Abstract
Soft tissue tumors are rare mesenchymal tumors with divergent differentiation. The diagnosis of soft tissue tumors is challenging for pathologists owing to the diversity of tumor types and histological overlap among the tumor entities. Present-day understanding of the molecular pathogenesis of soft tissue tumors has rapidly increased with the development of molecular genetic techniques (e.g., next-generation sequencing). Additionally, immunohistochemical markers that serve as surrogate markers for recurrent translocations in soft tissue tumors have been developed. This review aims to provide an update on recently described molecular findings and relevant novel immunohistochemical markers in selected soft tissue tumors.
Keywords: soft tissue tumor, sarcoma, molecular pathology, translocation, immunohistochemistry
1. Introduction
Soft tissue tumors comprise a heterogeneous group of tumors with a wide spectrum of differentiation. Soft tissue sarcomas represent less than 1% of all malignant neoplasms [1]. They present diagnostic challenges for pathologists due to the large number of tumor types, the rarity of each tumor type, their considerable morphologic diversity and overlap, and their intrinsic and technological complexity [2]. The classical diagnosis of soft tissue tumors is based on histological findings and ancillary tissue-based tests such as immunohistochemistry. Recent progress in molecular genetics in soft tissue tumors has improved diagnostic precision and refined the classification of these tumors [3].
Molecular genetics and immunohistochemistry are rapidly advancing areas in the diagnosis of soft tissue tumors. Immunohistochemistry plays a crucial role in providing genetic information on tumors. Various types of molecular alterations, including (1) specific chromosomal translocations, (2) specific mutations, (3) gene deletions, (4) gene amplifications, and (5) epigenetic alterations, are efficiently detectable via immunohistochemistry [4]. The classification of soft tissue tumors continues to evolve as new molecular genetic abnormalities are identified [5].
Herein, we review recently described molecular findings and relevant novel immunohistochemical markers in selected soft tissue tumors that can help with diagnosis.
2. Etiology
The etiology of most benign and malignant soft tissue tumors is unknown. Minorities are associated with germline mutations in tumor suppressor genes and occur in familial cancer syndromes, such as neurofibromatosis type 1, Gardner syndrome, Li–Fraumeni syndrome, Osler–Weber Rendu syndrome, etc. In rare cases (<10%), genetic and environmental factors, immunodeficiency, irradiation, and viral infections have been linked to the development of malignant soft tissue tumors [1].
Unlike carcinomas, most sarcomas do not arise from well-defined precursor lesions. Events comprising multistage tumorigenesis with progressively accumulated genetic alterations have not yet been clearly identified in most soft tissue tumors [1]. Some sarcomas recapitulate a recognizable mesenchymal lineage (e.g., skeletal muscle). However, they are believed to arise from pluripotent mesenchymal stem cells, which acquire somatic “driver” mutations in oncogenes and tumor suppressor genes [5].
3. Classification of Sarcomas Based on Karyotypic Complexity
Genetically, sarcomas can be separated into two major genetic groups [3,6]. One group of sarcomas (20%) is characterized by specific genetic changes and typically simple karyotypes, such as specific chromosomal translocations (e.g., FUS::DDIT3 in myxoid liposarcoma [MLPS]) and oncogenic mutations (e.g., KIT mutation in gastrointestinal stromal tumor [GIST]). A majority of sarcomas presenting with unique chromosomal translocations occur predominantly in young patients and tend to have a monomorphic microscopic appearance. Tumors with simple cytogenetic features often have distinctive molecular findings that may be diagnostically useful [7]. Another group of sarcomas (80%) is characterized by non-specific genetic alterations and complex unbalanced karyotypes that are characteristic of severe chromosomal and genetic instability. Sarcomas with more complex cytogenetic features are more common in adults and tend to be microscopically diverse, with a range of cell sizes and shapes within a single tumor [7]. The critical genetic events driving the biology of these sarcomas are largely unknown [8]. Moreover, it is important to recognize that even neoplasms with specific genetic alterations can progress and develop complex karyotypes with tumor progression.
4. Molecular Tests
Molecular tests have become increasingly important in diagnosing soft tissue tumors [9,10]. Common molecular methods include conventional cytogenetics, reverse-transcriptase polymerase chain reaction (RT-PCR), and fluorescence in situ hybridization (FISH). Conventional cytogenetics requires fresh tissue and is used to evaluate the entire karyotype. In contrast, FISH and RT-PCR are applied to identify specific translocations/amplifications associated with a given tumor type [11,12]. RT-PCR and FISH are considered complementary; the choice of one over the other is largely dictated by the expertise of the laboratory. Particularly, FISH is highly desirable in the evaluation of round cell sarcomas, spindle cell tumors, well-differentiated adipocytic tumors, and myxoid tumors [11].
More recently, next-generation sequencing (NGS) (massively parallel sequencing or deep sequencing) has emerged as a major tool for identifying known or novel molecular alterations in a wide array of soft tissue tumors [13,14,15,16]. NGS is a highly sensitive method for detecting genetic alterations and can help to diagnose more precisely and characterize more detailed genetic alterations. Additionally, NGS will provide further insight into the pathogenesis of soft tissue tumors and the basis for the development of targeted therapies. Current European Society for Medical Oncology (ESMO) guidelines [17] suggest that the morphologic and immunohistochemical analyses should be complemented by molecular pathology: (1) when the specific histologic diagnosis is uncertain, (2) when the clinicopathologic presentation is unusual, or (3) when the genetic information may have prognostic or predictive relevance. If the molecular analysis is not available, it is recommended that you send it to a reference with all molecular analysis equipment.
5. Immunohistochemistry
Over the last decade, molecular genetic findings have led to the development of novel, inexpensive, and quick diagnostic tests with immunohistochemical stains [18]. Recently described immunohistochemical markers are classified into three general categories: (1) protein correlates of molecular genetic alterations (e.g., β-catenin, MDM2, CDK4, H3K27me3, MYC, PDGFRA, RB1, SDHB, SMARCB1 [INI1], and SMARCA4 [BRG1]), (2) protein products of gene fusion (e.g., ALK, BCOR, CCNB3, CAMTA1, DDIT3, FOSB, SS18::SSX, TFE3, and pan-TRK), and (3) diagnostic markers identified by gene expression profiling (e.g., DOG1, ETV4, MUC4, NKX2-2, SATB2, and TLE1).
6. Practical Diagnostic Approach to Soft Tissue Tumors
The diagnosis of a soft tissue lesion requires both comprehensive clinical information and adequately processed tissue [9]. The first and most important step toward a correct diagnosis is the careful examination of conventionally stained sections at low magnification. In general, light microscopic assessment of morphology remains the cornerstone of diagnosing soft tissue tumors [19]. Usage of immunostains in the most effective and cost-efficient way requires an algorithmic approach and utilization of the reagents in panels [9]. The selection of a particular molecular test should be based on a specific differential diagnosis and relevant pretest probabilities [7]. Pathologists must exercise caution in interpreting cases because many tumors involve the same gene or even the same translocation [3]. The final diagnosis should be based on a coordinated interpretation of a reasonable morphological impression, clinical and radiologic data, and immunohistochemical and molecular findings that confirm the morphological impression [7].
7. Adipocytic Tumors
7.1. Spindle Cell Lipoma and Pleomorphic Lipoma
Spindle cell lipoma (SCL) is a benign adipocytic tumor composed of a variable admixture of bland spindle cells, mature adipocytes, and ropy collagen fibers [20]. Pleomorphic lipoma (PL) consists of mature adipocytes with pleomorphic stromal cells and multinucleated floret-like giant cells. SCLs and PLs are considered morphological variations of a single neoplasm. Genetically, SCL and PL are characterized by the deletion of chromosome 13q14, often in combination with the loss of chromosome 16q [21,22,23]. Immunohistochemistry shows that RB1 protein expression is lost in almost all SCL and PL [22]. CD34 is strongly expressed in spindle, pleomorphic, and floret-like giant cells.
Interestingly, loss of 13q14, including RB1, is seen in myofibroblastoma and cellular angiofibroma, which are morphologically similar to SCL and PL. The overlapping morphologic and genetic features support the hypothesis that these tumors are related entities, the so-called 13q/RB1 family of tumors [24,25,26].
7.2. Atypical Spindle Cell/Pleomorphic Lipomatous Tumor
In the 2020 WHO classification of tumors of soft tissue and bone, atypical spindle cell/pleomorphic lipomatous tumor (ASPLT) was described for the first time as a single entity. ASPLT is a benign adipocytic neoplasm characterized by mild to moderately atypical spindle cells, pleomorphic cells, mature adipocytes, and lipoblasts within a collagenous or myxoid stroma and ill-defined tumor margins [27]. Genetically, ASPLTs commonly harbor deletions or losses of 13q14, which includes RB1 and its flanking genes RCBTB2, DLEU1, and ITM2B [28,29,30,31]. Immunohistochemistry shows that the expression of RB1 in the nucleus is lost in approximately 50–70% of cases [28,29,30].
ASPLTs show no MDM2 or CDK4 amplification, which distinguishes them from atypical lipomatous tumor (ALT)/well-differentiated liposarcomas (WDLPS), and dedifferentiated liposarcomas (DDLPS). ASPLTs show molecular differences from SCL and PL, with typically more complex deletions of the 13q region and further deletions/losses of genes flanking RB1. However, there is a possibility that these features may represent a disease continuum [29,31,32,33]. ASPLT remains an evolving entity with a need for further understanding.
7.3. Atypical Lipomatous Tumor/Well-Differentiated Liposarcoma
Atypical lipomatous tumor/well-differentiated liposarcoma (ALT/WDLPS) is a locally aggressive mesenchymal neoplasm showing adipocytic differentiation with at least focal nuclear atypia in both adipocytes and stromal cells [34]. “ALT” and “WDLPS” are synonyms for explaining morphologically and genetically identical lesions. Genetically, amplification of MDM2 in 12q15 is almost always present. In addition, several other genes located in the 12q13–q15 region, including CDK4, TSPAN31, HMGA2, YEATS4, CPM, and FRS2, are commonly coamplified with MDM2 [35,36]. Immunohistochemically, MDM2 and/or CDK4 nuclear immunopositivity is present in most cases [36].
Immunohistochemistry for MDM2 and CDK4 has now become commonly used. However, these antibodies are not exclusively specific since they demonstrate positive staining in malignant peripheral nerve sheath tumors (MPNSTs) [37] and endometrial stromal sarcomas [38]. Nuclear expression of MDM2 in histiocytes in fat necrosis represents another major pitfall. Moreover, MDM2 is also amplified in intimal sarcoma, low-grade central osteosarcoma, and parosteal osteosarcoma [39,40,41]. Molecular testing for MDM2 amplification should be considered in recurrent lipomas, lipomatous tumors with equivocal cytologic atypia, large lipomatous tumors (>15 cm) without cytologic atypia, and lipomatous tumors lacking cytologic atypia in the retroperitoneum, pelvis, and abdomen [42].
7.4. Dedifferentiated Liposarcoma
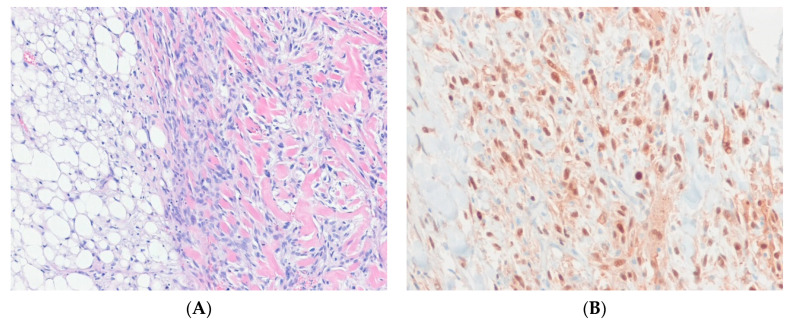
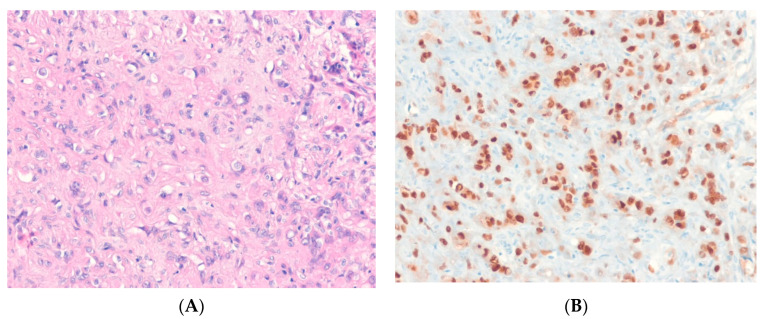
Dedifferentiated liposarcoma (DDLPS) is an ALT/WDLPS that shows an abrupt transition to non-lipogenic sarcoma of variable histological grade, either in the primary disease or in a recurrence [43]. It should be noted that a well-differentiated component may not be identifiable. The dedifferentiated component may show lipoblastic differentiation [44]. Genetically, DDLPS overlaps with ALT/WDLPS and is characterized by the amplification of MDM2 and CDK4 [35,45]. As in ALT/WDLPS, several other genes from the 12q13-q21 region and other chromosomal regions are variably coamplified with MDM2. The karyotypes and quantitative genomic profiles of DDLPS are often more complicated than those of ALT/WDLPS. Immunohistochemically, nuclear expression of MDM2 and/or CDK4 is observed in the majority of DDLPS cases [46] (Figure 1).
Figure 1.
Dedifferentiated liposarcoma. (A) The tumor shows an abrupt transition from a well-differentiated liposarcoma component (left) to a non-lipogenic sarcoma component (right), MDM2 (H&E stain, ×100). (B) The dedifferentiated part shows nuclear positivity for MDM2, indicating the presence of underlying MDM2 amplification (immunohistochemical stain for MDM2, ×200).
MDM2 immunohistochemistry and MDM2 amplification can help confirm a diagnosis of DDLPS and distinguish DDLPS from other undifferentiated sarcomas in the relevant clinical context [47]. It has been reported that undifferentiated pleomorphic sarcomas with MDM2 amplification are in fact DDLPS even in the absence of a well-differentiated LPS component [48].
7.5. Myxoid Liposarcoma
Myxoid liposarcoma (MLPS) is a malignant tumor composed of uniform, round to ovoid cells with a variable number of small lipoblasts. These cells are set in a myxoid stroma with a branching capillary vasculature [49]. Genetically, most MLPSs (>90%) are characterized by the t(12;16)(q13;p11) translocation, resulting in the FUS::DDIT3 fusion gene. In approximately 3% of MLPSs, t(12;22)(q13;q12) translocation results in EWSR1::DDIT3 fusion [50]. DDIT3 is a DNA-binding transcription factor involved in adipocytic differentiation [51,52]. The chimeric oncoprotein alters transcription and blocks adipocytic differentiation [53]. More than 50% of MLPS cases carry TERT promoter mutations [54], and approximately 25% have mutations that activate the PI3K/mTOR signaling pathway [55]. Recent findings have suggested that immunohistochemistry for DDIT3 is highly sensitive and specific for MLPS, including high-grade (round cell) MLPS cases [56].
The presence of FUS::DDIT3 or EWSR1::DDIT3 fusion helps to distinguish MLPS from other myxoid sarcomas and high-grade MLPS from various round cell sarcomas [57]. FUS and EWSR1 can replace each other and occur in other sarcomas, while DDIT3 is unique to MLPS. Thus, FISH break-apart probes directed at DDIT3 serve as a sensitive and specific strategy for MLPS diagnosis [58]. Furthermore, immunohistochemistry for DDIT3 could replace molecular genetic testing in many cases, although limited positivity can be observed in several other tumor types [56].
8. Fibroblastic and Myofibroblastic Tumors
8.1. Desmoid Fibromatosis
Desmoid fibromatosis is a locally aggressive but non-metastasizing deep-seated (myo)fibroblastic neoplasm with infiltrative growth and a high tendency to local recurrence [59]. The majority (90–95%) of sporadic desmoid tumors result from point mutations of the CTNNB1 gene on 3p21, which encodes β-catenin [60,61]. A minority of desmoid tumors occur in Gardner syndrome and harbor germline mutations of the APC gene on 5q21–q22 [62,63,64]. The activating mutations in CTNNB1 or inactivating mutations in APC interfere with the proteasomal degradation of β-catenin, resulting in the accumulation of β-catenin in the nucleus [65]. Immunohistochemically, nuclear expression of β-catenin is present in ~80% of tumors [66].
Importantly, although aberrant nuclear β-catenin is a helpful finding to support the diagnosis, nuclear β-catenin expression is also seen in superficial fibromatoses and some sarcomas and is consequently neither specific nor fully sensitive for desmoid fibromatosis [67,68]. CTNNB1 mutation analysis may be useful in small biopsy specimens and/or in cases of equivocal immunostaining for β-catenin [69,70]. Moreover, desmoid fibromatosis with specific mutations in exon 3 of CTNNB1, particularly S45F, has a greater tendency to local recurrence [71].
8.2. Solitary Fibrous Tumor
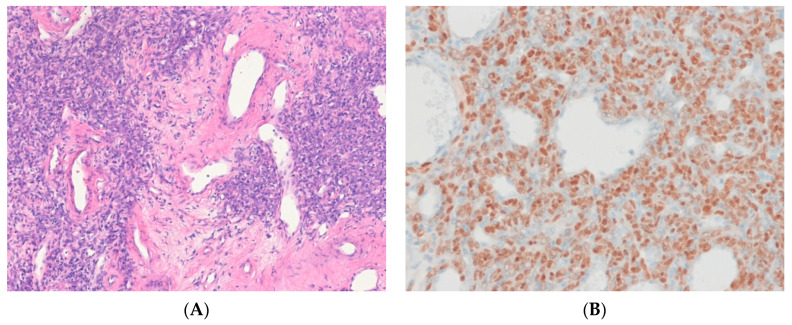
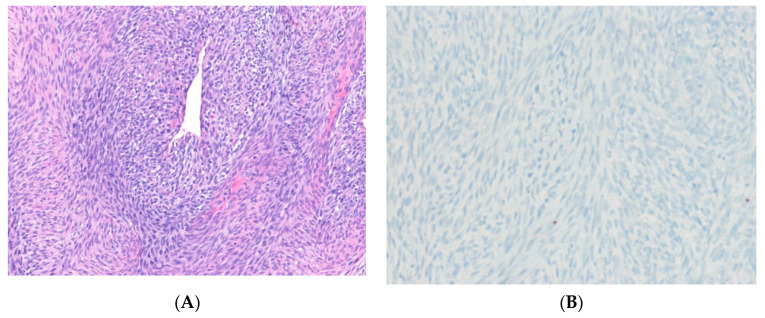
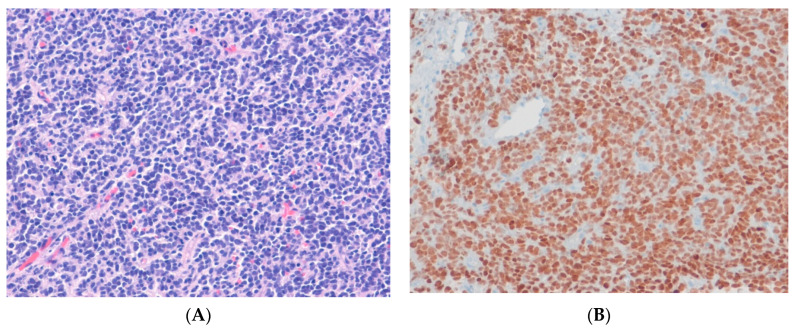
Solitary fibrous tumor (SFT) is a fibroblastic tumor with prominent, branched, thin-walled, dilated (staghorn) vasculature [72]. The genetic hallmark of SFT is a paracentric inversion involving chromosome 12q, resulting in the fusion of the NAB2 and STAT6 genes [73,74]. Overexpression of the NAB2::STAT6 gene fusion has been reported to induce proliferation in cultured cells and activate the expression of EGR-responsive genes [75]. These results establish NAB2::STAT6 as the defining driver mutation of SFT. Overexpression of ALDH1A1 (ALDH1), EGFR, JAK2, histone deacetylases, and retinoic acid receptors may also contribute to tumorigenesis [76,77]. In addition, TERT promoter mutations [78] and deletions or mutations of TP53 [79] are associated with aggressive behavior and dedifferentiation. Immunohistochemically, the NAB2::STAT6 fusion leads to the nuclear expression of STAT6 [80] (Figure 2). Thus, STAT6 immunohistochemistry is a sensitive and specific surrogate for all fusions [72]. CC34 is typically positive.
Figure 2.
Solitary fibrous tumor. (A) The tumor shows spindle to ovoid cells with hypocellular and hypercellular areas and a hemangiopericytoma-like vascular pattern (H&E stain, ×100). (B) The tumor cells show diffuse nuclear positivity for STAT6, suggesting the presence of an underlying NAB2::STAT6 fusion gene (immunohistochemical stain for STAT6, ×200).
IGF2 overexpression is consistently detected in SFTs, regardless of anatomical location, and may be associated with triggering hypoglycemia in some patients [76]. STAT6 gene is located on the long arm of chromosome 12 in the same region as MDM2 and is, therefore, coamplified and overexpressed in a subset of DDLPS [81]. Dedifferentiated SFTs show an abrupt transformation into an indistinguishable pleomorphic appearance. Nuclear expression of STAT6 may be lost in dedifferentiated SFT [82]. The behavior of SFTs has been challenging to predict. A new risk stratification model based on patient age, tumor size, necrosis, and mitotic activity has been shown to more accurately predict the prognosis of SFTs [83].
8.3. Inflammatory Myofibroblastic Tumor
Inflammatory myofibroblastic tumor (IMT) is a characteristic, rarely metastasizing neoplasm composed of myofibroblastic and fibroblastic spindle cells accompanied by an inflammatory infiltrate of plasma cells, lymphocytes, and/or eosinophils [84]. Genetically, IMTs are heterogeneous. In 50–60% of cases of IMT in children and young adults, the tumors fuse the ALK gene on 2p23 with various partner genes, including TPM3, TPM4, CLTC, and others [85,86]. ALK rearrangement is uncommon in IMTs diagnosed in older adults. ROS1 and NTRK3 gene rearrangements are each found in 5–10% of IMTs [87,88,89]. Very rare cases have RET or PDGFRB gene rearrangements [90]. Epithelioid inflammatory myofibroblastic sarcoma (EIMS) is a rare, aggressive variant dominated by epithelioid cells with amphophilic cytoplasm. EIMS has often been associated with RANBP2::ALK or RRBP1::ALK gene rearrangements [90,91]. Immunohistochemically, ALK expression is detectable in 50–60% of IMT cases (Figure 3). The ALK immunostaining pattern varies depending on the ALK fusion partner; for example, RANBP2::ALK is correlated with a nuclear membranous pattern, RRBP1::ALK with a perinuclear accentuated cytoplasmic pattern, and CLTC::ALK with a granular cytoplasmic pattern. This is while several other ALK fusion variants illustrate a diffuse cytoplasmic pattern (most commonly seen in IMT) [90].
Figure 3.
Inflammatory myofibroblastic tumor. (A) The tumor shows spindle cells with vesicular nuclei and eosinophilic cytoplasm admixed with lymphocytes and plasma cells (H&E stain, ×200). (B) The tumor cells show cytoplasmic positivity for ALK, indicating the presence of an underlying ALK rearrangement (immunohistochemical stain for ALK, ×200).
In ALK-negative cases, IHC for ROS1 and/or molecular assays for non-ALK gene fusions (e.g., NTRK3) may be useful [87,90]. ROS1-rearranged IMT typically shows a cytoplasmic expression of ROS1 [87]. Highly sensitive ALK antibody clones (5A4 and D5F3) can enhance the detection of the ALK protein in IMT [87]. A persistent partial response to the ALK inhibitor crizotinib has been reported in a patient with ALK-translocated IMT [92].
8.4. Low-Grade Fibromyxoid Sarcoma
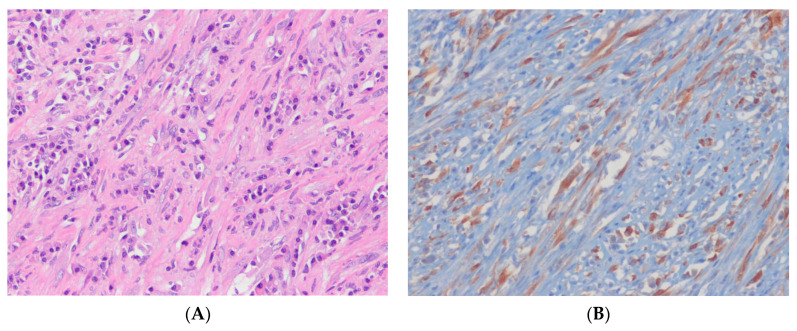
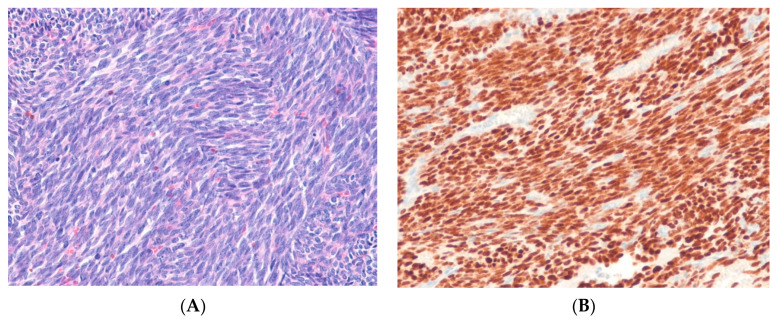
Low-grade fibromyxoid sarcoma (LGFMS) is a malignant fibroblastic neoplasm with bland spindle cells that grow in whorling growth pattern, a matrix that can be either collagenous or myxoid, and arcades of small blood vessels [93]. Genetically, LGFMSs exhibit a characteristic t(7;16)(q33;p11) translocation that results in the FUS::CREB3L2 fusion oncogene in > 90% of cases [94,95,96,97]. The FUS::CREB3L2 chimeric protein acts as an aberrant transcription factor, causing deregulated expression of CREB3L2 target genes [97]. Rare cases of LGFMS show the presence of FUS::CREB3L1 or EWSR1::CREB3L1 fusion genes [94,98]. The MUC4 gene on the long arm of chromosome 3 (3q29) is upregulated in LGFMS [97]. Immunohistochemically, tumor cells show strong, diffuse cytoplasmic expression of MUC4 in 100% of LGFMS cases [99,100] (Figure 4). EMA is also expressed in 80% of cases.
Figure 4.
Low-grade fibromyxoid sarcoma. (A) The tumor shows bland-appearing spindle cells in the myxoid and fibrous stroma, with short fascicles or a storiform pattern (H&E stain, ×100). (B) The tumor cells show cytoplasmic expression for MUC4, indicating the presence of underlying MUC4 upregulation (immunohistochemical stain for MUC4, ×200).
MUC4 is a high-molecular-weight transmembrane glycoprotein expressed on the cell membrane of many epithelial cells. It is a highly sensitive and specific marker for LGFMS and can help distinguish LGFMS from histologic mimics, such as soft tissue perineurioma [93]. If MUC4 is negative or unavailable to confirm the diagnosis, identification of FUS::CREB3L2 (or other uncommon variants) offers molecular genetic support for the diagnosis of LGFMS [93].
8.5. Sclerosing Epithelioid Fibrosarcoma
Sclerosing epithelioid fibrosarcoma (SEF) is a rare malignant fibroblastic neoplasm with epithelioid fibroblastic tumor cells arranged in cords and nests with dense sclerotic hyaline stroma. A subset of SEFs is morphologically and molecularly related to LGFMS [101]. Genetically, most cases of pure SEF harbor the EWSR1::CREB3L1 gene fusion [102,103,104]. In rare cases, EWSR1 is exchanged for FUS or PAX5, and/or CREB3L1 for CREB3L2, CREB3L3, or CREM [105,106]. SEF and LGFMS also have overlapping gene expression profiles. For instance, both tumors demonstrate high expression of MUC4 and CD24 [98]. Immunohistochemically, MUC4 is expressed in 80–90% of SEF cases with strong, diffuse, and cytoplasmic staining [107].
Cases showing hybrid features of SEF and LGFMS are now well described. Most cases of hybrid SEF/LGFMS show FUS::CREB3L2 gene fusion and MUC4 immunopositivity [96]. A small subset of SEFs lacks MUC4 expression. Recently, recurrent YAP1 and KMT2A gene rearrangements have been identified in MUC4-negative SEFs [108]. SEF has been shown to exhibit more aggressive behavior than LGFMS [109].
8.6. Infantile Fibrosarcoma
Conventional fibrosarcoma falls into two main categories: adult and infantile types. Infantile fibrosarcoma (IFS) is a malignant fibroblastic tumor that occurs most frequently in infancy [110]. It is a locally aggressive and rapidly growing tumor that rarely metastasizes. Most of these tumors harbor the ETV6::NTRK3 fusion resulting from t(12;15)(p13;q25). NTRK3 is a receptor tyrosine kinase. The ETV6::NTRK3 fusion gene encodes a constitutively activated chimeric tyrosine kinase that signals via the RAS-MAPK and PI3K signaling pathways to drive cellular transformation and oncogenesis [111]. In addition, EML4::NTRK3 fusion has been reported in rare cases [112]. Alternative gene fusions have been reported in a subset of cases, including NTRK1, NTRK2, BRAF, and MET [113,114]. Immunohistochemically, IFSs with NTRK gene rearrangements are often positive for a pan-TRK antibody [115].
The ETV6::NTRK3 fusion is found in a variety of neoplasms, such as cellular congenital mesoblastic nephroma [111], secretory carcinomas of the breast [116], and salivary gland-type secretory carcinoma [117]. It has been demonstrated that pan-TRK antibodies are not entirely specific for tumors with NTRK rearrangements [115]. Hence, FISH is most often used to confirm the diagnosis. Adult fibrosarcoma is an uncommon sarcoma composed of relatively monomorphic fibroblastic tumor cells with variable collagen production and often herringbone architecture. Its diagnosis is based on the principle of exclusion; true examples are exceedingly rare [118].
9. Vascular Tumors
9.1. Epithelioid Hemangioma
Epithelioid hemangioma is a benign vascular neoplasm consisting of well-formed blood vessels lined by plump, epithelioid (histiocytoid) endothelial cells with abundant eosinophilic cytoplasm and a variable infiltrate of eosinophils [119]. Genetically, epithelioid hemangiomas are characterized by recurrent fusion genes affecting the FOS or FOSB gene in approximately 50% of the cases. The gene partners for FOS include LMNA, MBNL1, VIM, and lincRNA [120,121], whereas FOSB is often fused to ZFP36, WWTR1, or ACTB [122,123]. The key event in the pathogenesis is the dysregulation of the FOS family (FOS, FOSB, FOSL1, and FOSL2) of transcription factors through chromosomal translocation. Immunohistochemically, a subset of cases shows FOS or FOSB expression, which may be diagnostically useful [121,123].
Interestingly, angiolymphoid hyperplasia with an eosinophilia subtype (cutaneous epithelioid hemangioma) may be non-neoplastic as they lack FOS or FOSB gene rearrangement [120]. Although FOSB fusions have not been found in cases of angiolymphoid hyperplasia with eosinophilia, the endothelial cells in this tumor are often positive for FOSB [120,124]. FOSB fusion-associated tumors, particularly ZFP36::FOSB, are more commonly cellular and solid, with some cytologic atypia and occasional necrosis [123].
9.2. Pseudomyogenic Hemangioendothelioma
Pseudomyogenic hemangioendothelioma (PHE) is a rarely metastasizing endothelial neoplasm commonly occurring in young adult males [125]. It often presents as multiple discontinuous nodules at different tissue levels and histologically mimics a myoid tumor or epithelioid sarcoma (EPS). PHEs have t(7;19)(q22;q13), resulting in SERPINE1::FOSB gene fusion [126,127]. An alternative ACTB::FOSB gene fusion was identified in half of the cases [122]. These fusions lead to the upregulation of FOSB, a member of the FOS family of transcription factors that encode leucine zipper proteins that interact with the JUN family to regulate cell proliferation, differentiation, angiogenesis, and survival. Immunohistochemically, nuclear staining for FOSB is present in almost all cases of PHE [124,128]. PHE also shows a nuclear expression of the endothelial transcription factors FLI1 and ERG [129,130]. Approximately 50% of cases are positive for CD31. In addition, SMA is predominantly expressed in one-third of tumors.
The clinicopathologic features and behavior do not differ between PHEs with SERPINE1::FOSB and ACTB::FOSB fusions are rare, although tumors with the ACTB variant occur more frequently as solitary lesions [122]. Recently, FOSB fusions with WWTR1 or CLTC have been described, the latter in a bone lesion in an adolescent [131,132].
9.3. Epithelioid Hemangioendothelioma
Epithelioid hemangioendothelioma (EHE) is a malignant vascular neoplasm composed of epithelioid endothelial cells within a characteristic myxohyaline stroma [133]. Genetically, EHE is characterized by a t(1;3)(p36;q25), resulting in a WWTR1::CAMTA1 fusion in 90% of cases [134,135,136]. Fusion of WWTR1 with CAMTA1 leads to dysregulation of the Hippo signaling pathway, such that WWTR1::CAMTA1 resides constitutively in the cell nucleus and drives oncogenic transformation [137]. The YAP1::TFE3 fusion due to a t(X;11)(p11;q13) translocation is observed in 5% of EHEs with a distinctly vasoformative morphology [138]. TFE3, an oncogenic transcription factor involved in other soft tissue tumor translocations, is upregulated as a consequence of the YAP1::TFE3 fusion. Immunohistochemically, EHE with WWTR1::CAMTA1 typically shows a diffusely strong nuclear expression of CAMTA1 [139] (Figure 5). Tumors with YAP1::TFE3 fusion show diffuse nuclear expression of TFE3 [138].
Figure 5.
Epithelioid hemangioendothelioma. (A) The tumor cells show epithelioid cells with oval or round nuclei and variable amounts of eosinophilic cytoplasm in myxohyaline stroma. Intracytoplasmic vacuoles are present (H&E stain, ×100). (B) The tumor cells show strong nuclear expression of CAMTA1, indicating the presence of an underlying WWTR1::CAMTA1 fusion gene (immunohistochemical stain for CAMTA1, ×200).
CAMTA1 is a highly sensitive and specific diagnostic marker and can help distinguish EHE with WWTR1::CAMTA1 fusion from cellular epithelioid hemangiomas and epithelioid angiosarcomas. TFE3-positive EHEs occasionally exhibit WWTR1::CAMTA1 gene fusions [140]. These findings show that two chromosomal alterations are not mutually exclusive but rather composable in EHEs.
10. Skeletal Muscle Tumors
10.1. Alveolar Rhabdomyosarcoma
Alveolar rhabdomyosarcoma (ARMS) is a malignant neoplasm composed of primitive monomorphic round cells with skeletal muscle differentiation [141]. Approximately 85% of ARMSs contain characteristic fusion genes. In fusion-positive ARMSs, PAX3::FOXO1 and PAX7::FOXO1 fusion genes are detected in 70–90% and 10–30% of the fusions, respectively [142,143]. PAX3 and PAX7 represent transcription factors that play an essential role in myogenesis [144]. The PAX::FOXO1 fusion proteins function as oncoproteins, affecting growth, survival, differentiation, and other signaling pathways by activating numerous downstream target genes, such as MET, ALK, FGFR4, MYCN, IGF1R, and MYOD1 [145,146,147]. Immunohistochemically, the nuclear expression of myogenin is strong and diffuse, unlike in embryonal rhabdomyosarcoma (RMS) and other RMS subtypes in which the staining pattern is focal [148,149]. MyoD1 is predominantly expressed in ARMS.
Approximately 20% of ARMS cases are fusion-negative [150]. Fusion-negative ARMS is genetically heterogeneous and may have alternative fusions with other genes (NCOA1 and INO80D). Histologic appearances of ARMS do not predict the presence or type of gene fusion. However, solid growth or mixed embryonal/alveolar patterns show a higher incidence of fusion negativity [150]. Fusion-positive ARMS patients have a poorer outcome than fusion-negative ARMS patients [151].
10.2. Spindle Cell/Sclerosing Rhabdomyosarcoma
Spindle cell/sclerosing rhabdomyosarcoma (RMS) is a type of RMS with fascicular spindle cells and/or primitive cells in a prominent hyaline collagenous stroma [152]. Spindle cell/sclerosing RMSs are categorized into three groups based on their genetic background. The first group, congenital/infantile spindle cell RMS, shows gene fusions involving VGLL2, SRF, TEAD1, NCOA2, and CITED2 [153,154]. The second group includes most of the spindle cell/sclerosing RMS in adolescents and young adults and a subset of tumors in older adults, showing the presence of the MYOD1 mutation [154,155]. The third group shows no recurrent, identifiable genetic alterations. Immunohistochemically, spindle cell/sclerosing RMS typically shows diffuse nuclear staining for MyoD1. Myogenin shows only limited expression in most spindle cells/sclerosing RMSs.
The recently described intraosseous spindle cell RMS shows two gene fusions, EWSR1/FUS::TFCP2 and MEIS1::NCOA2 [155,156]. Congenital/infantile spindle cell/sclerosing RMSs with gene fusions show a favorable clinical progression [157]. MYOD1-mutant spindle cells/sclerosing RMSs follow an aggressive clinical course despite multimodality therapy [158].
11. Gastrointestinal Stromal Tumor
A gastrointestinal stromal tumor (GIST) is a mesenchymal neoplasm characterized by differentiation to the interstitial cells of Cajal with variable behavior [159]. Approximately 75% of GISTs harbor activating mutations of KIT, most commonly in exon 11 (66% overall) or exon 9 (6%); mutations in exons 13 and 17 are rare (~1% each) [160,161]. Approximately 10% of GISTs harbor PDGFRA-activating mutations (most frequently in the stomach) [162,163]. The KIT or PDGFRA oncogene is located on chromosome 4 (4q12) and encodes type III receptor tyrosine kinases [162]. Downstream oncogenic signaling involves the RAS/MAPK and PI3K/AKT/mTOR signaling pathways [161,164]. Many KIT/PDGFRA wild-type GISTs have alterations in SDH subunit genes (5–10% overall) [165,166]. Almost all pediatric GISTs are SDH-deficient [165,167]. SDHA is the most commonly mutated subunit gene (~35% of SDH-deficient GISTs), followed by SDHB, SDHC, and SDHD [168]. Immunophenotypically, most GISTs show strong and diffuse expression of KIT (CD117). DOG1 (ANO1) is highly sensitive and specific for GIST and is useful in diagnosing KIT-negative GISTs [169]. SDH-deficient GISTs show a loss of SDHB protein expression, regardless of which SDH gene is mutated [168]. SDHA loss is specific to SDHA-mutant tumors [168].
Patients with PDGFRA-mutant tumors have a lower risk of metastasis than patients with KIT-mutant tumors [170]. GISTs harboring the PDGFRA D842V mutation have been shown to respond to avapritinib—a novel KIT and PDGFRA inhibitor [171]. Recently, the use of SDHB by immunohistochemistry has been used to stratify GIST into an SDHB-retained and an SDHB-deficient group, regardless of whether the responsible mutation was acquired or inherited. This widely available screening approach can facilitate decisions about further molecular testing strategies [172].
12. Peripheral Nerve Sheath Tumors
12.1. Malignant Peripheral Nerve Sheath Tumor
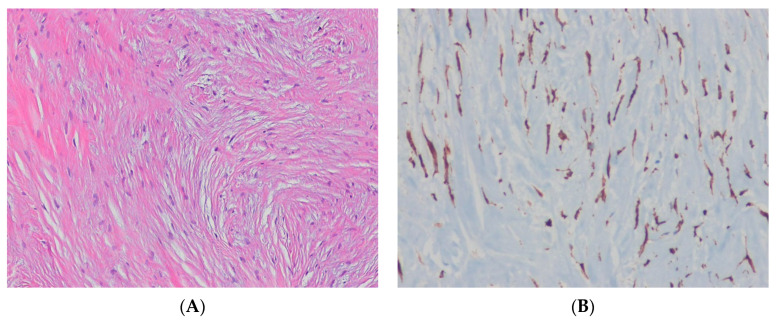
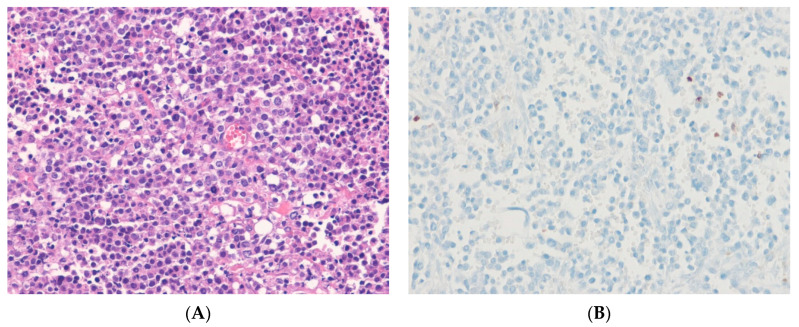
A malignant peripheral nerve sheath tumor (MPNST) is a malignant spindle cell tumor that shows evidence of nerve sheath differentiation. It often arises from a peripheral nerve, a pre-existing benign nerve sheath tumor, or in a patient with neurofibromatosis type 1 [173]. Genetically, conventional MPNSTs have complex karyotypes. Irrespective of whether they are NF1-associated, sporadic, or radiotherapy-associated, the majority of MPNSTs demonstrate highly recurrent and specific inactivating mutations of polycomb repressive complex 2 (PRC2) components (EED or SUZ12), NF1, and CDKN2A/CDKN2B [174,175,176]. Inactivation of the PRC2 results in the loss of histone H3K27 trimethylation (H3K27me3) [177,178]. Immunohistochemically, loss of H3K27me3 expression is more common in high-grade tumors (70–80%) than in low-grade tumors (20–30%) [178,179] (Figure 6). Thus, H3K27me3 can serve as a useful diagnostic marker for MPNST.
Figure 6.
Malignant peripheral nerve sheath tumor. (A) The tumor shows spindle cells in a fascicular pattern. Perivascular accentuation of cellularity is present (H&E stain, ×100). (B) The tumor cells show loss of H3K27me3 expression, indicating the presence of underlying inactivating mutations of the polycomb repressive complex 2 components (SUZ12 or EED) (immunohistochemical stain for H3K27me3, ×200).
At the molecular level, epithelioid MPNSTs differ from conventional MPNSTs. Approximately 75% of epithelioid MPNST cases show SMARCB1 gene inactivation, resulting in SMARCB1 loss by immunohistochemistry [180]. In contrast to conventional MPNSTs, nuclear staining for H3K27me3 is retained in epithelioid MPNSTs (i.e., normal). MPNSTs showing complete heterologous rhabdomyoblastic differentiation mimic spindle cell RMS. Immunohistochemistry for H3K27me3 reliably distinguishes MPNST with complete heterologous rhabdomyoblastic differentiation from spindle cell RMS [181].
12.2. Malignant Melanotic Nerve Sheath Tumor
A malignant melanotic nerve sheath tumor (MMNST) is a rare peripheral nerve sheath tumor composed of tumor cells with features of both Schwann cell and melanocytic differentiation [182]. It is most commonly associated with spinal or autonomic nerves near the midline. MMNSTs are frequently associated with the Carney complex. The majority of MMNSTs have inactivating mutations of the PRKAR1A gene on 17q24.2 [183]. PRKAR1A plays a central role in the development of MMNST. Immunohistochemically, PRKAR1A expression is typically lost in MMNST [183,184]. MMNSTs strongly express S100 protein, SOX10, and various melanocytic markers, including HMB45, Melan-A, and tyrosinase.
Psammoma bodies are found in approximately 50% of cases [182]. There are no clinical distinctions between psammomatous and non-psammomatous MMNSTs [183,184]. In addition, their histologic features do not correlate well with their clinical behavior. MMNSTs often show aggressive behavior [185,186]. It is critical to distinguish MMNST from malignant melanoma. The paravertebral location, heavy melanin pigmentation, psammoma bodies, and loss of PRKAR1A expression suggest the diagnosis of MMNST [184].
13. Tumors of Uncertain Differentiation
13.1. Synovial Sarcoma
Synovial sarcoma (SS) is a monomorphic spindle cell mesenchymal neoplasm with variable epithelial differentiation [187]. SS harbors a unique t(X;18)(p11.2;q11.2) translocation [188], by which one of the SSX genes (SSX1, SSX2, or SSX4) on the X chromosome fuses to SS18 on chromosome 18 [189]. Approximately two-thirds of SS cases harbor an SS18::SSX1 fusion, one-third harbor an SS18::SSX2 fusion, and uncommon cases harbor an SS18::SSX4 fusion [189,190]. SS18::SSX functions as an oncogene, and its expression is necessary to maintain the transformed phenotype of SS cells [191,192]. Immunohistochemically, a novel SS18-SSX fusion-specific antibody is highly sensitive (95%) and specific (100%) for SS [193] (Figure 7). Thus, SS18-SSX immunohistochemistry is a useful tool to confirm SS diagnosis, and it can replace molecular testing in most cases [194]. Moderate or strong nuclear staining for the transcriptional corepressor TLE1 is present in the majority of SS cases [195]. However, TLE1 staining is not specific to SS because it can also exist in histological mimics of SS, particularly MPNST and SFT [195].
Figure 7.
Monophasic synovial sarcoma. (A) The tumor shows fascicles of monomorphic spindle cells (H&E stain, ×100). (B) The tumor cells show strong nuclear expression for SS18-SSX antibody, consistent with the presence of an underlying SS18::SSX fusion gene (immunohistochemical stain for SS18-SSX antibody, ×200).
Interestingly, SS shows correlations between fusion type, tumor histology, and patient sex. Almost all SS18::SSX2 fusion cases show monophasic histology. In contrast, SS18::SSX1 fusion cases show an approximate 2:1 ratio of monophasic to biphasic SS [191]. Males show a 3:2 ratio of SS18::SSX1 to SS18::SSX2, whereas in females, the ratio is close to 1:1. Molecular confirmation (if available) of an SS18::SSX1/2/4 fusion should ideally be performed for optimal diagnostic accuracy [57]. Recently, novel and rare SSX1 fusions to non-SS18 genes have been reported in SS [196].
13.2. Epithelioid Sarcoma
Epithelioid sarcoma (EPS) is a malignant mesenchymal neoplasm exhibiting partial or complete epithelioid cytomorphology and evidence of epithelial differentiation [197]. Two clinicopathological subtypes are recognized in EPSs: (1) the classic (or distal) form, characterized by its propensity for acral sites and pseudogranulomatous growth pattern, and (2) the proximal-type (large cell) subtype, occurring mainly in proximal/truncal regions and consisting of nests and sheets of large epithelioid cells. Genetically, approximately 90% of both classic and proximal-type patients have SMARCB1 (INI1) deletion. The SMARCB1 gene (also called BAF47, INI1, or SNF5) at chromosome 22q11.2 encodes a protein part of the SWI/SNF chromatin-remodeling complex present in normal cells. Loss of expression of SWI/SNF chromatin-remodeling complex proteins plays an essential role in tumorigenesis [198]. Immunohistochemically, loss of SMARCB1 expression occurs in the majority of EPS cases [199,200,201]. ERG expression is commonly observed in EPSs, which can lead to confusion with endothelial tumors [202,203,204]. Most EPS cases are positive for cytokeratins and EMA. CD34 is expressed in 50–60% of EPS cases, which helps distinguish EPS from carcinomas.
An extreme minority of EPSs retain SMARCB1 (INI1) protein expression. The biological behavior of SMARCB1 (INI1)-preserved EPS is more aggressive than that of EPS with complete loss of SMARCB1 expression [198].
13.3. Extrarenal Rhabdoid Tumor
An extrarenal rhabdoid tumor (ERT) is a highly malignant soft tissue neoplasm composed of characteristic rounded or polygonal rhabdoid cells with glassy eosinophilic cytoplasm containing hyaline-like inclusion bodies, eccentric nuclei, and macronucleoli [205]. It mainly affects infants and children. Morphologically and genetically identical tumors also occur in the kidney and brain. Most ERTs are characterized by biallelic alterations of the SMARCB1 gene, resulting in a loss of expression of SMARCB1 (INI1). Immunohistochemically, the tumors show loss of SMARCB1 (INI1) expression [206,207]. In addition, SALL4 and glypican-3 expressions are frequently observed in ERTs [208,209].
A loss of SMARCB1 expression is also present in EPS, epithelioid MPNST, and myoepithelial carcinoma. When a tumor histologically similar to ERT occurs in adults, pathologists should first consider the possibility of malignant melanoma or other tumor types. Familial cases are typically associated with germline mutations in the SMARCB1 gene [210,211]. Mutations and/or loss of the SMARCA4 gene in 19p13.2 have been reported in rare rhabdoid tumors with retention of SMARCB1 expression [212].
13.4. Alveolar Soft Part Sarcoma
Alveolar soft part sarcoma (ASPS) is a rare tumor of uncertain histogenesis predominantly affecting the deep soft tissues of the extremities [213]. ASPS features variably discohesive epithelioid cells arranged in nests, resulting in a distinct alveolar growth pattern. It is characterized by a specific translocation, der(17)t(X;17)(p11.2;q25), resulting in ASPSCR1::TFE3 gene fusion. The ASPSCR1::TFE3 fusion protein activates c-Met signaling [214,215,216]. Immunohistochemically, ASPS shows nuclear immunoreactivity for TFE3 [217,218]. The immunopositivity for cathepsin K (100%) is also typical. In addition, calretinin (46%) [219,220] and focal desmin (50%) are also expressed in ASPS cases [221].
Although the ASPSCR1::TFE3 fusion in sarcomas appears highly specific and sensitive for ASPS, the same gene fusion is also found in a small subset of TFE3-rearranged renal cell carcinomas that affect young patients and have a morphology similar to ASPS [222]. In a clinical study with the c-Met inhibitor crizotinib in ASPS, disease stabilization was reported in most TFE3-rearranged ASPS MET-altered patients [223]. Therefore, c-MET could be a potential therapeutic target in ASPS.
13.5. Desmoplastic Small Round Cell Tumor
Desmoplastic small round cell tumor (DSRCT) is a malignant mesenchymal neoplasm of primitive small round tumor cells associated with prominent desmoplastic stroma and polyphenotypic differentiation [224]. It is characterized by a recurrent chromosomal translocation t(11;22)(p13;q12), resulting in the fusion of the EWSR1 gene on 22q12.2 and the WT1 gene on 11p13 [225,226,227,228]. The aberrant transcription of EWSR1::WT1 regulates the expression of various genes and activates the neural reprogramming factor ASCL1 to induce partial neural differentiation [229,230]. Immunohistochemically, DSRCT shows a characteristic polyphenotypic profile expressing epithelial, muscular, and neural markers. A polyclonal antibody to the carboxy (C)-terminus of WT1 is reactive and useful for diagnosis [231].
Recently, there was a report of three unusual tumors affecting the female genital tract with EWSR1::WT1 gene fusion lacking features of DSRCT [232]. These findings suggest the pleiotropy of the EWSR1::WT1 fusion is possible and not restricted to DSRCT. Detection of the EWSR1::WT1 gene fusion can be particularly useful in cases with unusual clinical or histological features [233]. DSRCT is an aggressive disease, despite multimodal therapies [234]. Hence, a better understanding of disease biology is necessary for identifying potential targets in the future [235].
13.6. Intimal Sarcoma
Intimal sarcomas are malignant mesenchymal tumors arising within the large blood vessels of the systemic and pulmonary circulations and in the heart [236]. The defining features are primarily intraluminal growth, obstruction of the lumen in the originating vessel, and seeding of tumor emboli in peripheral organs. Genetically, frequent amplifications/gains in the 12q13–q14 region (which contains MDM2 and CDK4) and (co)amplification/gains of PDGFRA, EGFR, and KIT are present [237,238,239,240]. MDM2 and PDGFR pathways may play a role in the pathogenesis of intimal sarcoma. Immunohistochemically, nuclear expression of MDM2 is observed in at least 70% of cases [237,240]. In addition, rare cases containing rhabdomyosarcomatous differentiation are positive for myogenin and MyoD1 [42].
Intimal sarcomas and undifferentiated cardiac sarcomas carry mutually exclusive MDM2, MDM4, and CDK6 amplifications and share a typical DNA methylation signature [241]. Many primary cardiac sarcomas with histological features of undifferentiated pleomorphic sarcoma are currently reported as intimal sarcomas, especially if there is MDM2 expression [242].
Table 1 is a summary of the recently described molecular genetic changes and related immunohistochemical markers in some soft tissue tumors.
Table 1.
Recently described molecular genetic alterations and immunohistochemical markers in selected soft tissue tumors.
| Tumor Category | Tumor Type | Cytogenetic Alterations | Molecular Alterations | Immunohistochemical Markers | Staining Pattern | References |
|---|---|---|---|---|---|---|
| Adipocytic tumors | Spindle cell/pleomorphic lipoma | Loss of 13q14 Loss of 16q |
RB1 deletion | RB1 | Loss | [20,21,22,23] |
| Atypical spindle cell/pleomorphic lipomatous tumor | Loss of 13q14 Monosomy 7 |
RB1 deletion (subset) | RB1 | Loss | [28,29,30,31] | |
| Atypical lipomatous tumor/well-differentiated liposarcoma | Gain of 12q13–15 region |
MDM2 amplification CDK4 amplification |
MDM2, CDK4 | Nuclear staining | [35,36] | |
| Dedifferentiated liposarcoma | Gain of 12q13–15 region |
MDM2 amplification CDK4 amplification |
MDM2, CDK4 | Nuclear staining | [45,46] | |
| Myxoid liposarcoma | t(12;16)(q13;p11) t(12;22)(q13;q12) |
FUS::DDIT3 (>90%) EWSR1::DDIT3 (3%) |
DDIT3 | Nuclear staining | [50,56] | |
| Fibroblastic and myofibroblastic tumors | Desmoid fibromatosis | Trisomy 8, trisomy 20 Loss of 5q21 |
CTNNB1 mutation APC mutation |
β-catenin | Nuclear staining | [60,61,65,66] |
| Solitary fibrous tumor | inv12(q13;q13) | NAB2::STAT6 | STAT6 | Nuclear staining | [72,73,74,80] | |
| Inflammatory myofibroblastic tumor | t(1;2)(q21;p23) t(2;19)(p23;p13) t(2;17)(p23;q23) t(6;17)(q22;p13) t(3;6)(q12;q22) |
TPM3::ALK TPM4::ALK CLTC::ALK ROS1::YWHAE ROS1::TFG1 |
ALK ROS1 |
Cytoplasmic staining | [85,86,87,88,89] | |
| Epithelioid inflammatory myofibroblastic sarcoma | t(2;2)(p23;q13) |
RANBP2::ALK RRBP1::ALK |
ALK | Nuclear membrane or perinuclear accentuation | [90,91] | |
| Low-grade fibromyxoid sarcoma | t(7:16)(q33;p11) t(11;16)(p11;p11) |
FUS::CREB3L2 (>90%) FUS::CREB3L1 EWSR1::CREB3L1 |
MUC4 | Cytoplasmic staining | [94,95,96,97,98,99,100] | |
| Sclerosing epithelioid fibrosarcoma | t(11;22)(p11;q12) t(11;16)(p11;p11) t(7;16)(q34;p11) |
EWSR1::CREB3L1 (80–90%) EWSR1::CREB3L2 FUS::CREB3L2 |
MUC4 | Cytoplasmic staining | [102,103,104,105,106,107] | |
| Infantile fibrosarcoma | t(12;15)(p13;q25) |
ETV6::NTRK3 EML4::NTRK3 |
Pan-TRK | Cytoplasmic or membranous staining |
[112,113,114,115] | |
| Vascular tumors | Epithelioid hemangioma | t(19;19)(q13;q13) t(7;19)(q22;q13) |
FOS::VIM, FOS::LMNA, ZFP36::FOSB, WWTR1::FOSB |
FOS (subset) FOSB (subset) |
Nuclear staining | [120,121,122,123] |
| Pseudomyogenic hemangioendothelioma | t(7;19)(q22;q13) |
SERPINE1::FOSB ACTB::FOSB |
FOSB | Nuclear staining | [126,127,128] | |
| Epithelioid hemangioendothelioma | t(1;3)(p36;q25) t(X;11)(p11;q13) |
WWTR1::CAMTA1 (85–90%) YAP1::TFE3 (5%) |
CAMTA1 TFE3 |
Nuclear staining | [134,135,136,137,138,139,140] | |
| Skeletal muscle tumors | Alveolar rhabdomyosarcoma | t(2;13)(q35;q14) t(1;13)(p36;q14) |
PAX3::FOXO1 (70–90%) PAX7::FOXO1 (10–30%) |
Myogenin MyoD1 |
Nuclear staining | [142,143,148,149] |
| Congenital/infantile spindle cell rhabdomyosarcoma | SRF::NCOA2, TEAD1::NCOA2, VGLL2::NCOA2, VGLL2::CITED2 | Myogenin MyoD1 |
Nuclear staining | [153,154] | ||
| MYOD1-mutant spindle cell/sclerosing rhabdomyosarcoma | MYOD1 mutations | Myogenin MyoD1 |
Nuclear staining | [154,155] | ||
| Intraosseous spindle cell rhabdomyosarcoma |
EWSR1/FUS::TFCP2 MEIS1::NCOA2 |
Myogenin MyoD1 |
Nuclear staining | [155,156] | ||
| Gastrointestinal stromal tumors | Gastrointestinal stromal tumor |
KIT mutations (75%) PDGFRA mutations (10%) |
KIT (CD117) DOG1 |
Cytoplasmic or membranous staining | [160,161,162,163,169] | |
| SDH-deficient gastrointestinal stromal tumor | SDH mutations | SDHB SDHA |
Loss | [165,166,168] | ||
| Peripheral nerve sheath tumor | MPNST | Complex karyotype with numerical and structural abnormalities | NF1 inactivation; PRC2 components (EED or SUZ12) inactivation | H3K27me3 | Loss | [177,178,179] |
| Epithelioid MPNST | 22q deletion | SMARCB1 inactivation | SMARCB1 | Loss | [180] | |
| MMNST | Mutation and/or loss of heterozygosity of 17q | PRKAR1A inactivation | PRKAR1A | Loss | [183,184] | |
| Tumors of uncertain differentiation | Synovial sarcoma | t(X;18)(p11.2;q11.2) |
SS18::SS1, SS2, or SS4 fusion; SS18L1::SSX1 fusion (rare) |
SS18-SSX | Nuclear staining | [189,190,193,194] |
| Epithelioid sarcoma | 22q11.2 deletion | SMARCB1 inactivation | SMARCB1 (INI1) | Loss | [197,199,200,201] | |
| Extrarenal rhabdoid tumor | 22q11.2 deletion | SMARCB1 inactivation | SMARCB1 (INI1) | Loss | [206,207] | |
| Alveolar soft part sarcoma | der(17)t(X;17)(p11;q25) | ASPSCR1::TFE3 | TFE3 | Nuclear staining | [217,218] | |
| DSRCT | t(11;22)(p13;q12) | EWSR1::WT1 | WT1 (C-erminus) | Nuclear staining | [225,226,227,228,231] | |
| Intimal sarcoma | 12q12-15 amplification |
MDM2 amplification PDGFRA amplification |
MDM2 | Nuclear staining | [237,238,239,240] |
MPNST, malignant peripheral nerve sheath tumor; PRC2, polycomb repressive complex 2; MMNST, malignant melanotic nerve sheath tumor; DSRCT, desmoplastic small round cell tumor; SDH, succinate dehydrogenase.
14. Undifferentiated Small Round Cell Sarcomas
14.1. Ewing Sarcoma
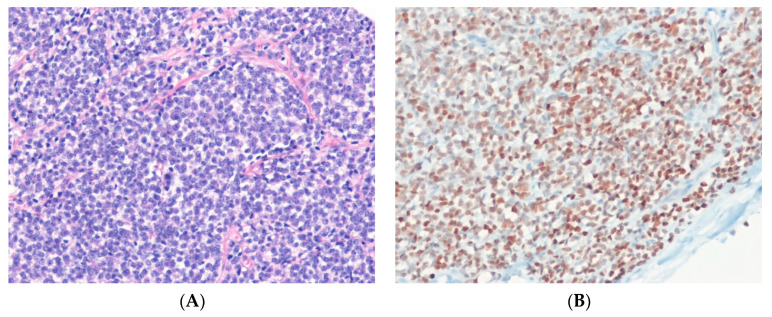
Ewing sarcoma (EWS) is a small round cell sarcoma characterized by a fusion of a FET gene family member (most commonly EWSR1) and an ETS gene family member [243]. Further mutations can occur in STAG2 (15–22%), CDKN2A (12%), and TP53 (7%) [244,245,246]. FET::ETS fusion genes encode chimeric transcription factors that function as master regulators to activate and repress thousands of genes. Expression of these aberrant transcription factors is required to develop EWS. The most common translocation (in 85–90% of cases) is t(11;22)(q24;q12), which results in the EWSR1::FLI1 fusion transcript and protein. The second most common is t(21;22)(q22;q12), which results in EWSR1::ERG in 5–10% of EWS cases. Immunohistochemically, strong, diffuse membranous expression of CD99 is observed in approximately 95% of EWSs. NKX2-2, a neuroendocrine/glial transcription factor, has a higher specificity than CD99 [247] (Figure 8). Strong nuclear ERG immunoreactivity is observed in EWS cases with EWSR1::ERG rearrangement [248]. FLI1 is expressed in the majority of EWS cases, regardless of the fusion variant. The adamantinoma-like variant of EWS consistently demonstrates diffuse cytokeratin, p63, and p40 positivity [249].
Figure 8.
Ewing sarcoma. (A) The tumor shows round cells with finely granular chromatin and clear cytoplasm (H&E stain, ×100). (B) The tumor cells show diffuse nuclear staining for NKX2-2, suggesting the presence of underlying NKX2-2 upregulation as a downstream target of the EWSR1::FLI1 fusion gene (immunohistochemical stain for NKX2-2, ×100).
Because EWSR1 fusions are identified in a diverse array of tumor types, FISH for EWSR1 is not specific for EWS; nonetheless, in the appropriate context, demonstration of EWSR1 rearrangement is sufficient to confirm the diagnosis [250]. A FISH-based approach using break-apart probes for EWSR1 and/or FUS uncovers most EWS cases. However, a minority of cases with complex inversion/insertion, structural rearrangements, or cryptic insertions may be negative by FISH and require RNA-based techniques for molecular diagnosis [251,252].
14.2. Round Cell Sarcoma with EWSR1-Non-ETS Fusions
Round cell sarcomas with EWSR1–non-ETS fusions are round, and spindle cell sarcomas with EWSR1 or FUS fusions involving partners irrelevant to the ETS gene family [253]. This category includes EWSR1/FUS::NFATC2 sarcomas [254] and EWSR1::PATZ1 sarcomas [255]. EWSR1/FUS::NFATC2 sarcomas have a preference for the bone and consist of round to spindle cells arranged in cords, nests, and trabeculae on a myxohyaline background. EWSR1::PATZ1 sarcomas usually occur in the deep soft tissue and have diverse morphologic features, with a small round to spindle cells in the fibrous stroma, variable necrosis, and mitoses. Immunohistochemically, CD99 is diffusely expressed in 50% of EWSR1/FUS::NFATC2 sarcoma cases. NKX2-2 and PAX7 may also be expressed [256,257]. AGGRECAN shows diffuse, cytoplasmic, and membranous staining [258]. NKX3-1 is frequently expressed in EWSR1::NFATC2 sarcomas [259,260]. EWSR1::PATZ1 sarcomas exhibit co-expression of myogenic markers (desmin, myogenin, MyoD1) and neural markers (S100 protein, SOX10, GFAP) [255].
The EWSR1/FUS::NFATC2 and EWSR1::PATZ1 fusions can be identified via diverse molecular approaches [261,262]. However, EWSR1::PATZ1 fusion is easily missed when using EWSR1 break-apart FISH [263]. Currently, NGS-based fusion panels are often applied to identify these gene rearrangements and confirm the diagnosis [250].
14.3. CIC-Rearranged Sarcoma
CIC-rearranged sarcoma is a high-grade undifferentiated round cell sarcoma defined by CIC-related gene fusions, most often CIC::DUX4 [264]. A CIC::DUX4 fusion is present in 95% of cases, resulting from either a t(4;19)(q35;q13) or a t(10;19)(q26;q13) translocation [265,266]. Rare cases are associated with non-DUX4 partner genes, including FOXO4, LEUTX, NUTM1, and NUTM2A [267,268,269]. The CIC::DUX4 fusion significantly enhances the CIC transcriptional activity and upregulates its targets, including CCND2, MUC5AC, and PEA3 family genes (e.g., ETV1, ETV4, and ETV5) [265,270]. In addition, the CIC::DUX sarcomas demonstrate frequent MYC amplification [271]. Immunohistochemically, DUX4 is a highly sensitive and specific marker for the differentiation of sarcoma with CIC::DUX4 fusion from its histologic mimics [272]. WT1 (anti-N-terminus monoclonal antibodies) (90–95%) and ETV4 (95–100%) are frequently positive and represent useful ancillary markers [273,274]. NKX2-2 is negative in CIC-rearranged sarcomas [275]. Sarcomas with CIC::NUTM1 fusions express NUT protein [267].
CIC-mutated or rearranged angiosarcomas represent a potential diagnostic pitfall [269]. CIC fusions have also been described in central nervous system tumors [276]. CIC-rearranged sarcomas are aggressive tumors with frequent metastases and poor outcomes. Their five-year survival rate ranges from 17–43%, and the response to standard EWS chemotherapy regimens is generally poor [267,277].
14.4. Sarcoma with BCOR Genetic Alteration
Sarcomas with BCOR genetic alterations are clinically distinct sarcomas arising in soft tissue and bone and divided into two main groups. The first group is characterized by sarcomas with BCOR-related gene fusions (BCOR-fusion sarcomas), most frequently BCOR::CCNB3. The second group shows internal tandem duplication in BCOR (BCOR-ITD sarcomas), described in infantile undifferentiated round cell sarcomas and primitive myxoid mesenchymal tumors of infancy [278,279]. BCOR-fusion and BCOR-ITD sarcomas show a similar gene expression signature [280,281,282,283]. Immunohistochemically, all tumors with various BCOR gene alterations show strong and diffuse nuclear positivity for BCOR (Figure 9). BCOR immunostaining may be diagnostically useful but is not specific [282,284]. Additionally, SATB2, TLE1, and cyclin D1 expressions are present in most sarcomas with BCOR genetic alterations. BCOR::CCNB3 sarcomas also express CCNB3 [284,285]. Pediatric soft tissue tumors with BCOR-ITD show membranous and cytoplasmic expression of EGFR [286].
Figure 9.
BCOR-rearranged sarcoma. (A) The tumor shows solid sheets of monomorphic round cells with round nuclei and scant eosinophilic cytoplasm, with delicate capillaries (H&E stain, ×100). (B) The tumor cells show diffuse nuclear staining for BCOR, indicating the presence of an underlying BCOR::CCNB3 fusion gene (immunohistochemical stain for BCOR, ×200).
BCOR family tumors share a morphologic spectrum with a similar immunoprofile and gene expression, suggesting a shared pathogenesis. Since immunohistochemistry for either BCOR or CCNB3 is not completely sensitive and specific, a molecular genetic approach is necessary for diagnosis [284]. BCOR::CCNB3 sarcomas often respond to EWS regimens and have a similar outcome [283]. The outcomes of the other BCOR family tumors need to be better defined.
Table 2 shows a summary of molecular genetic alterations and immunohistochemical markers in undifferentiated small round cell sarcomas.
Table 2.
Molecular genetic alterations and immunohistochemical markers in undifferentiated small round cell sarcomas.
| Tumor Type | Cytogenetic Alterations | Molecular Alterations | Immunohistochemical Markers | Staining Pattern | References |
|---|---|---|---|---|---|
| Ewing sarcoma | t(11;22)(q24;q12) t(21;22)(q22;q12) t(2;22)(q33;q12) t(7;22)(p22;q12) t(17;22)(q21;q12) |
EWSR1::FLI1 (85–90%) EWSR1::ERG (5–10%) EWSR1::FEV EWSR1::ETV1 EWSR1::ETV4 |
NKX2-2 FLI1 ERG CD99 |
NKX2-2, FLI1, ERG, nuclear staining; CD99, membranous staining |
[244,245,246,247,248] |
| Round cell sarcoma with EWSR1–non-ETS fusions |
EWSR1/FUS::FATC2 sarcoma; t(20;22)(q13;q12) t(16;20)(p11;q13) EWSR1::PATZ1 sarcoma; t(22;22)(q12;q12) |
EWSR1/FUS::FATC2 sarcoma; EWSR1::NFATC2 FUS::NFATC2 EWSR1::PATZ1 sarcoma; EWSR1::PATZ1 |
EWSR1/FUS::FATC2 sarcoma; AGGRECAN NKX3-1 EWSR1::PATZ1 sarcoma; Myogenic (myogenin, MyoD1) and neurogenic markers (S100 protein) |
AGGRECAN, cytoplasmic staining; NKX3-1, myogenin, myoD1, nuclear staining; S100 protein, nuclear and cytoplasmic staining |
[230,255,259] |
| CIC-rearranged sarcoma | t(4;19)(q35;q13) t(10;19)(q26;q13) t(X;19)(q13;q13) t(15;19)(q14;q13) |
CIC::DUX4 (95%) CIC::FOXO4 CIC::LEUTX CIC::NUTM1 CIC::NUTM2A |
DUX4 ETV4 WT1 (N-terminus) NUT |
Nuclear staining | [265,266,267,268,269,272,273,274] |
| Sarcoma with BCOR genetic alterations | inv(X)(p11.4p11.22) t(X;22)(p11q13) t(10;17)(q22;p13) |
BCOR::CCNB3, BCOR::MAML3, BCOR::ZC3H7B; BCOR internal tandem duplications |
BCOR CCNB3 SATB2 |
Nuclear staining | [282,283,284,285] |
15. Emerging Entities
15.1. EWSR1::SMAD3–Positive Fibroblastic Tumor
EWSR1::SMAD3–positive fibroblastic tumor (ESFT) is a benign neoplasm defined by a fusion of exon 7 of EWSR1 with exon 5 of SMAD3 [287,288,289]. It is characterized by small dermal and subcutaneous acral nodules and histological zonation with an acellular hyalinized center and peripheral fascicular spindle cell growth [290]. SMAD3 is a critical signal transducer in the TGF-β/SMAD signaling pathway involved in extracellular matrix synthesis by fibroblasts. Immunohistochemically, the fibroblastic tumor cells consistently show diffuse ERG nuclear expression, which correlates with a significant ERG mRNA upregulation [289].
At the transcriptional level, ESFTs also show overexpression of FN1 (fibronectin) [289]. Based on molecular data and morphologic and clinical similarities, ESFTs have been suggested to be associated with calcifying aponeurotic fibroma, lipofibromatosis, and lipofibromatosis-like neural tumors [289,290]. The diagnosis is primarily based on the detection of EWSR1::SMAD3 fusion [291]. The terminology of this tumor is provisional while the specific features await further determination.
15.2. NTRK-Rearranged Spindle Cell Neoplasm
NTRK-rearranged spindle cell neoplasms (other than infantile fibrosarcomas) are an emerging family of rare spindle cell neoplasms defined by NTRK fusions [292]. The tumors are characterized by haphazardly arranged monomorphic spindle cells, infiltrative growth in adipose tissue resembling lipofibromatosis, and characteristic stromal and perivascular keloid collagen. Most tumors harbor NTRK1 fusions with various partners, including LMNA, TPR, or TPM3 [293,294,295]. Rare cases with NTRK2 and NTRK3 fusions have also been reported [296]. The NTRK fusions lead to the activation of the oncogenic signaling pathway via chimeric proteins containing the tropomyosin receptor kinase domains of TRK-A, TRK-B, and TRK-C [297]. Immunohistochemically, most tumors with NTRK fusions are reactive with a monoclonal anti–pan-TRK antibody [117]. The staining can be either cytoplasmic or nuclear [115,298]. Most tumors show frequent coexpression of S100 protein and CD34. TRK-A immunohistochemistry is useful for the detection of NTRK1-rearranged tumors [299].
NTRK fusions have been reported at high frequency in various cancers, such as IFS and secretory breast carcinoma, and at a low frequency in other well-established tumors, such as GISTs [300,301]. Importantly, pan-TRK and TRK-A immunoreactivity are not completely specific, and additional molecular genetic testing is often required for a conclusive diagnosis. Hence, molecular detection of NTRK fusions can be useful in such cases [294,302]. Recently, NTRK-rearranged spindle cell neoplasms are ubiquitous tumors of myofibroblastic lineage with a distinct methylation class [303]. Our understanding of this tumor type is rapidly evolving. Additional genetic alterations will be discovered in further studies.
15.3. SWI/SNF Complex-Deficient Neoplasms
The SWItch/sucrose non-fermentable (SWI/SNF) complexes are a family of multi-subunit complexes that use the energy of adenosine triphosphate hydrolysis to remodel nucleosomes [304]. Chromatin remodeling processes mediated by the SWI/SNF complexes are critical to the modulation of gene expression across various cellular processes, including stemness, differentiation, and proliferation. Recently, mutations in the genes encoding different subunits of the SWI/SNF complex (SMARCB1, SMARCA4, ARID1A, ARID1B, ARID1, and PBRM) have been identified in many adult malignancies, which include encompassing epithelial and mesenchymal tumors [305,306]. Several SMARCA4-deficient poorly differentiated and undifferentiated carcinomas/sarcomas delineated primarily based on BRG1 protein loss have been characterized in various anatomical locations [307].
Thoracic SMARCA4-deficient undifferentiated tumor is a high-grade malignant neoplasm significantly affecting the thorax in adults. It shows an undifferentiated or rhabdoid phenotype and a lack of SMARCA4 (BRG1), a vital member of the SWI/SNF chromatin-remodeling complex [308]. The tumor is driven by biallelic inactivation of SMARCA4, including mainly nonsense and frameshift mutations. Histologically, the tumor consists of diffuse sheets of monomorphic, undifferentiated epithelioid cells with vesicular nuclei and prominent nucleoli, and frequent rhabdoid features. Immunohistochemically, complete loss of SMARCA4 (BRG1) expression is typical (Figure 10). Additionally, there is a family of tumors defined by SMARCA4 loss, including small cell carcinoma of the ovary, hypercalcemic type [309], and large cell malignancies originating in the uterus [310]. Recently, SMARCA4-deficient sinonasal carcinomas have also been reported [311].
Figure 10.
Thoracic SMARCA4-deficient undifferentiated tumor. (A) The tumor shows diffuse sheets of epithelioid tumor cells with uniform round nuclei and prominent nucleoli (H&E stain, ×200). (B) Most tumor cells show complete loss of SMARCA4, indicating the presence of underlying SMARCA4 inactivation (immunohistochemical stain for SMARCA4, ×200).
15.4. DICER1-Associated Sarcomas
The DICER1 gene is located on chromosome 14q32.13 and encodes an endoribonuclease in the ribonuclease (RNase) III family required for processing microRNA (miRNA) [312]. Dysregulation of microRNA by DICER1 mutations causes activation of oncogenes and underlies developmental and neoplastic disorders. A wide variety of DICER1-associated sarcomas have been reported in the literature [313]. DECER1-associated sarcomas, regardless of their site of origin, exhibit characteristic morphology that resembles pleuropulmonary blastoma [314]. The morphologic features include a subepithelial layer of malignant mesenchymal cells (cambium layer), areas of rhabdomyoblastic differentiation with positive staining for myogenin and myoD1, cellular/immature and occasionally malignant cartilage, bone/osteoid foci, and areas of anaplasia [315]. If a mesenchymal neoplasm is reported that has a combination of rhabdomyoblastic, cartilaginous, and neuroectodermal elements, a DICER1-associated neoplasm should be considered.
Kommoss et al. [316] reported the clinicopathological and molecular features of DICER1-mutant and DICER1-wild-type embryonal RMS in a series of genitourinary tumors. They suggested that DICER1-mutant ERMS might represent a distinct subtype in the future classification of RMS. Unsupervised hierarchical clustering of array-based whole-genome methylation data of a subset of DICER1-mutant sarcomas has revealed that they cluster together [317]. Further methylation studies are warranted to determine whether DICER1-mutant sarcomas constitute a distinct, identifiable subclass of sarcomas.
Table 3 shows a summary of molecular genetic alterations and immunohistochemical markers in emerging entities.
Table 3.
Molecular genetic alterations and immunohistochemical markers in emerging entities.
| Tumor Type | Cytogenetic Alterations | Molecular Alterations |
Immunohistochemical Markers | Staining Pattern | References |
|---|---|---|---|---|---|
| EWSR1::SMAD3–positive fibroblastic tumor | t(15;22)(q22.33;q12.2) | EWSR1::SMAD3 fusion | ERG | Nuclear staining | [289] |
| NTRK-rearranged spindle cell neoplasm |
NTRK1 fusions with LMNA, TPR, or TPM3; NTRK2, NTRK3 fusions |
Pan-TRK TRK-A |
Cytoplasmic or nuclear staining | [115,117,293,294,295,296,298], | |
| Thoracic SMARCA4-deficient undifferentiated tumor | Biallelic inactivation of SMARCA4 | SMARCA4 (BRG1) | Loss | [307] | |
| DICER1-associated sarcoma | DICER1 mutations | Myogenin MyoD1 |
Nuclear staining | [315] |
16. Conclusions
Advances in molecular techniques have refined the classification of soft tissue tumors. This review summarizes newly recognized and emerging entities, focusing on molecular alterations and antibodies as surrogate markers for molecular genetic techniques. In addition, already well-defined entities with recently described molecular changes are discussed. The critical genetic events driving the biology of soft tissue tumors are still largely unknown. Further studies with careful genomic-morphologic correlation are required to understand soft tissue pathology comprehensively.
Author Contributions
Conceptualization, J.H.C. and J.Y.R.; methodology, J.H.C. and J.Y.R.; resources, J.H.C. and J.Y.R.; writing—original draft preparation, J.H.C.; writing—review and editing, J.Y.R.; supervision, J.Y.R. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the 2022 Yeungnam University Research Grant.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Fletcher C.D.M., Baldini E.H., Blay J.Y., Gronchi A., Lazar A.J., Messiou C., Pollock R.E., Singer S. Soft tissue tumours: Introduction. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 6–12. [Google Scholar]
- 2.Sbaraglia M., Bellan E., Dei Tos A.P. The 2020 WHO classification of soft tissue tumours: News and perspectives. Pathologica. 2021;113:70–84. doi: 10.32074/1591-951X-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonescu C.R. The role of genetic testing in soft tissue sarcoma. Histopathology. 2006;48:13–21. doi: 10.1111/j.1365-2559.2005.02285.x. [DOI] [PubMed] [Google Scholar]
- 4.Chan J.K., Ip Y.T., Cheuk W. The utility of immunohistochemistry for providing genetic information on tumors. Int. J. Surg. Pathol. 2013;21:455–475. doi: 10.1177/1066896913502529. [DOI] [PubMed] [Google Scholar]
- 5.Horvai A. Bone, joints, and soft tissue tumors. In: Kumar V., Abbas A.K., Aster J.C., editors. Robbins and Cotran Pathologic Basis of Disease. 10th ed. Elsevier; Philadelphia, PA, USA: 2021. pp. 1208–1209. [Google Scholar]
- 6.Helman L.J., Meltzer P. Mechanisms of sarcoma development. Nat. Rev. Cancer. 2003;3:685–694. doi: 10.1038/nrc1168. [DOI] [PubMed] [Google Scholar]
- 7.Wang W.L., Lazar A.J. Applications of molecular testing to differential diagnosis. In: Hornick J.L., editor. Practical Soft Tissue Pathology. 2nd ed. Elsevier; Philadelphia, PA, USA: 2019. pp. 513–550. [Google Scholar]
- 8.Gibault L., Pérot G., Chibon F., Bonnin S., Lagarde P., Terrier P., Coindre J.M., Aurias A. New insights in sarcoma oncogenesis: A comprehensive analysis of a large series of 160 soft tissue sarcomas with complex genomics. J. Pathol. 2011;223:64–71. doi: 10.1002/path.2787. [DOI] [PubMed] [Google Scholar]
- 9.Golblum J.R., Folpe A.L., Weiss S.E., editors. Enzinger & Weiss’s Soft Tissue Tumors. 7th ed. Elsevier; Philadelphia, PA, USA: 2020. Approach to the diagnosis of soft tissue tumors; pp. 84–110. [Google Scholar]
- 10.Tanas M.R., Rubin B.P., Tubbs R.R., Billings S.D., Downs-Kelly E., Goldblum J.R. Utilization of fluorescence in situ hybridization in the diagnosis of 230 mesenchymal neoplasms: An institutional experience. Arch. Pathol. Lab. Med. 2010;134:1797–1803. doi: 10.5858/2009-0571-OAR.1. [DOI] [PubMed] [Google Scholar]
- 11.Tanas M.R., Goldblum J.R. Fluorescence in situ hybridization in the diagnosis of soft tissue neoplasms: A review. Adv. Anat. Pathol. 2009;16:383–391. doi: 10.1097/PAP.0b013e3181bb6b86. [DOI] [PubMed] [Google Scholar]
- 12.Oda Y., Yamamoto H., Kohashi K., Yamada Y., Iura K., Ishii T., Maekawa A., Bekki H. Soft tissue sarcomas: From a morphological to a molecular biological approach. Pathol. Int. 2017;67:435–446. doi: 10.1111/pin.12565. [DOI] [PubMed] [Google Scholar]
- 13.Groisberg R., Roszik J., Conley A., Patel S.R., Subbiah V. The role of next-generation sequencing in sarcomas: Evolution from light microscope to molecular microscope. Curr. Oncol. Rep. 2017;19:78. doi: 10.1007/s11912-017-0641-2. [DOI] [PubMed] [Google Scholar]
- 14.Cote G.M., He J., Choy E. Next-generation sequencing for patients with sarcoma: A single center experience. Oncologist. 2018;23:234–242. doi: 10.1634/theoncologist.2017-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szurian K., Kashofer K., Liegl-Atzwanger B. Role of next-generation sequencing as a diagnostic tool for the evaluation of bone and soft-tissue tumors. Pathobiology. 2017;84:323–338. doi: 10.1159/000478662. [DOI] [PubMed] [Google Scholar]
- 16.Al-Zaid T., Wang W.L., Somaiah N., Lazar A.J. Molecular profiling of sarcomas: New vistas for precision medicine. Virchows Arch. 2017;471:243–255. doi: 10.1007/s00428-017-2174-3. [DOI] [PubMed] [Google Scholar]
- 17.ESMO/European Sarcoma Network Working Group Soft tissue and visceral sarcomas: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014;25((Suppl. S3)):iii102–iii112. doi: 10.1093/annonc/mdu254. [DOI] [PubMed] [Google Scholar]
- 18.Hornick J.L. Novel uses of immunohistochemistry in the diagnosis and classification of soft tissue tumors. Mod. Pathol. 2014;27((Suppl. S1)):S47–S63. doi: 10.1038/modpathol.2013.177. [DOI] [PubMed] [Google Scholar]
- 19.Fletcher C.D.M. Tumors of soft tissue. In: Fletcher C.D.M., editor. Diagnostic Histopathology of Tumors. 5th ed. Elsevier; Philadelphia, PA, USA: 2021. pp. 1919–1984. [Google Scholar]
- 20.Billing S.D., Ud Din N. Spindle cell lipoma and pleomorphic lipoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 29–30. [Google Scholar]
- 21.Fletcher C.D., Akerman M., Dal Cin P., de Wever I., Mandahl N., Mertens F., Mitelman F., Rosai J., Rydholm A., Sciot R., et al. Correlation between clinicopathological features and karyotype in lipomatous tumors. A report of 178 cases from the Chromosomes and Morphology (CHAMP) Collaborative Study Group. Am. J. Pathol. 1996;148:623–630. [PMC free article] [PubMed] [Google Scholar]
- 22.Chen B.J., Mariño-Enríquez A., Fletcher C.D., Hornick J.L. Loss of retinoblastoma protein expression in spindle cell/pleomorphic lipomas and cytogenetically related tumors: An immunohistochemical study with diagnostic implications. Am. J. Surg. Pathol. 2012;36:1119–1128. doi: 10.1097/PAS.0b013e31825d532d. [DOI] [PubMed] [Google Scholar]
- 23.Dahlén A., Debiec-Rychter M., Pedeutour F., Domanski H.A., Höglund M., Bauer H.C., Rydholm A., Sciot R., Mandahl N., Mertens F. Clustering of deletions on chromosome 13 in benign and low-malignant lipomatous tumors. Int. J. Cancer. 2003;103:616–623. doi: 10.1002/ijc.10864. [DOI] [PubMed] [Google Scholar]
- 24.Magro G., Righi A., Casorzo L., Antonietta T., Salvatorelli L., Kacerovská D., Kazakov D., Michal M. Mammary and vaginal myofibroblastomas are genetically related lesions: Fluorescence in situ hybridization analysis shows deletion of 13q14 region. Hum. Pathol. 2012;43:1887–1893. doi: 10.1016/j.humpath.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Howitt B.E., Fletcher C.D. Mammary-type myofibroblastoma: Clinicopathologic characterization in a series of 143 cases. Am. J. Surg. Pathol. 2016;40:361–367. doi: 10.1097/PAS.0000000000000540. [DOI] [PubMed] [Google Scholar]
- 26.Flucke U., van Krieken J.H., Mentzel T. Cellular angiofibroma: Analysis of 25 cases emphasizing its relationship to spindle cell lipoma and mammary-type myofibroblastoma. Mod. Pathol. 2011;24:82–89. doi: 10.1038/modpathol.2010.170. [DOI] [PubMed] [Google Scholar]
- 27.Creytens D., Marino-Enriquez A. Atypical spindle cell/pleomorphic lipomatous tumour. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 34–35. [Google Scholar]
- 28.Mariño-Enriquez A., Nascimento A.F., Ligon A.H., Liang C., Fletcher C.D. Atypical spindle cell lipomatous tumor: Clinicopathologic characterization of 232 cases demonstrating a morphologic spectrum. Am. J. Surg. Pathol. 2017;41:234–244. doi: 10.1097/PAS.0000000000000770. [DOI] [PubMed] [Google Scholar]
- 29.Creytens D., Mentzel T., Ferdinande L., Lecoutere E., van Gorp J., Atanesyan L., de Groot K., Savola S., Van Roy N., Van Dorpe J., et al. “Atypical” pleomorphic lipomatous tumor: A clinicopathologic, immunohistochemical and molecular study of 21 cases, emphasizing its relationship to atypical spindle cell lipomatous tumor and suggesting a morphologic spectrum (atypical spindle cell/pleomorphic lipomatous tumor) Am. J. Surg. Pathol. 2017;41:1443–1455. doi: 10.1097/PAS.0000000000000936. [DOI] [PubMed] [Google Scholar]
- 30.Bahadır B., Behzatoğlu K., Hacıhasanoğlu E., Koca S.B., Sığırcı B.B., Tokat F. Atypical spindle cell/pleomorphic lipomatous tumor: A clinicopathologic, immunohistochemical, and molecular study of 20 cases. Pathol. Int. 2018;68:550–556. doi: 10.1111/pin.12719. [DOI] [PubMed] [Google Scholar]
- 31.Mentzel T., Palmedo G., Kuhnen C. Well-differentiated spindle cell liposarcoma (‘atypical spindle cell lipomatous tumor’) does not belong to the spectrum of atypical lipomatous tumor but has a close relationship to spindle cell lipoma: Clinicopathologic, immunohistochemical, and molecular analysis of six cases. Mod. Pathol. 2010;23:729–736. doi: 10.1038/modpathol.2010.66. [DOI] [PubMed] [Google Scholar]
- 32.Thway K. What’s new in adipocytic neoplasia? Histopathology. 2022;80:76–97. doi: 10.1111/his.14548. [DOI] [PubMed] [Google Scholar]
- 33.Demicco E.G. Molecular updates in adipocytic neoplasms. Semin. Diagn. Pathol. 2019;36:85–94. doi: 10.1053/j.semdp.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Sbaraglia M., Dei Tos A.P., Pedeutour F. Atypical lipomatous tumour/well-differentiated liposarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 36–38. [Google Scholar]
- 35.Italiano A., Bianchini L., Gjernes E., Keslair F., Ranchere-Vince D., Dumollard J.M., Haudebourg J., Leroux A., Mainguené C., Terrier P., et al. Clinical and biological significance of CDK4 amplification in well-differentiated and dedifferentiated liposarcomas. Clin. Cancer Res. 2009;15:5696–5703. doi: 10.1158/1078-0432.CCR-08-3185. [DOI] [PubMed] [Google Scholar]
- 36.Binh M.B., Sastre-Garau X., Guillou L., de Pinieux G., Terrier P., Lagacé R., Aurias A., Hostein I., Coindre J.M. MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well-differentiated and dedifferentiated liposarcoma subtypes: A comparative analysis of 559 soft tissue neoplasms with genetic data. Am. J. Surg. Pathol. 2005;29:1340–1347. doi: 10.1097/01.pas.0000170343.09562.39. [DOI] [PubMed] [Google Scholar]
- 37.Makise N., Sekimizu M., Kubo T., Wakai S., Hiraoka N., Komiyama M., Fukayama M., Kawai A., Ichikawa H., Yoshida A. Clarifying the distinction between malignant peripheral nerve sheath tumor and dedifferentiated liposarcoma: A critical reappraisal of the diagnostic utility of MDM2 and H3K27me3 Status. Am. J. Surg. Pathol. 2018;42:656–664. doi: 10.1097/PAS.0000000000001014. [DOI] [PubMed] [Google Scholar]
- 38.Schoolmeester J.K., Sciallis A.P., Greipp P.T., Hodge J.C., Dal Cin P., Keeney G.L., Nucci M.R. Analysis of MDM2 amplification in 43 endometrial stromal tumors: A potential diagnostic pitfall. Int. J. Gynecol. Pathol. 2015;34:576–583. doi: 10.1097/PGP.0000000000000187. [DOI] [PubMed] [Google Scholar]
- 39.He X., Pang Z., Zhang X., Lan T., Chen H., Chen M., Yang H., Huang J., Chen Y., Zhang Z., et al. Consistent amplification of FRS2 and MDM2 in low-grade osteosarcoma: A genetic study of 22 cases with clinicopathologic analysis. Am. J. Surg. Pathol. 2018;42:1143–1155. doi: 10.1097/PAS.0000000000001125. [DOI] [PubMed] [Google Scholar]
- 40.Wunder J.S., Eppert K., Burrow S.R., Gokgoz N., Bell R.S., Andrulis I.L. Co-amplification and overexpression of CDK4, SAS and MDM2 occurs frequently in human parosteal osteosarcomas. Oncogene. 1999;18:783–788. doi: 10.1038/sj.onc.1202346. [DOI] [PubMed] [Google Scholar]
- 41.Neuville A., Collin F., Bruneval P., Parrens M., Thivolet F., Gomez-Brouchet A., Terrier P., de Montpreville V.T., Le Gall F., Hostein I., et al. Intimal sarcoma is the most frequent primary cardiac sarcoma: Clinicopathologic and molecular retrospective analysis of 100 primary cardiac sarcomas. Am. J. Surg. Pathol. 2014;38:461–469. doi: 10.1097/PAS.0000000000000184. [DOI] [PubMed] [Google Scholar]
- 42.Zhang H., Erickson-Johnson M., Wang X., Oliveira J.L., Nascimento A.G., Sim F.H., Wenger D.E., Zamolyi R.Q., Pannain V.L., Oliveira A.M. Molecular testing for lipomatous tumors: Critical analysis and test recommendations based on the analysis of 405 extremity-based tumors. Am. J. Surg. Pathol. 2010;34:1304–1311. doi: 10.1097/PAS.0b013e3181e92d0b. [DOI] [PubMed] [Google Scholar]
- 43.Dei Tos A.P., Marino-Enriquez A., Pedeutour F. Dedifferentiated liposarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 39–41. [Google Scholar]
- 44.Mariño-Enríquez A., Fletcher C.D., Dal Cin P., Hornick J.L. Dedifferentiated liposarcoma with “homologous” lipoblastic (pleomorphic liposarcoma-like) differentiation: Clinicopathologic and molecular analysis of a series suggesting revised diagnostic criteria. Am. J. Surg. Pathol. 2010;34:1122–1131. doi: 10.1097/PAS.0b013e3181e5dc49. [DOI] [PubMed] [Google Scholar]
- 45.Saâda-Bouzid E., Burel-Vandenbos F., Ranchère-Vince D., Birtwisle-Peyrottes I., Chetaille B., Bouvier C., Château M.C., Peoc’h M., Battistella M., Bazin A., et al. Prognostic value of MGA2, CDK4, and JUN amplification in well-differentiated and dedifferentiated liposarcomas. Mod. Pathol. 2015;28:1404–1414. doi: 10.1038/modpathol.2015.96. [DOI] [PubMed] [Google Scholar]
- 46.Thway K., Flora R., Shah C., Olmos D., Fisher C. Diagnostic utility of p16, CDK4, and MDM2 as an immunohistochemical panel in distinguishing well-differentiated and dedifferentiated liposarcomas from other adipocytic tumors. Am. J. Surg. Pathol. 2012;36:462–469. doi: 10.1097/PAS.0b013e3182417330. [DOI] [PubMed] [Google Scholar]
- 47.Sirvent N., Coindre J.M., Maire G., Hostein I., Keslair F., Guillou L., Ranchere-Vince D., Terrier P., Pedeutour F. Detection of MDM2-CDK4 amplification by fluorescence in situ hybridization in 200 paraffin-embedded tumor samples: Utility in diagnosing adipocytic lesions and comparison with immunohistochemistry and real-time PCR. Am. J. Surg. Pathol. 2007;31:1476–1489. doi: 10.1097/PAS.0b013e3180581fff. [DOI] [PubMed] [Google Scholar]
- 48.Le Guellec S., Chibon F., Ouali M., Perot G., Decouvelaere A.V., Robin Y.M., Larousserie F., Terrier P., Coindre J.M., Neuville A. Are peripheral purely undifferentiated pleomorphic sarcomas with MDM2 amplification dedifferentiated liposarcomas? Am. J. Surg. Pathol. 2014;38:293–304. doi: 10.1097/PAS.0000000000000131. [DOI] [PubMed] [Google Scholar]
- 49.Thway K., Nielsen T.O. Myxoid liposarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 42–44. [Google Scholar]
- 50.Powers M.P., Wang W.L., Hernandez V.S., Patel K.S., Lev D.C., Lazar A.J., López-Terrada D.H. Detection of myxoid liposarcoma-associated FUS-DDIT3 rearrangement variants including a newly identified breakpoint using an optimized RT-PCR assay. Mod. Pathol. 2010;23:1307–1315. doi: 10.1038/modpathol.2010.118. [DOI] [PubMed] [Google Scholar]
- 51.Han J., Murthy R., Wood B., Song B., Wang S., Sun B., Malhi H., Kaufman R.J. ER stress signalling through eIF2α and CHOP, but not IRE1α, attenuates adipogenesis in mice. Diabetologia. 2013;56:911–924. doi: 10.1007/s00125-012-2809-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brenner S., Bercovich Z., Feiler Y., Keshet R., Kahana C. Dual regulatory role of polyamines in adipogenesis. J. Biol. Chem. 2015;290:27384–27392. doi: 10.1074/jbc.M115.686980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pérez-Mancera P.A., Bermejo-Rodríguez C., Sánchez-Martín M., Abollo-Jiménez F., Pintado B., Sánchez-García I. FUS-DDIT3 prevents the development of adipocytic precursors in liposarcoma by repressing PPARgamma and C/EBPalpha and activating eIF4E. PLoS ONE. 2008;3:e2569. doi: 10.1371/journal.pone.0002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koelsche C., Renner M., Hartmann W., Brandt R., Lehner B., Waldburger N., Alldinger I., Schmitt T., Egerer G., Penzel R., et al. TERT promoter hotspot mutations are recurrent in myxoid liposarcomas but rare in other soft tissue sarcoma entities. J. Exp. Clin. Cancer Res. 2014;33:33. doi: 10.1186/1756-9966-33-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trautmann M., Cyra M., Isfort I., Jeiler B., Krüger A., Grünewald I., Steinestel K., Altvater B., Rossig C., Hafner S., et al. Phosphatidylinositol-3-kinase (PI3K)/Akt signaling is functionally essential in myxoid liposarcoma. Mol. Cancer Ther. 2019;18:834–844. doi: 10.1158/1535-7163.MCT-18-0763. [DOI] [PubMed] [Google Scholar]
- 56.Baranov E., Black M.A., Fletcher C.D.M., Charville G.W., Hornick J.L. Nuclear expression of DDIT3 distinguishes high-grade myxoid liposarcoma from other round cell sarcomas. Mod. Pathol. 2021;34:1367–1372. doi: 10.1038/s41379-021-00782-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Italiano A., Di Mauro I., Rapp J., Pierron G., Auger N., Alberti L., Chibon F., Escande F., Voegeli A.C., Ghnassia J.P., et al. Clinical effect of molecular methods in sarcoma diagnosis (GENSARC): A prospective, multicentre, observational study. Lancet Oncol. 2016;17:532–538. doi: 10.1016/S1470-2045(15)00583-5. [DOI] [PubMed] [Google Scholar]
- 58.Narendra S., Valente A., Tull J., Zhang S. DDIT3 gene break-apart as a molecular marker for diagnosis of myxoid liposarcoma--assay validation and clinical experience. Diagn. Mol. Pathol. 2011;20:218–224. doi: 10.1097/PDM.0b013e3182107eb9. [DOI] [PubMed] [Google Scholar]
- 59.Fritchie K.J., Crago A.M., van de Rijn M. Desmoid fibromatosis. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 93–95. [Google Scholar]
- 60.Amary M.F., Pauwels P., Meulemans E., Roemen G.M., Islam L., Idowu B., Bousdras K., Diss T.C., O’Donnell P., Flanagan A.M. Detection of beta-catenin mutations in paraffin-embedded sporadic desmoid-type fibromatosis by mutation-specific restriction enzyme digestion (MSRED): An ancillary diagnostic tool. Am. J. Surg. Pathol. 2007;31:1299–1309. doi: 10.1097/PAS.0b013e31802f581a. [DOI] [PubMed] [Google Scholar]
- 61.Crago A.M., Chmielecki J., Rosenberg M., O’Connor R., Byrne C., Wilder F.G., Thorn K., Agius P., Kuk D., Socci N.D., et al. Near universal detection of alterations in CTNNB1 and Wnt pathway regulators in desmoid-type fibromatosis by whole-exome sequencing and genomic analysis. Genes Chromosomes Cancer. 2015;54:606–615. doi: 10.1002/gcc.22272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giarola M., Stagi L., Presciuttini S., Mondini P., Radice M.T., Sala P., Pierotti M.A., Bertario L., Radice P. Screening for mutations of the APC gene in 66 Italian familial adenomatous polyposis patients: Evidence for phenotypic differences in cases with and without identified mutation. Hum. Mutat. 1999;13:116–123. doi: 10.1002/(SICI)1098-1004(1999)13:2<116::AID-HUMU3>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 63.Sturt N.J., Gallagher M.C., Bassett P., Philp C.R., Neale K.F., Tomlinson I.P., Silver A.R., Phillips R.K. Evidence for genetic predisposition to desmoid tumours in familial adenomatous polyposis independent of the germline APC mutation. Gut. 2004;53:1832–1836. doi: 10.1136/gut.2004.042705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang W.L., Nero C., Pappo A., Lev D., Lazar A.J., López-Terrada D. CTNNB1 genotyping and APC screening in pediatric desmoid tumors: A proposed algorithm. Pediatr. Dev. Pathol. 2012;15:361–367. doi: 10.2350/11-07-1064-OA.1. [DOI] [PubMed] [Google Scholar]
- 65.Barker N. The canonical Wnt/beta-catenin signalling pathway. Methods Mol. Biol. 2008;468:5–15. doi: 10.1007/978-1-59745-249-6_1. [DOI] [PubMed] [Google Scholar]
- 66.Bhattacharya B., Dilworth H.P., Iacobuzio-Donahue C., Ricci F., Weber K., Furlong M.A., Fisher C., Montgomery E. Nuclear beta-catenin expression distinguishes deep fibromatosis from other benign and malignant fibroblastic and myofibroblastic lesions. Am. J. Surg. Pathol. 2005;29:653–659. doi: 10.1097/01.pas.0000157938.95785.da. [DOI] [PubMed] [Google Scholar]
- 67.Carlson J.W., Fletcher C.D. Immunohistochemistry for beta-catenin in the differential diagnosis of spindle cell lesions: Analysis of a series and review of the literature. Histopathology. 2007;51:509–514. doi: 10.1111/j.1365-2559.2007.02794.x. [DOI] [PubMed] [Google Scholar]
- 68.Ng T.L., Gown A.M., Barry T.S., Cheang M.C., Chan A.K., Turbin D.A., Hsu F.D., West R.B., Nielsen T.O. Nuclear beta-catenin in mesenchymal tumors. Mod. Pathol. 2005;18:68–74. doi: 10.1038/modpathol.3800272. [DOI] [PubMed] [Google Scholar]
- 69.Colombo C., Bolshakov S., Hajibashi S., Lopez-Terrada L., Wang W.L., Rao P., Benjamin R.S., Lazar A.J., Lev D. ‘Difficult to diagnose’ desmoid tumours: A potential role for CTNNB1 mutational analysis. Histopathology. 2011;59:336–340. doi: 10.1111/j.1365-2559.2011.03932.x. [DOI] [PubMed] [Google Scholar]
- 70.Le Guellec S., Soubeyran I., Rochaix P., Filleron T., Neuville A., Hostein I., Coindre J.M. CTNNB1 mutation analysis is a useful tool for the diagnosis of desmoid tumors: A study of 260 desmoid tumors and 191 potential morphologic mimics. Mod. Pathol. 2012;25:1551–1558. doi: 10.1038/modpathol.2012.115. [DOI] [PubMed] [Google Scholar]
- 71.Colombo C., Miceli R., Lazar A.J., Perrone F., Pollock R.E., Le Cesne A., Hartgrink H.H., Cleton-Jansen A.M., Domont J., Bovée J.V., et al. CTNNB1 45F mutation is a molecular prognosticator of increased postoperative primary desmoid tumor recurrence: An independent, multicenter validation study. Cancer. 2013;119:3696–3702. doi: 10.1002/cncr.28271. [DOI] [PubMed] [Google Scholar]
- 72.Demicco E.G., Fritchie K.J., Han A. Solitary fibrous tumour. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 104–106. [Google Scholar]
- 73.Chmielecki J., Crago A.M., Rosenberg M., O’Connor R., Walker S.R., Ambrogio L., Auclair D., McKenna A., Heinrich M.C., Frank D.A., et al. Whole-exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat. Genet. 2013;45:131–132. doi: 10.1038/ng.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mohajeri A., Tayebwa J., Collin A., Nilsson J., Magnusson L., von Steyern F.V., Brosjö O., Domanski H.A., Larsson O., Sciot R., et al. Comprehensive genetic analysis identifies a pathognomonic NAB2/STAT6 fusion gene, nonrandom secondary genomic imbalances, and a characteristic gene expression profile in solitary fibrous tumor. Genes Chromosomes Cancer. 2013;52:873–886. doi: 10.1002/gcc.22083. [DOI] [PubMed] [Google Scholar]
- 75.Robinson D.R., Wu Y.M., Kalyana-Sundaram S., Cao X., Lonigro R., Sung Y.S., Chen C.L., Zhang L., Wang R., Su F., et al. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat. Genet. 2013;45:180–185. doi: 10.1038/ng.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hajdu M., Singer S., Maki R.G., Schwartz G.K., Keohan M.L., Antonescu C.R. IGF2 over-expression in solitary fibrous tumours is independent of anatomical location and is related to loss of imprinting. J. Pathol. 2010;221:300–307. doi: 10.1002/path.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bertucci F., Bouvier-Labit C., Finetti P., Metellus P., Adelaide J., Mokhtari K., Figarella-Branger D., Decouvelaere A.V., Miquel C., Coindre J.M., et al. Gene expression profiling of solitary fibrous tumors. PLoS ONE. 2013;8:e64497. doi: 10.1371/journal.pone.0064497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Demicco E.G., Wani K., Ingram D., Wagner M., Maki R.G., Rizzo A., Meeker A., Lazar A.J., Wang W.L. TERT promoter mutations in solitary fibrous tumour. Histopathology. 2018;73:843–851. doi: 10.1111/his.13703. [DOI] [PubMed] [Google Scholar]
- 79.Kurisaki-Arakawa A., Akaike K., Hara K., Arakawa A., Takahashi M., Mitani K., Yao T., Saito T. A case of dedifferentiated solitary fibrous tumor in the pelvis with TP53 mutation. Virchows Arch. 2014;465:615–621. doi: 10.1007/s00428-014-1625-3. [DOI] [PubMed] [Google Scholar]
- 80.Doyle L.A., Vivero M., Fletcher C.D., Mertens F., Hornick J.L. Nuclear expression of STAT6 distinguishes solitary fibrous tumor from histologic mimics. Mod. Pathol. 2014;27:390–395. doi: 10.1038/modpathol.2013.164. [DOI] [PubMed] [Google Scholar]
- 81.Doyle L.A., Tao D., Mariño-Enríquez A. STAT6 is amplified in a subset of dedifferentiated liposarcoma. Mod. Pathol. 2014;27:1231–1237. doi: 10.1038/modpathol.2013.247. [DOI] [PubMed] [Google Scholar]
- 82.Schneider N., Hallin M., Thway K. STAT6 loss in dedifferentiated solitary fibrous tumor. Int. J. Surg. Pathol. 2017;25:58–60. doi: 10.1177/1066896916650257. [DOI] [PubMed] [Google Scholar]
- 83.Demicco E.G., Wagner M.J., Maki R.G., Gupta V., Iofin I., Lazar A.J., Wang W.L. Risk assessment in solitary fibrous tumors: Validation and refinement of a risk stratification model. Mod. Pathol. 2017;30:1433–1442. doi: 10.1038/modpathol.2017.54. [DOI] [PubMed] [Google Scholar]
- 84.Yamamoto H. Inflammatory myofibroblastic tumour. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 109–111. [Google Scholar]
- 85.Bridge J.A., Kanamori M., Ma Z., Pickering D., Hill D.A., Lydiatt W., Lui M.Y., Colleoni G.W., Antonescu C.R., Ladanyi M., et al. Fusion of the ALK gene to the clathrin heavy chain gene, CLTC, in inflammatory myofibroblastic tumor. Am. J. Pathol. 2001;159:411–415. doi: 10.1016/S0002-9440(10)61711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lawrence B., Perez-Atayde A., Hibbard M.K., Rubin B.P., Dal Cin P., Pinkus J.L., Pinkus G.S., Xiao S., Yi E.S., Fletcher C.D., et al. TPM3-ALK and TPM4-ALK oncogenes in inflammatory myofibroblastic tumors. Am. J. Pathol. 2000;157:377–384. doi: 10.1016/S0002-9440(10)64550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hornick J.L., Sholl L.M., Dal Cin P., Childress M.A., Lovely C.M. Expression of ROS1 predicts ROS1 gene rearrangement in inflammatory myofibroblastic tumors. Mod. Pathol. 2015;28:732–739. doi: 10.1038/modpathol.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alassiri A.H., Ali R.H., Shen Y., Lum A., Strahlendorf C., Deyell R., Rassekh R., Sorensen P.H., Laskin J., Marra M., et al. ETV6-NTRK3 is expressed in a subset of ALK-negative inflammatory myofibroblastic tumors. Am. J. Surg. Pathol. 2016;40:1051–1061. doi: 10.1097/PAS.0000000000000677. [DOI] [PubMed] [Google Scholar]
- 89.Antonescu C.R., Suurmeijer A.J., Zhang L., Sung Y.S., Jungbluth A.A., Travis W.D., Al-Ahmadie H., Fletcher C.D., Alaggio R. Molecular characterization of inflammatory myofibroblastic tumors with frequent ALK and ROS1 gene fusions and rare novel RET rearrangement. Am. J. Surg. Pathol. 2015;39:957–967. doi: 10.1097/PAS.0000000000000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mariño-Enríquez A., Wang W.L., Roy A., Lopez-Terrada D., Lazar A.J., Fletcher C.D., Coffin C.M., Hornick J.L. Epithelioid inflammatory myofibroblastic sarcoma: An aggressive intra-abdominal variant of inflammatory myofibroblastic tumor with nuclear membrane or perinuclear ALK. Am. J. Surg. Pathol. 2011;35:135–144. doi: 10.1097/PAS.0b013e318200cfd5. [DOI] [PubMed] [Google Scholar]
- 91.Lee J.C., Li C.F., Huang H.Y., Zhu M.J., Mariño-Enríquez A., Lee C.T., Ou W.B., Hornick J.L., Fletcher J.A. ALK oncoproteins in atypical inflammatory myofibroblastic tumours: Novel RRBP1-ALK fusions in epithelioid inflammatory myofibroblastic sarcoma. J. Pathol. 2017;241:316–323. doi: 10.1002/path.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yamamoto H., Yoshida A., Taguchi K., Kohashi K., Hatanaka Y., Yamashita A., Mori D., Oda Y. ALK, ROS1 and NTRK3 gene rearrangements in inflammatory myofibroblastic tumours. Histopathology. 2016;69:72–83. doi: 10.1111/his.12910. [DOI] [PubMed] [Google Scholar]
- 93.Doyle L.A., Mertens F. Low-grade fibromyxoid sarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 127–129. [Google Scholar]
- 94.Mertens F., Fletcher C.D., Antonescu C.R., Coindre J.M., Colecchia M., Domanski H.A., Downs-Kelly E., Fisher C., Goldblum J.R., Guillou L., et al. Clinicopathologic and molecular genetic characterization of low-grade fibromyxoid sarcoma, and cloning of a novel FUS/CREB3L1 fusion gene. Lab. Investig. 2005;85:408–415. doi: 10.1038/labinvest.3700230. [DOI] [PubMed] [Google Scholar]
- 95.Matsuyama A., Hisaoka M., Shimajiri S., Hayashi T., Imamura T., Ishida T., Fukunaga M., Fukuhara T., Minato H., Nakajima T., et al. Molecular detection of FUS-CREB3L2 fusion transcripts in low-grade fibromyxoid sarcoma using formalin-fixed, paraffin-embedded tissue specimens. Am. J. Surg. Pathol. 2006;30:1077–1084. doi: 10.1097/01.pas.0000209830.24230.1f. [DOI] [PubMed] [Google Scholar]
- 96.Guillou L., Benhattar J., Gengler C., Gallagher G., Ranchère-Vince D., Collin F., Terrier P., Terrier-Lacombe M.J., Leroux A., Marquès B., et al. Translocation-positive low-grade fibromyxoid sarcoma: Clinicopathologic and molecular analysis of a series expanding the morphologic spectrum and suggesting potential relationship to sclerosing epithelioid fibrosarcoma: A study from the French Sarcoma Group. Am. J. Surg. Pathol. 2007;31:1387–1402. doi: 10.1097/PAS.0b013e3180321959. [DOI] [PubMed] [Google Scholar]
- 97.Möller E., Hornick J.L., Magnusson L., Veerla S., Domanski H.A., Mertens F. FUS-CREB3L2/L1-positive sarcomas show a specific gene expression profile with upregulation of CD24 and FOXL1. Clin. Cancer Res. 2011;17:2646–2656. doi: 10.1158/1078-0432.CCR-11-0145. [DOI] [PubMed] [Google Scholar]
- 98.Lau P.P., Lui P.C., Lau G.T., Yau D.T., Cheung E.T., Chan J.K. EWSR1-CREB3L1 gene fusion: A novel alternative molecular aberration of low-grade fibromyxoid sarcoma. Am. J. Surg. Pathol. 2013;37:734–738. doi: 10.1097/PAS.0b013e31827560f8. [DOI] [PubMed] [Google Scholar]
- 99.Cowan M.L., Thompson L.D., Leon M.E., Bishop J.A. Low-grade fibromyxoid sarcoma of the head and neck: A clinicopathologic series and review of the literature. Head Neck Pathol. 2016;10:161–166. doi: 10.1007/s12105-015-0647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Doyle L.A., Möller E., Dal Cin P., Fletcher C.D., Mertens F., Hornick J.L. MUC4 is a highly sensitive and specific marker for low-grade fibromyxoid sarcoma. Am. J. Surg. Pathol. 2011;35:733–741. doi: 10.1097/PAS.0b013e318210c268. [DOI] [PubMed] [Google Scholar]
- 101.Doyle L.A., Mertens F. Sclerosing epithelioid fibrosarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 130–132. [Google Scholar]
- 102.Arbajian E., Puls F., Magnusson L., Thway K., Fisher C., Sumathi V.P., Tayebwa J., Nord K.H., Kindblom L.G., Mertens F. Recurrent EWSR1-CREB3L1 gene fusions in sclerosing epithelioid fibrosarcoma. Am. J. Surg. Pathol. 2014;38:801–808. doi: 10.1097/PAS.0000000000000158. [DOI] [PubMed] [Google Scholar]
- 103.Prieto-Granada C., Zhang L., Chen H.W., Sung Y.S., Agaram N.P., Jungbluth A.A., Antonescu C.R. A genetic dichotomy between pure sclerosing epithelioid fibrosarcoma (SEF) and hybrid SEF/low-grade fibromyxoid sarcoma: A pathologic and molecular study of 18 cases. Genes Chromosomes Cancer. 2015;54:28–38. doi: 10.1002/gcc.22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Doyle L.A., Hornick J.L. EWSR1 rearrangements in sclerosing epithelioid fibrosarcoma. Am. J. Surg. Pathol. 2013;37:1630–1631. doi: 10.1097/PAS.0b013e3182a05a6b. [DOI] [PubMed] [Google Scholar]
- 105.Dewaele B., Libbrecht L., Levy G., Brichard B., Vanspauwen V., Sciot R., Debiec-Rychter M. A novel EWS-CREB3L3 gene fusion in a mesenteric sclerosing epithelioid fibrosarcoma. Genes Chromosomes Cancer. 2017;56:695–699. doi: 10.1002/gcc.22474. [DOI] [PubMed] [Google Scholar]
- 106.Arbajian E., Puls F., Antonescu C.R., Amary F., Sciot R., Debiec-Rychter M., Sumathi V.P., Järås M., Magnusson L., Nilsson J., et al. In-depth genetic analysis of sclerosing epithelioid fibrosarcoma reveals recurrent genomic alterations and potential treatment targets. Clin. Cancer Res. 2017;23:7426–7434. doi: 10.1158/1078-0432.CCR-17-1856. [DOI] [PubMed] [Google Scholar]
- 107.Doyle L.A., Wang W.L., Dal Cin P., Lopez-Terrada D., Mertens F., Lazar A.J., Fletcher C.D., Hornick J.L. MUC4 is a sensitive and extremely useful marker for sclerosing epithelioid fibrosarcoma: Association with FUS gene rearrangement. Am. J. Surg. Pathol. 2012;36:1444–1451. doi: 10.1097/PAS.0b013e3182562bf8. [DOI] [PubMed] [Google Scholar]
- 108.Kao Y.C., Lee J.C., Zhang L., Sung Y.S., Swanson D., Hsieh T.H., Liu Y.R., Agaram N.P., Huang H.Y., Dickson B.C., et al. Recurrent YAP1 and KMT2A gene rearrangements in a subset of MUC4-negative sclerosing epithelioid fibrosarcoma. Am. J. Surg. Pathol. 2020;44:368–377. doi: 10.1097/PAS.0000000000001382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Warmke L.M., Meis J.M. Sclerosing epithelioid fibrosarcoma: A distinct sarcoma with aggressive features. Am. J. Surg. Pathol. 2021;45:317–328. doi: 10.1097/PAS.0000000000001559. [DOI] [PubMed] [Google Scholar]
- 110.Davis J.L., Antonescu C.R., Bahrami A. Infantile fibrosarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 119–121. [Google Scholar]
- 111.Tognon C., Garnett M., Kenward E., Kay R., Morrison K., Sorensen P.H. The chimeric protein tyrosine kinase ETV6-NTRK3 requires both Ras-Erk1/2 and PI3-kinase-Akt signaling for fibroblast transformation. Cancer Res. 2001;61:8909–8916. [PubMed] [Google Scholar]
- 112.Church A.J., Calicchio M.L., Nardi V., Skalova A., Pinto A., Dillon D.A., Gomez-Fernandez C.R., Manoj N., Haimes J.D., Stahl J.A., et al. Recurrent EML4-NTRK3 fusions in infantile fibrosarcoma and congenital mesoblastic nephroma suggest a revised testing strategy. Mod. Pathol. 2018;31:463–473. doi: 10.1038/modpathol.2017.127. [DOI] [PubMed] [Google Scholar]
- 113.Wegert J., Vokuhl C., Collord G., Del Castillo Velasco-Herrera M., Farndon S.J., Guzzo C., Jorgensen M., Anderson J., Slater O., Duncan C., et al. Recurrent intragenic rearrangements of EGFR and BRAF in soft tissue tumors of infants. Nat. Commun. 2018;9:2378. doi: 10.1038/s41467-018-04650-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Flucke U., van Noesel M.M., Wijnen M., Zhang L., Chen C.L., Sung Y.S., Antonescu C.R. TFG-MET fusion in an infantile spindle cell sarcoma with neural features. Genes Chromosomes Cancer. 2017;56:663–667. doi: 10.1002/gcc.22470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hung Y.P., Fletcher C.D.M., Hornick J.L. Evaluation of pan-TRK immunohistochemistry in infantile fibrosarcoma, lipofibromatosis-like neural tumour and histological mimics. Histopathology. 2018;73:634–644. doi: 10.1111/his.13666. [DOI] [PubMed] [Google Scholar]
- 116.Del Castillo M., Chibon F., Arnould L., Croce S., Ribeiro A., Perot G., Hostein I., Geha S., Bozon C., Garnier A., et al. Secretory breast carcinoma: A histopathologic and genomic spectrum characterized by a joint specific ETV6-NTRK3 gene fusion. Am. J. Surg. Pathol. 2015;39:1458–1467. doi: 10.1097/PAS.0000000000000487. [DOI] [PubMed] [Google Scholar]
- 117.Skalova A., Michal M., Simpson R.H. Newly described salivary gland tumors. Mod. Pathol. 2017;30((Suppl. S1)):S27–S43. doi: 10.1038/modpathol.2016.167. [DOI] [PubMed] [Google Scholar]
- 118.Yoshida A., Folpe A.L. Adult fibrosarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 122–123. [Google Scholar]
- 119.Bovée J.V.M.G., Huang S.C., Wang J. Epithelioid haemangioma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 152–153. [Google Scholar]
- 120.Huang S.C., Zhang L., Sung Y.S., Chen C.L., Krausz T., Dickson B.C., Kao Y.C., Agaram N.P., Fletcher C.D., Antonescu C.R. Frequent FOS gene rearrangements in epithelioid hemangioma: A molecular study of 58 cases with morphologic reappraisal. Am. J. Surg. Pathol. 2015;39:1313–1321. doi: 10.1097/PAS.0000000000000469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.van IJzendoorn D.G., de Jong D., Romagosa C., Picci P., Benassi M.S., Gambarotti M., Daugaard S., van de Sande M., Szuhai K., Bovée J.V. Fusion events lead to truncation of FOS in epithelioid hemangioma of bone. Genes Chromosomes Cancer. 2015;54:565–574. doi: 10.1002/gcc.22269. [DOI] [PubMed] [Google Scholar]
- 122.Agaram N.P., Zhang L., Cotzia P., Antonescu C.R. Expanding the spectrum of genetic alterations in pseudomyogenic hemangioendothelioma with recurrent novel ACTB-FOSB gene fusions. Am. J. Surg. Pathol. 2018;42:1653–1661. doi: 10.1097/PAS.0000000000001147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Antonescu C.R., Chen H.W., Zhang L., Sung Y.S., Panicek D., Agaram N.P., Dickson B.C., Krausz T., Fletcher C.D. ZFP36-FOSB fusion defines a subset of epithelioid hemangioma with atypical features. Genes Chromosomes Cancer. 2014;53:951–959. doi: 10.1002/gcc.22206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hung Y.P., Fletcher C.D., Hornick J.L. FOSB is a useful diagnostic marker for pseudomyogenic hemangioendothelioma. Am. J. Surg. Pathol. 2017;41:596–606. doi: 10.1097/PAS.0000000000000795. [DOI] [PubMed] [Google Scholar]
- 125.Hornick J.L., Agaram N.P., Bovée J.V.M.G. Pseudomyogenic haemangioendothelioma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 169–171. [Google Scholar]
- 126.Trombetta D., Magnusson L., von Steyern F.V., Hornick J.L., Fletcher C.D., Mertens F. Translocation t(7;19)(q22;q13)−a recurrent chromosome aberration in pseudomyogenic hemangioendothelioma? Cancer Genet. 2011;204:211–215. doi: 10.1016/j.cancergen.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 127.Walther C., Tayebwa J., Lilljebjörn H., Magnusson L., Nilsson J., von Steyern F.V., Øra I., Domanski H.A., Fioretos T., Nord K.H., et al. A novel SERPINE1-FOSB fusion gene results in transcriptional up-regulation of FOSB in pseudomyogenic haemangioendothelioma. J. Pathol. 2014;232:534–540. doi: 10.1002/path.4322. [DOI] [PubMed] [Google Scholar]
- 128.Sugita S., Hirano H., Kikuchi N., Kubo T., Asanuma H., Aoyama T., Emori M., Hasegawa T. Diagnostic utility of FOSB immunohistochemistry in pseudomyogenic hemangioendothelioma and its histological mimics. Diagn. Pathol. 2016;11:75. doi: 10.1186/s13000-016-0530-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mirra J.M., Kessler S., Bhuta S., Eckardt J. The fibroma-like variant of epithelioid sarcoma. A fibrohistiocytic/myoid cell lesion often confused with benign and malignant spindle cell tumors. Cancer. 1992;69:1382–1395. doi: 10.1002/1097-0142(19920315)69:6<1382::AID-CNCR2820690614>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 130.Hornick J.L., Fletcher C.D. Pseudomyogenic hemangioendothelioma: A distinctive, often multicentric tumor with indolent behavior. Am. J. Surg. Pathol. 2011;35:190–201. doi: 10.1097/PAS.0b013e3181ff0901. [DOI] [PubMed] [Google Scholar]
- 131.Panagopoulos I., Lobmaier I., Gorunova L., Heim S. Fusion of the genes WWTR1 and FOSB in pseudomyogenic hemangioendothelioma. Cancer Genomics Proteomics. 2019;16:293–298. doi: 10.21873/cgp.20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bridge J.A., Sumegi J., Royce T., Baker M., Linos K. A novel CLTC-FOSB gene fusion in pseudomyogenic hemangioendothelioma of bone. Genes Chromosomes Cancer. 2021;60:38–42. doi: 10.1002/gcc.22891. [DOI] [PubMed] [Google Scholar]
- 133.Rubin B.P., Deyrup A.T., Doyle L.A. Epithelioid haemangioendothelioma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 172–175. [Google Scholar]
- 134.Errani C., Zhang L., Sung Y.S., Hajdu M., Singer S., Maki R.G., Healey J.H., Antonescu C.R. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosomes Cancer. 2011;50:644–653. doi: 10.1002/gcc.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mendlick M.R., Nelson M., Pickering D., Johansson S.L., Seemayer T.A., Neff J.R., Vergara G., Rosenthal H., Bridge J.A. Translocation t(1;3)(p36.3;q25) is a nonrandom aberration in epithelioid hemangioendothelioma. Am. J. Surg. Pathol. 2001;25:684–687. doi: 10.1097/00000478-200105000-00019. [DOI] [PubMed] [Google Scholar]
- 136.Tanas M.R., Sboner A., Oliveira A.M., Erickson-Johnson M.R., Hespelt J., Hanwright P.J., Flanagan J., Luo Y., Fenwick K., Natrajan R., et al. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci. Transl. Med. 2011;3:98ra82. doi: 10.1126/scitranslmed.3002409. [DOI] [PubMed] [Google Scholar]
- 137.Tanas M.R., Ma S., Jadaan F.O., Ng C.K., Weigelt B., Reis-Filho J.S., Rubin B.P. Mechanism of action of a WWTR1(TAZ)-CAMTA1 fusion oncoprotein. Oncogene. 2016;35:929–938. doi: 10.1038/onc.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Antonescu C.R., Le Loarer F., Mosquera J.M., Sboner A., Zhang L., Chen C.L., Chen H.W., Pathan N., Krausz T., Dickson B.C., et al. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer. 2013;52:775–784. doi: 10.1002/gcc.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Doyle L.A., Fletcher C.D., Hornick J.L. Nuclear expression of CAMTA1 distinguishes epithelioid hemangioendothelioma from histologic mimics. Am. J. Surg. Pathol. 2016;40:94–102. doi: 10.1097/PAS.0000000000000511. [DOI] [PubMed] [Google Scholar]
- 140.Lee S.J., Yang W.I., Chung W.S., Kim S.K. Epithelioid hemangioendotheliomas with TFE3 gene translocations are compossible with CAMTA1 gene rearrangements. Oncotarget. 2016;7:7480–7488. doi: 10.18632/oncotarget.7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kohashi K., Bode-Lesniewska B. Alveolar rhabdomyosarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 205–208. [Google Scholar]
- 142.Barr F.G., Galili N., Holick J., Biegel J.A., Rovera G., Emanuel B.S. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nat. Genet. 1993;3:113–117. doi: 10.1038/ng0293-113. [DOI] [PubMed] [Google Scholar]
- 143.Davis R.J., D’Cruz C.M., Lovell M.A., Biegel J.A., Barr F.G. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- 144.Buckingham M., Relaix F. PAX3 and PAX7 as upstream regulators of myogenesis. Semin. Cell Dev. Biol. 2015;44:115–125. doi: 10.1016/j.semcdb.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 145.Cao L., Yu Y., Bilke S., Walker R.L., Mayeenuddin L.H., Azorsa D.O., Yang F., Pineda M., Helman L.J., Meltzer P.S. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res. 2010;70:6497–6508. doi: 10.1158/0008-5472.CAN-10-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gryder B.E., Yohe M.E., Chou H.C., Zhang X., Marques J., Wachtel M., Schaefer B., Sen N., Song Y., Gualtieri A., et al. PAX3-FOXO1 establishes myogenic super enhancers and confers BET bromodomain vulnerability. Cancer Discov. 2017;7:884–899. doi: 10.1158/2159-8290.CD-16-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Skapek S.X., Ferrari A., Gupta A.A., Lupo P.J., Butler E., Shipley J., Barr F.G., Hawkins D.S. Rhabdomyosarcoma. Nat. Rev. Dis. Primers. 2019;5:1. doi: 10.1038/s41572-018-0051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hostein I., Andraud-Fregeville M., Guillou L., Terrier-Lacombe M.J., Deminière C., Ranchère D., Lussan C., Longavenne E., Bui N.B., Delattre O., et al. Rhabdomyosarcoma: Value of myogenin expression analysis and molecular testing in diagnosing the alveolar subtype: An analysis of 109 paraffin-embedded specimens. Cancer. 2004;101:2817–2824. doi: 10.1002/cncr.20711. [DOI] [PubMed] [Google Scholar]
- 149.Rekhi B., Gupta C., Chinnaswamy G., Qureshi S., Vora T., Khanna N., Laskar S. Clinicopathologic features of 300 rhabdomyosarcomas with emphasis upon differential expression of skeletal muscle specific markers in the various subtypes: A single institutional experience. Ann. Diagn. Pathol. 2018;36:50–60. doi: 10.1016/j.anndiagpath.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 150.Lindberg M.R., editor. Diagnostic Pathology. Soft Tissue Tumors. 3rd ed. Elsevier; Philadelphia, PA, USA: 2019. Alveolar rhabdomyosarcoma; pp. 385–391. [Google Scholar]
- 151.Williamson D., Missiaglia E., de Reyniès A., Pierron G., Thuille B., Palenzuela G., Thway K., Orbach D., Laé M., Fréneaux P., et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J. Clin. Oncol. 2010;28:2151–2158. doi: 10.1200/JCO.2009.26.3814. [DOI] [PubMed] [Google Scholar]
- 152.Agaram N.P., Szuhai K. Spindle cell/sclerosing rhabdomyosarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 211–213. [Google Scholar]
- 153.Mosquera J.M., Sboner A., Zhang L., Kitabayashi N., Chen C.L., Sung Y.S., Wexler L.H., LaQuaglia M.P., Edelman M., Sreekantaiah C., et al. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosomes Cancer. 2013;52:538–550. doi: 10.1002/gcc.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Alaggio R., Zhang L., Sung Y.S., Huang S.C., Chen C.L., Bisogno G., Zin A., Agaram N.P., LaQuaglia M.P., Wexler L.H., et al. A molecular study of pediatric spindle and sclerosing rhabdomyosarcoma: Identification of novel and recurrent VGLL2-related fusions in infantile cases. Am. J. Surg. Pathol. 2016;40:224–235. doi: 10.1097/PAS.0000000000000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Agaram N.P., Chen C.L., Zhang L., LaQuaglia M.P., Wexler L., Antonescu C.R. Recurrent MYOD1 mutations in pediatric and adult sclerosing and spindle cell rhabdomyosarcomas: Evidence for a common pathogenesis. Genes Chromosomes Cancer. 2014;53:779–787. doi: 10.1002/gcc.22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Agaram N.P., LaQuaglia M.P., Alaggio R., Zhang L., Fujisawa Y., Ladanyi M., Wexler L.H., Antonescu C.R. MYOD1-mutant spindle cell and sclerosing rhabdomyosarcoma: An aggressive subtype irrespective of age. A reappraisal for molecular classification and risk stratification. Mod. Pathol. 2019;32:27–36. doi: 10.1038/s41379-018-0120-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dashti N.K., Wehrs R.N., Thomas B.C., Nair A., Davila J., Buckner J.C., Martinez A.P., Sukov W.R., Halling K.C., Howe B.M., et al. Spindle cell rhabdomyosarcoma of bone with FUS-TFCP2 fusion: Confirmation of a very recently described rhabdomyosarcoma subtype. Histopathology. 2018;73:514–520. doi: 10.1111/his.13649. [DOI] [PubMed] [Google Scholar]
- 158.Rekhi B., Upadhyay P., Ramteke M.P., Dutt A. MYOD1 (L122R) mutations are associated with spindle cell and sclerosing rhabdomyosarcomas with aggressive clinical outcomes. Mod. Pathol. 2016;29:1532–1540. doi: 10.1038/modpathol.2016.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dei Tos A.P., Hornick J.L., Miettinen M., Wanless I.R., Wardelmann E. Gastrointestinal stromal tumour. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 216–221. [Google Scholar]
- 160.Emile J.F., Brahimi S., Coindre J.M., Bringuier P.P., Monges G., Samb P., Doucet L., Hostein I., Landi B., Buisine M.P., et al. Frequencies of KIT and PDGFRA mutations in the MolecGIST prospective population-based study differ from those of advanced GISTs. Med. Oncol. 2012;29:1765–1772. doi: 10.1007/s12032-011-0074-y. [DOI] [PubMed] [Google Scholar]
- 161.Joensuu H., Hohenberger P., Corless C.L. Gastrointestinal stromal tumour. Lancet. 2013;382:973–983. doi: 10.1016/S0140-6736(13)60106-3. [DOI] [PubMed] [Google Scholar]
- 162.Heinrich M.C., Corless C.L., Duensing A., McGreevey L., Chen C.J., Joseph N., Singer S., Griffith D.J., Haley A., Town A., et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 163.Corless C.L., Schroeder A., Griffith D., Town A., McGreevey L., Harrell P., Shiraga S., Bainbridge T., Morich J., Heinrich M.C. PDGFRA mutations in gastrointestinal stromal tumors: Frequency, spectrum and in vitro sensitivity to imatinib. J. Clin. Oncol. 2005;23:5357–5364. doi: 10.1200/JCO.2005.14.068. [DOI] [PubMed] [Google Scholar]
- 164.Corless C.L., Barnett C.M., Heinrich M.C. Gastrointestinal stromal tumours: Origin and molecular oncology. Nat. Rev. Cancer. 2011;11:865–878. doi: 10.1038/nrc3143. [DOI] [PubMed] [Google Scholar]
- 165.Boikos S.A., Pappo A.S., Killian J.K., LaQuaglia M.P., Weldon C.B., George S., Trent J.C., von Mehren M., Wright J.A., Schiffman J.D., et al. Molecular subtypes of KIT/PDGFRA wild-type gastrointestinal stromal tumors: A report from the National Institutes of Health Gastrointestinal Stromal Tumor Clinic. JAMA Oncol. 2016;2:922–928. doi: 10.1001/jamaoncol.2016.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Janeway K.A., Kim S.Y., Lodish M., Nosé V., Rustin P., Gaal J., Dahia P.L., Liegl B., Ball E.R., Raygada M., et al. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proc. Natl. Acad. Sci. USA. 2011;108:314–318. doi: 10.1073/pnas.1009199108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Miettinen M., Wang Z.F., Sarlomo-Rikala M., Osuch C., Rutkowski P., Lasota J. Succinate dehydrogenase-deficient GISTs: A clinicopathologic, immunohistochemical, and molecular genetic study of 66 gastric GISTs with predilection to young age. Am. J. Surg. Pathol. 2011;35:1712–1721. doi: 10.1097/PAS.0b013e3182260752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wagner A.J., Remillard S.P., Zhang Y.X., Doyle L.A., George S., Hornick J.L. Loss of expression of SDHA predicts SDHA mutations in gastrointestinal stromal tumors. Mod. Pathol. 2013;26:289–294. doi: 10.1038/modpathol.2012.153. [DOI] [PubMed] [Google Scholar]
- 169.Espinosa I., Lee C.H., Kim M.K., Rouse B.T., Subramanian S., Montgomery K., Varma S., Corless C.L., Heinrich M.C., Smith K.S., et al. A novel monoclonal antibody against DOG1 is a sensitive and specific marker for gastrointestinal stromal tumors. Am. J. Surg. Pathol. 2008;32:210–218. doi: 10.1097/PAS.0b013e3181238cec. [DOI] [PubMed] [Google Scholar]
- 170.Joensuu H., Rutkowski P., Nishida T., Steigen S.E., Brabec P., Plank L., Nilsson B., Braconi C., Bordoni A., Magnusson M.K., et al. KIT and PDGFRA mutations and the risk of GI stromal tumor recurrence. J. Clin. Oncol. 2015;33:634–642. doi: 10.1200/JCO.2014.57.4970. [DOI] [PubMed] [Google Scholar]
- 171.Heinrich M.C., Jones R.L., von Mehren M., Schöffski P., Serrano C., Kang Y.K., Cassier P.A., Mir O., Eskens F., Tap W.D., et al. Avapritinib in advanced PDGFRA D842V-mutant gastrointestinal stromal tumour (NAVIGATOR): A multicentre, open-label, phase 1 trial. Lancet Oncol. 2020;21:935–946. doi: 10.1016/S1470-2045(20)30269-2. [DOI] [PubMed] [Google Scholar]
- 172.Brčić I., Argyropoulos A., Liegl-Atzwanger B. Update on molecular genetics of gastrointestinal stromal tumors. Diagnostics. 2021;11:194. doi: 10.3390/diagnostics11020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nielsen G.P., Chi P. Malignant peripheral nerve sheath tumour. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 254–257. [Google Scholar]
- 174.Lee W., Teckie S., Wiesner T., Ran L., Prieto Granada C.N., Lin M., Zhu S., Cao Z., Liang Y., Sboner A., et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat. Genet. 2014;46:1227–1232. doi: 10.1038/ng.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang M., Wang Y., Jones S., Sausen M., McMahon K., Sharma R., Wang Q., Belzberg A.J., Chaichana K., Gallia G.L., et al. Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat. Genet. 2014;46:1170–1172. doi: 10.1038/ng.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pemov A., Hansen N.F., Sindiri S., Patidar R., Higham C.S., Dombi E., Miettinen M.M., Fetsch P., Brems H., Chandrasekharappa S.C., et al. Low mutation burden and frequent loss of CDKN2A/B and SMARCA2, but not PRC2, define premalignant neurofibromatosis type 1-associated atypical neurofibromas. Neuro-Oncol. 2019;21:981–992. doi: 10.1093/neuonc/noz028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Prieto-Granada C.N., Wiesner T., Messina J.L., Jungbluth A.A., Chi P., Antonescu C.R. Loss of H3K27me3 expression is a highly sensitive marker for sporadic and radiation-induced MPNST. Am. J. Surg. Pathol. 2016;40:479–489. doi: 10.1097/PAS.0000000000000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schaefer I.M., Fletcher C.D., Hornick J.L. Loss of H3K27 trimethylation distinguishes malignant peripheral nerve sheath tumors from histologic mimics. Mod. Pathol. 2016;29:4–13. doi: 10.1038/modpathol.2015.134. [DOI] [PubMed] [Google Scholar]
- 179.Pekmezci M., Cuevas-Ocampo A.K., Perry A., Horvai A.E. Significance of H3K27me3 loss in the diagnosis of malignant peripheral nerve sheath tumors. Mod. Pathol. 2017;30:1710–1719. doi: 10.1038/modpathol.2017.97. [DOI] [PubMed] [Google Scholar]
- 180.Schaefer I.M., Dong F., Garcia E.P., Fletcher C.D.M., Jo V.Y. Recurrent SMARCB1 inactivation in epithelioid malignant peripheral nerve sheath tumors. Am. J. Surg. Pathol. 2019;43:835–843. doi: 10.1097/PAS.0000000000001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hornick J.L., Nielsen G.P. Beyond “Triton”: Malignant peripheral nerve sheath tumors with complete heterologous rhabdomyoblastic differentiation mimicking spindle cell rhabdomyosarcoma. Am. J. Surg. Pathol. 2019;43:1323–1330. doi: 10.1097/PAS.0000000000001290. [DOI] [PubMed] [Google Scholar]
- 182.Folpe A.L., Hameed M. Malignant melanotic nerve sheath tumour. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 258–260. [Google Scholar]
- 183.Wang L., Zehir A., Sadowska J., Zhou N., Rosenblum M., Busam K., Agaram N., Travis W., Arcila M., Dogan S., et al. Consistent copy number changes and recurrent PRKAR1A mutations distinguish Melanotic Schwannomas from Melanomas: SNP-array and next generation sequencing analysis. Genes Chromosomes Cancer. 2015;54:463–471. doi: 10.1002/gcc.22254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Torres-Mora J., Dry S., Li X., Binder S., Amin M., Folpe A.L. Malignant melanotic schwannian tumor: A clinicopathologic, immunohistochemical, and gene expression profiling study of 40 cases, with a proposal for the reclassification of “melanotic schwannoma”. Am. J. Surg. Pathol. 2014;38:94–105. doi: 10.1097/PAS.0b013e3182a0a150. [DOI] [PubMed] [Google Scholar]
- 185.Vallat-Decouvelaere A.V., Wassef M., Lot G., Catala M., Moussalam M., Caruel N., Mikol J. Spinal melanotic schwannoma: A tumour with poor prognosis. Histopathology. 1999;35:558–566. doi: 10.1046/j.1365-2559.1999.00786.x. [DOI] [PubMed] [Google Scholar]
- 186.Khoo M., Pressney I., Hargunani R., Tirabosco R. Melanotic schwannoma: An 11-year case series. Skelet. Radiol. 2016;45:29–34. doi: 10.1007/s00256-015-2256-8. [DOI] [PubMed] [Google Scholar]
- 187.Suumeijer A.J.H., Ladanyi M., Nielsen T.O. Synovial sarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 290–293. [Google Scholar]
- 188.Ladanyi M., Antonescu C.R., Leung D.H., Woodruff J.M., Kawai A., Healey J.H., Brennan M.F., Bridge J.A., Neff J.R., Barr F.G., et al. Impact of SYT-SSX fusion type on the clinical behavior of synovial sarcoma: A multi-institutional retrospective study of 243 patients. Cancer Res. 2002;62:135–140. [PubMed] [Google Scholar]
- 189.dos Santos N.R., de Bruijn D.R., van Kessel A.G. Molecular mechanisms underlying human synovial sarcoma development. Genes Chromosomes Cancer. 2001;30:1–14. doi: 10.1002/1098-2264(2000)9999:9999<::AID-GCC1056>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 190.Amary M.F., Berisha F., Bernardi Fdel C., Herbert A., James M., Reis-Filho J.S., Fisher C., Nicholson A.G., Tirabosco R., Diss T.C., et al. Detection of SS18-SSX fusion ranscripts in formalin-fixed paraffin-embedded neoplasms: Analysis of conventional RT-PCR, qRT-PCR and dual color FISH as diagnostic tools for synovial sarcoma. Mod. Pathol. 2007;20:482–496. doi: 10.1038/modpathol.3800761. [DOI] [PubMed] [Google Scholar]
- 191.Jones K.B., Barrott J.J., Xie M., Haldar M., Jin H., Zhu J.F., Monument M.J., Mosbruger T.L., Langer E.M., Randall R.L., et al. The impact of chromosomal translocation locus and fusion oncogene coding sequence in synovial sarcomagenesis. Oncogene. 2016;35:5021–5032. doi: 10.1038/onc.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Barrott J.J., Illum B.E., Jin H., Hedberg M.L., Wang Y., Grossmann A., Haldar M., Capecchi M.R., Jones K.B. Paracrine osteoprotegerin and β-catenin stabilization support synovial sarcomagenesis in periosteal cells. J. Clin. Investig. 2018;128:207–218. doi: 10.1172/JCI94955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Baranov E., McBride M.J., Bellizzi A.M., Ligon A.H., Fletcher C.D.M., Kadoch C., Hornick J.L. A novel SS18-SSX fusion-specific antibody for the diagnosis of synovial sarcoma. Am. J. Surg. Pathol. 2020;44:922–933. doi: 10.1097/PAS.0000000000001447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Perret R., Velasco V., Le Guellec S., Coindre J.M., Le Loarer F. The SS18-SSX antibody has perfect specificity for the SS18-SSX fusion protein: A validation study of 609 neoplasms including 2 unclassified tumors with SS18-Non-SSX fusions. Am. J. Surg. Pathol. 2021;45:582–584. doi: 10.1097/PAS.0000000000001628. [DOI] [PubMed] [Google Scholar]
- 195.Foo W.C., Cruise M.W., Wick M.R., Hornick J.L. Immunohistochemical staining for TLE1 distinguishes synovial sarcoma from histologic mimics. Am. J. Clin. Pathol. 2011;135:839–844. doi: 10.1309/AJCP45SSNAOPXYXU. [DOI] [PubMed] [Google Scholar]
- 196.Yoshida A., Arai Y., Satomi K., Kubo T., Ryo E., Matsushita Y., Hama N., Sudo K., Komiyama M., Yatabe Y., et al. Identification of novel SSX1 fusions in synovial sarcoma. Mod. Pathol. 2022;35:228–239. doi: 10.1038/s41379-021-00910-x. [DOI] [PubMed] [Google Scholar]
- 197.Oda Y., Dal Cin P., Le Loarer F., Nielsen T.O. Epithelioid sarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 294–296. [Google Scholar]
- 198.Hornick J.L., Dal Cin P., Fletcher C.D. Loss of INI1 expression is characteristic of both conventional and proximal-type epithelioid sarcoma. Am. J. Surg. Pathol. 2009;33:542–550. doi: 10.1097/PAS.0b013e3181882c54. [DOI] [PubMed] [Google Scholar]
- 199.Kohashi K., Izumi T., Oda Y., Yamamoto H., Tamiya S., Taguchi T., Iwamoto Y., Hasegawa T., Tsuneyoshi M. Infrequent SMARCB1/INI1 gene alteration in epithelioid sarcoma: A useful tool in distinguishing epithelioid sarcoma from malignant rhabdoid tumor. Hum. Pathol. 2009;40:349–355. doi: 10.1016/j.humpath.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 200.Sakharpe A., Lahat G., Gulamhusein T., Liu P., Bolshakov S., Nguyen T., Zhang P., Belousov R., Young E., Xie X., et al. Epithelioid sarcoma and unclassified sarcoma with epithelioid features: Clinicopathological variables, molecular markers, and a new experimental model. Oncologist. 2011;16:512–522. doi: 10.1634/theoncologist.2010-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Miettinen M., Wang Z., Sarlomo-Rikala M., Abdullaev Z., Pack S.D., Fetsch J.F. ERG expression in epithelioid sarcoma: A diagnostic pitfall. Am. J. Surg. Pathol. 2013;37:1580–1585. doi: 10.1097/PAS.0b013e31828de23a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Stockman D.L., Hornick J.L., Deavers M.T., Lev D.C., Lazar A.J., Wang W.L. ERG and FLI1 protein expression in epithelioid sarcoma. Mod. Pathol. 2014;27:496–501. doi: 10.1038/modpathol.2013.161. [DOI] [PubMed] [Google Scholar]
- 203.Kohashi K., Yamada Y., Hotokebuchi Y., Yamamoto H., Taguchi T., Iwamoto Y., Oda Y. ERG and SALL4 expressions in SMARCB1/INI1-deficient tumors: A useful tool for distinguishing epithelioid sarcoma from malignant rhabdoid tumor. Hum. Pathol. 2015;46:225–230. doi: 10.1016/j.humpath.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 204.Kohashi K., Yamamoto H., Yamada Y., Kinoshita I., Taguchi T., Iwamoto Y., Oda Y. SWI/SNF chromatin-remodeling complex status in SMARCB1/INI1-preserved epithelioid sarcoma. Am. J. Surg. Pathol. 2018;42:312–318. doi: 10.1097/PAS.0000000000001011. [DOI] [PubMed] [Google Scholar]
- 205.Oda Y., Biegel J.A., Pfister S.M. Extrarenal rhabdoid tumour. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 309–311. [Google Scholar]
- 206.Schneppenheim R., Frühwald M.C., Gesk S., Hasselblatt M., Jeibmann A., Kordes U., Kreuz M., Leuschner I., Martin Subero J.I., Obser T., et al. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am. J. Hum. Genet. 2010;86:279–284. doi: 10.1016/j.ajhg.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hoot A.C., Russo P., Judkins A.R., Perlman E.J., Biegel J.A. Immunohistochemical analysis of hSNF5/INI1 distinguishes renal and extra-renal malignant rhabdoid tumors from other pediatric soft tissue tumors. Am. J. Surg. Pathol. 2004;28:1485–1491. doi: 10.1097/01.pas.0000141390.14548.34. [DOI] [PubMed] [Google Scholar]
- 208.Hollmann T.J., Hornick J.L. INI1-deficient tumors: Diagnostic features and molecular genetics. Am. J. Surg. Pathol. 2011;35:e47–e63. doi: 10.1097/PAS.0b013e31822b325b. [DOI] [PubMed] [Google Scholar]
- 209.Yoshida A., Asano N., Kawai A., Kawamoto H., Nakazawa A., Kishimoto H., Kushima R. Differential SALL4 immunoexpression in malignant rhabdoid tumours and epithelioid sarcomas. Histopathology. 2015;66:252–261. doi: 10.1111/his.12460. [DOI] [PubMed] [Google Scholar]
- 210.Kohashi K., Nakatsura T., Kinoshita Y., Yamamoto H., Yamada Y., Tajiri T., Taguchi T., Iwamoto Y., Oda Y. Glypican 3 expression in tumors with loss of SMARCB1/INI1 protein expression. Hum. Pathol. 2013;44:526–533. doi: 10.1016/j.humpath.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 211.Versteege I., Sévenet N., Lange J., Rousseau-Merck M.F., Ambros P., Handgretinger R., Aurias A., Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 212.Eaton K.W., Tooke L.S., Wainwright L.M., Judkins A.R., Biegel J.A. Spectrum of SMARCB1/INI1 mutations in familial and sporadic rhabdoid tumors. Pediatr. Blood Cancer. 2011;56:7–15. doi: 10.1002/pbc.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jambhekar N.A., Landanyi M. Alveolar soft part sarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 297–299. [Google Scholar]
- 214.Tsuda M., Davis I.J., Argani P., Shukla N., McGill G.G., Nagai M., Saito T., Laé M., Fisher D.E., Ladanyi M. TFE3 fusions activate MET signaling by transcriptional up-regulation, defining another class of tumors as candidates for therapeutic MET inhibition. Cancer Res. 2007;67:919–929. doi: 10.1158/0008-5472.CAN-06-2855. [DOI] [PubMed] [Google Scholar]
- 215.Kobos R., Nagai M., Tsuda M., Merl M.Y., Saito T., Laé M., Mo Q., Olshen A., Lianoglou S., Leslie C., et al. Combining integrated genomics and functional genomics to dissect the biology of a cancer-associated, aberrant transcription factor, the ASPSCR1-TFE3 fusion oncoprotein. J. Pathol. 2013;229:743–754. doi: 10.1002/path.4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Mukaihara K., Tanabe Y., Kubota D., Akaike K., Hayashi T., Mogushi K., Hosoya M., Sato S., Kobayashi E., Okubo T., et al. Cabozantinib and dastinib exert anti-tumor activity in alveolar soft part sarcoma. PLoS ONE. 2017;12:e0185321. doi: 10.1371/journal.pone.0185321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Pang L.J., Chang B., Zou H., Qi Y., Jiang J.F., Li H.A., Hu W.H., Chen Y.Z., Liu C.X., Zhang W.J., et al. Alveolar soft part sarcoma: A bimarker diagnostic strategy using TFE3 immunoassay and ASPL-TFE3 fusion transcripts in paraffin-embedded tumor tissues. Diagn. Mol. Pathol. 2008;17:245–252. doi: 10.1097/PDM.0b013e31815d68d7. [DOI] [PubMed] [Google Scholar]
- 218.Williams A., Bartle G., Sumathi V.P., Meis J.M., Mangham D.C., Grimer R.J., Kindblom L.G. Detection of ASPL/TFE3 fusion transcripts and the TFE3 antigen in formalin-fixed, paraffin-embedded tissue in a series of 18 cases of alveolar soft part sarcoma: Useful diagnostic tools in cases with unusual histological features. Virchows Arch. 2011;458:291–300. doi: 10.1007/s00428-010-1039-9. [DOI] [PubMed] [Google Scholar]
- 219.Martignoni G., Gobbo S., Camparo P., Brunelli M., Munari E., Segala D., Pea M., Bonetti F., Illei P.B., Netto G.J., et al. Differential expression of cathepsin K in neoplasms harboring TFE3 gene fusions. Mod. Pathol. 2011;24:1313–1319. doi: 10.1038/modpathol.2011.93. [DOI] [PubMed] [Google Scholar]
- 220.Chamberlain B.K., McClain C.M., Gonzalez R.S., Coffin C.M., Cates J.M. Alveolar soft part sarcoma and granular cell tumor: An immunohistochemical comparison study. Hum. Pathol. 2014;45:1039–1044. doi: 10.1016/j.humpath.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 221.Folpe A.L., Deyrup A.T. Alveolar soft-part sarcoma: A review and update. J. Clin. Pathol. 2006;59:1127–1132. doi: 10.1136/jcp.2005.031120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Argani P., Antonescu C.R., Illei P.B., Lui M.Y., Timmons C.F., Newbury R., Reuter V.E., Garvin A.J., Perez-Atayde A.R., Fletcher J.A., et al. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: A distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am. J. Pathol. 2001;159:179–192. doi: 10.1016/S0002-9440(10)61684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Schöffski P., Wozniak A., Kasper B., Aamdal S., Leahy M.G., Rutkowski P., Bauer S., Gelderblom H., Italiano A., Lindner L.H., et al. Activity and safety of crizotinib in patients with alveolar soft part sarcoma with rearrangement of TFE3: European Organization for Research and Treatment of Cancer (EORTC) phase II trial 90101 ‘CREATE’. Ann. Oncol. 2018;29:758–765. doi: 10.1093/annonc/mdx774. [DOI] [PubMed] [Google Scholar]
- 224.Agaram N.P., Antonescu C.R., Ladanyi M. Desmoplastic small round cell tumour. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft tissue and bone tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 306–308. [Google Scholar]
- 225.Sawyer J.R., Tryka A.F., Lewis J.M. A novel reciprocal chromosome translocation t(11;22)(p13;q12) in an intraabdominal desmoplastic small round-cell tumor. Am. J. Surg. Pathol. 1992;16:411–416. doi: 10.1097/00000478-199204000-00010. [DOI] [PubMed] [Google Scholar]
- 226.Biegel J.A., Conard K., Brooks J.J. Translocation (11;22)(p13;q12): Primary change in intra-abdominal desmoplastic small round cell tumor. Genes Chromosomes Cancer. 1993;7:119–121. doi: 10.1002/gcc.2870070210. [DOI] [PubMed] [Google Scholar]
- 227.Ladanyi M., Gerald W. Fusion of the EWS and WT1 genes in the desmoplastic small round cell tumor. Cancer Res. 1994;54:2837–2840. [PubMed] [Google Scholar]
- 228.Gerald W.L., Rosai J., Ladanyi M. Characterization of the genomic breakpoint and chimeric transcripts in the EWS-WT1 gene fusion of desmoplastic small round cell tumor. Proc. Natl. Acad. Sci. USA. 1995;92:1028–1032. doi: 10.1073/pnas.92.4.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Gerald W.L., Haber D.A. The EWS-WT1 gene fusion in desmoplastic small round cell tumor. Semin. Cancer Biol. 2005;15:197–205. doi: 10.1016/j.semcancer.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 230.Kang H.J., Park J.H., Chen W., Kang S.I., Moroz K., Ladanyi M., Lee S.B. EWS-WT1 oncoprotein activates neuronal reprogramming factor ASCL1 and promotes neural differentiation. Cancer Res. 2014;74:4526–4535. doi: 10.1158/0008-5472.CAN-13-3663. [DOI] [PubMed] [Google Scholar]
- 231.Barnoud R., Sabourin J.C., Pasquier D., Ranchère D., Bailly C., Terrier-Lacombe M.J., Pasquier B. Immunohistochemical expression of WT1 by desmoplastic small round cell tumor: A comparative study with other small round cell tumors. Am. J. Surg. Pathol. 2000;24:830–836. doi: 10.1097/00000478-200006000-00008. [DOI] [PubMed] [Google Scholar]
- 232.Schoolmeester J.K., Folpe A.L., Nair A.A., Halling K., Sutton B.C., Landers E., Karnezis A.N., Dickson B.C., Nucci M.R., Kolin D.L. EWSR1-WT1 gene fusions in neoplasms other than desmoplastic small round cell tumor: A report of three unusual tumors involving the female genital tract and review of the literature. Mod. Pathol. 2021;34:1912–1920. doi: 10.1038/s41379-021-00843-5. [DOI] [PubMed] [Google Scholar]
- 233.Gerald W.L., Ladanyi M., de Alava E., Cuatrecasas M., Kushner B.H., LaQuaglia M.P., Rosai J. Clinical, pathologic, and molecular spectrum of tumors associated with t(11;22)(p13;q12): Desmoplastic small round-cell tumor and its variants. J. Clin. Oncol. 1998;16:3028–3036. doi: 10.1200/JCO.1998.16.9.3028. [DOI] [PubMed] [Google Scholar]
- 234.Subbiah V., Lamhamedi-Cherradi S.E., Cuglievan B., Menegaz B.A., Camacho P., Huh W., Ramamoorthy V., Anderson P.M., Pollock R.E., Lev D.C., et al. Multimodality treatment of desmoplastic small round cell tumor: Chemotherapy and complete cytoreductive surgery improve patient survival. Clin. Cancer Res. 2018;24:4865–4873. doi: 10.1158/1078-0432.CCR-18-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Mello C.A., Campos F.A.B., Santos T.G., Silva M.L.G., Torrezan G.T., Costa F.D., Formiga M.N., Nicolau U., Nascimento A.G., Silva C., et al. Desmoplastic small round cell tumor: A review of main molecular abnormalities and emerging therapy. Cancers. 2021;13:498. doi: 10.3390/cancers13030498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Bode-Lesniewska B., Debiec-Rychter M., Tavora F. Intimal sarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours. Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 315–317. [Google Scholar]
- 237.Bode-Lesniewska B., Zhao J., Speel E.J., Biraima A.M., Turina M., Komminoth P., Heitz P.U. Gains of 12q13-14 and overexpression of mdm2 are frequent findings inintimal sarcomas of the pulmonary artery. Virchows Arch. 2001;438:57–65. doi: 10.1007/s004280000313. [DOI] [PubMed] [Google Scholar]
- 238.Zhang H., Macdonald W.D., Erickson-Johnson M., Wang X., Jenkins R.B., Oliveira A.M. Cytogenetic and molecular cytogenetic findings of intimal sarcoma. Cancer Genet. Cytogenet. 2007;179:146–149. doi: 10.1016/j.cancergencyto.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 239.Zhao J., Roth J., Bode-Lesniewska B., Pfaltz M., Heitz P.U., Komminoth P. Combined comparative genomic hybridization and genomic microarray for detection of gene amplifications in pulmonary artery intimal sarcomas and adrenocortical tumors. Genes Chromosomes Cancer. 2002;34:48–57. doi: 10.1002/gcc.10035. [DOI] [PubMed] [Google Scholar]
- 240.Dewaele B., Floris G., Finalet-Ferreiro J., Fletcher C.D., Coindre J.M., Guillou L., Hogendoorn P.C., Wozniak A., Vanspauwen V., Schöffski P., et al. Coactivated platelet-derived growth factor receptor α and epidermal growth factor receptor are potential therapeutic targets in intimal sarcoma. Cancer Res. 2010;70:7304–7314. doi: 10.1158/0008-5472.CAN-10-1543. [DOI] [PubMed] [Google Scholar]
- 241.Koelsche C., Benhamida J.K., Kommoss F.K.F., Stichel D., Jones D.T.W., Pfister S.M., Heilig C.E., Fröhling S., Stenzinger A., Buslei R., et al. Intimal sarcomas and undifferentiated cardiac sarcomas carry mutually exclusive MDM2, MDM4, and CDK6 amplifications and share a common DNA methylation signature. Mod. Pathol. 2021;34:2122–2129. doi: 10.1038/s41379-021-00874-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Burke A.P. Cardiac undifferentiated pleomorphic sarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours Thoracic Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 262–264. [Google Scholar]
- 243.de Álava E., Lessnick S.L., Stamenkovic I. Ewing sarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 323–325. [Google Scholar]
- 244.Brohl A.S., Solomon D.A., Chang W., Wang J., Song Y., Sindiri S., Patidar R., Hurd L., Chen L., Shern J.F., et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 2014;10:e1004475. doi: 10.1371/journal.pgen.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Crompton B.D., Stewart C., Taylor-Weiner A., Alexe G., Kurek K.C., Calicchio M.L., Kiezun A., Carter S.L., Shukla S.A., Mehta S.S., et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014;4:1326–1341. doi: 10.1158/2159-8290.CD-13-1037. [DOI] [PubMed] [Google Scholar]
- 246.Tirode F., Surdez D., Ma X., Parker M., Le Deley M.C., Bahrami A., Zhang Z., Lapouble E., Grossetête-Lalami S., Rusch M., et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014;4:1342–1353. doi: 10.1158/2159-8290.CD-14-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Machado I., Yoshida A., Morales M.G.N., Abrahão-Machado L.F., Navarro S., Cruz J., Lavernia J., Parafioriti A., Picci P., Llombart-Bosch A. Review with novel markers facilitates precise categorization of 41 cases of diagnostically challenging, “undifferentiated small round cell tumors”. A clinicopathologic, immunophenotypic and molecular analysis. Ann. Diagn. Pathol. 2018;34:1–12. doi: 10.1016/j.anndiagpath.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 248.Wang W.L., Patel N.R., Caragea M., Hogendoorn P.C., López-Terrada D., Hornick J.L., Lazar A.J. Expression of ERG, an Ets family transcription factor, identifies ERG-rearranged Ewing sarcoma. Mod. Pathol. 2012;25:1378–1383. doi: 10.1038/modpathol.2012.97. [DOI] [PubMed] [Google Scholar]
- 249.Folpe A.L., Goldblum J.R., Rubin B.P., Shehata B.M., Liu W., Dei Tos A.P., Weiss S.W. Morphologic and immunophenotypic diversity in Ewing family tumors: A study of 66 genetically confirmed cases. Am. J. Surg. Pathol. 2005;29:1025–1033. doi: 10.1097/01.pas.0000167056.13614.62. [DOI] [PubMed] [Google Scholar]
- 250.Hornick J.L. Soft tissues. In: Longacre T.A., Greenson J.K., Hornick J.L., Reuter V.E., editors. Mills and Sternberg’s Diagnostic Surgical Pathology. 7th ed. Wolters Kluwer; Philadelphia, PA, USA: 2022. pp. 220–223. [Google Scholar]
- 251.Chen S., Deniz K., Sung Y.S., Zhang L., Dry S., Antonescu C.R. Ewing sarcoma with ERG gene rearrangements: A molecular study focusing on the prevalence of FUS-ERG and common pitfalls in detecting EWSR1-ERG fusions by FISH. Genes Chromosomes Cancer. 2016;55:340–349. doi: 10.1002/gcc.22336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Newby R., Rowe D., Paterson L., Farquharson M.A., MacDuff E., Coupe A., Hale J., Dildey P., Bown N. Cryptic EWSR1-FLI1 fusions in Ewing sarcoma: Potential pitfalls in the diagnostic use of fluorescence in situ hybridization probes. Cancer Genet. Cytogenet. 2010;200:60–64. doi: 10.1016/j.cancergencyto.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 253.Le Loarer F., Szuhai K., Tirode F. Round cell sarcoma with EWSR1–non-ETS fusions. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 326–329. [Google Scholar]
- 254.Diaz-Perez J.A., Nielsen G.P., Antonescu C., Taylor M.S., Lozano-Calderon S.A., Rosenberg A.E. EWSR1/FUS-NFATc2 rearranged round cell sarcoma: Clinicopathological series of 4 cases and literature review. Hum. Pathol. 2019;90:45–53. doi: 10.1016/j.humpath.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Chougule A., Taylor M.S., Nardi V., Chebib I., Cote G.M., Choy E., Nielsen G.P., Deshpande V. Spindle and round cell sarcoma with EWSR1-PATZ1 gene fusion: A sarcoma with polyphenotypic differentiation. Am. J. Surg. Pathol. 2019;43:220–228. doi: 10.1097/PAS.0000000000001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Toki S., Wakai S., Sekimizu M., Mori T., Ichikawa H., Kawai A., Yoshida A. PAX7 immunohistochemical evaluation of Ewing sarcoma and other small round cell tumours. Histopathology. 2018;73:645–652. doi: 10.1111/his.13689. [DOI] [PubMed] [Google Scholar]
- 257.Charville G.W., Wang W.L., Ingram D.R., Roy A., Thomas D., Patel R.M., Hornick J.L., van de Rijn M., Lazar A.J. EWSR1 fusion proteins mediate PAX7 expression in Ewing sarcoma. Mod. Pathol. 2017;30:1312–1320. doi: 10.1038/modpathol.2017.49. [DOI] [PubMed] [Google Scholar]
- 258.Perret R., Escuriol J., Velasco V., Mayeur L., Soubeyran I., Delfour C., Aubert S., Polivka M., Karanian M., Meurgey A., et al. NFATc2-rearranged sarcomas: Clinicopathologic, molecular, and cytogenetic study of 7 cases with evidence of AGGRECAN as a novel diagnostic marker. Mod. Pathol. 2020;33:1930–1944. doi: 10.1038/s41379-020-0542-z. [DOI] [PubMed] [Google Scholar]
- 259.Yoshida K.I., Machado I., Motoi T., Parafioriti A., Lacambra M., Ichikawa H., Kawai A., Antonescu C.R., Yoshida A. NKX3-1 is a useful immunohistochemical marker of EWSR1-NFATC2 sarcoma and mesenchymal chondrosarcoma. Am. J. Surg. Pathol. 2020;44:719–728. doi: 10.1097/PAS.0000000000001441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Yoshida A., Hashimoto T., Ryo E., Yoshida K.I., Motoi T., Yatabe Y., Mori T. Confirmation of NKX3-1 expression in EWSR1-NFATC2 sarcoma and mesenchymal chondrosarcoma using monoclonal antibody immunohistochemistry, RT-PCR, and RNA in situ hybridization. Am. J. Surg. Pathol. 2021;45:578–582. doi: 10.1097/PAS.0000000000001627. [DOI] [PubMed] [Google Scholar]
- 261.Wang G.Y., Thomas D.G., Davis J.L., Ng T., Patel R.M., Harms P.W., Betz B.L., Schuetze S.M., McHugh J.B., Horvai A.E., et al. EWSR1-NFATC2 translocation-associated sarcoma clinicopathologic findings in a rare aggressive primary bone or soft tissue tumor. Am. J. Surg. Pathol. 2019;43:1112–1122. doi: 10.1097/PAS.0000000000001260. [DOI] [PubMed] [Google Scholar]
- 262.Bode-Lesniewska B., Fritz C., Exner G.U., Wagner U., Fuchs B. EWSR1-NFATC2 and FUS-NFATC2 gene fusion-associated mesenchymal tumors: Clinicopathologic correlation and literature review. Sarcoma. 2019;2019:9386390. doi: 10.1155/2019/9386390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Bridge J.A., Sumegi J., Druta M., Bui M.M., Henderson-Jackson E., Linos K., Baker M., Walko C.M., Millis S., Brohl A.S. Clinical, pathological, and genomic features of EWSR1-PATZ1 fusion sarcoma. Mod. Pathol. 2019;32:1593–1604. doi: 10.1038/s41379-019-0301-1. [DOI] [PubMed] [Google Scholar]
- 264.Antonescu C.R., Yoshida A. CIC-rearranged sarcoma. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 330–332. [Google Scholar]
- 265.Kawamura-Saito M., Yamazaki Y., Kaneko K., Kawaguchi N., Kanda H., Mukai H., Gotoh T., Motoi T., Fukayama M., Aburatani H., et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum. Mol. Genet. 2006;15:2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- 266.Italiano A., Sung Y.S., Zhang L., Singer S., Maki R.G., Coindre J.M., Antonescu C.R. High prevalence of CIC fusion with double-homeobox (DUX4) transcription factors in EWSR1-negative undifferentiated small blue round cell sarcomas. Genes Chromosomes Cancer. 2012;51:207–218. doi: 10.1002/gcc.20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Le Loarer F., Pissaloux D., Watson S., Godfraind C., Galmiche-Rolland L., Silva K., Mayeur L., Italiano A., Michot A., Pierron G., et al. Clinicopathologic features of CIC-NUTM1 sarcomas, a new molecular variant of the family of CIC-fused sarcomas. Am. J. Surg. Pathol. 2019;43:268–276. doi: 10.1097/PAS.0000000000001187. [DOI] [PubMed] [Google Scholar]
- 268.Sugita S., Arai Y., Tonooka A., Hama N., Totoki Y., Fujii T., Aoyama T., Asanuma H., Tsukahara T., Kaya M., et al. A novel CIC-FOXO4 gene fusion in undifferentiated small round cell sarcoma: A genetically distinct variant of Ewing-like sarcoma. Am. J. Surg. Pathol. 2014;38:1571–1576. doi: 10.1097/PAS.0000000000000286. [DOI] [PubMed] [Google Scholar]
- 269.Huang S.C., Zhang L., Sung Y.S., Chen C.L., Kao Y.C., Agaram N.P., Singer S., Tap W.D., D’Angelo S., Antonescu C.R. Recurrent CIC gene abnormalities in angiosarcomas: A molecular study of 120 cases with concurrent investigation of PLCG1, KDR, MYC, and FLT4 Gene Alterations. Am. J. Surg. Pathol. 2016;40:645–655. doi: 10.1097/PAS.0000000000000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Specht K., Sung Y.S., Zhang L., Richter G.H., Fletcher C.D., Antonescu C.R. Distinct transcriptional signature and immunoprofile of CIC-DUX4 fusion-positive round cell tumors compared to EWSR1-rearranged Ewing sarcomas: Further evidence toward distinct pathologic entities. Genes Chromosomes Cancer. 2014;53:622–633. doi: 10.1002/gcc.22172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Smith S.C., Buehler D., Choi E.Y., McHugh J.B., Rubin B.P., Billings S.D., Balzer B., Thomas D.G., Lucas D.R., Goldblum J.R., et al. CIC-DUX sarcomas demonstrate frequent MYC amplification and ETS-family transcription factor expression. Mod. Pathol. 2015;28:57–68. doi: 10.1038/modpathol.2014.83. [DOI] [PubMed] [Google Scholar]
- 272.Siegele B., Roberts J., Black J.O., Rudzinski E., Vargas S.O., Galambos C. DUX4 immunohistochemistry is a highly sensitive and specific marker for CIC-DUX4 fusion-positive round cell tumor. Am. J. Surg. Pathol. 2017;41:423–429. doi: 10.1097/PAS.0000000000000772. [DOI] [PubMed] [Google Scholar]
- 273.Hung Y.P., Fletcher C.D., Hornick J.L. Evaluation of ETV4 and WT1 expression in CIC-rearranged sarcomas and histologic mimics. Mod. Pathol. 2016;29:1324–1334. doi: 10.1038/modpathol.2016.140. [DOI] [PubMed] [Google Scholar]
- 274.Le Guellec S., Velasco V., Pérot G., Watson S., Tirode F., Coindre J.M. ETV4 is a useful marker for the diagnosis of CIC-rearranged undifferentiated round-cell sarcomas: A study of 127 cases including mimicking lesions. Mod. Pathol. 2016;29:1523–1531. doi: 10.1038/modpathol.2016.155. [DOI] [PubMed] [Google Scholar]
- 275.Yoshida A., Goto K., Kodaira M., Kobayashi E., Kawamoto H., Mori T., Yoshimoto S., Endo O., Kodama N., Kushima R., et al. CIC-rearranged sarcomas: A study of 20 cases and comparisons with Ewing sarcomas. Am. J. Surg. Pathol. 2016;40:313–323. doi: 10.1097/PAS.0000000000000570. [DOI] [PubMed] [Google Scholar]
- 276.Sturm D., Orr B.A., Toprak U.H., Hovestadt V., Jones D.T.W., Capper D., Sill M., Buchhalter I., Northcott P.A., Leis I., et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164:1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Antonescu C.R., Owosho A.A., Zhang L., Chen S., Deniz K., Huryn J.M., Kao Y.C., Huang S.C., Singer S., Tap W., et al. Sarcomas with CIC-rearrangements are a distinct pathologic entity with aggressive outcome: A clinicopathologic and molecular study of 115 cases. Am. J. Surg. Pathol. 2017;41:941–949. doi: 10.1097/PAS.0000000000000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Antonescu C.R., Puls F., Tirode F. Sarcoma with BCOR genetic alterations. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 333–335. [Google Scholar]
- 279.Kao Y.C., Sung Y.S., Zhang L., Huang S.C., Argani P., Chung C.T., Graf N.S., Wright D.C., Kellie S.J., Agaram N.P., et al. Recurrent BCOR internal tandem duplication and YWHAE-NUTM2B fusions in soft tissue undifferentiated round cell sarcoma of infancy: Overlapping genetic features with clear cell sarcoma of kidney. Am. J. Surg. Pathol. 2016;40:1009–1020. doi: 10.1097/PAS.0000000000000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Roy A., Kumar V., Zorman B., Fang E., Haines K.M., Doddapaneni H., Hampton O.A., White S., Bavle A.A., Patel N.R., et al. Recurrent internal tandem duplications of BCOR in clear cell sarcoma of the kidney. Nat. Commun. 2015;6:8891. doi: 10.1038/ncomms9891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Specht K., Zhang L., Sung Y.S., Nucci M., Dry S., Vaiyapuri S., Richter G.H., Fletcher C.D., Antonescu C.R. Novel BCOR-MAML3 and ZC3H7B-BCOR gene fusions in undifferentiated small blue round cell sarcomas. Am. J. Surg. Pathol. 2016;40:433–442. doi: 10.1097/PAS.0000000000000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Kao Y.C., Sung Y.S., Zhang L., Jungbluth A.A., Huang S.C., Argani P., Agaram N.P., Zin A., Alaggio R., Antonescu C.R. BCOR overexpression is a highly sensitive marker in round cell sarcomas with BCOR genetic abnormalities. Am. J. Surg. Pathol. 2016;40:1670–1678. doi: 10.1097/PAS.0000000000000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Kao Y.C., Owosho A.A., Sung Y.S., Zhang L., Fujisawa Y., Lee J.C., Wexler L., Argani P., Swanson D., Dickson B.C., et al. BCOR-CCNB3 fusion positive sarcomas: A clinicopathologic and molecular analysis of 36 cases with comparison to morphologic spectrum and clinical behavior of other round cell sarcomas. Am. J. Surg. Pathol. 2018;42:604–615. doi: 10.1097/PAS.0000000000000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Matsuyama A., Shiba E., Umekita Y., Nosaka K., Kamio T., Yanai H., Miyasaka C., Watanabe R., Ito I., Tamaki T., et al. Clinicopathologic diversity of undifferentiated sarcoma with bcor-ccnb3 fusion: Analysis of 11 cases with a reappraisal of the utility of immunohistochemistry for BCOR and CCNB3. Am. J. Surg. Pathol. 2017;41:1713–1721. doi: 10.1097/PAS.0000000000000934. [DOI] [PubMed] [Google Scholar]
- 285.Shibayama T., Okamoto T., Nakashima Y., Kato T., Sakurai T., Minamiguchi S., Kataoka T.R., Shibuya S., Yoshizawa A., Toguchida J., et al. Screening of BCOR-CCNB3 sarcoma using immunohistochemistry for CCNB3: A clinicopathological report of three pediatric cases. Pathol. Int. 2015;65:410–414. doi: 10.1111/pin.12319. [DOI] [PubMed] [Google Scholar]
- 286.Salgado C.M., Zin A., Garrido M., Kletskaya I., DeVito R., Reyes-Múgica M., Bisogno G., Donofrio V., Alaggio R. Pediatric soft tissue tumors with BCOR ITD express EGFR but not OLIG2. Pediatr. Dev. Pathol. 2020;23:424–430. doi: 10.1177/1093526620945528. [DOI] [PubMed] [Google Scholar]
- 287.Antonescu C.R., Suurmeijer A.J.H. EWSR1-SMAD3–positive fibroblastic tumour (emerging) In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 76–77. [Google Scholar]
- 288.Michal M., Berry R.S., Rubin B.P., Kilpatrick S.E., Agaimy A., Kazakov D.V., Steiner P., Ptakova N., Martinek P., Hadravsky L., et al. EWSR1-SMAD3–rearranged fibroblastic tumor: An emerging entity in an increasingly more complex group of fibroblastic/myofibroblastic neoplasms. Am. J. Surg. Pathol. 2018;42:1325–1333. doi: 10.1097/PAS.0000000000001109. [DOI] [PubMed] [Google Scholar]
- 289.Kao Y.C., Flucke U., Eijkelenboom A., Zhang L., Sung Y.S., Suurmeijer A.J.H., Antonescu C.R. Novel EWSR1-SMAD3 gene fusions in a group of acral fibroblastic spindle cell neoplasms. Am. J. Surg. Pathol. 2018;42:522–528. doi: 10.1097/PAS.0000000000001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Armstrong S.M., Demicco E.G. What’s new in fibroblastic tumors? Virchows Arch. 2020;476:41–55. doi: 10.1007/s00428-019-02682-x. [DOI] [PubMed] [Google Scholar]
- 291.Lam S.W., Silva T.M., Bovée J.V.M.G. New molecular entities of soft tissue and bone tumors. Curr. Opin. Oncol. 2022;34:354–361. doi: 10.1097/CCO.0000000000000844. [DOI] [PubMed] [Google Scholar]
- 292.Suurmeijer A.J.H., Antonescu C.R. NTRK-rearranged spindle cell neoplasm (emerging) In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours Soft Tissue and Bone Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 287–289. [Google Scholar]
- 293.Agaram N.P., Zhang L., Sung Y.S., Chen C.L., Chung C.T., Antonescu C.R., Fletcher C.D. Recurrent NTRK1 gene fusions define a novel subset of locally aggressive lipofibromatosis-like neural tumors. Am. J. Surg. Pathol. 2016;40:1407–1416. doi: 10.1097/PAS.0000000000000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Suurmeijer A.J.H., Dickson B.C., Swanson D., Zhang L., Sung Y.S., Cotzia P., Fletcher C.D.M., Antonescu C.R. A novel group of spindle cell tumors defined by S100 and CD34 co-expression shows recurrent fusions involving RAF1, BRAF, and NTRK1/2 genes. Genes Chromosomes Cancer. 2018;57:611–621. doi: 10.1002/gcc.22671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Haller F., Knopf J., Ackermann A., Bieg M., Kleinheinz K., Schlesner M., Moskalev E.A., Will R., Satir A.A., Abdelmagid I.E., et al. Paediatric and adult soft tissue sarcomas with NTRK1 gene fusions: A subset of spindle cell sarcomas unified by a prominent myopericytic/haemangiopericytic pattern. J. Pathol. 2016;238:700–710. doi: 10.1002/path.4701. [DOI] [PubMed] [Google Scholar]
- 296.Yamazaki F., Nakatani F., Asano N., Wakai S., Sekimizu M., Mitani S., Kubo T., Kawai A., Ichikawa H., Yoshida A. Novel NTRK3 fusions in fibrosarcomas of adults. Am. J. Surg. Pathol. 2019;43:523–530. doi: 10.1097/PAS.0000000000001194. [DOI] [PubMed] [Google Scholar]
- 297.Cocco E., Scaltriti M., Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 2018;15:731–747. doi: 10.1038/s41571-018-0113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Hechtman J.F., Benayed R., Hyman D.M., Drilon A., Zehir A., Frosina D., Arcila M.E., Dogan S., Klimstra D.S., Ladanyi M., et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am. J. Surg. Pathol. 2017;41:1547–1551. doi: 10.1097/PAS.0000000000000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Rudzinski E.R., Lockwood C.M., Stohr B.A., Vargas S.O., Sheridan R., Black J.O., Rajaram V., Laetsch T.W., Davis J.L. Pan-Trk immunohistochemistry identifies NTRK rearrangements in pediatric mesenchymal tumors. Am. J. Surg. Pathol. 2018;42:927–935. doi: 10.1097/PAS.0000000000001062. [DOI] [PubMed] [Google Scholar]
- 300.Antonescu C.R. Emerging soft tissue tumors with kinase fusions: An overview of the recent literature with an emphasis on diagnostic criteria. Genes Chromosomes Cancer. 2020;59:437–444. doi: 10.1002/gcc.22846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Lam S.W., Briaire-de Bruijn I.H., van Wezel T., Cleven A.H.G., Hogendoorn P.C.W., Cleton-Jansen A.M., Bovée J.V.M.G. NTRK fusions are extremely rare in bone tumours. Histopathology. 2021;79:880–885. doi: 10.1111/his.14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Davis J.L., Lockwood C.M., Stohr B., Boecking C., Al-Ibraheemi A., DuBois S.G., Vargas S.O., Black J.O., Cox M.C., Luquette M., et al. Expanding the spectrum of pediatric NTRK-rearranged mesenchymal tumors. Am. J. Surg. Pathol. 2019;43:435–445. doi: 10.1097/PAS.0000000000001203. [DOI] [PubMed] [Google Scholar]
- 303.Tauziède-Espariat A., Duchesne M., Baud J., Le Quang M., Bochaton D., Azmani R., Croce S., Hostein I., Kesrouani C., Guillemot D., et al. NTRK-rearranged spindle cell neoplasms are ubiquitous tumors of myofibroblastic lineage with a distinct methylation class. Histopathology. 2023;82:596–607. doi: 10.1111/his.14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Masliah-Planchon J., Bièche I., Guinebretière J.M., Bourdeaut F., Delattre O. SWI/SNF chromatin remodeling and human malignancies. Annu. Rev. Pathol. 2015;10:145–171. doi: 10.1146/annurev-pathol-012414-040445. [DOI] [PubMed] [Google Scholar]
- 305.Agaimy A. Moving from “single gene” concept to “functionally homologous multigene complex”: The SWI/SNF paradigm. Semin. Diagn. Pathol. 2021;38:165–166. doi: 10.1053/j.semdp.2021.02.003. [DOI] [PubMed] [Google Scholar]
- 306.Schaefer I.M., Hornick J.L. SWI/SNF complex-deficient soft tissue neoplasms: An update. Semin. Diagn. Pathol. 2021;38:222–231. doi: 10.1053/j.semdp.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Nambirajan A., Jain D. Recent updates in thoracic SMARCA4-deficient undifferentiated tumor. Semin. Diagn. Pathol. 2021;38:83–89. doi: 10.1053/j.semdp.2021.06.001. [DOI] [PubMed] [Google Scholar]
- 308.Yoshida A., Boland J.M., Jain D., Le Loarer F., Rekhtman N. Thoracic SMARCA4-deficient undifferentiated tumor. In: The WHO Classification of Tumours Editorial Board, editor. WHO Classification of Tumours Thoracic Tumours. 5th ed. IARC Press; Lyon, France: 2020. pp. 111–114. [Google Scholar]
- 309.Jelinic P., Mueller J.J., Olvera N., Dao F., Scott S.N., Shah R., Gao J., Schultz N., Gonen M., Soslow R.A., et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat. Genet. 2014;46:424–426. doi: 10.1038/ng.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Lin D.I., Allen J.M., Hecht J.L., Killian J.K., Ngo N.T., Edgerly C., Severson E.A., Ali S.M., Erlich R.L., Ramkissoon S.H., et al. SMARCA4 inactivation defines a subset of undifferentiated uterine sarcomas with rhabdoid and small cell features and germline mutation association. Mod. Pathol. 2019;32:1675–1687. doi: 10.1038/s41379-019-0303-z. [DOI] [PubMed] [Google Scholar]
- 311.Agaimy A., Jain D., Uddin N., Rooper L.M., Bishop J.A. SMARCA4-deficient sinonasal carcinoma: A series of 10 cases expanding the genetic spectrum of SWI/SNF-driven sinonasal malignancies. Am. J. Surg. Pathol. 2020;44:703–710. doi: 10.1097/PAS.0000000000001428. [DOI] [PubMed] [Google Scholar]
- 312.Foulkes W.D., Priest J.R., Duchaine T.F. DICER1: Mutations, microRNAs and mechanisms. Nat. Rev. Cancer. 2014;14:662–672. doi: 10.1038/nrc3802. [DOI] [PubMed] [Google Scholar]
- 313.McCluggage W.G., Foulkes W.D. DICER1-associated sarcomas: Towards a unified nomenclature. Mod. Pathol. 2021;34:1226–1228. doi: 10.1038/s41379-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 314.Warren M., Hiemenz M.C., Schmidt R., Shows J., Cotter J., Toll S., Parham D.M., Biegel J.A., Mascarenhas L., Shah R. Expanding the spectrum of dicer1-associated sarcomas. Mod. Pathol. 2020;33:164–174. doi: 10.1038/s41379-019-0366-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.McCluggage W.G., Foulkes W.D. DICER1-associated sarcomas at different sites exhibit morphological overlap arguing for a unified nomenclature. Virchows Arch. 2021;479:431–433. doi: 10.1007/s00428-021-03087-5. [DOI] [PubMed] [Google Scholar]
- 316.Kommoss F.K.F., Stichel D., Mora J., Esteller M., Jones D.T.W., Pfister S.M., Brack E., Wachtel M., Bode P.K., Sinn H.P., et al. Clinicopathologic and molecular analysis of embryonal rhabdomyosarcoma of the genitourinary tract: Evidence for a distinct DICER1-associated subgroup. Mod. Pathol. 2021;34:1558–1569. doi: 10.1038/s41379-021-00804-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.McCluggage W.G., Foulkes W.D. DICER1-sarcoma: An emerging entity. Mod. Pathol. 2021;34:2096–2097. doi: 10.1038/s41379-021-00935-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.