Abstract
High grade (grade 3) neuroendocrine neoplasms (G3 NENs) have poor survival outcomes. From a clinical standpoint, G3 NENs are usually grouped regardless of primary site and treated similarly. Little is known regarding the underlying genomics of these rare tumors, especially when compared across different primary sites. We performed whole transcriptome (n = 46), whole exome (n = 40) and gene copy number (n = 43) sequencing on G3 NEN FFPE samples from diverse organs (in total 17 were lung, 16 were gastroenteropancreatic, 13 other). G3 NENs despite arising from diverse primary sites did not have gene expression profiles that were easily segregated by organ of origin. Across all G3 NENs, TP53, APC, RB1 and CDKN2A were significantly mutated. The CDK4/6 cell cycling pathway was mutated in 95% of cases, with upregulation of oncogenes within this pathway. G3 NENs had high tumor mutation burden (mean 7.09 mutations/MB), with 20% having >10 mutations/MB. Two somatic copy number alterations were significantly associated with worse prognosis across tissue types: focal deletion 22q13.31 (HR, 7.82; p = 0.034) and arm amplification 19q (HR, 4.82; p = 0.032). This study is among the most diverse genomic study of high-grade neuroendocrine neoplasms. We uncovered genomic features previously unrecognized for this rapidly fatal and rare cancer type that could have potential prognostic and therapeutic implications.
Keywords: grade 3, high grade, neuroendocrine carcinoma, neuroendocrine neoplasm, neuroendocrine tumor, CDKN2A, CDK4, TP53, RB1, APC, genomics, sequencing, cell cycling, tumor mutation burden
INTRODUCTION
Neuroendocrine neoplasms (NENs) commonly originate from the gastroenteropancreatic (GEP) organs and the lungs, but can also arise from most other organs in the body, including the genitourinary tract, head and neck, the gynecologic organs, and breast.(Kunz 2015) In contrast to other solid tumors, the most important factor that determines prognosis and treatment of NENs is not the stage, but the pathologic grade based on the World Health Organization (WHO) classification system (Table S1).(Klimstra DS 2019) Unlike lower grade (grade 1 and 2, G1/2) neuroendocrine tumors (NETs) which have a median overall survival (OS) of 16 and 8 years respectively, high grade (grade 3, G3) NENs have an OS of only about 7 months.(Dasari, et al. 2018; Dasari, et al. 2017) These G3 NENs are rare, with an annual incidence of 10.52 per 100,000 worldwide.(Leoncini, et al. 2017) They are most often poorly differentiated but can rarely be well-differentiated; if poorly differentiated, they are subdivided by large cell or small cell morphology.
As a distinct, highly aggressive entity that does not respond to treatments for lower grade NENs or to conventional treatments for adenocarcinoma or squamous cell carcinomas, G3 NENs pose unique challenges. It is still unknown whether G3 NENs arising from different organs are biologically similar or distinct, yet in the metastatic setting, they are currently all treated with the same first-line chemotherapy regimen (platinum and etoposide) borrowed from experience with small cell lung cancer.(Oronsky B 2018; Sorbye, et al. 2013) Whether this uniform treatment strategy, which is only effective at prolonging survival by a number of months, is biologically sound and should continue to be pursued would depend on a deeper understanding of G3 NENs across tissue types. While past sequencing studies each focused on a specific type of G3 NEN have reported a variety of mutations,(Uccella, et al. 2021) there is also evidence to suggest that high grade NENs across organs may show greater homogeneity than previously thought.(Balanis, et al. 2019)
Prior comprehensive genomic studies on grade 3 NENs have not evaluated the features of multi-site tissue origins,(Busico, et al. 2019; George, et al. 2018b; Girardi, et al. 2017; Kim, et al. 2016; Raj, et al. 2018; Rekhtman, et al. 2016; Vijayvergia, et al. 2016; Wang, et al. 2019; Wong, et al. 2018; Yachida, et al. 2012) and in the case of GEP NENs whole exome and genome studies were mostly limited to grade 1 and 2 NENs, with sample sizes of only 10 and 102 patients which illustrate the rarity of NENs in general.(Jiao, et al. 2011; Scarpa, et al. 2017)
Addressing the lack of comparative genomic profiling in grade 3 NENs, we leveraged whole genome (WGS), exome (WES) and transcriptome sequencing (WTS) on matched tumor and normal tissues arising from diverse organs with paired clinical outcome data. Our analysis revealed that G3 NENs had gene expression profiles that did not easily segregate by organ, that they shared mutations in TP53, RB1, APC, CDKN2A, and the CDK4/6 cell cycling pathway, and harbored two somatic copy number alterations (SCNAs) that could have prognostic potential across tissue types.
METHODS
Patient Cohort and Study Procedures
A retrospective review was conducted of all patients diagnosed with a grade 3 neuroendocrine neoplasm regardless of tissue origin from 2000 until January 2018 at our institution, under an approved protocol by the Stanford University Institutional Review Board. Waiver of consent was obtained for sequencing under the IRB protocol. Clinical records including treatment data were obtained from chart review.
Tumor samples were independently reviewed by an additional pathologist with expertise in NETs and NENs to confirm diagnosis. Macro-dissection was performed if required to isolate neoplastic tissue. All samples were determined to have greater than 20% tumor content on pathology review. Additional matched normal tissue from pathology specimens were also sequenced when available. Any samples mixed with a non-neuroendocrine neoplasm were excluded.
A grade 3 neuroendocrine neoplasm was defined per WHO classification as >20% Ki67 index or >20 mitoses/10 high powered field (for lung: >10 mitoses/10 high powered field). See Table S1.
WGS and WES Sequencing
Forty pairs of tumor and normal tissue samples were sequenced. Each tumor and normal tissue sample had 2 FFPE sections (each 10 micron thick) available for DNA extraction. DNA was extracted using an automated fluidics system, MagnaPure 96 instrument (Roche). From each sample, DNA was taken for library construction with unique dual sequencing adaptors (IDT) using the Kapa Hyper Prep Kit (Roche, Basel, Switzerland), following standard manufacturer guidelines. The libraries were sequenced on an iSeq (Illumina Inc.) to normalize and pool the samples based on their relative number of sequencing reads. A small in was sequenced on a high-output S2 flowcell (Illumina Inc.) to 22 million reads across all samples resulting in an average coverage of 1x. The sequencing files were converted to Fastq files with the BCL-FASTQ Tool (Illumina Inc.) and adapters were removed, the sequencing metrics were generated by PICARD tools (Broad Institute) and duplication rates by FASTP.(Chen, et al. 2018) Reads were aligned and filtered for duplication using Sentieon tools (Sentieon Inc., Mountain View, CA).(Kendig, et al. 2019) Copy number was estimated by CNVkit, per 100kbp bins, and a pooled reference of all the available normal tissue data. We ran Gistic2 to estimate the significant and recurrent CNVs with the following arguments: “-refgene hg38.UCSC.add_miR.160920.refgene.mat -maxspace 10000 -ta 0.1 -td 0.1 -qvt 0.25 -broad 1 -brlen 0.7 -twoside 1 -conf 0.99 -genegistic 1 -armpeel 1 -savegene 1 -res 0.05 -smallmem 1 -js 4”. We set the noise cut-off for both deletion and amplification to 0.1. Coupling CNVkit with Gistic2 enabled us to identify recurrent arm- and focal-level CNAs with statistical significance. The remaining library of each sample was pooled per 8 and enriched for exonic regions by hybrid capture using xGen Exome Research Panel 1.0 (IDT). The capture pools were again sequenced on an iSeq to re-normalize each pool before loading on a second S2 flowcell for WES to an average of 76 million reads. The sequencing files and metrics were again generated using the BCL-FASTQ and PICARD tools, with duplication rates by FASTP. Final sequencing depth was a mean of 240x for tumor samples and 182x for normal tissue. Duplication rate for low-pass WGS data was 31.7%, while for WES data was 11.5%. WGS and WES data were mapped to the human genome NCBI Assembly GRCh38 using BWA-Mem and processed by Sentieon (version: v201808.03); duplicate reads were marked by using MarkDuplicates. Somatic variants were extracted by pairing the tumor and normal Fastq files using Sentieon’s TNHaplotyper (Sentieon Inc., Mountain View, CA). We removed variants with significant strand orientation bias as previously described.(Diossy, et al. 2019) Variants with an allele fraction below 20% were further filtered to reduce false positives from FFPE processing, including insertion artifact. Maftools (v2.7.10) was used to visualize variant data.(Mayakonda, et al. 2018) Significantly mutated genes were identified using dNdScv with q<0.2.(Martincorena, et al. 2017) Analysis was adjusted for multiple testing using Benjamini-Hochberg’s false discovery rate.
RNA sequencing
Forty-six tumor and 21 normal samples were available for RNA sequencing. RNA was extracted using the RNAstorm kit (Cell Data Sciences, Fremont CA). Library preparation was performed using the KapaRNA Hyper Kit (Roche). The libraries were normalized and pooled in groups of 8 or 16, depending on library concentration, and enriched for exonic sequences by hybrid capture, using the xGen Exome Research Panel 1.0 (IDT). All captured pools were then sequenced on a NovaSeq 6000 instrument (Illumina Inc.) to 3.5 billion reads in total or 52.7 million reads per sample. The coverage range was similar for tumor and normal samples (mean PF reads for tumor samples was 54.5 × 106 and for normal samples was 48.9 × 106, and insert size was 204.0 bp and 196.0 bp respectively). The exome capture resulted in close to 80% of reads mapping to coding sequences and there was uniform coverage across the transcript, without any outlier samples. PCA analysis did not reveal any technical bias from extraction batch, capture pools or gender.
RNA-Seq Analysis
The raw sequencing data were mapped to the human genome (GRCh38) using STAR (v2.5). Uniquely mapped reads were used for generating a gene expression matrix. Principal component analysis (PCA) and differential gene expression analysis were performed using DESeq2.(Anders and Huber 2010) Ontology and pathway enrichment analysis were done using the R package Piano (v2.0.0),(Väremo, et al. 2013) with the version 7.1 annotated gene sets (C2 and C5) downloaded from the MSigDB database.(Subramanian, et al. 2005) Cell type enrichment analysis was performed using xCell.(Aran, et al. 2017)
Immunohistochemical Staining
A commercially available primary Rabbit monoclonal antibody directed against cyclin D1/bcl-1 (SP4, Thermo Fisher Scientific, Waltham, MA, catalog # RM-9104-S) was used at a dilution of 1:400. Formalin-fixed, paraffin-embedded (FFPE) sections of tumor and non-neoplastic tissue were cut. Standard automated methods were used for immunohistochemistry that include deparaffinization, antigen retrieval, peroxidase blocking, primary and secondary antibody incubation, detection and counterstaining with hematoxylin. Staining was performed on a Ventana Ultra instrument (Roche Tissue Diagnostics, Tucson, AZ) using Ventana CC1 antigen retrieval solution at pH 8.5.
Integrated Analysis with Machine Learning
CNTools was used to map the segmented copy number data to genes. Mutation MAF files as generated per above were aggregated into a gene-by-patient matrix, with a value of 1 if mutation is found, and a zero otherwise. We used the methods described by Zhao et al. for generating a sample-by-sample similarity matrix with three omics data types: copy number, mutation, and mRNA expression.(Zhao and Yan 2019) In brief, each data type was first transformed into a pairwise patient affinity matrix by unsupervised Random Forest (RF) learning. Random walk was subsequently employed to fuse these matrices into one similarity matrix. Consensus clustering consisted of 1000 iterations of hierarchical clustering, with 0.9 subsampling ratio, and agglomerative average linkage and Pearson correlation was employed to cluster these 40 patients.(Monti, et al. 2020) The optimal cluster number (k=3) was determined using the Gap statistic.(Tibshirani, et al. 2020)
Mutational Signature Analysis
Mutational signatures as established per prior publication was analysed using Maftools and compared to COSMIC v2 signatures, with cosine similarity calculated to determine the best match of samples to established signatures.(Alexandrov, et al. 2013) The optimal number of signatures was determined by Cophenetic correlation.
Clonal Population Estimation
EXPANDS (version 2.0.0) with default parameters estimated the clonal subpopulations of tumor cells in each sample.(Andor, et al. 2014) We used only SNV found in canonical chromosomes excluding sex chromosomes and mitochondrial chromosome. To have balanced clonal landscape, only SNVs derived from the regions overlapped with 50 or more reads in both tumor and normal samples were used. Furthermore, we excluded any SNVs that could not be explained by a sub-population present in 10% or more of the sample. All SNVs within segments with estimated copy number variation were used to predict tumor phylogeny with the following information: i) the number of sub-populations that were present in a tumor sample, ii) the size of each sub-population, and iii) the list of mutations for each sub-population. Founder mutations were defined by subpopulations exceeding 70% of total tumor cells.
Tumor Mutation Burden
Tumor mutation burden was calculated by dividing the total number of nonsynonymous mutations by the size of the coding region. The comparison groups used were TCGA cohorts from the MC3 project. Analysis was done using Maftools with 39 (size of the coding region) as the capture size.
Survival Analysis and Statistics
Clinical data including date of diagnosis, date of death and treatment data were obtained for each patient through the institutional electronic medical record system. Date of data cutoff was 12/31/2018. Survival was estimated using the Kaplan-Meier method with log-rank test for statistical significance. Hazard ratio was estimated using multivariate Cox proportional hazard regression model, controlling for the variables of age, stage, primary site, number of lines of treatment and resection status. Statistical analysis was performed using R package survival (v3.2–7) and Prism (v8, Graphpad). To isolate SCNAs which predicted death risk, the lasso method for variable selection in the Cox model was used as a first screen, with the R package glmnet (v4.1). Mutational enrichment analysis was done with pairwise and groupwise Fisher exact test. Gene expression analysis was done with Bonferroni-Holm method to account for multiple comparisons.
RESULTS
Patient samples and clinical characteristics
We identified 46 tumor samples and 40 matched normal tissue samples from patients with grade 3 NENs (Table S2, Supplemental Data). All samples underwent formal pathology review to confirm the diagnosis, including grade. The cohort included several primary sites: gastroenteropancreatic (GEP; n = 16), lung (n = 17) and other/miscellaneous (gynecologic, head and neck, breast, bladder and unknown primary; n = 13). Overall, 43 samples underwent low coverage whole genome sequencing; 40 tumor-normal pairs had whole exome sequencing and 46 tumors along with 21 additional normal samples had RNA sequencing. In total, 40 samples had data from all three platforms.
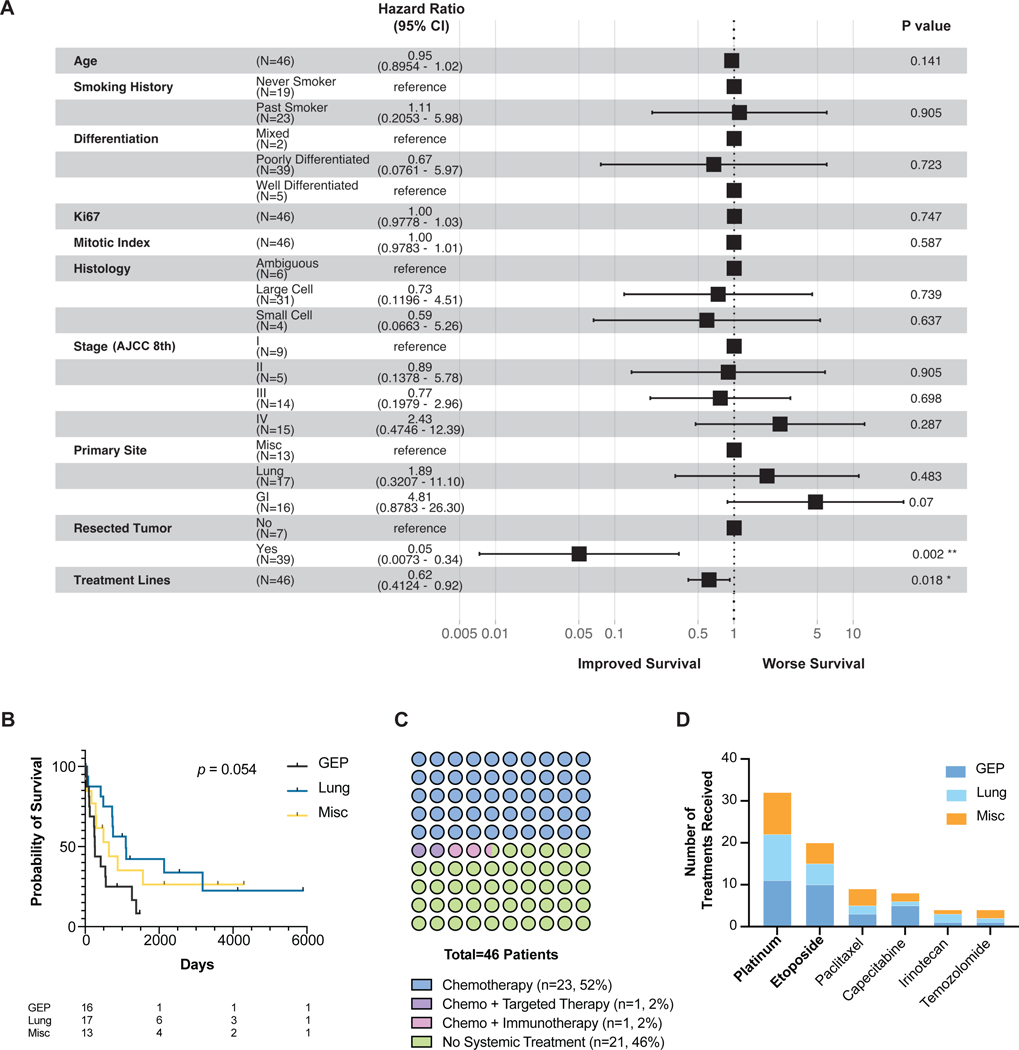
In comparing the overall survival (OS) from patients with different G3 NENs, a Cox multivariate proportional hazard regression analysis accounting for factors including age, smoking history and stage showed that neoplasms from a GEP site trended towards the worst survival (hazard ratio HR 4.81, p = 0.07; Fig. 1a–b), consistent with other studies.(Dasari, et al. 2018) Ki67 staining, mitotic index, and degree of differentiation (well vs. poorly differentiated) are the conventional pathologic metrics used to characterize NENs, and did not correlate with a survival difference (Fig. 1a). The variables that did significantly correlate with OS were prior number of lines of treatment (HR 0.62; p = 0.018), and resection of the primary tumor (HR 0.05; p = 0.002).
Figure 1. Clinical characteristics.
(a) Forest plot of Cox multivariate analysis of 46 patients.
(b) Kaplan-Meier survival curve of patients separated by tissue origin. Table lists the numbers at risk.
(c) Dot plot of systemic therapies received per patient. In the No Systemic Treatment group, surgery, radiation and/or hospice services were administered.
(d) Number of treatments of each chemotherapy received. Some patients received the same chemotherapy more than once. Only the most frequent chemotherapies were listed.
Abbreviations: GEP, gastroenteropancreatic. Misc, miscellaneous. AJCC, American Joint Committee on Cancer.
Half of the patients did not receive any systemic therapy (n = 21, 46%), often due to poor performance status (Fig. 1c). In patients who received therapy (n = 25, 54%), all received chemotherapy with platinum and etoposide being the most common first-line regimen regardless of tumor origin (Fig. 1d). One patient with a lung primary received targeted therapy (erlotinib) and another patient with a lung primary received immunotherapy (nivolumab).
Gene expression of G3 NENs did not easily segregate by organ
To date, the gene expression profiles of G3 NENs have not been thoroughly characterized. Such an investigation could shed light on the extent of homogeneity or heterogeneity among G3 NENs arising from different organs and has significant implications for whether our current “one-size fits all” treatment approach is validated. Platinum and etoposide is currently the recommended first line therapy for all high grade NENs regardless of primary site.(Network 2020)
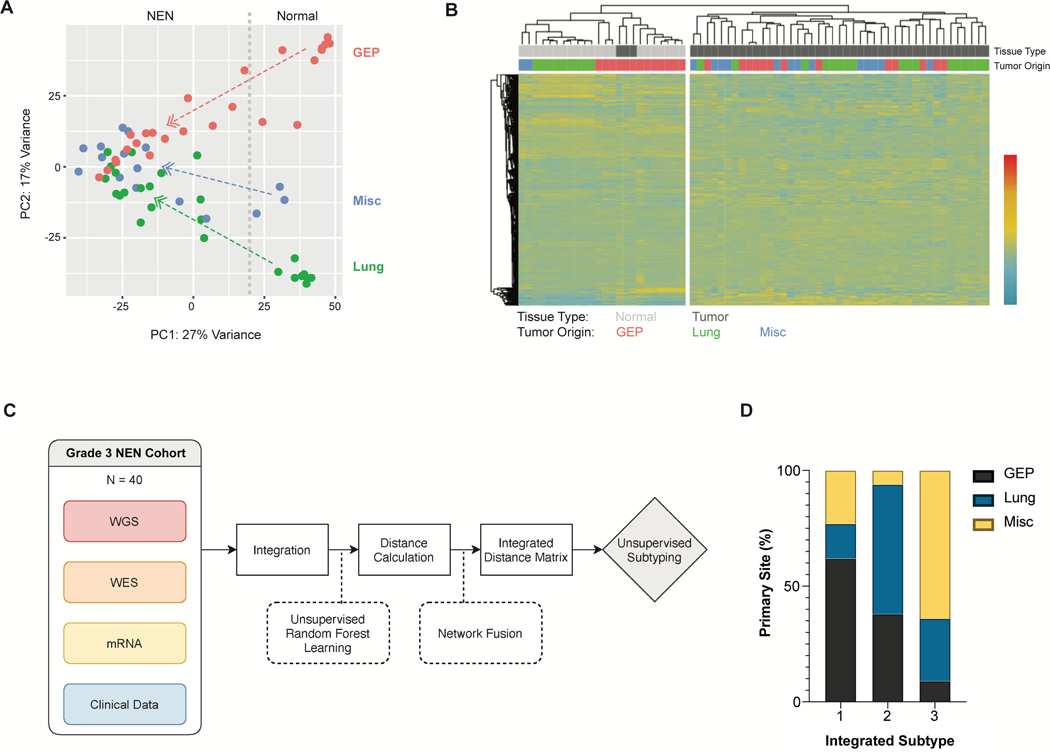
We conducted RNA-Seq analysis on 46 tumor samples along with 21 normal samples. An unsupervised principal-component analysis (PCA) showed that while the normal samples self-segregated based on organ, the G3 NENs formed a single cluster suggestive of a possible convergent expression signature (Fig. 2a). Unsupervised hierarchical clustering similarly showed no clear separation of G3 NENs by organ, in contrast to the clear separation seen with normal tissue (Fig. 2b).
Figure 2. Transcriptomic profiles.
(a) Principal Component Analysis (PCA) of adjacent normal tissue (n = 21), grade 3 neuroendocrine neoplasms (NENs) from gastroenteropancreatic (GEP) (n = 16), lung (n = 17) and miscellaneous primary sites (n = 13; gynecologic, head and neck, breast, bladder and unknown primary). Divided line marks separation between normal and tumor samples.
(b) Unsupervised clustering heatmap of above samples based on the top 30,000 differentially expressed genes. Clustering method was complete linkage with Pearson correlation.
(c) Schematic of machine learning and network analysis algorithm to integrate 4 layers of data.
(d) Bar graph showing subtypes from integrated genomic and transcriptomic analysis (n = 40). Gap statistic determined the optimal number of subtypes was three.
Abbreviations: GEP, gastroenteropancreatic. WGS, whole-genome sequencing. WES, whole-exome sequencing. PC1, principal component 1. PC2, principal component 2.
To further verify that the genomic profile of a G3 NEN was not necessarily tied to its originating organ, we used machine learning and network analysis to integrate low-pass WGS, WES and WTS data for 40 samples using a method we had previously established (Fig. 2c).(Zhao and Yan 2019) Unsupervised subtyping of such integrated data using Gap statistic showed that these G3 NENs were best divided into three subtypes. The subtypes were not clearly delineated by organ, but rather each subtype comprised a mixture of primary sites, further suggesting that the genomic profile of G3 NENs from different organs could be similar (Fig. 2d). Survival analysis showed that subtype 1 exhibited the least intra-group survival variability and had the worst outcomes (Fig. S1).
TP53, RB1, APC, CDKN2A are significantly mutated genes
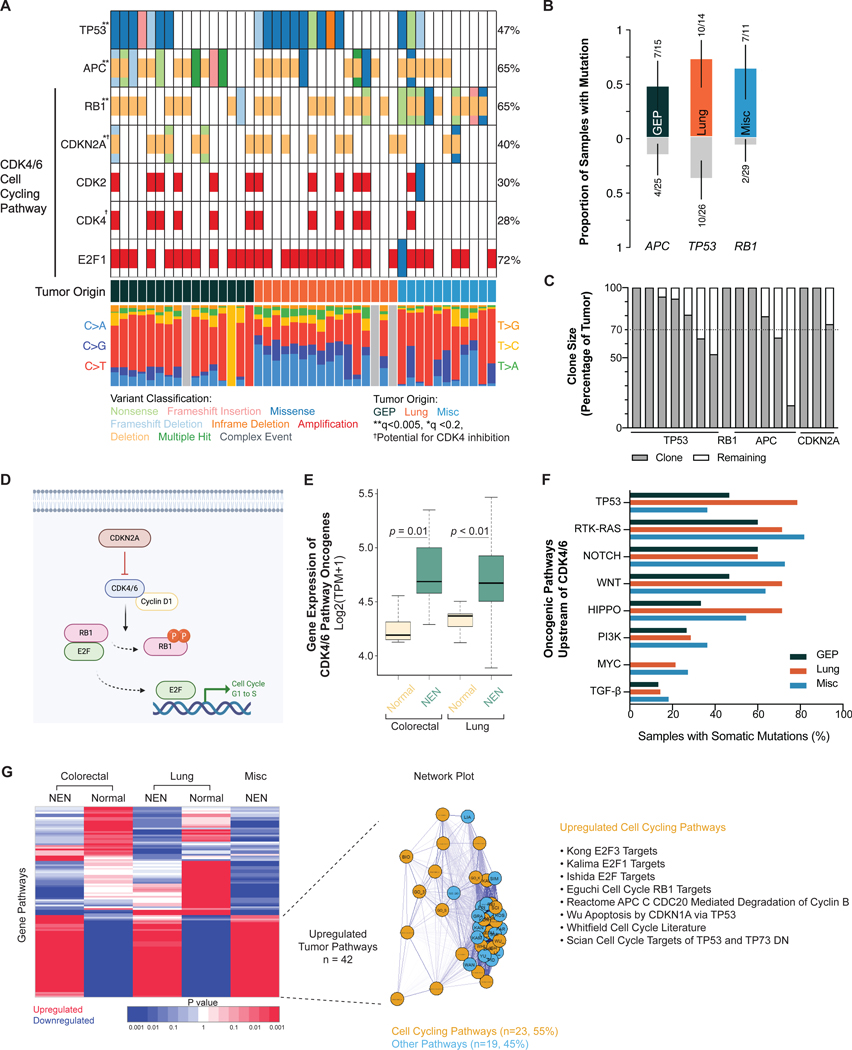
Previous whole-exome and whole-genome studies have focused on grades 1 and 2 NETs of the pancreas, and have identified mutations in telomere maintenance, chromatin remodeling, mTOR signaling, and DNA damage repair pathways.(Jiao, et al. 2011; Scarpa, et al. 2017) More recent studies in G3 NENs of the GI tract have identified mutations in TP53 and RB1, among other genes.(Venizelos, et al. 2021; Yachida, et al. 2022) Using the ratio of nonsynonymous to synonymous mutations (dNdScv method as previously established),(Martincorena, et al. 2017) we identified TP53, RB1, APC and CDKN2A as significantly mutated genes (q-value <0.005 except CDKN2A with q-value <0.2; Fig. 3a, Supplemental Data) across all samples. Subclonal composition analysis with EXPANDS further showed these genes to be likely founder mutations, with their clones exceeding 70% of total tumor cells (Fig. 3c).(Andor, et al. 2014) While these mutations were found across a variety of organ sites (GEP, lung and misc.), specific mutations were preferentially associated with a particular organ group. Mutation enrichment analysis (pairwise and groupwise Fisher exact test) determined that TP53 was more often found in lung NENs, APC in GEP and RB1 in non-lung/non-GEP tissues (p <0.05) (Fig. 3b).
Figure 3. Significantly mutated genes and CDK4/6 cell cycling pathway dysregulation.
(a) Oncoplot showing significantly mutated genes marked by asterisk(s) per dNdScv method. Low-pass WGS n = 43; WES n = 40.
(b) TP53 mutations were enriched in lung, APC in gastroenteropancreatic, and RB1 in non-lung/non-GEP neoplasms (p < 0.05, pairwise and groupwise Fisher exact test). N = 40.
(c) EXPANDS analysis showing the clone size of each significantly mutated gene. Dotted line shows the 70% threshold often used to define a founder clone.
(d) Schematic of CDK4/6 cell cycling pathway.
(e) Boxplot showing gene expression levels of the CDK4/6 pathway oncogenes (CCND1, CCDN2, CCND3, CCNE1, CDK2, CDK4, CDK6, E2F1, E2F3) between neoplasms and normal tissue. Mean ± SD (colorectal normal: 4.26 ± 0.15, n = 7; colorectal tumor: 4.77 ± 0.35, n = 8; lung normal: 4.34 ± 0.11, n = 9; lung tumor: 4.71 ± 0.48, n = 17). Statistical test: Holm-Bonferroni method.
(f) Frequency of mutations in canonical oncogenic pathways upstream of CDK4/6 pathway.
(g) Unsupervised clustering heatmap of gene pathways. Gene pathways universally upregulated by all neoplasms compared to normal tissue (n = 42) comprised mostly of cell cycling pathways (n = 23).
Abbreviations: WGS, whole genome sequencing. WES, whole exome sequencing. GEP, gastroenteropancreatic. NEN, neuroendocrine neoplasm. Misc, miscellaneous.
While APC, RB1 and CDKN2A all had frequent genomic deletions, TP53 had mutations exclusively. Most TP53 mutations were missense, and almost all (90%) were within the DNA-binding domain, consistent with prior literature showing the localization of hotspot mutations in a range of human cancers within this domain (Fig. S2).(Baugh, et al. 2018) KRAS and BRAF, although did not meet threshold for significant mutation, were altered in 8 and 5% of tumor samples respectively.
In plotting the frequency of DNA substitution mutations (Fig. 3a), there appeared to be organ-specific profiles, e.g. an abundance of C>A transversions for G3 lung neoplasms, suggestive of different incipient mutational processes. Using the mathematical algorithm established by Alexandrov et al. to characterize mutational signatures,(Alexandrov, et al. 2013) we isolated three signatures: deamination in the case of GEP NENs likely as a result of DNA repair error (93%),(Alexandrov, et al. 2013) smoking in lung NENs (93%) and ultraviolet light exposure in some tumors from miscellaneous other primary sites (Fig. S3). The finding of a deamination mutagenesis signature has been reported for lower grade GEP NENs.(Scarpa, et al. 2017) The observed smoking signature is consistent with prior research, including presence in high grade lung NENs,(Alexandrov, et al. 2016; George, et al. 2018a) and absence in lower grade lung NETs (pulmonary carcinoids).(Fernandez-Cuesta, et al. 2014)
CDK4/6 cell cycling pathway is frequently altered
Whereas prior targeted sequencing studies had found frequent mutations in TP53 and RB1 in G3 NENs, the involvement of CDKN2A is less frequently reported.(Peifer, et al. 2012; Roy, et al. 2018; Yachida, et al. 2012) CDKN2A (cyclin-dependent kinase inhibitor 2A) is a tumor suppressor gene that encodes p16 and p14, regulates cell cycle progression by inhibiting the cyclin-dependent kinases CDK4/6 from phosphorylating RB1 and initiating cell cycling thru the transcription factor E2F (Fig. 3d).(Sanchez-Vega, et al. 2018) CDKN2A was deleted in 40% of samples (all hemizygous losses), spanning across G3 NENs from different organs (Fig. 3a). When accounting for other components of the CDK4/6 pathway to include CDK2/4/6 amplification, E2F1 amplification, and RB1 deletion, the total percentage of affected samples reached 95% (n = 41/43).
Gene expression analysis confirmed that established oncogenes in the pathway (CCND1, CCDN2, CCND3, CCNE1, CDK2, CDK4, CDK6, E2F1, E2F3)(Sanchez-Vega, et al. 2018) showed statistically significant higher levels of expression in G3 colorectal (p = 0.01) and lung neoplasms (p = 0.009) when compared to their normal tissue respectively (Fig. 3e). Immunohistochemical staining with a CCND1 antibody in tumors with higher CCND1 gene expression than paired normal controls (7 colorectal tumors) confirmed the aberrant expression of this protein in tumors (86% of tumors positive vs 0% of normal controls positive, p = 0.0014; Fig. S4). Unsupervised hierarchical clustering of differentially expressed genes between G3 neoplasms and normal tissue showed 42 commonly shared upregulated pathways among all G3 neoplasms: 23 (55%) of which were also related to cell cycling (Fig. 3g, Supplemental Data). Canonical pathways upstream of CDK4/6 cell cycling including TP53, NOTCH, RTK-RAS, WNT, HIPPO, PI3K, MYC and TGF-β were also frequently mutated across G3 neoplasms from diverse organs (Fig. 3f, S5).(Choi and Anders 2014; Du, et al. 2020; George, et al. 2018a)
Grade 3 neuroendocrine neoplasms harbor high tumor mutation burden
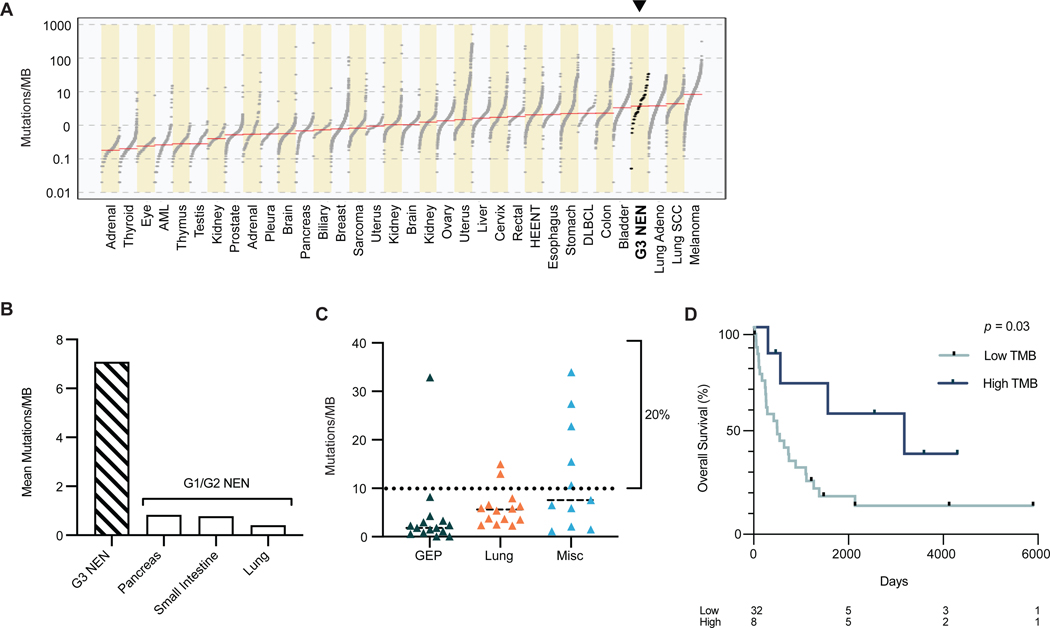
Concomitant with the recent success in treating solid tumors with checkpoint inhibitors, there is increasing data that show improved treatment efficacy in tumors with a higher tumor mutation burden (TMB).(Assi and Padda 2020; Lu, et al. 2020; Weber and Fottner 2018) When comparing the TMB of G3 NENs in our cohort to published data from 33 TCGA cohorts,(Ellrott, et al. 2018) acknowledging differences in sequencing techniques, G3 NENs had the fourth highest TMB levels, just behind melanoma, lung squamous cell carcinoma and lung adenocarcinoma (Fig. 4a). The mean number of mutations per megabase (MB) was 7.09, similar to prior reports.(George, et al. 2018a; George, et al. 2015; Peifer, et al. 2012; Puccini, et al. 2020) The high TMB of G3 NENs is significantly different from the lower TMB levels of grade 1 and grade 2 NETs from various organs, including pancreas, small intestine and the lungs, each with <1 mutations/MB as previously reported (Fig. 4b).(Fernandez-Cuesta, et al. 2014; Mafficini and Scarpa 2019; Yao, et al. 2019)
Figure 4. Tumor mutation burden.
(a) Grade 3 neuroendocrine neoplasms (NEN) rank 4th in order of highest tumor mutation burden (TMB) when compared to 33 tumor types in The Cancer Genome Atlas project. Red line indicates median TMB. N = 40.
(b) Mean TMB of grade 3 NENs (7.08, SD = 8.5, n = 40) compared to lower grade tumors previously reported (pancreas 0.82, small intestine 0.77, lung 0.4). (Fernandez-Cuesta, et al. 2014; Mafficini and Scarpa 2019; Yao, et al. 2019)
(c) TMB among grade 3 NENs separated by tissue origin. Dotted line marks 10 mutations/MB, a known predictor of response to immunotherapy as validated in other cancers. The miscellaneous group has the highest TMB (vs GEP: p = 0.04), with head and neck primaries in this group showing the highest TMB (21.5 mutations/MB). Mean ± SD: GEP (4.28 ± 8.17, n = 15), Lung (6.01 ± 3.82, n = 14), Misc (12.28 ± 11.24, n = 11). One-way ANOVA/Tukey’s multiple comparison test.
(d) Patients with high TMB had better overall survival than patients with lower TMB. Multivariate analysis accounted for age, stage, primary site, resected tumor and number of treatment lines, p = 0.03.
Abbreviations: GEP, gastroenteropancreatic. NEN, neuroendocrine neoplasm. TMB, tumor mutation burden. SD, standard deviation.
Research on other cancers has indicated that higher TMB, especially more than 10 mutations/MB, is associated with an improved response to checkpoint inhibitors.(Lu, et al. 2020; Melendez, et al. 2018; Samstein, et al. 2019) Pembrolizumab, a PD-1 inhibitor, is FDA approved for all solid tumors with more than 10 mutations/MB.(FDA 2020) In our cohort, as much as 20% of samples had more than 10 mutations/MB, with head and neck primary tumors showing the highest TMB (mean 21.5 mutations/MB), and GEP NENs showing the lowest (4.28 mutations/MB; Fig. 4c). Patients with any G3 NEN with more than 10 mutations/MB had better overall survival compared to those with fewer mutations (multivariate analysis p = 0.03, Fig. 4d). This phenomenon has been observed in numerous other cancer types, although the mechanism remains unclear.(Klebanov, et al. 2019)
Select copy number alterations increase the risk of death multifold
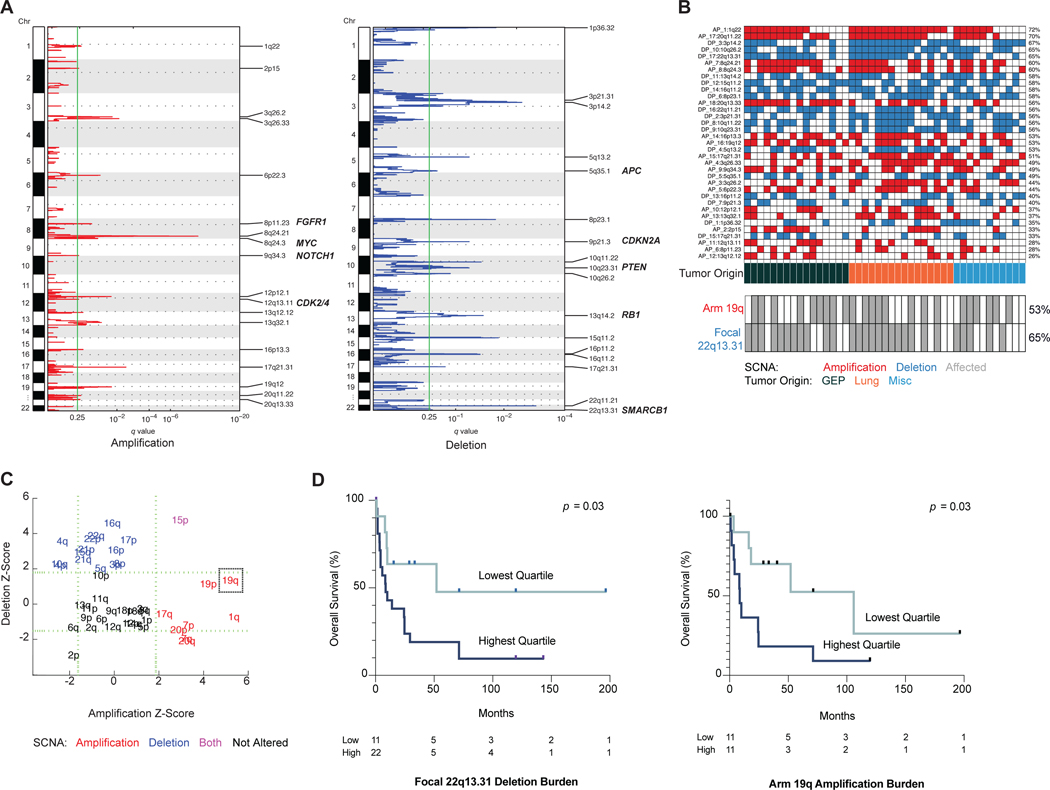
Low grade (G1 and G2) pulmonary NETs have no significant copy number alterations (SCNAs), while those from the gastroenteropancreatic system have numerous SCNAs.(Boons, et al. 2022; Fernandez-Cuesta, et al. 2014; Hashemi, et al. 2013; Karpathakis, et al. 2016) The extent of copy number changes in high grade NENs and their clinical associations remain unclear. From the results of whole genome sequencing, we discovered that G3 NENs had frequent chromosomal instability: 18 focal amplifications, 17 focal deletions, 18 arm amplifications and 23 arm deletions (Fig. 5a,c). No specific pattern was noted based on organ type (Fig. 5b). The most common arm level alterations were gain of arm 1q (58% samples affected), 19q (53%), 7p (53%) and 20q (53%); loss of arm 16q (60%), 22p (60%), 22q, 21p, 15p and 4q (each 55%). The most frequent focal alterations were: gain of 1q22 (72%), 20q11.22 (70%) and 8q24.21 (60%); loss of 3p14.2 (67%), 10q26.2 (65%) and 22q13.31 (65%; Supplemental Data).
Figure 5. Two somatic copy number alterations confer increased risk of death.
(a) Focal somatic copy number alterations per GISTIC analysis, q > 0.25. N = 43.
(b) Oncoplot of focal somatic copy number alterations. Frequency of arm 19q amplification and focal 22q13.31 deletion are mapped (gray = affected).
(c) Arm level somatic copy number alterations.
(d) Kaplan-Meier survival curves for differing burdens of copy number alteration. Focal deletion in 22q13.31 was associated with worse survival (HR 7.82, p = 0.034 by multivariate Cox analysis). Arm amplification of 19q was associated with worse survival (HR 4.82, p = 0.032 by multivariate Cox).
Abbreviations: GEP, gastroenteropancreatic. NEN, neuroendocrine neoplasm; SCNA, somatic copy number alteration.
After multiple test correction, a lasso-based model and Cox multivariate analysis showed that two SCNAs were associated with significantly worse prognosis: focal deletion of 22q13.31 (HR, 7.82; p = 0.034), and arm amplification of 19q (HR, 4.82; p = 0.032) (Fig. 5d, S6). Among 22q13 deletion carriers, the top quartile of those with the highest copy change had worst median OS compared to the rest: 9.9 months vs. 24 months (p = 0.03), and for 19q amplification carriers the top quartile versus the rest was 8.7 months vs. 36.7 months (p = 0.03). These two SCNAs occurred frequently across all neoplasms irrespective of primary site: focal deletion 22q13.31 was present in 65% of tumor samples, and arm amplification 19q in 53% of tumor samples (Fig. 5b). Deletion of 22q13.31 has been identified in cancers of the lung, ovary, head and neck, brain, and insulinomas, a form of pancreatic NET.(Bertonha, et al. 2015; Felicio, et al. 2018; Jonkers, et al. 2006; Liu, et al. 2015; Nakamura, et al. 2005; Reis, et al. 2002; Sondka, et al. 2018) Known tumor suppressor genes that were deleted in the wide peak of 22q13.31 were SMARCB1, CHEK2, NF2 and EP300, while known oncogenes in arm 19q were BCL3, CCNE1, AKT2, CD79A, CNOT3 (Supplemental Data). SMARCB1 and EP300 are both involved in chromatin remodeling. SMARCB1 is a known tumor suppressor located within this region. The protein is involved in chromatin remodeling, as a core subunit of the SWI/SNF chromatin remodeling complex.
DISCUSSION
High grade NENs are rapidly fatal with median survival of only 7 months, and can arise from a wide range of organs.(Dasari, et al. 2018) They are currently all treated with the same first-line chemotherapy (platinum and etoposide) regardless of primary site, owing largely to past experience with small cell lung cancer.(Oronsky B 2018) Whether this one-size fits all approach in treating all G3 NENs regardless of origin may have a biological basis is still unclear.
While past studies investigating G3 NENs from specific tissues have reported TP53 and RB1 to be the most common mutations, site-characteristic mutations have also been reported, for example APC in GEP tumors and STK11/KEAP1 in lung tumors.(Uccella, et al. 2021) Some prior research has suggested that G3 NENs were more similar to non-NENs from the same sites than to other G3 NENs.(Chen, et al. 2020) However, many of these studies only compared G3 NENs with non-NENs from the same site, and not to G3 NENs from other sites.(Chen, et al. 2020; Furlan, et al. 2013; Jesinghaus, et al. 2017; Sahnane, et al. 2015; Takizawa, et al. 2015; Woischke, et al. 2017) They also examined a small number of genes (ranging from 3 to <200) as points of comparison.
As whole transcriptome sequencing is often a more comprehensive assessment tool than select gene sequencing to reveal true underlying tumor behavior, we compared G3 NENs from different sites using this technology. We found that, interestingly, the gene expression profiles of NENs did not easily segregate by organ of origin. An integrated analysis combining WGS, WES and WTS data again showed no clear segregation of tumors by organ. These data taken together are consistent with a prior study which found the small cell subtype of G3 NENs from the prostate and bladder to be transcriptomically similar to that from the lung.(Balanis, et al. 2019) Together, these findings help inform our current uniform treatment approach to G3 neoplasms regardless of tumor origin,(Strosberg, et al. 2010) and is consistent with clinical experience in which platinum/etoposide therapy is more effective for G3 NENs than site-specific therapy normally employed for adenocarcinoma or squamous cell carcinoma for any particular organ. Further studies will have to be done, especially with larger sample sizes, to further investigate the similarities and differences between G3 NENs, which is important to guide both future clinical trial eligibility and therapy development.
With WES, we found that TP53, RB1 and APC were the most frequently implicated genes. These findings are consistent with prior studies on both lung and GEP NENs which showed these mutations to be among the most common.(George, et al. 2018a; Karlsson, et al. 2015; Peifer, et al. 2012; Puccini, et al. 2020) Dysregulation of the CDK4/6 cell cycling pathway, including alterations of RB1 and CDKN2A, was found in 95% of cases across G3 NENs from different organs. Transcriptome data confirmed significantly higher levels of oncogene expression in this pathway compared to normal tissue, as well as other signaling networks related to cell cycling. Other studies in G3 NENs of the lung have also reported genomic alterations resulting in loss of CDKN2A and upregulation of cell cycle and mitosis, though at lower frequencies.(George, et al. 2018a; Jones, et al. 2004; Peifer, et al. 2012; Rekhtman, et al. 2016) One pancreatic study found loss of CDKN2A present in 25% of G3 NENs, and when examined among G1–3 NENs it was associated with a higher tumor grade, increased risk of metastasis and shorter survival.(Roy, et al. 2018)
To aid new therapeutic discoveries, our finding that the CDK4/6 cell cycling pathway was involved in most cases may provide an opportunity for therapy with CDK4/6 inhibitors. CDK inhibition has not been clinically tested in G3 NENs, though there is pre-clinical data showing response to ribociclib in a NET cell line, and a case report of a dramatic response to palbociclib in a patient with breast G3 NEN harboring amplification of cyclin D1.(Aristizabal Prada, et al. 2018; Shanks, et al. 2018) As canonical oncogenic pathways such as PI3K that are upstream of CDK4/6 were frequently altered as well, dual therapies may be efficacious. Combinations of a CDK4/6 inhibitor with a RAS or PI3K inhibitor have already been tested in the preclinical setting with success.(Du, et al. 2020) Additionally, 20% of cases harbored high TMB (≥10 mutations/Mb), which raises the potential for therapy with pembrolizumab, which is now FDA-approved to treat any solid tumor that meets this threshold.(FDA 2020)
G3 neoplasms showed frequent chromosomal instability with focal and arm level copy number alterations, in contrast to lower grade NETs.(Fernandez-Cuesta, et al. 2014; Hashemi, et al. 2013; Karpathakis, et al. 2016) We did not detect specific CNV patterns based on tissue type, but identified two SCNAs found in over 50% of patients—deletion 22q13.31 and arm amplification 19q—that were common across all organs and had negative correlation with survival, with hazard ratios of 4–7. As there still exists a range of survival times with G3 NENs, and currently no reliable marker to predict survival within this group, these two SCNAs could potentially be used as prognostic markers but would require future prospective validation. Amplification of 19q includes known oncogenes that include CCNE1, Cyclin E1, which is involved in cell cycling. Amplification of this region, frequent loss of 3p (67%) involving RB1 and 9p involving CDKN2A in our cohort further illustrate the theme of cell cycling dysregulation as a primary oncogenic pathway in G3 NENs. These three CNV alterations have similarly been reported before for G3 lung NENs (small cell and large cell).(George, et al. 2018a; Peifer, et al. 2012)
One limitation of our study was the use of FFPE samples, which can cause relatively more sequencing artifacts when compared to fresh frozen (FF) samples.(Do and Dobrovic 2015) While large gene panels numbering hundreds of genes are now routinely ordered on FFPE tumor samples, there is less experience with performing whole genome sequencing on such samples.(Frampton, et al. 2013) However, an increasing number of studies are demonstrating the reliability of FFPE sequencing in such circumstances, with some studies showing >97–99% concordance between FFPE and FF samples.(Ahn, et al. 2016; Astolfi, et al. 2015; Carrick, et al. 2015; Munchel, et al. 2015; Spencer, et al. 2013; Van Allen, et al. 2014; Xia, et al. 2020) A second limitation of our study is the limited number of samples sequenced due to the rarity of G3 NENs. However, to our knowledge, this study is one of the first to discern differences between multiple G3 NEN primary sites. A third caveat of this study is the inclusion of poorly-differentiated (n = 39), well differentiated (n = 5), and mixed well and poorly-differentiated tumors (n = 2). While G3 GEP NENs are newly categorized into well-differentiated G3 NET and poorly-differentiated neuroendocrine carcinoma in the most recent version of the WHO classification system, NENs from other primary sites have not been similarly categorized.(Klimstra DS 2019) For example, the recent 2021 WHO classification for lung NENs group continues to establish well and poorly-differentiated G3 NENs as one entity, citing insufficient evidence for doing otherwise.(Board 2021) For NENs from an unknown primary site, it is further unclear which classification schema should be used. As our study aimed to compare G3 NENs across organs, to mitigate the non-uniformity of these classification systems, we employed a common inclusion criteria consisting of either ki-67 above 20% or mitotic index above 10/high power field. The few well-differentiated tumors in this study behaved similarly to the poorly-differentiated tumors, with high markers of proliferation, similar transcriptomic profiles, as well as highly aggressive clinical course.
Despite the above limitations, our exploratory study found genomic features common to all G3 NENs and may provide additional insight into future prognostic marker and therapy development.
Supplementary Material
Acknowledgements and Funding
This study was made possible by a generous donation from The Helen B. Rodde Fund, by the Stanford Cancer Institute Research Database, Stanford Data Coordinating Center, NCI Cancer Center Support Grant 5P30CA124435 and Stanford NIH/NCRR CTSA Award Number UL1 RR025744.
P.L.K. has received research funding from Novartis (Advanced Accelerator Applications); she has served as an advisor to Novartis (Advanced Accelerator Applications), Amgen, Genentech, Crinetics, RayzeBio, Ipsen Rising Star, Natera, HutchMed.
Footnotes
DECLARATIONS
Availability of Data and Materials
Whole exome sequencing data (n = 33) and transcriptome sequencing data (n = 36) permitted for sharing by IRB are deposited at the NIH database of genotypes and phenotypes (dbGaP) under accession number: phs002070.
Conflicts of Interest
The remaining authors declare no conflicts of interest.
REFERENCES
- Ahn D, Ozer H, Hancioglu B, Lesinski G, Timmers C & Bekaii-Saab T 2016. Whole-exome tumor sequencing study in biliary cancer patients with a response to MEK inhibitors. Oncotarget 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov L, Ju Y, Haase K, Van Loo P, Martincorena I, Nik-Zainal S, Totoki Y, Fujimoto A, Nakagawa H, Shibata T, et al. 2016. Mutational signatures associated with tobacco smoking in human cancer. Science 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. 2013. Signatures of mutational processes in human cancer. Nature 500 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S & Huber W 2010. Differential Expression Analysis for Sequence Count Data. Genome biology 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andor N, Harness JV, Müller S, Mewes HW & Petritsch C 2014. EXPANDS: expanding ploidy and allele frequency on nested subpopulations. In Bioinformatics, pp 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D, Hu Z & Butte AJ 2017. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 18 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aristizabal Prada ET, Nolting S, Spoettl G, Maurer J & Auernhammer CJ 2018. The Novel Cyclin-Dependent Kinase 4/6 Inhibitor Ribociclib (LEE011) Alone and in Dual-Targeting Approaches Demonstrates Antitumoral Efficacy in Neuroendocrine Tumors in vitro. Neuroendocrinology 106 58–73. [DOI] [PubMed] [Google Scholar]
- Assi HA & Padda SK 2020. Latest advances in management of small cell lung cancer and other neuroendocrine tumors of the lung. Cancer Treat Res Commun 23 100167. [DOI] [PubMed] [Google Scholar]
- Astolfi A, Urbini M, Indio V, Nannini M, Genovese C, Santini D, Saponara M, Mandrioli A, Ercolani G, Brandi G, et al. 2015. Whole exome sequencing (WES) on formalin-fixed, paraffin-embedded (FFPE) tumor tissue in gastrointestinal stromal tumors (GIST). BMC genomics 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanis N, Sheu K, Esedebe F, Patel S, Smith B, Park J, Alhani S, Gomperts B, Huang J, Witte O, et al. 2019. Pan-cancer Convergence to a Small-Cell Neuroendocrine Phenotype that Shares Susceptibilities with Hematological Malignancies. Cancer cell 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh E, Ke H, Levine A, Bonneau R & Chan C 2018. Why are there hotspot mutations in the TP53 gene in human cancers? Cell death and differentiation 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertonha FB, Barros Filho Mde C, Kuasne H, Dos Reis PP, da Costa Prando E, Munoz JJ, Roffe M, Hajj GN, Kowalski LP, Rainho CA, et al. 2015. PHF21B as a candidate tumor suppressor gene in head and neck squamous cell carcinomas. Mol Oncol 9 450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Board WCoTE 2021. Thoracic Tumours. Lyon, France: International Agency for Research on Cancer. [Google Scholar]
- Boons G, Vandamme T, Mariën L, Lybaert W, Roeyen G, Rondou T, Papadimitriou K, Janssens K, Op de Beeck B, Simoens M, et al. 2022. Longitudinal Copy-Number Alteration Analysis in Plasma Cell-Free DNA of Neuroendocrine Neoplasms is a Novel Specific Biomarker for Diagnosis, Prognosis, and Follow-up. Clin Cancer Res 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busico A, Maisonneuve P, Prinzi N, Pusceddu S, Centonze G, Garzone G, Pelligrinelli A, Giacomelli L, Mangogna A, Paolino C, et al. 2019. Gastroenteropancreatic High-Grade Neuroendocrine Neoplasms (H-NENs): histology and molecular analysis, two sides of the same coin. Neuroendocrinology. [DOI] [PubMed] [Google Scholar]
- Carrick D, Mehaffey M, Sachs M, Altekruse S, Camalier C, Chuaqui R, Cozen W, Das B, Hernandez B, Lih C, et al. 2015. Robustness of Next Generation Sequencing on Older Formalin-Fixed Paraffin-Embedded Tissue. PloS one 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Liu M, Zhang Y, Guo Y, Chen M & Chen J 2020. Genetic Characteristics of Colorectal Neuroendocrine Carcinoma: More Similar to Colorectal Adenocarcinoma. Clinical colorectal cancer. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y & Gu J 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics (Oxford, England) 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y & Anders L 2014. Signaling through cyclin D-dependent kinases. Oncogene 33. [DOI] [PubMed] [Google Scholar]
- Dasari A, Mehta K, Byers LA, Sorbye H & Yao JC 2018. Comparative study of lung and extrapulmonary poorly differentiated neuroendocrine carcinomas: A SEER database analysis of 162,983 cases. Cancer 124 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasari A, Shen C, Halperin D, Zhao B, Zhou S, Xu Y, Shih T & Yao J 2017. Trends in the Incidence, Prevalence, and Survival Outcomes in Patients With Neuroendocrine Tumors in the United States. JAMA oncology 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diossy M, Sztupinszki Z, Krzystanek M, Borcsok J, Eklund AC, Csabai I, Pedersen AG & Szallasi Z 2019. Strand Orientation Bias Detector (SOBDetector) to remove FFPE sequencing artifacts. [DOI] [PubMed] [Google Scholar]
- Do H & Dobrovic A 2015. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clinical chemistry 61. [DOI] [PubMed] [Google Scholar]
- Du Q, Guo X, Wang M, Li Y, Sun X & Li Q 2020. The application and prospect of CDK4/6 inhibitors in malignant solid tumors. Journal of hematology & oncology 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, Hess J, Ma S, Chiotti KE, McLellan M, et al. 2018. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst 6 271–281.e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA 2020. FDA approves pembrolizumab for adults and children with TMB-H solid tumors | FDA. [Google Scholar]
- Felicio PS, Bidinotto LT, Melendez ME, Grasel RS, Campacci N, Galvao HCR, Scapulatempo-Neto C, Dufloth RM, Evangelista AF & Palmero EI 2018. Genetic alterations detected by comparative genomic hybridization in BRCAX breast and ovarian cancers of Brazilian population. Oncotarget 9 27525–27534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Cuesta L, Peifer M, Lu X, Sun R, Ozretic L, Seidal D, Zander T, Leenders F, George J, Muller C, et al. 2014. Frequent mutations in chromatin-remodelling genes in pulmonary carcinoids. Nat Commun 5 3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton G, Fichtenholtz A, Otto G, Wang K, Downing S, He J, Schnall-Levin M, White J, Sanford E, An P, et al. 2013. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nature biotechnology 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan D, Sahnane N, Mazzoni M, Pastorino R, Carnevali I, Stefanoli M, Ferretti A, Chiaravalli A, La Rosa S & Capella C 2013. Diagnostic utility of MS-MLPA in DNA methylation profiling of adenocarcinomas and neuroendocrine carcinomas of the colon-rectum. Virchows Archiv : an international journal of pathology 462. [DOI] [PubMed] [Google Scholar]
- George J, Walter V, Peifer M, Alexandrov LB, Seidel D, Leenders F, Maas L, Müller C, Dahmen I, Delhomme TM, et al. 2018a. Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nature Communications 9 1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Walter V, Peifer M, Alexandrov L, Seidel D, Leenders F, Maas L, Müller C, Dahmen I, Delhomme T, et al. 2018b. Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nature Communications 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Lim J, Jang S, Cun Y, Ozretić L, Kong G, Leenders F, Lu X, Fernández-Cuesta L, Bosco G, et al. 2015. Comprehensive genomic profiles of small cell lung cancer. Nature 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi DM, Silva ACB, Rego JFM, Coudry RA & Riechelmann RP 2017. Unraveling molecular pathways of poorly differentiated neuroendocrine carcinomas of the gastroenteropancreatic system: A systematic review. Cancer Treat Rev 56 28–35. [DOI] [PubMed] [Google Scholar]
- Hashemi J, Fotouhi O, Sulaiman L, Kjellman M, Hoog A, Zedenius J & Larsson C 2013. Copy number alterations in small intestinal neuroendocrine tumors determined by array comparative genomic hybridization. BMC Cancer 13 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesinghaus M, Konukiewitz B, Keller G, Kloor M, Steiger K, Reiche M, Penzel R, Endris V, Arsenic R, Hermann G, et al. 2017. Colorectal mixed adenoneuroendocrine carcinomas and neuroendocrine carcinomas are genetically closely related to colorectal adenocarcinomas. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 30. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, et al. 2011. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M, Virtanen C, Honjoh D, Miyoshi T, Satoh Y, Okumura S, Nakagawa K, Nomura H & Ishikawa Y 2004. Two prognostically significant subtypes of high-grade lung neuroendocrine tumours independent of small-cell and large-cell neuroendocrine carcinomas identified by gene expression profiles. Lancet (London, England) 363. [DOI] [PubMed] [Google Scholar]
- Jonkers YM, Claessen SM, Feuth T, van Kessel AG, Ramaekers FC, Veltman JA & Speel EJ 2006. Novel candidate tumour suppressor gene loci on chromosomes 11q23–24 and 22q13 involved in human insulinoma tumourigenesis. J Pathol 210 450–458. [DOI] [PubMed] [Google Scholar]
- Karlsson A, Brunnström H, Lindquist KE, Jirström K, Jönsson M, Rosengren F, Reuterswärd C, Cirenajwis H, Borg Å, Jönsson P, et al. 2015. Mutational and gene fusion analyses of primary large cell and large cell neuroendocrine lung cancer. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpathakis A, Dibra H, Pipinikas C, Feber A, Morris T, Francis J, Oukrif D, Mandair D, Pericleous M, Mohmaduvesh M, et al. 2016. Prognostic Impact of Novel Molecular Subtypes of Small Intestinal Neuroendocrine Tumor. Clin Cancer Res 22 250–258. [DOI] [PubMed] [Google Scholar]
- Kendig K, Baheti S, Bockol M, Drucker T, Hart S, Heldenbrand J, Hernaez M, Hudson M, Kalmbach M, Klee E, et al. 2019. Sentieon DNASeq Variant Calling Workflow Demonstrates Strong Computational Performance and Accuracy. Frontiers in genetics 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Lee SJ, Park SH, Park JO, Lim HY, Kang WK, Lee J & Park YS 2016. Genomic Profiling of Metastatic Gastroenteropancreatic Neuroendocrine Tumor (GEP-NET) Patients in the Personalized-Medicine Era. J Cancer 7 1044–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebanov N, Artomov M, Goggins WB, Daly E, Daly MJ & Tsao H 2019. Burden of unique and low prevalence somatic mutations correlates with cancer survival. Sci Rep 9 4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimstra DS KG, La Rosa S, Rindi G 2019. Classification of neuroendocrine neoplasms of the digestive system. In WHO Classification of Tumours: Digestive System Tumours, edn 5th, p 16. Ed WCoTEB (Ed). Lyon: International Agency for Research on Cancer. [Google Scholar]
- Kunz PL 2015. Carcinoid and neuroendocrine tumors: building on success. J Clin Oncol 33 1855–1863. [DOI] [PubMed] [Google Scholar]
- Leoncini E, Boffetta P, Shafir M, Aleksovska K, Boccia S & Rindi G 2017. Increased incidence trend of low-grade and high-grade neuroendocrine neoplasms. Endocrine 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Stucker I, Chen C, Goodman G, McHugh MK, D’Amelio AM Jr., Etzel CJ, Li S, Lin X & Christiani DC 2015. Genome-wide Gene-Asbestos Exposure Interaction Association Study Identifies a Common Susceptibility Variant on 22q13.31 Associated with Lung Cancer Risk. Cancer Epidemiol Biomarkers Prev 24 1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Zhang P, Zhang Y, Li Z, Gong JF, Li J, Li Y, Zhang X, Lu Z, Wang X, et al. 2020. Efficacy, safety and biomarkers of toripalimab in patients with recurrent or metastatic neuroendocrine neoplasms: a multiple-center phase Ib trial. Clin Cancer Res. [DOI] [PubMed] [Google Scholar]
- Mafficini A & Scarpa A 2019. Genetics and Epigenetics of Gastroenteropancreatic Neuroendocrine Neoplasms. Endocr Rev 40 506–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, Davies H, Stratton MR & Campbell PJ 2017. Universal Patterns of Selection in Cancer and Somatic Tissues. In Cell, pp 1029–1041 e1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayakonda A, Lin D, Assenov Y, Plass C & Koeffler H 2018. Maftools: Efficient and Comprehensive Analysis of Somatic Variants in Cancer. Genome research 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez B, Van Campenhout C, Rorive S, Remmelink M, Salmon I & D’Haene N 2018. Methods of measurement for tumor mutational burden in tumor tissue. Transl Lung Cancer Res 7 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti S, Tamayo P, Mesirov J & Golub T 2020. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Machine Learning 52 91–118. [Google Scholar]
- Munchel S, Hoang Y, Zhao Y, Cottrell J, Klotzle B, Godwin A, Koestler D, Beyerlein P, Fan J, Bibikova M, et al. 2015. Targeted or whole genome sequencing of formalin fixed tissue samples: potential applications in cancer genomics. Oncotarget 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Ishida E, Shimada K, Kishi M, Nakase H, Sakaki T & Konishi N 2005. Frequent LOH on 22q12.3 and TIMP-3 inactivation occur in the progression to secondary glioblastomas. Lab Invest 85 165–175. [DOI] [PubMed] [Google Scholar]
- Network NCC 2020. Neuroendocrine and Adrenal Tumors. Ed Shah M: National Comprehensive Cancer Network. [DOI] [PubMed] [Google Scholar]
- Oronsky B ea 2018. Nothing But NET: A Review of Neuroendocrine Tumors and Carcinomas. - PubMed - NCBI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T, et al. 2012. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 44 1104–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccini A, Poorman K, Salem M, Soldato D, Seeber A, Goldberg R, Shields A, Xiu J, Battaglin F, Berger M, et al. 2020. Comprehensive Genomic Profiling of Gastroenteropancreatic Neuroendocrine Neoplasms (GEP-NENs). Clin Cancer Res 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj N, Shah R, Stadler Z, Mukherjee S, Chou J, Untch B, Li J, Kelly V, Saltz LB, Mandelker D, et al. 2018. Real-Time Genomic Characterization of Metastatic Pancreatic Neuroendocrine Tumors Has Prognostic Implications and Identifies Potential Germline Actionability. JCO Precis Oncol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis PP, Rogatto SR, Kowalski LP, Nishimoto IN, Montovani JC, Corpus G, Squire JA & Kamel-Reid S 2002. Quantitative real-time PCR identifies a critical region of deletion on 22q13 related to prognosis in oral cancer. Oncogene 21 6480–6487. [DOI] [PubMed] [Google Scholar]
- Rekhtman N, Pietanza MC, Hellmann MD, Naidoo J, Arora A, Won H, Halpenny DF, Wang H, Tian SK, Litvak AM, et al. 2016. Next-Generation Sequencing of Pulmonary Large Cell Neuroendocrine Carcinoma Reveals Small Cell Carcinoma-like and Non-Small Cell Carcinoma-like Subsets. Clin Cancer Res 22 3618–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, LaFramboise WA, Liu TC, Cao D, Luvison A, Miller C, Lyons MA, O’Sullivan RJ, Zureikat AH, Hogg ME, et al. 2018. Loss of Chromatin Remodeling Proteins and/or CDKN2A Associates With Metastasis of Pancreatic Neuroendocrine Tumors and Reduced Patient Survival Times. Gastroenterology 154 2060–2063 e2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahnane N, Furlan D, Monti M, Romualdi C, Vanoli A, Vicari E, Solcia E, Capella C, Sessa F & La Rosa S 2015. Microsatellite unstable gastrointestinal neuroendocrine carcinomas: a new clinicopathologic entity. Endocrine-related cancer 22. [DOI] [PubMed] [Google Scholar]
- Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ, Omuro A, et al. 2019. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 51 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vega F, Mina M, Armenia J, Chatila W, Luna A, La K, Dimitriadoy S, Liu D, Kantheti H, Saghafinia S, et al. 2018. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpa A, Chang DK, Nones K, Corbo V, Patch AM, Bailey P, Lawlor RT, Johns AL, Miller DK, Mafficini A, et al. 2017. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 543 65–71. [DOI] [PubMed] [Google Scholar]
- Shanks A, Choi J & Karur V 2018. Dramatic response to cyclin D–dependent kinase 4/6 inhibitor in refractory poorly differentiated neuroendocrine carcinoma of the breast. In Proc (Bayl Univ Med Cent), pp 352–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I & Forbes SA 2018. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer 18 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorbye H, Welin S, Langer SW, Vestermark LW, Holt N, Osterlund P, Dueland S, Hofsli E, Guren MG, Ohrling K, et al. 2013. Predictive and prognostic factors for treatment and survival in 305 patients with advanced gastrointestinal neuroendocrine carcinoma (WHO G3): the NORDIC NEC study. Ann Oncol 24 152–160. [DOI] [PubMed] [Google Scholar]
- Spencer D, Sehn J, Abel H, Watson M, Pfeifer J & Duncavage E 2013. Comparison of clinical targeted next-generation sequence data from formalin-fixed and fresh-frozen tissue specimens. J Mol Diagn 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strosberg JR, Coppola D, Klimstra DS, Phan AT, Kulke MH, Wiseman GA & Kvols LK 2010. The NANETS consensus guidelines for the diagnosis and management of poorly differentiated (high-grade) extrapulmonary neuroendocrine carcinomas. Pancreas 39 799–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha V, Mukherjee S, Ebert B, Gillette M, Paulovich A, Pomeroy S, Golub T, Lander E, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa N, Ohishi Y, Hirahashi M, Takahashi S, Nakamura K, Tanaka M, Oki E, Takayanagi R & Oda Y 2015. Molecular characteristics of colorectal neuroendocrine carcinoma; similarities with adenocarcinoma rather than neuroendocrine tumor. Human pathology 46. [DOI] [PubMed] [Google Scholar]
- Tibshirani R, Walther G & Hastie T 2020. Estimating the number of clusters in a data set via the gap statistic - Tibshirani - 2001 - Journal of the Royal Statistical Society: Series B (Statistical Methodology) - Wiley Online Library. Journal of the Royal Statistical Society. [Google Scholar]
- Uccella S, La Rosa S, Metovic J, Marchiori D, Scoazec J, Volante M, Mete O & Papotti M 2021. Genomics of High-Grade Neuroendocrine Neoplasms: Well-Differentiated Neuroendocrine Tumor with High-Grade Features (G3 NET) and Neuroendocrine Carcinomas (NEC) of Various Anatomic Sites. Endocrine pathology 32. [DOI] [PubMed] [Google Scholar]
- Van Allen E, Wagle N, Stojanov P, Perrin D, Cibulskis K, Marlow S, Jane-Valbuena J, Friedrich D, Kryukov G, Carter S, et al. 2014. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nature medicine 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Väremo L, Nielsen J & Nookaew I 2013. Enriching the Gene Set Analysis of Genome-Wide Data by Incorporating Directionality of Gene Expression and Combining Statistical Hypotheses and Methods. Nucleic acids research 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venizelos A, Elvebakken H, Perren A, Nikolaienko O, Deng W, Lothe I, Couvelard A, Hjortland G, Sundlöv A, Svensson J, et al. 2021. The molecular characteristics of high-grade gastroenteropancreatic neuroendocrine neoplasms. Endocrine-related cancer 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayvergia N, Boland PM, Handorf E, Gustafson KS, Gong Y, Cooper HS, Sheriff F, Astsaturov I, Cohen SJ & Engstrom PF 2016. Molecular profiling of neuroendocrine malignancies to identify prognostic and therapeutic markers: a Fox Chase Cancer Center Pilot Study. Br J Cancer 115 564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Sun L, Bao H, Wang A, Zhang P, Wu X, Tong X, Wang X, Luo J, Shen L, et al. 2019. Genomic dissection of gastrointestinal and lung neuroendocrine neoplasm. Chin J Cancer Res 31 918–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MM & Fottner C 2018. Immune Checkpoint Inhibitors in the Treatment of Patients with Neuroendocrine Neoplasia. Oncol Res Treat 41 306–312. [DOI] [PubMed] [Google Scholar]
- Woischke C, Schaaf C, Yang H, Vieth M, Veits L, Geddert H, Märkl B, Stömmer P, Schaeffer D, Frölich M, et al. 2017. In-depth mutational analyses of colorectal neuroendocrine carcinomas with adenoma or adenocarcinoma components. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 30. [DOI] [PubMed] [Google Scholar]
- Wong HL, Yang KC, Shen Y, Zhao EY, Loree JM, Kennecke HF, Kalloger SE, Karasinska JM, Lim HJ, Mungall AJ, et al. 2018. Molecular characterization of metastatic pancreatic neuroendocrine tumors (PNETs) using whole-genome and transcriptome sequencing. Cold Spring Harb Mol Case Stud 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Van Hummelen P, Kubit M, Lee H, Bell J, Grimes S, Wood-Bouwens C, Greer S, Barker T, Haslem D, et al. 2020. Whole genome analysis identifies the association of TP53 genomic deletions with lower survival in Stage III colorectal cancer. Scientific reports 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S, Vakiani E, White CM, Zhong Y, Saunders T, Morgan R, de Wilde RF, Maitra A, Hicks J, Demarzo AM, et al. 2012. Small cell and large cell neuroendocrine carcinomas of the pancreas are genetically similar and distinct from well-differentiated pancreatic neuroendocrine tumors. Am J Surg Pathol 36 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S, Totoki Y, Noë M, Nakatani Y, Horie M, Kawasaki K, Nakamura H, Saito-Adachi M, Suzuki M, Takai E, et al. 2022. Comprehensive Genomic Profiling of Neuroendocrine Carcinomas of the Gastrointestinal System. Cancer Discov 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Garg A, Chen D, Capdevila J, Engstrom P, Pommier R, Van Cutsem E, Singh S, Fazio N, He W, et al. 2019. Genomic profiling of NETs: a comprehensive analysis of the RADIANT trials. Endocr Relat Cancer 26 391–403. [DOI] [PubMed] [Google Scholar]
- Zhao L & Yan H 2019. MCNF: A novel method for cancer subtyping by integrating multi-omics and clinical data. IEEE/ACM transactions on computational biology and bioinformatics. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.