Abstract
Atopic dermatitis (AD) is a multifactorial disease with underlying barrier disruption and altered microbial flora, resulting in dry skin, eczematous inflammation with persistent pruritis. Mouse models have been heavily utilized to investigate AD pathophysiology. Among various AD mouse models, AD-like inflammation induced by topical calcipotriol, a vitamin D3 analog referred to as MC903 in experimental settings, is a versatile model that can be applied to any strains of mice, that can be used for immunological and morphological studies. Herein, we provide basic protocols for the topical application of MC903 and approaches to assess phenotypes. After inducing AD-like inflammation, the skin is harvested for flow cytometry analysis, as well as histological and immunofluorescence microscopy analyses. The combination of these approaches will enable accurate characterization of the degree of inflammation, type of inflammatory infiltrate, and localization of immune infiltrates.
Basic Protocol 1:
Application of MC903 and gross phenotype assessment
Basic Protocol 2:
Processing the skin for flow cytometry analysis
Support Protocol 1:
Skin immune cell surface staining and flow cytometry analysis
Basic Protocol 3:
Harvesting skin for histological analysis
Basic Protocol 4:
Immunofluorescence staining to identify immune cell infiltrates
Keywords: Atopic dermatitis, MC903, flow cytometry, histological analysis
INTRODUCTION:
AD is a multifactorial disease that involves genetic factors, the microbiome, and immune responses that lead to overt inflammation. There is still much to be learned about the pathophysiology has not been fully understood and mouse models have been utilized as the most important research tool to investigate its pathophysiology in vivo. Various AD mouse models have been established, which we can be divided into three categories: 1) Inbred models, 2) Genetically-engineered models, and 3) models induced by epicutaneous application of exogenous agents (Kim, Kobayashi, & Nagao, 2019). Understanding the validity, benefits, and limitations of each model carefully would allow researchers to the best matched mouse model based on what they want to investigate. MC903 is calcipotriol, which is widely used to induce atopic dermatitis-like inflammation (M. Li et al., 2009; Wang et al., 2021). Compared with inbred or genetically engineered models, the MC903 model can be applied to any strains of mice without the need to crossbreed a certain mouse line to another background. For example, studying the roles of T cells in the filaggrin-mutated Flaky tail mice requires it to be crossed to a lymphocyte-deficient background, such as the Rag2−/− mice. However, MC903 can be directly applied to Rag2−/− mice, thereby providing huge time, logistic and financial advantages.
Here, we provide a practical protocol in which atopic skin inflammation can be induced by topically applying the vitamin D3 analogue, MC903 to both mouse ears and trunk skin and on how phenotypes are evaluated. We also provide protocols for flow cytometry, and histological analyses of skin lesions that will enable immunophenotyping of the immune cell infiltrates and their localization.
BASIC PROTOCOL 1: Application of MC903 and gross phenotype assessment
MC903 is used topically to treat psoriasis in humans. Although this compound does not induce AD in humans, the finding that topical application of MC903 in mice induces skin inflammation with a bias towards type 2 immunity has established it as a convenient model to induce atopic dermatitis-like skin inflammation in various contexts. This model relies neither on epidermal barrier defect nor dysbiosis and can be used to assess immune responses directly in various genetically engineered models, in both C57BL/6 and BALB/c genetic backgrounds (Malhotra et al., 2018). The following protocol describes the preparation and application of MC903 in C57BL/6 mice and how to analyze gross skin phenotype. Application of MC903 is commonly done on ear skin in part because it facilitates quantification of ear thickness as a measure of disease activity. In this protocol, we also apply MC903 to the back skin, given the limited amount of tissue that can be obtained for flow cytometry from the ear skin. We also apply MC903 in two different concentrations. Researchers may utilize the optimal concentration depending on their needs. For example, if a genetically engineered mouse model is expected to display exacerbated phenotype as compared to controls, one might start by using 40 μM concentration. Conversely, if the mouse model of interest is expected to show attenuated phenotype, then one might start using 80 μM concentration.
Materials:
Mice: C57BL/6, 8 weeks old, female (The Jackson Laboratory, cat. No. 000664)
Isoflurane
O2 source
Hair clipper
Microfoil shaver
MC903, 40 or 80 μM concentration (Tocris Bioscience, cat. 2700) (see REAGENTS AND SOLUTIONS)
100% ethanol (EtOH)
200 μl pipet
Digital caliper (Mitsutoyo, cat MDC-25MJ)
- Two days before the MC903 application, shave dorsal skin
- Anesthetize mice using isoflurane.
- Apply ear tags to the right ear to enable monitoring of individual mice.
-
Shave an area of roughly 2 cm x 2 cm with a hair clipper. This eliminates the terminal hairs.Then finely shave remaining hair using a micro foil shaver. This eliminates the stubs so that skin surface can be better observed.
- Carefully smear MC903 solutions or EtOH (vehicle control) directly onto one ear and shaved skin using a pipette.
- Apply 10 uL of MC903 solution each to ventral and dorsal sides of the left ear of experimental mice and ethanol to control mice.
- Apply 90 uL of MC903 or ethanol to the shaved patch.
- Typically, the total amount of MC903 is displayed in nmol, where 0.25 to 1 nmol are applied to mouse ears (C. Li et al., 2017; M. Li et al., 2009; Wang et al., 2021). 20 μl of 40 μM concentration would equate to 0.8 nmol.
Repeat step 4 daily for 11 days.
Take photos of the skin phenotype and measure ear thicknesses with a digital caliper on Days 0, 4, 8, and 12, immediately prior to each application (see Fig.1A).
Mice are euthanized immediately after checking gross phenotype and measurement ear thickness on Day 12 to be processed for the next protocol.
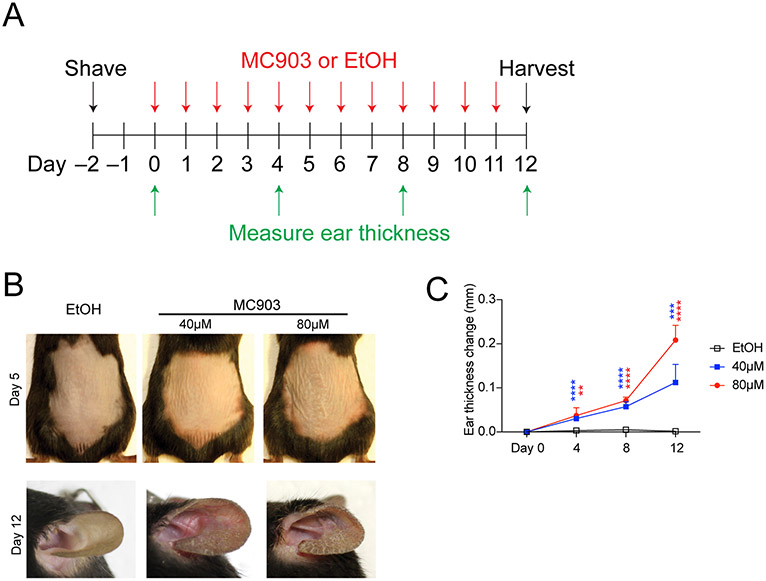
Figure 1. Experimental scheme and gross phenotype.
A: Experimental schedule of MC903 topical application. B. Representative images of gross phenotype of back and ear at day 5 and day 12, respectively. MC903 treated mice begin to show gross signs of inflammation at around day 5 on either skin site. C: Ear thickness typically begins to increase at around day 4. Statistics are performed between EtOH group and each MC903 group. EtOH (n=5), 40 μM MC903 (n=5) and 80 μM MC903 (n=6).
After shaving the back skin with micro foil shavers, anagen (hair growth) is induced around at day 6 to 9 and the skin becomes pigmented in C57BL/6 mice, masking the skin inflammation in the central areas. However, erythema can be observed in the periphery of anagen patches. Using BALB/c mice may facilitate evaluation of the gross phenotype, if applicable.
BASIC PROTOCOL 2 : Processing the skin for flow cytometry analysis
Introductory paragraph:
Histological analyses, immunofluorescence microscopy, and immunohistochemistry are useful for studying the spatial distribution of inflammation in skin sections. However, quantification of immune cell subsets may be challenging in sections. The use of flow cytometry will enable an unbiased analysis of the whole lesion and detailed immune cell subsetting can be performed using various panels of antibodies. The generation of single-cell suspensions from whole skin (epidermis and dermis) involves a combination of mechanical dissociation and enzymatic digestion. Liberase, which is a mixture of collagenase blend and thermolysin, is critical for the latter process. The type of Liberase that we have used has evolved from CI (Nagao et al., 2009), TL (Adachi et al., 2015) and now T-Flex (Sakamoto et al., 2021). We have not observed notable changes in the efficiency of skin digestion.
The following protocol describes how to make skin cell suspension for flow cytometry analysis.
Materials:
CO2 chamber
37°C incubator
Refrigerated centrifuge
Ice
Ophthalmic scissors
2 forceps with watchmaker forceps with curved tips
Size 15 scalpel (e.g. Kai medical, #515-A)
Kimwipe®
100 mm TC-treated Cell Culture Dish (e.g. BD Falcon)
60 mm TC-treated Culture Dish (e.g. Corning)
6 well plate (e.g. Corning)
50 ml conical centrifuge tubes (e.g., BD Falcon)
40 μm cell Strainer (e.g. BD Falcon)
100 μm cell Strainer (e.g. BD Falcon)
12ml syringe (e.g. Corning)
5% FACS buffer (see REAGENTS AND SOLUTIONS)
Whole skin digestion solution (see REAGENTS AND SOLUTIONS)
0.25 % Trypsin-EDTA
96-well Clear V-Bottom Not Treated Polypropylene Storage Microplate (Falcon®, cat#353263)
Sealing Tape (Themo Scientific, cat# 15036)
Automated cell counter (Invitrogen™ Countess™ Automated Cell Counter)
Mice from basic protocol 1
Add 5 ml of whole skin digestion solution per well of 6-well plate and place on ice until use.
-
Euthanize control (EtOH) and experimental (MC903) mice on Day 12 in a CO2 chamber.
The number of mice one can euthanize at once will depend on the speed at which each researcher is able to process the subsequent steps. Cell viability may decrease if it takes longer time to process samples.
Shave the back skin gently to get rid of excess hair (optional).
Excise the ears and dorsal skin where MC903 or ethanol was applied to using scissors and forceps.
Place the skin sample on the lid of a 10 cm petri dish and excise a square piece of 0.5 cm x 0.5 cm skin with a #15 scalpel. Float them on cold PBS and set aside for H&E and frozen sections (refer to Basic Protocols 3 and 4). Identical anatomical sites should be taken from control and experimental mice.
Float remaining skin on 10 ml of cold PBS in 10 cm petri dishes while other samples are processed.
Transfer the remaining skin samples onto lids of 10 cm petri dishes. Scrape off hypodermis and extra connective tissues by using 2 forceps with watchmaker forceps with curved tips.
Put the skin samples back into each petri dish and rinse off debris with cold PBS.
Remove extra PBS by briefly placing the skin samples on Kimwipe® and then transfer them into 6 cm petri dishes. Take 1 ml of whole skin digestion solution from the 6-well plates prepared earlier and add to the skin samples.
Mince skin samples well using scissors. This mincing process is the most critical part of skin digestion. Mince until the skin samples become paste-like in appearance. To minimize tissue damage, this procedure should be on ice.
Transfer minced skin into 6-well plates containing 4 ml of whole skin digestion solution.
Incubate cells for 2 hours at 37°C in an incubator.
10 min before the incubation is done, add 1 ml of 0.25% of Trypsin-EDTA into each well.
At the end of 2 hours, add 4ml of 5% FACS buffer into each well to stop enzymatic reaction.
Mechanically dissociate cells with 12 ml syringes. Pump the cell suspension 10 times.
Filter the cell suspension through 100 μm Cell Strainers set on 50 ml conical centrifuge tubes.
Centrifuge for 8 min at 400 x g. Make sure pellets are formed. Discard the supernatant.
Resuspend cells with 10 ml of 5% FACS buffer and pass the solution through 40 μm Cell Strainers set on 50 ml conical centrifuge tubes.
Centrifuge for 8 min at 400 x g and discard the supernatant. Resuspend cells with 5% FACS buffer. The volume should be under 300 μl.
Count cells with an automated cell counter. About 5 x 105 cells per cm2 can be expected.
Transfer cell suspensions into each well of 96-well clear V-Bottom plate.
Centrifuge the plate for 3 min at 400 x g and discard the supernatant.
Resuspend and wash the cells with 200 μl of cold PBS and centrifuge the plate for 3 min at 400 x g. Discard supernatant. Proceed immediately to the first step of Support Protocol 1.
SUPPORT PROTOCOL 1: Cell surface staining and flow cytometry analysis
Flow cytometry analysis enables the distinction of immune cell subsets and to study functional characteristics of each subset. In this protocol, we introduce a basic staining protocol for skin single-cell suspensions with which CD4 and CD8 T cells can be identified. If detection of cytokine expression or transcription factors are needed, additional cytokine and intracellular staining is required.
Materials:
Zombie Yellow™ Fixable Viability Kit
TruStain FcX™ (anti-mouse CD16/32) Antibody, clone:93
Antibodies (Table 1), fluorescently labeled based on the laser setting of flow cytometer
Table1.
Antibodies used for analysis of skin cells by flow cytometry
| Antibody markers |
Company | Catalog # | Clone | Fluorophore | Final concentration |
|---|---|---|---|---|---|
| CD45 | Biolegend | 103134 | 30F-11 | Brilliant Violet 421 | 1/200 |
| CD90.2 (Thy1.2) | Biolegend | 105305 | 30H-12 | FITC | 1/200 |
| CD3ε | Biolegend | 100330 | 145-2C11 | APC-Cy7 | 1/200 |
| CD4 | BD Horizon | 562285 | RM4-5 | PE-CF594 | 1/200 |
| CD8a | Biolegend | 100722 | 53-6.7 | PE-Cy7 | 1/100 |
| CD103 | Biolegend | 121414 | 2E7 | APC | 1/100 |
| CD2 | Biolegend | 100108 | RM2-5 | PE | 1/100 |
Refrigerated centrifuge
5 ml polystyrene round-bottom tube with cell-strainer cap (Falcon®, cat#352235)
LSR Fortessa (BD Biosciences) or equivalent
FlowJo software (FlowJo, LLC)
Single-Cell Suspension (see basic protocol 2)
Dilute Zombie Yellow™ Fixable Viability Kit with PBS at 1:200 dilution. Add 100 μl of this solution into each well of spun-down cells (Basic Protocol 2) and incubate for 15 min covered from light at room temperature (RT). During incubation time, prepare antibody master mix (Table 1) so that the cell surface staining process can be executed in a timely manner.
Centrifuge plate for 3 min at 400 x g and discard supernatant.
Dilute Fc block at 1:200 dilution with 5% FACS buffer. Add 100 μl into each well and incubate for 5 min in dark on ice.
Centrifuge for 3 min at 400 x g and discard supernatant.
Add 100 μl of antibody master mix (see table 1) into each well and incubate for 25 min in dark on ice.
Centrifuge for 3 min at 400 x g and discard the supernatant.
Resuspend and wash the cells with 200 μl of 5% FACS buffer.
Centrifuge for 3 min at 400 x g and discard the supernatant.
Repeat 7-8 steps once.
Resuspend cells in 200 μl of 5% FACS buffer and pass through the filter caps of 5 ml polystyrene round-bottom tube immediately before flow cytometry analysis.
If cell acquisition for flow cytometry cannot be done on the same day, fix the cells with FluoroFix™ Buffer (Biolegend cat 422101). After step 9, add 200 μl of FluoroFix™ Buffer into each well and incubate for 30 min at RT. Centrifuge for 3 min at 400 x g and discard the supernatant. Resuspend and wash the cells with 200 μl of 5% FACS buffer. Centrifuge for 3 min at 400 x g and discard the supernatant. Repeat this washing step once and resuspend the cells with 200 μl of 5% FACS buffer. Cover the plate with aluminum foil and store at 4 °C. Fluorescent dyes are usually stable up to 3 days.
Reference a figure/discuss the expected results of the flow data obtained in step 10.
BASIC PROTOCOL 3: Harvesting skin for histological analysis
Hematoxylin and eosin (H&E) stain is the most common staining method in histology, which not only allows us to study the morphology and spatial distribution of immune infiltrates but also to study tissue responses.
Materials:
Formalin solution, neutral buffered, 10% (Sigma, HT501128-4L)
Histology cassette and Foam Biopsy Pads (Thomas scientific, 1219D48 and 1150P62)
NPV.view 2 (HAMAMATSU PHOTONICS)
Excised skin sample from Basic Protocol 2
Place the 0.5 x 0.5 cm skin sample taken from Basic Protocol 2 (step 5) in a dish. Split the samples into two rectangular pieces, where the long axis of the rectangle is parallel to the direction of hair. One piece is used for H&E and the other for frozen sections (refer to Basic Protocol 4).
To prevent rolling up of the skin, we place the skin sample in a histology cartridge with sponges, which is immediately immersed in a bottle of 10% formalin and are outsourced to a histology service for sectioning and staining.
Provide instructions to the histology service to cut the skin sections along the long axis of the rectangle to generate vertical sections that expose the entirety of the hair follicle structures. Exposing the hair follicles will allow for better understanding the orientation in skin.
If applicable, scan the H&E slides and observe using the NPV.view 2 software.
BASIC PROTOCOL 4: Immunofluorescence staining
Immunofluorescence (IF) staining enables us to visualize molecules of interest, thereby enabling analysis of the distribution of specific immune cell subsets. Like flow cytometry, IF is ideal for detecting marker molecules expressed by a cell type of interest at the protein level. Here, we stained for CD4 and CD8 T cells.
Materials:
Cryostat
Tissue-Tek® OCT compound (SAKKURA, 4583)
Tissue-Tek® Cryomold® Intermediate (SAKKURA, 4566)
PARAFILM® (Bemis, HD234526A)
Cytology Adhesive Microscope Slides (Superfrost® Plus Gold Slides, Themo scientific, #6685L60)
Cover glasses
Acetone
Glass Staining Dish (Fisher scientific, #02-913-306)
PAP Pen (RESERCH PRODUCTS INTERNATIONAL, #195505)
TruStain FcX™ (anti-mouse CD16/32 antibody, clone:93)
Skim milk (Morinaga)
Phosphate Buffered Saline (PBS)
ProLong™ Gold antifade reagent with DAPI (Invitrogen #P36931)
Leica TCS SP8 confocal microscopy
Skin tissue from Basic Protocol 3 step 1
Rat anti-CD4 antibody (Table 2)
Table2.
Antibodies used for analysis of IF
| Antibody markers |
Company | Catalog # | Clone | Fluorophore | Final concentration |
|---|---|---|---|---|---|
| CD8 | Biolegend | 100723 | 53-6.7 | Alexa Fluor 488 | 1/100 |
| CD4 | Biolegend | 100506 | RM4-5 | Alexa Fluor 568* | 1/100 |
Conjugated in-house
Alexa Fluor™ 568 Antibody Labeling Kit (Thermo Fisher Scientific, A20184)
Rat anti-CD8 antibody (Table 2)
Embed the bisected half of skin tissues from Basic Protocol 3 step 1 in Tissue-Tek® O.C.T. compound and store air-tight at −80°C until use.
Cut frozen skin sections using a cryostat at thicknesses of 8 μm. Use adhesive slides to avoid the detachment of the skin sections during subsequent steps. After air-drying, store the slides in a slide box sealed with PARAFILM® and store at −20°C until use.
- Prepare primary antibodies (Table 2) and label the purified antibody if it is necessary.
- In this protocol, a rat anti-CD4 antibody (0.5mg/ml) was labeled in-house with the Alexa Fluor™ 568 Antibody Labeling Kit following the manufacturer’s instructions. This labeling should be done on a different date and the optimal concentration for staining should be confirmed by each user. The labeled anti-CD4 antibody was used to stain sections together with rat anti-CD8 antibody pre-labeled with Alexa Fluor™ 488. Using fluorescent dye-labeled primary antibodies enable the simultaneous use of two or more antibodies generated in the same species.
Fix slides in acetone in Glass Staining Dish at −20°C for 2 min.
Take out slides from acetone and air dry.
Rehydrate the slide in PBS 5min at RT. Do not allow sections to dry in subsequent steps.
Circle the section with PAP pen to create hydrophobic barriers. It helps to prevent blocking buffer and antibody solution from spreading or running off the slides.
Block the section with PBS containing 3% skim milk and Fc block (1:200) for 1hr at RT.
Rinse the slide in PBS.
Dilute primary antibodies with blocking buffer and add antibody solution on each section (see Table 2).
Incubate overnight at 4°C.
Wash the slide in PBS for 5min. Repeat 2 times.
Mount on Prolong gold with DAPI and allow the mounting media to harden over night at RT. If the mounting media is not dry on the next day, let dry for another day.
Scan the slides with confocal microscopy at the appropriate excitation wavelength of fluorophore. Slides may be stored protected from light at 4°C for several months should re-acquiring of the images be necessary.
REAGENTS AND SOLUTIONS:
MC903
Dissolve MC903 (Tocoris, purity ≥98%) with 100% ethanol (EtOH) and make 1 mM stock solution. For example, dissolve 10mg MC903 with 24.24 ml EtOH to make 1mM. Make 200 μl/tube aliquots stock solution and store at −20°C.
Right before application, dilute stock solution with 100% EtOH to make 40 μM and 80 μM solutions. For example, to make 40 μM solution, make 1:25 dilution of 1mM stock solution with 100% EtOH.
5% FACS Buffer
Add 5% volume of fetal calf serum into PBS and store at 4°C.
Liberase stock solution
Liberase T-Flex Research Grade (ROCHE, cat 05989132001) contains one bottle of 500mg collagenase blend and 2 vials of 15mg thermolysin. All procedure should be done on ice.
Reconstitute the collagenase vial with 20ml of cold PBS. Pipette gently until it fully dissolves.
Reconstitute each thermolysin vial with 15ml of cold PBS.
Add the two thermolysin solution into the collagenase bottle (total 50ml) to make Liberase stock solution. This contains 10 mg/ml of collagenase and 0.6 mg/ml of thermolysin.
Aliquot 1ml of Liberase stock solution into Eppendorf tubes.
Store the aliquots at −20°C for up to 6 months.
DNase stock solution
Dissolve 1 mg Deoxyribonuclease I from bovine pancreas (Sigma, cat DN25) in 1 mL of 0.15 M NaCl to make 1 mg/ml DNase stock solution.
Whole skin digestion solution
Supplement 40 ml of Roswell Park Memorial Institute (RPMI) 1640 medium with 1ml of Liberase stock solution and 40 μl of DNase stock solution.
COMMENTARY:
Background Information:
Atopic dermatitis (AD) is a common chronic inflammatory skin disease characterized by impaired epidermal barrier that results in chronic, eczematous skin inflammation. Loss-of-function mutation in FLG, a gene encoding the structural protein filaggrin that is degraded into natural moisturizing factors, has been identified as a major predisposition factor in AD patients (Palmer et al., 2006). Although FLG mutation is relatively prevalent in the Caucasian population with up to 50% of AD patients harboring the mutation, it is less prevalent in other ethnic populations (Brunner & Guttman-Yassky, 2019). Nevertheless, this finding established that genetically impaired epidermal barrier underlies AD pathophysiology. More recently, Genome-Wide Association Studies have identified various genes that are associated with AD not restricted to genes that regulate barrier function but also those that regulate immunity (Sandilands, Sutherland, Irvine, & McLean, 2009).
Loss of microbial diversity (dysbiosis) in the skin bacterial flora that likely arise due to impaired barrier functions may also play an important role in driving skin inflammation in AD(Kobayashi et al., 2015). It has been identified over 50 years ago that the pathobiont, Staphylococcus aureus, can be frequently isolated from AD skin. 16S microbiome analyses have demonstrated that S. aureus predominates the microbiome of active AD lesions, and metagenomic analyses suggest that different S. aureus strains may have distinct potentials in promoting immune responses in the skin (Kong et al., 2012; Paller et al., 2019). Whereas the exact roles of S. aureus in driving atopic skin inflammation remains to be elucidated, the findings that atopic skin inflammation can be mitigated via antibiotics in AD mouse models with spontaneous dysbiosis suggest that dysbiosis plays an important role in driving pathology in AD.
Historically, immunopathophysiology of AD has been compared to that of psoriasis(Nomura et al., 2003). Psoriatic skin inflammation is enriched in IL-17, as well as interferon gamma-producing T cells. In contrast, T cells in AD skin is Th2-biased and produce the type 2 cytokines IL-13. Interestingly, AD skin is also enriched in CD8 T cells that produce IL-13, highlighting both CD4 and CD8 T cell subsets as important sources of this cytokine(Hijnen et al., 2013). Mouse as well as human studies have also identified group 2 innate lymphoid cells (ILCs) as important sources of IL-13 (Alkon et al., 2022; Leyva-Castillo et al., 2020). The efficacy of the anti-IL4/IL13 receptor blocking antibodies in human AD points to the importance of this cytokine-cytokine receptor signaling axis in driving AD pathology(Beck et al., 2014). The MC903-induced AD-like inflammation model can be utilized to study aspects of these immune responses. It is a versatile model in that it can be applied to any mouse strains or genetically-engineered mouse models, in the absence of other AD factors such as barrier disruption or dysbiosis.
Critical Parameters:
Hair follicles are unique structures in skin that undergo cyclical bouts of growth that generates hair. The growing phase is called the anagen and the resting phase is telogen. Mice usually exit from a uniform phase of anagen by 8 weeks of age, after which spontaneous anagen patches can be observed after 12 weeks of age. We recommend starting MC903 application between 8-10 weeks of age to initiate the experiment in a uniform, telogen skin. The shaving of hair and the inflammation caused by MC903 does induce anagen, which may render it difficult to observe gross phenotypes due to darkening of skin in mouse strains with black fur coats, requiring histological analysis for disease activity evaluation. This is not an issue in BALB/c background with white fur coats, provided that this background is appropriate for the particular research question. It is also recommended to use female mice for stable acquisition of single cell suspensions for flow cytometry. Male mice naturally develop thicker skin which is challenging to digest, and the physical trauma from fighting may induce secondary inflammatory changes.
Ear skin has been historically used to induce MC903 mediated atopic dermatitis-like inflammation. Ear skin is useful MC903 can be applied on one ear and vehicle control on the other. This site is also convenient to quantify ear thickness as a measure of disease activity. However, ear skins lack the hypodermis (typical ILC2s reside in the adipose tissue, including the hypodermis in the trunk skin (Kobayashi et al., 2019)), have non-cycling hair follicles, and sparsely harbor resident memory T cells (Hirai et al., 2021). The advantage of using back skin is that it represents most of the body and more cells may be retrieved, enabling microscopy and multiple flow cytometry analyses from one mouse. Researchers should consider these differences when they determine the site of MC903 application. In both the ears and the back skin, the cartilage layer and the hypodermal layer are scraped off prior to preparation for flow cytometry, since these layers may interact with enzymatic digestions.
Troubleshooting:
Table 3.
Troubleshooting Guide for making whole skin cell suspension
| Problem | Possible Cause | Solution |
|---|---|---|
| Cells to be used for flow cytometry are lost after the first centrifuge | Skin digestion was not sufficient. | Make sure to mince the skin well, until it become paste-like. It is also important to use sharp scissors each time. |
| Insufficient enzyme activity | A common mistake is adding FBS into the enzyme mix, which can result in decreased enzymatic activity. If the above is not the case, consider changing to a new batch of Liberase. | |
| Insufficient filtration | Remaining debris may lead to a loss of cells. If clumps of debris are observed in the cell suspension, filter again. | |
| Positive signals are not observed in flow cytometry | Compensation issue | Re-run compensation again |
| Forgetting to add all antibodies | Make sure all antibodies are added | |
| High background signal in immunofluorescence images | Insufficient blocking | Blocking non-specific signals are an important step in immunofluorescence microscopy. Make sure to block for at least an hour at room temperature. |
| Concentration of the primary antibody is too high | High concentration of the primary antibodies may lead to high background, especially in the epidermis. Optimize the concentration of each antibody. Staining overnight at 4°C may help decrease background. It should be noted that polyclonal rabbit antibodies are known to provide high background in the epidermis. | |
| Cross-reactivity of secondary antibodies to Fc block | If secondary antibodies are used, the blocking buffer should not contain Fc block, which is made in rats. An anti-rat secondary antibody would recognize not only the primary antibody, but also Fc block antibody. |
Statistical Analysis:
Student’s t test was used to measure significance between two groups and ANOVA with Tukey’s multiple comparison test was used to measure significance when comparing multiple groups. Significance is indicated as follows in all figures: ns = not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Understanding Results:
In this protocol MC903 or EtOH was topically applied for a total of 12 consecutive days and was harvested on Day 12 (Figure 1A). Skin erythema (redness) and increases in ear thickness was observed on around day 4, where 80 μM MC903 concentration induced more erythema and ear thickness increases as compared to 40 μM concentration (Figure 1B, 1C). Ear thickness continued to increase over the course of the experiment. If mice experience signs of stress or deterioration of health, they should be euthanized immediately.
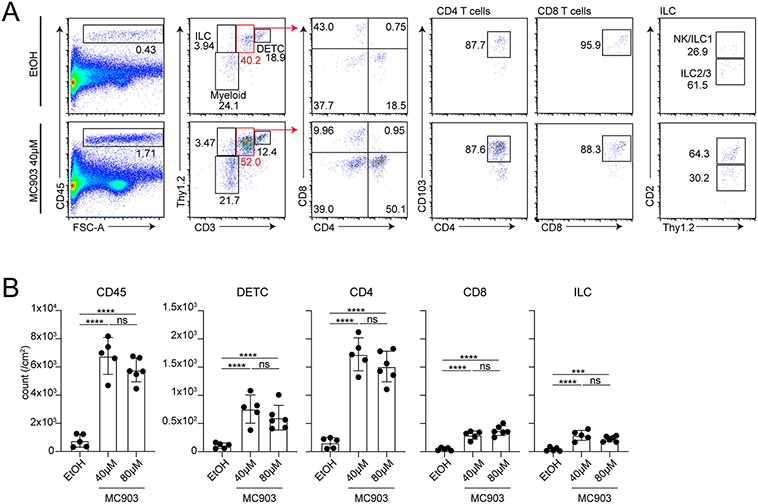
Flow cytometry analysis of MC903- or EtOH-treated skin revealed increased proportions of CD45+ immune cells as demonstrated in the CD45 versus forward scatter area plot (Figure 2A). Plotting the CD45+ gate for Thy1.2 versus CD3 enables the distinction of several immune cell subsets. Thy1.2+ CD3high population represents dendritic epidermal T cells, CD3mid population contain dermal γδ delta T cells, CD4, and CD8 T cells, Thy1.2+ CD3− population represents innate lymphoid cells, and Thy1.2− CD3− population represents myeloid cells. CD3mid population can be further distinguished for CD4 and CD8 T cells with markers of interest, in this case with a resident memory T cell marker, CD103. Thy1.2+ CD3− ILCs are a very minor population, which can be divided into CD2+ and CD2− populations, where the latter represents a mixture of group 2 and 3 ILCs. Further detection of lineage-determining transcription factors such as GATA3 and RORgt will enable their distinction. CD2+ ILCs are not well-characterized, but it likely represents a subset of ILC1 (Sakamoto et al., 2021). As quantified in Figure 2B, all lymphoid lineages analyzed increase in number upon MC903 application.
Figure. 2. Gating strategy for flow cytometry analysis.
A: Representative plots for immune cells from MC903 (40 μM)- or EtOH-treated skin. After excluding doublets and viability dye (Zombie yellow+ cells)-positive cells, CD45+ cells were plotted for CD3 vs Thy1.2. Thy1.2high CD3high cells are dendritic epidermal T cells (DETC). Thy1.2high CD3− cells represent ILCs. Thy1.2−CD3− cells contain a mixture of myeloid cells. Thy1.2+CD3mid contain dermal γδ T cells and αβ T cells. This gate can further be shown for CD4 T cells and CD8 T cells based on CD4 and CD8 expression. Most of CD4 and CD8 T cells express resident marker CD103. Thy1.2high CD3− ILC gate was further divided into NK/ILC1 and ILC2/3 based on CD2 expression. B: Each graph shows cell numbers of indicated populations per cm2. EtOH (n=5), 40 μM MC903 (n=5) and 80 μM MC903 (n=6).
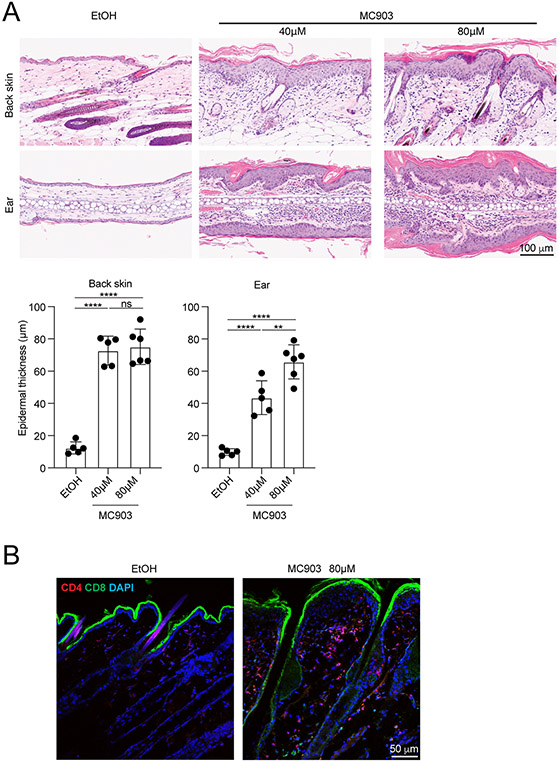
Histological analysis by H&E staining enables detailed studies on what tissue responses occur. It can be appreciated in Figure 3A that MC903 induces thickening of the cornified layer, epidermis, and dermis. Mononuclear infiltrates are observed predominantly in the dermis. Quantification of epidermal thickness is also a good measure for diseases activity, which can be measured on scanned images using NDP2.view 2.
Figure. 3. Histological and immunofluorescence analysis of skin sections.
A. H&E staining of back skin (upper panel) and ear skin (lower panel) from formalin-fixed paraffin-embedded skin samples. Epidermal thickness from scanned H&E images were measured in the NDP.view 2 software. B. CD4 and CD8 T cells were visualized by immunofluorescence microscopy by staining frozen skin sections with primary-labeled antibodies directed against CD4 (red) and CD8 (green). DAPI (blue) staining facilitates visualization of the skin structure.
Immunofluorescence microscopy visualizing CD4 and CD8 demonstrates a predominance of CD4 T cell infiltration (Figure 3B). ILCs are still challenging to demonstrate in sections because they require a combination of multiple markers to identify.
Time Considerations:
BASIC PROTOCOL 1: 14 days to complete MC903 application. 30 to 60 min per day.
BASIC PROTOCOL 2: 3-4 hours.
SUPPORT PROTOCOL: 3 hours. 24 hours would be necessary if additional cytokine and intracellular staining is to be performed.
BASIC PROTOCOL 3: 1 hour.
BASIC PROTOCOL 4: 1 to 3 hours for cutting frozen sections. 2-3 days to complete staining steps.
ACKNOWLEDGEMENTS:
This work was supported by the intramural research programs of NIAMS (1 ZIA AR041226-01)
K.S. was supported in part by KAKENHI (22K08427) and by Hamamatsu University School of Medicine Grant-in-Aid (655009, 711441).
Footnotes
CONFLICT OF INTEREST STATEMENT:
Authors declare no conflicts of interest.
DATA AVAILABILITY STATEMENT:
The data presented in this protocol are available from the corresponding author upon reasonable request.
LITERATURE CITED:
- Adachi T, Kobayashi T, Sugihara E, Yamada T, Ikuta K, Pittaluga S, Nagao K (2015). Hair follicle-derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nat Med, 21(11), 1272–1279. doi: 10.1038/nm.3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkon N, Bauer WM, Krausgruber T, Goh I, Griss J, Nguyen V, Stingl G (2022). Single-cell analysis reveals innate lymphoid cell lineage infidelity in atopic dermatitis. J Allergy Clin Immunol, 149(2), 624–639. doi: 10.1016/j.jaci.2021.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck LA, Thaçi D, Hamilton JD, Graham NM, Bieber T, Rocklin R, Radin AR (2014). Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N Engl J Med, 371(2), 130–139. doi: 10.1056/NEJMoa1314768 [DOI] [PubMed] [Google Scholar]
- Brunner PM, & Guttman-Yassky E (2019). Racial differences in atopic dermatitis. Ann Allergy Asthma Immunol, 122(5), 449–455. doi: 10.1016/j.anai.2018.11.015 [DOI] [PubMed] [Google Scholar]
- Hijnen D, Knol EF, Gent YY, Giovannone B, Beijn SJ, Kupper TS, Clark RA (2013). CD8(+) T cells in the lesional skin of atopic dermatitis and psoriasis patients are an important source of IFN-γ, IL-13, IL-17, and IL-22. J Invest Dermatol, 133(4), 973–979. doi: 10.1038/jid.2012.456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai T, Yang Y, Zenke Y, Li H, Chaudhri VK, De La Cruz Diaz JS, Kaplan DH (2021). Competition for Active TGFβ Cytokine Allows for Selective Retention of Antigen-Specific Tissue-Resident Memory T Cells in the Epidermal Niche. Immunity, 54(1), 84–98.e85. doi: 10.1016/j.immuni.2020.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kobayashi T, & Nagao K (2019). Research Techniques Made Simple: Mouse Models of Atopic Dermatitis. J Invest Dermatol, 139(5), 984–990.e981. doi: 10.1016/j.jid.2019.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Glatz M, Horiuchi K, Kawasaki H, Akiyama H, Kaplan DH, Nagao K (2015). Dysbiosis and Staphylococcus aureus Colonization Drives Inflammation in Atopic Dermatitis. Immunity, 42(4), 756–766. doi: 10.1016/j.immuni.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Voisin B, Kim DY, Kennedy EA, Jo JH, Shih HY, Nagao K (2019). Homeostatic Control of Sebaceous Glands by Innate Lymphoid Cells Regulates Commensal Bacteria Equilibrium. Cell, 176(5), 982–997.e916. doi: 10.1016/j.cell.2018.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Segre JA (2012). Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res, 22(5), 850–859. doi: 10.1101/gr.131029.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva-Castillo JM, Galand C, Mashiko S, Bissonnette R, McGurk A, Ziegler SF, Geha RS (2020). ILC2 activation by keratinocyte-derived IL-25 drives IL-13 production at sites of allergic skin inflammation. J Allergy Clin Immunol, 145(6), 1606–1614.e1604. doi: 10.1016/j.jaci.2020.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Maillet I, Mackowiak C, Viala C, Di Padova F, Li M, Ryffel B (2017). Experimental atopic dermatitis depends on IL-33R signaling via MyD88 in dendritic cells. Cell Death Dis, 8(4), e2735. doi: 10.1038/cddis.2017.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hener P, Zhang Z, Ganti KP, Metzger D, & Chambon P (2009). Induction of thymic stromal lymphopoietin expression in keratinocytes is necessary for generating an atopic dermatitis upon application of the active vitamin D3 analogue MC903 on mouse skin. J Invest Dermatol, 129(2), 498–502. doi: 10.1038/jid.2008.232 [DOI] [PubMed] [Google Scholar]
- Malhotra N, Leyva-Castillo JM, Jadhav U, Barreiro O, Kam C, O'Neill NK, Geha RS (2018). RORα-expressing T regulatory cells restrain allergic skin inflammation. Sci Immunol, 3(21). doi: 10.1126/sciimmunol.aao6923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao K, Ginhoux F, Leitner WW, Motegi S, Bennett CL, Clausen BE, Udey MC (2009). Murine epidermal Langerhans cells and langerin-expressing dermal dendritic cells are unrelated and exhibit distinct functions. Proc Natl Acad Sci U S A, 106(9), 3312–3317. doi: 10.1073/pnas.0807126106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura I, Goleva E, Howell MD, Hamid QA, Ong PY, Hall CF, Leung DY (2003). Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J Immunol, 171(6), 3262–3269. doi: 10.4049/jimmunol.171.6.3262 [DOI] [PubMed] [Google Scholar]
- Paller AS, Kong HH, Seed P, Naik S, Scharschmidt TC, Gallo RL, Irvine AD (2019). The microbiome in patients with atopic dermatitis. J Allergy Clin Immunol, 143(1), 26–35. doi: 10.1016/j.jaci.2018.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, McLean WH (2006). Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet, 38(4), 441–446. doi: 10.1038/ng1767 [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Jin SP, Goel S, Jo JH, Voisin B, Kim D, Nagao K (2021). Disruption of the endopeptidase ADAM10-Notch signaling axis leads to skin dysbiosis and innate lymphoid cell-mediated hair follicle destruction. Immunity, 54(10), 2321–2337.e2310. doi: 10.1016/j.immuni.2021.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandilands A, Sutherland C, Irvine AD, & McLean WHI (2009). Filaggrin in the frontline: role in skin barrier function and disease. Journal of Cell Science, 122(9), 1285–1294. doi: 10.1242/jcs.033969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Trier AM, Li F, Kim S, Chen Z, Chai JN, Kim BS (2021). A basophil-neuronal axis promotes itch. Cell, 184(2), 422–440.e417. doi: 10.1016/j.cell.2020.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this protocol are available from the corresponding author upon reasonable request.