Abstract
Cancer is a societal burden demanding innovative approaches. A major problem with the conventional chemotherapeutic agents is their strong toxicity and other side effects due to their poor selectivity. Uncontrolled proliferation of cancer cells is due to mutations, deletions, or amplifications in genes (oncogenes) encoding for proteins that regulate cell growth and division, such as transcription factors, for example, c-MYC. The direct targeting of the c-MYC protein has been attempted but so far unsuccessfully, as it lacks a definite binding site for the modulators. Meanwhile, another approach has been explored since the discovery that G-quadruplex secondary DNA structures formed in the guanine-rich sequences of the c-MYC promoter region can downregulate the transcription of this oncogene. Here, we will overview the major achievements made in the last decades towards the discovery of a new class of anticancer drugs targeting G-quadruplexes in the c-MYC promoter of cancer cells.
Keywords: G-quadruplexes, c-MYC, cancer, small molecules, G4 stabilizers
1. Introduction
Cancer is one of the most prevalent diseases in the world, with nearly 18 million new cases and 10 million deaths in 2020 [1]. These numbers clearly indicate that, despite the existence of many different ways to treat cancer, such as surgery, radiation therapy, chemotherapy, immunotherapy, and targeted therapies, these treatments do not lead to a cure in most cases [2]. This can happen for different reasons, for example, due to the presence of metastasis, cancer heterogeneity, and resistance to traditional chemotherapy [3]. For these reasons, and also due to the side effects of radiation and chemotherapy, it is of the utmost importance to find more efficient therapies with fewer side effects.
Cancer encompasses a large group of disorders resulting from the alteration in the expression of several genes, which leads to abnormal cell growth [4]. One of such oncogenes is c-MYC, which is amplified in many solid tumors, including almost all serous ovarian, breast, and lung carcinomas [5]. In addition, the chromosomal translocation of c-MYC can occur in leukemias and lymphomas [6], and the activity of c-MYC can also be deregulated at the level of expression and stability of c-MYC mRNA and proteins [7]. Thus, for the above mentioned reasons, and also due to the fact that c-MYC encodes for a transcription factor that controls the transcription of many genes involved in cell cycle progression, cell growth, differentiation, apoptosis, metabolism, DNA replication, and mRNA maturation [7,8,9,10], c-MYC has for a long time been considered to be an attractive drug target [11]. The direct pharmacological inhibition of the c-MYC protein has not yet been achieved, probably because it lacks a definite binding site due to its flat structure. However, many other indirect strategies to target c-MYC have been attempted [12]. These strategies include blocking c-MYC transcription with BRD4, CDK7, and CDK9 inhibitors, blocking MYC mRNA translation by targeting mTOR and CPEB, and also targeting the regulators of c-MYC stability such as some kinases and/or deubiquitinases (USP28, USP36, USP7, PLK1, and AURKA). Moreover, targeting MYC-MAX complexes by using a decoy (OmoMYC), or targeting genes that exhibit synthetic lethality with the overexpression of MYC (for example, CDK1 and GLS) have also been attempted [13,14].
Meanwhile, in the last two decades, another strategy to target c-MYC has been explored: to directly target the c-MYC gene at the transcriptional level. Here, we will summarize the achievements made so far to control the expression of this oncogene and inhibit cancer cell proliferation, with G-quadruplex-interactive small molecules.
2. G-Quadruplex Nucleic Acids as Drug Targets
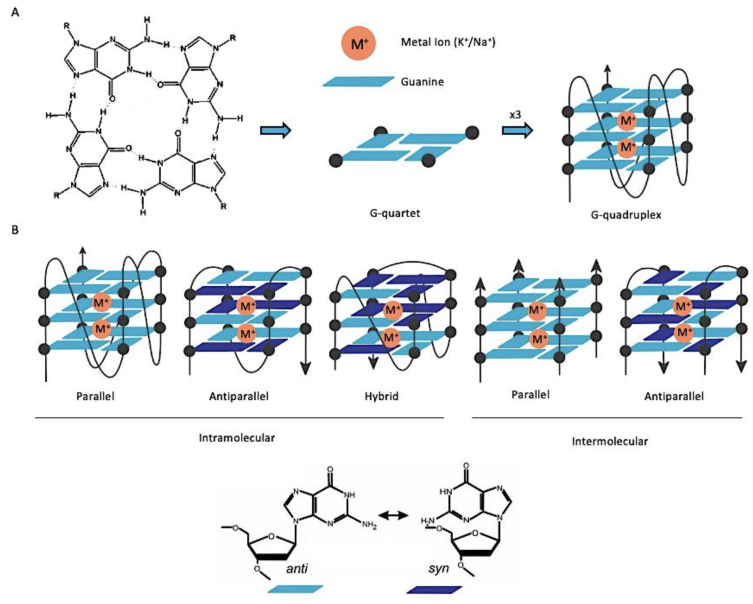
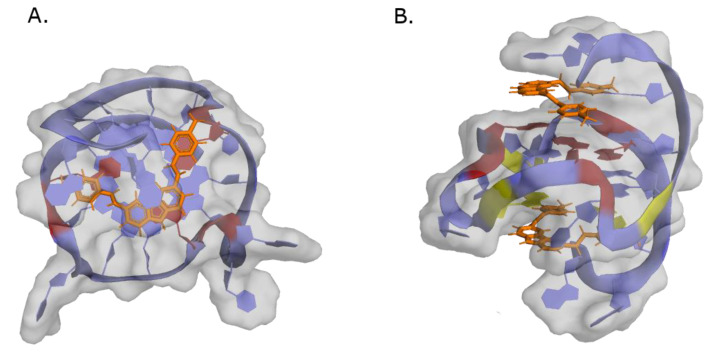
G-quadruplexes (G4) are secondary structures formed from DNA or RNA sequences with at least four runs of consecutive guanines [15,16]. Hydrogen-bonding between four guanines forms planar square structures called G-tetrad or G-quartet, which in turn can stack on top of each other, in an arrangement stabilized by π-π interactions and monovalent cations, called G-quadruplexes (Figure 1A). G4s can be formed by single-stranded DNA (intramolecular) or by two or four different strands (intermolecular) [17]. The orientation of the glycosidic bonds (syn and anti), and the variations in the loop size and sequence, determine the topology (parallel, anti-parallel, or hybrid) and stability of the intramolecular quadruplex (Figure 1B) [17].
Figure 1.
General structure and topologies of G-quadruplexes (G4). (A) Representation of a G-quartet stabilized by Hoogsteen hydrogen bonds (left). Schematic representation of a G-quadruplex additionally stabilized by π-π interactions and monovalent cations (right). (B) Schematic illustration of different intra- and intermolecular G4 topologies with different guanosine conformations (syn and anti).
G4-forming sequences are not randomly distributed in the human genome. Computer analysis has revealed a prevalence of this type of sequence in the telomeres and promoter regions of proto-oncogenes and the involvement of G4s in the regulation of epigenetic, replication, transcription, and translation processes has been demonstrated by several studies [16,18,19,20]. These observations, combined with the fact that G4s can assume different topologies depending on the sequence and environmental factors inducing the folding [21], soon led researchers to hypothesize that G4 structures could be selectively targeted by small molecules. Over the years, many G4 ligands have been developed and studied in vitro and in vivo for their potential as drug leads to treat not only cancer but also other pathologies [22,23,24,25,26].
3. Regulation of c-MYC Transcription by G-Quadruplexes in Promoter Region
In normal circumstances, MYC expression is highly controlled, but in many cancer cells it is overexpressed, leading to tumor progression [13]. Consequently, changes in MYC’s expression are directly correlated to cancer development [9,27,28].
The c-MYC gene is part of a family composed of two other genes, MYCN and MYCL, that, like with c-MYC, encode for the proteins involved in cancer development [13,29]. Interestingly, this family includes two additional genes, b-MYC and s-MYC, that differ from the previous ones because they encode for proteins that suppress tumor growth [30,31].
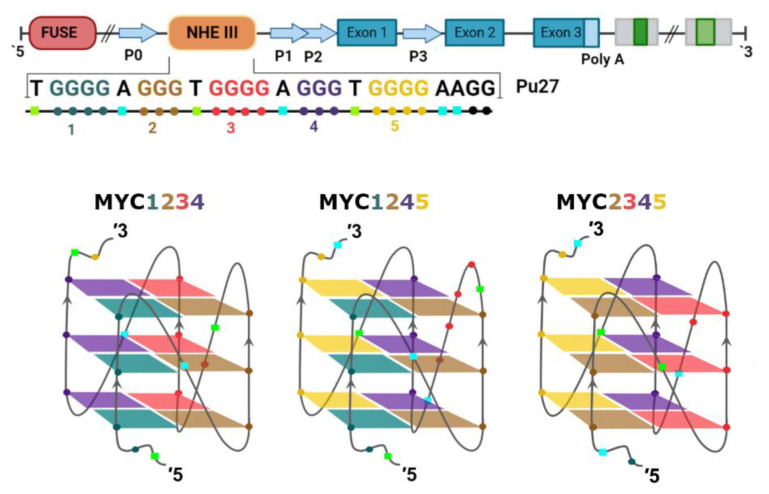
The c-MYC is regulated at transcriptional and post-transcriptional levels by several mechanisms [13]. This transcriptional regulation is complex and involves different elements, such as two transcription start locations, four promoters (P0, P1, P2, and P3), and the interaction with several DNA-binding proteins and regulatory elements. The most important promoters are P1 and P2 because they perform 75–90% of the transcriptions [32,33]. Upstream of the P1 promoter is a nuclease hypersensitivity element (NHE III1) that is the recognition region of transcription factor SP1, which activates c-MYC transcription. However, this is a GC-rich sequence called Pu-27, which can form intramolecular G4s that have been shown to silence c-MYC transcription [34,35,36,37]. This 27-nucleotide sequence has five runs of consecutive guanines, thus being able to fold into three different parallel G4s (Figure 2). However, the G4 formed by Runs 2, 3, 4, and 5 (MYC2345) was shown to be the more stable one [38], and for this reason it was considered to be the most relevant in the control of c-MYC transcription.
Figure 2.
Pu27 c-MYC sequence (top) and representation of different guanine combinations to form different G-quadruplexes (bottom).
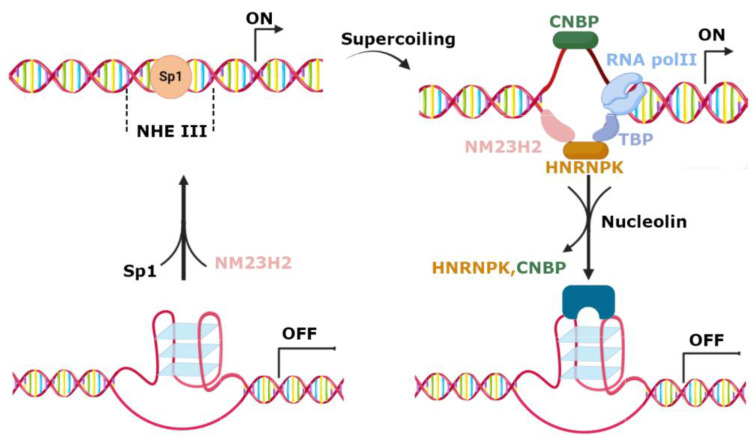
Active and silenced DNA secondary structures and consequent activation and deactivation of c-MYC transcription can be controlled by several proteins (Figure 3) [39]. Nucleolin regulates c-MYC transcription by promoting G4 formation and stabilization, leading to transcriptional arrest [40]. On the contrary, transcriptional activation is induced by the nucleoside diphosphate kinase NM23H2 protein [41] and poly ADP-ribose polymerase [42] that bind and unfold the c-MYC quadruplex. Single-strand DNA can be stabilized via the binding of heterogeneous nuclear ribonucleoprotein K (HNRNPK) and the cellular nucleic acid binding protein (CNBP), leading to the activation of c-MYC transcription [43].
Figure 3.
Schematic representation of c-MYC transcription regulation. G4 can downregulate the transcription by preventing the recognition of transcription factors. NM23H2 can unfold G4 leading to the transcriptional active DNA form. Single-strand DNA formation is induced by negative supercoiling, allowing for the binding of heterogeneous nuclear ribonucleoprotein K (HNRNPK) and the cellular nucleic acid binding protein (CNBP), and the posterior activation of c-MYC transcription. A negative regulation of this transcription occurs via the stabilization of G4 with nucleolin. Adapted from [24].
4. c-MYC G4 Stabilizing Small Molecules with Anticancer Activity
Due to the interest in G4 structures as potential anticancer drug targets, a great number of small molecules has been studied to assess them as quadruplex stabilizing compounds [22]. These compounds typically consist of an aromatic core (usually polyaromatic or macrocyclic) with positively charged side chains, that can interact, respectively, with the G4 tetrads, their phosphate backbone, and/or with water molecules that are present in the grooves [22,44,45]. Many small molecules with antiproliferative activity in cancer cell lines also bind and strongly stabilize in in vitro DNA G4 structures of oncogene-promoter regions, suggesting a potential use of them as new anticancer drugs [46]. In this section, we will focus on the small molecules for which the anticancer mechanism of action has been suggested to be by targeting and stabilizing G4 in the promoter region of c-MYC.
4.1. Macrocycles
Porphyrins are a family of N-heterocyclic molecules in which four pyrrole rings are connected, forming a macrocyclic compound. They provide a versatile base for several applications [47,48,49], such as chemosensors, photosensitizers [47], and antimicrobial agents [50], and as drugs in photodynamic therapy [51]. However, because of their photoactive properties, they can also induce harmful side effects.
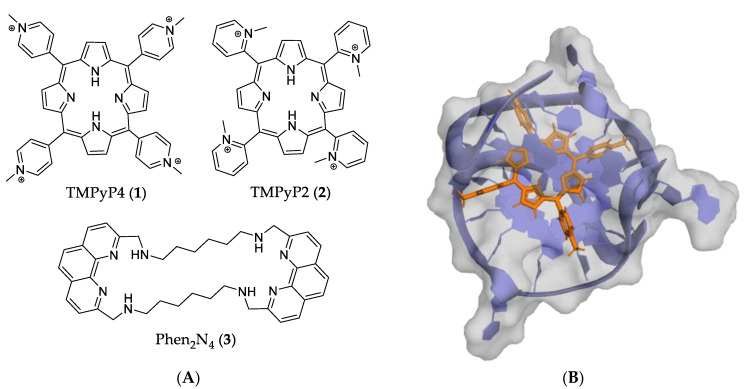
One of the most studied G4 ligands of this type is 5,10,15,20-tetrakis(N-methyl-4-pyridyl) porphyrin (TMPyP4) (1). TMPyP4 and its structural isomer TMPyP2 (2) (Figure 4A) can stabilize DNA G4 in different ways. The first is via external stacking with the outer layer of the guanine tetrads (Figure 4B) and the second is via binding to the diagonal and middle TTA loops [52,53,54]. Although both can stabilize the G4, Compound 2 has restricted rotation around the meso bond, which decreases its binding strength with the G-tetrads, justifying its low capacity to stabilize the G4 [37,54].
Figure 4.
(A) Structures of porphyrin macrocycles 1 and 2 and macrocycle 3. (B) PDB 2A5R representing porphyrin 1 in complex with a two-quartet c-MYC G4.
Furthermore, in vitro studies have shown that 1 can reduce telomerase activity, decrease c-MYC and hTERT expression, and inhibit cancer cell growth [55,56]. There are also a few in vivo studies that have focused on these compounds. These studies have shown that Compound 1 could decrease tumor growth and prolong animal survival compared with untreated controls and with Compound 2 [55]. However, 1 has shown poor selectivity for G4 structures compared to duplex DNA [57]. Moreover, additional studies have also shown that 1 is not able to discriminate between different G4 topologies [22].
Another class of macrocyclic compounds were studied by Carvalho et al. [58]. Different side chain lengths and NH content macrocyclic phenanthrolines were designed. The dimer phen2N4 (3) (Figure 4A) showed a good ability to stabilize both c-MYC G4 and telomeric G4 (21G), with a ΔTm of 17.2 and 20.3 °C, respectively, in a FRET melting assay. This experiment also showed that Compound 3 has selectivity towards G4 over duplex DNA. Moreover, the authors performed a fluorescent intercalator displacement (FID) assay, in which Compound 3 exhibited low values of EC50 (0.87 µM), which suggests a high binding affinity to c-MYC G4. Interestingly, an in vitro study demonstrated that Compound 3 can decrease the unwinding activity of Pif1 helicase. However, cytotoxicity assays resulted in no significant effect in the growth of HeLa cancer cells at low concentrations [58]. In another study, Compound 3 was shown to be highly cytotoxic (IC50 < 0.01 µM) for MCF-7 breast cancer cells [59].
4.2. Ligands with Four or More Fused Aromatic Rings
Indoloquinolines are tetracyclic aromatic compounds that have been extensively studied and have been shown to bind to several secondary structures of nucleic acids [60]. Moreover, several studies have shown that their selectivity and binding affinity toward G4s can change according to the side chains [23].
Taking this into consideration, Liu et al. [61] designed and analyzed four series of disubstituted indoloquinolines, with paired substitutions in Positions 7 and 11, 8 and 11, and 9 and 11, aiming to improve the anticancer activity of the lead SYUIQ-5 (4—Figure 5). Indoloquinoline 4 is a known binder of c-MYC G4, which intercalates between the 3′-outer tetrad of the G4 and a CG base pair (Figure 6A). Additionally, it establishes strong electrostatic interactions with guanine carbonyl groups, due to the positively charged protonated quinoline nitrogen of the tetracyclic indoloquinoline [62]. Moreover, this compound presented strong antiproliferative activity against several cancer cell lines, with IC50 values between 0.24 and 4.8 µM. It also reduced c-MYC transcription in cancer cells, which was suggested to occur by interfering with the interaction between NM23-H2 and c-MYC G4 [63].
Figure 5.
Structures of tetracyclic compounds: indoloquinolines 4, 6, 7, and 11, bioisosters 5 and 8–10, and indoloisoquinoline 12.
Figure 6.
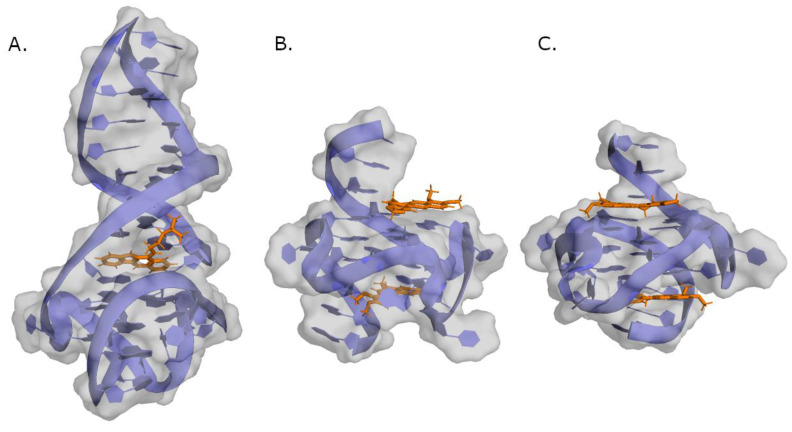
Representation of different compounds (colored sticks) in complex with a DNA structure (represented as cartoon and transparent surface). (A) Indoloquinoline 4 in complex with c-MYC G4 (PDB ID: 7PNG). (B) Conformer B of berberine in complex with c-MYC G4 (PDB ID: 7N7E). (C) Conformer A of berberine in complex with c-MYC G4 (PDB ID: 7N7D).
After analyzing the di-substituted derivatives of 4, Compound 5, an indoloquinoline bioisoster with two tertiary amines in the side chains (Figure 5), was found to be the most promising one. In a FRET melting assay, the compound increased the melting temperature of c-MYC G4 in 26.6 °C and in a microscale thermophoresis (MST) assay showed a higher affinity to c-MYC G4 than to duplex DNA (KDduplex/KDc-MYC = 8.0). Moreover, the compound can inhibit c-MYC transcription and cancer cell proliferation (IC50 of 4.7 µM) in Burkitt’s lymphoma (RAJI) cell line. Additionally, in vivo studies were performed in a RAJI xenograft model, and Compound 5 with a dosage of 30 mg/Kg for 2 weeks inhibited tumor growth, inducing a reduction in tumor volume of 27.4% [61]. Another study by Hurley et. al. analyzed indoloquinolines with a 11-piperazinyl substitution. The most promising compounds were 6 and 7 (Figure 5), which could stabilize the c-MYC G4, presenting ΔTm values of 7 and 17 °C in a CD melting assay. Additionally, the cytotoxicity in RAJI cell lines was tested, and both compounds also presented good IC50 values (2.3 and 3.1 µM, respectively) [64]. However, further experiments have suggested that the anticancer activity shown by these compounds cannot only be attributed to their capacity to stabilize c-MYC G4.
A different study demonstrated that three hybrids of triazole and indoloquinoline’s bioisoster (Compounds 8, 9, and 10—Figure 5) that increased the melting temperature of c-MYC G4 between 13 and 22 °C could also downregulate c-MYC transcription and expression. They also showed high cytotoxicity for different cancer cell lines (IC50 = 0.02–5.53 µM) and lower cytotoxicity against non-malignant human cells. Additionally, Compound 10, in an in vivo assay using a human lung cancer xenograft (A549 cell line), induced the significant inhibition of cancer cell proliferation (38%) and a decrease in tumor volume [65].
The N5-methylated indoloquinoline, PIQ-4m (11) (Figure 5), was shown to be a strong binder of c-MYC G4, with an EC50 of 1.7 µM in the thiazole orange (TO) displacement assay, and was also able to form a 1:2 ligand/G4 complex [66,67]. GQC-05 (12), an indoloisoquinoline derivative (Figure 5), is a well-known binder of c-MYC G4, inducing a ΔTm of 21 °C in a FRET melting assay. Moreover, it can downregulate the expression of this gene alone or in combination with other drugs, such as Navitoclax, a BCL-2 inhibitor [68,69,70,71].
Another class of compounds that can target DNA are fluoroquinolones [72]. Quarfloxin (CX-3543) (13) and Pidnarulex (CX-5461) (14) (Figure 7) are two fluoroquinolone derivatives that have been developed to preferentially target G4 nucleic acids, both inducing ΔTm > 15 °C in a FRET melting assay, for different G4s [73]. They entered into human clinical trials for the treatment of cancer [74,75,76]. Compound 13 reached a Phase II clinical study for the treatment of Neuroendocrine Carcinoma (NCT00780663), but it was abandoned due to poor clinical outcomes [76,77,78]. More recently, 14 entered in Phase I clinical trial for solid tumors withBRCA1/2, PALB2 or homologous recombination deficiency mutations (NCT04890613) [75]. The results of this Phase I study were recently released, demonstrating that the compound is well tolerated and that the observed reversion of PALB2 and BRCA2 mutations confirmed the proposed mechanism of action [79].
Figure 7.
Structures of c-MYC G4 ligands with four or more fused aromatic rings.
Compound 13 has higher selectivity to parallel G4 structures when compared to porphyrin 1 [24] and shows low IC50 values (around 2 µM) in cell viability assays with different cancer cell lines. Initially, the mechanism of action of 13 was thought to be through the binding to the G4 of the c-MYC promoter, but later it was discovered that it probably acts through binding to ribosomal DNA G4, inhibiting RNA-polymerase I activity and the formation of nucleolin/G4 complexes. This mechanism generates the redistribution of nucleolin into the nucleoplasm and, consequently, the inhibition of c-MYC transcription and cancer cell apoptosis [78]. There are several proposed mechanisms of action for Compound 14, with the most acknowledged being the synthetic lethality due to the stabilization of G4 structures in DNA. This causes a loss of function of the replication fork and induces DNA breaks, leading to apoptosis, particularly in cells with deficiencies in the DNA damage repair mechanisms [79,80]. Moreover, another recent study has indicated that 14 also inhibits topoisomerase II activity [81].
It has been shown that phenanthroimidazole derivatives can act as anticancer agents through different mechanisms, including by binding to c-MYC G4. Wu et al. [82] analyzed six compounds with different substitutions in the benzene ring linked to the imidazole. The most promising derivatives were those with chlorine atoms. It was shown that Compound 15 (Figure 7) can downregulate the expression of c-MYC and stabilize the G4 in the promoter region of this gene (ΔTm = 4.4 °C in a FRET melting assay). This ligand also showed good antiproliferative activity against cancer cells (IC50 values around 1 µM) and moderate toxicity for non-cancer human keratinocyte HaCaT cells (IC50 = 16.8 µM). APTO-253 (16—Figure 7) is also a phenanthroimidazole derivative that entered Phase I clinical trials for patients with acute myelogenous leukemia (AML) or myelodysplastic syndrome (MDS) (NCT02267863) [83]. The first outcomes have been published, revealing that APTO-253 has been well tolerated at doses up to 150 mg/m2 [84]. Compound 16 is known to present antiproliferative activity against human colon leukemia, the non-small cell lung, and renal and prostate cancer cell lines by inducing Krüppel-like factor (KLF) tumor suppressors [85]. In a FRET melting assay, 16 showed time-dependent stabilization of G4s, with propensity to stabilize c-MYC G4, and it was more selective to G4 than Compounds 1 and 14 that were used as controls. Compound 16 is also able to induce cytotoxicity in AML and in different lymphoma cell lines, with IC50 values between 57 nM and 1.75 µM. It also decreases c-MYC expression at mRNA and protein levels [86].
Compound BMH-21 (17) and its congener 18 (Figure 7) are examples of other compounds with four fused aromatic rings that can efficiently bind to c-MYC G4, as was shown by NMR studies (ΔTm ~10 °C). They bind to the Pu22, a 22-mer sequence of Pu27, and inhibit lymphoma cell (SUDHL-4) growth with IC50 values below 1 µM. Compound 18 forms a 2:1 ligand/G4 complex, in which each molecule binds to a different part of G4 [87].
Another polyaromatic compound is EMICORON (19—Figure 7) , a perylene diimide derivative that causes telomere uncapping and binds to G4s through π-π interactions with the terminal G-tetrads. In addition, its side chains can bind to the grooves of G4 due to their positive charges. Studies have demonstrated that this compound can bind to the c-MYC and BCL-2 oncogene promoter G4s (ΔTm = 16.4 and 15.4 °C, respectively, in a FRET melting assay) [88] and downregulate the expression of these oncogenes in cancer cells. Compound 19 presented good in vitro anticancer activity and in vivo studies with patient-derived xenografts of a metastatic lymph node of a colon carcinoma in a nude mouse showing a 64% inhibition of tumor growth when treated with 19 [89].
Berberine (20—Figure 7) is a natural product known to bind to G4 in a 2:1 stoichiometry as seen in the pdb structures of both conformers (7N7D and 7N7E)—Figure 6 [90]. This compound increased the melting temperature of c-MYC G4 by more than 6 °C in a CD study and it can induce cancer cell apoptosis in a dose-dependent manner. Additionally, in an in vivo assay with mice implanted with colon cancer cells (CT26), Compound 20 led to a significant decrease in tumor size after 14 days of treatment [91]. Berberine derivatives were also studied as c-MYC G4 ligands. One study showed that a 9-N-substituted derivative (21—Figure 7) could stabilize c-MYC G4 in a FRET melting assay, increasing the ΔTm of the complex to 29 °C. This compound could also inhibit the amplification of Pu27 in a PCR polymerase assay, with IC50 = 2 µM. Moreover, 21 promoted the total inhibition of HL60 cell proliferation at 5 and 10 µM and in an MTT assay presented IC50 = 4 µM in the NCI cell line [92].
4.3. Ligands with Three Fused Aromatic Rings
Phenoxazine is a tricyclic molecule that is part of known antibiotic and antitumor agents [93,94]. Phenoxazone B5 (22—Figure 8) is a phenoxazine derivative discovered in an in silico study as being a promising G4 ligand. This compound showed antiproliferative activity and a larger inhibitory effect on c-MYC transcription (by 25–40%) in the Ramos cell line than in the CA46 cell line, in which the c-MYC transcription is no longer regulated by the NHEIII1 G4-forming sequence in the promoter. These observations strongly suggest that the compound targets the promoter c-MYC G4. Moreover, 22 also showed better inhibitory activity of c-MYC promoter amplification via Taq polymerase than of BCl2, VEGF, and HIF-1α promoters, which also have G4-forming sequences. However, by using CD and a molar equivalent of the ligand, the compound was unable to increase the melting temperature of c-MYC G4 [95]. In another study, focused on the lead 5pro (Compound 23—Figure 8), two other phenoxazines with different side chains were studied. However, these new compounds did not induce better variations in Tm of the G4 in a FRET melting assay (ΔTm= 0 and 9 °C, respectively), when compared to the lead compound (ΔTm = 18 °C) [93].
Figure 8.
Structures of c-MYC G4 ligands with three fused aromatic rings.
Another imidazole chemotype, imidazole-purines, that can bind to c-MYC G4 was studied by Pelliccia et al. [96]. They compared several compounds to determine which one had a better selectivity to G4s in c-MYC and BCL2 gene promoters. Compound 24 (Figure 8) was the one with the ability to stabilize both G4s in a FRET melting assay (ΔTm = 12.8 °C for c-MYC G4 and 6.7 °C for BCL2 G4) and to downregulate c-MYC and BCL2 expression by 66% and 67%, respectively, after the treatment of Jurkat cells (human leukemia cell line) with 25 µM of the compound for 24 h.
Based on the structure of IZCZ-0 (25—Figure 8), four compounds were synthesized and the most promising derivative was the tetra-aryl derivative IZCZ-3 (26—Figure 8). It exhibited stronger affinity to c-MYC G4 (ΔTm of 20 °C) in comparison to the other G4s. Additionally, it showed low IC50 values of cytotoxicity to SiHa, HeLa, Huh7, and A375 cancer cells (2.1–4.2 µM), and higher IC50 values to normal BJ fibroblasts and mesangial cell lines (IC50 = 15.6–15.9 µM). Furthermore, it showed anticancer activity in vivo against human cervical squamous cancer cells, inducing a reduction in tumor weight of 69, 64, and 57% after treatment with 20, 10, and 5 mg/kg of the compound, respectively [97].
Another class of compounds, the pyrazolo[1,2-a]benzo[1,2,3,4]tetrazin-3-one derivatives (PBTs), was studied to evaluate the interaction with c-MYC G4 and the regulation of this oncogene transcription. This study showed that the substitution in C8 and C9 (27) and only in C8 (28) with chlorine atoms (Figure 8) improves the G4 stabilizing effect of these derivatives (ΔTm = 4.0 and 1.9 °C, respectively). Moreover, molecular docking studies have predicted that these chlorine atoms interact with adenine 3 of the G4 [98]. These compounds also presented good antiproliferative activity against different cancer cell lines in an MTT assay, with IC50 values between 13.5 and 13.9 µM for Compound 26 and 17.7 and 20.5 µM for Compound 27 [99].
Due to their planar aromatic ring system, carbazoles are a good choice for quadruplex recognition. Several studies have analyzed the binding of carbazole derivatives to G4s. In one of these studies, a derivative named Cz-1 (29—Figure 9) induced the highest increase in melting temperature of c-MYC G4 permitted in the FRET melting assay (15.8 °C). In other words, 29 showed a good ability to stabilize c-MYC G4 [100]. Furthermore, this compound was shown to be selective to c-MYC G4 over duplex DNA, with KD = 0.21 µM, and showed low values of IC50 in a cell proliferation assay with HeLa and HCT116 cells (3.4 µM and 3.2 µM, respectively) [100]. Another study conducted by Gluszynska et al. [101] also showed that carbazoles are potent G4 stabilizers. In this case, the carbazoles were also combined with triazoles and other pharmacophores. Interestingly, the most promising compound, 30 (Figure 9), was the only one with the carbazole and the benzothiazole moieties.
Figure 9.
Structure of Carbazoles showing cancer cell antiproliferative activity and high capacity to stabilize c-MYC G4.
Several other hybrid molecules were developed, for example, carbazole-triazole 31, 32, and 33 (Figure 9). Compound 31 presented the best selectivity and a higher affinity to c-MYC G4 than the other two hybrids, with KD = 0.17 µM. In a proliferation assay with HCT116 colon cancer cells, this compound showed an IC50 value of 2.1 µM and it was also proven that it was able to downregulate c-MYC transcription and to induce apoptosis in a dose-dependent manner [102]. Overall, Compound 31 was also better when compared to compound 29, both in stabilizing the G4 and in the antiproliferative effect over HCT116 cancer cells.
In an additional study, a series of carbazole derivatives were designed and analyzed using a FRET melting assay to determine their selectivity for G4. The most promising was Compound 34 (Figure 9), showing a relatively high value of ∆Tm for G4-DNA (23.4 °C) and a lower value for double-stranded DNA. Compounds 35 and 36 (Figure 9) also presented good values of ∆Tm, but lower than those of 34. Furthermore, 35 and 36 displayed significant cytotoxicity for cancer cell lines (IC50 = 2–6 µM) and insignificant values of cytotoxicity for human normal cells (IC50 > 50 µM) [103].
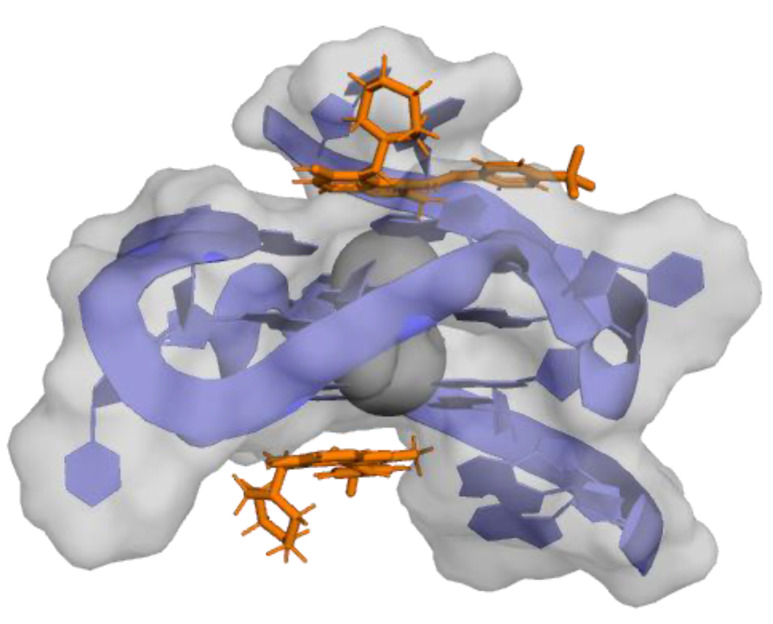
3,6-bis(1-methyl-4-vinylpyridinium carbazole diiodide (BMVC—37—Figure 9) is another carbazole that binds to c-MYC G4, forming 1:1 and 2:1 ligand/G4 complexes (Figure 10). BMVC changes its conformation to perform better binding to G4 and it perfectly matches three bases of the G-tetrads (Figure 10). Furthermore, Compound 36 showed KD = 36 nM for its complex with c-MYC G4 and induced a significant increase in the thermal stability of this G4 in a CD assay. This compound was also shown to repress c-MYC expression in MCF-7 breast cancer cells at 4 and 10 µM concentrations [104].
Figure 10.
Representation of Compound 37 (colored sticks) in complex with a c-MYC G4 structure (cartoon). Guanines interacting with BMVC in top G-quartet at 5′ are colored red, and in yellow are the ones at 3′ G-tetrad. (A) represents the structure of the 1:1 complex (PDB ID: 6JJ0). (B) represents the structure of the 2:1 complex (PDB ID: 6O2L).
4.4. Ligands with Two Fused Aromatic Rings
Quinazolines are N-containing heterocyclic compounds that are widely used as therapeutic agents [105,106]. Sysu12d (38) (Figure 11) is a 2,4-disubstituted quinazoline derivative that can stabilize the c-MYC G4 in the Pu27 sequence [107], and downregulate RNA polymerase I through the disruption of the nucleolin–rDNA interaction. This compound also presented good antiproliferative activity against several cancer cell lines, with IC50 values between 3.1 and 6.3 µM [107]. Another study by Li et al. [108] showed that Compound 39 (Figure 11) could stabilize c-MYC G4 in a FRET melting assay (ΔTm = 23.7 °C) and reduce tumor growth by 49% (10 mg/kg) and 58% (20 mg/kg) in an in vivo assay using human liver cancer cell lines transplanted in a nude mouse. Sysu-ID-01 (40—Figure 11) is another quinazoline derivative that showed the ability to bind to NM23-H2, down- regulating the transcription and translation of c-MYC, but its binding to c-MYC G4 is not as potent (ΔTm = 9 °C) as to NM23-H2 [109]. Because of these results, more studies were conducted to improve compounds’ bioavailability and to study the effects of different substituents. The most promising compounds in this study, 41 and 42 (Figure 11), showed high and specific binding, as well as a stabilizing effect on c-MYC G4 (ΔTm = 12.1 and 12.9 °C, respectively), accompanied by the inhibition of this gene transcription. Additionally, in an in vivo anticancer assay, both compounds inhibited the proliferation of SiHa cells (squamus cell carcinoma) in a dose-dependent manner [110]. Another study by Wang et al. [111] found another isaindigotone derivative (Compound 43—Figure 11) that could downregulate c-MYC transcription by disrupting the interaction between NM23-H2 and c-MYC G4. This compound presents weak binding to c-MYC G4 (ΔTm = 0.54 °C) but strong binding to the protein. Compound 43 can also induce cell cycle arrest, apoptosis, and SiHa cell proliferation arrest in a dose-dependent manner. In vivo studies in a SiHa xenograft mouse model were also performed. Compound 43 inhibited tumor growth by 48 and 65% at 2.5 and 5.0 mg/kg, respectively.
Figure 11.
Structure of c-MYC G4 ligands with two fused aromatic rings.
Quinoxaline or benzopyrazine compounds are naphthalene derivatives. Their main biological activities are antibacterial, antiviral, and antiparasitic, but they also present anticancer activity via different mechanisms [112,113]. Recently, Hu et al. [114] reported a new quinoxaline that can downregulate c-MYC transcription by targeting the G4s in the gene promoter. In this study, they synthesized 10 different quinoxalines with different side chains and several different electron-donating groups. Through structure–activity relationship studies, they concluded that the amino side chains were essential to the compounds binding to the G4 and extra positively charged amino substituents strengthened the interactions with c-MYC G4. The most promising compound was QN-1 (44) (Figure 11). It bound strongly to c-MYC G4 (KD = 1.3 µM) and was selective to this G4 over other G4s with different topologies. Additionally, its structure is more “drug-like” than the other G4 ligands. It also presented low values of IC50 (0.7–0.9 µM) in a cell proliferation assay with different cancer cell lines and higher values compared to normal fibroblast cells (IC50 = 4.6 µM). Compound 44 also showed the ability to inhibit c-MYC transcription in a concentration-dependent manner and had weaker effects in other oncogenes’ transcriptions, which shows the preference to silence the c-MYC gene in the breast cancer 4T1 cell line. Moreover, they conducted an in vivo study to evaluate the ability of Compound 44 to inhibit tumor growth in a triple-negative breast cancer (TNBC) mouse model. At 2.5, 5, and 10 mg/kg, QN-1 significantly inhibited the cancer cell growth (TGI = 42, 47, and 60%) comparable to classical chemotherapeutics for TNBC, but with lower side effects [114].
Imidazole-benzothiazole is a promising G4-binding motif. Taking this into consideration, IZTZ-1 (45—Figure 11) was developed and studied. It showed, in vitro, strong binding to c-MYC G4 (ΔTm = 15 °C) and inhibitory activity regarding melanoma cell growth (IC50 = 2.2 µM). Furthermore, a dual luciferase reporter assay and a flow cytometry assay showed that 45 could downregulate c-MYC transcription and expression and induce apoptosis. Moreover, it also inhibited tumor growth in a breast cancer xenograft mouse model [115].
The indolizinone 46 in Figure 11 was shown via fluorescence titration to selectively recognize MYC G4 (Ka = 9.9 × 105). This compound can also downregulate the transcription of c-MYC in two different cancer cell lines with different translocation break points within the c-MYC (human non-small cell line A549 and human laryngeal epithelial cell line—Hep2) [116]. The 7-azaindole-1-carboxamide derivative 47 (Figure 11) was investigated as a dual G4 binder/PARP inhibitor. The study showed that the compound stacks on the G4-tetrad trough π-π interactions, forming a 2:1 ligand/G4 complex with Ka = 106.1 M−1 [117]. Moreover, this compound also presents antiproliferative activity in HCC1937 cell lines, with an IC50 of 19.4 µM. In an in vivo assay, Compound 47 reduced the tumor volume by 35% in a MX1 cell line through the administration of 100 mg/kg every 2 days for 2 weeks [118].
Compound D089 (48—Figure 11) is a benzofuran derivative discovered through an investigation using a focused library of “drug-like” small molecules binding c-MYC G4 sequences [119]. It has a preference to bind to c-MYC G4 over other G4s in oncogenes promoters and duplex DNA, and it binds through a reversible interaction, as observed using a sensorgram. Furthermore, this compound showed better antiproliferative activity against myeloma cells (IC50 = 5.8 µM) than the well-studied G4 binder BRACO-19 (IC50 = 15.3 µM) [120]. Another study discovered DC-34 (9—Figure 11), a benzofuran derivative very similar to 48 that can also selectively inhibit c-MYC transcription by binding to the G4 in its promoter. It exhibited that a ΔTm of 7.5 °C forms a 2:1 complex, stacking over two guanines at each terminal G-tetrad plane, with reconfigured segments capping the ligand, as shown in Figure 12. This compound presents better antiproliferative activity against myeloma cells (IC50 of 3.4 µM) than Analog 48 [121].
Figure 12.
Schematic representation of Compound 49 (colored sticks) in a 2:1 complex with a c-MYC G4 structure (PDB ID: 5W77).
4.5. Flexible G4 Ligands
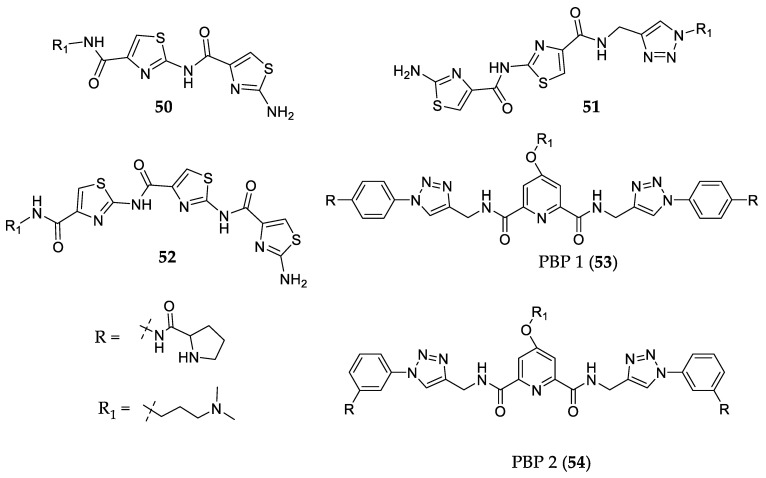
Thiazoles are present in several anticancer compounds [122] such as bleomycin and tiazofurin [123]. The thiazole moiety can be used in polyamide compounds as shown by Dutta et al. [122]. These researchers synthesized three thiazole polyamides (50, 51, and 52—Figure 13), and the latter compound (52) was shown to be more selective to c-MYC G4 than to other gene promoter G4s and duplex DNA, binding strongly when compared to the other two polyamide derivatives of 50 and 51. Compound 53 also showed the best antiproliferative activity against cancer cells compared to Compound 50 (IC50 values of 3.8 µM and 17.6 µM, respectively), but both showed insignificant cytotoxicity for non-malignant cells (IC50 > 50 µM).
Figure 13.
Structure of flexible c-MYC G4 ligands 50–54.
Debnath et al. [124] developed two peptidomimetic congeners that were able to bind to and stabilize G4 structures. A FRET melting assay showed that PBP1 (53) and PBP2 (54) (Figure 13) have high selectivity for c-MYC G4 over duplex DNA and a thiazole orange displacement assay demonstrated that 53 and 54 have good affinities to different G4s, with EC50 values regarding c-MYC G4 of 8.5 µM and 1.3 µM, respectively. They also investigated the growth-inhibiting activities of compounds using an MTT assay. Compound 53 showed higher values of IC50 than 54 (17.9 and 3.3 µM, respectively). Afterwards, other authors also studied these compounds and noticed that 53 presented a higher dissociation constant (11.2 µM) than 54 (1.42 µM) via isothermal titration calorimetry studies, indicating therefore that 54 is a better binder of c-MYC G4. These authors also analyzed the preference of binding to c-MYC G4 and to BCL2 G4, concluding that 54 is selective toward c-MYC G4 whereas 53 does not show a significant preference. Both compounds presented potent cytotoxic activity in MCF-7 breast cancer cells (IC50 = 3.8 and 7.1 µM), moderate cytotoxicity for other cancer cell lines, and low cytotoxicity for normal cells [125].
4.6. Metal Complexes
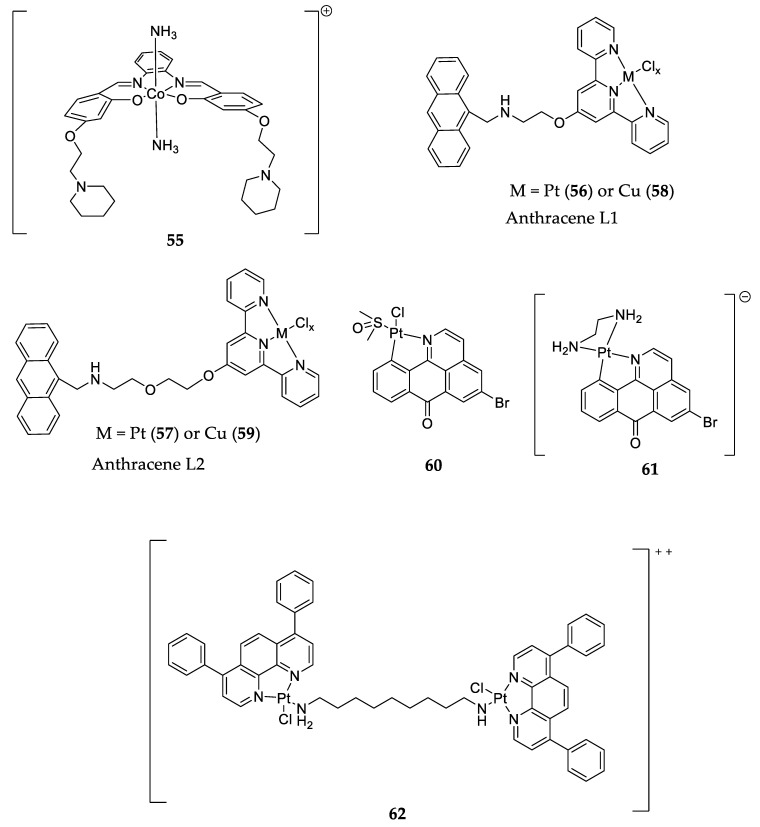
At the beginning of this century, the first G4 binders based on square planar metal complexes were reported. Since then, hundreds of new metal complex derivatives binding to G4 have been identified. Square-planar complexes with Ni II, Cu II, Co III, and Pt II are selective toward G4 over duplex DNA [24,126,127]. Co III compounds with NH3 as the coordinated ligand (55—Figure 14) have a higher affinity to c-MYC G4 than other complexes. The proposed binding mechanism considers that the NH3 groups establish hydrogen bond interactions with guanine’s oxygen atoms and that the salphen ligand displays π-π stacking interactions with the G-tetrad [126].
Figure 14.
Structure of metal complexes able to bind to and stabilize c-MYC G4.
The complexes of Cu II and Pt II anthracene-containing terpyridine ligands (PtL1,2—56 and 57; and CuL1,2—58 and 59; Figure 14) are selective toward G4 over duplex DNA. In one study, the Pt complexes showed better G4 stabilization values in a FRET melting assay when compared to the other complexes or the ligands without complexation with metals. Furthermore, Pt complexes presented a higher affinity to c-MYC G4 than the Cu complexes. The authors also concluded that the compounds with bigger linkers (L2—57 and 59) had more affinity to bind to G4 than the smaller ones (56 and 58), as can be seen by the ΔTm values in Table 1 [128].
Table 1.
Summary of best c-MYC G4 ligands’ characteristics.
| Binding to c-MYC G4 | Anticancer Activity | |||||
|---|---|---|---|---|---|---|
| Group | N° | Strength (Kd, Ka, ΔTm) |
Preference for c-MYC G4 (Yes/No) |
In Vitro | In Vivo | Ref. |
| Macrocycles | 1 | No |
|
|
[55,56] | |
| 3 | ΔTm = 17.2 °C | No |
|
[58,59] | ||
| Ligands with four or more fused aromatic rings | 4 | Yes |
|
[63] | ||
| 5 | ΔTm = 26.6 °C | __ |
|
|
[61] | |
| 6 7 |
ΔTm = 7 °C ΔTm = 17 °C |
No |
|
[64] | ||
| 8 9 10 |
ΔTm = 22.0 °C ΔTm = 16.6 °C ΔTm = 13.7 °C |
Yes |
|
|
[65] | |
| 12 | ΔTm = 21 °C | __ |
|
[69] | ||
| 13 | ΔTm > 15 °C | __ |
|
Reached Phase II clinical trial. | [73,76] | |
| 14 | ΔTm > 15 °C | No |
|
|
[81,131] | |
| 15 |
ΔTm = 4.4 °C | __ |
|
|
[82] | |
| 16 | - | No |
|
|
[83,85,86] | |
| 17 18 |
ΔTm ~ 10 °C | Yes |
|
[87] | ||
| 19 | ΔTm = 16.4 °C | No |
|
|
[88,89] | |
| 20 | ΔTm > 6 °C | No |
|
|
[91] | |
| 21 | ΔTm = 29 °C | Yes |
|
[92] | ||
| Ligands with three fused aromatic rings | 22 | Yes |
|
- | [95] | |
| 24 | ΔTm = 12.8 °C | No |
|
[96] | ||
| 26 | ΔTm = 20 °C | Yes |
|
|
[97] | |
| 27 28 |
ΔTm = 4 °C ΔTm = 1.9 °C |
Yes |
|
[98,99] | ||
| 29 | ΔTm= 15.8 °C | Yes |
|
[100] | ||
| 31 | Kd = 0.17 µM | __ |
|
[102] | ||
| 34 | ΔTm = 23.4 °C | __ |
|
[103] | ||
| 37 | Kd = 36 nM | Yes |
|
[104] | ||
| 38 | ΔTm = 15.0 °C | __ |
|
[107] | ||
| Ligands with two fused aromatic rings | 39 | ΔTm = 23.7 °C | __ |
|
|
[108] |
| 40 | ΔTm = 9 °C | __ |
|
[109] | ||
| 41 42 |
ΔTm = 12.1 °C ΔTm = 12.9 °C |
__ |
|
|
[110] | |
| 43 | Binding affinity to NM23-H2 protein (KD = 3.1 μM) | No |
|
|
[111] | |
| 44 | KD = 1.3 μM | Yes |
|
|
[114] | |
| 45 | ΔTm = 15 °C | Yes |
|
|
[115] | |
| 46 | Ka = 9.9 × 105 M−1 | Yes |
|
[116] | ||
| 47 | Ka = 106.1 M−1 | No |
|
|
[117,118] | |
| 48 | Yes |
|
[119,120] | |||
| 49 | ΔTm = 7.5 °C | Yes |
|
[121] | ||
| Flexible ligands | 52 | ΔTm = 22 °C | Yes |
|
[122] | |
| 53 54 |
ΔTm = 15.0 °C ΔTm = 6.0 °C |
No |
|
- | [124,125] | |
| Metal complexes | 56 57 58 59 |
ΔTm = 10.2 °C ΔTm = 15.6 °C ΔTm = 2.0 °C ΔTm = 7.3 °C |
No |
|
[128] | |
| 60 61 |
__ |
|
[129] | |||
Other platinum complexes [Pt(L)(DMSO)Cl] (60) and [Pt(L)(pn)]Cl (61)) with 5-bromo-oxoisoaporphine as ligands (Figure 14) were studied for their ability to downregulate c-MYC expression through binding to its promoter G4. Complex 62 inhibited 99.9% of c-MYC protein levels and it also showed better selectivity to c-MYC in an FID assay than Complex 60. Their cytotoxicity was evaluated via an MTT assay, and it was reported that the IC50 values varied between 5.1 and 31.1 µM for different cancer cell lines, but they showed very low cytotoxicity for non-malignant cells [129].
Dinuclear platinum complexes like 62 (Figure 14) were also shown to be able to stabilize G4 structures and bind more selectively to c-MYC G4 than to duplex DNA, with a ΔTm of 8.5 °C in a FRET melting assay. These complexes were able to interact with G-tetrads via π stacking and cross-linking with purines in G4 through their alkyl chains [130].
5. Conclusions
The c-MYC proto-oncogene encodes for a transcription factor whereby expression is deregulated in many cancer types. For this reason, several strategies have been used in the past years to, directly or indirectly, target c-MYC but, so far, these attempts have been unsuccessful. Another approach being studied aims to target the expression of c-MYC in cancer cells via the stabilization of the G4 structures present in its promoter region.
Table 1 summarizes the major achievements made so far. It can be concluded that several small molecules with different chemotypes and shapes have been investigated, and these compounds have a core with differently organized aromatic rings to better interact with G-quartets and stabilize the G4. Interestingly, all of the molecules reaching clinical trials (13, 14, and 16) have four fused aromatic rings and are potent G4 stabilizers, but they are not selective toward c-MYC G4. On the other hand, some c-MYC G4 ligands with two fused aromatic rings and consequently being more flexible were shown to have a preference of binding to this G4, and also showed good anticancer activity in vitro and in vivo (44 and 45). Moreover, it is among this class of compounds that we can find G4 ligands that are believed to act in cancer cells by targeting the G4-binding proteins or the interaction between the c-MYC G4 and the protein, such as the quinazolines 42 and 43. In fact, research into the G4-protein interactome is emerging and it is believed that targeting these interactions with small molecules may be the solution to achieve the desired selectivity toward a certain G4 structure.
Overall, this review has put into evidence all of the efforts that have been made to target c-MYC G4 and the new approaches being explored, and has highlighted several lead compounds that can be used in further investigations.
Abbreviations
AURKA: Aurora Kinase A; BRD4: Bromodomain-containing protein 4; CDK: Cyclin-dependent kinase 7; CPEB: cytoplasmic polyadenylation element binding; GLS: glutaminase; mTOR: mammalian target of rapamycin; PLK1: Serine/threonine protein kinase; USP: ubiquitin-specific peptidase.
Author Contributions
Conceptualization, A.P.; writing—original draft preparation, B.B., I.M.A., and R.E.; writing—review and editing, E.M. and A.P. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
Israa M. Aljnadi acknowledges the Global Platform for Syrian Students and the University of Lisbon for a doctoral fellowship. All authors acknowledge the financial support from Fundação para a Ciência e Tecnologia (FCT, Portugal) under projects UIDB/04138/2020, UIDP/04138/2020 (iMed.ULisboa), and 2022.06099.PTDC.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.World Health Organization Estimated Number of New Cases in 2020. [(accessed on 7 October 2022)]. Available online: https://gco.iarc.fr/today/online-analysis-table?v=2020&mode=cancer&mode_population=continents&population=900&populations=900&key=asr&sex=0&cancer=39&type=0&statistic=5&prevalence=0&population_group=0&ages_group%5B%5D=0&ages_group%5B%5D=17&group_cancer=1&include_nmsc=0&include_nmsc_other=1.
- 2.Yahya E.B., Alqadhi A.M. Recent Trends in Cancer Therapy: A Review on the Current State of Gene Delivery. Life Sci. 2021;269:119087. doi: 10.1016/j.lfs.2021.119087. [DOI] [PubMed] [Google Scholar]
- 3.Mansoori B., Mohammadi A., Davudian S., Shirjang S., Baradaran B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017;7:339–348. doi: 10.15171/apb.2017.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization Cancer. [(accessed on 7 October 2022)]. Available online: https://www.who.int/health-topics/cancer#tab=tab_1.
- 5.Ciriello G., Miller M.L., Aksoy B.A., Senbabaoglu Y., Schultz N., Sander C. Emerging Landscape of Oncogenic Signatures across Human Cancers. Nat. Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalla-Favera R., Bregni M., Erikson J., Patterson D., Gallo R.C., Croce C.M. Human C-Myc Onc Gene Is Located on the Region of Chromosome 8 That Is Translocated in Burkitt Lymphoma Cells. Proc. Natl. Acad. Sci. USA. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wahlström T., Henriksson M.A. Impact of MYC in Regulation of Tumor Cell Metabolism. Biochim. Biophys. Acta. 2015;1849:563–569. doi: 10.1016/j.bbagrm.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Rohban S., Campaner S. Myc Induced Replicative Stress Response: How to Cope with It and Exploit It. Biochim. Biophys. Acta. 2015;1849:517–524. doi: 10.1016/j.bbagrm.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Bretones G., Delgado M.D., León J. Myc and Cell Cycle Control. Biochim. Biophys. Acta. 2015;1849:506–516. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 10.Tu W.B., Helander S., Pilstål R., Hickman K.A., Lourenco C., Jurisica I., Raught B., Wallner B., Sunnerhagen M., Penn L.Z. Myc and Its Interactors Take Shape. Biochim. Biophys. Acta. 2015;1849:469–483. doi: 10.1016/j.bbagrm.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Chen B.-J., Wu Y.-L., Tanaka Y., Zhang W. Small Molecules Targeting C-Myc Oncogene: Promising Anti-Cancer Therapeutics. Int. J. Biol. Sci. 2014;10:1084–1096. doi: 10.7150/ijbs.10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf E., Eilers M. Targeting MYC Proteins for Tumor Therapy. Annu. Rev. Cancer Biol. 2020;4:61–75. doi: 10.1146/annurev-cancerbio-030518-055826. [DOI] [Google Scholar]
- 13.Chen H., Liu H., Qing G. Targeting Oncogenic Myc as a Strategy for Cancer Treatment. Signal Transduct. Target. Ther. 2018;3 doi: 10.1038/s41392-018-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duffy M.J., O’Grady S., Tang M., Crown J. MYC as a Target for Cancer Treatment. Cancer Treat. Rev. 2021;94:102154. doi: 10.1016/j.ctrv.2021.102154. [DOI] [PubMed] [Google Scholar]
- 15.Nakanishi C., Seimiya H. G-Quadruplex in Cancer Biology and Drug Discovery. Biochem. Biophys. Res. Commun. 2020;531:45–50. doi: 10.1016/j.bbrc.2020.03.178. [DOI] [PubMed] [Google Scholar]
- 16.Robinson J., Raguseo F., Nuccio S.P., Liano D., Di Antonio M. DNA G-Quadruplex Structures: More than Simple Roadblocks to Transcription? Nucleic Acids Res. 2021;49:8419–8431. doi: 10.1093/nar/gkab609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S. Quadruplex DNA: Sequence, Topology and Structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spiegel J., Adhikari S., Balasubramanian S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020;2:123–136. doi: 10.1016/j.trechm.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosiol N., Juranek S., Brossart P., Heine A., Paeschke K. G-Quadruplexes: A Promising Target for Cancer Therapy. Mol. Cancer. 2021;20:40. doi: 10.1186/s12943-021-01328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan J., Lan L. The DNA Secondary Structures at Telomeres and Genome Instability. Cell Biosci. 2020;10:47. doi: 10.1186/s13578-020-00409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Y., Iida K., Nagasawa K. Topologies of G-Quadruplex: Biological Functions and Regulation by Ligands. Biochem. Biophys. Res. Commun. 2020;531:3–17. doi: 10.1016/j.bbrc.2019.12.103. [DOI] [PubMed] [Google Scholar]
- 22.Mendes E., Aljnadi I.M., Bahls B., Victor B.L., Paulo A. Major Achievements in the Design of Quadruplex-Interactive Small Molecules. Pharmaceuticals. 2022;15:300. doi: 10.3390/ph15030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendes E., Bahls B., Aljnadi I.M., Paulo A. Indoloquinolines as Scaffolds for the Design of Potent G-Quadruplex Ligands. Bioorg. Med. Chem. Lett. 2022;72:128862. doi: 10.1016/j.bmcl.2022.128862. [DOI] [PubMed] [Google Scholar]
- 24.Francisco A.P., Paulo A. Oncogene Expression Modulation in Cancer Cell Lines by DNA G-Quadruplex-Interactive Small Molecules. Curr. Med. Chem. 2017;24:4873–4904. doi: 10.2174/0929867323666160829145055. [DOI] [PubMed] [Google Scholar]
- 25.Awadasseid A., Ma X., Wu Y., Zhang W. G-Quadruplex Stabilization via Small-Molecules as a Potential Anti-Cancer Strategy. Biomed. Pharmacother. 2021;139:111550. doi: 10.1016/j.biopha.2021.111550. [DOI] [PubMed] [Google Scholar]
- 26.Ruggiero E., Zanin I., Terreri M., Richter S.N. G-Quadruplex Targeting in the Fight against Viruses: An Update. Int. J. Mol. Sci. 2021;22:10984. doi: 10.3390/ijms222010984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C., Zhang J., Yin J., Gan Y., Xu S., Gu Y., Huang W. Alternative Approaches to Target Myc for Cancer Treatment. Signal Transduct. Target. Ther. 2021;6:117. doi: 10.1038/s41392-021-00500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burton R.A., Mattila S., Taparowsky E.J., Post C.B. B-Myc: N-Terminal Recognition of Myc Binding Proteins. Biochemistry. 2006;45:9857–9865. doi: 10.1021/bi060379n. [DOI] [PubMed] [Google Scholar]
- 29.Dhanasekaran R., Deutzmann A., Mahauad-Fernandez W.D., Hansen A.S., Gouw A.M., Felsher D.W. The MYC Oncogene - the Grand Orchestrator of Cancer Growth and Immune Evasion. Nat. Rev. Clin. Oncol. 2022;19:23–36. doi: 10.1038/s41571-021-00549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryan K.M., Birnie G.D. Myc Oncogenes: The Enigmatic Family. Biochem. J. 1996;314:713–721. doi: 10.1042/bj3140713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noguchi K., Yamana H., Kitanaka C., Mochizuki T., Kokubu A., Kuchino Y. Differential Role of the JNK and P38 MAPK Pathway in C-Myc- and s-Myc-Mediated Apoptosis. Biochem. Biophys. Res. Commun. 2000;267:221–227. doi: 10.1006/bbrc.1999.1952. [DOI] [PubMed] [Google Scholar]
- 32.González V., Hurley L.H. The C-MYC NHE III(1): Function and Regulation. Annu. Rev. Pharmacol. Toxicol. 2010;50:111–129. doi: 10.1146/annurev.pharmtox.48.113006.094649. [DOI] [PubMed] [Google Scholar]
- 33.Wang W., Hu S., Gu Y., Yan Y., Stovall D.B., Li D., Sui G. Human MYC G-Quadruplex: From Discovery to a Cancer Therapeutic Target. Biochim. Biophys. Acta Rev. Cancer. 2020;1874:188410. doi: 10.1016/j.bbcan.2020.188410. [DOI] [PubMed] [Google Scholar]
- 34.Islam M.A., Thomas S.D., Murty V.V., Sedoris K.J., Miller D.M. C-Myc Quadruplex-Forming Sequence Pu-27 Induces Extensive Damage in Both Telomeric and Nontelomeric Regions of DNA. J. Biol. Chem. 2014;289:8521–8531. doi: 10.1074/jbc.M113.505073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simonsson T., Pecinka P., Kubista M. DNA Tetraplex Formation in the Control Region of C-Myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.You H., Wu J., Shao F., Yan J. Stability and Kinetics of C-MYC Promoter G-Quadruplexes Studied by Single-Molecule Manipulation. J. Am. Chem. Soc. 2015;137:2424–2427. doi: 10.1021/ja511680u. [DOI] [PubMed] [Google Scholar]
- 37.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct Evidence for a G-Quadruplex in a Promoter Region and Its Targeting with a Small Molecule to Repress c-MYC Transcription. Proc. Natl. Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phan A.T., Modi Y.S., Patel D.J. Propeller-Type Parallel-Stranded G-Quadruplexes in the Human c-Myc Promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brázda V., Hároníková L., Liao J.C.C., Fojta M. DNA and RNA Quadruplex-Binding Proteins. Int. J. Mol. Sci. 2014;15:17493–17517. doi: 10.3390/ijms151017493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González V., Guo K., Hurley L., Sun D. Identification and Characterization of Nucleolin as a C-Myc G-Quadruplex-Binding Protein. J. Biol. Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Postel E.H., Berberich S.J., Flint S.J., Ferrone C.A. Human C-Myc Transcription Factor PuF Identified as Nm23-H2 Nucleoside Diphosphate Kinase, a Candidate Suppressor of Tumor Metastasis. Science. 1993;261:478–480. doi: 10.1126/science.8392752. [DOI] [PubMed] [Google Scholar]
- 42.Fekete A., Kenesi E., Hunyadi-Gulyas E., Durgo H., Berko B., Dunai Z.A., Bauer P.I. The Guanine-Quadruplex Structure in the Human c-Myc Gene’s Promoter Is Converted into B-DNA Form by the Human Poly(ADP-Ribose)Polymerase-1. PLoS ONE. 2012;7:e42690. doi: 10.1371/journal.pone.0042690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cashman D.J., Buscaglia R., Freyer M.W., Dettler J., Hurley L.H., Lewis E.A. Molecular Modeling and Biophysical Analysis of the C-MYC NHE-III1 Silencer Element. J. Mol. Model. 2008;14:93–101. doi: 10.1007/s00894-007-0254-z. [DOI] [PubMed] [Google Scholar]
- 44.Ohnmacht S.A., Neidle S. Small-Molecule Quadruplex-Targeted Drug Discovery. Bioorg. Med. Chem. Lett. 2014;24:2602–2612. doi: 10.1016/j.bmcl.2014.04.029. [DOI] [PubMed] [Google Scholar]
- 45.Neidle S. Structured Waters Mediate Small Molecule Binding to G-Quadruplex Nucleic Acids. Pharmaceuticals. 2021;15:7. doi: 10.3390/ph15010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paulo A., Castillo C.C., Neidle S. Comprehensive Medicinal Chemistry III. Elsevier; Amsterdam, The Netherlands: 2017. Targeting Promoter Quadruplex Nucleic Acids for Cancer Therapy; pp. 308–340. [Google Scholar]
- 47.Badran H.M., Eid K.M., Ammar H.Y. DFT and TD-DFT Studies of Halogens Adsorption on Cobalt-Doped Porphyrin: Effect of the External Electric Field. Results Phys. 2021;23:103964. doi: 10.1016/j.rinp.2021.103964. [DOI] [Google Scholar]
- 48.Chou J.-H., Kosal M.E., Nalwa H.S., Rakow N.A., Suslick K.S. Applications of Porphyrins and Metalloporphyrins to Materials Chemistry. ChemInform. 2003;34 doi: 10.1002/chin.200317263. [DOI] [Google Scholar]
- 49.Auwärter W., Écija D., Klappenberger F., Barth J.V. Porphyrins at Interfaces. Nat. Chem. 2015;7:105–120. doi: 10.1038/nchem.2159. [DOI] [PubMed] [Google Scholar]
- 50.Stojiljkovic I., Evavold B.D., Kumar V. Antimicrobial Properties of Porphyrins. Expert Opin. Investig. Drugs. 2001;10:309–320. doi: 10.1517/13543784.10.2.309. [DOI] [PubMed] [Google Scholar]
- 51.Huang H., Song W., Rieffel J., Lovell J.F. Emerging Applications of Porphyrins in Photomedicine. Front. Phys. 2015;3:23. doi: 10.3389/fphy.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phan A.T., Kuryavyi V., Gaw H.Y., Patel D.J. Small-Molecule Interaction with a Five-Guanine-Tract G-Quadruplex Structure from the Human MYC Promoter. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nielsen M.C., Ulven T. Macrocyclic G-Quadruplex Ligands. Curr. Med. Chem. 2010;17:3438–3448. doi: 10.2174/092986710793176320. [DOI] [PubMed] [Google Scholar]
- 54.Han F.X., Wheelhouse R.T., Hurley L.H. Interactions of TMPyP4 and TMPyP2 with Quadruplex DNA. Structural Basis for the Differential Effects on Telomerase Inhibition. J. Am. Chem. Soc. 1999;121:3561–3570. doi: 10.1021/ja984153m. [DOI] [Google Scholar]
- 55.Grand C.L., Han H., Muñoz R.M., Weitman S., Von Hoff D.D., Hurley L.H., Bearss D.J. The Cationic Porphyrin TMPyP4 Down-Regulates c-MYC and Human Telomerase Reverse Transcriptase Expression and Inhibits Tumor Growth in Vivo. Mol. Cancer Ther. 2002;1:565–573. [PubMed] [Google Scholar]
- 56.Mikami-Terao Y., Akiyama M., Yuza Y., Yanagisawa T., Yamada O., Yamada H. Antitumor Activity of G-Quadruplex-Interactive Agent TMPyP4 in K562 Leukemic Cells. Cancer Lett. 2008;261:226–234. doi: 10.1016/j.canlet.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 57.Ruan T.L., Davis S.J., Powell B.M., Harbeck C.P., Habdas J., Habdas P., Yatsunyk L.A. Lowering the Overall Charge on TMPyP4 Improves Its Selectivity for G-Quadruplex DNA. Biochimie. 2017;132:121–130. doi: 10.1016/j.biochi.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 58.Carvalho J., Quintela T., Gueddouda N.M., Bourdoncle A., Mergny J.-L., Salgado G.F., Queiroz J.A., Cruz C. Phenanthroline Polyazamacrocycles as G-Quadruplex DNA Binders. Org. Biomol. Chem. 2018;16:2776–2786. doi: 10.1039/C8OB00247A. [DOI] [PubMed] [Google Scholar]
- 59.Cruz C., Cairrao E., Silvestre S., Breitenfeld L., Almeida P., Queiroz J.A. Targeting of Mitochondria-Endoplasmic Reticulum by Fluorescent Macrocyclic Compounds. PLoS ONE. 2011;6:e27078. doi: 10.1371/journal.pone.0027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Riechert-Krause F., Weisz K. Indoloquinolines as DNA Binding Ligands. Heterocycl. Comm. 2013;19:145–166. doi: 10.1515/hc-2013-0034. [DOI] [Google Scholar]
- 61.Liu H.-Y., Chen A.-C., Yin Q.-K., Li Z., Huang S.-M., Du G., He J.-H., Zan L.-P., Wang S.-K., Xu Y.-H., et al. New Disubstituted Quindoline Derivatives Inhibiting Burkitt’s Lymphoma Cell Proliferation by Impeding c-MYC Transcription. J. Med. Chem. 2017;60:5438–5454. doi: 10.1021/acs.jmedchem.7b00099. [DOI] [PubMed] [Google Scholar]
- 62.Vianney Y.M., Weisz K. Indoloquinoline Ligands Favor Intercalation at Quadruplex-Duplex Interfaces. Chemistry. 2022;28:e202103718. doi: 10.1002/chem.202103718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ou T.-M., Lin J., Lu Y.-J., Hou J.-Q., Tan J.-H., Chen S.-H., Li Z., Li Y.-P., Li D., Gu L.-Q., et al. Inhibition of Cell Proliferation by Quindoline Derivative (SYUIQ-05) through Its Preferential Interaction with c-Myc Promoter G-Quadruplex. J. Med. Chem. 2011;54:5671–5679. doi: 10.1021/jm200062u. [DOI] [PubMed] [Google Scholar]
- 64.Boddupally P.V.L., Hahn S., Beman C., De B., Brooks T.A., Gokhale V., Hurley L.H. Anticancer Activity and Cellular Repression of C-MYC by the G-Quadruplex-Stabilizing 11-Piperazinylquindoline Is Not Dependent on Direct Targeting of the G-Quadruplex in the c-MYC Promoter. J. Med. Chem. 2012;55:6076–6086. doi: 10.1021/jm300282c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng D.-Y., Kuang G.-T., Wang S.-K., Peng W., Lin S.-L., Zhang Q., Su X.-X., Hu M.-H., Wang H., Tan J.-H., et al. Discovery of Novel 11-Triazole Substituted Benzofuro[3,2-b]Quinolone Derivatives as c-Myc G-Quadruplex Specific Stabilizers via Click Chemistry. J. Med. Chem. 2017;60:5407–5423. doi: 10.1021/acs.jmedchem.7b00016. [DOI] [PubMed] [Google Scholar]
- 66.Funke A., Dickerhoff J., Weisz K. Towards the Development of Structure-Selective G-Quadruplex-Binding Indolo[3,2-b]Quinolines. Chemistry. 2016;22:3170–3181. doi: 10.1002/chem.201504416. [DOI] [PubMed] [Google Scholar]
- 67.Funke A., Karg B., Dickerhoff J., Balke D., Müller S., Weisz K. Ligand-Induced Dimerization of a Truncated Parallel MYC G-Quadruplex. Chembiochem. 2018;19:505–512. doi: 10.1002/cbic.201700593. [DOI] [PubMed] [Google Scholar]
- 68.Montoya J.J., Turnidge M.A., Wai D.H., Patel A.R., Lee D.W., Gokhale V., Hurley L.H., Arceci R.J., Wetmore C., Azorsa D.O. In Vitro Activity of a G-Quadruplex-Stabilizing Small Molecule That Synergizes with Navitoclax to Induce Cytotoxicity in Acute Myeloid Leukemia Cells. BMC Cancer. 2019;19:1251. doi: 10.1186/s12885-019-6464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brown R.V., Danford F.L., Gokhale V., Hurley L.H., Brooks T.A. Demonstration That Drug-Targeted down-Regulation of MYC in Non-Hodgkins Lymphoma Is Directly Mediated through the Promoter G-Quadruplex. J. Biol. Chem. 2011;286:41018–41027. doi: 10.1074/jbc.M111.274720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mohamad Anuar N.N., Nor Hisam N.S., Liew S.L., Ugusman A. Clinical Review: Navitoclax as a pro-Apoptotic and Anti-Fibrotic Agent. Front. Pharmacol. 2020;11:564108. doi: 10.3389/fphar.2020.564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaiser C.E., Gokhale V., Yang D., Hurley L.H. Gaining Insights into the Small Molecule Targeting of the G-Quadruplex in the c-MYC Promoter Using NMR and an Allele-Specific Transcriptional Assay. Top. Curr. Chem. 2013;330:1–21. doi: 10.1007/128_2012_333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Drlica K. Mechanism of Fluoroquinolone Action. Curr. Opin. Microbiol. 1999;2:504–508. doi: 10.1016/S1369-5274(99)00008-9. [DOI] [PubMed] [Google Scholar]
- 73.Xu H., Di Antonio M., McKinney S., Mathew V., Ho B., O’Neil N.J., Santos N.D., Silvester J., Wei V., Garcia J., et al. CX-5461 Is a DNA G-Quadruplex Stabilizer with Selective Lethality in BRCA1/2 Deficient Tumours. Nat. Commun. 2017;8:14432. doi: 10.1038/ncomms14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Teng F.-Y., Jiang Z.-Z., Guo M., Tan X.-Z., Chen F., Xi X.-G., Xu Y. G-Quadruplex DNA: A Novel Target for Drug Design. Cell. Mol. Life Sci. 2021;78:6557–6583. doi: 10.1007/s00018-021-03921-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Knox J.J., Oza A.M., Provencher D.M., Soong J., McCormick D., Chen J., Chen J., Chang A. Phase 1b Expansion Study of CX-5461 in Patients with Solid Tumors and BRCA2 and/or PALB2 Mutation. J. Clin. Oncol. 2022;40:TPS622. doi: 10.1200/JCO.2022.40.4_suppl.TPS622. [DOI] [Google Scholar]
- 76.Pharmaceuticals C. Quarfloxin in Patients with Low to Intermediate Grade Neuroendocrine Carcinoma. [(accessed on 10 July 2022)]; Available online: https://clinicaltrials.gov/ct2/show/NCT00780663.
- 77.Singh M., Gupta R., Comez L., Paciaroni A., Rani R., Kumar V. BCL2 G Quadruplex-Binding Small Molecules: Current Status and Prospects for the Development of next-Generation Anticancer Therapeutics. Drug Discov. Today. 2022;27:2551–2561. doi: 10.1016/j.drudis.2022.06.002. [DOI] [PubMed] [Google Scholar]
- 78.Drygin D., Siddiqui-Jain A., O’Brien S., Schwaebe M., Lin A., Bliesath J., Ho C.B., Proffitt C., Trent K., Whitten J.P., et al. Anticancer Activity of CX-3543: A Direct Inhibitor of RRNA Biogenesis. Cancer Res. 2009;69:7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- 79.Hilton J., Gelmon K., Bedard P.L., Tu D., Xu H., Tinker A.V., Goodwin R., Laurie S.A., Jonker D., Hansen A.R., et al. Results of the Phase I CCTG IND.231 Trial of CX-5461 in Patients with Advanced Solid Tumors Enriched for DNA-Repair Deficiencies. Nat. Commun. 2022;13:3607. doi: 10.1038/s41467-022-31199-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu H., Hurley L.H. A First-in-Class Clinical G-Quadruplex-Targeting Drug. The Bench-to-Bedside Translation of the Fluoroquinolone QQ58 to CX-5461 (Pidnarulex) Bioorg. Med. Chem. Lett. 2022;77:129016. doi: 10.1016/j.bmcl.2022.129016. [DOI] [PubMed] [Google Scholar]
- 81.Bruno P.M., Lu M., Dennis K.A., Inam H., Moore C.J., Sheehe J., Elledge S.J., Hemann M.T., Pritchard J.R. The Primary Mechanism of Cytotoxicity of the Chemotherapeutic Agent CX-5461 Is Topoisomerase II Poisoning. Proc. Natl. Acad. Sci. USA. 2020;117:4053–4060. doi: 10.1073/pnas.1921649117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu Q., Song Y., Liu R., Wang R., Mei W., Chen W., Yang H., Wang X. Synthesis, Docking Studies and Antitumor Activity of Phenanthroimidazole Derivatives as Promising c-Myc G-Quadruplex DNA Stabilizers. Bioorg. Chem. 2020;102:104074. doi: 10.1016/j.bioorg.2020.104074. [DOI] [PubMed] [Google Scholar]
- 83.Aptose Biosciences Inc. A Study of APTO-253 in Patients with Relapsed or Refractory AML or MDS. [(accessed on 10 July 2022)]; Available online: https://clinicaltrials.gov/ct2/show/NCT02267863.
- 84.Ohanian M., Arellano M.L., Levy M.Y., O’Dwyer K., Babiker H., Mahadevan D., Zhang H., Rastgoo N., Jin Y., Marango J., et al. A Phase 1a/b Dose Escalation Study of the MYC Repressor Apto-253 in Patients with Relapsed or Refractory AML or High-Risk MDS. Blood. 2021;138:3411. doi: 10.1182/blood-2021-150049. [DOI] [Google Scholar]
- 85.Cercek A., Wheler J., Murray P.E., Zhou S., Saltz L. Phase 1 Study of APTO-253 HCl, an Inducer of KLF4, in Patients with Advanced or Metastatic Solid Tumors. Investig. New Drugs. 2015;33:1086–1092. doi: 10.1007/s10637-015-0273-z. [DOI] [PubMed] [Google Scholar]
- 86.Local A., Zhang H., Benbatoul K.D., Folger P., Sheng X., Tsai C.-Y., Howell S.B., Rice W.G. APTO-253 Stabilizes G-Quadruplex DNA, Inhibits MYC Expression, and Induces DNA Damage in Acute Myeloid Leukemia Cells. Mol. Cancer Ther. 2018;17:1177–1186. doi: 10.1158/1535-7163.MCT-17-1209. [DOI] [PubMed] [Google Scholar]
- 87.Musso L., Mazzini S., Rossini A., Castagnoli L., Scaglioni L., Artali R., Di Nicola M., Zunino F., Dallavalle S. C-MYC G-Quadruplex Binding by the RNA Polymerase I Inhibitor BMH-21 and Analogues Revealed by a Combined NMR and Biochemical Approach. Biochim. Biophys. Acta Gen. Subj. 2018;1862:615–629. doi: 10.1016/j.bbagen.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 88.Micheli E., Altieri A., Cianni L., Cingolani C., Iachettini S., Bianco A., Leonetti C., Cacchione S., Biroccio A., Franceschin M., et al. Perylene and Coronene Derivatives Binding to G-Rich Promoter Oncogene Sequences Efficiently Reduce Their Expression in Cancer Cells. Biochimie. 2016;125:223–231. doi: 10.1016/j.biochi.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 89.Porru M., Zizza P., Franceschin M., Leonetti C., Biroccio A. EMICORON: A Multi-Targeting G4 Ligand with a Promising Preclinical Profile. Biochim. Biophys. Acta Gen. Subj. 2017;1861:1362–1370. doi: 10.1016/j.bbagen.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 90.Dickerhoff J., Brundridge N., McLuckey S.A., Yang D. Berberine Molecular Recognition of the Parallel MYC G-Quadruplex in Solution. J. Med. Chem. 2021;64:16205–16212. doi: 10.1021/acs.jmedchem.1c01508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wen L., Han Z., Li J., Du Y. C-MYC and HIF1α Promoter G-Quadruplexes Dependent Metabolic Regulation Mechanism of Berberine in Colon Cancer. J. Gastrointest. Oncol. 2022;13:1152–1168. doi: 10.21037/jgo-22-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ma Y., Ou T.-M., Hou J.-Q., Lu Y.-J., Tan J.-H., Gu L.-Q., Huang Z.-S. 9-N-Substituted Berberine Derivatives: Stabilization of G-Quadruplex DNA and down-Regulation of Oncogene c-Myc. Bioorg. Med. Chem. 2008;16:7582–7591. doi: 10.1016/j.bmc.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 93.Tsvetkov V.B., Varizhuk A.M., Lizunova S.A., Nikolenko T.A., Ivanov I.A., Severov V.V., Belyaev E.S., Shitikov E.A., Pozmogova G.E., Aralov A.V. Phenoxazine-Based Scaffold for Designing G4-Interacting Agents. Org. Biomol. Chem. 2020;18:6147–6154. doi: 10.1039/D0OB00983K. [DOI] [PubMed] [Google Scholar]
- 94.Tsvetkov V.B., Turaev A.V., Petrunina N.A., Melnik D.M., Khodarovich Y.M., Pozmogova G.E., Zatsepin T.S., Varizhuk A.M., Aralov A.V. Phenoxazine Pseudonucleotides in DNA I-Motifs Allow Precise Profiling of Small Molecule Binders by Fluorescence Monitoring. Analyst. 2021;146:4436–4440. doi: 10.1039/D1AN00660F. [DOI] [PubMed] [Google Scholar]
- 95.Kang H.-J., Park H.-J. In Silico Identification of Novel Ligands for G-Quadruplex in the c-MYC Promoter. J. Comput. Aided Mol. Des. 2015;29:339–348. doi: 10.1007/s10822-014-9826-z. [DOI] [PubMed] [Google Scholar]
- 96.Pelliccia S., Amato J., Capasso D., Di Gaetano S., Massarotti A., Piccolo M., Irace C., Tron G.C., Pagano B., Randazzo A., et al. Bio-Inspired Dual-Selective BCL-2/c-MYC G-Quadruplex Binders: Design, Synthesis, and Anticancer Activity of Drug-like Imidazo[2,1-i]Purine Derivatives. J. Med. Chem. 2020;63:2035–2050. doi: 10.1021/acs.jmedchem.9b00262. [DOI] [PubMed] [Google Scholar]
- 97.Hu M.-H., Wang Y.-Q., Yu Z.-Y., Hu L.-N., Ou T.-M., Chen S.-B., Huang Z.-S., Tan J.-H. Discovery of a New Four-Leaf Clover-like Ligand as a Potent c-MYC Transcription Inhibitor Specifically Targeting the Promoter G-Quadruplex. J. Med. Chem. 2018;61:2447–2459. doi: 10.1021/acs.jmedchem.7b01697. [DOI] [PubMed] [Google Scholar]
- 98.Mulliri S., Laaksonen A., Spanu P., Farris R., Farci M., Mingoia F., Roviello G.N., Mocci F. Spectroscopic and in Silico Studies on the Interaction of Substituted Pyrazolo[1,2-a]Benzo[1,2,3,4]Tetrazine-3-One Derivatives with c-Myc G4-DNA. Int. J. Mol. Sci. 2021;22:6028. doi: 10.3390/ijms22116028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mingoia F., Di Sano C., Di Blasi F., Fazzari M., Martorana A., Almerico A.M., Lauria A. Exploring the Anticancer Potential of Pyrazolo[1,2-a]Benzo[1,2,3,4]Tetrazin-3-One Derivatives: The Effect on Apoptosis Induction, Cell Cycle and Proliferation. Eur. J. Med. Chem. 2013;64:345–356. doi: 10.1016/j.ejmech.2013.03.046. [DOI] [PubMed] [Google Scholar]
- 100.Das T., Panda D., Saha P., Dash J. Small Molecule Driven Stabilization of Promoter G-Quadruplexes and Transcriptional Regulation of c-MYC. Bioconjug. Chem. 2018;29:2636–2645. doi: 10.1021/acs.bioconjchem.8b00338. [DOI] [PubMed] [Google Scholar]
- 101.Głuszyńska A., Juskowiak B., Kuta-Siejkowska M., Hoffmann M., Haider S. Carbazole Ligands as C-Myc G-Quadruplex Binders. Int. J. Biol. Macromol. 2018;114:479–490. doi: 10.1016/j.ijbiomac.2018.03.135. [DOI] [PubMed] [Google Scholar]
- 102.Panda D., Saha P., Das T., Dash J. Target Guided Synthesis Using DNA Nano-Templates for Selectively Assembling a G-Quadruplex Binding c-MYC Inhibitor. Nat. Commun. 2017;8:16103. doi: 10.1038/ncomms16103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jana S., Panda D., Saha P., Pantos G.D., Dash J. Dynamic Generation of G-Quadruplex DNA Ligands by Target-Guided Combinatorial Chemistry on a Magnetic Nanoplatform. J. Med. Chem. 2019;62:762–773. doi: 10.1021/acs.jmedchem.8b01459. [DOI] [PubMed] [Google Scholar]
- 104.Liu W., Lin C., Wu G., Dai J., Chang T.-C., Yang D. Structures of 1:1 and 2:1 Complexes of BMVC and MYC Promoter G-Quadruplex Reveal a Mechanism of Ligand Conformation Adjustment for G4-Recognition. Nucleic Acids Res. 2019;47:11931–11942. doi: 10.1093/nar/gkz1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang D., Gao F. Quinazoline Derivatives: Synthesis and Bioactivities. Chem. Cent. J. 2013;7:95. doi: 10.1186/1752-153X-7-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Theivendren P. &.; Kumar, P. Quinazoline Marketed Drugs—A Review. Res. Pharm. 2011;1:1–21. [Google Scholar]
- 107.Su L., Zheng H., Li Z., Qiu J., Chen S., Liu J., Ou T.-M., Tan J.-H., Gu L.-Q., Huang Z.-S., et al. Mechanistic Studies on the Anticancer Activity of 2,4-Disubstituted Quinazoline Derivative. Biochim. Biophys. Acta. 2014;1840:3123–3130. doi: 10.1016/j.bbagen.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 108.Li Z., Liu C., Huang C., Meng X., Zhang L., He J., Li J. Quinazoline Derivative QPB-15e Stabilizes the c-Myc Promoter G-Quadruplex and Inhibits Tumor Growth in Vivo. Oncotarget. 2016;7:34266–34276. doi: 10.18632/oncotarget.9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shan C., Lin J., Hou J.-Q., Liu H.-Y., Chen S.-B., Chen A.-C., Ou T.-M., Tan J.-H., Li D., Gu L.-Q., et al. Chemical Intervention of the NM23-H2 Transcriptional Programme on c-MYC via a Novel Small Molecule. Nucleic Acids Res. 2015;43:6677–6691. doi: 10.1093/nar/gkv641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shan C., Yan J.-W., Wang Y.-Q., Che T., Huang Z.-L., Chen A.-C., Yao P.-F., Tan J.-H., Li D., Ou T.-M., et al. Design, Synthesis, and Evaluation of Isaindigotone Derivatives to Downregulate c-Myc Transcription via Disrupting the Interaction of NM23-H2 with G-Quadruplex. J. Med. Chem. 2017;60:1292–1308. doi: 10.1021/acs.jmedchem.6b01218. [DOI] [PubMed] [Google Scholar]
- 111.Wang Y.-Q., Huang Z.-L., Chen S.-B., Wang C.-X., Shan C., Yin Q.-K., Ou T.-M., Li D., Gu L.-Q., Tan J.-H., et al. Design, Synthesis, and Evaluation of New Selective NM23-H2 Binders as c-MYC Transcription Inhibitors via Disruption of the NM23-H2/G-Quadruplex Interaction. J. Med. Chem. 2017;60:6924–6941. doi: 10.1021/acs.jmedchem.7b00421. [DOI] [PubMed] [Google Scholar]
- 112.Pereira J.A., Pessoa A.M., Cordeiro M.N.D.S., Fernandes R., Prudêncio C., Noronha J.P., Vieira M. Quinoxaline, Its Derivatives and Applications: A State of the Art Review. Eur. J. Med. Chem. 2015;97:664–672. doi: 10.1016/j.ejmech.2014.06.058. [DOI] [PubMed] [Google Scholar]
- 113.Tariq S., Somakala K., Amir M. Quinoxaline: An Insight into the Recent Pharmacological Advances. Eur. J. Med. Chem. 2018;143:542–557. doi: 10.1016/j.ejmech.2017.11.064. [DOI] [PubMed] [Google Scholar]
- 114.Hu M.-H., Wu T.-Y., Huang Q., Jin G. New Substituted Quinoxalines Inhibit Triple-Negative Breast Cancer by Specifically Downregulating the c-MYC Transcription. Nucleic Acids Res. 2019;47:10529–10542. doi: 10.1093/nar/gkz835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu T.-Y., Huang Q., Huang Z.-S., Hu M.-H., Tan J.-H. A Drug-like Imidazole-Benzothiazole Conjugate Inhibits Malignant Melanoma by Stabilizing the c-MYC G-Quadruplex. Bioorg. Chem. 2020;99:103866. doi: 10.1016/j.bioorg.2020.103866. [DOI] [PubMed] [Google Scholar]
- 116.Yang F., Sun X., Wang L., Li Q., Guan A., Shen G., Tang Y. Selective Recognition of C-Myc Promoter G-Quadruplex and down-Regulation of Oncogene c-Myc Transcription in Human Cancer Cells by 3,8a-Disubstituted Indolizinone. RSC Adv. 2017;7:51965–51969. doi: 10.1039/C7RA09870G. [DOI] [Google Scholar]
- 117.Dallavalle S., Musso L., Artali R., Aviñó A., Scaglioni L., Eritja R., Gargallo R., Mazzini S. G-Quadruplex Binding Properties of a Potent PARP-1 Inhibitor Derived from 7-Azaindole-1-Carboxamide. Sci. Rep. 2021;11:3869. doi: 10.1038/s41598-021-83474-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cincinelli R., Musso L., Merlini L., Giannini G., Vesci L., Milazzo F.M., Carenini N., Perego P., Penco S., Artali R., et al. 7-Azaindole-1-Carboxamides as a New Class of PARP-1 Inhibitors. Bioorg. Med. Chem. 2014;22:1089–1103. doi: 10.1016/j.bmc.2013.12.031. [DOI] [PubMed] [Google Scholar]
- 119.Gaikwad S.M., Phyo Z., Arteaga A.Q., Gorjifard S., Calabrese D.R., Connors D., Huang J., Michalowski A.M., Zhang S., Liu Z.-G., et al. A Small Molecule Stabilizer of the MYC G4-Quadruplex Induces Endoplasmic Reticulum Stress, Senescence and Pyroptosis in Multiple Myeloma. Cancers. 2020;12:2952. doi: 10.3390/cancers12102952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Felsenstein K.M., Saunders L.B., Simmons J.K., Leon E., Calabrese D.R., Zhang S., Michalowski A., Gareiss P., Mock B.A., Schneekloth J.S., Jr. Small Molecule Microarrays Enable the Identification of a Selective, Quadruplex-Binding Inhibitor of MYC Expression. ACS Chem. Biol. 2016;11:139–148. doi: 10.1021/acschembio.5b00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Calabrese D.R., Chen X., Leon E.C., Gaikwad S.M., Phyo Z., Hewitt W.M., Alden S., Hilimire T.A., He F., Michalowski A.M., et al. Chemical and Structural Studies Provide a Mechanistic Basis for Recognition of the MYC G-Quadruplex. Nat. Commun. 2018;9:4229. doi: 10.1038/s41467-018-06315-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dutta D., Debnath M., Müller D., Paul R., Das T., Bessi I., Schwalbe H., Dash J. Cell Penetrating Thiazole Peptides Inhibit C-MYC Expression via Site-Specific Targeting of c-MYC G-Quadruplex. Nucleic Acids Res. 2018;46:5355–5365. doi: 10.1093/nar/gky385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Meunier B. Hybrid Molecules with a Dual Mode of Action: Dream or Reality? Acc. Chem. Res. 2008;41:69–77. doi: 10.1021/ar7000843. [DOI] [PubMed] [Google Scholar]
- 124.Debnath M., Ghosh S., Chauhan A., Paul R., Bhattacharyya K., Dash J. Preferential Targeting of I-Motifs and G-Quadruplexes by Small Molecules. Chem. Sci. 2017;8:7448–7456. doi: 10.1039/C7SC02693E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Paul R., Das T., Debnath M., Chauhan A., Dash J. G-Quadruplex-Binding Small Molecule Induces Synthetic Lethality in Breast Cancer Cells by Inhibiting c-MYC and BCL2 Expression. Chembiochem. 2020;21:963–970. doi: 10.1002/cbic.201900534. [DOI] [PubMed] [Google Scholar]
- 126.Vilar R. Medicinal Chemistry. Elsevier; Amsterdam, The Netherlands: 2020. Interaction of Metal Complexes with G-Quadruplex DNA; pp. 425–445. Advances in Inorganic Chemistry. [Google Scholar]
- 127.Palma E., Carvalho J., Cruz C., Paulo A. Metal-Based G-Quadruplex Binders for Cancer Theranostics. Pharmaceuticals. 2021;14:605. doi: 10.3390/ph14070605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gama S., Rodrigues I., Mendes F., Santos I.C., Gabano E., Klejevskaja B., Gonzalez-Garcia J., Ravera M., Vilar R., Paulo A. Anthracene-Terpyridine Metal Complexes as New G-Quadruplex DNA Binders. J. Inorg. Biochem. 2016;160:275–286. doi: 10.1016/j.jinorgbio.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 129.Wei Z.-Z., Qin Q.-P., Meng T., Deng C.-X., Liang H., Chen Z.-F. 5-Bromo-Oxoisoaporphine Platinum(II) Complexes Exhibit Tumor Cell Cytotoxcicity via Inhibition of Telomerase Activity and Disruption of c-Myc G-Quadruplex DNA and Mitochondrial Functions. Eur. J. Med. Chem. 2018;145:360–369. doi: 10.1016/j.ejmech.2017.12.092. [DOI] [PubMed] [Google Scholar]
- 130.He L., Meng Z., Xu D., Shao F. Dual Functional Dinuclear Platinum Complex with Selective Reactivity towards C-Myc G-Quadruplex. Sci. Rep. 2018;8:767. doi: 10.1038/s41598-017-19095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Haddach M., Schwaebe M.K., Michaux J., Nagasawa J., O’Brien S.E., Whitten J.P., Pierre F., Kerdoncuff P., Darjania L., Stansfield R., et al. Discovery of CX-5461, the First Direct and Selective Inhibitor of RNA Polymerase I, for Cancer Therapeutics. ACS Med. Chem. Lett. 2012;3:602–606. doi: 10.1021/ml300110s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.