Abstract
The juvenile-to-adult phase transition during vegetative development is a critical decision point in a plant’s life cycle. This transition is mediated by a decline in levels of miR156/157 and an increase in the activities of its direct targets, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) proteins. In Arabidopsis, the juvenile-to-adult transition is characterized by an increase in the length to width ratio of the leaf blade (a change in the distal region of a leaf), but what mediates this change in lamina shape is not known. Here, we show that ectopic expression of SPL9 and SPL13 produces enlarged and elongated leaves, resembling leaves from the blade-on-petiole1 (bop1) bop2 double mutant. The expression of BOP1/BOP2 is down-regulated in successive leaves, correlating with the amount of miR156 and antagonistic to the expression of SPL9 and SPL13 in leaves. SPL9 and SPL13 bind to the promoters of BOP1/BOP2 directly to repress their expression, resulting in delayed establishment of proliferative regions in leaves, which promotes more blade outgrowth (the distal region of a leaf) and suppresses petiole development (the proximal region of a leaf). Our results reveal a mechanism for leaf development along the proximal–distal axis, a heteroblastic character between juvenile leaves and adult leaves.
Keywords: Blade, BOP1, BOP2, petiole, SPL9, SPL13, vegetative phase change
Repression of BOP1/2expression by miR156-targeted SPL transcription factors controls establishment of proliferative regions in Arabidopsis leaves, thus controlling the blade and petiole development along the proximal–distal axis.
Introduction
After germination, flowering plants go through a juvenile vegetative phase and an adult vegetative phase before they flower. During the juvenile phase in Arabidopsis, the levels of miR156 and miR157 are high. As the shoot matures, the levels of miR156 and miR157 decline, and the activities of a group of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors increase (Wu et al., 2009; Xu et al., 2016b; He et al., 2018). The juvenile-to-adult phase transition (vegetative phase change) is largely controlled by the miR156–SPL module, and changes in miR156 and SPL activities resulted in changes in a plant’s immune response, plant–herbivore interactions, temperature stress responses, reproductive competence, and grain yield (Jiao et al., 2010; Stief et al., 2014; Mao et al., 2017; Wang et al., 2018; Hyun et al., 2019). Therefore, vegetative phase change is essential for the survival and reproductive fitness of a plant, and it is important to understand the mechanisms of vegetative phase change and how vegetative phase change traits are regulated.
In the juvenile phase, the Arabidopsis shoot produces small round leaves without abaxial trichomes. As the miR156/157 levels decline and the activities of their direct targets (SPLs) increase, the shoot produces leaves with more serrations at the blade margin, abaxial trichomes, and a more elongated blade (a change of the distal region of a leaf). Blade margin serration is largely controlled by CUP-SHAPED COTYLEDON (CUC) and TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors, with increased CUC activities inducing blade margin serration (Palatnik et al., 2003; Bilsborough et al., 2011; Hasson et al., 2011). Increased activities of the SPL proteins (such as SPL9) in adult leaves destabilize the CUC–TCP complex by physically interacting with TCP, releasing CUC proteins to induce leaf margin serration (Rubio-Somoza et al., 2014). The occurrence of abaxial trichomes is controlled by the interaction between the miR172 targeted TARGET-OF EAT1 (TOE1)/TOE2 and the adaxial/abaxial polarity regulator KANADI1 (KAN1) and their action on the trichome initiation gene GLABRA1 (GL1) (Wang et al., 2019; Xu et al., 2019). SPL9 and SPL15 directly activate the expression of MIR172b, leading to the down-regulation of its direct targets, TOE1/2 (Wu et al., 2009; Hyun et al., 2016). In juvenile leaves where the activities of miR156-targeted SPL proteins are low, TOE1/2 levels are high, and they physically interact with the abaxial identity protein KAN1 to repress the expression of the trichome initiation gene GL1 to suppress trichome production on the abaxial side of the juvenile leaves. In adult leaves where the activities of miR156-targeted SPL proteins are high, TOE1/2 levels are low and GL1 is de-repressed, resulting in production of trichomes on the abaxial side (Kerstetter et al., 2001; Wang et al., 2019; Xu et al., 2019). Together, the spatial and temporal interaction between KAN1 and TOE1/2 controls the spatial and temporal expression of GL1 to control the abaxial trichome production in juvenile and adult leaves. Overexpression of MIR172B (35S::MIR172B) greatly accelerates abaxial trichome production in 35S::MIR156A (from leaf 90 to leaf 11). The lamina shape of 35S::MIR156A, however, cannot be restored by 35S::MIR172B (Wu et al., 2009). Blade margin serration and abaxial trichome production during vegetative phase change have been well studied, but the mechanisms that regulate leaf shape during vegetative phase change have yet to be elucidated.
BLADE-ON-PETIOLE 1 (BOP1) and BOP2 genes encode the BTB-POZ domain of NPR1 subfamily transcription co-factors and have several roles during vegetative and reproductive development (Hepworth et al., 2005; Norberg et al., 2005; Ha et al., 2007; Jun et al., 2010; Khan et al., 2012). BOP1/2 are expressed in leaf primordia and floral primordia, in petioles of leaves, at the base of floral organs, and at the pedicel axis connecting the pedicel to the primary inflorescence (Hepworth et al., 2005; Norberg et al., 2005; Ha et al., 2007, 2010; Jun et al., 2010; Xu et al., 2010; Khan et al., 2012). Correspondingly, BOP1/2 have roles in leaf patterning along the proximal–distal axis, floral meristem identity, floral organ abscission, and fruit phyllotaxy (Hepworth et al., 2005; Norberg et al., 2005; Xu et al., 2010; Khan et al., 2012). In Arabidopsis, mutations in BOP1/2 simultaneously result in blade outgrowth on the petiole (Hepworth et al., 2005; Norberg et al., 2005; Ha et al., 2007; Jun et al., 2010). Orthologs of BOP1/2 in Medicago truncatula (NOOT), pea (COCH), and Lotus japonicus (NOOT-BOP-COCH-LIKE) promote proximal–distal patterning in leaves and floral organ patterning, similar to the roles of BOP1/2 in Arabidopsis. NOOT and NOOT-BOP-COCH-LIKE also promote identity of nodules, an organ that is found in Medicago truncatula and Lotus japonicus but not in Arabidopsis (Couzigou et al., 2012; Magne et al., 2018). Orthologs of BOP1/2 in rice were reported to control leaf sheath development (proximal region of a leaf, comparable to petiole in Arabidopsis) and to be repressed by the miR156-SPL module; mutations in OsBOP1/2/3 and the miR156-SPL module resulted in changed blade to sheath ratio and blade length to whole leaf length ratio (Toriba et al., 2019, 2020). The relative length of sheath (the proximal region of a leaf) has been a major heteroblastic character in rice. However, one of the major heteroblastic characters in Arabidopsis is the relative blade shape (the distal region of a leaf). Here, we analysed Arabidopsis leaf development along the proximal–distal axis spatially and temporally. We found that the more elongated blade is accompanied by suppression of petiole development and prolonged cell proliferation activities in the blade. The more elongated blade in adult leaves is probably caused by increased activities of miR156-targeted SPLs, which directly repress BOP1/2 expression, leading to the suppression of petiole development and continued outgrowth of blade.
Materials and methods
Plant material and growth conditions
All stocks used in this study were in the Col-0 genetic background. bop1-3 bop2-1, BOP1::GUS, and BOP2::GUS were gifts from Dr S. R. Hepworth (Xu et al., 2010). SPL9::sSPL9-GUS, SPL13::sSPL13-GUS, SPL9::rSPL9-GUS, and SPL13::rSPL13-GUS were gifts from Dr R. S. Poethig (Xu et al., 2016b). miR156-resistant SPL13 (rSPL13), rSPL13-HA, rSPL9-HA, and rSPL9-GFP were constructed in the Golden Gate system (Engler et al., 2014). Primers for making the constructs with the Golden Gate system are listed in Supplementary Table S1. The constructs were transformed into Arabidopsis Col-0 or spl9 spl13 using the floral dipping method (Clough and Bent, 1998). Primers for genotyping bop1, bop2, spl2, spl9, spl10, spl11, spl13, and spl15 were described previously (Xu et al., 2010, Xu et al., 2016b). Seeds were sown on Sunshine #8 potting soil, stratified at 4 °C for 2–4 d, and then transferred into Conviron growth chambers maintained at constant 22 °C in either long days (LDs; 16 h light: 8 h dark) or short days (SDs; 10 h light:14 h dark). Unless otherwise specified, all gene expression and chromatin immunoprecipitation (ChIP) analyses were performed with plants grown in LDs. Leaf length and width were measured by ImageJ software (imagej.nih.gov).
RT–quantitative PCR
Twelve-day-old seedlings of Col-0, spl9/13, rSPL13-GUS, rSPL13 Col-0, and rSPL13 spl9/13 plants or specific leaves as indicated in each figure, were harvested in liquid nitrogen. Tissues were ground into fine powder in liquid nitrogen, and total RNA was extracted using TRIzol (Thermo Fisher Scientific) followed by Turbo DNase (Thermo Fisher Scientific) treatment, according to the manufacturer’s instructions. cDNA was reverse transcribed from 1 μg of RNA with SuperScript III reverse transcriptase (Thermo Fisher Scientific), and quantitative PCR (qPCR) was performed using a Bio-Rad CFX96 real-time system. Primers used for qPCR are listed in Supplementary Table S1. All RT-qPCR experiments were performed in three biological replicates, and two reference genes, ACTIN2 (ACT2, AT3G18780) and EUKARYOTIC TRANSLATION INITIATION FACTOR 4A1 (EIF4A1, AT3G13920), were used in quantification analysis.
β-Glucuronidase staining analysis
Seedlings at different developmental stages as indicated in figures were pre-fixed in 90% acetone on ice for 10–20 min, followed by washing in β-glucuronidase (GUS) washing buffer (100 mM potassium phosphate buffer, pH 7.0, with 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, and 0.2% Triton X-100), then exchanged in GUS staining buffer. The tissue was vacuumed for 10 min and then incubated in GUS staining solution (100 mM potassium phosphate buffer, pH 7.0, with 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, 0.2% Triton X-100, and 2 mM X-Gluc) at 37 °C overnight. The chlorophyll was removed by washing stained tissues with 70% ethanol three times.
Chromatin immunoprecipitation
Three grams of 2-week-old Col-0 or transgenic plants grown in LDs were harvested and cross-linked in 1% formaldehyde under vacuum for 15 min. Tissues were ground in liquid nitrogen and suspended in extraction buffer 1 (0.4 M sucrose, 10 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 5 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 0.1% Triton X-100). Pellets were washed with extraction buffer 2 (0.25 M sucrose, 10 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 5mM β-mercaptoethanol, 1 mM PMSF, and 1% TritonX-100), and resuspended in nuclei lysis buffer (50 mM Tris–HCl, pH 8.0, 10 mM EDTA, and 1% SDS). DNA was then diluted in buffer (1.2 mM EDTA, 16.7 mM Tris–HCl, pH 8.0, 167 mM NaCl, and 0.01% SDS) and sonicated using a Covaris ultrasonicator M220. The hemagglutinin (HA) antibody and green fluorescent protein (GFP) antibody used in this study were from Sigma (cat. no. 11666606001) and Thermo Fisher Scientific (cat. no. A11122), respectively. The transposon TA3 was used as a negative control for non-specific binding. Primers used in ChIP analysis are listed in Supplementary Table S1.
Results
SPL13 promotes vegetative phase change and delays petiole development
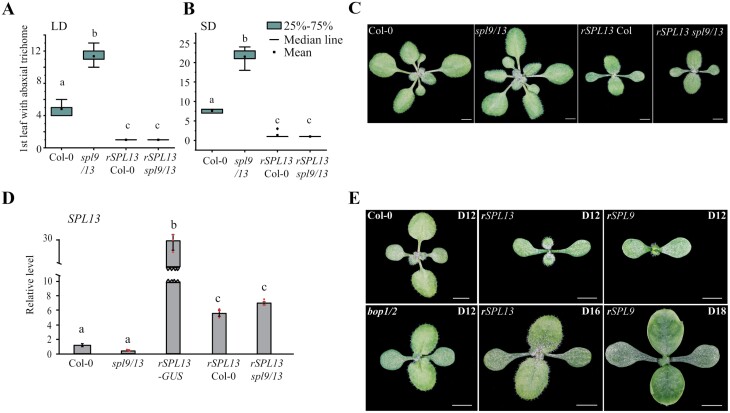
Genetic and molecular analysis have shown that miR156 and miR157 function redundantly to repress a group of SPL transcription factors that promote the adult vegetative phase and flowering (Wang et al., 2009; Xu et al., 2016b; He et al., 2018; Hyun et al., 2019). Among the 10 miR156/157-targeted SPLs, SPL13 plays essential roles in promoting phase transitions (Xu et al., 2016b). To further investigate the role of SPL13 in plant development, we made a construct containing a miR156-resistant SPL13 (SPL13::rSPL13) and transformed this construct into both Columbia wild type (Col-0) and the spl9 spl13 double mutant. Fourteen out of 18 (77.8%) T1 plants in a Col-0 background produced abaxial trichomes earlier than Col-0, whereas 18 out of 26 (69.2%) transgenics in an spl9 spl13 background produced abaxial trichomes earlier than spl9 spl13. We selected a representative line from each group and analysed its phenotype and the expression level of SPL13. Col-0 produced abaxial trichomes on leaf 4.8 ± 0.8 in LDs and 7.6 ± 0.5 in SDs, whereas the spl9/l3 double mutant produced abaxial trichomes on leaf 11.4 ± 0.9 in LDs and 21.5 ± 1.8 in SDs. In contrast, SPL13::rSPL13 Col-0 and SPL13::rSPL13 spl9/13 produced abaxial trichomes on leaf 1 ± 0.0 in LDs and leaf 1.3 ± 0.7 in SDs (Fig. 1A, B). This indicates that SPL13::rSPL13 complements the mutant phenotype of spl9/13, but also suggests that rSPL13 is overexpressed in both genetic backgrounds.
Fig. 1.
Plants ectopically expressing SPL13 and SPL9 are accelerated in vegetative phase change and mimic bop1 bop2 double mutant at seedling stage. (A, B) Ectopic expression of SPL13 in Col-0 and spl9/13 accelerated abaxial trichome production in both LDs (A) and SDs (B). (C) Sixteen-day-old Col-0, spl9 spl13, and rSPL13 plants in Col-0 or spl9 spl13 (spl9/13) plants growing in LDs. Scale bars: 3 mm. (D) RT-qPCR analysis of SPL13 transcripts in 12-day-old Col-0, spl9/13, rSPL13-GUS, rSPL13 Col-0, and rSPL13 spl9/13 seedlings. SPL13 is highly expressed in rSPL13-GUS, rSPL13 Col-0 and rSPL13 spl9/13 seedlings. Values are relative to Col-0 and represent the mean ±SEM from three biological replicates (red dots). Shared letters indicate not significantly different groups, different letters indicate significantly different groups; P<0.001, one-way ANOVA. (E) Twelve-day-old Col-0, rSPL13 plants, rSPL9 plants, and bop1 bop2 (bop1/2), 16-day-old rSPL13 plants, and 18-day-old rSPL9 plants growing in LDs. Note that the 16-day-old rSPL13 and 18-day-old rSPL9 plants mimic 12-day-old bop1/2. Scale bars: 3 mm.
Leaf emergence was delayed in both SPL13::rSPL13 Col-0 and SPL13::rSPL13 spl9/13 plants. At day 16, when petioles were clearly visible in Col-0 and spl9/13, leaf 1 and leaf 2 of the transgenic lines did not have visible petioles (Fig. 1C), resembling the bop1 bop2 (bop1/2) double mutant (Hepworth et al., 2005; Norberg et al., 2005; Ha et al., 2007; Xu et al., 2010). We grew the SPL13::rSPL13 spl9/13 plant (rSPL13 hereafter), spl9/13, bop1/2, and rSPL13-GUS plants together with Col-0 to examine their development (Supplementary Fig. S1). The spl9/13 double mutant had a faster rate of leaf initiation than Col-0, while rSPL13 plants, rSPL13-GUS plants, and bop1/2 had a slower rate of leaf initiation than Col-0 (Supplementary Fig. S1A, B). Similarly, the spl9/13 double mutant was delayed in producing abaxial trichomes, while the rSPL13 and rSPL13-GUS plants were markedly accelerated in producing abaxial trichomes, and bop1/2 was slightly accelerated in producing abaxial trichomes (Supplementary Fig. S1C, D). Because the phenotypes of rSPL13 plants were more severe than rSPL13-GUS, we examined the relative abundance of SPL13 in these plants by RT-qPCR. These results showed that SPL13 transcripts were elevated 5.7-fold in rSPL13 Col-0 and 7.1-fold in rSPL13 spl9/13 and were elevated about 30-fold in rSPL13-GUS plants (Fig. 1D). This suggests that the GUS tag at the 3ʹ end interferes with the activity of SPL13.
To confirm that the less severe phenotype in rSPL13-GUS plants is caused by the relatively large GUS tag, we transformed SPL13::rSPL13-HA and SPL9::rSPL9-HA constructs into the spl9/13 double mutant. The SPL13::rSPL13-HA spl9/13 plants looked very much like the SPL13::rSPL13 spl9/13 plants, while SPL9::rSPL9-HA spl9/13 plants had enlarged leaves 1 and 2 in which the boundary between blade and petiole was indistinct (Fig. 1E). Like the rSPL13 plants, the SPL9::rSPL9-HA spl9/13 (rSPL9 hereafter) plants produced abaxial trichomes on leaf 1, significantly earlier than wild type (WT) (5.4 ± 0.5) and bop1/2 (4.4 ± 0.5) (Supplementary Fig. S1E). These results suggest that having a small HA tag at the C-terminus of a protein does not interfere with the activities of the protein significantly, and ectopic expression of either SPL9 or SPL13 protein suppresses petiole development.
BOP1 and 2 act redundantly in vegetative phase change
BOP1 and BOP2 have been reported to function redundantly in regulating leaf and flower development while BOP2 functions by itself to promote photo-morphogenesis (Norberg et al., 2005; Ha et al., 2007; Jun et al., 2010; Xu et al., 2010; Khan et al., 2012; Zhang et al., 2017). To determine if BOP1 and BOP2 function redundantly in vegetative phase change, we examined the bop1 and bop2 single mutants as well as the bop1/2 double mutant. The bop1 and bop2 single mutants did not have a significant effect on two major vegetative phase change characters: the first leaf with abaxial trichome and the leaf blade length: blade width ratio. However, the bop1/2 double mutant was slightly accelerated in abaxial trichome production and had a larger blade length: blade width ratio (Supplementary Fig. S2A, B), indicating functional redundancy between BOP1 and BOP2 during vegetative phase change. The early abaxial trichome production in rSPL9 and rSPL13 plants could be attributed to the down-regulation of TOE1/2 genes, which then de-repress the trichome initiation gene GL1 (Wu et al., 2009; Wang et al., 2019; Xu et al., 2019). Our RT-qPCR analysis of bop1/2 leaves 1 and 4 showed that TOE1 and TOE2 were down-regulated in leaf 4 of the bop1/2 double mutant, suggesting that the accelerated trichome production in bop1/2 is probably caused by the down-regulation of TOE1/2 (Supplementary Fig. S2C, D) (Khan et al., 2015).
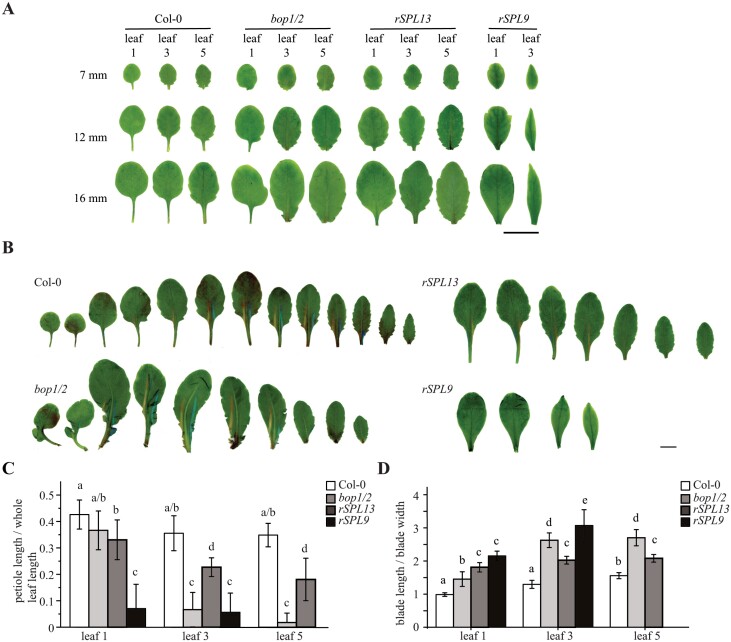
Petiole development is suppressed in rSPL13, rSPL9, and bop1/2 double mutant plants
Although the bop1/2 double mutant did not produce abaxial trichomes as early as rSPL9 plants and rSPL13 plants, the leaves of all three genotypes were very much delayed in developing a distinct petiole (Fig. 1E). We then compared leaf development in these genotypes, with a focus on petiole development. In LD conditions when the leaves are about 7 mm long, Col-0 has developed a visible and distinct petiole in leaf 1 and leaf 3 (juvenile leaves), and the petiole is just about to become visible in leaf 5 (normally an adult leaf) (Fig. 2A; Supplementary Fig. S3). However, in bop1/2, rSPL13, and rSPL9 plants, the petiole was not visible in 7 mm-long primordia of leaf 1, 3, or 5 (Fig. 2A; Supplementary Fig. S3A). This suggests that BOP1/2 promote petiole development while SPL9 and SPL13 suppress petiole development. When leaf 3 and leaf 5 are about 12 mm long, Col-0 has developed distinct petioles while bop1/2, rSPL13, and rSPL9 plants had not developed or just began to develop the petiole (Fig. 2A; Supplementary Fig. S3B). When leaf 5 was about 16 mm long, the petioles of Col-0 had elongated substantially, while bop1/2 has not yet developed any petioles, and rSPL13 plants had just begun to develop petioles (Fig. 2A; Supplementary Fig. S3C). Next, we examined petiole and blade development in mature leaves (when the inflorescence was about 1 cm long, and rosette leaves did not obviously grow). Overall, leaves 1 and 2 in bop1/2, rSPL13, and rSPL9 plants were bigger than Col-0 (Fig. 2B). Studies in rice showed that leaf sheath development is disrupted and the sheath length: whole leaf length ratio is shifted in Osbop mutants and mSPL14 plants (Toriba et al., 2019). As the examination of leaf development showed that petiole development was suppressed in bop1/2, rSPL13, and rSPL9 plants, we examined the petiole length: whole leaf length ratio in leaf 1, leaf 3, and leaf 5 (rSPL9 plants did not produce more than four leaves most of the time). Our results showed that the petiole length: whole leaf length ratio in bop1/2, rSPL13, and rSPL9 plants was significantly smaller than corresponding leaves in Col-0, except for leaf 1 in bop1/2 (Fig. 2C). As the blade length: blade width ratio (lamina shape) is one of the key characteristic traits of adult leaves in Arabidopsis, we analysed the lamina shape as well. Our data showed that the blade length: blade width ratio was significantly higher in all the corresponding leaves of bop1/2, rSPL13, and rSPL9 plants (Fig. 2D), suggesting a more elongated lamina. Together, our results suggest that petiole development and blade development are associated: BOP1/2 suppress and SPL9/13 promote blade development, while BOP1/2 promote and SPL9/13 suppress petiole development in Arabidopsis.
Fig. 2.
BOP1/2 promote while SPL9 and SPL13 suppress petiole development. (A) Morphology of Col-0, bop1/2, rSPL13, and rSPL9 leaf 1, leaf 3, and leaf 5 when they are about 7, 12, and 16 mm long. (B) Heteroblasty of Col-0, bop1/2, rSPL13, and rSPL9 plants. Plants were grown in LDs and leaves were dissected when inflorescence is about 1 cm long. (C, D) Petiole length: whole leaf length ratio (C) and blade length: blade width ratio (D) in leaves 1, 3, and 5 of Col-0, bop1/2, rSPL13, and rSPL9 plants. Shared letters above groups indicate not significantly different groups, and different letters above groups indicate significantly different groups; P<0.05, two-way ANOVA. Scale bars in (A, B): 1 cm.
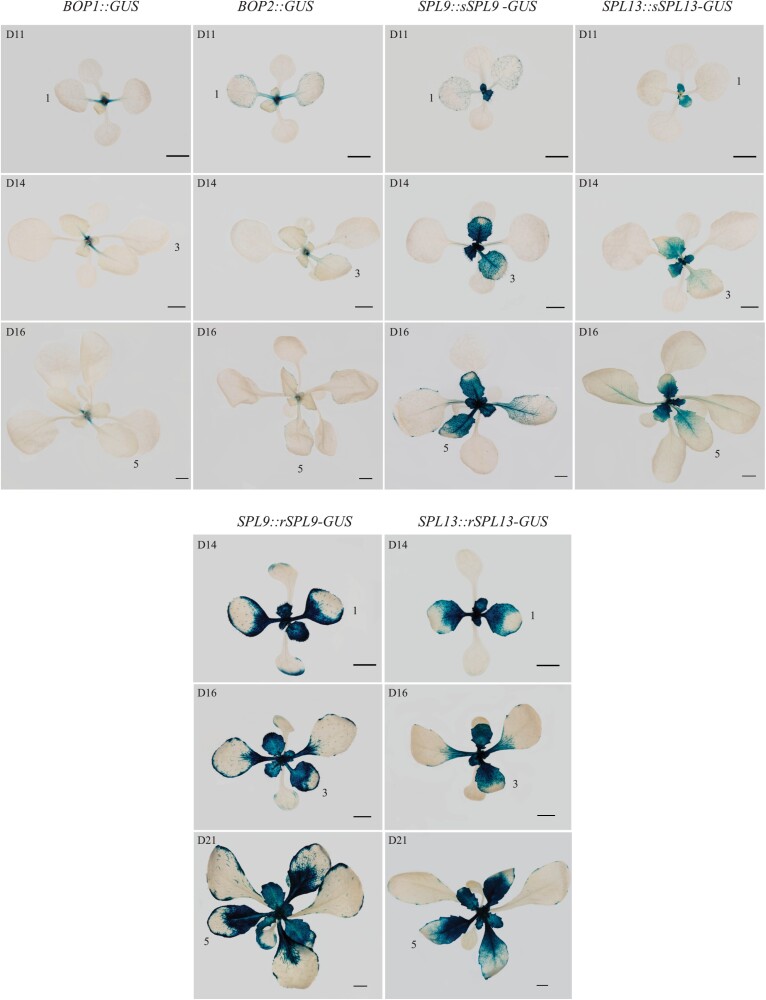
SPL9 and SPL13 repress BOP1 and BOP2
The results described above suggest that BOP1/2 and SPL9/13 function in opposite directions to regulate blade and petiole development. To investigate if they act antagonistically, we compared the expression pattern of BOP1, BOP2, miR156-sensitive SPL9 (sSPL9), miR156-resistant SPL9 (rSPL9), sSPL13, and rSPL13 genes using their GUS reporters (Fig. 3). In LDs, GUS expression driven by the BOP1 promoter (BOP1::GUS) was mainly detected in the petiole, whereas BOP2::GUS was detected in the petiole, midvein, and veins within the lamina of leaf 1 and 2 (Fig. 3). The expression of BOP1/2 was lower in the petiole (BOP1) or both the petiole and the blade (BOP2) of leaf 3 and 5 compared with leaf 1 and 2 (Fig. 3). sSPL9 protein detected from the SPL9::sSPL9-GUS plants was slightly expressed in leaf 1 and 2, whereas sSPL13 protein was excluded from leaf 1 and 2 (Fig. 3). Consistent with previous observations (Xu et al., 2016b; He et al., 2018), sSPL9 and sSPL13 proteins were expressed at higher levels in developing leaf 5 and 6 and were expressed at relatively lower levels in leaf 3 and 4, with the highest expression in the proximal region of the blade and petiole (Fig. 3). rSPL9 and rSPL13 proteins were highly expressed throughout the leaf when leaves were small, when petioles were not visible or just visible. They were localized to the proximal region of the blade and the petiole in larger leaves, when petioles were well developed (Fig. 3). Thus, BOP1/2 are highly expressed in the petiole of leaf 1 and 2, whereas SPL9 and SPL13 proteins are barely expressed there. In contrast, SPL9 and SPL13 proteins are more highly expressed in the petiole of leaf 5 than BOP1/2, suggesting that BOP1/2 and SPL9/13 interact antagonistically.
Fig. 3.
Expression of BOP1::GUS, BOP2::GUS, SPL9::sSPL9-GUS (sensitive to miR156), SPL13::sSPL13-GUS, SPL9::rSPL9-GUS (resistant to miR156), and SPL13::rSPL13-GUS. Plants were grown in LDs. All transgenic plants were in Col-0 background, and plants were harvested for GUS staining analysis when their leaf 1, leaf 3, and leaf 5 were at similar developmental stages. Scale bars: 2 mm.
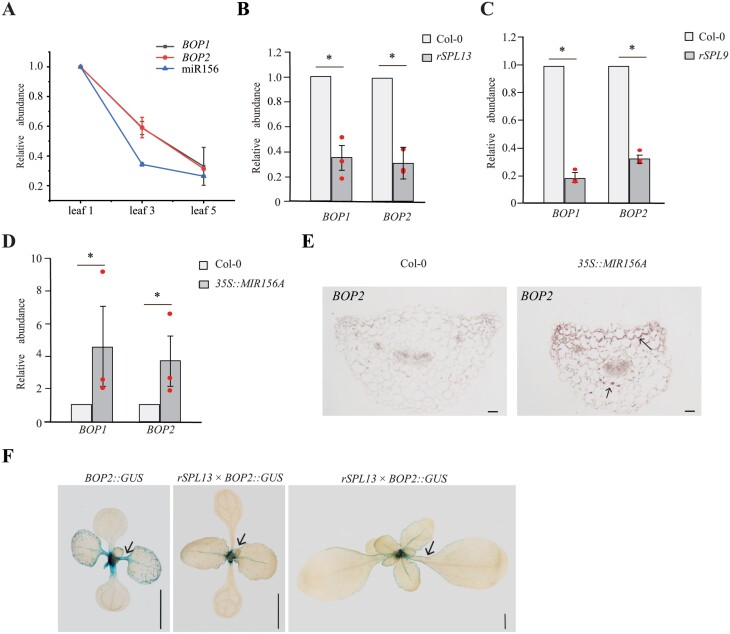
To confirm these observations, we measured the abundance of these transcripts by RT-qPCR (Fig. 4A). The amount of mature miR156 decreased about 70% from leaf 1 to leaf 3, whereas transcripts of BOP1 and BOP2 decreased about 40%. The amount of miR156 continued to drop in leaf 5, as did the transcripts of BOP1/2 (Fig. 4A). Thus, miR156 levels are positively correlated with the transcripts of BOP1/2, possibly through the repression of BOP1/2 by the SPL proteins targeted by miR156. To test this hypothesis, we examined the expression of BOP1/2 in the first two leaves of rSPL13 plants and rSPL9 plants. BOP1/2 mRNA levels were reduced about 60% in rSPL13 plants and 70% in rSPL9 plants (Fig. 4B, C). Conversely, BOP1/2 mRNA levels were elevated in 35S::MIR156A leaf 5 (Fig. 4D). To examine how miR156-targeted SPL proteins regulate BOP2 expression we used in situ hybridization and reporter genes to examine the effect of SPL proteins on the expression of BOP2. BOP2 was not detected in the petiole of Col-0 leaf 5 by in situ hybridization but was detected in the cortical cells underneath the upper epidermis and in a few cells surrounding the vascular bundle of 35S::MIR156A leaf 5 (Fig. 4E). BOP2::GUS was expressed at high levels in the petiole and veins of the first two leaves of Col-0 but was expressed at a much lower level in the petioles of the first two leaves in rSPL13 plants (Fig. 4F). Together, the RT-qPCR analysis and expression analysis suggest that SPL9 and SPL13 repress BOP1/2 to prolong blade development and delay petiole development in Arabidopsis.
Fig. 4.
SPL9 and SPL13 repress BOP1/2 in leaves. (A) RT-qPCR analysis of BOP1, BOP2, and miR156 in leaf 1, leaf 3, and leaf 5 in Col-0 wild-type. The amount of transcript of BOP1, BOP2 and miR156 in leaf 1 was set to 1. (B) RT-qPCR analysis of BOP1 and BOP2 in leaf 1 and 2 of Col-0 and rSPL13 plants. (C) RT-qPCR analysis of BOP1 and BOP2 in leaf 1 and 2 of Col-0 and rSPL9 plants. (D) RT-qPCR analysis of BOP1 and BOP2 in leaf 5 of Col-0 and 35S::MIR156A. The transcripts of BOP1 or BOP2 in Col-0 were set to 1. Values are means ±SEM from three biological replicates (red dots). *Significant difference between Col-0 and transgenic plants; P<0.001, one-way ANOVA. (E) In situ hybridization analysis of BOP2 in the fifth leaf petiole of Col-0 and 35S::MIR156A. BOP2 is not expressed in the fifth leaf petiole of Col-0, but is expressed in the cortical cells underneath the adaxial epidermis and in a few cortical cells around the vascular bundle in 35S::MIR156A. Arrows indicate expression of BOP2. Scale bars: 50 μm. (F) Expression of BOP2::GUS in Col-0 and rSPL13 plants. Note that BOP2 was strongly expressed in the petiole and veins of Col-0 leaf 1 and 2, while BOP2 expression is reduced in rSPL13 veins when its total leaf length is the same as the Col-0 (petiole was not yet visible), and BOP2 is expressed at much lower levels at the margin of petiole when rSPL13’s petiole is visible. Arrows indicate expression of BOP2. Scale bars: 3 mm.
Genetic interaction between miR156-targeted SPLs and BOP1/2
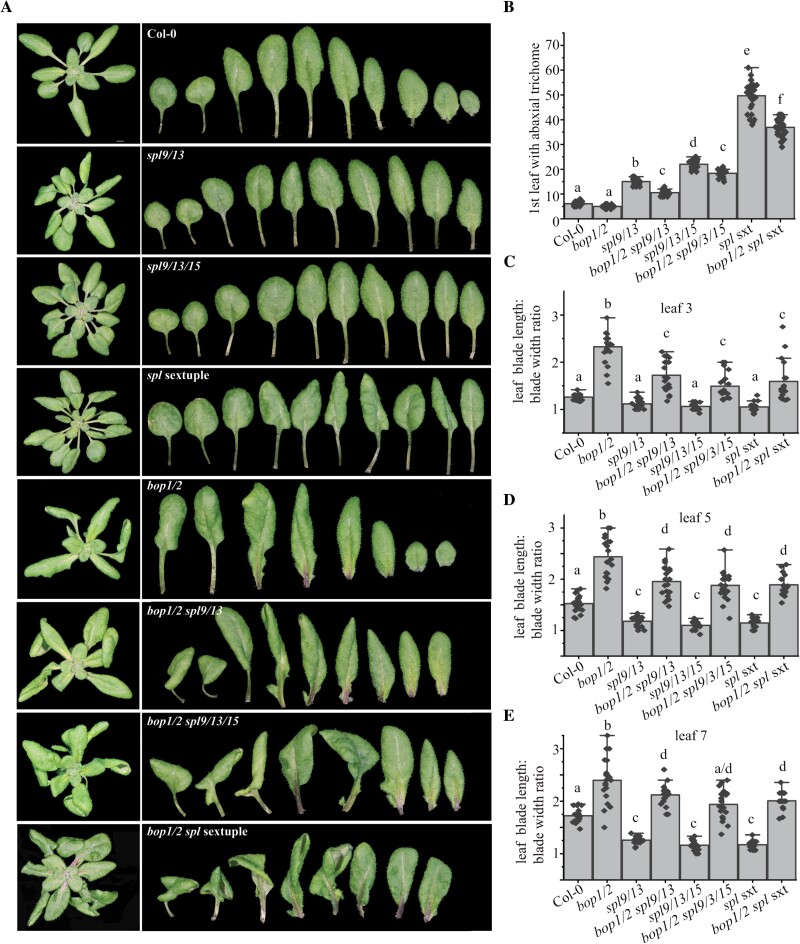
To investigate the genetic interaction between BOP1/2 and miR156-targeted SPLs, we crossed bop1/2 to loss-of-function spl mutants. Our previous analysis showed that SPL9 and SPL13 have the most significant effect on vegetative phase change, although SPL2, SPL10, SPL11, and SPL15 also contribute to this process (Xu et al., 2016b). We next introduced bop1/2 into spl9/13, spl9/13/15, and spl2/9/10/11/13/15 (spl sextuple, spl sxt) mutants. The rosettes of the bop1/2 spl9/13, bop1/2 spl9/13/15, and bop1/2 spl sextuple (bop1/2 spl sxt) mutants look like the bop1/2 rosette (Fig. 5A). Their abaxial trichome production was intermediate between bop1/2 and the corresponding spl9/13, spl9/13/15, and the spl sxt mutants, but much closer, respectively, to the trichome production in spl9/13, spl9/13/15, and the spl sxt than to bop1/2 (Fig. 5B). Quantitative examination of the lamina shape in leaf 3, leaf 5, and leaf 7 of the mutants showed that leaves of bop1/2 have more elongated lamina than Col-0, while spl9/13, spl9/13/15, and the spl sxt have a rounder lamina than Col-0 (Fig. 5C–E). The lamina shape in bop1/2 spl9/13/15, and bop1/2 spl sxt mutants was intermediate between their bop1/2 and spl9/13/15 or spl sxt parents, but much closer to the lamina shape in bop1/2 (Fig. 5C–E). Together, these results suggest that BOP1/2 and additional targets of the miR156-targeted SPLs are involved in determining lamina shape in Arabidopsis. The abaxial trichome production, on the other hand, is largely controlled by factors other than BOP1/2 (Wu et al., 2009; Wang et al., 2019; Xu et al., 2019).
Fig. 5.
Genetic interaction between bop1/2 and spl mutants. (A) Whole plant and heteroblasty of Col-0, spl9/13, bop1/2 spl9/13, spl9/13/15, bop1/2 spl9/13/15, spl2/9/10/11/13/15 (spl sextuple), and bop1/2 spl sextuple. (B) The first leaf with abaxial trichome in Col-0, bop1/2, spl mutants, and bop1/2 spl mutants. (C–E) The blade length/width ratio in leaf 3 (C), leaf 5 (D), and leaf 7 (E) of Col-0, bop1/2, spl mutants, and bop1/2 spl mutants. Shared letters above groups indicate no significant difference between them, while different letters above groups indicate the groups that are significantly different; P<0.01 (B–E), one-way ANOVA.
SPL9 and SPL13 binds to BOP1 and BOP2 directly
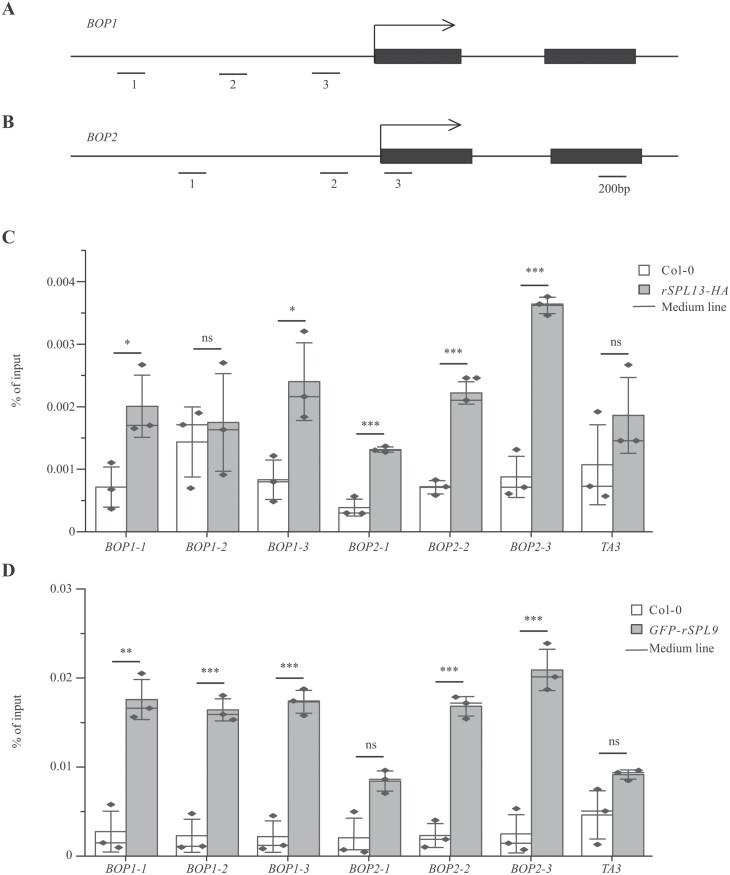
To investigate if miR156-targeted SPL9 and SPL13 represses BOP1/2 directly, we used chromatin immunoprecipitation followed by qPCR (ChIP-qPCR) to examine the binding of SPL9 and SPL13 to BOP1 and BOP2. We used a homozygous line from SPL13::rSPL13-HA spl9/13 (rSPL13-HA hereafter) transgenic plants, and made new SPL9::GFP-rSPL9 spl9/13 transgenic plants as the SPL9::rSPL9-HA spl9/13 we made earlier (Fig. 1) could not be maintained as homozygotes. We chose a SPL9::GFP-rSPL9 spl9/13 line (GFP-rSPL9 hereafter) that produces abaxial trichomes on leaf 1(Supplementary Fig. S4), similar to the rSPL9-HA plants we generated previously (Supplementary Fig. S1E). We collected vegetative tissues for ChIP-qPCR analysis. We identified potential SPL binding sites in BOP1 and BOP2 using ATHAMAP (athamap.de) and examined the abundance of these sites in chromatin immunoprecipitated with antibodies to HA and GFP. We found that both SPL9 and SPL13 proteins bind to the BOP1 promoter 1200–1400bp (BOP1-1) upstream of its transcription start site (TSS) and near its TSS (BOP1-3) (Fig. 6). SPL9 and SPL13 proteins bind to BOP2 promoter and right after its TSS (BOP2-2 and BOP2-3). SPL9 and SPL13 proteins also have unique binding sites at BOP1 and BOP2: SPL9 binds to BOP1-2 whereas SPL13 does not bind to this site, and SPL13 associates with BOP2-1 whereas SPL9 does not (Fig. 6). Together, our ChIP-qPCR analysis suggests that SPL9 and SPL13 proteins have overlapping and distinctive binding sites in BOP1 and BOP2 genomic DNA, which may explain the similar and distinctive phenotypes of rSPL9 and rSPL13 plants.
Fig. 6.
SPL9 and SPL13 bind to BOP1 and BOP2 directly. (A, B) Schematic diagram of the genomic structure of BOP1 (A) and BOP2 (B). Black boxes indicate exons, arrows indicate transcription start site. 1, 2, and 3 represent potential SPL binding sites to BOP1/2. (C) ChIP-qPCR analysis of rSPL13-HA occupancy at BOP1 and BOP2. (D) ChIP-qPCR analysis of GFP–rSPL9 occupancy at BOP1 and BOP2. Values are means ±SEM from three biological replicates (black dots). *P<0.05, **P<0.01, ***P<0.001; ns denotes not significantly different, P>0.05; one-way ANOVA. TA3 is a negative control.
AN3 activity is prolonged in leaves of bop1/2 and rSPL13
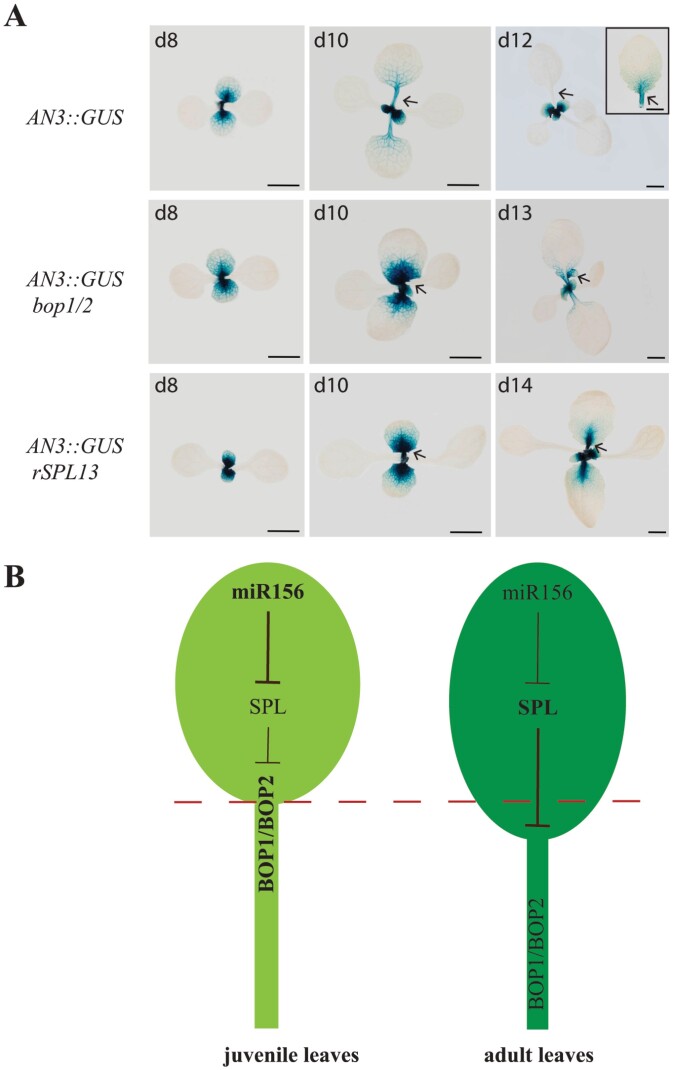
Next, we investigated the basis for the increased blade outgrowth and suppressed development of the petiole in bop1/2, rSPL9, and rSPL13 plants. Studies of Arabidopsis leaf development suggest that establishment of a proliferative region at the junction between blade and petiole is important for blade and petiole development (Ichihashi et al., 2011). This proliferative region is highly marked by the cell division marker pCYCB1;1::Dbox::GUS and AN3::GUS (Ichihashi et al., 2011). Leaves of the angustifolia3-4 (an3-4) mutant are smaller than WT, and the epidermal cell number in an3-4 is proportionally smaller than WT, suggesting that AN3 regulates cell proliferation in leaves (Kawade et al., 2013). Analysis of AN3 expression in developing leaves using an AN3::GUS reporter showed that AN3 is widely expressed in developing leaf primordia, is localized to the proliferative region when the blade-petiole boundary is just established, and its expression is almost gone when the leaf matures (Ichihashi et al., 2011). To investigate if the establishment of the proliferative region is affected in bop1/2 and rSPL13 leaves, we amplified a 4-kb AN3 promoter and fused it to GUS and transformed this construct into Col-0. We selected a line that showed a similar expression pattern to the one examined by Ichihashi et al. and crossed this line to the bop1/2 double mutant and the rSPL13 transgenic line. As leaf initiation is severely delayed in rSPL13 homozygotes (Fig. 1B), we selected plants that were homozygous for AN3::GUS and heterozygous for rSPL13 for comparison. We found that at day 8 when petioles are just about to develop in Col-0, AN3::GUS was localized to the proliferative region (Fig. 7A). At day10 when both the leaf blade and leaf petiole had elongated in Col-0, AN3::GUS was detected in the whole petiole and the proximal region of the blade, at much lower levels than its levels at day 8. At day 12, AN3::GUS could no longer be detected in leaf 1 and 2 of Col-0. The fifth leaf of Col-0 is normally an adult leaf in LD conditions and AN3::GUS could be detected in the petiole and the proximal region of the blade when its overall length was the same as its first leaf (Fig. 7A, inset). Petioles were not visible in 8-day-old bop1/2 nor rSPL13 plants, and AN3::GUS was more widely detected in their blades than Col-0 at this stage, with a higher concentration in the proximal region of the blade. Petioles were just visible in 10-day-old bop1/2 and rSPL13 plants, and their AN3::GUS expression was persistently detected in the proliferation zone at higher levels than its expression in Col-0 at day 10. At day 13 and day14, when leaf 1 and 2 of bop1/2 and rSPL13 plants were developed into the same length as Col-0 leaf 1 and 2, respectively, AN3::GUS could still be detected in the proximal region of the blade (arrows), indicating that cell proliferation persisted longer in the blades of bop1/2 and rSPL13 plants than in Col-0. These results indicate that the establishment of the proliferative region is delayed in bop1/2 and rSPL13 plants and the more elongated lamina in them may be a result of prolonged AN3 activity in their blade.
Fig. 7.
Mechanism for leaf blade development. (A) Expression of AN3::GUS in Col-0 and mutants. Top: expression of AN3::GUS in Col-0 at day (d) 8, d10, and d12 and the expression of AN3::GUS in leaf 5 (inset in image of d12) when it is at similar length to leaf 1 at d12. Middle: expression of AN3::GUS in bop1/2 at similar developmental stages to Col-0. Bottom: expression of AN3::GUS in rSPL13 plants at similar developmental stages to Col-0. Scale bars: 2 mm. Arrows indicate the expression of AN3::GUS in Col-0, bop1/2 double mutant, and rSPL13 plants at the proximal region of the blade. (B) Model for juvenile leaf and adult leaf development. In juvenile leaves where miR156 levels are high, miR156-targeted SPL levels are low, and BOP1/2 are activated, resulting in early establishment of the blade–petiole boundary and shorter blades. In adult leaves where miR156 levels are down-regulated, miR156-targeted SPLs are up-regulated and BOP1/2 are down-regulated, resulting in delayed establishment of the blade–petiole boundary and longer blades.
Discussion
Up-regulation of miR156-targeted SPLs resulted in changes of heteroblastic traits in vegetative leaves (Wu et al., 2009; Xu et al., 2016b; He et al., 2018). Lamina shape is one of the heteroblastic traits between juvenile leaves and adult leaves in Arabidopsis. Here, we show that a more elongated lamina in Arabidopsis adult leaves is caused by down-regulation of BOP1/2. SPL9 and SPL13 directly repress BOP1/2 to delay petiole development, resulting in more blade outgrowth and more elongated lamina.
The shape of the lamina and abaxial trichome production are controlled by different mechanisms
Although the bop1/2 double mutant produces abaxial trichomes slightly earlier than WT, its lamina is much more elongated than that of WT, suggesting that BOP1/2 have a larger role in controlling the lamina shape than in trichome production. Several lines of evidence suggest that trichome production and lamina shape are controlled by different mechanisms. Trichome production is largely controlled by the trichome initiation gene GL1, which is repressed by the miR172-targeted TOE1/2 genes (Wang et al., 2019; Xu et al., 2019). miR156-targeted SPLs directly activate MIR172b, which represses TOE1/2 genes (Wu et al., 2009). In the GL1 dominant mutant gl1-D, whose TOE1/2 binding site is mutated, abaxial trichome production is accelerated but the lamina shape is not significantly different from WT, suggesting that lamina shape and abaxial trichome production are controlled by different mechanisms (Xu et al., 2019). Consistent with this, abaxial trichome production in the 35S::MIR172B 35S::MIR156A double mutant is close to that of 35S::MIR172B while the lamina shape in the double mutant is close to that of 35S::MIR156A (Wu et al., 2009). In the bop1/2 spl sextuple mutant, trichome production is closer to that of the spl sextuple mutant. However, its lamina shape is closer to that of the bop1/2 double mutant (Fig. 5). Together, these results suggest that abaxial trichome production in leaves is mainly controlled by the miR172–TOE module, while lamina shape is controlled by BOP1/2 and other unknown SPL targets.
SPL controls leaf development along the proximal–distal axis by repressing BOP1/2
Previous analysis showed that the establishment of a proliferative region at the junction between petiole and blade is essential for leaf development along the proximal–distal axis in Arabidopsis (Ichihashi et al., 2011). A proliferative region is established when the petiole has just developed from the proximal region of a leaf. AN3 is highly expressed in this proliferative region and its expression decreases as the leaf matures (Ichihashi et al., 2011). The spatial and temporal examination of blade and petiole development in Col-0, bop1/2, and rSPL13 plants suggested that leaf development along the proximal–distal axis is largely dependent on the occurrence of the petiole: when petiole development is significantly delayed in both the bop1/2 double mutant and rSPL13 plants, the establishment of the proliferative region is delayed in them, resulting in longer AN3 activity and more blade outgrowth than in Col-0 (Fig. 7). In WT juvenile leaves, where activities of miR156-targeted SPLs are low, BOP1/2 are highly expressed, and the proliferative region is established early. AN3 diminishes sooner in juvenile lamina, resulting in a rounder lamina. In adult leaves, where activities miR156-targeted SPLs are increased, BOP1/2 are down-regulated, and the establishment of the proliferative region is delayed. AN3 activity then persists longer in adult leaves than in juvenile leaves, resulting in a more elongated lamina (Fig. 7B). Leaf 1 and 2 in rSPL13 plants and bop1/2 are bigger than leaf 1 and 2 in Col-0, and it is quite possible that the increase in size is caused by down-regulation of BOP1/2. It is, however, not clear if this is connected to the activity of AN3, as expression of AN3 is not expanded laterally in rSPL13 plants and bop1/2 leaves.
Heteroblastic trait in Arabidopsis and rice
In Arabidopsis, the proximal region of a leaf develops as a petiole, and the distal region develops as a blade. In rice, the proximal region of a leaf develops as a sheath, and the distal region develops as a blade (Ichihashi et al., 2011; Toriba et al., 2019). Although the petiole and sheath develop at different stages in Arabidopsis and rice leaves, their development is controlled by similar genes. Sheath development is severely suppressed in the rice osbop1/2/3 triple mutant, and petiole development is severely suppressed in the Arabidopsis bop1/2 double mutant (Hepworth et al., 2005; Norberg et al., 2005; Jun et al., 2010; Toriba et al., 2019), suggesting conserved roles for BOP in monocots and dicots. Studies in rice found that miR156-targeted SPLs repress OsBOP1/2/3 expression to control sheath development. Here, we found that miR156-targeted SPL9 and SPL13 repress BOP1/2 directly in leaves to control petiole and blade development (Figs 4, 6).
The lamina shape is a heteroblastic trait in Arabidopsis, while the relative sheath length is a heteroblastic trait in rice (Wu et al., 2009; Yang et al., 2012; Xu et al., 2016a, b; Toriba et al., 2019; Cheng et al., 2021). Here, our data showed that the lamina shape is correlated to the relative petiole length in juvenile leaves and adult leaves (Fig. 2). Together, studies in Arabidopsis and rice suggest that changes in the three ratios blade length: blade width, petiole length: whole leaf length, and leaf sheath: blade are caused by disrupted development at the proximal region of leaves, petiole, or sheath. This suggests that the development of the proximal region of a leaf may be a key factor for heteroblastic traits given the conserved roles of BOP1/2 and SPLs in higher plants.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Plants ectopically expressing SPL13 mimic bop1 bop2 double mutant.
Fig. S2. BOP1 and BOP2 act redundantly to promote petiole development and suppress blade outgrowth.
Fig. S3. BOP1/2 promote while SPL9 and SPL13 suppress petiole development.
Fig. S4. Ectopic expression of GFP-rSPL9 spl9/13 accelerated abaxial trichome production in both LDs and SDs.
Fig. S5. RT-qPCR analysis of gene expression normalized to EIF4A1.
Table S1. Primers used in this study.
Acknowledgements
We thank Dr Scott Poethig and Dr Beth Krizek for critical reading of the manuscript.
Contributor Information
Tieqiang Hu, Department of Biological Sciences, University of South Carolina, Columbia, SC 29208, USA.
Darren Manuela, Department of Biological Sciences, University of South Carolina, Columbia, SC 29208, USA.
Mingli Xu, Department of Biological Sciences, University of South Carolina, Columbia, SC 29208, USA.
Rainer Melzer, University College Dublin, Ireland.
Author contributions
TH and MX designed experiments, performed the experiments, and analysed data. DM made the SPL9::GFP-rSPL9 transgenic plants. MX wrote the manuscript. DM and MX revised the manuscript.
Conflict of interest
The authors declare no conflict of interest.
Funding
This work was supported by a grant from the National Science Foundation (IOS 1947274) to MX and startup funds from University of South Carolina to MX.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary data published online.
References
- Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M.. 2011. Model for the regulation of Arabidopsis thaliana leaf margin development. Proceedings of the National Academy of Sciences, USA 108, 3424–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YJ, Shang GD, Xu ZG, Yu S, Wu LY, Zhai D, Tian SL, Gao J, Wang L, Wang JW.. 2021. Cell division in the shoot apical meristem is a trigger for miR156 decline and vegetative phase transition in Arabidopsis. Proceedings of the National Academy of Sciences, USA 118, e2115667118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF.. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Couzigou JM, Zhukov V, Mondy S, et al. 2012. NODULE ROOT and COCHLEATA maintain nodule development and are legume orthologs of Arabidopsis BLADE-ON-PETIOLE genes. The Plant Cell 24, 4498–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert TM, Werner S, Jones JD, Patron NJ, Marillonnet S.. 2014. A golden gate modular cloning toolbox for plants. ACS Synthetic Biology 3, 839–843. [DOI] [PubMed] [Google Scholar]
- Ha CM, Jun JH, Fletcher JC.. 2010. Control of Arabidopsis leaf morphogenesis through regulation of the YABBY and KNOX families of transcription factors. Genetics 186, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CM, Jun JH, Nam HG, Fletcher JC.. 2007. BLADE-ON-PETIOLE 1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. The Plant Cell 19, 1809–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson A, Plessis A, Blein T, Adroher B, Grigg S, Tsiantis M, Boudaoud A, Damerval C, Laufs P.. 2011. Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. The Plant Cell 23, 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Xu M, Willmann MR, McCormick K, Hu T, Yang L, Starker CG, Voytas DF, Meyers BC, Poethig RS.. 2018. Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana. PLoS Genetics 14, e1007337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Zhang Y, McKim S, Li X, Haughn GW.. 2005. BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. The Plant Cell 17, 1434–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Richter R, Vincent C, Martinez-Gallegos R, Porri A, Coupland G.. 2016. Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Developmental Cell 37, 254–266. [DOI] [PubMed] [Google Scholar]
- Hyun Y, Vincent C, Tilmes V, Bergonzi S, Kiefer C, Richter R, Martinez-Gallegos R, Severing E, Coupland G.. 2019. A regulatory circuit conferring varied flowering response to cold in annual and perennial plants. Science 363, 409–412. [DOI] [PubMed] [Google Scholar]
- Ichihashi Y, Kawade K, Usami T, Horiguchi G, Takahashi T, Tsukaya H.. 2011. Key proliferative activity in the junction between the leaf blade and leaf petiole of Arabidopsis. Plant Physiology 157, 1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, et al. 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature Genetics 42, 541–544. [DOI] [PubMed] [Google Scholar]
- Jun JH, Ha CM, Fletcher JC.. 2010. BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2. The Plant Cell 22, 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawade K, Horiguchi G, Usami T, Hirai MY, Tsukaya H.. 2013. ANGUSTIFOLIA3 signaling coordinates proliferation between clonally distinct cells in leaves. Current Biology 23, 788–792. [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS.. 2001. KANADI regulates organ polarity in Arabidopsis. Nature 411, 706–709. [DOI] [PubMed] [Google Scholar]
- Khan M, Ragni L, Tabb P, et al. 2015. Repression of lateral organ boundary genes by PENNYWISE and POUND-FOOLISH is essential for meristem maintenance and flowering in Arabidopsis. Plant Physiology 169, 2166–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Xu M, Murmu J, Tabb P, Liu Y, Storey K, McKim SM, Douglas CJ, Hepworth SR.. 2012. Antagonistic interaction of BLADE-ON-PETIOLE1 and 2 with BREVIPEDICELLUS and PENNYWISE regulates Arabidopsis inflorescence architecture. Plant Physiology 158, 946–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magne K, Couzigou JM, Schiessl K, et al. 2018. MtNODULE ROOT1 and MtNODULE ROOT2 are essential for indeterminate nodule identity. Plant Physiology 178, 295–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YB, Liu YQ, Chen DY, Chen FY, Fang X, Hong GJ, Wang LJ, Wang JW, Chen XY.. 2017. Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nature Communications 8, 13925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norberg M, Holmlund M, Nilsson O.. 2005. The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 132, 2203–2213. [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D.. 2003. Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I, Zhou CM, Confraria A, Martinho C, von Born P, Baena-Gonzalez E, Wang JW, Weigel D.. 2014. Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Current Biology 24, 2714–2719. [DOI] [PubMed] [Google Scholar]
- Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Bäurle I.. 2014. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. The Plant Cell 26, 1792–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriba T, Tokunaga H, Nagasawa K, Nie F, Yoshida A, Kyozuka J.. 2020. Suppression of leaf blade development by BLADE-ON-PETIOLE orthologs is a common strategy for underground rhizome growth. Current Biology 30, 509–516.e3. [DOI] [PubMed] [Google Scholar]
- Toriba T, Tokunaga H, Shiga T, Nie F, Naramoto S, Honda E, Tanaka K, Taji T, Itoh JI, Kyozuka J.. 2019. BLADE-ON-PETIOLE genes temporally and developmentally regulate the sheath to blade ratio of rice leaves. Nature Communications 10, 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhou L, Shi H, et al. 2018. A single transcription factor promotes both yield and immunity in rice. Science 361, 1026–1028. [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D.. 2009. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhou CM, Mai YX, et al. 2019. A spatiotemporally regulated transcriptional complex underlies heteroblastic development of leaf hairs in Arabidopsis thaliana. The EMBO Journal 38, e100063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS.. 2009. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hu T, McKim SM, Murmu J, Haughn GW, Hepworth SR.. 2010. Arabidopsis BLADE-ON-PETIOLE1 and 2 promote floral meristem fate and determinacy in a previously undefined pathway targeting APETALA1 and AGAMOUS-LIKE24. The Plant Journal 63, 974–989. [DOI] [PubMed] [Google Scholar]
- Xu M, Hu T, Smith MR, Poethig RS.. 2016a. Epigenetic regulation of vegetative phase change in Arabidopsis. The Plant Cell 28, 28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hu T, Zhao J, Park MY, Earley KW, Wu G, Yang L, Poethig RS.. 2016b. Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genetics 12, e1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Qian Z, Zhou B, Wu G.. 2019. Age-dependent heteroblastic development of leaf hairs in Arabidopsis. New Phytologist 224, 741–748. [DOI] [PubMed] [Google Scholar]
- Yang L, Wu G, Poethig RS.. 2012. Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proceedings of the National Academy of Sciences, USA 109, 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Holmlund M, Lorrain S, Norberg M, Bako L, Fankhauser C, Nilsson O.. 2017. BLADE-ON-PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. eLife 6, e26759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary data published online.