Abstract
The world is slowly recovering from the Coronavirus disease 2019 (COVID-19) pandemic; however, humanity has experienced one of its According to work by Mishra et al. (2020), the study’s first phase included a cohort of 5,262 subjects, with 3,325 Fitbit users constituting the majority. However, among this large cohort of 5,262 subjects, most significant trials in modern times only to learn about its lack of preparedness in the face of a highly contagious pathogen. To better prepare the world for any new mutation of the same pathogen or the newer ones, technological development in the healthcare system is a must. Hence, in this work, PCovNet+, a deep learning framework, was proposed for smartwatches and fitness trackers to monitor the user’s Resting Heart Rate (RHR) for the infection-induced anomaly. A convolutional neural network (CNN)-based variational autoencoder (VAE) architecture was used as the primary model along with a long short-term memory (LSTM) network to create latent space embeddings for the VAE. Moreover, the framework employed pre-training using normal data from healthy subjects to circumvent the data shortage problem in the personalized models. This framework was validated on a dataset of 68 COVID-19-infected subjects, resulting in anomalous RHR detection with precision, recall, F-beta, and F-1 score of 0.993, 0.534, 0.9849, and 0.6932, respectively, which is a significant improvement compared to the literature. Furthermore, the PCovNet+ framework successfully detected COVID-19 infection for 74% of the subjects (47% presymptomatic and 27% post-symptomatic detection). The results prove the usability of such a system as a secondary diagnostic tool enabling continuous health monitoring and contact tracing.
Keywords: COVID-19, Wearables, Long short-term memory, Convolutional neural network, Variational autoencoder, Resting heart rate, Anomaly detection
1. Introduction
COVID-19 posed one of the most severe threats to modern healthcare systems in decades. According to the World Health Organization (WHO), over 614 million confirmed cases of COVID-19, along with 6.5 million casualties, were reported until October 2022 (WHO, 2022). Although effective vaccines have been available and the global pandemic situation has improved, the overall scenario pointed out the lack of preparedness in the healthcare systems around the world (Haldane et al., 2021, Mazumder et al., 2020). During the pandemic, researchers from different disciplines contributed their expertise to tackle the disease. As a result, a plethora of publications in different domains introduced the use of new tools and redefined the existing ones to tackle the healthcare crisis better (Gianola et al., 2020). Likewise, this work focuses on the use of smartwatches and fitness trackers to combat COVID-19 infection. After repeated mutations and waves of infections for two years, according to the WHO, the end of the pandemic is in sight now but yet to be fully achieved (UN News, 2022). The consequences of this work have significance that goes beyond just managing the existing COVID-19 outbreak, and can also be useful for fighting against other comparable pandemics that may arise in the future. Moreover, the technique and the model developed here will be highly beneficial in many other healthcare applications as well.
Among the family of coronaviruses, Severe Acute Respiratory Syndrome Coronavirus-2, or in short, SARS-CoV-2, is the pathogen responsible for the latest global coronavirus pandemic. Being part of the same family, it has similar characteristics, namely incubation period and reproduction number, as its predecessors — Middle East Respiratory Syndrome Coronavirus (MARS) and SARS-CoV. However, unlike its predecessors, SARS-CoV-2 caused a global pandemic due to its duration of viral shedding, asymptomatic infection rate, and rapid mutations in different global regions (Wu et al., 2021, Abdelrahman et al., 2020, Gandhi et al., 2020). One of the most effective methods to combat the asymptomatic COVID-19 infection can be to frequently test individuals in high-risk zones with active detection tools; ideally, the reverse transcription polymerase chain reaction (RT-PCR). However, due to the required resources and the cost to perform RT-PCRs, the widespread use of RT-PCR in active and frequent monitoring of such pathogens on a global scale is still in question (Augustine et al., 2020a, Augustine et al., 2020b). On the other hand, a more feasible approach can be to deploy continuous monitoring passively via devices such as smartwatches and fitness trackers (Alyafei et al., 2022).
In the domain of passive COVID-19 monitoring systems, several studies showed promising results. Alyafei et al. surveyed different COVID-19 detection systems, from laboratory-based to wearable ones (Alyafei et al., 2022). Several studies have shown that wearable sensor-based systems, as a part of the digital infrastructure, can be an effective mode for remote patient monitoring and virtual assessment (Seshadri et al., 2020, Dunn et al., 2018, Roblyer, 2020, Channa et al., 2021, Amft et al., 2020, Quer et al., 2021). In this regard, Buchhorn et al. showed a correlation between heart rate (HR) and heart rate variability (HRV) in an experiment on a 58-year-old male subject during the whole period of COVID-19 infection (Buchhorn et al., 2020). In another study, Ponomarev et al. showed a significant correlation between COVID-19 infection symptoms and HRV (Ponomarev et al., 2021). While clinical wearable devices have been used in numerous experimental settings, non-clinical commercial off-the-shelf (COTS) devices like smartwatches and fitness trackers have also been used in some studies for COVID-19 detection. The COTS devices contain sensors to effectively monitor HR, HRV, activity (step count), burned calories, sleep stages, and durations (Piwek et al., 2016, Mahajan et al., 2020). Mishra et al. demonstrated a correlation between resting heart rate (RHR) and COVID-19 infection using wearable smartwatch data (Mishra et al., 2020). Moreover, Mitratza et al. presented a systematic review on the use of wearable sensors for detecting SARS-CoV-2 symptoms (Mitratza et al., 2022). Their findings reiterated the results of Mishra et al. for using HR as a potential indicator for Covid-19 infection. Additionally, Radin et al. experimented on the correlation between sleep duration and RHR pattern with COVID-19 infection (Radin et al., 2020). Since the number of smartwatches and fitness tracker users has increased drastically over the past few years, this mode of passive COVID-19 detection system can have a crucial effect on the overall healthcare system (Vogels, 2022).
Like all other fields of technology, artificial intelligence (AI) has made notable contributions in the field of healthcare and wearables at the same time. During the COVID-19 pandemic, AI-based research has increased exponentially from wearables-based early COVID-19 detection (Liu et al., 2021, Bogu and Snyder, 2021, Abir et al., 2022) to chest X-ray (Wang et al., 2021, Arias-Londoño et al., 2020, Tahir et al., 2021, Yamaç et al., 2021) or computed tomography (CT)-based (Chen et al., 2020, Qiblawey et al., 2021) COVID-19 diagnosis. Among the wearables paradigm, Liu et al. introduced contrastive loss with a convolutional autoencoder to detect abnormality in heart rate. Moreover, Mishra et al. led a study on a cohort of over 5000 subjects to collect HR, steps, and sleep information using different smartwatches (Mishra et al., 2020). Cho et al. improved upon that study by proposing a One Class-Support Vector Machine (OC-SVM) for pre-symptomatic COVID-19 detection (Cho et al., 2022). Their method improved the presymptomatic detection performance over the statistical method proposed by Mishra et al. In another work, Bogu et al. used a long short-term memory (LSTM)-based autoencoder model to identify abnormal HR in that data (Bogu and Snyder, 2021). Merrill and Althoff proposed a self-supervised pretraining approach using healthy subject data and employed attention layers (Merrill and Althoff, 2022).
At the beginning of wearables-based COVID-19 detection research, there was an acute shortage of reliable data and a state-of-the-art method established for such studies. Hence, several concepts from other domains were adopted. Due to the nature of the available data, abnormal vital detection due to COVID-19 or similar respiratory diseases is mainly designed as an anomaly detection problem where abnormal data is rare compared to normal data. Autoencoders and variational autoencoders are often used in such scenarios across different domains (Kiran et al., 2018).
In the earlier work, a Long Short-term Memory (LSTM)-based Variational Autoencoder (VAE) model, PCovNet, was proposed to detect anomalous RHR from the smartwatch data (Abir et al., 2022). The framework was validated on 25 COVID-19-infected users’ data and showed promising results in early detection and precision score. Moreover, it was designed to make predictions based on a sequence of 8 RHR data points (1 data point per hour). In short, there were two particular shortcomings in the study which set the primary objectives of this work.
-
(i)
An anomaly detection system that takes weeks to make a prediction is not practical in a real-world setting; thus, a longer sequence was not employed. On the other hand, incorporating a longer RHR sequence could enhance the model’s robustness. Hence, the first objective of this work was to establish a balance between model robustness and real-world usability.
-
(ii)
Although precision and F-beta scores were very promising in the previous work, recall, and F-1 needed some improvements to achieve model reliability. Therefore, the second objective of this work was to achieve improvements in the performance metrics.
To achieve these objectives, PCovNet+, an improved hybrid anomaly detection pipeline, was proposed that will work at the backend of smartwatches to detect anomalous RHR considering 16 days of RHR data. This work employed Convolutional Neural Network (CNN)-based Variational Autoencoder (VAE) architecture as the primary anomaly detector and a Long Short-term Memory (LSTM)-based network to generate temporal-aware embeddings in the latent layer of the VAE based on past RHR information. The architecture was motivated by the work of Lin et al. (2020). Moreover, this work employed pretraining using Healthy group data to solve the low availability of personalized models. In a nutshell, the contributions of this work are as follows:
Firstly, a new CNN-VAE-based anomaly detection framework was proposed including a separate LSTM network to generate temporal-aware embeddings of the latent vector of the primary model.
Secondly, a pretrained model was generated using the data from the Healthy group to create base models for the CNN-VAE and the LSTM networks. These base models were finetuned with each subject’s baseline data to achieve a personalized version of the base model.
Thirdly, the model was validated on 68 COVID-19-infected individuals’ data.
2. Material and methods
2.1. Dataset
As a part of a multi-phase study to determine the correlation of COVID-19 infection with human vitals, namely heart rate, sleep, and activity profile, a research group from the Department of Genetics of Stanford University published two datasets (COVID-19, 2020, COVID-19, 2021). They used Research Electronic Data Capture (REDCap) survey web platform to track the symptom onset date, symptom severity, and diagnosis, along with other demographic and clinical data (Mishra et al., 2020, Alavi et al., 2022). The vitals data were primarily collected using smartwatches, such as Fitbit (Fitbit, 2022), Apple Watch (Apple Inc., 2022), and Garmin (Garmin, 2022), in both retrospective and prospective studies. They developed a cross-platform smartphone application named MyPHD to collect smartwatch data and synchronize it with the survey information (Mishra et al., 2020, Alavi et al., 2022).
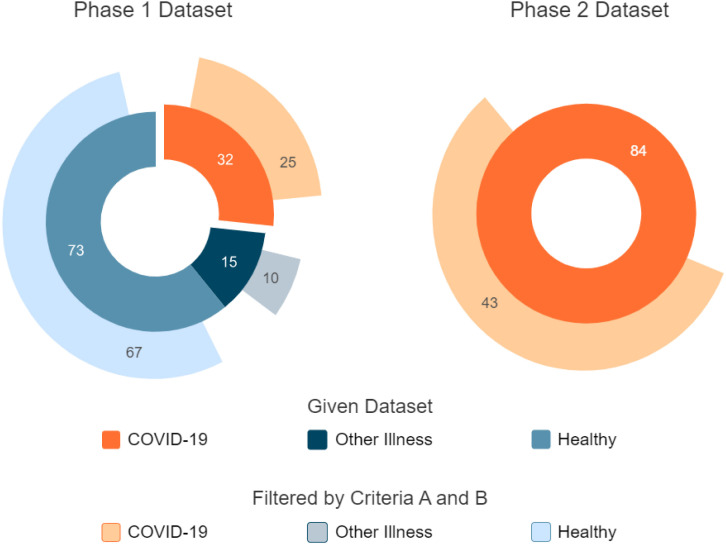
In this work, publicly available data from their study’s first and second phases was used. Fig. 1 illustrates the dataset distribution.
Fig. 1.
Dataset distribution. The left donut chart denotes the phase-1 dataset and the left one represents the phase-2 dataset.
2.1.1. First phase data
According to work by Mishra et al. (2020), the study’s first phase included a cohort of 5262 subjects, with 3325 Fitbit users constituting the majority. However, among this large cohort of 5262 subjects, only 32 had smartwatch data, namely heart rate, steps, and sleep duration during the COVID-19 infection. Hence, the given dataset included these 32 COVID-19 infected subjects, with 73 healthy and 15 patients with other respiratory diseases, whereas sleep data were only provided for the COVID-19 group
2.1.2. Second phase data
In the second phase study, Alavi et al. (2022) enrolled 3318 subjects between November 2020 and July 2021. Of these subjects, 278 were positively diagnosed with COVID-19; however, only 84 (49 Fitbit and 35 Apple Watch users) had recorded wearable data, namely heart rate, and step count during their infection. Moreover, 34 of them were diagnosed after enrollment, and 50 were diagnosed before that. Although the second phase dataset provided over 2000 users’ vital data, they only added the symptom information for those 84 subjects (COVID-19, 2021).
2.1.3. Combined dataset
To formulate the anomaly detection problem, the baseline data were needed which was not explicitly annotated in the provided dataset. Hence, this work relied on the literature to determine the number of days after infection till symptom onset (incubation period) and the number of days after the infection when the host releases live particles (viral shedding) of SARS-CoV-2. According to several research groups, this pathogen’s median incubation period ranges from 3 days to 5 days (COVID-19, 2021, Alavi et al., 2022). Although the evidence of viral shedding for up to 83 days was reported in a study (Cevik et al., 2021), after 21 days of the symptom onset, the amount of viral pathogen in the host fluid is reduced to the detection limit (He et al., 2020). Based on these findings, two criteria were set for qualifying a subject’s data for this study.
A. The wearables data must contain heart rate and steps during the same timestamps.
B. The provided data must range from at least 20 days before the symptom onset to 21 days afterward.
In this work, the PCovNet+ framework was developed and validated on the combined data from both phases that satisfied the abovementioned criteria. Afterward, the combined dataset consisted of 68 subjects from the COVID-19 group, 10 from the Other Illness group, and 67 from the Healthy group.
2.2. Data preprocessing
The combined dataset primarily comprised the heart rate and steps information. However, the targeted data channel for this work was the resting heart rate (RHR) that could be derived from the two given data channels. Moreover, before feeding the data into the anomaly detection model, the train and test sets needed to be determined appropriately, which entailed some extra layer of preprocessing for this study.
2.2.1. Resting heart rate calculation
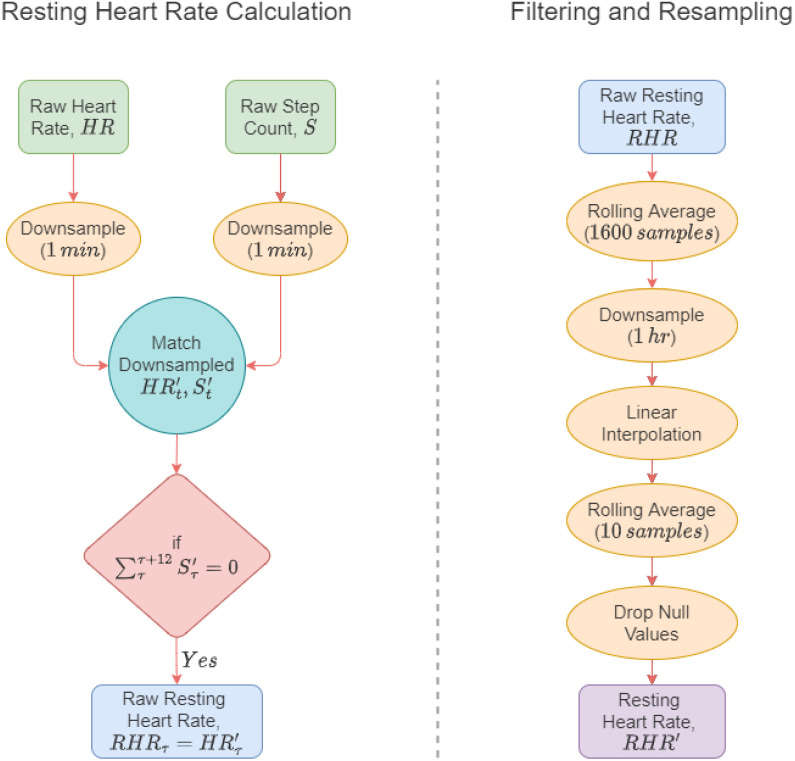
In this work, the RHR calculation algorithm by Mishra et al. (2020) and Bogu and Snyder (2021) was adopted where the authors considered the heart rate as RHR when the step count was zero for 12 consecutive minutes. The process is illustrated in the left flow diagram of Fig. 2.
Fig. 2.
Data preparation. At the left, raw HR and step counts are used to extract the raw resting heart rate. At the right, the raw resting heart rate undergoes several steps of filtering and resampling, resulting in the final resting heart rate.
As the timestamps were not the same for the given heart rate and steps channels, at first, the channels were downsampled to 1 min. Afterward, the two channels were merged and the heart rates were taken where the step count value was zero. Lastly, the heart rates were further filtered based on the timestamps where the step counts were not zero for at least 12 min. The resultant heart rate was considered the raw RHR for the experiment.
2.2.2. Filtering and resampling
The raw RHR, albeit not including the effects of any physical activity, is not devoid of any stress or mental activity that might not represent a complete resting state. As a result, the raw RHR value contains significant fluctuations in the data, which was filtered with a moving average window of 1600 data points.
As RHR does not change every minute, the filtered RHR was downsampled to 1 h as the next step of preprocessing. However, the resampled RHR contained missing values for some subjects due to the merging operation of RHR calculation in Section 2.2.1. To circumvent this issue, linear interpolation was used to impute the missing values.
After the missing value imputation, however, there were spikes in the imputed data due to linear interpolation. Hence, another moving average filter was used with only 10 data points and null values were dropped to produce the final RHR data channel for the experiment. The flow of these steps is shown on the right side of Fig. 2.
2.2.3. Dataset split
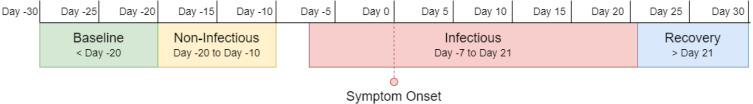
As discussed in Section 2.1, this work used prior literature (Cevik et al., 2021, Guan et al., 2020, Li et al., 2020) to determine the incubation period and viral shedding of SARS-CoV-2. Moreover, Mishra et al. (2020) and Bogu and Snyder (2021) divided the user dataset into four distinct regions, which were also adopted in this work. Fig. 3 represents these four regions.
Fig. 3.
COVID-19 infection stages — baseline period, non-infectious period, infectious period, and recovery period. In general, the baseline period contains normal data and the infectious period contains anomalous data. The other two may or may not have any anomaly.
1. Baseline Period: This is considered the ‘normal’ region of the data. This region includes all the RHR data 20 days before the symptom onset.
2. Non-infectious Period: It starts right after the baseline period and ends 10 days before the symptom onset. This region is equivalent to the baseline period for most of the subjects. However, according to He et al. (2020), a few with a prolonged incubation period might show some viral shedding during this time.
3. Infectious Period: This ranges from 7 days before the symptom onset to 21 days after that. Most viral shedding occurs during this period; hence, the anomalous RHR is most likely to occur during these days.
4. Recovery Period: Generally, after the infectious period, an infected individual’s vitals gradually return to the baseline. Hence, the period after 21 days of the symptom onset is regarded as the recovery period. However, the retainment of the baseline RHR varies in time for different individuals. For some individuals, the prolonged effect of COVID-19 infection can span months.
Train, validation, and test set were assigned based on the properties of these four periods.
Train Set: As this task was approached as an anomaly detection problem, the model was trained with normal data. Hence, only the baseline period was used in the train set.
Validation Set: 5% of the train set was used for validation during the model training. Hence, it also included only the normal data from the baseline period.
Test Set: To test the performance, both anomalous and normal data were needed in the test set. Although the anomalous RHR regions were approximated based on literature in different populations, the actual infection region is much harder to determine. Therefore, data from only Day 0 to Day 7 of infection was annotated as the anomalous test set.
2.2.4. Standardization
Standardization is a common preprocessing step to prepare the dataset for training the models efficiently. Moreover, the RHR values had different ranges for different subjects in the dataset. Hence, we used StandardScaler from the Scikit-Learn (Pedregosa et al., 2011) Python package.
At first, the mean () and standard deviation () of the train set were calculated and the mean was converted to zero and scaled by dividing the standard deviation value. For the test set, the same process was followed with the mean and standard deviation of the train set. It is given in Equation 1.
| (1) |
2.2.5. Segmentation
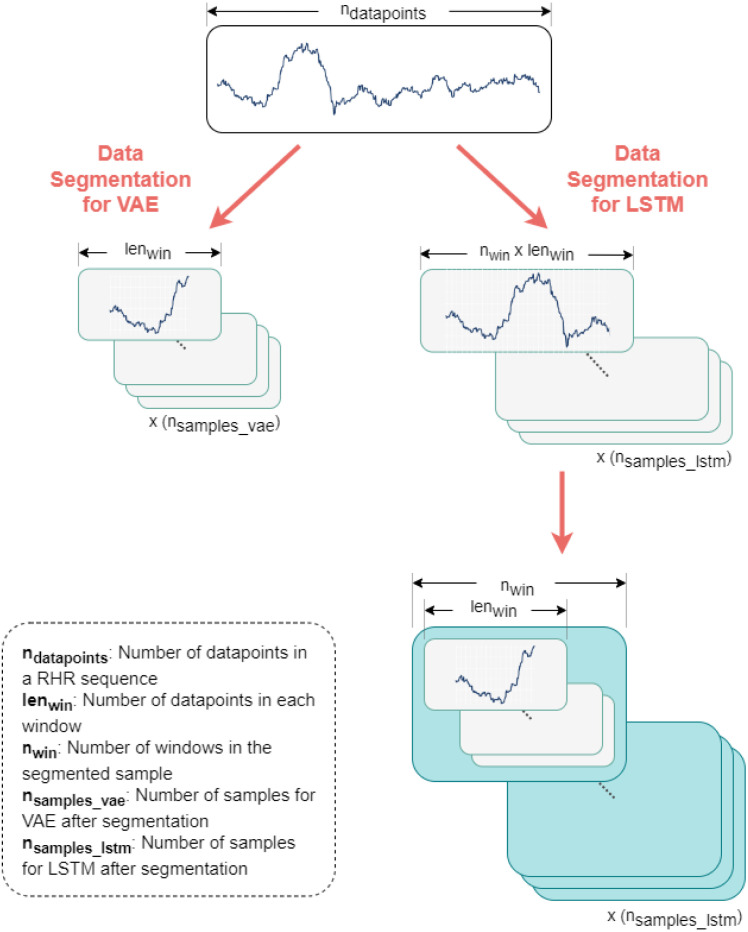
In this framework, two separate models were used — a CNN-VAE model and an LSTM network which needed two separate segmentation methods for training. Fig. 4 illustrates these segmentation methods.
Fig. 4.
Data segmentation process for CNN-VAE and LSTM networks. Data segmentation for the VAE is illustrated in the left-hand sequence where the entire sequence is converted into a three-dimensional matrix (, , ). On the other hand, data segmentation for the LSTM network is shown. At first, the data sequence is segmented into a similar three-dimensional matrix with an increased window length (, , ) and later segmented into a four-dimensional matrix (, , , ).
For CNN-VAE, the length of the window is denoted as . Both the training and testing datasets were segmented into data points per window by moving 1 data point per sample. The total number of samples for CNN-VAE can be represented by Eq. (2) ,
| (2) |
where denotes the total number of samples after segmentation for the CNN-VAE model. and respectively denote the number of data points in the segmented dataset and the unsegmented one. After segmentation, the dataset shape was (, , ) where is the number of data channels which is 1 as only RHR was used in this experiment.
On the other hand, the LSTM model took multiple numbers of windows, denoted as , from the CNN-VAE. Hence, for each training sample of the LSTM, the dataset had windows which constitutes a total number of data points in total. The segmentation for this case was done in two steps. At first, the original data were segmented in the same way as the CNN-VAE one but with a longer window of which was only for the CNN-VAE. Afterward, these segmented windows are sliced into smaller windows in a separate dimension. The total number of samples for the LSTM model can be calculated with Eq. (3),
| (3) |
where represents the total number of samples in the segmented dataset for the LSTM model. and denote the same properties as in Eq. (2). The segmented dataset shape for the LSTM model was (, , , ) where is 1 since RHR was only used in this work.
2.2.6. Augmentation
Data augmentation includes the techniques that are often used in the preprocessing stage of a deep learning experiment to artificially generate new data points from the existing ones to increase the volume of the overall dataset. Although this process is more common for image domain problems, the benefits can be exploited for time series problems (Iwana and Uchida, 2021). Um et al. showed that using such data augmentation methods on smartwatch sensors improved the overall deep learning model prediction (Um et al., 2017) and Bogu et al. used the same methods in their work on the same dataset as this study (Bogu and Snyder, 2021). Moreover, the previous study showed evidence of carefully chosen augmentation techniques to improve results (Abir et al., 2022). Hence, the same seven augmentation techniques were adopted here.
1. Scaling: The signal’s amplitude was multiplied with a random value of a Gaussian distribution of mean 1 and standard deviation 0.1.
2. Rotation: Horizontal mirroring was applied by randomly rotating the signal.
3. Permutation: The signal was sliced into 1 to 4 segments and permutated randomly to create perturbation.
4. Magnitude Warping: The signal was multiplied with a cubic spline having four knots that are determined randomly from a Gaussian distribution of mean 1 and standard deviation 0.2.
5. Time Warping : The temporal locations of the data points were perturbed based on a cubic spline.
6. Window Warping: A portion of the signal window was chosen randomly, and the frequency was altered by a factor of 0.5 or 2.
7. Window Slicing: The signal window was sliced randomly by 90%.
2.3. Anomaly detection model
In general, for both autoencoder and variational autoencoder architectures, the models learn crucial features of the normal input data and output a reconstructed version of the input. When test data with the anomaly is passed to the model, it also tries to reconstruct it. However, since it was only trained with normal data, the reconstruction loss for anomalous data was much higher. As a result, simple thresholding was able to differentiate the normal and anomalous data based on the model’s reconstruction loss.
After the first phase study, Bogu et al. from the Stanford Group proposed a Long Short-Term Memory Networks (LSTM)-based autoencoder for anomaly detection (LAAD) framework for anomalous RHR detection (Bogu and Snyder, 2021), which established the possibility of using a deep learning-based anomaly detection system with smartwatch data. The PCovNet framework used LSTM-based variational autoencoders and achieved better presymptomatic detection and F-beta score compared to the LAAD framework (Abir et al., 2022).
Both architectures adopted LSTM layers; however, the most distinctive features of these architectures are the bottleneck layer in the middle, which is often called the latent layer. For autoencoder architecture, the latent layer represents the fundamental features that are required to reconstruct the original signal. However, such a latent layer generally represents a discrete latent space of the original data, which cannot reconstruct data effectively for all possible values of the latent vectors. On the other hand, VAE resorts to variational inference for generating the latent layer, which is achieved by sampling the latent vector into a Gaussian-like distribution. This technique results in a continuous latent space that ensures effective reconstruction in the decoder.
The RHR change takes place over several days; however, making a single prediction taking several days to weeks of data is not feasible for a real-world scenario. Hence, this study used CNN-VAE at first to learn the local patterns of RHR and a separate LSTM network to create embeddings considering a more extended sequence (weeks) of past RHR for the final prediction. To clarify, the trained LSTM network works on the latent space and converts it based on a more extended sequence of past data. The method of using a separate LSTM network for transforming the latent space was motivated by the work of Lin et al. (2020) where the authors validated this process on several anomaly detection problems.
In this case, the continuous nature of the latent space is essential since the LSTM embeddings convert the encoder-generated latent space. This is the primary reason for choosing VAE architecture over the autoencoder one.
2.3.1. CNN-VAE architecture
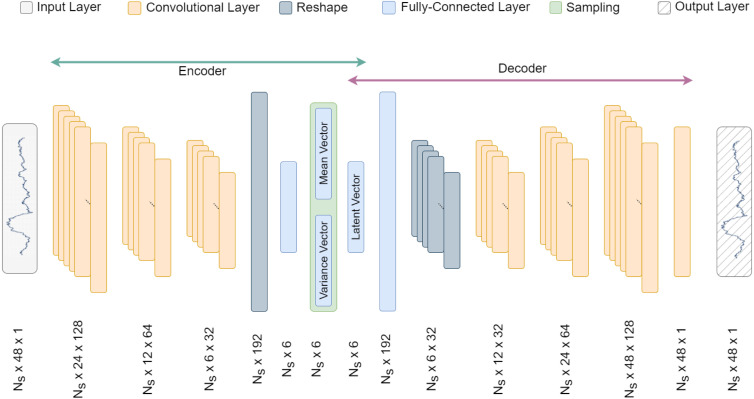
As per the VAE architecture, CNN-VAE is divided into two blocks — encoder and decoder. In both blocks, convolutional layers were used, since time series data is regarded as 1-dimensional data. Fig. 5 shows the detailed CNN-VAE architecture.
Fig. 5.
Detailed CNN-VAE architecture. denotes the number of samples. Here, three CNN layers are used in the encoder and the decoder blocks.
Given an input , the reconstructed data , and latent vector , the overall CNN-VAE process can be expressed by Eqs. (4), (5).
| (4) |
| (5) |
Encoder. The encoder consists of three convolutional layers, each with kernel size 3, stride 2, rectified linear unit (ReLU) activation function, and decreasing number of filters: 128, 64, and 32. These three 1D convolutional layers extract the features from each layer and pass them to the next. Afterward, the output of the third convolutional layer is flattened and fed into a dense layer with the size of the latent layer and the same ReLU activation function. According to the VAE architecture, the last dense layer outputs two separate layers, which describe the mean and variance of the latent space distribution and are sampled using the sampling function in Eq. (6).
| (6) |
Here, is the sampled latent layer which represents the latent space distribution. , , and denote the latent distribution mean, variance, and a randomly sampled unit Gaussian ()). The is scaled by the variance and shifted by the mean to constitute the latent space distribution. This process of introducing is often called the reparameterization trick, which allows the error to backpropagate through the network during the training process (Kingma and Welling, 2022).
Decoder. The decoder takes the latent layer as input and outputs the reconstructed signal of the original input. This block consists of a dense layer and four convolutional layers. At first, the latent vector is fed to a dense layer of nodes, which is reshaped to 6 × 32 array. Afterward, the reshaped array is passed through four convolutional layers of 32, 64, 128, and 1 filter. The first three convolutional layers use the ReLU activation function, and the last one employs the sigmoid activation function. The last layer’s output is the same as the input data.
Loss calculation. Due to the variational inference method used in the VAE architecture, the latent layer distribution is converted to a Gaussian-like one as the training progresses. Hence, apart from the reconstruction loss, the Kullback–Leibler (KL) Divergence of the latent layer distribution and a Gaussian was also calculated.
For input vector, the model outputs a corresponding prediction vector, and is a normal distribution with the mean vector and variance vector The overall loss calculation for CNN-VAE can be expressed by Eqs. (7), (8), and (9).
| (7) |
| (8) |
| (9) |
Here, , and denote the reconstruction loss, KL divergence loss, and total loss, respectively.
2.3.2. LSTM architecture
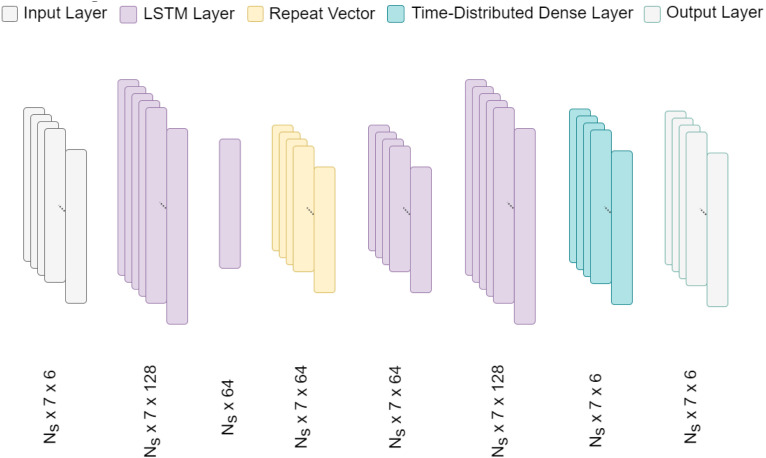
This framework adopted a simple LSTM autoencoder architecture for the embedding generation, which is depicted in Fig. 6. As mentioned before, after the separate training process, the LSTM layer works amidst the encoder and decoder of CNN-VAE. It takes the CNN-VAE latent layer as input and creates embeddings of the same shape that are more aware of the long-term RHR traits. The input and output of this network are of the same shape as the latent layer.
Fig. 6.
Detailed LSTM architecture. denotes the number of samples. In total four LSTM layers are used in the LSTM network.
The LSTM network, in total, consists of four LSTM layers. The first two layers have 128 and 64 LSTM units, respectively; however, the second one does not return a sequence. The condensed output is passed through two more LSTM layers with 64 and 128 units, respectively. At last, a time-distributed dense layer was used to retain the same shape as the input. Additionally, mean-squared error (MSE) was used as a loss function for the LSTM network.
2.3.3. Training
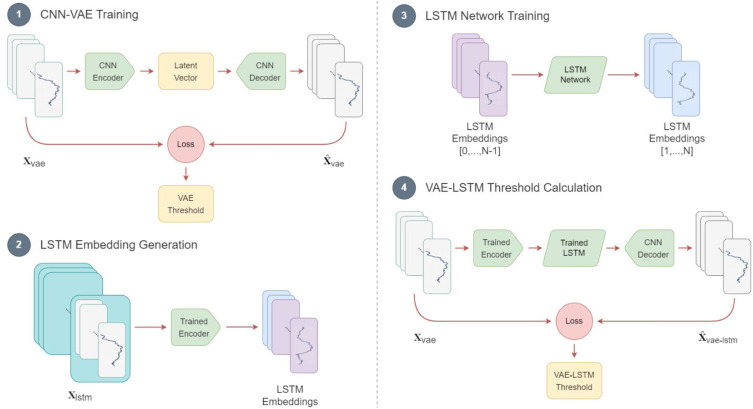
The combined dataset consists of both healthy and COVID-19-infected subject data. Like most other anomaly detection frameworks, the CNN-VAE and LSTM networks are trained using normal data. Since COVID-19-infected subjects do not often have a very long baseline (normal) data sequence, both models were pretrained using the healthy subjects’ data. Afterward, the baseline data of the COVID-19-infected subjects was used to fine-tune the pretrained model and make them personalized for each subject. Fig. 7 depicts the training sequence of the CNN-VAE and LSTM, along with the threshold calculation.
Fig. 7.
Diagram of training sequence of PCovNet+ framework.
CNN_VAE. The network was trained in the traditional way where the three-dimensional (, , ) segmented dataset was passed through the network as input and used the same dataset to compare with the reconstructed version. Reconstruction and KL divergence losses are calculated and minimized in each training step over the training sequence.
LSTM_VAE. The LSTM network training was done differently than the CNN-VAE. For this task, the dataset was segmented into a four-dimensional array (, , , ) according to Section 2.2.5.
For each LSTM training sample, first windows were passed to the trained CNN-VAE encoder to create the correspondent latent vector. These latent vectors (, , ) were used as the input of the LSTM network. On the other hand, for each LSTM training sample, the last windows were passed to the trained CNN_VAE encoder to create the correspondent latent vector to pass as the target of the LSTM network.
Hence, during the training process, the network, over time, learned to predict the immediate next window based on a more extended sequence of data compared to the CNN-VAE.
Threshold Calculation. After training, the combined network was used to generate the training loss. Since the training samples only included the normal data, the threshold was set at the maximum of the training loss.
2.3.4. Prediction
In this framework, the CNN-VAE was the primary anomaly detector that took a shorter data sequence into account compared to the LSTM network. Although the CNN-VAE could reconstruct the given input on its own, the LSTM network was added between the encoder and decoder to create embeddings of the latent vector.
After training the two networks, for predictions, the data only needed to be segmented for the CNN-VAE. The segmented data were passed through the CNN-VAE encoder to generate the latent vector. Afterward, the latent vector is passed through the LSTM network to generate embeddings which are then passed through the CNN-VAE decoder to make the final output. Data were labeled normal or anomalous by comparing the total loss with a threshold.
3. Experiments
3.1. Experimental setup
For all the experiments, the Adam optimizer (Kingma and Ba, 2014) was used along with batch size , learning rate , and epochs with early stopping criteria (patience ). This framework used Python programming language for this work along with Pandas (The pandas development team, 2020, McKinney, 2010) and Numpy (Harris et al., 2020) packages for data processing and array handling. Also, MatplotLib (Hunter, 2007) and Seaborn (Waskom, 2021) packages were used for data visualization. For the deep learning part, this work employed the Keras (Chollet et al., 2015) and TensorFlow (Abadi et al., 2016) packages. For the experiments, the hardware specification was — Intel Core i5 11th generation (2.4 GHz) processor, 16 GB RAM, and NVIDIA MX330 GPU. The shapes of the train and test sets for CNN-VAE and the LSTM networks are shown in Supplementary Appendix 1.
3.2. Evaluation metrics
Similar to the previous work, two types of evaluation metrics were considered for this study as well (Abir et al., 2022).
-
Statistical metrics: Precision, recall, F-1, and F-beta () scores were considered for this study. These metrics can be expressed by Eqs. (10), (11), (12), and (13) (Jabir et al., 2021).
(10) (11) (12) (13) -
Problem-specific metric: For this problem setting, early, late, and failed detections are three metrics to gain insight of the model’s performance across different subjects.
3.3. LSTM embeddings
In this experiment, the effectiveness of adding LSTM networks to the CNN-VAE was tested, along with the performance difference in three aspects. Note that the augmented train data described in Section 2.2.6 were used for this experiment.
At first, the experiment considered the numeric metrics of PCovNet+ for Phase-1, Phase-2, and combined datasets with and without the LSTM embeddings. The metrics give a quantitative evaluation of the framework between the two settings. Here, the test data from Day 0 (Symptom Onset) to Day 14 of infection we considered.
The second aspect involved examining the training and testing loss distributions. In this case, the test set from Day −20 to the last data in the recovery period (> Day 21) was considered, which resulted in both normal and anomalous data in the test set.
Finally, the experiment analyzed anomaly plots before and after using LSTM embeddings for a subject in which the CNN-VAE failed. The test data for this part was also taken from Day −20 to the end of the recovery period (> Day 21).
3.4. Test data regions
The datasets do not have labels for the HR data as normal or anomalous. Instead, the label is deduced passively using the given symptom onset and the relevant literature as described in Section 2.2.3. The earlier research put forward the notion that the metrics may not accurately indicate the detection of anomalies because the abnormal RHR does not appear at the same time in the infection region for every individual (Abir et al., 2022). Here, the issue was quantitively examined further by changing the data regions for the test set.
Here, only the infectious period was indicated as the anomalous region and the rest as normal. Moreover, the LSTM embeddings were used with CNN-VAE along with data augmentation on the train set. For evaluation, the baseline period for training was considered and five different regions were set as test data –
-
(i)
From Day −20 to Day −10 (non-infectious period) and from Day −7 to the rest (both infectious period and recovery period)
-
(ii)
From Day −20 to Day −10 (non-infectious period) and from Day 0 to Day 14 (partial infectious period)
-
(iii)
From Day 0 to Day 14 (partial infectious period) and the recovery period
-
(iv)
From Day 0 to Day 14 (partial infectious period)
-
(v)
From Day 0 to Day 7 (partial infectious period)
4. Results
This section presents the findings from the experiments in Sections 3.1, 3.2.
4.1. Impact of LSTM network
Table 1 shows that the statistical metrics for each dataset improve with the LSTM embeddings. In particular, the recall shows a significant improvement which signifies that the model can recognize more anomalous RHR data points compared to the CNN-VAE alone. Moreover, with the increase of recall, along with the improvement in already high precision, the F-1 improves by over 21% for Phase-1 and over 11% for the Phase-2 dataset. Regarding the problem-specific metrics, the failed cases are reduced for both datasets. On top of that, the model recognizes more subjects’ infections before symptom onset, resulting in better presymptomatic detection performance.
Table 1.
VAE vs. VAELSTM model performance.
| Dataset | Total subject | Model | Precision | Recall | F-beta | F-1 | Early detection | Delayed detection | Failed detection |
|---|---|---|---|---|---|---|---|---|---|
| Phase-1 | 25 | VAE | 0.9898 | 0.3887 | 0.9749 | 0.5582 | 36% | 40% | 24% |
| VAE LSTM | 0.9939 | 0.5848 | 0.9870 | 0.7363 | 65% | 10% | 25% | ||
| Phase-2 | 43 | VAE | 0.9916 | 0.2753 | 0.9667 | 0.4310 | 28% | 40% | 32% |
| VAE LSTM | 0.9930 | 0.4832 | 0.9827 | 0.6501 | 38% | 36% | 26% | ||
| Combined | 68 | VAE | 0.9916 | 0.2753 | 0.9667 | 0.4310 | 31% | 40% | 29% |
| VAE LSTM | 0.9930 | 0.4832 | 0.9827 | 0.6501 | 47% | 27% | 26% | ||
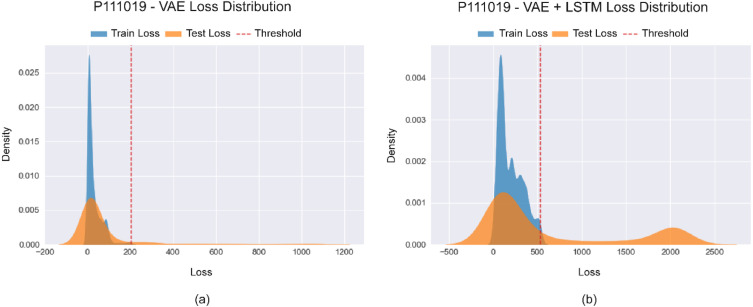
From Fig. 8(a), it can be observed that the train and test loss distributions are both sharper and mostly overlapped. Based on the thresholding method of separating the anomalous losses from the normal ones, only a thin tail is separated from the test distribution. However, the separation is more distinct in Fig. 8(b), which represents the losses after employing LSTM embeddings. Here, the test loss distribution has two peaks at a significant distance from one another. The left peak signifies the normal data in the test set can be easily deduced by the overlapping train loss distribution. On the other hand, the right peak of the test loss distribution signifies the anomalous data points in the test set. Moreover, the large separation ensures ease of separation between the normal and anomalous data.
Fig. 8.
Difference between the loss distributions of (a) CNN-VAE and (b) CNN-VAE with LSTM embeddings for subject id P111019.
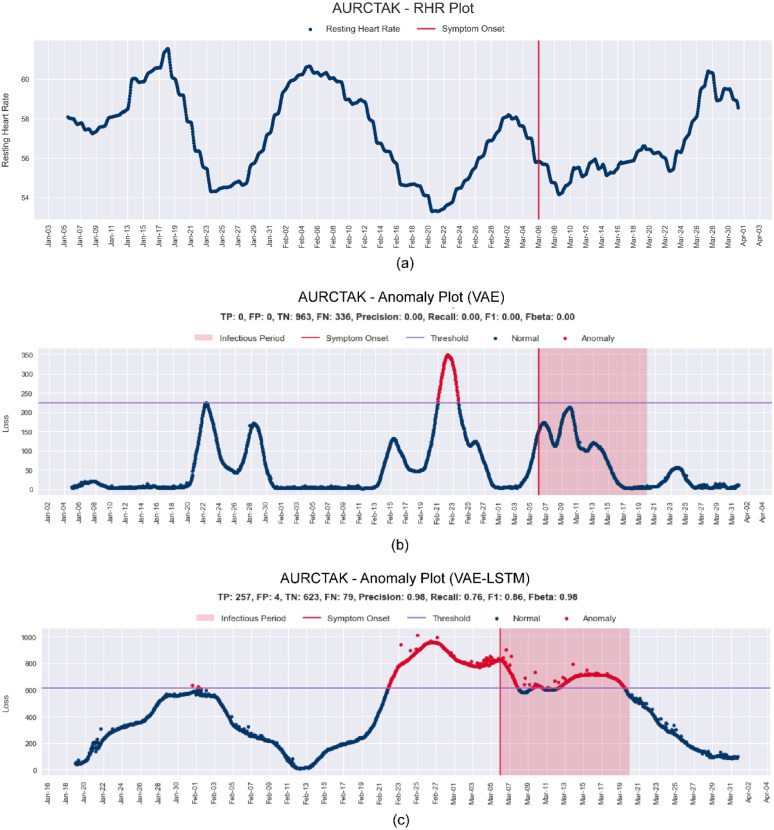
Fig. 9(a) shows RHR data of the subject id AURCTAK from the Phase-1 dataset. Here, no prominent elevation in RHR was found near the symptom onset compared to the baseline region. As both the baseline and the infectious region have an elevation in RHR, the CNN-VAE fails to make any prediction based on the elevation but rather makes an anomaly prediction for the lowest RHR trough. On the contrary, in Fig. 9(c), it is evident that after using the LSTM embeddings, the subtle trends in elevations and decreases of RHR are considered. As a result, the anomaly predictions are well clustered around the symptom onset.
Fig. 9.
(a) RHR Plot and the difference between anomaly plots (b) before and (c) after LSTM embeddings for subject id AURCTAK from the Phase-1 dataset.
4.2. Performance on different test set regions
The results of PCovNet+ are shown here based on the experiment settings described in Section 3.2. Fig. 10 illustrates the regions of the test set for this experiment and Table 2 shows the statistical metrics for the corresponding test set data region. The previous work Abir et al. (2022) and also, Bogu and Snyder (2021) used region (a) for the test set. However, as shown in Table 2, this region has the lowest precision, F-beta, and F-1 than the others.
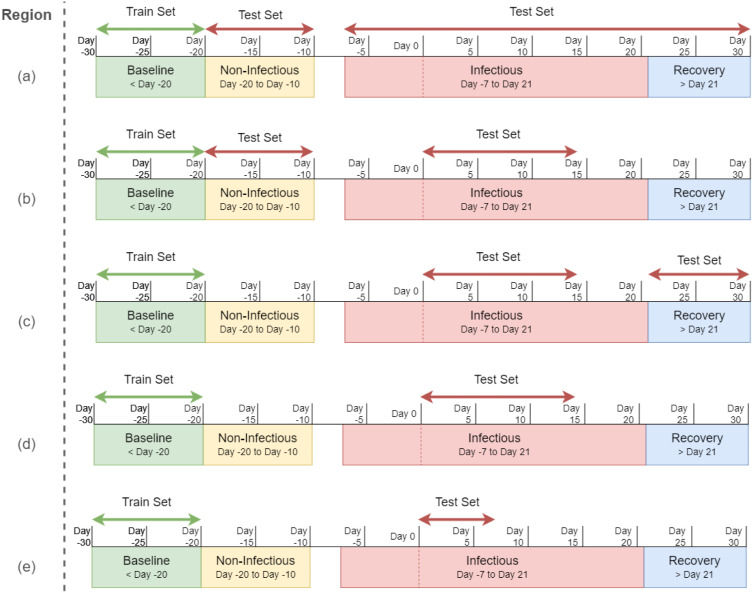
Fig. 10.
Different test sets from the infection periods, where the test set contains (a) all the data after the baseline region, (b) non-infectious period and Day 0 to Day 14, (c) only Day 0 to Day 14 and the recovery period, (d) Day 0 to Day 14, and (d) only Day 0 to Day 7.
Table 2.
CNN-VAE with LSTM embeddings results for different test set data regions.
| Region | Test set day range | Precision | Recall | F-beta | F-1 |
|---|---|---|---|---|---|
| (a) | Day −20 to Day −10 and Day −7 to Day 14 | 0.3180 | 0.4990 | 0.3189 | 0.3784 |
| (b) | Day −20 to Day −10 and Day 0 to Day 14 | 0.7013 | 0.5340 | 0.6987 | 0.5992 |
| (c) | Day 0 to Day 14 and Day 21 | 0.3938 | 0.5340 | 0.3947 | 0.4494 |
| (d) | Day 0 to Day 14 | 0.9930 | 0.4832 | 0.9827 | 0.6501 |
| (e) | Day 0 to Day 7 | 0.9855 | 0.4606 | 0.9744 | 0.6276 |
Region (b) leaves Day −7 to Day 0 and the recovery region in the test set. A significant increase in precision (about 30%), recall (3.5%), F-beta (about 38%), and F-1 (about 22%) can be noted. However, this performance increase can be either for the removal of recovery period data or Day −7 to Day 0 data. This is clarified in region (c), where only Day −20 to Day 0 data is dropped but recovery period data are kept. There is a performance increase in each metric compared to region (a); however, the metrics are far below region (b). So, it can be concluded that the non-infectious period and the Day −7 to Day 0 data include misclassifications.
To further assess the impact of the recovery region, it was removed in region (d). This time, the performance surpassed that of region (b) in precision, F-beta, and F-1 by about 28%. Therefore, it can be deduced that the recovery period data led to significant misclassification. The test data region was further analyzed by only including data from Day 0 to Day 7 in region (e). However, in this case, the metrics slightly decreased compared to those of region (d).
Based on the results, it can be concluded that the non-infectious, recovery, and infectious periods before symptom onset had significant misclassification. However, as previously mentioned, these regions do not have any reliable ground truth and were labeled based on literature. Moreover, due to the nature of the problem, it is highly unlikely to obtain reliable ground truth through laboratory testing. Each subject would need to be tested for COVID-19 infection daily to obtain such data; however, during the non-infectious period, patients are often unaware of their infection.
5. Discussion
The primary object of this work was to detect anomalous RHR using smartwatch data. This framework first processed the HR and steps data to generate RHR of the subjects. After the preprocessing steps, the train data, containing only the normal RHR was used to train the CNN-VAE network. The CNN-VAE network encoder block was used to generate latent vectors to train the LSTM network. The LSTM worked as an embedding generator for the CNN-VAE decoder that takes the latent vector and generates temporal-aware embeddings of the same shape as the latent vector. These embeddings were used to generate a reconstructed signal by the CNN-VAE decoder.
5.1. Comparison with previous study
As mentioned earlier, the PCovNet+ framework was a continuation of the previous work, PCovNet (Abir et al., 2022). However, compared to the previous study, this work only used the min–max threshold estimation (MTE) which decreased the model’s sensitivity but increased the robustness (Abir et al., 2022). To clarify, the model resulted in better statistical metrics but a lower detection rate than MTE. The rationale behind choosing robustness over sensitivity is the false alarms which might cause widespread panic among the users.
Moreover, in this study, the test set range was changed for evaluation compared to the earlier works. As the infectious period varies for each subject, a fixed range might result in some mislabeling, which is explored in detail in Section 4.
In Table 3, the comparison of PCovNet+ performance against two previous works on the same cohort data is presented. However, for PCovNet+, the available dataset is larger than the other one, which is shown in the ‘Number of Subjects’ column of Table 3. A notable increase in performance can be identified from the statistical metrics. In this work, the precision, recall, F-beta, and F-1 are ahead of the second-best result by about 5%, 20%, 7%, and 21%, respectively. This output shows that PCovNet+ surpassed its predecessors by a large margin in terms of robustness.
Table 3.
Comparison of PCovNet+ with the previous work.
| Framework | Precision | Recall | F-beta | F-1 | Early detection | Delayed detection | Failed detection | Number of subjects |
|---|---|---|---|---|---|---|---|---|
| LAAD (Bogu and Snyder, 2021) | 0.894 | 0.331 | 0.895 | 0.483 | 56% | 36% | 8% | 25 |
| PCovNet (STE) (Abir et al., 2022) | 0.904 | 0.274 | 0.883 | 0.421 | 80% | 20% | 0% | 25 |
| PCovNet (MTE) (Abir et al., 2022) | 0.946 | 0.234 | 0.918 | 0.375 | 44% | 44% | 12% | 25 |
| PCovNet+ | 0.993 | 0.534 | 0.985 | 0.693 | 47% | 27% | 26% | 68 |
However, the detection metric is lower than the other ones due to two factors. Firstly, prioritizing robustness over sensitivity by choosing the MTE method to calculate the threshold can be responsible for the decrease in detection rate which was shown in the previous study (Abir et al., 2022). Secondly, the increase in the dataset might contribute to the decrease in the detection rate as well.
5.2. Real-world usability of PCovNet+
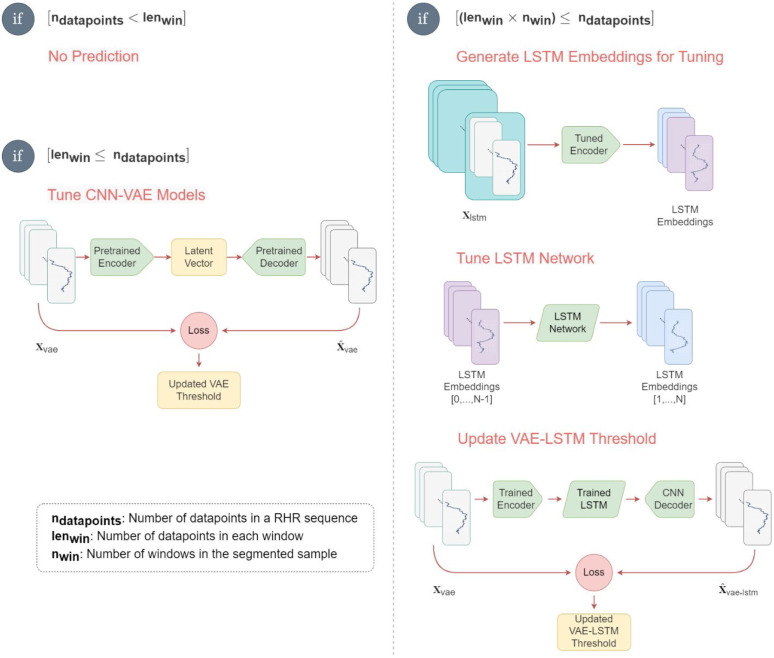
PCovNet+ used healthy subjects’ data to pre-train both the CNN-VAE and the LSTM networks. The loss curves of the pretraining are illustrated in Supplementary Appendix 2. Afterward, the model was fine-tuned on each infected subject’s baseline data to make the model personalized for that subject. The real-world implementation of the PCovNet+ is shown in Fig. 11.
Fig. 11.
Implementation steps of PCovNet+ framework for online learning. Different steps are shown in the diagram based on the availability of data during real-time implementation.
Let denote the number of available RHR data points for a particular subject. and respectively denote the number of data points per window and the number of windows for LSTM embedding generation.
Firstly, while , which is the case at the beginning for any subject, There are not enough RHR data points yet to make any prediction. Since each hour generates an RHR datapoint, for , the model does not give any prediction for the first 2 days.
Secondly, when , there is enough data to make a prediction and at the same time, fine-tune the CNN-VAE network. After each RHR datapoint, a new window is generated at this stage. The threshold will be updated if the user marks his health situation as normal.
Thirdly, the LSTM network is fine-tuned while . At first, the trained CNN-VAE encoder is used to generate LSTM embeddings from all the previous data. Then the embeddings are used to fine-tune the LSTM network. Lastly, the updated VAE-LSTM network is used to generate a loss. Like before, if the subject labels his current health condition as normal, the threshold will be updated.
This anomaly detection framework was trained and fine-tuned only with the normal (baseline) data from the users. If a user feels unwell for a certain range of time during the online learning, RHR during that time must not be used for the finetuning. Hence, user feedback is necessary in this regard.
5.3. Caveats
This study was performed on a cohort size of a few thousand, and only a few had available smartwatch data during their COVID-19 and other respiratory infections. Hence, to obtain a reliable ground truth of their actual infection date, the group would have needed to predict the infected individuals successfully beforehand and perform RT-PCR on each one of them every day. It was an improbable task from a prospective study with resource-constraint settings. On the other hand, only the symptoms and symptom onset date for a retrospective study can be known, but not the actual infection date, at least not without laboratory testing.
To circumvent the ground truth issue, like previous works (Mishra et al., 2020, Bogu and Snyder, 2021, Abir et al., 2022), this work relied on the literature to approximate different regions of infection lifetime. One fatal flaw with this approach is that each infected subject has a different incubation period, viral shedding, and infection type (for some subjects, the anomalous RHR persists long after recovery). Hence, this approximation cannot be regarded as a ground truth.
Another caveat of this work is the dataset diversity. As the study in this field is comparatively new, the volume of the dataset with diversity is still an issue. Hence, the framework developed in this work is not guaranteed to show similar performance on subjects from different regions of the world.
5.4. Implications and future directions
Although the model performance was improved significantly compared to the previous work, there is still room for improvement.
Dataset volume and diversity. In this work, the datasets are generated using some of the most advanced smartwatches and fitness trackers, namely Fitbit, Apple Watch, and Garmin, which incorporate industry-leading wearable sensors. More datasets with diverse wearable devices should be built and explored to achieve a generalized model. Also, having multiple big datasets with diverse populations and devices would create scopes for effective cross-validation of the models as well. Another aspect of achieving generalization is to work on larger datasets. In this regard, this study employs a larger dataset than the previous study; however, it is still not enough in deep learning. The future work includes the creation of a dataset with subjects of diverse nationalities.
Real-world implementation. Since the start of COVID-19, smartwatch-based COVID-19 detection has received special attention from a number of research groups worldwide. Although the COVID-19 infection rate has decreased all over the world after vaccination, we are still prone to respiratory diseases. As shown in the previous work, these anomaly detection systems can work similarly for other respiratory illnesses apart from COVID-19 (Abir et al., 2022). Moreover, any AI system’s usability in the real world often varies from the theoretical results. Hence, the next priority is to deploy the system in the real world and verify the performance to have a complete picture of the framework.
Collaboration. The objective of this work is to help the COVID-19 diagnosis process. However, at the same time, these systems can work as excellent tools for contact tracing for COVID-19 and other respiratory diseases. However, more collaboration of the leading research groups and industry to share data and research findings is needed to approach the goal of making smartwatches and fitness trackers personal healthcare companions.
6. Conclusion
In this work, PCovNet+, a framework for anomalous RHR detection using smartwatch data, was presented which was an improvement of the previous work on the same topic. This study involved a larger dataset of 68 subjects and introduced a new anomaly detection model introducing several new steps from other domains, e.g., LSTM embeddings and model pretraining using Healthy group data. From the experiments and presented data, a significant improvement in the statistical metrics was seen — approximately 5%, 30%, 7%, and 32% increase in precision, recall, F-beta, and F-1, respectively. Moreover, this study further explored the lack of ground truth issue in this work which might impact both the statistical metrics and detection rate significantly. In this regard, the two core research objectives have been addressed.
Overall, this work explores the COVID-19 prediction using smartwatch data further and improves the robustness of the previous work. Even though this system cannot replace laboratory-based active detection systems, it can be used as a secondary diagnostic tool. However, this study shows its crucial usability in regular human life combating respiratory diseases like COVID-19.
CRediT authorship contribution statement
Farhan Fuad Abir: Conceptualization, Methodology, Software, Validation, Original draft preparation. Muhammad E.H. Chowdhury: Supervision, Writing – original draft, Writing – review & editing, Funding acquisition. Malisha Islam Tapotee: Investigation, Writing – original draft. Adam Mushtak: Investigation, Writing – original draft. Amith Khandakar: Investigation, Writing – original draft. Sakib Mahmud: Software, Writing – review & editing. Anwarul Hasan: Supervision, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors would like to show gratitude to Mishra et al. (2020) for providing their study data.
Funding
This work was supported by the Qatar National Research Grant: UREP28-144-3-046. The statements made herein are solely the responsibility of the authors.
Code availability
The working code of the work is provided in the following GitHub repository: https://github.com/farhanfuadabir/PCovNet
Ethical consideration
Since the dataset is collected by a research group from the Department of Genetics of Stanford University and published publicly (Yamaç et al., 2021, Chen et al., 2020), there is no ethical consideration applies for this work.
Footnotes
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.engappai.2023.106130.
Appendix A. Supplementary data
The following is the Supplementary material related to this article.
Supplementary materials for PCovNet+ model.
Data availability
The first phase raw data and symptom onset information was provided by Mishra et al. (2020) which is available in the following link. https://storage.googleapis.com/gbsc-gcp-project-ipop_public/COVID-19/COVID-19-Wearables.zip
Moreover, the second phase data and symptom onset information was given by Alavi et al. and can be downloaded for the following link (Alavi et al., 2022). https://storage.googleapis.com/gbsc-gcp-project-ipop_public/COVID-19-Phase2/COVID-19-Phase2-Wearables.zip
References
- Abadi M., Agarwal A., Barham P., Brevdo E., Chen Z., Citro C., Corrado G.S., Davis A., Dean J., Devin M., Ghemawat S., Goodfellow I., Harp A., Irving G., Isard M., Jia Y., Jozefowicz R., Kaiser L., Kudlur M., Levenberg J., Mane D., Monga R., Moore S., Murray D., Olah C., Schuster M., Shlens J., Steiner B., Sutskever I., Talwar K., Tucker P., Vanhoucke V., Vasudevan V., Viegas F., Vinyals O., Warden P., Wattenberg M., Wicke M., Yu Y., Zheng X. 2016. TensorFlow: Large-scale machine learning on heterogeneous distributed systems. arXiv. [DOI] [Google Scholar]
- Abdelrahman Z., Li M., Wang X. Comparative review of SARS-CoV-2, SARS-CoV, MERS-CoV, and influenza a respiratory viruses. Front. Immunol. 2020;11:2309. doi: 10.3389/fimmu.2020.552909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abir F.F., Alyafei K., Chowdhury M.E.H., Khandaker A., Ahmed R., Hossain M.S., Mahmud S., Rahman A., Abbas T.O., Zughaier S.M., Naji K.K. PCovNet: A presymptomatic COVID-19 detection framework using deep learning model using wearables data. Comput. Biol. Med. 2022 doi: 10.1016/j.compbiomed.2022.105682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavi A., Bogu G.K., Wang M., Rangan E.S., Brooks A.W., Wang Q., Higgs E., Celli A., Mishra T., Metwally A.A., Cha K., Knowles P., Alavi A.A., Bhasin R., Panchamukhi S., Celis D., Aditya T., Honkala A., Rolnik B., Hunting E., Dagan-Rosenfeld O., Chauhan A., Li J.W., Bejikian C., Krishnan V., McGuire L., Li X., Bahmani A., Snyder M.P. Real-time alerting system for COVID-19 and other stress events using wearable data. Nature Med. 2022;28:175–184. doi: 10.1038/s41591-021-01593-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alyafei K., Ahmed R., Abir F.F., Chowdhury M.E.H., Naji K.K. A comprehensive review of COVID-19 detection techniques: From laboratory systems to wearable devices. Comput. Biol. Med. 2022;149 doi: 10.1016/j.compbiomed.2022.106070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amft O., Lopera González L.I., Lukowicz P., Bian S., Burggraf P. Wearables to fight COVID-19: From symptom tracking to contact tracing. IEEE Pervasive Comput. 2020;19:53–60. doi: 10.1109/MPRV.2020.3021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apple Inc. Apple; 2022. Apple Watch. https://www.apple.com/shop/buy-watch/apple-watch (accessed April 19, 2022) [Google Scholar]
- Arias-Londoño J.D., Gómez-García J.A., Moro-Velázquez L., Godino-Llorente J.I. Artificial intelligence applied to chest X-ray images for the automatic detection of COVID-19. A thoughtful evaluation approach. Ieee Access. 2020;8:226811–226827. doi: 10.1109/ACCESS.2020.3044858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine R., Das S., Hasan A., S A., Abdul Salam S., Augustine P., Dalvi Y.B., Varghese R., Primavera R., Yassine H.M., Thakor A.S., Kevadiya B.D. Rapid antibody-based COVID-19 mass surveillance: Relevance, challenges, and prospects in a pandemic and post-pandemic world. J. Clin. Med. 2020;9:3372. doi: 10.3390/jcm9103372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine R., Hasan A., Das S., Ahmed R., Mori Y., Notomi T., Kevadiya B.D., Thakor A.S. Loop-mediated isothermal amplification (LAMP): A rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology. 2020;9:182. doi: 10.3390/biology9080182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogu G.K., Snyder M.P. Deep learning-based detection of COVID-19 using wearables data. MedRxiv. 2021 [Google Scholar]
- Buchhorn R., Baumann C., Willaschek C. Heart rate variability in a patient with coronavirus disease 2019. Int. Cardiovasc. Forum J. 2020 [Google Scholar]
- Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channa A., Popescu N., Skibinska J., Burget R. The rise of wearable devices during the COVID-19 pandemic: A systematic review. Sensors. 2021;21:5787. doi: 10.3390/s21175787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., Chen Q., Huang S., Yang M., Yang X., et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.R., Kim J.H., Yoon H.R., Han Y.S., Kang T.S., Choi H., Lee S. Machine learning-based optimization of pre-symptomatic COVID-19 detection through smartwatch. Sci. Rep. 2022;12:7886. doi: 10.1038/s41598-022-11329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet F., et al. 2015. Keras. https://github.com/fchollet/keras. [Google Scholar]
- 2020. COVID-19 wearables data, phase-1. https://storage.googleapis.com/gbsc-gcp-project-ipop_public/COVID-19/COVID-19-Wearables.zip. [Google Scholar]
- 2021. COVID-19 wearables data, phase-2. https://storage.googleapis.com/gbsc-gcp-project-ipop_public/COVID-19-Phase2/COVID-19-Phase2-Wearables.zip. [Google Scholar]
- Dunn J., Runge R., Snyder M. Wearables and the medical revolution. Pers. Med. 2018;15:429–448. doi: 10.2217/pme-2018-0044. [DOI] [PubMed] [Google Scholar]
- Fitbit . 2022. Fitbit official site for activity trackers and more. https://www.fitbit.com/global/uk/home (accessed April 19, 2022) [Google Scholar]
- Gandhi M., Yokoe D.S., Havlir D.V. Asymptomatic transmission, the achilles’ heel of current strategies to control Covid-19. N. Engl. J. Med. 2020;382:2158–2160. doi: 10.1056/NEJMe2009758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmin . 2022. Smartwatches with fitness and health tracking Garmin. https://www.garmin.com/en-US/c/sports-fitness/activity-fitness-trackers/ (accessed April 19, 2022) [Google Scholar]
- Gianola S., Jesus T.S., Bargeri S., Castellini G. Chracteristics of academic publications, preprints, and registered clinical trials on the COVID-19 pandemic. PLOS ONE. 2020;15 doi: 10.1371/journal.pone.0240123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S., et al. Clinical characteristics of 2019 novel coronavirus infection in China. MedRxiv. 2020 [Google Scholar]
- Haldane V., De Foo C., Abdalla S.M., Jung A.-S., Tan M., Wu S., Chua A., Verma M., Shrestha P., Singh S., et al. Health systems resilience in managing the COVID-19 pandemic: lessons from 28 countries. Nat. Med. 2021:1–17. doi: 10.1038/s41591-021-01381-y. [DOI] [PubMed] [Google Scholar]
- Harris C.R., Millman K.J., van der Walt S.J., Gommers R., Virtanen P., Cournapeau D., Wieser E., Taylor J., Berg S., Smith N.J., Kern R., Picus M., Hoyer S., van Kerkwijk M.H., Brett M., Haldane A., del Río J.F., Wiebe M., Peterson P., Gérard-Marchant P., Sheppard K., Reddy T., Weckesser W., Abbasi H., Gohlke C., Oliphant T.E. Array programming with NumPy. Nature. 2020;585:357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Lau E.H., Wu P., Deng X., Wang J., Hao X., Lau Y.C., Wong J.Y., Guan Y., Tan X., et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- Hunter J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]
- Iwana B.K., Uchida S. Time series data augmentation for neural networks by time warping with a discriminative teacher. 2020 25th International Conference on Pattern Recognition; ICPR; IEEE; 2021. pp. 3558–3565. [Google Scholar]
- Jabir B., Falih N., Rahmani K. Accuracy and efficiency comparison of object detection open-source models. Int. J. Online Biomed. Eng. 2021;17 [Google Scholar]
- Kingma D.P., Ba J. 2014. Adam: A method for stochastic optimization. ArXiv Preprint arXiv:1412.6980. [Google Scholar]
- Kingma D.P., Welling Max. 2022. Auto-encoding variational Bayes. [DOI] [Google Scholar]
- Kiran B.R., Thomas D.M., Parakkal R. An overview of deep learning based methods for unsupervised and semi-supervised anomaly detection in videos. J. Imaging. 2018;4:36. doi: 10.3390/jimaging4020036. [DOI] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus–Infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Clark R., Birke R., Schönborn S., Trigoni N., Roberts S. Anomaly detection for time series using VAE-LSTM hybrid model. ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing; ICASSP; 2020. pp. 4322–4326. [DOI] [Google Scholar]
- Liu S., Han J., Puyal E.L., Kontaxis S., Sun S., Locatelli P., Dineley J., Pokorny F.B., Dalla Costa G., Leocani L., et al. Fitbeat: COVID-19 estimation based on wristband heart rate using a contrastive convolutional auto-encoder. Pattern Recognit. 2021 doi: 10.1016/j.patcog.2021.108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A., Pottie G., Kaiser W. Transformation in healthcare by wearable devices for diagnostics and guidance of treatment. ACM Trans. Comput. Healthc. 2020;1:1–12. [Google Scholar]
- Mazumder H., Hossain M.M., Das A. Geriatric care during public health emergencies: lessons learned from novel corona virus disease (COVID-19) pandemic. J. Gerontol. Soc. Work. 2020;63:257–258. doi: 10.1080/01634372.2020.1746723. [DOI] [PubMed] [Google Scholar]
- McKinney, W., 2010. Data Structures for Statistical Computing in Python. Austin, TX, pp. 51–56.
- Merrill M.A., Althoff T. 2022. Self-supervised pretraining and transfer learning enable flu and COVID-19 predictions in small mobile sensing datasets. [DOI] [Google Scholar]
- Mishra T., Wang M., Metwally A.A., Bogu G.K., Brooks A.W., Bahmani A., Alavi A., Celli A., Higgs E., Dagan-Rosenfeld O., et al. Pre-symptomatic detection of COVID-19 from smartwatch data. Nat. Biomed. Eng. 2020;4:1208–1220. doi: 10.1038/s41551-020-00640-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitratza M., Goodale B.M., Shagadatova A., Kovacevic V., van de Wijgert J., Brakenhoff T.B., Dobson R., Franks B., Veen D., Folarin A.A., Stolk P., Grobbee D.E., Cronin M., Downward G.S. The performance of wearable sensors in the detection of SARS-CoV-2 infection: a systematic review. Lancet Digit. Health. 2022;4:e370–e383. doi: 10.1016/S2589-7500(22)00019-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- Piwek L., Ellis D.A., Andrews S., Joinson A. The rise of consumer health wearables: promises and barriers. PLoS Med. 2016;13 doi: 10.1371/journal.pmed.1001953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev A., Tyapochkin K., Surkova E., Smorodnikova E., Pravdin P. Heart rate variability as a prospective predictor of early COVID-19 symptoms. MedRxiv. 2021 [Google Scholar]
- Qiblawey Y., Tahir A., Chowdhury M.E., Khandakar A., Kiranyaz S., Rahman T., Ibtehaz N., Mahmud S., Maadeed S.A., Musharavati F., et al. Detection and severity classification of COVID-19 in CT images using deep learning. Diagnostics. 2021;11:893. doi: 10.3390/diagnostics11050893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quer G., Radin J.M., Gadaleta M., Baca-Motes K., Ariniello L., Ramos E., Kheterpal V., Topol E.J., Steinhubl S.R. Wearable sensor data and self-reported symptoms for COVID-19 detection. Nat. Med. 2021;27:73–77. doi: 10.1038/s41591-020-1123-x. [DOI] [PubMed] [Google Scholar]
- Radin J.M., Wineinger N.E., Topol E.J., Steinhubl S.R. Harnessing wearable device data to improve state-level real-time surveillance of influenza-like illness in the USA: a population-based study. Lancet Digit. Health. 2020;2:e85–e93. doi: 10.1016/S2589-7500(19)30222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roblyer D.M. Perspective on the increasing role of optical wearables and remote patient monitoring in the COVID-19 era and beyond. JBO. 2020;25 doi: 10.1117/1.JBO.25.10.102703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri D.R., Davies E.V., Harlow E.R., Hsu J.J., Knighton S.C., Walker T.A., Voos J.E., Drummond C.K. Wearable sensors for COVID-19: A call to action to harness our digital infrastructure for remote patient monitoring and virtual assessments. Front. Digit. Health. 2020;2 doi: 10.3389/fdgth.2020.00008. https://www.frontiersin.org/articles/10.3389/fdgth.2020.00008 (accessed October 2, 2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir A., Qiblawey Y., Khandakar A., Rahman T., Khurshid U., Musharavati F., Islam M., Kiranyaz S., Chowdhury M. 2021. Deep learning for reliable classification of COVID-19, MERS, and SARS from chest X-ray images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The pandas development team . 2020. Pandas-dev/pandas: Pandas. [DOI] [Google Scholar]
- Um T.T., Pfister F.M., Pichler D., Endo S., Lang M., Hirche S., Fietzek U., Kulić D. Proceedings of the 19th ACM International Conference on Multimodal Interaction. 2017. Data augmentation of wearable sensor data for parkinson’s disease monitoring using convolutional neural networks; pp. 216–220. [Google Scholar]
- 2022. The end of the COVID-19 pandemic is in sight: WHO, UN News. https://news.un.org/en/story/2022/09/1126621 (accessed October 2, 2022) [Google Scholar]
- Vogels E.a. Pew Research Center; 2022. About One-in-Five Americans Use a Smart Watch or Fitness Tracker. https://www.pewresearch.org/fact-tank/2020/01/09/about-one-in-five-americans-use-a-smart-watch-or-fitness-tracker/ (accessed October 2, 2022) [Google Scholar]
- Wang Z., Xiao Y., Li Y., Zhang J., Lu F., Hou M., Liu X. Automatically discriminating and localizing COVID-19 from community-acquired pneumonia on chest X-rays. Pattern Recognit. 2021;110 doi: 10.1016/j.patcog.2020.107613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskom M.L. Seaborn: statistical data visualization. J. Open Source Softw. 2021;6:3021. [Google Scholar]
- 2022. WHO coronavirus (COVID-19) dashboard. https://covid19.who.int (accessed October 2, 2022) [Google Scholar]
- Wu Z., Harrich D., Li Z., Hu D., Li D. The unique features of SARS-CoV-2 transmission: Comparison with SARS-CoV, MERS-CoV and 2009 H1N1 pandemic influenza virus. Rev. Med. Virol. 2021;31 doi: 10.1002/rmv.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaç M., Ahishali M., Degerli A., Kiranyaz S., Chowdhury M.E., Gabbouj M. Convolutional sparse support estimator-based COVID-19 recognition from X-ray images. IEEE Trans. Neural Netw. Learn. Syst. 2021;32:1810–1820. doi: 10.1109/TNNLS.2021.3070467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials for PCovNet+ model.
Data Availability Statement
The first phase raw data and symptom onset information was provided by Mishra et al. (2020) which is available in the following link. https://storage.googleapis.com/gbsc-gcp-project-ipop_public/COVID-19/COVID-19-Wearables.zip
Moreover, the second phase data and symptom onset information was given by Alavi et al. and can be downloaded for the following link (Alavi et al., 2022). https://storage.googleapis.com/gbsc-gcp-project-ipop_public/COVID-19-Phase2/COVID-19-Phase2-Wearables.zip