Abstract
Simple Summary
Transposable elements (TEs) are genetic parasites that mobilize themselves from one locus to another in the host genome. DDD/E transposase gene is the most abundant gene in nature and its protein product catalyzes DNA transposition. The IS481 family is a group of prokaryotic TEs that encode a DDD/E transposase. Here, we report a group of eukaryotic TEs that shows close affinity to some of the prokaryotic IS481 family members, and designate them IS481EU. IS481EU was found in palabasalids including Trichomonas vaginalis. Although most TEs with DDD/E transposase generate direct repeats of fixed or similar lengths (target site duplications, TSDs) at both ends of insertion, IS481EU generates TSDs of discrete lengths (~4 bps, ~15 bps, or ~25 bps). The unique characteristics of IS481EU in protein sequences and the distribution of TSD lengths support its placement as a new superfamily of eukaryotic DNA transposons. The transmission of prokaryotic IS481 to a eukaryotic lineage likely resulted in the birth of IS481EU, and may have contributed to lineage-specific evolutionary trajectories.
Abstract
DDD/E transposase gene is the most abundant gene in nature and many DNA transposons in all three domains of life use it for their transposition. A substantial number of eukaryotic DNA transposons show similarity to prokaryotic insertion sequences (ISs). The presence of IS481-like DNA transposons was indicated in the genome of Trichomonas vaginalis. Here, we surveyed IS481-like eukaryotic sequences using a bioinformatics approach and report a group of eukaryotic IS481-like DNA transposons, designated IS481EU, from parabasalids including T. vaginalis. The lengths of target site duplications (TSDs) of IS481EU are around 4 bps, around 15 bps, or around 25 bps, and strikingly, these discrete lengths of TSDs can be observed even in a single IS481EU family. Phylogenetic analysis indicated the close relationships of IS481EU with some of the prokaryotic IS481 family members. IS481EU was not well separated from IS3EU/GingerRoot in the phylogenetic analysis, but was distinct from other eukaryotic DNA transposons including Ginger1 and Ginger2. The unique characteristics of IS481EU in protein sequences and the distribution of TSD lengths support its placement as a new superfamily of eukaryotic DNA transposons.
Keywords: DNA transposon, insertion sequence, DDD/E transposase, IS481, IS481EU, target site duplications (TSDs), parabasalids
1. Introduction
Transposable elements (TEs), transposons or mobile DNA, include a wide variety of DNA segments that can, in a process called transposition, move or duplicate themselves from one location to another [1]. TEs are traditionally grouped into two groups: retrotransposons and DNA transposons [2]. DDD/E transposase/integrase is the most abundant gene in nature [3], and is the most common transposase in both eukaryotic and prokaryotic DNA transposons [4]. DDD/E transposases constitute the RNase H fold [5] and contain three catalytic residues (DDD or DDE) for DNA strand transfer [4]. Upon integration, a short DNA segment is duplicated at both sides of DNA transposons and they are called target site duplications (TSDs). Integrases encoded by long terminal repeat (LTR) retrotransposons and retroviruses are also a type of DDD/E transposases [6]. Recombination-activating gene 1 (RAG1), the catalytic component for the V(D)J recombination system of jawed vertebrates, is a domesticated DDD/E transposase derived from a DNA transposon [7,8].
An insertion sequence (IS) was originally defined as a short DNA segment encoding only the enzymes necessary for its transposition [9,10,11]. Around 30 IS “families” are designated at ISFinder (https://www-is.biotoul.fr/, accessed on 23 May 2022), and the majority of them encode a DDD/E transposase or its variety. Sharing additional conserved residues apart from the DDE motif, some IS families are more closely related to each other than to others. The transposases of the IS3 family, the IS481 family, and the IS1202 group show a certain level of similarity to one another. Currently the IS1202 group is classified inside the ISNCY (IS not classified yet) family. Known eukaryotic DNA transposons are classified into 23 “superfamilies” in Repbase (https://www.girinst.org/repbase/, accessed on 24 May 2022) [12,13]. Among them, 21 superfamilies encode a DDD/E transposase. Like prokaryotic IS families, different eukaryotic transposon superfamilies sometimes share conserved motifs. For example, Dada, hAT, Kolobok, MuDR, and P share the signature motif “C/DxxH” between the second D and the last E conserved residues [14,15].
Despite the ancient splitting of eukaryotes and prokaryotes, a notable level of similarity is observed between a number of eukaryotic DDD/E transposases and their prokaryotic counterparts. IS630 in prokaryotes and Mariner, Tc1, pogo, and many related DNA transposons in eukaryotes have related DDD/E transposases, and are altogether called ITm [16]. Besides the sequence similarity of transposases, they generate 2 bp TSDs of TA in general. Multiple horizontal transfers from prokaryotes to eukaryotes have been suggested in this superfamily [17]. Prokaryotic IS1380 and eukaryotic piggyBac are distantly related, and they share 4 bp TSDs and the 5′-CC..GG-3′ termini [18,19]. Eukaryotic Merlin DNA transposons show a clear relationship with the prokaryotic IS1016 group inside of the IS1595 family [20]. Both generate 8 bp TSDs. Eukaryotic Zator shows a similarity to the prokaryotic ISAzo13 family and both of them generate 3 bp TSDs [21,22]. The relationship between prokaryotic IS256 and eukaryotic MuDR was reported and they share a signature of conserved residues in addition to the catalytic core with other eukaryotic DNA transposons [14,15,23,24]. They generate 4 bp to 10 bp TSDs. Eukaryotic Harbinger and ISL2EU superfamilies, as well as their relatives, called Spy, Nuwa, Pangu, and ISL2PR show a similarity to prokaryotic ISL2 and IS5 and together constitute PHIS [25,26,27]. They generate no TSDs or 1 bp to 4 bp TSDs. The horizontal transfer of prokaryotic IS5 from bacteria to the genome of bdelloid rotifer was also reported [26]. The transposase similarity and the similar lengths of TSDs suggest a shared mechanism of transposition, even between eukaryotic and prokaryotic TEs.
In our previous article, we reported three proteins that show similarity to the prokaryotic IS481 transposases from the genome of Trichomonas vaginalis, which was designated as IS481EU [28]. The presence of DNA transposons encoding an IS481-like transposase was also indicated and designated as Banshee [29]. However, these sequences have not been analyzed further and few characteristics of these putative transposons have been reported to date. Here, we report a group of eukaryotic DNA transposons showing similarity to the prokaryotic IS481 family from the multiple genomes of palabasalids, including T. vaginalis. One of the distinctive features of these IS481EU DNA transposons is the generation of three distinct lengths of TSDs: around 4 bp, around 15 bp or around 25 bp TSDs. The phylogenetic analysis suggested the close affinity of IS481EU with a sublineage of the IS481 family. IS481EU was not separated from the IS3EU/GingerRoot superfamily of eukaryotic DNA transposons in our phylogenetic analysis, but the different features of proteins and TSDs support its position as a new superfamily of eukaryotic DNA transposons.
2. Materials and Methods
2.1. Characterization of IS481EU Families
Censor [30] searches were performed against the genome of T. vaginalis with the protein sequences reported in [28]. Censor hits were extracted and clustered with BLASTCLUST 2.2.25 in the NCBI BLAST package with the thresholds at 75% length coverage and 75% sequence identity. The consensus sequence for each cluster was generated with the 50% majority rule applied and the help of homemade scripts. Censor searches were performed with the consensus sequence of each cluster against the genome. Up to 10 Censor hits were extracted with 5000 bp flanking sequences at both sides. Consensus sequences were regenerated to be elongated to find both termini. The termini were determined based on the terminal TG..CA signatures and the presence of TSDs.
Censor searches were performed with the protein sequences of IS481EU from T. vaginalis against the genomes of palabasalids (Table 1). Genome sequences were downloaded from NCBI Assembly (https://www.ncbi.nlm.nih.gov/assembly, accessed on 18 May 2022). The characterization of complete IS481EU sequences was conducted similarly to the cases of IS481EU from T. vaginalis. All characterized consensus sequences are available as Supplementary Materials (Data S1) and have also been submitted to Repbase (https://www.girinst.org/repbase/, accessed on 24 May 2022) [12].
Table 1.
The distribution of IS481EU in parabasalids.
| Phylum | Order | Family | Species | Genome Assembly | IS481EU Family |
|---|---|---|---|---|---|
| Parabasalia | Trichomonadida | Trichomonadidae | Trichomonas vaginalis | ASM289133v1 | IS481EU-1_TV to 10_TV, IS481EU-N1_TV to N3_TV, 4N1_TV, 5N1_TV, 8N1_TV, and 10N1_TV. |
| Trichomonas tenax | PRJEB22701 | IS481EU-1_TrTenax | |||
| Trichomonas gallinae | MiGF1c1.0 | IS481EU-1_TrGallinae to IS481EU-5_TrGallinae | |||
| Tritrichomonadida | Tritrichomonadidae | Tritrichomonas foetus | TF_PacBio | IS481EU-1_TrFoetus | |
| Dientamoebidae | Histomonas meleagridis | ASM2018611v1 | IS481EU-1_HisMel to 5_HisMel |
2.2. Protein Dataset and Phylogenetic Analysis
All protein sequences of IS481EU families from T. vaginalis and Histomonas meleagridis were used for the phylogenetic analysis. All protein sequences of the IS481 family and the IS1202 group were extracted from ISFinder (https://www-is.biotoul.fr/, accessed on 24 May 2022). Representatives of the IS3 family were chosen based on [11], and were extracted from ISFinder (https://www-is.biotoul.fr/, accessed on 23 May 2022) [9]. The protein sequences of all autonomous IS3EU (17 entries), Ginger1 (23 entries), and Ginger2 (30 entries) were extracted from Repbase (https://www.girinst.org/repbase/, accessed on 24 May 2022) [12]. The protein sequences of GingerRoot were extracted from the dataset reported in the original article [31]. Representatives of LTR retrotransposons and endogenous retroviruses were obtained from Repbase (https://www.girinst.org/repbase/, accessed on 24 May 2022) [12]. The sequences of HIV-1 and HTLV-1 were extracted from the dataset available at GyDB (https://gydb.org/, accessed on 24 May 2022) [32]. They were aligned with the help of MAFFT v.7.407 [33] with the linsi option or Clustal Omega [34] in the Seaview package. The conserved transposase domains were extracted and realigned with Probcons version 1.12 [35]. Protein sequences with a large deletion were removed.
A maximum likelihood tree was generated on the PhyML 3.0 server (http://www.atgc-montpellier.fr/phyml/, accessed on 27 May 2022) [36] with 100 bootstrapping supports and 2 datasets. One dataset was composed of the IS481EU and IS481 proteins. The substitution model Q.pfam +G+I was chosen based on the Bayesian information criterion (BIC). The other dataset was composed of the representatives of IS481EU, IS481, IS1202, IS3, IS3EU, Ginger1, Ginger2, GingerRoot, LTR retrotransposons, and retroviruses. The substitution model Q.pfam +G+I+F was chosen based on the BIC. The phylogenetic tree was rooted at the midpoint and visualized with FigTree v.1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 27 May 2022).
3. Results
3.1. Eukaryotic TEs Related to the Prokaryotic IS481 Family
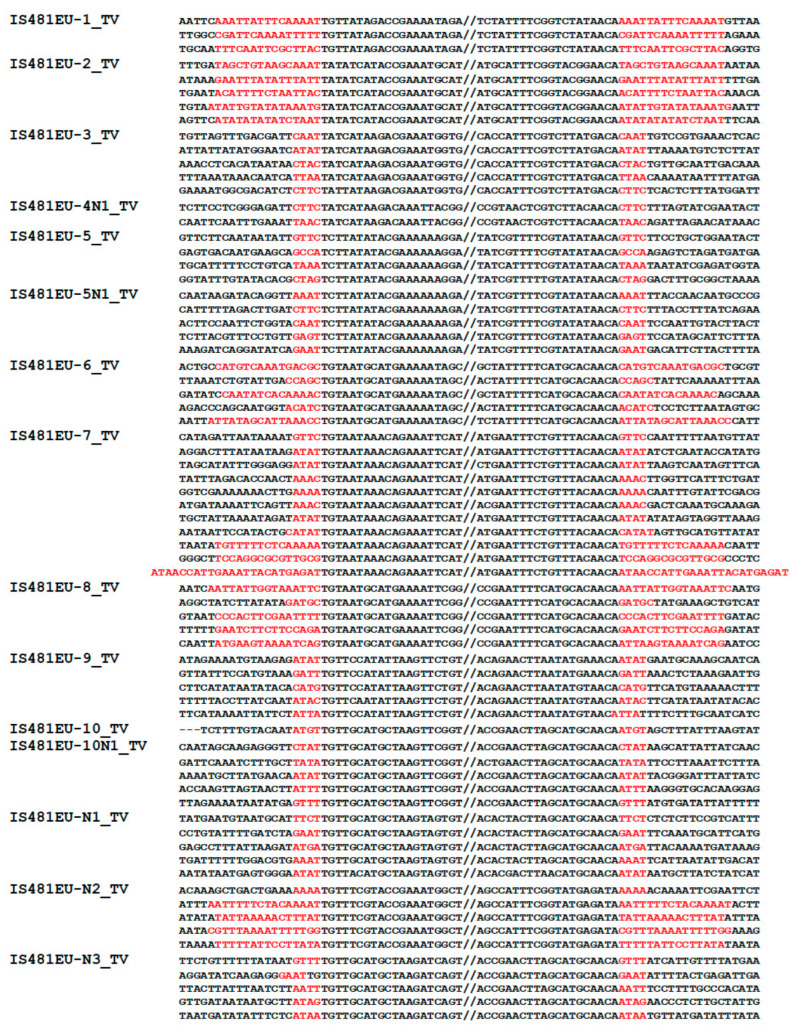
In the survey of TEs encoding a protein similar to the reported IS481EU transposases in the genome of a protist T. vaginalis, we were able to characterize 10 autonomous IS481EU families and 7 non-autonomous families (Table 1 and Table S1). The letter N in the family name indicates a non-autonomous family. While we could characterize the autonomous counterparts of four non-autonomous families (for example, IS481EU-4_TV for IS481EU-4N1_TV), we could not find the autonomous counterparts of the remaining three non-autonomous families (IS481EU-N1_TV to IS481EU-N3_TV). These IS481EU transposons from T. vaginalis show the terminal signature TNT..AYA (Figure 1). All IS481EU families have terminal inverted repeats (TIRs), mostly shorter than 50 bps. All autonomous families encode a single protein containing a DDD/E transposase. The most abundant IS481EU family is IS481EU-7_TV and it has 376 copies which retain both termini. One of the interesting characteristics of IS481EU is the distinct lengths of TSDs. Several families of IS481EU, such as IS481EU-1_TV and IS481EU-2_TV generate 15 bp or 16 bp TSDs, while other families such as IS481EU-3_TV and IS481EU-5_TV generate 4 bp TSDs. IS481EU-6_TV and IS481EU-8_TV show both 4 bp TSDs and around 15 bp TSDs. The majority of IS481EU-7_TV copies are flanked with 4 bp TSDs; however, two copies are flanked with 15 bp TSDs, and one copy with 24 bp TSDs. Non-autonomous IS481EU families show the same features in the lengths of TSDs. Banshee was reported to generate 4 bp or 15 bp TSDs and starts with TGT [29], so Banshee is likely a family of IS481EU.
Figure 1.
Termini and boundaries of IS481EU families from T. vaginalis. If there are fewer than five copies with both termini, all copies with TSDs are shown with flanking sequences. If there are more than five copies with both termini, only the 5 representative copies with TSDs are shown, except for IS481EU-7_TV, for which 10 representative copies are shown. TSDs are colored in red.
Homology search at the NCBI BLAST website (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 18 May 2022) with the transposases of IS481EU families revealed the presence of similar transposases in the genomes of Trichomonas tenax, Trichomonas gallinae, Tritrichomonas foetus, and Histomonas meleagridis (Table 1 and Table S1). Although most of these transposases were found as a single copy in the respective genome, we were able to reconstruct five complete DNA transposon consensus sequences from the genome of H. meleagridis. Trichomonas and Histomonas belong to the different orders inside palabasalids. We investigated the TSDs of IS481EU from H. meleagridis to check whether they share the same features as IS481EU from T. vaginalis (Figure 2). All IS481EU families from H. meleagridis show TGT…ACA termini, and many of their copies generate 15 bp or 16 bp TSDs upon integration. IS481EU-5_HisMel has a non-autonomous family (IS481EU-5N1_HisMel) and one of its copies is flanked by degenerated 24/25 bp TSDs.
Figure 2.
Termini and boundaries of IS481EU families from H. meleagridis. All copies with TSDs are shown with flanking sequences. TSDs are colored in red.
3.2. The Phylogenetic Relationship between IS481EU and IS481
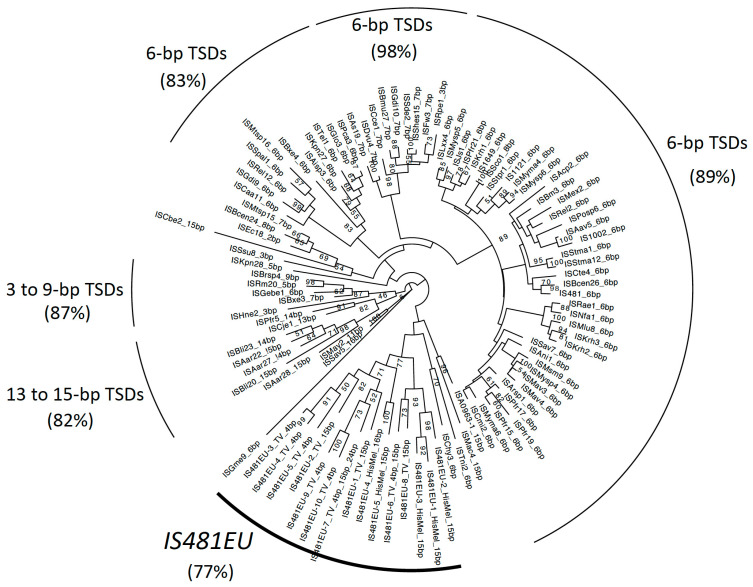
We performed a phylogenetic analysis of IS481EU with all prokaryotic IS481 elements available on ISFinder (Figure 3). The IS481 family is distributed among both bacteria and archaea. The IS481 family generates 2 to 15 bp TSDs, but the majority of IS481s generate 6 bp TSDs (ISFinder, https://www-is.biotoul.fr/, accessed on 23 May 2022). Three large groups composed of prokaryotic IS481 elements generating 6 bp TSDs were statistically supported (Bootstrap values: 89%, 98%, and 83%). The other two prokaryotic lineages were supported statistically and were composed of ISs with different lengths of TSDs.
Figure 3.
Phylogenetic tree based on the transposase domains of IS481EU and IS481 elements. The length of TSDs is shown at each node after the element name. Lineages with ample bootstrap supports (over 70%) are indicated.
Families of IS481EU clustered together with ample statistical support (77% bootstrap value). Based on the phylogeny, the ancestral IS481EU family seems to have generated 15 bp TSDs. Two sublineages of IS481EU show 4 bp TSDs. One is the sublineage composed of three families: IS481EU-3_TV, IS481EU-4_TV, and IS481EU-5_TV. The other is composed of IS481EU-9_TV and IS481EU-10_TV. Its sister family, IS481EU-7_TV, is an exceptional family that generates 4 bp, 15 bp, and 24 bp TSDs. Another family, IS481EU-6_TV generates both 4 bp and 15 bp TSDs, but 4 bp TSDs are dominant. The phylogeny indicates that the change in TSD length has occurred multiple times in the evolution of IS481EU. The mixed phylogenetic positions of IS481EU from T. vaginalis and H. meleagridis suggests the presence of multiple lineages of IS481EU in the evolution of parabasalids, although the current data cannot exclude the possibility of the horizontal transfer of IS481EU between parabasalids.
Although the statistical support is not high enough, two lineages of prokaryotic IS481 elements are positioned close to the IS481EU lineage. ISMac4 and ISA0963-1 are from archaea and generate 15 bp TSDs. ISTni2 and ISChy3 generate 6 bp TSDs and are found in bacteria. Unfortunately, the current data cannot clarify the origin of IS481EU.
3.3. The Origins of IS3EU/GingerRoot and IS481EU in Eukaryotes
The BLASTP search on ISFinder (https://www-is.biotoul.fr/, accessed on 23 May 2022) with the transposase of IS481EU-1_TV found its similarity to many IS3 family members, as well as IS481 family members. IS481 was originally considered a member of IS3 [10] and, thus, the sequence similarity between these two families is expected. Besides these two groups, three ISs (ISArsp14, ISBli29, and ISHahy13) were hit. All of them belong to the ISNCY (IS not classified yet) family. ISHahy13 is a compact IS and belongs to the IS1202 group inside ISNCY. IS1202 is reported to be distantly related to IS481 [10]. ISArsp14 and ISBli29 are large ISs with passenger genes. These three ISs terminate with 5′-TG and CA-3′, as with IS3 and IS481. We included the three groups of ISs (IS481, IS3, and IS1202) for the phylogenetic analysis.
Our previous analysis supported the independent origin of IS481EU from Ginger1, Ginger2, or Polinton [28]. Since the report, two groups of eukaryotic TEs have been reported: IS3EU and GingerRoot. As a designated superfamily name, IS3EU have been listed in Repbase (https://www.girinst.org/repbase/, accessed on 24 May 2022) since 2013. Now, there are 45 IS3EU families stored in Repbase as of 24 May 2022, 17 of which are autonomous [12]. The complete IS3EU families are always associated with TA..TA termini. TSDs are usually 6 bp long. GingerRoot was characterized as a new group of TEs showing similarity to Ginger1 and Gigner2 from the clubmoss Selaginella lepidophylla, and related TEs are supposed to be present in animals too [31]. GingerRoot families generate 6 bp TSDs, and their termini are mostly TA..TA and occasionally TG..CA. Since there is a certain level of sequence similarity between IS3 and IS481, the relationship between IS481EU, IS3EU, and GingerRoot was investigated (Figure 4).
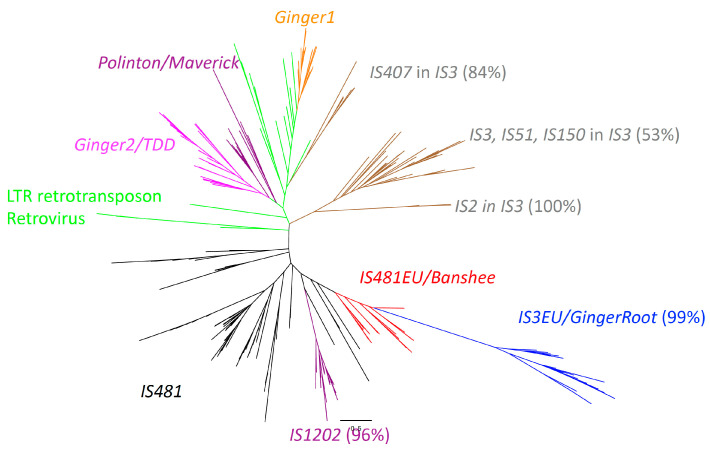
Figure 4.
Phylogenetic tree based on the transposase domains of eukaryotic IS481EU, IS3EU/GingerRoot, Ginger1, Ginger2/TDD, Polinton/Maverick superfamilies, and prokaryotic IS481, IS3, IS1202 families, and the integrases of LTR retrotransposons and retroviruses. Lineages with ample bootstrap supports (over 50%) are indicated. IS2, IS3, IS51, IS150, and IS407 are subfamilies inside the IS3 family. The colors of lineages are as follows: orange, Ginger1; magenta, Ginger2/TDD; purple, Polinton/Maverick; red, IS481EU/Banshee; blue, IS3EU/GingerRoot; black, IS481, grey, IS3; purple, IS1202; green, LTR retrotransposons and retroviruses.
IS481EU, IS3EU, GingerRoot, Polinton, IS3, IS481, and IS1202 families do not have an HHCC motif upstream of the transposase core unlike the integrase of HIV-1 [28]. The N-terminal domain of integrase, which includes an HHCC motif, coordinates the binding of a zinc ion to form a structure similar to those of helix-turn-helix DNA-binding domains, and stimulates catalytic activity [37]. The first catalytic D is located 100 to 300 residues downstream from the N-terminus in IS481EU, IS3EU, IS3, IS481, and IS1202, and it is possible that the N-terminal region of these transposases is functionally equivalent to the N-terminal domain of integrase. The N-terminal regions of IS481EU proteins show a weak similarity to helix-turn-helix DNA-binding proteins such as TetR transcriptional regulators, according to the results of HHpred, protein homology detection, and structure prediction software [38]. Only the transposase core regions were able to be aligned reliably (Figure S1).
Due to the low sequence conservation and the short length of transposases, only a few lineages were statistically supported. First, IS3EU and GingerRoot belong to the same lineage with strong statistical support (Bootstrap value: 99%), and thus this lineage is designated as IS3EU/GingerRoot hereafter. Second, the monophyly of the entire IS1202 family is supported (Bootstrap value: 96%). Third, the IS3 family is separated into three lineages, and two of them are well supported. The monophyly of the IS2 subgroup is highly supported (Bootstrap value: 100%), and the monophyly of the IS407 subgroup is supported too (Bootstrap value: 84%). The clustering of IS3, IS51, and IS150 subgroups is moderately supported (53%).
As we have no obvious outgroup, the root of this phylogenetic tree cannot be determined. The presence of the N-terminal HHCC domain of integrase in LTR retrotransposons, retroviruses, and Ginger1 [28], suggests the common origin of these transposases and integrases, and so, the basal positions of some LTR retrotransposons could be artifacts. Although there is no statistical support, the distinct positions of two lineages of eukaryotic TEs indicate at least two independent origins of eukaryotic TEs. One is composed of IS481EU and IS3EU/GingerRoot, and the other is of Ginger1, Ginger2, Polinton, LTR retrotransposons, and retroviruses. IS3EU/GingerRoot and IS481EU are not separated. The close affinity of ISTni2, ISA0963_1, and ISMac4 with IS481EU is again seen in this phylogenetic analysis, although there is no significant statistical support.
4. Discussion
Here, we report IS481EU, a new group of eukaryotic DNA transposons encoding a DDD/E transposase. The origin of IS481EU could not be distinguished from that of IS3EU/GingerRoot but is likely distinct from the origin of Ginger1, Ginger2, Polinton, LTR retrotransposons, and retroviruses. Although there is still a possibility that IS481EU and IS3EU/GingerRoot share a eukaryotic ancestral DNA transposon, based on the distinct characteristics of these two groups, we propose IS481EU as a new superfamily of eukaryotic DNA transposons. The close relationship of IS481EU to some of prokaryotic IS481 members suggests the horizontal transfer of an IS481 family member to a certain eukaryote contributed to the birth of IS481EU. This is the third superfamily of eukaryotic transposons designated based on the similarity to the prokaryotic ISs, following ISL2EU and IS3EU [12]. We can expect that as more genomes are sequenced, there will be a greater chance that new ISxEUs would be identified. Thus, it is tempting to introduce a new term MILET (Minor IS-like Eukaryotic Transposon Superfamilies) to refer to these “minor” superfamilies which may have spread at a later evolutionary time or only achieved limited success in their transition from prokaryotes to eukaryotes.
The relationship between IS481EU and Banshee should be mentioned. Although there is no sequence information publicly available for Banshee, it is reported to generate 4 or 15 bp TSDs and started with 5′-TGT [29]. Banshee was found only in T. vaginalis [10]. We could not find any other TE families related to IS481 other than IS481EU from T. vaginalis. These results strongly suggest that the reported Banshee is a member of IS481EU.
TEs belonging to the same superfamily tend to generate TSDs of similar lengths, which could be an indicator of the associated superfamily [13,39]. In the case of IS3 family, there are three types of TSD length and they correlate with the subgroups inside the IS3 family [9]. The IS407 subgroup is associated with 4 bp TSDs, and the IS2 subgroup is associated with 5 bp TSDs. Other IS3 family members are flanked with 3 bp TSDs. The characteristic TSD lengths are 4 bp for Ginger1, 4 or 5 bp for Ginger2, 6 bp for Polinton, and 3–6 bp for LTR retrotransposons and retroviruses [28,39,40].
Retroviral integrase recognizes scissile phosphodiester bonds in target DNA which are separated by 4 to 6 bp for strand transfer [41]. The target DNA is significantly kinked to optimally position the catalytic site for the pairwise strand transfer events in the complex. Severe target DNA bending is also observed in transpososomes of various superfamilies of DNA transposons including bacterial Mu, IS21, and eukaryotic Mos1 [42,43,44]. The single-stranded gaps are repaired by cellular enzymes and result in TSDs. Thus, different degrees of target DNA bending along with the unique distance of two catalytic sites would lead to the characteristic lengths of TSDs. Conserved lengths of TSDs in each superfamily of DNA transposons and LTR retrotransposons suggest the conservative nature of transposase–DNA interaction.
Contrary to the relatively conserved TSD lengths among the IS3 family and the superfamilies of eukaryotic DNA transposons related to them, the IS481 and IS1202 families show discrete lengths of TSDs among members. The majority of IS481 family members generate 6 bp TSDs, but not a few IS481 family members generate ~15 bp TSDs (ISFinder: https://www-is.biotoul.fr/, accessed on 23 May 2022). The IS1202 group members show three separate ranges of the length of TSDs: 5 or 6 bp, 15 to 17 bp, and 24 to 29 bp (ISFinder: https://www-is.biotoul.fr/, accessed on 23 May 2022). The IS1202 group is located inside the IS481 family in the phylogeny (Figure 4). The three separate length ranges of TSDs for the IS1202 group is indicative of the conserved mechanisms to generates TSDs. Even in this circumstance, IS481EU is still unusual. One characteristic of IS481EU is the two or more discrete lengths of TSDs, which can be observed even among the copies of the same family. The extreme case is observed in IS481EU-7_TV. Among the 11 full-length copies, 7 were flanked with 4 bp TSDs, 1 with 5 bp TSDs, 2 with 15 bp TSDs, and 1 with 24 bp TSDs (Figure 1). We can speculate that the three discrete lengths of TSDs seen for IS481EU, IS481, and IS1202 are generated through similar mechanisms.
How are such largely discrete lengths of TSDs generated? The situation is different from the two different lengths of TSDs seen among the Academ superfamily [45]. In the Academ superfamily, AcademH generates 9 or 10 bp TSDs, while AcademX generates 3 or 4 bp TSDs. The difference in the TSD lengths between these two Academ lineages seems related to the different enzymes, helicase and nuclease, encoded by them. Here, as seen in the case of IS481EU-7_TV, the enzymes functioning during transposition are the same that generate two or more different lengths of TSDs. Notably, the three distinct ranges of TSD lengths observed in the groups of IS1202, IS481, and IS481EU are close to 10 bps apart, a measurement that equals one full turn of the B-DNA double helix.
The transposases of IS481EU show clear similarity to retroviral integrases (Figure S1), the structure and function of which are well-studied [41]. Among the three discrete ranges of TSD lengths seen among IS481EU, IS481, and IS1202, it is likely that the shortest 4 to 6 bp TSDs would correspond to the TSDs of retroviruses, which are 4 to 6 bps in length. Retroviral integrase works as a tetramer, with a dimer-of-dimers architecture. The inner subunits of each dimer are responsible for interaction with DNA, while the outer subunits attach to the inner subunits but not to the target DNA. Thus, the length of TSDs is dictated by the distance between the two catalytic cores of inner subunits and the bendability of target DNA. The 4 bp TSDs observed upon IS481EU integration are likely to be generated similarly to the cases of retroviral integrases, by two inner subunits of IS481EU transposase tetramer.
We have currently no clue for how the longer TSDs are generated upon transposition of IS481EU. It can be speculated that the two ranges of longer TSDs (~16 bp and ~25 bp) are generated when the target DNA is cleaved by one inner subunit and one outer subunit, or by two outer subunits, with different DNA-binding surfaces. Another possibility is that IS481EU has different multimer conformations and different conformations result in different lengths of TSDs. Alternatively, DNA might form a loop between two binding sites. Structural and biochemical analysis is necessary to clarify the mechanisms generating different lengths of TSDs.
5. Conclusions
IS481EU is characterized from the unicellular eukaryotic lineage parabasalids, and here is proposed as a new superfamily of eukaryotic DNA transposons. It shows a new link between prokaryotic and eukaryotic TEs via the sequence similarity in their termini and transposases, as well as the TSD length variations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology12030365/s1, Figure S1: Multiple alignment of DDD/E transposase cores; Data S1: Consensus or representative sequences of IS481EU. Data S2: Multiple alignment of DDD/E transposase domains of IS481EU and IS481; Data S3: Multiple alignment of transposase domains of eukaryotic IS481EU, IS3EU/GingerRoot, Ginger1, Ginger2/TDD, Polinton/Maverick superfamilies, and prokaryotic IS481, IS3, IS1202 families, and the integrases of LTR retrotransposons and retroviruses. Table S1: The characteristics of IS481EU families.
Author Contributions
Conceptualization, K.K.K.; methodology, K.K.K. and W.B.; validation, K.K.K.; formal analysis, K.K.K. and W.B.; investigation, K.K.K. and W.B.; data curation, K.K.K. and W.B.; writing—original draft preparation, K.K.K.; writing—review and editing, K.K.K. and W.B.; visualization, K.K.K.; supervision, K.K.K.; project administration, K.K.K. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available as Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Curcio M.J., Derbyshire K.M. The outs and ins of transposition: From mu to kangaroo. Nat. Rev. Mol. Cell Biol. 2003;4:865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- 2.Finnegan D.J. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5:103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- 3.Aziz R.K., Breitbart M., Edwards R.A. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 2010;38:4207–4217. doi: 10.1093/nar/gkq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hickman A.B., Chandler M., Dyda F. Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit. Rev. Biochem. Mol. Biol. 2010;45:50–69. doi: 10.3109/10409230903505596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majorek K.A., Dunin-Horkawicz S., Steczkiewicz K., Muszewska A., Nowotny M., Ginalski K., Bujnicki J.M. The RNase H-like superfamily: New members, comparative structural analysis and evolutionary classification. Nucleic Acids Res. 2014;42:4160–4179. doi: 10.1093/nar/gkt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyda F., Hickman A.B., Jenkins T.M., Engelman A., Craigie R., Davies D.R. Crystal structure of the catalytic domain of HIV-1 integrase: Similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 7.Kapitonov V.V., Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang S., Tao X., Yuan S., Zhang Y., Li P., Beilinson H.A., Zhang Y., Yu W., Pontarotti P., Escriva H., et al. Discovery of an Active RAG Transposon Illuminates the Origins of V (D) J Recombination. Cell. 2016;166:102–114. doi: 10.1016/j.cell.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siguier P., Gourbeyre E., Varani A., Ton-Hoang B., Chandler M. Everyman’s Guide to Bacterial Insertion Sequences. Microbiol. Spectr. 2015;3:MDNA3-0030-2014. doi: 10.1128/microbiolspec.MDNA3-0030-2014. [DOI] [PubMed] [Google Scholar]
- 11.Ross K., Varani A.M., Snesrud E., Huang H., Alvarenga D.O., Zhang J., Wu C., McGann P., Chandler M. TnCentral: A Prokaryotic Transposable Element Database and Web Portal for Transposon Analysis. mBio. 2021;12:e0206021. doi: 10.1128/mBio.02060-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao W., Kojima K.K., Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kojima K.K. Structural and sequence diversity of eukaryotic transposable elements. Genes Genet. Syst. 2020;94:233–252. doi: 10.1266/ggs.18-00024. [DOI] [PubMed] [Google Scholar]
- 14.Kojima K.K., Jurka J. A superfamily of DNA transposons targeting multicopy small RNA genes. PLoS ONE. 2013;8:e68260. doi: 10.1371/journal.pone.0068260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan Y.W., Wessler S.R. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl. Acad. Sci. USA. 2011;108:7884–7889. doi: 10.1073/pnas.1104208108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tellier M., Bouuaert C.C., Chalmers R. Mariner and the ITm Superfamily of Transposons. Microbiol. Spectr. 2015;3:MDNA3-0033-2014. doi: 10.1128/microbiolspec.MDNA3-0033-2014. [DOI] [PubMed] [Google Scholar]
- 17.Shi S., Puzakov M., Guan Z., Xiang K., Diaby M., Wang Y., Wang S., Song C., Gao B. Prokaryotic and Eukaryotic Horizontal Transfer of Sailor (DD82E), a New Superfamily of IS630-Tc1-Mariner DNA Transposons. Biology. 2021;10:1005. doi: 10.3390/biology10101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yusa K. piggyBac Transposon. Microbiol. Spectr. 2015;3:MDNA3-0028-2014. doi: 10.1128/microbiolspec.MDNA3-0028-2014. [DOI] [PubMed] [Google Scholar]
- 19.Sarkar A., Sim C., Hong Y.S., Hogan J.R., Fraser M.J., Robertson H.M., Collins F.H. Molecular evolutionary analysis of the widespread piggyBac transposon family and related "domesticated" sequences. Mol. Genet. Genom. 2003;270:173–180. doi: 10.1007/s00438-003-0909-0. [DOI] [PubMed] [Google Scholar]
- 20.Feschotte C. Merlin, a new superfamily of DNA transposons identified in diverse animal genomes and related to bacterial IS1016 insertion sequences. Mol. Biol. Evol. 2004;21:1769–1780. doi: 10.1093/molbev/msh188. [DOI] [PubMed] [Google Scholar]
- 21.Bao W., Jurka M.G., Kapitonov V.V., Jurka J. New superfamilies of eukaryotic DNA transposons and their internal divisions. Mol. Biol. Evol. 2009;26:983–993. doi: 10.1093/molbev/msp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arkhipova I.R. Using bioinformatic and phylogenetic approaches to classify transposable elements and understand their complex evolutionary histories. Mob. DNA. 2017;8:19. doi: 10.1186/s13100-017-0103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hua-Van A., Capy P. Analysis of the DDE motif in the Mutator superfamily. J. Mol. Evol. 2008;67:670–681. doi: 10.1007/s00239-008-9178-1. [DOI] [PubMed] [Google Scholar]
- 24.Eisen J.A., Benito M.I., Walbot V. Sequence similarity of putative transposases links the maize Mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res. 1994;22:2634–2636. doi: 10.1093/nar/22.13.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han M.J., Xiong C.L., Zhang H.B., Zhang M.Q., Zhang H.H., Zhang Z. The diversification of PHIS transposon superfamily in eukaryotes. Mob. DNA. 2015;6:12. doi: 10.1186/s13100-015-0043-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gladyshev E.A., Arkhipova I.R. A single-copy IS5-like transposon in the genome of a bdelloid rotifer. Mol. Biol. Evol. 2009;26:1921–1929. doi: 10.1093/molbev/msp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han M.J., Xu H.E., Zhang H.H., Feschotte C., Zhang Z. Spy: A new group of eukaryotic DNA transposons without target site duplications. Genome Biol. Evol. 2014;6:1748–1757. doi: 10.1093/gbe/evu140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bao W., Kapitonov V.V., Jurka J. Ginger DNA transposons in eukaryotes and their evolutionary relationships with long terminal repeat retrotransposons. Mob. DNA. 2010;1:3. doi: 10.1186/1759-8753-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feschotte C., Pritham E.J. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohany O., Gentles A.J., Hankus L., Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinform. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerbin S., Wai C.M., VanBuren R., Jiang N. GingerRoot: A Novel DNA Transposon Encoding Integrase-Related Transposase in Plants and Animals. Genome Biol. Evol. 2019;11:3181–3193. doi: 10.1093/gbe/evz230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Llorens C., Futami R., Covelli L., Dominguez-Escriba L., Viu J.M., Tamarit D., Aguilar-Rodríguez J., Vicente-Ripolles M., Fuster G., Bernet G.P., et al. The Gypsy Database (GyDB) of mobile genetic elements: Release 2.0. Nucleic Acids Res. 2011;39:D70–D74. doi: 10.1093/nar/gkq1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh K., Kuma K., Toh H., Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Do C.B., Mahabhashyam M.S., Brudno M., Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 37.Engelman A., Cherepanov P. Retroviral Integrase Structure and DNA Recombination Mechanism. Microbiol. Spectr. 2014;2:1–22. doi: 10.1128/microbiolspec.MDNA3-0024-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zimmermann L., Stephens A., Nam S.Z., Rau D., Kubler J., Lozajic M., Gabler F., Söding J., Lupas A.N., Alva V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018;430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 39.Kapitonov V.V., Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat. Rev. Genet. 2008;9:411–412. doi: 10.1038/nrg2165-c1. [DOI] [PubMed] [Google Scholar]
- 40.Kapitonov V.V., Jurka J. Self-synthesizing DNA transposons in eukaryotes. Proc. Natl. Acad. Sci. USA. 2006;103:4540–4545. doi: 10.1073/pnas.0600833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lesbats P., Engelman A.N., Cherepanov P. Retroviral DNA Integration. Chem. Rev. 2016;116:12730–12757. doi: 10.1021/acs.chemrev.6b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris E.R., Grey H., McKenzie G., Jones A.C., Richardson J.M. A bend, flip and trap mechanism for transposon integration. Elife. 2016;5:e15537. doi: 10.7554/eLife.15537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arias-Palomo E., Berger J.M. An Atypical AAA+ ATPase Assembly Controls Efficient Transposition through DNA Remodeling and Transposase Recruitment. Cell. 2015;162:860–871. doi: 10.1016/j.cell.2015.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montano S.P., Pigli Y.Z., Rice P.A. The mu transpososome structure sheds light on DDE recombinase evolution. Nature. 2012;491:413–417. doi: 10.1038/nature11602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kojima K.K. AcademH, a lineage of Academ DNA transposons encoding helicase found in animals and fungi. Mob. DNA. 2020;11:15. doi: 10.1186/s13100-020-00211-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available as Supplementary Materials.