Abstract
High Risk Human Papilloma Viruses (HR-HPV) persistently infect women with Human Immunodeficiency Virus-1 (HIV-1). HPV-16 escapes immune surveillance in HIV-1 positive women receiving combined antiretroviral therapy (cART). HIV-1 Tat and HPV E6/E7 proteins exploit Notch signaling. Notch-1, a developmentally conserved protein, influences cell fate from birth to death. Notch-1 and its downstream targets, Hes-1 and Hey-1 contribute to invasive and aggressive cancers. Cervical cancer cells utilize Notch-1 and hyper-express CXCR4, a co-receptor of HIV-1. Accumulating evidence shows that HIV-1 affects cell cycle progression in pre-existing HPV infection. Additionally, Tat binds Notch-1 receptor for activation and influences cell proliferation. Oncogenic viruses may interfere or converge together to favor tumor growth. The molecular dialogue during HIV-1/HPV-16+ co-infections in the context of Notch-1 signaling has not been explored thus far. This in vitro study was designed with cell lines (HPV-ve C33A and HPV-16+ CaSki) which were transfected with plasmids (pLEGFPN1 encoding HIV-1 Tat and pNL4-3 encoding HIV-1 [full HIV-1 genome]). HIV-1 Tat and HIV-1 inhibited Notch-1expression, with differential effects on EGFR. Notch-1 inhibition nullified Cyclin D expression with p21 induction and increased G2-M cell population in CaSki cells. On the contrary, HIV-1 infection shuts down p21 expression through interaction of Notch-1 downstream genes Hes-1-EGFR and Cyclin D for G2-M arrest, DDR response and cancer progression. This work lays foundations for future research and interventions, and therefore is necessary. Our results describe for the first time how HIV-1 Tat cancers have an aggressive nature due to the interplay between Notch-1 and EGFR signaling. Notch-1 inhibitor, DAPT used in organ cancer treatment may help rescue HIV-1 induced cancers.
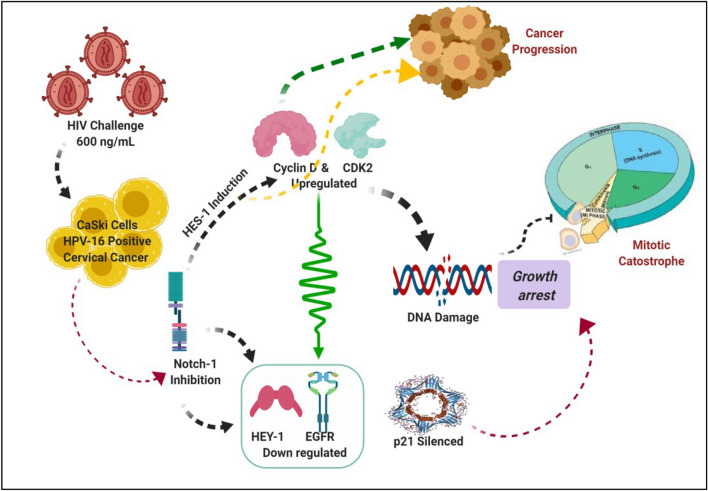
Graphical abstract
The illustration shows how HIV interacts with HPV-16 to induce Notch 1 suppression for cancer progression (Created with BioRender.com).
Supplementary Information
The online version contains supplementary material available at 10.1007/s13337-023-00809-y.
Keywords: HIV-1, HIV-HPV co-infections, HIV-Tat, Notch-1, Cell cycle genes
Introduction
There is an increased risk of co-infections with Human papillomavirus (HPV) with persistence of many carcinogenic subtypes in Human Immunodeficiency Virus (HIV-1) positive women [1]. They are more prone to invasive cervical cancers too [2, 3]. Epidemiological studies from a cohort of HIV-1 positive women established an incidence of 35.3% carcinogenic HPV genotypes. Oncogenic HPV-16 also escapes immune surveillance in HIV-1 positive women during combined Antiretroviral Therapy (cART) [4]. HPV vaccines have not yet been examined for the recurrence of cervical intraepithelial lesions in HIV-1 positive women [5].
HPV lifecycle orchestrates growth and differentiation events through HPV trans-forming genes-E6/E7 facilitating tumorigenesis [6]. The viral trans activator protein Tat, molecular weight 14–16 KDa, transcriptionally activates the expression of 10 kb RNA genome HIV-1 [7]. During HIV-1 infection, Tat and HPV E6/E7 proteins exploit Notch signaling to exacerbate HPV-associated disease pathogenesis [6, 8]. The transmembrane Notch-1 protein is essential for proliferation, differentiation and apoptosis at all stages of life. In vitro studies in TZM-bl-based reporter gene assays performed at Tat concentration (1–5 ng/mL) in the human brain inferred that Tat functions as a Notch ligand during neurogenesis whilst binding to the Notch receptor, thereby activating Notch signaling cascade [9]. Notch-1 activation, through ligand binding from neighboring cell encounter, induces proteolysis and release of Notch-1 Intracellular Domain (NICD) into the nucleus, the main gene processing centre [8]. Concomitantly, Numb, an evolutionary conserved protein enhances Notch-1 signaling by repressing NICD ubiquitination [10]. NICD also binds with DNA binding transcription factor protein, CSL to initiate Notch-1 downstream targets, Hes-1 and Hey-1 gene transcription associated with carcinogenesis and progression of cervical carcinoma [11]. Alternative spliced Numb iso variants, short (Numb S) and long (Numb L) also influence Hes-1 expression [12].
Notch-1, Numb and EGFR have similar roles in cell fate specifications including their involvement in cancers. In vitro studies in breast cancer cell lines demonstrated a positive link between Notch-1 and EGFR when transfected with HIV-1 [13]. Notch-1 regulates growth and differentiation via the Akt pathway through cyclin D, CDK2 and p21 in T-cell acute lymphoblastic leukemia cell line [14, 15]. Cervical cancer cells simultaneously utilize Notch-1 [16] as well as hyper-express CXCR4, a co-receptor of HIV-1. The cross interference of intricate pathways converge together viz CXCR4/Notch-1, Wnt/Notch-1, Notch-1/Snail/Numb, Notch-1/EGFR are implicated in cancer progression, survival and chemotaxis via intracellular signaling /downstream mediators [17, 18].
Recurrent, invasive cervical carcinomas due to HIV-1 infection and interference with cell cycle marker expression, prompted us to examine the effects of HIV-1 Tat and full HIV-1 genome on HPV-ve C33A cells and HPV-16+ CaSki cells using transient transfection. While this is an in vitro study, it lays the foundations for future research and interventions, and therefore important. We demonstrated here for the first time that both HIV-1 Tat and HIV-1 suppresses Notch-1 expression in CaSki (HPV-16+) cells through significant Hes-1induction and increased CDK2 expression for G0-G1 accumulation at the expense of S phase. HIV-1 Tat amplifies EGFR and p21 expression though Cyclin D is depleted, whereas HIV-1 transfection decreases EGFR and p21 expression concomitant with over-active Cyclin D. Elevated Cyclin D preserves proliferation for unbridled mitosis, G2-M arrest, genomic instability and cancer progression [19, 20]. Taken together, HIV-1 Tat shows an inverse relationship between Notch-1 and EGFR which favors an aggressive phenotype whilst HIV-1 has a crosstalk between Notch-1 and EGFR (Graphical abstract) during HIV-1/HPV-16+ co-infections.
Materials and methods
Media and experimental reagents
The primary antibodies for Notch-1 (ab65297), Hes-1 (ab49170), EGFR (ab2430), CDK2 (ab235941,) and GAPDH (sc32233) were purchased from Abcam, UK and SantaCruz, USA. The DNA transfection reagent, Jet PEI was procured from GeneX, India. The Notch-1 inhibitor-N-[N-(3, 5-difluorophenacetyl)-Lalanyl]-S-phenylglycine t-butyl ester (DAPT) was obtained from Abcam, UK. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), Dimethyl sulfoxide (DMSO), Antibiotic antimycotic solution (100X) [A5955], and Ethanol, 2-Mercaptoethanol and 2-Propanol (Isopropanol) were obtained from Sigma, USA. Roswell Park Memorial Institute (RPMI) 1640, Dulbecco's Modified Eagle Medium (DMEM) and N-2-hydroxyethylpiperazine-N-2-ethane sulfonic acid (HEPES-1M) were obtained from HiMedia Laboratories, India. The Fetal Bovine Serum (FBS) was obtained from Moregate, Australia. DNA was extracted using GF-1 Tissue DNA Extraction Kit from Vivantis Technologies, Malaysia. RNA isolation kit, Plasmid Miniprep Plus Purification Kit were procured from Gene Mark, USA. The Cycletest™ Plus DNA kit was procured from BD Biosciences, USA. Power SYBR® Green RNA to CT TM 1-Step Kit was procured from Thermofisher, USA.
Cell lines
Adherent cervical tumor derived cell lines HPV-Negative C33A cells (HPV-ve for HPV DNA and RNA having cervical cancer phenotype) and HPV-16+ CaSki cells (with inherent HPV-16+ and HPV-18+ sequences and endogenous Notch-1), were used for transfection.
Cell culture
CaSki and C33A cell lines were procured from National Centre for Cell Sciences (NCCS), Pune, India. CaSki cell line was propagated in RPMI-1640 with sodium bicarbonate without l-Glutamine supplemented with 10% fetal bovine serum and antibiotic, antimycotic solution (An antibiotic-antifungal cocktail having 10,000 units penicillin, 10 mg/mL streptomycin and 25 μg/mL Amphotericin B, which is solubilized in proprietary citrate buffer). C33A cells were grown DMEM medium consisting of l-Glutamine, Sodium pyruvate, Glucose and Sodium bicarbonate. Anti-mycotic solution was used at a concentration of 10 ml/L of media.
Plasmids
The pLEGFPN1-(RV-Tat 86) plasmid was a gift from Dr. Francesca Peruzzi, Associate Professor, LSU Health Sciences, New Orleans, LA, USA. The pNL4.3, a full-length replication competent, infectious HIV-1 subtype B, was obtained through National Institutes of Health (NIH), AIDS Research and Reference Rea-gent Program (ARRRP), USA. The pmigR1 encoding Intracellular Notch-1(ICN1) was kindly presented by Dr. Warren Pear, Professor of Pathology, Laboratory Medicine and Molecular Genetics, University of Pennsylvania, Philadelphia PA, USA. This plasmid was used as a positive control for Notch-1 activation.
Cell proliferation
Briefly, CaSki and C33A cells (104/well) were seeded in 96 micro-well culture plates and maintained at 37 °C in a CO2 incubator. Cells were exposed to serial dilutions of plasmid DNA/DAPT and incubated overnight for 24 h. Later, 20 µL of MTT solution (5 mg/mL) was added to each well and then the plates were incubated for 4 h. Supernatant was replaced with DMSO, and the plates were incubated for additional 1 h. The absorbance was measured at 550/630 nm. The viability was calculated in percentage with reference to control sets.
Recombinant cloning
Plasmids were transformed using DH-5α through heat shock (42–45 °C) and rapid chilling on ice. After heat shock treatment, the plasmid DH-5α mixture was added to pre-warmed SOC or LB without antibiotics. LB agar plate containing ampicillin was used to grow the transformed bacteria, at 32 °C. Colonies were further inoculated in 15 mL LB mixed with 65% glycerol for stock preparation and plasmid amplification.
Plasmid purification
Briefly, bacterial cells were pelleted, resuspended in 250 µL of Solution I (as per the manufacturer's protocol, Gene Mark plasmid miniprep purification kit). With subsequent addition of 250 μL Solution II, and following 4–5 times mixing, Solution III (250 µL) was added. The spin columns containing lysate/s were centrifuged, rinsed and purged of purified DNA with preheated elution buffer. The DNA was quantified for each plasmid viz HIV-1 Tat, HIV-1 and migR1 encoding Intracellular Notch-1 (ICN1) and stored at − 20 °C for transfection.
Transient transfection
Transfections were carried according to the manufacturer’s instructions (jetPEI® DNA transfection reagent, USA). Briefly, CaSki and C33A cells were seeded in T-25 flasks (1 × 106) with 5 mL medium containing FCS and antibiotics. FCS in culture medium enhances transfection with DNA. Hence, we were used media containing FCS for transfections of both CaSki and C33A cell lines. Quantified DNA (600 ng/mL) was used for transfecting one million cells [21]. The media was replenished the next day and cell lines maintained overnight in a CO2 incubator. Cells were detached for further analysis.
Cell cycle analysis
Transfected cells were scraped gently, washed twice in cold PBS and processed using BD Cycletest™ Plus DNA Kit as per the manufacturer's protocol. Briefly, cells were pelleted, mixed with Solution A (trypsin buffer) for 10 min followed by incubation with Solution B (trypsin inhibitor and RNase buffer) for 10 min. Finally, the cells were stained with 200 µL of cold Solution C (PI stain solution) kept for 10 min in the dark at 4 °C and analyzed for DNA content by BD FACSAria™ Fusion Flow Cytometer (BD, USA).
DNA isolation
DNA from transfected cells was isolated using the manufacturer’s protocol (Vivantis Technologies, Malaysia). Briefly, the cell pellet (5 × 106 ) was resuspended in PBS, treated with Proteinase K and lysis enhancer in Tris buffer. After incubation at 65 °C for 10 min the cells were transferred to the column, followed by column washing with wash buffer. DNA was eluted with preheated elution buffer and quantified using spectrophotometer.
Polymerase chain reaction (PCR)
Notch-1 PCR was optimized with DNA isolated from human fetal buccal mucosal (FBM) cell line procured from ACTREC, Mumbai, India. Twenty µL reaction mix (1.2 µL DNA template, primer mix and PCR dye master mix (5Xia)) was subjected to the following conditions; 95 °C for 4 min, 45 cycles at 95 °C for 20 s, 60 °C for 20 s, two extension steps, − 72 °C for 20 s, and 2 min. Notch-1 PCR protocol was also used for CaSki cells.
Tat PCR was performed with DNA isolated from CaSki and C33A cells using the following conditions; 95 °C for 9 min followed by 34 cycles at 95 °C for 1 min, 53 °C for 2 min, two extension steps at 72 °C for 1 min and 7 min respectively. PCR products were resolved on a 2% agarose gel and visualized on a UV transilluminator after staining with ethidium bromide.
Western blot analysis
C33A and CaSki cells were harvested, washed, and lysed with RIPA buffer supplemented with phenylmethylsulfonyl fluoride (PMSF). Protein extracts (30 μg) per sample were subjected to 8% SDS-PAGE and transferred to polyvinylidene difluoride membranes (EMD Millipore, USA). Membranes were blocked with 5% nonfat skim milk for 1 h at room temperature, probed with the appropriate primary antibodies (Notch-1, Hes-1, CDK2, EGFR and GAPDH a endogenous protein) at 1:1000 dilution, washed, and then incubated with the corresponding goat anti‑rabbit and anti-mouse secondary antibody (1:5000 dilution). The chemiluminescent signal was detected using ECL system (Intron, Seongnam, Korea). Semi‑quantitative analyses of the intensities of the protein bands were analyzed by using ImageJ software (NIH, USA).
RNA isolation
RNA was isolated from transfected cell lines as per the manufacturer’s protocol (GeneMark Biolab, Thailand). Briefly, transfected culture cells were lysed with lysis methanol cocktail in 70% ethanol. The lysate was loaded on the spin column and washed. After DNAse treatment, the columns were washed again with wash buffer. Purified RNA (30–50 µL) was collected, quantified by a NanoDrop 2000c (ThermoFisher Scientific, USA) and processed for qPCR.
Quantitative real-time PCR (qRT-PCR)
The qRT-PCR was conducted using Applied Biosystems™ Power SYBR™ Green RNA-to-CT™ One-Step Kit and ABI 7500 FAST v2.3 (ThermoFisher, Singapore) as per the manufacturer’s instructions. The qRT-PCR master mix (10 μL) consisted of a cocktail of 5 μL of 2X SYBR Green Real-time PCR Master, 2 μL primers -forward and reverse, 0.08 μL, RNA template (up to 100 ng), 2 μL, RNAse free water and 0.92 μL, RT Enzyme (125X). The thermal cycling conditions were 48 °C for 30 min, followed by 1 cycle at 95 °C for 10 min, 45 cycles at 95 °C for 15 s, 60 °C for 10 s, and 72 °C for 30 s. Experiments were run in triplicates with β-actin as housekeeping gene. Relative concentrations were calculated using 7500 FAST software v2.3. Cycle threshold (CT) values of transfected C33A cell line (an HPV-ve cell line) served as the internal control for normalizing sample variations. The forward and reverse primers used for qRT-PCR were human Notch-1, Hes-1, Hey-1, cyclin D, CDK2, p21, EGFR, and β-actin. (Supplementary, Table S1).
Densitometry and statistical analysis
The results were analyzed by GraphPad Prism 5 for Windows (GraphPad Software, USA). Student t-test was used to determine the statistical significance of the data expressed as mean ± standard deviation (SD). *p values ≤ 0.05 were considered statistically significant.
Results
Effect of HIV-1 Tat transfection on CaSki cells
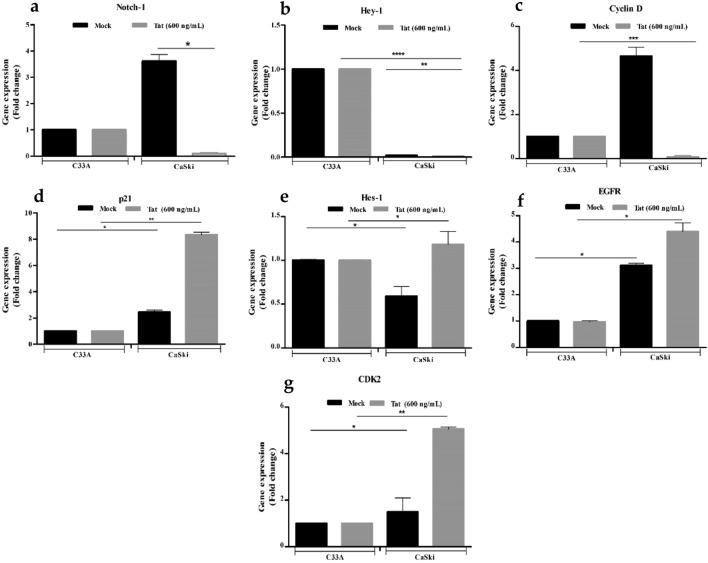
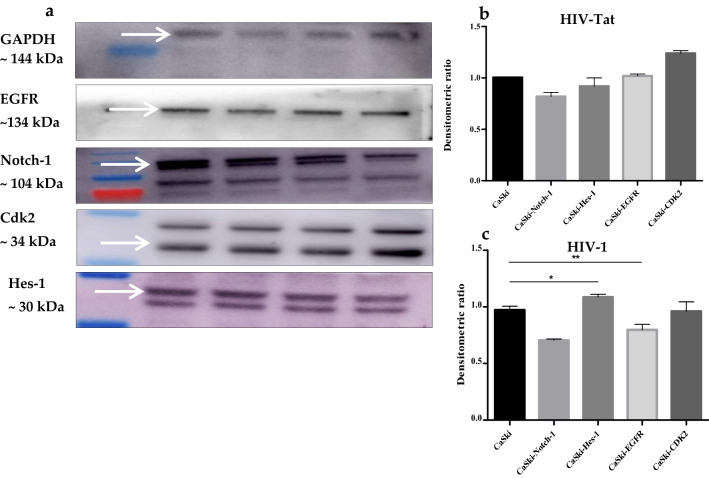
HIV-1 Tat maintained the viability of C33A and CaSki cells at 37 °C. HIV-1 Tat significantly inhibited Notch-1 activity as observed by the downstream transcriptional activator, Hes-1 hyperexpression in CaSki cells (Fig. 1e,*p ≤ 0.05). For WB analysis, four proteins (Notch-1, Hes-1, EGFR and CDK2) were normalized with the GAPDH protein. Protein levels were plotted as the densitometric ratio of CaSki + HIV-1 Tat (600 ng/mL) versus CaSki only (Fig. 3b). Hey-1, (Fig. 1b), the downstream Notch-1 transcriptional re-pressor was significantly suppressed following transfection with Tat (C33A mock vs. CaSki mock *p ≤ 0.05; C33A 600 ng/mL vs. CaSki 600 ng/mL, *p ≤ 0.05). Although HIV-1 Tat suppressed Notch-1 expression, EGFR was significantly upregulated when C33A and CaSki cell lines were compared (Fig. 1f,*p ≤ 0.05). CDK2 induction (Fig. 1g, *p ≤ 0.05) in Tat transfected CaSki cells, showed concomitant G0/G1 phase accumulation (60.8%; 66.20%; Supplemental, Table 2) when compared to HPV-ve C33A cells (55.9%: 81.55%, *p ≤ 0.05). Cyclin D expression was significantly shut down in Caski (HPV-16+) cells (Fig. 1c) which coincided with a significant p21 induction (Fig. 1d) as well as increased cell numbers in CaSki G2-M (12.3%; Supplemental, Table S2) phase cells.
Fig. 1.
Relative gene expression of HPV-16+ CaSki Cells normalized with HPV negative C33A cells after transfection with Tat DNA. The x-axis shows transfected cell lines with Tat DNA. Mock represents solvent control. Fold change for the following genes was calculated using HPV-16+ CaSki cells, a Notch-1 and its transcription factors, b Hes-1, c Hey-1, d EGFR, e Cyclin D, f CDK2, and g p21. (* denotes p ≤ 0.05, the significant difference level)
Fig. 3.
a Western blotting analysis to assess band density using Image J software for four proteins. GAPDH served as the house keeping gene. Intracellular Notch-1 (ICN1) was taken as positive control, b Densitometric ratio of HIV-1 Tat transfected CaSki cells (600 ng/mL) compared with CaSki mock, c Densitometric ratio of HIV-1 transfected CaSki cells (600 ng/mL) compared with CaSki mock. (* denotes p ≤ 0.05, the significant difference level)
Notch-1 activation explains aberrant mitosis in CaSki cells during HIV-1 transfection
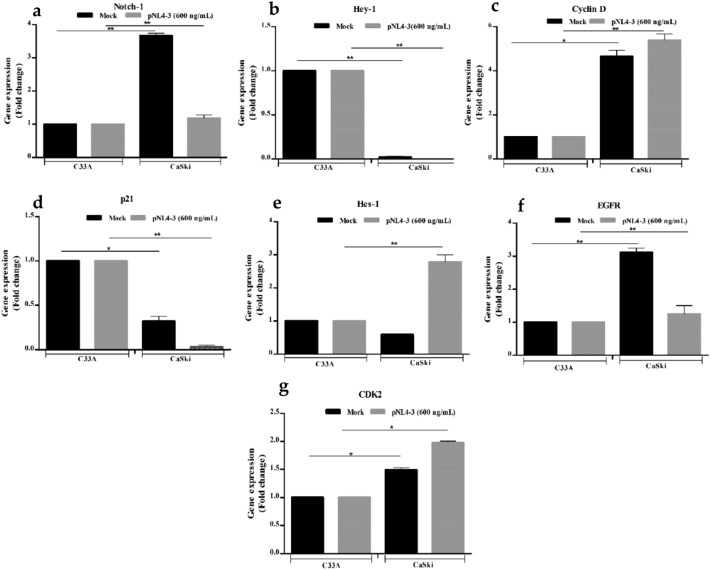
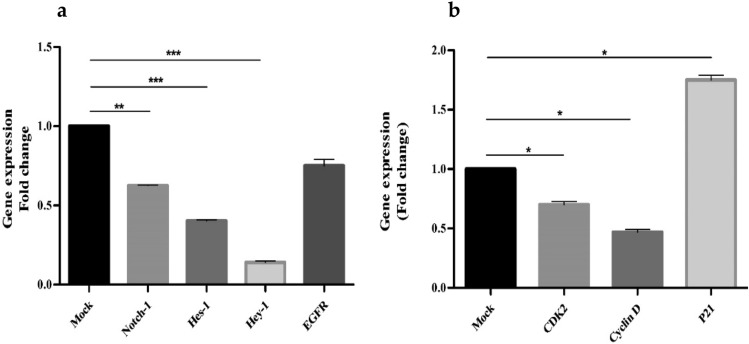
HIV-1 maintained the viability in C33A and HPV-16+ CaSki cells at 37 °C. Figure 2a and b show significant inhibition of Notch-1 expression, though its downstream target Hes-1 was overexpressed in HPV-16+ CaSki cells (*p ≤ 0.05) indicative of Notch-1 signaling.
Fig. 2.
Relative gene expression of HPV-16+ CaSki Cells normalized with HPV negative C33A cells after transfection with HIV-1 DNA. The x-axis shows transfected cell lines with HIV-1 DNA. Mock represents solvent control. Fold change for the following genes was calculated using HPV-16+ CaSki cells, a Notch-1 and its transcription factors, b Hes-1, c Hey-1, d EGFR, e Cyclin D, f CDK2, and g p21. (* denotes p ≤ 0.05, the significant difference level)
Notch-1 and Hes-1 proteins decreased and matched with the qPCR profile (Fig. 3a). HIV-1 also significantly inhibited Notch-1 transcriptional repressor Hey-1 (Fig. 3c). Interestingly, HIV-1 induced reverse expressions of EGFR (Fig. 3c) and Cyclin D (Fig. 3c) as that of HIV-1 Tat. EGFR inhibition affects G1 phase of the cell cycle, whereas overactive Cyclin D permits cellular differentiation and cancer progression. Additionally, overactive Cyclin D showed hyper CDK2 (Fig. 2g) expression corroborating increased G0/G1 cell accumulation. EGFR and CDK2 (Fig. 3b) protein levels overlap with the qPCR data.HIV-1 transfection of CaSKi cells showed significant G0-G1 phase cell accumulation (*p ≤ 0.05; 78.35%; Supplemental, Table S3) compared with C33A cells (60.5%). A significant decrease in the G2-M cell population (4.65%; *p < 0.05; Supplemental, Table S3) was observed in CaSki cells as compared to HPV-ve C33A cells (*p ≤ 0.05; Supplemental, Table S3). Consequent to G2-M arrest (Supplemental, Table S3) which collates with p21 (*p ≤ 0.05; Fig. 2d) shut down, HIV-1 transfection confirmed transcriptional Hes-1 mediated ~ p21 repression in a bHLH domain dependent manner plausibly promoting Cyclin D induction. Excess Cyclin D forming complexes with CDK2, DNA Damaged Response (DDR), genomic instability, subsequent DNA repair, unscheduled aberrant mitosis and cancer progression.
Incubation with Notch-1 inhibitor rescued p21 expression
CaSki cells were incubated with 62.5 mM/L DAPT to assess the effects of Notch-1 blockade on proliferation of HPV-16+ CaSki cells. MTT assay data were showed suppression of Notch-1 which affected the growth and proliferation of CaSki cells (Supplemental, Fig. S5). qPCR data demonstrated a significant decreased expression for Notch-1, Hes-1, Hey-1 and EGFR gene 24 h post incubation of DAPT with CaSki cells (*p ≤ 0.05; Fig. 4a) as compared with solvent Control. Cell cycle analysis also showed G0/G1 arrest in DAPT treated CaSki cells (Supplemental, Table S4). Consistent with G0-G1 arrest, cyclin D (*p ≤ 0.05; Fig. 4b) was maintained, with significant p21 restoration (*p ≤ 0.05; Fig. 4b), and significant appreciable CDK2 expression. (*p ≤ 0.05; Fig. 4b).
Fig. 4.
qPCR analysis of HPV-16+ CaSki cells treated with Notch-1 inhibitor, DAPT for 24 h; a Mock represents solvent control used in preparation of 62.5 mM/L DAPT, a Notch-1 inhibitor. X-axis represents the relative gene expression (fold change) for mock, Notch-1, Hes-1, Hey-1 and EGFR, b Mock represents solvent control used in preparation of 62.5 mM/L DAPT, a Notch-1 inhibitor. X-axis represents the relative gene expression (fold change) for mock, CDK2, cyclin D and p21. (* denotes p ≤ 0.05, the significant difference level)
Discussion
In this study, we used an in vitro concept, transient transfection, to establish Notch-1 signaling in HPV-16 + CaSki cells taking lessons learnt from Notch-1 signaling in human breast cancer cell lines [13]. Though this is an in vitro study, it lays foundations for future research and interventions, and therefore is necessary. Notch family genes, besides normal embryogenesis are also implicated in cancers [22]. Studies in differentiated HIV-1 infected macrophage cell line (U1) when incubated with HPV-infected CaSki culture supernatant demonstrated a significant increase in HIV-1 replication which was associated with increased cytochrome P450, superoxide dismutase 1 (SOD-1) and HPV oncoprotein E6 expression [23]. Earlier studies in human cervical tissues elaborated an altered expression of regulatory and cell cycle proteins in the cervix due to HIV-1 infection occurring in the context of a co-existing HPV infection. Whilst HIV-1 alone may subvert cell cycle progression to enhance cervical carcinogenesis, Tat, besides disturbing cell cycle progression, also has a super advantage over cellular proliferation [8, 24].
HPV associated cancers require Notch-1 inhibition to induce malignant transformation [6, 22]. Tat significantly elicited Notch-1 inhibition in HPV-16+ CaSki cells through activation of Notch-1 signaling pathway, characterized by its downstream target genes Hes-1 and Hey-1 [25]. Hey-1 appears to be depleted in CaSki cells a possible characteristic of transformed cell lines. Based on our findings, we propose that HIV-1 Tat plausibly binds to EGF repeats on the extracellular Notch-1 domain, consequently suppressing Notch-1 expression [26, 27]. Hes-1 and Hey-1 genes are involved in adhesion, invasion, angiogenesis and proliferative events during carcinogenesis [28, 29]. In T-cell leukemia, Hes-1 binds to the nuclear Cyclin D promoter to elicit Notch-1 signaling. Concomitantly, Hes-1 induction and Cyclin D repression are obligatory events for sustenance and cancer progression in T-cell leukemia [30]. Our observations in HIV-1 Tat transfected HPV-16+ CaSki cells corroborate with the above reported findings [28, 30]. HIV-1 Tat significantly induces Hes-1 expression for Cyclin D shut down (*p ≤ 0.05), with suppression of EGFR. Now, EGFR, the transcriptional target of Notch-1 [31], cooperates with downstream targets like Cyclin D and CDK2 for growth, proliferation and cell cycle progression. Additionally, Cyclin D, affiliated as downstream target of multiple oncogenic pathways, is also functional in G1 stage of the cell cycle [32, 33]. Cell cycle analysis demonstrate that HIV-1 Tat significantly activates EGFR with consequent significant G0/G1 phase cell accumulation (*p ≤ 0.05; Supplementary, Table S2) in HPV-16+ CaSki cells as compared with the HPV-ve, C33A cells. Recent evidences highlight that Cyclin D depletion disturbs oxidative balance in cancer cells. Subsequently, cancer cells succumb to high lethal oxidative stress inducing irreversible senescence [33]. Hence, HIV-1 Tat adjusts the tumor microenvironment to maintain the proliferative state to evade irreversible senescence through Hes-1 activation, EGFR and p21 amplification drives cell migration and invasion for an aggressive less therapeutic phenotype in HPV-16+ CaSki cells [34]. The above qRT-PCR data significantly collates with cell cycle analysis of G2-M phase population of HPV-16+ CaSki cells (*p ≤ 0.05).
HIV-1, significantly (**p ≤ 0.005) inhibits Notch-1 and Hey-1 expression though its downstream transcriptional activator Hes-1 (*p ≤ 0.05). Concomitantly, an oscillatory gene expression between Hes-1 and EGFR following HIV-1 infection indicates a plausible crosstalk between Notch-1 and EGFR pathways [13, 31]. As reported earlier, Hes-1 and Cyclin D gene expressions manipulate cancer maintenance and progression [30]. EGFR suppression and significant hyperactive CDK2 expression corroborate with cell cycle data showing significant G0/G1 phase cellular accumulation/s at the expense of S phase (*p ≤ 0.05; Supplemental, Table S3) for cancer progression. Excess CDK2 forms nuclear complexes with Cyclin D, disturbing cell cycle progression [33]. Although, there is significant CDK2 hyper expression (*p ≤ 0.05), yet CDK2 is insufficient to phosphorylate p21 such that p21 subsequently shuts down proliferation (*p ≤ 0.05; Fig. 2d, g), decline in polyploid G2-M phase HPV-16+ CaSki cells (4.65%; (*p ≤ 0.05; Supplemental, Table S3) was observed (data not shown) [33, 34]. HIV-1 Vpr (Viral Protein R) transcription causes G2-M of the cell cycle arrest inhibiting cell proliferation [35]. Additive effects of HIV-1 Vpr and Notch-1/CXCR4 [17] partner together to inhibit p21 and juxtapose aberrant mitosis establishing G2-M arrest, replication restart for metastasis and cancer progression [35, 36].
Our results with DAPT, a Notch-1 inhibitor and γ-secretase blocker revealed significant recovery in the G0/G1 and S phase HPV-16+ CaSki cells (Supplemental, Table S4). Notch-1 inhibition was approximately 35% (*p ≤ 0.05; Fig. 4a). Downstream targets Hes-1 & Hey-1 were significantly inhibited (*p ≤ 0.05; Fig. 4a). There was a marginal recovery in p21 expression (*p ≤ 0.05; Fig. 4b). DAPT rescues G0/G1 (*p ≤ 0.05; Supplemental, Table S4) and S phase (*p ≤ 0.05; Supplemental, Table S4) during HPV carcinogenesis.
Conclusions
Our study highlights for the first time that Tat inhibits Notch-1 expression in HPV-16+ CaSki cells. Concomitantly, Tat hyper expresses Hes-1 and EGFR cumulatively amplifying p21 expression. Taken together, Tat favors an invasive and aggressive phenotype for cancer metastasis. On the contrary, HIV-1 infection exploits Hes-1 expression such that the overactive cyclin D shuts down p21 expression triggering unbridled mitosis (giant cell formation), G2-M arrest, damaged DNA response (DDR) for cancer growth and progression. However, treatment of CaSki cells with a Notch-1 inhibitor, DAPT provides protection against disease progression in HIV-1/HPV-16+ cervical cancers and offers a therapeutic promise with a marginal recovery in p21 G0/G1 and S phase.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors gratefully acknowledge Drs VM Katoch and Soumya Swaminathan, Former-Director General/s ICMR, New Delhi, India, for extramural support. Constructive discussions from Dr. Pradeep Seth, Chairman, Limited Department Competitive Examination (LDCE) committee and Dr. Madhu Chhanda Das, Scientist D, ECD-II, ICMR, New Delhi, India, are appreciated. Dr. SM Shahabuddin and Mr. Ajit Patil, Technical officer B, ICMR-NARI, Pune, India, are thanked for help with the cross-referencing and initial laboratory work respectively.
Funding
This research work was funded by an extramural Grant from the Indian Council of Medical Research (ICMR), New Delhi, India (No. 5/2–3/LDCE/2015-ECD-II).
Declarations
Conflict of interest
The authors have no conflict of interest to declare.
Consent for publication
All authors have read the manuscript and approved the final manuscript.
Institutional review board
ICMR-NARI institutional review board approved this research study. Waiver of consent (vide NARI/EC Protocol No.: 2015–15) for C33A and CaSki cell lines was also granted.
Informed consent
Written informed consent has been obtained from both participating patient(s).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Godbole SV, Nandy K, Gauniyal M, et al. HIV and cancer registry linkage identifies a substantial burden of cancers in persons with HIV in India. Medicine (Baltimore) 2016;95(37):e4850. doi: 10.1097/MD.0000000000004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debeaudrap P, Sobngwi J, Tebeu PM, Clifford GM. Residual or recurrent precancerous lesions after treatment of cervical lesions in human immunodeficiency virus-infected women: a systematic review and meta-analysis of treatment failure. Clin Infect Dis. 2019;69(9):1555–1565. doi: 10.1093/cid/ciy1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryu A, Nam K, Kwak J, Kim J, Jeon S. Early human papillomavirus testing predicts residual/recurrent disease after LEEP. J Gynecol Oncol. 2012;23(4):217–225. doi: 10.3802/jgo.2012.23.4.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mane A, Sahasrabuddhe VV, Nirmalkar A, et al. Rates and determinants of incidence and clearance of cervical HPV genotypes among HIV-seropositive women in Pune, India. J Clin Virol. 2017;88:26–32. doi: 10.1016/j.jcv.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menon S, Rossi R, Kariisa M, Callens S. Determining the HPV vaccine schedule for a HIV-infected population in sub Saharan Africa, a commentary. Virol J. 2018;15(1):129. doi: 10.1186/s12985-018-1039-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weijzen S, Zlobin A, Braid M, Miele L, Kast WM. HPV16 E6 and E7 oncoproteins regulate Notch-1 expression and cooperate to induce transformation. J Cell Physiol. 2003;194(3):356–362. doi: 10.1002/jcp.10217. [DOI] [PubMed] [Google Scholar]
- 7.Li L, Dahiya S, Kortagere S, et al. Impact of Tat genetic variation on HIV-1 disease. Adv Virol. 2012 doi: 10.1155/2012/123605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nyagol J, Leucci E, Onnis A, et al. The effects of HIV-1 Tat protein on cell cycle during cervical carcinogenesis. Cancer Biol Ther. 2006;5(6):684–690. doi: 10.4161/cbt.5.6.2907. [DOI] [PubMed] [Google Scholar]
- 9.Kovall RA, Gebelein B, Sprinzak D, Kopan R. The canonical notch signaling pathway: structural and biochemical insights into shape, sugar, and force. Dev Cell. 2017;41(3):228–241. doi: 10.1016/j.devcel.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan Y, Gao X, Chen J, Liu Y, He JJ. HIV Tat impairs neurogenesis through functioning as a Notch ligand and activation of Notch signaling pathway. J Neurosci. 2016;36(44):11362–11373. doi: 10.1523/JNEUROSCI.1208-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo Z, Mu L, Zheng Y, et al. NUMB enhances Notch signaling by repressing ubiquitination of NOTCH1 intracellular domain. J Mol Cell Biol. 2020;12(5):345–358. doi: 10.1093/jmcb/mjz088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rong C, Feng Y, Ye Z. Notch is a critical regulator in cervical cancer by regulating Numb splicing. Oncol Lett. 2017;13(4):2465–2470. doi: 10.3892/ol.2017.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tripathi R, Rath G, Sharma V, et al. HES1 protein modulates human papillomavirus-mediated carcinoma of the uterine cervix. J Glob Oncol. 2019;5:1–10. doi: 10.1200/JGO.18.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai J, Ma D, Zang S, et al. Cross-talk between Notch and EGFR signaling in human breast cancer cells. Cancer Invest. 2009;27(5):533–540. doi: 10.1080/07357900802563036. [DOI] [PubMed] [Google Scholar]
- 15.Raja R, Ronsard L, Lata S, Trivedi S, Banerjea AC. HIV-1 Tat potently stabilises Mdm2 and enhances viral. Biochem J. 2017;474(14):2449–2464. doi: 10.1042/BCJ20160825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo D, Ye J, Dai J, et al. Notch-1 regulates Akt signaling pathway and the expression of cell cycle regulatory proteins cyclin D1, CDK2 and p21 in T-ALL cell lines. Leuk Res. 2009;33(5):678–685. doi: 10.1016/j.leukres.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 17.Colombo M, Mirandola L, Chiriva-Internati M, et al. Cancer cells exploit Notch signaling to redefine a supportive cytokine milieu. Front Immunol. 2018;9:1823. doi: 10.3389/fimmu.2018.01823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu C, Zhao H, Chen H, Yao Q. CXCR4 in breast cancer: oncogenic role and therapeutic targeting. Drug Des Dev Ther. 2015;9:4953–4964. doi: 10.2147/DDDT.S84932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsaouli G, Ferretti E, Bellavia D, Vacca A, Felli MP. Notch/CXCR4 partnership in acute lymphoblastic leukemia progression. J Immunol Res. 2019 doi: 10.1155/2019/5601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi B, Sharifi HJ, DiGrigoli S, et al. Inhibition of HIV early replication by the p53 and its downstream gene p21. Virol J. 2018;15(1):53. doi: 10.1186/s12985-018-0959-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iijima K, YukihitoIshizaka Y. DNA unwinding by viral protein R initializes complicated cellular responses in HIV-1 infection: defining the viper’s first bite. J Emerg Dis Virol. 2018;4(1):1–11. doi: 10.16966/247. [DOI] [Google Scholar]
- 22.Ajiro M, Zheng ZM. E6^E7, a novel splice isoform protein of human papillomavirus 16, stabilizes viral E6 and E7 oncoproteins via HSP90 and GRP78. mBio. 2015;6(1):e02068-14. doi: 10.1128/mBio.02068-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabina R, et al. Extracellular vesicles from human papilloma virus-infected cervical cancer cells enhance HIV-1 replication in differentiated U1 cell line. Viruses. 2020;12(2):239. doi: 10.3390/v12020239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicol Alcina F, et al. Cell-cycle and suppressor proteins expression in uterine cervix in HIV/HPV co-infection: comparative study by tissue micro-array (TMA) BMC Cancer. 2008;8:289. doi: 10.1186/1471-2407-8-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aster JC, Pear WS, Blacklow SC. The varied roles of notch in cancer. Annu Rev Pathol. 2017;12:245–275. doi: 10.1146/annurev-pathol-052016-100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barillari G, Palladino C, Bacigalupo I, Leone P, Falchi M, Ensoli B. Entrance of the Tat protein of HIV-1 into human uterine cervical carcinoma cells causes upregulation of HPV-E6 expression and a decrease in p53 protein levels. Oncol Lett. 2016;12(4):2389–2394. doi: 10.3892/ol.2016.4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuster-Gossler K, et al. Context-dependent sensitivity to mutations disrupting the structural integrity of individual EGF repeats in the mouse notch ligand DLL1. Genetics. 2016;202(3):1119–1133. doi: 10.1534/genetics.115.184515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Cao Y, Li M, Jin F. Upregulation of HES1 promotes cell proliferation and invasion in breast cancer as a prognosis marker and therapy target via the AKT pathway and EMT process. J Cancer. 2018;9(4):757–766. doi: 10.7150/jca.22319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z, Sanders AJ, Liang G, Song E, Jiang WG, Gong C. Hey factors at the crossroad of tumorigenesis and clinical therapeutic modulation of hey for anticancer treatment. Mol Cancer Ther. 2017;16(5):775–786. doi: 10.1158/1535-7163.MCT-16-0576. [DOI] [PubMed] [Google Scholar]
- 30.D'Altri T, Gonzalez J, Aifantis I, Espinosa L, Bigas A. Hes1 expression and CYLD repression are essential events downstream of Notch1 in T-cell leukemia. Cell Cycle. 2011;10(7):1031–1036. doi: 10.4161/cc.10.7.15067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purow BW, Sundaresan TK, Burdick MJ, et al. Notch-1 regulates transcription of the epidermal growth factor receptor through p53. Carcinogenesis. 2008;29(5):918–925. doi: 10.1093/carcin/bgn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laphanuwat P, Likasitwatanakul P, Sittithumcharee G, et al. Cyclin D1 depletion interferes with oxidative balance and promotes cancer cell senescence. J Cell Sci. 2018;131(12):jcs214726. doi: 10.1242/jcs.214726. [DOI] [PubMed] [Google Scholar]
- 33.Ronchini C, Capobianco AJ. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic) Mol Cell Biol. 2001;21(17):5925–5934. doi: 10.1128/MCB.21.17.5925-5934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dash BC, El-Deiry WS. Phosphorylation of p21 in G2/M promotes cyclin B-Cdc2 kinase activity. Mol Cell Biol. 2005;25(8):3364–3387. doi: 10.1128/MCB.25.8.3364-3387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chowdhury IH, Wang XF, Landau NR, et al. HIV-1 Vpr activates cell cycle inhibitor p21/Waf1/Cip1: a potential mechanism of G2/M cell cycle arrest. Virology. 2003;305(2):371–377. doi: 10.1006/viro.2002.1777. [DOI] [PubMed] [Google Scholar]
- 36.Topacio BR, Zatulovskiy E, Cristea S, et al. Cyclin D-Cdk 4,6 drives cell-cycle progression via the retinoblastoma protein's C-terminal helix. Mol Cell. 2019;74(4):758–770.e4. doi: 10.1016/j.molcel.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.