Abstract
Background:
Intracerebral hemorrhage (ICH) has an estimated heritability of 29%. We developed a genomic risk score for ICH and determined its predictive power in comparison to standard clinical risk factors.
Methods:
We combined genome-wide association data from individuals of European ancestry for ICH and related traits in a meta-genomic risk score (metaGRS) (2.6 million variants). We tested associations with ICH and its predictive performance in addition to clinical risk factors in a held-out validation dataset (842 cases and 796 controls). We tested associations with risk of incident ICH in the population-based UK Biobank cohort (486,784 individuals, 1,526 events, median follow-up 11.3 years).
Results:
One SD increment in the metaGRS was significantly associated with 31% higher odds for ICH (95%CI: 1.16-1.48) in age-, sex- and clinical risk factor-adjusted models. The metaGRS identified individuals with almost 5-fold higher odds for ICH in the top score percentile (OR: 4.83, 95%CI: 1.56-21.2). Predictive models for ICH incorporating the metaGRS in addition to clinical predictors showed superior performance compared to clinical risk factors alone (c-index: 0.695 vs 0.686). The metaGRS showed similar associations for lobar and non-lobar ICH, independent of the known APOE risk locus for lobar ICH. In the UK Biobank, the metaGRS was associated with higher risk of incident ICH (HR: 1.15, 95%CI: 1.09-1.21). The associations were significant within both a relatively high-risk population of antithrombotic medications users, as well as among a relatively low-risk population with a good control of vascular risk factors and no use of anticoagulants.
Conclusions:
We developed and validated a genomic risk score that predicts lifetime risk of ICH beyond established clinical risk factors among individuals of European ancestry. Whether implementation of the score in risk prognostication models for high-risk populations, such as patients under antithrombotic treatment, could improve clinical decision making should be explored in future studies.
Keywords: intracerebral hemorrhage, hemorrhagic stroke, genetics, genomic risk score
Graphical Abstract
INTRODUCTION
Intracerebral hemorrhage (ICH) is the most devastating type of stroke. Although it accounts for 10-20% of all acute cerebrovascular events, it is responsible for almost 50% of stroke-related morbidity and mortality.1 Given the lack of effective treatments and the devastating outcome of ICH, primary and secondary prevention is critical. As with many complex human diseases, ICH risk is comprised of both environmental and genetic factors. Early genome-wide association studies (GWAS) of ICH estimated a heritability of 29% and revealed a polygenic architecture,2,3 whereas large-scale GWAS for established risk factors for ICH, such as hypertension and smoking, have identified hundreds of associated genomic loci that cumulatively explain a large proportion of the variance of these traits.4-9
Beyond defining heritability and providing insight into biological mechanisms, GWAS findings have begun to show promise for disease risk prediction. Genomic risk scores (GRS), biomarkers representing the aggregated effect of many genetic variants on a given trait, have been proposed as powerful tools for identifying individuals at high risk for complex traits, with a predictive performance at times comparable to that of rare monogenic mutations.10 A GRS for ICH could have clinical utility in decision making algorithms, such as for complementing risk-benefit calculation tools in patients prescribed antithrombotic medications. Despite acknowledged limitations, pertaining mainly to translation of the accuracy of GRS findings from the cohort- to the individual-based level, as well as sex- and ancestry-specific predictive differences,11,12 if constructed according to best practices,11-14 an ICH GRS could have important implications for patient selection for clinical trials. However, as opposed to other vascular diseases,10 efforts to construct a GRS for ICH lag behind, likely because of the lower statistical power in available datasets due to the relative rarity of the disease.
Recent advances in analytical approaches offer more efficient alternative methods for GRS construction that allow to utilize data from multiple GWAS in order to overcome power limitations of genomic datasets of rare diseases.15,16 Based on the assumption that the majority of genetic variants exert their effects on a given disease by affecting intermediate traits, constructing a GRS based on genomic data of traits in the causal pathway of the disease of interest can improve both power and prediction.15 In this study, we sought to investigate whether combining genetic liability for possible ICH risk factors and traits reflecting pathologies underlying ICH into a meta-Genomic Risk Score (metaGRS) could improve our ability to predict ICH events among individuals of European ancestry. To further explore the potential utility of such a genomic score for clinical risk prediction, we also assessed whether the metaGRS improves ICH risk prediction beyond established clinical risk factors.
METHODS
Data availability statement
The data that support the findings of this study will be available from the corresponding author upon reasonable request. MetaGRS single nucleotide polymorphism (SNP)-specific weights will be made publicly available at The Polygenic Score (PGS) Catalog.
Study design and participating studies
As our primary data source, we used genotype and phenotype data from 1,861 ICH cases and 1,722 ICH-free controls from three independent GWAS datasets: the North American (USA) multi-center Genetics of Cerebral Hemorrhage on Anticoagulation (GOCHA) study, a prospectively collected case-control study of European ancestry subjects aged > 55 years with primary ICH17; the European member sites contributing ICH cases and controls to the International Stroke Genetics Consortium (EUR/ISGC); and the Genetic and Environmental Risk Factors for Hemorrhagic Stroke (GERFHS) study, a prospectively collected case-control study of subjects > 18 years of age with spontaneous ICH in the Greater Cincinnati region18. GOCHA and EUR/ISGC were the training datasets, whereas GERFHS was our primary validation dataset. Furthermore, we performed an external validation of the derived score for incident ICH events in the UK Biobank (UKBB) cohort, over a median follow-up of 11.3 years among 486,623 individuals aged 40-69 years at recruitment without a prior history of ICH.19 All studies were approved by the Institutional Review Board or Ethics Committee at each participating institution. We followed the GRS reporting guidelines as outlined in the Polygenic Risk Score Reporting Standards developed by the Clinical Genome Resource (ClinGen) Complex Disease Working Group and the PGS Catalog14. Details on ascertainment of cases and controls and on imputation of the GWAS datasets are provided in Supplemental Material.
Trait-specific GRS and ICH metaGRS construction
We used GOCHA (436 ICH cases and 405 controls) and EUR/ISGC (577 ICH cases and 523 controls) as training datasets to develop GRS for 21 traits associated with ICH risk. We leveraged publicly available GWAS summary-level data from international consortia, as detailed in Table S1.4,20-32 For all traits, we used data from European-only populations and excluded duplicate and ambiguous AT/GC SNPs and SNPs with MAF≤1%. No GOCHA or EUR/ISGC cases or controls were included in the selected studies. For each trait, we generated a range of candidate trait-specific GRS and selected the best-performing ones based on the highest area under the receiving operating characteristics curve of a logistic regression model for ICH. Following a standardized meta-analytic approach, we constructed an ICH metaGRS creating a weighted-average of the trait-specific GRS. We used GERFHS as our primary validation dataset. Details are provided in Supplemental Methods, Tables S2-S23.
ICH metaGRS performance and clinical evaluation in GERFHS and UKBB
We explored the metaGRS performance in the validation dataset and its comparison with clinical predictors. The primary metaGRS and alternative versions were entered as linear predictors in logistic regression models for ICH in the primary validation dataset. We performed complementary analyses to investigate the association between the metaGRS and ICH. We evaluated the potential clinical utility of the metaGRS exploring its associations with ICH independently of clinical risk factors in GERFHS and performed external validation in a population-based setting in the UKBB (Supplemental Methods).
RESULTS
Construction of a metaGRS for ICH in the training dataset
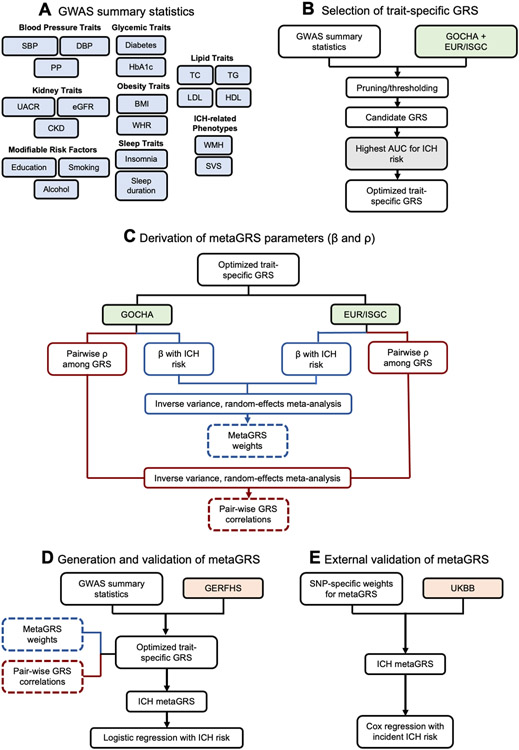
A schematic of our study design is provided in Figure 1. Following quality checks, we developed 21 optimized GRS for ICH-associated traits on the basis of associations with ICH in GOCHA and EUR/ISGC. The numbers of variants included in these GRS ranged from 213 to 1,148,192 (Table S23). Detailed results are provided in the Supplemental Results, Tables S24-S25, and Figures S1-S4.
Figure 1. Study design.
(A) Individual genomic risk scores (GRS) were derived for intracerebral hemorrhage (ICH)-related traits from publicly available summary statistics. (B) The GRS were optimized in the combined training dataset of GOCHA and EUR/ISGC. (C) MetaGRS parameters were determined on the basis of association with ICH in the training datasets. (D) The metaGRS was compiled in the GERFHS validation dataset and associations with ICH were explored using logistic regression. (E) The metaGRS was externally validated in the general population-based UKBB cohort using Cox proportional hazards regression analyses.
Associations between the metaGRS and ICH in the validation dataset
We explored associations between the derived metaGRS and ICH risk in GERFHS. Characteristics of ICH cases and ICH-free controls in GERFHS are presented in Table S26. After adjusting for age, sex, and two PCs, one SD increase in the metaGRS was associated with 45% higher odds of ICH (OR 1.45; 95% CI: 1.30-1.63; p=6.2x10−11). Patients in higher thresholds of the metaGRS distribution were at progressively higher risk for ICH (Figure 2). Notably, patients in the top 2.5% and 1% had substantially increased odds of ICH, respectively, compared to the rest of sample. We observed an expected gradual change in ICH odds when moving to either the higher or lower ends of the metaGRS distribution from the middle decile (Figure S5). Modeling risk of ICH relative to the bottom 10% of the metaGRS loading did not reveal any significant non-linear effects (Figure S6).
Figure 2. Odds for intracerebral hemorrhage (ICH) across the metaGRS distribution.
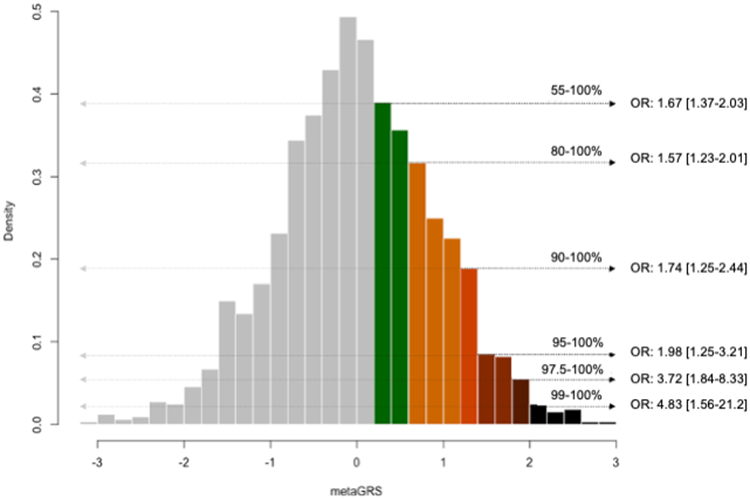
Depicted is the metaGRS distribution (centered around a mean of 0 and a standard deviation of 1) in the GERFHS validation dataset and the odds ratios (OR) for ICH per percentile group, relative the rest of the sample, as derived from logistic regression models adjusted for age, sex and the first 2 principal components of the population structure.
In analyses comparing ICH odds between the primary and the alternative metaGRS, we overall found that the primary metaGRS achieved the best performance (Figure S7). However, these metaGRS with penalized and adjusted weights still demonstrated significant associations with ICH risk, albeit with weaker effect size estimates and lower predictive performances (Figure S7, Table S27).
Predictive performance of metaGRS for ICH in comparison with clinical risk factors
We next explored the performance of the metaGRS with established clinical risk factors for predicting ICH in GERFHS. History of ischemic stroke, hypertension, diabetes, high cholesterol, heavy alcohol use, anticoagulant use, and education less than high school were most strongly associated with ICH risk (Table S28). After adjusting for these clinical risk factors, the metaGRS continued to be independently associated with ICH risk (OR = 1.31 per one standard deviation of the metaGRS; 95% CI 1.16 – 1.48 p < 0.0001). Adding the metaGRS to a model including the clinical risk factors significantly improved the model fit (AIC of clinical predictors = 1989.65, AIC of clinical predictors + metaGRS = 1972.86, LRT = 4x10−5).
The metaGRS showed comparable predictive performance to hypertension, and higher than the remaining clinical risk factors apart from education (Figure 3). Importantly, the c-index of a model including the entire set of clinical risk factors (C: 0.686, 95%CI: 0.663-0.718) increased after including the metaGRS in the model (C: 0.695, 95%CI: 0.673 – 0.727, Figure 3). Similar results were observed for the alternative metaGRS versions (Figure S8, Table S29). Stratified analyses showed significant associations of the metaGRS with both lobar and non-lobar ICH (Tables S30-S34). When exploring in the same model the metaGRS and APOE genotype, a known risk locus for ICH, we found that both were independently associated with the odds of ICH (Table S35).
Figure 3. Performance of clinical risk factors, metaGRS, and their combination for predicting odds for intracerebral hemorrhage (ICH).
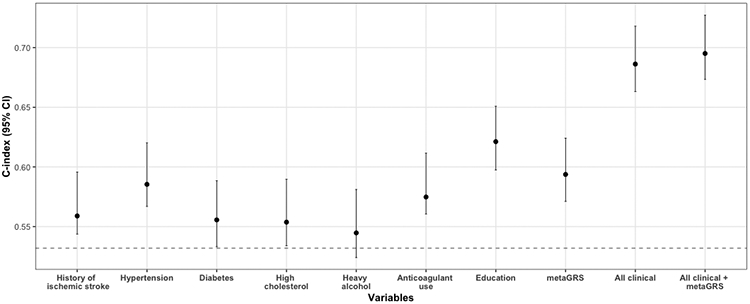
Depicted are the c-indices derived from logistic regression models including age, sex, 2 principal components of population structure (baseline model, dotted line) and additionally in successive models the reported clinical variables (history of ischemic stroke, hypertension, diabetes, high cholesterol, heavy alcohol use, anticoagulant medication use, and education) and the metaGRS in the validation GERFHS dataset. Percentile confidence intervals of the c-indices were calculated after bootstrapping over 1000 iterations.
Validation of the metaGRS in the UKBB population
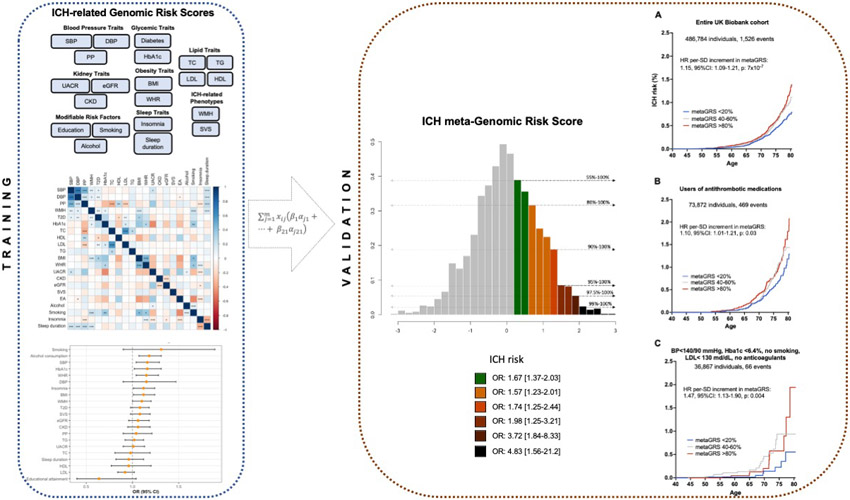
As a final step, we explored associations between the metaGRS and incident ICH risk in a general population sample. In the prospective population-based UKBB cohort, a total of 486,623 participants without a history of ICH, were followed-up for a median of 11.3 years (IQR: 10.6-11.1 years) (Table S36). We again found the metaGRS to be associated with a higher risk of incident ICH (HR per SD increment: 1.15, 95%CI: 1.09-1.21, p=7x10−7) after adjusting for age, sex, the first 10 PCs of population structure, kinship, genotyping chip, and genetic ancestry. Accounting for death as a competing risk with the subdistribution hazard approach did not substantially alter the results (HR per SD increment of metaGRS: 1.14, 95%CI: 1.08-1.20, p=4x10−6). Figure 4A presents the Kaplan-Meier curves with age as the time variable for individuals at upper, median, and lower quantiles of the metaGRS. Within both high-risk individuals using antithrombotic medications at baseline, as well as low-risk individuals with well-controlled vascular risk factors at baseline and no antithrombotic medications, the metaGRS retained association with incident ICH risk (Figures 4B, C). We again found the metaGRS to be associated with a higher risk of incident ICH (HR per SD increment: 1.15, 95%CI: 1.09-1.21, p=7x10−7, concordance: 0.705, 95%CI: 0.693-0.717) in a model adjusted for age, sex, the first 10 PCs of population structure, kinship, genotyping chip, and genetic ancestry.
Figure 4. MetaGRS and cumulative risk of incident intracerebral hemorrhage (ICH) in the population-based UK Biobank sample.
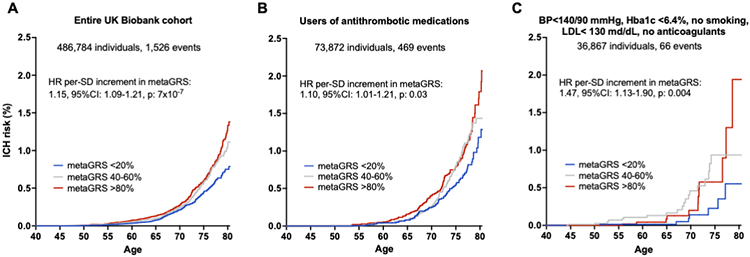
The results are derived from (A) entire UKBB population, (B) users of antithrombotic medications at baseline, and (C) low-risk individuals with conventional vascular risk factors under control and no use of anticoagulant medications after excluding prevalent cases of ICH. Depicted are the Kaplan-Meier curves for different metaGRS quantiles, as well as the hazard ratios (HR) per standard deviation (SD) increment in metaGRS, as derived from Cox proportional hazards regression models adjusted for baseline age, sex, the first 10 principal components of the population structure, genetic ancestry, genotyping chip, and kinship.
DISCUSSION
We developed a genomic risk score for ICH in a training dataset of 1,013 cases and 928 controls based on GWAS data for 21 ICH-related traits. We found the derived metaGRS to be significantly associated with the odds of ICH in an independent validation dataset of 842 ICH cases and 796 ICH-free controls. The metaGRS was independent of traditional clinical risk factors of ICH and improved model performance in prediction of ICH. Furthermore, the score was significantly associated with incident ICH risk in a population-based cohort study of 480,000 individuals followed-up for a median of 11 years (1,500 incident ICH events). Our results provide important insights into genomic prediction for ICH and could have implications for clinical practice.
First, the metaGRS identified individuals at very high risk for ICH. For example, individuals at the top percentile had almost 5-fold increased odds for ICH, as compared to the rest of the population. While it remains to be clarified how these individuals would benefit from potential primary preventive interventions, this information could be useful both for screening for hypertension, the main clinical risk factor for ICH, and early initiation of antihypertensive treatment, as well as for decision making when considering initiation of antiplatelet or anticoagulation treatments that might increase ICH risk. These risk stratification strategies based on genomic information are increasingly important as millions of persons in the US and around the world have been genotyped by direct-to-consumer genotyping companies.
Second, the metaGRS improved risk discrimination for ICH when compared to classical clinical predictors. Specifically, it was associated with ICH risk independently of vascular risk factors and was found to have a predictive value superior to all predictors except for education. The predictive power of the metaGRS was comparable to that of hypertension, the clinical risk factor for ICH that explains the most variance in the trait.33 These findings support the incorporation of genetic information into clinical tools aiming to quantify ICH risk within specific patient subgroups. A post-hoc analysis of trial data showed that among patients with atrial fibrillation and a CHA2-DS2-VASc score of 2, a high genomic risk score for ischemic stroke led to an absolute ischemic stroke risk equivalent to those with a higher score.34 Whether integration of a genomic risk score for ICH in such analyses could lead to a more precise assessment of the risk-benefit ratio for specific patients remains to be determined. Along these lines, the several clinical trials currently evaluating anticoagulation as a secondary prevention strategy after ICH constitute a unique opportunity for genomic-based risk-stratification, as a portion of them have built-in biobanks that are collecting DNA samples.
Third, despite its rarity, we found the metaGRS to be significantly associated with prospective ICH risk in the general population. As expected, the strength of association was attenuated in the relatively healthy population of UKBB, compared to the disease-focused case-control setting where the metaGRS was developed. The metaGRS was associated with a higher risk of ICH even among individuals with evidence-based control of relevant risk factors, who were not actively smoking, had blood pressure of 140/90 mmHg or less, no evidence of diabetes, normal BMI, and who reported no use of anticoagulants. While such analyses are restricted by lack of power, our results suggest that for individuals with a high genetic risk, the recommended treatment targets for modifiable risk factors might not be sufficient for primary ICH prevention. The importance of this observation lies on the fact that the genomic information is available long before risk factors are present and could thus be used for earlier risk stratification in otherwise low-risk individuals. Concomitantly, the metaGRS was also associated with a higher risk of ICH even among a high-risk group of individuals using antithrombotic medications, indicating its potential utility among a relevant group of patients for whom bedside calculation of ICH risk might be particularly relevant to clinical care.
Our study has limitations. First, the sample sizes of the available genetic datasets for ICH are limited, as compared to other clinical endpoints. This introduces uncertainty to the association estimates between the genetic variants and ICH risk, which were used to construct the metaGRS, and thus impacts negatively on its predictive performance. Indeed, when compared with metaGRS for other traits that have been developed in larger datasets, such as coronary artery disease and ischemic stroke,15,16 the association with incident ICH events is weaker. Additionally, while previous studies exist exploring the genetic architecture of ICH and cerebral small vessel disease that could potentially be used for construction and comparison of our genomic risk score, these were either derived from the same population sample as the one we use in the current study, are ICH GWAS in ancestrally diverse populations, or derived from candidate gene studies2,3,35. As such, utilizing the above studies to construct a GRS would result in significant over-estimation of association estimates13 and limit the GRS accuracy partly due to linkage disequilibrium and allele frequency differences across populations36, as well as by including a limited number of non-replicated loci37. Second, ICH is a phenotypically heterogeneous disease, with the most common etiologies being hypertensive small vessel disease (typically in non-lobar locations) and cerebral amyloid angiopathy (typically in lobar locations). To maximize the power of our approach, we have pooled cases, which could have negatively impacted the predictive performance for specific ICH etiologies. While our score was predictive for both non-lobar and lobar ICHs, developing etiology-specific scores might be of more relevance for specific clinical scenarios. Furthermore, in the training and the validation datasets, we did not account for dose of risk factors, due to either suboptimal phenotyping in the datasets, or to avoid including a larger number of clinical variables than already included, which would likely contribute to model instability, respectively. Third, while the metaGRS showed significant associations with risk of incident ICH in the UKBB, we could not explore its effects in concert with other clinical predictors, because the metaGRS was generated using datasets including data from the UKBB. Therefore, independent validation either of a score trained in an entirely UKBB-independent dataset or of the described metaGRS in another external cohort would be necessary. Fourth, the metaGRS was constructed solely based on data from individuals of European genetic ancestry, and may thus not be applicable for individuals of other ancestries. Larger multi-ethnic GWAS studies of ICH currently underway will facilitate the generation of ancestry-specific GWAS datasets. Furthermore, because our metaGRS is derived from genetic susceptibility to common vascular risk factors, it is likely to carry associations with other cardiovascular traits as well, despite being trained on ICH. This is a fundamental limitation of the technique, and our metaGRS is therefore not intended as a tool to identify risk of ICH at the exclusion of other endpoints, such as coronary artery disease. However, this expected pleiotropic effect of the metaGRS allows for pursuing follow-up research, focused on composite outcomes that share common biological underpinnings. Last, overarching limitations and challenges still exist on the generation and validation of GRS across disease states, which apply to our metaGRS as well. Some of them include the possible differences in sex-specific predictive performances, the translation of GRS estimates from the cohort- to the individual-specific level which has been suggested to introduce additional variability, as well as the heterogeneity of the different methods for GRS construction which could ultimately hinder clinical application.11,12 Towards that end, efforts are currently underway to standardize and delineate procedures surrounding GRS construction and reporting, such that prediction models incorporating GRS-based estimations can be leveraged in a consistent and reproducible manner.14
In conclusion, our study represents the first comprehensive attempt to develop and validate a genomic risk score for ICH. Our results demonstrate that the incorporation of genomic information in clinical prediction models for ICH could enhance predictive performance. As such, it lays the groundwork for future analyses in larger genetic datasets for ICH to optimally combine genomic information to maximize predictive benefit. Exploration of the performance of genomic risk scores for ICH in clinical trials of patients receiving antithrombotic medications could offer useful insights in risk prediction of ICH in this high-risk population with potential relevance for clinical decision making.
Supplementary Material
Acknowledgements
Funding Sources:
MKG is supported by a Walter-Benjamin fellowship from the German Research Foundation (Deutsche Forschungsgemeinschaft [DFG], GZ: GE 3461/1-1) and the FöFoLe program of LMU Munich (FöFoLe-Forschungsprojekt Reg.-Nr. 1120). HIH is suported by NIH R01HL138423, R01HL156024, R01HL138423-05S1 and 1R01AG072592. GJF is supported by the NIH (K76AG059992, R03NS112859), the American Heart Association (AHA) (18IDDG34280056 and 817874), and a pilot grant from the Claude D. Pepper Older Americans Independence Center at Yale (P30AG021342). JR and CDA are supported by NIH R01NS103924, U01NS069673, AHA 18SFRN34250007, and AHA-Bugher 21SFRN812095. Additional sources of funding for GOCHA, GERFHS, and EUR/ISGC are provided in the Supplemental Material.
Non-standard Abbreviations and Acronyms
- ICH
intracerebral hemorrhage
- GWAS
genome-wide association study
- GRS
genomic risk score
- SNP
single nucleotide polymorphism
- GOCHA
Genetics of Cerebral Hemorrhage on Anticoagulation
- GERFHS
Genetic and Environmental Risk Factors for Hemorrhagic Stroke
- EUR/ISGC
European member sites contributing to the International Stroke Genetics Consortium
- UKBB
UK Biobank
- QC
quality control
- PC
principal component
- HRC
Haplotype Reference Consortium
- MAF
minor allele frequency
- LD
linkage disequilibrium
- AUC
area under the receiving-operating characteristics curve
- CAD
coronary artery disease
- AIC
Akaike Information Criterion
- HES
hospital episode statistics
- SBP
systolic blood pressure
- DBP
diastolic blood pressure
- PP
pulse pressure
- WMH
white matter hyperintensities
- CKD
chronic kidney disease
- eGFR
estimated glomerular filtration rate
- UACR
urine albumin-to-creatinine ratio
- TC
total cholesterol
- TG
triglycerides
- LDL
low-density lipoprotein
- HDL
high-density lipoprotein
- T2D
type 2 diabetes mellitus
- HbA1c
hemoglobin A1c
- BMI
body mass index
- WHR
waist-to-hip ratio
- SVS
small vessel stroke
- EA
educational attainment
Footnotes
Disclosures: HIH has consulted for Acuta Capital, Novartis and Nutriglobal. GJF reports grants from NIH and grants from AHA. CDL reports grants from NIH. JR reports grants from NIH; compensation from National Football League for expert witness services; compensation from Takeda Development Center Americas, Inc. for consultant services; and grants from AHA. CDA has consulted for ApoPharma and Invitae; receives Sponsored Research Support from Bayer AG, AHA, and Massachusetts General Hospital; and reports grants from NIH. The other authors have nothing to disclose.
REFERENCES
- 1.Biffi A, Kuramatsu JB, Leasure A, Kamel H, Kourkoulis C, Schwab K, Ayres AM, Elm J, Gurol ME, Greenberg SM, et al. Oral Anticoagulation and Functional Outcome after Intracerebral Hemorrhage. Annals of neurology. 2017;82:755–765. doi: 10.1002/ana.25079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woo D, Falcone GJ, Devan WJ, Brown WM, Biffi A, Howard TD, Anderson CD, Brouwers HB, Valant V, Battey TW, et al. Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. Am J Hum Genet. 2014;94:511–521. doi: 10.1016/j.ajhg.2014.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung J, Marini S, Pera J, Norrving B, Jimenez-Conde J, Roquer J, Fernandez-Cadenas I, Tirschwell DL, Selim M, Brown DL, et al. Genome-wide association study of cerebral small vessel disease reveals established and novel loci. Brain. 2019;142:3176–3189. doi: 10.1093/brain/awz233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, Ntritsos G, Dimou N, Cabrera CP, Karaman I, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50:1412–1425. doi: 10.1038/s41588-018-0205-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giri A, Hellwege JN, Keaton JM, Park J, Qiu C, Warren HR, Torstenson ES, Kovesdy CP, Sun YV, Wilson OD, et al. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nature genetics. 2019;51:51–62. doi: 10.1038/s41588-018-0303-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann TJ, Ehret GB, Nandakumar P, Ranatunga D, Schaefer C, Kwok PY, Iribarren C, Chakravarti A, Risch N. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat Genet. 2017;49:54–64. doi: 10.1038/ng.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Surendran P, Drenos F, Young R, Warren H, Cook JP, Manning AK, Grarup N, Sim X, Barnes DR, Witkowska K, et al. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat Genet. 2016;48:1151–1161. doi: 10.1038/ng.3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wain LV, Vaez A, Jansen R, Joehanes R, van der Most PJ, Erzurumluoglu AM, O'Reilly PF, Cabrera CP, Warren HR, Rose LM, et al. Novel Blood Pressure Locus and Gene Discovery Using Genome-Wide Association Study and Expression Data Sets From Blood and the Kidney. Hypertension. 2017. doi: 10.1161/hypertensionaha.117.09438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warren HR, Evangelou E, Cabrera CP, Gao H, Ren M, Mifsud B, Ntalla I, Surendran P, Liu C, Cook JP, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017;49:403–415. doi: 10.1038/ng.3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nature Genetics. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert SA, Abraham G, Inouye M. Towards clinical utility of polygenic risk scores. Hum Mol Genet. 2019;28:R133–R142. doi: 10.1093/hmg/ddz187 [DOI] [PubMed] [Google Scholar]
- 12.Ding Y, Hou K, Burch KS, Lapinska S, Privé F, Vilhjálmsson B, Sankararaman S, Pasaniuc B. Large uncertainty in individual polygenic risk score estimation impacts PRS-based risk stratification. Nat Genet. 2022;54:30–39. doi: 10.1038/s41588-021-00961-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi SW, Mak TS, O'Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc. 2020;15:2759–2772. doi: 10.1038/s41596-020-0353-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wand H, Lambert SA, Tamburro C, Iacocca MA, O'Sullivan JW, Sillari C, Kullo IJ, Rowley R, Dron JS, Brockman D, et al. Improving reporting standards for polygenic scores in risk prediction studies. Nature. 2021;591:211–219. doi: 10.1038/s41586-021-03243-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abraham G, Malik R, Yonova-Doing E, Salim A, Wang T, Danesh J, Butterworth AS, Howson JMM, Inouye M, Dichgans M. Genomic risk score offers predictive performance comparable to clinical risk factors for ischaemic stroke. Nat Commun. 2019;10:5819. doi: 10.1038/s41467-019-13848-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inouye M, Abraham G, Nelson CP, Wood AM, Sweeting MJ, Dudbridge F, Lai FY, Kaptoge S, Brozynska M, Wang T, et al. Genomic Risk Prediction of Coronary Artery Disease in 480,000 Adults: Implications for Primary Prevention. J Am Coll Cardiol. 2018;72:1883–1893. doi: 10.1016/j.jacc.2018.07.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biffi A, Sonni A, Anderson CD, Kissela B, Jagiella JM, Schmidt H, Jimenez-Conde J, Hansen BM, Fernandez-Cadenas I, Cortellini L, et al. Variants at APOE influence risk of deep and lobar intracerebral hemorrhage. Ann Neurol. 2010;68:934–943. doi: 10.1002/ana.22134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo D, Sauerbeck LR, Kissela BM, Khoury JC, Szaflarski JP, Gebel J, Shukla R, Pancioli AM, Jauch EC, Menon AG, et al. Genetic and environmental risk factors for intracerebral hemorrhage: preliminary results of a population-based study. Stroke. 2002;33:1190–1195. doi: 10.1161/01.str.0000014774.88027.22 [DOI] [PubMed] [Google Scholar]
- 19.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler E, Leong A, Liu C-T, Hivert M-F, Strawbridge RJ, Podmore C, Li M, Yao J, Sim X, Hong J, et al. Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLOS Medicine. 2017;14:e1002383. doi: 10.1371/journal.pmed.1002383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pulit SL, Stoneman C, Morris AP, Wood AR, Glastonbury CA, Tyrrell J, Yengo L, Ferreira T, Marouli E, Ji Y, et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet. 2019;28:166–174. doi: 10.1093/hmg/ddy327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, Payne AJ, Steinthorsdottir V, Scott RA, Grarup N, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nature Genetics. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teumer A, Li Y, Ghasemi S, Prins BP, Wuttke M, Hermle T, Giri A, Sieber KB, Qiu C, Kirsten H, et al. Genome-wide association meta-analyses and fine-mapping elucidate pathways influencing albuminuria. Nature Communications. 2019;10:4130. doi: 10.1038/s41467-019-11576-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wuttke M, Li Y, Li M, Sieber KB, Feitosa MF, Gorski M, Tin A, Wang L, Chu AY, Hoppmann A, et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nature Genetics. 2019;51:957–972. doi: 10.1038/s41588-019-0407-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Persyn E, Hanscombe KB, Howson JMM, Lewis CM, Traylor M, Markus HS. Genome-wide association study of MRI markers of cerebral small vessel disease in 42,310 participants. Nat Commun. 2020;11:2175. doi: 10.1038/s41467-020-15932-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese A-K, van der Laan SW, Gretarsdottir S, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nature Genetics. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, Nguyen-Viet TA, Bowers P, Sidorenko J, Karlsson Linnér R, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nature Genetics. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–244. doi: 10.1038/s41588-018-0307-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wootton RE, Richmond RC, Stuijfzand BG, Lawn RB, Sallis HM, Taylor GMJ, Hemani G, Jones HJ, Zammit S, Davey Smith G, et al. Evidence for causal effects of lifetime smoking on risk for depression and schizophrenia: a Mendelian randomisation study. Psychol Med. 2019:1–9. doi: 10.1017/s0033291719002678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dashti HS, Jones SE, Wood AR, Lane JM, van Hees VT, Wang H, Rhodes JA, Song Y, Patel K, Anderson SG, et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nature Communications. 2019;10:1100. doi: 10.1038/s41467-019-08917-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane JM, Jones SE, Dashti HS, Wood AR, Aragam KG, van Hees VT, Strand LB, Winsvold BS, Wang H, Bowden J, et al. Biological and clinical insights from genetics of insomnia symptoms. Nat Genet. 2019;51:387–393. doi: 10.1038/s41588-019-0361-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Donnell MJ, Chin SL, Rangarajan S, Xavier D, Liu L, Zhang H, Rao-Melacini P, Zhang X, Pais P, Agapay S, et al. Global and regional effects of potentially modifiable risk factors associated with acute stroke in 32 countries (INTERSTROKE): a case-control study. Lancet. 2016;388:761–775. doi: 10.1016/S0140-6736(16)30506-2 [DOI] [PubMed] [Google Scholar]
- 34.Marston NA, Patel PN, Kamanu FK, Nordio F, Melloni GM, Roselli C, Gurmu Y, Weng LC, Bonaca MP, Giugliano RP, et al. Clinical Application of a Novel Genetic Risk Score for Ischemic Stroke in Patients With Cardiometabolic Disease. Circulation. 2021;143:470–478. doi: 10.1161/CIRCULATIONAHA.120.051927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wahab KW, Tiwari HK, Ovbiagele B, Sarfo F, Akinyemi R, Traylor M, Rotimi C, Markus HS, Owolabi M. Genetic risk of Spontaneous intracerebral hemorrhage: Systematic review and future directions. J Neurol Sci. 2019;407:116526. doi: 10.1016/j.jns.2019.116526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vilhjálmsson BJ, Yang J, Finucane HK, Gusev A, Lindström S, Ripke S, Genovese G, Loh PR, Bhatia G, Do R, et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am J Hum Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosand J Genetic Risk Factors for Medication-Related Hemorrhagic Stroke. Appendix 1d. In; Version 6, November/26/2010, Amendment: 5. [Google Scholar]
- 39.Gomis M, Ois A, Rodriguez-Campello A, Cuadrado-Godia E, Jiménez-Conde J, Subirana I, Dávalos A, Roquer J. Outcome of intracerebral haemorrhage patients pre-treated with statins. European journal of neurology. 2010;17:443–448. [DOI] [PubMed] [Google Scholar]
- 40.Domingues-Montanari S, Hernandez-Guillamon M, Fernandez-Cadenas I, Mendioroz M, Boada M, Munuera J, Rovira A, Maisterra O, Parés M, Gutierrez M. ACE variants and risk of intracerebral hemorrhage recurrence in amyloid angiopathy. Neurobiology of aging. 2011;32:551. e513–551. e522. [DOI] [PubMed] [Google Scholar]
- 41.Pera J, Slowik A, Dziedzic T, Pulyk R, Wloch D, Szczudlik A. Glutathione peroxidase 1 C593T polymorphism is associated with lobar intracerebral hemorrhage. Cerebrovascular Diseases. 2008;25:445–449. [DOI] [PubMed] [Google Scholar]
- 42.Hallstroöm Br, Joönsson A-C, Nerbrand C, Norrving B, Lindgren A. Stroke incidence and survival in the beginning of the 21st century in southern Sweden: comparisons with the late 20th century and projections into the future. Stroke. 2008;39:10–15. [DOI] [PubMed] [Google Scholar]
- 43.Seifert T, Lechner A, Flooh E, Schmidt H, Schmidt R, Fazekas F. Lack of association of lobar intracerebral hemorrhage with apolipoprotein E genotype in an unselected population. Cerebrovasc Dis. 2006;21:266–270. doi: 10.1159/000091225 [DOI] [PubMed] [Google Scholar]
- 44.Anderson CD, Falcone GJ, Phuah CL, Radmanesh F, Brouwers HB, Battey TW, Biffi A, Peloso GM, Liu DJ, Ayres AM, et al. Genetic variants in CETP increase risk of intracerebral hemorrhage. Ann Neurol. 2016;80:730–740. doi: 10.1002/ana.24780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O'Connell J, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG, Donnelly P, Eichler EE, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raîche G, Walls TA, Magis D, Riopel M, Blais J-G. Non-graphical solutions for Cattell’s scree test. Methodology: European Journal of Research Methods for the Behavioral and Social Sciences. 2013;9:23–29. doi: 10.1027/1614-2241/a000051 [DOI] [Google Scholar]
- 49.Falcone GJ, Kirsch E, Acosta JN, Noche RB, Leasure A, Marini S, Chung J, Selim M, Meschia JF, Brown DL, et al. Genetically Elevated LDL Associates with Lower Risk of Intracerebral Hemorrhage. Annals of Neurology. 2020;88:56–66. doi: 10.1002/ana.25740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Georgakis MK, Gill D, Webb AJS, Evangelou E, Elliott P, Sudlow CLM, Dehghan A, Malik R, Tzoulaki I, Dichgans M. Genetically determined blood pressure, antihypertensive drug classes, and risk of stroke subtypes. Neurology. 2020;95:e353–e361. doi: 10.1212/wnl.0000000000009814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harshfield EL, Georgakis MK, Malik R, Dichgans M, Markus HS. Modifiable lifestyle factors and risk of stroke: a Mendelian randomization analysis. medRxiv. 2020:2020.2003.2017.20037549. doi: 10.1101/2020.03.17.20037549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marini S, Georgakis MK, Chung J, Henry JQA, Dichgans M, Rosand J, Malik R, Anderson CD. Genetic overlap and causal inferences between kidney function and cerebrovascular disease. Neurology. 2020;94:e2581–e2591. doi: 10.1212/wnl.0000000000009642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marini S, Merino J, Montgomery BE, Malik R, Sudlow CL, Dichgans M, Florez JC, Rosand J, Gill D, Anderson CD, et al. Mendelian Randomization Study of Obesity and Cerebrovascular Disease. Annals of Neurology. 2020;87:516–524. doi: 10.1002/ana.25686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venables WN, Ripley BD. Modern Applied Statistics with S. Springer Publishing Company, Incorporated; 2010. [Google Scholar]
- 55.Friedman JH, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. 2010. 2010;33:22. doi: 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Falcone Guido J, Biffi A, Devan William J, Jagiella Jeremiasz M, Schmidt H, Kissela B, Hansen Björn M, Jimenez-Conde J, Giralt-Steinhauer E, Elosua R, et al. Burden of Risk Alleles for Hypertension Increases Risk of Intracerebral Hemorrhage. Stroke. 2012;43:2877–2883. doi: 10.1161/STROKEAHA.112.659755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kittner SJ, Sekar P, Comeau ME, Anderson CD, Parikh GY, Tavarez T, Flaherty ML, Testai FD, Frankel MR, James ML, et al. Ethnic and Racial Variation in Intracerebral Hemorrhage Risk Factors and Risk Factor Burden. JAMA Netw Open. 2021;4:e2121921. doi: 10.1001/jamanetworkopen.2021.21921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lioutas VA, Beiser AS, Aparicio HJ, Himali JJ, Selim MH, Romero JR, Seshadri S. Assessment of Incidence and Risk Factors of Intracerebral Hemorrhage Among Participants in the Framingham Heart Study Between 1948 and 2016. JAMA Neurol. 2020;77:1252–1260. doi: 10.1001/jamaneurol.2020.1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marini S, Crawford K, Morotti A, Lee MJ, Pezzini A, Moomaw CJ, Flaherty ML, Montaner J, Roquer J, Jimenez-Conde J, et al. Association of Apolipoprotein E With Intracerebral Hemorrhage Risk by Race/Ethnicity: A Meta-analysis. JAMA Neurol. 2019;76:480–491. doi: 10.1001/jamaneurol.2018.4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. Journal of the American statistical association. 1999;94:496–509. [Google Scholar]
- 61.Team RC. R: A language and environment for statistical computing. 2013.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study will be available from the corresponding author upon reasonable request. MetaGRS single nucleotide polymorphism (SNP)-specific weights will be made publicly available at The Polygenic Score (PGS) Catalog.