Abstract
Prognostic analysis for patient survival often employs gene expressions obtained from high-throughput screening for tumor tissues from patients. When dealing with survival data, a dependent censoring phenomenon arises, and thus the traditional Cox model may not correctly identify the effect of each gene. A copula-based gene selection model can effectively adjust for dependent censoring, yielding a multi-gene predictor for survival prognosis. However, methods to assess the impact of various types of dependent censoring on the multi-gene predictor have not been developed. In this article, we propose a sensitivity analysis method using the copula-graphic estimator under dependent censoring, and implement relevant methods in the R package “compound.Cox”. The purpose of the proposed method is to investigate the sensitivity of the multi-gene predictor to a variety of dependent censoring mechanisms. In order to make the proposed sensitivity analysis practical, we develop a web application. We apply the proposed method and the web application to a lung cancer dataset. We provide a template file so that developers can modify the template to establish their own web applications.
Keywords: copula, Cox regression, dependent censoring, gene expression, high-dimensional data, Kendall’s tau, lung cancer, prognostic prediction, survival analysis, survival prediction
1. Introduction
Prognostic analysis for survival often employs gene expressions obtained from high-throughput screening for tumor tissues from patients [1,2,3,4,5,6]. Here, the primary interest is often to find a set of genes that are strong predictors of survival. The simplest and yet most commonly used approach is to select a small number of significant genes found in a sequence of univariate Cox models fitted to patients’ survival data [5,6,7,8,9].
A linear predictor using multiple genes has been shown to be useful for survival prediction. Usually, a multi-gene predictor is more accurate than a single-gene predictor as the prognostic ability of a single gene is limited. A predictor optimally constructed from the selected genes becomes a useful biomarker for predicting survival in breast cancer [10,11,12,13,14,15], lung cancer [6,7,8,9,16,17], gastric cancer [18,19], ovarian cancer [20,21,22,23,24], skin cancer [25], liver cancer [26,27], bladder cancer [28], head and neck cancer [29,30], glioma [31], myeloproliferative neoplasms [32], kidney cancer [33], and cancers of mixed types [34,35]. These analyses were performed mostly based on univariate Cox regression with the significance scaling of p-values, such as 0.05, 0.01, and 0.001, followed by cross-validation and/or external validation. The primary reason for applying Cox regression is that it can easily handle censored survival times, i.e., survival time may not be observable for all patients under investigation.
However, Cox regression critically relies on the independent censoring assumption: survival time and censoring time need to be statistically independent. This assumption is easily violated when patients drop out from the study due to the worsening of his/her health condition or patients are removed for transplantation [36,37]. Dependent censoring also arises due to unobserved factor [38,39,40,41]. In such cases, survival time is censored dependently on their health status. If the independent censoring assumption is violated, Cox regression analyses may not correctly identify the effect of each gene and thus may fail to select truly effective genes [41]. Therefore, the resultant predictor based on selected genes may have reduced ability to predict survival.
A copula-based gene selection approach [41] can effectively adjust for dependent censoring, yielding a multi-gene predictor. Besides the development of a multi-gene predictor, copula-based dependent censoring models have gained popularity in recent years for dealing with survival data [42,43,44,45,46,47,48,49,50]. However, methods to assess the impact of various types of dependent censoring on the multi-gene predictor have not been developed. In this article, we propose a sensitivity analysis method using the copula-graphic estimator [47,51,52,53,54,55,56] under dependent censoring, and implement relevant methods in the R package compound.Cox [9]. While the CG estimator has mostly been discussed for estimation of survival functions, its adaptation to the context of survival prognostic prediction remains unclear. This article aims to fill this gap by proposing detailed statistical methodologies; see Section 3 for details.
Another important goal of this article is to establish a Shiny-based web application (in Section 4) that can be manipulated by any user online without installing any software. This effectively connects the development of a sensitivity analysis and its implementation in order to carry out a patient’s prognosis. The application is written by R packages Shiny [57] and compound.Cox [9]. While this application is developed using data on lung cancer patients, the computational framework can be extended to other survival data from other patients. We provide the R code in Supplementary Materials so that users can easily edit the code to be adapted to their own data analyses.
The article is constructed as follows: Section 2 reviews the backgrounds of this research. Section 3 introduces the proposed sensitivity analysis method. Section 4 describes the development of the software and web application. Section 5 provides the results of applying the proposed method to the lung cancer data. Section 6 concludes with discussion.
2. Backgrounds
In this section, we shall first review gene selection methods [1,9,41] based on survival data. Gene selection is a required step before building a prognostic prediction method. Next, we shall review prognostic prediction methods based on selected genes.
We define basic notations as follows: is survival time and is a -vector of genes from individuals . The conditional survival function given is denoted by , and the conditional hazard function given is defined by . Similarly, denotes the conditional hazard function given the j-th gene .
2.1. Classical Gene Selection Method
The approach called univariate selection [1,9] is performed as follows: In the initial step, a significance of each gene is examined by univariate Cox regression one-by-one. Then, a subset of significant genes is selected with a p-value threshold, such as 0.05, 0.01, and 0.001. This is the most standard approach for gene selection in medical research.
In the univariate selection, a significance of gene on survival time is measured by fitting a Cox proportional hazards model [58] given by
| (1) |
for each gene . Here, the parameter measures the association between survival and the gene, and the baseline hazard function is a nuisance parameter. Cox’s partial likelihood estimator [58], denoted by , is used to obtain the p-value for the Wald test for vs. without specifying . One selects genes that exhibit smaller p-values than a threshold value. While the Cox’s approach is simple and widely accepted, the validity of the gene selection method relies on an important assumption.
To describe the assumption for censoring mechanisms, we define as random censoring time. The (training) samples consist of , , where and , where is the indicator function. The samples are often referred to as training samples since they are used to train the model (1). For the estimator to capture the true in Equation (1), one needs to impose the following assumption [37,41]:
Independent censoring assumption: Survival time and censoring time are independent conditionally on . That is,
If this assumption is violated, Cox’s estimator is biased for the true value of [37,41]. This assumption is violated when is dropout time from the study due to the worsening of his/her health status or is time at removal for transplantation [36,37].
2.2. Copula-Based Gene Selection Method
Copulas provide a general model to relax the assumption of independent censoring. Copula-based dependent censoring models have gained popularity in recent years for dealing with survival data [41,42,43,44,45,46,47,48,49,50,55,56]. For the purpose of gene selection, [41] introduced the following copula model:
| (2) |
where is a copula [59] with a parameter that specifies the strength of dependence.
The copula model (2) can specify a variety of dependent censoring structures. For implementation, a copula with a simple form, such as the Clayton, Gumbel, or Frank copula, is suggested. Furthermore, a copula is better parameterized so that gives the independence copula . An example is the Clayton copula of the form
This copula accommodates the independence copula as its limiting case by as . As the value of departs from zero, the level of dependence increases. To measure the degree of dependence, it is convenient to use Kendall’s tau, defined as under the Clayton copula. The value of ranges from independence () to perfect positive dependence (). While the Clayton copula permits only positive dependence, other copulas (e.g., the Frank copula) give negative dependence.
Under the Clayton copula model, a significance of gene on survival time is measured by fitting the Cox model (1), namely,
Here, measures the association between survival and a gene. The significance of a gene is measured by the estimator proposed by [41], denoted as . Accordingly, a subset of significant genes is constructed via a p-value threshold for testing vs. . The value of can be estimated by that maximizes the cross-validated c-index [37,41]. The computation is implemented by the R package, compound.Cox [9], and explained by the book [37].
2.3. Prognostic Prediction Method
Once genes are selected based on survival data, one often wishes to assess their prognostic ability for future patients. In many cases, the metric of interest is the ability of the genes to separate good and poor prognosis groups. For this purpose, researchers typically use a multi-gene predictor, defined as a weighted sum of gene expressions [1,5,6,7,8,9].
To formulate a multi-gene predictor, suppose that selected genes are measured for a future patient for a prognostic prediction purpose, denoted as . Survival prediction can be formulated by the prognostic index { XE “prognostic index:prognostic index”}(PI) defined as
The PI is a weighted sum of genes whose weights reflect the degree of univariate association. A high (low) value of the PI gives a high (low) risk of death. Thus, a patient may be classified into either a low-risk group or a high-risk group using a cut-off value (e.g., the median of the PI values calculated from the training samples).
If were assumed in , the PI would be the classical predictor based on univariate Cox regression under the independent censoring assumption [1,5,6,7,8,9]. As the value of is estimated by maximizing the cross-validated c-index [37,41], the resultant may have an improved predictive ability over the traditional PI. In medical research, it is the standard to display two Kaplan–Meier (KM) estimates for survival functions calculated for the two groups to see if there is a clear separation between them. However, if dependent censoring exists in test samples, the KM estimators are biased [52]. Therefore, we shall propose a method to examine a sensitivity of the PI to dependent censoring.
3. Proposed Methods
In this section, we propose a sensitivity analysis method using the CG estimator to assess the impact of dependent censoring on survival prognostic prediction.
This section deals with test samples that are different from the training samples used to select genes and develop the PI (Section 2). In our proposed method, the primary role of the test samples is to examine the sensitivity of the prognostic ability of the PI to dependent censoring. In many medical studies, random multiple splits for training/test sets are applied to develop and validate a PI. However, the goal of the proposed sensitivity analysis is different. A sensitivity analysis is performed to examine the potential effect of dependent censoring that may be present in the test samples.
Define as survival time, as censoring time, and as a -vector of genes from individuals in the test samples. The test samples consist of , , where and , where the sample size is possibly different from the size of the training samples. Let be the PI for the i-th patient in the test samples, where is computed by the training samples (Section 2.2).
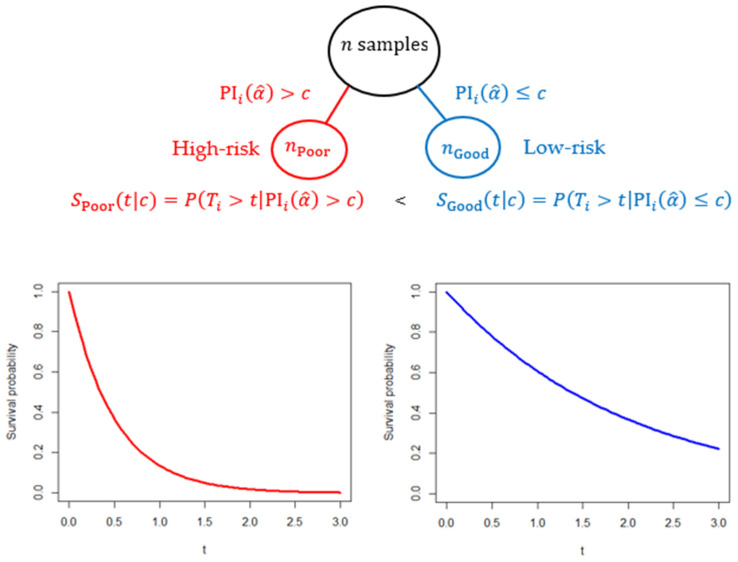
Figure 1 shows how the PI is applied to classify patients into a high-risk (low-risk) group if his/her PI is greater (less) than a cut-off value . Concretely, is a good prognosis (low-risk) group, and is a poor prognosis (high-risk) group. If the classification is successful, the conditional survival function is higher than the conditional survival function , namely, .
Figure 1.
Good and poor prognosis group separated by the PI applied for the testing sample of size n. Concretely, is a good prognosis (low-risk) group, and is a poor prognosis (high-risk) group. If the classification is successful, survival functions exhibit . The cut-off value can be the median of the PI values calculated from the training samples.
The cut-off value may be chosen to be the 50th percentile (median) of the PIs that are computed by the training samples. The median was chosen to ensure a desirable statistical power to detect the difference of the two groups and to achieve robust groupings by eliminating the instability due to unevenly allocated sample sizes. Additionally, this rule has been commonly employed in the analysis of survival prognostic prediction with a multi-gene predictor [6,20,25,26].
3.1. Sensitivity Analysis via the Copula-Graphic Estimator
This section describes the proposed method of assessing the sensitivity of the PI by using the CG estimator.
Let be a good prognosis (low-risk) group, and be a poor prognosis (high-risk) group, where is a cut-off value. Let and be the sizes of the groups. To assess the sensitivity of the prognostic ability of the PI, one needs to understand how well the two survival probabilities are differentiated between the two groups (Figure 1). Accordingly, one needs to estimate two conditional survival functions:
We suggest estimating them by the CG estimator of [52] that adjusts for the effect of dependent censoring { XE “Kendall’s tau:Kendall’s tau”}. In order to apply the CG estimator, it is necessary to impose the following Archimedean copula models:
| (3) |
| (4) |
where is a generator function that specifies the dependence structure. It is a continuous and decreasing function such that and [59]. The value of in Equations (3) and (4) can be chosen arbitrarily and can be different from the value estimated by the training samples.
If the models (3) and (4) are assumed, one can estimate and consistently by the following CG estimators:
where and are the numbers at risk at .
Note that the CG estimator reduces to the KM estimator by , and hence, the resultant estimator reduces to the KM estimator, e.g.,
For instance, for the Clayton copula, one has with . The copula generated by is the independence copula:
This means that the KM estimator is the CG estimator under the independent censoring assumption.
To compute the CG estimators, a parametric form of the copula must be specified (examples of parametric copulas will be shown in Section 3.2). That is, both the form of and the parameter value of must be specified. However, both of them are difficult to estimate with test samples since they provide modest information about the copula [51,52,60].
Our strategy to tackle this difficulty is a sensitivity analysis that is performed under a variety of copula models [52,53,54,55,56]. More precisely, we suggest plotting and for the three forms of and many different parameter values of . If and are clearly separated for a variety of and , we conclude that the prognostic ability the PI is shown to be robust against dependent censoring. This strategy is in line with previous sensitivity analyses [52,53,54,55,56], yet it is a new method in the context of prognostic survival analysis.
An objective metric of “clear separation” between and is obtained via a significance test proposed by Emura and Chen [37,41]. Formally, one can reject the null hypothesis against for a large value of , where
where is the largest time point where survivors are seen for both good and poor groups.
The p-value of the test is computed by , where is a random permutation for based on for a random permutation . To stabilize, we suggest a large number, say . Note that is known as the restricted mean survival time difference (RMSTD).
The actual implementation of the proposed sensitivity analysis is technical. Thus, the development of the software implementation and the online web application are desirable for clinicians and patients to use. We leave this topic to Section 4. The following subsection provides specific examples of parametric copulas that are currently implemented in our developed R functions and web application.
3.2. Examples of Parametric Copulas
This subsection describes some examples of parametric copulas in order to perform a sensitivity analysis.
If the generator is for , it yields the Clayton copula
Kendall’s tau for measuring dependency is given by . The resultant Clayton CG estimator is
If the generator is for , it yields the Gumbel copula
Kendall’s tau for measuring dependency is given by . The resultant Gumbel CG estimator is
Finally, one can obtain the Frank CG estimator by the generator , .
By letting for the Gumbel copula, one has , and hence, the resultant CG estimators reduce to the KM estimator, namely, . For the Clayton copula, one has with . Hence, as . The case of the Frank copula is similar.
4. Software and Web Application
This section describes the development of software to establish a Shiny-based web application for the practical implementation of the proposed sensitivity analysis.
In order to implement the aforementioned sensitivity analyses (Section 3.1), we developed computational/graphical facilities for the CG estimators. First, we refined the computational routines for the CG estimators in the R package compound.Cox [9]. The package now includes the functions CG.Clayton(.), CG.Frank(.), and CG.Gumbel(.), which can compute the CG estimators under the three copulas. These functions compute the CG estimators and graphically display the plot possibly colored by an option (the S.col option). This is useful for visually distinguishing the good prognosis group (specify S.col=“blue”) and poor prognosis group (specify S.col=“red”). Second, we devise a new function, CG.test(.), for testing the prognostic difference for the good and poor prognosis groups based on the difference statistic between two CG estimates. The p-value is computed by the permutation test (Section 3.1). This is useful for objectively measuring the prognostic ability of the PI. While is recommended, this number may be reduced to to produce a web application that works reasonably fast.
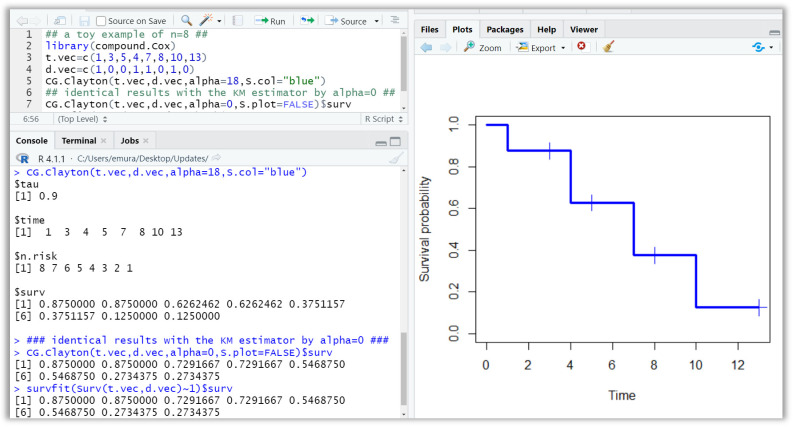
An example of the CG estimator under the Clayton copula is given in Figure 2. Users may run the R code (upper-left panel) after installing the compound.Cox package. Then, the plot of the CG estimates is displayed in the right panel. The numerical values of the estimates (survival probabilities at ) are shown in the lower-left panel. This example uses data 1, 3, 5, 4, 7, 8, 10, 13; 1, 0, 0, 1, 0, 1, 0. The data are artificial data, which may be regarded as patients from a good prognosis group (hence, we specify the S.col =“blue” option to make the plot blue-colored). The Clayton copula’s parameter is specified as (Kendall’s tau is 0.9). If one specifies the copula parameter as , this yields the KM estimator. We see that the numerical results are identical to the results computed by the survfit(.) function (lower-left panel).
Figure 2.
The R console showing an example of the CG estimator under the Clayton copula. The upper-left panel is the R code. The right panel is the plot of the Clayton CG estimates computed by data 1, 3, 5, 4, 7, 8, 10, 13; 1, 0, 0, 1, 0, 1, 0. The lower-left panel shows the numerical values of the estimates (survival probabilities). The Clayton copula’s parameter is specified as (Kendall’s tau is 0.9). If one specifies the copula parameter as , this yields the KM estimator. We see that the numerical results are identical to the results computed by the survfit(.) function (lower-left panel).
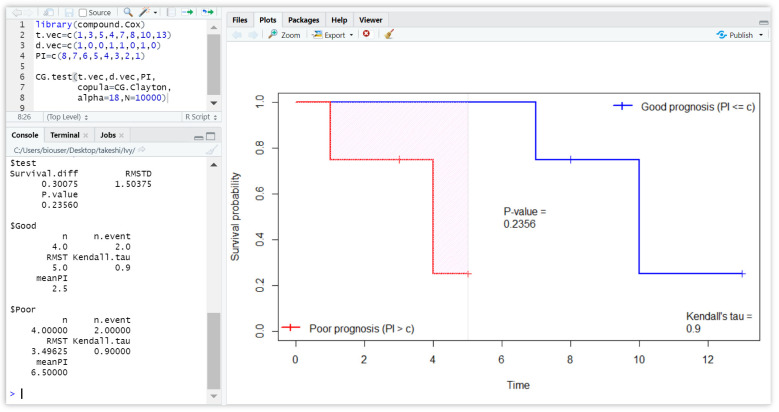
An example for testing the difference for the good and poor prognosis is given in Figure 3. This example uses prognostic indices 8, 7, 6, 5, 4, 3, 2, 1 to predict the survival outcomes and already defined. The figure displays the plots of the Clayton CG estimates that are separated between good and poor prognoses group. In addition, the figure shows the p-value computed by the permutation test. More detailed results, such as the difference () and the RMSTD (), are given in the left panel.
Figure 3.
The R console showing the test results for the difference for the good and poor prognoses. The right panel gives the plots of the Clayton CG estimates computed by data 1, 3, 5, 4, 7, 8, 10, 13; 1, 0, 0, 1, 0, 1, 0; 8, 7, 6, 5, 4, 3, 2, 1. The Clayton copula’s parameter is specified as . More detailed results, such as the difference () and the RMSTD (), are given in the left panel.
While we developed the infrastructures of the computational/graphical facilities for the CG estimators, the actual implementation of the sensitivity analysis is still complex. This is because the sensitivity analysis requires the understanding of the R programming and the proposed methods of Section 3. To further make our methodologies accessible to users, we propose a method to produce an interactive web application that can be manipulated by users with minimal knowledge (e.g., clinicians and patients).
We suggest an R package Shiny to make an online web application that can display the plots of and given a user-specified form of and a value of . With Shiny, one can easily convert R programs into a web application. This process can be carried out by creating the “app.R” file, a program file written in the R language. We develop a template file (available in Supplementary Materials) so that developers can modify the template for their own prediction settings. Once the template is appropriately tuned by developers, the web application is immediately built and can be made publicly available through a platform shinyapps.io (https://www.shinyapps.io/ (accessed on 10 January 2023). A concrete implementation of the web application shall be described in the next section.
5. Results
The purpose of this section is to demonstrate the proposed methods of Section 3 by using the lung cancer data originally reported in [6] and now available in the Lung object in the compound.Cox package [9]. We first introduce the dataset.
5.1. Lung Cancer Data
Briefly, the Lung data are a survival dataset containing training samples and test samples of 125 surgically treated non-small-cell lung cancer patients [6]. The endpoint of interest is overall survival { XE “overall survival:overall ”}, i.e., months from surgery to death. Overall survival time and censoring status (death or alive) were recorded. Additionally, available are high-dimensional gene expressions that may predict survival. All the gene expressions were coded as 1, 2, 3, or 4 according to the quartile levels following the original study [6].
In the following analysis, we selected the 16 genes and then constructed the multi-gene predictor as described in Section 2.2–Section 2.3 (also reported in [37,41]). The resultant predictor takes the form for the i-th patient in the test samples, where is computed by the training samples given the copula parameter (Kendall’s tau = 0.90), where are gene expression{ XE “gene expression:gene expression ”}s taking values 1, 2, 3, and 4. In terms of gene symbols, the 16-gene predictor is
| PI = (0.51 × MMP16) + (0.51 × ZNF264) + (0.50 × HGF) + (−0.49 × HCK) + (0.47 × NF1) + (0.46 × ERBB3) + (0.57 × NR2F6) + (0.77 × AXL) + (0.51 × CDC23) + (0.92 × DLG2) + (−0.34 × IGF2) + (0.54 × RBBP6) + (0.51 × COX11) + (0.40 × DUSP6) + (−0.37 × ENG) + (−0.41 × IHPK1). |
In order to examine the sensitivity of the PI under a variety of dependent censoring scenarios, we shall apply the proposed sensitivity analysis methods.
5.2. Sensitivity Analysis via the Web App
As prescribed in Section 3, we formed a good prognosis group and a poor prognosis group , where the cut-off value was set to be the 50th percentile (median) of the PIs in the training samples (as we proposed in Section 3). The test samples were then separated into two groups. Then, we evaluated the prognostic performance of the PI by displaying the estimated survival functions and given a copula and a value of .
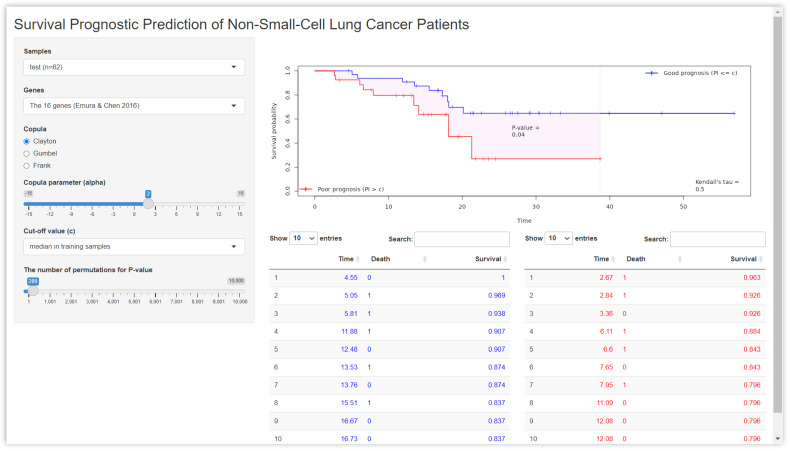
We constructed a web application (Figure 4) by the methods of Section 4. The interactive web version is available in https://takeshi.shinyapps.io/lung/ (accessed on 10 January 2023). The main goal of this web application is to visualize how the two prognosis groups are separated by the PI for a user-specified choice of copulas and any value of .
Figure 4.
The web application for the sensitivity analysis based on the lung cancer data. The interactive web version is available in https://takeshi.shinyapps.io/lung/ (accessed on 10 January 2023). The main goal of this web application is to visualize how the two prognosis groups are separated by the 16-gene PI for a user-specified choice of (copula) and any value of (copula parameter).
Using this web application, we tried the Clayton, Gumbel, and Frank copulas under several values of . For the Clayton copula and , one can see a good separation between the plots of and as shown in the right panel of Figure 4; one can also see the p-value (=0.04 based on N = 200 permutations).
Table 1 summarizes the p-values under the three copulas with selected values of whose Kendall’s measures of correlation (tau) take −0.75, −0.46, 0, 0.33, 0.46, 0.50, and 0.75. The p-values were obtained based on N = 10,000 permutations (by setting the left panel of Figure 4). One can observe that the p-values obtained are less than or around 0.05 for positive dependence (Kendall’s tau = 0.75 or 0.50), while they are greater than 0.10 for negative dependence and independence (tau = −0.75, −0.46, 0). Therefore, the PI would lose the ability of prognostic prediction when censoring in the test data is negatively dependent on survival. Therefore, our analysis reveals a potential drawback for the PI developed by Emura and Chen [41] when negatively dependent censoring arises. On the other hand, the PI gains its improved prognostic ability when censoring is positively dependent on survival. The p-value reaches nearly 1% level for strongly positive dependence (tau = 0.88 or 0.91). This is reasonable since the PI is developed under the Clayton copula with strongly positive dependence (, tau = 0.90).
Table 1.
p-values for testing the difference of survival prognosis (good vs. poor) under the three copulas with selected values of α (Kendall’s tau takes −0.75, −0.46, 0, 0.33, 0.46, 0.50, 0.75).
| Prognostic Index (PI) | |||
|---|---|---|---|
| Emura and Chen [41] | Chen et al. [6] | ||
| Clayton copula | (tau = 0.33) | 0.065 | 0.036 |
| (tau = 0.50) | 0.036 | 0.036 | |
| (tau = 0.75) | 0.021 | 0.036 | |
| (tau = 0.88) | 0.011 | 0.029 | |
| Gumbel copula | (tau = 0.00) | 0.171 | 0.040 |
| (tau = 0.50) | 0.058 | 0.029 | |
| (tau = 0.75) | 0.028 | 0.029 | |
| (tau = 0.91) | 0.011 | 0.025 | |
| Frank copula | (tau = −0.75) | 0.360 | 0.059 |
| (tau = −0.46) | 0.307 | 0.054 | |
| (tau = 0.46) | 0.054 | 0.034 | |
| (tau = 0.75) | 0.018 | 0.032 | |
Recall that the cut-off value was set to be the median of ’s. If we change the cut-off value to be the first or third quartile (25% or 75% points) of ’s, then the separation between the plots of and is not clear. This confirms our suggestion to apply the median as the cut-off value.
5.3. Comparison with the 16-Gene Pedictor Developed by Chen et al. [6]
The proposed sensitivity analysis is useful for any type of established predictor, not restricted to the predictor developed by the copula-based gene selection method. Furthermore, the proposed web application allows one to compare different predictors in a user-friendly manner. In order to demonstrate this feature, we performed a sensitivity analysis for the PI based on Chen et al. [6] defined as
| PI = (−1.09 × ANXA5) + (1.32 × DLG2) + (0.55 × ZNF264) + (0.75 × DUSP6) + (0.59 × CPEB4) + (−0.84 × LCK) + (−0.58 × STAT1) + (0.65 × RNF4) + (0.52 × IRF4) + (0.58 × STAT2) + (0.51 × HGF) + (0.55 × ERBB3) + (0.47 × NF1) + (−0.77 × FRAP1) + (0.92 × MMD) + (0.52 × HMMR). |
Table 1 shows the p-values testing the difference of survival prognosis (good vs. poor) made by the PI. We observe that the p-values are mostly less than 0.05 for the three copulas with selected values of α (Kendall’s tau takes −0.75, −0.46, 0, 0.33, 0.46, 0.50, 0.75). Therefore, the predictive ability of the PI is justified for a variety of dependent censoring models. This extremely confirms the validity and robustness of the PI developed by Chen et al. [6], even though they are developed under the independent censoring model. On the other hand, the ability of the PI under strong positive dependence is reduced compared to the PI developed by Emura and Chen [41]. If strong positive dependence exists in the test samples, the latter PI better prognosticates survival. However, we do not suggest determining the single best copula and the single value of α in the sensitivity analysis based on the CG estimators [52,53,54,55,56]. Instead, we suggest tying different copulas and their parameters to understand the sensitivity of the PI.
6. Conclusions and Discussion
In this article, we propose a novel sensitivity analysis method for a multi-gene predictor for survival. The primary tool for the proposed method is the CG estimator [52], which can effectively adjust for the impact of dependent censoring on survival data. Consequently, a unique feature of the proposed method is the ability to handle dependent censoring, which has been of increasing concern to medical statisticians in a variety of survival analysis methods [41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56]. The lung cancer data analysis demonstrates the implementation of the proposed method on a web application (Figure 4; https://takeshi.shinyapps.io/lung/ (accessed on 10 January 2023)). A user of this application can easily perform a sensitivity analysis on survival prognostic models using a smartphone.
From a methodological point of view, the proposed method is a novel implementation of the CG estimator [52] to the context of prognostic prediction. So far, the CG estimator has been applied to a single population setting [44,51,52,53,54], two-sample comparison [55], simple regression [43], multiple regression [60,61], factorial designs [56], and survival forests [47]. The objective of survival forests is to classify patients into several groups, which is similar to the goal of the proposed method. The difference is that survival forests use a decision tree while ours uses a multi-gene predictor. However, survival forests have not been adapted to deal with high-dimensional gene expressions. Hence, it is of great interest to extend survival forests to handle both dependent censoring and high-dimensional covariates as in a survival tree [62].
In this article, we employed the R package Shiny to transfer our sensitivity analysis formulas to a user-friendly web application. Some authors have used the Shiny package in other contexts. A series of works by Fournier et al. [63], Asar et al. [64], and Lenain et al. [65] made a Shiny-based web application for the dynamic prediction of long-term kidney graft failure. Similarly, the dynamic prediction method of Emura et al. [21] (see also the updated works of [66]) was transferred to web applications [67]. Clearly, this type of survival prognostic prediction tool can promote the development of personalized medicine, allowing clinicians to utilize powerful prognostic prediction models.
We note that the issue of dependent censoring in survival prognostic prediction is not restricted to medical research, which can be seen in the reliability analysis of mechanical items or systems [68,69,70,71,72,73]. In the reliability prediction of mechanical equipment, the survival probability is usually modeled by parametric distributions, such as the Weibull distribution. The analysis of a prognostic prediction model in engineering settings poses additional challenges tailored to parametric analyses. Recall that the CG estimator is a semiparametric estimator that does not assume any parametric form of the survival function. The semiparametric CG estimator [52] is not easily modified to parametric models since the estimator may require some numerical integrations.
We focus on the three Archimedean copulas: the Clayton, Gumbel, and Frank copulas, which have been extensively used in survival analysis [74,75,76,77,78] and other fields [79,80,81]. Owing to their remarkable popularity, our choices of copulas are natural. Furthermore, these Archimedean copulas allow simple formulas to compute the CG estimators [52]. However, there are a large number of non-Archimedean copulas popular in a variety of applications, such as the FGM copula, Gaussian copula, trigonometric copula, and Celebioglu–Cuadras copula [82,83,84,85,86,87]. As the CG estimators have not been considered for these non-Archimedean copulas, it is of great interest to develop computational tools for them.
Acknowledgments
We thank the Guest Editor and the three reviewers for their helpful comments that improved this manuscript.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/biomedicines11030797/s1, the “app.R” file, a program file written in the R language. Readers may run this program (after installing the Shiny R package) to see how our web application was created.
Author Contributions
Conceptualization, T.E.; Formal analysis, C.-T.Y. and T.E.; Funding acquisition, G.-Y.L.; Methodology, C.-T.Y. and T.E.; Supervision, G.-Y.L. and T.E.; Writing—original draft, C.-T.Y. and T.E. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the results in this article are reproducible by the code available in Supplementary Materials.
Conflicts of Interest
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This work was financially supported by JSPS KAKENHI (22K11948) and Chang Gung Memorial Hospital (BMRP745).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Witten D.M., Tibshirani R. Survival analysis with high-dimensional covariates. Stat. Methods Med. Res. 2010;19:29–51. doi: 10.1177/0962280209105024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newcombe P.J., Ali H.R., Blows F.M., Provenzano E., Pharoah P.D., Caldas C.M., Richardson S.T. Weibull regression with Bayesian variable selection to identify prognostic tumour markers of breast cancer survival. Stat. Methods Med. Res. 2017;26:414–436. doi: 10.1177/0962280214548748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Q., Wang J., Liu M., Zhu Q., Li Q., Xie C., Han C., Wang Y., Gao M., Liu J. Weighted correlation gene network analysis reveals a new stemness index-related survival model for prognostic prediction in hepatocellular carcinoma. Aging. 2020;12:13502. doi: 10.18632/aging.103454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharjee A. Big Data Analytics in Oncology with R. CRC Press; New York, NY, USA: 2022. [Google Scholar]
- 5.Matsui S. Statistical issues in clinical development and validation of genomic signatures. In: Matsui S., Buyse M., Simon R., editors. Design and Analysis of Clinical Trials for Predictive Medicine. CRC Press; Boca Raton, FL, USA: 2015. pp. 207–226. [Google Scholar]
- 6.Chen H.-Y., Yu S.-L., Chen C.-H., Chang G.-C., Chen C.-Y., Yuan A., Cheng C.-L., Wang C.-H., Terng H.-J., Kao S.-F., et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N. Engl. J. Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 7.Beer D.G., Kardia S.L., Huang C.-C., Giordano T.J., Levin A.M., Misek D.E., Lin L., Chen G., Gharib T.G., Thomas D.G., et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 8.Emura T., Chen Y.-H., Chen H.-Y. Survival prediction based on compound covariate under Cox proportional hazard models. PLoS ONE. 2012;7:e47627. doi: 10.1371/journal.pone.0047627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emura T., Matsui S., Chen H.-Y. compound.Cox: Univariate feature selection and compound covariate for predicting survival. Comput. Methods Programs Biomed. 2019;168:21–37. doi: 10.1016/j.cmpb.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 10.Jenssen T.K., Kuo W.P., Stokke T., Hovig E. Association between gene expressions in breast cancer and patient survival. Hum. Genet. 2002;111:411–420. doi: 10.1007/s00439-002-0804-5. [DOI] [PubMed] [Google Scholar]
- 11.Van’T Veer L.J., Dai H., Van De Vijver M.J., He Y.D., Hart A.A.M., Mao M., Peterse H.L., Van Der Kooy K., Marton M.J., Witteveen A.T., et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 12.Li D., Hu X.J., Wang R. Evaluating Association between Two Event Times with Observations Subject to Informative Censoring. J. Am. Stat. Assoc. 2021 doi: 10.1080/01621459.2021.1990766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sotiriou C., Wirapati P., Loi S., Harris A., Fox S., Smeds J., Nordgren H., Farmer P., Praz V., Haibe-Kains B., et al. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 14.Haibe-Kains B., Desmedt C., Loi S., Culhane A., Bontempi G., Quackenbush J., Sotiriou C. A Three-Gene Model to Robustly Identify Breast Cancer Molecular Subtypes. J. Natl. Cancer Inst. 2012;104:311–325. doi: 10.1093/jnci/djr545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng M., Xiang L. Correlation-based joint feature screening for semi-competing risks outcomes with application to breast cancer data. Stat. Methods Med. Res. 2021;30:2428–2446. doi: 10.1177/09622802211037071. [DOI] [PubMed] [Google Scholar]
- 16.Li F., Niu Y., Zhao W., Yan C., Qi Y. Construction and validation of a prognostic model for lung adenocarcinoma based on endoplasmic reticulum stress-related genes. Sci. Rep. 2022;12:19857. doi: 10.1038/s41598-022-23852-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding H., Shi L., Chen Z., Lu Y., Tian Z., Xiao H., Deng X., Chen P., Zhang Y. Construction and evaluation of a prognostic risk model of tumor metastasis-related genes in patients with non-small cell lung cancer. BMC Med. Genom. 2022;15:187. doi: 10.1186/s12920-022-01341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neto C., Brito M., Lopes V., Peixoto H., Abelha A., Machado J. Application of Data Mining for the Prediction of Mortality and Occurrence of Complications for Gastric Cancer Patients. Entropy. 2019;21:1163. doi: 10.3390/e21121163. [DOI] [Google Scholar]
- 19.Zhang H., Lin Y., Zhuang M., Zhu L., Dai Y., Lin M. Screening and identification of CNIH4 gene associated with cell proliferation in gastric cancer based on a large-scale CRISPR-Cas9 screening database DepMap. Gene. 2023;850:146961. doi: 10.1016/j.gene.2022.146961. [DOI] [PubMed] [Google Scholar]
- 20.Yoshihara K., Tajima A., Yahata T., Kodama S., Fujiwara H., Suzuki M., Onishi Y., Hatae M., Sueyoshi K., Fujiwara H., et al. Gene Expression Profile for Predicting Survival in Advanced-Stage Serous Ovarian Cancer across Two Independent Datasets. PLoS ONE. 2010;5:e9615. doi: 10.1371/journal.pone.0009615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emura T., Nakatochi M., Matsui S., Michimae H., Rondeau V. Personalized dynamic prediction of death according to tumour progression and high-dimensional genetic factors: Meta-analysis with a joint model. Stat. Methods Med. Res. 2018;27:2842–2858. doi: 10.1177/0962280216688032. [DOI] [PubMed] [Google Scholar]
- 22.Waldron L., Haibe-Kains B., Culhane A.C., Riester M., Ding J., Wang X.V., Ahmadifar M., Tyekucheva S., Bernau C., Risch T., et al. Comparative Meta-analysis of Prognostic Gene Signatures for Late-Stage Ovarian Cancer. J. Natl. Cancer Inst. 2014;106:049. doi: 10.1093/jnci/dju049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emura T., Nakatochi M., Murotani K., Rondeau V. A joint frailty-copula model between tumour progression and death for meta-analysis. Stat. Methods Med Res. 2017;26:2649–2666. doi: 10.1177/0962280215604510. [DOI] [PubMed] [Google Scholar]
- 24.Gao L., Jiang W., Yue Q., Ye R., Li Y., Hong J., Zhang M. Radiomic model to predict the expression of PD-1 and overall survival of patients with ovarian cancer. Int. Immunopharmacol. 2022;113:109335. doi: 10.1016/j.intimp.2022.109335. [DOI] [PubMed] [Google Scholar]
- 25.Arora C., Kaur D., Lathwal A., Raghava G.P. Risk prediction in cutaneous melanoma patients from their clinico-pathological features: Superiority of clinical data over gene expression data. Heliyon. 2020;6:e04811. doi: 10.1016/j.heliyon.2020.e04811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu P., Dong C., Shi H., Yan Z., Zhang J., Liu J. Constructing and validating of m7G-related genes prognostic signature for hepatocellular carcinoma and immune infiltration: Potential biomarkers for predicting the overall survival. J. Gastrointest. Oncol. 2022;13:3169–3182. doi: 10.21037/jgo-22-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z., Wang J., Li S., Li L., Li L., Li D., Guo H., Gao D., Liu S., Ruan C., et al. Prognostic prediction and immune infiltration analysis based on ferroptosis and EMT state in hepatocellular carcinoma. Front. Immunol. 2022;13:1076045. doi: 10.3389/fimmu.2022.1076045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang X., Guo Y., Chen Z., Zhang F., Huang J., Qin Y. A prognostic risk prediction model based on ferroptosis-related long non-coding RNAs in bladder cancer: A bulk RNA-seq research and scRNA-seq validation. Medicine. 2022;101:e32558. doi: 10.1097/MD.0000000000032558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou L., Cheng Q., Hu Y., Tan H., Li X., Wu S., Zhou T., Zhou J. Cuproptosis-related LncRNAs are potential prognostic and immune response markers for patients with HNSCC via the integration of bioinformatics analysis and experimental validation. Front. Oncol. 2022;12:1030802. doi: 10.3389/fonc.2022.1030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J., Xu Z., Yuan Z., Teh B.M., Zhou C., Shen Y. Identification of a cuproptosis-related lncRNA signature to predict the prognosis and immune landscape of head and neck squamous cell carcinoma. Front. Oncol. 2022;12:983956. doi: 10.3389/fonc.2022.983956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He Y., Lin Z., Tan S. Identification of prognosis-related gene features in low-grade glioma based on ssGSEA. Front. Oncol. 2022;12:1056623. doi: 10.3389/fonc.2022.1056623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vannucchi A.M., Guglielmelli P. Molecular prognostication in Ph-negative MPNs in 2022. Hematology. 2022;1:225–234. doi: 10.1182/hematology.2022000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z., Xia Z., Yu Y., Cai L., Jian W., Wang T., Xue W., Wang X., Wang B., Zhang P., et al. A pyroptosis-associated signature plays a role in prognosis prediction in clear cell renal cell carcinoma. BMC Med. Genom. 2022;15:204. doi: 10.1186/s12920-022-01339-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi J., Oh I., Seo S., Ahn J. G2Vec: Distributed gene representations for identification of cancer prognostic genes. Sci. Rep. 2018;8:13729. doi: 10.1038/s41598-018-32180-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim M., Oh I., Ahn J. An Improved Method for Prediction of Cancer Prognosis by Network Learning. Genes. 2018;9:478. doi: 10.3390/genes9100478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staplin N.D., Kimber A.C., Collett D., Roderick P.J. Dependent censoring in piecewise exponential survival models. Stat. Methods Med. Res. 2015;24:325–341. doi: 10.1177/0962280214544018. [DOI] [PubMed] [Google Scholar]
- 37.Emura T., Chen Y.H. Analysis of Survival Data with Dependent Censoring: Copula-Based Approaches. Springer; Singapore: 2018. [Google Scholar]
- 38.Schneider S., Demarqui F., Colosimo E.A., Mayrink V.D. An approach to model clustered survival data with dependent censoring. Biom. J. 2020;62:157–174. doi: 10.1002/bimj.201800391. [DOI] [PubMed] [Google Scholar]
- 39.Schneider S., Demarqui F., de Freitas Costa E. Free-ranging dogs’ lifetime estimated by an approach for long-term survival data with dependent censoring. Environ. Ecol. Stat. 2002;29:869–911. doi: 10.1007/s10651-022-00549-0. [DOI] [Google Scholar]
- 40.Bhattacharjee A., Vishwakarma G.K., Banerjee S., Ong S.H. A modified risk detection approach of biomarkers by frailty effect on multiple time to event data. J. Comput. Appl. Math. 2023;419:114681. doi: 10.1016/j.cam.2022.114681. [DOI] [Google Scholar]
- 41.Emura T., Chen Y.-H. Gene selection for survival data under dependent censoring: A copula-based approach. Stat. Methods Med. Res. 2016;25:2840–2857. doi: 10.1177/0962280214533378. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y.-H. Semiparametric marginal regression analysis for dependent competing risks under an assumed copula. J. R. Stat. Soc. Ser. B. 2010;72:235–251. doi: 10.1111/j.1467-9868.2009.00734.x. [DOI] [Google Scholar]
- 43.Braekers R., Veraverbeke N. A copula-graphic estimator for the conditional survival function under dependent censoring. Can. J. Stat. 2005;33:429–447. doi: 10.1002/cjs.5540330308. [DOI] [Google Scholar]
- 44.Emura T., Michimae H. A copula-based inference to piecewise exponential models under dependent censoring, with application to time to metamorphosis of salamander larvae. Environ. Ecol. Stat. 2017;24:151–173. doi: 10.1007/s10651-017-0364-4. [DOI] [Google Scholar]
- 45.Xu J., Ma J., Connors M.H., Brodaty H. Proportional hazard model estimation under dependent censoring using copulas and penalized likelihood. Stat. Med. 2018;37:2238–2251. doi: 10.1002/sim.7651. [DOI] [PubMed] [Google Scholar]
- 46.Czado C., Van Keilegom I. Dependent censoring based on parametric copulas. Biometrika. 2022:asac067. doi: 10.1093/biomet/asac067. [DOI] [Google Scholar]
- 47.Moradian H., Larocque D., Bellavance F. Survival forests for data with dependent censoring. Stat. Methods Med. Res. 2019;28:445–461. doi: 10.1177/0962280217727314. [DOI] [PubMed] [Google Scholar]
- 48.Deresa N.W., Van Keilegom I., Antonio K. Copula-based inference for bivariate survival data with left truncation and dependent censoring. Insur. Math. Econ. 2022;107:1–21. doi: 10.1016/j.insmatheco.2022.07.011. [DOI] [Google Scholar]
- 49.Huang X.W., Wang W., Emura T. A copula-based Markov chain model for serially dependent event times with a dependent terminal event. Jpn. J. Stat. Data Sci. 2021;4:917–951. doi: 10.1007/s42081-020-00087-8. [DOI] [Google Scholar]
- 50.Lo S.M., Wilke R.A. A copula model for dependent competing risks. J. R. Stat. Soc. Ser. C (Appl. Stat.) 2010;59:359–376. doi: 10.1111/j.1467-9876.2009.00695.x. [DOI] [Google Scholar]
- 51.Zheng M., Klein J.P. Estimates of marginal survival for dependent competing risks based on an assumed copula. Biometrika. 1995;82:127–138. doi: 10.1093/biomet/82.1.127. [DOI] [Google Scholar]
- 52.Rivest L.P., Wells M.T. A martingale approach to the copula-graphic estimator for the survival function under dependent censoring. J. Multivar. Anal. 2001;79:138–155. doi: 10.1006/jmva.2000.1959. [DOI] [Google Scholar]
- 53.de Uña-Álvarez J., Veraverbeke N. Generalized copula-graphic estimator. Test. 2013;22:343–360. doi: 10.1007/s11749-012-0314-2. [DOI] [Google Scholar]
- 54.de Uña-Álvarez J., Veraverbeke N. Copula-graphic estimation with left-truncated and right-censored data. Statistics. 2013;51:387–403. doi: 10.1080/02331888.2016.1274898. [DOI] [Google Scholar]
- 55.Emura T., Hsu J.H. Estimation of the Mann–Whitney effect in the two-sample problem under dependent censoring. Comput. Stat. Data Anal. 2020;150:106990. doi: 10.1016/j.csda.2020.106990. [DOI] [Google Scholar]
- 56.Emura T., Ditzhaus M., Dobler D., Murotani K. Factorial survival analysis for treatment effects under dependent censoring. arXiv. 2023 doi: 10.1177/09622802231215805.2302.01617 [DOI] [PubMed] [Google Scholar]
- 57.Winston Chang J.C., Allaire J.J., Xie Y., McPherson J. Shiny: Web Application Framework for R. CRAN. 2021. [(accessed on 1 January 2023)]. Available online: https://CRAN.R-project.org/package=shiny.
- 58.Cox D.R. Regression models and life-tables. J. R. Stat. Soc. Ser. B (Methodol.) 1972;34:187–202. doi: 10.1111/j.2517-6161.1972.tb00899.x. [DOI] [Google Scholar]
- 59.Nelsen R.B. An Introduction to Copulas. Springer; New York, NY, USA: 2007. [Google Scholar]
- 60.Lo S.M., Wilke R.A. A regression model for the copula-graphic estimator. J. Econom. Methods. 2014;3:21–46. doi: 10.1515/jem-2012-0016. [DOI] [Google Scholar]
- 61.Sujica A., Van Keilegom I. The copula-graphic estimator in censored nonparametric location-scale regression models. Econom. Stat. 2018;7:89–114. doi: 10.1016/j.ecosta.2017.07.002. [DOI] [Google Scholar]
- 62.Emura T., Hsu W.C., Chou W.C. A survival tree based on stabilized score tests for high-dimensional covariates. J. Appl. Stat. 2023;50:264–290. doi: 10.1080/02664763.2021.1990224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fournier M.-C., Foucher Y., Blanche P., Legendre C., Girerd S., Ladrière M., Morelon E., Buron F., Rostaing L., Kamar N., et al. Dynamic predictions of long-term kidney graft failure: An information tool promoting patient-centred care. Nephrol. Dial. Transplant. 2019;34:1961–1969. doi: 10.1093/ndt/gfz027. [DOI] [PubMed] [Google Scholar]
- 64.Asar Ö., Fournier M.-C., Dantan E. Dynamic predictions of kidney graft survival in the presence of longitudinal outliers. Stat. Methods Med. Res. 2021;30:185–203. doi: 10.1177/0962280220945352. [DOI] [PubMed] [Google Scholar]
- 65.Lenain R., Dantan E., Giral M., Foucher Y., Asar Ö., Naesens M., Hazzan M., Fournier M.-C. External Validation of the DynPG for Kidney Transplant Recipients. Transplantation. 2021;105:396–403. doi: 10.1097/TP.0000000000003209. [DOI] [PubMed] [Google Scholar]
- 66.Kawakami R., Michimae H., Lin Y.-H. Assessing the numerical integration of dynamic prediction formulas using the exact expressions under the joint frailty-copula model. Jpn. J. Stat. Data Sci. 2021;4:1293–1321. doi: 10.1007/s42081-021-00133-z. [DOI] [Google Scholar]
- 67.Emura T., Michimae H., Matsui S. Dynamic Risk Prediction via a Joint Frailty-Copula Model and IPD Meta-Analysis: Building Web Applications. Entropy. 2022;24:589. doi: 10.3390/e24050589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kordestani M., Saif M., Orchard M.E., Razavi-Far R., Khorasani K. Failure Prognosis and Applications—A Survey of Recent Literature. IEEE Trans. Reliab. 2021;70:728–748. doi: 10.1109/TR.2019.2930195. [DOI] [Google Scholar]
- 69.Hong Y., Meeker W.Q., McCalley J.D. Prediction of remaining life of power transformers based on left truncated and right censored lifetime data. Ann. Appl. Stat. 2009;3:857–879. doi: 10.1214/00-AOAS231. [DOI] [Google Scholar]
- 70.Mitra D., Kundu D., Balakrishnan N. Likelihood analysis and stochastic EM algorithm for left truncated right censored data and associated model selection from the Lehmann family of life distributions. Jpn. J. Stat. Data Sci. 2021;4:1019–1048. doi: 10.1007/s42081-021-00115-1. [DOI] [Google Scholar]
- 71.Zheng R., Najafi S., Zhang Y. A recursive method for the health assessment of systems using the proportional hazards model. Reliab. Eng. Syst. Saf. 2022;221:108379. doi: 10.1016/j.ress.2022.108379. [DOI] [Google Scholar]
- 72.Emura T., Michimae H. Left-truncated and right-censored field failure data: Review of parametric analysis for reliability. Qual. Reliab. Eng. Int. 2022;38:3919–3934. doi: 10.1002/qre.3161. [DOI] [Google Scholar]
- 73.Zuo Z., Wang L., Lio Y. Reliability Estimation for Dependent Left-Truncated and Right-Censored Competing Risks Data with Illustrations. Energies. 2023;16:62. doi: 10.3390/en16010062. [DOI] [Google Scholar]
- 74.Peng M., Xiang L., Wang S. Semiparametric regression analysis of clustered survival data with semi-competing risks. Comp. Stat. Data Anal. 2018;124:53–70. doi: 10.1016/j.csda.2018.02.003. [DOI] [Google Scholar]
- 75.Burzykowski T., Molenberghs G., Buyse M., Geys H., Renard D. Validation of surrogate end points in multiple randomized clinical trials with failure time end points. J. R. Stat. Soc. Ser. C Appl. Stat. 2001;50:405–422. doi: 10.1111/1467-9876.00244. [DOI] [Google Scholar]
- 76.Rotolo F., Paoletti X., Michiels S. surrosurv: An R package for the evaluation of failure time surrogate endpoints in individual patient data meta-analyses of randomized clinical trials. Comput. Methods Programs Biomed. 2018;155:189–198. doi: 10.1016/j.cmpb.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 77.Emura T., Sofeu C.L., Rondeau V. Conditional copula models for correlated survival endpoints: Individual patient data meta-analysis of randomized controlled trials. Stat. Methods Med. Res. 2021;30:2634–2650. doi: 10.1177/09622802211046390. [DOI] [PubMed] [Google Scholar]
- 78.Orenti A., Boracchi P., Marano G., Biganzoli E., Ambrogi F. A pseudo-values regression model for non-fatal event free survival in the presence of semi-competing risks. Stat. Methods Appl. 2022;31:709–727. doi: 10.1007/s10260-021-00612-3. [DOI] [Google Scholar]
- 79.Ghosh S., Sheppard L.W., Holder M.T., Loecke T.D., Reid P.C., Bever J.D., Reuman D.C. Advances in Ecological Research. Volume 62. Academic Press; Cambridge, MA, USA: 2020. Copulas and their potential for ecology; pp. 409–468. [Google Scholar]
- 80.Zhou Y., Lu Z., Shi Y., Cheng K. The copula-based method for statistical analysis of step-stress accelerated life test with dependent competing failure modes. Proc. Inst. Mech. Eng. O J. Risk Reliab. 2019;233:401–418. doi: 10.1177/1748006X18793251. [DOI] [Google Scholar]
- 81.Shih J.-H., Konno Y., Chang Y.-T., Emura T. Copula-Based Estimation Methods for a Common Mean Vector for Bivariate Meta-Analyses. Symmetry. 2022;14:186. doi: 10.3390/sym14020186. [DOI] [Google Scholar]
- 82.Chesneau C. Theoretical study of some angle parameter trigonometric copulas. Modelling. 2022;3:140–163. doi: 10.3390/modelling3010010. [DOI] [Google Scholar]
- 83.Susam S.O. A multi-parameter Generalized Farlie-Gumbel-Morgenstern bivariate copula family via Bernstein polynomial. Hacet. J. Math. Stat. 2022;51:618–631. doi: 10.15672/hujms.993698. [DOI] [Google Scholar]
- 84.Ota S., Kimura M. Effective estimation algorithm for parameters of multivariate Farlie–Gumbel–Morgenstern copula. Jpn. J. Stat. Data Sci. 2021;4:1049–1078. doi: 10.1007/s42081-021-00118-y. [DOI] [Google Scholar]
- 85.Zhang Z., Charalambous C., Foster P. A Gaussian copula joint model for longitudinal and time-to-event data with random effects. Comp. Stat. Data Anal. 2023;181:107685. doi: 10.1016/j.csda.2022.107685. [DOI] [Google Scholar]
- 86.Chesneau C. Theoretical Contributions to Three Generalized Versions of the Celebioglu–Cuadras Copula. Analytics. 2023;2:31–54. doi: 10.3390/analytics2010003. [DOI] [Google Scholar]
- 87.Bekrizadeh H., Parham G.A., Jamshidi B. A new asymmetric class of bivariate copulas for modeling dependence. Commun. Stat.-Simul. Comput. 2017;46:5594–5609. doi: 10.1080/03610918.2016.1169292. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the results in this article are reproducible by the code available in Supplementary Materials.