin oligosaccharides by keratan sulfate Gal-6-sulfotransferase. Glycobiology 10, 203–211. 10.1093/glycob/10.2.203 [DOI] [PubMed] [Google Scholar]
- Uchimura K., Rosen S. D. (2006). Sulfated L-selectin ligands as a therapeutic target in chronic inflammation. Trends Immunol. 27, 559–565. 10.1016/j.it.2006.10.007 [DOI] [PubMed] [Google Scholar]
- Weyers A., Yang B., Solakyildirim K., Yee V., Li L., Zhang F., et al. (2013). Isolation of bovine corneal keratan sulfate and its growth factor and morphogen binding. FEBS J. 280, 2285–2293. 10.1111/febs.12165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuoka Y., Kobayashi M., Kurokawa D., Akasaka K., Saiga H., Taira M. (2009). Evolutionary origins of blastoporal expression and organizer activity of the vertebrate gastrula organizer gene lhx1 and its ancient metazoan paralog lhx3. Development 136, 2005–2014. 10.1242/dev.028530 [DOI] [PubMed] [Google Scholar]
- Yasuoka Y., Matsumoto M., Yagi K., Okazaki Y. (2020). Evolutionary history of GLIS genes illuminates their roles in cell reprograming and ciliogenesis. Mol. Biol. Evol. 37, 100–109. 10.1093/molbev/msz205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuoka Y. (2020). Morphogenetic mechanisms forming the notochord rod: The turgor pressure‐sheath strength model. Dev. Growth Differ. 62, 379–390. 10.1111/dgd.12665 [DOI] [PubMed] [Google Scholar]
- Yasuoka Y., Suzuki Y., Takahashi S., Someya H., Sudou N., Haramoto Y., et al. (2014). Occupancy of tissue-specific cis-regulatory modules by Otx2 and TLE/Groucho for embryonic head specification. Nat. Commun. 5, 4322. 10.1038/ncomms5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. K. S., Holland L. Z. (2009). Amphioxus whole-mount in situ hybridization. Cold Spring Harb. Protoc. 4, pdb.prot5286. 10.1101/pdb.prot5286 [DOI] [PubMed] [Google Scholar]
- Yusa A., Kitajima K., Habuchi O. (2006). N-linked oligosaccharides on chondroitin 6-sulfotransferase-1 are required for production of the active enzyme, golgi localization, and sulfotransferase activity toward keratan sulfate. J. Biol. Chem. 281, 20393–20403. 10.1074/jbc.M600140200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.