Abstract
The outbreak of novel Coronavirus, an enduring pandemic declared by WHO, has consequences to an alarming ongoing public health menace which has already claimed several million human lives. In addition to numerous vaccinations and medications for mild to moderate COVID-19 infection, lack of promising medication or therapeutic pharmaceuticals remains a serious concern to counter the ongoing coronavirus infections and to hinder its dreadful spread. Global health emergencies have called for urgency for potential drug discovery and time is the biggest constraint apart from the financial and human resources required for the high throughput drug screening. However, computational screening or in-silico approaches appeared to be an effective and faster approach to discover potential molecules without sacrificing the model animals. Accumulated shreds of evidence on computational studies against viral diseases have revealed significance of in-silico drug discovery approaches especially in the time of urgency. The central role of RdRp in SARS-CoV-2 replication makes it promising drug target to curtain on going infection and its spread. The present study aimed to employ E-pharmacophore-based virtual screening to reveal potent inhibitors of RdRp as potential leads to block the viral replication. An energy-optimised pharmacophore model was generated to screen the Enamine REAL DataBase (RDB). Then, ADME/T profiles were determined to validate the pharmacokinetics and pharmacodynamics properties of the hit compounds. Moreover, High Throughput Virtual Screening (HTVS) and molecular docking (SP & XP) were employed to screen the top hits from pharmacophore-based virtual screening and ADME/T screen. The binding free energies of the top hits were calculated by conducting MM-GBSA analysis followed by MD simulations to determine the stability of molecular interactions between top hits and RdRp protein. These virtual investigations revealed six compounds having binding free energies of −57.498, −45.776, −46.248, −35.67, −25.15 and −24.90 kcal/mol respectively as calculated by the MM-GBSA method. The MD simulation studies confirmed the stability of protein ligand complexes, hence, indicating as potent RdRp inhibitors and are promising candidate drugs to be further validated and translated into clinics in future.
Keywords: COVID-19, RNA-dependent RNA polymerase MM-GBSA, REAL DataBase – Enamine, ADME/T
1. Introduction
Coronavirus outbreak since 2019 in China has taken the world by storm. An ongoing pandemic has caused serious global public health threats with millions of people reported to have contracted coronavirus worldwide [1], [2]. Studies have reported this positive-sense, single-stranded RNA virus as a novel beta-coronavirus or dubbed SARS-CoV-2. It belongs to the betacoronavirus genus of the coronaviridae family comprising the alpha- and beta-coronaviruses [3], [4], [5]. The biggest genome (26.4–31.7 kb) among all known RNA viruses is found in this enclosed virus [6]. Its genome exhibits striking resemblances with the already known members of the family MERS (Middle East Respiratory Syndrome) and SARS and their non-structural proteins were reported to exhibit a significant role in the transcription and virus replication process. These non-structural proteins comprising RNA-dependent RNA polymerase (RdRp), main protease (Mpro) and papain-like protease [7], [8].
RNA dependent RNA polymerase (RdRp), plays an important role in the RNA replication within the life cycle of the severely acute respiratory coronavirus-2 (SARS-CoV-2), causing the lethal respiratory illness i.e. COVID-19 [9]. RdRp also known as nsp12, remains a highly conserved drug target against SARS-CoV-2 over the past years. Nsp12 is an indispensable and critical component of the corona viral replication/transcription machinery complex as it catalyses the Corona virus RNA synthesis [10]. RdRp reportedly does not exhibit host cell homology that nominates it as a providential antiviral drug target with minimum toxicity to human cells. Moreover, it does not possess a proofreading activity [11]. These disclosures have paved the way to conduct drug repurposing of mutagenic nucleoside analog inhibitors or chain terminator-based therapeutic drugs like favipiravir and remdesivir. Such nucleoside analogs have been proved to be potent drugs in antiviral therapies due to their decisive role in terminating viral RNA synthesis [12]. The prime targets of these drugs have been non-structural proteins due to the least probability of getting mutations. Although several mutations and resultant mutants have been reported for the coronaviruses, highly conserved proteins are rarely expected to get mutated during their life cycle. The underlying reason remained the deadly effects on the virus itself due to mutation in these highly conserved proteins [13]. Having rare chances to be mutated makes the main protease and RdRp the promising drug targets for prospective therapeutic strategies.
New oral coronavirus medications are still being researched till today. Molnupiravir, fluvoxamine and Paxlovid are three noval oral antiviral medicines that are successful in lowering the mortality and hospitalization rates in COVID-19 patients. Paxlovid, recently released by Pfizer, is an oral antiviral drug candidate for SARS-CoV-2 protease inhibitors. In Paxlovid, Ritonavir and PF-07321332 are combined. Paxlovid prevents the coronavirus to prudence new virions by inhibiting the SARS-COV-2-3Cl protease which is required to produce individual viral proteins from polypeptide. The combination of low dose of ritonavir along with Paxlovid helps in slowing down the metabolism of PF-07321332 and hence helping to fight against the virus by maintaining high concentration for long in body. Apart from limited effectiveness of Paxlovid and few other drugs [14], [15], the unavailability of effective drug, the current medication relies on management and supportive therapies. The occurrence of frequent recombination events has made the discovery of potent therapeutic drugs against corona viruses even more onerous than ever, which requires more exhaustive insight into the molecular aspect of corona viruses and their life cycle [16]. The computational screening techniques accelerate the drug designing and discovery process by the screening large libraries of molecules against specific proteins [17]. The target proteins may include both structural and nonstructural proteins of SARS-COV-2 as alternate targets apart from the previously investigated viral main protease [18], [19]. These new target proteins include Nsp1 [20], Nsp15 [21], Nsp16 [22], RdRp [23], and host protein such as ACE2 [24] used to investigate the potential bioactive compounds from various plant sources.
Moreover, drug repurposing holds a significant potential in identifying and discovering the potential drug for COVID-19 from the previously designed and approved drugs [19], [25]. Mostly, the drugs being investigated for COVID-19 due to their efficacy and efficiency are repurposed and clinically approved known drugs including the ritonavir and lopinavir for HIV protease inhibitors [26], [27], oseltamivir, remdesivir [28], [29], [30], and hydroxychloroquine [31]. The adverse effects of the clinically approved antimalarial drug i.e. hydroxychloroquine have been reported that suggested halting its use for COVID patients worldwide. These drugs are reported to have severe consequences such as tissue damage and worsen organ injury due to autophagy inhibition [32]. Among other known drugs, the potential advantages of antiviral drugs, including remdesivir and ritonavir have been reported for COVID-19 [25]. Numerous computational researches including drug repurposing have been reported over a period of the ongoing pandemic. The drug targets of these studies include structural [21], [33], [34] as well nonstructural proteins [25] including RdRp, Envelope and protease [35], [36].
In-silico approach has been extensively endorsed by researchers and scientists from around the world. Numerous studies have been projected over the past months to identify effective inhibitors or repurposed drugs against non-structural proteins and RdRp of SARS-CoV-2. To understand the mode of action of these drugs, molecular modelling provides substantial insight into the important interactions between ligands and chosen drug targets [37], [38], [39]. The importance of natural products also has overshadowed the current research studies to reveal their potential as potent inhibitors against the coronavirus proteases including cyanin, chlorogeninwere, amentoavone, catechin-7-o-gallate, agathisavone [40], green tea polyphenols [41], and taraxerol [42]. Immense work has been done on various phytochemicals to reveal their antiviral and RdRp-inhibiting activities including phytochemicals hesperidin from the Andean region [43], neoandrographolide (AGP3) from Andrographis paniculata [44], flavonoids (usararotenoid A, and 12α-epi-millettosin) and alkaloids (10′-hydroxyusambarensine, cryptospirolepine, strychnopentamine) from African phytochemicals [45], and plenty of others. The phytochemicals and their derivatives have been identified as prospective inhibitors of RNA-dependent RNA polymerase as they exhibit substantial antiviral properties [38].
The current study aims to implement the computationally efficient E-pharmacophore based virtual screening strategy to discover anti-SARS-CoV-2 compounds from REAL Database – Enamine. The potent inhibitors of SARS-CoV-2 RdRp from this approach will impair the viral replication and hence curtain the COVID-19. The high throughput virtual screening based on E-pharmacophore model was used to screen the library of compounds against a specific target followed by molecular docking to analyse the interaction between the RdRp protein and potent inhibitors. Subsequently, ADME/T properties were assessed and free binding energies were calculated using the MM-GBSA approach. Eventually, MD simulations were employed on the top docked protein-ligand complexes to comprehend the dynamic behaviour and interactions of the complexes. This identification will introduce crucial understanding of how small molecules bind to RdRp and support our efforts to identify and create in silico anti-SARS-CoV-2 RdRp therapeutics.
2. Material and methods
2.1. Preparation of the protein
The crystal structure of the RdRp complex with remdesivir (PDB ID: 7BV2) having resolution of 2.5 Å was retrieved from protein data bank (PDB) [46]. To correct and refine the protein and to rectify the structural artifacts, the protein preparation wizard of Schrödinger was used [47]. This ensured high-confidence structural correctness by transforming the protein structure from the raw state to refined state and prepared adequately for molecular docking and molecular dynamic studies. The process involved assigning the corrected bond orders, omitting water molecules and other non-specific chemical components from the crystal structure, and addition of hydrogen atoms to the protein structure for remodelling the tautomeric and ionization states of amino acid residues. ([48]; Schrodinger Release 2018-1). Protein preparation wizard was utilised to pre-process the RdRp-inhibitor complex by removing the water molecules from the complex, positioned within 5 Å from the het-groups. Subsequent addition of missing hydrogen atoms along with hydrogen bond optimisation was followed by restrained energy minimization of RdRp structure by employing the OPLS4 force field to achieve high accuracy [49].
2.2. Molecular docking
Schrödinger's GLIDE module was used to dock the prepared ligands with RdRp which produced different poses of protein-ligand complexes to identify their binding mode and affinity [50]. XP descriptors helped in analysing the docking results providing significant details of various non-bonding interactions [51]. The energy for each pose was identified by Glide XP descriptor information, to optimise the ligand conformation of the ligand-protein complex. Before proceeding to the docking procedure, Schrödinger's Glide Grid Generation was used to prepare grid with Remdesivir (PDB ID: 7BV2) inhibitor as a reference ligand, resulting in the development of a boundary box around the active site of RdRp [52]. Grid generation was preceded by the docking using the prepared ligand. Subsequently, a protein-ligand complex was used for generation of E-pharmacophore model.
2.3. Energy-optimised pharmacophore (E-pharmacophore)
The intrinsic flexibility of the target active site was incorporated into Energy-optimised pharmacophore (E-pharmacophore) model generation to generate energetically efficient pharmacophores which could screen millions of molecules computationally in a short amount of time. The E-Pharmacophore methods combined the features of ligands with structure-based approaches to study the intrinsic flexibility of active sites and ligand binding [53]. An E-pharmacophore model was developed by using the protein-ligand complex of RdRp in Phase module. The default six pharmacophore features including: i) aromatic ring (R), ii) negatively ionizable region (N), iii) hydrogen bond acceptor (A), iv) hydrogen bond donor (D), v) hydrophobic group (H) and vi) positively ionizable region (P) were employed in Phase module to generate E-pharmacophore model.
2.4. Preparation of the ligand
The LigPrep module packaged by Schrödinger was used to prepare ligands by applying certain filters and generating a customized ligand library. The preparation of the ligands is carried out by adjusting the torsion of the ligand and then assigning the protonation states [54]. Subsequent refinement of 3D structures by addition of hydrogen atoms, generation of stereoisomers and identification of ionization states, led to ligand refinement. Furthermore, the OPLS4 force field incorporated in PHASE was used for energy minimization and to optimise the low-energy 3D structure of the ligand [55].
2.5. Virtual screening based on E-pharmacophore hypothesis
Virtual screening based on E-pharmacophore model was performed by utilising the Phase module of the Schrodinger suite for over 1 billion conformers of molecules from the Enamine REAL DataBase (RDB) database.
2.6. ADME/T analysis and Lipinski parameters
Schrödinger's QikProp module was employed to analyse the ADME/T profile of the potential compounds found by E-pharmacophore based virtual screen. To filter these compounds according to the Lipinski rule [56], the ADME/T properties analysis was conducted. Properties assessed involved molecular weight, brain/blood partition coefficient, lipophilicity (log Po/w), hydrophobicity and hydrophilicity (QPlogPo/w, QPlogS, QPPCaco, QPlogBB, and QPPMDCK), and human oral absorption (HOA%).
2.7. Virtual screening by molecular docking
After the initial screening of all the compounds in REAL Database – Enamine, based on E-pharmacophore and then filtering of revealed hits based on their ADME/T profiles, the best hits were selected for further processing by performing molecular docking with RdRp. GLIDE (Grid-based Ligand Docking with Energetics) module of Schrödinger was employed for stepwise molecular docking studies to screen the top hits. Firstly, the High throughput Virtual Screening (HTVS) and then standard precision docking (SP) were performed for all the molecules obtained by performing E-pharmacophore based screen and ADME/T analysis. Eventually, extra precision (XP) docking was performed using the resultant high-scoring compounds from SP docking studies to further specify the top hits.
2.8. Calculation of binding free energy by using MM-GBSA
To quantitatively measure the binding affinity between the protein and small molecules identified by performing XP docking, the molecular mechanics-generalized born surface area (MM-GBSA) approach was adopted. The prime module of Schrödinger was used to estimate the theoretical binding free energies of the hits obtained from XP docking, based on the glide scores, were utilised for MM-GBSA (Molecular Mechanical-Generalized Born Surface Area) analysis [57]. The energy properties of free ligand, free protein, and protein-ligand complex were utilised for the estimation of binding free energies [58].
ΔGbind calculations are executed using the following equations:
| (1) |
Using Eq. (1), ΔGbind is obtained by the difference between total of the free energies of the ligand (Glig) and the receptors (Grec) and the free energy of protein-ligand complex (Gcom).
ΔGbind is supposed to comprise enthalpy (H) [59] and entropy (TΔS) as presented in Eq. (2).
| (2) |
2.9. Molecular dynamics simulations
The Desmond module of Schrodinger was employed to conduct MD simulation studies to determine the dynamic binding behaviour and binding stability of the protein-inhibitor complexes using their docked poses [55]. Solvation of the complexes was done with the simple point-charge (SPC) water model, the system being neutralized with Na+/Cl− ions. Furthermore, potential energy of the protein-ligand complexes was minimized and relaxed under NPT ensemble to eliminate high energies in the predicted model. The molecular simulation was performed at 1 atm pressure and 300 K temperature for 100 ns and NPT production run under the OPLS4 force field. Finally, the stability of the protein-ligand complex was determined by calculating RMSD and for determination of fluctuation in structure RMSF was calculated.
3. Results
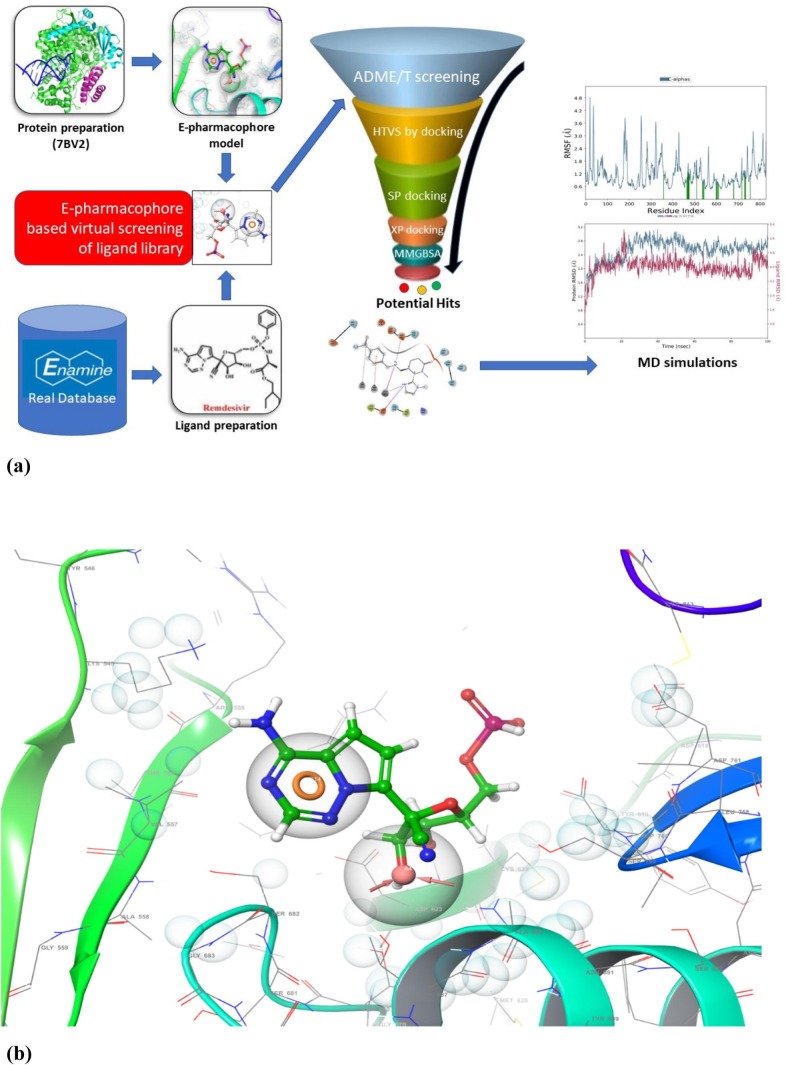
The work flow followed during the current drug screening process is depicted in Fig. 1a. This mainly includes selection of compound data base, energy optimised E-pharmacophore model, virtual screening for selection of top hit compounds and MD simulation to validate those compounds.
Fig. 1.
(a) Schematic representation of virtual screening. REAL Database – Enamine was screened for E-phamacophore developed from RdRp-Remdesivir complex and then obtained hits were prioritised based on their E-phamacophore features and ADME/T profiles. These hits were docked using SP and XP docking with RdRp followed the evaluation of stability of docked-complexes via MD simulations.
(b) E-pharmacophore model of RdRP of SARS-CoV-2 with remdesivir (RTP) inhibitor.
3.1. Energy-optimised pharmacophore (e-pharmacophore)
The E-pharmacophore hypothesis of the selected target i.e., RdRp was generated to develop an energetically optimised pharmacophore model by Schrödinger's PHASE module. An energy-optimised pharmacophore hypothesis (AR) was generated based on the protein-ligand complex. The hypothesis assisted to derive electrostatic and steric features of the ligands with binding sites of RdRp. The generated Energy-based pharmacophore model was consisting of one hydrogen bond acceptors (A) and one aromatic rings (R) (Fig. 1b). The hypothesis was used for the screening of more than 1 billion conformers from the REAL Database - Enamine to predict compound with favourable pharmacophore features.
3.2. ADME/T analysis and Lipinski parameters
ADME/T analysis and Lipinski parameters were assessed for the molecules to determine their pharmacokinetic profile. Pharmacokinetic profiles of hits and the findings of the ADME/T studies are shown in Table 1 and Table 2 . The most significant descriptors taken into consideration includes QPlogPo/w, QPlogS, QPlogBB, and QPPMDCK (determining hydrophobicity and hydrophilicity), #star (showing number of descriptors that fall beyond the recommended range), and Rule of Five (indicating total violations of Lipinski's rule of five). Molecules were selected on the basis of ADME/T properties and having zero (0) violation of Rule of Five. Total no of 0.354224 million molecules were selected for further processing.
Table 1.
ADME/T properties of all six compounds selected after HTVS.
| S. no. | QPlogPo/w (predicted octanol/water partition coefficient) | QPlogBB | Mol. MW (Da) | Percent human-oral absorption | FISA (hydrophilic component of the SASA (SASA on N, O, and H on heteroatoms). | PISA (carbon and attached hydrogen) component of the SASA) | QPlogHERG | QPlogS (mol dm−3) |
|---|---|---|---|---|---|---|---|---|
| Comp 1 | 0.899 | −1.116 | 355.439 | 88.173 | 175.542 | 229.680 | −5.027 | −2.256 |
| Comp 2 | −0.765 | −0.626 | 324.465 | 73.495 | 194.381 | 200.000 | −5.556 | 1.087 |
| Comp 3 | 1.370 | −1.585 | 292.294 | 84.717 | 194.381 | 82.737 | −5.141 | −3.505 |
| Comp 4 | 2.448 | 0.216 | 341.4 | 91.614 | 61.213 | 229.087 | −5.988 | −2.817 |
| Comp 5 | 3.341 | −0.841 | 372.8 | 100.000 | 119.249 | 239.892 | −5.312 | −4.813 |
| Comp 6 | 2.680 | −0.465 | 301.3 | 100.00 | 82.091 | 282.432 | −5.337 | −4.069 |
Table 2.
The XP Gscore, RO5 violations and MMGBSA energies before and after MD simulations of top six hit compounds. The compounds are ranked in ascending order of their MMGBSA energies after MD simulations.
| Compound | XP GScore | MMGBSA (before MD) | mol MW | Docking score | Rule of Five violation | Prime energy | MMGBSA (after MD) |
|---|---|---|---|---|---|---|---|
| Comp 1 | −7.986 | −57.498 | 355.439 | −7.986 | 0 | −34,026.0 | −70.498 |
| Comp 2 | −8.029 | −45.776 | 324.465 | −8.029 | 0 | −34,041.1 | −63.776 |
| Comp 3 | −7.168 | −46.248 | 292.294 | −7.168 | 0 | −33,983.2 | −60.248 |
| Comp 4 | −6.257 | −35.67 | 342.827 | −5.643 | 0 | −33,679.9 | −55.67 |
| Comp 5 | −6.333 | −25.15 | 343.359 | −6.333 | 0 | −33,720.8 | −44.01 |
| Comp 6 | −7.686 | −24.90 | 303.323 | −5.124 | 0 | −33,719.8 | −38.34 |
3.3. Receptor-based high throughput virtual screening
The top hits retrieved as a result of E-pharmacophore-dependent screening combined with ADME/T profiling were processed for further filtering for HTVS (539440). Standard precision (SP) docking was performed on 53,944 molecules. Outputs were filtered out after each of these processes based on glide scores and then filtered molecules were employed for the next docking process. In the next step, extra precision (XP) docking analysis was performed for the selected molecules. From the output of SP docking, 4821 compounds were designated for XP docking, based on the glide scores. Six compounds have good XP GScore and having good interactions with the residues of binding pocket (Table 2). Thymoquinone showed low binding affinity towards RdRp and had been used as negative control in previous studies [60]. In the current study, the thymoquinone XP Gscore (−2.80 kcal/mol) was twofold lower in comparison to that of our lowest hit compound (Comp. 5: −6.333 kcal/mol, Table 2). These top hit molecules (Table 3 ) were further proceeded to the MMGBSA analysis.
Table 3.
The IUPAC names and smiles of the top six hit compounds.
| Compound no. | Name | Smiles |
|---|---|---|
| Comp-1 | (4-[({[(2S,3R)-1-Methyl-2-(1-methyl-1H-imidazol-2-yl)-6-oxo-3-piperidinyl]methyl}amino)methyl]benzamide) | NC( O)c1ccc(cc1)C[NH2+]C[C@@H](CCC2 O)[C@H](N2C)c3n(C)cc[nH+]3 |
| Comp-2 | (N2-(2-{1-[(1-Methyl-4-piperidinyl)acetyl]-3-piperidinyl}ethyl)glycinamide) | NC( O)C[NH2+]CC[C@H]1CCCN(C1)C( O)C[C@H]2CC[N@H+](C)CC2 |
| Comp-3 | (5-(3,4,5-trimethoxyphenyl)-1H-pyrazole-3-carbohydrazide) | NNC( O)c1cc([nH]n1)-c2cc(OC)c(OC)c(c2)OC |
| Comp-4 | (N-[(2,1,3-benzothiadiazol-4-yl)methyl]-5-[(morpholin-4-yl)methyl]pyridin-2-amine) | n1snc(c12)cccc2CNc(nc3)ccc3CN4CCOCC4 |
| Comp-5 | (2-Chloro-3-isopropoxy-N-[(1-oxo-1,2,3,4-tetrahydro-4-isoquinolinyl)methyl]benzamide) | CC(C)Oc(ccc1)c(Cl)c1C( O)NC[C@@H](CNC2 O)c(c23)cccc3 |
| Comp-6 | (N-[(5-Fluoro-1H-benzimidazol-4-yl)methyl]-2-(methoxymethyl)-6-methyl-4-pyrimidinamine) | COCC1 NC( CC( [NH+]1)C)NCC2 C3[NH]C NC3 CC C2F |
3.4. Binding free energy calculations
MM-GBSA method was used to estimate the Binding free energies. Compounds projecting high glide scores were subjected to MM-GBSA analysis to calculate binding energies and identify the most promising inhibitors. Six compounds, also identified as high g-scoring hits during the molecular docking, were revealed to have binding energy. The Comp 1–6 showed binding free energies of −57.498, −45.776, −46.248, −35.67, −25.15 and −24.90 kcal/mol respectively as calculated by the MM-GBSA method. The values of MMGBSA and glide scores of the top six hits are enlisted in Table 2. However, the binding free energies from MMGBSA analysis for remdesivir (PDB ID: 7BV2) as a positive control and thymoquinone as negative control, were −6.61 and −12.41 kcal/mol respectively.
3.5. Interaction profiles of hit molecules with RdRp
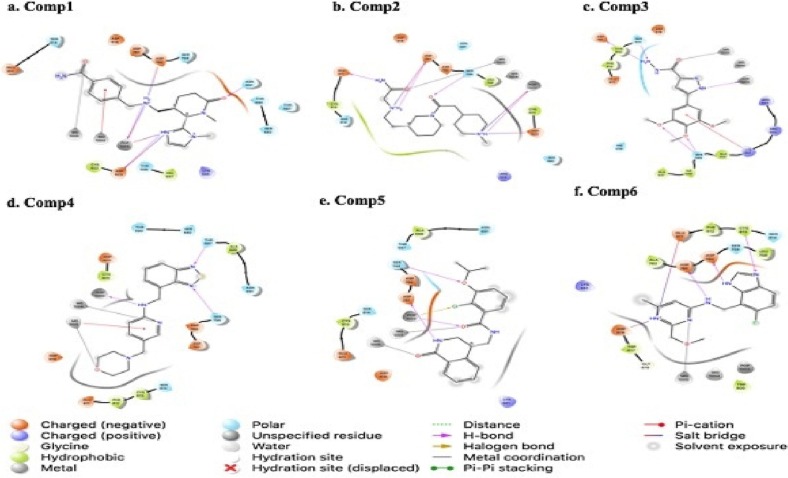
Intermolecular interaction patterns of the RdRp structure were determined using Schrödinger's ligand interaction diagram that displays hydrogen bonds, pi-pi stacking and salt bridges etc. as intermolecular interactions (Laskowski and Swindells [67]). The hydrogen bonds play a pivotal role in stabilizing the protein-ligand complex (Patil et al. [68]). For RdRp protein, the compound 1 made salt bridges with ASP 760 and ASP 623 with the distance of 4.69 and 1.86 Å respectively, hydrogen bond with ASN 691 (1.86 Å) and ASP 623(1.89 Å). Compound 1 also established hydrogen bond and salt bridge with phosphate ion and pi-cation and metal coordination with Mg+2. Compound 2 made hydrogen bonds with ASP 761, SER 759, GLU 811 with the distance of 2.61, 2.68 and 2.10 Å respectively and phosphate ion and made salt bridges with ASP 761 (2.16 Å), ASP 623 (3.87 Å) and phosphate ion. Compound 3 showed hydrogen bonding with SER 549, ASP 761, SER 814 with the distance of 1.98, 2.11, 2.02 and 2.09 Å respectively and with phosphate ion while made pi-cation interactions with LYS 551(5.54 Å) and metal coordination with magnesium ion (Fig. 2a–d). The amino acid residues that exhibited hydrogen bonding with compound 4 include SER 759 with the distance of 2.22 Å, and THR 687 with the distance of 2.20 Å. Compound 4 also made pi-cation and other interaction magnesium ion and hydrogen bond with phosphate ion as indicated in Fig. 2d. The compound 5 made 3 hydrogen bonds with SER 759 with the distance of 2.22 Å, ASP 761 with the distance of 1.96 Å and with phosphate ion, metal coordination with magnesium and halogen bond with phosphate ion (Fig. 2e). Compound 6 formed 2 salt bridge with GLU 811 with the distance of 2.98 Å and Asp 618 with the distance of 4.04 Å, metal coordination with magnesium and 3 hydrogen bonds with ASP 760, ASP 761 and CYS 813 with the distance of 1.66, 2.27 and 2.53 Å respectively. (Fig. 2f). In accordance to the previous in silico drug screens exploiting the same binding pocket, the current E-pharmacophore based approach have revealed similar interacting residues in addition to the novel interacting residues. Hence, these interactions play important role in stabilizing the ligands with in the binding pocket of receptor and resultantly block the RdRp from replicating the virus.
Fig. 2.
Intermolecular interaction in 2D between the top six hit compounds and RdRp of SARS-CoV2. Type of interactions is color coded as shown in figure.
3.6. Molecular dynamics simulations
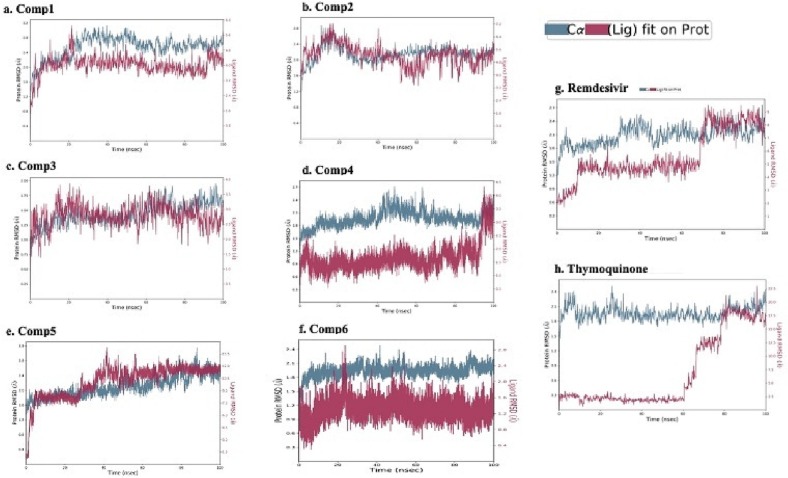
MD simulations studies were performed on protein-ligand complexes to determine the dynamic stability of the RdRp complexes with the hit compounds. High binding free energies-exhibiting top hits selected based on MM-GBSA analysis, were subjected to MD simulations. These top hits included comp 1, comp 2, comp 3, comp 4, comp 5 and comp 6 with binding free energies of −57.498, −45.776, −46.248, −35.67, −25.15, and −24.90 kcal/mol respectively. RdRp in complex with each hit compound i.e. RdRp-hits complexes were employed and the projected changes in their conformation from the initial structure over the simulation period of 100 ns were expressed as RMSD. Moreover, RMSF values were also calculated to analyse the structural stability, atomic mobility, and flexibility of residues during protein-hit interaction. The RMSF values were estimated to be less than 3 Å that confirmed the status of the stable protein. The RdRp-hits complexes showing small deviations i.e. RMSD below 3 Å owing to slight conformational changes compared to the initial structures. MD simulation studies indicated the RMSD values for Compound 1 were within range of 3 Å. It showed a small deviation of almost 0.4 Å at almost 20 ns and after that the simulation was converged and no larger deviation was observed throughout the simulation. The Compound 2 also showed stable interaction as there was a small deviation at around 18 ns and after that no large deviation was observed throughout the simulation. The Compound 3 was stable throughout the simulation as there was no large deviation observed (Fig. 3a–c). The RMSD of Compound 4 fell within 3 Å revealing the absence of large deviations and confirmed the binding stability of Compound 4 with RdRp. No significant deviation was observed in the Ligand RMSD during the first 95 ns. However, in the very end of simulation (after 95 ns) there was a jump in the ligand RMSD deviations. This deviation might be due to conformational changes after which the ligand RMSD was equilibrated (Fig. 3d). The interacting amino acid residues before and after the jump were analysed and compared for possible changes in the interactions. Most of the interacting residues were same except for LYS 551, CYS 622 THR 687, ALA 688, lost their interactions and LYS 545 made new interactions. This was probably due to single bond rotation at the amine group in compound 4. For Compound 4-RdRp complex, the protein backbone was also found to remain consistent during the MD simulation and found no significant deviation in comparison to the native structure and confirmed the binding stability of Compound 4 with RdRp. More precisely, the backbone RMSD was consistent till the end (100 ns) of simulation and deviation ranges from 1.5 to 2.4 Å. For Compound 5-RdRp and Compound 6-RdRp complexes the backbone RMSD was also consistent throughout the simulation (Fig. 3e–f). For Compound 5 the deviations were observed at 5 ns and then almost 30 to 40 ns and then it was stabilised. For Compound 6 there was no significant deviation and the interactions were consistent throughout the simulation (Fig. 3e–f). Higher deviation during the above duration might be due to conformational changes. As a positive control RMSD deviations for RdRp complex with Remdesivir (PDB ID: 7BV2) were also analysed. The RMSD showed there was a flip at almost 10 ns and then at 70 ns after that the simulation was converged and system was equilibrated. (Fig. 3g). Similarly, Thymoquinone, used as negative control, also show large deviation of 2.5 Å to 22.5 Å in RMSD during MD simulation which revealed weak binder (Fig. 3h).
Fig. 3.
Stability of ligand-protein complexes determined by molecular dynamic simulation for 100 ns. Root Mean Square Deviation plots of top six hit compounds (a–f), Remdesivir (g) and Thymoquinone (h) complexes with SARS-CoV-2RdRp.
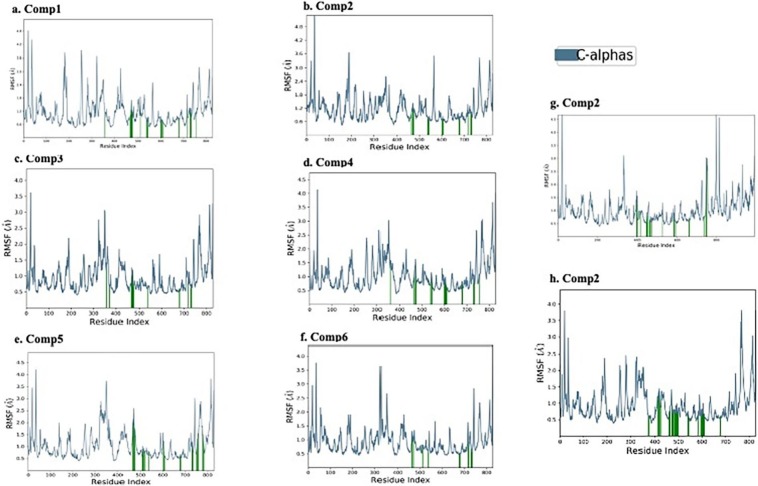
Furthermore, the catalytic region of RdRp exhibited stability for all the complexes with RMSF values. The calculated RMSF values for comp 1–6 (Fig. 4a–f), Remdesivir (Fig. 4g) and Thymoquinone (Fig. 4h), from the MD simulation trajectories are represented and no considerable fluctuations were observed. The green vertical lines in the plots represent the interacting residues of RdRp. The results indicating these compounds have strong binding affinity with the RdRp of SARS-Cov-2 for 100 ns MD simulation. Resultantly, these compounds have to ability to block the enzyme responsible for replication of virus in body.
Fig. 4.
Stability of the ligand-protein complexes determined by molecular dynamic simulation for 100 ns. Root Mean Square Fluctuation plots of top six hit compounds (a–f), Remdesivir (g) and Thymoquinone (h) complexes with SARS-CoV-2RdRp.
4. Discussion
In spite of extensive vaccination campaigns, new SARS-CoV-2 variations and reinfection cases are still being reported globally, raising major concerns about the COVID-19. In this regard, a multi-pronged strategy that includes social withdrawal, working from home, routine testing, vaccination, and immunity development was used to treat SARS-CoV-2 viral infection. The concurrent use of medications such as Remdesivir and 2-deoxy-d-glucose aids in the fight against COVID-19 [61], [62]. To improve the efficacy and accessibility of the illness treatment, new medications must be discovered. In the current study, energy-optimised pharmacophore modelling and structure-based virtual screening were used to find potential inhibitors of the RNA-dependent RNA polymerase (RdRp). Pharmacophore is a geometrical depiction of the chemical functionalities [63]. E-Pharmacophore model-based HTVS is a significant computational approach to advance and expedite the process of drug designing and discovery. This highlights the importance of in-silico analysis in the modern scientific community as it efficiently allows for the rapid screening of large libraries for promising hits without time-intensive processes. To find the most favourable pharmacophore features-containing hit molecules, more than 1 billion conformers (inhibitors/compounds) from the REAL Database-Enamine were screened. The resultant molecules were subjected to subsequent molecular docking including HTVS, SP and XP to identify the top hits. The docking study intended to assess the molecular interactions of distinct poses with the residues of RdRp active site and arrange in order of their binding scores. Following the HTVS and SP docking processes, the retrieved hits were filtered out based on the glide scores. The top selected hits with the glide scores were employed to conduct XP docking. Six compounds, viz., Comps 1–6 showed −7.986, −8.029, −7.168, −6.257, −6.333, −7.686 kcal/ mol respectively were identified as top XP GScore, thus exhibiting favourable pharmacological properties. Current E-pharmacophore based approach have revealed similar interacting residues such as 623, 759 and 760 as found in previous in silico investigations in the same binding pocket [20], [64]. However, several new interacting residues including SER 549, LYS 551, ASP 761 and GLU 811 were also revealed. Similar interactions were been previously reported, and they demonstrated good activity [65]. The six compounds will be included to the COVID-19 drug discovery process. Moreover, Kumar et al. [66] identified potential inhibitors of SARSCoV-2Mpro from available natural products databases through a combination of computational approaches, such as pharmacophore based virtual screening, molecular docking, MD simulation, and MMGBSA. Their results revealed that the top-ranked molecules SN00293542, and SN00382835 occupied the active site of the target, the main protease like that of the co-crystal ligand. These molecules may emerge as promising ligands against SARS-CoV-2 and therefore further investigation is required.
The hit molecules were subjected to the MM-GBSA analysis to calculate binding free energies. The inhibitors that had high glide score and binding energies resulted in promising candidate against SARS-CoV-2. These top hits include Comp-1 and -6 with binding free energies of −57.498, −45.776, −46.248, −35.67, −25.15, and −24.90 kcal/mol respectively, revealed as the potent inhibitors of RNA-dependent RNA polymerase (RdRp). Moreover, analysis of ADME/T properties and Lipinski's rule of five also revealed favourable pharmacokinetic properties. By using high-throughput virtual screening (HTVS) against the extensive ZINC compound database combined with extensive molecular dynamics (MD) simulations, Brunt et al. [9] discovered four non-nucleoside small molecules that bind to SARS-CoV-2 RdRp more favourably than the active form of the well-known drug remdesivir (RTP) and adenosine triphosphate (ATP).
Molecular dynamics (MD) simulation is a computational approach to studying biomolecules structural stability and molecular behaviour (Rather et al. [69]). MD simulation studies were conducted to reveal the binding status of the RdRp-hit complexes to identify the most stable binding complex that indicate its potential to strongly bind with the RdRp active site to inhibit its activity. Binding stability was assessed by determining the RMSD values of the complexes. In the current study, the MD simulation studies confirmed the stability of protein ligand complexes. Earlier, Karthic et al. [61] reported that MD simulation results revealed that RMSD variations were more pronounced in the unbound root mean square deviations (RdRp) than in the complexes. The conformational alterations in RdRp in the presence of the inhibitor ligand occurred prior to the final structural rearrangements to achieve the optimum posture, as shown by the fluctuating RMSD of the ligand-bound SARS-CoV-2 RdRp complexes until 30 ns, at which point it plateaued. The ligands that were compared, REM and FP, had larger fluctuations than Comp-1 and EMC-1. This demonstrated that the TP ligands stabilised before REM and FP. The findings of the present study deduced that these hits hold the promising prospect to be a candidate lead compound against RdRp of SARS-CoV-2.
5. Conclusion
Millions of people have died as a result of the COVID-19 pandemic brought on by the SARS-CoV-2 virus. Although the current immunization campaign offers some hope, the virus is likely to develop a resistance to the vaccinations that are now in use. Hence, there is a great need for new therapies that target viral replication. In this study, we aimed to use the remdesivir to SARS-CoV-2 RdRp complex. Using the model constructed from PDB 7BV2, as remdesivir, an antiviral drug that targets the RdRp of SARS-CoV-2, was one of the first antiviral drugs to be approved for COVID-19 clinical trials. Most of the current studies, including structural, cell-based, with animal models, or with human subjects, have concentrated their efforts on the effect of RDV on the WT-RdRp of SARS-CoV-2. In current study E-pharmacophore and structure-based virtual screening followed by MD simulations have evolved as the most efficient computational approach to expedite the drug discovery process. Computational drug discovery holds the promising potential to screen large libraries of virtual hit molecules or compounds to identify the potent inhibitor against a specific target. The present study intended and thus succeeded in identifying the effective inhibitor against SARS-CoV-2 RdRp. Comp-1 and -6 were identified as the high XP Gscoring thereby high binding free energy-containing hit exhibiting strong and stable binding affinity, validated by MD simulation, towards the RdRp active site. Moreover, a favourable pharmacokinetic profile and Lipinski properties evinced the promising potential of these compounds as an effective inhibitors of RNA-dependent RNA polymerase delineating its prospective utilization against SARS-CoV-2. However, a rational assessment regarding the benefits and possible side effects of the proposed hit compound is mandatory to ascertain its reliability and efficacy against COVID-19.
Funding
This research received no external funding.
CRediT authorship contribution statement
Hafiz Muzzammel Rehman: Original drafting, review, editing.: Muhammad Sajjad: Original drafting, Conceptualization; review, editing.: Muhammad Akhtar Ali: Original drafting, review, editing.: Roquyya Gul: Original drafting, review.: Muhammad Naveed: Drafting, editing.: Muhammad Shahbaz Aslam: Review, editing, Khyber Shinwari: Original drafting, Conceptualization, review.: Munir Ahmad Bhinder: Drafting, editing, review.: Muhammad Usman Ghani: Original drafting, editing.: Mahjabeen Saleem: Original drafting, conceptualization, editing, review.: Mohd Ashraf Rather: Drafting, editing, review.: Ishtiyaq Ahmad: Editing, review.: Adnan Amin: Editing, review.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
No data was used for the research described in the article.
References
- 1.Lu H., Stratton C.W., Tang Y.W. Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J. Med. Virol. 2020;92(4):401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agarwal D., Zafar I., Ahmad S.U., Kumar S., Sundaray J.K., Rather M.A. Structural, genomic information and computational analysis of emerging coronavirus (SARS-CoV-2) Bull.Natl.Res.Cent. 2022;46(1):1–16. doi: 10.1186/s42269-022-00861-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A., Neuman B.W., et al. 2020. Severe acute respiratory syndrome-related coronavirus: the species and its viruses–a statement of the Coronavirus Study Group. bioRxiv 2020.02. 07.937862. [Google Scholar]
- 5.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su S., Wong G., Shi W., Liu J., Lai A.C., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elmezayen A.D., Al-Obaidi A., Şahin A.T., Yelekçi K. Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J. Biomol. Struct. Dyn. 2021;39(8):2980–2992. doi: 10.1080/07391102.2020.1758791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalita P., Padhi A.K., Zhang K.Y., Tripathi T. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microb. Pathog. 2020;145 doi: 10.1016/j.micpath.2020.104236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunt D., Lakernick P.M., Wu C. Discovering new potential inhibitors to SARS-CoV-2 RNA dependent RNA polymerase (RdRp) using high throughput virtual screening and molecular dynamics simulations. Sci. Rep. 2022;12:19986. doi: 10.1038/s41598-022-24695-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campagnola G., Gong P., Peersen O.B. High-throughput screening identification of poliovirus RNA-dependent RNA polymerase inhibitors. Antivir. Res. 2011;91(3):241–251. doi: 10.1016/j.antiviral.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19(3):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 13.Zhang D., Chen J., Deng L., Mao Q., Zheng J., Wu J., Zeng C., Li Y. Evolutionary selection associated with the multi-function of overlapping genes in the hepatitis B virus. Infect. Genet. Evol. 2010;10(1):84–88. doi: 10.1016/j.meegid.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Wang L., Berger N.A., Davis P.B., Kaelber D.C., Volkow N.D., Xu R. 2022. COVID-19 rebound after Paxlovid and Molnupiravir during January-June 2022. medRxiv. 2022 Jun 22. [Google Scholar]
- 15.Wen W., Chen C., Tang J., Wang C., Zhou M., Cheng Y., Zhou X., Wu Q., Zhang X., Feng Z., Wang M. Efficacy and safety of three new oral antiviral treatment (molnupiravir, fluvoxamine and Paxlovid) for COVID-19:a meta-analysis. Ann.Med. 2022;54(1):516–523. doi: 10.1080/07853890.2022.2034936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prajapat M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H., Kumar S., Bhattacharyya A., Kumar H., Bansal S., et al. Drug targets for corona virus: a systematic review. Indian J. Pharmacol. 2020;52(1):56. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sliwoski G., Kothiwale S., Meiler J., Lowe E.W. Computational methods in drug discovery. Pharmacol. Rev. 2014;66(1):334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhardwaj V.K., Singh R., Sharma J., et al. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2021;39:3449–3458. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhardwaj V.K., Singh R., Das P., Purohit R. Evaluation of acridinedione analogs as potential SARS-CoV-2 main protease inhibitors and their comparison with repurposed anti-viral drugs. Comput. Biol. Med. 2021;128 doi: 10.1016/j.compbiomed.2020.104117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh R., Bhardwaj V.K., Das P., Purohit R. A computational approach for rational discovery of inhibitors for non-structural protein 1 of SARS-CoV-2. Comput. Biol. Med. 2021;135 doi: 10.1016/j.compbiomed.2021.104555. 104555-104555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh R., Bhardwaj V.K., Purohit R. Inhibition of nonstructural protein 15 of SARS-CoV-2 by golden spice: a computational insight. Cell Biochem. Funct. 2022;40:926–934. doi: 10.1002/cbf.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh R., Bhardwaj V.K., Sharma J., et al. In-silico evaluation of bioactive compounds from tea as potential SARS-CoV-2 nonstructural protein 16 inhibitors. J. Tradit.Complement.Med. 2022;12:35–43. doi: 10.1016/j.jtcme.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh R., Bhardwaj V.K., Purohit R. Potential of turmeric-derived compounds against RNA-dependent RNA polymerase of SARS-CoV-2: an in-silico approach. Comput. Biol. Med. 2021;139 doi: 10.1016/j.compbiomed.2021.104965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh R., Bhardwaj V.K., Sharma J., et al. Identification of potential plant bioactive as SARS-CoV-2 spike protein and human ACE2 fusion inhibitors. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arun K.G., Sharanya C.S., Abhithaj J., Francis D., Sadasivan C. Drug repurposing against SARS-CoV-2 using E-pharmacophore based virtual screening, molecular docking and molecular dynamics with main protease as the target. J. Biomol. Struct. Dyn. 2021;39(13):4647–4658. doi: 10.1080/07391102.2020.1779819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baden L.R., Rubin E.J. COVID-19-the search for effective therapy. N. Engl. J. Med. 2020;382(19):1851–1852. doi: 10.1056/NEJMe2005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., et al. A trial of lopinavir–ritonavir in adults hospitalized with severe COVID-19. N. Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arya A., Dwivedi V.D. Synergistic effect of vitamin D and remdesivir can fight COVID-19. J. Biomol. Struct. Dyn. 2020;39(11):4198–4199. doi: 10.1080/07391102.2020.1773929. [DOI] [PubMed] [Google Scholar]
- 29.Hendaus M.A. Remdesivir in the treatment of coronavirus disease 2019 (COVID-19): a simplified summary. J. Biomol. Struct. Dyn. 2020;39(10):3787–3792. doi: 10.1080/07391102.2020.1767691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muralidharan N., Sakthivel R., Velmurugan D., Gromiha M.M. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J. Biomol. Struct. Dyn. 2021;39(7):2673–2678. doi: 10.1080/07391102.2020.1752802. [DOI] [PubMed] [Google Scholar]
- 31.Mitjà O., Clotet B. Use of antiviral drugs to reduce COVID-19 transmission. Lancet Glob. Health. 2020;8(5):e639–e640. doi: 10.1016/S2214-109X(20)30114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edelstein C.L., Venkatachalam M.A., Dong Z. Autophagy inhibition by chloroquine and hydroxychloroquine could adversely affect acute kidney injury and other organ injury in critically ill patients with COVID-19. Kidney Int. 2020;98(1):234–235. doi: 10.1016/j.kint.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Oliveira O.V., Rocha G.B., Paluch A.S., Costa L.T. Repurposing approved drugs as inhibitors of SARS-CoV-2 S-protein from molecular modeling and virtual screening. J. Biomol. Struct. Dyn. 2021;39(11):3924–3933. doi: 10.1080/07391102.2020.1772885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta M.K., Vemula S., Donde R., Gouda G., Behera L., Vadde R. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J. Biomol. Struct. Dyn. 2021;39(7):2617–2627. doi: 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elfiky A.A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silicoperspective. J. Biomol. Struct. Dyn. 2021;39(9):3204–3212. doi: 10.1080/07391102.2020.1761882. Jun. Epub 2020 May 6. PMID: 32338164; PMCID: PMC7222627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elfiky A.A., Azzam E.B. Novel guanosine derivatives against MERS CoV polymerase: an in-silico perspective. J. Biomol. Struct. Dyn. 2021;39(8):2923–2931. doi: 10.1080/07391102.2020.1758789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Battisti V., Wieder O., Garon A., Seidel T., Urban E., Langer T. A computational approach to identify potential novel inhibitors against the coronavirus SARS-CoV-2. Mol. Inform. 2020;39(10) doi: 10.1002/minf.202000090. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gajjar N.D., Dhameliya T.M., Shah G.B. In search of RdRp and mpro inhibitors against SARS CoV-2: molecular docking, molecular dynamic simulations and ADMET analysis. J. Mol. Struct. 2021;1239 doi: 10.1016/j.molstruc.2021.130488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian L., Qiang T., Liang C., Ren X., Jia M., Zhang J., Li J., Wan M., YuWen X., Li H., et al. RNA-dependent RNA polymerase (RdRp) inhibitors: the current landscape and repurposing for the COVID-19 pandemic. Eur. J. Med. Chem. 2021;213 doi: 10.1016/j.ejmech.2021.113201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nallusamy S., Mannu J., Ravikumar C., Angamuthu K., Nathan B., Nachimuthu K., Ramasamy G., Muthurajan R., Subbarayalu M., Neelakandan K. Shortlisting phytochemicals exhibiting inhibitory activity against major proteins of SARS-CoV-2 through virtual screening. Res. Square. 2020 doi: 10.21203/rs.3.rs-31834/v1. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors–an in-silico docking and molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2020;39(12):4362–4374. doi: 10.1080/07391102.2020.1779818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kar P., Sharma N.R., Singh B., Sen A., Roy A. Natural compounds from Clerodendrum spp. as possible therapeutic candidates against SARS-CoV-2: an in silico investigation. J. Biomol. Struct. Dyn. 2020;39(13):4774–4785. doi: 10.1080/07391102.2020.1780947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mosquera-Yuqui F., Lopez-Guerra N., Moncayo-Palacio E.A. Targeting the 3CLpro and RdRp of SARS-CoV-2 with phytochemicals from medicinal plants of the Andean Region: molecular docking and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1835716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murugan N.A., Pandian C.J., Jeyakanthan J. Computational investigation on Andrographis paniculata phytochemicals to evaluate their potency against SARS-CoV-2 in comparison to known antiviral compounds in drug trials. J. Biomol. Struct. Dyn. 2021;39(12):4415–4426. doi: 10.1080/07391102.2020.1777901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogunyemi O.M., Gyebi G.A., Elfiky A.A., Afolabi S.O., Ogunro O.B., Adegunloye A.P., Ibrahim I.M. Alkaloids and flavonoids from African phytochemicals as potential inhibitors of SARS-Cov-2 RNA-dependent RNA polymerase: an in silico perspective. Antiviral Chem Chemother. 2020;28 doi: 10.1177/2040206620984076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin W., Mao C., Luan X., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Falco F., Di Giovanni C., Cerchia C., De Stefano D., Capuozzo A., Irace C., Iuvone T., Santamaria R., Carnuccio R., Lavecchia A. Novel non-peptide small molecules preventing IKKβ/NEMO association inhibit NF-κB activation in LPS-stimulated J774 macrophages. Biochem. Pharmacol. 2016;104:83–94. doi: 10.1016/j.bcp.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 49.Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J.Y., Wang L., Lupyan D., Dahlgren M.K., Knight J.L., et al. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2016;12(1):281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 50.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein− ligand complexes. J. Med. Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 51.Yoo J., Medina-Franco J.L. Homology modeling, docking and structure-based pharmacophore of inhibitors of DNA methyltransferase. J. Comput. Aided Mol. Des. 2011;25(6):555–567. doi: 10.1007/s10822-011-9441-1. [DOI] [PubMed] [Google Scholar]
- 52.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., Shaw D.E. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 53.Salam N.K., Nuti R., Sherman W. Novel method for generating structure-based pharmacophores using energetic analysis. J. Chem. Inf. Model. 2009;49(10):2356–2368. doi: 10.1021/ci900212v. [DOI] [PubMed] [Google Scholar]
- 54.Banerjee K., Gupta U., Gupta S., Wadhwa G., Gabrani R., Sharma S.K., Jain C.K. Molecular docking of glucosamine-6-phosphate synthase in Rhizopus oryzae. Bioinformation. 2011;7(6):285–290. doi: 10.6026/007/97320630007285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Den Driessche G., Fourches D. Adverse drug reactions triggered by the common HLA-B* 57: 01 variant: a molecular docking study. J Cheminform. 2017;9(1):13. doi: 10.1186/s13321-017-0202-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaddaguti V., Rao T.V., Rao A.P. Potential mosquito repellent compounds of Ocimum species against 3N7H and 3Q8I of Anophelesgambiae. 3 Biotech. 2016;6(1):26. doi: 10.1007/s13205-015-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cappel D., Hall M.L., Lenselink E.B., Beuming T., Qi J., Bradner J., Sherman W. Relative binding free energy calculations applied to protein homology models. J. Chem. Inf. Model. 2016;56(12):2388–2400. doi: 10.1021/acs.jcim.6b00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abdullah J.A., Aldahham B.J., Rabeea M.A., Asmary F.A., Alhajri H.M., Islam M.A. Synthesis, characterization and in-silico assessment of novel thiazolidinone derivatives for cyclin-dependent kinases-2 inhibitors. J. Mol. Struct. 2021;1223 [Google Scholar]
- 59.Onufriev A., Bashford D., Case D.A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 2004;55(2):383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 60.Elfiky A.A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in-silico perspective. J. Biomol. Struct. Dyn. 2021;39(9):3204–3212. doi: 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karthic A., Kesarwani V., Singh R.K., Yadav P.K., Chaturvedi N., Chauhan P., Yadav B.S., Kushwaha S.K. Computational analysis reveals monomethylated triazolopyrimidine as a novel inhibitor of SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) Molecules. 2022;27(3):801. doi: 10.3390/molecules27030801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verma A., Adhikary A., Woloschak G., Dwarakanath B.S., Papineni R.V. A combinatorial approach of a polypharmacological adjuvant 2-deoxy-D-glucose with low dose radiation therapy to quell the cytokine storm in COVID-19 management. Int. J. Radiat. Biol. 2020;96:1323–1328. doi: 10.1080/09553002.2020.1818865. [DOI] [PubMed] [Google Scholar]
- 63.Proekt A., Hemmings H.C., Jr. In: Pharmacology And Physiology for Anesthesia: Foundations And Clinical Application. 2nd ed. Hemmings H.C. Jr., Egan T.D., editors. Elsevier Inc.; 2019. Mechanisms of drug action; pp. 2–19. [Google Scholar]
- 64.Bhardwaj V.K., Singh R., Sharma J., et al. Bioactive molecules of tea as potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2. Front.Med. 2021;8 doi: 10.3389/fmed.2021.684020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mohammad A., Al-Mulla F., Wei D.-Q., Abubaker J. Remdesivir MD simulations suggest a more favourable binding to SARS-CoV-2 RNA dependent RNA polymerase mutant P323L than wild-type. Biomolecules. 2021;11(7):919. doi: 10.3390/biom11070919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar B.K., Faheem Sekhar, Kondapalli V.G.C., Ojha R., Prajapati V.K., Pai A., Murugesan S. Pharmacophore based virtual screening, molecular docking, molecular dynamics and MM-GBSA approach for identification of prospective SARS-CoV-2 inhibitor from natural product databases. J. Biomol. Struct. Dyn. 2020:1–24. doi: 10.1080/07391102.2020.1824814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laskowski R.A., Swindells M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 68.Patil R., Das S., Stanley A., Yadav L., Sudhakar A., Varma A.K. Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PloS one. 2010;5(8) doi: 10.1371/journal.pone.0012029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rather M.A., Dutta S., Guttula P.K., Dhandare B.C., Yusufzai S.I., Zafar M.I. Structural analysis, molecular docking and molecular dynamics simulations of G-protein-coupled receptor (kisspeptin) in fish. J. Biomol. Struct. Dyn. 2020;38(8):2422–2439. doi: 10.1080/07391102.2019.1633407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.