OBJECTIVES:
Perturbed host metabolism is increasingly recognized as a pillar of sepsis pathogenesis, yet the dynamic alterations in metabolism and its relationship to other components of the host response remain incompletely understood. We sought to identify the early host-metabolic response in patients with septic shock and to explore biophysiological phenotyping and differences in clinical outcomes among metabolic subgroups.
DESIGN:
We measured serum metabolites and proteins reflective of the host-immune and endothelial response in patients with septic shock.
SETTING:
We considered patients from the placebo arm of a completed phase II, randomized controlled trial conducted at 16 U.S. medical centers. Serum was collected at baseline (within 24 hr of the identification of septic shock), 24-hour, and 48-hour postenrollment. Linear mixed models were built to assess the early trajectory of protein analytes and metabolites stratified by 28-day mortality status. Unsupervised clustering of baseline metabolomics data was conducted to identify subgroups of patients.
PATIENTS:
Patients with vasopressor-dependent septic shock and moderate organ dysfunction that were enrolled in the placebo arm of a clinical trial.
INTERVENTIONS:
None.
MEASUREMENTS AND MAIN RESULTS:
Fifty-one metabolites and 10 protein analytes were measured longitudinally in 72 patients with septic shock. In the 30 patients (41.7%) who died prior to 28 days, systemic concentrations of acylcarnitines and interleukin (IL)-8 were elevated at baseline and persisted at T24 and T48 throughout early resuscitation. Concentrations of pyruvate, IL-6, tumor necrosis factor-α, and angiopoietin-2 decreased at a slower rate in patients who died. Two groups emerged from clustering of baseline metabolites. Group 1 was characterized by higher levels of acylcarnitines, greater organ dysfunction at baseline and postresuscitation (p < 0.05), and greater mortality over 1 year (p < 0.001).
CONCLUSIONS:
Among patients with septic shock, nonsurvivors exhibited a more profound and persistent dysregulation in protein analytes attributable to neutrophil activation and disruption of mitochondrial-related metabolism than survivors.
Keywords: biomarkers, chemokines, cytokines, metabolomics, precision medicine
KEY POINTS
Question: What is the relationship between metabolites and their changes over time with mortality and the host response in patients with septic shock?
Findings: Acylcarnitines were elevated at baseline and throughout the first 48 hours of shock in patients who died prior to 28 days. Greater inflammation was associated with metabolic dysfunction, and metabolic subgroups had distinct patterns of organ function and differential mortality.
Meaning: Persistent disruption in mitochondrial-related metabolism early in the course of septic shock is associated with poor patient outcomes, but further work with longer follow-up is needed to fully explore the metabolic underpinnings in sepsis.
Sepsis is a syndrome characterized by a dysregulated host response to an infection, which manifests as life-threatening multiorgan dysfunction (1). The clinical trajectory and outcomes for patients with sepsis is highly heterogeneous, with mortality in septic shock exceeding 40% (2). The mechanisms underlying this variability in patient outcomes remain poorly understood, threatening the development of novel pharmacotherapy and improvement of care for patients with sepsis (3). Identification of the biological mechanisms underlying sepsis heterogeneity holds promise in personalizing care for patients with sepsis through predictive and prognostic enrichments, disease subphenotyping, and rational drug discovery (4).
Altered metabolism and host bioenergetics are increasingly recognized as cornerstones of sepsis pathophysiology and sepsis heterogeneity (5). The Surviving Sepsis Campaign recently listed an improved understanding of sepsis-induced metabolic disruptions as one of five key translational research questions in the field, and elevated lactate concentrations are associated with mortality and are formally codified in the definition of septic shock (1, 6). Clearance of lactate has also been proposed as a metabolic biomarker and physiologic endpoint of randomized controlled clinical trials (7–9), yet the kinetics and bioenergetics of lactate are complex, and remediation of elevated concentrations has not consistently translated to improved survival (10–13). This stresses the need for additional prognostic and predictive metabolic biomarkers beyond lactate in patients with sepsis.
Metabolomics is a discovery science that seeks to identify and quantify small molecules in a given biospecimen (14). In sepsis and other critical illnesses, metabolomics studies have consistently identified differences among patients stratified by mortality and organ function (15–17), and shown promise in understanding variable response to treatment (18, 19). These studies have implicated mitochondrial dysfunction, protein catabolism, and perturbed lipid metabolism as potential drivers of poor patient outcomes (20). However, most studies in critical care metabolomics have been cross-sectional, leveraging single blood samples of convenience, and included patients with highly variable illness severity and/or diagnosis at presentation.
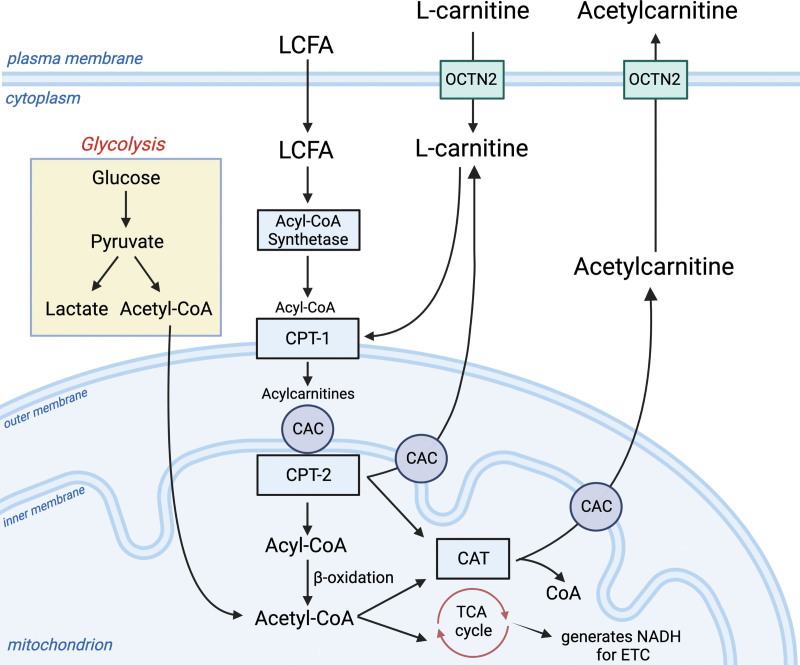
In this study, we analyzed longitudinal serum samples from patients in the placebo arm of the Rapid Administration of Carnitine in sEpsis (RACE) clinical trial (21). Our primary goal was to describe the early host-metabolic response in patients with vasopressor-dependent shock using existing metabolomics data and obtained inflammatory protein measurements. Since RACE assessed an intervention with supplemental L-carnitine, the metabolomics data set included measurements of acylcarnitines (fatty-acid esters that are formed when fatty acids conjugate with L-carnitine in the mitochondria as shown in Fig. 1). We and others have previously shown that sepsis nonsurvivors have elevated blood levels of acylcarnitines compared with survivors and that blood levels of acylcarnitines are associated with sepsis-induced organ dysfunction (15, 17, 19, 22, 23). This signal is attributed to a disruption in mitochondrial fatty-acid metabolism (24). As such, blood levels of acylcarnitines along with abundant polar compounds (e.g., amino acids) could provide mechanistic insights to sepsis-induced perturbations in metabolism. In a secondary analysis, we also sought to determine if metabolomics data alone, agnostic to any clinical or demographic patient characteristics, could identify clusters or subphenotypes of septic shock and assess the relationship between metabolites and protein biomarkers of the host response.
Figure 1.
Glycolysis and mitochondrial-related metabolism. L-carnitine is required for the transport of long-chain fatty acids (LCFA) into the mitochondrion. This process enlists the carnitine shuttle, which begins with the entrance of L-carnitine into the cell from the blood through the organic cation transporter (OCTN2). LCFAs enter the cell and are converted to long-chain acyl-CoAs by acyl-CoA synthetase (EC 6.2.1.3). The L-carnitine shuttle apparatus uses L-carnitine via carnitine palmitoyl transferase 1 (CPT1; EC 2.3.1.21) to convert L-carnitine and LCFA-CoAs to acylcarnitines. The transporter, carnitine-acylcarnitine carrier (CAC; SLC25A20), moves the newly formed long-chain acylcarnitines into the mitochondrial matrix in exchange for free carnitine. Here, long-chain acyl groups are transferred back to CoA by CPT2 (EC 2.3.1.21), which regenerates Acyl-CoA for use in β-oxidation (sepsis-induced dysfunction of β-oxidation can lead to elevations in acylcarnitines). This process also generates L-carnitine. Here, L-carnitine can either be transported out of the mitochondrion by CAC or used as a substrate for carnitine acetyl-transferase (CAT; EC2.3.1.7), which converts it and Acetyl-CoA to acetylcarnitine. Acetylcarnitine moves through CAC and OCTN2 back into the bloodstream. This process may be enhanced during sepsis and times of metabolic stress, serving as a crucial sink for excess acetyl groups that may be toxic to the cell. The Acetyl-CoA produced by β-oxidation sources the tricarboxylic acid (TCA) cycle as does the Acetyl-CoA produced from the oxidative decarboxylation of pyruvate in the mitochondrial matrix. NADH = nicotinamide adenine dinucleotide (NAD) + hydrogen (H); CoA = coenzyme A; ETC = electron transport chain. This figure was created using Biorender.com.
MATERIALS AND METHODS
Study Population and Design
We performed a secondary analysis of patients who were randomized to the saline placebo arm of the RACE clinical trial (21). Abbreviated inclusion criteria for the parent trial included: 1) enrollment within 24 hours of the identification of septic shock, 2) a blood lactate level exceeding 18 mg/dL, 3) a Sequential Organ Failure Assessment (SOFA) score of at least six, and 4) receipt of high-dose vasopressors within 4 hours of enrollment (25). Patients in the placebo arm received a 20-mL bolus of 0.9% normal saline followed by a 1-liter 12-hour infusion, rather than an equivalent volume of L-carnitine. Patients in this analysis were also required to have at least one serum sample available for metabolomics. We excluded patients who were allocated to receive intravenous L-carnitine to focus on the metabolic trajectory of the disease course without any interference that may be introduced by the study drug. Furthermore, we have previously identified phenotypes of L-carnitine response (19). The trial protocol was approved by the institutional review board (IRB) of all 16 centers and registered with clinicaltrials.gov (NCT01665092) prior to patient enrollment. All patients or their legally authorized representative provided informed consent prior to randomization. Procedures were followed in accordance with the ethical standards of the responsible institutional committees on human experimentation at each clinical site and with the Helsinki Declaration of 1975. Data generated as part of the ancillary study to the parent clinical trial was done using deidentified biospecimens that cannot be linked to a specific individual by the investigator(s) directly or indirectly through a coding system. As such, the study protocol (HUM00104311) was deemed “not regulated” by the University of Michigan’s IRB, and IRB approval was not required; a letter of waiver was issued on July 14, 2015.
Serum Sampling and Assays
Blood samples were collected at enrollment (baseline or day 0), 24 hours (day 1), and 48 hours (day 2) after saline initiation (Supplementary Fig. 1, http://links.lww.com/CCX/B155). Serum was obtained from whole blood, frozen at –80°C, and shipped on dry ice to the University of Michigan College of Pharmacy Nuclear Magnetic Resonance (NMR) Metabolomics Laboratory to await metabolomics assays. Further details regarding sample processing from this trial have been reported elsewhere (19, 26).
For our analysis, we used existing metabolomics data that were generated as part of an ancillary metabolomics study to the RACE trial (19). We employed the same metabolomics strategy as our prior work, which included the measurement of low-molecular-weight, polar metabolites as measured by NMR spectroscopy using previously described methods (18). Briefly, spectra were acquired on a Varian (now Agilent, Santa Clara, CA) 11.74 Tesla (500 MHz) NMR spectrometer (27). Spectra were acquired at room temperature using a 1 H,1 H- nuclear overhauser effect spectroscopy (NOESY) pulse sequence and analyzed using the Processor and Profiler modules of the Chenomx NMR Suite 8.2 (Chenomx, Edmonton, AB, Canada) software. Acylcarnitines, which are esters formed by the conjugation of fatty acids with L-carnitine (24), were measured by reverse-phase liquid chromatography with tandem mass spectrometry (LC-MS/MS) (23). Internal standards (NSK-B, Cambridge Isotope Laboratories, Tewksbury, MA) were used to allow for absolute quantification of L-carnitine, acetylcarnitine (C2), and several acylcarnitines (C3, C4, C5, C8, C14, and C16), whereas other acylcarnitine species were measured using relative quantification by peak area.
Residual serum from the same samples was used to measure inflammatory and immune-related cytokines of the host response on a Milliplex magnetic bead immunoassay panel run on a Luminex 200 Instrument (Thermo Fisher Scientific, Waltham, MA). Angiopoietin-2 (ANG-2) was measured using a commercially available enzyme-linked immunoassay (Invitrogen, Thermo Fisher Scientific) in accordance with the manufacturer’s instructions.
Statistical Analyses and Outcomes
Missing data for metabolites quantified by NMR and protein markers were assumed to be left-censored and below the limit of detection (28). For each analyte, we imputed missing observations as the minimum concentration observed divided by two. Following imputation, concentrations were log-transformed and z-scaled to have a mean of zero and sd of one. There were no missing data for L-carnitine, acetylcarnitine, or the acylcarnitines quantified by LC-MS.
For the primary analysis, we fit a series of general linear mixed models to assess metabolite or protein concentrations over time in patients stratified by their mortality status at 28 days. Similar approaches have been successfully used in the analysis of multiomics biomarker data in other cohorts of patients with critical illness (29, 30). For each metabolite, we built: 1) a null model consisting of a patient-level (random) intercept, 2) a fixed-effect model, which added “Time” (baseline, day 1, and day 2) and “Mortality Status” (survivor vs nonsurvivor at 28 d), and 3) an interaction model, which further added a “Time*Mortality” interaction. All models contained patient age, sex, and baseline SOFA Score as covariates. Equations for each analyte concentration at time (t) in patient (i) were modeled as such (please note each model also contained fixed-effect parameters for age, sex, and baseline SOFA):
Null Model: Analytet,i = β0 + b0,i
Fixed-Effect Model: Analytet,i = β0 + βTime * Timepointt + βMortality * Mortalityi + b0,i
Interaction Model: Analytet,i = β0 + βTime * Timepointt + βMortality * Mortalityi + βinteraction * Timepointt * Mortalityi + b0,i
Models were compared based on the approximate F test according to the Kenward-Roger approach with the “pbkrtest” R package (31, 32). We selected between the fixed-effect and interaction models, opting for the latter when the interaction p value was less than 0.05. The overall p value was then determined by comparing the selected model to the null model. The resulting p values were rank-ordered to determine the most perturbed signals and corrected for multiple comparison according to the false discovery procedure of Benjamini and Hochberg (33). Analyte concentrations of sepsis survivors and nonsurvivors were visualized on the log10-scale, with differences at each time point assessed by the Mann-Whitney U test. We assessed the correlation between metabolites and protein biomarkers over time using repeated-measure correlation analysis using the “Rmcorr” R package (34).
We also sought to identify if there were clusters or metabolic subtypes of patients with septic shock at trial enrollment. We employed an unsupervised clustering approach (i.e., naïve to patient demographics, outcomes, and clinical status) using baseline metabolomics data and the K-means algorithm, with Euclidean distance (35). To prevent overfitting, clustering was done on the first five principal components of the metabolomics data, representing ~70% of the variance in the dataset. The optimal number of clusters was selected using the Silhouette method and the “NbClust” R package (36). We tested clustering performance by maximizing the average Silhouette width with k ranging from one to 10 clusters. We also assessed the stability of the optimal number of clusters and the assignment of individual patients to subgroups using the full metabolomics dataset, given recent calls to consider the use of principal component analysis (PCA) and dimensionality reduction (37). Individual metabolites driving cluster separation were visualized using the “ComplexHeatMap” R package (38).
We assessed for patient differences in demographics, comorbidities, and organ dysfunction between metabolic clusters using linear mixed models, the Mann-Whitney U test, or chi-square test, as appropriate. We also tested the impact of metabolic cluster assignment on mortality status at 28 days using logistic regression and on mortality out to 1 year using Kaplan-Meir analysis and the log-rank test. All data analysis and figure generation were completed using the statistical programming language R (Version 4.1.0) (39). All raw data and the code used to conduct our analysis are available on Github (https://github.com/UMichNMR-Metabolomics, accessed June 22, 2022).
RESULTS
Patient Characteristics, Metabolites, and Protein Analytes of the Host Response
Seventy-two patients from the RACE trial were randomized to receive saline placebo and had at least one serum sample available for metabolomics (Supplementary Fig. 1, http://links.lww.com/CCX/B155). Of these, 30 patients (41.7%) died within 28 days of trial enrollment. Patients who died had greater organ dysfunction but were similar with respect to age, sex, and baseline comorbidities as measured by a modified version of the Charlson comorbidity index (40, 41) (Table 1).
TABLE 1.
Patient Demographics, Laboratory Values, and Physiologic Parameters Stratified by 28-d Mortality
| Patient Characteristic | 28-d Survivor, n = 42a | 28-d Nonsurvivor, n = 30a | P b |
|---|---|---|---|
| Age (yr) | 62 (54–70) | 62 (47–72) | 0.8 |
| Sex | |||
| Female | 22 (52%) | 13 (43%) | 0.4 |
| Male | 20 (48%) | 17 (57%) | |
| Self-reported race | |||
| African American | 13 (31%) | 9 (30%) | > 0.9 |
| Caucasian | 29 (69%) | 21 (70%) | |
| Total Sequential Organ Failure Assessment score | 11 (7–13) | 13 (9–15) | 0.016 |
| Charlson comorbidity index | 4.00 (2.25–4.75) | 5.00 (3.00–5.75) | 0.4 |
| Clinical lactate (mmol/L) | 3.15 (2.58–6.20) | 4.20 (2.90–6.50) | 0.9 |
| Unknown (n) | 6 | 5 | |
| Creatinine (mg/dL) | 1.90 (1.47–2.70) | 1.81 (1.12–2.88) | 0.6 |
| Platelet count (cells/mm3) | 165 (101–227) | 146 (75–215) | 0.4 |
| Unknown (n) | 0 | 1 | |
| Total bilirubin (mg/dL) | 0.95 (0.40–1.99) | 2.40 (0.70–4.20) | 0.005 |
| Unknown (n) | 0 | 1 | |
| WBC Count (cells/mm3) | 25 (14–31) | 18 (12–25) | 0.7 |
| Unknown (n) | 9 | 9 | |
| Body mass index | 29 (23–37) | 24 (21–35) | 0.14 |
| Unknown (n) | 0 | 1 | |
| Respiratory rate (breaths/min) | 20 (17–24) | 20 (16–26) | 0.7 |
| Heart rate (beats/min) | 101 (94–113) | 104 (92–114) | > 0.9 |
| Cumulative vasopressor index | 4 (3–8) | 6 (4–8) | 0.092 |
| Unknown (n) | 2 | 1 | |
Described as median (interquartile range) for continuous variables and n (%) for categorical variables
Calculatd by one-way analysis of variance for continuous variables or Pearson χ2 test for categorical variables.
A total of 51 metabolites and 10 protein biomarkers of the host response were measured at baseline, 24 hours, and 48 hours. Metabolites included L-carnitine, acetylcarnitine, and 22 acylcarnitines measured by LC-MS and 27 small, predominately polar, molecules by NMR spectroscopy. Protein analytes included markers of the host-immune and inflammatory response and endothelial cell activation. A full list of the measured analytes and the analytical platform used are provided in the first two columns of Supplementary Table 1 (http://links.lww.com/CCX/B155).
Early Host Response Over Time Stratified by 28-Day Mortality Status
We fit linear mixed models for each protein analyte and metabolite and found the inclusion of fixed effects for time and mortality status improved model fit across 39 (76%) metabolites and 10 (100%) protein markers of the host response (overall false discovery rate < 0.05). This indicates the concentration of analytes and metabolites indicative of the host response significantly changed throughout the early course of shock and/or by a patient’s mortality status. In addition, the linear trajectory of 14 of these analytes and metabolites varied based on mortality status, as indicated by a significant “Time*Mortality” interaction (p < 0.05).
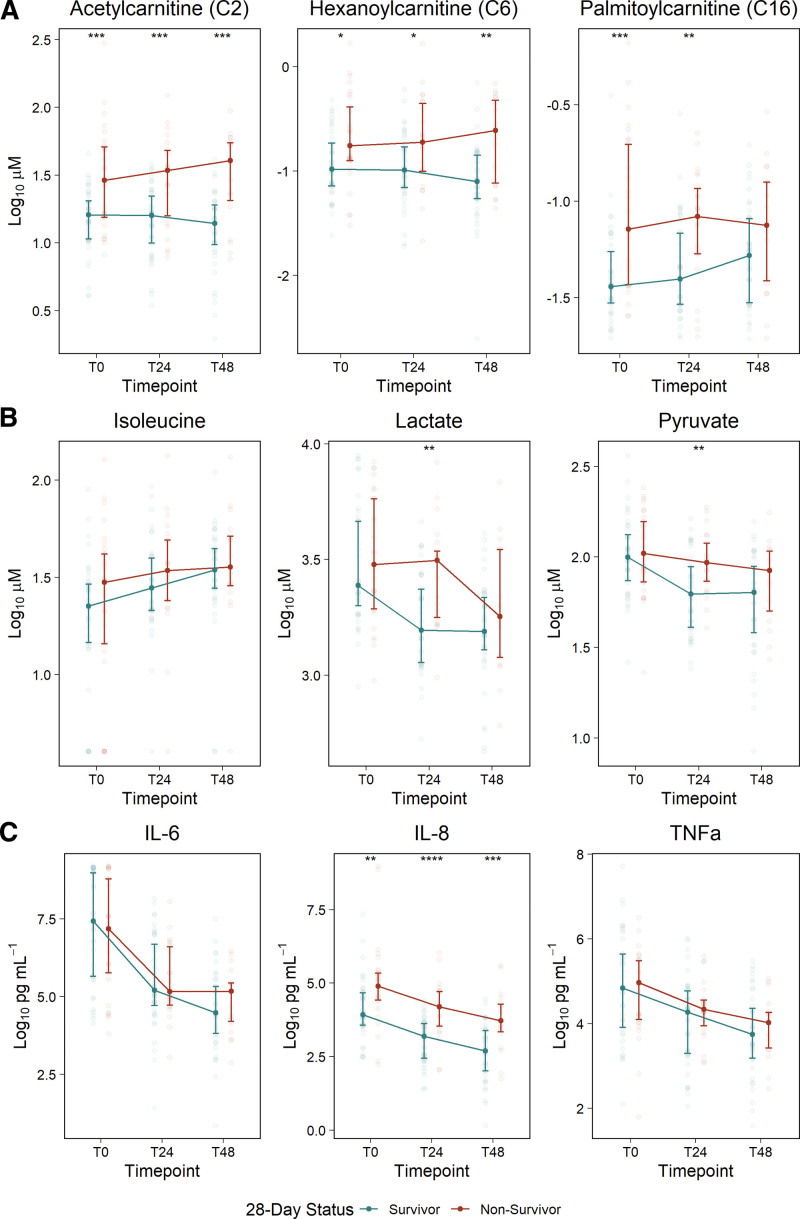
The most perturbed signals across analytical platforms are highlighted in Figure 2 and included acetylcarnitine (C2) the medium-chain acylcarnitine, C6, and the long-chain acylcarnitine, C16 (LC-MS; Fig. 2A); lactate, pyruvate, and isoleucine (NMR; Fig. 2B); and interleukin (IL)-6, IL-8, and tumor necrosis factor (TNF)-α (protein immunoassays; Fig. 2C). Patients who died experienced greater inflammation and disruption in mitochondrial-related metabolism that were sustained over the 2 days postenrollment, as evidenced by persistent elevations in IL-8 and acylcarnitines, regardless of chain length. IL-6, TNF-α, and pyruvate concentrations were similar at individual time points (Mann-Whitney U test; p > 0.05), but decreased, or were cleared, at a much slower rate in patients who died, as indicated by a significant and positive “Time*Mortality” interaction (βTime*Mortality > 0; p < 0.05). Lactate levels tended to steadily decline in both groups (βTime = –0.456; 95% CI, –0.583 to –0.33), whereas isoleucine levels increased (βTime = 0.346; 95% CI, 0.223–0.469).
Figure 2.
Temporal changes in metabolites and inflammatory cytokines in 28-d septic shock survivors and non-survivors. The most perturbed analytes across time and 28-d mortality status measured using three analytical platforms: A, acylcarnitines measured by liquid chromatography-mass spectrometry; B, metabolites measured by nuclear magnetic resonance; and C, Proteins measured by immunoassays. Analyte concentrations at each time point are visualized with the median ± 25th and 75th percentiles, with differences between mortality groups assessed by the Mann-Whitney U test. Analytes presented here were chosen according to the rank-ordered overall p value from linear mixed modeling as described in the Materials and Methods section. IL = interleukin.
The full linear mixed modeling results for all protein analytes and metabolites are shown in Table S1 (http://links.lww.com/CCX/B155). Numerous metabolites were elevated in patients who did not survive past 28 days, indicated by a βMortality coefficient significantly greater than zero. This included many acylcarnitines of varying chain length, 2-hydroxybutyrate and 3-hydroxybutyrate (a ketone body), propylene glycol, and amino acids (methionine, glycine, alanine, tyrosine, and glutamine). Among patients who died, concentrations of ANG-2 decreased more slowly (βTime*Mortality > 0), whereas concentrations of nine long-chain acylcarnitines decreased more rapidly (βTime*Mortality < 0) than in survivors.
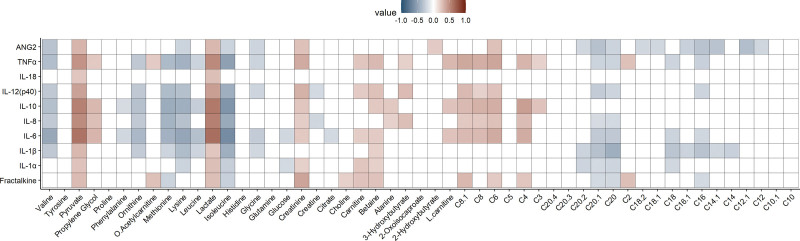
Repeated-measure correlations between protein analytes and metabolites are shown in Figure 3. Notably, strong positive correlations included those between numerous inflammatory cytokines and markers of endothelial activation (ANG-2 and fractalkine) with lactate and pyruvate. In contrast, branched-chain and numerous other amino acids tended to be negatively correlated with inflammatory cytokines. Similarly, long-chain acylcarnitines were negatively correlated with endothelial markers.
Figure 3.
Repeated-measure correlations between protein and metabolite analytes. The Rmcorr R package was used to determine the repeated-measure correlation coefficient between proteins and metabolites measured across three time points. The ggcorrplot R package was used to visualize the results. Positive correlations are indicated in red, while negative correlations are shown in blue, and only significant (p < 0.05) correlation pairs are included. ANG2 = angiopoietin-2; TNFα = tumor necrosis factor-alpha; IL = interleukin.
Baseline Metabolic Clustering
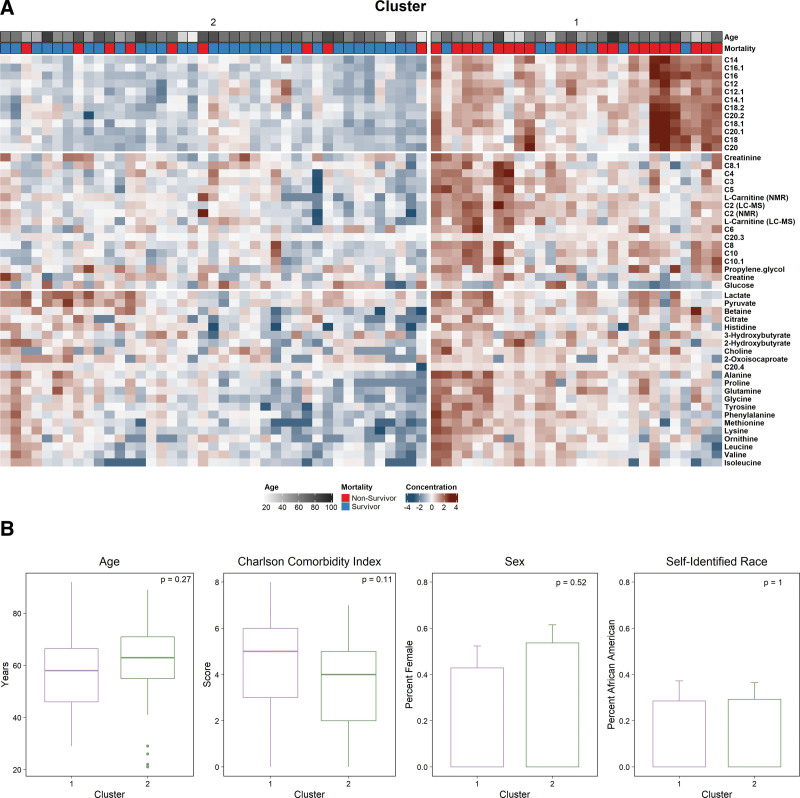
Unsupervised clustering of baseline metabolomics data revealed two distinct groups of patients, with 28 patients assigned to cluster 1 and 41 to cluster 2. Two clusters were selected as the optimal number for k in both the PCA dimensionality-reduced dataset and in the full metabolomics data (Supplementary Figs. 2, A and B, http://links.lww.com/CCX/B155). Patients were assigned to the same cluster regardless of the method used (Supplementary Fig. 2C, http://links.lww.com/CCX/B155). The heat map of metabolites driving separation demonstrates significant metabolic heterogeneity among patients with septic shock at trial enrollment (Fig. 4A). Clusters differed most dramatically across medium- and long-chain acylcarnitines. Across clusters, patient age, sex, self-identified race, and baseline comorbidities were similar (Fig. 4B).
Figure 4.
Metabolite concentration and patient characteristics stratified by cluster assignment. A, Heat map comparison of metabolites stratified by cluster assignment. Concentrations of metabolites were log-transformed and z-scaled as described in the methods. Patients were clustered after principal component analysis of the baseline metabolomics data, with two groups best separating the data (cluster 1: n = 28 and cluster 2: n = 41). Patient age and 28-d mortality are shown as annotation above the heat map. B, Patient demographics, comorbidities, sex, and self-identified race stratified by metabolic cluster assignment. p values reported are from the Mann-Whitney U test or χ2, as appropriate.
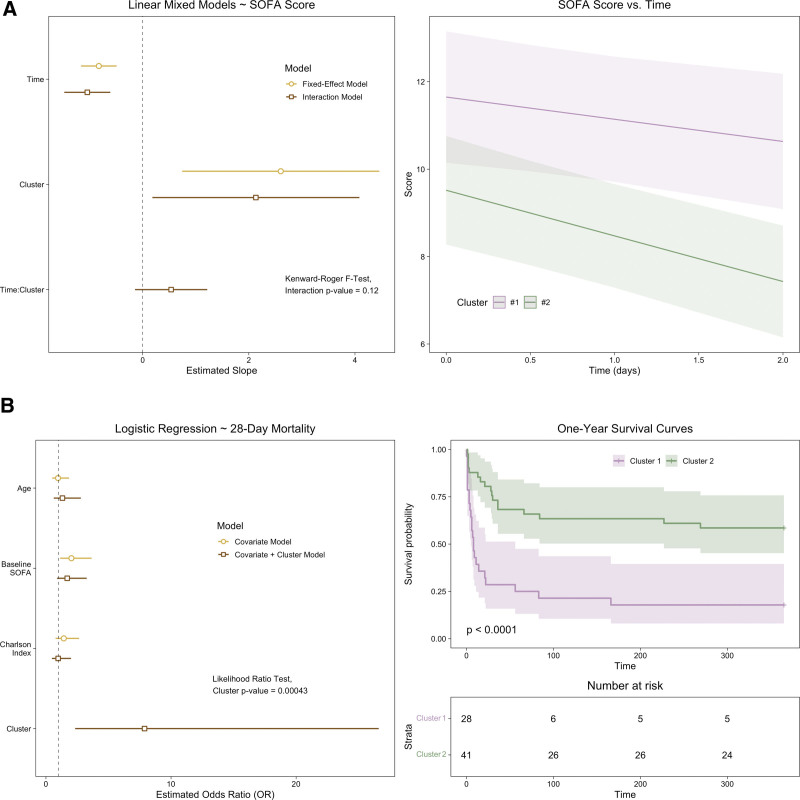
We assessed the trajectory of organ dysfunction between groups using linear mixed models with a patient-level intercept and time, cluster, and their interaction as fixed effects (Fig. 5A). The linear SOFA score trajectory was not different between metabolic clusters (βTime*Cluster1 = 0.536; 95% CI, –0.15 to 1.22; p = 0.12); however, patients in cluster one had worse organ function at baseline (Mann-Whitney U test; p < 0.05), which was sustained over the early course of their illness (Mann-Whitney U test at T24 and T48; p < 0.01).
Figure 5.
Patient outcomes stratified by metabolic cluster assignment. A, Model coefficients (left) and predictions (right) from linear mixed models, with Sequential Organ Failure Assessment (SOFA) score as the outcome variable. The fixed-effect model included the Time when SOFA was measured and Cluster assignment. The interaction model also included the Time*Cluster interaction, and its inclusion was assessed with the Kenward-Roger F test. A, The SOFA score predictions for each cluster (cluster 1 in purple, n = 28, cluster 2 in green, n = 41) from the interaction model are shown with their 95% CI. The median and interquartile range are also plotted for each time point, with between-cluster differences assed by the Mann-Whitney U test. B, Model coefficients from logistic regression, with 28-day mortality (left) and survival curves out to 1 year (right) between clusters. The probability of 28-day mortality was modeled with a covariate model that included age, baseline SOFA score, and the Charlson Comorbidity Index. A second model added cluster assignment as a predictor variable, and the likelihood ratio test was used to determine the impact of its inclusion. Survival curves for each metabolic cluster and the number at risk were plotted and assessed with the log-rank test. Survival curves were censored at 1 yr.
We also assessed differences in mortality between the metabolic clusters. In a logistic regression model adjusted for age, baseline SOFA score, and comorbidities, assignment to cluster 1 was associated with a greater probability of 28-day mortality (Fig. 5B; odds ratio = 7.8; 95% CI, 2.45–28.62). Addition of assigned cluster as a covariate to a logistic regression model of 28-day mortality with age, baseline SOFA score, and Charlson Comorbidity Index was informative based on the likelihood ratio test (p = 0.0004; Fig. 5B).One-year survival curves between the two metabolic clusters were also significantly different (log-rank test; p < 0.001).
DISCUSSION
In this study, we found a distinct metabolic and inflammatory signature in the early host response in 28-day nonsurvivor patients with septic shock. This signature was derived from measurements of serum acylcarnitines, small polar metabolites, and protein biomarkers of inflammation and endothelial activation. Specifically, patients who died had sustained elevations in IL-8 and acylcarnitines throughout the first 48 hours after the identification of septic shock. In addition, levels of pyruvate, IL-6, TNF-α, and ANG-2 declined at a slower rate in patients who died. Importantly, we also found a metabolic pattern of inflammation, whereby inflammatory cytokines were correlated with glycolysis products (e.g., lactate) and branched-chain amino acid (BCAA) catabolism. Taken together, our work further corroborates a role of disrupted mitochondrial-related metabolism and host inflammation in directing clinical outcomes (15, 17, 19, 22, 23). These disruptions continue early during septic shock and persist in patients who die by 28 days.
In addition to our main findings, unsupervised clustering revealed two distinct metabolic groups. These subgroups of patients with septic shock had similar demographics and comorbidities; however, patients in the first cluster were characterized by greater organ dysfunction and had a greater likelihood of 1 year mortality. Our findings demonstrate the substantial metabolic heterogeneity of patients within a well-defined cohort of septic shock, highlighting the syndromic nature of the disease and the potential for multiomic data to inform biologically driven endotyping of sepsis.
Metabolic perturbations are a well-established aspect of sepsis pathophysiology and have reliably been linked to acute illness severity and patient mortality (3). Described disruptions include a hypermetabolic state that results in catabolism of protein and fat, a glycolytic shift with a subsequent upregulation of the TCA cycle, and mitochondrial dysfunction (42–44). Our work contributes toward the Surviving Sepsis Campaign’s goal to better understand sepsis-induced metabolic disruptions (6) by providing a more comprehensive mapping of the dynamic metabolic changes in sepsis and its relationship to the host-immune response. This information is needed to direct the rationale design of targeted, metabolic pharmacotherapy and to inform biologic mechanisms underlying sepsis phenotypes and heterogeneity.
By leveraging the serially collected blood samples of the RACE trial, we show that elevations in acylcarnitines and IL-6, IL-8, and TNF-α persist early in the course of illness despite shock resuscitation treatment. The most perturbed signals in the early host response over time included acetylcarnitine (C2), the medium-chain acylcarnitine, C6, and the long-chain acylcarnitine, C16. Acetylcarnitine is the most abundant acylcarnitine in the blood and reflects the acetyl “pool” of the host, which is critically important in energy production (e.g., acetyl-CoA; Fig. 1) and the synthesis of the neurotransmitter, acetylcholine. Medium-chain acylcarnitines are derived from medium-chain fatty acids that are conjugated with L-carnitine in preparation for beta-oxidation, but they can also be produced by peroxisomal metabolism (24). In sepsis nonsurvivors, elevations in IL-8 and acetylcarnitine (C2) and other short-chain (C3-C5) acylcarnitines persisted to 48 hours. Short-chain acylcarnitines (C3-C5) can be derived from BCAA, leucine, isoleucine, and valine via mitochondrial enzymes that occur independent of fatty-acid metabolism and may reflect BCAA catabolism. From our baseline analysis, clusters differed most dramatically across medium (C6–C12) and long-chain (C13–C20) acylcarnitines (24) indicative of dysfunction in β-oxidation.
In aggregate, our findings are consistent with prior studies but add the important element of time-series data. Using an integrated metabolomic and proteomic analysis, Langley et al (15) found alterations in fatty-acid oxidation in sepsis nonsurvivors. Namely, patients who died had consistent elevations in acylcarnitines of various chain lengths and down regulation of fatty-acid transport proteins, suggesting a decreased capacity for β-oxidation of fatty acids by the mitochondria. Subsequent studies have been largely consistent with these findings and demonstrated that elevations in acylcarnitines are related to not only differential mortality and organ dysfunction (16, 45, 46) but also to systemic levels of mitochondrial DNA (47) and concentrations of inflammatory cytokines (17). The enhancement of analytical platforms and the growth of metabolomics as a field will likely continue to add promise to addressing these questions of variable patient outcomes in sepsis and other critical illness (48, 49).
Endothelial dysfunction and hyperinflammation are also well characterized components of sepsis pathophysiology, associated with patient outcomes, and represent potential avenues for targeted therapeutic development. Elevated concentrations of IL-8 have been associated with patient mortality in multiple cohorts of patients with sepsis and have been proposed as a prognostic biomarker for clinical trial enrichment (50–52). Similar findings have been reported in sepsis or sepsis-induced acute respiratory distress syndrome (ARDS) for IL-6 (53–55) and ANG-2 (56–58). Consistent with these studies, we found that concentrations of IL-8 were elevated in sepsis nonsurvivors over the first 2 days. Although we did not observe the same differences for IL-6 or ANG-2, the rate of decline for these protein analytes was slower in patients who died. These differences may be attributed to the fact that this cohort was comprised exclusively of patients with septic shock, presenting at a more advanced stage of infection.
Our study also further informs details of the interaction between the host metabolic and immune response, finding a positive correlation between numerous hyperinflammatory cytokines and the glycolysis end products, pyruvate and lactate (Fig. 1). In contrast, the BCAA tended to be lower in patients with a more hyperinflamed state. Alanine, known to be released from skeletal muscle following oxidation of BCAA (59), was positively correlated with both IL-8 and IL-10. Our findings here are consistent with a signature of BCAA catabolism that may result from a shifting preference for different energy substrates and a variable metabolic response that tracks with the host-immune response to infection (46). Conditions of chronic, low-grade inflammation such as obesity and type 2 diabetes are associated with increased systemic levels of BCAA (e.g., less catabolism) (60), whereas the inverse or normal levels have been reported in sepsis (61, 62). Although the precise mechanism for this has not been elucidated, we hypothesize that accelerated oxidation of BCAA results from a heightened acute inflammatory response.
The failure of numerous clinical trials and the lack of any targeted therapy beyond antibiotics have led to calls to reevaluate the approach to defining and treating sepsis and other critical care syndromes (4, 63). This shift toward disease subphenotyping (or endotyping) and defining treatable traits leverages individual patient characteristics and laboratory values, omic-based descriptions of an individual’s biological response, and unsupervised statistical or machine learning methods to cluster similar patients. These efforts have recently been reviewed (64), but notably include the identification of distinct patient clusters based on electronic health record data (65–67) and gene expression data (68, 69). In ARDS, similar approaches have defined hyperinflammatory and hypoinflammatory subphenotypes, with dramatic differences in clinical outcomes and response to therapy (70–74). Rogers et al (75) recently used metabolomics data from patients with sepsis and found distinct groups separated by concentrations of plasma lipids and with variable organ dysfunction and mortality. Here, we employed a similar approach on a smaller, but well-defined, cohort of patients exclusively with septic shock. Using metabolomics and a widely used clustering algorithm, we found two distinct groups of patients driven primarily by increased systemic concentrations of long-chain acylcarnitines, suggesting excess fatty-acid supply and/or incomplete β-oxidation (76). Patients assigned to cluster one tended to have significantly more organ dysfunction at baseline and postresuscitation and a greater risk of death. Furthermore, assignment to cluster 1 was associated with mortality even when added to a model containing SOFA score, age, and Charlson comorbidity index. Although it would be tempting to make direct comparisons between our and the Rogers et al (75) cohort, this is likely not to be meaningful because of the use of different biofluids and analytical platforms (77). We used quantitative, targeted assays for the measurement of metabolites in serum, whereas Rogers et al (75) assayed plasma using a broad analytical platform (relative quantification) that was enriched for lipids. Nevertheless, collectively, these findings demonstrate that metabolomics data are beneficial to defining subclasses of sepsis and directing future work that seeks to redefine critical illness based on biological underpinnings of disease pathobiology.
Our work has several limitations that are worthy of consideration. Sepsis heterogeneity is multifaceted. The sample size of the cohort limited our ability to assess the impact of variability in the site of infection and to account for treatments with specific pharmacologic agents (antibiotics, corticosteroids, and specific sedative agents or vasopressors). We acknowledge that we cannot account for all potential confounding variables and that our analysis only permits the introduction of metabolic subgroups as hypothesis-generating and that a larger cohort of patients will be needed for reproducibility and validation. As part of future validation, assessment of plasma analyte measurements may be warranted since we used residual serum volume for our analyte assays (78). Furthermore, in our analysis, we used an existing metabolomics data set that was generated as part of an ancillary study to the parent clinical trial (19). The biospecimens for this work was limited to serum samples, which did not permit cell- or tissue-level measurements of mitochondrial metabolism. Furthermore, we were not able to specifically measure fatty acid mobilization or identify the origins of the acylcarnitines we measured, and therefore, cannot pinpoint the specific mechanism by which the sepsis-induced elevation in acylcarnitines occurs. Notably, although peroxisomal metabolism may contribute to these signals, the abundance of acylcarnitines is produced by mitochondria (24). In addition, our approach was directed by our prior work; these assays were targeted and relatively limited in scope, measuring only acylcarnitines and a modest number of polar molecules (18). Use of larger cohorts of patients using untargeted metabolic profiling will likely find additional signals related to patient outcomes and may result in additional clusters of metabolically distinct patients. These studies should ideally integrate other patient characteristics and measures of the host-biological response to provide a more comprehensive understanding of the biological changes during sepsis. Finally, metabolomic and other omic data are not currently readily available within clinical practice. Point-of-care testing and thoughtful assay development and implementation will be required for the translation of metabolomics and subphenotyping findings.
CONCLUSIONS
In summary, we found that early concentrations of acylcarnitines and IL-8 are persistently elevated in patients with septic shock who do not survive. Unsupervised clustering of baseline metabolomics data also revealed two groups of patients with differentiating organ function and mortality. These findings reinforce that metabolic derangements of the host, particularly those related to a disruption in mitochondrial-related metabolism (e.g., fatty acid metabolism), continue postresuscitation and may be useful for prognostic and/or predictive enrichment to combat sepsis heterogeneity.
Supplementary Material
Footnotes
Supported, in part, by the National Institute of General Medical Sciences via R01GM103799 (to Dr. Jones), K23GM113041 (to Dr. Puskarich), and R01GM111400 and R35GM136312 (to Dr. Stringer).
Dr. Jennaro is supported by the American Foundation of Pharmaceutical Education. The remaining authors have disclosed that they do not have any potential conflicts of interest. The content is solely the responsibility of the authors and does not necessarily represent the official views of National Institute of General Medical Sciences or the National Institutes of Health.
The parent clinical trial (NCT01665092) was conducted from March 5, 2013, to February 5, 2018 at 16 large medical centers in the United States. The research protocol was approved by the local institutional review boards in accordance with National Institutes of Health guidelines and requirements.
All patients or their legally authorized representatives signed informed consent prior to participation. The metabolomics analysis of the collected deidentified serum samples was deemed “not regulated” by the University of Michigan’s Institutional Review Board.
The datasets generated by this work are available in the National Institutes of Health’s Metabolomics Workbench repository, https://www.metabolomicsworkbench.org/; accession number ST001319. All raw data and the code used to conduct our analysis are available on Github (https://github.com/UMichNMR-Metabolomics, accessed June 22, 2022).
Drs. Jennaro, Puskarich, Jones, and Stringer designed the study concept and analysis plan. Drs. Puskarich and Jones performed the clinical study and ensured the collection of blood samples. Dr. Jennaro, Dr. Evans, Dr. Karnovsky, Mr. Flott, and Ms. McLellan generated data. Dr. Jennaro performed the data analysis, and Dr. Jennaro and Stringer interpreted the data’s results and wrote the first draft of the article. All authors critically read and reviewed the article and approved of the submitted version.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccejournal).
REFERENCES
- 1.Singer M, Deutschman CS, Seymour CW, et al. : The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016; 315:801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vincent J-L, Jones G, David S, et al. : Frequency and mortality of septic shock in Europe and North America: A systematic review and meta-analysis. Crit Care 2019; 23:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavaillon J-M, Singer M, Skirecki T: Sepsis therapies: Learning from 30 years of failure of translational research to propose new leads. EMBO Mol Med 2020; 12:e10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maslove DM, Tang B, Shankar-Hari M, et al. : Redefining critical illness. Nat Med 2022; 28:1141–1148 [DOI] [PubMed] [Google Scholar]
- 5.Leligdowicz A, Matthay MA: Heterogeneity in sepsis: New biological evidence with clinical applications. Crit Care 2019; 23:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deutschman CS, Hellman J, Ferrer Roca R, et al. : The surviving sepsis campaign: Basic/translational science research priorities*. Crit Care Med 2020; 48:1217–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones AE: Lactate clearance for assessing response to resuscitation in severe sepsis. Acad Emerg Med 2013; 20:844–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puskarich MA, Trzeciak S, Shapiro NI, et al. : Whole blood lactate kinetics in patients undergoing quantitative resuscitation for severe sepsis and septic shock. Chest 2013; 143:1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vincent JL, Quintairos ESA, Couto L, Jr, et al. : The value of blood lactate kinetics in critically ill patients: A systematic review. Crit Care 2016; 20:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones AE, Shapiro NI, Trzeciak S, et al. ; Emergency Medicine Shock Research Network (EMShockNet) Investigators: Lactate clearance vs central venous oxygen saturation as goals of early sepsis therapy: A randomized clinical trial. JAMA 2010; 303:739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jansen TC, van Bommel J, Schoonderbeek FJ, et al. ; LACTATE study group: Early lactate-guided therapy in intensive care unit patients: A multicenter, open-label, randomized controlled trial. Am J Respir Crit Care Med 2010; 182:752–761 [DOI] [PubMed] [Google Scholar]
- 12.Hernandez G, Bellomo R, Bakker J: The ten pitfalls of lactate clearance in sepsis. Intensive Care Med 2019; 45:82–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernández G, Ospina-Tascón GA, Damiani LP, et al. ; The ANDROMEDA SHOCK Investigators and the Latin America Intensive Care Network (LIVEN): Effect of a resuscitation strategy targeting peripheral perfusion status vs serum lactate levels on 28-day mortality among patients with septic shock: The ANDROMEDA-SHOCK Randomized Clinical Trial. JAMA 2019; 321:654–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serkova NJ, Standiford TJ, Stringer KA: The emerging field of quantitative blood metabolomics for biomarker discovery in critical illnesses. Am J Respir Crit Care Med 2011; 184:647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langley RJ, Tsalik EL, van Velkinburgh JC, et al. : An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med 2013; 5:195ra95–195ra95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogers AJ, McGeachie M, Baron RM, et al. : Metabolomic derangements are associated with mortality in critically ill adult patients. PLoS One 2014; 9:e87538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung KP, Chen GY, Chuang TY, et al. : Increased plasma acetylcarnitine in sepsis is associated with multiple organ dysfunction and mortality: A multicenter cohort study. Crit Care Med 2019; 47:210–218 [DOI] [PubMed] [Google Scholar]
- 18.Puskarich MA, Finkel MA, Karnovsky A, et al. : Pharmacometabolomics of l-carnitine treatment response phenotypes in patients with septic shock. Ann Am Thorac Soc 2015; 12:46–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puskarich MA, Jennaro TS, Gillies CE, et al. : Pharmacometabolomics identifies candidate predictor metabolites of an L-carnitine treatment mortality benefit in septic shock. Clin Transl Sci 2021; 14:2288–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Englert JA, Rogers AJ: Metabolism, metabolomics, and nutritional support of patients with sepsis. Clin Chest Med 2016; 37:321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones AE, Puskarich MA, Shapiro NI, et al. : Effect of levocarnitine vs placebo as an adjunctive treatment for septic shock: The Rapid Administration of Carnitine in Sepsis (RACE) randomized clinical trial. JAMA Netw Open 2018; 1:e186076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mickiewicz B, Tam P, Jenne CN, et al. : Integration of metabolic and inflammatory mediator profiles as a potential prognostic approach for septic shock in the intensive care unit. Crit Care 2015; 19:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puskarich MA, Evans CR, Karnovsky A, et al. : Septic shock nonsurvivors have persistently elevated acylcarnitines following carnitine supplementation. Shock 2018; 49:412–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dambrova M, Makrecka-Kuka M, Kuka J, et al. : Acylcarnitines: Nomenclature, biomarkers, therapeutic potential, drug targets, and clinical trials. Pharmacol Rev 2022; 74:506–551 [DOI] [PubMed] [Google Scholar]
- 25.Vincent JL, Moreno R, Takala J, et al. : The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the working group on sepsis-related problems of the European Society of Intensive Care Medicine. Intensive Care Med 1996; 22:707–710 [DOI] [PubMed] [Google Scholar]
- 26.Gillies CE, Jennaro TS, Puskarich MA, et al. : A multilevel Bayesian approach to improve effect size estimation in regression modeling of metabolomics data utilizing imputation with uncertainty. Metabolites 2020; 10:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McHugh CE, Flott TL, Schooff CR, et al. : Rapid, reproducible, quantifiable NMR metabolomics: Methanol and methanol: Chloroform precipitation for removal of macromolecules in serum and whole blood. Metabolites 2018; 8:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antonelli J, Claggett BL, Henglin M, et al. : Statistical workflow for feature selection in human metabolomics data. Metabolites 2019; 9:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Vught LA, Wiewel MA, Hoogendijk AJ, et al. : The host response in patients with sepsis developing intensive care unit-acquired secondary infections. Am J Respir Crit Care Med 2017; 196:458–470 [DOI] [PubMed] [Google Scholar]
- 30.Faust HE, Oniyide O, Wang Y, et al. : Early plasma nuclear DNA, mitochondrial DNA, and nucleosome concentrations are associated with acute kidney injury in critically ill trauma patients. Crit Care Explor 2022; 4:e06–63-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kenward MG, Roger JH: Small sample inference for fixed effects from restricted maximum likelihood. Biometrics 1997; 53:983–997 [PubMed] [Google Scholar]
- 32.Halekoh U, Højsgaard S: A Kenward-Roger approximation and parametric bootstrap methods for tests in linear mixed models – the R package pbkrtest. J Stat Softw 2014; 59:1–3226917999 [Google Scholar]
- 33.Benjamini Y, Hochberg Y: Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol) 1995; 57:289–300 [Google Scholar]
- 34.Bakdash JZ, Marusich LR: Repeated measures correlation. Front Psychol 2017; 8:456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lucas C, Wong P, Klein J, et al. ; Yale IMPACT Team: Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020; 584:463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charrad M, Ghazzali N, Boiteau V, et al. : NbClust: An R package for determining the relevant number of clusters in a data set. J Stat Softw 2014; 61:1–36 [Google Scholar]
- 37.Elhaik E: Principal component analyses (PCA)-based findings in population genetic studies are highly biased and must be reevaluated. Sci Rep 2022; 12:14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu Z, Eils R, Schlesner M: Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016; 32:2847–2849 [DOI] [PubMed] [Google Scholar]
- 39.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria, R Foundation for Statistical Computing, 2021 [Google Scholar]
- 40.Charlson ME, Pompei P, Ales KL, et al. : A new method of classifying prognostic comorbidity in longitudinal studies: Development and validation. J Chronic Dis 1987; 40:373–383 [DOI] [PubMed] [Google Scholar]
- 41.Jennaro T, Puskarich M, Karnovsky A, et al. : Early metabolic profiles of patients with septic shock reveal distinct patient clustering characterized by ongoing metabolic dysfunction and differential mortality. D24 SEPSIS BIOMARKERS AND OUTCOMES: WHAT CAN WE PREDICT? Am Thorac Soc 2022;205:A5155–A515A [Google Scholar]
- 42.Hussain H, Vutipongsatorn K, Jiménez B, et al. : Patient stratification in sepsis: Using metabolomics to detect clinical phenotypes, sub-phenotypes and therapeutic response. Metabolites 2022; 12:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Preau S, Vodovar D, Jung B, et al. : Energetic dysfunction in sepsis: A narrative review. Ann Intensive Care 2021; 11:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singer M: The role of mitochondrial dysfunction in sepsis-induced multi-organ failure. Virulence 2014; 5:66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mickiewicz B, Duggan GE, Winston BW, et al. ; Alberta Sepsis Network: Metabolic profiling of serum samples by 1H nuclear magnetic resonance spectroscopy as a potential diagnostic approach for septic shock. Crit Care Med 2014; 42:1140–1149 [DOI] [PubMed] [Google Scholar]
- 46.Mickiewicz B, Tam P, Jenne CN, et al. : Integration of metabolic and inflammatory mediator profiles as a potential prognostic approach for septic shock in the intensive care unit. Crit Care Med 2015; 19:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johansson PI, Nakahira K, Rogers AJ, et al. : Plasma mitochondrial DNA and metabolomic alterations in severe critical illness. Crit Care 2018; 22:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eckerle M, Ambroggio L, Puskarich MA, et al. : Metabolomics as a driver in advancing precision medicine in sepsis. Pharmacotherapy 2017; 37:1023–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evangelatos N, Bauer P, Reumann M, et al. : Metabolomics in sepsis and its impact on public health. Public Health Genomics 2017; 20:274–285 [DOI] [PubMed] [Google Scholar]
- 50.Anderson BJ, Calfee CS, Liu KD, et al. : Plasma sTNFR1 and IL8 for prognostic enrichment in sepsis trials: A prospective cohort study. Crit Care 2019; 23:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mikacenic C, Price BL, Harju-Baker S, et al. : A two-biomarker model predicts mortality in the critically ill with sepsis. Am J Respir Crit Care Med 2017; 196:1004–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong HR, Lindsell CJ, Pettilä V, et al. : A multibiomarker-based outcome risk stratification model for adult septic shock*. Crit Care Med 2014; 42:781–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mat-Nor MB, Md Ralib A, Abdulah NZ, et al. : The diagnostic ability of procalcitonin and interleukin-6 to differentiate infectious from noninfectious systemic inflammatory response syndrome and to predict mortality. J Crit Care 2016; 33:245–251 [DOI] [PubMed] [Google Scholar]
- 54.Jekarl DW, Lee S-Y, Lee J, et al. : Procalcitonin as a diagnostic marker and IL-6 as a prognostic marker for sepsis. Diagn Microbiol Infect Dis 2013; 75:342–347 [DOI] [PubMed] [Google Scholar]
- 55.Miguel-Bayarri V, Casanoves-Laparra EB, Pallás-Beneyto L, et al. : Prognostic value of the biomarkers procalcitonin, interleukin-6 and C-reactive protein in severe sepsis. Med Intensiva 2012; 36:556–562 [DOI] [PubMed] [Google Scholar]
- 56.Parikh SM, Mammoto T, Schultz A, et al. : Excess circulating angiopoietin-2 may contribute to pulmonary vascular leak in sepsis in humans. PLoS Med 2006; 3:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ricciuto DR, dos Santos CC, Hawkes M, et al. : Angiopoietin-1 and angiopoietin-2 as clinically informative prognostic biomarkers of morbidity and mortality in severe sepsis. Crit Care Med 2011; 39:702–710 [DOI] [PubMed] [Google Scholar]
- 58.Reilly JP, Wang F, Jones TK, et al. : Plasma angiopoietin-2 as a potential causal marker in sepsis-associated ARDS development: Evidence from Mendelian randomization and mediation analysis. Intensive Care Med 2018; 44:1849–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holeček M: Branched-chain amino acids in health and disease: Metabolism, alterations in blood plasma, and as supplements. Nutr Metab (Lond) 2018; 15:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang S, Zeng X, Ren M, et al. : Novel metabolic and physiological functions of branched chain amino acids: A review. J Anim Sci Biotechnol 2017; 8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Freund H, Atamian S, Holroyde J, et al. : Plasma amino acids as predictors of the severity and outcome of sepsis. Ann Surg 1979; 190:571–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Freund HR, Ryan JA, Jr, Fischer JE: Amino acid derangements in patients with sepsis: Treatment with branched chain amino acid rich infusions. Ann Surg 1978; 188:423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prescott HC, Calfee CS, Thompson BT, et al. : Toward smarter lumping and smarter splitting: rethinking strategies for sepsis and acute respiratory distress syndrome clinical trial design. Am J Respir Crit Care Med 2016; 194:147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, Markal A, Balch JA, et al. : Methods for phenotyping adult patients in sepsis and septic shock: A scoping review. Crit Care Explor 2022; 4:e0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z, Ho KM, Gu H, et al. : Defining persistent critical illness based on growth trajectories in patients with sepsis. Crit Care 2020; 24:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Z, Zhang G, Goyal H, et al. : Identification of subclasses of sepsis that showed different clinical outcomes and responses to amount of fluid resuscitation: A latent profile analysis. Crit Care 2018; 22:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seymour CW, Kennedy JN, Wang S, et al. : Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. JAMA 2019; 321:2003–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maslove DM, Tang BM, McLean AS: Identification of sepsis subtypes in critically ill adults using gene expression profiling. Crit Care 2012; 16:R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sweeney TE, Azad TD, Donato M, et al. : Unsupervised analysis of transcriptomics in bacterial sepsis across multiple datasets reveals three robust clusters. Crit Care Med 2018; 46:915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Calfee CS, Delucchi K, Parsons PE, et al. ; NHLBI ARDS Network: Subphenotypes in acute respiratory distress syndrome: Latent class analysis of data from two randomised controlled trials. Lancet Respir Med 2014; 2:611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Calfee CS, Janz DR, Bernard GR, et al. : Distinct molecular phenotypes of direct vs indirect ARDS in single-center and multicenter studies. Chest 2015; 147:1539–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Famous KR, Delucchi K, Ware LB, et al. ; ARDS Network: Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am J Respir Crit Care Med 2017; 195:331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Calfee CS, Delucchi KL, Sinha P, et al. ; Irish Critical Care Trials Group: Acute respiratory distress syndrome subphenotypes and differential response to simvastatin: Secondary analysis of a randomised controlled trial. Lancet Respir Med 2018; 6:691–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sinha P, Delucchi KL, Thompson BT, et al. ; NHLBI ARDS Network: Latent class analysis of ARDS subphenotypes: A secondary analysis of the statins for acutely injured lungs from sepsis (SAILS) study. Intensive Care Med 2018; 44:1859–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rogers AJ, Leligdowicz A, Contrepois K, et al. : Plasma metabolites in early sepsis identify distinct clusters defined by plasma lipids. Crit Care Explor 2021; 3:e0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McCann MR, George De la Rosa MV, Rosania GR, et al. : L-carnitine and acylcarnitines: mitochondrial biomarkers for precision medicine. Metabolites 2021; 11:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kennedy AD, Ford L, Wittmann B, et al. : Global biochemical analysis of plasma, serum and whole blood collected using various anticoagulant additives. PLoS One 2021; 16:e0249797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rosenberg-Hasson Y, Hansmann L, Liedtke M, et al. : Effects of serum and plasma matrices on multiplex immunoassays. Immunol Res 2014; 58:224–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.