Abstract
SARS-CoV-2, the causative agent of COVID-19 disease has resulted in the death of millions worldwide since the beginning of the pandemic in December 2019. While much progress has been made to understand acute manifestations of SARS-CoV-2 infection, less is known about post-acute sequelae of COVID-19 (PASC). We investigated the levels of both Spike protein (Spike) and viral RNA circulating in patients hospitalized with acute COVID-19 and in patients with and without PASC. We found that Spike and viral RNA were more likely to be present in patients with PASC. Among these patients, 30% were positive for both Spike and viral RNA; whereas, none of the individuals without PASC were positive for both. The levels of Spike and/or viral RNA in the PASC-positive patients were found to be increased or remained the same as in the acute phase; whereas, in the PASC-negative group, these viral components decreased or were totally absent. Additionally, this is the first report to show that part of the circulating Spike is linked to extracellular vesicles without any presence of viral RNA in these vesicles. In conclusion, our findings suggest that Spike and/or viral RNA fragments persist in the recovered COVID-19 patients with PASC up to 1 year or longer after acute SARS-CoV-2 infection.
Keywords: Extracellular vesicles, SARS-CoV-2, COVID-19, Spike, Viral RNA, plasma
INTRODUCTION
The COVID-19 pandemic caused by SARS-CoV-2 continues to have devastating global consequences1. Recognized now is also a plethora of symptoms that persist long after acute infection, known as Post-acute Sequelae of COVID-19 or PASC in 10–30% of patients after 12 weeks2,3. While the definition and scope of PASC remain to be determined, it is currently defined as new or persistent symptoms of COVID-19 beyond 4 weeks of acute infection not explained by other underlying etiologies4,5. If only a small fraction of patients go on to develop PASC, it will have an enormous impact for years. It is imperative to understand the mechanism(s) of acute and post-acute COVID-19 disease pathogenesis, identify those most at risk of developing PASC, and investigate targets for future treatments.
Recent studies suggest that SARS-CoV-2 RNA disseminates to extra-pulmonary tissues in asymptomatic/mild infections and that viral RNA can reside in tissues for over 7 months6. SARS-CoV-2 Spike protein (Spike) mediates lung injury and vascular damage by inducing endothelial dysfunction and inflammation7,8. We previously reported significant apoptosis of microvascular endothelial cells on exposure to plasma-derived extracellular vesicles (EVs) from hospitalized COVID-19 patients9. In this study, we examined if the circulation of SARS-CoV-2 RNA or Spike is associated with acute disease severity and if its persistence correlated with manifestations of PASC.
METHODS
Sample and data collection
Plasma samples (n=116) were obtained from COVID-19 patients (Acute) admitted to the University of Kansas Health System (TUKHS) from May 2020 through December 20219. Samples from individuals never exposed to SARS-CoV-2 with matched co-morbidities (n=15) were obtained from the TUKHS Biospecimen Repository Core Facility (BRCF). PASC patients were recruited from our Long-COVID clinic at TUKHS (N=33) with acute infections ranging from March 2020 to May 2021. This group was defined as having one or more symptoms greater than 8–12 weeks post-infection, reported via a Research Electronic Data Capture (REDCap) survey or to a provider. Biospecimens were obtained at initial enrollment at least 8–12 weeks post-acute infection. PASC-negative patients were identified through the Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV) −2 trial (a study for outpatient treatment of COVID-19) participants after they had completed 24 weeks of the trial and did not have any persistent symptoms (N=14). We were also able to follow 12 patients that were included in the Acute COVID Biorepository as well as enrolled in our PASC Biorepository.
These studies were approved by the IRB at the University of Kansas Medical Center. Demographic information and all other clinical data were collected via electronic medical records or survey data and stored in a secure database (Table 1). Surveys were conducted at initial enrollment and subsequently, every 3 months through REDCap utilizing the standardized World Health Organization-established Global COVID-19 and Clinical Platform Case Report Form for Post-COVID conditions.
Table 1:
Demographics & Characteristics of Acute COVID-19 and PASC Patients Included in Analysis
| Acute (n=116) | PASC Negative (n=14) | PASC Positive (n=33) | |
|---|---|---|---|
| Age (median, IQR) | 57 (43.8–71.0) | 57 (43.7 – 62.7) | 49 (42.0 – 63.0) |
| Sex (total, %) | |||
| Male | 65 (56.0%) | 8 (57.1%) | 13 (39.4%) |
| Female | 50 (43.1%) | 6 (42.9%) | 20 (60.6%) |
| Race (total, %) | |||
| Asian | 1 (0.8%) | 1 (7.1%) | 1 (3.0%) |
| Black or African American | 21 (18.1%) | 2 (14.3%) | 2 (6.1%) |
| White | 64 (55.2%) | 8 (57.1%) | 24 (72.7%) |
| Other | 29 (25.0%) | 1 (7.1%) | 4 (12.1%) |
| Unknown | 1 (0.8%) | 2 (14.3%) | 2 (6.1%) |
| Ethnicity (total, %) | |||
| Hispanic | 29 (25.0%) | 1 (7.1%) | 4 (12.1%) |
| Non-Hispanic | 85 (73.3%) | 9 (64.3%) | 26 (78.8%) |
| Unknown | 2 (1.72%) | 4 (28.5%) | 3 (9.1%) |
| BMI (median, IQR) * | 31.9 (26.6–39.4) | 33.35 (29.9 – 36.6) | 30.1 (26.9 – 39.9) |
| Length of hospitalization (days) (median, IQR) * | 8.5 (4.0 – 15.7) | 3.5 (2 – 7) | 0 (0 – 7) |
| Days between symptom onset/last positive test and study enrollment (median, IQR) * | 5.0 (3.0–8.0) | 235 (144 – 263.0) | 201 (183.0 – 266.0) |
| 28 – 100 days | --- | 3 (21.4%) | 3 (9.1%) |
| 101 – 200 days | --- | 3 (21.4%) | 13 (39.4%) |
| 201 – 300 days | --- | 5 (35.7%) | 12 (36.4%) |
| 301 – 400 days | --- | 1 (7.1%) | 2 (6.1%) |
| 401 – 500 days | --- | 1 (7.1%) | 2 (6.1%) |
| 500 + days | --- | 0 (0.0%) | 1 (3.0%) |
BMI data was only available on 31/33 PASC positive patients and 8/14 PASC negative patients; length of hospitalization available on 29/33 PASC-positive patients and 8/14 PASC negative patients.
Measurement of SARS-CoV-2 RNA Using Droplet Digital PCR
RNA was isolated using 200μl plasma as previously reported by others10. After centrifugation of plasma at approximately 21,000 × g for 2h at 4 °C, the supernatant was removed and 750 μL of TRIzol-LS™ Reagent (ThermoFisher) was added to the pellet and incubated on ice for 10 minutes. Following incubation, 200 μL of chloroform (Millipore-Sigma) was added, vortexed and centrifuged at 21,000 × g for 15 min at 4 °C. Subsequently, the aqueous layer was removed and treated with an equal volume of isopropanol (Sigma) followed by addition of GlycoBlue™ Coprecipitant (ThermoFisher) and 100 μL 3 M Sodium Acetate (Life Technologies) and incubation on dry ice until frozen. RNA was later pelleted by centrifugation at 21,000 × g for 45 mins at 4 °C. The supernatant was then discarded, and the RNA pellet was washed with cold 70% ethanol followed by resuspension in diethyl pyrocarbonate-treated water (ThermoFisher). SARS-CoV-2 RNA copies were quantified using the QX200TM Droplet Digital TM PCR System (ddPCR) and BioRad SARS-CoV-2 ddPCR Kit according to the manufacturer’s instructions. Data was analyzed using the QuantaSoftTM 1.7.4 Software (Bio-Rad) and SARS-CoV-2 quantification was expressed in number copies/μl. Also, to check if the plasma derived viral RNA is from EVs, virions and /or is present as free RNA, plasma was subjected to RNase A (0.5 μg/μl) treatment at 37°C for 20 minutes. The reaction was immediately stopped by adding TRIzol-LS followed by RNA isolation11 12. The quality of RNA was determined using Nanodrop followed by quantification of SARS-CoV-2 RNA copies using ddPCR.
Isolation of Extracellular Vesicles
Extracellular vesicles (EVs) were isolated from EDTA plasma using Ultracentrifugation. Hemolysis of blood was ruled out by visually inspecting plasma samples for any pink discoloration indicative of hemolysis or by measuring the absorbance of hemoglobin at 414nm. About 1ml of platelet-free plasma (PFP) was centrifuged at 20,000 × g for 15 minutes at 4°C to remove large-sized EVs. The supernatants were then centrifuged at 100,000 × g for 70 minutes at 4°C, washed with PBS, and spun again at 100,000 × g for 70 minutes to isolate small EVs. The abundance, size distribution and purity of small EVs was compared among different groups using the NanoSight LM10 system, TEM and Western blot analysis of EV markers using CD9 (Cat# 13174T/ 13174S, Cell Signaling Technology, MA, USA), CD81 (Cat# NB100-65805SS, Novus Biologicals, LLC), TSG101 (Cat# NBP2-77452SS, Novus Biologicals, LLC) and Alix (Cat# 2171T, Cell Signaling Technology, MA, USA) antibodies as mentioned in our previous publication9.
Spike protein ELISA
The levels of SARS-CoV-2 Spike protein in the plasma (1:5 dilution) and EV lysate (50 μg EV protein) were determined using the SAR-CoV-2 (2019-nCoV) Spike protein ELISA Kit according to the manufacturer instructions (SinoBiological). The sensitivity of the kit is 10.28 pg/mL and previous studies have shown that this sensitivity is enough to detect Spike protein in plasma13 14. To check the levels of Spike protein in EVs treated with proteinase K, 100μg of Spike protein-positive COVID-19 EVs were treated with and without 50 μg/ml of proteinase K (Exiqon) for 30 min at 37 °C. Then the reaction was inhibited by incubation with 5mM phenylmethylsulfonyl fluoride (PMSF) for 10 min at 37 °C (Mol. Pharmaceutics 2018, 15, 3, 1073–1080). EVs were also treated with and without heparinase II (0.9 mIU/mL, New England Biolabs) for 3 h at 37 °C and then incubated again with fresh enzyme overnight at 37°C 15. After proteinase K or heparinase treatment, EVs were washed with PBS and concentrated using Amicon Ultra-4 centrifugal filters (10 kDa, Millipore Sigma, USA).
Statistical Analysis
Differences in the continuous variables among the groups were assessed using Kruskal Wallis or Wilcoxon rank sum Test. Categorical outcomes among the groups were assessed using Fisher exact test or test of proportions. Spearman’s rank correlation analyses were carried out to assess the relationship between each of the plasma Spike, EV-Spike protein and viral RNA with Age, BMI, etc. (Table 2). All tests were considered statistically significant if the p-values were less than 0.05. The statistical analyses were carried out using the statistical software R version 4.0.0 16.
Table 2:
Correlation Analyses of Clinical Characteristics with Viral RNA, Spike level in Plasma and Plasma-derived EVs in COVID-19 Individuals
| Viral RNA [Correlation (p-value)] | Plasma spike [Correlation (p-value)] | SEVs spike [Correlation (p-value)] | |
|---|---|---|---|
| Acutely -infected COVID-19 patients | |||
| Age | 0.0981 (0.2948) | 0.0398 (0.6728) | −0.0061 (0.9597) |
| BMI | 0.04 (0.6695) | 0.0809 (0.3898) | −0.2108 (0.0776) |
| Peak WHO Score | 0.0497 (0.6026) | 0.2002 (0.0351) | 0.0716 (0.5646) |
| Length of hospitalization | 0.1379 (0.2549) | 0.253 (0.0359) | 0.1579 (0.324) |
| WBC Count | 0.1009 (0.2856) | 0.0567 (0.5508) | 0.1446 (0.2325) |
| Lymphocyte Count | −0.133 (0.1806) | −0.0676 (0.4995) | −0.2072 (0.1031) |
| Creatinine | 0.1477 (0.1153) | 0.0055 (0.9536) | 0.0509 (0.6758) |
| Lactate Dehydrogenase | 0.2532 (0.0273) | 0.1904 (0.0994) | 0.1781 (0.2309) |
| Ferritin | 0.1543 (0.1586) | 0.0627 (0.5689) | 0.1888 (0.1892) |
| D.Dimer | 0.0863 (0.4136) | 0.2338 (0.0249) | 0.3544 (0.0092) |
| C-Reactive Protein | 0.1335 (0.2149) | 0.1451 (0.1774) | −0.1266 (0.3713) |
| PASC- positive patients | |||
| Age | 0.0322 (0.8586) | 0.0044 (0.9807) | −0.1102 (0.6343) |
| Days between last positive test and study enrollment | −0.1219 (0.4993) | 0.0391 (0.8291) | −0.1635 (0.4789) |
| BMI | −0.0126 (0.9464) | −0.1261 (0.4992) | −0.1465 (0.5377) |
| Days of Hospitalization | 0.4197 (0.0234) | −0.0428 (0.8253) | 0.442 (0.0581) |
Correlation analyses with BMI on n=31 and with length of hospitalization on n= 29 PASC-positive patients.
RESULTS
Presence of Viral RNA, soluble Spike and/or EV-linked Spike in the plasma of COVID-19 patients with acute disease
The reverse transcription-droplet digital polymerase chain reaction (RT-ddPCR) analysis revealed the presence of viral RNA in 35% of hospitalized COVID-19 patients (Figure 1A (i)). To check if positivity was due to the presence of free viral RNA in circulation, RNAse treatment was given to a small set of samples (n=10) before ddPCR analysis (Figure 1A (ii), and viral RNA levels dropped in most of the samples (p<0.001). Some samples still showed high levels of viral RNA possibly due to the existence of free virions in the plasma or because the incubation time with RNAse was too short to completely degrade free RNA in samples with higher viral RNA levels, to begin with.
Figure 1:
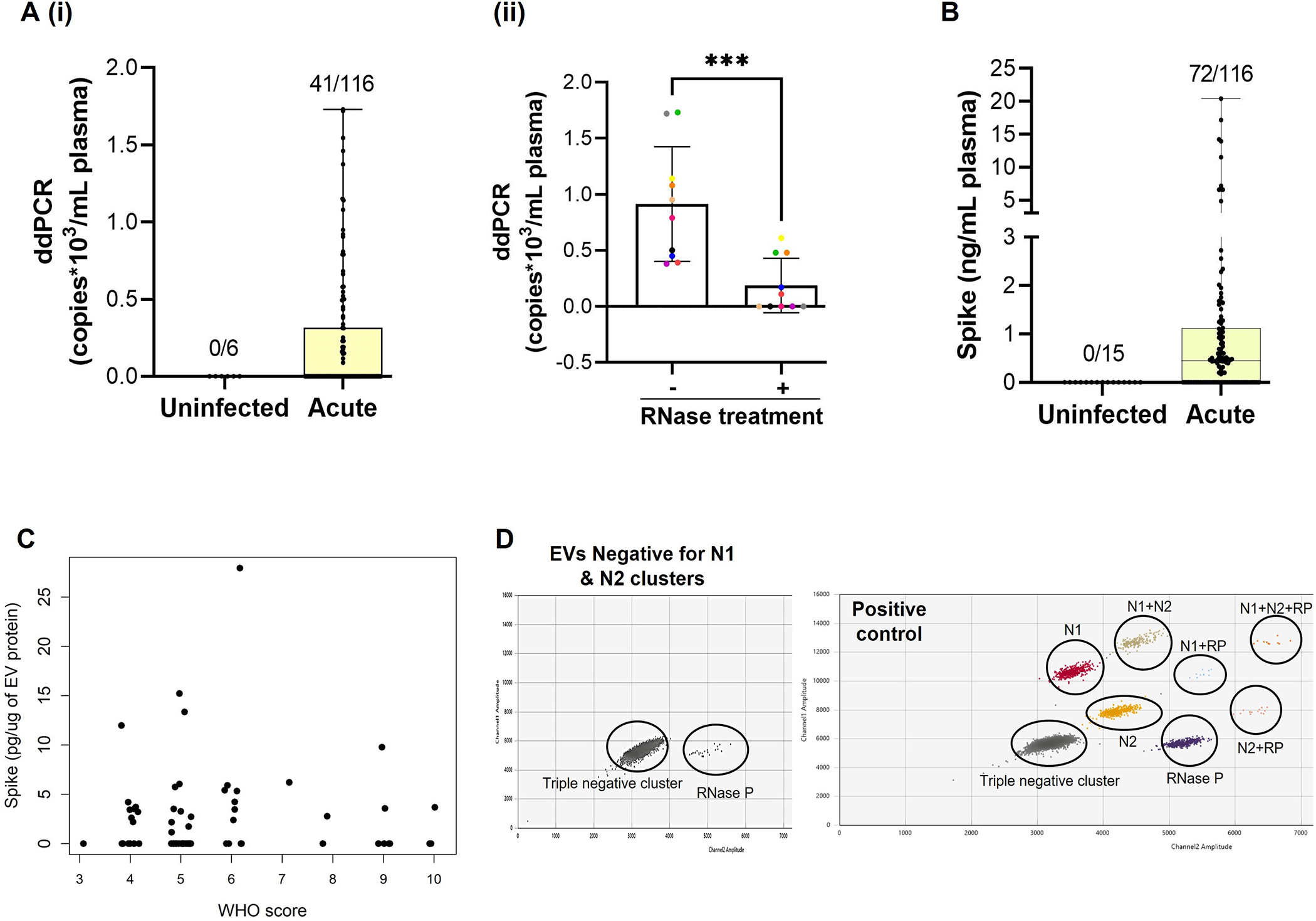
The presence of SARS-CoV-2 RNA (A) and Spike protein (B) in the plasma of symptomatic hospitalized COVID-19 patients (Acute Disease). SARS-CoV-2 RNA copies were measured by dd-PCR (A) and Spike protein by ELISA (B) in the plasma from Acute COVID-19 patients and never infected with COVID-19 controls. Aii) Viral RNA copies the plasma after RNase treatment (n=10). C) Spike ELISA in the plasma derived small EVs from the subjects that showed positivity for Spike protein in total plasma (n=71). D) Representative 2D plot of dd-PCR showing absence of viral RNA in EVs as detected by the presence of internal control (RP) but absence of Nucleocapsid cluster. EVs from n=11 subjects that showed positivity for viral RNA in the corresponding plasma were analyzed. Also shown is the 2D plot of positive control (synthetic RNA transcript containing five gene targets: E, N, ORF1ab, RdRP and S Genes of SARS-CoV-2) showing the presence of nucleocapsid (N) cluster. *** p ≤ 0.001.
Plasma analysis for Spike by ELISA showed the presence of Spike in 62% of Acute COVID-19 patients (Figure 1B). The level of Spike showed a significant positive correlation with peak WHO score (p=0.03) along with D-Dimer (p=0.02) and length of hospitalization (p=0.03) (Table 2). Some of the samples that had circulating Spike did not have detectable levels of viral RNA, while other samples that had viral RNA did not have detectable Spike. While 34% of acute COVID-19 patients were positive for only Spike, positivity for viral RNA alone was observed in only 6% of the total samples analyzed. This suggests that Spike may be freely circulating independently of circulating virions and circulating viral RNA may include free viral RNA or its fragments during acute infection with SARS-CoV-2.
We previously demonstrated an increased number of plasma-derived small EVs and its altered cargo to be associated with disease severity of hospitalized COVID-19 patients 9. To see if circulating Spike is linked to EVs, we analyzed EVs from the individuals which showed positivity for Spike in total plasma. Results indicated that 44% of Spike-positive hospitalized patients showed the presence of Spike in EVs (Figure 1C). The level of Spike-linked EVs significantly correlated with D-Dimer (p=0.009) in Acute COVID-19 patients (Table 2). We also checked for the presence of viral RNA using ddPCR in EVs from Acute patients. Interestingly, we were not able to detect viral RNA in any of the EV samples tested (Figure 1D).
Persistent circulation of Viral RNA and EV-linked /soluble Spike is associated with PASC
We grouped PASC patients into PASC-positive (n=33) and PASC-negative (n=14) based on self-reported symptoms. PASC symptoms included primarily fatigue (81.8%), shortness of breath (66.7%), brain fog (57.6%) along with sleep disturbances, mood changes, loss of taste/smell, fever, myalgias, headaches, chest pain, post-exertional malaise and cough etc. There was no difference in vaccination status between PASC-positive and negative groups. We screened to ensure that administrations of mRNA vaccines were not confounding the levels of Spike detected in PASC patients. The earliest time between a recent vaccination and blood draw for this study was 21 days in one patient, and this patient was PASC-negative and did not have detectable levels of Spike. All other vaccinations were more than 30 days before consent/biospecimens were obtained. Patients with PASC were more likely to have a pre-existing pulmonary disease which in most cases was mild-to-moderate asthma or obstructive sleep apnea while PASC negative were more likely to have diabetes, coronary artery disease or kidney disease given the nature of the ACTIV-2 cohort.
Examination of the viral RNA in the plasma from PASC-positive and PASC-negative individuals showed that 28% of PASC-negative samples had viral RNA copies ranging from 0.08–2.13 copies/ul; whereas, 59% of PASC-positive patients had viral RNA in circulation (0.07–12.74 copies/ul) (Figure 2Ai). RNase treatment of plasma before RNA isolation resulted in the drop of viral RNA copies in most of the samples from the PASC-positive group (Figure 2Aii). Furthermore, viral RNA in plasma showed a strong positive correlation with days of hospitalization (p=0.043).
Figure 2:
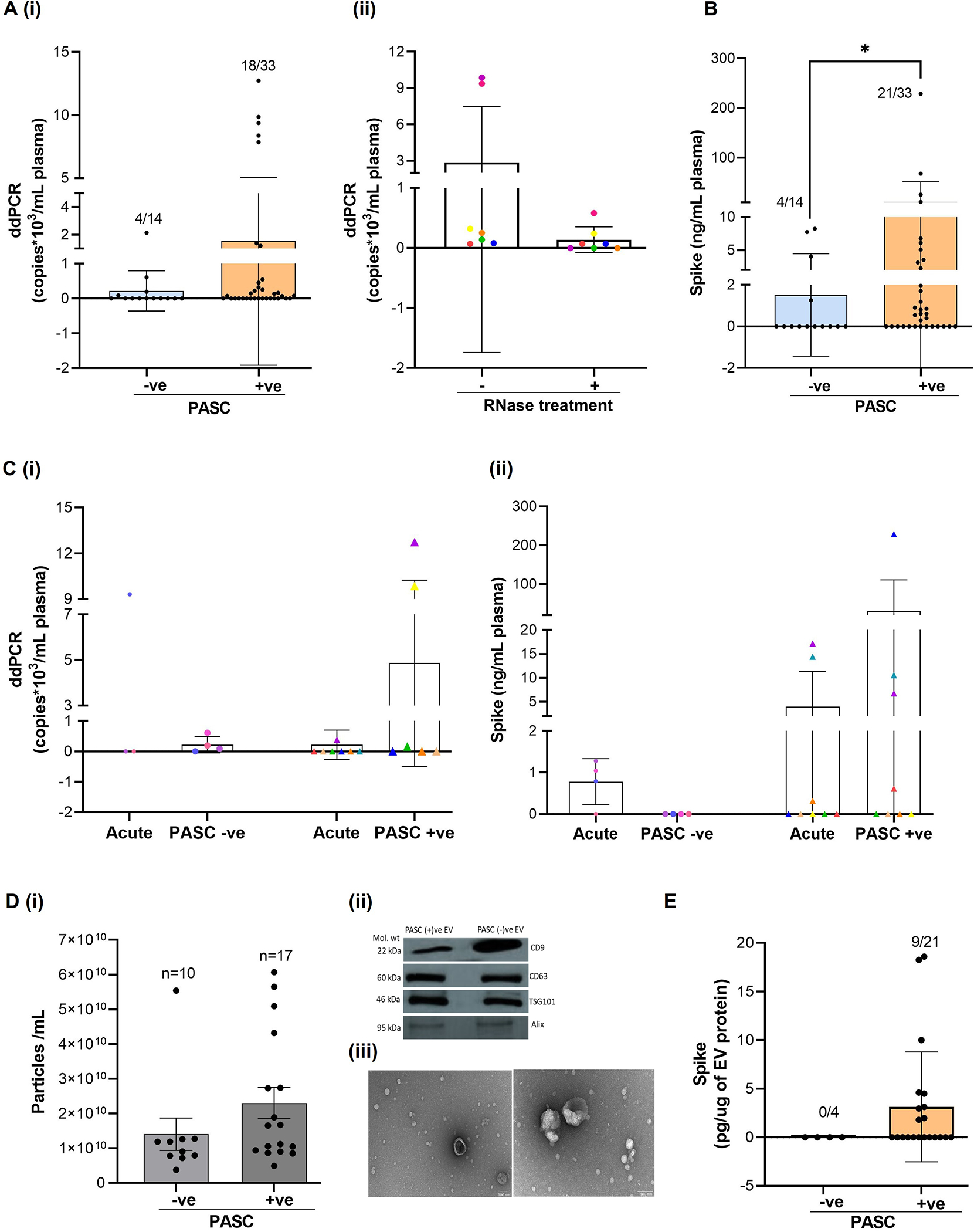
Viral RNA and/or Spike protein persistence in circulation is associated with PASC. Viral RNA (A) and Spike protein (B) levels were analyzed in the plasma from COVID -19 patients with PASC (persistent symptoms of COVID-19 at 8–12 weeks or more) (PASC +ve,N=33) and COVID-19 recovered patients that did not have any persistent symptoms (PASC-ve,N=14). SARS-CoV-2 RNA copies were measured using droplet digital -PCR and Spike protein by ELISA. C) Longitudinal analysis of viral RNA (C(i) and Spike protein (C(ii) in individuals (N=12) at acute and post- acute phase of COVID-19 disease with and without PASC. D) Comparison of small EV numbers in the plasma from individuals with and without PASC as measured by Nanoparticle Tracking Analysis (i). Characterization of plasma derived small EVs using western blot (ii) and Transmission electron microscope (TEM) (iii). Representative TEM micrograph showing EVs at 30,000X and 50000X magnification. E) Spike protein in the plasma derived small EVs from individuals with and without PASC as measured by ELISA. * p ≤ 0.05.
Parallel evaluation of Spike in the plasma found the presence of Spike in 4 of 14 PASC-negative individuals and in 21 out of 33 PASC-positive (64%) samples (Figure 2B) (p<0.05). Additionally, we found that 33% (11/33) of the PASC-positive group showed positivity for both Spike and viral RNA. None of the samples in the PASC-negative group showed positivity for both Spike and viral RNA. Comparison of the levels of viral RNA and Spike during the acute COVID-19 and post-recovery phase of COVID-19 in the same patients (n=12), showed that Spike and/or viral RNA in the patients from the PASC-positive group increased or remained the same as in the acute phase; whereas, in the PASC-negative group, Spike was found to be totally absent and viral RNA either decreased to very low copy numbers or was undetectable (Figure 2 C).
Next, we isolated small EVs from the plasma of PASC-ve and PASC+ve samples that showed positivity for Spike protein and/or viral RNA. Nanoparticle tracking analysis revealed a trend towards an increase in the total number of EVs in the PASC+ve group as compared to the PASC-ve group (p=0.203) (Figure 2D,i). The lack of significant difference was potentially due to one of the PASC-ve individuals that showed a much higher number of EVs compared to the rest of the subjects and had multiple underlying comorbidities, including diabetes, heart failure, and kidney disease. In addition, EVs were characterized using western blot that confirmed the presence of EV-linked tetraspanins and other markers of exosomes (Figure 2Dii); and TEM analysis that indicated the presence of EVs in the range of 25–150 nm in SEVs (Figure 2D iii). Examination of the Spike levels in the EVs from the plasma samples that showed positivity for Spike revealed the presence of EV-linked Spike in 43% (9/21) of samples from the PASC-positive group (Figure 2E). Interestingly, EVs from the PASC-negative group did not show any positivity for Spike ELISA.
Presence of Spike protein on the surface of EVs
To test if Spike is on the surface of EVs, we first compared the levels of Spike in the EVs treated with or without proteinase K. To have enough protein for the analysis, we pooled two patient samples each for a set of n=3. As shown in Figure 3A, a significant decrease was observed in the levels of Spike in the EV preparation from ‘Severe’ group patients after the proteinase K treatment (p<0.05). Further, cytokine and growth factors have been found to be linked to the EV surface through the heparan sulfate proteoglycans and Clausen et al. have reported that Spike interacts with heparan sulfate through the receptor binding domain17. Therefore, to assess if Spike is associated on EV surfaces by binding to heparan sulfate, we examined the levels of Spike in heparinase II-treated EVs. Heparinase II treatment of COVID-19 EVs showed decreased levels of Spike when compared to untreated EVs (Figure 3B, n=3, each a pool of EVs from 2 patients) We confirmed the removal of membrane proteins by proteinase K treatment by examining the levels of tetraspanins in EV preparations before and after proteinase K treatment. As expected proteinase K treatment resulted in the degradation of CD81 and CD9 from the EV membrane whereas both tetraspanins were still present on EVs after heparinase II treatment.
Figure 3:
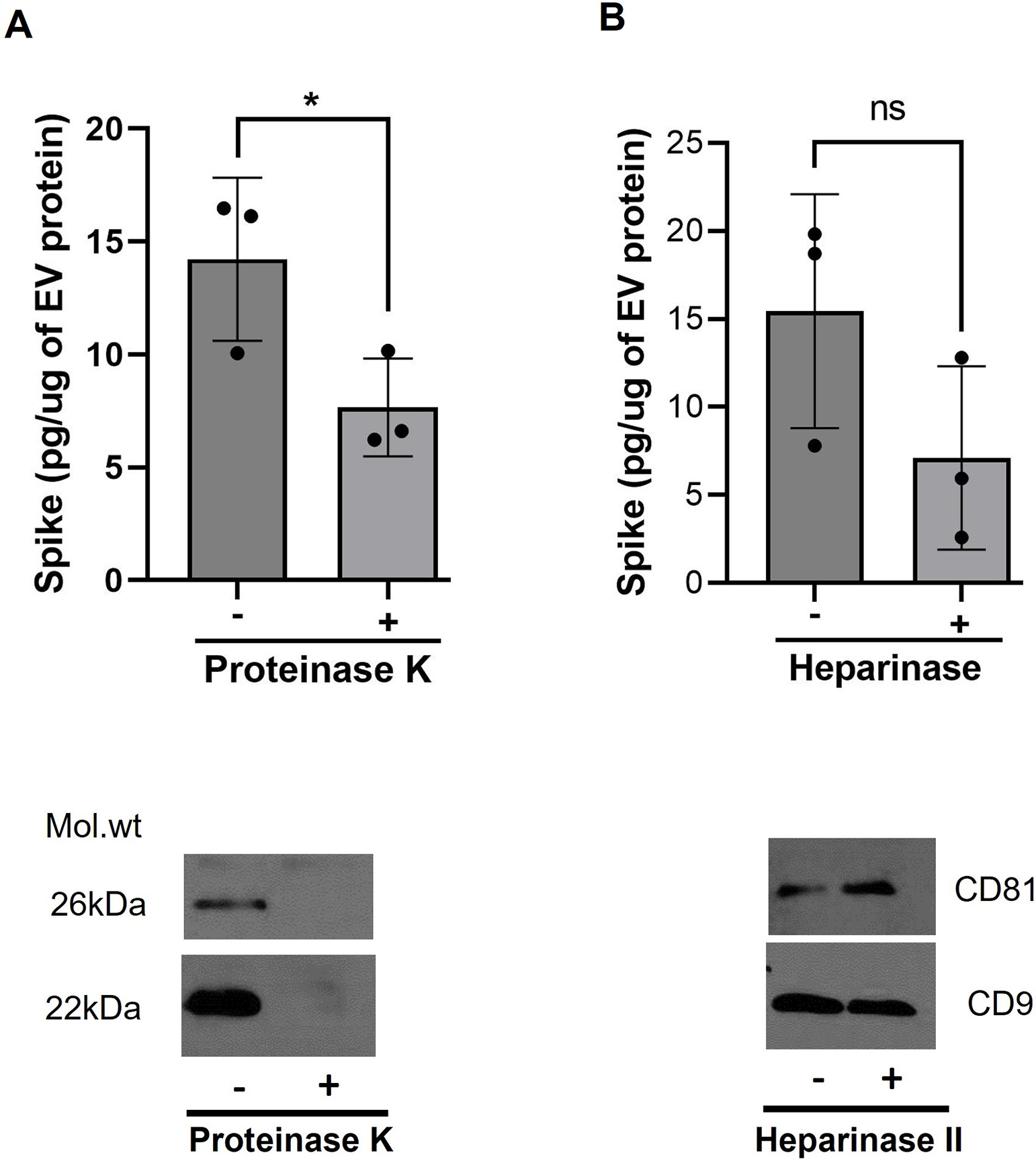
Spike protein is partly associated on the surface of EVs. Decrease in levels of Spike in EVs treated with proteinase K (A) and heparinase II (B) in comparison to untreated SEVs, as detected by ELISA. EVs from 2 patients were pooled in a set of n=3 for the analysis. * p≤0.05. Western blots below showing loss of tetraspanins from the surface of un-infected EVs confirmed the activity of proteinase K while the heparinase II treatment didn’t show any effect on these surface markers.
DISCUSSION
We report that Spike and viral RNA can persist for up to 1 year or longer after acute SARS-CoV-2 infection and are associated with PASC. Our observations reveal that levels of Spike and/or viral RNA in the PASC-positive patients increase or remain the same as in the acute phase; whereas, in the PASC-negative group, these viral components decreased or were totally absent. Interestingly, we also report the persistent circulation of Spike-linked EVs in PASC-positive patients.
Mechanisms of how SARS-CoV-2 or its components persist and cause prolonged symptoms long after the infection has yet to be elucidated. Proposed theories include that parts of the SARS-CoV-2 genome can be incorporated into the DNA of infected cells and fragments of the SARS-CoV-2 genome may be transcribed at random 18,19. This could be due to endogenous Long interspersed nuclear element-1 (LINE-1) retrotransposon mediated reverse transcription and integration into host genome 20 21. Alternative explanations for SARS-CoV-2 persistence include mechanisms used by other RNA viruses to persist in host cells: evasion of host immune response, infection of long-lived cells, and/or infection of immune-privileged tissues such as the brain, eyes, or testes, or diminished cell-mediated immunityt2,22. Additionally, SARS-CoV-2 may persist due to an insufficient immune response during acute COVID-1923,24.
We did not observe any correlation of viral RNA in plasma with acute disease severity as observed in previous reports 25,26. However, some positive correlation of Spike with peak WHO score during hospitalization and length of hospitalization was observed. This could be explained due to lack of clearance by cell-mediated immunity23,24 and/or decreased production of early functional, neutralizing antibodies both of which are considered a predictor of COVID-19 severity. While we didn’t perform whole RNA sequencing, we may be detecting viral RNA fragments because we observed a decrease in viral RNA in most of the samples after RNAse treatment. Further our results revealed that only 33% of PASC patients were positive for both plasma Spike and viral RNA.
In our recently published study, we found that levels of EN-RAGE, IL-18R1, and tissue factor (TF) in EVs were especially correlated with COVID-19 severity and length of hospitalization. We also found that EVs from hospitalized COVID-19 patients induce apoptosis of endothelial cells in vitro9. Here, we observed that EVs carry Spike in both acute COVID-19 and PASC patients. Given that SARS-CoV-2 replicates in cytosol using endosomal pathway27 and we found Spike to be associated with EVs, it is possible that SARS-CoV-2 can usurp EV biogenesis pathways and conceal itself within EVs for protection against neutralizing antibodies. In addition to the traditional complete viral particles that are secreted into the extracellular space, the viral biogenesis pathway can overlap with EV pathways in a number of ways, producing a continuum of particles like naked virions, EVs containing infective viral genomes, and quasi-enveloped viruses28 29. We did not observe viral RNA in EVs in post-COVID patients, though a very low copy number may persist below the limit of detection of ddPCR. Removal of surface proteoglycans from EVs with Heparinase II or treatment with Proteinase K showed a decrease in Spike levels suggesting Spike to be partly present on the surface of EVs via heparin sulfate interactions. However, the lack of total loss of Spike after treatment suggests that some of the Spike proteins may also be encapsulated in the EVs. Numerous studies shed light on the key role of Heparan sulfate proteoglycans with the SARS-CoV-2 binding at the cell membrane. Heparan sulfate interacts with the adjacent residues of the ACE2-binding site at the receptor binding domain (RBD) of the S1 subunit of the SARS-CoV-2 trimeric S-protein30,31.
We must acknowledge the limitations of our study. PASC has been difficult to study due to the heterogeneity of clinical presentations and lack of a uniform definition. Our PASC-positive patients were defined as those with self-reported, subjective symptoms which may result in an error in cohort designation. Many of the included patients had comorbidities making it difficult to ascertain if reported symptoms were worsening chronic conditions or from PASC. Another limitation is that our Acute cohort were all hospitalized patients and received standard-of-care therapy for their COVID-19 infection, which has evolved over the pandemic. Patients enrolled in the PASC cohort did not all have moderate/severe initial infection and most only had mild acute infections. Thus the PASC cohort could not be compared directly to our acutely infected cohort which lacked individuals with mild infection. We also did not have access to the type of variant each patient was infected with or their viral load due to institutional laboratory constraints and acknowledge that we could not discount re-infection with COVID during long-term follow-up routinely; however, samples were drawn 8–12 weeks after known past infection. An earlier study reports that PASC patients with mild symptoms during acute infection have higher levels of viral RNA 32 which could be why we observed increased positivity of viral RNA in the PASC-positive group compared to Acute patients. Nevertheless, in a small subset of patients (N=12) where we were able to follow hospitalized patients from their moderate to severe acute illness into the PASC cohort, we found an increase in viral copy numbers in a few individuals compared to a decrease in viral RNA in PASC-negative group. We also acknowledge the small sample size of PASC-negative patients who were recruited from ACTIV-2 had possibly received treatments that could have impacted our results. Despite these limitations, our study reveals intriguing findings that the persistence of Spike protein and viral RNA fragments from SARS-CoV-2 is associated with PASC and may contribute to its development.
Acknowledgements:
We acknowledge Aaron Lehman, Department of Internal Medicine, KUMC, for his help in processing blood samples and; Luigi Boccardi and Maggie Chen for their assistance in maintaining clinical data and COVID-19 biorepository.
Funding Statement:
The funds to carry out the study were supported by National Institute of Health (NIH) grants R01 HL129875 awarded to N.K.D.
Footnotes
Conflict of Interest Disclosure:
The authors report no conflict of interest.
Ethics Approval Statement:
The study was approved by the IRB at the University of Kansas Medical Center.
Data availability statement:
Data will be available upon a reasonable request
REFERENCES
- 1.(WHO) WHO. WHO Coronavirus (COVID-19) Dashboard. In:2022.
- 2.Griffin DE. Why does viral RNA sometimes persist after recovery from acute infections? PLoS Biol. 2022;20(6):e3001687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groff D, Sun A, Ssentongo AE, et al. Short-term and Long-term Rates of Postacute Sequelae of SARS-CoV-2 Infection: A Systematic Review. JAMA Netw Open. 2021;4(10):e2128568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfaff ER, Girvin AT, Bennett TD, et al. Identifying who has long COVID in the USA: a machine learning approach using N3C data. Lancet Digit Health. 2022;4(7):e532–e541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC. Long COVID or Post-COVID Conditions. 2022.
- 6.Daniel C, Sydney S, Sabrina R, et al. Nature Portfolio. 2022.
- 7.Kumar N, Zuo Y, Yalavarthi S, et al. SARS-CoV-2 Spike Protein S1-Mediated Endothelial Injury and Pro-Inflammatory State Is Amplified by Dihydrotestosterone and Prevented by Mineralocorticoid Antagonism. Viruses. 2021;13(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang R, Mamun A, Dominic A, Le NT. SARS-CoV-2 Mediated Endothelial Dysfunction: The Potential Role of Chronic Oxidative Stress. Front Physiol. 2020;11:605908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishnamachary B, Cook C, Kumar A, Spikes L, Chalise P, Dhillon NK. Extracellular vesicle-mediated endothelial apoptosis and EV-associated proteins correlate with COVID-19 disease severity. J Extracell Vesicles. 2021;10(9):e12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fajnzylber J, Regan J, Coxen K, et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020;11(1):5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bracht JWP, Gimenez-Capitan A, Huang CY, et al. Analysis of extracellular vesicle mRNA derived from plasma using the nCounter platform. Sci Rep. 2021;11(1):3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutierrez Garcia G, Galicia Garcia G, Zalapa Soto J, et al. Analysis of RNA yield in extracellular vesicles isolated by membrane affinity column and differential ultracentrifugation. PLoS One. 2020;15(11):e0238545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao Y, Su B, Guo X, et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182(1):73–84 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, Lee A, Grigoryan L, et al. Mechanisms of innate and adaptive immunity to the Pfizer-BioNTech BNT162b2 vaccine. Nat Immunol. 2022;23(4):543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christianson HC, Svensson KJ, van Kuppevelt TH, Li JP, Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc Natl Acad Sci U S A. 2013;110(43):17380–17385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; [computer program]. 2020. [Google Scholar]
- 17.Clausen TM, Sandoval DR, Spliid CB, et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell. 2020;183(4):1043–1057 e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Richards A, Barrasa MI, Hughes SH, Young RA, Jaenisch R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc Natl Acad Sci U S A. 2021;118(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smits N, Rasmussen J, Bodea GO, et al. No evidence of human genome integration of SARS-CoV-2 found by long-read DNA sequencing. Cell Rep. 2021;36(7):109530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet. 2009;10(1):19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanciano S, Cristofari G. Measuring and interpreting transposable element expression. Nat Rev Genet. 2020;21(12):721–736. [DOI] [PubMed] [Google Scholar]
- 22.Paz-Bailey G, Rosenberg ES, Doyle K, et al. Persistence of Zika Virus in Body Fluids - Final Report. N Engl J Med. 2018;379(13):1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakraborty S, Gonzalez JC, Sievers BL, et al. Early non-neutralizing, afucosylated antibody responses are associated with COVID-19 severity. Sci Transl Med. 2022;14(635):eabm7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Beltran WF, Lam EC, Astudillo MG, et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184(2):476–488 e411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs JL, Bain W, Naqvi A, et al. Severe Acute Respiratory Syndrome Coronavirus 2 Viremia Is Associated With Coronavirus Disease 2019 Severity and Predicts Clinical Outcomes. Clin Infect Dis. 2022;74(9):1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagman K, Hedenstierna M, Gille-Johnson P, et al. Severe Acute Respiratory Syndrome Coronavirus 2 RNA in Serum as Predictor of Severe Outcome in Coronavirus Disease 2019: A Retrospective Cohort Study. Clin Infect Dis. 2021;73(9):e2995–e3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shang C, Zhuang X, Zhang H, et al. Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice. Virol J. 2021;18(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altan-Bonnet N Extracellular vesicles are the Trojan horses of viral infection. Curr Opin Microbiol. 2016;32:77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longatti A, Boyd B, Chisari FV. Virion-independent transfer of replication-competent hepatitis C virus RNA between permissive cells. J Virol. 2015;89(5):2956–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L, Chopra P, Li X, et al. Heparan Sulfate Proteoglycans as Attachment Factor for SARS-CoV-2. ACS Cent Sci. 2021;7(6):1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalra RS, Kandimalla R. Engaging the spikes: heparan sulfate facilitates SARS-CoV-2 spike protein binding to ACE2 and potentiates viral infection. Signal Transduct Target Ther. 2021;6(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batra A, Clark JR, Kang AK, et al. Persistent viral RNA shedding of SARS-CoV-2 is associated with delirium incidence and six-month mortality in hospitalized COVID-19 patients. Geroscience. 2022;44(3):1241–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be available upon a reasonable request


