Summary
Background
Accurate identification of pCR to neoadjuvant chemotherapy (NAC) is essential for determining appropriate surgery strategy and guiding resection extent in breast cancer. However, a non-invasive tool to predict pCR accurately is lacking. Our study aims to develop ensemble learning models using longitudinal multiparametric MRI to predict pCR in breast cancer.
Methods
From July 2015 to December 2021, we collected pre-NAC and post-NAC multiparametric MRI sequences per patient. We then extracted 14,676 radiomics and 4096 deep learning features and calculated additional delta-value features. In the primary cohort (n = 409), the inter-class correlation coefficient test, U-test, Boruta and the least absolute shrinkage and selection operator regression were used to select the most significant features for each subtype of breast cancer. Five machine learning classifiers were then developed to predict pCR accurately for each subtype. The ensemble learning strategy was used to integrate the single-modality models. The diagnostic performances of models were evaluated in the three external cohorts (n = 343, 170 and 340, respectively).
Findings
A total of 1262 patients with breast cancer from four centers were enrolled in this study, and pCR rates were 10.6% (52/491), 54.3% (323/595) and 37.5% (66/176) in HR+/HER2−, HER2+ and TNBC subtype, respectively. Finally, 20, 15 and 13 features were selected to construct the machine learning models in HR+/HER2−, HER2+ and TNBC subtypes, respectively. The multi-Layer Perception (MLP) yields the best diagnostic performances in all subtypes. For the three subtypes, the stacking model integrating pre-, post- and delta-models yielded the highest AUCs of 0.959, 0.974 and 0.958 in the primary cohort, and AUCs of 0.882–0.908, 0.896–0.929 and 0.837–0.901 in the external validation cohorts, respectively. The stacking model had accuracies of 85.0%–88.9%, sensitivities of 80.0%–86.3%, and specificities of 87.4%–91.5% in the external validation cohorts.
Interpretation
Our study established a novel tool to predict the responses of breast cancer to NAC and achieve excellent performance. The models could help to determine post-NAC surgery strategy for breast cancer.
Funding
This study is supported by grants from the National Natural Science Foundation of China (82171898, 82103093), the Deng Feng project of high-level hospital construction (DFJHBF202109), the Guangdong Basic and Applied Basic Research Foundation (grant number, 2020A1515010346, 2022A1515012277), the Science and Technology Planning Project of Guangzhou City (202002030236), the Beijing Medical Award Foundation (YXJL-2020-0941-0758), and the Beijing Science and Technology Innovation Medical Development Foundation (KC2022-ZZ-0091-5). Funding sources were not involved in the study design, data collection, analysis and interpretation, writing of the report, or decision to submit the article for publication.
Keywords: Breast cancer, Multi-parametric MRI, Neoadjuvant chemotherapy, Pathological complete response, Longitudinal radiomics, Deep learning
Research in context.
Evidence before this study
Primary tumor often shows different response to neoadjuvant chemotherapy (NAC) in patients with breast cancer. Different outcome after NAC could benefit from different surgery strategy. Predicting pCR to NAC is significant for identifying candidates suitable for more limited operations such as breast-conserving surgery (BCS) even without surgery. However, tools predicting pCR in non-invasive way are limited. Radiomics and deep learning, are powerful tools for quantitative analysis of medical images. They might capture visually unrecognizable tumor heterogeneity from multi-parametric magnetic resonance images (prior to NAC and after NAC) and predict pCR based on precise analysis to specific molecular subtypes of breast cancer. We searched PubMed and Web of Science for the keywords (“Breast cancer”, “Breast tumor”, “Neoadjuvant chemotherapy”, “Pathological complete response”, “Magnetic resonance image”, “Radiomics”, “Texture analysis”, “Machine learning”, “Longitudinal Magnetic resonance image”, “Ensemble learning” or “Deep learning”) up to February 7, 2023, with no language restrictions. No study has investigated the efficacy of longitudinal MRI-based radiomics and deep learning analysis for predicting pCR in breast cancer treated with NAC in each molecular subtype.
Added value of this study
To our best knowledge, this is the first multicenter cohorts radiomics and deep learning study using the longitudinal MRI for predicting pCR. We constructed ensemble learning models using pre-NAC and post-NAC MRI and accurately predicted pCR for each molecular subtype of breast cancer. For breast cancer patients, this model can help surgeons decide on breast-conserving surgery and sentinel lymph node biopsy after NAC treatment. Importantly, the model is robust across different centers even MRI scanners changed.
Implications of all the available evidence
Our ensemble models reinforce the knowledge of using radiomics and deep learning tools to predict pCR to NAC in breast cancer by precise analysis based on molecular subtypes. Our non-invasive models can serve as a supplement to current surgical risk stratification strategies and require no additional expense since MRI examination is routinely performed in clinical practice for breast cancer before and after NAC treatment.
Introduction
Breast cancer has become the most common cancer and causes the most cancer-related death among women, which is experiencing a gradual increase.1 Neoadjuvant chemotherapy (NAC) has been a standard treatment to downstage tumor for breast cancer. Pathological complete response (pCR) is an effective surrogate endpoint to predict prognosis in breast cancer.2, 3, 4 The pCR rates were tremendously different among various molecular subtypes due to tumor heterogeneity.5, 6, 7, 8 Only about 30–50% of breast cancer reached pCR (defined as ypT0/isypN0) after the completion of NAC. Moreover, about 29% of breast cancers failed to respond to NAC and 7.9% even upstaged after NAC.9 For surgery after NAC, patients who achieved pCR could benefit from breast-conserving surgery (BCS), even omitting surgery instead of breast mastectomy.10, 11, 12 However, the gold standard of pCR assessment depends on the pathological results of surgical specimens after NAC. There still needs to be more consensus in identifying appropriate patients eligible for breast-conserving surgery. Thus, it is clinically significant to predict the pCR to improve surgical risk stratification and patient management.
There have been several methods to monitor tumor response to NAC, such as physical examination, ultrasonography (US), computed tomography (CT), mammography, magnetic resonance imaging (MRI) and positron emission tomography/computed tomography (PET/CT).13, 14, 15, 16 Those methods provide helpful information on the tumor size and extent.17, 18, 19 The response evaluation criteria in solid tumors (RECIST) was applied to classify breast cancer response into four types: (1) complete response, (2) partial response, (3) stable disease and (4) progressive disease according to their volumetric changes.20 However, in clinical practice, the complete response assessed by RECIST is insufficient to indicate pCR. The ACRIN 6657/I-SPY Trial found that pCR could be predicted in the early stage of NAC according to tumor size change monitoring by MRI examination.21 MRI performs better in determining tumor extent and morphology prior to and after NAC than mammography and US, as it provides comprehensive imaging information of tumors.22 Dynamic contrast-enhanced (DCE) MRI is considered an accurate tool in monitoring the residual tumor. The apparent diffusion coefficient (ADC) mapping derived by diffusion-weighted imaging (DWI) also provides useful quantitative information.23,24 More recently, investigators have suggested that using tumor stage, hormone receptor status, HER2 status and MRI radiological features has shown promise for predicting pCR to NAC.13,14 However, a meta-analysis found that MRI only had a sensitivity of 64% for predicting pCR.25 Besides, for detecting the axillary lymph node metastasis after NAC, a study reported that the sensitivity was 77% and the specificity was 54% of MRI. In contrast, the diagnostic indexes of US are 50% and 72%, respectively.26
Radiomics and deep learning are emerging interdisciplinary combining medical imaging and the computer field, which extracts lots of quantitative information from medical images and shows great potential to assist in clinical diagnosis and treatment.27, 28, 29, 30, 31 In 2019, we reported a MRI-based radiomics model to predict pCR in breast cancer, and the RMM model had excellent performances with AUCs of 0.71–0.80 in multicenter validation.13 However, breast tumors are spatially and temporally heterogeneous in different molecular subtypes, which results in diverse imaging-derived characteristics. MRI imaging could reflect a comprehensive view of the entire tumor and monitor longitudinal tumor change during NAC. We hypothesize that radiomics and deep learning could acquire more quantitative features from longitudinal multi-parametric MRI in different subtypes of breast cancer to predict pCR better. There is no deep learning radiomics model integrating the pre-NAC and post-NAC MRI to predict pCR to NAC. The feasibility of radiomics and deep learning tools to predict pCR based on longitudinal MRI remains to be tested in multicenter datasets. Therefore, we aim to construct and validate different artificial intelligence models for predicting pCR using longitudinal multi-parametric MRI based on various molecular subtypes.
Methods
Study participants
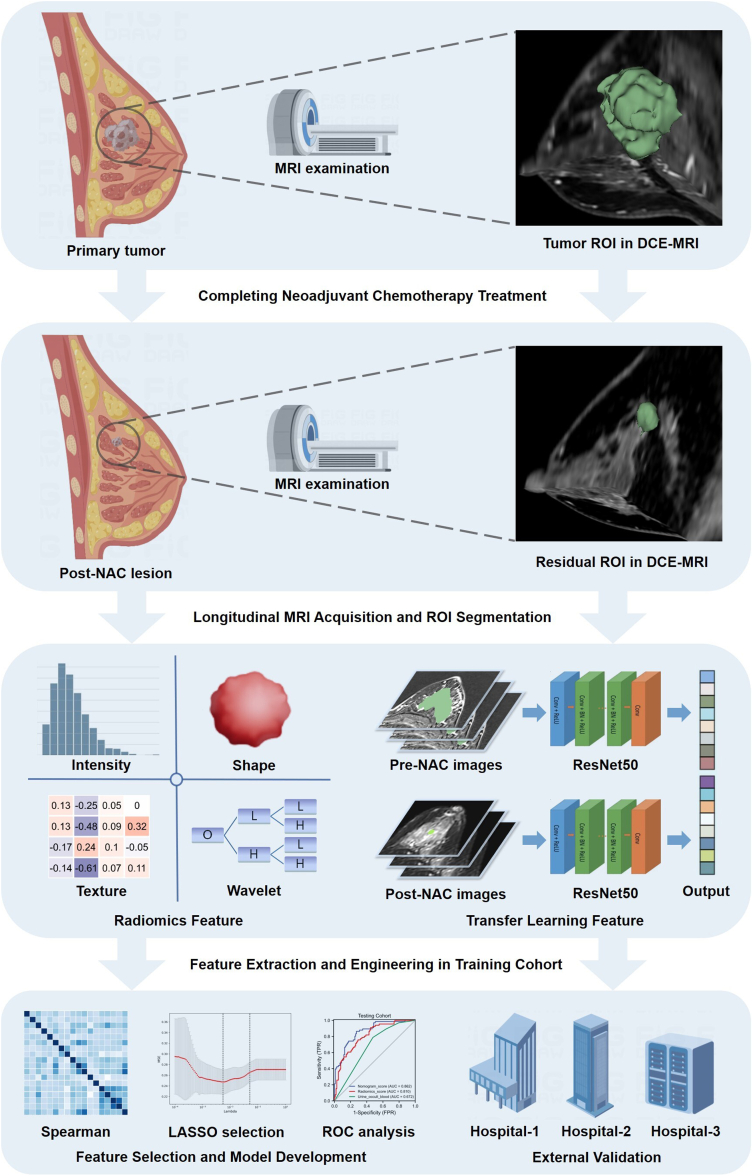
From July 2015 to December 2021, the patients with breast cancer were retrospectively recruited from the Guangdong Provincial People's Hospital and used as the primary cohort to develop machine learning models. Three external validation cohorts were also consecutively enrolled from the First People's Hospital of Foshan, the Shantou Central Hospital and the First Affiliated Hospital of Sun Yat-sen University. The eligibility criteria were as follows: (i) invasive breast cancer; (ii) completing NAC treatment and following surgery; (iii) acquisition of MRI data prior to and after NAC treatment; (iv) complete clinical and pathological data. Exclusion criteria were: (i) bilateral breast cancer; (ii) incomplete or non-standard NAC treatment or surgery; (iii) inadequate MRI quality or lack of MRI data; (iv) metastatic disease or another malignancy. Clinical data were reviewed and collected from the electronic medical record system. The study design and pipeline are shown as Fig. 1.
Fig. 1.
The study design and workflow of longitudinal MRI-based radiomics deep learning in predicting pCR to neoadjuvant chemotherapy.
Ethics statement
This study has received approval from the Institutional Ethics Review Board of all the involved hospitals. As this was a retrospective study, the requirement for informed consent was waived.
Treatment strategies and pathological assessment
All patients received 6 or 8 cycles of NAC treatment. The regimens were based on either taxane or taxane combined with anthracycline. All human epidermal growth factor receptor 2 positive (HER2+) patients also received trastuzumab or a combination of trastuzumab and pertuzumab drugs. After systemic NAC treatment, breast-conserving surgery or mammectomy was performed. Sentinel lymph node biopsy (SLNB) with/without axillary lymph node dissection (ALND) was performed to determine axillary lymph node staging.
The tumor type and receptor status were confirmed by immunohistochemistry (IHC) of US-guided core biopsies. HR was defined as positive, with ≥1% of nuclear staining of estrogen receptor (ER) or progesterone receptor (PR). Tumors were considered HER2- when IHC 0 and 1+ grades were observed, while HER2+ was determined with IHC 3+ grade.32,33 The HER2 gene amplification was determined by gene amplification by fluorescence in situ hybridization (FISH) when HER2 expression graded 2+ was obtained by IHC. Depending on the receptor status, all the patients were classified into three subtypes depending on receptor status as follows: (i) HR+/HER2−; (ii) HER2+; (iii) TN (triple-negative). To evaluate the degree of Ki-67 expression, we set the cutoff index as 20%, with <20% indicating low expression and ≥20% indicating high expression. The pCR was defined as ypT0/is/ypN0 according to the pathological examination of surgical specimen.
MRI acquisition and post-processing
All the MRI examinations were performed with 1.5- or 3.0- Tesla scanners within two weeks before initiation of NAC and after completing NAC. The imaging sequences included T2-weighted images (T2WI), dynamic contrast-enhanced (DCE) images, and diffusion-weighted imaging (DWI). After intravenous injection of gadolinium contrast agent (0.2 ml/kg) within 2 min, the first post-contrast images were acquired, and then five subsequent post-contrast images were acquired. Axial DWI images were acquired with two b-values (0 and 1000 s/mm2). Detailed information about MRI acquisition could be seen in Supplementary materials.
Tumor segmentation and radiomics analysis
Three breast-specialized radiologists delineated the tumor regions of interest (ROI) and 5-mm peri-tumor regions with the 3D Slicer software (version 4.10.2, www. slicer.org).34,35 The regions of necrosis, calcifications, or hemorrhage were carefully avoided. Detailed methods for the ROI segmentation and registration can be seen in the Supplementary material. Then we performed the feature extraction using Pyradiomics Module (https://github.com/Radiomics/pyradiomics).29,36 Filters (Laplacian of Gaussian filters and wavelet) were used to get more derived images. All the radiomics features could be classified into 7 types: (i) 14 shape-based features; (ii) 234 first-order features; (iii) 182 gray-level dependence matrix (GLDM) features; (iv) 208 gray-level size zone matrix (GLSZM) features; (v) 65 neighboring gray-tone difference matrix (NGTDM) features; (vi) 208 gray-level run-length matrix (GLRLM) features; (vii) 312 gray-level co-occurrence matrix (GLCM) features. A total of 1223 features were extracted from each ROI and its corresponding MRI sequence, totaling 14,676 features from the twelve ROIs (tumor region and peri-tumor region) and MRI sequences (pre-NAC DCE, T2WI and DWI; post-NAC DCE, T2WI and DWI) were obtained per patient. Furthermore, to reflect the longitudinal change of tumors, the delta-radiomics features were calculated as the relative net change from the pre-NAC radiomics feature value to the post-NAC radiomics feature value. A total of 21,924 radiomics features could be obtained per patient, including pre-NAC, post-NAC and delta-NAC radiomics feature sets. The different features and their detailed explanation can be seen in the Supplementary material.
Deep learning analysis
The deep learning model was trained using the ResNet50 framework.37 Detailed network architecture is reported in the Supplementary material. The deep learning model received multiple inputs, including the pre-NAC and post-NAC DCE MRI images. We chose the slice of the largest section of breast tumor for each DCE MRI sequence per patient. The input ROI images contained the whole tumor region and its border region, which were manually cropped from the raw MRI images. The original images of DCE MRI were input and the pixels in any image were normalized to 0–1000. The image box included the lesion was resampled to 448 × 448 pixels. The training Adam optimizer was 0.001 and the batch size was 64. L2 regularization and early stopping were used to prevent over-fitting. The loss rate was used to evaluate the model performance. The deep learning process was developed and each slice was an independent input. After the deep learning model training was finished, the features in the full connecting layer were extracted as the deep learning features (DLF). For external validation cohorts, the ROI images were input into the deep learning model and analyzed layer by layer. The feature values in the full connecting layer were extracted as well. A total of 4096 features were obtained from the pre-NAC and post-NAC ROI images. Then the relative net change from the pre-NAC deep learning features to the post-NAC deep learning features were also calculated to get delta-deep learning features.
Feature selection
We considered that radiomics and deep learning features had diverse importance degrees in different molecular subtypes, so we performed feature selection steps for each subtype (HR+/HER2−, HER2+ and TNBC). To ensure the stability of features extracted from ROI, 100 patients were randomly selected and the ROIs segmentations were performed twice by different radiologists. Then the inter-class correlation coefficient of each feature was calculated. To select the features most correlated with the pCR outcome, we used the U test to select the features with significant differences between pCR group and non-pCR group. The Boruta method was used to calculate each feature's Shapley value and the max shadow value. When a Shapley value was higher than the max shadow value, the corresponding feature was selected for further analysis. Furthermore, we used the least absolute shrinkage and selection operator (LASSO) logistic regression supported by Onekey AI platform to reduce the number of features. The Spearman correlation analysis was performed, and the correlation coefficients among features were calculated to evaluate their multi-collinearity. If there is any coefficient value ≥0.8 or ≤−0.8 of a pair of features, then only the feature with a better diagnostic performance was retained. Then we used the final feature set to construct machine learning models for each molecular subtype. A random forest model was develop using all the selected features as to compare the feature contributions on a same model. Detailed information about feature selection could be seen in Supplementary materials.
Development and assessment of models
We performed the model construction and evaluation with the scikit-learn package (version: 0.18) in Python 3.70. The primary cohort was used for model construction with repeated cross-validation. The external validation cohorts were used for the final evaluation of models. We used the selected radiomics and deep learning features to construct three single-modality models (pre-, post- and delta-). The model construction was a supervised task based on the pCR and non-pCR label, and five robust classification algorithms supported by Onekey AI platform, including Logistic Regression, Random Forest, XGBoost, SVM and Multi-Layer Perception (MLP) neural network were used. Each classified model was trained to accurately fit the labeled data and predict the testing cohorts. In our study, we performed ensemble learning integrating features from different images to predict pCR more accurately. The single-modality model output scores were obtained and used to develop a stacking model with Support Vector Machine (SVM). To choose the best model hyper parameters fitting the model, the grid searching method and 5-fold cross-validation were performed. Four folds (80% of the patients) were used to train the model and the rest (20% of the patients) were used to select the best hyperparameters. To ensure the robustness of the model, we repeated 1000 times the whole construction process with the bootstrap method. The best performing models in the primary cohort were then used to test the external validation cohorts. The receiver operating characteristic (ROC) curve, calibration curve, and decision curve were used to demonstrate the prediction ability of the models visually. The diagnostic indexes of pre-, post-, delta- and stacking models, including AUC (with the 95% CI), specificity, sensitivity, accuracy, positive predictive value (PPV) and negative predictive value (NPV) were also calculated. Detailed information about machine learning process could be seen in Supplementary materials.
Statistical analysis
The patient's baseline data were evaluated using SPSS (version 20.0) and statistics packages (python version). Continuous variables were described as mean ± standard deviation, and the categorical variables were described as frequencies and percentages. The normal or non-normal distribution of continuous variables was determined by the Kolmogorov–Smirnov (KS) test. The homogeneity of continuous variance was tested by the Levene test. The Mann–Whitney U test or Student's t-test was used to compare their inter-group differences. The Chi-squared test or Fisher exact test was used to compare their difference. A two-sided p < 0.05 indicated statistical significance. The 95% confidence interval (CI) of AUC was calculated using the bootstrap method (1000 intervals). The DeLong testing method was performed to compare the AUCs of different models.38
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. All investigators read, discussed, and approved the final version of this manuscript. All investigators had full access to the dataset and took responsibility for the authenticity and integrity of the dataset as well as the decision to submit for publication.
Results
Baseline characteristics of patients
Between July 2015 and December 2021, 54 (11.7%) of 463 patients in the primary cohort, 42 (11.2%) of 385 patients in the validation cohort 1, 30 (15.0%) of 200 patients in the validation cohort 2, and 46 (11.9%) of 386 patients in the validation cohort 3 were excluded due to tumor progression or intolerance during NAC. Finally, we retrieved 1262 patients with breast cancer in this study. The characteristics of all 1262 patients (the mean interval between two MRI examinations, 171.5 days, and the range, 127–206 days) were described in Table 1. In total, 441 patients achieved pCR (34.9%) and 821 patients still remained residual invasive breast cancer or axillary lymph node metastasis (65.1%). For each cohort, the pCR rates were 38.4%, 36.2%, 29.4% and 32.4% in the primary cohort, validation cohort 1, validation cohort 2 and validation cohort 3, respectively. For each molecular subtype, the HR+/HER2- subtype had the lowest pCR rate of a total of 10.6% (52/491) in all cohorts compared to the other two subtypes (HER2+ subtype, 54.3% [323/595]; and TNBC subtype, 37.5% [66/176]). For each cohort, significant differences were observed in baseline characteristics of ER, PR, HER2 and Ki-67 between patients with pCR and patients with non-pCR. No significant differences were observed in age and clinical stage between patients with pCR and patients with non-pCR in all the cohorts, except for a significant difference in the clinical stage (p < 0.01) in primary cohort.
Table 1.
Characteristics of the patients in different cohorts.
| Characteristics | Primary cohort (N = 409) |
Validation cohort 1 (N = 343) |
Validation cohort 2 (N = 170) |
Validation cohort 3 (N = 340) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pCR (n = 157) | non-pCR (n = 252) | p value | pCR (n = 124) | non-pCR (n = 219) | p value | pCR (n = 50) | non-pCR (n = 120) | p value | pCR (n = 110) | non-pCR (n = 230) | p value | |
| Age (year)a | 48.34 ± 10.12 | 48.74 ± 10.33 | 0.457 | 48.91 ± 10.82 | 49.19 ± 9.16 | 0.651 | 50.17 ± 10.72 | 49.24 ± 10.21 | 0.392 | 49.13 ± 10.58 | 48.73 ± 10.22 | 0.478 |
| Clinical stage (%)b | <0.01 | 0.612 | 0.298 | 0.399 | ||||||||
| I | 5 (3.18%) | 11 (4.37%) | 2 (1.61%) | 2 (0.91%) | 0 (0.00%) | 1 (0.83%) | 2 (1.82%) | 2 (0.87%) | ||||
| II | 127 (80.89%) | 167 (66.27%) | 80 (64.52%) | 134 (61.19%) | 16 (32.00%) | 53 (44.17%) | 55 (50.00%) | 103 (44.78%) | ||||
| III | 25 (15.93%) | 74 (29.36%) | 42 (33.87%) | 83 (37.90%) | 34 (68.00%) | 67 (55.83%) | 53 (48.18%) | 125 (54.35%) | ||||
| ER status (%)b | <0.01 | <0.01 | <0.01 | 0.014 | ||||||||
| Positive | 94 (59.87%) | 197 (78.17%) | 62 (50.00%) | 148 (67.58%) | 18 (36.00%) | 90 (75.00%) | 57 (51.82%) | 152 (66.09%) | ||||
| Negative | 63 (40.13%) | 55 (21.83%) | 62 (50.00%) | 71 (32.42%) | 32 (64.00%) | 30 (25.00%) | 53 (48.18%) | 78 (33.91%) | ||||
| PR status (%)b | <0.01 | <0.01 | <0.01 | 0.01 | ||||||||
| Positive | 90 (57.32%) | 189 (75.00%) | 60 (48.39%) | 145 (66.21%) | 15 (30.00%) | 89 (74.17%) | 53 (48.18%) | 147 (63.91%) | ||||
| Negative | 67 (42.68%) | 63 (25.00%) | 64 (51.61%) | 74 (33.79%) | 35 (70.00%) | 31 (25.83%) | 57 (51.82%) | 83 (36.09%) | ||||
| HER-2 status (%)b | <0.01 | <0.01 | <0.01 | <0.01 | ||||||||
| Positive | 114 (72.61%) | 72 (28.57%) | 94 (75.81%) | 66 (30.14%) | 36 (72.00%) | 59 (49.17%) | 79 (71.82%) | 75 (32.61%) | ||||
| Negative | 43 (27.39%) | 180 (71.43%) | 30 (24.19%) | 153 (69.86%) | 14 (28.00%) | 61 (50.83%) | 31 (28.18%) | 155 (67.39%) | ||||
| Ki-67 status (%)b | 0.035 | 0.115 | 0.032 | 0.016 | ||||||||
| Positive | 130 (82.80%) | 186 (73.81%) | 100 (80.65%) | 160 (73.06%) | 40 (80.00%) | 110 (91.67%) | 88 (80.00%) | 155 (67.39%) | ||||
| Negative | 27 (17.20%) | 66 (26.19%) | 24 (19.35%) | 59 (26.94%) | 10 (20.00%) | 10 (8.33%) | 22 (20.00%) | 75 (32.61%) | ||||
| Cancer subtype (%)b | <0.01 | <0.01 | <0.01 | <0.01 | ||||||||
| HR+/HER2− | 19 (12.10%) | 154 (61.11%) | 14 (11.29%) | 120 (54.79%) | 7 (14.00%) | 48 (40.00%) | 12 (10.91%) | 117 (50.87%) | ||||
| HER2+ | 114 (72.61%) | 72 (28.57%) | 94 (75.81%) | 66 (30.14%) | 36 (72.00%) | 59 (49.17%) | 79 (71.82%) | 75 (32.61%) | ||||
| TN | 24 (15.29%) | 26 (10.32%) | 16 (12.90%) | 33 (15.07%) | 7 (14.00%) | 13 (10.83%) | 19 (17.27%) | 38 (16.52%) | ||||
p value < 0.05 was considered statistically signifificance.
pCR, pathologic complete response; ER, ertrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor-2; HR, hormone receptor.
Normally distributed continuous variables, expressed as mean ± standard deviation, use independent t-test to observe inter-group difference.
Categorical variables, expressed as frequencies (proportions), use χ2 test to observe inter-group difference.
Feature extraction and selection
A total of 14,676 radiomics features and 4096 deep learning features were extracted. Furthermore, another 7338 delta-radiomics and 2048 delta-deep learning features were calculated by the extracted features. To ensure the reproducibility of the features, a total of 2842 ineligible features with an ICC <0.75 were excluded. The Mann–Whitney U test showed that 5482 features were significantly associated with pCR outcome. Then we performed independent Boruta and LASSO feature selection pipeline for each subtype. The Boruta method was used to select the important and robust features with higher Shapley values than the max shadow value by 1000 internals bootstrap. To further decrease the feature dimension and simplify model, we used LASSO to determine the optimal feature numbers for each subtype. Finally, we extracted 15, 20 and 13 the most optimal features for HR+/HER2− subtype, HER2+ subtype and TNBC subtype, respectively. The feature sets were as follows: (i) HR+/HER2− subtype, two features (GLDM), three features (GLRLM), one feature (NGTDM), one feature (GLCM), and eight features (DLF). (ii) HER2+ subtype, three features (GLDM), three features (GLRLM), two feature (NGTDM), two feature (GLCM), and ten features (DLF). (iii) TNBC subtype, one feature (GLDM), two features (GLRLM), three feature (NGTDM), one feature (GLCM), and six features (DLF).
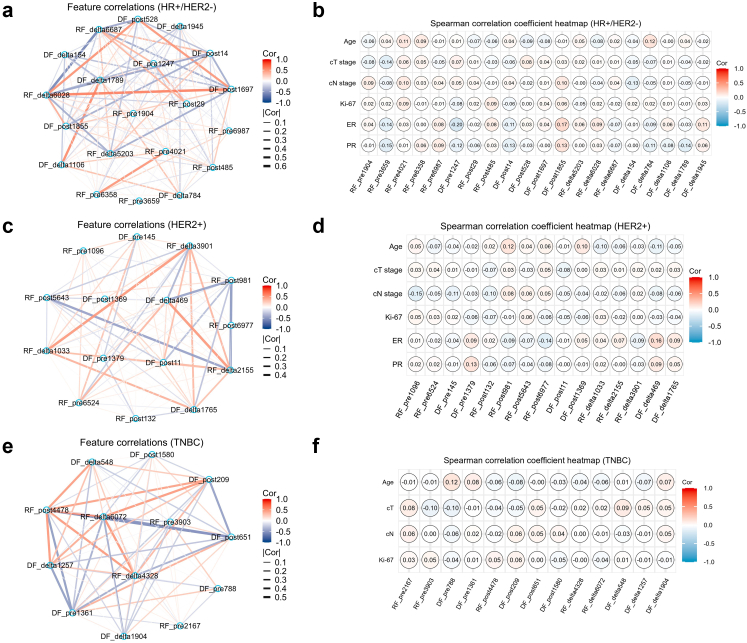
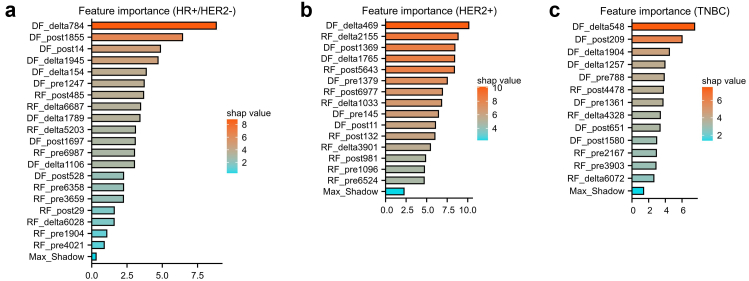
All the selected features showed high ICCs (>0.75) between different radiologists and significant differences (p < 0.05) using the Mann–Whitney U test between the pCR group and non-pCR group in the primary cohort. The Spearman correlation coefficient network diagrams showed the associations of the selected features. All the selected features showed no high correlation to the other features with a correlation coefficient value between −0.6 and 0.6 (Fig. 2; a, c, e). Besides, we also evaluate the relationship between imaging features and clinical characteristics (Fig. 2; b, d, f). The feature importance was evaluated by the Boruta model and calculated the Shapley values of each feature (Fig. 3).
Fig. 2.
The Spearman correlation coefficient network diagrams showed the relations between each pair of selected features in HR+/HER2− (a), HER2+ (c) and TNBC subtype (e). the Spearman correlation coefficient heat maps showed the relations between selected features and clinical characteristics in HR+/HER2− (b), HER2+ (d) and TNBC subtype (f). Each feature was independent predictor as there was no correlation coefficient >0.8 in each subtype. And all the imaging-derived features were independent from clinical characteristics as no correlation coefficient >0.8 was observed in each subtype.
Fig. 3.
The horizontal bar charts showed the feature importance of the selected radiomics and deep learning features in HR+/HER2− (a), HER2+ (b) and TNBC subtype (c). In the three random forest models, all the Shapley values of selected features were higher than the corresponding max shadow value in each subtype. It indicated that all the features contributed to develop the models.
Development and performance of models
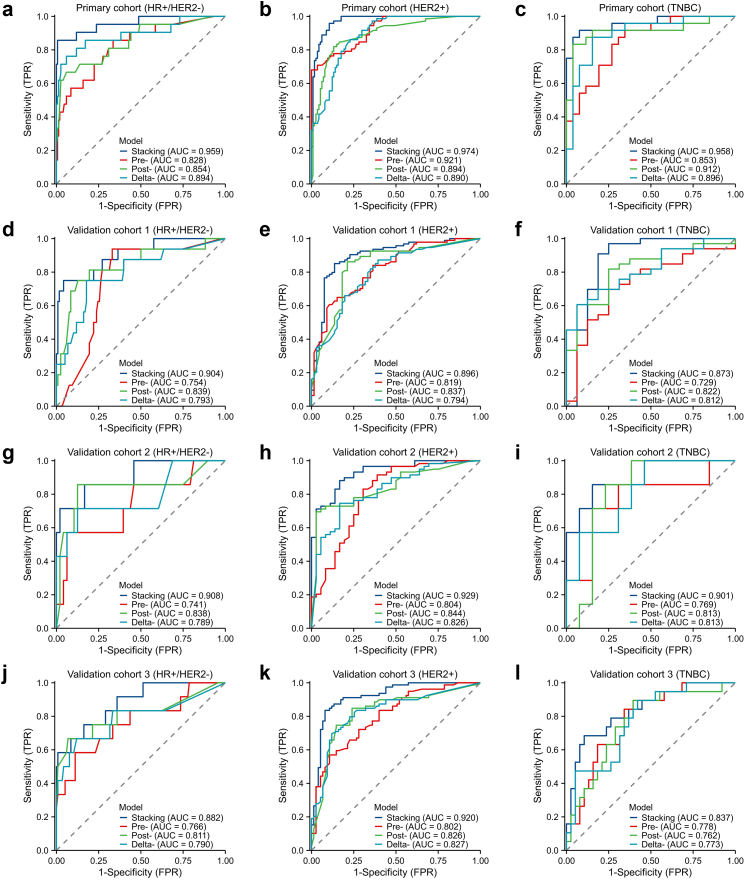
For each molecular subtype, we developed five robust supervised models to predict pCR and compared their performance to determine the most optimal model. MLP neural network performed better than other classifiers in all subtypes and feature sets in the model training step. Then it was used to construct machine learning models. According to the Youden index testing, the threshold of the best score to identify pCR was set in each subtype. To compare the models developed by different feature sets (pre-, post- and delta-), we selected the best model for further analysis and used ensemble learning to integrate the single-modality models' outputs. Table 2 described the diagnostic indexes of the stacking models for each subtype in the primary cohort and validation cohorts. In the primary cohort, three stacking models yielded AUCs of 0.959 (95% CI: 0.908–1.0), 0.974 (95% CI: 0.955–0.993) and 0.958 (95% CI: 0.906–1.0) in HR+/HER2−, HER2+ and TNBC subtype, respectively. Compared with the single-modality prediction models, the stacking model performed better than the pre-NAC models (AUCs: 0.828 [HR+/HER2−], 0.921 [HER2+] and 0.853 [TNBC]), the post-NAC models (AUCs: 0.854 [HR+/HER2−], 0.894 [HER2+] and 0.912 [TNBC]) and the delta-model (AUCs: 0.894 [HR+/HER2−], 0.890 [HER2+] and 0.896 [TNBC]). Fig. 4 showed all the models’ AUCs for each subtype. The Delong test showed that the ensemble learning significantly improve the model performance to predict pCR (all p < 0.05). The sensitivities (85.7% [HR+/HER2−], 95.8% [HER2+] and 91.7% [TNBC]) of stacking models also outperformed the three single-modality models (66.7%–85.7% [HR+/HER2−], 68.1%–86.1% [HER2+] and 83.3%–91.7% [TNBC]). The specificities (99.3% [HR+/HER2−], 89.5% [HER2+] and 92.3% [TNBC]) of stacking models were also high enough to identify the non-pCR patients. All the performance results are based on the 1000-round 5-fold cross-validation.
Table 2.
Performances of SVM stacking models for predicting pCR to NAC in various molecular subtypes and cohorts.
| Molecular subtype | Cohort | AUC (95% CI) | ACC (%) | SEN (%) | SPE (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|
| HR+/HER2− | PC | 0.959 (0.908–1.00) | 97.68 | 85.71 | 99.34 | 94.73 | 98.05 |
| VC-1 | 0.904 (0.817–0.991) | 93.28 | 75.00 | 95.76 | 70.58 | 96.58 | |
| VC-2 | 0.908 (0.776–1.00) | 94.54 | 71.42 | 97.91 | 83.33 | 95.91 | |
| VC-3 | 0.882 (0.779–0.985) | 82.94 | 75.00 | 83.76 | 32.14 | 97.02 | |
| HER2+ | PC | 0.974 (0.955–0.993) | 91.93 | 95.83 | 89.47 | 85.18 | 97.14 |
| VC-1 | 0.896 (0.842–0.949) | 85.62 | 85.10 | 86.36 | 89.88 | 80.28 | |
| VC-2 | 0.929 (0.881–0.978) | 86.31 | 88.13 | 83.33 | 89.65 | 81.08 | |
| VC-3 | 0.920 (0.876–0.965) | 87.66 | 83.54 | 92.00 | 91.66 | 84.14 | |
| TNBC | PC | 0.958 (0.906–1.00) | 92.00 | 91.66 | 92.30 | 91.66 | 92.30 |
| VC-1 | 0.873 (0.735–1.00) | 87.75 | 90.90 | 81.25 | 90.90 | 81.25 | |
| VC-2 | 0.901 (0.755–1.00) | 85.00 | 85.71 | 84.61 | 75.00 | 91.66 | |
| VC-3 | 0.837 (0.725–0.949) | 82.45 | 68.42 | 89.47 | 76.47 | 85.00 |
PC, primary cohort; VC, validation cohort; AUC, the area under curve; ACC, accuracy; SEN, sensitivity; SPE, specificity; NPV, negative predictive value; PPV, positive predictive value; 95% CI, 95% confidence interval; HR, hormone receptor; HER2, human epidermal growth factor receptor 2; TNBC, triple negative breast cancer; pCR, pathological complete response.
Fig. 4.
Predictive performances of the different models in the primary and external validation cohorts (a–l). Plots show the ROC curves of stacking model, pre-model, post-model and delta-model, in HR+/HER2− (a), HER2+ (b) and TNBC subtype (c), respectively, in the primary cohort. Plots show the ROC curves of stacking model, pre-model, post-model and delta-model, in HR+/HER2− (d), HER2+ (e) and TNBC subtype (f), respectively, in the validation cohort 1. Plots show the ROC curves of stacking model, pre-model, post-model and delta-model, in HR+/HER2− (g), HER2+ (h) and TNBC subtype (i), respectively, in the validation cohort 2. Plots show the ROC curves of the stacking model, pre-model, post-model and delta-model, in HR+/HER2− (j), HER2+ (k) and TNBC subtype (l), respectively, in the validation cohort 3.
After model construction, the three stacking models based on molecular subtypes were tested in external validation cohorts. Table 3 described the diagnostic indexes of the different models for total patients in the primary cohort and validation cohorts. The models accurately predict the pCR in validation cohort 1 (AUCs: 0.904 [HR+/HER2−], 0.896 [HER2+] and 0.873 [TNBC]), validation cohort 2 (AUCs: 0.908 [HR+/HER2−], 0.929 [HER2+] and 0.901 [TNBC]) and validation cohort 3 (AUCs: 0.882 [HR+/HER2−], 0.920 [HER2+] and 0.837 [TNBC]). In all patients, the specificities were noticeably high in external validation cohorts (91.5%, 90.7% and 87.4%). In comparison, the sensitivities were moderately high (85.3%, 86.3% and 80.0%). The stacking models also showed higher performances than single-modality models in all subtypes. The post-models had higher mean AUCs (0.835, 0.838 and 0.809) than the other two single-modality models (pre-: 0.780, 0.779 and 0.784; delta: 0.796, 0.812 and 0.803) in all the validation cohorts. They were all lower than stacking models by the DeLong test (all p < 0.05). The specificities (91.5%, 90.7% and 87.4%) and NPV (89.7%, 89.8% and 90.1%) of stacking models were also significantly high to identify non-pCR patients in the validation cohorts. However, the sensitivities (85.3%, 86.3% and 80.0%) and PPV (87.8%, 87.6% and 75.2%) were lower than the specificities in the validation cohorts. Detailed information about model results could be seen in Supplementary materials.
Table 3.
Performances of combining different machine learning models for predicting pCR to NAC in different cohorts.
| Cohort | Model | Mean AUC | ACC (%) | SEN (%) | SPE (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|
| PC | Stacking | 0.965 | 94.37 | 93.16 | 94.86 | 87.90 | 97.19 |
| Pre- | 0.873 | 78.48 | 76.06 | 79.45 | 59.73 | 89.23 | |
| Post- | 0.879 | 88.01 | 79.48 | 91.43 | 78.81 | 91.75 | |
| Delta- | 0.892 | 86.06 | 84.61 | 86.64 | 71.73 | 93.35 | |
| VC-1 | Stacking | 0.895 | 88.92 | 85.31 | 91.50 | 87.76 | 89.70 |
| Pre- | 0.780 | 71.42 | 66.43 | 75.00 | 65.51 | 75.75 | |
| Post- | 0.835 | 83.67 | 83.91 | 83.50 | 78.43 | 87.89 | |
| Delta- | 0.796 | 77.55 | 79.72 | 76.00 | 70.37 | 83.97 | |
| VC-2 | Stacking | 0.918 | 88.82 | 86.30 | 90.72 | 87.50 | 89.79 |
| Pre- | 0.779 | 82.94 | 86.30 | 80.41 | 76.82 | 88.63 | |
| Post- | 0.838 | 82.35 | 72.60 | 89.69 | 84.12 | 81.30 | |
| Delta- | 0.812 | 79.41 | 76.71 | 81.44 | 75.67 | 82.29 | |
| VC-3 | Stacking | 0.891 | 85.00 | 80.00 | 87.39 | 75.21 | 90.13 |
| Pre- | 0.784 | 77.35 | 62.72 | 84.34 | 65.71 | 82.55 | |
| Post- | 0.809 | 82.94 | 82.72 | 83.04 | 70.00 | 90.95 | |
| Delta- | 0.803 | 79.70 | 72.72 | 83.04 | 67.22 | 86.42 |
PC, primary cohort; VC, validation cohort; AUC, the area under curve; ACC, accuracy; SEN, sensitivity; SPE, specificity; NPV, negative predictive value; PPV, positive predictive value; pCR, pathological complete response.
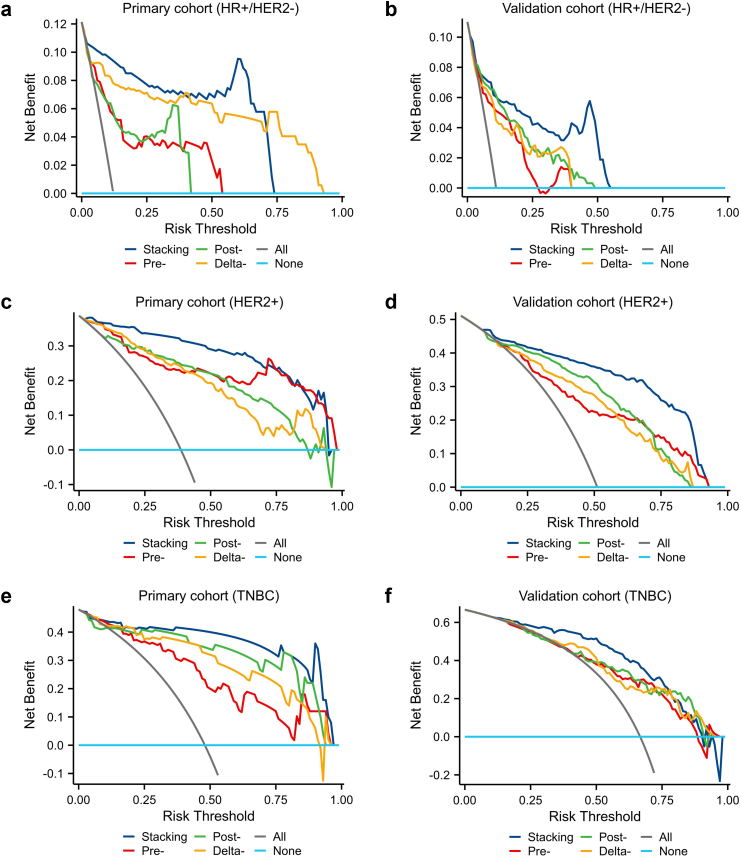
To evaluate the clinical benefit value, we used decision curve analysis to identify the model score interval that could benefit patients from model suggestions. For the HER2+ subtype and TNBC subtype, when the threshold was set at 0.08–0.91 (HER2+) and 0.11–0.89 (TNBC), their clinical net benefits were higher than 0 in the validation cohorts. However, for the HR+/HER2− subtype, only the threshold was set at the interval of 0.06–0.56, the clinical net benefits were higher than 0. That might be due to the relatively high proportion of non-pCR patients after NAC. Fig. 5 showed the decision curves of all models for each subtype.
Fig. 5.
Predictive performances of the different models in the primary and external validation cohorts (a–f). Plots show the decision curves of stacking model, pre-model, post-model and delta-model, in HR+/HER2− (a), HER2+ (c) and TNBC subtype (e), respectively, in the primary cohort. Plots show the decision curves of the stacking model, pre-model, post-model and delta-model, in HR+/HER2− (b), HER2+ (d) and TNBC subtype (f), respectively, in the validation cohorts.
Discussion
Accurate assessment of pCR is essential as an urgent need for de-escalation surgery instead of mastectomy in patients with breast cancer. The implementation of surgical risk stratification of breast cancer after completing NAC can be promoted by using ensemble learning on the longitudinal MRI. However, few previous studies reported radiomics and deep learning biomarkers based on molecular subtypes to predict pCR in breast cancer. In our study, to meet individualized treatment need, we developed more precise models based on specific molecular subtypes of breast cancer.8 Our stacking model discriminated pCR and residual invasive cancer with mean AUC values of 0.891–0.918 in all the external validation cohorts. More importantly, the result indicated that longitudinal MRI-based ensemble learning models could assist in assessing breast cancer response to NAC.
The pCR rate in our study was 34.9%, which is in keeping with rates reported in the ACOSOG Z1071 trial (34%) and the NSABP B-27 trial (26%).39, 40, 41 For patients expected to achieve pCR after NAC, a limited surgery strategy can be performed to reduce surgical complications and the economic burden. To monitor the efficacy of NAC, the present clinical tool for evaluating tumor size change is the RECIST, which is applied widely.20 Some studies showed that breast tumor response to NAC was associated with clinical TNM stage, hormone receptor status, HER2 status, and tumor Ki-67 index.13,42,43 However, RECIST and conventional clinicopathologic characteristics cannot accurately predict pCR to NAC. Several studies have suggested that MRI had an outstanding diagnosis performance in breast response to NAC.24,26 Loo et al. found that pCR was more easily achieved in TNBC and HER2+ subtypes than HR+/HER2-subtype and MRI helped monitor tumor change during NAC.44 But MRI had a favorable specificity of 91% and a poor sensitivity of 63% in predicting pCR to NAC in breast cancer.45 To get more information from MRI images, we constructed a radiomics machine learning model using three MRI sequences. It achieved an excellent performance in predicting pCR to NAC with an AUC of 0.71–0.80.13 Some researchers also reported that radiomics and deep learning classifiers had promising potential in predicting pCR to NAC with AUC values in testing cohorts.15,16,46,47 These previous studies demonstrated the challenges of accurate assessment of pCR to NAC. Previous studies showed that molecular subtypes had different pCR rates, which indicated that tumor heterogeneity in medical images might influence model performance when mixing them to analyze together.3 Due to the heterogeneity of breast cancer, radiomics and deep learning to MRI need more precise analysis on specific subtypes, and clinical applicability needs to be validated in multicenter cohorts with large sample sizes. We also investigated the potential of longitudinal multi-parametric MRI in assessing tumor response to NAC and performed ensemble learning using MRI-based radiomics and deep learning features in our study.
Radiomics and deep learning offer the potential to identify who will achieve pCR, whereas clinical factors are limited in the predicting pCR. It is challenging to meet with the success of limited breast surgery after NAC due to lacking outperforming methods in evaluating pCR. Our study had several notable advantages compared to previous studies, such as US-based deep learning analysis on HER2+ subtype, or radiomics analysis on mammograms.31,48 First, our study constructed MRI-based ensemble learning models to predict pCR based on molecular subtypes. In contrast, previous studies only used conventional clinical and radiologic characteristics and did not perform precise subtypes analysis, or only used radiomics method.49, 50, 51, 52 Secondly, we collected longitudinal MRI data, including pre-NAC and post-NAC sequences, which contained the tumor change information during NAC, rather than single-modality models.53 Previous study also reported that delta-radiomics had higher performance.54 We found that pre-, post- and delta-features were all crucial for predicting pCR. In each subtype, delta-features always contributed more according to the feature Shapley analysis (total value [delta-] > total value [post-] > total value [pre-]). The correlations among those imaging-derived features were assessed and no association was found (all Spearman correlation coefficient <0.54, p > 0.05), which indicated that each feature was an independent predictor. Thirdly, our study conducted the largest study (n = 1262) of breast cancer treated with NAC in multicenter cohorts. In all subtypes, pre-model achieved mean AUCs of 0.780, 0.779 and 0.784 in three validation cohorts, and post-model and delta-model improved the performance (mean AUCs of post-model, 0.835, 0.838 and 0.809; mean AUCs of delta-model, 0.796, 0.812 and 0.803) after completing NAC. Notably, the ensemble learning of several feature sets provided the best diagnostic performance (mean AUCs, 0.895, 0.918 and 0.891; DeLong test, all p < 0.05). The phenomenon revealed that more dramatic changes of tumors during NAC were reflected on MRI images, which enabled machine learning model to predict pCR powerfully.
Our models could integrate heterogenous radiomics and deep features of tumor and are worthy of further study. We also found that all selected imaging-derived features had no significant correlation with age, cT stage, cN stage, Ki-67, ER, PR and HER2 status (spearman correlation coefficient <0.54, all p > 0.05). According to the pathological results of biopsy, pre-NAC and post-NAC MRI data and ROI subjected to the corresponding stacking model would output a risk score. Patients expected to reach pCR by the stacking models would benefit from breast-conversing operations. However, surgical clearance should be considered when the residual lesion was predicted. Finally, 89.7%, 89.8% and 90.1% of non-pCR patients benefited from our models, while 87.8%, 87.5% and 75.2% of pCR patients could be determined before surgery. Moreover, MRI examination had already been routinely used during NAC, and our model could provide additional information on tumor response without extra spending. We considered that the scanning field strength of different MRI scanners might influence the model performance, so we collected the MRI data acquired with 3.0 T scanners in validation cohort 3, to validate the models developed by 1.5 T MRI data in the primary cohort. After image post-processing, including normalization and resampling, the same features were selected and input into the models. The stacking models performed great prediction results in validation cohorts 3, with AUCs of 0.882 (95% CI, 0.779–0.985), 0.920 (95% CI, 0.876–0.965) and 0.837 (95% CI, 0.725–0.949) in HR+/HER2−, HER2+ and TNBC subtype. Although MRI scanning field strengths were significantly different, high specificities (83.8% [HR+/HER2−], 92.0% [HER2+] and 89.5% [TNBC]) and NPVs (97.0% [HR+/HER2−], 84.1% [HER2+] and 85.0% [TNBC]) were observed to determine non-pCR patients.
Despite encouraging findings, our study has several limitations. First, the deep learning and radiomics model was built using retrospective data. Prospective data from more clinical trials would improve the clinical evidence of our model. Second, imbalanced ratios of molecular subtypes might influence the clinical implementation of the model, especially in the HR+/HER2− subtype due to its relatively low pCR rate. Third, only the pre-NAC and post-NAC MRI sequences were used to develop the models, and in the future, we will further study the predictive potential of mid-NAC MRI in predicting pCR. Fourth, we only used the MRI data to develop models, and more medical images, including pathological whole slide images and ultrasound images, might improve our models.
To conclude, we constructed ensemble learning models using pre-NAC MRI and post-NAC MRI to accurately predict pCR for each molecular subtype of breast cancer. For breast cancer patients, this model can help surgeons decide on breast-conserving surgery and SLNB after NAC treatment. Prospective studies with external validation could provide strong clinical evidence for our diagnostic model and explore the clinical application value for tailored treatment in wider regions and populations.
Contributors
YHH contributed to the conception design, collection and analysis of data, and manuscript writing. TZ contributed to the data analysis, data interpretation and manuscript writing. XLZ contributed to the collection and analysis of data, and manuscript writing. WL contributed to the conception design. XXZ contributed to the analysis of pathology. MYC contributed to the provision of study materials of patients. FJ, LLZ and CQY contributed to the provision of study materials of patients and data proofreading. ZYW, GLY and YL contributed to the administrative support, provision of study materials of patients and manuscript revision. KW contributed to the conception design, funding acquisition and manuscript revision. All authors read and approved the final manuscript. All authors had full access to the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis as well as the decision to submit for publication.
Data sharing statement
The datasets generated during and analyzed during the current study are available by the corresponding author Kun Wang, upon reasonable request.
Declaration of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Funding sources were not involved in the study design, data collection, analysis and interpretation, writing of the report, or decision to submit the article for publication.
Acknowledgements
This study is supported by grants from the National Natural Science Foundation of China (82171898, 82103093), the Deng Feng project of high-level hospital construction (DFJHBF202109), the Guangdong Basic and Applied Basic Research Foundation (grant number, 2020A1515010346, 2021A1515011570, 2022A1515012277), the Guangzhou Science and Technology Project (202002030236, 202102021055), the Beijing Medical Award Foundation (YXJL-2020-0941-0758), and the Beijing Science and Technology Innovation Medical Development Foundation (KC2022-ZZ-0091-5). Funding sources were not involved in the study design, data collection, analysis and interpretation, writing of the report, or decision to submit the article for publication.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2023.101899.
Contributor Information
ZhiYong Wu, Email: stwuzy@163.com.
GuoLin Ye, Email: 13902816950@139.com.
Ying Lin, Email: Linying3@mail.sysu.edu.cn.
Kun Wang, Email: gzwangkun@126.com.
Appendix A. Supplementary data
References
- 1.Curigliano G., Burstein H.J., Winter E.P., et al. De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the primary therapy of early breast cancer 2017. Ann Oncol. 2019;30(7):1181. doi: 10.1093/annonc/mdy537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cortazar P., Zhang L., Untch M., et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384(9938):164–172. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 3.von Minckwitz G., Untch M., Blohmer J.U., et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol. 2012;30(15):1796–1804. doi: 10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 4.Spring L.M., Bar Y., Isakoff S.J. The evolving role of neoadjuvant therapy for operable breast cancer. J Natl Compr Canc Netw. 2022;20(6):723–734. doi: 10.6004/jnccn.2022.7016. [DOI] [PubMed] [Google Scholar]
- 5.Goorts B., van Nijnatten T.J., de Munck L., et al. Clinical tumor stage is the most important predictor of pathological complete response rate after neoadjuvant chemotherapy in breast cancer patients. Breast Cancer Res Treat. 2017;163(1):83–91. doi: 10.1007/s10549-017-4155-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J.H., Bahri S., Mehta R.S., et al. Impact of factors affecting the residual tumor size diagnosed by MRI following neoadjuvant chemotherapy in comparison to pathology. J Surg Oncol. 2014;109(2):158–167. doi: 10.1002/jso.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Nonneville A., Houvenaeghel G., Cohen M., et al. Pathological complete response rate and disease-free survival after neoadjuvant chemotherapy in patients with HER2-low and HER2-0 breast cancers. Eur J Cancer. 2022;176:181–188. doi: 10.1016/j.ejca.2022.09.017. [DOI] [PubMed] [Google Scholar]
- 8.Wolf D.M., Yau C., Wulfkuhle J., et al. Redefining breast cancer subtypes to guide treatment prioritization and maximize response: predictive biomarkers across 10 cancer therapies. Cancer Cell. 2022;40(6):609–623.e6. doi: 10.1016/j.ccell.2022.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fayanju O.M., Ren Y., Thomas S.M., et al. The clinical significance of breast-only and node-only pathologic complete response (pCR) after neoadjuvant chemotherapy (NACT): a review of 20,000 breast cancer patients in the national cancer data base (NCDB) Ann Surg. 2018;268(4):591–601. doi: 10.1097/SLA.0000000000002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pilewskie M., Morrow M. Axillary nodal management following neoadjuvant chemotherapy: a review. JAMA Oncol. 2017;3(4):549–555. doi: 10.1001/jamaoncol.2016.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goetz M.P., Gradishar W.J., Anderson B.O., et al. NCCN guidelines insights: breast cancer, version 3.2018. J Natl Compr Canc Netw. 2019;17(2):118–126. doi: 10.6004/jnccn.2019.0009. [DOI] [PubMed] [Google Scholar]
- 12.Kuerer H.M., Smith B.D., Krishnamurthy S., et al. Eliminating breast surgery for invasive breast cancer in exceptional responders to neoadjuvant systemic therapy: a multicentre, single-arm, phase 2 trial. Lancet Oncol. 2022;23(12):1517–1524. doi: 10.1016/S1470-2045(22)00613-1. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z., Li Z., Qu J., et al. Radiomics of multiparametric MRI for pretreatment prediction of pathologic complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Clin Cancer Res. 2019;25(12):3538–3547. doi: 10.1158/1078-0432.CCR-18-3190. [DOI] [PubMed] [Google Scholar]
- 14.Goorts B., Dreuning K.M.A., Houwers J.B., et al. MRI-based response patterns during neoadjuvant chemotherapy can predict pathological (complete) response in patients with breast cancer. Breast Cancer Res. 2018;20(1):34. doi: 10.1186/s13058-018-0950-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li P., Wang X., Xu C., et al. 18F-FDG PET/CT radiomic predictors of pathologic complete response (pCR) to neoadjuvant chemotherapy in breast cancer patients. Eur J Nucl Med Mol Imaging. 2020;47(5):1116–1126. doi: 10.1007/s00259-020-04684-3. [DOI] [PubMed] [Google Scholar]
- 16.Zhuang X., Chen C., Liu Z., et al. Multiparametric MRI-based radiomics analysis for the prediction of breast tumor regression patterns after neoadjuvant chemotherapy. Transl Oncol. 2020;13(11) doi: 10.1016/j.tranon.2020.100831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dialani V., Chadashvili T., Slanetz P.J. Role of imaging in neoadjuvant therapy for breast cancer. Ann Surg Oncol. 2015;22(5):1416–1424. doi: 10.1245/s10434-015-4403-9. [DOI] [PubMed] [Google Scholar]
- 18.Han S., Choi J.Y. Prognostic value of (18)F-FDG PET and PET/CT for assessment of treatment response to neoadjuvant chemotherapy in breast cancer: a systematic review and meta-analysis. Breast Cancer Res. 2020;22(1):119. doi: 10.1186/s13058-020-01350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber J.J., Jochelson M.S., Eaton A., et al. MRI and prediction of pathologic complete response in the breast and axilla after neoadjuvant chemotherapy for breast cancer. J Am Coll Surg. 2017;225(6):740–746. doi: 10.1016/j.jamcollsurg.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz L.H., Litière S., de Vries E., et al. RECIST 1.1-Update and clarification: from the RECIST committee. Eur J Cancer. 2016;62:132–137. doi: 10.1016/j.ejca.2016.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hylton N.M., Blume J.D., Bernreuter W.K., et al. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy--results from ACRIN 6657/I-SPY TRIAL. Radiology. 2012;263(3):663–672. doi: 10.1148/radiol.12110748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobbes M.B., Prevos R., Smidt M., et al. The role of magnetic resonance imaging in assessing residual disease and pathologic complete response in breast cancer patients receiving neoadjuvant chemotherapy: a systematic review. Insights Imaging. 2013;4(2):163–175. doi: 10.1007/s13244-013-0219-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park S.H., Moon W.K., Cho N., et al. Diffusion-weighted MR imaging: pretreatment prediction of response to neoadjuvant chemotherapy in patients with breast cancer. Radiology. 2010;257(1):56–63. doi: 10.1148/radiol.10092021. [DOI] [PubMed] [Google Scholar]
- 24.Partridge S.C., Zhang Z., Newitt D.C., et al. Diffusion-weighted MRI findings predict pathologic response in neoadjuvant treatment of breast cancer: the ACRIN 6698 multicenter trial. Radiology. 2018;289(3):618–627. doi: 10.1148/radiol.2018180273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu Y.L., Pan S.M., Ren J., Yang Z.X., Jiang G.Q. Role of magnetic resonance imaging in detection of pathologic complete remission in breast cancer patients treated with neoadjuvant chemotherapy: a meta-analysis. Clin Breast Cancer. 2017;17(4):245–255. doi: 10.1016/j.clbc.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 26.You S., Kang D.K., Jung Y.S., An Y.S., Jeon G.S., Kim T.H. Evaluation of lymph node status after neoadjuvant chemotherapy in breast cancer patients: comparison of diagnostic performance of ultrasound, MRI and 18F-FDG PET/CT. Br J Radiol. 2015;88(1052) doi: 10.1259/bjr.20150143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambin P., Rios-Velazquez E., Leijenaar R., et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yip S.S., Aerts H.J. Applications and limitations of radiomics. Phys Med Biol. 2016;61(13):R150–R166. doi: 10.1088/0031-9155/61/13/R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zwanenburg A., Vallières M., Abdalah M.A., et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295(2):328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matikas A., Johansson H., Grybäck P., et al. Survival outcomes, digital TILs, and on-treatment PET/CT during neoadjuvant therapy for HER2-positive breast cancer: results from the randomized PREDIX HER2 trial. Clin Cancer Res. 2023;29(3):532–540. doi: 10.1158/1078-0432.CCR-22-2829. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y., Wang Y., Wang Y., et al. Early prediction of treatment response to neoadjuvant chemotherapy based on longitudinal ultrasound images of HER2-positive breast cancer patients by Siamese multi-task network: a multicentre, retrospective cohort study. eClinicalMedicine. 2022;52 doi: 10.1016/j.eclinm.2022.101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammond M.E., Hayes D.F., Dowsett M., et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28(16):2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolff A.C., Hammond M.E., Hicks D.G., et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31(31):3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 34.Fedorov A., Beichel R., Kalpathy-Cramer J., et al. 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging. 2012;30(9):1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng G.Z., San Jose Estepar R., Folch E., Onieva J., Gangadharan S., Majid A. Three-dimensional printing and 3D slicer: powerful tools in understanding and treating structural lung disease. Chest. 2016;149(5):1136–1142. doi: 10.1016/j.chest.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Griethuysen J.J.M., Fedorov A., Parmar C., et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kocak M.A., Ramirez D., Erkip E., Shasha D.E. SafePredict: a meta-algorithm for machine learning that uses refusals to guarantee correctness. IEEE Trans Pattern Anal Mach Intell. 2021;43(2):663–678. doi: 10.1109/TPAMI.2019.2932415. [DOI] [PubMed] [Google Scholar]
- 38.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 39.Boughey J.C., Ballman K.V., Le-Petross H.T., et al. Identification and resection of clipped node decreases the false-negative rate of sentinel lymph node surgery in patients presenting with node-positive breast cancer (T0-T4, N1-N2) who receive neoadjuvant chemotherapy: results from ACOSOG Z1071 (alliance) Ann Surg. 2016;263(4):802–807. doi: 10.1097/SLA.0000000000001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bear H.D., Anderson S., Smith R.E., et al. Sequential preoperative or postoperative docetaxel added to preoperative doxorubicin plus cyclophosphamide for operable breast cancer: National Surgical Adjuvant Breast and Bowel Project Protocol B-27. J Clin Oncol. 2006;24(13):2019–2027. doi: 10.1200/JCO.2005.04.1665. [DOI] [PubMed] [Google Scholar]
- 41.Yam C., Abuhadra N., Sun R., et al. Molecular characterization and prospective evaluation of pathologic response and outcomes with neoadjuvant therapy in metaplastic triple-negative breast cancer. Clin Cancer Res. 2022;28(13):2878–2889. doi: 10.1158/1078-0432.CCR-21-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy B.L., Hoskin T., Heins C.D.N., Habermann E.B., Boughey J.C. Preoperative prediction of node-negative disease after neoadjuvant chemotherapy in patients presenting with node-negative or node-positive breast cancer. Ann Surg Oncol. 2017;24(9):2518–2525. doi: 10.1245/s10434-017-5872-9. [DOI] [PubMed] [Google Scholar]
- 43.Kantor O., Sipsy L.M., Yao K., James T.A. A predictive model for axillary node pathologic complete response after neoadjuvant chemotherapy for breast cancer. Ann Surg Oncol. 2018;25(5):1304–1311. doi: 10.1245/s10434-018-6345-5. [DOI] [PubMed] [Google Scholar]
- 44.Loo C.E., Straver M.E., Rodenhuis S., et al. Magnetic resonance imaging response monitoring of breast cancer during neoadjuvant chemotherapy: relevance of breast cancer subtype. J Clin Oncol. 2011;29(6):660–666. doi: 10.1200/JCO.2010.31.1258. [DOI] [PubMed] [Google Scholar]
- 45.Yuan Y., Chen X.S., Liu S.Y., Shen K.W. Accuracy of MRI in prediction of pathologic complete remission in breast cancer after preoperative therapy: a meta-analysis. AJR Am J Roentgenol. 2010;195(1):260–268. doi: 10.2214/AJR.09.3908. [DOI] [PubMed] [Google Scholar]
- 46.Antunovic L., De Sanctis R., Cozzi L., et al. PET/CT radiomics in breast cancer: promising tool for prediction of pathological response to neoadjuvant chemotherapy. Eur J Nucl Med Mol Imaging. 2019;46(7):1468–1477. doi: 10.1007/s00259-019-04313-8. [DOI] [PubMed] [Google Scholar]
- 47.Li Y., Fan Y., Xu D., et al. Deep learning radiomic analysis of DCE-MRI combined with clinical characteristics predicts pathological complete response to neoadjuvant chemotherapy in breast cancer. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.1041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skarping I., Larsson M., Förnvik D. Analysis of mammograms using artificial intelligence to predict response to neoadjuvant chemotherapy in breast cancer patients: proof of concept. Eur Radiol. 2022;32(5):3131–3141. doi: 10.1007/s00330-021-08306-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu C., Jarrett A.M., Zhou Z., et al. MRI-based digital models forecast patient-specific treatment responses to neoadjuvant chemotherapy in triple-negative breast cancer. Cancer Res. 2022;82(18):3394–3404. doi: 10.1158/0008-5472.CAN-22-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng Q., Ke M., Zhong L., et al. Radiomics based on dynamic contrast-enhanced MRI to early predict pathologic complete response in breast cancer patients treated with neoadjuvant therapy. Acad Radiol. 2022;S1076-6332(22) doi: 10.1016/j.acra.2022.11.006. [DOI] [PubMed] [Google Scholar]
- 51.Khan N., Adam R., Huang P., Maldjian T., Duong T.Q. Deep learning prediction of pathologic complete response in breast cancer using MRI and other clinical data: a systematic review. Tomography. 2022;8(6):2784–2795. doi: 10.3390/tomography8060232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joo S., Ko E.S., Kwon S., et al. Multimodal deep learning models for the prediction of pathologic response to neoadjuvant chemotherapy in breast cancer. Sci Rep. 2021;11(1) doi: 10.1038/s41598-021-98408-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng Y., Cheng Z., Gong C., et al. Pretreatment DCE-MRI-based deep learning outperforms radiomics analysis in predicting pathologic complete response to neoadjuvant chemotherapy in breast cancer. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.846775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu S., Du S., Gao S., Teng Y., Jin F., Zhang L. A delta-radiomic lymph node model using dynamic contrast enhanced MRI for the early prediction of axillary response after neoadjuvant chemotherapy in breast cancer patients. BMC Cancer. 2023;23(1):15. doi: 10.1186/s12885-022-10496-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.