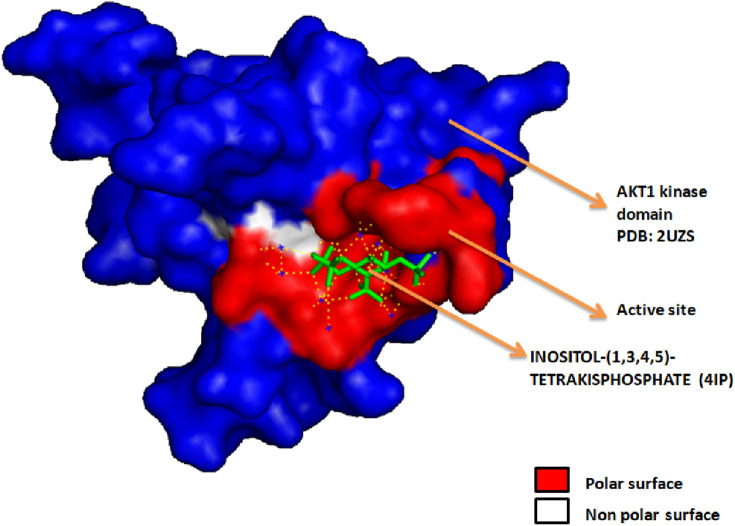
Abstract
Prostate cancer is the second most fatal malignancy in men after lung cancer, and the fifth leading cause of death. Piperine has been utilized for its therapeutic effects since the time of Ayurveda. According to traditional Chinese medicine, piperine has a wide variety of pharmacological effects, including anti-inflammatory, anti-cancer, and immune-regulating properties. Based on the previous study, Akt1 (protein kinase B) is one of the targets of piperine, it belongs to the group of oncogenes and the mechanism of the Akt1 is an interesting approach for anticancer drug design. From the peer-reviewed literature, five piperine analogs were identified altogether, and a combinatorial collection was formed. However, may not be entirely clear how piperine analogs work to prevent prostate cancer. In the present study, serine-threonine kinase domain Akt1 receptor was employed to analyze the efficacy of piperine analogs against standards using in silico methodologies. Additionally, their drug-likeness was evaluated utilizing online servers like Molinspiration and preADMET. Using AutoDock Vina, the interactions of five piperine analogs and two standards with Akt1 receptor was investigated. Our study reveals that piperine analog-2 (pip2) shows highest binding affinity (− 6.0 kcal/mol) by forming 6 hydrogen bonds with more hydrophobic interactions compared to other four analogs and standards. In conclusion, the piperine analog pip2, which shows strong inhibition affect in Akt1-cancer pathway, may be employed as chemotherapeutic drugs.
Graphical abstract
Keywords: Piperine, Akt1, preADMET, pip2, In silico methods, AutoDock Vina
Introduction
Prostate cancer is the second most fatal malignancy in men after lung cancer and the fifth leading cause of death. The primary cause of cancer in males is prostate cancer (PCa), which is brought on by the uncontrolled proliferation of abnormal cells in the prostate gland (Cai Jian et al. 2022). As per a World Health Organization estimate, there will be 10.0 million cancer-related fatalities in 2020. Around 70% of fatalities in low- and middle-income nations are thought to be caused by cancer (Mathers 2020; Sung et al. 2021). PCa is a complex and varied disease that can be classified as either aggressive or non-aggressive cancer (Nguyen et al. 2016). The typical age of diagnosis for males with PCa is 66, and approximately 6 in 10 receive therapy beyond the age of 65 (Siegel et al. 2020). There is a huge demand for innovative anticancer medications with minimal side effects, and spices are a promising source and may be utilized to treat chemotherapeutics (Atanasov et al. 2015). Researchers have discovered encouraging findings in this field, and new research is being carried out to find novel drugs (Krzyszczyk et al. 2018).
Piperine (1-[5-[1,3-benzodioxol-5-yl]-1-oxo-2,4-pentadienyl]) also known as Black pepper (Piper nigrum), of the cinnamamides group is the source of piperidine, a nitrogenous natural plant alkaloid (Salehi et al. 2019). The anti-microbial, anti-fungal, antioxidant, anti-apoptotic, anti-depressant, anti-oxidant, and anti-inflammatory effects of this substance are significant physiological and phytoconstituents ones. Piperine has a methylenedioxyphenyl (MDP) ring, a side chain containing conjugated double bonds, and a piperidine moiety attached to a side chain by a carbonyl amide linkage (Kumar et al. 2017; Li et al. 2007; Shrivastava et al. 2013; Hammad et al. 2017). Among the several specific ligands under investigation, we concentrated on piperine in the current work due to its superior pharmacokinetics and five Piperine analogs that demonstrated stronger binding affinity, and it was thought to be the most promising drug. Based on the literature, focusing on the Piperine core scaffold, we chose 5 Piperine analogs. Except for pip1, all of the piperine analogs employed in this study had two benzene rings in their structure, as well as alkynes, amino, sulfonamide, hydroxyl, nitro, and methyl groups (Zarai et al. 2013; Mittal et al. 2000; Yu et al. 2021).
Literature studies, however, point to the critical role that aberrant Akt activation plays in improving survival and preventing apoptosis in a number of cancer model systems (Cheung et al. 2013). Likewise, several experimental and clinical investigations on PCa have found a clear correlation between elevated Akt activity and the development of malignant phenotypes (Bedolla et al. 2007; Makhov et al. 2012). In addition, it is understood that the Akt family consists of three members, Akt1, Akt2, and Akt3, each of which possesses a highly conserved serine-threonine kinase domain. One of these, Akt1, is thought to be crucial in controlling the signaling pathway that drives the growth of malignant tumors, including PCa (Zinda et al. 2001; Pandya et al. 2021), the same has been proved in our previous study also (Prakash et al. 2022). The effect of Akt1 action on piperine analogs and the prostate cancer target therapy is discussed in this paper utilizing an in silico technique.
Drug development requires much time, money, and involves multidisciplinary research. Thus, in silico investigations have been included to the process of assessing a drug candidate in order to save time and expenses. Nowadays, effective methods for studying the crucial variables needed to assess a compound’s chemical and physical qualities are known as in silico methods (Sattarinezhad et al. 2015). As these factors affect pharmacokinetic characteristics, the primary goal of the investigations is to prevent irrational expenditures related to compound biological tests. In this study, the structural and physicochemical parameters are compared with a vast number of experimental findings. Using these characteristics, in silico computer-assisted screenings may be carried out (Brito et al. 2011). The ligand-macromolecule interactions involving structurally particular medicines are the focus of the pharmacodynamic investigation. The pharmacodynamic properties of the ligand molecules are extremely useful in determining the biological efficacy of piperine analogs against prostate cancer. Virtual screening was used to carry out a molecular docking strategy in search of the best piperine analogue against the standard and serine-threonine kinase domain Akt1 protein. All piperine analogs were examined for effective protein–ligand interactions with the Akt1 receptor and compared to commonly prescribed medications of PCa. This aids in the subsequent research of potential lead compounds that result in powerful new PCa inhibitors that may prevent PCa.
In ligand-based pharmacophore modeling, 3D molecules of two or more ligands are aligned on aspects of the training molecules common pharmacophore. It is necessary to acknowledge that each of the pharmacophore shared chemical characteristics is crucial (Prabhavathi et al. 2021). A pharmacophore model is checked for compounds that map to it and meet its criteria as well as being highly likely to be active during experimental testing. In the current work, pharmacophore modeling is used to uncover powerful drug that act as PCa inhibitor.
This research purpose is to identify possible hits for piperine analogs against serine-threonine kinase domain of Akt1 protein while also carry out the necessary in silico pharmacokinetic and pharmacological analysis.
Methods
Preparation of ligands
Piperine analogs chemical structures were obtained from peer-reviewed literature. The chemical structures of Piperine analogs were drawn by using ACD/chemSketch (Freeware) (https://www.acdlabs.com/resources/free-chemistry-software-apps/) and the PDB file of ligand were generated using OpenBabel software (O’Boyle et al. 2011). The SPDBV software was used to do energy reduction, and Auto Dock Vina was used to convert PDB files of inhibitor-containing piperine analogs into PDBQT files for further analysis. In order to analyze the binding relationship between the selected piperine analogs and the PCa target, the FDA-approved and clinically used PCa inhibitors Flutamide (standard 1) and cabazitaxel (standard 2) was used (https://www.cancer.gov/about-cancer/treatment/drugs/prostate).
Preparation of receptor
From the RCSB PDB, the atomic coordinates of the Akt1 kinase domain were obtained (https://www.rcsb.org/pdb/). For the docking investigation, the co-crystallized structure of Akt1 (PDB ID: 2UZS, resolution: 2.46Å) was acquired. In order to prepare the structure for docking analysis, co-crystallized heteroatoms and water molecules were taken out using SPDBV software, and then polar hydrogen and Gasteiger charges were added using Auto Dock Vina. Then, structures were saved as PDBQT files for extensive analysis.
Pharmacokinetic phase
Lipinski’s Rule of Five and PreADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) features were used to predict the drug-likeness of the compounds. The pharmacokinetics of medicine is determined by the molecular characteristics of a specific drug, which are primarily defined by Rule of Five (Lipinski 2004).
Drug-likeness prediction
According to the chemical and structural characteristics of the drug molecule, drug-likeness determines if the studied molecule is similar to a known drug. The key pharmacophore aspects that influence a molecules behavior in terms of bioavailability, transportation, toxicity, reactivity, and other properties on living organisms include hydrophobicity, hydrogen bonding, electron distribution, and molecular size. Utilizing the cheminformatics service Molinspiration, the chemical characteristics and biological activity of piperine analogs were assessed in the current investigation (https://www.molinspiration.com/). The server includes a broad range of tools for processing and manipulating molecules, such as tautomer synthesis, molecule fragmentation, normalisation of molecules, computation of different chemical characteristics used in QSAR studies, and fragment-based virtual screening. The server estimates the bioactivity score for the most significant therapeutic targets, such as GPCR ligand, kinase inhibitors, ion channel modulators, enzymes, and nuclear receptors, based on the Lipinski Rule of Five (Venkatesh et al. 2017).
ADMET prediction
PreADMET is a web-based tool for predicting ADMET (absorption, distribution, metabolism, excretion, and toxicity) (https://preadmet.webservice.bmdrc.org/) of piperine analogs processing excellent binding affinity with receptor protein based on lowest binding energy. Since the characteristics of the molecules have a significant influence in the pre-clinical and clinical phases, the ADMET is the most crucial metric in drug discovery research and is taken into account when creating the medicine. The human intestinal absorption (HIA) of pharmaceuticals may be predicted using the Caco2 cell (human colon adenocarcinoma) and MDCK (Madin-Darby canine kidney) cell models. Blood brain barrier (BBB) and plasma protein binding (PPB) are important factors in the drug distribution phase. Based on the value of BBB penetration, PreADMET forecasts the proportion of drug bound in plasma protein to simulate in vitro data on humans and anticipate in vivo data. Drug effectiveness and distribution are influenced directly or indirectly by how well a drug interacts with target molecules when it is unbound. The activity of a drug is dependent on how it interacts with plasma proteins. Based on the findings of the Ames test, PreADMET predicts the level of toxicity, and the value of the prediction results is either “positive” or “negative” PreADMET, based on the National Toxicology Program and containing the findings of the in vivo carcinogenicity tests performed on mice and rats over a two-year period, was finally used to hypothesize carcinogenicity (Yamashita et al. 2000; Singh et al. 2016).
Computer-aided drug design of piperine analogs for the treatment of prostate cancer
Pharmacodynamic phase
The pharmacodynamic phase of a structurally specific drug is focused on drug-macromolecule interactions. In other aspects, it involves altering one drug by the use of another drug and additional circumstances. For the most part, a ligand-receptor complex is created when ligand interacts with a receptor during pharmacodynamics research. The drugs interaction with a receptor determines how it behaves when it is present at the site of action.
Molecular docking studies
Using the AutoDock Vina platform, the binding mechanism and interaction of Akt1 with piperine analogs were studied (Trott et al. 2010). This programme needs a grid box that has already been generated. It uses XYZ coordinates to serve as the frontier for active pocket amino acids in the receptor. By utilizing the PDBsum server (http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/) to compare the Akt1 (PDB ID: 2UZS) with other Akt1 crystallographic structures that are available in the PDB, the active pocket amino acid residues was discovered. PDBsum predicts the exact ligand for unbound structures and also allows for quick examination of binding sites that are comparable across the structures with various global folds as well as related folds. Using the Autogrid tool, the grid optimization was completed, and the grid box was centered to encompass all of the identified active pocket amino acid residues (Arg86, Lys14, Asn53, Arg25, Arg23, Lys17, Tyr18, Ile19, and Gly16). The grid box size was set at 20, 20 and 22 and on mass center 15.371, 6.581 and 15.557 for x, y and z co-ordinates respectively, with space separated by 1.0 Å (grid-point spacing). Exhaustiveness 10 was used for the molecular docking to get more realistic value. Ten alternative forms of confirmations were created with their corresponding binding affinities, once docking was completed using AutoDock software. Results with the lowest binding energy or highest binding affinity were taken into consideration for additional post-docking investigations. LigPlot v.1.4.5 was used to build two-dimensional docking for the investigation of hydrogen bond and hydrophobic interactions after the docked complex of Akt1 protein and piperine analogs with excellent binding affinity was shown in PyMOL viewer.
Pharmacophore study
To determine the optimum interactions with a particular molecular target and to activate or inhibit its biological activities, a pharmacophore model is a collection of electronic and electrostatic interactions structure. A pharmacophore model was created utilizing the PharmaGist web platform to build the new leads for biological activity against prostate cancer. The pharmacophore model, which consists of a collection of 3D features required for the bioactive ligand, was developed using a minimum of three pharmacophoric attributes (Ashtekar et al. 2019).
Result and discussion
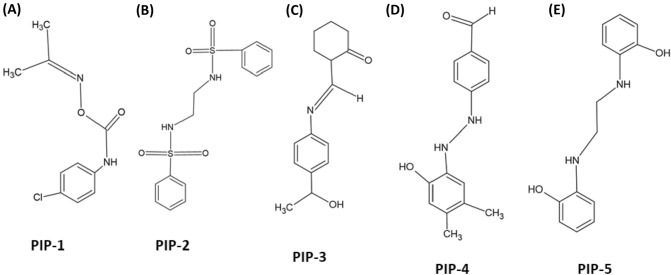
The development of new drugs is justified by the identification of creative and powerful inhibitors derived from piperine analogs that prevent PCa. The five piperine analogs are filtered using the Lipinski rule, physicochemical concepts, molecular characteristics, and drug-likeness. Based on absorption in the human body and physicochemical parameters, suitable compounds with membrane permeability were chosen; their chemical structures are shown in Fig. 1 with their complete information, including LogP value, given in Table 1. For in silico experiments, the current assessment of five known piperine analogs is taken into account. Using the previously specified filtering criteria, so finally one analog were discovered that might be employed to treat prostate cancer.
Fig. 1.
Chemical structure of Piperine analog molecules a–e
Table 1.
ChemSpider ID, IUPAC name, and LogP value of the Piperine analogs and standards
| Symbol | ChemSpider ID | IUPAC name of the compound | LogP |
|---|---|---|---|
| Pip1 | 190,885 | [(4-chlorophenyl)amino][(propan-2-ylideneamino)oxy]methanone | 2.67 ± 0.58 |
| Pip2 | 65,724 | N-{2-[(phenylsulfonyl)amino]ethyl}benzenesulfonamide | 2.04 ± 0.45 |
| Pip3 | – | 2-[(E)-{[4-(1-hydroxyethyl)phenyl]imino}methyl]cyclohexan-1-one | 1.20 ± 0.56 |
| Pip4 | – | 4-[2-(2-hydroxy-4,5-dimethylphenyl)hydrazinyl]benzaldehyde | 3.42 ± 0.39 |
| Pip5 | 387,590 | 2,2’-(1,2-Ethanediyldiimino)diphenol | 2.05 ± 0.27 |
| Standard 1 | 3280 | 2-Methyl-N-[4-nitro-3-(trifluoromethyl)phenyl]propanamide | 3.72 ± 0.40 |
| Standard 2 | 8,029,779 | [(1S,2S,3R,4S,7R,9S,10S,12R,15S)-4-acetyloxy-1-hydroxy-15-[(2R,3S)-2-hydroxy-3-[(2-methylpropan-2yl)oxycarbonylamino]-3-phenylpropanoyl]oxy-9,12-dimethoxy-10,14,17,17-tetramethyl-11-oxo-6-oxatetracyclo[11.3.1.03,10.04,7]heptadec-13-en-2-yl] benzoate | 7.56 ± 0.74 |
Drug-likeness properties
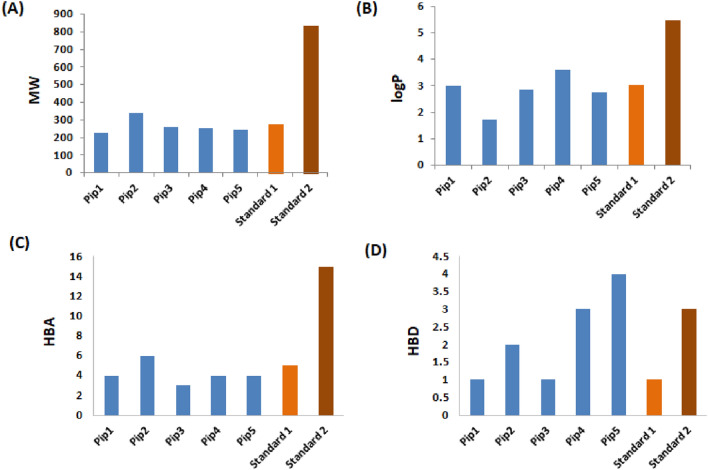
In Table 2, the characteristics of five piperine analogs compared with standards that are retrieved through the Molinspiration server are given. The fact that every molecule has a molecular weight within the permissible range (MW ≤ 500) and every molecule has embraced Lipinski’s rule of five is significant, since it directly affects the identification of drugs that affect the target function. The range of Lipinski’s limit is less than 10 and 5 respectively, since the number of hydrogen bond donors (OH and NH atoms) and acceptors (O and N atoms) in piperine analogs was 0 to 7 and 0 to 5 respectively. In the phase of drug distribution, the hydrophobicity and lipophilicity of the molecule are assessed using the MLogP value (octagonal/water partition co efficient) (Clark 1999). All of the piperine analogs MLogP values fell within the permitted range (Lipinski’s rule of five) as shown in Fig. 2. The Topological Polar Surface Area (TPSA), which is based on the sum of all polar atoms such oxygen, nitrogen, and bonded hydrogen, is another significant physicochemical parameter utilised to forecast drug distribution characteristics. Intestinal absorption, hydrogen bonding potential, bioavailability, blood brain barrier penetration (BBB), and Caco-2 cell permeability are all well-represented by TPSA. The quantity of rotatable bonds (≤ 10) and TPSA score (140) suggest high bioavailability. All piperine analogs were discovered to have less than 10 rotatable bonds. The value of TPSA of all piperine analogs discovered to be in the range of 0.00–140 plainly exhibiting strong oral bioavailability shows that rotatable polar atomic bonds improve the flexibility of molecules for more adaptive and efficient interaction with the enzyme active site.
Table 2.
Physicochemical properties of piperine analogs and standards
| Compound | MW | miLogP | TPSA (Å) | nON | nOHNH | nrotb | natoms | Volume |
|---|---|---|---|---|---|---|---|---|
| Pip1 | 226.66 | 2.98 | 50.70 | 4 | 1 | 3 | 15 | 194.33 |
| Pip2 | 340.43 | 1.70 | 92.34 | 6 | 2 | 7 | 22 | 276.72 |
| Pip3 | 259.35 | 2.84 | 49.66 | 3 | 1 | 3 | 19 | 257.45 |
| Pip4 | 256.31 | 3.58 | 61.35 | 4 | 3 | 4 | 19 | 240.38 |
| Pip5 | 244.29 | 2.74 | 64.51 | 4 | 4 | 5 | 18 | 229.90 |
| Standard 1 | 276.21 | 3.02 | 74.92 | 5 | 1 | 4 | 19 | 220.011 |
| Standard 2 | 835.94 | 5.48 | 202.4 | 15 | 3 | 15 | 60 | 758.90 |
Fig. 2.
Plots of percentage performance of piperine analogs and FDA prostate cancer drugs for oral bioavailability parameters according to Ro5, A Molecular weight (MW), B Partition coefficient (Log P), C Hydrogen bond acceptor (HBA), D Hydrogen bond donor (HBD)
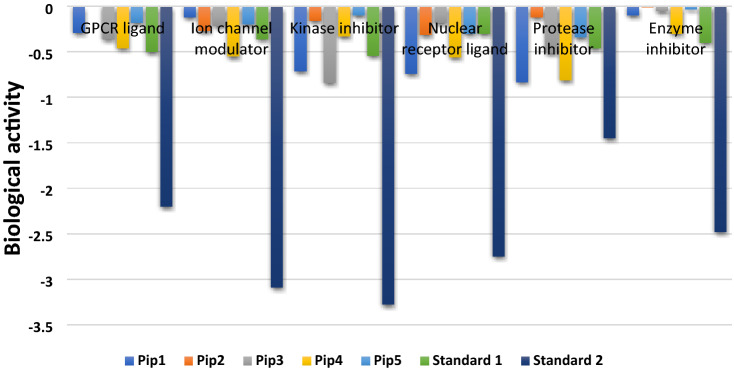
The piperine analogs were tested for their biological activity against GPCR ligands, kinase inhibitors, nuclear receptor ligands, Ion channel modulators, protease inhibitor, and enzyme inhibitory action (Table 3). The current work shows that piperine analogs are physiologically active and have physiological effects when combined with enzymes such as nuclear receptor ligands, protease inhibitors, GPCR ligands, and others. The bioactivity score for pip2 and pip5 molecules was determined to be 0.50 and Pip1, pip4, and pip3 was ≤ 0.50 for GPCR ligand, Ion channel modulator, Kinase inhibitor, nuclear receptor ligand, protease inhibitor, and enzyme inhibitor. Together, the findings demonstrate that pip2 and pip5 piperine analogs have considerable physicochemical features that are consistent with higher biological activity as shown in Fig. 3.
Table 3.
Biological activity of piperine analogs and standards
| Compound | GPCR ligand | Ion channel modulator | Kinase inhibitor | Nuclear receptor ligand | Protease inhibitor | Enzyme inhibitor |
|---|---|---|---|---|---|---|
| Pip1 | − 0.29 | − 0.12 | − 0.71 | − 0.74 | − 0.83 | − 0.10 |
| Pip2 | 0.00 | − 0.27 | − 0.16 | − 0.31 | − 0.12 | − 0.01 |
| Pip3 | − 0.36 | − 0.21 | − 0.84 | − 0.18 | − 0.51 | − 0.05 |
| Pip4 | − 0.46 | − 0.55 | − 0.33 | − 0.56 | − 0.81 | − 0.30 |
| Pip5 | − 0.18 | − 0.20 | − 0.10 | − 0.30 | − 0.34 | − 0.03 |
| Standard 1 | − 0.50 | − 0.36 | − 0.54 | − 0.30 | − 0.46 | − 0.40 |
| Standard 2 | − 2.20 | − 3.09 | − 3.28 | − 2.75 | − 1.45 | − 2.48 |
Fig. 3.
Bioactivities of piperine analogs and standards are calculated using Molinspiration tool. Note: GPCR ligand–protein coupled receptor ligand; Ion channel modulator; nuclear receptor legend; Kinase inhibitor; Protease inhibitor; Enzyme inhibitor. If the bioactivity scores is (≥ 0.00) considered biological active, if the bioactivity scores (− 0.50 to 0.0) it is moderately active and finally if the bioactivity scores (< − 0.50) it is inactive
Pharmacokinetics and toxicity
Using preADMET tool, the pharmacokinetic characteristics of five piperine analogs with higher binding energies were further assessed. Physicochemical characteristics that impact drug molecule absorption and bioavailability, such as lipophilicity (clogP), polar surface area, molecular weight (MW), and water solubility (logS), were previously assessed using Molinspiration server. A crucial factor in determining a molecule’s oral availability is lipophilicity (logP). The piperine analogs TPSA (Topological Polar Surface Area) is determined to be 140, suggesting that they have a good bioavailability (Veber et al. 2002).
Piperine analogs are more flexible than standard flutamide due to the presence of additional rotatable bonds, and they show high binding potential in docking studies. The piperine counterparts have a hydrogen bond acceptor and donor count that falls within the optimal ranges of 0–6 and 0–4, respectively. All five piperine analogs and known inhibitors have HIA (human intestinal absorption) values between 85 and 100%. This suggests that the digestive system can effectively absorb all piperine equivalents. Additionally, average permeability to Caco2 cell (4–70) and MDCK cell model is demonstrated using piperine analogs (predicted value in the range 4–70). With the exception of pip1, the PPB binding evaluation of piperine analogs in the distribution phase showed considerable binding energy with plasma proteins (predicted value ≥ 80%). The most of the time, weak plasma protein binding substances can readily traverse cell membranes and interact with targets. The pip1, pip2, and pip3 exhibited minimal absorption in the CNS (predicted value less than 0.4), whereas pip4 and pip5 showed intermediate absorption, according to BBB penetration (predicted value between 2.0 and 1.0). As opposed to pip1, pip2, and pip3, standards have strong CNS absorption whereas piperine analogs have less adverse effects. As indicated in Table 4, the pip2 molecule exhibited the lowest skin-permeability when compared to the standard, which is another key risk assessment feature of the compounds after accidental contact with the skin. The effects of piperine analogs on cancer and mutagenesis were assessed during the toxicology phase. The Ames test is an easy way to determine, if five piperine analogs are mutagenic, with pip1, pip3, pip4, and pip5 showing that they are mutagens and pip2 showing that they are not. Finally, the drug-likeness and ADMET values of the chosen piperine analogs pip1, pip2, pip3, pip4, and pip5 suggest that they have the potential to function as effective inhibitors of the Akt1 receptor. Pip2 is a more effective inhibitor of the Akt1 receptor in piperine analogs.
Table 4.
ADMET properties of piperine analogs and standards against Akt1 receptor
| Compoud | ADMET | Toxicity | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HIA (%) | PPB (%) | BBB (%) | Caco2 | Skin Permeability | Ames test | MDCK(nm/sec) | Rodent Carcinogenicity | ||
| Carcino Mouse | Carcino Rat |
||||||||
| Pip1 | 95.090 | 65.143 | 0.130 | 22.743 | − 2.231 | Mutagen | 197.36 | Positive | Negative |
| Pip2 | 95.920 | 88.311 | 0.340 | 1.471 | − 1.635 | Non-mutagen | 4.054 | Negative | Negative |
| Pip3 | 95.330 | 82.175 | 0.172 | 24.618 | − 2.806 | Mutagen | 34.558 | Negative | Positive |
| Pip4 | 88.843 | 83. 093 | 1.748 | 19.771 | − 3.685 | Mutagen | 6.645 | Positive | Positive |
| Pip5 | 85.754 | 81.241 | 2.440 | 6.873 | − 3.71 | Mutagen | 277.09 | Negative | Negative |
| Standard 1 | 89.516 | 85.603 | 0.921 | 3.854 | − 2.063 | Mutagen | 46.775 | Positive | Negative |
| Standard 2 | 92.19 | 82.401 | 0.236 | 23.023 | − 1.467 | Non-mutagen | 0.0434 | Positive | Negative |
Molecular docking studies
All five piperine analogs were docked alongside the standards flutamide and cabazitaxel against the kinase domain of Akt1, and the active site of Akt1 domain of PDB 2UZS as shown in Fig. 4. The ligand was categorised and contrasted with recognised inhibitors according to binding affinity, hydrogen bonding and hydrophobic interactions. The results show that the binding affinities of pip2, pip3, and pip4 give acceptable antagonists with the discovered standards and docking affinities as above as − 5.4 kcal/mol.
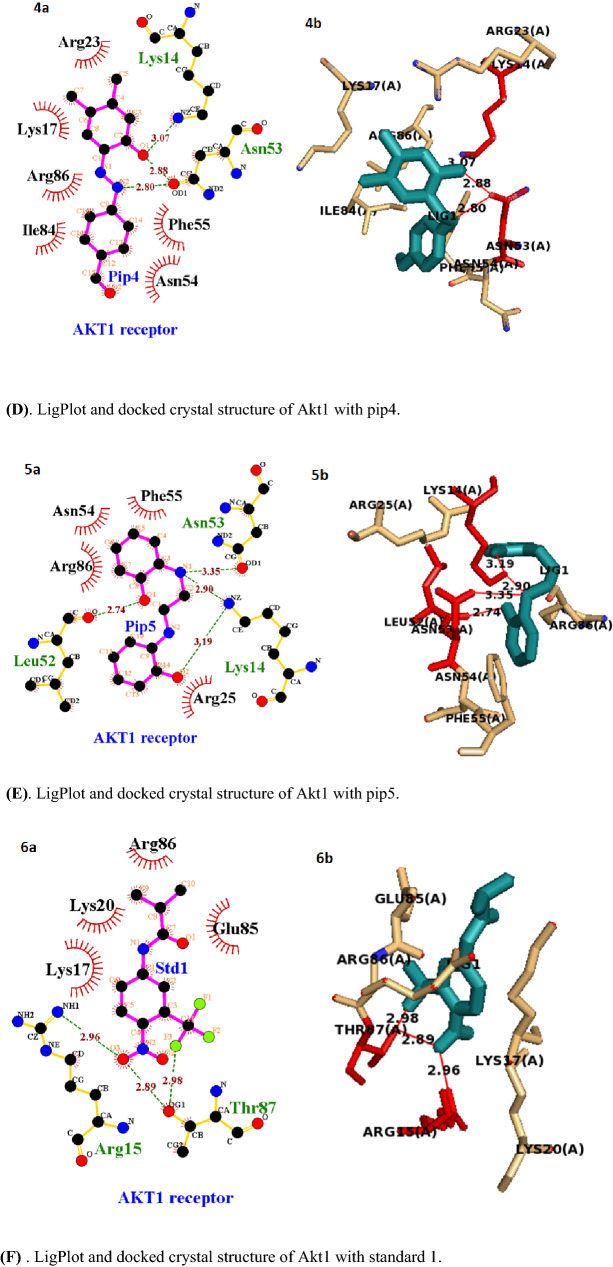
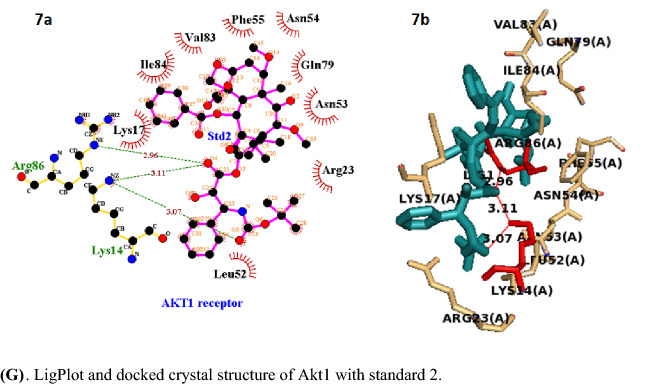
Fig. 4.
Active site of Akt1 domain of PDB: 2UZS. A LigPlot and docked crystal structure of Akt1 with pip1. B LigPlot and docked crystal structure of Akt1 with pip2. C. LigPlot and docked crystal structure of Akt1 with pip3. D. LigPlot and docked crystal structure of Akt1 with pip4. E. LigPlot and docked crystal structure of Akt1 with pip5. F. LigPlot and docked crystal structure of Akt1 with standard 1. G. LigPlot and docked crystal structure of Akt1 with standard 2
The four piperine analogs, with exception of pip1and pip5, theoretically have good binding affinity in comparison to the standard drug flutamide and cabazitaxel. The active pocket amino acid residues Arg86, Lys14, Asn53, Lys17, Tyr18, Ile19, Leu52, and Asn54 of 2UZS form hydrogen bonds with the piperine analogs pip1, pip2, pip3, pip4, and pip5, and the bond lengths range from 2.81 to 3.34. Hydrogen bond and affinity of piperine analogs and standards against Akt1 receptor and are tabulated in Table 5, and their 2D and 3D structures are shown in Fig. 5 [(A), (B), (C), (D), (E), (F), (G)]. In piperine analog, pip2 shows higher binding affinity by forming 6 hydrogen bonds with more hydrophobic interactions compared to other four analogs and two standards.
Table 5.
Molecular docking analysis of piperine analogs and standards against Akt1 receptor
| Ligands | Affinity (Kcal/mol) | H bonds | H bond length(Å) | H bond with | Hydrophobic interactions |
|---|---|---|---|---|---|
| Pip1 | − 5.2 | 1 | 2.91 | 2UZS:Arg86::PIP1:O2 | Phe55,Asn53, Arg23, Lys14, Lys17, Gly16, Tyr18 |
| Pip2 | -6.0 | 6 |
3.11 2.88 2.81 3.18 3.08 2.91 |
2UZS:Lys14::PIP2:O1 2UZS:Lys14::PIP2:O1 2UZS:Arg86::PIP2:O2 2UZS:Arg86::PIP2:O3 2UZS:Arg86::PIP2:O3 2UZS:Asn54::PIP2:O4 |
Phe55, Ile84, Leu52, Lys17, Gly16, Tyr18, Arg23, Asn53 |
| Pip3 | − 5.6 | 4 |
3.34 3.32 3.23 2.89 |
2UZS:Arg86::PIP3:N 2UZS:Lys14::PIP3:N 2UZS:Tyr18::PIP3:O2 2UZS:Lys17::PIP3:O2 |
Gly16, Asn53, Ile84, Phe55 |
| Pip4 | − 5.7 | 3 |
3.07 2.88 2.80 |
2UZS:Lys14::PIP4:O1 2UZS:Asn53::PIP4:N2 2UZS:Asn53::PIP4:N2 |
Arg23, Lys17, Arg86, Ile84 Asn54, Phe55 |
| Pip5 | − 5.4 | 4 |
3.35 3.19 2.90 2.74 |
2UZS:Asn53::PIP5:N1 2UZS:Lys14::PIP5:O2 2UZS:Lys14::PIP5:N1 2UZS:Leu52::PIP5:O1 |
Phe55, Asn54, Arg86, Arg25 |
| Standard 1 | − 5.4 | 3 |
2.89 2.96 2.98 |
2UZS:Thr87::STD1:O3 2UZS:Arg15::STD1:O3 2UZS:Thr87::STD1:F3 |
Arg86, Lys17, Lys20, Glu85 |
| Standard 2 | − 5.2 | 3 |
3.11 3.07 2.96 |
2UZS:Lys14::STD2:O4 2UZS:Lys14::STD2:O5 2UZS:Arg86::STD2:O4 |
Asn54, Phe55, Val83, Ile84, Lys17,Leu52, Arg23, Asn53, Gln79 |
Fig. 5.
Docking results of screened piperine analogs and standards against prostate cancer targeting Akt1 using AutoDock Vina tool
Pharmacophore model
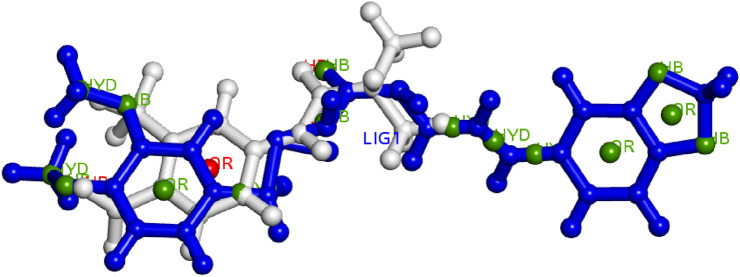
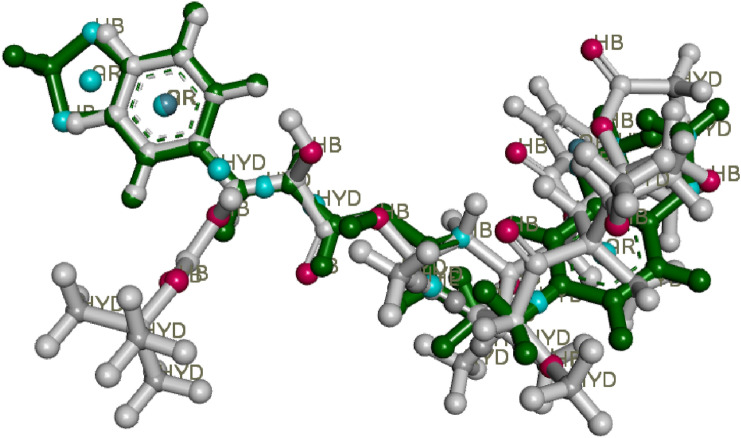
There is a collection of pharmacophore with characteristics (1 aromatic, 1donar, 2 acceptors) but not all of the input compounds could include all of the features since they come from distinct structural components. So, we employed models created from various combinations of the input molecules structure. The chosen models, along with the best score values, features, and molecules implicated, are given in Table 6. The cumulative total of the matching key pharmacophore points produced the best score in a pairwise match. PharmaGist looks as many critical selected features for various input compounds that are connected by significant pairwise alignments as it possible. The model 1 was built using 3 chemicals; pip2, standard1 and standard2, however it took into account pip2 and standard1 for the best pairwise score. The model 1 has a score of 7.523 and 4 features, as shown in Fig. 6. The model 2 was built using 3 chemicals; pip2, standard1 and standard2, however it took into account pip2 and standard2 for the best pairwise score, and the model 2 has a score of 6.021 and 3 features, as shown in Fig. 7. Due to their comparable pharmacophore properties, the screened molecule can function as PCa inhibitor.
Table 6.
The best pairwise features of pharmacophore model
| Pharmacophore model | Score | Number of features | Features | Molecules involved |
|---|---|---|---|---|
| 1 | 7.523 | 4 | 1Ar, 1D, 2A | Pip2 and standard 1 |
| 2 | 6.021 | 3 | 1Ar, 2A | Pip2 and standard 2 |
Fig. 6.
Pharmacophore model-1 aligned on three compounds pip2, standard1 and standard2, HYD-Hydrophobic, AR- Aromatic features
Fig. 7.
Pharmacophore model-2 aligned on three compounds pip2, standard 1 and standard 2, HYD-Hydrophobic, AR- Aromatic features
Conclusion
pip1, pip2, pip3, pip4, and pip5 are five piperine analogs that have shown potential as Akt1 inhibitors in the current study. They have great binding affinities and considerable drug-likeness properties. One of these pip2 validates the overall drug design parameters by demonstrating a significant binding interaction with the conserved amino acids in the receptor molecule by demonstrating higher binding affinity than the standards flutamide and cabazitaxel. As a result of these findings, the chosen piperine analogs were investigated in further depth and through structural alteration to create potential new, more potent drugs with improved therapeutic action against prostate cancer.
Acknowledgements
The author is thankful to Kuvempu University, Shivamogga, and Karnataka, India for providing the necessary facility to carry out the work and also the Government of India—Ministry of Tribal Affairs for the financial support.
Abbreviations
- PCa
Prostate cancer
- Akt1
Serine-threonine kinase
- ADMET
Absorption, Distribution, Metabolism, Excretion, and Toxicity
- Pip
Piperine analog
Author contribution
PN designed and carried out the experiments; contributed to the interpretation of the results, conceptualization the work and wrote the entire manuscript.
Data availability
All the data we generated in this paper is available in the body of the manuscript as supporting figures and tables. We do not have any ethical or legal considerations for not making our data publicly available.
Declarations
Conflict of interest
The author declares that they have no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ashtekar SS, Bhatia NM, Bhatia MS. Exploration of leads from natural domain targeting HER2 in breast cancer: an in-silico approach. Int J Pept Res Ther. 2019;25(2):659–667. doi: 10.1007/s10989-018-9712-y. [DOI] [Google Scholar]
- Atanasov AG, Waltenberger B, Pferschy-Wenzig EM, Linder T, Wawrosch C, Uhrin P, Temml V, Wang L, Schwaiger S, Heiss EH, Rollinger JM, Schuster D, Breuss JM, Bochkov V, Mihovilovic MD, Kopp B, Bauer R, Dirsch VM, Stuppner H. Discovery and resupply of pharmacologically active plant-derived natural products: a review. Biotechnol Adv. 2015;33(8):15821614. doi: 10.1016/j.biotechadv.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedolla R, Prihoda TJ, Kreisberg JI, Malik SN, Krishnegowda NK, Troyer DA, Ghosh PM. Determining risk of biochemical recurrence in prostate cancer by immunohistochemical detection of PTEN expression and Akt activation. Clin Cancer Res. 2007;13(13):3860–3867. doi: 10.1158/1078-0432.CCR-07-0091. [DOI] [PubMed] [Google Scholar]
- Brito MAD. Pharmacokinetic study with computational tools in the medicinal chemistry course. Braz J Pharm Sci. 2011;47:797–805. doi: 10.1590/S1984-82502011000400017. [DOI] [Google Scholar]
- Cai J, Zhao J, Gao P, Xia Y. Patchouli alcohol suppresses castration-resistant prostate cancer progression by inhibiting NF-κB signal pathways. Translat Androl Urol. 2022;11(4):528–542. doi: 10.21037/tau-22-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung M, Testa JR. Diverse mechanisms of AKT pathway activation in human malignancy. Curr Cancer Drug Targets. 2013;13:2340–2344. doi: 10.2174/1568009611313030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DE. Rapid calculation of polar molecular surface area and its application to the prediction of transport phenomena. 2. Prediction of blood-brain barrier penetration. J Pharm Sci. 1999;88(8):815–821. doi: 10.1021/js980402t. [DOI] [PubMed] [Google Scholar]
- Hammad AS, Ravindran S, Khalil A, Munusamy S. Structure-activity relationship of piperine and its synthetic amide analogs for therapeutic potential to prevent experimentally induced ER stress in vitro. Cell Stress Chaperones. 2017;22:417–428. doi: 10.1007/s12192-017-0786-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyszczyk P, Acevedo A, Davidoff EJ, Timmins LM, Marrero-Berrios I, Patel M, White C, Lowe C, Sherba JJ, Hartmanshenn C, O'Neill KM, Balter ML, Fritz ZR, Androulakis IP, Schloss RS, Yarmush ML. The growing role of precision and personalized medicine for cancer treatment. Technology. 2018;6(3–4):79–100. doi: 10.1142/S2339547818300020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dobos GJ, Rampp T. The significance of ayurvedic medicinal plants. J Evid Based Complement Altern Med. 2017;22:494–501. doi: 10.1177/2156587216671392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang C, Li W, Koike K, Nikaido T, Wang MW. Antidepressant-like effects of piperine and its derivative, antiepilepsirine. J Asian Nat Prod Res. 2007;9(3–5):421–430. doi: 10.1080/10286020500384302. [DOI] [PubMed] [Google Scholar]
- Lipinski CA. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Makhov PB, Golovine K, Kutikov A, Teper E, Canter DJ, Simhan J, Uzzo RG, Kolenko VM. Modulation of Akt/mTOR signaling overcomes sunitinib resistance in renal and prostate cancer cells. Mol Cancer Ther. 2012;11(7):1510–1517. doi: 10.1158/1535-7163.MCT-11-0907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers CD. History of global burden of disease assessment at the World Health Organization. Arch Public Health. 2020;78:77. doi: 10.1186/s13690-020-00458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal R, Gupta RL. In vitro antioxidant activity of piperine. Methods Find Exp Clin Pharmacol. 2000;22:271–274. doi: 10.1358/mf.2000.22.5.796644. [DOI] [PubMed] [Google Scholar]
- Monique B. Pharmacokinetic study with computational tools in the medicinal chemistry course. Braz J Pharm Sci. 2011;47:797–805. doi: 10.1590/S1984-82502011000400017. [DOI] [Google Scholar]
- Prakash N, Hanumanthappa M, Preema KA, Vijayalaksmi V. Investigating drug-target interactions of piperine in prostate cancer using network pharmacology and docking studies. Res J Biotech. 2022;17(10):32–42. doi: 10.25303/1710rjbt32042. [DOI] [Google Scholar]
- Nguyen-Nielsen M, Borre M. Diagnostic and therapeutic strategies for prostate cancer. Semin Nucl Med. 2016;46(6):484–490. doi: 10.1053/j.semnuclmed.2016.07.002. [DOI] [PubMed] [Google Scholar]
- O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open babel: an open chemical toolbox. J Cheminformatics. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya N, Kumar A. Piperine analogs arrest c-myc gene leading to downregulation of transcription for targeting cancer. Sci Rep. 2021;11(1):22909. doi: 10.1038/s41598-021-01529-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhavathi H, Dasegowda KR, Renukananda KH, Lingaraju K, Naika HR. Exploration and evaluation of bioactive phytocompounds against BRCA proteins by in silico approach. J Biomol Struct Dyn. 2021;39(15):5471–5485. doi: 10.1080/07391102.2020.1790424. [DOI] [PubMed] [Google Scholar]
- Salehi B, Zakaria ZA, Gyawali R, Ibrahim SA, Rajkovic J, Shinwari ZK, Khan T, Sharifi-Rad J, Ozleyen A, Turkdonmez E, Valussi M, Tumer TB, Monzote Fidalgo L, Martorell M, Setzer WN. Piper species: a comprehensive review on their phytochemistry, biological activities and applications. Molecules (basel, Switzerland) 2019;24(7):1364. doi: 10.3390/molecules24071364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattarinezhad E, Bordbar AK, Fani N. Piperine derivatives as potential inhibitors of Survivin: an in silico molecular docking. Comput Biol Med. 2015;63:219–227. doi: 10.1016/j.compbiomed.2015.05.016. [DOI] [PubMed] [Google Scholar]
- Shrivastava P, Vaibhav K, Tabassum R, Khan A, Ishrat T, Khan MM, Ahmad A, Islam F, Safhi MM, Islam F. Anti-apoptotic and anti-inflammatory effect of Piperine on 6-OHDA induced Parkinson's rat model. J Nutr Biochem. 2013;24(4):680–687. doi: 10.1016/j.jnutbio.2012.03.018. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- Singh S, Das T, Awasthi M, Pandey VP, Pandey B, Dwivedi UN. DNA topoisomerase-directed anticancerous alkaloids: ADMET-based screening, molecular docking, and dynamics simulation. Biotechnol Appl Biochem. 2016;63(1):125–137. doi: 10.1002/bab.1346. [DOI] [PubMed] [Google Scholar]
- Sung H, Ferlay J, Siegel RL. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Venkatesh SSL, Krishna V, Jayabaskaran C. In Silico Analysis of ADME-T properties of Amentoflavone. Bioinform Proteomics. 2017;1(3):000118. [Google Scholar]
- Yamashita S, Furubayashi T, Kataoka M, Sakane T, Sezaki H, Tokuda H. Optimized conditions for prediction of intestinal drug permeability using Caco-2 cells, European Journal of Pharmaceutical Sciences. Eur J Pharm Sci. 2000;10(3):195–204. doi: 10.1016/S0928-0987(00)00076-2. [DOI] [PubMed] [Google Scholar]
- Yu JW, Yuan HW, Bao LD, Si LG. Interaction between piperine and genes associated with sciatica and its mechanism based on molecular docking technology and network pharmacology. Mol Diversity. 2021;25(1):233–248. doi: 10.1007/s11030-020-10055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarai Z, Boujelbene E, Salem N, Gargouri Y, Sayari A. Antioxidant and antimicrobial activities of various solvent extracts, piperine and piperic acid from Piper nigrum. LWT Food Sci Technol. 2013;50:634–641. doi: 10.1016/j.lwt.2012.07.036. [DOI] [Google Scholar]
- Zinda MJ, Johnson MA, Paul JD, Horn C, Konicek BW, Lu ZH, Sandusky G, Thomas JE, Neubauer BL, Lai MT, Graff JR. AKT-1, -2, and -3 are expressed in both normal and tumor tissues of the lung, breast, prostate, and colon. Clin Cancer Res. 2001;7(8):2475–2479. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data we generated in this paper is available in the body of the manuscript as supporting figures and tables. We do not have any ethical or legal considerations for not making our data publicly available.