Abstract
The dataset provided with this article describes a targeted lipidomics analysis performed on the serum of COVID-19 patients characterized by different degree of severity. As the ongoing pandemic has posed a challenging threat for humanity, the data here presented belong to one of the first lipidomics studies carried out on COVID-19 patients’ samples collected during the first pandemic waves. Serum samples were obtained from hospitalized patients with a molecular diagnosis of SARS-CoV-2 infection detected after nasal swab, and categorized as mild, moderate, or severe according to pre-established clinical descriptors. The MS-based targeted lipidomic analysis was performed by MRM using a Triple Quad 5500+ mass spectrometer, and the quantitative data were acquired on a panel of 483 lipids. The characterization of this lipidomic dataset has been outlined using multivariate and univariate descriptive statistics and bioinformatics tools.
Keywords: Lipidomics, COVIDomics, COVID-19 research, Serum lipidome, Lipid metabolism, Lipids mass spectrometry, Targeted metabolomics, Multiple reaction monitoring
Specifications Table
| Subject | Omics: lipidomics Biochemistry |
| Specific subject area | Lipidomics analysis of COVID-19 serum samples. |
| Type of data | Chart Graph Figure |
| How the data were acquired | The data were acquired by FIA-MS/MS in the MRM mode using a Triple Quad 5500+ System. |
| Data format | Raw Analyzed Filtered |
| Description of data collection | Serum samples were collected from COVID-19 patients with different degree of severity, categorized as mild (n = 20), moderate (n = 16), or severe (n = 17). Mass spectrometry data were acquired using a targeted lipidomics platform and quantitative analysis of lipids was performed using internal standards. The presence of sample or analyte outliers has been verified using statistical methods. Data were normalized by calculating the sum of original concentrations per lipid class over the number of subjects, or using the log10-transformation and the Pareto scaling method. |
| Data source location | CEINGE–Biotecnologie Avanzate Franco Salvatore s.c.ar.l, Naples 80145, Italy |
| Data accessibility | The raw MS files have been deposited to the MetaboLights repository with the study identifier MTBLS6844. The files are available at the following link: www.ebi.ac.uk/metabolights/MTBLS6844. |
| Related research article | M. Caterino, M. Gelzo, S. Sol, R. Fedele, A. Annunziata, C. Calabrese, G. Fiorentino, M. D'Abbraccio, C. Dell'Isola, F.M. Fusco, R. Parrella, G. G. Fabbrocini, I. Gentile, I. Andolfo, M. Capasso, M. Costanzo, A. Daniele, E. Marchese, R. Polito, R. Russo, C. Missero, M. Ruoppolo, G. Castaldo. Dysregulation of lipid metabolism and pathological inflammation in patients with COVID-19. Sci Rep (2021). 11(1):2941. doi:10.1038/s41598-021-82,426-7. |
Value of the Data
-
•
This dataset refers to one of the first mass spectrometry-based targeted lipidomics analyses on serum samples from COVID-19 patients
-
•
The lipidome analyzed here is related to patients with different degree of severity
-
•
Researchers studying the impact of COVID-19 on lipid metabolism can benefit from these data
-
•
These data can help improving the classification system of COVID-19 disease and the identification of putative therapeutic targets
1. Objective
The pandemic of coronavirus disease 2019 (COVID-19) caused by infection of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been recognized as a global health threat, affecting several million individuals worldwide.
As part of the original research previously conducted by Caterino et al. [1], the need for the identification of metabolic processes perturbed upon SARS-CoV-2 infection in humans led to the acquisition of the present lipidomics dataset, among many others for COVIDomics research [2,3]. While several attempts to sustain the severe effects of the disease on human health have been made, including drug repurposing and vaccines development, effective therapeutic options are seriously needed [4]. In fact, disturbed pathways or dysregulated molecules can be pharmacologically targeted to prevent serious sequelae in affected patients.
This data article aims at providing a deeper description of the serum lipidome of COVID-19 patients at different stages of severity, showing raw and normalized concentrations data and including visual quantitative comparisons of lipids between the three groups of patients that were categorized according to several clinical descriptors as mild, moderate, and severe.
2. Data Description
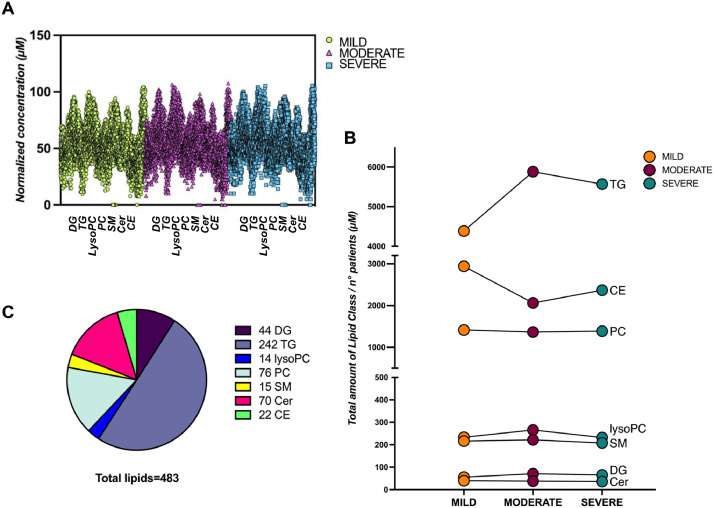
The data here reported are related to a study conducted by Caterino et al. on the lipidome of serum samples obtained from hospitalized COVID-19 patients [1]. After molecular diagnosis of SARS-CoV-2 infection, patients were categorized as mild (n = 20), moderate (n = 16), or severe (n = 17) and their sera were analyzed by mass spectrometry (MS)-based targeted lipidomics. The data were acquired on a triple quadrupolar mass spectrometer, and quantitative insights were obtained by calculating lipids concentrations after targeted analysis by Multiple Reaction Monitoring (MRM). The MS-targeted analysis allowed the identification and quantification of a panel of 483 lipids, classified as diacylglycerols (DG), triacylglycerols (TG), lysophosphatidylcholines (lysoPC), phosphatidylcholines (PC), sphingomyelins (SM), ceramides (Cer), and cholesterol esters (CE). The raw concentration values (µM) of the lipids measured by MS in each replicate of the experimental conditions analyzed are available as supplementary material of the related research article [1]. Further, the raw MS files have been made publicly available into the MetaboLights repository with the study identifier MTBLS6844 (www.ebi.ac.uk/metabolights/MTBLS6844) [5]. The distribution of lipid concentrations per class, obtained by averaging the replicates and normalizing the means in each analyzed condition (mild, moderate, severe), did not show any particular outlier, with comparable concentration ranges for all the lipid classes analyzed (Fig. 1a). The total amount of each lipid class was calculated as the sum of the lipid concentrations within that class, and normalized with respect to the number of subjects included in each experimental condition. These results show the trend of abundance of each lipid class in the mild, moderate, and severe patients’ serum lipidomes (Fig. 1b).
Fig. 1.
Visualization of the distribution of COVID-19 serum lipidomics data. A) The concentration of lipids was normalized by averaging the replicates and normalizing the means in each analyzed condition (mild, moderate, severe) for each lipid class. B) The total amount of each lipid class was calculated as the sum of the lipid concentrations within that class, and divided by the number of patients included in each experimental condition. The increasing or decreasing trend for each lipid class can be observed in mild, moderate and severe patients. C) Pie chart describing the composition of the lipid panel analyzed by MS-MRM-based lipidomics, with detail of the number of analytes for each class.
The distribution of each lipid class over the total of 483 lipid molecules measured shows the following numerosity assignment: DG=44 molecules, TG=242 molecules, lysoPC=14 molecules, PC=76 molecules, SM=15 molecules, Cer=70 molecules, CE=22 molecules (Fig. 1c).
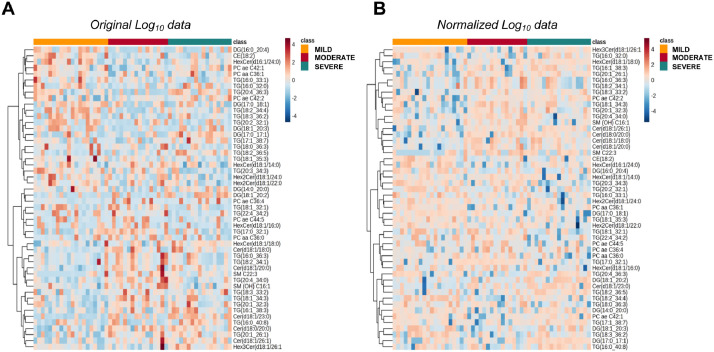
Lipid concentrations were employed to build heatmaps within MetaboAnalyst 5.0 software to visualize the distribution of analytes abundance within the replicates of the same condition class and between the three classes. Fig. 2a shows the distribution of the top-50 lipids with original log10-transformed concentrations, while the log10-transformed and normalized data were used for visualization in Fig. 2b.
Fig. 2.
Heatmaps showing the serum abundance of the top-50 lipids in the mild, moderate, and severe COVID-19 patients using (A) original log10 concentrations and (B) normalized log10 concentrations.
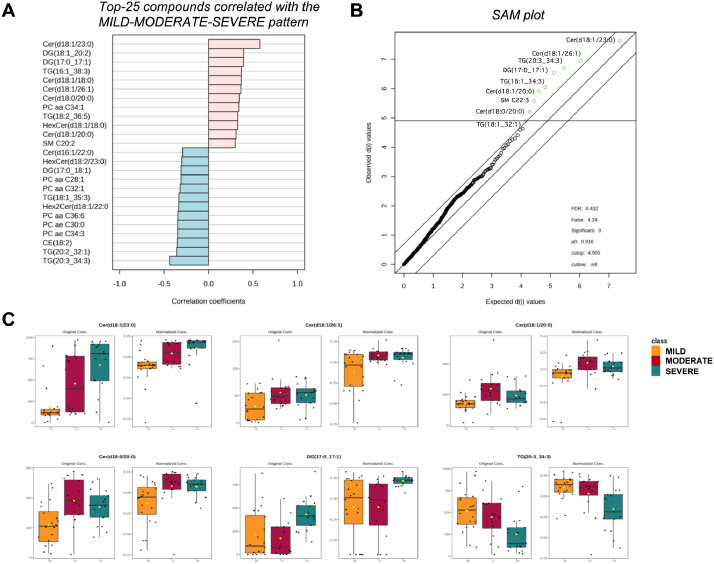
The lipidomic data were further used for pattern search analysis to select the top-25 compounds with a pattern of increasing concentration from mild-to moderate-to severe COVID-19, and of decreasing concentration from severe-to moderate-to mild COVID-19, for which the Cer(d18:1/23:0) showed a significant positive correlation (correlation coefficient >0.5) (Fig. 3a). Significance Analysis of Metabolomics (SAM) identified 9 significant features that are the ones that mostly deviate from the “observed-expected d line” (Fig. 3b). SAM analysis confirmed that six out of the nine molecules overlapped with those found in the pattern analysis, namely Cer(d18:1/23:0), Cer(d18:1/26:1) Cer(d18:1/20:0), Cer(d18:0/20:0), DG(17:0_17:1), TG(20:3_34:3). The plots relative to these six molecules were reported for the mild, moderate, and severe conditions using both original and normalized concentration values (Fig. 3c).
Fig. 3.
Correlation analysis of significant lipids across the mild, moderate, and severe COVID-19 patients. A) The PatternHunter tool of MetaboAnalyst was used to search a crescent pattern of concentration in the order mild-to moderate-to severe and viceversa, with significant correlation coefficients >0.5 or <–0.5. B) Significance Analysis of Metabolites (SAM) plot depicts the observed relative differences versus the expected relative differences of advanced significance metabolites estimated by data permutation. The solid (central) diagonal line indicates where these two measures are the same. The dotted (lateral) lines are drawn at a distance of a delta from the solid line. With a given delta of 0.7, nine metabolites highlighted as green dots were identified to be significant (q-value <0.432). C) Six selected significant metabolites in common between pattern correlation and SAM analyses were plotted as function of their original and normalized concentration.
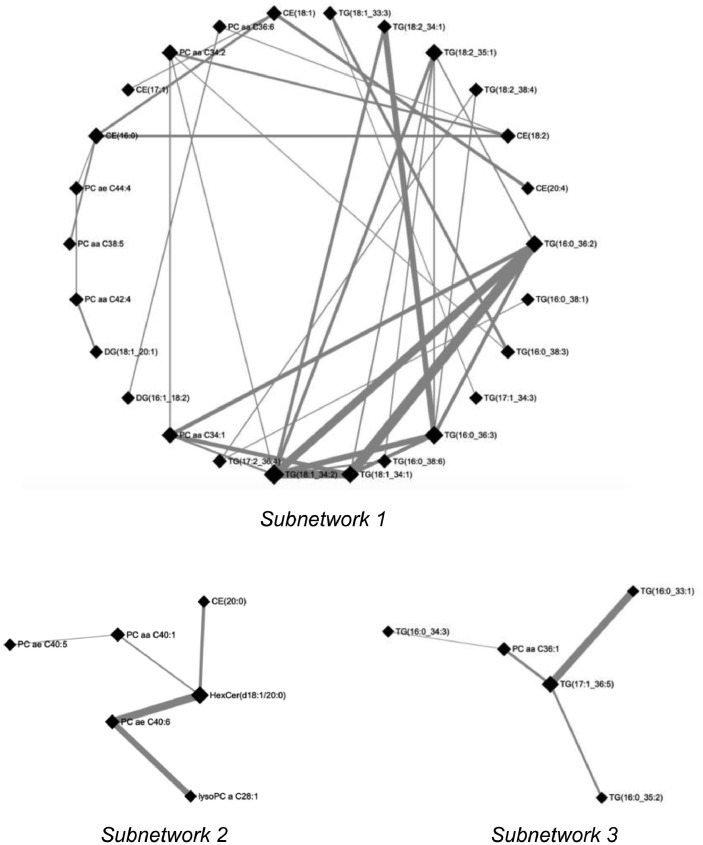
Finally, a lipid-lipid correlation network was built using the Debiased Sparse Partial Correlation (DSPC) network function within MetaboAnalyst 5.0 software. In particular, the three most dense subnetworks were selected and reported in Fig. 4, as Subnetwork 1 (26 nodes, 36 edges), Subnetwork 2 (6 nodes, 5 edges), Subnetwork 3 (5 nodes, 4 edges). In the figure, the nodes are input metabolites, and the edges represent measures of their association, whereas thicker edges are characterized by lower p-values associated within those connections.
Fig. 4.
The Debiased Sparse Partial Correlation (DSPC) network function was used to select the three most dense lipid-lipid connection subnetworks.
3. Experimental Design, Materials and Methods
3.1. Patients recruitment
Hospitalized COVID-19 patients (n = 53) were enrolled and categorized in three severity groups as mild (n = 20), moderate (n = 16), and severe (n = 17), on the basis of a seven-point ordinal scale that includes signs and clinical descriptors [1]. The study protocol (n°191/20) was approved by Ethics Committee at the University of Naples Federico II. All methods and experimental procedures were performed in accordance with the relevant guidelines and regulations included in the above-mentioned protocol. The presented work was carried out in agreement with the Declaration of Helsinki. This study did not involve human subjects under the age of 18 years. Each patient (and/or legal guardian) provided fully informed consent to the participation and the use of their biological samples for research purposes.
3.2. Targeted lipidomics analysis
The serum lipidome of COVID-19 patients was analyzed using a MS-based metabolomics platform with the application of a targeted approach [6], [7], [8]. The lipidomic characterization of sera was performed employing the standardized protocols of the MxP Quant 500 kit (Biocrates Life Sciences Innsbruck, Austria) coupled with MS analysis [9]. Blanks, seven calibration standards, three levels of quality control samples (human-based QCs), and patients’ sera were processed, run, and analyzed by direct flow injection analysis–tandem mass spectrometry (FIA-MS/MS) in the MRM mode to target and quantify analytes from the different available lipid classes [10]. The MS platform used was composed by a SCIEX 5500+ QTRAP (AB Sciex, Framingham, MA, USA) mass spectrometer coupled with a 1260 Infinity II HPLC (Agilent Technologies, Santa Clara, CA, USA). COVID-19 samples were prepared and run in triplicates to reduce the analytical variability.
In brief, the analytical procedure required that 10 µL of the samples were directly added onto a 96-well plate, and dried under nitrogen for 30 min. A sample derivatization step of 1 hour was then performed using 50 µL of 5% phenyl isothiocyanate (PITC), followed by another drying step of 1 hour. Further, 300 µL of extraction solvent (5 mM ammonium acetate/methanol) was added and the plate was shaken for 30 min (450 rpm). The extract content was eluted by centrifugation and 10 µL of each extract were diluted with 490 µL of FIA solvent.
Sample volumes of 20 µL were injected into the system within the mobile phase (FIA solvent) at a flow rate of 0.03 mL/min until 1.6 min, followed by flow rates of 0.20 mL/min for 1.6 min and 0.02 mL/min for 0.20 min. The electrospray ionization (ESI) source was set in positive ion mode with the following parameters: spray voltage=5.5 kV, temperature=450 °C, GS1=20 psi, GS2=40 psi, CUR=30 psi, CAD=8 psi. For quantitation, FIA data were analyzed using the MetIDQ Oxygen software (Biocrates Life Sciences Innsbruck, Austria). The final dataset was composed of concentration values (µM) for the 483 lipids targeted by MRM.
3.3. Features selection, bioinformatics, data visualization
Several multiomics and bioinformatics tools can provide information regarding the distribution and quality of acquired omics data. In this case, the serum concentration values of 483 lipids from COVID-19 patients were processed by univariate and multivariate statistics and bioinformatics using GraphPad Prism 9.0 and MetaboAnalyst 5.0 platforms [11], [12], [13], [14], [15]. In Prism, the replicates of each analyzed condition (mild, moderate, severe) were averaged and their means normalized. In addition, the total amount of each lipid class was obtained by calculating the sum of original lipid concentrations divided by the number of subjects representative of that condition.
In MetaboAnalyst, the lipidomic dataset underwent imputation to replace the missing values (missing values being replaced by 1/5 of minimum positive values of their corresponding variables), data were log10-transformed and scaled using the Pareto scaling method. Heatmaps were obtained using both original concentrations and normalized values as data source, with the setting of Euclidean as distance measure and Ward as clustering method.
The PatternHunter function was used to select a predefined pattern of concentration in the three conditions, and find significant correlations between analytes and the pattern chosen. In this case, the pattern mild-moderate-severe was selected with the Spearman rank correlation as distance measure. Other significant analytes were selected using the Significance Analysis of Microarrays or Significance Analysis of Metabolomics (SAM) [16] set with a default delta score=0.7. Finally, the Debiased Sparse Partial Correlation (DSPC) network function was employed to build a functional lipid-lipid connection network that provides association between several lipids [17]. With respect to the present lipidome dataset, the three most dense lipid-lipid interaction subnetworks were extracted.
Ethics Statements
Fully informed consent was obtained from the subjects participating in this study after approval of the Ethics Committee at the University of Naples Federico II (protocol n°191/20). The research was carried out in accordance with the Declaration of Helsinki.
CRediT authorship contribution statement
Michele Costanzo: Conceptualization, Methodology, Software, Writing – review & editing. Marianna Caterino: Conceptualization, Methodology, Software, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by the project “CEINGE TASK-FORCE COVID19”, code D64I200003800 by Regione Campania for the fight against Covid-19 (DGR n.140, 17–03–2020).
Data Availability
Dysregulation of lipid metabolism and pathological inflammation in patients with COVID-19 (Original data) (Supplementary Information 3).
References
- 1.Caterino M., Gelzo M., Sol S., Fedele R., Annunziata A., Calabrese C., Fiorentino G., D'Abbraccio M., Dell'Isola C., Fusco F.M., Parrella R., Fabbrocini G., Gentile I., Andolfo I., Capasso M., Costanzo M., Daniele A., Marchese E., Polito R., Russo R., Missero C., Ruoppolo M., Castaldo G. Dysregulation of lipid metabolism and pathological inflammation in patients with COVID-19. Sci. Rep. 2021;11:2941. doi: 10.1038/s41598-021-82426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costanzo M., Caterino M., Fedele R., Cevenini A., Pontillo M., Barra L., Ruoppolo M. COVIDomics: the proteomic and metabolomic signatures of COVID-19. Int. J. Mol. Sci. 2022;23:2414. doi: 10.3390/ijms23052414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis H.-M., Liu Y., Frampas C.F., Longman K., Spick M., Stewart A., Sinclair E., Kasar N., Greener D., Whetton A.D., Barran P.E., Chen T., Dunn-Walters D., Skene D.J., Bailey M.J. Metabolomics markers of COVID-19 are dependent on collection wave. Metabolites. 2022;12:713. doi: 10.3390/metabo12080713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costanzo M., De Giglio M.A.R., Roviello G.N. Anti-Coronavirus vaccines: past investigations on SARS-CoV-1 and MERS-CoV, the approved vaccines from BioNTech/Pfizer, Moderna, Oxford/AstraZeneca and others under development against SARS-CoV-2 infection. Curr. Med. Chem. 2021:28. doi: 10.2174/0929867328666210521164809. [DOI] [PubMed] [Google Scholar]
- 5.Haug K., Cochrane K., Nainala V.C., Williams M., Chang J., Jayaseelan K.V., O'Donovan C. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costanzo M., Caterino M., Ruoppolo M. Targeted metabolomics, in: Metabolomics Perspect. From Theory to Pract. Appl. 2022:219–236. doi: 10.1016/B978-0-323-85062-9.00006-4. [DOI] [Google Scholar]
- 7.Zhang N.R., Hatcher N.G., Ekroos K., Kedia K., Kandebo M., Marcus J.N., Smith S.M., Bateman K.P., Spellman D.S. Validation of a multiplexed and targeted lipidomics assay for accurate quantification of lipidomes. J. Lipid Res. 2022;63 doi: 10.1016/j.jlr.2022.100218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sethi S., Brietzke E. Recent advances in lipidomics: analytical and clinical perspectives. Prostaglandins Other Lipid Mediat. 2017:128–129. doi: 10.1016/j.prostaglandins.2016.12.002. 8–16. [DOI] [PubMed] [Google Scholar]
- 9.Viallon V., His M., Rinaldi S., Breeur M., Gicquiau A., Hemon B., Overvad K., Tjønneland A., Rostgaard-Hansen A.L., Rothwell J.A., Lecuyer L., Severi G., Kaaks R., Johnson T., Schulze M.B., Palli D., Agnoli C., Panico S., Tumino R., Ricceri F., Verschuren W.M.M., Engelfriet P., Onland-Moret C., Vermeulen R., Nøst T.H., Urbarova I., Zamora-Ros R., Rodriguez-Barranco M., Amiano P., Huerta J.M., Ardanaz E., Melander O., Ottoson F., Vidman L., Rentoft M., Schmidt J.A., Travis R.C., Weiderpass E., Johansson M., Dossus L., Jenab M., Gunter M.J., Bermejo J.Lorenzo, Scherer D., Salek R.M., Keski-Rahkonen P., Ferrari P. A new pipeline for the normalization and pooling of metabolomics data. Metabolites. 2021;11:631. doi: 10.3390/metabo11090631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue X., Liu W., Liu Y., Shen M., Zhai Y., Ma Z., Cao Z. Development, validation, and clinical application of an FIA-MS/MS method for the quantification of lysophosphatidylcholines in dried blood spots. J. Clin. Lab. Anal. 2022;36 doi: 10.1002/jcla.24099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costanzo M., Caterino M., Salvatori I., Manganelli V., Ferri A., Misasi R., Ruoppolo M. Proteome data of neuroblastoma cells overexpressing Neuroglobin. Data Br. 2022;41 doi: 10.1016/j.dib.2022.107843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costanzo M., Fiocchetti M., Ascenzi P., Marino M., Caterino M., Ruoppolo M. Proteomic and bioinformatic investigation of altered pathways in neuroglobin-deficient breast cancer cells. Molecules. 2021;26:2397. doi: 10.3390/molecules26082397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia J., Psychogios N., Young N., Wishart D.S. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez Melo M., Remacle N., Cudré-Cung H.P., Roux C., Poms M., Cudalbu C., Barroso M., Gersting S.W., Feichtinger R.G., Mayr J.A., Costanzo M., Caterino M., Ruoppolo M., Rüfenacht V., Häberle J., Braissant O., Ballhausen D. The first knock-in rat model for glutaric aciduria type I allows further insights into pathophysiology in brain and periphery. Mol. Genet. Metab. 2021;133:157–181. doi: 10.1016/j.ymgme.2021.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Costanzo M., Caterino M., Cevenini A., Jung V., Chhuon C., Lipecka J., Fedele R., Guerrera I.C., Ruoppolo M. Dataset of a comparative proteomics experiment in a methylmalonyl-CoA mutase knockout HEK 293 cell model. Data Br. 2020;33 doi: 10.1016/j.dib.2020.106453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alakwaa F.M., Savelieff M.G. Bioinformatics analysis of metabolomics data unveils association of metabolic signatures with methylation in breast cancer. J. Proteome Res. 2020;19:2879–2889. doi: 10.1021/acs.jproteome.9b00755. [DOI] [PubMed] [Google Scholar]
- 17.Pang Z., Chong J., Zhou G., de Lima Morais D.A., Chang L., Barrette M., Gauthier C., Jacques P.-É., Li S., Xia J. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Dysregulation of lipid metabolism and pathological inflammation in patients with COVID-19 (Original data) (Supplementary Information 3).