Abstract
Antibiotic resistance (AR) remains one of the greatest threats to global health, and Aeromonas species have the potential to spread AR in the aquatic environment. The spread of resistance to antibiotics important to human health, such as third-generation cephalosporins (3GCs) and carbapenems, is of great concern. We isolated and identified 15 cefotaxime (3GC)- and 51 carbapenem-resistant Aeromonas spp. from untreated hospital and treated municipal wastewater in January 2020. The most common species were Aeromonas caviae (58%), A. hydrophila (17%), A. media (11%), and A. veronii (11%). Almost all isolates exhibited a multidrug-resistant phenotype and harboured a diverse plasmidome, with the plasmid replicons ColE, IncU, and IncR being the most frequently detected. The most prevalent carbapenemase gene was the plasmid-associated blaKPC-2 and, for the first time, the blaVIM-2, blaOXA-48, and blaIMP-13 genes were identified in Aeromonas spp. Among the 3GC-resistant isolates, the blaGES-5 and blaMOX genes were the most prevalent. Of the 10 isolates examined, three were capable of transferring carbapenem resistance to susceptible recipient E. coli. Our results suggest that conventionally treated municipal and untreated hospital wastewater is a reservoir for 3GC- and carbapenem-resistant, potentially harmful Aeromonas spp. that can be introduced into aquatic systems and pose a threat to both the environment and public health.
Keywords: Aeromonas, hospital wastewater, municipal wastewater, ESBL, AmpC, carbapenemase, plasmid transfer
1. Introduction
Antibiotic resistance (AR) is undoubtedly one of the greatest threats to global health [1]. In addition to the issue of AR in the clinical setting, there has been increasing interest in recent years in the role that the environment, particularly wastewater, plays in the maintenance and spread of AR. Due to the multi-faceted nature of AR, a One Health approach that incorporates interactions between the human, animal, and environmental microbiome is needed to fully understand the spread of AR [1]. Continuous identification, assessment and monitoring of AR hotspots is incredibly important for controlling and preventing the spread of AR. Hospital wastewater assessment is critical because hospital sewage is a major contributor of antibiotic-resistant bacteria (ARB) and antibiotic-resistance genes (ARGs) to wastewater systems and, subsequently, to the environment [2,3]. Another well-known AR hotspot is wastewater treatment plants (WWTPs), where bacterial populations are reduced many times over, but which still discharge large amounts of ARB into water bodies on a daily basis [4]. One of the predominant genera in treated wastewater is Aeromonas [5,6], which are used as bacterial indicators of environmental AR [6,7,8].
Members of the genus Aeromonas are gram-negative bacilli that are ubiquitous in aquatic environments, including freshwater, seawater, and sewage [9,10,11]. Some Aeromonas species, particularly Aeromonas salmonicida, Aeromonas hydrophila, and Aeromonas veronii are known fish pathogens [11]. However, Aeromonas species are also considered as potential human pathogens causing various diseases such as gastroenteritis, respiratory, blood and skin infections [12]. These diseases are mainly caused by four species, Aeromonas caviae, Aeromonas dhakensis, A. hydrophila, and A. veronii [13].
The rapid increase of AR in Aeromonas spp. is of serious concern [5,6]. Of particular concern is the increasing resistance of Aeromonas species to antibiotics of critical importance to human health, such as third-generation cephalosporins (3GCs) and carbapenems [3,14,15]. The 3GCs are generally used to treat infections caused by gram-negative bacteria. However, the increasing prevalence of resistance to these antibiotics mediated by extended-spectrum β-lactamases (ESBLs) has led to the increasing use of carbapenems as last-resort antibiotics for the treatment of these infections. The most common mechanism of resistance to carbapenems in gram-negative bacteria is the production of carbapenemases, which confer resistance to virtually all β-lactams. Carbapenem resistance in Aeromonas spp. is usually mediated by the “carbapenem-hydrolyzing Aeromonas” metallo-β-lactamase (CphA), which is encoded by the cphA gene on the chromosome [16,17]. Other clinically relevant and plasmid-associated carbapenemase genes such as blaKPC-2, blaVIM-1, blaIMP-4, blaNDM-1 and blaOXA-830 have also been detected in Aeromonas spp. both in the chromosome and on plasmids [3,18,19,20,21,22,23,24,25,26]. In addition, several plasmid-associated ESBL genes have also been found in ESBL-producing Aeromonas spp. such as blaCTX-M-15, blaSHV-12, blaPER-1, blaFOX-2, blaVEB-9 and blaGES-5, [10,14,27,28,29]. Therefore, Aeromonas spp. in the environment may serve as a reservoir for genes conferring resistance to antibiotics, which are vital in human medicine. Characterization of environmental Aeromonas strains is, therefore, critical for understanding AR from a One Health perspective.
To date, few studies have investigated the characteristics of 3GC- and carbapenem-resistant Aeromonas spp. from wastewater matrices [3,14,30]. Therefore, the aim of this study was to isolate and characterize carbapenemase- and ESBL-producing Aeromonas spp. from untreated hospital and treated municipal wastewater from the Zagreb WWTP. The AR profile against a number of clinically important antibiotics and the presence of priority ARGs (ESBL, carbapenemase) and plasmid replicon types in these isolates were analyzed by phenotypic and genotypic tests. In addition, we investigated the ability of selected carbapenemase-producing isolates to transfer their carbapenem-resistant phenotype to a β-lactam-susceptible Escherichia coli, and transferable plasmids were analyzed for carbapenemase genes and plasmid replicon types.
2. Results
2.1. Identification, AR Profiles and ß-Lactamase Production in Aeromonas isolates
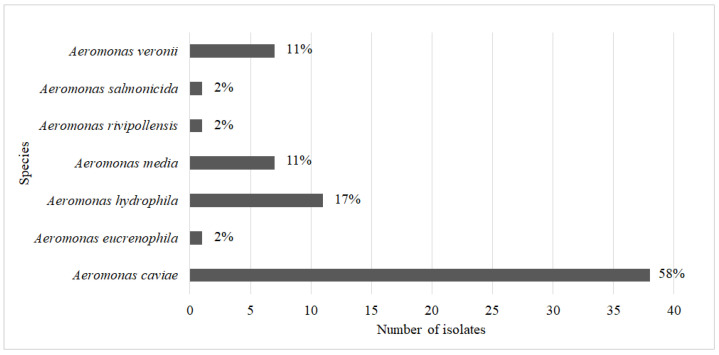
A total of 66 isolates of the genus Aeromonas were isolated from municipal wastewater (n = 59) and hospital wastewater (n = 7). Of these, fifteen isolates were selected on selective media with cefotaxime (representative of 3GCs) and fifty one isolates were selected on media with carbapenems. Fifty isolates were identified to the species level using matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS), and the other sixteen isolates, which could only be identified to the genus level using MALDI-TOF MS, were identified to the species level using their 16S rRNA gene sequences. The most prevalent species were A. caviae with 38 isolates (58%), followed by A. hydrophila with 11 isolates (17%), Aeromonas media and A. veronii with 7 isolates each (11%), and Aeromonas eucrenophila, Aeromonas rivipollensis, and A. salmonicida with only 1 isolate each (Figure 1).
Figure 1.
Number of isolates of each specific Aeromonas species.
To compare the accuracy of the MALDI-TOF MS method in identifying Aeromonas at the species level, 16S rRNA sequencing of 35 Aeromonas spp. carrying carbapenemase genes was also performed (Table S3). Thirty-one isolates had the same species identification as MALDI-TOF MS, and four isolates showed a discrepancy between MALDI-TOF MS and 16S rRNA sequencing. Three isolates of A. caviae were identified as A. hydrophila by 16S sequencing, and A. salmonicida was identified as A. media by 16S sequencing.
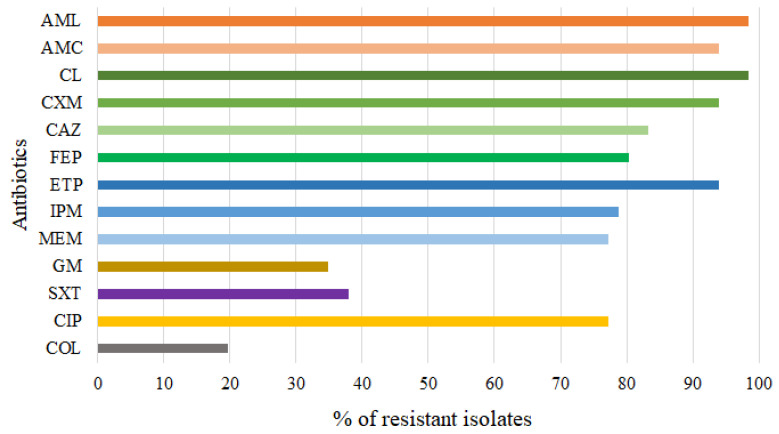
All Aeromonas isolates were further subjected to antibiotic susceptibility testing against a range of clinically relevant antibiotics. More than 93% of the isolates were resistant to penicillins (amoxicillin (AML) and amoxicillin/clavulanic acid (AMC)) and first and second generation cephalosporins (cephalexin (CL) and cefuroxime (CXM)) (Figure 2). In addition, more than 80% of the isolates were resistant to third and fourth generation cephalosporins (ceftazidime (CAZ) and cefepime (FEP)) and carbapenems (ertapenem (ETP), imipenem (IPM), and meropenem (MEM)), and more than 77% were resistant to ciprofloxacin (CIP). On the other hand, the isolates studied showed lower resistance to gentamicin (GM) (35%), sulfamethoxazole–trimethoprim (SXT) (38%), and colistin (COL) (20%) (Figure 2).
Figure 2.
Antibiotic resistance patterns of 66 Aeromonas isolates. AML = amoxicillin, AMC = amoxicillin/clavulanic acid, CL = cephalexin, CXM = cefuroxime, CAZ = ceftazidime, FEP = cefepime, ETP = ertapenem, IPM = imipenem, MEM = meropenem GM = gentamicin, SXT = trimethoprim/sulfamethoxazole, CIP = ciprofloxacin, COL = colistin.
Different levels of resistance to carbapenem antibiotics were found in different Aeromonas spp. Almost all A. caviae, A. hydrophila, A. media, and A. veroni were resistant to ertapenem (Table S1). In addition, almost all A. media and A. veroni were also resistant to imipenem and meropenem. In contrast, the prevalence of resistance of A. caviae and A. hydrophila to imipenem and meropenem averaged 75% and 82%, respectively (Table S1). Colistin resistance was detected in single isolates of A. veronii and A. eucrenophila, three isolates of A. hydrophila and eight isolates of A. caviae (Table S1). With the exception of A. eucrenophila, all of these colistin-resistant isolates were co-resistant to at least one of the three carbapenem antibiotics.
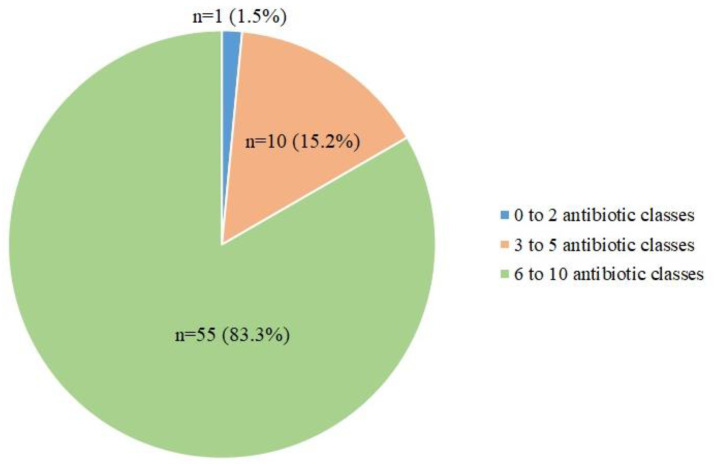
Almost all Aeromonas isolates (65/66) were multidrug-resistant (Figure 3), with 10 isolates showing co-resistance to 3–5 antibiotic classes and 55 isolates (majority A. caviae) showing co-resistance to 6–10 antibiotic classes (Figure 3).
Figure 3.
A pie chart showing the distribution of Aeromonas isolates resistant to a different number of antibiotic classes.
We also phenotypically screened 15 Aeromonas isolates resistant to cefotaxime (3GC-resistant) for the production of ESBLs and plasmid-mediated AmpC (pAmpC) using the double-disc synergy test (DDST) and the combined disc test with phenylboronic acid (pAmpC test). The ESBL test identified all 3GC-resistant isolates (n = 15) as ESBL-positive, and the pAmpC test identified 11/15 positive isolates. We also tested all isolates (n = 66) for carbapenemase production using the Carba-NP test and identified 45/66 isolates as phenotypic carbapenemase producers.
2.2. Detection of ESBL, AmpC and Carbapenemase Genes
Polymerase chain reaction (PCR) of genomic DNA for the five clinically relevant carbapenemase genes (blaKPC, blaNDM, blaIMP, blaVIM, and blaOXA-48) and Sanger sequencing of the obtained amplicons were performed for all 66 Aeromonas isolates (Table 1). The most frequently detected carbapenemase gene was the blaKPC-2 gene, which was found in 41 isolates (62%), 6 of which co-occurred with blaVIM-2 (in A. caviae), and 1 with blaIMP-13 (in A. media). Two A. caviae isolates were positive for the blaNDM-1 gene and one for the blaOXA-48 gene. Isolates identified as ESBL producers (n = 15) were screened for ESBL genes (blaCTX-M groups 1, 2, 9, blaTEM, blaSHV, blaPER, blaVEB, blaGES and blaSME) by PCR and Sanger sequencing of the obtained amplicons (Table 1). Ten isolates were shown to carry at least one of the ESBL genes targeted. The most frequently detected ESBL gene was blaGES-5, which was found in 8 of 10 isolates (in 5 A. caviae and 3 A. media). In two isolates (A. caviae and A. media), blaGES-5 co-occurred with blaTEM-1 and in one isolate with blaSHV-12 (in A. media). One A. caviae isolate carried blaGES-5+TEM-1+CTX-M-15 and one carried blaGES-5+TEM-1+OXA-48. The gene blaVEB-9 was detected in one A. hydrophila isolate (Table 1). Of the 11 strains that produced pAmpC, 6 carried blaMOX (5 A. caviae, 1 A. hydrophila), 1 of them together with blaCIT (A. caviae), 1 with blaFOX (A. caviae), and 1 with both blaCIT and blaFOX (A. caviae). Two isolates carried only blaCIT, one carried only blaFOX, and two contained blaACC. None of the mcr genes were detected in the colistin-resistant strains (n = 13).
Table 1.
Diversity, ESBL, pAmpC, and carbapenemase genes and incompatibility (Inc) groups of plasmids of resistant Aeromonas isolates and their respective transconjugants. Bold letters indicate isolates used for plasmid transfer. The asterisk * indicates isolate that successfully transferred carbapenemase resistance to the E. coli CV601 recipient. TWW = treated municipal wastewater, H = hospital wastewater.
| Species | Isolates | Transconjugants | |||||
|---|---|---|---|---|---|---|---|
| ID | Source | ARGs Detected on Genomic DNA | ARGs Detected on Plasmid DNA | Plasmid Replicons Detected |
ARGs Detected on Plasmid DNA |
Plasmid Replicons Detected |
|
| A. caviae | A36 | TWW | KPC-2 | KPC-2 | ColE, | ||
| A1 | TWW | KPC-2 | KPC-2 | ColE, R | |||
| A4 | TWW | KPC-2 | KPC-2 | ColE, R | |||
| A9 | TWW | KPC-2 | KPC-2 | ColE, R | |||
| A11 | TWW | KPC-2 | KPC-2 | ColE, R | |||
| A23 | TWW | KPC-2 | KPC-2 | ColE, R | |||
| A3 | TWW | KPC-2 | KPC-2 | ColE, R, P | |||
| A20 | TWV | KPC-2 | KPC-2 | ColE, R | |||
| A31 | TWW | KPC-2 | KPC-2 | ColE, R, HI1 | |||
| A6 | TWW | KPC-2 | KPC-2 | ColE, U, HI1, L/M, FrepB, K | |||
| A19 | TWW | KPC-2 | KPC-2 | ColE, U | |||
| A30 | TWW | KPC-2 | KPC-2 | ColE | |||
| A2 * | TWW | KPC-2 | KPC-2 | ColE, N, Y | KPC-2 | ColE, N | |
| A15 | TWW | KPC-2 | KPC-2 | ColE, K | |||
| A32 | TWW | KPC-2 | KPC-2 | ColE, K | |||
| A38 | TWW | KPC-2 | KPC-2 | ColE, K | |||
| A8 | TWW | KPC-2 | KPC-2 | K | |||
| A18 | H | KPC-2 | KPC-2 | - | |||
| A21 | TWW | KPC-2 | KPC-2 | P | |||
| A12 | TWW | KPC-2, VIM-2 | KPC-2, VIM-2 | - | |||
| A35 | TWW | KPC-2, VIM-2 | KPC-2, VIM-2 | ColE, R, HI1 | |||
| A37 | TWW | KPC-2, VIM-2 | KPC-2, VIM-2 | ColE, HI1, N | |||
| A29 | TWW | KPC-2, VIM-2 | KPC-2, VIM-2 | ColE, | |||
| A28 | TWW | KPC-2, VIM-2 | KPC-2, VIM-2 | ColE, U | |||
| A10 | TWW | KPC-2, VIM-2 | KPC-2, VIM-2 | U | |||
| A24 * | TWW | NDM-1 | NDM-1 | ColE | - | ColE | |
| A14 | TWW | CTX-M-15, TEM-1, GES-5, ACC | CTX-M-15, TEM-1 | W, P | |||
| A22 | TWW | TEM-1, GES-5, FOX, CIT, MOX | TEM-1, FOX, CIT, MOX | ColE, I1-1y, FIA, W, Y, FrepB | |||
| A26 | TWW | TEM-1, MOX | TEM-1, MOX | ColE, W | |||
| A34 | TWW | OXA-48; TEM-1, GES-5, MOX | TEM-1, MOX | ColE, Y, FrepB | |||
| A5 | TWW | KPC-2 | - | - | |||
| A13 | TWW | VIM-2, ACC | - | - | |||
| A17 | TWW | GES-5, CIT, MOX | CIT, MOX | - | |||
| A33 | TWW | GES-5, MOX, FOX | MOX | - | |||
| A27 | TWW | NDM-1, FOX | - | - | |||
| A7 | TWW | - | - | - | |||
| A16 | TWW | - | - | - | |||
| A25 | TWW | - | - | - | |||
| A. hydrophila | A39 | TWW | KPC-2 | KPC-2 | ColE, U, K, FrepB | ||
| A40 | TWW | KPC-2 | KPC-2 | ColE, U, K, FrepB | |||
| A42 | TWW | KPC-2 | KPC-2 | ColE, R | |||
| A45 | TWW | KPC-2 | KPC-2 | ColE, R | |||
| A48 | TWW | KPC-2 | KPC-2 | ColE, U | |||
| A49 | TWW | KPC-2 | KPC-2 | ColE | |||
| A41 | TWW | KPC-2 | KPC-2 | - | |||
| A46 | TWW | VEB-9 | - | - | |||
| A47 | TWW | MOX | - | - | |||
| A44 | TWW, | - | - | - | |||
| A43 | H | - | - | - | |||
| A. media | A51 | TWW | KPC-2 | KPC-2 | ColE, | ||
| A50 | TWW | KPC-2 | KPC-2 | ColE, R, U | |||
| A56 | TWW | KPC-2 | KPC-2 | ColE, R, U | |||
| A55 | TWW | KPC-2, IMP-13 | KPC-2 | ColE, R, U | |||
| A54 | TWW | KPC-2, GES-5 | KPC-2, GES-5 | - | |||
| A52 | TWW | TEM-1, GES-5 | TEM-1, GES-5 | ColE, | |||
| A53 | TWW | SHV-12, GES-5 | SHV-12 | ColE, FIC | |||
| A. veronii | A63 | TWW | KPC-2 | KPC-2 | ColE, U, K, FrepB | ||
| A60 | H | KPC-2 | KPC-2 | R, U, L/M, FrepB | |||
| A62 | H | - | - | - | |||
| A57 | H | - | - | - | |||
| A58 | H | - | - | - | |||
| A59 | H | - | - | - | |||
| A61 | TWW | - | - | - | |||
| A. salmonicida | A64 * | TWW | KPC-2 | KPC-2 | ColE, K | KPC-2 | ColE |
| A. rivipollensis | A65 | TWW | CIT | - | - | ||
| A. eucrenophila | A66 | TWW | - | - | - | ||
To determine whether the ESBL and carbapenemase genes were placed on plasmids, plasmid DNA was subjected to target PCR for the corresponding genes. Out of 15 ESBL-producing isolates, 2 A. media isolates carried blaGES-5 and 1 carried blaSHV-12 on plasmids. The blaCTX-M-15 gene was found on plasmid DNA from one A. caviae. All 5 isolates carrying blaTEM-1 were confirmed to carry it on plasmids. Regarding the carbapenemase genes, 40/41 isolates carried blaKPC-2 on plasmids. The gene blaVIM-2 was found on plasmids of 6/7 A. caviae isolates, and 1 A. caviae carried blaNDM-1 on the plasmid. The blaOXA-48 and blaIMP-13 genes were not detected on the plasmids.
Isolates that had ESBL and/or carbapenemase genes on a plasmid (n = 47) were analyzed for the presence of plasmid replicon types by PCR-based replicon typing (PBRT) (Table 1). No plasmid replicons were detected in 4 Aeromonas isolates that were positive for carbapenemase genes by regular PCR (Table 1). In the remaining 43 isolates, 17 different plasmid replicon types were detected, with 10 isolates having one plasmid type and 33 isolates having more than one plasmid. The most common plasmid replicon type detected in Aeromonas isolates was ColE (n = 38). The combination of two plasmid replicon types (ColE + IncR, ColE + IncK, ColE + IncU, ColE + IncW, ColE + IncFIC, and IncW + IncP) was detected in 18 isolates, while the combination of three (ColE + IncR + IncHI1, ColE + IncR + IncP, ColE + IncN + IncY, ColE + IncHI1 + IncN, ColE + IncY + FrepB, and ColE + IncR + IncU) and four replicon types (ColE + IncU + IncK + IncFrepB, and IncR + IncU + IncL/M + IncFrepB) were detected in 9 and 4 isolates, respectively. Two A. caviae isolates had six plasmids (ColE + IncU + IncHI1 + IncL/M + IncFrepB + IncK, and ColE + IncI1-1y + IncFIA + IncW + IncY + IncFrepB) (Table 1).
2.3. Enterobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction (ERIC-PCR) Fingerprinting
Genetic similarity of all 66 Aeromonas spp. was assessed by ERIC-PCR analysis (Figure S1A–D). The fingerprints of the isolates consisted of 1 to 13 amplification bands ranging in size from 100 bp to 10,000 bp. Overall, the examined isolates exhibited great intraspecies diversity as they were assigned to 50 different ERIC-PCR profiles (27/38 for A. caviae, 9/11 for A. hydrophila, 7/7 for A. media, and 7/7 for A. veronii).
2.4. Conjugal Transfer of Carbapenem Resistance and Genotypic Characterization of Captured Plasmids
Conjugation experiments were performed to determine the possible transfer of carbapenem resistance from Aeromonas spp. to other bacteria. These experiments were performed using 10 randomly selected carbapenem-resistant Aeromonas isolates carrying carbapenemase genes on a plasmid (4 A. caviae, 2 A. hydrophila, 1 A. media, 2 A. veronii, and 1 A. salmonicida) as donors and the susceptible E. coli CV601 as the recipient. On LB plates containing meropenem (0.12 mg/L), we selected transconjugants that matched the phenotype of the donors. Transfer of the meropenem resistance phenotype was successful in three of ten isolates tested, namely, one KPC-2-carrying A. caviae, one NDM-1-carrying A. caviae, and one KPC-2-carrying A. salmonicida (Table 1). The transfer frequency of meropenem-resistant transconjugants was 6.41 × 10−8 and 7.52 × 10−5 per recipient in KPC-2-carrying A. caviae and A. salmonicida, respectively, and 8.59 × 10−7 per recipient in NDM-1-carrying A. caviae.
Plasmid DNA was extracted from all three transconjugants and analysed by PCR for the presence of carbapenemase genes and plasmid replicon types (Table 1). The carbapenemase gene blaKPC-2 was detected in transconjugants from blaKPC-2-PCR positive A. caviae and A. salmonicida, whereas blaNDM-1 was not detected in transconjugant from blaNDM-1-PCR positive A. caviae. In transconjugants from blaNDM-1-PCR positive A. caviae and blaKPC-2-PCR positive A. salmonicida, a plasmid was detected that was assigned to the ColE replicon type. In contrast, a combination of the replicon types of ColE and IncN plasmids was detected in transconjugant from blaKPC-2-PCR positive A. caviae (Table 1).
3. Discussion
The present study aimed to characterize Aeromonas spp. from treated municipal and untreated hospital wastewater that exhibited resistance to the antibiotics 3GCs and carbapenems, which are important for human health, using culture-dependent and culture-independent approaches. Aeromonas spp. are ubiquitous bacteria found in various aquatic habitats worldwide [9,11]. Considering this and the fact that they can easily acquire and exchange ARGs [30], research needs to focus more on the impact of these potentially harmful bacteria on human health.
In this study, 66 resistant Aeromonas strains were successfully isolated and identified to the species level by MALDI-TOF MS and/or 16S rRNA gene sequencing. These isolates could be assigned to 7 different species, with A. caviae (58%) dominating, followed by A. hydrophila, (17%), A. media (11%) and A. veronii (11%), while the prevalence of A. eucrenophila, A. rivipollensis and A. salmonicida was 2%. Of note, MALDI-TOF MS showed very good correlation with the results of 16S rRNA sequencing, with the exception of three cases of A. caviae identified as A. hydrophila and one case of A. salmonicida identified as A. media by 16S rRNA sequencing. In a previous study [31], MALDI-TOF MS was shown to be a more accurate method than 16S sequencing for distinguishing Aeromonas spp., particularly A. caviae and A. hydrophila and, therefore, we relied on the results from MALDI-TOF MS as the correct identification.
In other studies on aeromonads from wastewater environments, A. caviae was identified as the most abundant species in Brazil [32], while A. veronii was the predominant species in wastewater in Portugal [5]. Further assessment of genetic relatedness within these species by ERIC-PCR revealed high intraspecies diversity, as 7 species were assigned to 50 different ERIC-PCR profiles. This is in agreement with other studies that also showed high heterogeneity in ERIC sequences of Aeromonas strains [33,34,35,36].
Although Aeromonas spp. are environmental bacteria found primarily in aquatic environments, some members of this genus are pathogenic [13]. These include A. caviae, A. dhakensis, A. hydrophila, and A. veronii, which have been associated with gastrointestinal and respiratory tract infections, soft tissue and skin infections, etc. [11,12,13]. These species have also been isolated from untreated and treated wastewater [5,32]. Therefore, the predominant isolation of these species from hospital and municipal wastewater in this study suggests that these sources serve as reservoirs for these pathogens and as potential pathways for their transmission to humans and animals.
Characterizing the AR profile of Aeromonas isolates from wastewater is critical to understanding AR from the perspective of the One Health approach. In our study, all but one isolate was multidrug-resistant (i.e., resistant to ≥3 antibiotic classes). As expected, of all antibiotics tested, resistance to cephalosporins (first–fourth generation) and carbapenems (80%) was most common, followed by fluoroquinolones (CIP) (77%). Interestingly, 18% of our Aeromonas isolates (mainly A. caviae and A. hydrophila) were resistant to both carbapenems and colistin, limiting treatment options for Aeromonas infections, as carbapenems and colistin play an important role as “last resort” in the treatment of infections caused by gram-negative MDR bacteria. Therefore, the discharge of untreated hospital wastewater and treated municipal wastewater could increase the prevalence of these opportunistic pathogens with clinically relevant AR phenotypes in environmental waters and pose a threat to human health.
The positive AmpC β-lactamase result in some Aeromonas isolates was not unexpected as Aeromonas spp. are known to harbor chromosomal AmpC β-lactamase genes [11]. Among these genes, blaMOX was the most prevalent. Previously, blaMOX had been described in A. caviae [27] and in Aeromonas sp. from treated wastewater [14]. In addition, Aeromonas sanarellii, A. caviae, and A. media have been identified as carriers of three different variants of blaMOX [37]. Many previous studies have shown that Aeromonas isolates from the environment were susceptible to the 3GCs [5,38,39]. However, in this study, 15 Aeromonas isolates were isolated on selective agar with cefotaxime (3GC), all of which were resistant to ceftazidime (3GC) and produced ESBLs. Four different ESBL genes (blaGES-5, blaSHV-12, blaCTX-M-15, and blaVEB-9) and one narrow spectrum β-lactamase gene (blaTEM-1) were found in these isolates, with blaGES-5 being the predominant gene and found mainly in A. caviae and A. media isolates. This is consistent with a previous study in which the blaGES-5 gene was also found in A. caviae isolated from hospital wastewater in Brazil [29]. Although this gene is mainly of chromosomal origin in A. caviae and A. media isolates, it was also detected in an A. media isolate on the plasmid replicon ColE which, to our knowledge, had not previously been associated with the carriage of the blaGES-5 gene. The other ESBL genes detected have also been found in environmental Aeromonas spp. in previous studies [10,28,40,41].
The majority of A. caviae (37/38), A. hydrophila (7/8), and A. media (6/7) that were resistant to at least one of the carbapenem antibiotics tested produced carbapenemases and mainly possessed the blaKPC-2 gene. Previous studies have demonstrated the presence of blaKPC-2 in Aeromonas strains from natural waters in Brazil [42,43] and from wastewater in Brazil [3], the United States [30], China [20], and Japan [18]. Besides the present study, only one previous study detected blaKPC-2 in Aeromonas sp. from activated sludge in Europe (Poland) [14]. In addition, in the present study, blaVIM-2, blaNDM-1, and blaOXA-48 were detected in A. caviae, while blaIMP-13 was found in A. media. To our knowledge, the blaVIM-2, blaOXA-48, and blaIMP-13 genes have never been detected in Aeromonas strains, and the blaNDM-1 gene was only recently discovered in two separate clinical cases of A. caviae in China [24,25]. Some of these genes (e.g., blaOXA-48, blaIMP-13) in the Aeromonas isolates studied in this work were mainly of chromosomal origin. However, most of our Aeromonas isolates carried carbapenemase genes on plasmids, especially blaKPC-2, blaVIM-2, and blaNDM-1, suggesting that they were acquired by HGT, probably under the selection pressure of antibiotics in hospital and municipal wastewater [2]. Interestingly, in six A. caviae isolates blaKPC-2 co-occurred with blaVIM-2 on a plasmid DNA, while in one A. media blaKPC-2 co-occurred with blaIMP-13 but was not present on plasmids. To our knowledge, this is the first report of the co-occurrence of both gene combinations (blaKPC-2 + blaVIM-2 and blaKPC-2 + blaIMP-13) in Aeromonas strains.
Plasmid-mediated multidrug resistance plays an important role in the worldwide spread of ARGs [44]. This study showed that Aeromonas isolates harbored various plasmid replicons, with ColE (n = 38), IncR (n = 15) and IncU (n = 11) being the most frequently detected. Our carbapenem-resistant Aeromonas isolates carrying these plasmids were also positive for blaKPC-2, blaVIM-2, or blaNDM-1 genes, suggesting possible mobilization of these genes by plasmid-mediated transfer. The plasmid replicons ColE and IncR have been associated with the carriage of blaKPC-2 in Enterobacterales isolates [45,46,47] while IncU (also known as IncP-6 [48]) has been associated with the transfer of blaKPC-2 from Aeromonas to E. coli [18]. In addition to blaKPC-2, other carbapenamase genes such as blaNDM-1 and blaOXA-48 [49,50] and ESBL genes such as blaCTX-M-15 [51] have also been found on the ColE plasmid in enterobacteria. To our knowledge, this is one of the first reports of Aeromonas isolates carrying blaKPC-2, blaVIM-2 or blaNDM-1 genes associated with conjugative ColE plasmids.
In the present study, transferable carbapenem (meropenem)-resistant plasmids were also captured from two A. caviae and one A. salmonicida strains to carbapenem-susceptible E. coli CV601. One ColE plasmid was captured from A. caviae (blaNDM-1 positive) and A. salmonicida (blaKPC-2 positive), and another one was captured from A. caviae (blaKPC-2 positive) in combination with replicon type IncN. The presence of blaKPC-2 in the ColE plasmid in transconjugant from A. salmonicida suggests that these plasmids may play an important role in the spread of Aeromonas carrying plasmid-localized blaKPC-2 genes. Moreover, the observation made in this study that two plasmids (ColE and IncN) were co-transferred from A. caviae (blaKPC-2 positive) to E. coli is also interesting in light of the findings of Barry et al. (2019) [52]. They demonstrated co-transfer of small cryptic plasmids such as Col440I along with larger plasmids carrying blaKPC-2 from blaKPC-positive Citrobacter freundii to E. coli. They found that the larger blaKPC-plasmids were never transferred alone and, therefore, hypothesized that these small plasmids are often transferred with conjugative plasmids carrying ARGs because they might play a helper role. The same could be true for our small (ColE) and larger (IncN) plasmids detected in blaKPC-positive transconjugant. The conjugative plasmid replicon IncN has already been associated with the carriage of blaKPC-2 in enterobacteria [53]. However, we cannot exclude the possibility that blaKPC-2 is present on both plasmids or only on ColE. In addition, blaNDM-1 was not detected in the transconjugant from NDM-1-positive A. caviae, suggesting that it may be localized on other mobile genetic element that is integrated into the host chromosome.
Aeromonas spp. are considered susceptible to colistin, with the exception of Aeromonas jandaei and A. hydrophila [54,55]. In our study, 3/11 A. hydrophila were resistant to colistin, one A. veronii and one A. eucrenophila, and most A. caviae (8/38). Although mcr genes (mcr-1, mcr-3, and mcr-5) have been found previously in Aeromonas spp. [56,57,58], our A. caviae, A. eucrenophila and A. hydrophila isolates were negative for all mcr genes tested (mcr1–5). Nevertheless, it is possible that these isolates have a novel variant of the mcr gene or another resistance mechanism, such as the addition of L-Ara-4N to the lipopolysaccharide layer [59]. Therefore, whole genome sequencing of these isolates would be required to fully elucidate the mechanism of colistin resistance.
4. Materials and Methods
4.1. Wastewater Sampling
Treated municipal wastewater (secondary effluent; 24-h composite samples) and untreated hospital wastewater (grab samples) from two hospitals (H1 and H2) in Zagreb, Croatia were collected on three consecutive days in winter 2020 (January). Municipal wastewater underwent stepwise treatment (primary and secondary-aerobic biodegradation) prior to discharge into the Sava River. Hospital wastewater was not treated in the hospital prior to discharge into the municipal wastewater system, as is common practice in all hospitals in Croatia. Samples were collected in sterile glass bottles (2.5 L), transported in cool boxes with ice blocks, and processed in the laboratory within 2 h.
4.2. Isolation of 3GC- and Carbapenem-Resistant Aeromonas
A series of dilutions of municipal and hospital wastewater samples were prepared in 0.85% NaCl (tenfold dilution up to 1:10,000). The dilutions were filtered in triplicate under vacuum through sterile mixed cellulose ester membrane filters (47 mm diameter, 0.22 µm pore size, Whatman, GE Healthcare, Life Science, Chicago, IL, USA) and the filters were placed on Rapid’ E. coli 2 (BioRad, Hercules, CA, USA) agar plates supplemented with 4 mg/L cefotaxime (CTX) and CHROMagar mSuperCARBA (CHROMagar, Paris, France) agar plates. The plates were incubated at 37 °C for 24 h. Different colonies of Rapid’ E. coli 2 + CTX and CHROMagar mSuperCARBA were selected for isolation of presumptive 3GC- or carbapenem-resistant Aeromonas spp. A total of 200 presumptive isolates were purified on the same medium and stored in a 20% glycerol stock at −80 °C.
4.3. Identification of Isolates
Isolates were identified to species level by MALDI-TOF MS. Isolates were streaked on Mueller–Hinton plates (Oxoid, Basingstoke, UK), incubated overnight for 18–24 h at 37 °C, and sent to the Laboratory of Mass Spectrometry and Functional Proteomics at the Ruđer Bošković Institute. Pure cultures were transferred directly to the spots of the MALDI-TOF MS target using a toothpick. A score value greater than 2.00 for the species level and a score value between 1.70 and 1.99 was indicated for successful identification. Isolates that could not be identified to species level by MALDI-TOF MS and those that carried carbapenemase gene(s) were additionally analyzed by sequencing of the 16S rRNA gene. The 1465 pb fragment was amplified with the universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) [60]. Thermocycling conditions were as follows: initial denaturation at 98 °C for 5 min, followed by 35 amplification cycles of denaturation for 10 s at 98 °C, annealing for 30 s at 60 °C and extension for 1:30 min at 72 °C, followed by a final extension at 72 °C for 5 min. PCR products were sent to Macrogen Europe (Amsterdam, The Netherlands) for purification and Sanger sequencing. Partial nucleotide sequences of the 16S rRNA genes were compared with the homologous sequences of the different Aeromonas species available in the GenBank database using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 20 February 2023). All sequences were identified to species level (≥99% sequence identity).
4.4. Antibiotic Susceptibility Testing and Phenotypic Identification of ESBLs, pAmpC and Carbapenemases
Antibiotic susceptibility testing was performed by the disk diffusion method using commercially available disks according to EUCAST guidelines [61]. Antibiotics tested included AML (25 µg), AMC (30 µg, BD BBL), CL (30 µg, OXOID), CXM (30 µg, BD BBL), CAZ (10 µg, OXOID), FEP (30 µg, BD BBL), ETP (10 µg, BD BBL), IPM (10 µg, BD BBL), MEM (10 µg, BD BBL), GM (10 µg, BD BBL), SXT (1.25/23.75 µg, BD BBL), and CIP (5 µg, BD BBL). Antibiotic disks AML, CL, and CAZ were purchased from OXOID (Basingstoke, UK) and the others from BD BBL (Franklin Lakes, NJ, USA). In addition, for isolates resistant to any of the carbapenems, the minimum inhibitory concentration (MIC) was determined by broth microdilution according to EUCAST guidelines. In addition, the susceptibility to COL was tested for all isolates using the MIC. Briefly, the initial concentration of 64 mg/L (COL, IPM, and MEM) or 16 mg/L (ETP) was serially double diluted in a sterile 96-well plate to a final concentration of 1 mg/L (COL, IPM, MEM) or 0.25 mg/L (ETP). Wells contained 90 μL Mueller–Hinton broth (Merck, Darmstadt, Germany) or, in the case of colistin, cation-adjusted Mueller–Hinton broth 2 (Sigma-Aldrich, Steinheim am Albuch, Germany) and serially diluted antibiotics. Each well was inoculated with 10 μL of an overnight bacterial culture diluted to a concentration of 5 × 105 CFU/mL. The plates were incubated overnight at 37 °C, and the lowest concentration at which no visible growth occurred was determined as the MIC of the isolates. Escherichia coli ATCC 25,922 and Escherichia coli NCTC 13,846 were used for quality control.
For phenotypic determination of ESBL and pAmpC production, 3GC-resistant isolates were subjected to DDST and pAmpC tests, respectively. For ESBL production, overnight cultures were diluted in 0.85% NaCl to a concentration of 0.5 McFarland and plated onto Mueller–Hinton agar using a sterile cotton swab. The CAZ (30 µg) and CTX (30 µg) discs were placed 20 mm and 30 mm (centre to centre) away from the amoxicillin-clavulanate (AMC, 20 + 10 µg) disc, respectively. If, after overnight incubation at 37 °C, synergy with clavulanate occurred with one of the 3GCs (enlargement of the zone of inhibition), this was considered a positive result for ESBL production (EUCAST guidelines).
The pAmpC production was determined using a cefoxitin disk (30 µg) alone and in combination with phenylboronic acid (300 µg) [62] applied to the inoculated Muller–Hinton plates. If there was an increase in the zone of inhibition of ≥5 mm after overnight incubation at 37 °C, the isolate was classified as a pAmpC producer.
All isolates that were resistant to carbapenems (both 3GC- and carbapenem-resistant) were subjected to an in-house CarbaNP assay to determine carbapenemase production [63]. Briefly, bacterial suspensions in Tris-HCL lysis buffer were mixed with 100 µL phenol red solution containing ZnSO4 × 7H2O (0.1 mM) and imipenem-cilastatin (12 mg/mL). After incubation at 37 °C for a maximum of 2 h, the bacterial strains that changed the color of the suspension from red to orange or yellow were classified as carbapenemase producers.
4.5. Genomic and Plasmid DNA Extraction
Genomic DNA was extracted from overnight cultures of all Aeromonas isolates and transconjugants using the Quick-DNATM Miniprep Plus Kit (Zymo, Irvine, CA, USA) according to the manufacturer’s instructions. Plasmid DNA was extracted from overnight cultures of ESBL-, pAmpC-, and carbapenemase-producing Aeromonas isolates and three carbapenem-resistant transconjugants using the ExtractNow Plasmid Mini Kit (Minerva Biolabs, Berlin, Germany) according to the manufacturer’s protocol. The extracted genomic and plasmid DNA was stored at −20 °C until further analysis.
4.6. Detection of Target ARGs by PCR and Sanger Sequencing of the Amplicons
Target ARGs in genomic and plasmid DNA extracted from Aeromonas isolates and transconjugants were detected by regular PCR with specific primers and conditions (Table 2), and the gene variant was identified by Sanger sequencing of the amplicons obtained. 3GC-resistant isolates (n = 15) were screened for ESBL genes by singleplex PCR (blaCTX-M groups 1, 2, and 9) and multiplex PCR (blaTEM, blaSHV, blaPER, blaVEB, blaGES, and blaSME) and for pAmpC genes (blaFOX, blaEBC, blaCIT, blaACC, blaMOX) by multiplex PCR (Table 2). All Aeromonas isolates (n = 66) and three transconjugants were screened for the presence of carbapenemase genes (blaKPC, blaNDM, blaOXA-48-like, blaIMP, and blaVIM). The colistin-resistant strains were screened for the mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5 genes (Table 2). All PCR products were separated by 1.5% gel electrophoresis at 100 V for 60 min, stained with ethidium bromide, and visualized in the UV transilluminator. All positive PCR products were sent to Macrogen (Amsterdam, The Netherlands) for purification and sequencing in forward direction. The resulting sequences were compared with the reference in the NCBI database using BLASTX search.
4.7. Plasmid Replicon Typing
PBRT was used to identify the replicon type of plasmids in Aeromonas isolates and transconjugants. This was achieved by PCR amplification with plasmid DNA of the strains and transconjugants using the specific primer sets for 22 replicons and conditions listed in Table S2 [64,65,66]. Reactions were performed using the Hot Start Core Kit Ab+ (Jena Bioscience, Jena, Germany). PCR products were separated by electrophoresis (100 V, 60 min) on a 1.5% agarose gel and stained with ethidium bromide.
Table 2.
Specific primers and conditions for polymerase chain reaction (PCR) of resistance genes.
| Target Gene | Primer Name | Primer Nucleotide Sequence (5′–3′) |
Amplicon Size (bp) | PCR Conditions | Reference | |
|---|---|---|---|---|---|---|
| Carbapenemases | bla KPC | KPC-F | AGTTCTGCTGTCTTGTCT | 793 | Initial denaturation step at 94 °C for 2:30 min; 30 cycles of 94 °C for 20 s, annealing temperature: 57 °C for blaKPC, blaVIM, blaOXA-48, 55 °C for blaIMP, 58 °C for blaNDM for 25 s; 72 °C for 45 s; final extension 72 °C for 2 min |
[67] |
| KPC-R | CTTGTCATCCTTGTTAGGC | |||||
| bla VIM | VIM MJ-F | GGTGAGTATCCGACAGTC | 442 | |||
| VIM-MJ-R | CAGCACCRGGATAGAAGAG | |||||
| bla IMP | IMP MJ-F1 | GGYGTTTATGTTCATACWTC | 235 | |||
| IMP MJ-R1 | GGATYGAGAATTAAGCCACTC | |||||
| bla OXA-48 | OXA-48A | TTGGTGGCATCGATTATCGG | 744 | [68] | ||
| OXA-48B | GAGCACTTCTTTTGTGATGGC | |||||
| bla NDM | NDM-F | TGGCAGCACACTTCCTATC | 813 | [69] | ||
| NDM-R | AGATTGCCGAGCGACTTG | |||||
| bla CTX-M | bla CTX-M-1 | M13U | GGTTAAAAAATCACTGCGTC | 864 | Initial denaturation step at 94 °C for 2:30 min; 30 cycles of 94 °C for 20 s, annealing temperature: 55 °C for 25 s; 72 °C for 45 s; final extension 72 °C for 2 min |
[70] |
| M13L | TTGGTGACGATTTTAGCCGC | |||||
| bla CTX-M-2 | M25U | ATGATGACTCAGAGCATTCG | 866 | |||
| M25L | TGGGTTACGATTTTCGCCGC | |||||
| bla CTX-M-9 | M9U | ATGGTGACAAAGAGAGTGCA | 870 | |||
| M9L | CCCTTCGGCGATGATTCTC | |||||
| ESBL-multiplex 1 | bla TEM | TEM-F | GCGGTAAGATCCTTGAGAGT | 620 | Initial denaturation step at 94 °C for 2:30 min; 35 cycles of 94 °C for 30 s, annealing temperature: 55 °C for 30 s; 72 °C for 45 s; final extension 72 °C for 2 min |
[67] |
| TEM-R | TACGATACGGGAGGGCTTA | |||||
| bla SHV | SHV-F | TTCGCCTGTGTATTATCTCC | 494 | |||
| SHV-R | CGCCTCATTCAGTTCCG | |||||
| bla PER | PER-F | CTGGGCTCCGATAATGA | 349 | |||
| PER-R | CTGGTCGCCWATGATGA | |||||
| ESBL-multiplex 2 | bla VEB | VEB-F | ATGCCAGAATAGGAGTAGC | 673 | Initial denaturation step at 94 °C for 2:30 min; 35 cycles of 94 °C for 30 s, annealing temperature: 58 °C for 30 s; 72 °C for 45 s; final extension 72 °C for 2 min |
|
| VEB-R | AATTGTCCATTCGGTAAAGTAAT | |||||
| bla GES | GES-F | CTAGCATCGGGACACAT | 504 | |||
| GES-R | GACAGAGGCAACTAATTCG | |||||
| bla SME | SME-F | GCTCAGGTATGACATTAGGT | 350 | |||
| SME-R | CCAATCAGCAGGAACACTA | |||||
| AmpC- multiplex | bla FOX | FOX-F | AACATGGGGTATCAGGGAGATG | 190 | Initial denaturation step at 94 °C for 2:30 min; 35 cycles of 94 °C for 30 s, annealing temperature: 55 °C for 30 s; 72 °C for 45 s; final extension 72 °C for 2 min |
[71] |
| FOX-R | CAAAGCGCGTAACCGGATTGG | |||||
| bla EBC | EBC-F | TCGGTAAAGCCGATGTTGCGG | 302 | |||
| EBC-R | CTTCCACTGCGGCTGCCAGTT | |||||
| bla CIT | CIT-F | TGGCCAGAACTGACAGGCAAA | 462 | |||
| CIT-R | TTTCTCCTGAACGTGGCTGGC | |||||
| bla ACC | ACC-F | AACAGCCTCAGCAGCCGGTTA | 346 | |||
| ACC-R | TTCGCCGCAATCATCCCTAGC | |||||
| bla DHA | DHA-F | AACTTTCACAGGTGTGCTGGGT | 405 | |||
| DHA-R | CCGTACGCATACTGGCTTTGC | |||||
| bla MOX | MOX-F | GCTGCTCAAGGAGCACAGGAT | 520 | |||
| MOX-R | CACATTGACATAGGTGTGGTGC | |||||
| mcr-multiplex | mcr-1 | mcr-1-F | AGTCCGTTTGTTCTTGTGGC | 320 | Initial denaturation step at 94 °C for 15 min; 25 cycles of 94 °C for 30 s, annealing temperature: 58 °C for 1:30 min; 72 °C for 1 min; final extension 72 °C for 10 min |
[72] |
| mcr-1-R | AGATCCTTGGTCTCGGCTTG | |||||
| mcr-2 | mcr-2-F | CAAGTGTGTTGGTCGCAGTT | 715 | |||
| mcr-2-R | TCTAGCCCGACAAGCATACC | |||||
| mcr-3 | mcr-3-F | AAATAAAAATTGTTCCGCTTATG | 929 | |||
| mcr-3-R | AATGGAGATCCCCGTTTTT | |||||
| mcr-4 | mcr-4-F | TCACTTTCATCACTGCGTTG | 1116 | |||
| mcr-4-R | TTGGTCCATGACTACCAATG | |||||
| mcr-5 | mcr-5-F | ATGCGGTTGTCTGCATTTATC | 1664 | |||
| mcr-5-R | TCATTGTGGTTGTCCTTTTCTG | |||||
4.8. ERIC-PCR
All Aeromonas were fingerprinted by ERIC-PCR using the primers ERIC-1R (5′-ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC-2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′) [73]. The temperature profiles for amplification were as follows: initial denaturation at 95 °C for 7 min, followed by 30 cycles of amplification with denaturation for 30 s at 90 °C, annealing for 1 min at 52 °C, and extension for 8 min at 65 °C, followed by a final extension at 65 °C for 16 min [33]. The amplification products were separated by 1.5% gel electrophoresis for 90 min at 100 V. The size of the amplified products was determined by comparison with a 1-kb DNA ladder (Promega, Fitchburg, WI, USA).
The pattern of DNA fingerprint bands generated by ERIC-PCR was analyzed, and dendrograms were generated with GelJ software v2.0 [74], using the Dice coefficient to calculate similarity between fingerprints and UPGMA (the unweighted pair-group method with average linkages) method for cluster analysis.
4.9. Conjugation Assay
To investigate the potential plasmid transfer of carbapenem resistance, in vitro conjugation experiments were performed using 10 different carbapenemase-producing Aeromonas strains (4 A. caviae, 2 A. hydrophila, 2 A. veronii, 1 A. media, 1 A. salmonicida) as donors and the rifampicin- and kanamycin-resistant E. coli strain CV601 as plasmid recipient. The filter conjugation assay was performed as previously described [75]. Briefly, donor and recipient strains were grown overnight at 28 °C in LB broth. A total of 500 μL of the overnight cultures of each donor and recipient strain were mixed, centrifuged, and the pellets were resuspended in 200 µL of physiological saline and then spotted onto a filter for mating. After overnight incubation at 28 °C, the filters were washed in physiological saline, and 100 μL of the conjugation mixture was spread on LB agar plates containing kanamycin (50 mg/L), rifampicin (50 mg/L), and MEM (0.12 mg/L) and incubated for 48 to 72 h at 28 °C. Transconjugants were determined by the green fluorescence of the green fluorescent protein. Putative transconjugants were verified by BOX-PCR as previously described [76] and tested for carbapenemase genes and plasmid types as described above. Transfer frequencies were calculated as the total number of transconjugants divided by the total number of recipients.
4.10. Data Accessibillity
The Sanger sequence data were submitted to NCBI GenBank, and the accession numbers for each gene are as follows: OQ348696-OQ348735 (blaKPC), OQ348736 (blaOXA-48), OQ348737 (blaIMP), OQ348738-OQ348739 (blaNDM), OQ348740 (blaCTX-M), OQ348741-OQ348745 (blaTEM), OQ348746-OQ348752 (blaGES), OQ348753-OQ348758 (blaVIM), OQ348759 (blaSHV), OQ348760 (blaVEB), and OQ532911-OQ532961 (16S rRNA gene, Table S3).
5. Conclusions
This study showed that hospital and municipal wastewater contains multidrug-resistant Aeromonas species, some of which are opportunistic pathogens of clinical importance. These Aeromonas isolates exhibited a diverse plasmidome and harbored ESBL and/or carbapenemase genes, which are frequently localized on plasmids. Successful plasmid-mediated transfer of the carbapenem resistance phenotype from A. caviae and A. salmonicida strains to susceptible E. coli recipients was also demonstrated. All these data suggest that Aeromonas spp. in the environment may serve not only as a reservoir for clinically important carbapenemase genes but also as a source for their transfer to other bacteria, including pathogens.
Acknowledgments
We thank the hospital and wastewater treatment plant staff for providing wastewater samples. We also warmly acknowledge Milena Milaković and Stela Križanović for assistance with antibiotic susceptibility testing.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics12030513/s1, Figure S1. Dendrogram showing genetic relatedness of (A) 38 Aeromonas caviae, (B) 11 Aeromonas hydrophila, (C) 7 Aeromonas media and (D) 7 Aeromonas veronii strains determined by analysis of ERIC-PCR fingerprints using the Dice similarity coefficient and UPGMA clustering method; Table S1: Antimicrobial susceptibility patterns of Aeromonas isolates; Table S2. Specific primers and conditions used in plasmid replicon typing; Table S3. Identification of Aeromonas strains by MALDI-TOF MS and/or 16S rRNA gene sequencing. Red letters indicate discrepancies between MALDI-TOF MS and 16S identification. ID represents the isolate designation; TWW and H refer to the source of bacterial isolation, treated municipal wastewater and hospital wastewater, respectively. ND, not determined.
Author Contributions
Conceptualization, N.U.-K.; methodology, N.U.-K., S.D. and A.P.; formal analysis, S.D. and A.P.; funding acquisition, N.U.-K.; resources, N.U.-K.; investigation: S.D., A.P. and M.D.; project administration: N.U.-K.; supervision, N.U.-K.; visualization: S.D. and A.P.; writing—original draft: N.U.-K., S.D. and A.P.; writing—review and editing: N.U.-K., S.D., A.P. and M.D. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data from this study have been deposited in NCBI GenBank, and the accession numbers for each gene are as follows: OQ348696–OQ348735 (blaKPC), OQ348736 (blaOXA-48), OQ348737 (blaIMP), OQ348738-OQ348739 (blaNDM), OQ348740 (blaCTX-M), OQ348741-OQ348745 (blaTEM), OQ348746-OQ348752 (blaGES), OQ348753-OQ348758 (blaVIM), OQ348759 (blaSHV), OQ348760 (blaVEB), and OQ532911-OQ532961 (16S rRNA gene).
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was funded by the Croatian Science Foundation under project number IP-2019-04-5539.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Hernando-Amado S., Coque T.M., Baquero F., Martínez J.L. Defining and Combating Antibiotic Resistance from One Health and Global Health Perspectives. Nat. Microbiol. 2019;4:1432–1442. doi: 10.1038/s41564-019-0503-9. [DOI] [PubMed] [Google Scholar]
- 2.Brown K.D., Kulis J., Thomson B., Chapman T.H., Mawhinney D.B. Occurrence of Antibiotics in Hospital, Residential, and Dairy Effluent, Municipal Wastewater, and the Rio Grande in New Mexico. Sci. Total Environ. 2006;366:772–783. doi: 10.1016/j.scitotenv.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Picão R.C., Cardoso J.P., Campana E.H., Nicoletti A.G., Petrolini F.V.B., Assis D.M., Juliano L., Gales A.C. The Route of Antimicrobial Resistance from the Hospital Effluent to the Environment: Focus on the Occurrence of KPC-Producing Aeromonas spp. and Enterobacteriaceae in Sewage. Diagn. Microbiol. Infect. Dis. 2013;76:80–85. doi: 10.1016/j.diagmicrobio.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Rizzo L., Manaia C., Merlin C., Schwartz T., Dagot C., Ploy M.C., Michael I., Fatta-Kassinos D. Urban Wastewater Treatment Plants as Hotspots for Antibiotic Resistant Bacteria and Genes Spread into the Environment: A Review. Sci. Total Environ. 2013;447:345–360. doi: 10.1016/j.scitotenv.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 5.Figueira V., Vaz-Moreira I., Silva M., Manaia C.M. Diversity and Antibiotic Resistance of Aeromonas spp. in Drinking and Waste Water Treatment Plants. Water Res. 2011;45:5599–5611. doi: 10.1016/j.watres.2011.08.021. [DOI] [PubMed] [Google Scholar]
- 6.Goñi-Urriza M., Capdepuy M., Arpin C., Raymond N., Caumette P., Quentin C. Impact of an Urban Effluent on Antibiotic Resistance of Riverine Enterobacteriaceae and Aeromonas spp. Appl. Environ. Microbiol. 2000;66:125–132. doi: 10.1128/AEM.66.1.125-132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grilo M.L., Sousa-Santos C., Robalo J., Oliveira M. The Potential of Aeromonas spp. from Wildlife as Antimicrobial Resistance Indicators in Aquatic Environments. Ecol. Indic. 2020;115:106396. doi: 10.1016/j.ecolind.2020.106396. [DOI] [Google Scholar]
- 8.Varela A.R., Nunes O.C., Manaia C.M. Quinolone Resistant Aeromonas spp. as Carriers and Potential Tracers of Acquired Antibiotic Resistance in Hospital and Municipal Wastewater. Sci. Total Environ. 2016;542:665–671. doi: 10.1016/j.scitotenv.2015.10.124. [DOI] [PubMed] [Google Scholar]
- 9.Figueras M.J., Guarro J., Martinez-Murcia A. Clinically Relevant Aeromonas Species. Clin. Infect. Dis. 2000;30:988–989. doi: 10.1086/313839. [DOI] [PubMed] [Google Scholar]
- 10.Maravić A., Skočibušić M., Šamanić I., Fredotović Ž., Cvjetan S., Jutronić M., Puizina J. Aeromonas spp. Simultaneously Harbouring blaCTX-M-15, blaSHV-12, blaPER-1 and blaFOX-2, in Wild-Growing Mediterranean Mussel (Mytilus galloprovincialis) from Adriatic Sea, Croatia. Int. J. Food Microbiol. 2013;166:301–308. doi: 10.1016/j.ijfoodmicro.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Janda J.M., Abbott S.L. The Genus Aeromonas: Taxonomy, Pathogenicity, and Infection. Clin. Microbiol. Rev. 2010;23:35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamy B., Kodjo A., Laurent F. Prospective Nationwide Study of Aeromonas Infections in France. J. Clin. Microbiol. 2009;47:1234–1237. doi: 10.1128/JCM.00155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernández-Bravo A., Figueras M.J. An Update on the Genus Aeromonas: Taxonomy, Epidemiology, and Pathogenicity. Microorganisms. 2020;8:129. doi: 10.3390/microorganisms8010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piotrowska M., Przygodzińska D., Matyjewicz K., Popowska M. Occurrence and Variety of β-Lactamase Genes among Aeromonas spp. Isolated from Urban Wastewater Treatment Plant. Front. Microbiol. 2017;8:863. doi: 10.3389/fmicb.2017.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Liu H., Zhang L., Sun B. Application of Modified Carbapenem Inactivation Method and Its Derivative Tests for the Detection of Carbapenemase-Producing Aeromonas. Infect. Drug Resist. 2021;14:3949–3960. doi: 10.2147/IDR.S330115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinclair H.A., Heney C., Sidjabat H.E., George N.M., Bergh H., Anuj S.N., Nimmo G.R., Paterson D.L. Genotypic and Phenotypic Identification of Aeromonas Species and CphA-Mediated Carbapenem Resistance in Queensland, Australia. Diagn. Microbiol. Infect. Dis. 2016;85:98–101. doi: 10.1016/j.diagmicrobio.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Wu C.-J., Chen P.-L., Wu J.-J., Yan J.-J., Lee C.-C., Lee H.-C., Lee N.-Y., Chang C.-M., Lin Y.-T., Chiu Y.-C., et al. Distribution and Phenotypic and Genotypic Detection of a Metallo-β-Lactamase, CphA, among Bacteraemic Aeromonas Isolates. J. Med. Microbiol. 2012;61:712–719. doi: 10.1099/jmm.0.038323-0. [DOI] [PubMed] [Google Scholar]
- 18.Sekizuka T., Inamine Y., Segawa T., Hashino M., Yatsu K., Kuroda M. Potential KPC-2 Carbapenemase Reservoir of Environmental Aeromonas hydrophila and Aeromonas caviae Isolates from the Effluent of an Urban Wastewater Treatment Plant in Japan. Environ. Microbiol. Rep. 2019;11:589–597. doi: 10.1111/1758-2229.12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu X., Yu X., Shang Y., Xu H., Guo L., Liang Y., Kang Y., Song L., Sun J., Yue F., et al. Emergence and Characterization of a Novel IncP-6 Plasmid Harboring blaKPC–2 and qnrS2 Genes in Aeromonas taiwanensis Isolates. Front. Microbiol. 2019;10:2132. doi: 10.3389/fmicb.2019.02132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Qiu Y., Fang C., Dai X., Zhang L. Genomic Characterization of a Multidrug-Resistant Aeromonas caviae Isolate Carrying a Novel blaKPC-2 -Harbouring Plasmid and an IMP-4-Encoding Phage-like Plasmid. Microbiol. Spectr. 2022;10:e00840-22. doi: 10.1128/spectrum.00840-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adler A., Assous M.V., Paikin S., Shulman A., Miller-Roll T., Hillel S., Aronov R., Carmeli Y., Schwaber M.J. Emergence of VIM-Producing Aeromonas caviae in Israeli Hospitals. J. Antimicrob. Chemother. 2014;69:1211–1214. doi: 10.1093/jac/dkt505. [DOI] [PubMed] [Google Scholar]
- 22.Bosch T., Schade R., Landman F., Schouls L., van Dijk K. A blaVIM-1 Positive Aeromonas hydrophila Strain in a near-Drowning Patient: Evidence for Interspecies Plasmid Transfer within the Patient. Future Microbiol. 2019;14:1191–1197. doi: 10.2217/fmb-2019-0091. [DOI] [PubMed] [Google Scholar]
- 23.Walsh T.R., Weeks J., Livermore D.M., Toleman M.A. Dissemination of NDM-1 Positive Bacteria in the New Delhi Environment and Its Implications for Human Health: An Environmental Point Prevalence Study. Lancet Infect. Dis. 2011;11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 24.Xu S., Tu J., Zhang L., Chen Y., Dong X., Chi X., Xu H. Detection of NDM-1-Positive Aeromonas caviae from Bacteremia by Using Whole-Genome Sequencing. Infect. Drug Resist. 2022;15:2835–2841. doi: 10.2147/IDR.S360353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo X., Mu K., Zhao Y., Zhang J., Qu Y., Hu D., Jia Y., Dai P., Weng J., Wang D., et al. Emergence of blaNDM– 1-Carrying Aeromonas caviae K433 Isolated from Patient with Community-Acquired Pneumonia. Front. Microbiol. 2022;13:825389. doi: 10.3389/fmicb.2022.825389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Q., Zhou W., Qian C., Shen K., Zhu X., Zhou D., Sun Z., Lu W., Liu H., Li K., et al. OXA-830, a Novel Chromosomally Encoded Extended-Spectrum Class D β-Lactamase in Aeromonas simiae. Front. Microbiol. 2019;10:2732. doi: 10.3389/fmicb.2019.02732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye Y., Xu X.-H., Li J.-B. Emergence of CTX-M-3, TEM-1 and a New Plasmid-Mediated MOX-4 AmpC in a Multiresistant Aeromonas caviae Isolate from a Patient with Pneumonia. J. Med. Microbiol. 2010;59:843–847. doi: 10.1099/jmm.0.016337-0. [DOI] [PubMed] [Google Scholar]
- 28.Girlich D., Poirel L., Nordmann P. Diversity of Clavulanic Acid-Inhibited Extended-Spectrum β-Lactamases in Aeromonas spp. from the Seine River, Paris, France. Antimicrob. Agents Chemother. 2011;55:1256–1261. doi: 10.1128/AAC.00921-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conte D., Palmeiro J.K., Bavaroski A.A., Rodrigues L.S., Cardozo D., Tomaz A.P., Camargo J.O., Dalla-Costa L.M. Antimicrobial Resistance in Aeromonas Species Isolated from Aquatic Environments in Brazil. J. Appl. Microbiol. 2021;131:169–181. doi: 10.1111/jam.14965. [DOI] [PubMed] [Google Scholar]
- 30.Mathys D.A., Mollenkopf D.F., Feicht S.M., Adams R.J., Albers A.L., Stuever D.M., Grooters S.V., Ballash G.A., Daniels J.B., Wittum T.E. Carbapenemase-Producing Enterobacteriaceae and Aeromonas spp. Present in Wastewater Treatment Plant Effluent and Nearby Surface Waters in the US. PLoS ONE. 2019;14:e0218650. doi: 10.1371/journal.pone.0218650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin H.B., Yoon J., Lee Y., Kim M.S., Lee K. Comparison of MALDI-TOF MS, Housekeeping Gene Sequencing, and 16S RRNA Gene Sequencing for Identification of Aeromonas Clinical Isolates. Yonsei Med. J. 2015;56:550. doi: 10.3349/ymj.2015.56.2.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martone-Rocha S., Piveli R.P., Matté G.R., Dória M.C., Dropa M., Morita M., Peternella F.A., Matté M.H. Dynamics of Aeromonas Species Isolated from Wastewater Treatment System. J. Water Health. 2010;8:703–711. doi: 10.2166/wh.2010.140. [DOI] [PubMed] [Google Scholar]
- 33.Szczuka E., Kaznowski A. Typing of Clinical and Environmental Aeromonas sp. Strains by Random Amplified Polymorphic DNA PCR, Repetitive Extragenic Palindromic PCR, and Enterobacterial Repetitive Intergenic Consensus Sequence PCR. J. Clin. Microbiol. 2004;42:220–228. doi: 10.1128/JCM.42.1.220-228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maiti B., Raghunath P., Karunasagar I., Karunasagar I. Typing of Clinical and Environmental Strains of Aeromonas spp. Using Two PCR Based Methods and Whole Cell Protein Analysis. J. Microbiol. Methods. 2009;78:312–318. doi: 10.1016/j.mimet.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 35.Latif-Eugenín F., Beaz-Hidalgo R., Silvera-Simón C., Fernandez-Cassi X., Figueras M.J. Chlorinated and Ultraviolet Radiation -Treated Reclaimed Irrigation Water Is the Source of Aeromonas Found in Vegetables Used for Human Consumption. Environ. Res. 2017;154:190–195. doi: 10.1016/j.envres.2016.12.026. [DOI] [PubMed] [Google Scholar]
- 36.Davin-Regli A., Bollet C., Chamorey E., Colonna D’Istria V., Cremieux A. A Cluster of Cases of Infections Due to Aeromonas hydrophila Revealed by Combined RAPD and ERIC-PCR. J. Med. Microbiol. 1998;47:499–504. doi: 10.1099/00222615-47-6-499. [DOI] [PubMed] [Google Scholar]
- 37.Ebmeyer S., Kristiansson E., Larsson D.G.J. CMY-1/MOX-Family AmpC β-Lactamases MOX-1, MOX-2 and MOX-9 Were Mobilized Independently from Three Aeromonas Species. J. Antimicrob. Chemother. 2019;74:1202–1206. doi: 10.1093/jac/dkz025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kämpfer P., Christmann C., Swings J., Huys G. In Vitro Susceptibilities of Aeromonas Genomic Species to 69 Antimicrobial Agents. Syst. Appl. Microbiol. 1999;22:662–669. doi: 10.1016/S0723-2020(99)80019-8. [DOI] [PubMed] [Google Scholar]
- 39.Aravena-Román M., Inglis T.J.J., Henderson B., Riley T.V., Chang B.J. Antimicrobial Susceptibilities of Aeromonas Strains Isolated from Clinical and Environmental Sources to 26 Antimicrobial Agents. Antimicrob. Agents Chemother. 2012;56:1110–1112. doi: 10.1128/AAC.05387-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harnisz M., Korzeniewska E. The Prevalence of Multidrug-Resistant Aeromonas spp. in the Municipal Wastewater System and Their Dissemination in the Environment. Sci. Total Environ. 2018;626:377–383. doi: 10.1016/j.scitotenv.2018.01.100. [DOI] [PubMed] [Google Scholar]
- 41.Amos G.C.A., Hawkey P.M., Gaze W.H., Wellington E.M. Waste Water Effluent Contributes to the Dissemination of CTX-M-15 in the Natural Environment. J. Antimicrob. Chemother. 2014;69:1785–1791. doi: 10.1093/jac/dku079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Araujo C.F.M., Silva D.M., Carneiro M.T., Ribeiro S., Fontana-Maurell M., Alvarez P., Asensi M.D., Zahner V., Carvalho-Assef A.P.D. Detection of Carbapenemase Genes in Aquatic Environments in Rio de Janeiro, Brazil. Antimicrob. Agents Chemother. 2016;60:4380–4383. doi: 10.1128/AAC.02753-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montezzi L.F., Campana E.H., Corrêa L.L., Justo L.H., Paschoal R.P., da Silva I.L.V.D., Souza M.d.C.M., Drolshagen M., Picão R.C. Occurrence of Carbapenemase-Producing Bacteria in Coastal Recreational Waters. Int. J. Antimicrob. Agents. 2015;45:174–177. doi: 10.1016/j.ijantimicag.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 44.Dolejska M., Papagiannitsis C.C. Plasmid-Mediated Resistance Is Going Wild. Plasmid. 2018;99:99–111. doi: 10.1016/j.plasmid.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 45.Chen L., Mathema B., Chavda K.D., DeLeo F.R., Bonomo R.A., Kreiswirth B.N. Carbapenemase-Producing Klebsiella pneumoniae: Molecular and Genetic Decoding. Trends Microbiol. 2014;22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reyes J., Cárdenas P., Tamayo R., Villavicencio F., Aguilar A., Melano R.G., Trueba G. Characterization of blaKPC-2 -Harboring Klebsiella pneumoniae Isolates and Mobile Genetic Elements from Outbreaks in a Hospital in Ecuador. Microbial. Drug Resist. 2021;27:752–759. doi: 10.1089/mdr.2019.0433. [DOI] [PubMed] [Google Scholar]
- 47.Kraftova L., Finianos M., Studentova V., Chudejova K., Jakubu V., Zemlickova H., Papagiannitsis C.C., Bitar I., Hrabak J. Evidence of an Epidemic Spread of KPC-Producing Enterobacterales in Czech Hospitals. Sci. Rep. 2021;11:15732. doi: 10.1038/s41598-021-95285-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haines A.S., Cheung M., Thomas C.M. Evidence That IncG (IncP-6) and IncU Plasmids Form a Single Incompatibility Group. Plasmid. 2006;55:210–215. doi: 10.1016/j.plasmid.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 49.Wu W., Feng Y., Tang G., Qiao F., McNally A., Zong Z. NDM Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings. Clin. Microbiol. Rev. 2019;32:e00115-18. doi: 10.1128/CMR.00115-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Findlay J., Perreten V., Poirel L., Nordmann P. Molecular Analysis of OXA-48-Producing Escherichia coli in Switzerland from 2019 to 2020. Eur. J. Clin. Microbiol. Infect. Dis. 2022;41:1355–1360. doi: 10.1007/s10096-022-04493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loconsole D., Accogli M., de Robertis A.L., Capozzi L., Bianco A., Morea A., Mallamaci R., Quarto M., Parisi A., Chironna M. Emerging High-Risk ST101 and ST307 Carbapenem-Resistant Klebsiella pneumoniae Clones from Bloodstream Infections in Southern Italy. Ann. Clin. Microbiol. Antimicrob. 2020;19:24. doi: 10.1186/s12941-020-00366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barry K.E., Wailan A.M., Sheppard A.E., Crook D., Vegesana K., Stoesser N., Parikh H.I., Sebra R., Mathers A.J. Don’t Overlook the Little Guy: An Evaluation of the Frequency of Small Plasmids Co-Conjugating with Larger Carbapenemase Gene Containing Plasmids. Plasmid. 2019;103:1–8. doi: 10.1016/j.plasmid.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 53.Chen L., Hu H., Chavda K.D., Zhao S., Liu R., Liang H., Zhang W., Wang X., Jacobs M.R., Bonomo R.A., et al. Complete Sequence of a KPC-Producing IncN Multidrug-Resistant Plasmid from an Epidemic Escherichia coli Sequence Type 131 Strain in China. Antimicrob. Agents Chemother. 2014;58:2422–2425. doi: 10.1128/AAC.02587-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Komeda T., Shrestha S., Sherchan J.B., Tohya M., Hishinuma T., Sherchand J.B., Tada T., Kirikae T. Emergence of a Highly Colistin-Resistant Aeromonas jandaei Clinical Isolate Harbouring Four Genes Encoding Phosphoethanolamine Transferases in Nepal. Int. J. Antimicrob. Agents. 2022;59:106544. doi: 10.1016/j.ijantimicag.2022.106544. [DOI] [PubMed] [Google Scholar]
- 55.Liu J., Xiao G., Zhou W., Yang J., Wang Y., Wu Y., Cheng X., Sun Z. Various Novel Colistin Resistance Mechanisms Interact to Facilitate Adaptation of Aeromonas hydrophila to Complex Colistin Environments. Antimicrob. Agents Chemother. 2021;65:e0007121. doi: 10.1128/AAC.00071-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yin W., Li H., Shen Y., Liu Z., Wang S., Shen Z., Zhang R., Walsh T.R., Shen J., Wang Y. Novel Plasmid-Mediated Colistin Resistance Gene mcr-3 in Escherichia coli. mBio. 2017;8:e00543-17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma S., Sun C., Hulth A., Li J., Nilsson L.E., Zhou Y., Börjesson S., Bi Z., Bi Z., Sun Q., et al. Mobile Colistin Resistance Gene mcr-5 in Porcine Aeromonas hydrophila. J. Antimicrob. Chemother. 2018;73:1777–1780. doi: 10.1093/jac/dky110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eichhorn I., Feudi C., Wang Y., Kaspar H., Feßler A.T., Lübke-Becker A., Michael G.B., Shen J., Schwarz S. Identification of Novel Variants of the Colistin Resistance Gene mcr-3 in Aeromonas spp. from the National Resistance Monitoring Programme GERM-Vet and from Diagnostic Submissions. J. Antimicrob. Chemother. 2018;73:1217–1221. doi: 10.1093/jac/dkx538. [DOI] [PubMed] [Google Scholar]
- 59.Gonzalez-Avila L.U., Loyola-Cruz M.A., Hernández-Cortez C., Bello-López J.M., Castro-Escarpulli G. Colistin Resistance in Aeromonas spp. Int. J. Mol. Sci. 2021;22:5974. doi: 10.3390/ijms22115974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weisburg W.G., Barns S.M., Pelletier D.A., Lane D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.European Committee on Antimicrobial Susceptibility Testing . The European Committee on Antimicrobial Susceptibility Testing. Universidade de Cabo Verde; Praia, Cabo Verde: 2020. Breakpoint tables for interpretation of MICs and zone diameters. Version 10.0. [Google Scholar]
- 62.Gupta G., Tak V., Mathur P. Detection of AmpC β Lactamases in Gram-Negative Bacteria. J. Lab. Physicians. 2014;6:001–006. doi: 10.4103/0974-2727.129082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nordmann P., Poirel L. Emerging Carbapenemases in Gram-Negative Aerobes. Clin. Microbiol. Infect. 2002;8:321–331. doi: 10.1046/j.1469-0691.2002.00401.x. [DOI] [PubMed] [Google Scholar]
- 64.Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K.L., Threlfall E.J. Identification of Plasmids by PCR-Based Replicon Typing. J. Microbiol. Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 65.García-Fernández A., Fortini D., Veldman K., Mevius D., Carattoli A. Characterization of Plasmids Harbouring qnrS1, qnrB2 and qnrB19 Genes in Salmonella. J. Antimicrob. Chemother. 2009;63:274–281. doi: 10.1093/jac/dkn470. [DOI] [PubMed] [Google Scholar]
- 66.Poirel L., Bonnin R.A., Nordmann P. Genetic Features of the Widespread Plasmid Coding for the Carbapenemase OXA-48. Antimicrob. Agents Chemother. 2012;56:559–562. doi: 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jelić M. Mechanisms of Antimicrobial Resistance in Carbapenem-Resistant Enterobacteriaceae. University of Zagreb; Zagreb, Croatia: 2018. [Google Scholar]
- 68.Poirel L., Héritier C., Tolün V., Nordmann P. Emergence of Oxacillinase-Mediated Resistance to Imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004;48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Revathi G., Siu L.K., Lu P.-L., Huang L.-Y. First Report of NDM-1-Producing Acinetobacter baumannii in East Africa. Int. J. Infect. Dis. 2013;17:e1255–e1258. doi: 10.1016/j.ijid.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 70.Saladin M., Cao V.T.B., Lambert T., Donay J.-L., Herrmann J.-L., Ould-Hocine Z., Verdet C., Delisle F., Philippon A., Arlet G. Diversity of CTX-M Î2-Lactamases and Their Promoter Regions from Enterobacteriaceae Isolated in Three Parisian Hospitals. FEMS Microbiol. Lett. 2002;209:161–168. doi: 10.1111/j.1574-6968.2002.tb11126.x. [DOI] [PubMed] [Google Scholar]
- 71.Pérez-Pérez F.J., Hanson N.D. Detection of Plasmid-Mediated AmpC β-Lactamase Genes in Clinical Isolates by Using Multiplex PCR. J. Clin. Microbiol. 2002;40:2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rebelo A.R., Bortolaia V., Kjeldgaard J.S., Pedersen S.K., Leekitcharoenphon P., Hansen I.M., Guerra B., Malorny B., Borowiak M., Hammerl J.A., et al. Multiplex PCR for Detection of Plasmid-Mediated Colistin Resistance Determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for Surveillance Purposes. Eurosurveillance. 2018;23:1–11. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Versalovic J., Koeuth T., Lupski J.R. Distribution of Repetitive DNA Sequences in Eubacteria and Application to Fingerpriting of Bacterial Genomes. Nucleic. Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heras J., Domínguez C., Mata E., Pascual V., Lozano C., Torres C., Zarazaga M. GelJ—A Tool for Analyzing DNA Fingerprint Gel Images. BMC Bioinform. 2015;16:270. doi: 10.1186/s12859-015-0703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gekenidis M.-T., Kläui A., Smalla K., Drissner D. Transferable Extended-Spectrum β-Lactamase (ESBL) Plasmids in Enterobacteriaceae from Irrigation Water. Microorganisms. 2020;8:978. doi: 10.3390/microorganisms8070978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martin B., Humbert O., Camara M., Guenzi E., Walker J., Mitchell T., Andrew P., Prudhomme M., Alloing G., Hakenbeck R., et al. A Highly Conserved Repeated DNA Element Located in the Chromosome of Streptococcus pneumoniae. Nucleic Acids Res. 1992;20:3479–3483. doi: 10.1093/nar/20.13.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data from this study have been deposited in NCBI GenBank, and the accession numbers for each gene are as follows: OQ348696–OQ348735 (blaKPC), OQ348736 (blaOXA-48), OQ348737 (blaIMP), OQ348738-OQ348739 (blaNDM), OQ348740 (blaCTX-M), OQ348741-OQ348745 (blaTEM), OQ348746-OQ348752 (blaGES), OQ348753-OQ348758 (blaVIM), OQ348759 (blaSHV), OQ348760 (blaVEB), and OQ532911-OQ532961 (16S rRNA gene).