Abstract
We report the spillover of highly pathogenic avian influenza A(H5N1) into marine mammals in the northeastern United States, coincident with H5N1 in sympatric wild birds. Our data indicate monitoring both wild coastal birds and marine mammals will be critical to determine pandemic potential of influenza A viruses.
Keywords: avian influenza, H5N1, influenza, viruses, zoonoses, virology, ecology, Caniformia, harbor seals, gray seals, New England, United States
Highly pathogenic avian influenza (HPAI) viruses are of concern because of their pandemic potential, socioeconomic impact during agricultural outbreaks, and risks to wildlife conservation. Since October 2020, HPAI A(H5N1) virus, belonging to the goose/Guangdong H5 2.3.4.4b clade, has been responsible for >70 million poultry deaths and >100 discrete infections in many wild mesocarnivore species (1). As of January 2023, H5N1 infections in mammals have been primarily attributed to consuming infected prey, without evidence of further transmission among mammals.
We report an HPAI A(H5N1) virus outbreak among New England harbor and gray seals that was concurrent with a wave of avian infections in the region, resulting in a seal unusual mortality event (UME); evidence of mammal adaptation existed in a small subset of seals. Harbor (Phoca vitulina) and gray (Halichoerus grypus) seals in the North Atlantic are known to be affected by avian influenza A virus and have experienced previous outbreaks involving seal-to-seal transmission (2–7). Those seal species represent a pathway for adaptation of avian influenza A virus to mammal hosts that is a recurring event in nature and has implications for human health.
The Study
The first detections of HPAI clade 2.3.4.4b viruses in North America were in wild and domestic birds in November 2021 in Canada and late December 2021 in the United States (8–11). Starting on January 20, 2022, avian oropharyngeal or cloacal samples were collected from wild birds by personnel in 4 wildlife clinics in Massachusetts. Additional opportunistic samples were collected in Maine and Massachusetts in response to suspicious avian deaths in seabird breeding colonies. We screened samples from 1,079 individual wild birds representing 78 avian species of concern for H5 influenza and identified 119 infected birds from 21 species (Appendix 1 Figure 1, panel A; Appendix 2). Wild birds in New England experienced 2 waves of influenza infections during 2022. The first wave peaked in March and was largely represented by raptor deaths (39.1% influenza-positive birds). A second wave began in June; gull (38% influenza-positive) and eider (26.8% influenza-positive) deaths were most frequently reported during the second wave. Mortality events affected seabird breeding colonies throughout the coastal region during the second wave; 8 islands had >1 bird test positive for H5 (Appendix 1 Table).
During January 20–July 31, 2022, opportunistic nasal, oral, conjunctival or rectal swab samples were collected from 132 stranded seals along the North Atlantic coast from Maine to Virginia (Appendix 2). HPAI virus was not detected in any of the 82 seals that were sampled through May 31, 2022. Concurrent with the second wave of avian infections, increased seal strandings in Maine led to a National Oceanic and Atmospheric Administration declaration of a UME beginning on June 1, 2022, that included 164 harbor and 11 gray seals in Maine during June and July (12). Swab samples were collected from 41 of those animals; 17/35 harbor and 2/6 gray seals were HPAI-positive and were within coastal regions of known and suspected HPAI outbreaks among terns, eiders, cormorants, and gulls (Appendix 1 Figure 1, panel B). Respiratory symptoms were observed with a subset of neurologic cases, although most stranded seals were deceased. The respiratory tract was the most consistent source of reverse transcription PCR–positive samples from affected seals (15/19 nasal, 16/19 oral, 6/19 conjunctiva, and 4/19 rectal samples).
We sequenced influenza A viruses from swab samples, resulting in 71 avian- and 13 seal-derived virus genomes from New England. We performed phylogenetic analysis of sequences from New England and the most closely related available virus sequences by using IQ-TREE (https://www.iqtree.org) (Figure 1; Appendix 1). We classified all but 1 virus as nonreassortant Eurasia 2.3.4.4b viruses and included those in further analyses (Appendix 1 Figures 2, 3). Sequences fell into 4 distinct clusters; 2 lineages were unique to New England. We found single-nucleotide polymorphisms (SNPs) (Appendix 1 Figures 4–7) and amino acid mutations (Figure 2) by using vSNP (https://github.com/USDA-VS/vSNP). Most sequences fell within a dominant New England–specific cluster that spanned the first and second waves (lineage A in this study). All second-wave viruses from lineage A exhibited the acquisition of new, shared mutations. That cluster spanned diverse species, including gulls, geese, eiders, raptors, and seals. A small number of raptor-derived sequences clustered with the primary lineage prevalent in North America at the time of sampling. All but 1 sample from the second wave of avian infections fell into either lineage A or a smaller, unique cluster primarily associated with terns (lineage B in this study).
Figure 1.
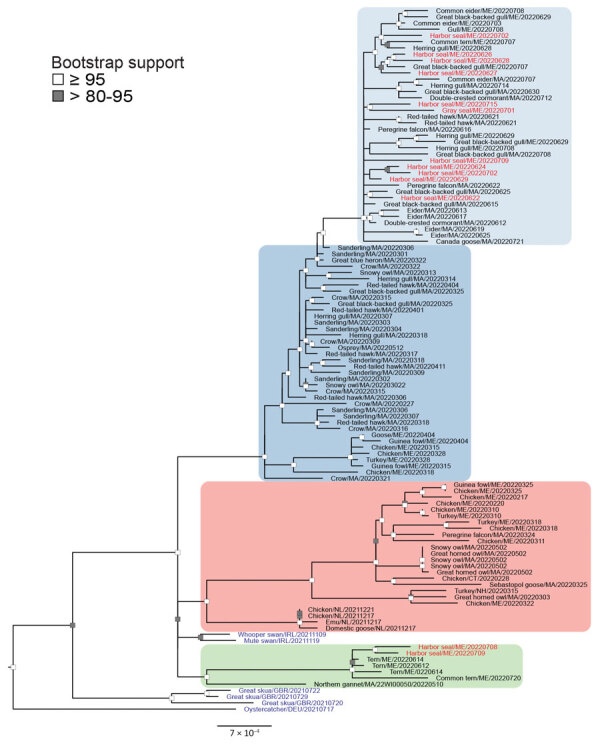
Phylogenetic analysis of highly pathogenic avian influenza A(H5N1) viruses from New England birds and seals, United States. Complete genomes of HPAI H5N1 viruses (GISAID database, https://www.gisaid.org) were compared by using IQ-TREE (https://www.iqtree.org) with the Ultrafast bootstrap (n = 10,000) option and A/chicken/NL/FAV-0033/2021 as a reference. Bootstrap support values >80 are shown at nodes. Red text indicates seal-derived sequences, black text avian-derived sequences from New England and Newfoundland, and blue text indicates avian-derived sequences from Europe . Branches are shaded on the basis of lineage groups: primary lineage from North America, pink; New England–specific lineage A, 1st wave blue, 2nd wave light blue; and New England–specific lineage B, green. All newly reported specimens were collected in the New England region during February–July 2022. Scale bar indicates nucleotide substitutions per site.
Figure 2.
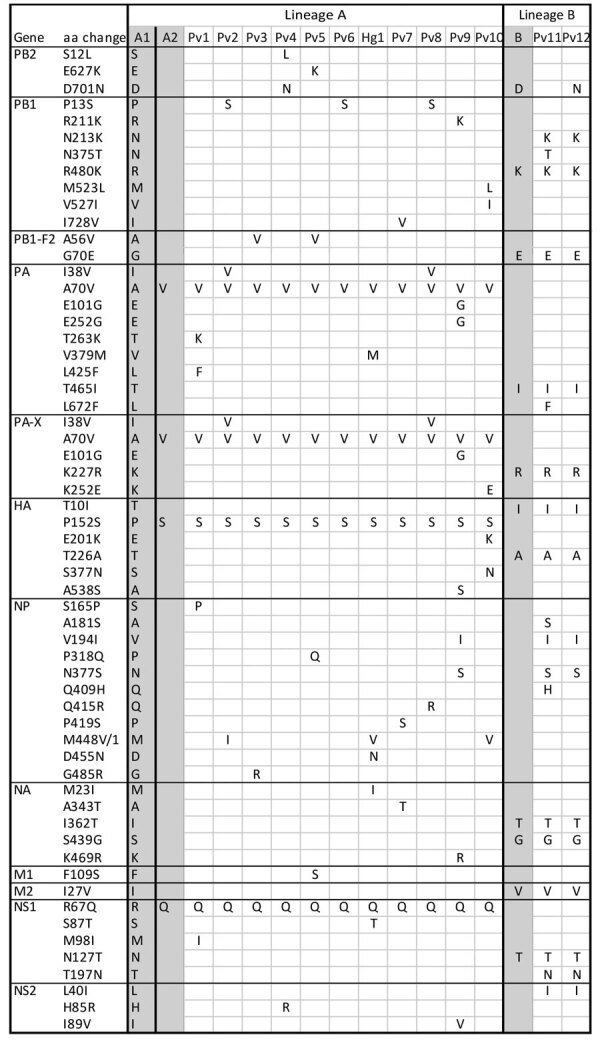
Amino acid changes in highly pathogenic avian influenza A(H5N1) viruses from New England birds and seals, United States. Each single-nucleotide polymorphism (SNP) that resulted in an amino acid change within ≥1 seal-derived sequence is shown. SNPs were observed in 12 H5N1 virus genes resulting in amino acid changes in corresponding proteins: PB2, PB1, PB1-F2, PA, PA-X, HA, NP, NA, M1, M2, NS1, and NS2. All avian virus reference sequences are shaded gray. A/Sanderling/MA/CW_22–112 (H5N1) (GISAID database, https://www.gisaid.org) (labeled A1) was used as a reference for first wave lineage A sequences; A/common eider/MA/TW_22–1400 (H5N1) (labeled A2) was used as a reference for second wave lineage A sequences; and A/common tern/MA/20220612_1 (H5N1) (labeled B) was used as a reference for lineage B sequences. Four aa differ between first and second wave lineage A viruses and 10 aa differ between first wave lineage A and lineage B. Second wave lineage A and lineage B seal-derived virus sequences, sampling date, and sampling location in Maine, USA, are indicated for each seal as follows: Pv1, MME22-112, 2022 Jun 22, Wells; Pv2, MME22-117, 2022 Jun 24, Yarmouth; Pv3, MME22-121, 2022 Jun 26, Georgetown; Pv4, MME22-122, 2022 Jun 27, New Harbor; Pv5, MME22-131, 2022 Jun 28, Harpswell; Pv6, MME22-133, 2022 Jun 29, S. Portland; Hg1, MME22-144, 2022 Jul 1, Phippsburg; Pv7, MME22-150, 2022 Jul 2, Westport; Pv8, MME22-155, 2022 Jul 2, Falmouth; Pv9, MME22-198, 2022 Jul 9, Wells; Pv10, MME22-230, 2022 Jul 15, Kennebunkport; Pv11, MME22-191, 2022 Jul 8, Harpswell; Pv12, MME22-195, 2022 Jul 9, Harpswell. HA, hemagglutinin; Hg, gray seal; M, matrix; NA, neuraminidase; NP, nucleoprotein; NS, nonstructural; PA, polymerase acidic; PB, polymerase basic; Pv, harbor seal.
We inferred that >2 spillover events occurred in the seal population during the second wave of avian infections. Of the sequences derived from seals, 11/13 clustered with second wave lineage A (Figure 1). We found 4 aa changes in specific proteins in both birds and seals that were distinct from the first wave of HPAI (polymerase acidic protein, A70V; polymerase acidic X protein, A62V; hemagglutinin protein, P152S; and nonstructural 1 protein, R67Q) (Figure 2). Within second wave lineage A, we found 37 aa changes in >1 seal sequence that were infrequent or absent from bird sequences. Most changes were unique; each occurred in only 1 animal. The polymerase basic 2 protein amino acid substitutions, E627K (in seal no. Pv/MME-22–131) and D701N (in seal no. Pv/MME-22–122), previously associated with mammalian adaptation were each present in 1 seal in second wave lineage A. An additional 2/13 seal-derived sequences clustered with lineage B; 10 aa mutations occurred in both bird and seal sequences (Figure 2). Another 10 aa changes occurred in at least 1 seal sequence that were infrequent or absent in the bird sequences. In contrast to lineage A, most amino acid changes were shared between the 2 seals in lineage B and were derived from animals stranded within the same town and sampled 1 day apart. The polymerase basic 2 protein substitution, D701N, was present in 1 seal from lineage B (Figure 2, seal no. PV12, MME-22–195).
Conclusions
Transmission from wild birds to seals was evident for >2 distinct HPAI H5N1 lineages in this investigation and likely occurred through environmental transmission of shed virus. Viruses were not likely acquired by seals through predation or scavenging of infected animals, because birds are not a typical food source for harbor or gray seals (13). Data do not support seal-to-seal transmission as a primary route of infection. If individual bird–seal spillover events represent the primary transmission route, the associated seal UME suggests that transmission occurred frequently and had a low seal species barrier. We observed novel amino acid changes throughout the virus genome in seals, including amino acid substitutions associated with mammal adaptation.
In contrast to outbreaks in agricultural settings, outbreaks of HPAI in wild populations can rarely be managed well through biosecurity measures or depopulation, which is particularly true for large, mobile marine species such as seals. Avian and mammalian colonial wildlife might be particularly affected by influenza A viruses, which could enable ongoing circulation between and within species, providing opportunities for reassortments of novel strains and study of mammalian virus adaptation. Migratory animals might further disseminate the viruses over broad geographic regions. Therefore, the interface of wild coastal birds and marine mammals is critical for monitoring the pandemic potential of influenza A viruses.
Additional information for highly pathogenic avian influenza A(H5N1) virus outbreak in New England seals, United States.
Samples and sequences used for study of highly pathogenic avian influenza A(H5N1) virus outbreak in New England seals, United States.
Acknowledgments
We thank the numerous staff and volunteers at Marine Mammals of Maine, College of the Atlantic, Seacoast Science Center, National Marine Life Center, Marine Mammal Alliance Nantucket, International Fund for Animal Welfare, Atlantic Marine Conservation Society, New York Marine Rescue Center, National Aquarium, Mystic Aquarium, Tufts Wildlife Clinic, New England Wildlife Center, New England Fisheries and Science Center, Wild Care Inc., Cape Wildlife Center, Linda Loring Nature Foundation, Maine Coastal National Wildlife Reservation, Rachel Carson National Wildlife Refuge, University of Massachusetts Nantucket Field Station, US Fish and Wildlife Services, and Massachusetts Audubon Society for providing critical expertise on marine and avian wildlife, obtaining samples from stranded animals, and providing essential insights on ecological contexts from the field; and the authors and originating and submitting laboratories of the sequences from GISAID’s EpiFlu Database (https://www.gisaid.org) on which this research is based.
The study was funded by Mount Sinai Center for Research in Influenza Pathogenesis and Transmission, a Center of Excellence in Influenza Research and Response established by the National Institute of Allergy and Infectious Disease, National Institutes of Health, US Department of Health and Human Services (contract no. 75N93021C00014). The scientific results and conclusions and any views or opinions expressed herein are those of the authors and do not necessarily reflect the views or policies of the US Government, its agencies, or any of the included organizations.
All sequencing data are available in the GISAID database (https://www.gisaid.org) (EPI_ISL_14098915, EPI_ISL_14098917–24, EPI_ISL_16632466, EPI_ISL_16632487–8, EPI_ISL_16632494–6, EPI_ISL_16632498–524, EPI_ISL_16632536–42, EPI_ISL_16641764–94, EPI_ISL_16641796–9) or as a single file on GitHub (https://github.com/ksawatzki/H5N1_EID). All additional data are available in the appendices.
Biography
Dr. Puryear is a virologist at The Cummings School of Veterinary Medicine at Tufts University in the Department of Infectious Disease and Global Health. Her research interests focus on epidemiology, evolution, and adaptation of wildlife diseases.
Footnotes
Suggested citation for this article: Puryear W, Sawatzki K, Hill N, Foss A, Stone JJ, Doughty L, et al. Highly pathogenic avian influenza A(H5N1) virus outbreak in New England seals, United States. Emerg Infect Dis. 2023 Apr [date cited]. https://doi.org/10.3201/eid2904.221538
These first authors contributed equally to this article.
References
- 1.World Organisation for Animal Health. World animal health information system [cited 2022 Jul 19]. https://wahis.woah.org/#/dashboards/qd-dashboard
- 2.Geraci JR, St Aubin DJ, Barker IK, Webster RG, Hinshaw VS, Bean WJ, et al. Mass mortality of harbor seals: pneumonia associated with influenza A virus. Science. 1982;215:1129–31. 10.1126/science.7063847 [DOI] [PubMed] [Google Scholar]
- 3.Callan RJ, Early G, Kida H, Hinshaw VS. The appearance of H3 influenza viruses in seals. J Gen Virol. 1995;76:199–203. 10.1099/0022-1317-76-1-199 [DOI] [PubMed] [Google Scholar]
- 4.Anthony SJ, St Leger JA, Pugliares K, Ip HS, Chan JM, Carpenter ZW, et al. Emergence of fatal avian influenza in New England harbor seals. MBio. 2012;3:e00166–12. 10.1128/mBio.00166-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodewes R, Bestebroer TM, van der Vries E, Verhagen JH, Herfst S, Koopmans MP, et al. Avian Influenza A(H10N7) virus-associated mass deaths among harbor seals. Emerg Infect Dis. 2015;21:720–2. 10.3201/eid2104.141675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodewes R, Zohari S, Krog JS, Hall MD, Harder TC, Bestebroer TM, et al. Spatiotemporal analysis of the genetic diversity of seal influenza A(H10N7) virus, northwestern Europe. J Virol. 2016;90:4269–77. 10.1128/JVI.03046-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puryear WB, Keogh M, Hill N, Moxley J, Josephson E, Davis KR, et al. Prevalence of influenza A virus in live-captured North Atlantic gray seals: a possible wild reservoir. Emerg Microbes Infect. 2016;5:e81. 10.1038/emi.2016.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canadian Food Inspection Agency, National Emergency Operation Centre, Geographic Information System Services. Highly pathogenic avian influenza—wild birds [cited 2022 Jul 19]. https://cfia-ncr.maps.arcgis.com/apps/dashboards/89c779e98cdf492c899df23e1c38fdbc
- 9.United States Department of Agriculture. USDA confirms highly pathogenic avian influenza in a wild bird in South Carolina [cited 2022 Jul 7]. https://www.aphis.usda.gov/aphis/newsroom/stakeholder-info/sa_by_date/sa-2022/hpai-sc
- 10.Bevins SN, Shriner SA, Cumbee JC Jr, Dilione KE, Douglass KE, Ellis JW, et al. Intercontinental movement of highly pathogenic avian influenza A(H5N1) clade 2.3.4.4 virus to the United States, 2021. Emerg Infect Dis. 2022;28:1006–11. 10.3201/eid2805.220318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caliendo V, Lewis NS, Pohlmann A, Baillie SR, Banyard AC, Beer M, et al. Transatlantic spread of highly pathogenic avian influenza H5N1 by wild birds from Europe to North America in 2021. Sci Rep. 2022;12:11729. 10.1038/s41598-022-13447-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Oceanic and Atmospheric Administration. 2018–2020 Pinniped unusual mortality event along the Northeast Coast [cited 2022 Jul 19]. https://www.fisheries.noaa.gov/new-england-mid-atlantic/marine-life-distress/2018-2020-pinniped-unusual-mortality-event-along
- 13.Bowen WD, Harrison GD. Comparison of harbour seal diets in two inshore habitats of Atlantic Canada. Can J Zool. 1996;74:125–35. 10.1139/z96-017 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information for highly pathogenic avian influenza A(H5N1) virus outbreak in New England seals, United States.
Samples and sequences used for study of highly pathogenic avian influenza A(H5N1) virus outbreak in New England seals, United States.


