SUMMARY
The mammalian spinal cord functions as a community of cell types for sensory processing, autonomic control, and movement. While animal models have advanced our understanding of spinal cellular diversity, characterizing human biology directly is important to uncover specialized features of basic function and human pathology. Here, we present a cellular taxonomy of the adult human spinal cord using single nucleus RNA-sequencing with spatial transcriptomics and antibody validation. We identified 29 glial clusters and 35 neuronal clusters, organized principally by anatomical location. To demonstrate the relevance of this resource to human disease, we analyzed spinal motoneurons, which degenerate in amyotrophic lateral sclerosis (ALS) and other diseases. We found that, compared with other spinal neurons, human motoneurons are defined by genes related to cell size, cytoskeletal structure, and ALS, suggesting a specialized molecular repertoire underlying their selective vulnerability. We include a web resource to facilitate further investigations into human spinal cord biology.
Graphical Abstract
eTOC blurb
Yadav, Matson et al. use single nucleus RNA-sequencing, spatial transcriptomics, and immunohistochemistry to profile the cell types of the adult human spinal cord, identifying sixty-four glial and neuronal populations. This resource reveals how the unique molecular environments of specific cell types could contribute to chronic pain or neurodegeneration.
INTRODUCTION
The mammalian spinal cord relays, processes, and transforms sensory inputs and descending cues from the brain into sensory, motor, respiratory, and autonomic outputs. These critical processes rely on a diverse array of spinal cord cell types, each with their own molecular repertoires, functions, and vulnerabilities to injury and disease. Most prominently, spinal motoneurons specifically degenerate in spinal muscular atrophy1 and in amyotrophic lateral sclerosis (ALS), though the molecular basis for this selective phenotype is not clear2,3. Spinal cord cell types have been extensively studied in model organisms, including molecular profiling at the single-cell level, to identify candidate cellular mechanisms for human pathophysiology including in chronic pain, neurodegeneration, and spinal cord injury4–12. However, technical obstacles and limited access to high quality tissue specimens have prevented the full application of single cell approaches to study human spinal cord biology directly. Thus, prior work has only been done on limited cell types or in human fetal tissue13–15.
To characterize the cell types of the adult human lumbar spinal cord, we used recently optimized tissue extraction methods on spinal cords from organ donor subjects and performed single nucleus RNA-sequencing (snRNA-seq) of over 50,000 nuclei. We identified 64 unique clusters, including 29 non-neuronal populations and 35 neuronal populations, and validated many of the expression patterns with spatial transcriptomics. We established a comprehensive taxonomy of the neuronal clusters, compared them with their mouse counterparts, and created a publicly available browsable interface as a resource for the field. Finally, we performed a focused analysis on the transcriptional profile of spinal motoneurons, identifying a molecular signature that could underlie their selective vulnerability in neurodegenerative disease.
RESULTS
We obtained post-mortem lumbar spinal cord tissue from 14 donor transplant cases (Supplemental Table S1), using neuroprotective conditions (see Methods). For snRNA-seq experiments, nuclei were isolated and profiled from seven donors (Fig. 1A), resulting in a dataset of 55,289 nuclei after quality control (Supplemental Fig. S1). Initial clustering of all nuclei clearly distinguished the major known cell classes, including oligodendrocytes and their precursors and progenitors, meningeal cells, astrocytes, endothelial and pericyte cells, microglia, and neurons; the latter included glutamatergic neurons, GABAergic/glycinergic neurons, and motoneurons (Fig. 1B). To determine whether the overall proportions of cell classes that we observed in the sequencing dataset reflected in vivo tissue composition, we analyzed the prevalence of oligodendrocytes, astrocytes, microglia, and neurons in adult human lumbar spinal cord tissue using antibody staining with classic cell class markers. We found similar proportions for neurons, astrocytes, microglia, and oligodendrocytes (Fig. 1C–D, p = 0.67, p = 0.33, p = 0.06, p = 0.06, Supplemental Fig. S1) in tissue compared with the snRNA-seq dataset. Overall, the major cell classes in the sequencing dataset showed clear segregation of previously reported markers for these cell types, each of these broad classes (Fig. 1B, Supplemental Fig. S1–S6), as described below.
Fig. 1: A single cell catalog of the human spinal cord reveals the gene expression signature of human motoneurons.
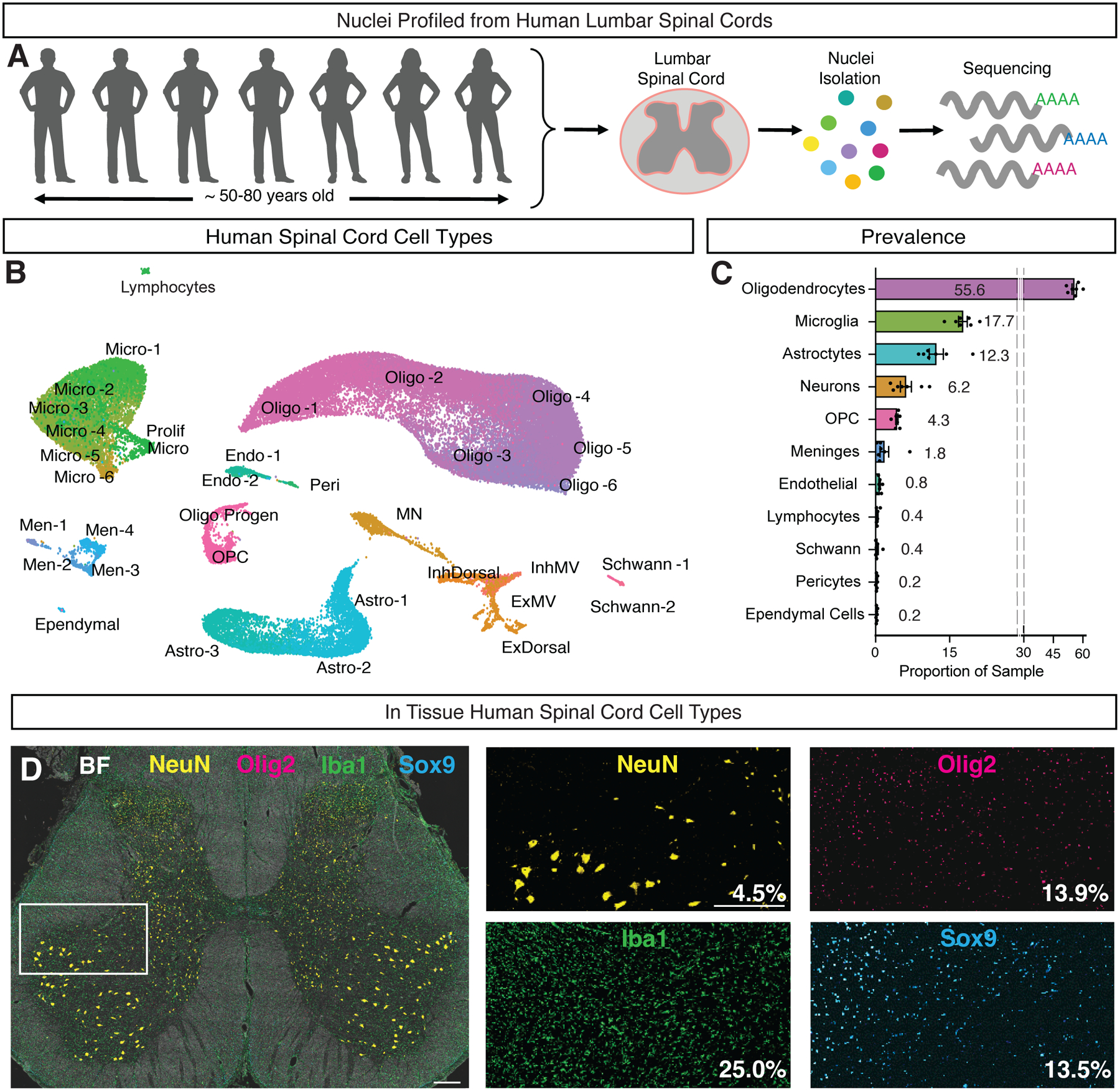
A, Lumbar spinal cord tissue was obtained from seven subjects and processed for single nucleus RNA-sequencing. B, UMAP plot showing the major cell classes of the human spinal cord. Cells of the oligodendrocyte lineage are shown in pink/purple and include two populations of Schwann cells (Schwann-1 and Schwann–2), oligodendrocyte precursor cells (OPC), progenitors (Oligo Progen), and six groups of oligodendrocytes (Oligo-1 through Oligo-6). Microglia are shown in green and include a putatively proliferating population (Prolif Micro), five groups of microglia (Micro-1 through Micro-4 and Perivascular Microglia) as well as a population of macrophages. Astrocytes are shown in turquoise and include three populations (WM Astro, GM Astro-1 and GM Astro-2). Meninges are shown in blue and include four populations (Men-1 through Men-4). Vascular cells are shown in teal and include two groups of endothelial cells (Venous/Capillary Endo and Arterial Endo) and pericytes/smooth muscle cells (Peri/SMC). Ependymal cells are shown in cyan. Neurons are shown in orange and include seven broad classes based on their neurotransmitter status and putative location: motoneurons (MN), excitatory dorsal neurons (ExDorsal), inhibitory dorsal neurons (InhDorsal), excitatory mid neurons (ExM), excitatory ventral neurons (EV), inhibitory mid neurons (InhM), and inhibitory ventral neurons (InhV). C, Bar plot showing the proportion of each cluster in each donor (N=7). Error bars are ± s.e.m. D, Multiplexed immunohistochemistry of the lumbar human spinal cord, stained for NeuN (yellow), IBA1 (green), SOX9 (turquoise), and OLIG2 (pink). Brightfield (BF) is shown in white. Mean percent of DAPI+ cells expressing NeuN, OLIG2, IBA1 and SOX9 are noted in the bottom right corner of each inset (N = 2). Scale bars are 500 μm. See also Supplemental Figures S1–S6.
Glial and Support Cell Populations of the Adult Human Lumbar Spinal Cord
We re-clustered non-neuronal cells and identified 29 subpopulations. We further characterized these groups by inspecting expression of known marker genes, and examined their spatial distribution using spatial transcriptomics on tissue from five donors (Fig. 2).
Fig. 2: Glial and support cell types in the human spinal cord.
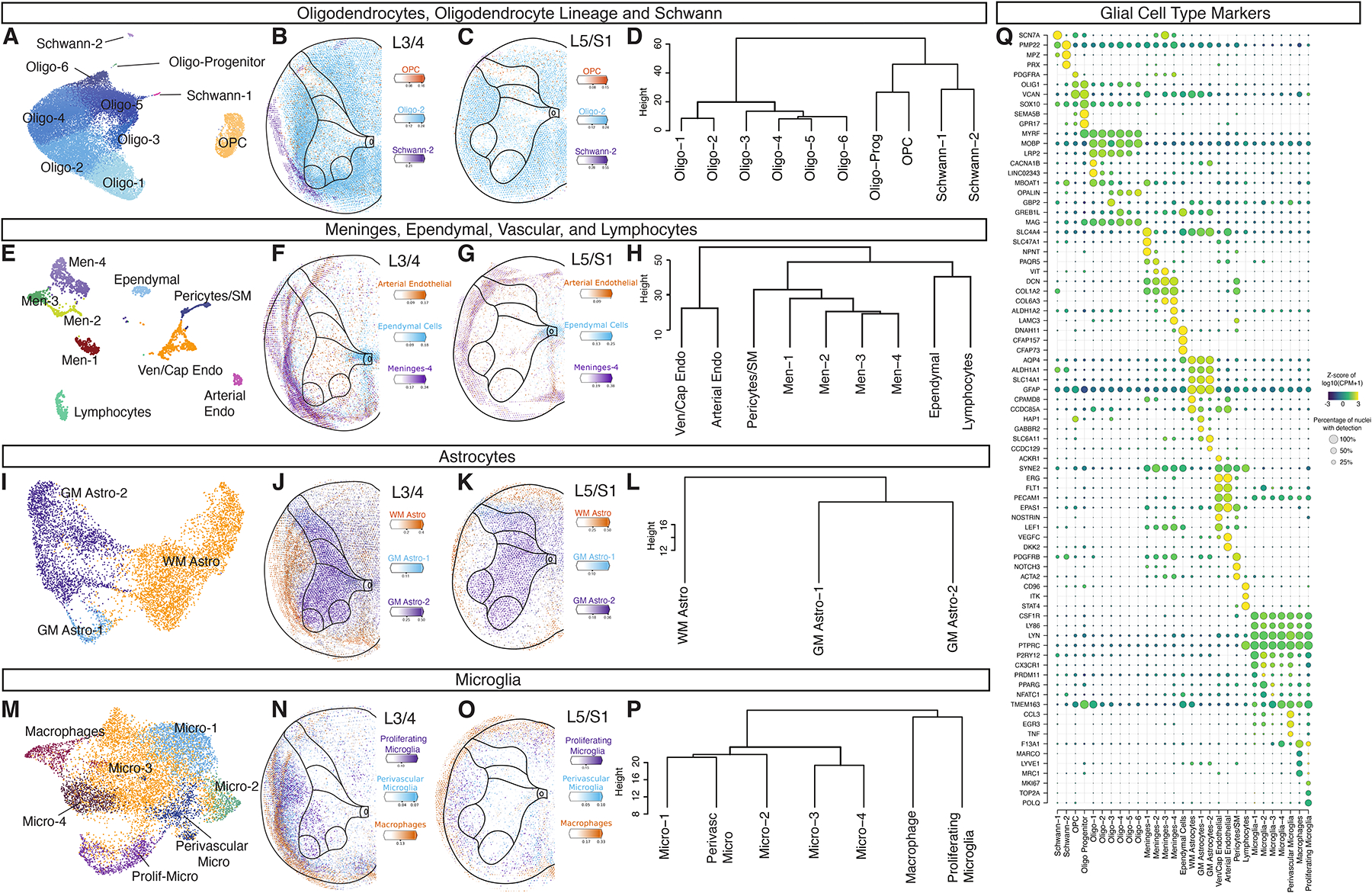
Glial cell types including Oligodendrocytes (A-D), Meninges, Ependymal, Vascular and Lymphocyte Cells (E-H), Astrocytes (I-L), Microglia and Macrophages (M-P). For each cell class, the UMAP shows the subtypes, the spatial feature plots show Cell2Location predictions, and the dendrogram depicts the relationships between the subtypes. Individual Cell2Location prediction for each cell type can be found in Supplemental Figure S7 and S8. Dendrograms were calculated using the top 2,000 highly variable genes from each population and Ward’s method. Q, Dot plot of markers for glial subtypes showing average expression (color) and percent expressed (dot size). See also Supplemental Figures S7–S9.
Amongst oligodendrocytes and related populations, we observed two groups of Schwann cells, a population of oligodendrocyte precursor cells and related progenitors, as well as six populations of oligodendrocytes that were distributed over the entire spinal cord tissue with a bias for the white matter, as expected (Fig. 2A–D, Fig. 2Q, Supplemental Fig. S7 and S8).
Amongst support cells, we identified four populations of meningeal-related cells that included putative meningeal fibroblasts (Meninges-1, SLC4A4) and perivascular fibroblasts (Meninges-3 and Meninges-4, expressing ABCA8, DCN, and COL1A1), and a group of ependymal cells (Fig. 2E–H, Fig. 2Q, Supplemental Fig. S7 and S8). Amongst astrocytes, we identified three populations, including one localized to the white matter (WM Astro) that expressed the fibrous astrocyte marker CD44. The other two astrocyte populations were localized to the gray matter (GM Astro-1 and GM Astro-2) and were enriched for genes involved in neural metabolism and signaling including the GABA transporter SLC6A11, the AMPA receptor regulator SHISA9, and the synaptic adhesion protein TENM2 (Fig. 2I–L, Fig. 2Q, Supplemental Fig. S7 and S8).
Amongst vascular cells, we identified two endothelial cell populations, one of which represented putative venous/capillary cells (Endothelial-1, expressing IL1R1, ACKR1, ABCG2, and MFSD2A), while the other represented putative arterial cells (Endothelial-2, expressing SEMA3G, BMX, VEGFC, and PLCG2). We also identified a putative pericyte population (expressing PDGFRB and NOTCH3) that likely included vascular smooth muscle cells as well (which are marked by SLIT3, ACTA2, and MYH11) and a cluster of lymphocytes (Fig. 2E–H, Fig. 2Q, Supplemental Fig. S7 and S8).
We observed six populations of microglia and one population of macrophages. A population of putative perivascular microglia were similar to a previously described aging-associated microglial population in mice16. This population showed enriched expression of inflammatory-related genes (CCL3, CCL4, IL1B, and ATF3), and were found just outside the main artery at the midline of the ventral horn of the spinal cord. Similarly, the cluster of macrophages (marked by MRC1, F13A1, LYVE1, and CD163) were found near, and within, the region of this main vessel (Fig. 2M–P, Fig. 2Q and Supplemental Fig. S7 and S8). We also observed a putative proliferative type of microglia characterized by expression of POLQ, TOP2A, and MKI67 (Fig. 2M–P, Fig. 2Q, Supplemental Fig. S7 and S8). In prior work on healthy adult mouse spinal cord proliferative microglia were not observed17. We therefore performed antibody staining on post-mortem tissue from three organ donor subjects not included in the snRNA-seq dataset to confirm the existence of this population in intact tissue. Indeed, we found that 23% of IBA1+ microglia in tissue co-expressed the proliferative marker Ki67 (Supplemental Fig. S9, 25% ± 0.5 of cells were IBA1+, with 5.8% ± 0.38 of cells double positive for IBA1 and Ki67). Although prior studies on post-mortem and surgically resected adult human brain tissue have also identified proliferative microglia18, it is yet to be determined whether this reflects normal adult human biology, is associated with the advanced age of the tissue donors, or is due to peri-mortem changes in spinal cord. Proliferative microglia were enriched in white matter, but also interestingly were found in the ventral horn near putative motoneurons (Supplemental Fig. S9).
Neuronal Atlas of the Adult Human Lumbar Spinal Cord
To characterize the neuronal populations of the adult human lumbar spinal cord, we sub-clustered the neuronal nuclei and identified 35 populations. These included a large population of spinal motoneurons (described below), a cluster defined by expression of immediate early response genes (IEG), and 33 main populations of spinal neurons (Fig. 3A, Supplemental Fig. S10). Each cluster contained nuclei from all seven donors, with the exception of Ex-Dorsal-1, Inh-Dorsal-4 and Inh-Dorsal-5 (Fig. 3A, Supplemental Fig. S11, Supplemental Table S2).
Fig. 3: Neuronal cell types in the human spinal cord.
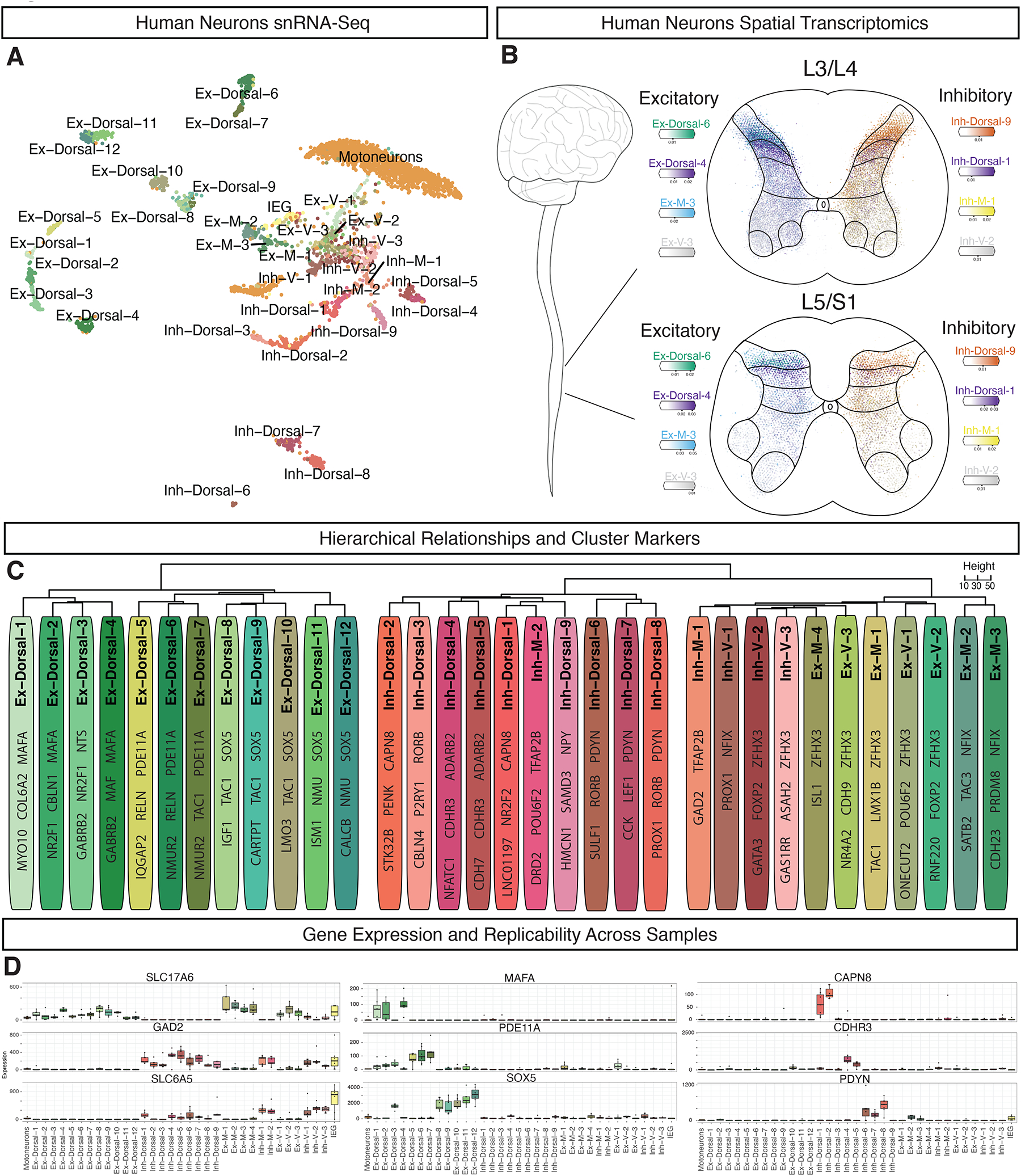
A, UMAP plot of human spinal neurons showing 35 populations. B, Cell2Location predictions on spatial transcriptomics data showing selected excitatory (left) and inhibitory (right) cell types at both L3/4 and L5/S1 segmental levels. C, Dendogram showing the relationship of neuronal subtypes, calculated using the top 2,000 highly variable genes and Ward’s method. For each cluster, 2–3 marker genes are listed. D, Neurotransmitter status markers SLC17A6, GAD2, and SLC6A5 (left column), dorsal excitatory markers MAFA, PDE11A, and SOX5 (middle column), and dorsal inhibitory markers CAPN8, CDHR3, and PDYN (right column). Box plots show the median expression of each gene in each cluster (Counts per Million of Unique Molecular Identifiers) per donor (N = 7), as well as the 25th and 75th percentile of expression, and whiskers show the most extreme point within 1.5 times the interquartile range. See also Supplemental Figures S10–S13.
The three main axes of gene expression variability amongst human spinal neurons were genes related to motoneuron identity, to spatial location, and to neurotransmitter status (Supplemental Fig. S12). To assign putative locations for each population, we used Cell2Location predictions based on spatial transcriptomics data, the spatial distribution of RNA expression for key marker genes, and comparison with data from macaque12 and mouse19, sorting clusters into general categories of dorsal, mid, and ventral cell types (Fig 3B–D). We next assigned putative neurotransmitter status to each population, identifying 19 glutamatergic (defined by the expression of SLC17A6), and 14 GABA/glycinergic populations (defined by expression of GAD1, GAD2, PAX2, and SLC6A5). A dendrogram of the overall cluster relationships confirmed that location and neurotransmitter status were the major organizational axes of spinal neurons, with dorsal excitatory clusters defining the first branch point, and then dorsal inhibitory clusters and mid-ventral clusters (of both general neurotransmitter types) defining subsequent branch points (Fig 3C). We therefore adopted a nomenclature for human spinal neuron populations that references both neurotransmitter status and location. These and other cell type features are summarized in Supplemental Table S3, with selected data in Fig 3 and Supplemental Fig. S10–S13.
Dorsal excitatory neurons were organized into 12 distinct clusters mainly localized to the superficial dorsal horn (lamina I/II), with the exception of Ex-Dorsal-4, which was localized to lamina III (Supplemental Fig. S7 and S8). Expression of LMX1B, as well as LBX1 and TLX3, in many of these populations suggested a dI5/dILB embryonic origin for these cells (Supplemental Fig. S13). These populations all expressed the vGLUT2 gene SLC17A6, with Ex-Dorsal-6 additionally expressing the vGLUT3 gene SLC17A8. One major group (Ex-Dorsal-1, Ex-Dorsal-2, and Ex-Dorsal-4) were likely nonpeptidergic and distinguished by the MAF/MAFA bZIP transcription factors, while the other clusters were likely peptidergic, distinguished by genes such as PAM, TAC1, TAC3, NMU, and GRP (Supplemental Fig. S10).
The dorsal inhibitory neurons were organized into 9 distinct cell types and were also mainly localized to the superficial dorsal horn, with the exceptions of Inh-Dorsal-1 and Inh-Dorsal-4 found in lamina III, and of Inh-Dorsal-5 which was found in both the superficial and deep dorsal horns (Supplemental Fig. S7 and S8). Expression of the inhibitory transcription factor PAX2, as well as GBX1 and minor expression of LBX1 suggested a dI4/dILA embryonic origin for these cells (Supplemental Fig. S13, Supplemental Table S3). Most populations expressed markers for both GABAergic (GAD1/2) and glycinergic (SLC6A5) cell types, with the exception of Inh-Dorsal-2 and Inh-Dorsal-3, which seemed exclusively GABAergic. The Inh-Dorsal-4 and Inh-Dorsal-5 cell types were putatively nonpeptidergic and marked by ADARB2, while most other dorsal inhibitory clusters were distinguished by neuropeptide genes such as NPPC, PENK, PDYN, and NPY (Supplemental Fig. S10).
Overall, mid and ventral cells were distinguished by expression of either the early-born markers ZFHX3/4 or the late-born markers NFIA/NFIB/NFIX and PROX1, similar to what has been reported recently in mouse20 (Supplemental Fig. S10). Ex-M-1, Ex-M-2, and Ex-M-3 were mid-excitatory cell types that expressed LMX1B and TLX3 suggesting a dI5/dILB origin (Supplemental Fig. S13). In contrast, Ex-M-4 and the ventral excitatory clusters Ex-V-1, Ex-V-2, and Ex-V-3 expressed very low but detectable levels of markers for the dI1–3 embryonic domains that are known to settle in the deep dorsal and ventral horn regions (BARHL1, BARHL2, LHX2, LHX9, ISL1, and OTP), as well as markers of the V0c (PITX2, CHAT), V2a (VSX2, SOX14), and V3 (SIM1, NKX2–2) domains (Supplemental Fig. S10 and S13). Amongst inhibitory mid and ventral clusters, Inh-M-2 were likely derived from the dI4/dILA domain, based on their expression of either GBX1 or LBX1. Inh-V-2 selectively expressed markers of the V2b embryonic lineage, including GATA2, GATA3, and MSX1. Spatial transcriptomics predictions from Cell2Location confirmed a mid/ventral location for each of these clusters (except Ex-M2) (Supplemental Fig S7 and S8).
Our characterization of neuronal populations suggested that dorsal clusters were readily distinguished by specific markers, whereas ventral clusters often displayed overlapping patterns of gene expression. Having previously observed this pattern in mouse spinal cord19, we next systematically assessed how distinct dorsal clusters were compared to ventral clusters in both human and mouse spinal cord. We examined two measures of distinctness: cluster separability (analogous to the tightness of gene expression distribution in a cluster) and distance between cluster centroids (analogous to the difference between means of gene expression).
To assess cluster separability, we used two approaches: silhouette scoring of each cluster (Fig. 4A–C, Supplemental Fig. S11) and a post-hoc machine learning approach to measure the proportion of nuclei from each cluster that could be unambiguously assigned (Supplemental Fig. S14A). Both of these approaches showed that dorsal clusters formed discrete groups that were well separated from each other, whereas mid and ventral clusters were substantially more overlapping (Fig. 4B,C). To assess how far apart cluster centroids were, we calculated pairwise correlations between cluster centroids over the 2000 most highly variable genes (Fig. 4D–F), and Euclidean distance between cluster centroids in principal component space (Fig. 3C and Russ et al19). We found that dorsal clusters have significantly lower correlations with each other and higher distances from each other, as compared to mid and ventral clusters, and this was also true for correlations at the individual cell level (Supplemental Fig. S14B). Overall, these analyses confirmed that dorsal neurons are organized into significantly more discrete groups than ventral neurons, pointing to fundamental differences in the overall organization of the dorsal and ventral regions of both the human and mouse spinal cords.
Fig. 4: Overall relationships among dorsal and ventral neuronal populations in human and mouse lumbar spinal cord.

A, Distributions of percell Silhouette scores in human and mouse spinal cord neurons, separated into dorsal and ventral groups. Higher silhouette scores indicate that cells belong to more clearly separated clusters. Cluster-level silhouette scores are shown in Supplemental Fig. S11. Two-way ANOVA, followed by Mann-Whitney tests for human and mouse dorsal vs ventral distributions were p < 0.0001, ****. B-C, UMAP of human neurons (B) and mouse neurons (C) colored by Silhouette score. D, Median gene expression correlation (Pearson’s R) of each cluster to other clusters, using the top 2,000 highly variable genes. Two-way ANOVA, followed by Mann-Whitney tests for human and mouse dorsal vs ventral distributions were p < 0.0001, ****. E-F, Heatmap of pairwise gene expression correlations (Pearson’s R) of human spinal cord clusters (E) and mouse spinal cord clusters (F, Russ et al.19). Correlation is colored from purple (low) to yellow (high). See also Supplemental Figure S14.
Having established similarities in the overall organization of dorsal and ventral neurons in the human expression patterns are shared or distinct between the two species. We thus integrated the neuronal data with prior harmonized datasets from postnatal mouse tissue (Fig. 5AB, Supplemental Fig. S15). We found that, overall, human neurons were enriched for KAZN, ROBO2 and DPP10, while mouse neurons were enriched for DCC, USP29, and ASIC2 (Fig. 5E, Supplemental Fig. S16). There was cluster-specific correspondence between the two datasets, with pairs of human-mouse dorsal clusters showing high correlations and specific relationships, while ventral clusters showed broader overall similarity (Fig 5C). We used a network analysis of cluster relatedness to identify human and mouse cell type pairs with high conservation (Fig. 5D). For example, human ExDorsal-4 is highly homologous to mouse Excit-05, a member of the MAF family located in lamina III-IV and associated with corrective reflexes and light touch processing (Fig. 5G)21. Both the human and mouse clusters are enriched for MAF, ADARB2, and RORA, while the human cluster is also enriched for MAFA (found in the spatial transcriptomics data in the deeper region of the dorsal horn) and the mechanosensitive protein PIEZO2, which may confer evolutionarily distinct functions in this population. In addition, human Inh-Dorsal-8 was highly homologous to mouse Inhib-11, a member of the Pdyn family located in lamina I-III and associated with mechanical allodynia pain symptoms and itch22–25 (Fig. 5L). Both clusters were enriched for the neuropeptides PDYN and PNOC, as well as PROX1 and TACR3. The human cluster was enriched for the neuropeptide NPPC while the mouse cluster was enriched for the neuropeptide Gal. In the future, such cross-species cell type relationships can be used to propose behavioral functions for a broad range of human neuronal populations.
Fig. 5: Relationships between human and mouse spinal cord neuronal populations.
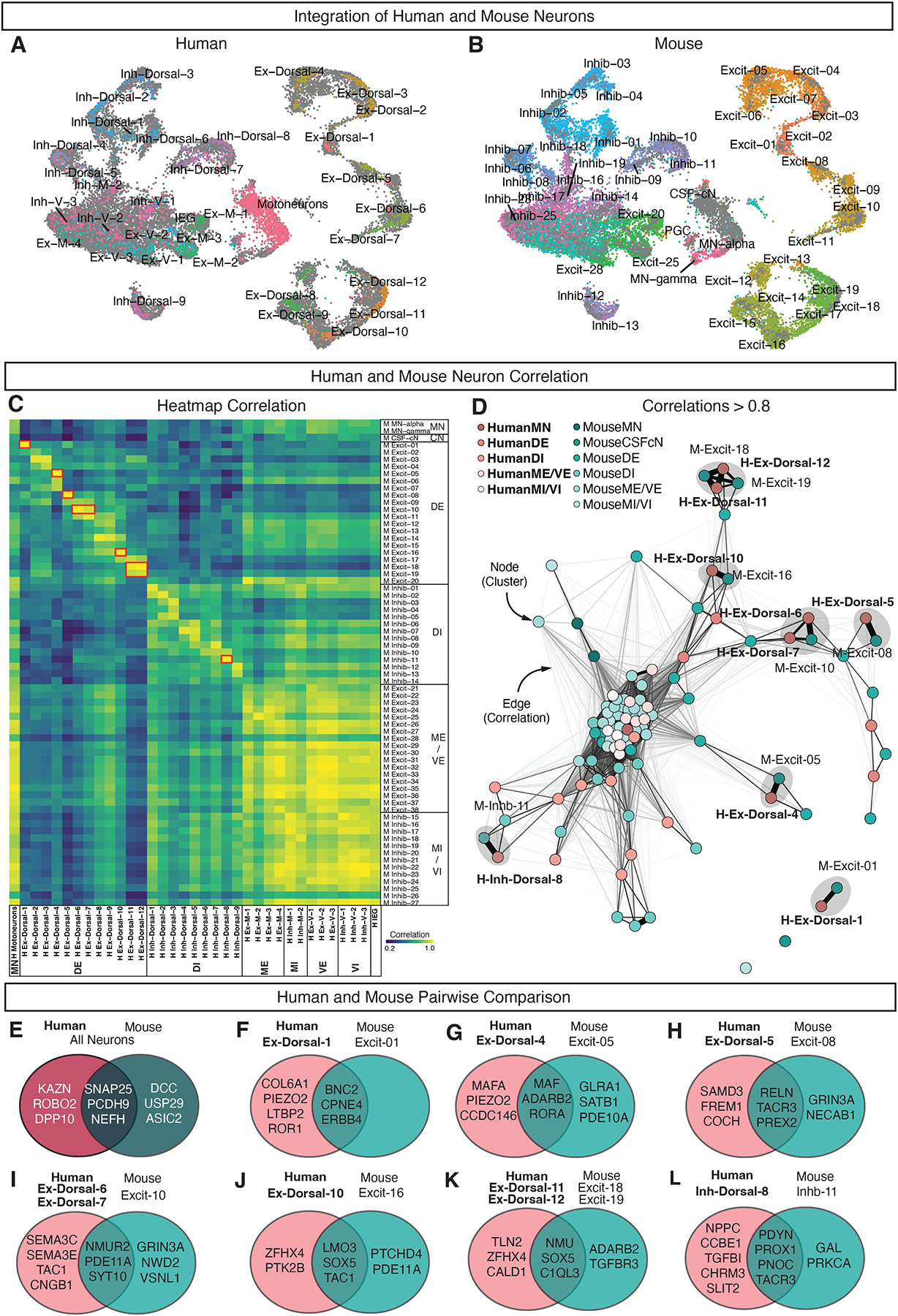
A-B, UMAP plots of integrated human and mouse spinal neuron data sets, colored by clusters from (A) human data set and (B) mouse data set (Russ et al.19). C, Heatmap of correlations values (Pearson’s R) between human clusters (columns) and mouse clusters (rows). Correlations were calculated using the top 2,000 highly variable genes from the integrated mouse-human data set. Red boxes highlight 7 pairs of clusters shown in E-L. Human clusters are bolded and mouse clusters are in regular font. D, Quotient graph showing neuronal clusters as nodes connected by edges. Edges represent correlations greater than 0.8 between human and mouse neuronal clusters. Edge thickness and length reflect correlation values, with greater correlations having thicker and shorter edges. Human clusters (bold, pink) and mouse clusters (teal) are shown. Grey circles highlight 7 pairs of clusters shown in E-L. E-L, Venn diagrams represent differentially expressed genes using the Wilcoxon Rank Sum test. The overlap in the two circles represents top genes enriched in both of the selected pair(s) of human and mouse neurons compared to all other human and mouse neurons, while human or mouse top enriched genes are shown in pink or teal circles, respectively. No differentially expressed genes were found for Mouse Excit-01. See also Supplemental Figures S15–S17.
As a resource, this cellular and gene expression atlas of the adult human spinal cord makes it possible to register known molecular correlates of pathophysiology with human spinal cell types. For example, GWAS studies have identified a number of human genomic loci associated with chronic pain26–28. We examined cell-type specific expression of the ten genes near loci that were significant in at least two independent cohorts for GWAS analysis (see Methods). Of these, CCDC26 was strongly enriched in microglia and macrophages while DCC, NOXA1, and SPOCK2 were enriched across the neuronal populations (Supplemental Fig. S16 and S17). Intriguingly, SOX5 was found in astrocytes, vascular cells, and in a group of related neuronal cell types (ExDorsal-8 through Ex-Dorsal-12) that are excellent candidates for a role in pathogenic mechanisms of chronic pain. Subsets of these dorsal excitatory peptidergic neurons selectively expressed the TAC1, TAC3, and CALCB genes for pain-related neuropeptides substance P and CGRP; they were enriched for the opioid receptor gene OPRK1, and they selectively expressed the opioid receptor gene OPRD1 (Supplemental Fig. S17). In addition, these cell types were putative homologues of mouse cell types Excit-14 through Excit-19 (Fig. 5C), which have been shown to play functional roles in mechanical nociception and pain coping mechanisms22,29–33. These data link population genetic studies with transcriptional profiling to propose specific cell types as central regulators of chronic pain in human patients.
Human motoneurons are defined by genes related to cell structure, cell size, and ALS
We next sought to use this cellular and molecular resource to study the gene expression profile of human motoneurons, and to determine whether their molecular repertoire relates to their selective vulnerability in diseases such as ALS. The human motoneuron cluster could not be divided into more refined discrete subtypes. This may reflect technical limits (these nuclei contained a relatively low number of genes per nucleus) or biological continua amongst adult human motoneuron features. Coclustering with mouse MNs from previously published datasets suggested a division into alpha/beta and gamma sub-types, but these were not clearly separated by human marker genes (Supplemental Fig. S24A–C). As a result, we analyzed human motoneurons as one group.
We examined the top 50 marker genes that distinguished the motoneuron cluster from other human spinal neurons (Fig. 6A). To determine whether these genes were enriched in motoneurons in spinal cord tissue, we assessed the distribution of the entire predicted gene signature in our spatial transcriptomics dataset. While a few genes may reflect background contamination (e.g. PLP1), we found that the predicted motoneuron Cell2Location distribution and the top motoneuron marker genes were strongly enriched in lamina IX in the ventral horn, confirming the overall expression pattern (Supplemental Fig. S18A). Motoneuron markers included those involved in acetylcholine synthesis and function (SLC5A7 and ACLY), as expected, but surprisingly were dominated by three partially overlapping sets of genes: (1) those involved in cytoskeletal structure, (2) neurofilament genes related to cell size, and (3) genes directly implicated in ALS pathogenesis (Fig. 6A).
Fig. 6. Human motoneurons are characterized by genes associated with ALS, cell structure, and increased cell size.
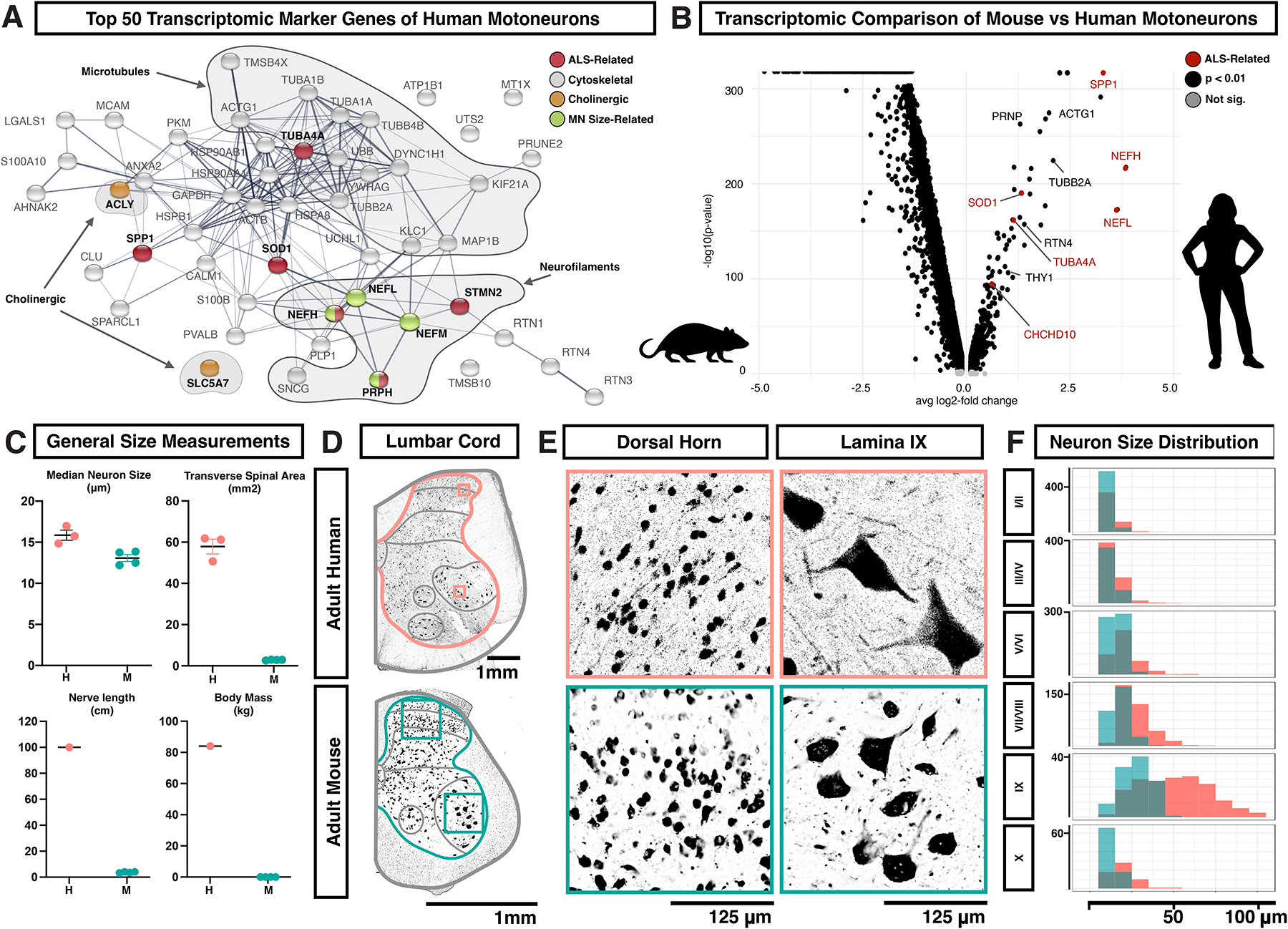
A, Association network plot constructed using the String protein database for the top 50 marker genes of human motoneurons, with selected categories highlighted (cholinergic transmission, orange; ALS, red; genes whose over-expression in mice causes enlargement and/or degeneration of motoneurons, green; cytoskeletal components, gray). B, Volcano plot showing genes enriched in either lumbar motoneurons from adult mice or lumbar motoneurons from adult humans. Genes are plotted by the average change in expression (avg log2-fold change) and by the statistical strength of the difference (-log10(p-value)) with significant genes in black and significant ALS-related genes in red. C, Gross anatomical and neuronal measurements of the human (H) and mouse (M) lumbar spinal cords, including median neuron size (μm), transverse area of the spinal cord (mm2), maximum nerve length (cm), and body mass (kg). D, Transverse sections of one side of the adult lumbar human (above) and mouse (below) spinal cords, with antibody labeling for NeuN. Images are representative of data from three subjects. Scale bars are 1 mm. Boxes indicate the regions shown in panel E. Gray lines indicate the laminar/regional boundaries used in panel F. E, Higher magnification view of NeuN labeled spinal neurons from panel D in the human (above) and mouse (below). The left-side images are from the dorsal horn and the right-side images are of putative motoneurons in lamina IX. Scale bars are 125 μm. F, Histogram showing the count distribution of neuron Feret distance in human (pink) and mouse (teal) across the different lamina regions of the adult lumbar spinal cord. Measurements are given in μm and the count scale is shown at the right of each plot. Bonferroni-adjusted Wilcoxon Rank Sum test p-values and Bhattacharyya Coefficients (BC) for human vs mouse distributions are as follows. I/II: p=7.5e-27, BC=0.93, III/IV: p=4.0e-12, BC=0.96, V/VI: p=3.2e-30, BC=0.89, VII/VIII: p=5.7e-49, BC=0.80, IX: p=1.6e-19, BC=0.71, X: p=9.5e-10, BC=0.92. See also Supplemental Figures S18–S23.
Cytoskeletal components were the most abundant category of motoneuron marker gene and the most enriched gene ontology (GO) terms, including GO annotation clusters related to microtubules (p=0.000009) and axon structure and neurofilaments (p=0.000018) (Supplemental Table S5). The marker genes that were structural components of neurofilaments (NEFL, NEFM, NEFH, and PRPH) have been directly linked to cell size, axon diameter, and degeneration34–39, providing a potential link between human motoneuron gene expression and cellular phenotype. Amongst ALS-related motoneuron marker genes, there were both cytoskeletal genes (NEFH, PRPH, TUBA4A, and STMN2), as well as genes that are not directly linked to cellular structure (SOD1, OPTN, and SPP1). Most of these markers showed enriched expression in lamina IX in the spatial transcriptomics dataset (as examples, Supplemental Fig. S18A).
We further examined the expression of a panel of ALS-related genes compiled from the literature40–47 across human spinal cord cell types. In addition to the genes above, CHCHD10 and KIF5A were enriched in spinal motoneurons, extending this signature profile (Supplemental Fig. S18B, S19, S20). We also observed enriched expression of SPP1, FUS, and C9ORF72 in microglia and STMN2, and TUBA4A in an excitatory midpopulation (Ex-M-1, Supplemental Fig. S18B, S19, S20). TARDBP was not detected at sufficient levels in the dataset to characterize its expression pattern. We next assessed the cell type distribution of co-varying gene modules of recently implicated in ALS pathogenesis based on spatial transcriptomic data48. In agreement with our findings above, the main module associated with disease progression in human tissue were enriched in microglia and motoneurons in our dataset (Supplemental Fig. S21). Given that the enriched expression of neurodegeneration-associated genes in human motoneuron transcriptomics may have been due to the age of the study donors, we also examined expression of ALS-related genes in a dataset of human embryonic spinal cord cell types13. We found patterns including low levels of gene expression (i.e. NEFH and TUBA4A), moderate but broad cell type expression (i.e. OPTN and PRPH), or high and ubiquitous cell type expression (i.e. SOD1 and STMN2) (Supplemental Fig. S22). Thus, the overall enrichment of ALS-related genes in human motoneurons was not apparent in newly formed motoneurons, but likely emerge at some point during motoneuron maturation or aging. To test whether this expression profile reflected a nonspecific enrichment of degeneration-associated genes in human motoneurons with age, we compared the expression of genes for multiple neurodegenerative diseases, including those with age-related associations, across human spinal cord cell types. This analysis revealed a specific association of ALS-related gene expression in human motoneurons (Supplemental Fig. S23)
To determine whether ALS-related genes are also enriched in motoneurons in mice, the major animal model for studying the genetic basis of neurodegenerative disease, we compared the human data to prior snRNA-seq data from lumbar skeletal motoneurons from adult mice4. We found that prominent ALS-related genes were enriched and were expressed at higher levels specifically in the human motoneurons as compared to mouse motoneurons (Fig. 6B and Supplemental Fig. S24C). To determine if this enrichment is unique to motoneurons, we examined the analysis of a recent study on conservation in human brain gene expression patterns49 and found that three genes of interest (SOD1, TUBA4A, OPTN) had a significantly higher mean human-to-mouse divergence score than other assayed genes (mean score of 0.587 ± 0.19 versus 1,426 other genes with mean 0.320 ± 0.123, p=0.0002).
Cell Size and Protein Expression in Human Lumbar Motoneurons
Why might human motoneurons be defined by genes related to cell size and structure, compared to other neurons? It is well established that human motoneurons are large, but to further investigate relative size differences, we analyzed neuron soma size across all laminae in human and mouse lumbar spinal cord tissue. Given the obvious differences in overall body size and anatomy, we expected that most classes of human neurons would be larger than mouse neurons. Surprisingly, human and mouse lumbar spinal neurons were approximately the same size, with a median Feret diameter (maximal caliper length) of 16.02 and 13.13 μm, respectively (human mean 20.3 ± 0.28 s.e.m; mouse mean 14.28 ± 0.12 s.e.m.) (Fig. 6C, Supplemental Fig. S24D–F, and Supplemental Table S6). By contrast, human lamina IX spinal neurons were approximately 2-fold larger than those in mouse and could be up to ~120 μm across compared to ~50 μm in mouse (Fig. 6D–F, Supplemental Fig. S24D–F, and Supplemental Table S6). These measurements are consistent with those previously reported for human and mouse spinal motoneuron soma50–52 and the same proportion that has been observed for human and mouse motoneuron axon caliber53–55. Assuming that human alpha motoneurons are within the higher end of this size distribution, then they are (1) much larger than other human spinal neurons, (2) increased in scale relative to mouse motoneurons, and (3) among the among the largest vertebrate neurons, including elephant motoneurons (~85 μm)56, human Betz corticospinal neurons (~60–100 μm)57, subsets of human dorsal root ganglion neurons (up to 100 μm)58 and salmon Mauthner cells (~87 μm)59. This notable size of human motoneurons may explain the specialized gene expression signature that we observed in this subclass of neurons.
To assess specific ALS-related features in tissue and in situ cell size, we next analyzed the protein expression of six ALS-related genes in post-mortem lumbar spinal cord from four donors using immunofluorescence. We found that neurons expressing NEFH, OPTN, PRPH, STMN2, and TUBA4A proteins were all enriched within the motoneuron region (lamina IX) of the lumbar spinal cord, with limited positive cells in other regions except for scattered, large cells in lamina III/IV of the dorsal horn (which may be projection neurons) and smaller neurons in medial lamina VII (Fig. 7A–D and Supplemental Table S7). SOD1 was present in lamina IX and throughout the spinal cord in a distinct peri-nuclear distribution, in contrast to the enriched RNA expression that we detected by snRNA-seq and by spatial transcriptomics. To ensure the accuracy of the SOD1 protein expression pattern, we validated the SOD1 antibody through targeted knockdown in human iPS neurons (Supplemental Fig. S25D). Overall, these data confirm the enriched expression of ALS-related proteins in human spinal motoneurons in tissue60,61.
Fig. 7. ALS-related proteins are enriched in human motoneurons.
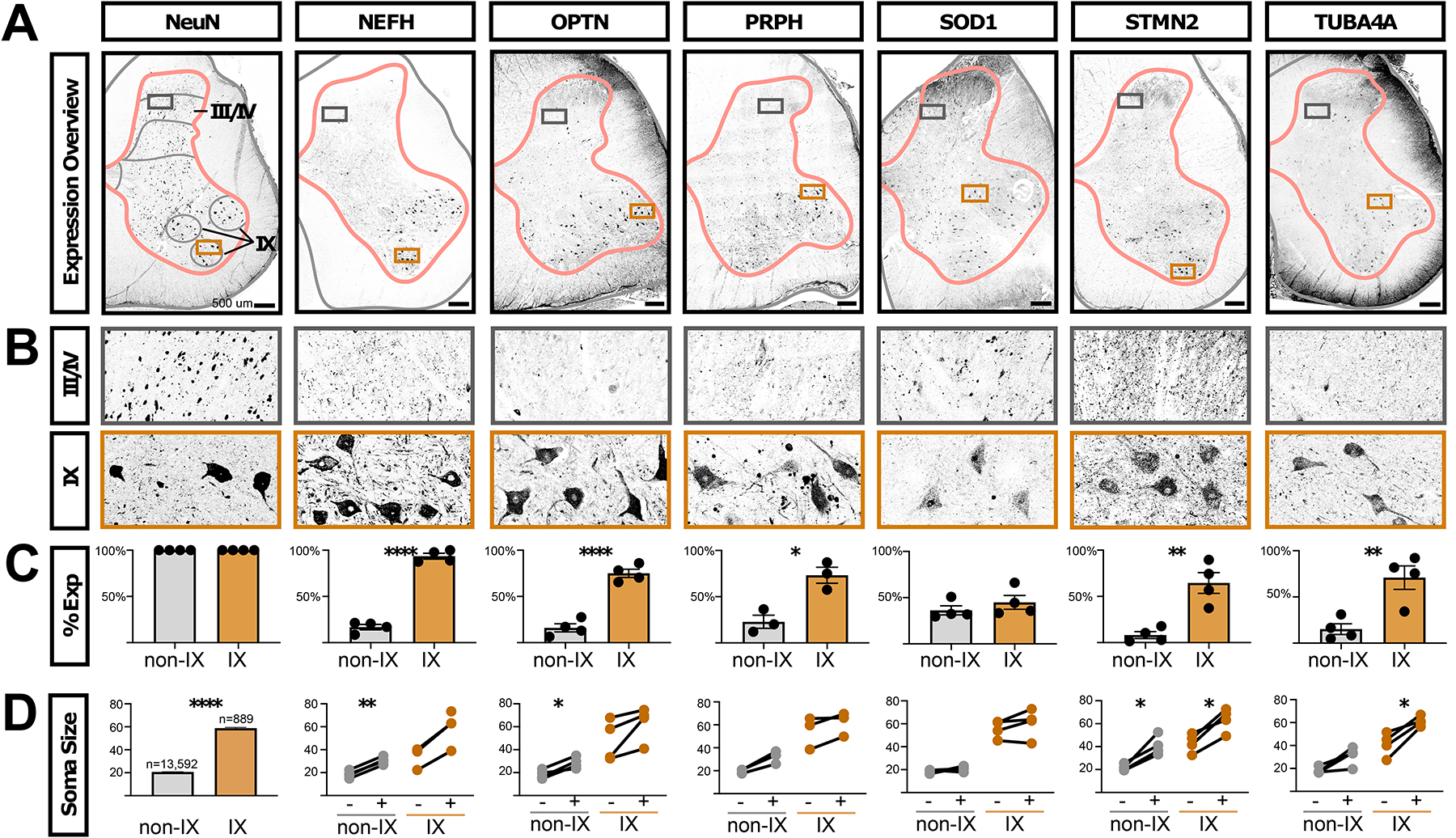
A, Antibody staining on adult human lumbar spinal cord against NeuN (RBFOX3 gene, general neural marker) and the ALS-related genes NEFH, OPTN, PRPH, SOD1, STMN2, and TUBA4A. Gray matter outlines are shown in pink and boundaries of lamina I/II, III/IV, V/VI, VII/VIII, IX, and X are shown in gray. Boxes indicate the enlarged images in panel. A, Images are representative of data from three subjects (two male and one female). Scale bars are 500 μm. B, Inset of the images in panel A, from the boxed region in laminae III/IV or lamina IX. The width of the insets is 500 μm. C, Quantification of the percent of NeuN+ neurons that co-expressed the indicated proteins in either all neurons not in lamina IX (non-IX) or those in lamina IX. The mean ± s.e.m. are shown. The plotted values and number of cells counted in each subject and category are available in Supplemental Table S7). Paired t-test results are shown where * indicates p < 0.05, ** indicates p < 0.005, **** indicates p < 0.0001. D, The sizes of NeuN+ neurons are shown for each indicated protein. For NeuN, 100% of cells were positive, by definition, and the total counts and sizes (mean ± s.e.m.) are shown for neurons not in lamina IX (non-IX) or those in lamina IX. For all other indicated proteins, the Feret distance sizes are shown for all neurons that did not (−) or did (+) express the indicated protein (mean Feret distance in μm). Each line joins values within one subject. There is an unpaired value for NEFH because we did not detect neurons in lamina IX that did not express NEFH. The plotted values and number of cells measured in each subject and category are available in Supplemental Table S7. Paired two-tailed t-test p-values, after Benjamini-Hochberg FDR correction, are shown where * indicates p < 0.05, ** indicates p < 0.005. **** indicates p < 0.0001. See also Supplemental Figures S24 and S25.
We also studied the expression of these proteins in the mouse spinal cord, using lumbar tissue from aged animals (11 months old) to approximate the advanced age of the human subjects in this study. We found that Nefh, Optn, Prph, Stmn2, and Tuba4a displayed enrichment in lamina IX, while Sod1 was expressed ubiquitously, similar to what has been previously described for Sod1 in mice (Supplemental Fig. S25A–C)61. Together with the comparative transcriptomic analysis above, this suggests that while human and mouse motoneurons are both enriched for expression of ALS-related genes, in human motoneurons the relative expression levels are higher and the enrichment of these genes as motoneuron-specific markers is greater.
Finally, we examined the relationship between expression of ALS-related genes and human spinal neurons size in tissue. We measured the Feret distances of human neurons expressing each ALS-related protein in comparison with non-expressing neurons. We found that neurons that expressed NEFH, OPTN, PRPH, STMN2, and TUBA4A were generally larger than non-expressing neurons, both within the motoneuron region of lamina IX and in other laminae (Fig. 7D, Supplemental Table S7). Within lamina IX, this likely reflects enrichment within the larger alpha motoneurons (versus gamma) and in other laminae, this may reflect expression within spinocerebellar projection neurons that degenerate in ALS62,63 or other large cell classes. Importantly, we found that the very largest lamina IX neurons – known to be most susceptible to degeneration in ALS2,53,54,64 – were the most likely to express these markers. For lamina IX neurons with a Feret distance greater than 70 μm, on average 100% expressed NEFH, 81% expressed OPTN, 88% expressed PRPH, 60% expressed SOD1, 90% expressed STMN2, and 95% expressed TUBA4A (Supplemental Table S7). These data further link motoneuron size and vulnerability to these cytoskeletal genes that have causative roles in motoneuron size and human disease.
DISCUSSION
The advent of single cell transcriptomic profiling approaches has transformed biology, with the potential to pinpoint therapeutic targets amidst the complexity of human disease. However, we still lack a comprehensive characterization of the human spinal cord that could provide crucial insights into chronic pain, spinal cord injury, and neurodegeneration. Here, we used snRNA-seq and spatial transcriptomics to create a cellular taxonomy of the adult human lumbar spinal cord. We identified dozens of cell types, including diverse glial and neuronal populations, and characterized their molecular repertoires and putative locations. We next used this atlas to examine cell-type specific mechanisms of pathophysiology, identifying a group of human dorsal horn neurons enriched for pain-related genes, and a specialized molecular signature in human motoneurons that links their extreme cell size with their vulnerability to degeneration in ALS. This atlas and an accompanying web-based resource (https://vmenon.shinyapps.io/humanspinalcord/) can serve as tools for further understanding human spinal cord biology.
There have been several recent studies on the molecular and cellular heterogeneity in the human spinal cord, particularly during development. Rayon and colleagues13, focused on first trimester spinal cord derived from four human embryos, identified diverse progenitor and neuronal populations, and performed a systematic comparison with the spinal cord cell types of the developing mouse spinal cord. Zhang and colleagues14 profiled the early and mid-stages of fetal development with an important focus on glial development and cell-cell communication. For the adult human spinal cord, Zhang and colleagues performed snRNA-seq on the spinal cord from two donors and identified coarse glial and neuronal cell types15. However, they did not characterize human neurons to the same degree as this study, especially with respect to motoneurons, nor did they validate predicted gene expression patterns in tissue. Here, we establish the first comprehensive taxonomy of the adult human spinal cord. With this broad view and comparison with similar work in mice, we found that the major axes of spinal neuron diversity are conserved across both species. In addition to neurotransmitter status, the primary factor in spinal neuron organization is dorsal-ventral location, with dorsal neurons forming robust and distinct clusters that each display specific marker genes while ventral clusters showed overlapping gene expression patterns. This is similar to what we and others have previously shown in adult mice16,17, but seems to be at odds with our knowledge of the many refined populations of ventral neurons within the cardinal V0, V1, V2, and V3 embryonic lineage domains65. There are multiple potential explanations for this discrepancy. First, it is possible that the dorsal-ventral pattern reflects multiple axes of neural diversity overlaid onto ventral neurons, effectively blurring the distinctions that would be apparent along any single axis. In addition to developmental lineage, it has recently been shown that transcriptional signatures of birthdate sub-divide spinal neurons during mouse and human development13,20,66–68. This includes a continuum of very early-born (ONECUT2), early-born (ZFHX3/4), and late-born (NFIB/NFIA/NEUROD2/6) factors and is most apparent amongst cell types that settle in the mid and ventral regions of the spinal cord. As birthdate is coupled to projection neuron versus local interneuron identity20, this level of transcriptional diversity may be sustained into adult stages to support cell-type specific functional requirements based on axon length. Together with other parameters, such as location within the ventral horn or electrophysiological specialization, these features may overlap each other to form broad ventral clusters that are less distinct from each other. Alternatively, it is possible that dorsal and ventral neurons in the adult require differential levels of ongoing gene expression related to their functions. Perhaps dorsal neurons (which express specific neuropeptides, neuropeptide receptors, and other genes involved in neuronal function) require specific transcriptional signatures to be sustained to perform more specialized computations. In contrast, ventral neurons may operate mainly based on their connectivity within the network and can therefore downregulate their lineage-based and embryonic molecular diversity once axon guidance has occurred and circuit structure is complete. These neurons may operate as a broad network whose tasks are carried out through global dynamic population activity69. In the future, relating transcriptional identity to connectivity, intrinsic electrophysiological parameters, and neural activity will help to resolve how spinal neural populations are organized for function.
An intriguing finding from our analysis is the enrichment of cytoskeletal gene expression in human motoneurons. All cells require a functional cytoskeleton, raising the question of why spinal motoneurons are particularly dependent on the proper expression and function of cytoskeletal-related genes. Interestingly, neurofilament genes that were enriched in human spinal motoneurons are precisely those structural components that drive increased axon caliber and cell size55,70–72. Over-expression of mouse NEFL, human NEFM, human NEFH, or mouse PRPH in transgenic mice can each cause enlargement and swellings of motoneuron somas and subsequent axon degeneration34,39, linking human motoneuron gene expression and potential degenerative phenotypes. Relatedly, these neurofilament genes are found in other large neurons in the nervous system, suggesting that they may be part of a common signature that permits increased cell size60,73–76. Large soma size and axon caliber may be required to sustain extensive dendritic trees and axons up to a meter long, to support cell energetics, or for firing rate and conduction parameters77–79. These large cells then rely critically on this protein network and are selectively vulnerable to its abnormal function. Human motoneurons were also distinguished by expression of the microtubule stability factors TUBA4A and STMN245,80, potentially highlighting a requirement for structural support in these peripherally projecting cells with long axons. Overall, these findings support a model of specific molecular repertoires for motoneuron cell structure that also confer selective vulnerability to degeneration43,80,81.
Although we captured all major cell types and most known subclasses of cells in this work, we foresee further advances as additional data sets of this type arise. Among motoneurons, we expected to observe well-established “alpha” and “gamma” subtypes based on transcriptional profiles recently described in mice but did not. This limitation may be experimental, reflecting the relatively low number of genes detected per motoneuron nucleus. In addition, we did not observe all known populations of neurons, such as cerebrospinal contacting neurons (CSF-cN), which represent less than one percent of mouse spinal neurons19. As technological advances allow for higher sensitivity transcriptomics on larger numbers of cells or improvements in in situ profiling methods, a clearer picture of refined neuronal populations and the heterogeneity within motoneurons will likely become apparent.
Overall, it is important to consider the spinal cord as a community of cell types that function together in normal health and disease. Here, we highlighted specific findings on a proliferative population of adult microglia, a group of dorsal excitatory neurons enriched for pain-related genes, and the molecular signature of motoneurons. But this work provides a comprehensive resource for transcriptional profiling of the dozens of cell types that make up the adult human lumbar spinal cord. As such, it will allow researchers to parse how genetic alterations could affect diverse cell-type specific molecular profiles in disease; how particular populations may respond to target molecular interventions and pharmacology; and how human spinal cell types may interact with each other through cell-cell signaling pathways. Thus, we hope this work, together with other ongoing efforts, will serve as a foundation for studying the wide range of cell types involved in human spinal cord function.
STAR★METHODS
Resource Availability
Lead Contact
Further information and requests for resources, reagents, or code should be directed to and will be fulfilled by the lead contact Ariel Levine (ariel.levine@nih.gov).
Materials Availability
The study did not generate new unique reagents.
Data and code availability
Anonymized raw sequencing data and counts tables are deposited in the Gene Expression Omnibus (GEO) with accession numbers GSE190442 and GSE222322 with associated metadata in Supplemental Table S2. In addition, visualization of expression data at the cluster and donor level are available through a searchable web resource at https://vmenon.shinyapps.io/humanspinalcord/.
Code for the evolutionary divergence analysis can be found at https://colab.research.google.com/drive/19Ty97LOwT3AmaVCJGKA8BXXNFYH_iSZ4?usp=sharing and https://colab.research.google.com/drive/19Ty97LOwT3AmaVCJGKA8BXXNFYH_iSZ4?usp=sharing Custom MATLAB-based code for quantification of cell counts is available at https://github.com/ArielLevineLabNINDS/CellCounter (https://doi.org/10.5281/zenodo.6967482).
Experimental Model and Subject Details
Lumbar spinal cords were obtained from neurologic determination of death organ-donor patients (all demographic data is listed in Supplemental Table S1) under the approval of the French institution for organ transplantation (Agence de la Biomédecine) or the Ottawa Health Science Network Research Ethics Board, following the template provided by the University of Ottawa and the Tri-Council Policy Statement Guidelines. Both approvals imply consent for using anonymized donor genetic information. Patients with neurological disease or major infections were excluded from the study. For mouse experiments, all mice were of 50:50 mixed background from strains C57BL/6J and BALB/CJ, housed in standard conditions. For basic anatomical experiments, two male and two female mice of approximately 24 weeks old were used. For ALS marker gene expression studies, two male and one female mice of approximately 11 months old were used. All procedures and experiments were approved by the Animal Care and Use Committee of NINDS (protocol #1384).
Method Details
Human spinal cord acquisition and preparation.
Human lumbar spinal cords were retrieved under chilled body and neuroprotective conditions as described previously82–84. The extraction procedure took 20–40 minutes and was done within three hours of cessation of circulation by aortic cross-clamp. Lumbar spinal cord tissue was flash frozen on liquid nitrogen in the operating room and stored at −80°C until nuclei isolation.For immunohistochemistry experiments, lumbar spinal cord tissue was isolated from organ-donor patients (~55–65 years old, 3 men, 1 woman). The tissue was immediately fixed in 4% paraformaldehyde for 24–48 hours, then washed in PBS, and placed in 30% sucrose for 2–4 days at 4°C before being embedded in OCT medium for sectioning. For Visium spatial transcriptomics, postmortem lumbar spinal cord from a non-neurological control subject (~75 years old, male) was acquired from the Target ALS Multicenter Postmortem Core as part of the New York Genome Center (NYGC) Amyotrophic Lateral Sclerosis (ALS) Consortium. Informed consent is acquired by each Target ALS member site through its own institutional review board (IRB) protocol and samples are transferred to the NYGC in accordance with all applicable foreign, domestic, federal, state, and local laws and regulations for processing, sequencing and analysis. The Biomedical Research Alliance of New York (BRANY) IRB serves as the central ethics oversight body for the NYGC ALS Consortium. Ethical approval for this study was given by the BRANY IRB.
Mouse work and spinal cord acquisition.
All procedures and experiments were approved by the Animal Care and Use Committee of NINDS (protocol #1384). Adult mice were of 50:50 mixed background from strains C57BL/6J and BALB/CJ, housed in standard conditions. For basic anatomical experiments, two male and two female mice of approximately 24 weeks old were used. For ALS marker gene expression studies, two male and one female mice of approximately 11 months old were used. To obtain spinal cord tissue, anesthetized mice were transcardially perfused with PBS followed by cold 4% paraformaldehyde (PFA). The spinal cords were harvested and post-fixed in cold 4% PFA overnight at 4°C, cryoprotected by immersion in 30% sucrose overnight at 4°C and embedded in OCT medium for sectioning.
Nuclei isolation.
Nuclei were isolated from fresh frozen human spinal cords using a triton-based protocol85. Briefly, after removing the dura, half a segment of spinal cord was placed in a Dounce homogenizer (Kontes Dounce Tissue Grinder) containing 500 μL of lysis buffer (0.32 M sucrose, 10 mM HEPES [pH 8.0], 5 mM CaCl2, 3 mM 586 MgAc, 0.1 mM ETDA, 1 mM DTT, 0.1% Triton X-100). After douncing with 5 strokes of pestle A and 5–10 strokes of pestle B, the lysate was diluted in 3 mL of sucrose buffer (0.32 M sucrose, 10 mM 588 HEPES [pH 8.0], 5 mM CaCl2, 3 mM MgAc, 0.1 mM ETDA, 1 mM DTT) and passed over a 70 μm strainer. The filtered lysate was centrifuged at 3,200 × g for 5 min at 4°C. After centrifugation, the pellet was resuspended in 3 mL of sucrose buffer and centrifuged again at 3,200 × g for 5 min at 4°C. After centrifugation, the pellet was resuspended in 3 mL sucrose buffer and incubated for 2 min on ice. The sample was transferred to an Oak Ridge tube and homogenized for 1 min using an Ultra-Turrax Homogenizer (IKA). Then, 12.5 mL of density sucrose buffer (1 M sucrose, 10 mM HEPES [pH 8.0], 3 mM MgAc, 1 mM DTT) was layered below the sample. The tube was centrifuged at 3,200 × g for 20 min and the supernatant immediately poured off. The nuclei on the side of the tube were resuspended with 100 μL of PBS with 0.04% BSA and 0.2 U/μL RNase inhibitor. Nuclei were inspected for visual appearance and quantified with a hemocytometer before proceeding with nuclei capture and sequencing.
Single-nucleus RNA sequencing.
Single nucleus RNA sequencing was carried out using Single-cell gene expression 3’ v3 kit on the Chromium platform (10X Genomics) according to manufacturer’s instructions with one modification. Following reversetranscription, an additional PCR cycle was added to the number of cycles for cDNA amplification to compensate for decreased cDNA abundance in nuclei compared to cells. Libraries were sequenced to a minimum depth of 20,000 reads per nucleus using an Illumina HiSeq 3000 (PE 26 – 8 – 98 bp). Raw sequencing reads were demultiplexed, aligned, and a count matrix was generated using CellRanger. For alignment, introns and exons were included in the reference genome (GRCh38).
Quality check analysis.
All the 10x runs for each human sample were initially filtered with an nUMI cutoff of >1000 and then nuclei with less than 5% mitochondrial gene contamination were retained. Next, the mitochondrial genes were also removed from the matrices. A total of 55,289 nuclei that passed quality control filtering, with mean detection of 2,187 genes per nucleus (Supplemental Fig. S1)
Top level UMAP and clustering.
The 7 human datasets were integrated using SCTransform normalization followed by CCA based integration using the Seurat 4.086,87 package. The integrated data sets were then jointly analyzed to identify the optimal number of principal components for downstream analysis, based on the ElbowPlot and PCheatmaps function. The number of PCs was set to 30 for clustering and UMAP visualization. The clusters, obtained using a value of 0.6 for Seurat’s resolution parameter, were then manually annotated based on the expression of marker genes for neurons, astrocytes, microglia, oligodendrocytes, OPCs, endothelial cells, pericytes, meningeal cells, Schwann cells, and lymphocytes.
Subclustering of major cell types.
Identification of subclusters within cell types was performed separately for three major cell types (neurons, microglia, astrocytes), with the rest being subclustered in two additional groups (Group 1- oligodendrocytes, OPCs, and Schwann cells; Group 2- endothelial cells, pericytes, meningeal cells, and lymphocytes). For each cell type/group, the subclustering was done in multiple rounds until no putative transcriptomic doublets or contamination of other cell types was observed, as described below.
For subclustering of major cell types, the raw counts were aggregated from all 7 datasets for each cell type, and then re-normalized (using log normalization) and scaled in order to prepare for integration. The integration of 7 datasets belonging to a particular cell type was performed based on the CCA-integration workflow from the Seurat 4.0 package. The optimal number of PCs was selected based on the ElbowPlot and PCheatmaps function for each cell type, in order to be used for subclustering and preparation of UMAP visualizations. Multiple resolutions were interrogated, depending on cell type, ranging from values of 0.08 to 3.
During each round, putative transcriptomic doublet clusters and contamination of other cell types was removed (based on co-expression of multiple major class genes) and the above steps were performed again. Doublets were identified by clusters that expressed markers for more than one cell type. All clusters were checked for doublets by their markers using the wilcox (Wilcoxon Rank Sum Test) and auroc (Area under the ROC curve) functions, as well as visually using the FeatureScatter option in Seurat.
Subclustering of neurons.
Neurons were clustered in 2 stages. During the first stage, log-normalization of raw counts and scaling (including regressing out the number of transcripts and the percentage of mitochondrial transcripts) of each dataset were performed, followed by integration based on the same steps described above. All highquality neuronal nuclei were clustered in an unsupervised manner testing a range of resolutions. This stage led to some clusters that were clearly distinct, together with a large group of nuclei (visualized in the central region of the UMAP) that did not subdivide well at lower resolutions, but were dispersed at higher resolutions. This prompted us to perform a focused re-clustering of this non-distinct group with new principal components based only on these nuclei. The spatial distribution of marker genes for this large group suggested a ventral identity, so we included all putative mid/ventral clusters into this step of the analysis. Raw counts were again extracted and normalized using SCTransform (to avoid dataset size related limitations), followed by the standard integration workflow in the Seurat 4.0 package. We also performed a focused sub-clustering of the motoneurons at this stage but could not identify robust sub-clusters within this cell type. During each clustering stage, cluster-specific genes were identified based on Wilcoxon Rank-Sum test and AUROC analysis within the FindMarkers function from Seurat 4.0. Based on these genes, distinct subpopulations based on expression of candidate markers were manually annotated.
In order to obtain a refined set of neuronal subpopulations, all the subclusters were interrogated for low gene detection, doublets, and other contamination from non-neuronal genes, and were subsequently removed from the analysis. All the refined clusters were then re-integrated to prepare a combined neuronal UMAP and mapped with refined subcluster annotations. Finally, we then analyzed each cluster independently to assign a “dorsal”, “mid”, “ventral” identity, guided by spatial distribution of marker genes, the predicted cell2location distribution of the cell type in spatial transcriptomics data, and relative similarity to previously described mouse or macaque cell types.
Cluster robustness assessment and silhouette scores.
We used two approaches to assess cluster robustness: a post-hoc machine learning-based classification approach, and a silhouette score approach.
For the post-hoc machine learning approach, we built a random forest classifier for every pair of neuronal clusters, trained on 80% of the nuclei. This classifier was then used to assign cluster membership for the remaining 20% of the cells, and the entire process repeated such that each cell in every pairwise cluster comparison was classified 100 times. A cell that was classified into its original cluster <90 times was deemed “misclassified”. For every pair of clusters, we then calculated the mean percentage of cells that were misclassified among the two clusters to generate pairwise cluster robustness scores. For visualization as a constellation diagram, we only connected cluster pairs with minimum misclassification percentage >3%, representing their connections with the mean misclassification percentage.
For silhouette score evaluation, we used the ‘silhouette’ function from the ‘cluster’ library in R (https://cran.r-project.org/web/packages/cluster/index.html), where the Euclidian distance matrix based on the first 25 PCs was used as input, together with the neuronal cell type annotations.
Tissue processing, Visium data generation, and Visium data preprocessing.
Frozen postmortem lumbar spinal cord from 5 non-neurological control subjects were embedded in Tissue Plus OCT Compound (Fisher Healthcare, catalog no. 4585) and cryosectioned at −16°C. Sections of 10 μm thickness were collected onto prechilled Visium Spatial Gene Expression Slides (10x Genomics, catalog no. 1000185) by warming the back of the slide to adhere the tissue. Four technical replicates for each subject were collected approximately 30 mm apart across two Visium slides that were processed in parallel to minimize technical batch effects. Where necessary due to tissue sectioning artifacts, additional technical replicates were collected and processed on a later date. All technical replicates for each subject were sequenced on the same flow cell.
Visium spatially resolved gene expression data was generated according to the Visium Spatial Gene Expression User Guide (10x Genomics, CG000239 Rev F). Briefly, tissue sections were fixed in chilled methanol and stained using hematoxylin and eosin. Brightfield histological images were acquired using an EC Plan-Neofluar 10x/0.3 M27 objective on a Zeiss Axio Observer Z1 fitted with a Zeiss Axiocam 506 mono (Carl Zeiss Microscopy, Germany). Raw CZI images were stitched using Zen 2012 (blue edition) (Carl Zeiss Microscopy, Germany) and exported as JPEGs. Tissue sections were permeabilized for 12 minutes which was selected as the optimal time based on tissue permeabilization time course experiments conducted using the Visium tissue optimization protocol. cDNA libraries were prepared and quantified according to the Visium Spatial Gene Expression User Guide (10x Genomics, CG000239 Rev F) and pooled at a concentration of 10 nM for sequencing.
Pooled spatial gene expression libraries were loaded at a concentration of 0.9 nM and sequenced on a NovaSeq 6000 System using a NovaSeq S4 Regent Kit v1.5 (200 cycles, Illumina, catalog no. 20027466) using the following recipe: read 1: 100 reads, i7 index read: 10 cycles, i5 index read: 10 cycles, read 2: 100 cycles. The average sequencing depth for each sample was approximately 200–280 × 106 reads.
Raw FASTQ files and histological images were processed using Space Ranger v.1.3.0, which uses a modified STAR v2.7.2a for genome alignment and performs barcode/UMI counting to generate feature-spot matrices. Reads were aligned to a GRCh38 reference genome filtered to exclude lncRNAs, pseudogenes and mitochondrially encoded genes.
Postprocessing and Computational analysis of Visium Spatial Transcriptomics data
Registration of Visium experiments was achieved using the OpenCV library (https://pypi.org/project/opencv-python-headless/). Briefly, histology images were annotated with 31 landmarks (14 points denoting features of the gray-white matter boundary for each side, and 3 points along the midline). These points were used to calculate a homography matrix from each experiment to a reference image using the findHomography function. Coordinates from each experiment were transformed to the resulting coordinate space using the perspectiveTransform function.
Cell type proportions comprising Visium experiments were estimated using the Cell2location package88 based on the following notebook (https://cell2location.readthedocs.io/en/latest/notebooks/cell2location_tutorial.html). The single-cell regression model was trained with the following parameters: max_epochs = 250 and lr = 0.002. The Cell2location model was initialized and trained with the following parameters: N_cells_per_location = 5, detection_alpha = 20, and max_epochs = 2000.
Cross-species analysis between human spinal cord vs mouse meta-analysis datasets.
Cross-species comparison between human and mouse meta-analysis19 spinal cord datasets were performed at two levels: 1. “Top-level”, which includes all major cell types and 2. Neurons only.
In both cases, the orthologous genes within mouse data matrix were converted to human homologs using biomaRt package89 from Bioconductor and in-house scripts. The raw counts from both human and mouse datasets were then split by different samples and then re-normalized, scaled and integrated. For the “top-level” analysis, SCTransform based integration was performed whereas for neurons only, log normalization-based integration was performed. Subsequently, UMAPs and correlation matrices were generated for further cross-species comparison of various cell types at top level and neuronal sub-clusters.
Cross-correlation of human and mouse cluster expression.
Cross-species cluster correlation measures were calculated from PCs in the integrated space (using 20 PCs for the top-level comparison of major cell classes), and Pearson correlation of the top 2,000 highly variable genes. Aggregate correlation values for each pair of clusters (one mouse, one human) were calculated as the mean correlation value across all human-mouse nuclei pairs from the respective clusters.
Quotient graphs using qgraph in R were used to show the correlations greater than 0.8 based on the top 2,000 highly variable genes between human and mouse spinal cord neurons (graph “cor”, layout “spring”).
GO analysis of human motoneuron marker genes.
The top markers (based on smallest adjusted p-value) of human spinal motoneurons were determined based on the Wilcoxon Rank Sum test and were analyzed using DAVID 6.8 GO enrichment analysis (https://david.ncifcrf.gov/summary.jsp). The general categories of GOTERM_BP_DIRECT, GOTERM_CC_DIRECT, and GOTERM_MF_DIRECT were analyzed and functional annotation clustering was performed using default parameters including medium classification stringency.
Focused comparison of mouse and human motor neurons.
Human motor neurons were compared to mouse lumbar skeletal motor neurons from a recent study4. Mouse MN genes were converted to human homologs using Homologene (https://CRAN.Rproject.org/package=homologene). Only genes with human homologs present in both datasets were included in the analysis (13,574). Raw counts were extracted from each original dataset, normalized using SCTransform, and integrated based on integration anchors. Clustering was performed as described above (resolution = 0.4), and differentially expressed genes were identified based on Wilcoxon Rank Sum test and ROC analysis within FindMarkers function from Seurat 4.0.
Analysis of evolutionarily convergence/divergence scores.
All available data on gene expression-based human:mouse divergence scores was downloaded from Pembroke et al49. Genes of interest were then extracted, yielding scores for three genes (SOD1, TUBA4A, OPTN) that overlapped with this data. We compared the mean and standard deviation of these three genes to the same metrics for the remainder of the assayed genes from the Pembroke report (N=1426 other genes) using a standard two-sided t-test.
Pain GWAS analysis.
All genes found in both Johnston et al26 and Suri et al27, or in Bortsov et al28 (which included separate discovery and replication cohorts) were examined: C8orf34, CCDC26, DCC, EXD3, MIPOL1, NOXA1, PSAP, SLC25A21, SOX5, SPOCK2.
Neurodegenerative disease gene analysis.
Post-QC scRNAseq count data was extracted for seven major cell classes of interest. For each gene per cell class, mean expression was calculated across all assayed cells of that class. These means were then z-scored to facilitate comparisons across multiple cell types. Additionally, genes that did not have any count based data available for that cell class were set to zero for z-scaled values. From this large dataframe of normalized counts per cell type, candidate genes for HSP, PD and ALS were extracted from Genomics England Expert Panel App genes audited at the “green” level of confidence [https://panelapp.genomicsengland.co.uk/]. AD genes were annotated by an expert panel and extracted90 and the ALS list was also supplemented with genes from the literature, as described in the main text. These extracted genes were then mapped to each cluster using the python package “seaborn”, with z-scores greater than 7 truncated to a value of 7 for display purposes. Code for the evolutionary divergence analysis can be found at https://colab.research.google.com/drive/19Ty97LOwT3AmaVCJGKA8BXXNFYH_iSZ4?usp=sharing and https://colab.research.google.com/drive/1BDJaiwhYnhMO9VJZNWUn9Iw_c87Y7mjr?usp=sharing.
SOD1 antibody validation in human iPS neurons with targeted knockdown.
Previously published human inducible pluripotent stem cells (hiPSCs) were used to knock down SOD191. A SOD1 or non-targeting control sgRNA was cloned into a mU6-sgRNA EF1a-puro-T2A-2XmycNLS-BFP vector (gift from Martin Kampmann’s lab; Addgene #127965). sgRNA sequences are as follows: SOD1: GAGGCACCACGACAGACCCG, non-targeting sgRNA: GAATATGTGCGTGCATGAAG. Lentivirus was produced via transduction of Lenti-X HEK 293T cells using Lipofectamine 3000 in DMEM high glucose GlutaMAX Supplement media containing 10% FBS. 24 hours post-transfection, media was replaced, including ViralBoost Reagent (ALSTEM, #VB100). 96 hours post-transfection, media was collected and concentrated 1:10 in 1xPBS using Lenti-X concentrator (Takara Bio, #631231), aliquoted, and stored at −80°C. 100 ml of these aliquots was used to transduce 100,000 hiPSCs to generate SOD1 KD and control lines. The cells were split and replated on Matrigel (Corning Incorporated #354277) coated coverslips with the viral concentrate in E8+Y-27632 ROCK Inhibitor and allowed to incubate for 24 hours at 37°C, 5% CO2. The media was replaced with E8 and the cells were allowed to grow for another 24 hours before fixation with 4% PFA in PBS for 10 mins at room temperature. Cells were washed with PBS 3 times and permeabilized in block (PBS + 3% donkey serum + 0.1% tritonX) for 30 mins at room temperature. Primary antibody targeting SOD1 (Sigma, HPA001401–100UL) was diluted at 1:500 in block and cells were incubated in primary overnight at 4°C on a rocker. The next day, cells were washed three times with PBST and incubated in block with secondary antibody (Jackson ImmunoResearch # 711-625-152) and Hoechst (Thermo Scientific #62249) for 1 hour at room temperature. Following 3 washes with PBST, the coverslips were mounted using ProLong Gold antifade reagent (Invitrogen #P36934). After curing, the coverslips were imaged using Nikon spinning disk confocal using laser wavelengths of 405 nm, 488 nm, 561 nm, and 640 nm at 100ms exposure and 75%, 25%, 25% and 100% power respectively. Images were edited using ImageJ.
Immunohistochemistry antibodies.
KI67 (Cell Signaling Tech, 9449S), IBA1 (Synaptic Systems, 234006), NeuN (Millipore Sigma, ABN90), SOX9 (Abcam, ab185966), OLIG2 (Millipore Sigma, MABN50), SOD1 (Sigma, HPA001401–100UL), OPTN (Proteintech, 10837–1-AP), Neurofilament H (Cell Signaling, 2836S), Chat (Millpore Sigma, AB144P), TUBA4A (Thermofisher, PA5–29546), Alexa Fluor® 647 Anti-alpha Tubulin (Abcam, ab190573), Stathmin-2/STMN2 (Novus, NBP1–49461), and Peripherin/PRPH (Millipore, AB1530).
Immunohistochemistry.
Immunohistochemistry for human and mouse spinal cords were performed as previously described92 with modifications for human spinal cords. For single-round immunohistochemistry, human spinal cords were cut at 14 μm, washed twice in TBS and placed in 0.05% sodium azide-TBS at 4°C for 3 days under a LED light to quench autofluorescence. This was done to quench the background autofluorescence and was based on a protocol optimized in brain tissue93. Human spinal cords were then placed in blocking buffer (1% IgG-free BSA, 10% normal donkey serum, in TBS) for one hour prior to incubation in blocking buffer and primary antibody for 48 hours at 4°C. Primary antibody was washed off three times in TBS with 0.025% triton before a 2-hour incubation in secondary antibody at room temperature. Secondary antibody was washed off three times in TBS with 0.025% triton before adding a coverslip. For multiplex immunohistochemistry, human spinal cords were cut at 10-μm and tissue was processed as previously described94.
We examined whether the human tissue underwent size changes during processing that could bias cell size measurements, but we found that processed and stained sections showed overall dimensions that were very similar to the cross-sectional dimensions of lumbar spinal cord from in vivo human MRI. Specifically, in sections from four donors over L3–L5, we measured 9.2 +/− 0.17 cm in width and 6.9 +/− 0.1 cm in height (mean +/− s.e.m.), while Toossi et al.95 showed that the L3–L5 spinal segments in MRI were ~8.5–9.3 cm in width and 7–7.5 cm in height and, in sections from four donors over L3–L5, we measured 9.2 +/− 0.17 cm in width and 6.9 +/− 0.1 cm in height (mean +/− s.e.m.).
Mouse spinal cords were cut at 50 μm and placed in blocking buffer (1% IgG-free BSA, 10% normal donkey serum, 0.1% Triton-X 100 in PBS) for one hour, then incubated in blocking buffer with primary antibody for 48 hours at 4°C. Primary antibody was washed off three times in PBS before a 2-hour incubation in secondary antibody at room temperature. Secondary antibody was washed off three times in PBS before adding a coverslip. There was less autofluorescence in mouse spinal cord tissue compared to human spinal cord tissue, given the ability to perfuse with PBS and PFA, timing of euthanasia and age of the mice. This rendered an LED quenching step unnecessary in mouse tissue.
Imaging.
Images of immunohistochemistry samples were imaged using a Zeiss 800 LSM confocal microscope.
Image analysis and quantification.
For quantification of neurons by laminae, the images were overlaid in Adobe Photoshop where borders between the gray and white matter and the lamina within the gray matter were drawn. These images were then exported to ImageJ for analysis. The cells were measured manually by outlining each cell using the selection tool and adding them to groups within the ROIManager in ImageJ based on lamina. Feret distance (maximum caliper, similar to diameter) measurements of all the ROIs for each section were saved in a spreadsheet. The white and gray matter of each subject were outlined in ImageJ and their areas were exported to a spreadsheet.
To identify colocalization of markers with NeuN, each neuron was first outlined with the selection tool in ImageJ and saved into different groups based on whether the cell was in lamina IX or not. Then, each cell that had co-occurrence of the markers were placed into separate groups (double positive in lamina IX and double positive outside lamina IX). Feret diameter measurements were then saved to a spreadsheet and the number of cells in each group were counted in Python.
For neuron and non-neuronal quantification in human tissue, all counts were done on single thin sections from each donor (10 um). DAPI was counted using a custom MATLAB-based code and only cells in which the nucleus (DAPI+) was present in the section were subsequently counted for antibody staining (NeuN, Oligo2, Iba1 and Sox9) which was done using the Count Tool in Adobe Photoshop. We ensured that cells on stitching boundaries were not double or miscounted. While a low level of background of antibody staining was present in some cases, we counted only cells which had a clear staining without high background.
Quantification and Statistical Analysis
Two-way ANOVA (repeated-measures) was used for assessing grouped data, such as the correlation and silhouette scores between human and mouse dorsal vs ventral neurons. Two-tailed t tests (unpaired) were used all for differences in silhouette scores and correlation between clusters as well as expression of protein and soma size, as indicated in figure legends. Bonferroni-adjusted Wilcoxon Rank Sum test p-values and Bhattacharyya Coefficients (BC) were used for comparison of human vs mouse cell diameter. Differences among groups were considered significant if p < 0.05. P values are denoted by asterisks: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; n.s – not significant. Data are represented as mean ± s.e.m. unless otherwise indicated. Statistical analyses were performed using GraphPad prism software and R.
Additional Resources
None.
Supplementary Material
Supplemental Table S1: Metadata on human subjects used in this study. Related to Figure 1.
Supplemental Table S2: Number of nuclei and average gene detection for each cluster.Related to Figure 1.
Supplemental Table S3: Marker expression and spatial location of clusters. Related to Figures 2 and 3.
Supplemental Table S4: Top 10 differentially expressed genes in each cluster. Related to Figures 2 and 3.
Supplemental Table S5: Gene Ontology (GO) analysis results for top 50 marker genes in motoneurons. Related to Figure 6.
Supplemental Table S6: Size measurements of neurons in adult human and mouse lumbar spinal cord. Related to Figure 7.
Supplemental Table S7: Quantification of protein expression. Related to Figure 7.
Supplemental Figures S1 through S25.
Supplemental Table S8: Genes used in disease enrichment analysis. Related to Figure 6.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| KI67 | Cell Signaling Tech | 9449S |
| IBA1 | Synaptic Systems | 234006 |
| NeuN | Millipore Sigma | ABN90P |
| SOX9 | Abcam | ab185966 |
| OLIG2 | Millipore Sigma | MABN50 |
| SOD1 | Sigma | HPA001401 |
| OPTN | Proteintech | 10837-1-AP |
| Neurofilament H | Cell Signaling | 2836S |
| Chat | Millpore Sigma | AB144P |
| TUBA4A | Thermofisher | PA5-29546 |
| Alexa Fluor® 647 Anti-alpha Tubulin | Abcam | ab190573 |
| Stathmin-2/STMN2 | Novus | NBP1-49461 |
| Peripherin/PRPH | Millipore | AB1530 |
| Biological samples | ||
| Human spinal cord | Ottawa Hospital Research Institute | N/A |
| Human spinal cord | Gui de Chauliac Hospital | N/A |
| Human spinal cord | Target ALS Multicenter Postmortem Core | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| ViralBoost Reagent | ALSTEM | #VB100 |
| Lenti-X concentrator | Takara Bio | #631231 |
| Matrigel | Corning Incorporated | #354277 |
| Hoechst | Thermo Scientific | #62249 |
| ProLong Gold antifade reagent | Invitrogen | #P36934 |
| Sucrose | Invitrogen | 15503-022 |
| 1 M HEPES (pH = 8.0) | Gibco | 15630-080 |
| CaCl2 | Sigma Aldrich | C1016-100G |
| MgAc | Sigma Aldrich | M5661-50G |
| 0.5 M EDTA (pH = 8.0) | Corning | MT-46034CI |
| Dithiothreitol (DTT) | Sigma Aldrich | 10197777001 |
| T riton-X | Sigma Aldrich | T8787 |
| 1 M Tris-HCl (pH = 7.4) | Sigma Aldrich | T2194 |
| 0.04% BSA | New England Biolabs | B9000S |
| 0.2 U/μL RNAse Inhibitor | Lucigen | 30281-1 |
| Trypan Blue Stain (0.4%) | Thermo Fisher Scientific | T10282 |
| Critical commercial assays | ||
| Chromium Single Cell 3’ GEM, Library & Gel Bead Kit V3 | 10X Genomics | PN-1000075 |
| Chromium Single Cell B Chip Kit | 10X Genomics | PN-1000074 |
| Chromium i7 Multiplex Kit | 10X Genomics | PN-120262 |
| Visium Spatial Gene Expression Slide & Reagent Kit | 10X Genomics | PN-1000184 |
| Visium Accessory Kit | 10X Genomics | PN-1000194 |
| Dual Index Kit TT Set A | 10X Genomics | PN-1000215 |
| Deposited data | ||
| Anonymized raw sequencing data | This paper | GEO: GSE190442 and GSE222322 |
| Raw mass spectrometry datasets | This paper | https://www.synapse.org/#!Synapse:syn27230711 |
| Experimental models: Cell lines | ||
| Human iPSC Line | Tian et al.91 | N/A |
| Experimental models: Organisms/strains | ||
| C57BL/6J | Jackson Laboratory | 000664 |
| BALB/cJ | Jackson Laboratory | 000651 |
| Recombinant DNA | ||
| mU6-sgRNA EF1 a-puro-T2A-2XmycNLS-BFP | Tian et al.91 | Addgene Plasmid pMK1334 #127965 |
| Software and algorithms | ||
| Seurat 4.0 | Hafemeister and Satija87; Stuart et al.86 | https://satiialab.org/seurat/index.html |
| biomaRt | Durinck et al.89 | https://bioconductor.org/packages/release/bioc/html/biomaRt.html |
| Homologene | National Center for Biotechnology Information | https://CRAN.R-proiect.org/package=homologene |
| Neurodegenerative disease gene analysis | This paper |
https://colab.research.google.com/drive/19Ty97LOwT3AmaVCJGKA8BXXNFYH_iSZ4?usp=sharing and https://colab.research.google.com/drive/1BDJaiwhYnhMO9VJZNWUn9Iw_c87Y7mjr?usp=sharing |
| Cell2Location | Kleshchevnikov et al.88 | https://cell2location.readthedocs.io/en/latest/ |
| OpenCV | opencv-python- headless 4.6.0.66 | https://pypi.org/proiect/opencv-python-headless/ |
| Fiji/ImageJ | Fiji v2.1.0 | https://imagei.net/software/fiii/ |
| Adobe Photoshop | Adobe Systems | https://www.adobe.com |
| Adobe Illustrator | Adobe Systems | |
| Other | ||
| Resource website | This paper | |
Highlights.
Single-nucleus RNA-sequencing and spatial transcriptomics of adult human spinal cord
Glial classes include proliferative microglia and white and gray matter astrocytes
Dorsal neuron types are more discrete and ventral neuron groups are more overlapping
The human motoneuron transcriptional profile is enriched for genes related to ALS
ACKNOWLEDGEMENTS
We gratefully acknowledge the gift of human tissue from the 11 donors included in this work and their families, whose contribution has been critical for this work. This work was supported by NINDS Intramural funds through 1ZIA NS003153 (KJEM, LL, IH, AJL), 1ZIANS003155 (SH and MEW) NS116350 (JP, KK, HP); by NICHD Intramural funds through 1ZIAHD008966 (MRA, CELP); by the Intramural Research Program of the NIA project ZO1 AG000535 (MAN); by R01 AG06683 and U54 AG076040 from NIA and the NIH Common Fund (AY, VM); by the Canadian Institutes of Health Research, the University of Ottawa Department of Surgery, the Ontario Neurotrauma Foundation, and the Praxis Spinal Cord Institute (AG, SA, JP, MMA, FAQ, SMA, APW, ECT, MEH); and by ANR-15-CE16-012, FRC-EET-2019 grants, CNRS/INSERM/Montpellier Hospital research supports (PFM, EB, LB) and a AL210154 grant (JP, KK, HP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
M.A.N.’s participation in this project was part of a competitive contract awarded to Data Tecnica International LLC by the National Institutes of Health to support open science research, he also currently serves on the scientific advisory board for Clover Therapeutics and is an advisor to Neuron23 Inc.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
REFERENCES
- 1.Arnold ES, and Fischbeck KH (2018). Spinal muscular atrophy. Handb Clin Neurol 148, 591–601. 10.1016/b978-0-444-64076-5.00038-7. [DOI] [PubMed] [Google Scholar]
- 2.Kawamura Y, Dyck PJ, Shimono M, Okazaki H, Tateishi J, and Doi H (1981). Morphometric comparison of the vulnerability of peripheral motor and sensory neurons in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol 40, 667–675. 10.1097/00005072-198111000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Ravits J, Appel S, Baloh RH, Barohn R, Brooks BR, Elman L, Floeter MK, Henderson C, Lomen-Hoerth C, Macklis JD, et al. (2013). Deciphering amyotrophic lateral sclerosis: what phenotype, neuropathology and genetics are telling us about pathogenesis. Amyotroph Lateral Scler Frontotemporal Degener 14 Suppl 1, 5–18. 10.3109/21678421.2013.778548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alkaslasi MR, Piccus ZE, Hareendran S, Silberberg H, Chen L, Zhang Y, Petros TJ, and Le Pichon CE (2021). Single nucleus RNA-sequencing defines unexpected diversity of cholinergic neuron types in the adult mouse spinal cord. Nat Commun 12, 2471. 10.1038/s41467-021-22691-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blum JA, Klemm S, Shadrach JL, Guttenplan KA, Nakayama L, Kathiria A, Hoang PT, Gautier O, Kaltschmidt JA, Greenleaf WJ, and Gitler AD (2021). Single-cell transcriptomic analysis of the adult mouse spinal cord reveals molecular diversity of autonomic and skeletal motor neurons. Nat Neurosci 24, 572–583. 10.1038/s41593-020-00795-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kathe C, Skinnider MA, Hutson TH, Regazzi N, Gautier M, Demesmaeker R, Komi S, Ceto S, James ND, Cho N, et al. (2022). The neurons that restore walking after paralysis. Nature 611, 540–547. 10.1038/s41586-022-05385-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milich LM, Choi JS, Ryan C, Cerqueira SR, Benavides S, Yahn SL, Tsoulfas P, and Lee JK (2021). Single-cell analysis of the cellular heterogeneity and interactions in the injured mouse spinal cord. J Exp Med 218. 10.1084/jem.20210040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tansley S, Uttam S, Ureña Guzmán A, Yaqubi M, Pacis A, Parisien M, Deamond H, Wong C, Rabau O, Brown N, et al. (2022). Single-cell RNA sequencing reveals time- and sex-specific responses of mouse spinal cord microglia to peripheral nerve injury and links ApoE to chronic pain. Nat Commun 13, 843. 10.1038/s41467-022-28473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun J, Qiu J, Yang Q, Ju Q, Qu R, Wang X, Wu L, and Xing L (2022). Single-cell RNA sequencing reveals dysregulation of spinal cord cell types in a severe spinal muscular atrophy mouse model. PLoS Genet 18, e1010392. 10.1371/journal.pgen.1010392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacLean M, López-Díez R, Vasquez C, Gugger PF, and Schmidt AM (2022). Neuronal-glial communication perturbations in murine SOD1(G93A) spinal cord. Commun Biol 5, 177. 10.1038/s42003-022-03128-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Häring M, Zeisel A, Hochgerner H, Rinwa P, Jakobsson JET, Lönnerberg P, La Manno G, Sharma N, Borgius L, Kiehn O, et al. (2018). Neuronal atlas of the dorsal horn defines its architecture and links sensory input to transcriptional cell types. Nat Neurosci 21, 869–880. 10.1038/s41593-018-0141-1. [DOI] [PubMed] [Google Scholar]
- 12.Arokiaraj CM, Kleyman M, Chamessian A, Shiers S, Kang B, Kennedy MM, Chen Y, Dum RP, Patterson R, Lewis DA, et al. (2022). Cellular and Molecular Organization of the Rhesus Macaque Dorsal Horn with Comparison to Mouse. bioRxiv, 2022.2004.2001.486135. 10.1101/2022.04.01.486135. [DOI] [Google Scholar]
- 13.Rayon T, Maizels RJ, Barrington C, and Briscoe J (2021). Single-cell transcriptome profiling of the human developing spinal cord reveals a conserved genetic programme with human-specific features. Development 148. 10.1242/dev.199711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q, Wu X, Fan Y, Jiang P, Zhao Y, Yang Y, Han S, Xu B, Chen B, Han J, et al. (2021). Single-cell analysis reveals dynamic changes of neural cells in developing human spinal cord. EMBO Rep 22, e52728. 10.15252/embr.202152728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D, Wei Y, Liu J, Chen H, Li J, Zhu T, and Zhou C (2021). Single-nucleus transcriptomic atlas of spinal cord neuron in human. bioRxiv, 2021.2009.2028.462103. 10.1101/2021.09.28.462103. [DOI] [Google Scholar]
- 16.Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, Walker AJ, Gergits F, Segel M, Nemesh J, et al. (2019). Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 50, 253–271.e256. 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matson KJE, Russ DE, Kathe C, Hua I, Maric D, Ding Y, Krynitsky J, Pursley R, Sathyamurthy A, Squair JW, et al. (2022). Single cell atlas of spinal cord injury in mice reveals a proregenerative signature in spinocerebellar neurons. Nat Commun 13, 5628. 10.1038/s41467-022-33184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olah M, Menon V, Habib N, Taga MF, Ma Y, Yung CJ, Cimpean M, Khairallah A, Coronas-Samano G, Sankowski R, et al. (2020). Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer’s disease. Nat Commun 11, 6129. 10.1038/s41467-020-19737-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russ DE, Cross RBP, Li L, Koch SC, Matson KJE, Yadav A, Alkaslasi MR, Lee DI, Le Pichon CE, Menon V, and Levine AJ (2021). A harmonized atlas of mouse spinal cord cell types and their spatial organization. Nat Commun 12, 5722. 10.1038/s41467-021-25125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osseward PJ 2nd, Amin ND, Moore JD, Temple BA, Barriga BK, Bachmann LC, Beltran F Jr., Gullo M, Clark RC, Driscoll SP, et al. (2021). Conserved genetic signatures parcellate cardinal spinal neuron classes into local and projection subsets. Science 372, 385–393. 10.1126/science.abe0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bourane S, Grossmann KS, Britz O, Dalet A, Del Barrio MG, Stam FJ, Garcia-Campmany L, Koch S, and Goulding M (2015). Identification of a spinal circuit for light touch and fine motor control. Cell 160, 503–515. 10.1016/j.cell.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan B, Cheng L, Bourane S, Britz O, Padilla C, Garcia-Campmany L, Krashes M, Knowlton W, Velasquez T, Ren X, et al. (2014). Identification of spinal circuits transmitting and gating mechanical pain. Cell 159, 1417–1432. 10.1016/j.cell.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brewer CL, Styczynski LM, Serafin EK, and Baccei ML (2020). Postnatal maturation of spinal dynorphin circuits and their role in somatosensation. Pain 161, 1906–1924. 10.1097/j.pain.0000000000001884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serafin EK, Paranjpe A, Brewer CL, and Baccei ML (2021). Single-nucleus characterization of adult mouse spinal dynorphin-lineage cells and identification of persistent transcriptional effects of neonatal hindpaw incision. Pain 162, 203–218. 10.1097/j.pain.0000000000002007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kardon AP, Polgár E, Hachisuka J, Snyder LM, Cameron D, Savage S, Cai X, Karnup S, Fan CR, Hemenway GM, et al. (2014). Dynorphin acts as a neuromodulator to inhibit itch in the dorsal horn of the spinal cord. Neuron 82, 573–586. 10.1016/j.neuron.2014.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston KJA, Adams MJ, Nicholl BI, Ward J, Strawbridge RJ, Ferguson A, McIntosh AM, Bailey MES, and Smith DJ (2019). Genome-wide association study of multisite chronic pain in UK Biobank. PLoS Genet 15, e1008164. 10.1371/journal.pgen.1008164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suri P, Palmer MR, Tsepilov YA, Freidin MB, Boer CG, Yau MS, Evans DS, Gelemanovic A, Bartz TM, Nethander M, et al. (2018). Genome-wide meta-analysis of 158,000 individuals of European ancestry identifies three loci associated with chronic back pain. PLoS Genet 14, e1007601. 10.1371/journal.pgen.1007601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bortsov AV, Parisien M, Khoury S, Martinsen AE, Lie MU, Heuch I, Hveem K, Zwart JA, Winsvold BS, and Diatchenko L (2022). Brain-specific genes contribute to chronic but not to acute back pain. Pain Rep 7, e1018. 10.1097/pr9.0000000000001018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gatto G, Bourane S, Ren X, Di Costanzo S, Fenton PK, Halder P, Seal RP, and Goulding MD (2021). A Functional Topographic Map for Spinal Sensorimotor Reflexes. Neuron 109, 91–104.e105. 10.1016/j.neuron.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peirs C, Williams SP, Zhao X, Walsh CE, Gedeon JY, Cagle NE, Goldring AC, Hioki H, Liu Z, Marell PS, and Seal RP (2015). Dorsal Horn Circuits for Persistent Mechanical Pain. Neuron 87, 797–812. 10.1016/j.neuron.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang T, Lin SH, Malewicz NM, Zhang Y, Zhang Y, Goulding M, LaMotte RH, and Ma Q (2019). Identifying the pathways required for coping behaviours associated with sustained pain. Nature 565, 86–90. 10.1038/s41586-018-0793-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petitjean H, Pawlowski SA, Fraine SL, Sharif B, Hamad D, Fatima T, Berg J, Brown CM, Jan LY, Ribeiro-da-Silva A, et al. (2015). Dorsal Horn Parvalbumin Neurons Are Gate-Keepers of Touch-Evoked Pain after Nerve Injury. Cell Rep 13, 1246–1257. 10.1016/j.celrep.2015.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith KM, Browne TJ, Davis OC, Coyle A, Boyle KA, Watanabe M, Dickinson SA, Iredale JA, Gradwell MA, Jobling P, et al. (2019). Calretinin positive neurons form an excitatory amplifier network in the spinal cord dorsal horn. Elife 8. 10.7554/eLife.49190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Z, Cork LC, Griffin JW, and Cleveland DW (1993). Increased expression of neurofilament subunit NF-L produces morphological alterations that resemble the pathology of human motor neuron disease. Cell 73, 23–33. 10.1016/0092-8674(93)90157-l. [DOI] [PubMed] [Google Scholar]
- 35.Xu Z, Cork LC, Griffin JW, and Cleveland DW (1993). Involvement of neurofilaments in motor neuron disease. J Cell Sci Suppl 17, 101–108. 10.1242/jcs.1993.supplement_17.15. [DOI] [PubMed] [Google Scholar]
- 36.Gama Sosa MA, Friedrich VL Jr., DeGasperi R, Kelley K, Wen PH, Senturk E, Lazzarini RA, and Elder GA (2003). Human midsized neurofilament subunit induces motor neuron disease in transgenic mice. Exp Neurol 184, 408–419. 10.1016/s0014-4886(03)00206-1. [DOI] [PubMed] [Google Scholar]
- 37.Côté F, Collard JF, and Julien JP (1993). Progressive neuronopathy in transgenic mice expressing the human neurofilament heavy gene: a mouse model of amyotrophic lateral sclerosis. Cell 73, 35–46. 10.1016/0092-8674(93)90158-m. [DOI] [PubMed] [Google Scholar]
- 38.Marszalek JR, Williamson TL, Lee MK, Xu Z, Hoffman PN, Becher MW, Crawford TO, and Cleveland DW (1996). Neurofilament subunit NF-H modulates axonal diameter by selectively slowing neurofilament transport. J Cell Biol 135, 711–724. 10.1083/jcb.135.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beaulieu JM, Nguyen MD, and Julien JP (1999). Late onset of motor neurons in mice overexpressing wild-type peripherin. J Cell Biol 147, 531–544. 10.1083/jcb.147.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregory JM, Fagegaltier D, Phatnani H, and Harms MB (2020). Genetics of Amyotrophic Lateral Sclerosis. Current Genetic Medicine Reports 8, 121–131. 10.1007/s40142-020-00194-8. [DOI] [Google Scholar]
- 41.Taylor JP, Brown RH Jr., and Cleveland DW (2016). Decoding ALS: from genes to mechanism. Nature 539, 197–206. 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown RH Jr., and Al-Chalabi A (2017). Amyotrophic Lateral Sclerosis. N Engl J Med 377, 1602. 10.1056/NEJMc1710379. [DOI] [PubMed] [Google Scholar]
- 43.Castellanos-Montiel MJ, Chaineau M, and Durcan TM (2020). The Neglected Genes of ALS: Cytoskeletal Dynamics Impact Synaptic Degeneration in ALS. Front Cell Neurosci 14, 594975. 10.3389/fncel.2020.594975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Theunissen F, Anderton RS, Mastaglia FL, Flynn LL, Winter SJ, James I, Bedlack R, Hodgetts S, Fletcher S, Wilton SD, et al. (2021). Novel STMN2 Variant Linked to Amyotrophic Lateral Sclerosis Risk and Clinical Phenotype. Front Aging Neurosci 13, 658226. 10.3389/fnagi.2021.658226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klim JR, Williams LA, Limone F, Guerra San Juan I, Davis-Dusenbery BN, Mordes DA, Burberry A, Steinbaugh MJ, Gamage KK, Kirchner R, et al. (2019). ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat Neurosci 22, 167–179. 10.1038/s41593-018-0300-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morisaki Y, Niikura M, Watanabe M, Onishi K, Tanabe S, Moriwaki Y, Okuda T, Ohara S, Murayama S, Takao M, et al. (2016). Selective Expression of Osteopontin in ALS-resistant Motor Neurons is a Critical Determinant of Late Phase Neurodegeneration Mediated by Matrix Metalloproteinase-9. Sci Rep 6, 27354. 10.1038/srep27354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto T, Murayama S, Takao M, Isa T, and Higo N (2017). Expression of secreted phosphoprotein 1 (osteopontin) in human sensorimotor cortex and spinal cord: Changes in patients with amyotrophic lateral sclerosis. Brain Res 1655, 168–175. 10.1016/j.brainres.2016.10.030. [DOI] [PubMed] [Google Scholar]
- 48.Maniatis S, Äijö T, Vickovic S, Braine C, Kang K, Mollbrink A, Fagegaltier D, Andrusivová Ž, Saarenpää S, Saiz-Castro G, et al. (2019). Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis. Science 364, 89–93. 10.1126/science.aav9776. [DOI] [PubMed] [Google Scholar]
- 49.Pembroke WG, Hartl CL, and Geschwind DH (2021). Evolutionary conservation and divergence of the human brain transcriptome. Genome Biol 22, 52. 10.1186/s13059-020-02257-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McHanwell S, and Biscoe TJ (1981). The sizes of motoneurons supplying hindlimb muscles in the mouse. Proc R Soc Lond B Biol Sci 213, 201–216. 10.1098/rspb.1981.0062. [DOI] [PubMed] [Google Scholar]
- 51.Ishihara A, Ohira Y, Tanaka M, Nishikawa W, Ishioka N, Higashibata A, Izumi R, Shimazu T, and Ibata Y (2001). Cell body size and succinate dehydrogenase activity of spinal motoneurons innervating the soleus muscle in mice, rats, and cats. Neurochem Res 26, 1301–1304. 10.1023/a:1014245417017. [DOI] [PubMed] [Google Scholar]
- 52.Kawamura Y, and Dyck PJ (1977). Lumbar motoneurons of man: III. The number and diameter distribution of large- and intermediate- diameter cytons by nuclear columns. J Neuropathol Exp Neurol 36, 956–963. 10.1097/00005072-197711000-00007. [DOI] [PubMed] [Google Scholar]
- 53.Sobue G, Matsuoka Y, Mukai E, Takayanagi T, and Sobue I (1981). Pathology of myelinated fibers in cervical and lumbar ventral spinal roots in amyotrophic lateral sclerosis. J Neurol Sci 50, 413–421. 10.1016/0022-510x(81)90153-2. [DOI] [PubMed] [Google Scholar]
- 54.Sobue G, Matsuoka Y, Mukai E, Takayanagi T, Sobue I, and Hashizume Y (1981). Spinal and cranial motor nerve roots in amyotrophic lateral sclerosis and X-linked recessive bulbospinal muscular atrophy: morphometric and teased-fiber study. Acta Neuropathol 55, 227–235. 10.1007/bf00691322. [DOI] [PubMed] [Google Scholar]
- 55.Nguyen MD, Larivière RC, and Julien JP (2000). Reduction of axonal caliber does not alleviate motor neuron disease caused by mutant superoxide dismutase 1. Proc Natl Acad Sci U S A 97, 12306–12311. 10.1073/pnas.97.22.12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hardesty I (1902). Observations on the medulla spinalis of the elephant with some comparative studies of the intumescentia cervicalis and the neurones of the columna anterior. Journal of Comparative Neurology 12, 125–182. 10.1002/cne.910120203. [DOI] [Google Scholar]
- 57.Braak H, and Braak E (1976). The pyramidal cells of Betz within the cingulate and precentral gigantopyramidal field in the human brain. A Golgi and pigmentarchitectonic study. Cell Tissue Res 172, 103–119. 10.1007/bf00226052. [DOI] [PubMed] [Google Scholar]
- 58.Haberberger RV, Barry C, Dominguez N, and Matusica D (2019). Human Dorsal Root Ganglia. Front Cell Neurosci 13, 271. 10.3389/fncel.2019.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zottoli SJ (1978). Comparison of Mauthner cell size in teleosts. J Comp Neurol 178, 741–757. 10.1002/cne.901780409. [DOI] [PubMed] [Google Scholar]
- 60.Tsang YM, Chiong F, Kuznetsov D, Kasarskis E, and Geula C (2000). Motor neurons are rich in non-phosphorylated neurofilaments: cross-species comparison and alterations in ALS. Brain Res 861, 45–58. 10.1016/s0006-8993(00)01954-5. [DOI] [PubMed] [Google Scholar]
- 61.Pardo CA, Xu Z, Borchelt DR, Price DL, Sisodia SS, and Cleveland DW (1995). Superoxide dismutase is an abundant component in cell bodies, dendrites, and axons of motor neurons and in a subset of other neurons. Proc Natl Acad Sci U S A 92, 954–958. 10.1073/pnas.92.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Averback P, and Crocker P (1982). Regular involvement of Clarke’s nucleus in sporadic amyotrophic lateral sclerosis. Arch Neurol 39, 155–156. 10.1001/archneur.1982.00510150025006. [DOI] [PubMed] [Google Scholar]
- 63.Williams C, Kozlowski MA, Hinton DR, and Miller CA (1990). Degeneration of spinocerebellar neurons in amyotrophic lateral sclerosis. Ann Neurol 27, 215–225. 10.1002/ana.410270302. [DOI] [PubMed] [Google Scholar]
- 64.McIlwain DL (1991). Nuclear and cell body size in spinal motor neurons. Adv Neurol 56, 67–74. [PubMed] [Google Scholar]
- 65.Gosgnach S, Bikoff JB, Dougherty KJ, El Manira A, Lanuza GM, and Zhang Y (2017). Delineating the Diversity of Spinal Interneurons in Locomotor Circuits. J Neurosci 37, 10835–10841. 10.1523/jneurosci.1829-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stam FJ, Hendricks TJ, Zhang J, Geiman EJ, Francius C, Labosky PA, Clotman F, and Goulding M (2012). Renshaw cell interneuron specialization is controlled by a temporally restricted transcription factor program. Development 139, 179–190. 10.1242/dev.071134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Francius C, Harris A, Rucchin V, Hendricks TJ, Stam FJ, Barber M, Kurek D, Grosveld FG, Pierani A, Goulding M, and Clotman F (2013). Identification of multiple subsets of ventral interneurons and differential distribution along the rostrocaudal axis of the developing spinal cord. PLoS One 8, e70325. 10.1371/journal.pone.0070325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delile J, Rayon T, Melchionda M, Edwards A, Briscoe J, and Sagner A (2019). Single cell transcriptomics reveals spatial and temporal dynamics of gene expression in the developing mouse spinal cord. Development 146. 10.1242/dev.173807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lindén H, Petersen PC, Vestergaard M, and Berg RW (2022). Movement is governed by rotational neural dynamics in spinal motor networks. Nature 610, 526–531. 10.1038/s41586-022-05293-w. [DOI] [PubMed] [Google Scholar]
- 70.Lee MK, and Cleveland DW (1996). Neuronal intermediate filaments. Annu Rev Neurosci 19, 187–217. 10.1146/annurev.ne.19.030196.001155. [DOI] [PubMed] [Google Scholar]
- 71.Hoffman PN, Griffin JW, and Price DL (1984). Control of axonal caliber by neurofilament transport. J Cell Biol 99, 705–714. 10.1083/jcb.99.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Friede RL, and Samorajski T (1970). Axon caliber related to neurofilaments and microtubules in sciatic nerve fibers of rats and mice. Anat Rec 167, 379–387. 10.1002/ar.1091670402. [DOI] [PubMed] [Google Scholar]
- 73.Bakken TE, Jorstad NL, Hu Q, Lake BB, Tian W, Kalmbach BE, Crow M, Hodge RD, Krienen FM, Sorensen SA, et al. (2021). Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature 598, 111–119. 10.1038/s41586-021-03465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e1022. 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nguyen MQ, von Buchholtz LJ, Reker AN, Ryba NJ, and Davidson S (2021). Single-nucleus transcriptomic analysis of human dorsal root ganglion neurons. Elife 10. 10.7554/eLife.71752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Limone F, Mordes D, Couto A, Pietiläinen O, Joseph BJ, Burberry A, Ghosh SD, Meyer D, Goldman M, Bortolin L, et al. (2021). Single-nucleus sequencing reveals enriched expression of genetic risk factors sensitises Motor Neurons to degeneration in ALS. bioRxiv, 2021.2007.2012.452054. 10.1101/2021.07.12.452054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schoenen J (1982). Dendritic organization of the human spinal cord: the motoneurons. J Comp Neurol 211, 226–247. 10.1002/cne.902110303. [DOI] [PubMed] [Google Scholar]
- 78.Manuel M, Chardon M, Tysseling V, and Heckman CJ (2019). Scaling of Motor Output, From Mouse to Humans. Physiology (Bethesda) 34, 5–13. 10.1152/physiol.00021.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perge JA, Niven JE, Mugnaini E, Balasubramanian V, and Sterling P (2012). Why do axons differ in caliber? J Neurosci 32, 626–638. 10.1523/jneurosci.4254-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Clark JA, Yeaman EJ, Blizzard CA, Chuckowree JA, and Dickson TC (2016). A Case for Microtubule Vulnerability in Amyotrophic Lateral Sclerosis: Altered Dynamics During Disease. Front Cell Neurosci 10, 204. 10.3389/fncel.2016.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hardy J, and Rogaeva E (2014). Motor neuron disease and frontotemporal dementia: sometimes related, sometimes not. Exp Neurol 262 Pt B, 75–83. 10.1016/j.expneurol.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 82.Galuta A, Sandarage R, Ghinda D, Auriat AM, Chen S, Kwan JCS, and Tsai EC (2020). A Guide to Extract Spinal Cord for Translational Stem Cell Biology Research: Comparative Analysis of Adult Human, Porcine, and Rodent Spinal Cord Stem Cells. Front Neurosci 14, 607. 10.3389/fnins.2020.00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bauchet L, Poulen G, Lonjon N, Vachiery-Lahaye F, Bourinet E, Perrin FE, and Hugnot JP (2022). Isolation and Culture of Precursor Cells from the Adult Human Spinal Cord. Methods Mol Biol 2389, 103–110. 10.1007/978-1-0716-1783-0_10. [DOI] [PubMed] [Google Scholar]
- 84.Dedek A, Xu J, Kandegedara CM, Lorenzo L, Godin AG, De Koninck Y, Lombroso PJ, Tsai EC, and Hildebrand ME (2019). Loss of STEP61 couples disinhibition to N-methyl-d-aspartate receptor potentiation in rodent and human spinal pain processing. Brain 142, 1535–1546. 10.1093/brain/awz105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Matson KJE, Sathyamurthy A, Johnson KR, Kelly MC, Kelley MW, and Levine AJ (2018). Isolation of Adult Spinal Cord Nuclei for Massively Parallel Single-nucleus RNA Sequencing. J Vis Exp. 10.3791/58413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e1821. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hafemeister C, and Satija R (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20, 296. 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kleshchevnikov V, Shmatko A, Dann E, Aivazidis A, King HW, Li T, Elmentaite R, Lomakin A, Kedlian V, Gayoso A, et al. (2022). Cell2location maps fine-grained cell types in spatial transcriptomics. Nat Biotechnol 40, 661–671. 10.1038/s41587-021-01139-4. [DOI] [PubMed] [Google Scholar]
- 89.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, and Huber W (2005). BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440. 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 90.Ramos DM, Skarnes WC, Singleton AB, Cookson MR, and Ward ME (2021). Tackling neurodegenerative diseases with genomic engineering: A new stem cell initiative from the NIH. Neuron 109, 1080–1083. 10.1016/j.neuron.2021.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tian R, Gachechiladze MA, Ludwig CH, Laurie MT, Hong JY, Nathaniel D, Prabhu AV, Fernandopulle MS, Patel R, Abshari M, et al. (2019). CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron 104, 239–255.e212. 10.1016/j.neuron.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sathyamurthy A, Johnson KR, Matson KJE, Dobrott CI, Li L, Ryba AR, Bergman TB, Kelly MC, Kelley MW, and Levine AJ (2018). Massively Parallel Single Nucleus Transcriptional Profiling Defines Spinal Cord Neurons and Their Activity during Behavior. Cell Rep 22, 2216–2225. 10.1016/j.celrep.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duong H, and Han M (2013). A multispectral LED array for the reduction of background autofluorescence in brain tissue. J Neurosci Methods 220, 46–54. 10.1016/j.jneumeth.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maric D, Jahanipour J, Li XR, Singh A, Mobiny A, Van Nguyen H, Sedlock A, Grama K, and Roysam B (2021). Whole-brain tissue mapping toolkit using large-scale highly multiplexed immunofluorescence imaging and deep neural networks. Nat Commun 12, 1550. 10.1038/s41467-021-21735-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Toossi A, Bergin B, Marefatallah M, Parhizi B, Tyreman N, Everaert DG, Rezaei S, Seres P, Gatenby JC, Perlmutter SI, and Mushahwar VK (2021). Comparative neuroanatomy of the lumbosacral spinal cord of the rat, cat, pig, monkey, and human. Sci Rep 11, 1955. 10.1038/s41598-021-81371-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1: Metadata on human subjects used in this study. Related to Figure 1.
Supplemental Table S2: Number of nuclei and average gene detection for each cluster.Related to Figure 1.
Supplemental Table S3: Marker expression and spatial location of clusters. Related to Figures 2 and 3.
Supplemental Table S4: Top 10 differentially expressed genes in each cluster. Related to Figures 2 and 3.
Supplemental Table S5: Gene Ontology (GO) analysis results for top 50 marker genes in motoneurons. Related to Figure 6.
Supplemental Table S6: Size measurements of neurons in adult human and mouse lumbar spinal cord. Related to Figure 7.
Supplemental Table S7: Quantification of protein expression. Related to Figure 7.
Supplemental Figures S1 through S25.
Supplemental Table S8: Genes used in disease enrichment analysis. Related to Figure 6.
Data Availability Statement
Anonymized raw sequencing data and counts tables are deposited in the Gene Expression Omnibus (GEO) with accession numbers GSE190442 and GSE222322 with associated metadata in Supplemental Table S2. In addition, visualization of expression data at the cluster and donor level are available through a searchable web resource at https://vmenon.shinyapps.io/humanspinalcord/.
Code for the evolutionary divergence analysis can be found at https://colab.research.google.com/drive/19Ty97LOwT3AmaVCJGKA8BXXNFYH_iSZ4?usp=sharing and https://colab.research.google.com/drive/19Ty97LOwT3AmaVCJGKA8BXXNFYH_iSZ4?usp=sharing Custom MATLAB-based code for quantification of cell counts is available at https://github.com/ArielLevineLabNINDS/CellCounter (https://doi.org/10.5281/zenodo.6967482).


