Abstract
Pancreatic cancer (PC) is considered to be the seventh most common cause of cancer-related deaths. The number of deaths caused by PC is estimated to increase in the future. An early diagnosis of PC is crucial for improving treatment outcomes. The most common histopathological subtype of PC is pancreatic ductal adenocarcinoma (PDAC). MicroRNAs (miRNAs)—which are endogenous non-coding RNAs involved in the posttranscriptional regulation of multiple gene expression—constitute useful diagnostic and prognostic biomarkers in various neoplasms, including PDAC. Circulating miRNAs detected in a patient’s serum or plasma are drawing more and more attention. Hence, this review aims at evaluating the clinical value of circulating miRNA in the screening, diagnosis, prognosis and monitoring of pancreatic ductal adenocarcinoma therapy.
Keywords: circulating miRNA, pancreatic cancer, diagnosis, screening, prognosis, therapy monitoring, pancreatic ductal adenocarcinoma
1. Introduction
Pancreatic cancer (PC)remains a leading cause of cancer-related deaths. It is estimated that PC was responsible for 466,003 deaths in 2020,which puts it as the seventhleading cause of cancer-related deaths [1]. The incidence of PC has been either stable or slightly increased, with the highest incidence in North America, Europe and Australia/New Zealand [2]. The 5year relative survival rate is 11.5%, and it is largely due to late diagnosis and asymptomatic course at early stages [3,4].The number of deaths caused by PC is predicted to increase to 111,500 by 2025 in the European Union. In that case, PC will become the third leading cause of cancer-related deaths in the EU [5].
The risk factors of PC are tobacco smoking, obesity, diabetes mellitus, a diet rich in processed meat and lack of physical activity. Moreover, several gene mutations contribute to an increased risk of pancreatic cancer (p53, K-ras, p16, PRSS1) [6,7].
The most common histopathological subtype of PC is pancreatic ductal adenocarcinoma (PDAC). It is estimated that it accounts for more than 90% of PC diagnoses. The other types of PC are considered to have an impact on prognosis but have a lesser impact on management decisions [8].
Early diagnosis of PC is crucial for improving treatment outcomes. Ultrasound (US), computed tomography (CT), magnetic resonance imaging (MRI) and endoscopic ultrasonography (EUS) with EUS-guided fine-needle aspiration (EUS-FNA) are conventional methods used in PC diagnosis [9]. The advantages and disadvantages of these diagnostic methods are presented in Supplementary Table S1 (S1). Novel biomarkers related to PC, such as circulating tumor extracellular vesicles, circulating microRNAs (miRNAs), circulating tumor cells (CTCs), cell-free circulating tumor DNAs (cfDNAs) and long noncoding RNAs (lncRNAs), emerged in recent years as markers that might be useful as diagnostics and prognostic factors [10,11].
MiRNAs are endogenous non-coding RNAs that are involved in posttranscriptional protein synthesis regulation and play a significant role in cancer initiation, progression, proliferation, metastasis and chemotherapy resistance. They bind to 3’ untranslated regions (3’ UTRs) of targeted transcripts causing transcript repression or degradation- through the formation of RNA-induced silencing complexes (RISCs) [12,13].The diagnostic and prognostic potential of different miRNAs has been proved in various neoplasms, such as cervical cancer, lung cancer, gastric cancer or colorectal cancer [14,15,16,17]. Apart from tissue samples, miRNA may be detected in bodily fluids, including saliva (miR-3679-5p, miR-940, miR-21, miR-23a, miR-23b and miR-29, for example) [18,19], pancreatic fluid [20], peritoneal lavage fluid [21], cerebrospinal fluid [22] or urine [23]. Furthermore, numerous studies provide evidence that blood-based miRNAs may play a role in PC diagnosis, prognosis and surveillance during treatment [24,25]. Therefore, circulating miRNAs have gained more attention in recent years, as minimally invasive cancer markers and an alternative and supplementary approach to more invasive diagnostic methods currently available.
2. Literature Search
The literature search and study selection followed PRISMA guidelines [26]. The articles dating from 2003 to 2022 that were included in this review were evaluated based on their titles and abstracts. The eligibility criteria were based on the PICOS framework: (1) Participants: patients with PDAC; (2) Interventions: the detection of circulating miRNAs (3) Comparisons: non-PC controls; (4) Outcomes: area under the curve (AUC) of receiver operating characteristic (ROC) analysis, diagnostic sensitivity (SEN) and specificity (SPE),prognostic value, screening value or surveillance value of miRNA and (5) Study design: diagnostic research.
A combination of entry terms, including “plasma”, “circulating”, “blood-based”, “free”, “exosomal”, “serum”, “miRNA”, “microRNA” and ”pancreatic cancer” was used to search the PubMed and EMBASE databases. We also performed a manual search of potentially eligible studies based on reference lists of extracted reviews. The search was performed from 5 to 16 September 2022.
The extracted data included: specimen type, the investigated miRNAs and their expression levels in PDAC patients, whether miRNAs were used as single markers or combined as the panel, normalization control, AUC of ROC in diagnostic utility analysis, diagnostic SEN and SPE of the test, test comparison to carbohydrate antigen 19-9 (CA19-9) SEN and SPE, usefulness of miRNA as a prognostic factor, screening performance of miRNA and disease surveillance with miRNA.
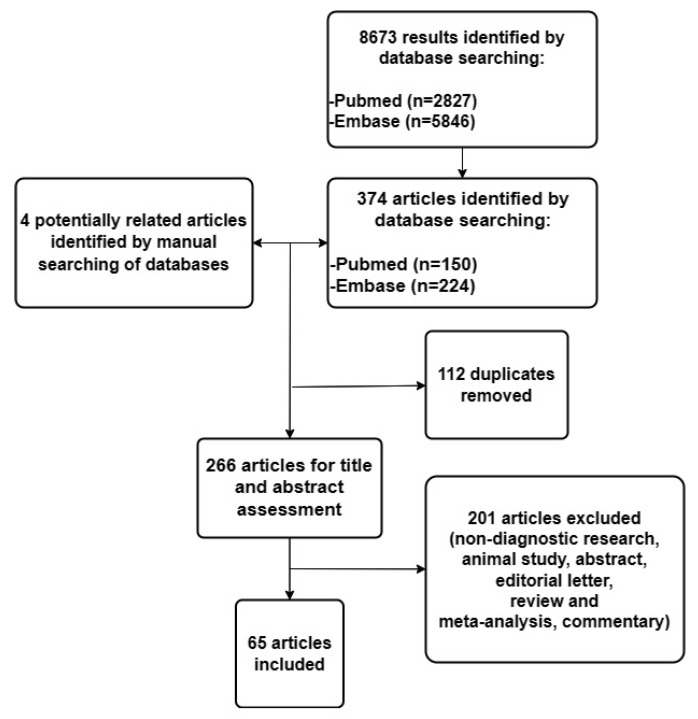
The flow diagram of the literature search and study selection process is presented in Figure 1.
Figure 1.
Flow diagram of the literature search and study selection process.
The aim of this study was to provide a literature review on the diagnostic, prognostic, predictive, screening and monitoring of the therapeutic value of circulating miRNA in patients diagnosed with PDAC.
The risk of bias was assessed with the QUADAS-2 tool, which assesses the risk in four domains: “patient selection”, “index test”, “reference standard” and “flow and timing” [27]. We have recognized that studies evaluated in this review exhibited a high risk of bias in the “patient selection” domain since they included patients with a definitive diagnosis. The risk of bias was intermediate in the “index test” domain because articles included in this review were trying to establish the threshold for the studied miRNAs. The risk of bias in “reference standard” and “flow and timing” was considered low.
3. Diagnostic Performance of Circulating miRNA
There were 44 studies included in this review that covered the topic of the diagnostic value of 110 single circulating miRNAs [28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70]. In those studies, miRNA expression was evaluated with real-time quantitative polymerase chain reaction (RT-Q-PCR). Details of the studies and their results, with predicted targeted genes, based on “miRBase: the microRNA database” are included in Supplementary Table S2. A hundred and ten different single miRNAs were tested 145 times in included the 44 studies. The diagnostic value determined with receiver operating characteristic curves (ROC) and their areas under the curve (AUC) varied within those studies from 0.618 to 1.0. In 17.93% (n = 26) of cases the AUC value reached or exceeded 0.9. Sensitivity (SEN) and specificity (SPE), if provided in the selected studies, ranged from 58% to 100% and 50% to 100%, respectively. The samples for tests were collected from blood plasma in 53.34% of cases and from blood serum in 49.66%. In 15.86% of cases, the tested miRNA was down-regulated compared to controls and, in 84.14% of cases, up-regulated compared to controls. In 42.76% of cases (n = 62), an endogenous control was made to normalize the data; in 51.03% of cases, an exogenous control was employed (n = 4). Both methods were used in 2.07% of cases (n = 3) In 4.14% of cases (n = 6), we did not find precise information about normalization methods (S2).
In 25 studies [28,30,31,36,38,39,43,44,45,49,50,51,54,55,57,58,62,64,65,67,70,71,72,73,74], miRNA was combined with other miRNAs, protein markers or CA19-9 into 33 diagnostic panels. The AUC value in ROC analyses was increased and ranged from 0.7207 to 1.0 (with 2 of them lacking data on AUC). SEN and SPE varied from 64% to 100% and 73% to 100%, respectively. In 51.52% (n = 17), cases the AUC value reached or exceeded 0.9. For details, see Supplementary Table S3.
4. Diagnostic Performance of Circulating Single miRNA Compared to CA19-9
We reviewed 16 studies that involved an analysis of the diagnostic performance of single miRNA compared to the diagnostic utility of CA19-9 [28,29,34,36,44,48,53,54,55,58,63,64,66,68,69,74]. In those studies,55 single circulating miRNAs were evaluated 59 times with 66.10% of them presenting better diagnostic value than CA19-9 alone and 23.73% worse than CA19-9 alone. In 6 cases there were no data to evaluate the diagnostic value compared to CA19-9 alone. In all cases with a worse diagnostic value of miRNA compared to CA19-9, an analysis of panels combining tested miRNA and CA19-9 was conducted. It proved that this combination improves the diagnostic performance of miRNA. Details are presented in Supplementary Table S4.
Three of the analyzed studies evaluated the expression of miRNA utilizing the droplet digital polymerase chain reaction quantification (ddPCR) [75,76,77]. The ddPCR is a method of nucleic acid amplification considered to be more sensitive than conventional real time quantitative PCR. Furthermore, it does not require any internal or external normalization, which improves its reliability [78,79].The results of those studies are presented in Table 1.
Table 1.
Diagnostic performance of single miRNAs using the ddPCR method for expression evaluation, CS–clinical stage, AUC–area under the curve, SEN–sensitivity and SPE-specificity.
| miRNA | Author | Source of Sample | Expression Level Compared to Non-Cancer Controls. | CS of PC Patients | Control | AUC | SEN | SPE | Diagnostic Performance Compared to CA19-9 | AUC of Tested miRNA Combined with CA19-9 | SEN with CA19-9 | SPE with CA19-9 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miR-1290 | Tavano et al. [75] | plasma | Up- regulated |
I-IV | Healthy subjects | 0.734 [0.678–0.789] | 56.3% | 89.5% | Worse results | 0.956 [0.933–0.979] | 88.6% | 96.6%. |
| miR-1273g-3p | Mazza et al. [76] | plasma | Up- regulated |
I-IV | Healthy subjects | 0.703 [0.639–0.768] | 77.7% | 57.3% | Worse results | 0.940 [0.909–0.972] | 90.2% | 87.3% |
| miR-122-5p | Mazza et al. [76] | plasma | Up- regulated |
I-IV | Healthy subjects | 0.658 [0.590–0.726] | 49.1% | 79.2% | Worse results | 0.933 (0.899–0.967) | 83.9% | 94.6% |
| let-7d | Suzuki et al. [77] | serum | Down- regulated |
I-IV | Non-tumour controls (chronic pancreatitis, billiary stone, others) | 0.83 [0.740–0.910] | 888.9% | 68.2% | Better results | - | - | - |
5. Prognostic Value of Circulating miRNA
We investigated 21 studies on the prognostic value of miRNA [28,32,35,36,37,46,48,52,53,61,64,65,66,72,74,80,81,82,83,84,85]. The prognostic value of different miRNAs was evaluated 30 times. In 24 cases, the prognostic value of the tested miRNA was confirmed with univariate analysis, which later was confirmed as an independent prognostic factor by multivariate Cox analysis in 17 cases. The most commonly tested circulating miRNA was miR-21, the up-regulated expression of which was associated with poor patient prognoses (5 studies). The same was also reported for miR-196a (2 studies). The details are provided in Supplementary Table S5. In a study by Gablo et al. [82], a panel consisting of down-regulated miR-99a-5p, miR-365a-3p and up-regulated miR-200c-3p had been proven to identify patients with poor prognosis after surgical resection with the sensitivity of 85% and specificity of 80%. According to the study by Vietsch et al. [86], two up-regulated miRNAs (miR-99a-5p and miR-125b-5p) are significantly associated with a shorter progression free survival (PFS).
6. Predictive Value of Circulating miRNA
The predictive value of circulating miRNA was evaluated in six studies covered here [46,83,84,85,87,88]. In a study by Miyamae et al. [53], an increased expression of plasma miR-744 indicated the tendency to worse PFS during gemcitabine therapy (p = 0.0533). Wang et al. [85] presented similar results for serum miR-21, the over-expression of which indicated a shortened time to progression (TTP) and overall survival (OS) among patients with PDAC stage III and IV during gemcitabine therapy (p = 0.029 for OS, CS III; p = 0.069 for OS, CS IV; p = 0.008 for TTP, CS III; p = 0.029 for TTP, CS IV). In a study by van der Sijde et al. [84], a high level of serum miR-373-3p before the start of treatment and a low level of serum miR-194-5p after one cycle of treatment were associated with early tumor progression during FOLFIRINOX therapy. In a study Demiray et al. [88], patients with low let-7c expression progressed significantly earlier than patients with high let-7c expression during FOLFIRINOX therapy. In study by Lu et al. [87], plasma miR-20a-5p expression was significantly lower in gemcitabine-resistant patients with PDAC than in non-resistant patients (p < 0.01). Kong et al. provided results that serum levels of miR-196a might be a useful marker for differentiating respectable and unresectable PDAC [83].
7. Screening Value of Circulating miRNA
In studies by Duell et al. [89] and Franklin et al. [38], the screening utility of circulating miRNA was investigated. In the nested case-control study by Duell et al. [89], six plasma miRNAs expression levels (miR-10a, -10b, -21-5p, -30c, -155 and -212) were statistically significantly associated with the risk of PDAC. However, in Franklin et al.’s. study, 15 miRNA candidate, that were differently expressed during initial screen (miR-574-3p, -885-5p, -144-3p, -130b-3p, -34a-5p, -24-3p, -106b-5p, -22-5p, -451a, -101-3p, -26a-5p, -197-3p, -423-3p, -122-5p, and let-7d-3p) were not altered in plasma before the diagnosis of PDAC, which left the authors with a conclusion that circulating miRNA expression levels change late after the diagnosis of PDAC [38].
8. Monitoring Therapies with Circulating miRNA
The usefulness of circulating miRNAs in therapy monitoring was addressed in 8 studies included in this review. Details can be found in Table 2. Most of the studies (7) evaluated the expression of miRNAs pre- and postoperatively, proving a change in expression levels. In one study, the expression of preoperatively down-regulated miRNA increased after a successful surgery [43] while in six studies the expression of preoperatively up-regulated miRNAs decreased after surgery [28,38,41,46,50,90,91]. The study by Meijer et al. [90] provided data on miR-181a-5p usefulness in therapy monitoring during systemic therapy with the FOLFIRINOX regimen.
Table 2.
Therapy monitoring with circulating miRNA.
| miRNA | Author | Source of Sample | Therapy Monitoring Utility | p-Value |
|---|---|---|---|---|
| miR-107 | Imamura et al. [43] | plasma | up-regulation after surgery | p = 0.0186 |
| miR-181a-5p | Meijer et al. [90] | plasma | down-regulation after treatment with FOLFIRINOX in non-progressive patients, compared with progressive patients | p = 0.003 |
| miR-18a | Morimura et al. [91] | plasma | down-regulation after surgery | p = 0.0077 |
| miR-196a | Slater et al. [57] | serum | down-regulation after surgery (9 cases) | - |
| miR-196b | Slater et al. [57] | serum | down-regulation after surgery (9 cases) | - |
| miR-221 | Kawaguchi et al. [46] | plasma | down-regulation after surgery | p = 0.018 |
| miR-223 | Komatsu et al. [53] | plasma | down-regulation after surgery | p = 0.0297 |
| miR-483-3p | Abue et al. [28] | plasma | down-regulation after surgery (2 cases) | - |
| miR-744 | Miyamae et al. [53] | plasma | down-regulation after surgery | p = 0.0063 |
9. Challenges to the Analysis of Circulating miRNA
Circulating miRNAs are good candidates for biomarkers. The relatively low risk of complications and minimally invasive and painless sampling procedure are what makes them potentially useful in cancer diagnosis, prognosis and therapy monitoring. However, the quantification of circulating miRNA might be challenging.
Although circulating miRNAs are considered rather stable molecules [92], proper measures should be taken during specimen sample collection and handling. Contamination with background cellular fraction of miRNAs caused by blood cell lysis remains a cause of severe preclinical bias. Collected samples should be processed within two hours. Plasma and serum could be placed at 4 °C for up to 24 h or preferably frozen at −80 °C for long-term storage [93]. What is more, the variability of circulating miRNA expression could also be influenced by BMI, sex and age. Thus, a proper experiment protocol is recommended [94].
Specimen control for hemolysis should involve a visual inspection or the spectrophotometric test of hemoglobin absorbance at 414 nm [95], with an absorbance cut-off value of >0.2 for a hemolyzed specimen. Samples that do not meet the criteria should be excluded from further analysis.
The spike-in control, which is a foreign small RNA molecule added to the sample before extraction, can be used for extraction efficiency control. The molecule is the same length as native miRNAs, but it lacks the sequence homology to endogenous miRNAs. It usually comesfrom an unrelated species, such as Caenorhabditis elegans. The use of such control can identify material losses [96]. Positive endogenous miRNAs controls are also used to estimate the extraction procedure. The miRNAs suitable for endogenous control should be present at stable and high levels in tested specimen and should not be affected by cellular material. This kind of control might be used alongside exogenous spike-in controls [97].
Among traditional high-throughput methods of miRNA analysis, such as microarrays or RNA sequencing (RNA-Seq), RT-Q-PCR with its high specificity and sensitivity is considered a gold standard. However, the main limitation of this approach is that only the targeted miRNAs can be quantified. It also requires predesigned RT-primers, the high costs of which might be a limiting factor as well [98].
It should be recommended for authors who plan future studies on clinical utility of circulating miRNAs to select an appropriate control group, set an independent intrinsic validation and perform an analysis of background factors influencing the levels of detected circulating miRNAs, most importantly age and sex, as presented in Takinawa et al.’s study [99].
10. Discussion
PDAC is surgically resectable in fewer than 20% of patients with PDAC [100]. A late diagnosis leads to a more serious clinical stage of the disease and a low 5-year survival rate [3]. The prognosis of PDAC has not improved over the past 20 years, with relatively stable mortality. Most patients are diagnosed in clinical stage IV of the disease, which means they are suitable for palliative treatment, with the median overall survival (mOS) estimated at 8 to 11 months. Since the overall survival depends mainly on the disease’s clinical stage, a search for methods with an efficient diagnosis of early PDAC is becoming more important.
Various miRNAs play important roles in the carcinogenesis and development of various cancers. Since circulating miRNAs emerged as a novel biomarker in various neoplasms [101,102,103] and are stable in bodily fluids in patients with PDAC, this review aims at providing an updated perspective on circulating miRNAs’ role in pancreatic cancer diagnosis. The single biomarker, the diagnostic utility of which was most frequently evaluated, was miR-21. Its diagnostic utility was examined in 7 studies reviewed here. The other commonly evaluated biomarkers were miR-155, miR-196a and miR-210, each of them present in 5 studies (S2).
The area under the curve (AUC) in receiver operating characteristics analysis (ROC) is used for evaluating the accuracy of diagnostic test. A result above 0.9 indicate a high diagnostic value. The diagnostic utility of circulating miRNA has been already established by multiple studies and meta-analyses [104,105] with miR-21 as a reliable biomarker in digestive system neoplasms with its 72% sensitivity, 82% specificity and an overall AUC of 0.86. However, the meta-analysis by Aalami et al. [106] demonstrated that single circulating miR-21 levels are not specific for subgroups of gastrointestinal cancers, such as esophageal squamous cell cancer, gastric cancer, pancreatic ductal adenocarcinoma or colorectal cancer. This meta-analysis’s limitations were high heterogeneity and publication bias. Most of the studies included in this review provided data of circulating miRNAs surpassing the commonly used CA19-9 as a biomarker in PDAC. However, some presented a poorer effect. To overcome the limitations of the diagnostic performance of single circulating miRNAs, they were combined into panels with CA19-9, other miRNAs or other useful biomarkers (S3 and S4)in several studies. The diagnostic panels reached high diagnostic value(AUC values above 0.9) in more cases than single miRNAs. In their meta-analysis, Pei et al. proved that multiple miRNAs combined into diagnostic panels provide a more accurate method of PDAC diagnosis than single circulating miRNAs. The diagnostic accuracy of panels of multiple miRNAs was verified in 19 studies. The sensitivity of such panels was 85% while the specificity was 89%. The AUC was determined to be 0.93. Compared to the diagnostic accuracy of a single miRNA in the 17 studies(sensitivity 78%, specificity 79% and AUC 0.84), combined miRNA panels provide a more accurate method of PDAC diagnosis. However, this meta-analysis had its limitations—most importantly, high heterogeneity among the reviewed studies caused by differences in miRNA profiling methods and specimen resources [107]. Moreover, the same miRNAs are deregulated in different types of neoplasm, meaning that they are not specific to any cancer type and are involved in various oncogenic pathways. For example, miRNA-21 is also involved in the carcinogenesis of colorectal cancer, breast cancer and glioblastomas [108]. Taking it into account, a combination of miRNAs can increase the accuracy of diagnostic panel regarding a specific cancer type. However, creating a complex diagnostic panel that could be implemented for clinical usage might be a costly and time-consuming procedure, which could be detrimental to its clinical utility.
Several prognostic factors are commonly used in pancreatic cancer therapy. Factors like histopathological type (adenosquamous carcinoma and undifferentiated carcinoma are considered pancreatic cancer types with a poorer prognosis than PDAC), high expression of CA 19-9 (in terms of survival after surgery, CA19-9 levels above 500 IU are related to a poor prognosis) or loss of SMAD 4 (mothers against decapentaplegic homolog 4) expression (poor prognosis) are implemented into treatment guidelines [109]. The search for other prognostic factors is important for the therapeutic process to determine which patients might benefit from specific therapies. For example, inflammation-related factors, such as neutrophil-to-lymphocyte ratio (NLR) play an increasing role in PDAC therapy since they are easy to obtain and relatively not expensive [110]. The prognostic value of circulating miRNA in PDAC is established in most studies as well. Similarly to circulating miRNAs’ diagnostic role, the miR-21 was the most frequently evaluated biomarker in terms of its prognostic performance. Three out of six studies reported miR-21 as an independent prognostic factor in multivariable regression analysis with p < 0.01.A similar effect was also detected for miR-196a (two studies, one of which included a multivariate analysis). In one study, the level of miR-21 was not associated with PDAC patients prognosis and, what is noteworthy, none of the six other tested miRNA expression levels was associated with PDAC prognosis [69] (S5). The authors suggest a longer time of observation and larger groups of enrolled patients could increase the value of further studies on this matter. The Guraya et al.’s meta-analysis infers that miR-21 constitutes a useful prognostic biomarker in digestive system neoplasms. The pooled hazard ratio (HR) of worse OS in patients with PDAC was 3.77 (1.63–8.73, p < 0.01). However, the meta-analysis has its limitations, with a high heterogeneity of the covered studies as the critical one [111].
The most intriguing role of miRNAs would be in screening patients with an increased risk of PDAC. Some of the markers in this review showed the potential for PDAC screening [89]. However, one study presented different results [38]. The number of studies evaluating this aspect of circulating miRNAs included in this review is low. It is probably due to the difficult methodology of such studies and a long time of patient follow-up. The differences between those studies are important. The study by Duell et al. was a prospective cohort study while in Franklin’s study, blood samples were retrieved from a biobank from retrospectively analyzed patients who had undergone pancreatic surgery. It seems that Duell’s study remains the only one evaluating the risk of pancreatic cancer among patients with an increased expression inmiRNA. With its prospective design and specimen collection many years before the diagnosis, it provides strong evidence for miRNAs screening value. However, the main qualities of the screening test should be its high specificity, safety and relatively low costs [112,113]. For now, circulating miRNAs testing seems to be unable to satisfy the cost criterion to be used as a population screening test. Therefore, the screening role of circulating miRNAs in PDAC remains uncertain and requires large, multicenter studies to provide reliable data.
The predictive value of circulating miRNAs indicates its utility to choose the most suitable therapy for the patient, either the surgical or systemic approach. The identification of patients that may respond to a certain therapy is crucial to improve treatment outcomes. This may also influence the decision to discontinue the therapy in patients who will no longer benefit from treatment. Studies included in this review provide data on this matter [46,83,84,85,87,88]. However, many factors contribute to the choice of therapy and the most significant of them is the patient’s performance status (PS) before the start of the treatment. Circulating miRNAs seem to be a promising factor. Still, further research is needed to determine their predictive role, especially in a prospective approach.
There are no specific biomarkers to monitor the response to treatment in PDAC. CA 19-9 expression has been suggested as a predictor for survival in patients with PDAC, but its decrease during systemic treatment is not associated with prolonged survival compared to patients who did not exhibit a significant decrease [114]. Therefore, the search for biomarkers, which may improve the decision-making process in PDAC treatment, is also relevant. Most of the studies covered in this analysis referred to circulating miRNAs changes in reference to surgical treatment. Expression of circulating miRNAs in postoperative specimen was significantly reduced compared to preoperative serum or plasma expression. These findings suggest that miRNAs are released by cancerous tissue and could be used to monitor tumor dynamics, such as the presence of a residual tumor or disease recurrence [28,38,41,46,50,91]. The study by Meijer et al. [90] demonstrated that miR-181a-5p is significantly down-regulated in patients with non-progressive PDAC compared to patients with progressive PDAC during FOLFIRINOX treatment. No such observation was made for patients treated with gemcitabine plus nab-paclitaxel. Notably, patients treated with the gemcitabine plus nab-paclitaxel regimen were older and had a worse PS, which could affect the result. Therefore, large-scale, prospective research is called for to establish circulating miRNA’s role in tumor response monitoring in PDAC.
Circulating miRNA constitutes a challenging biomarker to be implemented in clinical practice. First of all, its expression may be influenced by different factors like diet, medications, patient sex or the time of sample collection during the day. What is more, its stability relies on sample handling and storage. The quality control of extracted miRNA may be performed with endogenous miRNA controls or exogenous spike-in controls [115,116]. The studies handled in this review employed various quality control methods, the most common being RNU6B and cel-miR-39 (S2). Further research is required to determine the optimal protocol for circulating miRNA detection so that study results can be compared more easily.
11. Conclusions and Future Research
This review provides general information about circulating miRNAs diagnostic, prognostic and therapy monitoring value, which seems to be well-established in numerous studies. The screening value of circulating miRNA in PDAC remains uncertain and has to be further investigated. Due to the high mortality rate of pancreatic cancer patients, it is crucial to search for early diagnostic biomarkers to improve the survival rate.
There are many obstacles to employing circulating miRNA in clinical practice. A proper study design, reducing the risk of preclinical variability of miRNAs expression and standardized sample processing seem to be crucial for reducing the bias and may increase the reproducibility of the method. Standardization of diagnostic procedures for circulating miRNA expression should be the future aim. It would facilitate not only an easier juxtaposition of results, but also precise cancer diagnosis and prognosis. What is more, combining miRNAs into diagnostic panels seems to yield a more accurate method than using single miRNAs. Introducing other biomarkers, such as CA19-9, CEA, cytokines or proteins, into miRNAs diagnostic panels might improve their diagnostic accuracy. In terms of screening value, further research is needed to determine the panel with the highest specificity. Large-scale, prospective studies are required to determine predictive and therapy monitoring utility of circulating miRNAs.
After developing standardized and universally accepted methods for circulating miRNA testing in the future, these markers could be the novel additional diagnostic, prognostic and disease monitoring factor useful in clinical practice. However, further research is required to determine the optimal methods for circulating miRNA testing.
Abbreviations
pancreatic cancer (PC), pancreatic ductal adenocarcinoma (PDAC), microRNAs (miRNAs), ultrasound (US), computed tomography (CT), magnetic resonance imaging (MRI), endoscopic ultrasonography (EUS) with EUS-guided fine-needle aspiration (EUS-FNA), circulating tumor cells (CTCs), cell-free circulating tumor DNAs (cfDNAs), long noncoding RNAs (lncRNAs), 3′ untranslated regions (3′ UTRs), RNA-induced silencing complexes (RISCs), area under the curve (AUC), receiver operating characteristic (ROC) analysis, diagnostic sensitivity (SEN), diagnostic specificity (SPE), carbohydrate antigen 19-9 (CA19-9), real-time quantitative polymerase chain reaction (RT-Q-PCR), digital polymerase chain reaction quantification (ddPCR), CS–clinical stage, time-to progression (TTP), overall survival (OS), RNA sequencing (RNA-Seq), SMAD 4 (mothers against decapentaplegic homolog 4), neutrophil-to-lymphocyte ratio (NLR), hazard ratio (HR), performance status (PS).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24065113/s1. References [117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,219,220,221,222,223,224,225,226,227,228,229,230,231,232,233,234,235,236,237,238,239,240,241,242,243,244,245,246,247] are cited in Supplementary Materials.
Author Contributions
Conceptualization, J.W. and J.K.S.; methodology, J.W. and J.K.S.; validation, J.W., J.K.S. and I.G.; formal analysis, J.W.; investigation, J.W.; resources, J.W.; data curation, J.W.; writing—original draft preparation, J.W.; writing—review and editing, J.K.S. and I.G.; visualization, J.W.; supervision J.K.S. and I.G.; project administration, J.W.; funding acquisition, J.W. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by Medical University of Silesia—PCN-CDN-640-2-1-819/22.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M., Abnet C.C., Neale R.E., Vignat J., Giovannucci E.L., McGlynn K.A., Bray F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology. 2020;159:335–349.e15. doi: 10.1053/j.gastro.2020.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.NCI Website.National Cancer Institute (NCI) Cancer Stat Facts: Pancreatic Cancer. [(accessed on 4 October 2022)]; Available online: https://seer.cancer.gov/statfacts/html/pancreas.htmlhttps://seer.cancer.gov/statfacts/html/pancreas.html.
- 4.Ghaneh P., Costello E., Neoptolemos J.P. Biology and management of pancreatic cancer. Gut. 2007;56:1134. doi: 10.1136/gut.2006.103333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferlay J., Partensky C., Bray F. More deaths from pancreatic cancer than breast cancer in the EU by 2017. Acta Oncol. 2016;55:1158–1160. doi: 10.1080/0284186X.2016.1197419. [DOI] [PubMed] [Google Scholar]
- 6.Ilic M., Ilic I. Epidemiology of pancreatic cancer. World J. Gastroenterol. 2016;22:9694. doi: 10.3748/wjg.v22.i44.9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGuigan A., Kelly P., Turkington R.C., Jones C., Coleman H.G., McCain R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018;24:4846. doi: 10.3748/wjg.v24.i43.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collisson E.A., Bailey P., Chang D.K., Biankin A.V. Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2019;16:207–220. doi: 10.1038/s41575-019-0109-y. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L., Sanagapalli S., Stoita A. Challenges in diagnosis of pancreatic cancer. World J. Gastroenterol. 2018;24:2047–2060. doi: 10.3748/wjg.v24.i19.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian L., Li Q., Baryeh K., Qiu W., Li K., Zhang J., Yu Q., Xu D., Liu W., Brand R.E., et al. Biosensors for early diagnosis of pancreatic cancer: A review. Transl. Res. 2019;213:67–89. doi: 10.1016/j.trsl.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 11.O’Neill R.S., Stoita A. Biomarkers in the diagnosis of pancreatic cancer: Are we closer to finding the golden ticket? World J. Gastroenterol. 2021;27:4045. doi: 10.3748/wjg.v27.i26.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 13.He B., Zhao Z., Cai Q., Zhang Y., Zhang P., Shi S., Xie H., Peng X., Yin W., Tao Y., et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int. J. Biol. Sci. 2020;2020:2628–2647. doi: 10.7150/ijbs.47203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X., Wang H.K., Li Y., Hafner M., Banerjee N.S., Tang S., Briskin D., Meyers C., Chow L.T., Xie X., et al. MicroRNAs are biomarkers of oncogenic human papillomavirus infections. Proc. Natl. Acad. Sci. USA. 2014;111:4262–4267. doi: 10.1073/pnas.1401430111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MiRNA-491-5p Inhibits Cell Proliferation, Invasion and Migration via Targeting JMJD2B and Serves as a Potential Biomarker in Gastric Cancer—PubMed. [(accessed on 6 November 2022)]; Available online: https://pubmed.ncbi.nlm.nih.gov/29511447/ [PMC free article] [PubMed]
- 16.Pan J., Zhou C., Zhao X., He J., Tian H., Shen W., Han Y., Chen J., Fang S., Meng X., et al. A two-miRNA signature (miR-33a-5p and miR-128-3p) in whole blood as potential biomarker for early diagnosis of lung cancer. Sci. Rep. 2018;8:16699. doi: 10.1038/s41598-018-35139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan Y., Liu W., Zhang Y., Zhang Y., Sun S. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem. Biophys. Res. Commun. 2018;503:870–875. doi: 10.1016/j.bbrc.2018.06.089. [DOI] [PubMed] [Google Scholar]
- 18.Xie Z., Yin X., Gong B., Nie W., Wu B., Zhang X., Huang J., Zhang P., Zhou Z., Li Z. Salivary microRNAs show potential as a noninvasive biomarker for detecting resectable pancreatic cancer. Cancer Prev. Res. 2015;8:165–173. doi: 10.1158/1940-6207.CAPR-14-0192. [DOI] [PubMed] [Google Scholar]
- 19.Humeau M., Vignolle-Vidoni A., Sicard F., Martins F., Bournet B., Buscail L., Torrisani J., Cordelier P. Salivary MicroRNA in Pancreatic Cancer Patients. PLoS ONE. 2015;10:e0130996. doi: 10.1371/journal.pone.0130996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ideno N., Mori Y., Nakamura M., Ohtsuka T. Early Detection of Pancreatic Cancer: Role of Biomarkers in Pancreatic Fluid Samples. Diagnostics. 2020;10:1056. doi: 10.3390/diagnostics10121056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hata T., Mizuma M., Masuda K., Chiba K., Ishida M., Ohtsuka H., Nakagawa K., Morikawa T., Kamei T., Unno M. MicroRNA-593-3p Expression in Peritoneal Lavage Fluid as a Prognostic Marker for Pancreatic Cancer Patients Undergoing Staging Laparoscopy. Ann. Surg. Oncol. 2021;28:2235–2245. doi: 10.1245/s10434-020-09440-3. [DOI] [PubMed] [Google Scholar]
- 22.Baraniskin A., Kuhnhenn J., Schlegel U., Chan A., Deckert M., Gold R., Maghnouj A., Zöllner H., Reinacher-Schick A., Schmiegel W., et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood. 2011;117:3140–3146. doi: 10.1182/blood-2010-09-308684. [DOI] [PubMed] [Google Scholar]
- 23.Pavlidis L., Spyropoulou G.A., Papas A., Demiri E. Urinary Excretion of MicroRNA-126 Is a Biomarker for Hemangioma Proliferation. Plast. Reconstr. Surg. 2018;141:319E–320E. doi: 10.1097/PRS.0000000000004078. [DOI] [PubMed] [Google Scholar]
- 24.Daoud A.Z., Mulholland E.J., Cole G., McCarthy H.O. MicroRNAs in Pancreatic Cancer: Biomarkers, prognostic, and therapeutic modulators. BMC Cancer. 2019;19:1130. doi: 10.1186/s12885-019-6284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartel D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 26.Page M.J., McKenzie J.E., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D., Shamseer L., Tetzlaff J.M., Akl E.A., Brennan S.E., et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whiting P.F., Rutjes A.W.S., Westwood M.E., Mallett S., Deeks J.J., Reitsma J.B., Leeflang M.M.G., Sterne J.A.C., Bossuyt P.M.M. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 28.Abue M., Yokoyama M., Shibuya R., Tamai K., Yamaguchi K., Sato I., Tanaka N., Hamada S., Shimosegawa T., Sugamura K., et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int. J. Oncol. 2015;46:539. doi: 10.3892/ijo.2014.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aita A., Millino C., Sperti C., Pacchioni B., Plebani M., De Pittà C., Basso D. Serum miRNA Profiling for Early PDAC Diagnosis and Prognosis: A Retrospective Study. Biomedicines. 2021;9:845. doi: 10.3390/biomedicines9070845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akamatsu M., Makino N., Ikeda Y., Matsuda A., Ito M., Kakizaki Y., Saito Y., Ishizawa T., Kobayashi T., Furukawa T., et al. Specific MAPK-Associated MicroRNAs in Serum Differentiate Pancreatic Cancer from Autoimmune Pancreatitis. PLoS ONE. 2016;11:e0158669. doi: 10.1371/journal.pone.0158669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alemar B., Izetti P., Gregório C., Macedo G.S., Castro M.A.A., Osvaldt A.B., Matte U., Ashton-Prolla P. miRNA-21 and miRNA-34a Are Potential Minimally Invasive Biomarkers for the Diagnosis of Pancreatic Ductal Adenocarcinoma. Pancreas. 2016;45:84–92. doi: 10.1097/MPA.0000000000000383. [DOI] [PubMed] [Google Scholar]
- 32.Bartsch D.K., Gercke N., Strauch K., Wieboldt R., Matthäi E., Wagner V., Rospleszcz S., Schäfer A., Franke F.S., Mintziras I., et al. The Combination of MiRNA-196b, LCN2, and TIMP1 is a Potential Set of Circulating Biomarkers for Screening Individuals at Risk for Familial Pancreatic Cancer. J. Clin. Med. 2018;7:295. doi: 10.3390/jcm7100295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Q., Yang L., Xiao Y., Zhu J., Li Z. Circulating microRNA-182 in plasma and its potential diagnostic and prognostic value for pancreatic cancer. Med. Oncol. 2014;31:225. doi: 10.1007/s12032-014-0225-z. [DOI] [PubMed] [Google Scholar]
- 34.Cote G.A., Gore A.J., McElyea S.D., Heathers L.E., Xu H., Sherman S., Korc M. A pilot study to develop a diagnostic test for pancreatic ductal adenocarcinoma based on differential expression of select miRNA in plasma and bile. Am. J. Gastroenterol. 2014;109:1942–1952. doi: 10.1038/ajg.2014.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng T., Yuan Y., Zhang C., Zhang C., Yao W., Wang C., Liu R., Ba Y. Identification of Circulating MiR-25 as a Potential Biomarker for Pancreatic Cancer Diagnosis. Cell. Physiol. Biochem. 2016;39:1716–1722. doi: 10.1159/000447872. [DOI] [PubMed] [Google Scholar]
- 36.Dittmar R.L., Liu S., Tai M.C., Rajapakshe K., Huang Y., Longton G., DeCapite C., Hurd M.W., Paris P.L., Kirkwood K.S., et al. Plasma MicroRNA Biomarkers in Limited Volume Samples for Detection of Early Stage Pancreatic Cancer. Cancer Prev. Res. 2021;14:729. doi: 10.1158/1940-6207.CAPR-20-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fathi M., Aghdaie H.A., Ghafouri-Fard S., Shams R. Evaluation of potential of miR-8073 and miR-642 as diagnostic markers in pancreatic cancer. Mol. Biol. Rep. 2022;49:6475–6481. doi: 10.1007/s11033-022-07476-0. [DOI] [PubMed] [Google Scholar]
- 38.Franklin O., Jonsson P., Billing O., Lundberg E., Öhlund D., Nyström H., Lundin C., Antti H., Sund M. Plasma Micro-RNA Alterations Appear Late in Pancreatic Cancer. Ann. Surg. 2018;267:775. doi: 10.1097/SLA.0000000000002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ganepola G.A., Rutledge J.R., Suman P., Yiengpruksawan A., Chang D.H. Novel blood-based microRNA biomarker panel for early diagnosis of pancreatic cancer. World J. Gastrointest. Oncol. 2014;6:22–33. doi: 10.4251/wjgo.v6.i1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hua Y., Chen H., Wang L., Wang F., Wang P., Ning Z., Li Y., Liu L., Chen Z., Meng Z. Low serum miR-373 predicts poor prognosis in patients with pancreatic cancer. Cancer Biomark. 2017;20:95–100. doi: 10.3233/CBM-170231. [DOI] [PubMed] [Google Scholar]
- 41.Huang W.T., Lin T.S., Wu J.Y., Hong J.M., Chen Y.L., Qiu F.N. Evaluation of miR-429 as a novel serum biomarker for pancreatic ductal adenocarcinoma and analysis its tumor suppressor function and target genes. Eur. Rev. Med. Pharmacol. Sci. 2022;26:4638–4653. doi: 10.26355/EURREV_202207_29186. [DOI] [PubMed] [Google Scholar]
- 42.Hussein N.A.E.M., Kholy Z.A.E., Anwar M.M., Ahmad M.A., Ahmad S.M. Plasma miR-22-3p, miR-642b-3p and miR-885-5p as diagnostic biomarkers for pancreatic cancer. J. Cancer Res. Clin. Oncol. 2017;143:83–93. doi: 10.1007/s00432-016-2248-7. [DOI] [PubMed] [Google Scholar]
- 43.Imamura T., Komatsu S., Ichikawa D., Miyamae M., Okajima W., Ohashi T., Kiuchi J., Nishibeppu K., Konishi H., Shiozaki A., et al. Depleted tumor suppressor miR-107 in plasma relates to tumor progression and is a novel therapeutic target in pancreatic cancer. Sci. Rep. 2017;7:5708. doi: 10.1038/s41598-017-06137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang D., Yuan X., Ni J., Shen L., Cai M., Xu L. Low Serum miR-607 Level as a Potential Diagnostic and Prognostic Biomarker in Patients of Pancreatic Ductal Adenocarcinoma: A Preliminary Study. Can. J. Gastroenterol. Hepatol. 2021;2021:8882129. doi: 10.1155/2021/8882129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karasek P., Gablo N., Hlavsa J., Kiss I., Vychytilova-Faltejskova P., Hermanova M., Kala Z., Slaby O., Prochazka V. Pre-operative Plasma miR-21-5p Is a Sensitive Biomarker and Independent Prognostic Factor in Patients with Pancreatic Ductal Adenocarcinoma Undergoing Surgical Resection. Cancer Genom. Proteom. 2018;15:321–327. doi: 10.21873/cgp.20090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawaguchi T., Komatsu S., Ichikawa D., Morimura R., Tsujiura M., Konishi H., Takeshita H., Nagata H., Arita T., Hirajima S., et al. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br. J. Cancer. 2013;108:361–369. doi: 10.1038/bjc.2012.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan I.A., Rashid S., Singh N., Rashid S., Singh V., Gunjan D., Das P., Dash N.R., Pandey R.M., Chauhan S.S., et al. Panel of serum miRNAs as potential non-invasive biomarkers for pancreatic ductal adenocarcinoma. Sci. Rep. 2021;11:2824. doi: 10.1038/s41598-021-82266-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komatsu S., Ichikawa D., Miyamae M., Kawaguchi T., Morimura R., Hirajima S., Okajima W., Ohashi T., Imamura T., Konishi H., et al. Malignant potential in pancreatic neoplasm; new insights provided by circulating miR-223 in plasma. Expert Opin. Biol. Ther. 2015;15:773–785. doi: 10.1517/14712598.2015.1029914. [DOI] [PubMed] [Google Scholar]
- 49.Li A., Omura N., Hong S.M., Vincent A., Walter K., Griffith M., Borges M., Goggins M. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 2010;70:5226–5237. doi: 10.1158/0008-5472.CAN-09-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin M.S., Chen W.C., Huang J.X., Gao H.J., Sheng H.H. Aberrant expression of microRNAs in serum may identify individuals with pancreatic cancer. Int. J. Clin. Exp. Med. 2014;7:5226–5234. [PMC free article] [PubMed] [Google Scholar]
- 51.Liu G., Shao C., Li A., Zhang X., Guo X., Li J. Diagnostic Value of Plasma miR-181b, miR-196a, and miR-210 Combination in Pancreatic Cancer. Gastroenterol. Res. Pract. 2020;2020:6073150. doi: 10.1155/2020/6073150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J., Gao J., Du Y., Li Z., Ren Y., Gu J., Wang X., Gong Y., Wang W., Kong X. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int. J. Cancer. 2012;131:683–691. doi: 10.1002/ijc.26422. [DOI] [PubMed] [Google Scholar]
- 53.Miyamae M., Komatsu S., Ichikawa D., Kawaguchi T., Hirajima S., Okajima W., Ohashi T., Imamura T., Konishi H., Shiozaki A., et al. Plasma microRNA profiles: Identification of miR-744 as a novel diagnostic and prognostic biomarker in pancreatic cancer. Br. J. Cancer. 2015;113:1467–1476. doi: 10.1038/bjc.2015.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qu K., Zhang X., Lin T., Liu T., Wang Z., Liu S., Zhou L., Wei J., Chang H., Li K., et al. Circulating miRNA-21-5p as a diagnostic biomarker for pancreatic cancer: Evidence from comprehensive miRNA expression profiling analysis and clinical validation. Sci. Rep. 2017;7:1692. doi: 10.1038/s41598-017-01904-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi W., Lu Y., Gong R., Sun J.J., Liu G. Serum miR-629 is a novel molecular marker for diagnosis and the prognosis of pancreatic cancer. Eur. Rev. Med. Pharmacol. Sci. 2018;22:5187–5193. doi: 10.26355/EURREV_201808_15715. [DOI] [PubMed] [Google Scholar]
- 56.Škrha P., Hořínek A., Pazourková E., Hajer J., Frič P., Škrha J., Anděl M. Serum microRNA-196 and microRNA-200 in pancreatic ductal adenocarcinoma of patients with diabetes mellitus. Pancreatology. 2016;16:839–843. doi: 10.1016/j.pan.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 57.Slater E.P., Strauch K., Rospleszcz S., Ramaswamy A., Esposito I., Klöppel G., Matthäi E., Heeger K., Fendrich V., Langer P., et al. MicroRNA-196a and -196b as Potential Biomarkers for the Early Detection of Familial Pancreatic Cancer. Transl. Oncol. 2014;7:464–471. doi: 10.1016/j.tranon.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stroese A.J., Ullerich H., Koehler G., Raetzel V., Senninger N., Dhayat S.A. Circulating microRNA-99 family as liquid biopsy marker in pancreatic adenocarcinoma. J. Cancer Res. Clin. Oncol. 2018;144:2377–2390. doi: 10.1007/s00432-018-2749-7. [DOI] [PubMed] [Google Scholar]
- 59.Michael Traeger M., Rehkaemper J., Ullerich H., Steinestel K., Wardelmann E., Senninger N., Abdallah Dhayat S. The ambiguous role of microRNA-205 and its clinical potential in pancreatic ductal adenocarcinoma. J. Cancer Res. Clin. Oncol. 2018;144:2419–2431. doi: 10.1007/s00432-018-2755-9. [DOI] [PubMed] [Google Scholar]
- 60.Vieira N.F., Serafini L.N., Novais P.C., Neto F.S.L., De Assis Cirino M.L., Kemp R., Ardengh J.C., Saggioro F.P., Gaspar A.F., Sankarankutty A.K., et al. The role of circulating miRNAs and CA19-9 in pancreatic cancer diagnosis. Oncotarget. 2021;12:1638–1650. doi: 10.18632/oncotarget.28038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vila-Navarro E., Duran-Sanchon S., Vila-Casadesús M., Moreira L., Gins A., Cuatrecasas M., Josè Lozano J., Bujanda L., Castells A., Gironella M. Novel Circulating miRNA Signatures for Early Detection of Pancreatic Neoplasia. Clin. Transl. Gastroenterol. 2019;10:e00029. doi: 10.14309/ctg.0000000000000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Z. Diagnostic performance for declined microRNA-133a in pancreatic cancer. J. Cell. Biochem. 2019;121:3882–3886. doi: 10.1002/jcb.29547. [DOI] [PubMed] [Google Scholar]
- 63.Wang J., Chen J., Chang P., LeBlanc A., Li D., Abbruzzesse J.L., Frazier M.L., Killary A.M., Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev. Res. 2009;2:807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei J., Yang L., Wu Y.N., Xu J. Serum miR-1290 and miR-1246 as Potential Diagnostic Biomarkers of Human Pancreatic Cancer. J. Cancer. 2020;11:1325–1333. doi: 10.7150/jca.38048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu J., Cao Z., Liu W., You L., Zhou L., Wang C., Lou W., Sun B., Miao Y., Liu X., et al. Plasma miRNAs Effectively Distinguish Patients With Pancreatic Cancer From Controls: A Multicenter Study. Ann. Surg. 2016;263:1173–1179. doi: 10.1097/SLA.0000000000001345. [DOI] [PubMed] [Google Scholar]
- 66.Xu L., Cai Y., Chen X., Zhu Y., Cai J. Circulating MiR-1290 as a potential diagnostic and disease monitoring biomarker of human gastrointestinal tumors. BMC Cancer. 2021;21:989. doi: 10.1186/s12885-021-08729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu Q., Xu C., Yuan W., Wang C., Zhao P., Chen L., Ma J. Evaluation of Plasma MicroRNAs as Diagnostic and Prognostic Biomarkers in Pancreatic Adenocarcinoma: miR-196a and miR-210 Could Be Negative and Positive Prognostic Markers, Respectively. BioMed Res. Int. 2017;2017:6495867. doi: 10.1155/2017/6495867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu Y., Tong Y., Zhong A., Wang Y., Lu R., Guo L. Identification of Serum microRNA-25 as a novel biomarker for pancreatic cancer. Medicine. 2020;99:e23863. doi: 10.1097/MD.0000000000023863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuan W., Tang W., Xie Y., Wang S., Chen Y., Qi J., Qiao Y., Ma J. New combined microRNA and protein plasmatic biomarker panel for pancreatic cancer. Oncotarget. 2016;7:80033–80045. doi: 10.18632/oncotarget.12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zou X., Wei J., Huang Z., Zhou X., Lu Z., Zhu W., Miao Y. Identification of a six-miRNA panel in serum benefiting pancreatic cancer diagnosis. Cancer Med. 2019;8:2810–2822. doi: 10.1002/cam4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johansen J.S., Calatayud D., Albieri V., Schultz N.A., Dehlendorff C., Werner J., Jensen B.V., Pfeiffer P., Bojesen S.E., Giese N., et al. The potential diagnostic value of serum microRNA signature in patients with pancreatic cancer. Int. J. Cancer. 2016;139:2312–2324. doi: 10.1002/ijc.30291. [DOI] [PubMed] [Google Scholar]
- 72.Liu R., Chen X., Du Y., Yao W., Shen L., Wang C., Hu Z., Zhuang R., Ning G., Zhang C., et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin. Chem. 2012;58:610–618. doi: 10.1373/clinchem.2011.172767. [DOI] [PubMed] [Google Scholar]
- 73.Schultz N.A., Dehlendorff C., Jensen B.V., Bjerregaard J.K., Nielsen K.R., Bojesen S.E., Calatayud D., Nielsen S.E., Yilmaz M., Holländer N.H., et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014;311:392–404. doi: 10.1001/jama.2013.284664. [DOI] [PubMed] [Google Scholar]
- 74.Zhou X., Lu Z., Wang T., Huang Z., Zhu W., Miao Y. Plasma miRNAs in diagnosis and prognosis of pancreatic cancer: A miRNA expression analysis. Gene. 2018;673:181–193. doi: 10.1016/j.gene.2018.06.037. [DOI] [PubMed] [Google Scholar]
- 75.Tavano F., Gioffreda D., Valvano M.R., Palmieri O., Tardio M., Latiano T.P., Piepoli A., Maiello E., Pirozzi F., Andriulli A. Droplet digital PCR quantification of miR-1290 as a circulating biomarker for pancreatic cancer. Sci. Rep. 2018;8:16389. doi: 10.1038/s41598-018-34597-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mazza T., Gioffreda D., Fontana A., Biagini T., Carella M., Palumbo O., Maiello E., Bazzocchi F., Andriulli A., Tavano F. Clinical Significance of Circulating miR-1273g-3p and miR-122-5p in Pancreatic Cancer. Front. Oncol. 2020:10–44. doi: 10.3389/fonc.2020.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suzuki R., Asama H., Waragai Y., Takagi T., Hikichi T., Sugimoto M., Konno N., Watanabe K., Nakamura J., Kikuchi H., et al. Fibrosis-related miRNAs as serum biomarkers for pancreatic ductal adenocarcinoma. Oncotarget. 2017;9:4451–4460. doi: 10.18632/oncotarget.23377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dingle T.C., Sedlak R.H., Cook L., Jerome K.R. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin. Chem. 2013;59:1670–1672. doi: 10.1373/clinchem.2013.211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hindson C.M., Chevillet J.R., Briggs H.A., Gallichotte E.N., Ruf I.K., Hindson B.J., Vessella R.L., Tewari M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ali S., Dubaybo H., Brand R.E., Sarkar F.H. Differential Expression of MicroRNAs in Tissues and Plasma Co-exists as a Biomarker for Pancreatic Cancer. J. Cancer Sci. Ther. 2015;7:336. doi: 10.4172/1948-5956.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ali S., Almhanna K., Chen W., Philip P.A., Sarkar F.H. Differentially expressed miRNAs in the plasma may provide a molecular signature for aggressive pancreatic cancer. Am. J. Transl. Res. 2010;3:28–47. [PMC free article] [PubMed] [Google Scholar]
- 82.Gablo N., Trachtova K., Prochazka V., Hlavsa J., Grolich T., Kiss I., Srovnal J., Rehulkova A., Lovecek M., Skalicky P., et al. Identification and Validation of Circulating Micrornas as Prognostic Biomarkers in Pancreatic Ductal Adenocarcinoma Patients Undergoing Surgical Resection. J. Clin. Med. 2020;9:2440. doi: 10.3390/jcm9082440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kong X., Du Y., Wang G., Gao J., Gong Y., Li L., Zhang Z., Zhu J., Jing Q., Qin Y., et al. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig. Dis. Sci. 2011;56:602–609. doi: 10.1007/s10620-010-1285-3. [DOI] [PubMed] [Google Scholar]
- 84.van der Sijde F., Homs M.Y.V., van Bekkum M.L., van den Bosch T.P.P., Bosscha K., Besselink M.G., Bonsing B.A., de Groot J.W.B., Karsten T.M., Koerkamp B.G., et al. Serum miR-373-3p and miR-194-5p Are Associated with Early Tumor Progression during FOLFIRINOX Treatment in Pancreatic Cancer Patients: A Prospective Multicenter Study. Int. J. Mol. Sci. 2021;22:10902. doi: 10.3390/ijms222010902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang P., Zhuang L., Zhang J., Fan J., Luo J., Chen H., Wang K., Liu L., Chen Z., Meng Z. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol. Oncol. 2013;7:334–345. doi: 10.1016/j.molonc.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vietsch E.E., Peran I., Suker M., van den Bosch T.P.P., van der Sijde F., Kros J.M., van Eijck C.H.J., Wellstein A. Immune-Related Circulating miR-125b-5p and miR-99a-5p Reveal a High Recurrence Risk Group of Pancreatic Cancer Patients after Tumor Resection. Appl. Sci. 2019;9:4784. doi: 10.3390/app9224784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu H., Lu S., Yang D., Zhang L., Ye J., Li M., Hu W. MiR-20a-5p regulates gemcitabine chemosensitivity by targeting RRM2 in pancreatic cancer cells and serves as a predictor for gemcitabine-based chemotherapy. Biosci. Rep. 2019;39:BSR20181374. doi: 10.1042/BSR20181374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Demiray A.G., Demiray A., Yaren A., Yapar Taşköylü B., Gököz Doğu G., Değirmencioğlu S., Çakıroğlu U., Özhan N., Karan C., Çakan Demirel B., et al. Evaluation of Serum microRNA Let-7c and Let-7d as Predictive Biomarkers for Metastatic Pancreatic Cancer. Turk. J. Gastroenterol. 2022;33:696–703. doi: 10.5152/tjg.2022.21829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Duell E.J., Lujan-Barroso L., Sala N., Deitz McElyea S., Overvad K., Tjonneland A., Olsen A., Weiderpass E., Busund L.T., Moi L., et al. Plasma microRNAs as biomarkers of pancreatic cancer risk in a prospective cohort study. Int. J. Cancer. 2017;141:905–915. doi: 10.1002/ijc.30790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meijer L.L., Garajová I., Caparello C., Le Large T.Y.S., Frampton A.E., Vasile E., Funel N., Kazemier G., Giovannetti E. Plasma miR-181a-5p Downregulation Predicts Response and Improved Survival After FOLFIRINOX in Pancreatic Ductal Adenocarcinoma. Ann. Surg. 2020;271:1137–1147. doi: 10.1097/SLA.0000000000003084. [DOI] [PubMed] [Google Scholar]
- 91.Morimura R., Komatsu S., Ichikawa D., Takeshita H., Tsujiura M., Nagata H., Konishi H., Shiozaki A., Ikoma H., Okamoto K., et al. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br. J. Cancer. 2011;105:1733–1740. doi: 10.1038/bjc.2011.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Balzano F., Deiana M., Giudici S.D., Oggiano A., Baralla A., Pasella S., Mannu A., Pescatori M., Porcu B., Fanciulli G., et al. Molecules miRNA Stability in Frozen Plasma Samples. Molecules. 2015;20:19030–19040. doi: 10.3390/molecules201019030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Khan J., Lieberman J.A., Lockwood C.M. Variability in, variability out: Best practice recommendations to standardize pre-analytical variables in the detection of circulating and tissue microRNAs. Clin. Chem. Lab. Med. 2017;55:608–621. doi: 10.1515/cclm-2016-0471. [DOI] [PubMed] [Google Scholar]
- 94.Ameling S., Kacprowski T., Chilukoti R.K., Malsch C., Liebscher V., Suhre K., Pietzner M., Friedrich N., Homuth G., Hammer E., et al. Associations of circulating plasma microRNAs with age, body mass index and sex in a population-based study. BMC Med. Genom. 2015;8:61. doi: 10.1186/s12920-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kirschner M.B., Kao S.C., Edelman J.J., Armstrong N.J., Vallely M.P., Van Zandwijk N., Reid G. Haemolysis during Sample Preparation Alters microRNA Content of Plasma. PLoS ONE. 2011;6:e24145. doi: 10.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li Y., Kowdley K.V. Method for microRNA isolation from clinical serum samples. Anal. Biochem. 2012;431:69–75. doi: 10.1016/j.ab.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Duy J., Koehler J.W., Honko A.N., Minogue T.D. Optimized microRNA purification from TRIzol-treated plasma. BMC Genom. 2015;16:95. doi: 10.1186/s12864-015-1299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gemmati D., Longo G., Gallo I., Silva J.A., Secchiero P., Zauli G., Hanau S., Passaro A., Pellegatti P., Pizzicotti S., et al. Host genetics impact on SARS-CoV-2 vaccine-induced immunoglobulin levels and dynamics: The role of TP53, ABO, APOE, ACE2, HLA-A, and CRP genes. Front. Genet. 2022;13:3145. doi: 10.3389/fgene.2022.1028081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takizawa S., Matsuzaki J., Ochiya T. Circulating microRNAs: Challenges with their use as liquid biopsy biomarkers. Cancer Biomark. 2022;35:1–9. doi: 10.3233/CBM-210223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kleeff J., Korc M., Apte M., La Vecchia C., Johnson C.D., Biankin A.V., Neale R.E., Tempero M., Tuveson D.A., Hruban R.H., et al. Pancreatic cancer. Nat. Rev. Dis. Prim. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 101.Lampignano R., Kloten V., Krahn T., Schlange T. Integrating circulating miRNA analysis in the clinical management of lung cancer: Present or future? Mol. Aspects Med. 2020;72:100844. doi: 10.1016/j.mam.2020.100844. [DOI] [PubMed] [Google Scholar]
- 102.Marcuello M., Vymetalkova V., Neves R.P.L., Duran-Sanchon S., Vedeld H.M., Tham E., van Dalum G., Flügen G., Garcia-Barberan V., Fijneman R.J., et al. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol. Aspects Med. 2019;69:107–122. doi: 10.1016/j.mam.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 103.Hamam R., Hamam D., Alsaleh K.A., Kassem M., Zaher W., Alfayez M., Aldahmash A., Alajez N.M. Circulating microRNAs in breast cancer: Novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017;8:e3045. doi: 10.1038/cddis.2017.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Peng C., Wang J., Gao W., Huang L., Liu Y., Li X., Li Z., Yu X. Meta-analysis of the Diagnostic Performance of Circulating MicroRNAs for Pancreatic Cancer. Int. J. Med. Sci. 2021;18:660–671. doi: 10.7150/ijms.52706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wei L., Yao K., Gan S., Suo Z. Clinical utilization of serum- or plasma-based miRNAs as early detection biomarkers for pancreatic cancer: A meta-analysis up to now. Medicine. 2018;97:e12132. doi: 10.1097/MD.0000000000012132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Aalami A.H., Abdeahad H., Mesgari M. Circulating miR-21 as a potential biomarker in human digestive system carcinoma: A systematic review and diagnostic meta-analysis. Biomarkers. 2021;26:103–113. doi: 10.1080/1354750X.2021.1875504. [DOI] [PubMed] [Google Scholar]
- 107.Pei Z., Liu S.M., Huang J.T., Zhang X., Yan D., Xia Q., Ji C., Chen W., Zhang X., Xu J., et al. Clinically relevant circulating microRNA profiling studies in pancreatic cancer using meta-analysis. Oncotarget. 2017;8:22616–22624. doi: 10.18632/oncotarget.15148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Singh A., Singh A.K., Giri R., Kumar D., Sharma R., Valis M., Kuca K., Garg N. The role of microRNA-21 in the onset and progression of cancer. Future Med. Chem. 2021;13:1885–1906. doi: 10.4155/fmc-2021-0096. [DOI] [PubMed] [Google Scholar]
- 109.Ducreux M., Cuhna A.S., Caramella C., Hollebecque A., Burtin P., Goéré D., Seufferlein T., Haustermans K., Van Laethem J.L., Conroy T., et al. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2015;26((Suppl. 5)):v56–v68. doi: 10.1093/annonc/mdv295. [DOI] [PubMed] [Google Scholar]
- 110.Pointer D.T., Roife D., Powers B.D., Murimwa G., Elessawy S., Thompson Z.J., Schell M.J., Hodul P.J., Pimiento J.M., Fleming J.B., et al. Neutrophil to lymphocyte ratio, not platelet to lymphocyte or lymphocyte to monocyte ratio, is predictive of patient survival after resection of early-stage pancreatic ductal adenocarcinoma. BMC Cancer. 2020;20:750. doi: 10.1186/s12885-020-07182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guraya S. Prognostic significance of circulating microRNA-21 expression in esophageal, pancreatic and colorectal cancers; a systematic review and meta-analysis. Int. J. Surg. 2018;60:41–47. doi: 10.1016/j.ijsu.2018.10.030. [DOI] [PubMed] [Google Scholar]
- 112.Herman C. What makes a screening exam “good”? Virtual Mentor. 2006;8:34–37. doi: 10.1001/VIRTUALMENTOR.2006.8.1.CPRL1-0601. [DOI] [PubMed] [Google Scholar]
- 113.Wentzensen N., Wacholder S. From differences in means between cases and controls to risk stratification: A business plan for biomarker development. Cancer Discov. 2013;3:148. doi: 10.1158/2159-8290.CD-12-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hess V., Glimelius B., Grawe P., Dietrich D., Bodoky G., Ruhstaller T., Bajetta E., Saletti P., Figer A., Scheithauer W., et al. CA 19-9 tumour-marker response to chemotherapy in patients with advanced pancreatic cancer enrolled in a randomised controlled trial. Lancet Oncol. 2008;9:132–138. doi: 10.1016/S1470-2045(08)70001-9. [DOI] [PubMed] [Google Scholar]
- 115.Valihrach L., Androvic P., Kubista M. Circulating miRNA analysis for cancer diagnostics and therapy. Mol. Aspects Med. 2020;72:100825. doi: 10.1016/j.mam.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 116.Glinge C., Clauss S., Boddum K., Jabbari R., Jabbari J., Risgaard B., Tomsits P., Hildebrand B., Kaèaèb S., Wakili R., et al. Stability of Circulating Blood-Based MicroRNAs—Pre-Analytic Methodological Considerations. PLoS ONE. 2017;12:e0167969. doi: 10.1371/journal.pone.0167969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee E.S., Lee J.M. Imaging diagnosis of pancreatic cancer: A state-of-the-art review. World J. Gastroenterol. 2014;20:7864. doi: 10.3748/wjg.v20.i24.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000063.
- 119.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000065.
- 120.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000680.
- 121.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000098.
- 122.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000099.
- 123.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000104.
- 124.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000254.
- 125.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004556.
- 126.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0005865.
- 127.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000421.
- 128.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0005898.
- 129.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000445.
- 130.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000444.
- 131.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0005929.
- 132.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0005880.
- 133.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000425.
- 134.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0026478.
- 135.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000427.
- 136.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000436.
- 137.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000646.
- 138.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004658.
- 139.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000069.
- 140.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004489.
- 141.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000070.
- 142.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000256.
- 143.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000270.
- 144.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000257.
- 145.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0022692.
- 146.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000258.
- 147.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004559.
- 148.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0002821.
- 149.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0026608.
- 150.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000260.
- 151.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000259.
- 152.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000072.
- 153.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0002891.
- 154.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0007892.
- 155.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000222.
- 156.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004767.
- 157.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0002819.
- 158.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000226.
- 159.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0001080.
- 160.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0009201.
- 161.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000227.
- 162.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000073.
- 163.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000074.
- 164.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0001620.
- 165.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000682.
- 166.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004571.
- 167.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000318.
- 168.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000266.
- 169.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0009197.
- 170.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004493.
- 171.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000075.
- 172.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000076.
- 173.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004494.
- 174.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0026475.
- 175.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000267.
- 176.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000269.
- 177.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0022695.
- 178.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000272.
- 179.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004568.
- 180.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000278.
- 181.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000279.
- 182.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004570.
- 183.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000280.
- 184.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000077.
- 185.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004495.
- 186.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0019043.
- 187.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000080.
- 188.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004498.
- 189.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000081.
- 190.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000082.
- 191.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000420.
- 192.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000244.
- 193.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004674.
- 194.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0018985.
- 195.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000510.
- 196.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0005792.
- 197.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004506.
- 198.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000255.
- 199.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004557.
- 200.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0018104.
- 201.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0019892.
- 202.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000725.
- 203.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000726.
- 204.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000728.
- 205.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0001340.
- 206.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0001536.
- 207.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0016889.
- 208.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0018993.
- 209.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0019053.
- 210.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0001631.
- 211.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0019722.
- 212.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0019749.
- 213.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0019775.
- 214.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0002173.
- 215.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0002177.
- 216.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0002812.
- 217.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0022259.
- 218.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0003239.
- 219.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0003275.
- 220.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0023714.
- 221.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0024598.
- 222.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0024599.
- 223.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004810.
- 224.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0003298.
- 225.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0018444.
- 226.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0003326.
- 227.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0027398.
- 228.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0027500.
- 229.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0027542.
- 230.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0027600.
- 231.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0027638.
- 232.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000252.
- 233.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004553.
- 234.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0028111.
- 235.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0028117.
- 236.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004945.
- 237.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004946.
- 238.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0031000.
- 239.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0033692.
- 240.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004947.
- 241.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000093.
- 242.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004981.
- 243.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004982.
- 244.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000097.
- 245.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004511.
- 246.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0000689.
- 247.“miRDB Search Result.” [Online] [(accessed on 26 February 2023)]. Available online: https://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&full=mirbase&searchBox=MIMAT0004678.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.