Abstract
Influenza virus infection remains a threat to human health since viral hemagglutinins are constantly drifting, escaping infection and vaccine-induced antibody responses. Viral hemagglutinins from different viruses display variability in glycan recognition. In this context, recent H3N2 viruses have specificity for α2,6 sialylated branched N-glycans with at least three N-acetyllactosamine units (tri-LacNAc). In this work, we combined glycan arrays and tissue binding analyses with nuclear magnetic resonance experiments to characterize the glycan specificity of a family of H1 variants, including the one responsible for the 2009 pandemic outbreak. We also analyzed one engineered H6N1 mutant to understand if the preference for tri-LacNAc motifs could be a general trend in human-type receptor-adapted viruses. In addition, we developed a new NMR approach to perform competition experiments between glycans with similar compositions and different lengths. Our results point out that pandemic H1 viruses differ from previous seasonal H1 viruses by a strict preference for a minimum of di-LacNAc structural motifs.
Keywords: influenza virus, N-glycan, recognition, glycan array, NMR
Introduction
Influenza A viruses remain a threat to human health. Globally, the World Health Organization (WHO) estimates 290 000 to 650 000 annual deaths. One barrier to the transmission of avian viruses in humans is receptor specificity. Hemagglutinins of avian and human viruses are characterized by their different preferences for α2,3 (avian-type receptor) and α2,6 (human-type receptor) galactose-linked sialic acids, respectively.1,2 However, other elements of the glycan sequence are also important factors of HA specificity that contribute to viral transmission between species.3 For instance, recent H3N2 viruses have evolved specificity for α(2-6)-linked N-glycans that terminate poly-N-acetyllactosamine chains (poly-LacNAc chains).4,5 Complex N-glycans in mammalian and higher eukaryotes contain poly-LacNAc chains with a different number of repeats of the disaccharide (Galβ1–4GlcNAc),6,7 which can be branched, further terminated with sialic acid, and decorated with fucose and sulfates.8 In this context, it has been described that sulfation has an impact on glycan recognition of H1 and H5 hemagglutinins.9−11
N-glycans containing poly-LacNAc chains terminating in α2-6 linked sialic acids are relatively rare in the human and ferret respiratory tracts.12−14 We recently showed that biological membranes only need a low number of glycan receptors to be bound by contemporary human H3N2 viruses.4 In particular, the presentation of poly-LacNAc N-glycans is essential for contemporary H3N2 influenza virus binding. However, this analysis has been lacking for H1N1 viruses. Interestingly, in contrast to the observations for the recent H3N2 variants, these viruses do still bind erythrocytes,4,15 strongly suggesting that they display binding capability for N-glycans containing two LacNAc repeats and terminating in α2-6 linked sialic acid.
Studies carried out with glycan arrays have shown that H3 Vic/11 prefers human-type branched N-glycans with at least three LacNAc repeats. The hypothesis is that these glycans can interact with two binding sites of the hemagglutinin trimer in a bidentate binding mode.5 Although we recently showed that the minimal receptor for recent H3N2 viruses is a single arm containing a tri-LacNAc with an α2-6 linked sialic acid,4 data at the molecular level for this specificity for both human H1 and H3 viruses is missing. This is due to the challenging nature of experimentally studying the structure and interactions of poly-LacNAc glycans. In fact, their structural characterization is hampered due to the inherent flexibility of the glycan that highly precludes crystallization studies. In this context, the X-ray structures of a variety of HA/glycan complexes have been reported, but the available electron density only allowed for defining very short-chain oligosaccharides located at the hemagglutinin receptor binding site.16
Herein, we show the power of NMR spectroscopy to derive key details of the recognition features of poly-LacNAc glycans when bound to hemagglutinins. One H6 and three different human H1 variants, including the one responsible for the 2009 pandemic outbreak, have been characterized. Fittingly, the NMR results allow the clear detection of differences in the binding epitope between different HAs, which are in good agreement with the binding preferences determined by glycan arrays. In addition, a new NMR methodology, based on the employment of a paramagnetic molecular probe, has been developed to perform competition experiments between glycans with a similar composition but a different chain length.
Results
In Contrast to Seasonal H1, Pandemic H1 Prefers N-Glycans Displaying di-LacNAc Repeats
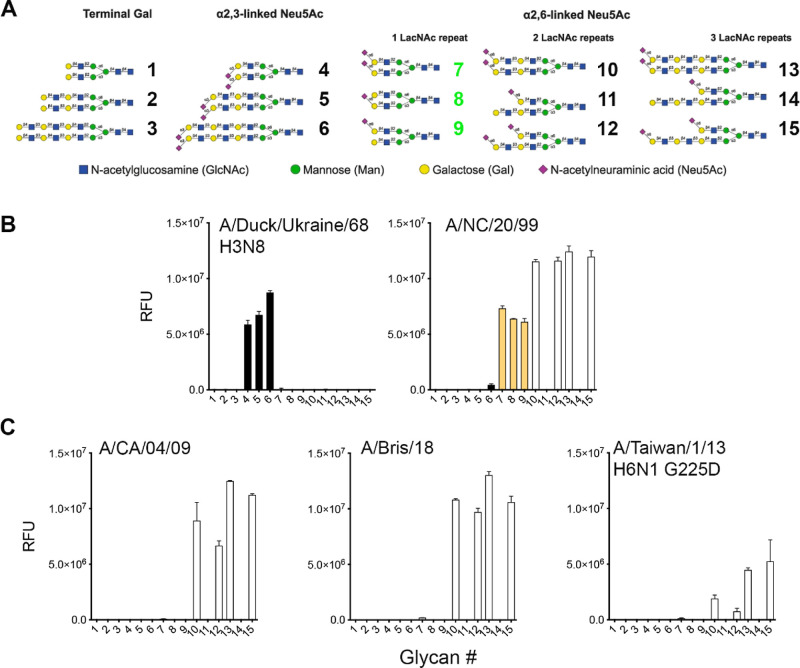
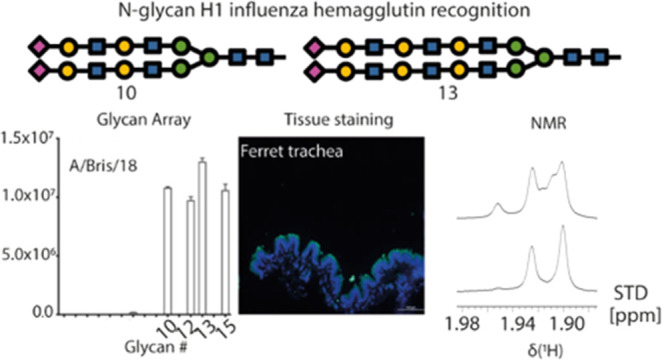
To determine if human H1 prefers to bind N-glycans presenting at least one tri-LacNAc arm with an α2,6 linked sialic acid, we employed a previously described glycan array (Figure 1A).4 Initially, A/Duck/Ukraine/68 H3N8 hemagglutinin was characterized to demonstrate the proper printing in the array of the α2,3 linked sialic acid displaying N-glycans (#4–6), Figure 1B left. Next, three human H1 proteins from distinct moments during their circulation in humans were analyzed. A/New Caledonia/20/99 (A/NC/20/99) represents seasonal H1s (Figure 1B right) circulating before they were replaced by the 2009 new pandemic lineage, represented here by A/CA/04/09 (Figure 1C left). This pandemic strain underwent antigenic changes in the last decade, and a recent vaccine strain A/Bris/18 (Figure 1C middle) was selected as a representative. Finally, an H6 G225D mutant protein (Figure 1C right) was also included since we previously established that this engineered mutant switched specificity from α2,3 to α2,6 sialylated glycans.17 This binding phenotype was highly similar to that of contemporary human H3N2 viruses.5 This protein was specifically employed to understand why zoonotic viruses isolated from humans and adapted to bind human receptors share this specificity.18−21 In this follow-up analysis, we clearly observe reduced responsiveness to the glycan array compared to the H1 isolates. Conversely, the specificity is very similar, with a slight preference for three LacNAc repeats.
Figure 1.
Glycan array analyses of recombinant influenza HA proteins. (A) Glycan structures imprinted on the array. (B) Reference HA of A/duck/Ukraine/68 H3N8 and a pre-pandemic H1 seasonal strain A/NC/20/99. (C) Receptor binding profiles of antigenically distinct pandemic H1 proteins, A/CA/04/09 versus A/Bris/18, and the H6 G225D mutant protein specific for human-type receptors. α2,3 linked structures are given in black bars, α2,6 linked structures with a single LacNAc are given in orange bars (green glycan numbers), and glycans with two and three LacNAc repeats are given in white bars. The Y-axes give the mean relative fluorescent units.
A/NC/20/99 H1 protein displayed a receptor binding profile similar to that of other pre-pandemic H1 proteins we had previously analyzed.22 This protein specifically binds the prototypical human-type receptor, defined as a single LacNAc terminating with an α2,6 linked sialic acid, as demonstrated with compounds #7–9 (Figure 1B right). Interestingly, compounds #11 and #14 are not bound, probably with the non-sialylated longer arm interfering. In contrast, A/CA/04/09 indeed prefers extended branched LacNAc repeats (Figure 1C left), as previously reported by others and us.5,23 The antigenically distant A/Bris/18 displayed an almost identical binding phenotype (Figure 1C middle). Importantly, both were able to bind α2,6 linked sialic acid on two LacNAc repeats, which is in line with their property to hemagglutinate turkey erythrocytes. Finally, the H6 G225D mutant protein displayed lower responsiveness overall (Figure 1C right).
Ferret Upper Respiratory Tract Is Bound by HA Proteins with Human-Type Receptor Specificity
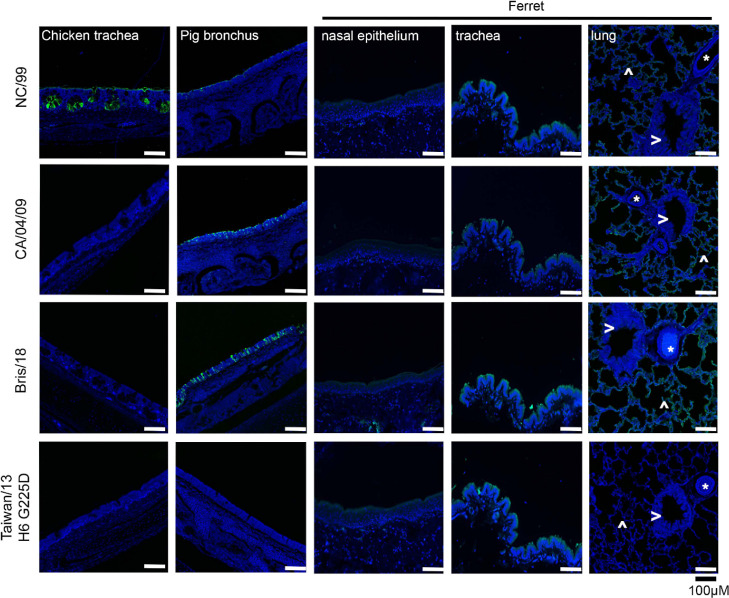
Once the differential receptor specificity of human H1 and human H6 was established, we analyzed the receptor binding profile on tissue sections from biologically relevant respiratory organs (see Figure 2). Tissues display organ and species-specific glycans. In this regard, the chicken trachea displays mainly avian-type receptors, whereas the pig lung expresses mainly human-type receptors (with different sialic acid modifications), and several parts of the ferret respiratory tract closely resemble the human respiratory glycome.24 Especially, the ferret tissues are of importance as they provide the animal model most similar to the human respiratory tract at the glycan level. The green fluorescent protein (GFP) was fused to the C-terminus of trimeric hemagglutinins, as previously described, to aid in the visualization of HA binding to tissues.25 Interestingly, the A/NC/20/99 H1 bound efficiently to the chicken trachea (Figure 2), which is counterintuitive as this is considered as an organ mainly displaying avian-type receptors. However, it is likely that very simple N-glycans with α2,6 linked sialic acids are also expressed but normally not bound by human strains. To confirm sialic acid dependency, we pretreated the chicken trachea with a sialidase, which abrogated binding completely, Figure S1. A binding experiment with the plant lectin SNA which binds α2,6-linked sialic acids further confirmed the presence of this structure. All tissues were bound by A/NC/20/99, albeit with different levels of fluorescent intensity. A/CA/04/09 and Bris/18 displayed almost identical binding profiles, as expected from the highly similar specificity determined by the glycan arrays. In contrast to A/NC/20/99, both A/CA/04/09 and Bris/18 failed to bind chicken trachea, while they showed increased fluorescence in the pig bronchus and the ferret lung. In the ferret lung, it is remarkable that mainly the alveoli are bound with higher intensity compared to A/NC/20/99. Clearly, ferret bronchi are not bound by any HA tested, which could indicate a discontinuity in glycan expression from the lung to the trachea.
Figure 2.
Tissue binding assays to relevant respiratory tissues, origin of tissue is given on top, and the staining HA on the left. Blue is nuclei staining by DAPI (4′,6-diamidino-2-phenylindole) and in green binding afforded by the recombinant HA proteins that are fused with a GFP. In the ferret tissues, lung alveoli are indicated with ^, blood vessels with *, and the bronchi with >.
Finally, the ferret trachea is bound by all HA tested. The H6N1 variant was only able to bind this tissue, perhaps due to its low and specific binding avidity.
Thus, the ferret upper respiratory tract does express complex N-glycans with multiple LacNAc repeats, confirming their suitability as an animal model for human influenza A viruses.
Harnessing the Power of NMR to Get a Molecular Grip on HA Specificity
Saturation transfer difference (STD) NMR experiments,26 were used to complement the glycan array and tissue binding data described above and provide the molecular recognition perspective at atomic resolution. In particular, the details of the interaction features of the sialylated tri-LacNAc biantennary N-glycan (13) with the four hemagglutinin variants specific for human-type receptors were scrutinized. STD NMR experiments have been widely employed to derive the binding epitope of diverse ligands, including glycans, to a variety of receptors.26 In this context, the STD experiment has been successfully used to analyze the interaction of sialylated trisaccharides with cells transfected with H5 and H1 variants of hemagglutinin.27 Nevertheless, the intricacy of the NMR spectra of complex glycans, especially in biantennary structures which display the same sugar repetitions in the two antennae, makes the analysis far from trivial.28
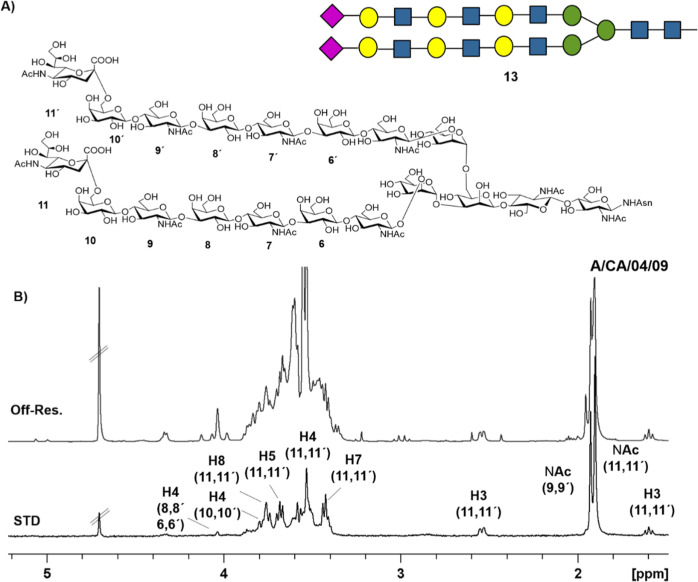
Fittingly, clear STD signals were observed for the tri-LacNAc-containing biantennary N-glycan 13 in the presence of the four hemagglutinin variants. Therefore, the experiments allowed the detection of the recognition event. In all cases, the higher STD intensities belong to the sialic acid signals (Figure 3 and 4).
Figure 3.
(A) Structure of the tri-LacNAc biantennary N-glycan employed in the NMR binding experiments. (B) Reference spectrum (off resonance) and STD NMR spectrum of the tri-LacNAc glycan in the presence of H1 A/CA/04/09 hemagglutinin. The signals of the hydrogens that appear in the STD spectrum are labeled with the proton number and the corresponding number of the glycan ring where they are located.
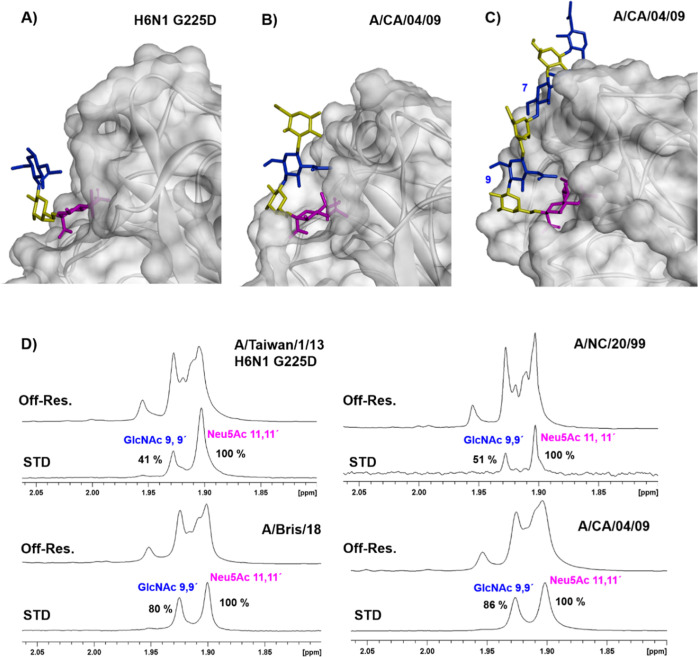
Figure 4.
(A) Expansion of the binding site of the X-ray crystallographic structure of H6N1 G225 D in complex with a trisaccharide, PDB code 5T0B (B) X-ray crystallographic structure of A/CA/04/09 bound to a pentasaccharide PDB code 3UBE (only four glycan units could be modeled in the electron density). (C) Computational model of the complex formed by the ligand displaying a tri-LacNAc motif with A/CA/04/09 based on the PDB code 3UBE. (D) Comparison of the STD NMR spectra (acetyl group region) recorded for the tri-LacNAc biantennary N-glycan 13 in the presence of the four hemagglutinins characterized in this work. The STD intensities are normalized to the highest signal that is the acetyl group of sialic acid.
However, there are marked differences in the STD pattern of the variants (Figures 4 and S4). For A/CA/04/09 and A/Bris/18, large STD effects were detected for the acetyl groups of GlcNAc 9 and 9′ and, although with a smaller intensity, the signals corresponding to H4 of the internal Gal units (8,8′,6,6′) were also observed (Figure S4). Therefore, it can be unambiguously concluded that the H1 A/CA/04/09 and A/Bris/18 variants interact not only with the exposed sialic acid moiety but also with the internal glycan units, which all together display an extended binding epitope (Figure S5).
In contrast for A/NC/20/99 and A/Taiwan/1/13 H6 G225D mutants, the STD NMR signal of the acetyl group of GlcNAcs 9 and 9′ is highly decreased with respect to that of A/CA/04/09 (Figure 4D), while the STD signal of H4 of the internal Gal moieties is very weak for A/NC/20/99 and cannot be even detected for the H6 G225D mutant (Figure S4). Therefore, these A/NC/20/99 and H6 G225D variants show significantly less interaction with the internal glycan units, and basically, only the terminal sialic acid is the interacting epitope. Interestingly, these NMR-based results are in full agreement with the X-ray structures described for A/Taiwan/1/13 H6 G225D17 and A/CA/04/0929 in complex with short-chain glycans (PDB codes: 5T0B and 3UBE, Figure 4A,B, respectively). The internal GlcNAc (blue color Figure 4B) is much closer to the protein surface in A/CA/04/09 (ca. distance between the 2-acetamido nitrogen to carboxyl oxygen of Asp190 is 3.0 Å) than in H6 G225D (ca. distance between the 2-acetamido nitrogen to the closer methyl group of V190 is 6.8 Å). Indeed, this closer proximity is due to the stabilizing interactions of the Asp190 carboxyl group with both 2-acetamido of GlcNAc and the 2-hydroxyl group of the adjacent internal Gal moiety.29 On the other hand, the H6 G225D variant cannot establish these interactions, since position 190 contains a valine residue. All these features are fully in line with the absence of the STD signal for the internal Gal unit.
Additional LacNAc Moieties Stabilize Receptor Binding
As a further step, to explore the effect of the number of LacNAc repeats in the recognition, the interactions of a di-LacNAc biantennary glycan with A/CA/04/09 and H6 G225D variants were characterized, and the STD intensities were compared to those measured for the tri-LacNAc analogue (Figure S6). For A/CA/04/09, the STD effect of the acetyl groups of the GlcNAc units 7 and 7′ (corresponding to 9, 9′ in the tri-LacNAc glycan) significantly decreases (from 86 to 48%, the values are normalized with respect to the highest signal that corresponds to the acetyl group of sialic acid that is considered 100%) in the di-LacNAc respect to the tri-LacNAc glycan. Thus, it can be deduced that the presence of the additional LacNAc repeat in the tri-LacNAc glycan increases the stabilizing interactions of the internal units.
It is tempting to hypothesize that a favorable bidentate binding (Figure S7) can only take place in the tri-LacNAc biantennary N-glycan since the length of the di-LacNAc repeat does not allow for simultaneous binding of two sialic acid moieties at their binding sites within the same hemagglutinin trimer.5 However, according to the array data discussed above, a high response is also observed for the recognition of the asymmetric glycan 15 (with just one arm containing three LacNAc repeats) by A/CA/04/09. Therefore, the tri-LacNAc glycans display a preferred interaction with this variant and do not require the existence of a bidentate binding event. In this context, the interaction of a tri-LacNAc chain to A/CA/04/09 (Figure 4C) was modeled using the available X-ray crystallographic coordinates for the lectin (PDB 3UBE) using computational chemistry tools. According to the obtained model, the glycan can establish interactions along the protein surface that are in complete agreement with the existence of an extended binding epitope in this variant in full agreement with the NMR results. Alternatively, this trend is not observed for the H6N1 variant, where only weak STD NMR signals were detected for the internal units, either using the tri-LacNAc or di-LacNAc biantennary glycans.
Using a Terminal Epitope as a Competitor Uncovers the Relative Binding Affinities
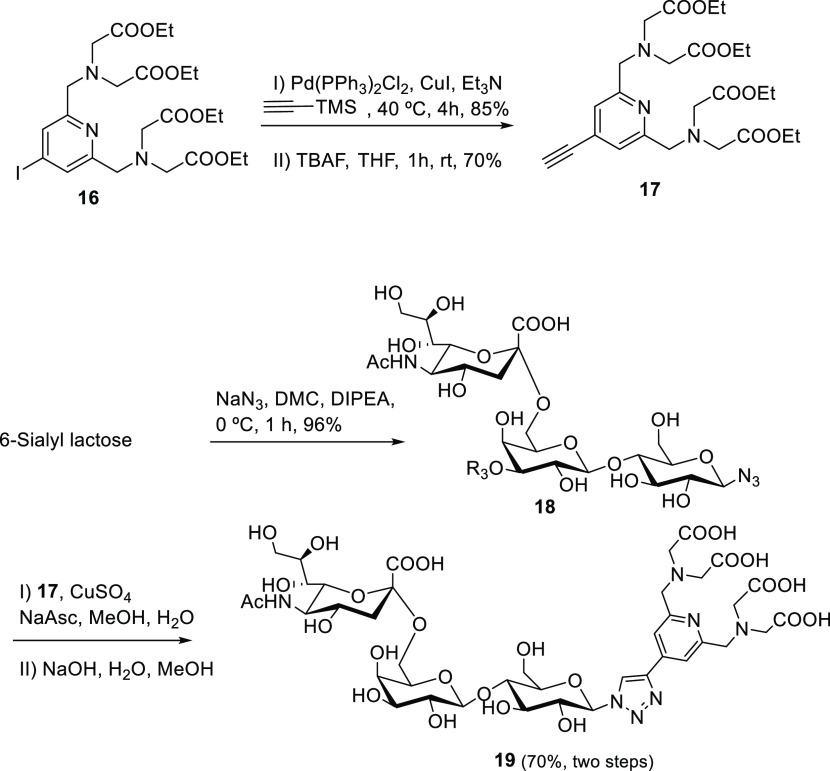
Finally, to further confirm the binding preferences and to derive the relative binding affinities of the different glycans, which contain the same monosaccharide units but within chains with different lengths, a new NMR protocol was devised. The major drawback of the standard NMR experiments is that the resonance signals of the monosaccharides within the glycans with different lengths appear at the same chemical shift, and therefore, it is not possible to detect individual signals for each of them. Thus, an α2-6 sialylated trisaccharide conjugated to a lanthanide binding tag was synthesized (Scheme 1) and used as a molecular probe to perform competition experiments.
Scheme 1. Synthesis of α2-6 Sialylated Trisaccharide Probe Bearing a Lanthanide Binding Tag.
The use of lanthanides in NMR-based structural studies started in the metalloprotein field by exchanging the natural metal for a paramagnetic lanthanide, such as Tb3+ or Dy3+.30 The method was further extended to other systems by employing diverse lanthanide binding tags that were covalently attached to the target molecule.31 The paramagnetic nucleus induces chemical shift changes (pseudo contact shifts, PCS) in the NMR signals of the molecule. These PCS contain valuable structural information since they depend on the distance between the NMR-active nuclei and the paramagnetic ion, as well as on the shape of the paramagnetic susceptibility tensor that is characteristic of each lanthanide.32 In the last years, this method has also been applied to the analysis of complex saccharides, including multiantenna glycans.28,33,34
Thus, following this concept, the target trisaccharide was synthesized as described in Scheme 1. In brief, compound 16 was obtained from chelidamic acid through six reaction steps as described by Qi et al.35 A Sonogashira coupling with trimethylsilylacetylene and further removal of the TMS group gave 17 in 60% yield for these two steps. On the other hand, 6-sialyl lactose was treated with DMC (2-chloro-1,3-dimethyl-1-H-imidazolium chloride), NaN3, and DIPEA in water at 0 °C to give rise to the corresponding β-azido derivative 18 (96%), without the need to protect the sugar moiety.36 Following a CuAAC coupling procedure, the 6-sialyl lactose azide derivative 18 was coupled with the PyMTA derivative 17. The synthesis was finished by deprotection of the carboxy groups with NaOH in MeOH and water, isolating 19 with a yield of 70% for the final two steps, Scheme 1.
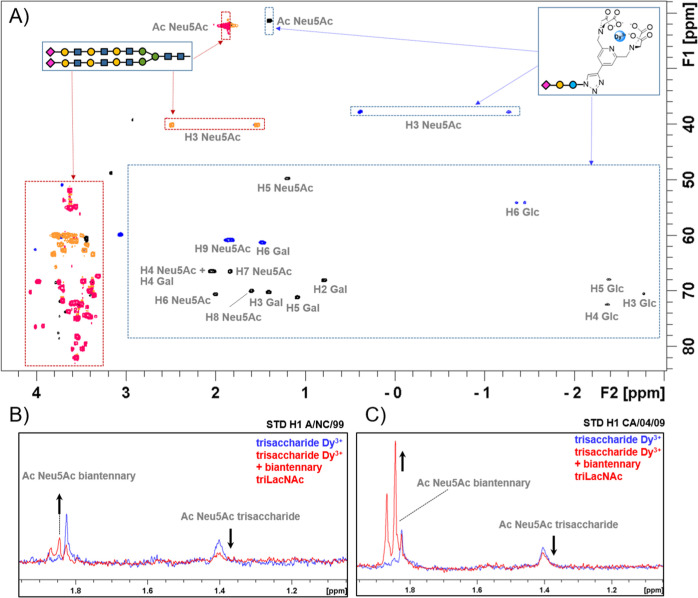
Thus, 1H and 1H–13C HSQC NMR spectra for trisaccharide 19 were recorded upon addition of dysprosium trichloride to the NMR tube (Figure 5). The standard 1D 1H NMR experiment showed a spectacular increase in signal dispersion, displaying the NMR signals of the Glc moiety attached to the lanthanide tag at negative chemical shifts (for instance, H3 Glc appears at δ −2.7 ppm). Actually, the overlaid of the 1H–13C HSQC spectra of a tri-LacNAc biantennary N-glycan with the trisaccharide derivative loaded with dysprosium shows the signals of both glycans in spectral regions that are clearly differentiated (Figure 5A). Therefore, it is possible to perform competition experiments under these experimental conditions. Thus, the STD NMR spectrum of the trisaccharide probe was acquired in the presence of two hemagglutinin variants H1 A/NC/20/99 and H1 A/CA/04/09. In both cases, STD signals were clearly detected, confirming the interaction (blue spectra, Figure 5B and 5C). In the next step, 0.5 equivalents of the tri-LacNAc biantennary glycan were added to each sample.
Figure 5.
(A) Overlaid of the edited 1H–13C HSQC spectrum of the tri-LacNAc biantennary glycan (red/orange colors) with the spectrum of the trisaccharide probe loaded with dysprosium (black/blue colors). (B,C) STD competition experiments of the trisaccharide probe with a biantennary tri-LacNAc glycan in the presence of H1 A/NC/20/99 and H1 A/CA/04/09, respectively.
The STD NMR signals of the trisaccharide probe showed a significant decrease in their intensities, while STD NMR signals for the tri-LacNAc glycan appeared. This evidence strongly suggests that both molecules compete for the same hemagglutinin binding site and that the longer glycan is replacing the probe. Given the use of sub-stoichiometric amounts of the tri-LacNAc moiety, it can be assessed that both HAs display higher affinity for the long-chain glycan than for the trisaccharide. The analysis of the data also permitted us to point out that STD signal intensities observed for the biantennary tri-LacNAc are stronger in the presence of H1 A/CA/04/09 than in the presence of A/NC/20/99 (Figure 5C). Indeed, A/CA/04/09 is the variant that also displays higher effects on the STD NMR experiments with the tri-LacNAc biantennary N-glycan described above (Figure S9). The STD experiment of compound 19 was also acquired in the absence of the metal to detect the signals of the tag that disappear in the presence of dysprosium due to the paramagnetic relaxation enhancement effect induced by the metal. No STD signals were detected for the protons of the chelating unit, pointing out that the lanthanide tag does not interact with the protein (Figure S8).
Discussion and Conclusions
Both NMR and glycan array analysis allow detecting the recognition of α(2-6) long chain glycans by the H1 and H6 hemagglutinins characterized in this work. The binding of these hemagglutinins to ferret respiratory tissues reveals that these glycans are present in the upper respiratory tract, probably enabling efficient viral transmission.
Interestingly, we detect differences in the recognition pattern of seasonal (A/NC/20/99) and pandemic (A/CA/04/09) hemagglutinin variants. The A/NC/20/99 strain interacts with short chains according to the array (Figure 1) (7–9) and tissue binding data, supporting previous data on the promiscuous specificity of A/NC/20/99.37 This result correlates with weak STD effects for the internal units of the tri-LacNAc glycan in NMR experiments that explains the lack of strong selectivity for long-chain glycans in this variant. In contrast, pandemic A/CA/04/09 displays a clear preference for long chains according to array data, and this result correlates with extended binding epitopes detected by NMR, where not only the sialic acid but also the GlcNAcs 9,9′ in the tri-LacNAc glycan display strong STD effects. Moreover, the interaction with the internal galactoses is also detected. A similar binding epitope was obtained for A/Bris/18 that results from the evolution of A/CA/04/09 during the last decade. Therefore, pandemic H1 maintains specificity for N-glycans with sialylated di-LacNAc and longer structures. NMR competition experiments confirmed the higher affinity of the tri-LacNAc structure than the short model compound, sialyl lactoside trisaccharide derivative 19, when bound to the A/CA/04/09 variant. In addition, both NMR and glycan array data show that A/CA/04/09 maintains binding to di-LacNAc glycans in contrast to H3N2 contemporary viruses in which this binding property is lost.4
H6N1 strain displays a lower response overall in array data and less interaction with the tri-LacNAc internal units than A/CA/04/09 according to STD NMR experiments. Therefore, this variant is less efficiently adapted to recognize long-chain glycans, and this is perhaps another barrier to sustained human-to-human transmission. If this is the case for other subtypes that can cause zoonotic infections, it remains to be explored. However, this H6 G225D protein can bind ferret upper respiratory tract tissues and is therefore primed to transmit between ferrets and therefore probably in humans.
Herein, the potential of combining NMR with glycan array data to characterize the binding preferences of hemagglutinins from different viruses is shown. Interestingly, pandemic H1 variants have evolved to recognize long-chain glycans with at least two LacNAc repeats. The HAs studied in this work bound to ferret upper respiratory tract tissue; therefore, this tissue expresses complex N-glycans with multiple LacNAc repeats, confirming its suitability as an animal model for human influenza A viruses.
In addition, a trisaccharide probe bearing a lanthanide binding tag has been designed to perform NMR competition experiments. This probe allows to discriminate the NMR signals, that otherwise will overlap, of common residues in sialylated glycans with different lengths toward HA, and it can be applied to explore the binding preferences of other lectins that recognized sialylated glycans.
Materials and Methods
Trimeric HA Expression Plasmid Generation
Recombinant trimeric influenza A virus hemagglutinin proteins were cloned using Gibson assembly from cDNAs encoding codon-optimized open reading frames. The pCD5 expression vector was adapted so that after the signal sequence, HA-encoding cDNAs are cloned in frame with a GCN4 trimerization motif (KQIEDKIEEIESKQKKIENEIARIKK), a tobacco etch virus (TEV) cleavage site, a fluorescent reporter open reading frame, and the Strep-tag II ((WSHPQFEKGGGSGGGSWSHPQFEK); IBA, Germany).25,38
Protein Expression and Purification
Expression vectors were transfected into HEK293S GnTI cells with polyethyleneimine I (PEI) in a 1:8 ratio (μg DNA:μg PEI) as previously described.25,38 The transfection mix was replaced after 6 h by 293 SFM II suspension medium (Invitrogen, 11686029) supplemented with glucose 2.0 g/L, sodium bicarbonate 3.6 g/L, primatone 3.0 g/L (Kerry), 1% glutaMAX (Gibco), 1.5% dimethyl sulfoxide, and 2 mM valproic acid. Culture supernatants were harvested 5 days post-transfection. Proteins were purified with sepharose Strep-Tactin beads (IBA Life Sciences) as previously described.
Glycan Microarray Binding of HA
The glycan microarray as earlier presented was already utilized.4 HAs were pre-complexed with mouse anti-Strep-tag-HRP and goat anti-mouse-Alexa555 antibodies in a 4:2:1 molar ratio. Respectively, in 50 μL phosphate-buffered saline (PBS) with 0.1% Tween-20. The mixture was incubated on ice for 15 min and then on the surface of the array for 90 min in a humidified chamber. Then, slides were rinsed successively with PBS-T (0.1% Tween-20), PBS, and deionized water. The arrays were dried by centrifugation and immediately scanned as described previously. Processing of the six replicates was performed by removing the highest and lowest replicate and subsequently calculating the mean value and standard deviation over the four remaining replicates.
Fluorescent Protein Histochemistry
Sections of formalin-fixed, paraffin-embedded chicken (Gallus gallus domesticus), pig bronchus, and ferret were obtained from the Department of Veterinary Pathobiology, Faculty of Veterinary Medicine, Utrecht University, the Netherlands. Protein histochemistry was performed as previously described.39 In short, tissue sections of 4 μm were deparaffinized and rehydrated, after which antigens were retrieved by heating the slides in 10 mM sodium citrate (pH 6.0) for 10 min. When indicated, neuraminidase treatment was performed for 18h at 37 °C with Vibrio cholerae neuraminidase (Roche, #11080725001) in 10 mM potassium acetate buffer, 0.1% Triton X-100, pH 4.2. Tissues were blocked overnight at 4 °C using 3% BSA (w/v) in PBS with 0.1% Tween-20 and subsequently stained using 5 μg/mL of pre-complexed HAs, as previously described for the glycan microarray. Lectin stains were performed with Erythrina cristagalli lectin (ECA, 1 μg/mL, B-1145-5, Vector Laboratories) and Sambucus nigra lectin (SNA, 2 μg/mL, B-1305-2, Vector Laboratories) precomplexed with the streptavidin-Alexa Fluor 488 conjugate (Invitrogen, #S11223, 0.2 and 0.4 μg/mL, respectively) and incubated for 90 min. Counterstain was performed with DAPI for 5 min at room temperature. The stained tissues were covered with cover slides using FluorSave (Merck Millipore).
NMR
STD NMR spectra were acquired in deuterated buffer TRIS-d11 50 mM, NaCl 100 mM pD 8.02 at 288 K using a Bruker 600 MHz spectrometer equipped with a cryoprobe. The glycan concentration was 0.3 mM, and the glycan/protein ratio was 40:1 for all HA variants. The experiments were performed with 2048 scans and 50 ms of saturation time. The on-resonance frequency was set to −0.3 ppm, and the off-resonance frequency was set at 100 ppm.
Competition experiments were acquired with samples of sialyllactose derivative 0.3 mM loaded with dysprosium. A sialyllactose/protein ratio of 40:1 was used for both A/CA/04/09 and A/NC/20/99 samples. tri-LacNAc biantennary N-glycan was added to these samples to a final concentration of 0.15 mM. STD experiments were acquired in the absence and in the presence of tri-LacNAc biantennary N-glycan with 4096 scans and a 50 ms saturation time. The on-resonance frequency was set at 6.5 ppm, and the off-resonance frequency was set at 100 ppm.
3D Models
The model of a TriLacNAc structural motif bound to H1 A/CA/04/09 was built by the superimposition of the coordinates of one of the arms of the triLacNAc biantennary structure obtained by molecular dynamics by Prof. Robert Woods’ group in complex with an H3N2 hemagglutinin variant5 (model coordinates kindly provided by Dr. Oliver Grant) onto the X-ray structure of A/CA/04/09, PDB code 3UBE, G chain.
Synthesis of the diLacNAc and triLacNAc Biantennary Glycans for NMR Studies
The synthesis of these glycans was carried out following the already described synthetic protocol by Nycholat et al.40
Synthesis of the Trisaccharide Probe
All chemicals were obtained from Aldrich/Merck (St. Louis, MO, USA), VWR (Radnor, PA, USA), and Fluorochem (Derbyshire, UK). TLC analyses were performed on Merck silica gel 60 F254 plates using phosphomolybdic acid or anisaldehyde and heat for detection. Silica gel NORMASIL 60 40–63 μm was used for flash chromatography. The detailed synthetic protocol and the compound characterization are given in the Supporting Information.
Acknowledgments
R.P.d.V. is a recipient of an ERC Starting grant from the European Commission (802780) and a Beijerinck Premium of the Royal Dutch Academy of Sciences. The glycan array setup was supported by the Netherlands Organization for Scientific Research (NWO, TOP-PUNT 718.015.003 to G.-J.P.H.B.). Dr. Lin Liu (CCRC) and Dr. Margreet A. Wolfert (Utrecht University) developed, printed, and validated the glycan microarray. We would like to thank Nikoloz Nemanichvili for technical assistance. A.C. acknowledges funding from Agencia Estatal de Investigación “Spanish Ministry of Science and Innovation” (MICINN) project PID2019-105237GB-I00. J.P.C. acknowledges funding by the Spanish MICINN, grant no. RTI2018-095588-B-I00 (co-funded by the European Regional Development Fund/European Social Fund, “Investing in your future”). JJB also thanks funding by the European Research Council (RECGLYCANMR, Advanced grant no. 788143), the Agencia Estatal de Investigación (Spain) for grants RTI2018-094751-B-C21 and C22 and PDI2021-1237810B–C21 and C22, and CIBERES, an initiative of the Instituto de Salud Carlos III (ISCIII), Madrid, Spain. The NMR spectra were acquired at the NMR service of CIB Margarita Salas and in the NMR facility of the UCM. We also acknowledge Prof. Robert Woods group for sending us the coordinates of a glycan-hemagglutinin model.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.2c00664.
Experimental procedures; characterization of new compounds; and additional NMR information (PDF)
Author Present Address
†† Poultry Diagnostic and Research Center, University of Georgia, Athens, United States
Author Present Address
‡‡ Charité-Univesitätsmedizin Berlin
Author Contributions
A.C.: conceptualization, NMR investigation and analysis, and writing original-draft; J.S.: synthesis of diLacNAc and triLacNAc glycans; J.M.O.: synthesis of the trisaccharide probe; J.P.C and G.D.: design and supervision of the synthesis of the trisaccharide probe; C.M.S. and K.M.B., R.vdW.: protein expression and tissue staining experiments; F.J.C.: supervision; C.M.N,: synthesis of diLacNAc and triLacNAc glycans; J.P.: methodology, supervision, and writing-review; G.J.B.: methodology and supervision; J.J.B.: methodology, supervision, and writing-review; R.P.dV.: conceptualization, glycan array investigation and analysis, tissue experiments, and writing original-draft. All authors have given approval to the final version of the manuscript. CRediT: Angeles Canales conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, visualization, writing-original draft, writing-review & editing; Javier Sastre investigation, methodology, resources, writing-review & editing; Jose M Orduña investigation, methodology, resources; Cindy Maria Spruit data curation, investigation; Javier Pérez-Castells data curation, formal analysis, investigation, methodology, writing-review & editing; Gema Domínguez investigation, methodology, resources; Kim M Bouwman data curation, methodology; Roosmarijn van der Woude data curation, investigation; Francisco Javier Cañada conceptualization, supervision, validation, writing-review & editing; Corwin M Nycholat methodology, resources; James C Paulson conceptualization, methodology, resources, supervision; Geert-Jan P. H. Boons conceptualization, funding acquisition, resources, supervision; Jesús Jiménez-Barbero conceptualization, funding acquisition, investigation, resources, supervision, validation, writing-original draft, writing-review & editing; Robert P. de Vries conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, visualization, writing-original draft, writing-review & editing.
The authors declare no competing financial interest.
Supplementary Material
References
- Thompson A. J.; de Vries R. P.; Paulson J. C. Virus recognition of glycan receptors. Curr. Opin. Virol. 2019, 34, 117–129. 10.1016/j.coviro.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G. N.; Paulson J. C. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 1983, 127, 361–373. 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Srinivasan A.; Viswanathan K.; Raman R.; Chandrasekaran A.; Raguram S.; Tumpey T. M.; Sasisekharan V.; Sasisekharan R. Quantitative biochemical rationale for differences in transmissibility of 1918 pandemic influenza A viruses. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 2800–2805. 10.1073/pnas.0711963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broszeit F.; van Beek R. J.; Unione L.; Bestebroer T. M.; Chapla D.; Yang J. Y.; Moremen K. W.; Herfst S.; Fouchier R. A. M.; de Vries R. P.; et al. Glycan remodeled erythrocytes facilitate antigenic characterization of recent A/H3N2 influenza viruses. Nat. Commun. 2021, 12, 5449. 10.1038/s41467-021-25713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W.; de Vries R. P.; Grant O. C.; Thompson A. J.; McBride R.; Tsogtbaatar B.; Lee P. S.; Razi N.; Wilson I. A.; Woods R. J.; et al. Recent H3N2 Viruses Have Evolved Specificity for Extended, Branched Human-type Receptors, Conferring Potential for Increased Avidity. Cell Host Microbe 2017, 21, 23–34. 10.1016/j.chom.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. L.; Kohler J. J.; Pfeffer S. R. Association of -1,3-N-acetylglucosaminyltransferase 1 and -1,4-galactosyltransferase 1, trans-Golgi enzymes involved in coupled poly-N-acetyllactosamine synthesis. Glycobiology 2009, 19, 655–664. 10.1093/glycob/cwp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujita M.; McAuliffe J.; Hindsgaul O.; Sasaki K.; Fukuda M. N.; Fukuda M. Poly-N-acetyllactosamine Synthesis in BranchedN-Glycans Is Controlled by Complemental Branch Specificity of i-Extension Enzyme and β1,4-Galactosyltransferase I. J. Biol. Chem. 1999, 274, 16717–16726. 10.1074/jbc.274.24.16717. [DOI] [PubMed] [Google Scholar]
- Stanley P.; Moremen K. W.; Lewis N. E.; Taniguchi N.; Aebi M.; et al. N-Glycans, 1-9. In Essentials of Glycobiology, 4 th ed.; Varki A., Cummings R. D., Esko J. D., Eds.; Cold Spring Harbor Laboratory Press: Long Island: NY, USA, 2022; Chapter 9. [Google Scholar]
- Xiong X.; Tuzikov A.; Coombs P. J.; Martin S. R.; Walker P. A.; Gamblin S. J.; Bovin N.; Skehel J. J. Recognition of sulphated and fucosylated receptor sialosides by A/Vietnam/1194/2004 (H5N1) influenza virus. Virus Res. 2013, 178, 12–14. 10.1016/j.virusres.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Childs R.; Palma A.; Wharton S.; Matrosovich T.; Liu Y.; Chai W.; Campanero-Rhodes M. A.; Zhang Y.; Eickmann M.; Kiso M.; Hay A.; Matrosovich M.; Feizi T. Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nat. Biotechnol. 2009, 27, 797–799. 10.1038/nbt0909-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Childs R. A.; Matrosovich T.; Wharton S.; Palma A. S.; Chai W.; Daniels R.; Gregory V.; Uhlendorff J.; Kiso M.; Klenk H. D.; Hay A.; Feizi T.; Matrosovich M. Altered Receptor Specificity and Cell Tropism of D222G Hemagglutinin Mutants Isolated from Fatal Cases of Pandemic A(H1N1) 2009 Influenza Virus. J. Virol. 2010, 84, 12069–12074. 10.1128/jvi.01639-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd-Leotis L.; Gao C.; Jia N.; Mehta A. Y.; Trost J.; Cummings S. F.; Heimburg-Molinaro J.; Cummings R. D.; Steinhauer D. A. Antigenic Pressure on H3N2 Influenza Virus Drift Strains Imposes Constraints on Binding to Sialylated Receptors but Not Phosphorylated Glycans. J. Virol. 2019, 93, 011788-19. 10.1128/jvi.01178-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N.; Barclay W. S.; Roberts K.; Yen H. L.; Chan R. W.; Lam A. K.; Air G.; Peiris J. S.; Dell A.; Nicholls J. M.; et al. Glycomic Characterization of Respiratory Tract Tissues of Ferrets. J. Biol. Chem. 2014, 289, 28489–28504. 10.1074/jbc.m114.588541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.; Karamanska R.; Chan R. W.; Chan M. C.; Jia N.; Air G.; Hopton C.; Wong M. P.; Dell A.; Malik Peiris J. S.; et al. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog. 2013, 9, e1003223 10.1371/journal.ppat.1003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby D. C.; Savanovica M.; Zhanga J.; Torellib A.; Jeeningac R. E.; Gagnond L.; Harrisa S. L. Interlaboratory Reproducibility of Standardized Hemagglutination Inhibition Assays. mSphere 2022, 7, e0095321 10.1128/msphere.00953-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A. J.; Paulson J. C. Adaptation of influenza viruses to human airway receptors. J. Biol. Chem. 2021, 296, 100017. 10.1074/jbc.REV120.013309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vries R. P.; Tzarum N.; Peng W.; Thompson A. J.; Ambepitiya Wickramasinghe I. N.; Pena A. T. T.; van Breemen M. J.; Zhu K. M.; McBride X.; Yu R.; Sanders W.; Verheije R. W.; Wilson M. H.; Paulson I. A.; Paulson J. C. A single mutation in Taiwanese H6N1 influenza hemagglutinin switches binding to human-type receptors. EMBO Mol. Med. 2017, 9, 1314–1325. 10.15252/emmm.201707726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R. P.; Peng W.; Grant O. C.; Thompson A. J.; Zhu X.; Bouwman K. M.; de la Pena A. T. T.; van Breemen M. J.; Ambepitiya Wickramasinghe I. N.; de Haan C. A. M.; et al. Three mutations switch H7N9 influenza to human-type receptor specificity. PLoS Pathog. 2017, 13, e1006390 10.1371/journal.ppat.1006390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W.; Bouwman K. M.; McBride R.; Grant O. C.; Woods R. J.; Verheije M. H.; Paulson J. C.; de Vries R. P. Enhanced Human-Type Receptor Binding by Ferret-Transmissible H5N1 with a K193T Mutation. J. Virol. 2018, 92, e02016–e02017. 10.1128/JVI.02016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzarum N.; de Vries R. P.; Peng W.; Thompson A. J.; Bouwman K. M.; McBride R.; Yu W.; Zhu X.; Verheije M. H.; Paulson J. C.; et al. The 150-Loop Restricts the Host Specificity of Human H10N8 Influenza Virus. Cell Rep. 2017, 19, 235–245. 10.1016/j.celrep.2017.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A. J.; Cao L.; Ma Y.; Wang X.; Diedrich J. K.; Kikuchi C.; Willis S.; Worth C.; McBride R.; Yates J. R. 3rd; et al. Human Influenza Virus Hemagglutinins Contain Conserved Oligomannose N-Linked Glycans Allowing Potent Neutralization by Lectins. Cell Host Microbe 2020, 27, 725–735. 10.1016/j.chom.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R.; de Vries R. P.; Zhu X.; Nycholat C. M.; McBride R.; Yu W.; Paulson J. C.; Wilson I. A. Preferential recognition of avian-like receptors in human influenza A H7N9 viruses. Science 2013, 342, 1230–1235. 10.1126/science.1243761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakdawala S. S.; Jayaraman A.; Halpin R. A.; Lamirande E. W.; Shih A. R.; Stockwell T. B.; Lin X.; Simenauer A.; Hanson C. T.; Vogel L.; et al. The soft palate is an important site of adaptation for transmissible influenza viruses. Nature 2015, 526, 122–125. 10.1038/nature15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P. S.; Böhm R.; Hartley-Tassell L. E.; Steen J. A.; Wang H.; Lukowski S. W.; Hawthorne P. L.; Trezise A. E.; Coloe P. J.; Grimmond S. M.; et al. Ferrets exclusively synthesize Neu5Ac and express naturally humanized influenza A virus receptors. Nat. Commun. 2014, 5, 5750. 10.1038/ncomms6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemanichvili N.; Tomris I.; Turner H. L.; McBride R.; Grant O. C.; van der Woude R.; Aldosari M. H.; Pieters R. J.; Woods R. J.; Paulson J. C.; et al. Fluorescent Trimeric Hemagglutinins Reveal Multivalent Receptor Binding Properties. J. Mol. Biol. 2019, 431, 842–856. 10.1016/j.jmb.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.; Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 2001, 123, 6108–6117. 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- Vasile F.; Panigada M.; Siccardi A.; Potenza D.; Tiana G. A combined NMR-computational study of the interaction between influenza virus hemagglutinin and sialic derivatives from human and avian receptors on the surface of transfected cells. Int. J. Mol. Sci. 2018, 19, 1267. 10.3390/ijms19051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez de Toro B.; Peng W.; Thompson A. J.; Dominguez G.; Canada F. J.; Perez-Castells J.; Paulson J. C.; Jimenez-Barbero J.; Canales A. Avenues to Characterize the Interactions of Extended N-Glycans with Proteins by NMR Spectroscopy: The Influenza Hemagglutinin Case. Angew. Chem., Int. Ed. Engl. 2018, 57, 15051–15055. 10.1002/anie.201807162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R.; McBride R.; Nycholat C. M.; Paulson J. C.; Wilson I. A. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J. Virol. 2012, 86, 982–990. 10.1128/jvi.06322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I.; Luchinat C.; Parigi G.; Pierattelli R. NMR spectroscopy of paramagnetic metalloproteins. Chembiochem 2005, 6, 1536–1549. 10.1002/cbic.200500124. [DOI] [PubMed] [Google Scholar]
- Miao Q.; Nitsche C.; Orton H.; Overhand M.; Otting G.; Ubbink M. Paramagnetic Chemical Probes for Studying Biological Macromolecules. Chem. Rev. 2022, 122, 9571–9642. 10.1021/acs.chemrev.1c00708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting G. Prospects for lanthanides in structural biology by NMR. J. Biomol. NMR 2008, 42, 1–9. 10.1007/s10858-008-9256-0. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T.; Sakae Y.; Zhang Y.; Yamamoto S.; Okamoto Y.; Kato K. Exploration of conformational spaces of high-mannose-type oligosaccharides by an NMR-validated simulation. Angew. Chem., Int. Ed. Engl. 2014, 53, 10941–10944. 10.1002/anie.201406145. [DOI] [PubMed] [Google Scholar]
- Canales A.; Boos I.; Perkams L.; Karst L.; Luber T.; Karagiannis T.; Domínguez G.; Cañada F. J.; Pérez-Castells J.; Häussinger D.; et al. Breaking the Limits in Analyzing Carbohydrate Recognition by NMR Spectroscopy: Resolving Branch-Selective Interaction of a Tetra-AntennaryN-Glycan with Lectins. Angew. Chem., Int. Ed. Engl. 2017, 56, 14987–14991. 10.1002/anie.201709130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M.; Hulsmann M.; Godt A. Synthesis and Hydrolysis of 4-Chloro-PyMTA and 4-Iodo-PyMTA Esters and Their Oxidative Degradation with Cu(I/II) and Oxygen. Synthesis 2016, 48, 3773–3784. 10.1055/s-0035-1561660. [DOI] [Google Scholar]
- Tanaka T.; Nagai H.; Noguchi M.; Kobayashi A.; Shoda S. One-step conversion of unprotected sugars to β-glycosyl azides using 2-chloroimidazolinium salt in aqueous solution. Chem. Commun. 2009, 23, 3378–3379. 10.1039/b905761g. [DOI] [PubMed] [Google Scholar]
- Chen L. M.; Rivailler P.; Hossain J.; Carney P.; Balish A.; Perry I.; Davis C. T.; Garten R.; Shu S.; Xu X.; Klimov A.; Paulson J. C.; Cox N. J.; Swenson S.; Stevens J.; Vincent A.; Gramer M.; Donis R. O. Receptor specificity of subtype H1 influenza A viruses isolated from swine and humans in the United States. Virology 2011, 412, 401–410. 10.1016/j.virol.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R. P.; de Vries E.; Bosch B. J.; de Groot R. J.; Rottier P. J.; de Haan C. A. The influenza A virus hemagglutinin glycosylation state affects receptor-binding specificity. Virology 2010, 403, 17–25. 10.1016/j.virol.2010.03.047. [DOI] [PubMed] [Google Scholar]
- Bouwman K. M.; Tomris I.; Turner H. L.; van der Woude R.; Shamorkina T. M.; Bosman G. P.; Rockx B.; Herfst S.; Snijder J.; Haagmans B. L.; et al. Multimerization- and glycosylation-dependent receptor binding of SARS-CoV-2 spike proteins. PLoS Pathog. 2021, 17, e1009282 10.1371/journal.ppat.1009282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nycholat C. M.; McBride R.; Ekiert D. C.; Xu R.; Rangarajan J.; Peng W.; Razi N.; Gilbert M.; Wakarchuk W.; Wilson I. A.; et al. Recognition of sialylated poly-N-acetyllactosamine chains on N- and O-linked glycans by human and avian influenza A virus hemagglutinins. Angew. Chem., Int. Ed. Engl. 2012, 51, 4860–4863. 10.1002/anie.201200596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.