Abstract
Simple Summary
HER2-equivocal cases represent around 15% of breast carcinomas, and 20–40% of them are HER2-amplified. The distinction between HER2-amplified and non-amplified cases is of great importance for patient management, and in this study, we investigated the performance of STRAT4 (a RT-qPCR platform) in the evaluation of HER2-equivocal cases. We compared this technique to the recommended methods (immunohistochemistry and in situ hybridization) for the evaluation of HER2-equivocal cases and non-equivocal HER2 cases. We found a 91.3% accuracy for the identification of HER2-positive tumors globally and 99.3% for that of non-equivocal HER2 cases, while the accuracy decreased to 80.7% for HER2-equivocal cases. Our results suggest that STRAT4 is not reliable for the evaluation of the HER2 amplification status in equivocal cases.
Abstract
Xpert Breast Cancer STRAT4 is a RT-qPCR platform that studies the mRNA expression of ESR1, PGR, MKI67 and ERBB2, providing a positive or negative result for each of these breast cancer biomarkers. Its concordance with immunohistochemistry (IHC) and in situ hybridization (ISH) has been previously demonstrated, but none of the previous works was focused on HER2-equivocal (2+) cases identified by IHC. Thus, we studied the concordance between IHC/ISH and STRAT4 results for 112 HER2 2+ IBC samples, using 148 HER2 0+, 1+ and 3+ (no-HER2 2+) samples for comparison. We found 91.3% accuracy for the determination of HER2 status globally, 99.3% for no-HER2 2+ samples and 80.7% for HER2 2+ samples. Regarding the other biomarkers, we obtained 96.4% accuracy for estrogen receptor, 84.1% for progesterone receptor and 58.2% for Ki67. Our results suggest that the use of ERBB2 mRNA for the evaluation of HER2 2+ cases is not a reliable reflex method to assess the ERBB2 amplification status.
Keywords: breast carcinoma, HER2, mRNA, immunohistochemistry
1. Introduction
The assessment of biomarkers expression in invasive breast carcinoma (IBC) is crucial for patient management. Currently, it is performed by evaluating the expression of the estrogen receptor (ER), progesterone receptor (PR), Ki67 and HER2. The expression of ER and PR is present in about 75% to 80% of IBC [1], especially in well and moderately differentiated tumors, and these patients are eligible for endocrine therapy. The recommended method for their evaluation is immunohistochemistry (IHC), assessing the percentage of positive tumoral cells and the staining intensity [1].
Ki67 is widely used in pathology to evaluate cell proliferation rate. In IBC, it is used as a prognostic factor and for treatment planning [2]. However, it is not widely accepted in clinical practice due to its variability because of preanalytical factors and low intra- and interobserver reproducibility, leading to different attempts to standardize its evaluation [3].
HER2 is overexpressed in approximately 15–20% of IBC, and only these tumors are eligible for anti-HER2 therapy. Its evaluation is usually performed with IHC, scoring the tumors as negative (score 0 or 1+), equivocal (score 2+) and positive (score 3+). In cases that show an equivocal HER2 immunohistochemical pattern (HER2 2+, according to 2018 ASCO-CAP guidelines [4]), additional testing must be performed using in situ hybridization (ISH) to determine its final positive (amplified) or negative (no amplified) status. Moreover, in some cases, unusual patterns of HER2 might be encountered, which also warrant further testing [5]. HER2 2+ represent around 15% of IBC [6], and HER2 amplification is detected in 20–40% of them [7,8,9].
Although it has been proven that patients with HER2 1+ and HER2 2+ tumors without amplification do not benefit from adjuvant trastuzumab therapy [10], recent studies have shown that the outcome of these patients may improve with HER2-directed antibody–drug conjugates [11]. Thus, the distinction between HER2 0+ and HER2 1+ is gaining relevance, as is the concept of “HER2-low” breast cancer, encompassing those HER2 1+ and HER2 2+ tumors without HER2 amplification [5,12].
As previously mentioned, HER2 2+ cases need additional testing with ISH, and this technique requires additional tissue and takes at least 2 days to be performed. However, new technologies are becoming available to assess IBC biomarkers. In this regard, Xpert® Breast Cancer STRAT4 mRNA (Cepheid, Sunnyvale, CA, USA) (STRAT4) is a real-time quantitative polymerase chain reaction (RT-qPCR) assay that studies the expression of ESR1, PGR, ERBB2 and MKI67 mRNA, providing a positive or negative result for each biomarker in approximately two hours [13]. Other advantages of this technique are its reproducibility and objectivity, as the software provides these results.
Previous studies reported the concordance of STRAT4 and immunohistochemistry for ER, PR, Ki67 and HER2, achieving good results [13,14,15,16,17,18,19]. We previously collaborated in a Europe-wide external quality assessment [14], achieving accuracies ≥90% for all biomarkers across five participating centers, highlighting the reproducibility of this technique. However, none of these STRAT4 studies were focused on the complex and salient group that comprises HER2 2+ cases.
Thus, the aim of this study was to investigate the performance of STRAT4 in the evaluation of HER2 2+ cases, using FISH amplification status as the gold standard for the final HER2 classification.
2. Materials and Methods
2.1. Case Selection
Consecutive HER2 2+ IBC samples diagnosed in our department between the years 2012 and 2018 were selected, all of them from female patients, for which there was enough tissue to perform the mRNA assay (see Section 2.3). In addition, consecutive HER2 0 + , 1+ and 3+ cases (from now on, these will be referred as no-HER2 2+ cases) diagnosed in our department between 2012 and 2018 were selected for comparison. This study was approved by the Ethics Committee of the Hospital Universitario Ramón y Cajal (reference number 361-20).
2.2. Immunohistochemistry
The antibodies used are shown in Table 1. A positive external control was placed on each immunohistochemical slide. The results were interpreted according to the last ASCO-CAP guidelines: 2020 ASCO-CAP guidelines [1] for ER and PR; and 2018 ASCO-CAP guidelines [4] for HER2. Ki67 was evaluated following the Updated Recommendations from the International Ki67 in Breast Cancer Working Group [3] and it was considered as indicative of a high-proliferative state when >20% of tumoral cells showed nuclear staining, as reported in previous studies of STRAT4 [13,14,17].
Table 1.
Details of the antibodies used.
| Biomarker | Antibody | Company | Concentration |
|---|---|---|---|
| ER | EP1 clone | Agilent | Ready to use |
| PR | PgR clone | Agilent | 1:50 |
| HER2 | HercepTest | Agilent | Ready to use |
| Ki67 | Mib-1 | Agilent | Ready to use |
All HER2 IHC slides were reviewed by two pathologists, and discordances were resolved by consensus. Additional HER2 testing was performed in HER2 2+ cases, defined as weak to moderate complete membrane staining observed in >10% of tumor cells [4]. HER2 fluorescent in situ hybridization (FISH) was performed for them, using the PathVysion HER-2 DNA Probe Kit (Abbot Laboratories, Chicago, IL, USA). The results were interpreted following the 2018 ASCO-CAP guidelines [4].
2.3. mRNA Expression with Xpert® Breast Cancer STRAT4
The samples were processed according to the manufacturers´ instructions, requiring at least 30% of cellularity. Briefly, each specimen was cut three times obtaining 4 μm samples that were placed in a 1.5 mL Eppendorf tube. This material was then mixed with 1.2 mL of lysis reagent and 20 µL of proteinase K. Subsequently, it was incubated at 80 °C for 30 min and mixed with 1.2 mL of ethanol ≥95%. For each sample, 520 µL of the solution was transferred to the sample chamber of a STRAT4 cartridge and placed into a GeneXpert module for RNA extraction, purification and RT-qPCR analysis.
The results were analyzed using the GeneXpert DX software. CYFIP1 was used as a reference for mRNA expression, and a delta cycle threshold (dCt = [CtCYFIP1] − [Cttarget]) was provided that uses a predefined cut-off value to classify the expression status of each biomarker (positive vs negative). In the case of ERBB2, the cut-off value is set to −1, for ESR1 to −1, for PGR to −3.5, and for MKI67 to −4 [13]. Thus, the expression of CYFIP1 was used both to normalize the biomarkers expression and as an mRNA quality control of the sample.
2.4. Statistical Analysis
All analyses were performed using R 4.1.0 [20]. The diagnosis accuracy (sensitivity, specificity, positive predictive value and negative predictive value) was studied. Comparisons of the means were performed using Wilcoxon signed-rank test, and correlation studies using the Spearman coefficient. The results were considered statistically significant if p < 0.05.
3. Results
3.1. Clinicopathological Features
A total of 260 cases were studied from 256 females (three multifocal and one bilateral), being 112 cases HER2 2+. The clinical and histopathological features are summarized in Table 2. Briefly, the median patient age was 58.9 years, 55.4% of the cases presented a histological grade 2, and most of them (81.2%) were IBC of no special type. Most expressed hormonal receptors (80.8% ER and 63.8% PR), 43% were HER2 2+, and 66.9% had a low-proliferative index.
Table 2.
Histopathological features of the series.
| Total | Percentage | ||
|---|---|---|---|
| 260 | 100 | ||
| Age, median (IQR) | 58.9 (15.9) | 256 | |
| Histological grade | 1 | 48 | 18.5 |
| 2 | 144 | 55.4 | |
| 3 | 68 | 26.2 | |
| Histological type | IBC, no special type | 211 | 81.2 |
| Lobular carcinoma | 32 | 12.3 | |
| Other | 17 | 6.5 | |
| ER | Positive | 210 | 80.8 |
| Low | 5 | 1.9 | |
| Negative | 45 | 17.3 | |
| PR | Positive | 166 | 63.8 |
| Negative | 94 | 36.2 | |
| HER2 | Positive (3+) | 31 | 11.9 |
| Equivocal 2+ (amplified) | 30 | 11.5 | |
| Equivocal 2+ (no amplified) | 82 | 31.5 | |
| Negative (1+) | 46 | 17.7 | |
| Negative (0+) | 71 | 27.3 | |
| Ki67 1 | High-proliferative | 86 | 33.1 |
| Low-proliferative | 173 | 66.9 |
1 In one no-HER2 2+ case Ki67 immunohistochemical status could not be retrieved.
3.2. STRAT4 Analysis
A summary of the STRAT4 results is shown in Table 3. Most cases were hormonal receptor-positive (80.4% for ESR1 and 73.5% for PGR), 73.5% were ERBB2-negative, and 27.3% rendered a negative MKI67 result. The histopathological features of the cases with a failed STRAT4 determination (n = 8, 3.2%) are shown in Table S1.
Table 3.
STRAT4 results of the series.
| Total | Percentage | ||
|---|---|---|---|
| ER | Positive | 209 | 80.4 |
| Negative | 43 | 16.5 | |
| Fail | 8 | 3.1 | |
| PR | Positive | 191 | 73.5 |
| Negative | 61 | 73.5 | |
| Fail | 8 | 3.1 | |
| HER2 | Positive | 61 | 23.5 |
| Negative | 191 | 73.5 | |
| Fail | 8 | 3.1 | |
| Ki67 | Positive | 181 | 69.6 |
| Negative | 71 | 27.3 | |
| Fail | 8 | 3.1 |
3.3. Diagnostic Accuracy Analysis
The eight cases with a failed STRAT4 determination were excluded in the subsequent analysis. The number of concordant and discordant cases for the different biomarkers is shown in Table 4 and a summary of the diagnostic accuracy statistics is shown in Table 5. The accuracy for HER2 determination reached 91.3% when studying all cases together, while it decreased to 80.7% for HER2 2+ cases and increased to 99.3% for no-HER2 2+ cases. The biomarkers results with both techniques of the discordant cases are shown in Table S2.
Table 4.
Number of concordant and discordant cases for the different biomarkers’ status.
| Biomarker | Group | Concordat | Discordant | Total | ||
|---|---|---|---|---|---|---|
| Positive | Negative | IHC Positive | IHC Negative | |||
| HER2 | Globally | 50 | 180 | 11 | 11 | 252 |
| HER2 2+ | 20 | 68 | 10 | 11 | 109 | |
| No-HER2 2+ | 30 | 111 | 1 | 0 | 143 | |
| HER2 0+ | 0 | 70 | 0 | 0 | 70 | |
| HER2 1+ | 0 | 42 | 0 | 0 | 42 | |
| HER2 3+ | 30 | 0 | 1 | 0 | 31 | |
| ER | 204 | 39 | 4 | 5 | 252 | |
| PR | 156 | 56 | 5 | 35 | 252 | |
| Ki67 | 78 | 68 | 3 | 102 | 251 | |
Table 5.
Diagnostic accuracy statics for each biomarker status.
| Biomarker | Group | Accuracy | Sensitivity | Specificity | Positive Predictive Value | Negative Predictive Value |
|---|---|---|---|---|---|---|
| HER2 | Globally | 91.3 | 82.0 | 94.2 | 82.0 | 94.2 |
| HER2 2+ | 80.7 | 66.7 | 86.1 | 64.5 | 87.2 | |
| No-HER2 2+ | 99.3 | 96.8 | 100 | 100 | 99.1 | |
| HER2 0+ | - | - | 100 | - | 100 | |
| HER2 1+ | - | - | 100 | - | 100 | |
| HER2 3+ | - | 96.8 | - | 100 | - | |
| ER | 96.4 | 98.1 | 88.6 | 97.6 | 90.7 | |
| PR | 84.1 | 96.9 | 61.5 | 81.7 | 91.8 | |
| Ki67 | 58.2 | 96.3 | 40.0 | 43.3 | 95.8 |
3.4. HER2 Results
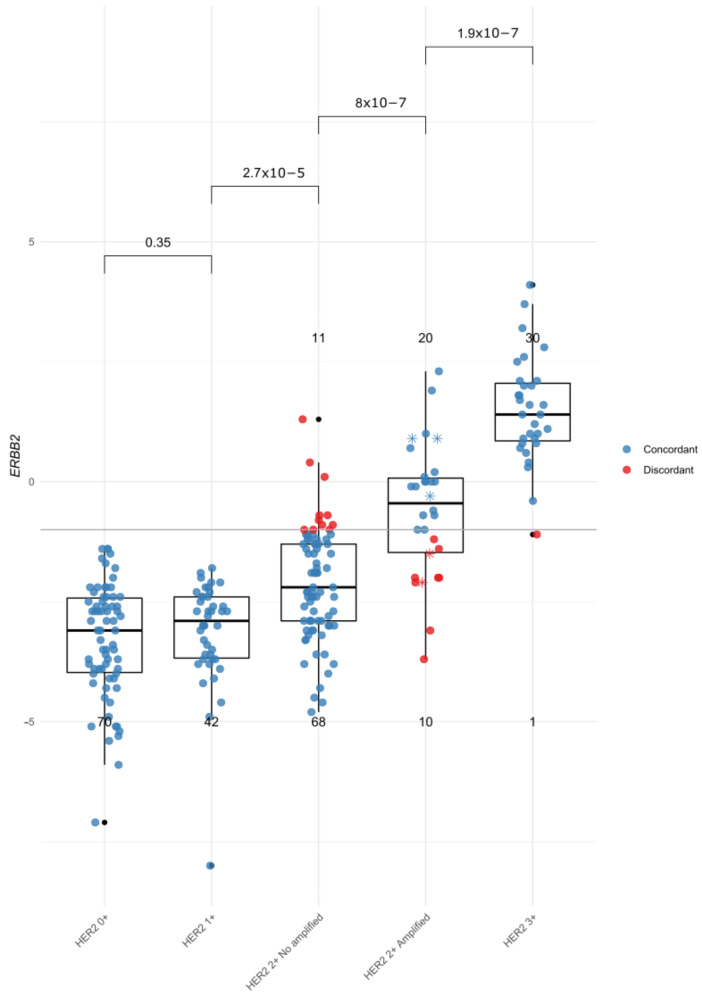
The STRAT4 ERBB2 dCt for each case according to their immunohistochemical and FISH HER2 status is shown in Figure 1. Twenty-one out of the 109 HER2 2+ cases (19.3%) and 1 out of the 31 HER2 3+ cases (3.2%) showed discordant results between mRNA expression and FISH amplification. All HER2 0+ and HER2 1+ cases where correctly categorized as HER2-negative by STRAT4. There were statistically significant differences between the ERBB2 dCt and the IHC/FISH results among all subgroups, except between HER2 0+ and HER2 1+.
Figure 1.
Boxplots of STRAT4 ERBB2 dCt by HER2 immunohistochemical (0+, 1+, 2+, 3+) and FISH (not amplified, amplified) results. Each individual case is represented with a colored point (blue for concordant cases, red for discordant cases). Asterisks (*) in the HER2 2+-amplified group represent the cases with signals amplified in clusters (n = 5). The horizontal gray line represents the cut-off value for STRAT4 ERBB2 dCt (−1). The bars above the boxplots represent the p-values of ERBB2 dCt comparison of the means.
Regarding the HER2 2+ cases without HER2 amplification analyzed by FISH, 11 out of 79 (13.9%) were classified as HER2-positive by STRAT4. All of them showed ≤2 HER2/CEP17 ratio and ≤4 mean HER2 copies (group 5, ISH-negative) according to the CAP-ASCO guidelines [4]). Eight of these cases scored near the STRAT4 cut-off for ERBB2 (dCt −0.7 to −1, being the cut-off for positivity ≥ −1), while three scored far above this point (dCt 0.1, 0.4 and 1.3).
Ten out of thirty (33.3%) HER2 2+ cases with HER2 amplification by FISH were classified as HER2-negative by STRAT4. Two of them showed ERBB2 signals amplified in clusters, with dCt values of −1.5 and −2.1. The remaining eight discordant cases showed ≥2 HER2/CEP17 ratio and ≥4 mean HER2 copies (group 1, ISH-positive) according to the CAP-ASCO guidelines [4]). As opposed to the non-amplified cases, all but one amplified sample (dCt −1.2) scored far below the cut-off point (dCt −1.4 to −3.7).
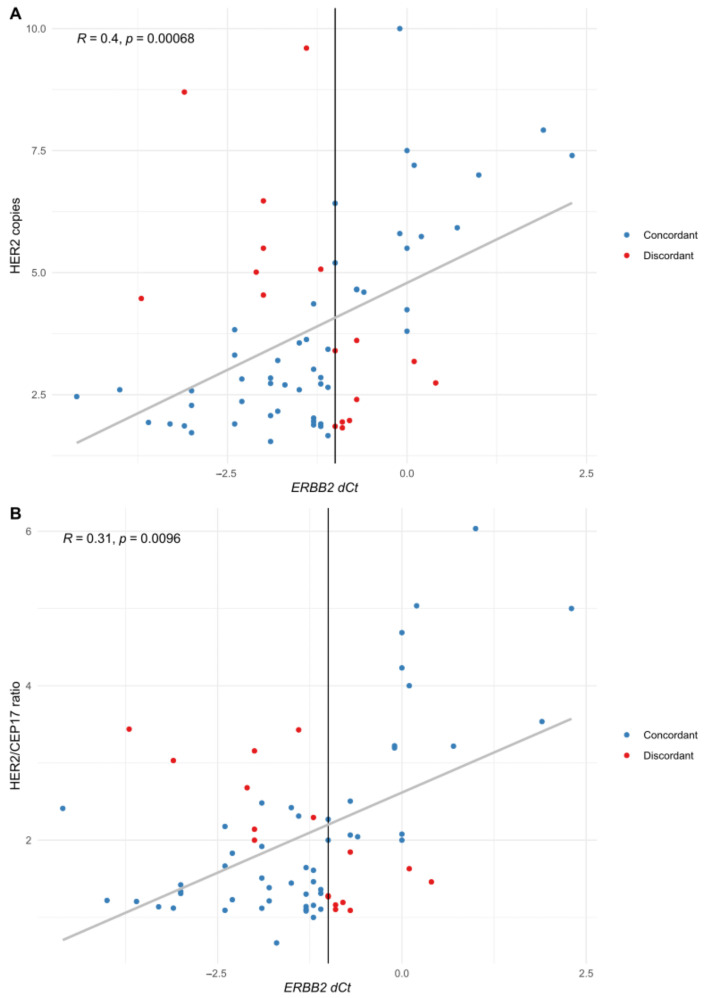
When studying the relationship between STRAT4 dCt and mean HER2 copies and HER2/CEP17 ratio, there was a positive correlation, as shown in Figure 2 (cases with HER2 signals amplified in clusters (n = 5) were excluded from these analyses because the number of HER2 signals could not be accurately counted).
Figure 2.
Correlation of HER2 copies (A) and HER2/CEP17 ratio (B) with ERBB2 expression. Each individual case is represented with a colored point (blue for concordant cases, red for discordant ones). The vertical line represents the cut-off value for STRAT4 ERBB2 dCt (−1).
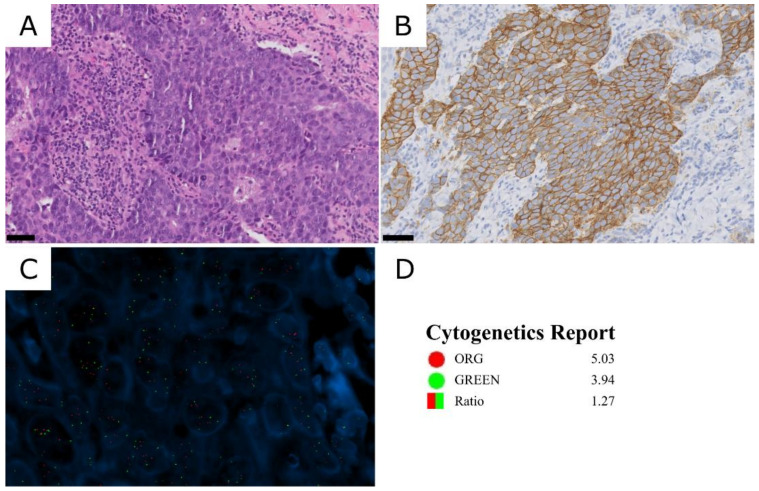
As illustrated previously, one HER2 3+ case showed discordant results between the HER2 immunohistochemical status and STRAT4, near the cut-off value of ERBB2 mRNA expression (dCt −1.1) (Figure 1). Due to this discordance, HER2 FISH was performed, which showed gains in both CEP17 and ERBB2 signals (ISH group 4 [4]) (Figure 3). This patient was treated with neoadjuvant therapy, including pertuzumab, trastuzumab and docetaxel, showing a pathological complete response in the subsequent surgical specimen.
Figure 3.
HER2 3+ case classified as HER2 negative by STRAT4 (case 279). (A) HE. (B) HER2 immunohistochemistry, showing intense and complete membrane staining in >10% of tumor cells. (C) HER2 FISH, showing gains for both probes. Blue: DAPI, green signals: CEP17 probe; red signals: ERBB2 probe. (D) Cytogenetics report. ORG: ERBB2 probe count; GREEN: CEP17 probe count. Scale bar: 50 µm.
3.5. Estrogen Receptor Results
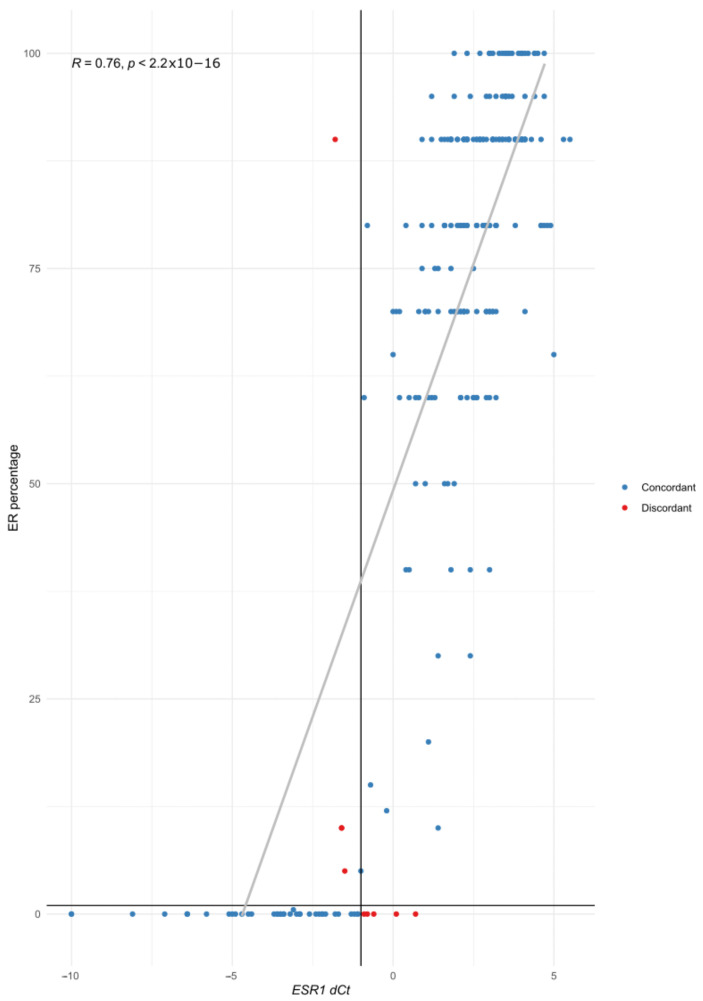
When comparing the ER results, nine cases (3.6%) showed discordant results. Three out of four discordant IHC-positive cases showed low ER expression (5% of ER-positive cells (n = 1) and 10% of ER-positive cells (n = 2)). The IHC ER percentage and ESR1 dCt showed a positive correlation (Figure 4).
Figure 4.
Correlation of ER IHC percentage (y axis) with ESR1 dCT (x axis). Each individual case is represented with a colored point (blue for concordant cases, red for discordant ones). The vertical line represents the cut-off value for STRAT4 ESR1 dCt (−1), and the horizontal line the cut-off value for ER (1%).
3.6. Progesterone Receptor Results
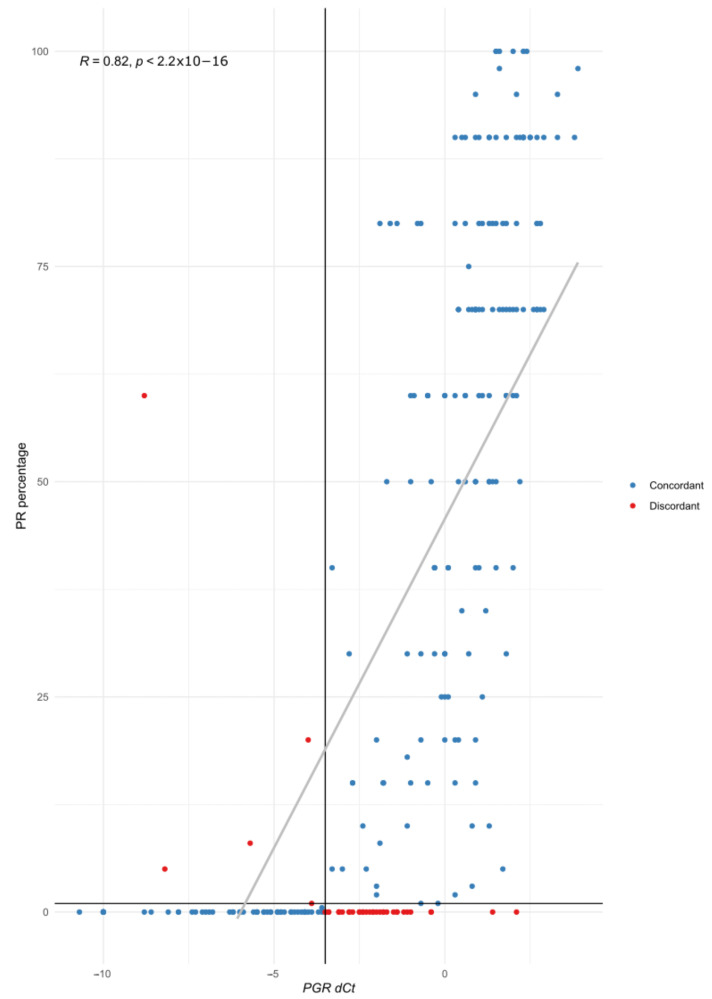
Regarding PR, 40 cases (15.9%) showed discordant results, and 35 were IHC-negative and STRAT4-positive. The IHC PR percentage and PGR dCt showed a positive correlation (Figure 5).
Figure 5.
Correlation of PR IHC percentage (y axis) with PGR dCT (x axis). Each individual case is represented with a colored point (blue for concordant cases, red for discordant ones). The vertical line represents the cut-off value for STRAT4 PGR dCt (−3.5), and the horizontal line the cut-off value for PR IHC (1%).
3.7. Ki67 Results
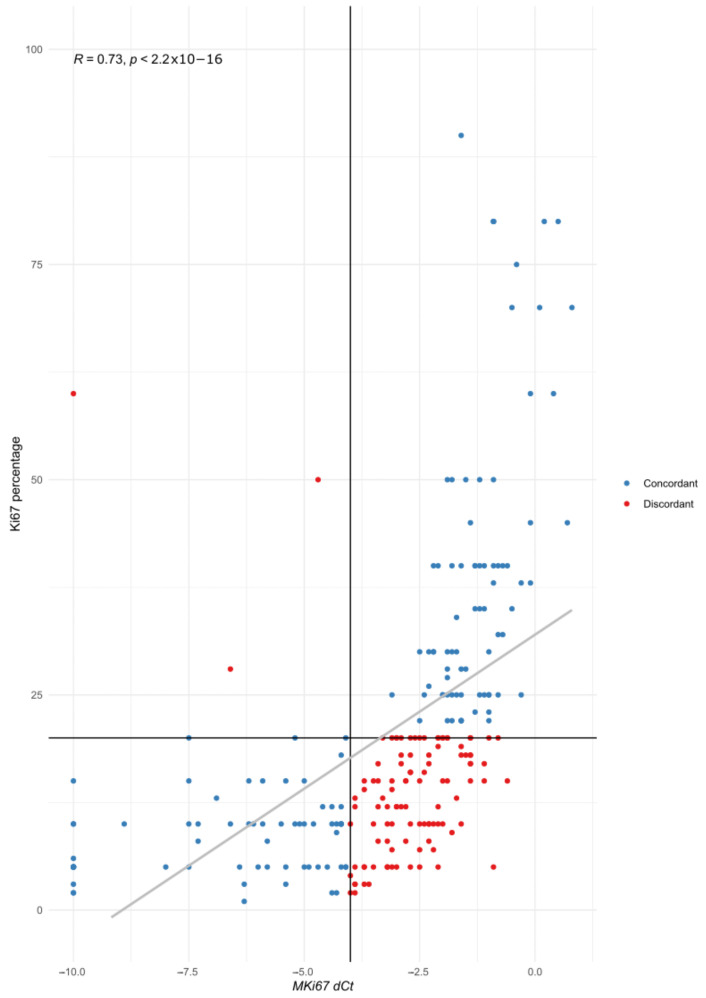
One hundred and five cases (41.7%) showed discordant results between IHC and STRAT4. The IHC Ki67 percentage and MKI67 dCt showed a positive correlation (Figure 6). Discordant cases with Ki67 expression <5% (n = 7) showed a STRAT4 dCt near the cut-off value (range −3.6 to −4).
Figure 6.
Correlation of Ki67 IHC percentage (y axis) with MKI67 dCT (x axis). Each individual case is represented with a colored point (blue for concordant cases, red for discordant ones). The vertical line represents the cut-off value for STRAT4 MKI67 dCt (−4), and the horizontal line the cut-off value for Ki67 IHC (20%).
4. Discussion
In this study, we evaluated the performance of STRAT4, a RT-qPCR platform, to assess breast cancer biomarker status in a cohort enriched in HER2 2+ cases. An acceptable concordance was observed between IHC and STRAT4 when evaluating HER2 globally and no-HER2 2+ cases. However, the concordance clearly decreased in HER2 2+ tumors. Thus, STRAT4 achieved a high concordance with IHC in HER2 determination in no-HER 2+ cases, with a 99.3% accuracy. However, HER2 2+ cases still represent a challenge, with an identification accuracy of 80.7%. This degree of discordance was not attributable to the specific series, since 26.8% of our HER2 2+ cases showed HER2 amplification by FISH, in accordance with the literature [7,8,9]. In addition, all cases rendered interpretable results for other STRAT4 markers (Supplementary Table S2), and the failure rate of STRAT4 (3.2%) in this series was similar to that reported in the literature [13,16,17].
Previous studies [13,14,15,17,18,19] reported around 90% accuracy for STRAT4, when studying globally the HER2 status (positive vs. negative), without distinguishing the HER2 2+ amplification status. A more recent study also reported a 90% concordance between the HER2 2+ FISH and STRAT4 results, although only 10 HER2 2+ cases were included [16], and the 2013 ASCO/CAP guidelines were followed [21].
The fact that two HER2 2+ cases that showed HER2 amplification with cluster signals were categorized as HER2-negative by STRAT4 is interesting. In these cases, we would expect to find high ERBB2 mRNA levels and HER2 protein levels, but the dCt value obtained was not near the positivity cut-off, and HER2 IHC was equivocal. Thus, this could be due to a transcription mechanism resulting in low mRNA and protein levels, even in the presence of gene amplification. In fact, several mechanisms play a role in regulating the mRNA and protein levels, such as transcription factors, promoters, micro-RNA or DNA methylation.
The discordant HER2 3+ case might be explained by the genetic alterations found, as we observed a high number of copies of both CEP17 and ERBB2 with the FISH assay. A polysomy affecting both chromosome 15 (reference gene CYPIP1) and 17 (ERBB2) could result in higher mRNA levels of both, thus resulting in a low dCt value. Nonetheless, it was suggested that increases in the number of copies of both ERBB2 and CEP17 are usually due to pericentromeric gains rather than to polysomies [22]. Downs-Kelly et al. [23] studied the effect of polysomy 17 on HER2 gene and protein expression and found that polysomy does not significantly contribute to a higher expression, although it has been reported that most polysomy cases that show 3+ by IHC are also amplified, as shown by FISH [24]. The FISH result in this discordant case, with a ERBB2/CEP17 ratio <2 and ≥4 and <6 ERBB2 signals/cell, would indicate it belonged to the ISH group 4. However, following the current ASCO/CAP guidelines [4], this sample should be categorized as HER2-positive, as the initial immunohistochemistry rendered a 3+ status (Figure 3) and in clinical practice it would not have been studied by FISH. In addition, the patient showed a great response to targeted therapy, confirming the HER2-positive status.
When the aforementioned ASCO/CAP 2013 guidelines [21] were applied, a poorer concordance between STRAT4 and FISH results was obtained, as six STRAT4-negative and non-amplified by FISH tumors (concordant cases using 2018 criteria) would have been classified as HER2-positive by the 2013 guidelines. In this regard, previous studies comparing both guidelines showed that following the 2018 criteria reduces the rate of HER2 positivity [25]. Moreover, it has been proven that the 2018 ASCO/CAP guidelines for HER2 [4] have a better concordance with HER2 mRNA expression assays [26].
There were statistically significant differences in ERBB2 dCt according to the HER2 status for all groups except the HER2 0+ compared to the HER2 1+ (Figure 1). Due to the recent development of HER2-directed antibody–drug conjugates [11], the correct classification of these cases is gaining importance, as it may change the clinical management of the patients. In this regard, a recent study by Atallah et al. [27] suggests new refined score patterns to distinguish HER2 0+ and HER2 1+ tumors, which correlates with mRNA expression.
Regarding the hormonal receptors, STRAT4 achieved the greatest accuracy in ER determination, in accordance with the literature [13,14,15,17], while for PR had worse results. Although it could be argued that some discrepancies could be attributed to the antibodies used, as they present different sensitivity and specificity, it has been shown that the Agilent/Dako platform has one of the strongest inter-observer agreement and accuracy compared to Leica and Ventana [28]. However, most studies that compared STRAT4 to IHC used Agilent/Dako, with accuracies ranging from 100% to 93.3% for ER (ours was 96.4%) and 89% to 96% for PR (ours was 84.1%) [14,15,16]. Only one study used Ventana, achieving 98.9% accuracy for ER and 89.8% for PR [17]. Another study reported 98.3% accuracy for ER and 86.7% for PR, without specifying the antibodies used [18].
Patients with higher ER and PR expression have a better response to endocrine therapy, and even those with as little as 1% positive staining still show a response [1]. Nevertheless, low ER expression (1% to 10% of weakly positive cells) must be reported, as recommended by the current guidelines [1], as these tumors are heterogenous, some of them with similar characteristics to ER-negative carcinomas [29,30], and do not benefit as much from endocrine treatments. Our study did not have sufficient power to evaluate low-ER tumors, and only five cases were classified in this group. However, three of them showed discordant results, highlighting the difficulty of assessing bordering values.
As shown before, previous studies also found a lower accuracy for PR when compared to ER [13,14,15,17,18], and these results are similar to ours. This may be explained by the heterogeneity of PR expression, as it is not as uniform as ER and usually presents a lower intensity, making its evaluation more difficult.
Ki67 expression led to the greatest discordance between the two techniques. In fact, it has the lowest concordance rates between all IHC biomarkers due to the difficulties in the standardization of its measurement [3]. Moreover, previous studies of the concordance of IHC and STRAT4 also found that Ki67 has the lowest accuracy, ranging from 73% to 90% [13,14,17]. However, we obtained even worse results, with 58.2% accuracy. This may be due to methodological differences, as in some studies, cases with intermediate IHC results were excluded from the analysis [17], or a small number of cases were analyzed [14]. There was significant scattering of the MKI67 dCt data for tumors with low proliferative rates, with Ki67 < 20% (Figure 6), in accordance with previous observations [13,15,17]. The best thresholds for Ki67 IHC and MKI67 dCt in our cohort according to the ROC curves were 15.5% and −3 dCt, respectively (Figures S1 and S2, versus 20% and −4 dCt used) [31].
The main limitation of our study is that the number of cases was low compared to those of other STRAT4 concordance studies [13,17].
5. Conclusions
Although the number of cases we studied was limited, we were able to analyze a large number of HER2 2+ cases, showing that they represent a complex group in which ERBB2 mRNA is not a reliable reflex method to assess ERBB amplification status, although it could be useful in areas where IHC and FISH are not available [16].
Further studies should evaluate the concordance of STRAT4 in other special IBC cases, such as those with ERBB2 mutations as well as HER2-low and low-ER cases, as it could provide useful information on biomarker status and patient management.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15061688/s1, Table S1: Histopathological features of the cases with a failed STRAT4 determination; Table S2: Biomarkers results of both techniques for the discordant cases; Figure S1: ROC curve for the best MKI67 dCt threshold in our series. Figure S2: ROC curve for the best Ki67 IHC threshold in our series.
Author Contributions
Conceptualization, J.P. and B.P.-M.; Data curation, I.C.-B.; Formal analysis, I.C.-B., D.S. and G.M.-B.; Funding acquisition, J.P.; Investigation, T.C.-C. and M.R.; Methodology, J.P. and B.P.-M.; Project administration, J.P. and B.P.-M.; Resources, J.P.; Supervision, J.P. and B.P.-M.; Visualization, I.C.-B.; Writing—original draft, I.C.-B., J.P. and B.P.-M.; Writing—review & editing, I.C.-B., T.C.-C., M.C.S., N.M.-J., M.C., D.S., G.M.-B., J.P. and B.P.-M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was approved by the Ethics Committee of the Hospital Universitario Ramón y Cajal (reference number 361–20).
Informed Consent Statement
Patient consent was waived by the Ethics Committee because little FFPE material would be used, patient management would not change irrespective of the findings, and due to the retrospective nature of the study.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding authors upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study has been funded by Instituto de Salud Carlos III (ISCIII) through the project “PI22/01892” and co-funded by the European Union. Developed with the financial support of Immune4ALL Project (PMP22/00054), with European funds of the Recovery, Transformation and Resilence Plan, and financed by the Instituto de Salud Carlos IIII. Funden by CIBERONC (grants CB16/12/00316 and CB16/12/00295) and the European Development Regional Fund “A way to achieve Europe” (FEDER). Cepheid provided material support for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Allison K.H., Hammond M.E.H., Dowsett M., McKernin S.E., Carey L.A., Fitzgibbons P.L., Hayes D.F., Lakhani S.R., Chavez-MacGregor M., Perlmutter J., et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J. Clin. Oncol. 2020;38:1346–1366. doi: 10.1200/JCO.19.02309. [DOI] [PubMed] [Google Scholar]
- 2.Yerushalmi R., Woods R., Ravdin P.M., Hayes M.M., Gelmon K.A. Ki67 in Breast Cancer: Prognostic and Predictive Potential. Lancet Oncol. 2010;11:174–183. doi: 10.1016/S1470-2045(09)70262-1. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen T.O., Leung S.C.Y., Rimm D.L., Dodson A., Acs B., Badve S., Denkert C., Ellis M.J., Fineberg S., Flowers M., et al. Assessment of Ki67 in Breast Cancer: Updated Recommendations from the International Ki67 in Breast Cancer Working Group. J. Natl. Cancer Inst. 2021;113:808–819. doi: 10.1093/jnci/djaa201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolff A.C., Hammond M.E.H., Allison K.H., Harvey B.E., Mangu P.B., Bartlett J.M.S., Bilous M., Ellis I.O., Fitzgibbons P., Hanna W., et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. Arch. Pathol. Lab. Med. 2018;142:1364–1382. doi: 10.5858/arpa.2018-0902-SA. [DOI] [PubMed] [Google Scholar]
- 5.Grassini D., Cascardi E., Sarotto I., Annaratone L., Sapino A., Berrino E., Marchiò C. Unusual Patterns of HER2 Expression in Breast Cancer: Insights and Perspectives. Pathobiology. 2022;89:278–296. doi: 10.1159/000524227. [DOI] [PubMed] [Google Scholar]
- 6.Egervari K., Szollosi Z., Nemes Z. Immunohistochemical Antibodies in Breast Cancer HER2 Diagnostics. Tumor Biol. 2008;29:18–27. doi: 10.1159/000132567. [DOI] [PubMed] [Google Scholar]
- 7.Hwang H.W., Hong S.A., Nam S.J., Kim S.W., Lee J.E., Yu J.-H., Lee S.K., Cho S.Y., Cho E.Y. Histologic Analysis According to HER2 Gene Status in HER2 2 + Invasive Breast Cancer: A Study of 280 Cases Comparing ASCO/CAP 2013 and 2018 Guideline Recommendations. Virchows Arch. 2022;480:749–758. doi: 10.1007/s00428-022-03280-0. [DOI] [PubMed] [Google Scholar]
- 8.Hoda R.S., Brogi E., Xu J., Ventura K., Ross D.S., Dang C., Robson M., Norton L., Morrow M., Wen H.Y. Impact of the 2018 American Society of Clinical Oncology/College of American Pathologists HER2 Guideline Updates on HER2 Assessment in Breast Cancer With Equivocal HER2 Immunohistochemistry Results With Focus on Cases With HER2/CEP17 Ratio <2.0 and Average HER2 Copy Number ≥4.0 and <6.0. Arch. Pathol. Lab. Med. 2019;144:597–601. doi: 10.5858/arpa.2019-0307-OA. [DOI] [PubMed] [Google Scholar]
- 9.Park S., Park H.S., Koo J.S., Yang W.I., Kim S.I., Park B.-W. Breast Cancers Presenting Luminal B Subtype Features Show Higher Discordant Human Epidermal Growth Factor Receptor 2 Results between Immunohistochemistry and Fluorescence in Situ Hybridization. Cancer. 2012;118:914–923. doi: 10.1002/cncr.26406. [DOI] [PubMed] [Google Scholar]
- 10.Fehrenbacher L., Cecchini R.S., Geyer Jr C.E., Rastogi P., Costantino J.P., Atkins J.N., Crown J.P., Polikoff J., Boileau J.-F., Provencher L., et al. NSABP B-47/NRG Oncology Phase III Randomized Trial Comparing Adjuvant Chemotherapy With or Without Trastuzumab in High-Risk Invasive Breast Cancer Negative for HER2 by FISH and With IHC 1+ or 2+ J. Clin. Oncol. 2020;38:444–453. doi: 10.1200/JCO.19.01455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Modi S., Jacot W., Yamashita T., Sohn J., Vidal M., Tokunaga E., Tsurutani J., Ueno N.T., Prat A., Chae Y.S., et al. Trastuzumab Deruxtecan in Previously Treated HER2-Low Advanced Breast Cancer. N. Engl. J. Med. 2022;387:9–20. doi: 10.1056/NEJMoa2203690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H., Katerji H., Turner B.M., Hicks D.G. HER2-Low Breast Cancers. Am. J. Clin. Pathol. 2022;157:328–336. doi: 10.1093/ajcp/aqab117. [DOI] [PubMed] [Google Scholar]
- 13.Wu N.C., Wong W., Ho K.E., Chu V.C., Rizo A., Davenport S., Kelly D., Makar R., Jassem J., Duchnowska R., et al. Comparison of Central Laboratory Assessments of ER, PR, HER2, and Ki67 by IHC/FISH and the Corresponding MRNAs (ESR1, PGR, ERBB2, and MKi67) by RT-QPCR on an Automated, Broadly Deployed Diagnostic Platform. Breast Cancer Res. Treat. 2018;172:327–338. doi: 10.1007/s10549-018-4889-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erber R., Hartmann A., Fasching P.A., Ruebner M., Stöhr R., Beckmann M.W., Zentgraf M., Popp V., Weidler J., Simon I., et al. Reproducibility of MRNA-Based Testing of ESR1, PGR, ERBB2, and MKI67 Expression in Invasive Breast Cancer-A Europe-Wide External Quality Assessment. Cancers. 2021;13:4718. doi: 10.3390/cancers13184718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janeva S., Parris T.Z., Nasic S., De Lara S., Larsson K., Audisio R.A., Olofsson Bagge R., Kovács A. Comparison of Breast Cancer Surrogate Subtyping Using a Closed-System RT-QPCR Breast Cancer Assay and Immunohistochemistry on 100 Core Needle Biopsies with Matching Surgical Specimens. BMC Cancer. 2021;21:439. doi: 10.1186/s12885-021-08171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mugabe M., Ho K.E., Ruhangaza D., Milner D., Rugwizangoga B., Chu V.C., Wu N.C., Rizo A., Weidler J.M., Wong W., et al. Use of the Xpert Breast Cancer STRAT4 for Biomarker Evaluation in Tissue Processed in a Developing Country. Am. J. Clin. Pathol. 2021;156:766–776. doi: 10.1093/ajcp/aqab016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filipits M., Rudas M., Singer C.F., Fitzal F., Bago-Horvath Z., Greil R., Balic M., Lax S.F., Halper S., Hulla W., et al. ESR1, PGR, ERBB2, and MKi67 MRNA Expression in Postmenopausal Women with Hormone Receptor-Positive Early Breast Cancer: Results from ABCSG Trial 6. ESMO Open. 2021;6:100228. doi: 10.1016/j.esmoop.2021.100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta S., Mani N.R., Carvajal-Hausdorf D.E., Bossuyt V., Ho K., Weidler J., Wong W., Rhees B., Bates M., Rimm D.L. Macrodissection Prior to Closed System RT-QPCR Is Not Necessary for Estrogen Receptor and HER2 Concordance with IHC/FISH in Breast Cancer. Lab. Investig. 2018;98:1076–1083. doi: 10.1038/s41374-018-0064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wasserman B.E., Carvajal-Hausdorf D.E., Ho K., Wong W., Wu N., Chu V.C., Lai E.W., Weidler J.M., Bates M., Neumeister V., et al. High Concordance of a Closed-System, RT-QPCR Breast Cancer Assay for HER2 MRNA, Compared to Clinically Determined Immunohistochemistry, Fluorescence in Situ Hybridization, and Quantitative Immunofluorescence. Lab. Investig. 2017;97:1521–1526. doi: 10.1038/labinvest.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Core Team R: A Language and Environment for Statistical Computing. [(accessed on 22 August 2022)]. Available online: https://www.R-project.org/
- 21.Wolff A.C., Hammond M.E.H., Hicks D.G., Dowsett M., McShane L.M., Allison K.H., Allred D.C., Bartlett J.M.S., Bilous M., Fitzgibbons P., et al. Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Update. J. Clin. Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 22.Hanna W.M., Rüschoff J., Bilous M., Coudry R.A., Dowsett M., Osamura R.Y., Penault-Llorca F., van de Vijver M., Viale G. HER2 in Situ Hybridization in Breast Cancer: Clinical Implications of Polysomy 17 and Genetic Heterogeneity. Mod. Pathol. 2014;27:4–18. doi: 10.1038/modpathol.2013.103. [DOI] [PubMed] [Google Scholar]
- 23.Downs-Kelly E., Yoder B.J., Stoler M., Tubbs R.R., Skacel M., Grogan T., Roche P., Hicks D.G. The Influence of Polysomy 17 on HER2 Gene and Protein Expression in Adenocarcinoma of the Breast: A Fluorescent In Situ Hybridization, Immunohistochemical, and Isotopic MRNA In Situ Hybridization Study. Am. J. Surg. Pathol. 2005;29:1221–1227. doi: 10.1097/01.pas.0000165528.78945.95. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y., Ma L., Liu D., Yang Z., Yang C., Hu Z., Chen W., Yang Z., Chen S., Zhang Z. Impact of Polysomy 17 on HER2 Testing of Invasive Breast Cancer Patients. Int. J. Clin. Exp. Pathol. 2013;7:163–173. [PMC free article] [PubMed] [Google Scholar]
- 25.Gordian-Arroyo A.M., Zynger D.L., Tozbikian G.H. Impact of the 2018 ASCO/CAP HER2 Guideline Focused Update. Am. J. Clin. Pathol. 2019;152:17–26. doi: 10.1093/ajcp/aqz012. [DOI] [PubMed] [Google Scholar]
- 26.McLemore L.E., Albarracin C.T., Gruschkus S.K., Bassett R.L., Wu Y., Dhamne S., Yim I., Lin K., Bedrosian I., Sneige N., et al. HER2 Testing in Breast Cancers: Comparison of Assays and Interpretation Using ASCO/CAP 2013 and 2018 Guidelines. Breast Cancer Res. Treat. 2021;187:95–104. doi: 10.1007/s10549-021-06208-5. [DOI] [PubMed] [Google Scholar]
- 27.Atallah N.M., Toss M.S., Green A.R., Mongan N.P., Ball G., Rakha E.A. Refining the Definition of HER2-Low Class in Invasive Breast Cancer. Histopathology. 2022;81:770–785. doi: 10.1111/his.14780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornaga E.N., Klimowicz A.C., Guggisberg N., Ogilvie T., Morris D.G., Webster M., Magliocco A.M. A Systematic Comparison of Three Commercial Estrogen Receptor Assays in a Single Clinical Outcome Breast Cancer Cohort. Mod. Pathol. 2016;29:799–809. doi: 10.1038/modpathol.2016.74. [DOI] [PubMed] [Google Scholar]
- 29.Najjar S., Allison K.H. Updates on Breast Biomarkers. Virchows Arch. 2022;480:163–176. doi: 10.1007/s00428-022-03267-x. [DOI] [PubMed] [Google Scholar]
- 30.Raghav K.P.S., Hernandez-Aya L.F., Lei X., Chavez-Macgregor M., Meric-Bernstam F., Buchholz T.A., Sahin A., Do K.-A., Hortobagyi G.N., Gonzalez-Angulo A.M. Impact of Low Estrogen/Progesterone Receptor Expression on Survival Outcomes in Breast Cancers Previously Classified as Triple Negative Breast Cancers. Cancer. 2012;118:1498–1506. doi: 10.1002/cncr.26431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.-C., Müller M. PROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding authors upon reasonable request.