Abstract
The appearance of persister cells with low metabolic rates are key factors leading to antibiotic treatment failure. Such persisters are multidrug tolerant and play a key role in the recalcitrance of biofilm-based chronic infections. Here, we present the genomic analyses of three distinct Pseudomonas aeruginosa Egyptian persister-isolates recovered from chronic human infections. To calculate the persister frequencies, viable counts were determined before and after treatment with levofloxacin. The susceptibilities of isolates to different antibiotics were determined using the agar-dilution method. To determine their recalcitrance, the levofloxacin persisters were further challenged with lethal concentrations of meropenem, tobramycin, or colistin. Furthermore, the biofilm formation of the persister strains was estimated phenotypically, and they were reported to be strong biofilm-forming strains. The genotypic characterization of the persisters was performed using whole genome sequencing (WGS) followed by phylogenetic analysis and resistome profiling. Interestingly, out of the thirty-eight clinical isolates, three isolates (8%) demonstrated a persister phenotype. The three levofloxacin-persister isolates were tested for their susceptibility to selected antibiotics; all of the tested isolates were multidrug resistant (MDR). Additionally, the P. aeruginosa persisters were capable of surviving over 24 h and were not eradicated after exposure to 100X-MIC of levofloxacin. WGS for the three persisters revealed a smaller genome size compared to PAO1-genome. Resistome profiling indicated the presence of a broad collection of antibiotic-resistance genes, including genes encoding for antibiotic-modifying enzymes and efflux pump. Phylogenetic analysis indicated that the persister isolates belong to a distinct clade rather than the deposited P. aeruginosa strains in the GenBank. Conclusively, the persister isolates in our study are MDR and form a highly strong biofilm. WGS revealed a smaller genome that belongs to a distinct clade.
Keywords: persister cells, Pseudomonas aeruginosa, multidrug-tolerant cells, killing curve, whole genome sequencing (WGS), resistome profiling, phylogenetic analysis
1. Introduction
Pseudomonas aeruginosa is an opportunistic pathogen that poses a threat in clinical settings due to its intrinsic and acquired resistance to a wide range of antibiotics and biocides [1,2,3]. During chronic infection, P. aeruginosa can be evolved toward a high-persister state, producing more antibiotic-tolerant cells [4]. Persister-cell formation is a phenomenon that contributes to the tolerance of a bacterial sub-population to antimicrobial agents. The presence of persister cells surviving high concentrations of antibiotics makes it virtually impossible to eradicate the chronic infection caused by P. aeruginosa. Although persisters constitute a small fraction of the population, they can be selected by surviving the treatment with high doses of bactericidal antibiotics [5]. Notably, this antibiotic tolerance of persister cells is distinct from genetically inherited resistance [6].
Although persister cells were discovered in Staphylococcus sp. as early as 1944 by Bigger [7], they were ignored for a long time. The first mutants identified to be altered in their persistence were the high persistence (hip) mutants in Escherichia coli that were discovered in 1983 [8]. Furthermore, persister cells have been described for other pathogens, including P. aeruginosa, Mycobacterium tuberculosis, Salmonella sp., and Streptococcus sp. [6,9]. The existence of persisters is believed to prolong and exacerbate the treatment of infectious diseases, such as tuberculosis and cystic fibrosis (CF)-associated lung infections [10].
Biofilm formation by bacteria is highly associated with increased resistance to conventional antibiotic therapy. Pseudomonas possesses a strong tendency to develop biofilm, which aids in its persistence and contributes to recalcitrant and/or recurrent infections after antibiotic therapy [11,12,13]. The biofilm growth mode creates micro-environment conditions that activate stringent response mechanisms and toxin–antitoxin (TA) systems that render the bacterial population dormant and hence highly tolerant to antibiotics [14].
Persister cells represent a stage of dormancy that protects them from complete eradication by antimicrobial substances, even in the presence of concentrations that vastly exceed the minimal inhibitory concentration (MIC). Persister cells are genetically identical to antibiotic-sensitive bacteria within a population but have a distinct phenotype in that they are tolerant to antibiotics [15,16]. The dormancy stage in persister cells is thought to be the underlying mechanism of antibiotic tolerance, since most antibiotics target bacterial components or pathways involved in replication [17]. Nevertheless, persister cells can switch from the dormant to the replicating stage. This ‘bet-hedging’ strategy is thought to be a survival strategy of microbial populations [18].
It has been reported that the level of persisters could be increased as a result of a heritable mutation in genes such as hipA (encodes the toxin entity of the toxin-antitoxin module hipBA) [8]. After 30 years of reporting hipA mutants, it was found that HipA inactivates glutamyl tRNA synthetase (GltX) by phosphorylation [19]. The inhibition of GltX stimulated the synthesis of guanosine pentaphosphate/tetraphosphate (ppGpp); ppGpp is a very important stress alarm, one that dramatically increases the persistence level [20,21].
Although persister cells can arise from stochastic events in growing cultures [22], evidence suggests that their formation can also be induced as a response to several environmental factors, such as nutrient and oxygen deprivation, oxidative stress, DNA damage, and antibiotics [23]. The addition of levofloxacin at a concentration of 10–100 times higher than its specific MIC was capable of inducing and separating persister cells [24]. Starvation, hypoxia, and antibiotic stress could be directly correlated to a reduced metabolic rate and decreased cellular activity. Reduced metabolic activity is related to increased persistence [25].
Previous reports have suggested that persister cells exhibit different phenotypes compared to the wild-type strains, including smaller colony sizes or changes in pigmentation [10,14,26]. Given that persister cells can enter a non-growing state, tolerate high concentrations of bactericidal antibiotics (for example by activating efflux pumps), and regrow once the treatment is ceased, these cells have already been linked to the relapse and recalcitrance of chronic infections [14].
This study aimed to examine the fraction of persister cells in the P. aeruginosa cultures of clinical isolates and to determine the susceptibility to different antimicrobials and the biofilm-forming capacity for P. aeruginosa persister isolates as compared to the reference strain PAO1. Moreover, whole genome sequencing, resistome profiling, and phylogenic analysis were performed for the three persister isolates compared to the first sequenced P. aeruginosa strain (PAO1 genome).
2. Materials and Methods
2.1. Bacterial Strains and Ethical Statement
Thirty-eight P. aeruginosa isolates were obtained from different sources (urine, sputum, and burn specimens) from patients admitted to El-Ahrar Educational Hospital and Zagazig University Hospital, Egypt. The standard strain of P. aeruginosa PAO1 was included in this study. It was provided from the stock culture collection of the Microbiology and Immunology Department, Faculty of Pharmacy, Zagazig University.
Experiments involving human samples were performed in accordance with the Declaration of Helsinki and were approved by the Zagazig University Institutional Review Board (Approval number: ZU-IRB-10360).
2.2. Antimicrobial Agents
The antibiotics used in the study were pharmaceutical grade products. Meropenem was obtained from the ADWIA Company, Egypt. Cefoperazone and tobramycin were obtained from the Egyptian Pharmaceutical Industries Company (EPICO), 10th of Ramadan city, Egypt. Cefepime was obtained from RAMEDA, Egypt. Levofloxacin and amikacin were obtained from the AMOUN Company, Egypt. Colistin and chloramphenicol were the products of SIGMA-TEC, 6th October City, Egypt. The batch numbers of the used antimicrobials were listed in Supplementary Table S1.
2.3. Determination of Viable Bacterial Count
Ten-fold serial dilutions (final volume 1000 μL) of the bacterial suspensions, from an overnight culture in a nutrient broth (Oxoid, Hampshire, England), were performed in Eppendorf tubes. Triplicates of 100 μL of the appropriate dilutions were plated on nutrient agar plates (Oxoid, Hampshire, England). The plates were incubated for 24–48 h at 37 °C. The average numbers of colonies on the appropriate plate were counted and the colony forming units (CFU)/mL were calculated [27].
2.4. Selection and Quantification of Levofloxacin Persister Cells
Levofloxacin was added to a growing nutrient broth culture of the isolate at a concentration of 10–100 times its MIC. The suspensions were incubated in a shaker incubator (200 rpm) at 37 °C for 24 h. Falcone tubes containing bacterial cultures were centrifuged for 5 min at 8000 rpm and the supernatant was removed without disrupting the pellet. The cells were washed by re-suspending them in 1 mL phosphate-buffered saline (PBS) and transferred to an Eppendorf tube for centrifugation for 5 min at 14,000 rpm. The supernatant was gently discarded and the pellets were re-suspended in 1 mL of Mueller–Hinton broth (MHB). The viable count was made as described previously [28].
2.5. Determination of the MIC of Tested Antibiotics
The MICs of antibiotics were determined using the agar dilution method, according to the clinical laboratory standard institute (CLSI) guideline [29]. The used antibiotics were levofloxacin, chloramphenicol, amikacin, cefoperazone, meropenem, tobramycin, cefepime, and colistin. The MICs were reported and interpreted as sensitive, intermediate, or resistant, according to the CLSI breakpoints indicated in Supplementary Table S2.
2.6. The Killing Curve of Bactericidal Antibiotics against Levofloxacin Persisters
Time-killing curves were performed for selected persister-forming strains with antibiotics, as described previously [30]. Three water-soluble antimicrobial agents (colistin, meropenem, and tobramycin) were selected for time-killing curve assay.
Briefly, the overnight cultures of P. aeruginosa cells, treated with 100X MIC of levofloxacin (~2–5 × 103 CFU/mL), were pelleted (at 6000 rpm for 20 min), washed, and resuspended in 1 mL of a double-strength broth. The stock solutions of antimicrobial agents were prepared with concentrations equivalent to their 2x MICs and added to the suspension of persister cells. Tubes were incubated in a shaker incubator (200 rpm) at 37 °C for up to 24 h. Viable counts were made at zero time and after that at 1, 3, 5 and 24 h. The experiments were performed in triplicate with three independent cultures.
2.7. Biofilm Formation Capacity Test
The quantitative assay of biofilm formation was performed according to Stepanovic et al. [31]. A suspension of tryptone soy broth (TSB, Oxoid, Hampshire, UK) was prepared from the overnight culture of each isolate and adjusted to 106 CFU/mL in the freshly prepared culture broth of TSB. Aliquots of 200 µL of the bacterial suspension were transferred to the wells of a flat-bottom microtiter plate and incubated at 37 °C for 48 h. The TSB was removed gently and distilled water was used to wash the plates to remove any planktonic cells with subsequent air drying. After 20 min of treatment with 200 µL of 99% fixing methanol, the biofilm was then stained with 200 µL of 1% crystal violet (CV) solution for 15 min. After washing of the plate, 33% glacial acetic acid was used as a solvent for CV and the optical density (OD) of the solubilized dye was measured at 570 nm with a spectrofluorimeter (Biotek, Winooski, VT, USA). Wells containing only media were included in the study to serve as a negative control. The experiment was performed in triplicate.
The cut-off optical density (ODc) was calculated as three times the standard deviations above the mean OD of the negative control. The tested strains were classified into: non-biofilm producer (OD ≤ ODc), weak biofilm producer (OD > ODc, but ≤2 × ODc), moderate biofilm producer (OD > 2 × ODc, but ≤ 4 × ODc), and strong biofilm producer (OD > 4 × ODc), as described previously [31].
2.8. Genomic DNA (gDNA) Extraction and Whole Genome Sequencing
The GeneJET™ Genomic DNA Purification Kit was used for gDNA extraction. The DNA purification protocol was performed according to the manufacturer’s instructions. The purified gDNA was used immediately or stored at −20 °C. For the whole-genome sequencing, genomic, short-read sequencing was performed on an Illumina NextSeq 500 sequencer (Illumina, The Netherlands) using a library prepared with MiSeq v3 (2 × 75 bp) reagent kit.
2.9. Genome Annotation and Phylogeny
The raw reads were filtered for adaptor sequences and low-quality bases using Trimmomatic v0.36 [32] and assembled de novo using Unicycler v0.4.8 [33]. For both tools, the default parameters were used. Only contigs > 500 bp were considered for further analysis. Genome-based annotation was carried out using PROKKA v1.11 [34]. Single nucleotide variants (SNVs) were identified by mapping filtered reads against the closed reference genome of P. aeruginosa PAO1 (NC_002516) using Snippy v4.3.6 (https://github.com/tseemann/snippy (accessed on 5 January 2023)). Furthermore, the isolates were identified using TYGC server (https://tygs.dsmz.de/ (accessed on 5 January 2023)), which compared the genomes to type-strain and calculated the in-silico DNA–DNA hybridization score [35].
All the available (as of June 2022) completely closed genomes of 445 isolates were downloaded from NCBI using the NCBI-genome-download script (https://github.com/kblin/ncbi-genome-download (accessed on 5 January 2023)). The phylogeny was constructed based on the MASH distance using Ondov et al. [36]. The multilocus sequence typing was carried out using MLST script (https://github.com/tseemann/mlst (accessed on 5 January 2023)). The pangenome atlas was created using gView Server (https://server.gview.ca/ (accessed on 5 January 2023)).
2.10. Resistome Analysis
The gene differences among the isolates were studied by calculating the core and pan-genome using the Panaroo tool v1.2.3 [37]. Virulence gene profiling was performed by aligning the amino-acid sequences of all genes (using 70% query coverage and 80% nucleotide identity) against the ‘virulence factor database (VFDB) using the Diamond v0.8.36.98 tool [38]. Antibiotic resistance gene profiling was carried out using NCBI’s AMRfinder (https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/AMRFinder/ (accessed on 8 January 2023)) as well as the ‘resistance gene identifier’ v5.05.5 against the CARD database (v3.0.3) [39]. The circular image was created using the BRIG tool v0.95 [40]. The heatmap for clustering AMR elements was constructed using ClustVis tool [41]. All the bioinformatics tools were run with default parameters, unless specified.
3. Results
3.1. Separation of P. aeruginosa Persister Cells and Determination of Their Antimicrobial Susceptibility
Thirty-eight P. aeruginosa isolates taken from chronically ill patients were studied for persister cells formation using 100X MIC of levofloxacin. From the thirty-eight P. aeruginosa isolates, only three isolates (8%) yielded persisters; these three isolates were isolated from respiratory tract infections and from patients with cystic fibrosis and were named P1, P2, and P3.
The resistance profile of these persisters showed increased resistance patterns to seven tested antimicrobials (Table 1). In comparison to the reference strain PAO1, all persister strains were resistant to meropenem, levofloxacin, chloramphenicol, and tobramycin (100%), while 90% of these strains were resistant to cefoperazone and colistin. Respective resistance was observed against cefepime (60%) and amikacin (70%). The three persister strains were found to be MDR.
Table 1.
Resistance profiles of the recovered persister strains and PAO1 standard strain.
| Strain Code | MICs (µg/mL) | MDR | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CHL | AK | LEV | CEP | FEP | MEM | CT | TOB | ||
| PAO1 | 2 (S) | 4 (S) | 0.5 (S) | 4 (S) | 1 (S) | 0.5 (S) | 1 (S) | 2 (S) | - |
| P1 | 256 (R) | 128 (R) | 8 (R) | 256 (R) | 64 (R) | 64 (R) | 16 (R) | 128 (R) | 5 |
| P2 | 512 (R) | 512 (R) | 8 (R) | 512 (R) | 128 (R) | 16 (R) | 32 (R) | 128 (R) | 5 |
| P3 | 256 (R) | 128 (R) | 16 (R) | 512 (R) | 64 (R) | 16 (R) | 16 (R) | 512 (R) | 5 |
S: Sensitive, R: Resistant, MDR: Multidrug Resistant, AK: Amikacin, CEP: Cefoperazone, FEP: Cefepime, MEM: Meropenem, CT: Colistin, TOB: Tobramycin, LEV: Levofloxacin, CHL: Chloramphenicol.
3.2. Survival Curve of Persisters in the Presence of Levofloxacin, Tobramycin, Meropenem, and Colistin
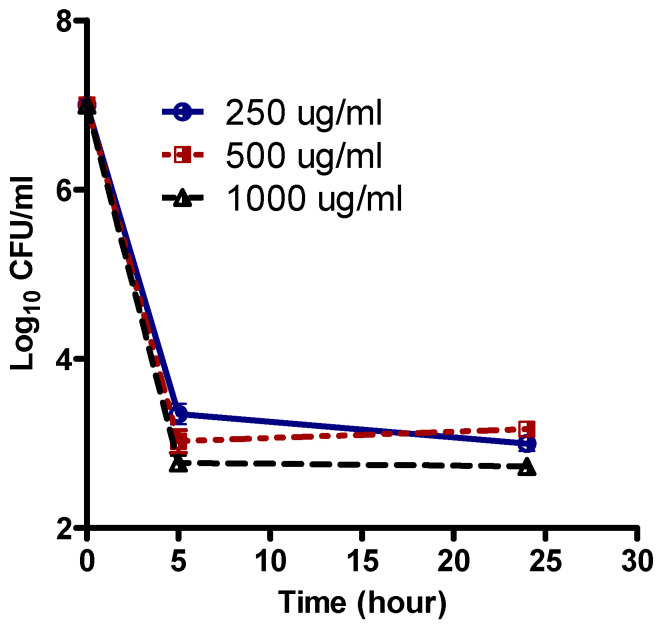
After overnight growth in Mueller–Hinton (MH) broth, (Oxoid, Hampshire, England), cells were exposed to increasing concentrations of levofloxacin (250, 500, 1000 μg/mL). The analysis of the number of survivor cells after 5 and 24 h (Figure 1) showed that the bulk of the population was effectively killed with 250 µg/mL levofloxacin. The killing curves were followed on the original strains.
Figure 1.
Population analysis curve of levofloxacin against P. aeruginosa persister (isolate P1). Levofloxacin was added at time zero at the indicated concentrations to a stationary-phase culture. Data are expressed as the Mean ± SEM of three independent experiments.
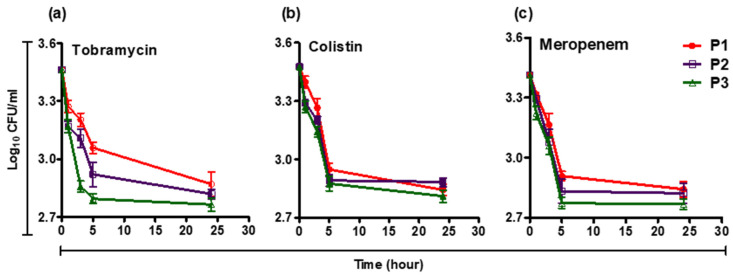
A small fraction of cells (~10−3) were tolerant for up to 1000 µg/mL levofloxacin (~100X MIC), The persisters after treatment with levofloxacin were exposed to lethal concentrations of colistin, meropenem, or tobramycin, and the survivor’s numbers were followed for 24 h (Figure 2).
Figure 2.
Biphasic time killing curves of (a) tobramycin, (b) colistin, and (c) meropenem against levofloxacin persisters of P. aeruginosa. After treatment with 100X-MIC of levofloxacin, P. aeruginosa strains (P1, P2, P3) were exposed to bactericidal concentrations of antibiotics and viable counts were determined at different time points (0, 1, 3, 5, and 24 h). Data are expressed as the Mean ± SEM of three independent experiments.
3.3. Biofilm Forming Capacity of Isolates
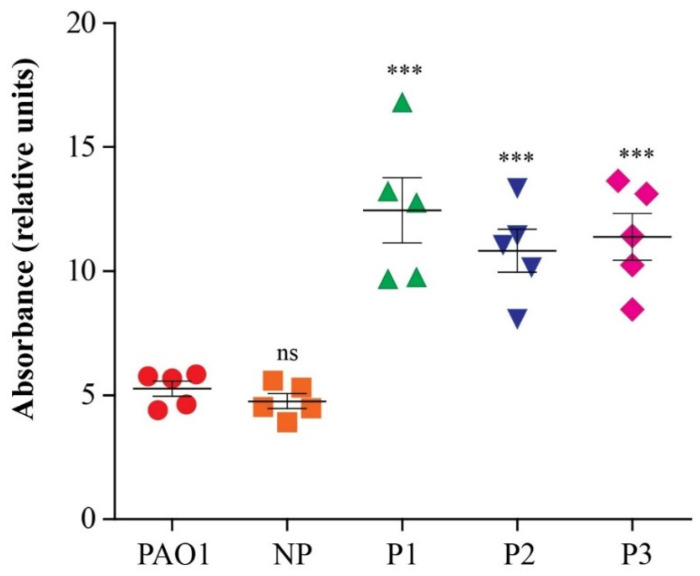
The biofilm-forming capacity of the three levofloxacin-persister isolates in addition to the PAO1 strain and a non-persister forming isolate (clinical isolate which had no survival cells after treatment with high concentration (100X MIC) of levofloxacin) was quantified using a spectrophotometric assay. All the tested isolates were strong biofilm producers, as the measured OD was greater than 4ODc (the ODc was 0.1). The persister-forming isolates showed the highest absorbance compared to PAO1, indicating their higher biofilm-forming capacity; the relative absorbance was calculated in comparison to the absorbance of ODc (Table 2 and Figure 3).
Table 2.
Biofilm forming capacity of persister strains compared to PAO1.
| Isolate | Absorbance (OD) at 570 nm | Biofilm Forming Capacity |
|---|---|---|
| P1 | 0.967 | S |
| P2 | 0.831 | S |
| P3 | 0.872 | S |
| PAO1 | 0.584 | S |
| NP | 0.547 | S |
| ODc | 0.1 | Control (no biofilm) |
S: strong biofilm forming, NP: non persister isolate.
Figure 3.
Biofilm formation by persister isolates compared to PAO1 standard strain and a non-persister (NP) clinical isolate. The quantitative assay of biofilm formation was performed using a spectrophotometric assay. All isolates formed strong biofilm with the highest strength observed with persister isolates. Data are expressed as the Mean ± SEM of three independent experiments. Two-tailed unpaired T-tests were employed to analyze significance; ns denotes no significance, *** p < 0.001.
3.4. Whole Genome Sequencing (WGS) Reveals Distinct Features of the Persisters
All the genome sequences were submitted to the public database under the project accession number PRJNA890046. Genomes of the P1, P2, and P3 isolates were deposited as 1 (SRR21888463), 2 (SRR21888462), and 4 (SRR21888461). Isolates were identified to be Pseudomonas aeruginosa, as all the three isolates showed more than 70% DDH to type strain. The genome sizes of the persister isolates were 4,471,320 bp, 4,951,902 bp, and 4,661,420 bp for the isolates P1, P2, and P3, respectively, compared to 6,264,404 bp the genome size of the PAO1_NC_002516 reference strain. The genomes were distinguished into 1547, 1688, and 1706 contigs, respectively. The numbers of protein-coding sequences within the genomes were 4606, 5130, and 4928 genes, for the persister isolates P1, P2, and P3, respectively, compared to the 5671 coding genes for the PAO1 reference strain. The GC contents of the genomes were 64.38, 63.91, and 64.14%, respectively, compared to 66.56% for the PAO1 genome. WGS confirmed species-level identification of all isolates with percent identity values of 98.5% for all assemblies to the reference strain PAO1.
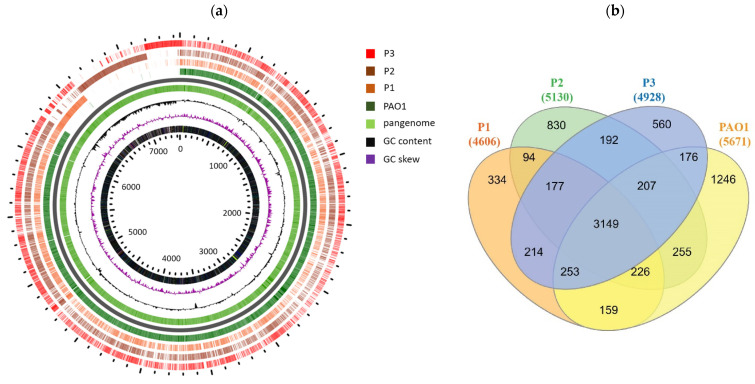
The circular map representing the entire genome assembly is shown in Figure 4. The circular image depicts the comparison of the genome of isolates P1, P2, and P3 to the reference genome of the PAO1 (NC_002516) strain. From the inner to outer circle is depicted the nucleotide position, GC content, GC skew (of the PAO1), and genes of isolates P1, P2, and P3, respectively. In addition, a comparison of the PAO1 genome with persister genomes revealed the presence of a number of genes (2401) in the persister genomes that were absent in the PAO1 genome (Supplementary Table S3).
Figure 4.
Whole genome sequencing (WGS) of the persister isolates. (a) Genomic comparison of the PAO1 and studied isolates. Using gView Server, the pan genome of the four isolates was constructed and the genes were mapped against it. (b) Venn diagram of genes in the three persister P. aeruginosa isolates and PAO1 strain. The four strains shared in 3149 genes, while 334, 830, and 560 genes are unique to P1, P2, and P3, respectively.
3.5. Antimicrobial Resistance Elements
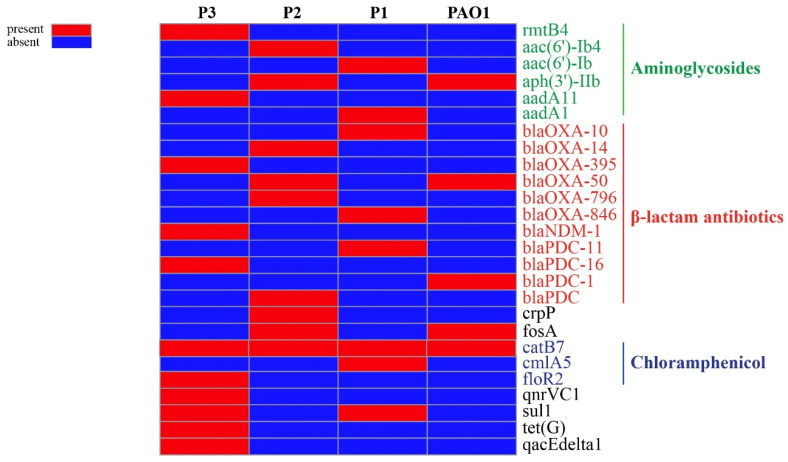
Resistome profiling was carried out to detect the resistance elements contributing to the MDR-pattern of these persisters forming isolates against anti-pseudomonas antibiotics. The analysis of antibiotics and antiseptic resistance elements pinpointed 26 resistance genes distributed among the isolates, 21 of them only exist in these persister isolates and 4 are shared with the PAO1 genome, while the blaPDC-1 gene is present in only the standard strain of PAO1 (Figure 5). These resistance elements represent a cocktail of resistance elements against most of the clinically used antibiotics, including the major aminoglycosides modifying enzymes acetyltransferases: aac(6′)-Ib and aac(6′)-Ib4, phosphotransferase: aph(3′)-IIb, nucleotidyltransferases: aadA1 and aadA11 beside the 16S-rRNA methylase rmtB4. Variants and sub-variants of different classes of β-lactamases were detected, including blaOXA-10, blaOXA-14, bla OXA-395, bla OXA-50, bla OXA-796, and bla OXA-864. Several pseudomonas-derived cephalosporinase (PDC) variants were also detected, i.e., bla PDC, blaPDC-1, blaPDC-11, and blaPDC-16 and finally the blaNDM-1 gene, which encodes for the New Delhi metallo-β-lactamase 1. Acquired resistance elements against chloramphenicol were also detected, including catB7, cmlA5, and floR2, while individual resistance factors against tetracycline (tetG), fosfomycin (fosA), sulphonamides (sul1), novel ciprofloxacin-modifying enzyme encoding gene crpP, quinolone resistance determinant (qnrVC1), and the antiseptic resistance gene (qacEΔ1) are mainly found in isolate number 3. The broad collection of antibiotic resistance genes found in the genomes are the reason for the MDR of these isolates.
Figure 5.
Resistome profiling of antimicrobial modifying enzymes in persister isolates compared to PAO1 standard strain. Antibiotic resistance gene profiling was carried out using NCBI’s AMRfinder as well as ‘resistance gene identifier’ v5.05.5 against the CARD database. Heatmap was constructed using Clustvis online tool.
Aside from the antibiotic modifying enzymes, the persisters’ genome harbored gene clusters that correspond to the efflux pump-mediated resistance. Genome analysis identified 24–30 genes shared among the three isolates. These genes code for membrane fusion proteins (MFP), resistance nodulation division proteins (RND), and outer membrane proteins (OMP) that constitute efflux pump systems (Table 3). Based on the identified gene clusters, nine multidrug resistance efflux pumps (MexAB-OprM, MexCD-OprJ, MexEF-OprN, MexGHI-OpmD, MexJK-OpmH, MexMN-OprM, MexPQ-OpmE, MexXY-OprM, and MexVW-OprM) were chromosomally encoded in the persister isolates. Moreover, mutagenic analysis of the MexAB-OprM efflux pump regulators recognized a few point mutations in the MexAB-OprM repressors (MexR, NalD, and NalC).
Table 3.
Genotypic characteristics of efflux pump systems in P. aeruginosa persister isolates.
| Persister Number | Efflux Pump Component | Efflux Pump | Mutation in MexAB-OprM Efflux Pump Regulatory Proteins | ||
|---|---|---|---|---|---|
| MFP | RND | OMP | |||
| P1 | MexA | MexB | OprM | MexAB-OprM | MexR (V126E), NalC (S209R, G71E) |
| MexC | MexD | OprJ | MexCD-OprJ | ||
| MexJ | MexK | OpmH | MexJK-OpmH | ||
| MexM | MexN | OprM | MexMN-OprM | ||
| MexP | MexQ | - | |||
| MexV | MexW | OprM | MexVW-OprM | ||
| P2 | MexA | MexB | OprM | MexAB-OprM | MexR (V126E), NalC (S209R, G71E) |
| MexC | MexD | OprJ | MexCD-OprJ | ||
| MexE | MexF | OprN | MexEF-OprN | ||
| MexG | MexH,I | - | |||
| MexJ MexM |
MexK MexN |
OpmH OprM |
MexJK-OpmH MexMN-OprM |
||
| MexP | MexQ | - | |||
| MexV | MexW | OprM | MexVW-OprM | ||
| P3 | MexA | MexB | OprM | MexAB-OprM | MexR (V126E), NalC (S209R, G71E) |
| MexC | MexD | OprJ | MexCD-OprJ | ||
| MexE | MexF | OprN | MexEF-OprN | ||
| MexG | MexH,I | OpmD | MexGHI-OpmD | ||
| MexJ | MexK | OpmH | MexJK-OpmH | ||
| MexM | MexN | OprM | MexMN-OprM | ||
| MexP | MexQ | - | |||
| MexV | MexW | OprM | MexVW-OprM | ||
3.6. Global Phylogenetic Analysis of the Persister Isolates
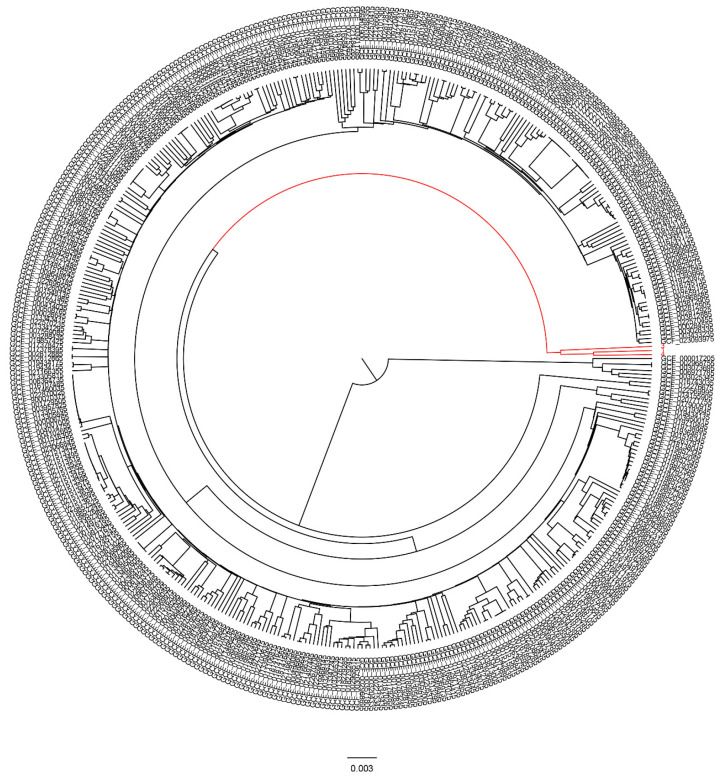
The phylogenomic location of the studied isolates was performed. Completely closed genomes of 445 P. aeruginosa isolates were downloaded from the public database and compared with these persister-forming isolates. When phylogenomically compared to previously known P. aeruginosa strains from different countries, the studied isolates were placed in a unified but separate clade (Figure 6 and Supplementary Table S4).
Figure 6.
Phylogenetic tree analysis of persister isolates compared to the genome of 445 P. aeruginosa isolates downloaded from the NCBI database. The phylogenetic tree was constructed based on the MASH distance using Ondov et al. [35]. Our persister isolates were placed in a unified but separate clade (indicated by the red color).
4. Discussion
Chronic infections represent a terrible therapeutic challenge and have a puzzling property. These infections are difficult and even impossible to eradicate. There is growing evidence that bacterial persisters are involved in the relapse of bacterial infections [42]. Th characterization of persister cells is important, not only in regarding the understanding of population dynamics but also concerning antibiotic tolerance in chronic-infections therapy [43]. Few studies have assayed persister formation in clinical isolates from complex host environments [44,45]. In the current study, the characterization of P. aeruginosa persisters isolated from chronic respiratory tract infections was performed using phenotypic and genotypic methods.
Initially, we isolated P. aeruginosa multidrug-tolerant persister forming strains. Although 100-fold MIC is unlikely to be achieved in the therapy of clinical infections, the application of high doses of antimicrobials would select for high persister mutants. The treatment with 100-fold MIC is a method of choice to identify highly drug-tolerant persister cells of P. aeruginosa [46]. Accordingly, in our study, three different P. aeruginosa isolates were identified as persister-forming strains. These isolates were recovered from 38 P. aeruginosa clinical isolates (8% persister-forming capacity). A low persister level, approaching 1% of the total population, was reported by Mulcahy et al. [44]. While a higher persister forming percentage (19%) was reported recently [45], their isolates were only obtained from CF infections, which is a deadly disease accompanied with a chronic respiratory infection.
Two of the three persister isolates (P2 and P3) formed both large colonies and small colonies on agar plates. Similarly, it was reported that S. aureus persisters induce a phenotypic change into small colony variants due to genomic inversion [47]. Furthermore, a recent study indicated that P. aeruginosa can persist for a long time in an acidic environment, through phenotypic alteration to a slow-growing colony phenotype [13].
The frequency of persisters was calculated for each isolate, and it ranged from 0.0017 to 0.0027 (approximately 10−3, meaning 1 out of 103 cells can form persister). Our result is quite consistent with the results of Salcedo-Sora and Kell [48], who reported a persister level within the range of 0.0012–0.0016. However, our results reveal much higher frequencies than the 10−5 and the 10–6 reported previously [24,49].
P. aeruginosa strains are intrinsically resistant to a variety of antimicrobials, and can further develop resistance during therapy [3]. In this study, the MIC results revealed that the persister-forming isolates were highly resistant to most of the tested antibiotics. Similarly, a recent study of Pseudomonas CF isolates revealed that more than 75% of the CF isolates were resistant to at least half of the antibiotics. Additionally, 25% of their isolates were found to be resistant to all the tested antibiotics, and only 36% of CF-isolates were resistant to tobramycin [50]. Furthermore, it was reported that persister (Hip) isolates were resistant to ciprofloxacin by 72.4% [45]. In this investigation, the time-killing curves using colistin, meropenem, and tobramycin revealed that the curves substantially differentiate not only between clinical isolates of P. aeruginosa but also between the different antibiotics. For example, isolate P1 showed a higher number of survival after exposure to tobramycin and meropenem, while isolate P2 had more survivor cells upon exposure to colistin.
The threat of P. aeruginosa is complicated due to its ability to form biofilms that provide a protected environment to bacterial cells which help in tolerating various stresses, including antimicrobials [51,52]. It was reported that the formation of P. aeruginosa persisters and biofilms are interrelated [53]. The biofilm formation was evaluated in the three persister-forming isolates, in addition to the PAO1 standard strain and a non-persister clinical isolate. All the tested isolates were strong biofilm formers; however, the persister isolates had stronger biofilms (as their OD was twice that of the PAO1 strain). In contrast, a previous study reported that the biofilm of PAO1 and their persister strain had the same strength [26].
Genomics has been a key element in the study of P. aeruginosa evolution. It has been reported that P. aeruginosa strains had a variable genome size of approximately 5–7 Mbp [54]. The omnipresence of P. aeruginosa may be related to its high genome plasticity. The genome of the P. aeruginosa PAO1 standard strain was sequenced approximately two decades ago [54]. However, the genetic mediators of persister formation for P. aeruginosa are poorly understood [4]. In the current study, genome sequencing of persister-forming isolates revealed that they had smaller genomes (4.66–4.9 Mbp), compared to PAO1-genome (6.3 Mbp) sequenced previously [55], and the genome of Pseudomonas CF-isolates (6.36 Mbp) recently sequenced [50]. The smaller genome size for our isolates could be due to the short-reads-based sequencing and assembly used in our study. Furthermore, it is documented that P. aeruginosa can customize its genome as individual strains have the ability to acquire or discard genomic segments to fit their needs for survival in virtually any environment [56].
Resistome profiling indicated the presence of a broad collection of antibiotic-resistance genes in our persisters’ genome, with 25 resistance genes distributed in persister-isolates, most of which confer resistance to β-lactams (11 genes), aminoglycoside resistance (6 genes), and chloramphenicol resistance (3 genes), while resistance to other antibiotic classes is mediated by one gene each. CARD analysis revealed that the CF-isolates genome contains a high number of resistance genes, including beta-lactam resistance genes (n = 6), efflux pumps (n = 37), antibiotic inactivation enzymes (n = 8), and only one aminoglycoside resistance gene [50].
Multidrug efflux pumps constitute a group of antibiotic resistance elements that are a core part of bacterial genomes [57]. The P. aeruginosa PAO1 genome contains several drug efflux systems, predominantly of the RND and MFS families [56]. In the current study, several multidrug efflux tripartite pumps that encoded in operon were recognized at the genome level in our persister isolates (6–8 classes of the RND multidrug efflux pump-operon in each genome), which explains their profound resistance against tested antibiotics. Consistent with our results, MexA/B pumps were reported to be closely associated with antimicrobial resistance and biofilm formation in clinical P. aeruginosa isolates recovered from respiratory infections [58].
It was reported previously that mutations in mexR, nalC, and nalD regulatory genes yield a MDR phenotype in P. aeruginosa [59,60]. Surprisingly, the identified point mutations within the MexAB-OprM efflux pump regulatory genes (MexR, NalC) have been previously described in clinical MDR–P. aeruginosa isolates [61,62]. In addition, an inactivating mutation in mexZ, encoding a repressor of the MexXY-OprM pump, was reported in P. aeruginosa persister isolates from CF patients [44]. All of these mutations favored the overexpression of these pump systems and caused increased resistance to different antimicrobials. Future work will focus on studying the transcriptional changes associated with persister formation to identify potential targets for the anti-persister drug.
Our results in resistome profiling of persister isolates allow the identification of genes that are likely to be involved in the antibiotic resistance of P. aeruginosa persister isolates. Knowledge of the complete genome sequence and encoded processes provides a wealth of information for the discovery of new antibiotic targets and for the development of more effective strategies to treat the life-threatening opportunistic infections caused by P. aeruginosa in humans.
Furthermore, phylogenetic analysis indicated that persister isolates belong to a unified but separate clade rather than the deposited P. aeruginosa strains in the public repositories. Further investigation is required to explore the validity and features of this clade. Similarly, the phylogenetic analysis of P. aeruginosa CF isolates performed by Datar et al. [50] revealed their unique clustering and indicate their location in different clades. To the best of our knowledge, our study is the first to study the genome sequence of persister isolates from local hospitals in Egypt.
5. Conclusions
The present study characterized, for the first time in Egypt, P. aeruginosa persister isolates. The persister isolates were strong biofilm producers and have a highly resistant phenotype when compared to the PAO1 standard strain. These results were confirmed by genome sequencing revealing a large number of resistance genes. Furthermore, WGS revealed a smaller genome that belong to a unified but separate clade compared to the genome of other P. aeruginosa strains deposited in the GenBank.
Acknowledgments
The authors would like to acknowledge the medical staff at clinical laboratories in Zagazig University Hospital and El-Ahrar Educational Hospital for providing clinical specimens.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens12030426/s1, Table S1: The batch number of antimicrobial agents used in the current study; Table S2: CLSI breakpoints of antimicrobial agents used in the current study; Table S3: The list of genes that found in persister strains genome and absent from PAO1 genome; Table S4: List of the 445 P. aeruginosa isolates downloaded from the NCBI database and used for Phylogenetic tree analysis
Author Contributions
F.E.S., A.A.K. and A.M.E.-G. contributed to the conception and design of the study and supervised the work; A.A.B. performed laboratory analysis; S.D., G.Y., A.M. and M.A.M. performed whole genome sequencing. All authors contributed to data analysis and interpretation, in addition to the writing and reviewing process of the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable as the specimens are provided by the hospital clinical laboratory and there was no direct contact with patients.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The work was supported by the Transregional Collaborative Research Centre SFB TR 84 (Innate Immunity of the Lung); TP A4 to M.A.M.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Kadry A.A., Serry F.M., El-Ganiny A.M., El-Baz A.M. Integron occurrence is linked to reduced biocide susceptibility in multidrug resistant Pseudomonas aeruginosa. Br. J. Biomed. Sci. 2017;74:78–84. doi: 10.1080/09674845.2017.1278884. [DOI] [PubMed] [Google Scholar]
- 2.Abbas H.A., El-Ganiny A.M., Kamel H.A. Phenotypic and genotypic detection of antibiotic resistance of Pseudomonas aeruginosa isolated from urinary tract infections. Afr. Health Sci. 2018;18:11–21. doi: 10.4314/ahs.v18i1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pang Z., Raudonis R., Glick B.R., Lin T.J., Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019;37:177–192. doi: 10.1016/j.biotechadv.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 4.Cameron D.R., Shan Y., Zalis E.A., Isabella V., Lewis K. A genetic determinant of persister cell formation in bacterial pathogens. J. Bacteriol. 2018;200:e00303–e00318. doi: 10.1128/JB.00303-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beharry Z., Palzkill T. Functional analysis of active site residues of the fosfomycin resistance enzyme FosA from Pseudomonas aeruginosa. J. Biol. Chem. 2005;280:17786–17791. doi: 10.1074/jbc.M501052200. [DOI] [PubMed] [Google Scholar]
- 6.Willenborg J., Willms D., Bertram R., Goethe R., Valentin-Weigand P. Characterization of multi-drug tolerant persister cells in Streptococcus suis. BMC Microbiol. 2014;14:120. doi: 10.1186/1471-2180-14-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bigger J.W. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet. 1944;244:497–500. doi: 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 8.Moyed H.S., Bertrand K.P. HipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher R.A., Gollan B., Helaine S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017;15:453–564. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 10.Pu Y., Zhao Z., Li Y., Zou J., Ma Q., Zhao Y., Ke Y., Zhu Y., Chen H., Baker M.A., et al. Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol. Cell. 2016;62:284–294. doi: 10.1016/j.molcel.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thi M.T.T., Wibowo D., Rehm B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020;21:8671. doi: 10.3390/ijms21228671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seleem N.M., Abd El Latif H.K., Shaldam M.A., El-Ganiny A.M. Drugs with new lease of life as quorum sensing inhibitors: For combating MDR Acinetobacter baumannii infections. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1687–1702. doi: 10.1007/s10096-020-03882-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed F., Mirani Z.A., Mirani P.N., Imdad M.J., Khan F.Z., Khan M.N., Khan A.B., Li Y., Zhao Y. Pseudomonas aeruginosa response to acidic stress and imipenem resistance. Appl. Sci. 2022;12:8357. doi: 10.3390/app12168357. [DOI] [Google Scholar]
- 14.Soares A., Alexandre K., Etienne M. Tolerance and persistence of Pseudomonas aeruginosa in biofilms exposed to antibiotics: Molecular mechanisms, antibiotic strategies and therapeutic perspectives. Front. Microbiol. 2020;11:2057. doi: 10.3389/fmicb.2020.02057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiuff C., Zappala R.M., Regoes R.R., Garner K.N., Baquero F., Levin B.R. Phenotypic tolerance: Antibiotic enrichment of noninherited resistance in bacterial populations. Antimicrob. Agents Chemother. 2005;49:1483–1494. doi: 10.1128/AAC.49.4.1483-1494.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwan B.W., Valenta J.A., Benedik M.J., Wood T.K. Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 2013;57:1468–1473. doi: 10.1128/AAC.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis K. Persister cells. Annu. Rev. Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 18.Kussell E., Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 19.Kaspy I., Rotem E., Weiss N., Ronin I., Balaban N.Q., Glaser G. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 2013;4:3001. doi: 10.1038/ncomms4001. [DOI] [PubMed] [Google Scholar]
- 20.Germain E., Castro-Roa D., Zenkin N., Gerdes K. Molecular mechanism of bacterial persistence by HipA. Mol. cell. 2013;52:248–254. doi: 10.1016/j.molcel.2013.08.045. [DOI] [PubMed] [Google Scholar]
- 21.Pacios O., Blasco L., Bleriot I., Fernandez-Garcia L., Ambroa A., López M., Bou G., Cantón R., Garcia-Contreras R., Wood T.K., et al. (p) ppGpp and its role in bacterial persistence: New challenges. Antimicrob. Agents Chemother. 2020;64:e01283-20. doi: 10.1128/AAC.01283-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maisonneuve E., Castro-Camargo M., Gerdes K. (p) ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2013;154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 23.Wang T., El Meouche I., Dunlop M.J. Bacterial persistence induced by salicylate via reactive oxygen species. Sci. Rep. 2017;7:43839. doi: 10.1038/srep43839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hazan R., Maura D., Que Y., Rahme L.G. Assessing Pseudomonas aeruginosa persister/antibiotic tolerant Cell. In: Filloux A., Ramos J.L., editors. Pseudomonas Methods and Protocols. Methods in Molecular Biology. Volume 1149. Humana; New York, NY, USA: 2014. pp. 699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood T.K., Knabel S.J., Kwan B.W. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 2013;79:7116–7121. doi: 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Žiemytė M., Carda-Diéguez M., Rodríguez-Díaz J.C., Ventero M.P., Mira A., Ferrer M.D. Real-time monitoring of Pseudomonas aeruginosa biofilm growth dynamics and persister cells’ eradication. Emerg. Microbes Infect. 2021;10:2062–2075. doi: 10.1080/22221751.2021.1994355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller J.H. Determination of viable cell counts: Bacterial growth curves. In: Miller J.H., editor. Experiments in Molecular Genetics. Cold Spring Harbor; New York, NY, USA: 1972. pp. 31–36. [Google Scholar]
- 28.De Groote V.N., Fauvart M., Kint C.I., Verstraeten N., Jans A., Cornelis P., Michiels J. Pseudomonas aeruginosa fosfomycin resistance mechanisms affect non-inherited fluoroquinolone tolerance. J. Med. Microbiol. 2011;60:329–336. doi: 10.1099/jmm.0.019703-0. [DOI] [PubMed] [Google Scholar]
- 29.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing; 27th Informational Supplement; M100–S28. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. [Google Scholar]
- 30.Narayanaswamy V.P., Keagy L.L., Duris K., Wiesmann W., Loughran A.J., Townsend S.M., Baker S. Novel Glycopolymer eradicates antibiotic- and CCCP-induced persister cells in Pseudomonas aeruginosa. Front. Microbiol. 2018;9:1724. doi: 10.3389/fmicb.2018.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stepanović S., Vuković D., Hola V., Di Bonaventura G., Djukić S., Cirković I., Ruzicka F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115:891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 32.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 35.Meier-Kolthoff J.P., Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ondov B.D., Treangen T.J., Melsted P., Mallonee A.B., Bergman N.H., Koren S., Phillippy A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tonkin-Hill G., MacAlasdair N., Ruis C., Weimann A., Horesh G., Lees J.A., Gladstone R.A., Lo S., Beaudoin C., Floto R.A., et al. Producing polished prokaryotic pangenomes with the panaroo pipeline. Genome Biol. 2020;21:180. doi: 10.1186/s13059-020-02090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 39.Jia B., Raphenya A.R., Alcock B., Waglechner N., Guo P., Tsang K.K., Lago B.A., Dave B.M., Pereira S., Sharma A.N., et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alikhan N.-F., Petty N.K., Ben Zakour N.L., Beatson S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metsalu T., Vilo J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen N.R., Lobritz M.A., Collins J.J. Microbial persistence and the road to drug resistance. Cell Host Microbe. 2013;13:632–642. doi: 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orman M.A., Brynildsen M.P. Establishment of a method to rapidly assay bacterial persister metabolism. Antimicrob. Agents Chemother. 2013;57:4398–4409. doi: 10.1128/AAC.00372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mulcahy L.R., Burns J.L., Lory S., Lewis K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 2010;192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartell J.A., Cameron D.R., Mojsoska B., Haagensen J.A., Pressler T., Sommer L.M., Lewis K., Molin S., Johansen H.K. Bacterial persisters in long-term infection: Emergence and fitness in a complex host environment. PLoS Pathogens. 2020;16:e1009112. doi: 10.1371/journal.ppat.1009112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lechner S., Lewis K., Bertram R. Staphylococcus aureus persisters tolerant to bactericidal antibiotics. J. Mol. Microbiol. Biotechnol. 2012;22:235–244. doi: 10.1159/000342449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guérillot R., Kostoulias X., Donovan L., Li L., Carter G.P., Hachani A., Vandelannoote K., Giulieri S., Monk I.R., Kunimoto M., et al. Unstable chromosome rearrangements in Staphylococcus aureus cause phenotype switching associated with persistent infections. Proc. Natl. Acad. Sci. USA. 2019;116:20135–20140. doi: 10.1073/pnas.1904861116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salcedo-Sora J.E., Kell D.B. A Quantitative survey of bacterial persistence in the presence of antibiotics: Towards antipersister antimicrobial discovery. Antibiotics. 2020;9:508. doi: 10.3390/antibiotics9080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coates J., Park B.R., Le D., Şimşek E., Chaudhry W., Kim M. Antibiotic-induced population fluctuations and stochastic clearance of bacteria. Elife. 2018;7:e32976. doi: 10.7554/eLife.32976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Datar R., Coello Pelegrin A., Orenga S., Chalansonnet V., Mirande C., Dombrecht J., Perry J.D., Perry A., Goossens H., van Belkum A. Phenotypic and genomic variability of serial peri-lung transplantation Pseudomonas aeruginosa isolates from cystic fibrosis patients. Front. Microbiol. 2021;12:604555. doi: 10.3389/fmicb.2021.604555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karatuna O., Yagci A. Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Ps. aeruginosa respiratory isolates. Clin. Microbiol. Infect. 2010;16:1770–1775. doi: 10.1111/j.1469-0691.2010.03177.x. [DOI] [PubMed] [Google Scholar]
- 52.Asbury J., Jazayeri J.A. The significance of Pseudomonas aeruginosa infection and biofilms to cystic fibrosis patients: The need for the development of new therapies. Adv. Biotechnol. Microbiol. 2018;8:555742. doi: 10.19080/AIBM.2018.08.555742. [DOI] [Google Scholar]
- 53.Drenkard E., Ausubel F.M. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature. 2002;416:740–743. doi: 10.1038/416740a. [DOI] [PubMed] [Google Scholar]
- 54.Jurado-Martín I., Sainz-Mejías M., McClean S. Pseudomonas aeruginosa: An audacious pathogen with an adaptable arsenal of virulence factors. Int. J. Mol. Sci. 2021;22:3128. doi: 10.3390/ijms22063128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M., et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 56.Mathee K., Narasimhan G., Valdes C., Qiu X., Matewish J.M., Koehrsen M., Rokas A., Yandava C.N., Engels R., Zeng E., et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl. Acad. Sci. USA. 2008;105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alcalde-Rico M., Olivares-Pacheco J., Alvarez-Ortega C., Cámara M., Martínez J.L. Role of the multidrug resistance efflux pump MexCD-OprJ in the Pseudomonas aeruginosa quorum sensing response. Front. Microbiolol. 2018;9:2752. doi: 10.3389/fmicb.2018.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhandari S., Adhikari S., Karki D., Chand A.B., Sapkota S., Dhungel B., Banjara M.R., Joshi P., Lekhak B., Rijal K.R. Antibiotic resistance, biofilm formation and detection of mexA/mexB efflux-pump genes among clinical isolates of Pseudomonas aeruginosa in a tertiary care hospital, Nepal. Front. Trop. Dis. 2022;2:810863. doi: 10.3389/fitd.2021.810863. [DOI] [Google Scholar]
- 59.Adewoye L., Sutherland A., Srikumar R., Poole K. The mexR repressor of the mexAB-oprM multidrug efflux operon in Pseudomonas aeruginosa: Characterization of mutations compromising activity. J. Bacteriol. 2002;184:4308–4312. doi: 10.1128/JB.184.15.4308-4312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sobel M.L., Hocquet D., Cao L., Plesiat P., Poole K. Mutations in PA3574 (nalD) lead to increased MexAB-OprM expression and multidrug resistance in laboratory and clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2005;49:1782–1786. doi: 10.1128/AAC.49.5.1782-1786.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suresh M., Nithya N., Jayasree P.R., Vimal K.P., Manish Kumar P.R. Mutational analyses of regulatory genes, mexR, nalC, nalD and mexZ of mexAB-oprM and mexXY operons, in efflux pump hyperexpressing multidrug-resistant clinical isolates of Pseudomonas aeruginosa. World J. Microbiol. Biotechnol. 2018;34:83. doi: 10.1007/s11274-018-2465-0. [DOI] [PubMed] [Google Scholar]
- 62.Amsalu A., Sapula S.A., De Barros Lopes M., Hart B.J., Nguyen A.H., Drigo B., Turnidge J., Leong L.E., Venter H. Efflux pump-driven antibiotic and biocide cross-resistance in Pseudomonas aeruginosa isolated from different ecological niches: A case study in the development of multidrug resistance in environmental hotspots. Microorganisms. 2020;8:1647. doi: 10.3390/microorganisms8111647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.