Abstract
Plasmodesmata are cytosolic bridges, lined by the plasma membrane and traversed by endoplasmic reticulum; plasmodesmata connect cells and tissues, and are critical for many aspects of plant biology. While plasmodesmata are notoriously difficult to extract, tissue fractionation and proteomic analyses can yield valuable knowledge of their composition. Here we have generated two novel proteomes to expand tissue and taxonomic representation of plasmodesmata: one from mature Arabidopsis leaves and one from the moss Physcomitrium patens, and leveraged these and existing data to perform a comparative analysis to identify evolutionarily conserved protein families that are associated with plasmodesmata. Thus, we identified β-1,3-glucanases, C2 lipid-binding proteins, and tetraspanins as core plasmodesmal components that probably serve as essential structural or functional components. Our approach has not only identified elements of a conserved plasmodesmal proteome, but also demonstrated the added power offered by comparative analysis for recalcitrant samples. Conserved plasmodesmal proteins establish a basis upon which ancient plasmodesmal function can be further investigated to determine the essential roles these structures play in multicellular organism physiology in the green lineages.
Keywords: Arabidopsis, cell to cell communication, phyloproteomics, Physcomitrium patens, plasmodesmata, proteomics
Plasmodesmata enable cell to cell communication for growth, development, and responses in plants. We used comparative analysis of plasmodesmal proteomes from divergent plants to identify conserved plasmodesmal elements.
Introduction
Plasmodesmata are membrane-lined connections that traverse the cell wall and interconnect the cytoplasm, plasma membrane, and endoplasmic reticulum (ER) between plant cells. The direct cytosol–cytosol contact enables the sharing of resources and information, underpinning growth, developmental, and response processes (Benitez-Alfonso, 2014; Sevilem et al., 2015; Brunkard and Zambryski, 2017; Cheval and Faulkner, 2018). Plasmodesmata are dynamic, responding to internal and external cues to create transient domains of connectivity within tissues. While it is established that their responses to a range of environmental signals are enabled by specialized signalling machinery (Cui and Lee, 2016; Cheval et al., 2020), the molecular machinery that brings about their biogenesis and structure is less well defined.
To obtain a comprehensive overview of proteins present at plasmodesmata, and ultimately build understanding of their role in physiology and development, proteomic characterization of plasmodesmata-enriched fractions has been performed on multiple occasions (Faulkner et al., 2005; Fernandez-Calvino et al., 2011; Park et al., 2017; Leijon et al., 2018; Brault et al., 2019). Such proteomes have provided valuable insights into plasmodesmal structure and function, identifying novel plasmodesmal machinery that has been leveraged to gain further understanding of plasmodesmata function in lateral root formation (Benitez-Alfonso et al., 2013) and immune signalling (Faulkner et al., 2013). Proteomic analyses also generate an expanding ‘parts list’ that allows us to ask whether recurrent protein classes are found at plasmodesmata in multiple plant tissues and species, and thus define a core protein complement of plasmodesmata (Kirk et al., 2022). However, sampling across differentiated tissues and taxonomic groups is hitherto poor, limiting the scope of such an approach. As plasmodesmata are understood to be a feature conserved across land plants (Brunkard and Zambryski, 2017), expanding our current knowledge relating to the plasmodesmata of flowering plants to extant species belonging to different taxonomic groups would give greater insight into core and conserved plasmodesmal components.
The bryophytes are a group of plants sister to the vascular plants (tracheophytes), with these clades diverging soon [~445 million years ago (Mya)] after the conquest of the land by the green kingdom (~490 Mya) (Morris et al., 2018). Electron microscopy has revealed that plasmodesmata across the tissues of bryophytes share architectural features, such as the outer plasma membrane lining and a central desmotubule (comprised of ER), with flowering plants (Ligrone and Duckett, 1994; Cook et al., 1997; Ligrone et al., 2000). These observations suggest that plasmodesmata are a trait present in the ancestor of all land plants and that elements of their structure observed across diverse extant species are essential to their function, being conserved or repeatedly recruited to plasmodesmata. Other than a limited analysis of the proteins present in plasmodesmata of the giant-celled green alga Chara corallina (Faulkner et al., 2005), molecular details about the composition of plasmodesmata outside the flowering plants are lacking, leaving questions of the molecular conservation of plasmodesmata unanswered.
A comparison of extant traits and molecular constituents between living ancestors would provide a powerful entry point towards establishing which plasmodesmal components are core, and which are derived. In recent years, extant species from Bryophyta such as Marchantia polymorpha and Physcomitrium (formerly Physcomitrella) patens have grown to be important models for plant research (Rensing et al., 2020; Naramoto et al., 2022). We took advantage of recent developments in methods for extracting plasmodesmata from differentiated green tissues to phylogenetically expand information of the molecular composition of plasmodesmata, generating new plasmodesmal proteomes from differentiated tissue of Arabidopsis thaliana and P. patens. Leveraging these and existing proteomes, we performed a comparative phylogenetic analysis, exploiting a Bayesian approach of repeated identification indicating conserved plasmodesmal association, thereby increasing the power in analysis of recalcitrant plasmodesmal samples. Thus, we identified core plasmodesmal proteins in consistently identified protein orthogroups validating members of β-1,3-glucanase, C2 lipid-binding protein, and tetraspanin families as conserved, core plasmodesmal proteins. Our approach, and new resources, has revealed essential features of plasmodesmata, with the potential to define basic rules and requirements of symplastic cell–cell communication in the multicellular green lineage.
Materials and methods
Plant material and growth conditions
For plasmodesmal extraction, A. thaliana Col-0 plants were grown on soil in short-day conditions (10 h light/14 h dark) at 22 °C. Leaves were harvested 5 weeks after germination. For stable transformation, A. thaliana plants were grown in long-day conditions (16 h light/8 h dark). Physcomitrium (Physcomitrella) patens tissues for generating plasmodesmal fractions were grown on BCD-AT medium in long-day conditions (16 h light/8 h dark) at 25 °C. Protonemal tissue was grown on top of nitrocellulose membrane for 1 week, whereas gametophore tissue was grown directly on the medium for 4 weeks. Routine P. patens culture for generating and maintaining transformants was performed under continuous light at 25 °C on BCD-AT medium. Nicotiana benthamiana plants were grown on soil with 16 h light/8 h dark at 23 °C.
Plasmodesmal purification
Plasmodesmata were extracted from expanded rosette leaves of 5-week-old Arabidopsis plants and a mix of P. patens protonemal and gametophore tissue. To fully homogenize differentiated tissue, we extracted plasmodesmata according to Cheval et al. (2020), with the key difference in approach from that in Faulkner and Bayer (2017) being the homogenization method and the use of Triton X-100 to disrupt chloroplasts. First, frozen mature tissue was ground into a powder in liquid nitrogen and suspended with extraction buffer [EB: 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1× cOmplete™ ULTRA protease inhibitors (Sigma), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1% (w/v) polyvinylpyrrolidone (PVP)-40 kDa (Sigma)], and ultrasonicated for 1 min in six 10 s pulses with a 5 s pause between each pulse (Soniprep 150 Plus, MSE). The sample was passed twice through a high-pressure homogenizer (EmulsiFlex™-B15, Avestin) at 80 psi. Triton X-100 (10% v/v) was added dropwise to the resultant homogenate to a final concentration of 0.5% (v/v) to disrupt residual chloroplasts, and cell walls were collected by centrifugation at 400 g. The cell wall pellet was washed three times (four for P. patens samples) with EB (15 ml) and centrifuged at 400 g. We validated the method for P. patens by calcofluor staining of cell walls at the different stages of fractionation (Supplementary Fig. S1), showing that the size of cell wall fragments generated by this approach is similar to those derived from A. thaliana suspension cells (30–100 µm) (Grison et al., 2015).
The cleaned cell wall pellet was incubated in an equal volume of cellulase buffer [CB: 20 mM MES-KOH pH 5.5, 100 mM NaCl, 2% w/v Cellulase R-10 (Yakult Pharmaceutical Co., Ltd, Japan), 1× cOmplete™ ULTRA protease inhibitors (Sigma), 1 mM PMSF] for 1 h at 37 °C, 200 rpm. Undigested cell wall was removed by centrifugation at 5000 g, and the supernatant was collected as the plasmodesmal membrane-containing fraction. The cell wall pellet was washed again with CB to extract residual plasmodesmal membranes and the soluble fraction was ultracentrifuged at 135 000 g for 1 h. The membrane pellet was resuspended in 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM DTT, 1×cOmplete™ ULTRA EDTA-free protease inhibitors (Sigma), 1 mM PMSF, 0.2% (v/v) IPEGAL®CA-630 (Sigma).
Mass spectrometry
Plasmodesmal samples were run 5 mm into a 1.5 mm thick 10% polyacrylamide Tris resolving gel (containing 0.1% SDS) without a stacking gel, in a glycine 0.1% SDS running buffer. The gel was washed in dH2O and then the band was excised. The bands were washed four times in 20% acetonitrile at 40 °C for 15 min to remove detergents, and then stored at 4 °C with 100 µl of dH2O.
MS analysis was performed by the Cambridge Centre of Proteomics. 1D gel bands were cut into 1 mm2 pieces, destained, reduced (DTT) and alkylated (iodoacetamide), and subjected to enzymatic digestion with trypsin overnight at 37 °C. Digested peptides were analysed by LC-MS/MS with a Dionex Ultimate 3000 RSLC nanoUPLC (Thermo Fisher Scientific Inc., Waltham, MA, USA) system and a Q Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific Inc.). Separation of peptides was performed by reverse-phase chromatography at a flow rate of 300 nl min–1 and a Thermo Scientific reverse-phase nano-Easy-spray column (Thermo Scientific PepMap C18, 2 µm particle size, 100 Å pore size, 75 µm i.d.×50 cm length). Peptides were loaded onto a pre-column (Thermo Scientific PepMap 100 C18, 5 µm particle size, 100 Å pore size, 300 µm i.d.×5 mm length) from the Ultimate 3000 autosampler with 0.1% formic acid for 3 min at a flow rate of 15 µl min–1. After this period, the column valve was switched to allow elution of peptides from the pre-column onto the analytical column. Solvent A was water+0.1% formic acid and solvent B was 80% acetonitrile, 20% water+0.1% formic acid. The linear gradient employed was 2–40% B in 90 min (the total run time including column washing and re-equilibration was 120 min).
The LC eluant was sprayed into the mass spectrometer by means of an Easy-spray source (Thermo Fisher Scientific Inc.). All m/z values of eluting ions were measured in an Orbitrap mass analyser, set at a resolution of 70 000, and scanned between m/z 380 and 1500. Data-dependent scans (Top 20) were employed to automatically isolate and generate fragment ions by higher energy collisional dissociation [HCD; normalized collision energy (NCE): 25%] in the HCD collision cell, and measurement of the resulting fragment ions was performed in the Orbitrap analyser, set at a resolution of 17 500. Singly charged ions and ions with unassigned charge states were excluded from being selected for MS/MS, and a dynamic exclusion of 20 s was employed.
Post-run, all MS/MS data were converted to mgf files and the files were then submitted to the Mascot search algorithm (Matrix Science, London UK, version 2.6.0) and searched against the Cambridge Centre of Proteomics database, including common contaminant sequences containing non-specific proteins such as keratins and trypsin. Variable modifications of oxidation (M) and deamidation (NQ) were applied, as well as a fixed modification of carbamidomethyl (C). The peptide and fragment mass tolerances were set to 20 ppm and 0.1 Da, respectively. A significance threshold value of P<0.05 and a peptide cut-off score of 20 were also applied. All data (DAT files) were then imported into the Scaffold program (Version 4.10.0, Proteome Software Inc., Portland, OR, USA). Proteins were classed as positively identified when the peptide and protein identification probability thresholds were >95% (Leijon et al., 2018) and proteins were identified in at least two replicates.
Gene Ontology analysis
Gene Ontology (GO) was used to test gene lists for cellular localization enrichment (Ashburner et al., 2000). A cellular localization GO term over-representation test was performed, using the Panther database (release 1 July 2022) (Thomas et al., 2003; Mi et al., 2019) and GO database (released 13 October 2022) with a Fisher’s exact test and false discovery rate (FDR) reported. Physcomitrium patens genes were annotated bioinformatically using phylogenetic backpropagation of GO terms via the Panther database (Gaudet et al., 2011). Graphs were drawn using ggplot2 in R (v4.0.0) (Wickham, 2016).
Bioinformatic analysis
HMMER v3.3 (hmmer.org) was used for sequence similarity searches (Eddy, 1998). The P. patens plasmodesmal proteome was downloaded as peptide sequences from UniProt and used as the reference database for a ‘phmmer’ search against which the A. thaliana UniProt proteome was run (UP000006548, accessed 24 April 2020) (Cheng et al., 2017). Protein matches were filtered at either E <1 × 10–100 or E <1 × 10–50 as stated in the text.
Orthofinder (v2.2.6) was used to create de novo orthogroups (Emms and Kelly, 2015, 2019). Plasmodesmal proteome protein sequences were downloaded using UniProt, TAIR (Araport11), and Phytozome v12.1 (Populus trichocarpa v3.1). Orthofinder was run on these sequences with default settings.
Phylogenetic analysis
A peptide sequence was downloaded from UniProt for each protein within an orthogroup. The protein FASTA sequences were aligned with Clustal Omega (v1.2.4; Sievers et al., 2011) to build a consensus sequence. The consensus sequence, in Stockholm format, was used as the basis for a hmmsearch (EBI, HmmerWeb version 2.41.1; Potter et al., 2018). A search was conducted against the EMBL Reference Proteomes database restricted to A. thaliana (taxon id: 3702), P. patens (taxon id: 3218), and P. trichocarpa (taxon id: 3694) species sequences with the sequence E-value cut-off 1 × 10–100, unless otherwise stated. Protein sequences were manually deduplicated for each gene.
The FASTA sequences for all identified homologues, from the hmmsearch, in all three species were downloaded and a bootstrapped non-rooted phylogenetic tree was generated using the ‘standard_trimmed_phyml_bootstrap’ ete workflow (v3.1.1; Huerta-Cepas et al., 2016). In this workflow, sequences are aligned with Clustal Omega, trimmed with TrimAI (Capella-Gutiérrez et al., 2009), and a phylogeny determined with 100 bootstraps using PhyML (Guindon et al., 2010). Trees were drawn using ggtree in R (v4.0.0) (Yu et al., 2017).
Molecular phylogeny for P. patens plasmodesmata-associated protein families (Supplementary Fig. S2) was determined using the maximum likelihood method (JTT matrix-based model) after sequences were aligned using MUSCLE run on MEGA (v7). A discrete Gamma distribution with five categories was used to model evolutionary rate differences among sites. All positions with <80% site coverage were eliminated.
Construct generation for protein tagging in moss
For mNeonGreen tagging of moss candidate plasmodesmal proteins, first a mNeonGreen tagging vector was generated. For this the mNeonGreen coding sequence was amplified using primers mNG-HindIII-F and mNG-stop-EcoRI-R (Supplementary Table S1) from pPY22 [Addgene plasmid #137082 (Yi and Goshima, 2020), introducing a GSGGSG-encoding linker before mNeonGreen in the process]. Next, using HindIII and EcoRI restriction sites, the Citrine fluorophore in pCTRN-nptII (Hiwatashi et al., 2008) was exchanged with the amplified mNeonGreen-encoding sequence, resulting in plasmid pmNG-nptII.
Moss mNeonGreen in-locus tagging constructs were assembled using InFusion recombination of PCR-amplified fragments. Four fragments were assembled: a vector backbone sequence amplified from pmNG-nptII using primers pBS-vec-PmeI-F and pBS-vec-PmeI-R; two gDNA-amplified homology arms of ~1 kb in length located upstream and downstream of the intended mNeonGreen integration site; and a mNeonGreen-encoding fragment (Supplementary Fig. S3) which, in the case of C-terminal fusions, was followed by a G418 resistance cassette (both amplified from pmNG-nptII using primers mNG-noStart-F+mNG-noStop-R or Link-mNG-F+Cassette-R, respectively). The resultant plasmids were verified by sequencing and linearized by PmeI digestion prior to transformation into P. patens.
Construct generation for N. benthamiana and A. thaliana expression
For localization analysis of putative plasmodesmal candidates by expression in N. benthamiana and A. thaliana tissues, binary vectors containing the coding sequence of the protein of interest fused to a fluorescent protein were generated. Typically, the coding sequence of a gene of interest was synthesized (Genewiz, China) as a Golden Gate L0 module in a pUC57 backbone, except for moss tetraspanin PpTET6 (A9TQE7; Pp3c4_3550V3), moss β-1,3-glucanase PpGHL17_18 (A0A2K1J8R8; Pp3c16_15860V3), and Arabidopsis β-1,3-glucanase AtBG_PPAP (Q9FHX5; AT5G42100) (see below). For synthesis, internal BsaI and BpiI restriction sites were removed via silent mutation, and appropriate 4 bp overhangs were added to enable Golden Gate cloning. Via a BsaI-mediated level 1 Golden Gate reaction, coding sequences were linked to an enhanced green fluorescent protein (eGFP)-, mCherry-, or mRuby3-encoding fragment with a 35S or ACT2 promoter and terminator regions placed upstream and downstream, respectively. Coding sequences for PpTET6 and PpGHL17_18 were amplified from moss cDNA and assembled into a Golden Gate level 0 acceptor plasmid, removing internal BsaI and BpiI sites in the process. For the β-1,3-glucanase, during fragment assembly an mNeonGreen-coding fragment was fused in-frame after the sequence coding for the catalytic domain. The tagging constructs for AtBG_PPAP and the N-terminal fusions of Q4A3V3 and AtGELP91 were generated by inserting mCitrine downstream of their predicted signal peptides and assembling the fusion in a Level 1 binary vector with a 35S promoter and either a 35S or heat shock protein terminator.
Plant transformation
Arabidopsis thaliana was transformed by floral dip (Clough and Bent, 1998). Nicotiana benthamiana was transformed by co-infiltration of Agrobacterium tumefaciens [GV3101 (pMP90)] strains harbouring either a binary plasmid coding for in planta expression of the transgene of interest or the p19 silencing suppressor. Leaves were imaged 2 d post-infiltration.
Physcomitrium patens was transformed using polyethylene glycol (PEG)-mediated protoplast transformation (Nishiyama et al., 2000). For constructs without a resistance cassette (i.e. those used for N-terminal or internal tagging of the protein of interest), the plasmid p35S-LoxP-HygR (pTN186; Genbank AB542059.1) was co-transformed in a 1:1 ratio such that a first selection step on hygromycin B-containing medium could be performed. Transformants with the correct single integration of the mNeonGreen expression constructs were identified using PCR.
Confocal imaging
Leaf tissue from N. benthamiana and A. thaliana was cut into 1 cm2 samples and mounted adaxially. Samples were imaged on a ZEISS LSM800 confocal microscope with a ×63/1.2 water immersion objective lens (C-APOCHROMAT ×63/1.2 water). GFP and mCitrine were excited at 488 nm with an argon laser and collected at 500–545 nm. mRuby was excited at 561 nm with a DPSS laser and collected at 590– 620 nm. Aniline blue [0.1% (w/v) in 1× phosphate-buffered saline (PBS) pH 7.4] was infiltrated adaxially and excited at 405 nm with a UV laser and collected at 430–470 nm. Wall fractions were stained with 0.01% Calcofluor white M2R (F3543, Sigma) and imaged by confocal microscopy with a ×20 objective (PLAN APOCHROMAT NA 0.8). Calcofluor white was excited at 405 nm with a UV laser and collected at 430–470 nm.
Moss protonemal cells were observed using a 39 mm diameter glass bottom dish, prepared with solidified BCD medium and grown for 4–6 d in a thin layer of the same medium except solidified with 0.7% (w/v) low melting agarose. For all moss fluorescence microscopy experiments, the second and third caulonemal cells relative to the tip of a protonemal filament were used. Imaging of endogenous moss proteins tagged with mNeonGreen was performed on a spinning disc confocal microscope consisting of a Nikon Ti-eclipse body equipped with a Yokogawa CSU-X1 spinning disc head and ×100 Plan Apo VC objective (NA 1.40). Image digitization was performed with a Photometrics Prime 95B cMOS camera with a ×1.2 post-magnification fitted in front of the camera. Typical exposures used were 500–3000 ms. For excitation of mNeonGreen, a 491 nm laser line was used and emitted light was filtered using a 527/60 bandpass emission filter. All microscope components were operated by MetaMorph software. Co-localization of aniline blue-stained callose deposits with mNeonGreen-tagged proteins of interest was performed on a Leica Stellaris 5 confocal microscope. Aniline blue prepared as a 1.6% (w/v) stock solution in 0.1 M phosphate buffer (pH 8.5) according to Muller et al. (2022) was diluted in water to a final concentration of 0.02% in water and then added to the imaging dishes for 48 h prior to observation [except for co-localization of β-1,3-glucanase PpGHL17_18 (A0A2K1J8R8) where a 2 h incubation was used]. Cells were imaged using a ×100 HC plan apo objective (NA 1.40). Excitation of aniline blue was done using a 405 nm solid state laser and emitted light was collected between 420 nm and 490 nm on a HyD detector with the pinhole set to 0.6 Airy units. Excitation of mNeonGreen was done using 505 nm light obtained from a pulsed white light laser, and emitted light was collected between 515 nm and 560 nm on a HyD detector, with the pinhole aperture set to 1 Airy unit. Frames of the two different probes were collected successively and a line-averaging factor of 8 was used.
Results
Generation of a plasmodesmal proteome from mature Arabidopsis leaves
There are currently two published plasmodesmata proteomes of A. thaliana (Fernandez-Calvino et al., 2011; Brault et al., 2019) that use suspension culture cells as biological material, as well as another from P. trichocarpa cell suspension cultures (Leijon et al., 2018). To define a novel A. thaliana plasmodesmata proteome that represented differentiated tissue, we extracted plasmodesmata from expanded leaves (assumed to be mature leaves, Kalve et al., 2014) of 5-week-old plants and characterized the proteome by MS. Proteins were considered positively identified in the same manner as in Leijon et al. (2018): if the protein (95% certainty; Searle, 2010) was represented in at least two of the three samples by at least one peptide (95% certainty; Keller et al., 2002). With these thresholds, 238 proteins were identified in the fraction (Supplementary Table S2).
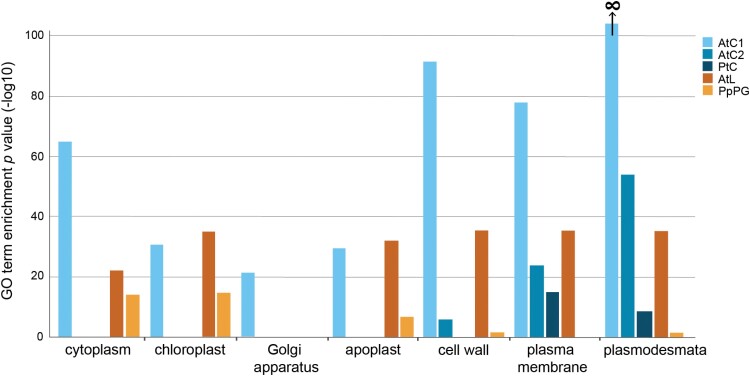
To assess if the mature leaf plasmodesmal fraction has sufficient purity to define a plasmodesmal proteome, we assessed whether it showed enrichment for the cellular localization ‘plasmodesma’ GO term. For ease of referencing, hereafter the proteomes are named ‘AtC1’ (Fernandez-Calvino et al., 2011), ‘AtC2’ (Brault et al., 2019), ‘PtC’ (Leijon et al., 2018), and ‘AtL’ (the proteome from mature leaves produced in this study). AtL was benchmarked against AtC2 and PtC for ‘plasmodesmal’ enrichment, noting that >50% of proteins in AtC1 are associated with the ‘plasmodesmata’ GO term, leading to the enrichment P-value of close to, and rounded to, zero. It was also benchmarked against AtC1, AtC2, and PtC for the proportion of putative contamination from other subcellular compartments. AtC2, PtC, and AtL were significantly enriched for ‘plasmodesmata’-labelled proteins (Fig. 1). Moreover, all flowering plant proteomes were significantly enriched for ‘cell wall’ and ‘plasma membrane’ proteins, which are both structural components of plasmodesmata. (Brault et al., 2019). The enrichment factor filtering used to define the AtC2 proteome worked extremely well, with other likely contaminant categories (e.g. ‘Golgi apparatus’ or ‘chloroplast’) not over-represented, unlike the unfiltered proteomes. However, the similarity between the representation of proteins in non-plasmodesmal cell components in AtC1 and AtL suggests that the latter is of comparable quality and defines a list of candidate plasmodesmal proteins from Arabidopsis leaves.
Fig. 1.
The Arabidopsis plasmodesmal fraction derived from expanded rosette leaves and the moss plasmodesmal fraction derived from protonema and gametophore tissue are enriched in plasmodesmal proteins. P-values for cell compartment GO term enrichment of plasmodesmal proteomes from cell suspension cultures (AtC1, AtC2, and PtC), expanded rosette leaves (AtL), and moss protonema and gametophore tissue (PpPG).
Generation of a plasmodesmal proteome from P. patens
In addition to proteomes from cell suspension cultures, a plasmodesmal proteome from N. benthamiana leaves (Park et al., 2017) exists, but none is available beyond dicotyledonous flowering plants. To expand the phylogenetic representation of plasmodesmal proteomes, we defined a novel plasmodesmal proteome from the moss P. patens (termed ‘PpPG’). We purified cell walls from a mix of protonema and gametophore tissue to produce wall fragments comparable in size with those generated during plasmodesmal fractionation from Arabidopsis cell culture (30–100 µm; Grison et al., 2015; Supplementary Fig. S1). We digested the cellulose in these fragments to release plasmodesmal membranes and analysed the proteins extracted from this fraction by MS. Proteins were identified in the same manner as for the Arabidopsis leaf proteome, generating a list of 215 candidate plasmodesmal proteins (Supplementary Table S3). We confirmed that this extraction protocol works in P. patens by checking for enrichment of proteins annotated with the plasmodesmal GO term. A total of 185 (86%) of the UniProt identifiers were mapped to the GO database, with plasmodesma-annotated proteins over-represented (seven proteins, P=3.19 × 10–5, 0.51 proteins expected) in the P. patens plasmodesmal fraction (Fig. 1). This value is reduced compared with the flowering plant plasmodesmal proteomes due to poor annotation of P. patens proteins within the ‘plasmodesmata’ GO term via phylogenetic backpropagation of Arabidopsis GO terms (Gaudet et al., 2011). Nonetheless, given the poor backpropagation of GO terms, we concluded that identification of several proteins with a plasmodesmata annotation suggests that the extraction protocol produced a protein fraction that probably contains a representative population of plasmodesmal proteins from P. patens.
Phylogenetic comparison of Arabidopsis, poplar, and moss plasmodesmal proteomes reveals orthogroups containing core proteins
To further characterize and compare the composition of the P. patens plasmodesmal proteome, we explored different bioinformatic approaches to find orthologous proteins. First, we used a one-to-one homologue database search approach. Using InParanoid 8.0 (pairwise BLAST, defining orthogroups from an ancestral protein sequence) and MetaPhOrs (defining orthogroups from a meta-analysis of many homologue databases), we converted the P. patens protein identifiers to their A. thaliana homologue identifier (Supplementary Table S4). Only 62 (InParanoid) and 52 (MetaPhOrs) P. patens proteins were matched to Arabidopsis proteins by this approach, but performing a GO term analysis on these two lists of Arabidopsis identifiers revealed enrichment of the plasmodesmata GO term (P=7.11 × 10–16 and 2.82 × 10–8 for InParanoid and MetaPhOrs, respectively). However, the low percentage of P. patens protein homologues identified (29% and 24%) by this method is too low to allow for the P. patens proteome to offer significant power in a comparative analysis.
Our next analysis involved comparing one with many, instead of relying on databases to convert P. patens proteins to A. thaliana homologues. To this end, we used HMMER (v3.3, profile hidden Markov models) to find the closest homologue for P. patens plasmodesmal proteins in A. thaliana. Using two arbitrary thresholds of E <1 × 10–50 and E <1 × 10–100, HMMER matched 147 (68%) and 80 (37%) P. patens proteins to A. thaliana proteins, respectively. Even at these conservative values, a HMMER search matched more proteins than database lookup tools. However, one to many mapping makes it difficult to translate the P. patens proteome members to specific A. thaliana proteins. One approach would be to take the most significant (i.e. most likely) homologue for each protein. However, taking P. patens A0A2K1JXU2 (‘X8 domain-containing protein’; Associated locus Pp3c10_5480V3) as an example, there are two almost indistinguishable top hits in A. thaliana: O49737 (E=4.2 × 10–101) and Q8L837 (E=6.3 × 10–101), suggesting that it is likely that the ancestral protein of A0A2K1JXU2 has undergone a duplication event in A. thaliana giving two equally likely homologues. In essence, this builds orthogroups restricted to one P. patens member.
Another consideration when using HMMER to assign homologues is that to find phylogenetically conserved proteins (i.e. to concurrently compare several lists among several species), one list would have to be chosen as the reference frame. Defining the P. patens proteome as the reference list allows the distribution of P. patens hits across the Arabidopsis proteomes to be compared, but any nuance from comparison between the A. thaliana proteomes is lost. Therefore, we tried a third, many to many approach by forming de novo orthogroups using the OrthoFinder software (Emms and Kelly, 2019).
OrthoFinder uses a pairwise BLAST approach to build orthogroups from an input set of protein sequences. We used OrthoFinder (v2.2.6) to define orthogroups (OGs) between five plasmodesmal proteomes: AtC1, AtC2, AtL, PpPG, and PtC. This analysis returned 992 orthogroups, of which 289 had more than one member and 288 contained proteins from multiple proteomes (Supplementary Fig. S4. Two orthogroups had members from all proteomes, and 17 had members from four of the five proteomes (Table 1; Supplementary Table S5). We noted that members of the IMK2 orthogroup (OG18) and OG9 both contain receptor-like kinases belonging to the LRR III group, and that the sole member of the calcium-dependent lipid-binding orthogroup (OG50) identified in the Arabidopsis proteomes shows similarity to members of the C2 lipid-binding orthogroup (OG3, phmmer search E=9.6 × 10–6). Therefore, OG18 and OG3 were not considered independently. Further, while OG19, representing DUF26-containing proteins that include the PDLPs, is represented in the proteomes from P. trichocarpa and A. thaliana, it does not have any P. patens homologues (Vaattovaara et al., 2019) and so we excluded it as a candidate core orthogroup. We defined the remaining 16 orthogroups as containing proteins that are ‘phylogenetically conserved plasmodesmal proteins’ (Table 1).
Table 1.
List of orthogroups identified in at least four of five proteomes
| Orthogroup | Protein class | No. of proteomes | No. of proteins | Localized at plasmodesmata |
|---|---|---|---|---|
| OG0 | β-1,3-Glucanase | 5 | 27 | Benitez-Alfonso et al. (2013); Levy et al. (2007) |
| OG1 | Peroxidase | 4 | 22 | No |
| OG3 | C2 lipid-binding | 4 | 16 | Brault et al. (2019) |
| OG4 | SKU5 | 4 | 13 | No |
| OG5 | GDSL esterase/lipase | 4 | 13 | No |
| OG6 | Tetraspanin | 4 | 12 | Fernandez-Calvino et al. (2011) |
| OG7 | ATP-binding cassette | 4 | 11 | No |
| OG8 | Aspartyl protease | 4 | 10 | No |
| OG9 | Leucine-rich repeat receptor-like kinase | 4 | 10 | Grison et al. (2019); Fernandez-Calvino et al. (2011) |
| OG10 | Leucine-rich repeat extensin-like | 4 | 10 | No |
| OG13 | Histone H2B | 4 | 9 | No |
| OG14 | Tubulin beta-7 | 4 | 9 | Blackman and Overall (1998) |
| OG16 | RNA-binding glycine-rich protein | 4 | 8 | No |
| OG18 | Inflorescence meristem receptor-like kinase 2 | 5 | 7 | No |
| OG19 | DUF26-containing protein | 4 | 7 | Thomas et al. (2008) |
| OG28 | Eukaryotic translation initiation factor 4A | 4 | 6 | No |
| OG40 | Subtilisin-like protease | 4 | 5 | No |
| OG50 | Calcium-dependent lipid-binding | 4 | 4 | No |
| OG63 | Ribosomal protein | 4 | 4 | No |
Moss core orthogroup members are plasmodesmal proteins
Rationalizing that plasmodesmata are defined by specialized membranes, we first considered orthogroups for which the representatives detected in the Arabidopsis proteomes have at least in silico support for membrane association [i.e. either predicted transmembrane helices or an omega site for glycosylphosphatidylinositol (GPI) anchor attachment]. This led us to refine our initial OGs of interest to: OG0 (β-1,3-glucanase), OG3 (C2 lipid-binding), OG6 (tetraspanin), OG7 (ATP-binding cassette), and OG9 (LRR RLK III). Proteins from OG0 (Benitez-Alfonso et al., 2013; Levy et al., 2007; Rinne et al., 2011), OG3 (Brault et al., 2019), OG6 (Fernandez-Calvino et al., 2011), and OG9 (Fernandez-Calvino et al., 2011; Grison et al., 2019) have already been validated as plasmodesmata associated in Arabidopsis by live imaging of fluorescent protein fusions. We selected OG0, OG3, OG6, and OG7, and identified P. patens homologues, all but one present in the P. patens plasmodesmal fraction, and further characterized their in vivo localization in the native tissues.
For OG0, three P. patens β-1,3-glucanases were present in the plasmodesmal fraction (Supplementary Tables S3, S5). We noted that the protein A0A2K1K5L9 (associated locus Pp3c8_940V3) had an incomplete catalytic domain, and therefore disregarded it for further analysis. We selected A0A2K1J8R8 (Pp3c16_15860V3.1, PpGHL17_18, Supplementary Table S6; Supplementary Figs S2, S3), a β-1,3-glucanase with a predicted GPI anchor similar to most known plasmodesmata-associated β-1,3-glucanases (Levy et al., 2007; Benitez-Alfonso et al., 2013; Gaudioso-Pedraza et al., 2018), as a moss representative of OG0. We generated a transgenic P. patens line that expresses a fluorescent protein fusion by inserting a mNeonGreen (mNG)-encoding sequence at the native genomic locus downstream of the predicted catalytic domain and before the predicted omega site for GPI anchor attachment (Supplementary Fig. S3). Live imaging of P. patens protonema shows that PpGHL17_18-mNG has a punctate localization at the cell junctions (Fig. 2A). Co-localization with aniline blue suggests that this fluorescence pattern is co-incident with staining of plasmodesmal callose (Fig. 2E) and therefore that PpGHL17_18 is a plasmodesmata-associated β-1,3-glucanase.
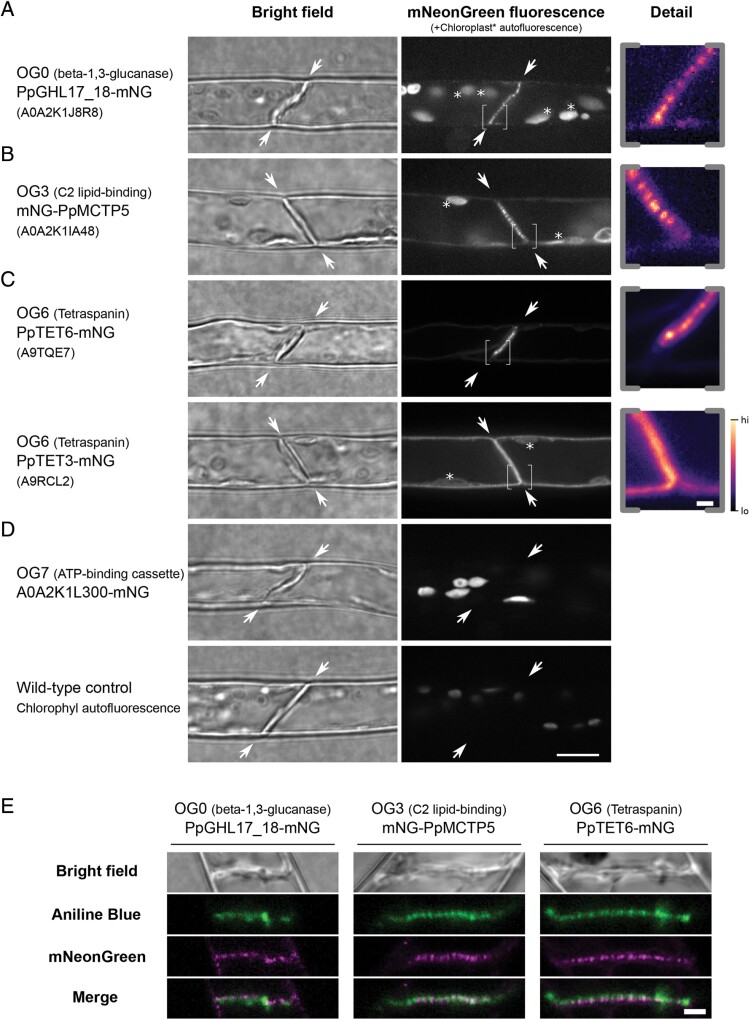
Fig. 2.
Localization of selected P. patens orthogroup members in moss protonemal cells reveals plasmodesmal association. (A–D) Micrographs of moss protonemal cells expressing the indicated protein fused to fluorescent protein mNeonGreen, taken under bright field (left) and fluorescence imaging conditions (right). Proteins belonging to the β-1,3-glucanase (A), C2 lipid-binding (B), tetraspanin (C), and ATP-binding cassette (D) orthogroups were localized. The dividing interface between two neighbouring cells where plasmodesmata are exclusively located in this tissue are highlighted by arrows. When the fusion protein was detected at this location, an expanded view of part of the dividing wall (indicated by brackets) is shown on the right in pseudocolour. Examples of autofluorescent chloroplasts are marked with an asterisk. The A0A2K1L300–mNG fusion protein localized to chloroplasts as levels of chloroplast autofluorescence under the same imaging and display conditions in wild-type tissue were vastly lower (bottom row). Scale bars are 10 µm in overview images, and 1 µm in expanded views. (E) Co-localization of the three mNeonGreen fusion proteins localizing to the cell interface (magenta) with the callose stain aniline blue (green). A single confocal plane is depicted showing co-occurrence of the callose and plasmodesmal protein fusion proteins (merge, bottom row). Scale bar is 1 µm.
For OG3, representing the C2 lipid-binding protein family that contains the plasmodesmata-associated MCTPs (Brault et al., 2019), no member was identified in the plasmodesmal fraction from P. patens (Supplementary Table S5). Therefore, we selected A0A2K1IA48 (Pp3c27_520V3.1, PpMCTP5, Supplementary Table S6; Supplementary Figs S2, S3) as a candidate P. patens plasmodesmal protein as it has the closest homology to Arabidopsis MCTP4 using a phmmer search and is most abundantly expressed in moss tissues (Ortiz-Ramírez et al., 2016; Fernandez-Pozo et al., 2020). We generated a fluorescent protein fusion by homologous recombination, inserting mNeonGreen at the N-terminus of PpMCTP5 and observed a punctate localization restricted to the cell junction (Fig. 2B). Again, aniline blue co-localization confirmed co-incidence of the signal with plasmodesmal callose, validating PpMCTP5 as a plasmodesmal C2 lipid-binding protein from P. patens (Fig. 2E). We also noted that PpMCTP5–mNG showed weak ER-associated fluorescence that was enriched at discrete foci at the periphery of the external surface of cells (Supplementary Fig. S5), possibly being points of connection between the ER and the plasma membrane as would be expected for proteins in membrane contact sites.
The tetraspanin group OG6 contained two members identified in the P. patens plasmodesmal fraction: A9RCL2 (Pp3c7_23740V3.1, PpTET3, Supplementary Table S6; Supplementary Figs S2, S3) and A9TQE7 (Pp3c4_3550V3.1, PpTET6, Supplementary Table S6, Supplementary Figs S2, S3). mNeonGreen fusions at the C-terminus of these two tetraspanins revealed two different patterns of localization. PpTET6 displayed a punctate pattern of localization at the cell periphery that co-localized with aniline blue staining of plasmodesmal callose (Fig. 2C, E). In contrast, PpTET3 showed even distribution in the periphery of the cell, suggesting that it is not enriched in plasmodesmata but present in the entire plasma membrane (Fig. 2C). Therefore, we validated only PpTET6 as a candidate plasmodesmata-associated protein.
OG7 represents ATP-binding cassette proteins that, in contrast to members from OG0, OG3, and OG6, have not been validated as plasmodesmata-associated proteins in any species. To test whether this group might represent novel core plasmodesmal proteins, we identified A0A2K1L300 in our purified plasmodesmal fraction (Supplementary Table S3) and inserted mNeonGreen by homologous recombination to generate an A0A2K1L300–mNG fusion. Live imaging shows that this protein fusion localizes to chloroplasts, suggesting that it is not enriched in plasmodesmata (Fig. 2D).
Plasmodesmal association of core orthogroup members is conserved in heterologous species
The validation of plasmodesmal association of P. patens proteins from orthogroups represented in plasmodesmal proteomes suggests that orthogroup analysis can identify core, conserved plasmodesmal proteins. We reasoned that such core plasmodesmal proteins would be recruited to plasmodesmata in any plant species and, to test this hypothesis, we expressed orthogroup representatives from Arabidopsis and P. patens in N. benthamiana leaf epidermal cells and used live-cell imaging to determine their association with plasmodesmata. For OG0, we inserted mCitrine downstream of the predicted signal peptide of Q9FHX5 (At5g42100, AtBG_PPAP) and mNeonGreen downstream of the catalytic domain of PpGHL17_18, and expressed the gene fusions transiently in N. benthamiana leaves. Both proteins showed punctate distribution across the cell periphery, with foci of fluorescence co-incident with aniline blue-stained plasmodesmal callose (Fig. 3A, B).
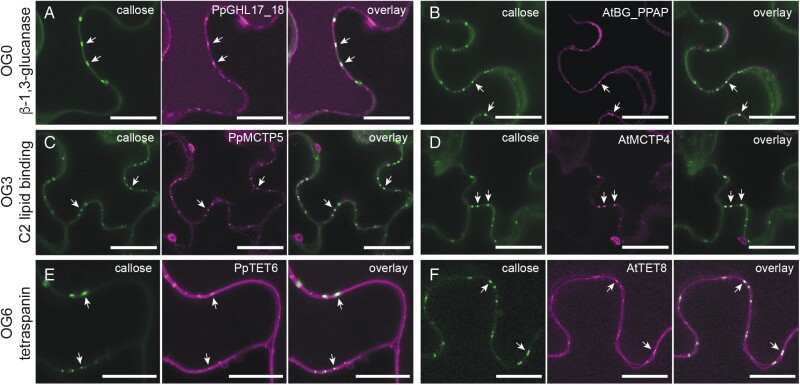
Fig. 3.
Proteins from OG0, OG3, and OG6 maintain plasmodesmal association in a heterologous species. Confocal micrographs of moss (A, C, E) and Arabidopsis (B, D, F) members of these orthogroups produced in N. benthamiana leaf epidermal cells. In each panel, aniline blue-stained plasmodesmal callose is green (left), the candidate–fluorescent protein fusion is magenta (centre), and the overlay is on the right. Members of OG0, representing β-1,3-glucanases, localize to the cell periphery and accumulate at plasmodesmata as indicated by aniline blue co-staining of plasmodesmal callose (arrows). (A) is PpGHL17_18–mNG and accumulates in the vacuole as well as at the cell periphery. (B) is AtBG_PPAP–mCitrine and is detected as diffuse labelling of the cell wall as well as at plasmodesmata. Members of OG3, representing C2 lipid-binding proteins, accumulate at plasmodesmata as indicated by aniline blue co-staining of plasmodesmal callose (arrows). (C) is PpMCTP5–GFP and (D) is AtMCTP4–GFP. Members of OG6, representing tetraspanins, localize to the plasma membrane at the cell periphery and accumulate in plasmodesmata as indicated by aniline blue co-staining of plasmodesmal callose (arrows). (E) is PpTET6–mNG and (F) is AtTET8–mRuby. Scale bars are 20 μm (A, E, F) or 25 μm (B, C, D).
Similarly, we generated C-terminal fusions of OG3 members Q9C8H3 (At1g51570, AtMCTP4) and PpMCTP5, and OG6 members Q8S8Q6 (AT2G23810, AtTET8) and PpTET6, with GFP or mRuby and observed punctate localization when expressed in N. benthamiana (Fig. 3C–F). These punctae co-localized with aniline blue-stained callose, confirming that these proteins can be recruited to plasmodesmata in a heterologous system. We further confirmed conservation of plasmodesmal association for C2 lipid-binding proteins by stable expression of a GFP fusion of PpMCTP5 in Arabidopsis. Again, this protein fusion localized in punctae at the cell periphery (Supplementary Fig. S6). Thus, β-1,3-glucanases (OG0), C2 lipid-binding proteins (OG3), and tetraspanins (OG6) show characteristics of core plasmodesmal proteins.
Screening of non-membrane proteins for plasmodesmal association in a heterologous system
Having observed that conserved plasmodesmal proteins maintain their localization in heterologous systems, we used this approach to test the plasmodesmal association of candidates from orthogroups for which members were not predicted to all have a membrane association. We chose Arabidopsis and P. patens representatives of OG5 (‘GDSL esterase/lipase’) and OG16 (glycine-rich RNA-binding proteins, GRPs), and screened for plasmodesmal association in N. benthamiana. For OG5, we noted that four members were identified in the P. patens plasmodesmal fraction. We selected P. patens Q4A3V3 (Pp3c18_1550V3.1, the member identified in plasmodesmal fractions with the highest number of peptide hits) and its closest homologue in Arabidopsis Q9LY84 (At5g14450, AtGELP91) for localization analysis. C-terminal protein fusions to GFP showed localization in a cellular reticulum suggestive of the ER (Fig. 4A, B; Supplementary Fig. S7). Co-localization of C-terminal protein fusions with aniline blue-stained plasmodesmal callose showed some reticulum aggregations overlaid with, or adjacent to, plasmodesmata. However, as the ER is continuous with the plasmodesmal desmotubule, and there were many sites where aniline blue signals did not overlay with AtGELP91 or Q4A3V3 fluorescence, we concluded that neither was specifically enriched at plasmodesmata relative to the rest of the ER. To test whether the location of the epitope tag interfered with protein localization, we generated N-terminal protein fusions by inserting mCitrine downstream of the predicted signal peptide of AtGELP91 and Q4A3V3. These localized to intracellular mobile bodies (possibly Golgi bodies) and showed faint diffuse localization at the cell periphery, suggesting that the proteins are secreted to the cell wall (Supplementary Fig. S7). While this infers that a C-terminal epitope tag might interfere with the proteins’ exit from the ER, we did not see these OG5 proteins accumulate at plasmodesmata when tagged at either terminus.
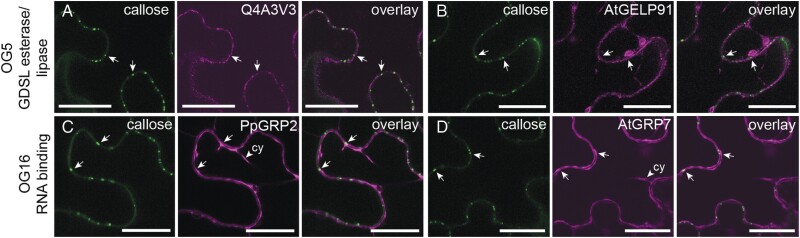
Fig. 4.
Proteins from OG5 and OG16 do not accumulate in plasmodesmata. Confocal micrographs of N. benthamiana leaf epidermal cells producing fusions of moss (A, C) and Arabidopsis (B, D) members of OG5 and OG16. Each panel shows aniline blue-stained callose on the left, the protein–fluorescent protein fusion in the centre, and the overlay of the two images on the right. The positions of two plasmodesmata-associated callose deposits are indicated by arrows in each panel. Members of OG5, containing GDSL esterase/lipases, show an uneven intracellular localization suggestive of a membrane reticulum such as the ER. (A) shows Q4A3V3–GFP and (B) shows AtGELP–GFP (Q9LY84). Members of OG16, containing RNA-binding proteins, show nucleo-cytosolic localization characteristic of soluble proteins. Arrowheads indicate cytoplasmic strands (Cy). (C) shows PpGRP2–mCherry (Q8LPB1) and (D) shows AtGRP7–GFP (Q03250). Scale bars are 25 μm.
For OG16 (GRPs), representatives were identified in both Arabidopsis and P. patens plasmodesmal proteomes. We selected Q03250 (At2g21660, AtGRP7) from Arabidopsis as it was represented in two of three Arabidopsis plasmodesmal proteomes, and Q8LPB1 (Pp3c11_19620V3.1, PpGRP2; Nomata et al., 2004) from P. patens as it had the highest number of unique peptides identified from our P. patens fraction. C-terminal fusions of both Arabidopsis and P. patens GRPs to GFP showed a nucleo-cytosolic localization in N. benthamiana leaves (Fig. 4C, D; Supplementary Fig. S7). Aniline blue staining of tissue producing AtGRP7–GFP and PpGRP2–mCherry suggests that neither GRP co-localizes with plasmodesmal callose. We further tested the localization of AtGRP7 and PpGRP2 by generating N-terminal protein fusions, but this produced an identical pattern of localization to the C-terminal fusions (Supplementary Fig. S7). Further, stable transformation of Arabidopsis with transgenes that encode C-terminal fusions of AtGRP7 and PpGRP2 produced similar localization patterns (Supplementary Fig. S8). Therefore, neither non-membrane-associated orthogroup showed specific plasmodesmal enrichment and association. Whether this arises because the plasmodesmal fraction of the ER and cytosol cannot be resolved from the cellular pool by light microscopy or because these proteins do not associate with plasmodesmata is unclear.
Phylogenetic analysis within orthogroups identifies different patterns of evolution of plasmodesmata association
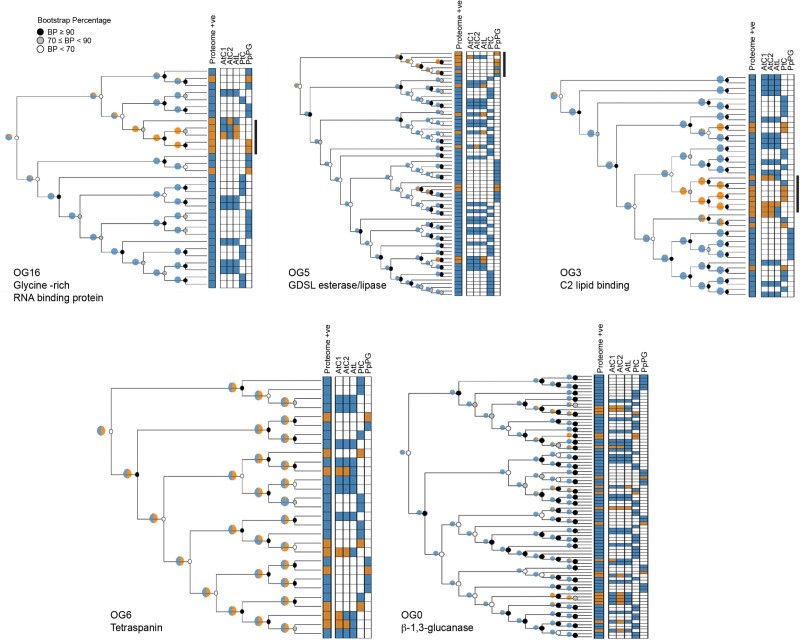
While live imaging can confirm plasmodesmal association of proteins that accumulate at plasmodesmata such that the fluorescence signal associated with plasmodesmata is greater than or separated from the surrounding pool, the approach is limited when plasmodesmal association is transient and accumulation is not a feature of protein behaviour. We could not confirm plasmodesmal association of OG5 members despite equally strong proteomic support for plasmodesmal association to those of OG6. Therefore, we explored whether protein family phylogenies could identify patterns that indicate a likelihood of conserved plasmodesmal association. We generated unrooted cladograms of the protein families that are represented by orthogroup members from Arabidopsis, poplar, and moss, overlaid the resulting trees with proteomic data, and assessed whether members identified in plasmodesmal fractions were distributed throughout a tree or were clustered in specific clades. OG3, OG5, and OG16, all show plasmodesmal association predominantly in a single branch of the tree (Fig. 5; Supplementary Figs S9–S11), suggesting that plasmodesmal association was gained once during evolution of the protein family. In contrast, plasmodesmal association in OG6 is dispersed across the whole tree, suggesting that each tetraspanin ancestor has an equal likelihood of being associated with plasmodesmata (Fig. 5; Supplementary Fig. S12). Similarly, OG0 (Fig. 5; Supplementary Fig. S13) shows no clear phylogenetic pattern associated with plasmodesmal association. As proteins validated as core plasmodesmal proteins are represented amongst trees that harbour single clades and whole-tree distribution of proteomic hits, this approach offers no further resolution in identifying core plasmodesmal proteins. However, for protein families with plasmodesmal association in specific clades, it offers potential to identify candidate plasmodesmal family members from species for which a proteome has not been generated.
Fig. 5.
Unrooted cladograms of orthogroup members from A. thaliana, P. trichocarpa, and P. patens as defined by a hmmsearch with a threshold of E <1 × 10–100 (OG0/OG3/OG5) or E <1 × 10–0 (OG6/OG16). Each tree has a heatmap of proteome matches for each protein in the tree, with orange indicating a proteome hit and blue indicating that the protein was not detected in the relevant proteome(s). For OG3, OG5, and OG16, plasmodesmal association appears to primarily correlate with a single clade within the tree, indicated by the black bar to the right of the proteome heatmap. Pie charts estimate the likely ancestral plasmodesmal localization (orange) by phylogenetic backpropagation. Node support is indicated by greyscale circles.
Discussion
Plasmodesmata are essential features of plant cells, but detailed molecular understanding of their structure and function has long been enigmatic. As membrane-rich structures embedded in the cell wall, they can be described as recalcitrant with respect to biochemical extraction and characterization, and knowledge of their composition has been revealed in a piecemeal fashion despite considerable research efforts. Despite technical challenges, proteomic strategies have underpinned major leaps in understanding of plasmodesmal function, yielding primary knowledge of plasmodesmal responses, as well as formulation of the current model for their core structure being a specialized membrane contact site (Tilsner et al., 2016). Recognizing the gains to be made by better understanding of the protein composition of plasmodesmata in different tissues and species, we used phyloproteomic comparison to define a more detailed atlas of plasmodesmal structure and function.
Defining a proteome is subject to sampling and technical variation that limits the depth of an analysis of samples from a single technical or biological context as subcellular fractionation and MS are inherently noisy techniques (Cargile et al., 2004). The caveat of this is that the most abundant proteins in the preparations will be the most consistently identified, and so some qualitative metric of abundance can be inferred from the repeated presence of a protein. This rationale also applies to a comparison of proteomes of different species in which consistent identification infers conservation and the approach can be used to identify core, essential, and conserved plasmodesmal proteins. Thus, comparative phylogenetic analysis of proteomes from different species gives power to identifying key plasmodesmal components from inherently noisy datasets. With the aim of increasing the analytical power of plasmodesmal proteomics, we generated two new plasmodesmal proteomes from differentiated tissues of Arabidopsis and the moss P. patens, and identified protein orthogroups that were represented across samples, hypothesizing that these contain proteins that are core to plasmodesmal structure and/or function.
The ER-derived desmotubule and apoplastic callose have been observed in plasmodesmata across the green lineage (Robards, 1976; Faulkner et al., 2009; Brecknock et al., 2011), suggesting the possibility that these, and other features of plasmodesmata employ families of conserved proteins. We reasoned that core, conserved proteins would associate with plasmodesmata in distantly related plant species, and proteins from β-1,3-glucanase (OG0), C2 lipid-binding protein (OG3), and tetraspanin (OG6) orthogroups demonstrated this behaviour. However, despite evidence of conservation, phylogenetic analysis of the relationships between the moss, poplar, and Arabidopsis protein families from which these orthogroups are derived does not reveal a single pattern of evolution of plasmodesmata association.
Our approach identified and confirmed β-1,3-glucanases and C2 lipid-binding proteins as core and conserved plasmodesmal components. For β-1,3-glucanases this aligns with their characterized role in callose homeostasis at plasmodesmata, with callose deposition detected at plasmodesmata in algae (Faulkner et al., 2009), moss (Fig. 2E; Tomoi et al., 2020; Muller et al., 2022), and flowering plants. Current structural models of plasmodesmata incorporate C2 lipid-binding domain proteins as connectors between the ER and the plasma membrane in specialized membrane contact sites (Brault et al., 2019). Consistent with the conservation of the plasma membrane and desmotubules in plasmodesmata, the conservation of C2 lipid-binding proteins in plasmodesmata suggests that they are a central and core element of plasmodesmata. We observed that the moss C2 lipid-binding protein PpMCTP5 localized at plasmodesmata in moss protonema, but also at other points where the ER sits at the cell periphery (Supplementary Fig. S5) as expected for proteins at membrane contact sites. This further supports the likelihood that there is functional conservation between Arabidopsis and moss C2 lipid-biding proteins and that membrane contact sites are an ancient feature of plasmodesmal structure.
Tetraspanins also showed conserved localization across different species; however, while they have been previously localized to plasmodesmata in Arabidopsis (Fernandez-Calvino et al., 2011), their functional relevance has not yet been established. Tetraspanins are associated with membrane compartmentalization in animals and function in the recruitment and activation of signalling components (Levy and Shoham, 2005; Kummer et al., 2020). For tetraspanins, plasmodesmal association is broadly represented across the cladogram (Fig. 5). As tetraspanins are found across different kingdoms of eukaryotic life, and as our trees are unrooted, it seems unlikely that tetraspanins were an evolutionary advance that specifically catalysed the formation of plasmodesmata. However, these proteins might be associated with the specialization of membrane function associated with the evolution of plasmodesmata. Indeed, as plasmodesmal membranes host localized and specialized signalling cascades, tetraspanins might serve to define the plasmodesmal membrane domain and require further investigation.
With callose deposition central to plasmodesmal function, we were surprised that our analysis did not detect callose synthases. While this might arise from our fractionation methods being suboptimal for their extraction, or from usage of non-quantitative MS methods, we found that if we reduced the stringency of protein identification in both our Arabidopsis leaf and moss plasmodesmal fractions, allowing an identification probability of >50% threshold for peptide and protein identification and a minimum of one sample, we identify an additional 12 orthogroups present in at least four of five proteomes, one of which represents callose synthases (Supplementary Tables S2, S3, S7). This low stringency analysis also reveals orthogroups containing heavy metal-associated isoprenylated proteins (HIPPs), which have been localized to plasmodesmata in Arabidopsis (Guo et al., 2021) and N. benthamiana (Cowan et al., 2018), and xyloglucan endotransglucosylase/hydrolase proteins, which have been confirmed as plasmodesmal proteins in a concomitant proteomic study (Gombos et al., 2023). In essence, by requiring the identification of a protein in multiple independent proteomes, we are increasing the a priori likelihood of protein identification within a sample. Taking this Bayesian idea, we can reduce the stringent identification criteria of known plasmodesmal proteins, as we are expecting them to appear in the samples. Moreover, proteins which are misidentified would not be classed as ‘core’.
This approach strengthens the confidence in identifying true plasmodesmal proteins by proteomic methods. However, despite their repeated identification in plasmodesmal proteomes, we were unable to resolve any association of proteins in the GRP and GDSL esterase/lipase families with plasmodesmata using confocal microscopy (Fig. 4; Supplementary Fig. S7). The limits of resolution of confocal microscopy suggest that this negative result might not exclude such proteins from having a transient or non-enriched (relative to the rest of the cell) association with plasmodesmata. Future work could use approaches with higher resolution such as immunolocalization by EM or super-resolution light microscopy to determine whether proteins are associated with plasmodesmata.
In addition to the increased power of analysis by comparative analysis, the data contained herein establish new knowledge of moss plasmodesmata. While moss genomes do not encode a family of PDLPs (Vaattovaara et al., 2019), which positively regulate callose deposition (Lee et al., 2011; Caillaud et al., 2014), the detection of stimulus-triggered callose deposition in bryophytes (Kitagawa et al., 2019) suggests that callose regulation of plasmodesmata is an ancestral feature. The absence of PDLPs from moss might indicate the possibility that moss has fewer, or less complex, regulatory processes for callose synthesis.
Cell to cell communication is a central feature of multicellularity. Therefore, a greater understanding of plasmodesmata promises to enhance our knowledge of a whole range of plant processes by resolving which cells and tissues coordinate and communicate to enable organism-level responses. The details of plasmodesmal structure and function are slowly being revealed, and we have demonstrated the benefit of enhancing the knowledge gained from technically difficult proteomic profiling by pooling new and existing information to identify conserved, core plasmodesmal components. Indeed, our approach offers further opportunity to define the core structure of plasmodesmata and expand our understanding across the evolutionary tree to which future efforts can add mechanistic and physiological understanding.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Validation of mature plant tissue fractionation of P. patens after successive disruption steps.
Fig. S2. Phylogenetic trees of protein families with plasmodesmata-associated members in P. patens inferred using the maximum likelihood method.
Fig. S3. Generation and validation of moss strains expressing candidate plasmodesmal proteins fused to fluorescent protein mNeonGreen.
Fig. S4. Venn diagram representing the overlap between orthogroups identified in each of the proteomes analysed in this study.
Fig. S5. Occurrence of labelled C2 lipid-binding protein-positive punctae at the cell periphery in addition to plasmodesmal association in moss.
Fig. S6. Stable expression of the P. patens C2 lipid-binding domain protein PpMCTP5–GFP in Arabidopsis.
Fig. S7. Members of the GDSL esterase/lipase OG5 and the glycine-rich RNA-binding protein OG16 do not associate with plasmodesmata.
Fig. S8. Stable expression of the RNA-binding proteins PpGRP2–GFP and AtGRP7–GFP in Arabidopsis.
Fig. S9. Unrooted cladograms of OG16 (glycine-rich RNA-binding protein) members from A. thaliana, P. trichocarpa, and P. patens with protein Uniprot identifiers listed.
Fig. S10. Unrooted cladograms of OG5 (GDSL esterase/lipase) members from A. thaliana, P. trichocarpa, and P. patens with protein Uniprot identifiers listed.
Fig. S11. Unrooted cladograms of OG3 (C2 lipid-binding) members from A. thaliana, P. trichocarpa, and P. patens with protein Uniprot identifiers listed.
Fig. S12. Unrooted cladograms of OG6 (tetraspanin) members from A. thaliana, P. trichocarpa, and P. patens with protein Uniprot identifiers listed.
Fig. S13. Unrooted cladograms of OG0 (β-1,3-glucanase) members from A. thaliana, P. trichocarpa, and P. patens with protein Uniprot identifiers listed.
Table S1. Primers used in this study.
Table S2. List of proteins identified in A. thaliana rosette leaf plasmodesmal fractions by MS, defining AtL.
Table S3. Filtered list of proteins identified in P. patens plasmodesmal fractions by MS, defining PpPG.
Table S4. One to one mapping of P. patens proteins identified in the plasmodesma-enriched fraction to Arabidopsis homologs using two methods.
Table S5. List of OG members identified in each proteome.
Table S6. Uniprot protein identifiers mapped to genome annotations (EnsemblPlants, Phypa_V3) and proposed names based on phylogenetic relationships.
Table S7. List of orthogroups identified in at least four of five proteomes when the stringency for protein identification in AtL and PpPG is reduced.
Acknowledgements
We thank Dr P.A.C. van Gisbergen for assistance with moss callose staining.
Contributor Information
Matthew G Johnston, Department of Cell and Developmental Biology, John Innes Centre, UK.
Andrew Breakspear, Department of Cell and Developmental Biology, John Innes Centre, UK.
Sebastian Samwald, Department of Cell and Developmental Biology, John Innes Centre, UK.
Dan Zhang, Department of Cell and Developmental Biology, John Innes Centre, UK.
Diana Papp, Department of Cell and Developmental Biology, John Innes Centre, UK.
Christine Faulkner, Department of Cell and Developmental Biology, John Innes Centre, UK.
Jeroen de Keijzer, Laboratory of Cell Biology, Wageningen University and Research, Wageningen, The Netherlands.
Karl-Josef Dietz, Bielefeld University, Germany.
Author contributions
MGJ, CF, and JdK, conceptualization, data curation, visualization, and writing—original draft; MGJ and JdK: formal analysis; CF, funding acquisition; MGJ, SS, DP, CF, and JdK: investigation; AB, DP, MGJ, JdK, and DZ: methodology; CF and JdK: project administration; MGJ, AB, SS, DP, MGJ, and JdK: resources; CF and JdK: supervision; MGJ and CF, validation; MGJ, SS, CF, and JdK: writing—review and editing.
Conflict of interest
The authors have no conflict of interest to declare.
Funding
This work was supported by the European Research Council (grant agreement 725459, ‘INTERCELLAR’); the Biotechnology and Biological Research Council (Institute Strategic Programme ‘Plant Health’ BBS/E/J/000PR9796); and the John Innes Foundation (Rotation Studentships to MGJ and SS).
Data availability
MS data generated for this study have been deposited in the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022) partner repository with the dataset identifier PXD038964. All other data supporting the findings of this study are available within the paper and within its supplementary data published online.
References
- Ashburner M, Ball CA, Blake JA, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y. 2014. Symplastic intercellular transport from a developmental perspective. Journal of Experimental Botany 65, 1857–1863. [DOI] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Faulkner C, Pendle A, Miyashima S, Helariutta Y, Maule A.. 2013. Symplastic intercellular connectivity regulates lateral root patterning. Developmental Cell 26, 136–147. [DOI] [PubMed] [Google Scholar]
- Blackman LM, Overall RL.. 1998. Immunolocalisation of the cytoskeleton to plasmodesmata of Chara corallina. The Plant Journal 14, 733–741. [Google Scholar]
- Brault ML, Petit JD, Immel F, et al. 2019. Multiple C2 domains and transmembrane region proteins (MCTPs) tether membranes at plasmodesmata. EMBO Reports 20, e47182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecknock S, Dibbayawan TP, Vesk M, Vesk PA, Faulkner C, Barton DA, Overall RL.. 2011. High resolution scanning electron microscopy of plasmodesmata. Planta 234, 749–758. [DOI] [PubMed] [Google Scholar]
- Brunkard JO, Zambryski PC.. 2017. Plasmodesmata enable multicellularity: new insights into their evolution, biogenesis, and functions in development and immunity. Current Opinion in Plant Biology 35, 76–83. [DOI] [PubMed] [Google Scholar]
- Caillaud M-C, Wirthmueller L, Sklenar J, Findlay K, Piquerez SJM, Jones AME, Robatzek S, Jones JDG, Faulkner C.. 2014. The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathogens 10, e1004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargile BJ, Bundy JL, Stephenson JL Jr. 2004. Potential for false positive identifications from large databases through tandem mass spectrometry. Journal of Proteome Research 3, 1082–1085. [DOI] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD.. 2017. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. The Plant Journal 89, 789–804. [DOI] [PubMed] [Google Scholar]
- Cheval C, Faulkner C.. 2018. Plasmodesmal regulation during plant–pathogen interactions. New Phytologist 217, 62–67. [DOI] [PubMed] [Google Scholar]
- Cheval C, Samwald S, Johnston MG, de Keijzer J, Breakspear A, Liu X, Bellandi A, Kadota Y, Zipfel C, Faulkner C.. 2020. Chitin perception in plasmodesmata characterizes submembrane immune-signaling specificity in plants. Proceedings of the National Academy of Sciences, USA 117, 9621–9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF.. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cook ME, Graham LE, Botha CEJ, Lavin CA.. 1997. Comparative ultrastructure of plasmodesmata of Chara and selected bryophytes: toward an elucidation of the evolutionary origin of plant plasmodesmata. American Journal of Botany 84, 1169–1178. [PubMed] [Google Scholar]
- Cowan GH, Roberts AG, Jones S, Kumar P, Kalyandurg PB, Gil JF, Savenkov EI, Hemsley PA, Torrance L.. 2018. Potato mop-top virus co-opts the stress sensor HIPP26 for long-distance movement. Plant Physiology 176, 2052–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Lee JY.. 2016. Arabidopsis callose synthases CalS1/8 regulate plasmodesmal permeability during stress. Nature Plants 2, 16034. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14, 755–763. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biology 16, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology 20, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C, Bayer EM.. 2017. Isolation of plasmodesmata. Methods in Molecular Biology 1511, 187–198. [DOI] [PubMed] [Google Scholar]
- Faulkner CR, Blackman LM, Cordwell SJ, Overall RL.. 2005. Proteomic identification of putative plasmodesmatal proteins from Chara corallina. Proteomics 5, 2866–2875. [DOI] [PubMed] [Google Scholar]
- Faulkner CR, Blackman LM, Collings DA, Cordwell SJ, Overall RL.. 2009. Anti-tropomyosin antibodies co-localise with actin microfilaments and label plasmodesmata. European Journal of Cell Biology 88, 357–369. [DOI] [PubMed] [Google Scholar]
- Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ.. 2013. LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proceedings of the National Academy of Sciences, USA 110, 9166–9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Calvino L, Faulkner C, Walshaw J, Saalbach G, Bayer E, Benitez-Alfonso Y, Maule A.. 2011. Arabidopsis plasmodesmal proteome. PLoS One 6, e18880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Pozo N, Haas FB, Meyberg R, et al. 2020. PEATmoss (Physcomitrella Expression Atlas Tool): a unified gene expression atlas for the model plant Physcomitrella patens. The Plant Journal 102, 165–177. [DOI] [PubMed] [Google Scholar]
- Gaudet P, Livstone MS, Lewis SE, Thomas PD.. 2011. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Briefings in Bioinformatics 12, 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudioso-Pedraza R, Beck M, Frances L, Kirk P, Ripodas C, Niebel A, Oldroyd GED, Benitez-Alfonso Y, de Carvalho-Niebel F.. 2018. Callose-regulated symplastic communication coordinates symbiotic root nodule development. Current Biology 28, 3562–3577. [DOI] [PubMed] [Google Scholar]
- Gombos S, Miras M, Howe V, et al. 2023. A high confidence Physcomitrium patens plasmodesmata proteome by iterative scoring and validation reveals diversification of cell wall proteins during evolution. New Phytologist 10.1111/nph.18730 [DOI] [PubMed] [Google Scholar]
- Grison MS, Fernandez-Calvino L, Mongrand S, Bayer EM.. 2015. Isolation of plasmodesmata from Arabidopsis suspension culture cells. Methods in Molecular Biology 1217, 83–93. [DOI] [PubMed] [Google Scholar]
- Grison MS, Kirk P, Brault ML, Wu XN, Schulze WX, Benitez-Alfonso Y, Immel F, Bayer EM.. 2019. Plasma membrane-associated receptor-like kinases relocalize to plasmodesmata in response to osmotic stress. Plant Physiology 181, 142–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Guo T, Weber H, Niemann MCE, Theisl L, Leonte G, Novák O, Werner T.. 2021. Arabidopsis HIPP proteins regulate endoplasmic reticulum-associated degradation of CKX proteins and cytokinin responses. Molecular Plant 14, 1918–1934. [DOI] [PubMed] [Google Scholar]
- Hiwatashi Y, Obara M, Sato Y, Fujita T, Murata T, Hasebe M.. 2008. Kinesins are indispensable for interdigitation of phragmoplast microtubules in the moss Physcomitrella patens. The Plant Cell 20, 3094–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Serra F, Bork P.. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Molecular Biology and Evolution 33, 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalve S, De Vos D, Beemster GTS.. 2014. Leaf development: a cellular perspective. Frontiers in Plant Science 5, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R.. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical Chemistry 74, 5383–5392. [DOI] [PubMed] [Google Scholar]
- Kirk P, Amsbury S, German L, Gaudioso-Pedraza R, Benitez-Alfonso Y.. 2022. A comparative meta-proteomic pipeline for the identification of plasmodesmata proteins and regulatory conditions in diverse plant species. BMC Biology 20, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Tomoi T, Fukushima T, Sakata Y, Sato M, Toyooka K, Fujita T, Sakakibara H.. 2019. Abscisic acid acts as a regulator of molecular trafficking through plasmodesmata in the moss Physcomitrella patens. Plant and Cell Physiology 60, 738–751. [DOI] [PubMed] [Google Scholar]
- Kummer D, Steinbacher T, Schwietzer MF, Thölmann S, Ebnet K.. 2020. Tetraspanins: integrating cell surface receptors to functional microdomains in homeostasis and disease. Medical Microbiology and Immunology 209, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Wang X, Cui W, et al. 2011. A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. The Plant Cell 23, 3353–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leijon F, Melzer M, Zhou Q, Srivastava V, Bulone V.. 2018. Proteomic analysis of plasmodesmata from populus cell suspension cultures in relation with callose biosynthesis. Frontiers in Plant Science 9, 1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Erlanger M, Rosenthal M, Epel BL.. 2007. A plasmodesmata-associated beta-1,3-glucanase in Arabidopsis. The Plant Journal 49, 669–682. [DOI] [PubMed] [Google Scholar]
- Levy S, Shoham T.. 2005. The tetraspanin web modulates immune-signalling complexes. Nature Reviews. Immunology 5, 136–148. [DOI] [PubMed] [Google Scholar]
- Ligrone R, Duckett JG.. 1994. Thallus differentiation in the marchantialean liverwort Asterella wilmsii (Steph.) with particular reference to longitudinal arrays of endoplasmic microtubules in the inner cells. Annals of Botany 73, 577–586. [Google Scholar]
- Ligrone R, Ducket JG, Renzaglia KS.. 2000. Conducting tissues and phyletic relationships of bryophytes. Philosophical Transactions of the Royal Society B: Biological Sciences 355, 795–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD.. 2019. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Research 47, D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JL, Puttick MN, Clark JW, Edwards D, Kenrick P, Pressel S, Wellman CH, Yang Z, Schneider H, Donoghue PCJ.. 2018. The timescale of early land plant evolution. Proceedings of the National Academy of Sciences, USA 115, E2274–e2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Fujita T, Coudert Y.. 2022. Callose detection and quantification at plasmodesmata in bryophytes. Methods in Molecular Biology 2457, 177–187. [DOI] [PubMed] [Google Scholar]
- Naramoto S, Hata Y, Fujita T, Kyozuka J.. 2022. The bryophytes Physcomitrium patens and Marchantia polymorpha as model systems for studying evolutionary cell and developmental biology in plants. The Plant Cell 34, 228–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Hiwatashi Y, Sakakibara I, Kato M, Hasebe M.. 2000. Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Research 7, 9–17. [DOI] [PubMed] [Google Scholar]
- Nomata T, Kabeya Y, Sato N.. 2004. Cloning and characterization of glycine-rich RNA-binding protein cDNAs in the moss Physcomitrella patens. Plant and Cell Physiology 45, 48–56. [DOI] [PubMed] [Google Scholar]
- Ortiz-Ramírez C, Hernandez-Coronado M, Thamm A, Catarino B, Wang M, Dolan L, Feijó JA, Becker JD.. 2016. A transcriptome atlas of Physcomitrella patens provides insights into the evolution and development of land plants. Molecular Plant 9, 205–220. [DOI] [PubMed] [Google Scholar]
- Park SH, Li F, Renaud J, Shen W, Li Y, Guo L, Cui H, Sumarah M, Wang A.. 2017. NbEXPA1, an α-expansin, is plasmodesmata-specific and a novel host factor for potyviral infection. The Plant Journal 92, 846–861. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y, Bai J, Bandla C, et al. 2022. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Research 50, D543–D552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD.. 2018. HMMER web server: 2018 update. Nucleic Acids Research 46, W200–W204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Goffinet B, Meyberg R, Wu SZ, Bezanilla M.. 2020. The moss Physcomitrium (Physcomitrella) patens: a model organism for non-seed plants. The Plant Cell 32, 1361–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne PL, Welling A, Vahala J, Ripel L, Ruonala R, KangasjärviJ, van der SchootC.. 2011. Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-beta-glucanases to reopen signal conduits and release dormancy in Populus. The Plant Cell 23, 130–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robards AW. 1976. Plasmodesmata in higher plants. In: Gunning BES, Robards AW, eds. Intercellular communication in plants: studies on plasmodesmata. Berlin, Heidelberg: Springer, 15–17. [Google Scholar]
- Searle BC. 2010. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 10, 1265–1269. [DOI] [PubMed] [Google Scholar]
- Sevilem I, Yadav SR, Helariutta Y.. 2015. Plasmodesmata: channels for intercellular signaling during plant growth and development. Methods in Molecular Biology 1217, 3–24. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CL, Bayer EM, Ritzenthaler C, Fernandez-Calvino L, Maule AJ.. 2008. Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biology 6, e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A.. 2003. PANTHER: a library of protein families and subfamilies indexed by function. Genome Research 13, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J, Nicolas W, Rosado A, Bayer EM.. 2016. Staying tight: plasmodesmal membrane contact sites and the control of cell-to-cell connectivity in plants. Annual Review of Plant Biology 67, 337–364. [DOI] [PubMed] [Google Scholar]
- Tomoi T, Kawade K, Kitagawa M, Sakata Y, Tsukaya H, Fujita T.. 2020. Quantitative imaging reveals distinct contributions of SnRK2 and ABI3 in plasmodesmatal permeability in Physcomitrella patens. Plant and Cell Physiology 61, 942–956. [DOI] [PubMed] [Google Scholar]
- Vaattovaara A, Brandt B, Rajaraman S, et al. 2019. Mechanistic insights into the evolution of DUF26-containing proteins in land plants. Communications Biology 2, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag. [Google Scholar]
- Yi P, Goshima G.. 2020. Rho of plants GTPases and cytoskeletal elements control nuclear positioning and asymmetric cell division during Physcomitrella patens branching. Current Biology 30, 2860–2868. [DOI] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y.. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution 8, 28–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MS data generated for this study have been deposited in the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022) partner repository with the dataset identifier PXD038964. All other data supporting the findings of this study are available within the paper and within its supplementary data published online.