Abstract
Myelination contributes not only to the rapid nerve conduction but also to axonal insulation and protection. In the central nervous system (CNS), the initial myelination features a multistep process where oligodendrocyte precursor cells undergo proliferation and migration before differentiating into mature oligodendrocytes. Mature oligodendrocytes then extend processes and wrap around axons to form the multilayered myelin sheath. These steps are tightly regulated by various cellular and molecular mechanisms, such as transcription factors (Olig family, Sox family), growth factors (PDGF, BDNF, FGF-2, IGF), chemokines/cytokines (TGF-β, IL-1β, TNFα, IL-6, IFN-γ), hormones (T3), axonal signals (PSA-NCAM, L1-CAM, LINGO-1, neural activity), and intracellular signaling pathways (Wnt/β-catenin, PI3 K/AKT/mTOR, ERK/MAPK). However, the fundamental mechanisms for initial myelination are yet to be fully elucidated. Identifying pivotal mechanisms for myelination onset, development, and repair will become the focus of future studies. This review focuses on the current understanding of how CNS myelination is initiated and also the regulatory mechanisms underlying the process.
Keywords: central nervous system, development, myelin sheath, microglia, oligodendrocyte
Introduction
Myelination is a well-coordinated program that generates myelin sheaths to wrap around axons. The ensheathment of axons results in an increased membrane resistance, a decreased membrane capacitance, and a specific molecular organization at the nodes of Ranvier (nodes, paranodes, and juxtaparanodes), thereby significantly facilitating a rapid saltatory impulse propagation along with the axons (Coman et al., 2006; Nave & Werner, 2014). As a result, action potentials can conduct along with myelinated axons up to 100-fold faster than unmyelinated ones with similar diameters (Monje, 2018). Apart from its role in speeding up nerve conduction, myelin also provides trophic axonal support by transferring pyruvate/lactate through monocarboxylate transporters (Philips & Rothstein, 2017), especially for longer axons whose segments are far (many centimeters or even meters in humans) away from the cell body but adjacent to local glia cells. Moreover, myelin also regulates axonal transport, the process whereby substances (such as organelles) are transferred between the cell body and the axon tip (Edgar & Garbern, 2004). Hence, central nervous system (CNS) myelination significantly benefits motor, sensory, and cognitive functions of the brain due to its essential roles in increasing the speed of action potential conduction and neuronal homeostasis.
In the CNS, the myelin sheath is formed as a multilamellar membrane structure by oligodendrocytes (OLs) (Nave & Werner, 2014; Simons & Nave, 2015), which occurs as a multistep process during development. These steps include oligodendrocyte precursor cell (OPC) proliferation, OPC migration, OPC differentiation, subsequent wrapping, and myelin compaction (Barateiro et al., 2016). Each step is efficiently orchestrated by both cellular and extracellular factors. However, the fundamental mechanism regulating initial myelination is not fully understood. Failure of CNS myelination would eventually lead to neurologic and neurodegenerative diseases, such as leukodystrophies (Kolodny, 1993). Hence, a comprehensive understanding of physiological behaviors in CNS myelination is required to identify potential therapeutic targets in pathological conditions.
In this review, we first summarize the key processes of initial myelination. Next, we highlight current progress in our understanding of mechanisms of initial CNS myelination from the aspects of transcription factors, extracellular factors, axonal regulation, and signaling pathways. Finally, we compare initial myelination with remyelination before concluding the review with open questions in this field.
Initial Events of CNS Myelination
OL Lineage Development
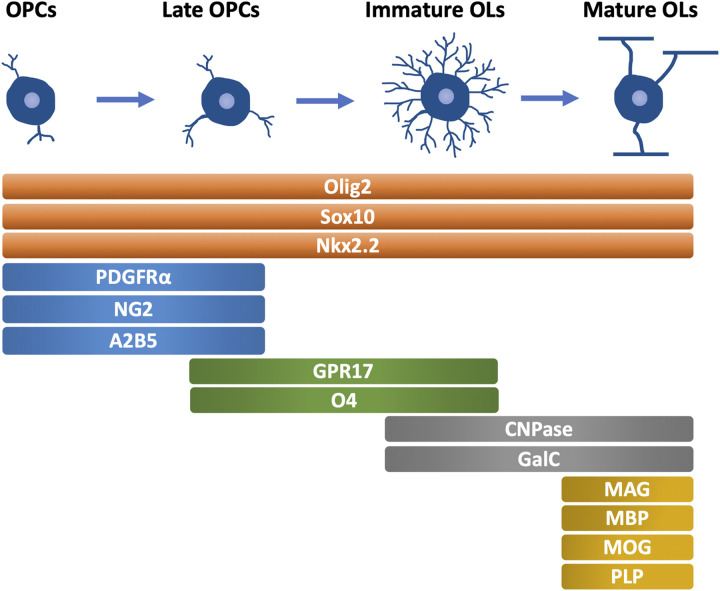
The maturation of OLs is a prerequisite for myelin sheath formation, highlighting the importance of OL lineage development for myelination. Generally, the OL lineage development can be divided into four stages, OPCs, late OPCs, immature (or premyelinating) OLs, and mature (or myelinating) OLs, which are characterized by the morphological changes and different expression patterns of specific markers (Figure 1) (Barateiro & Fernandes, 2014).
Figure 1.
Oligodendrocyte Lineage Development and Specific Markers in Each Stage (mammals). OPCs, oligodendrocyte precursor cells. OLs, oligodendrocyte. PDGFRα, platelet-derived growth factor receptor α. CNPase, 2′,3′-cyclic nucleotide 3′-phosphodiesterase. GalC, galactocerebroside C. MAG, myelin associated glycoprotein. MBP, myelin basic protein. MOG, myelin oligodendrocyte glycoprotein. PLP, proteolipid protein.
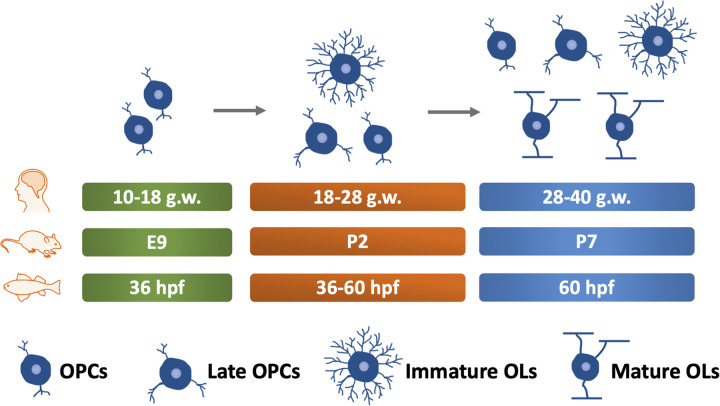
In the mouse brain, OPCs are first observed in the ventricular germinal layer of the lateral basal plate of the diencephalon at around embryonic day (E) 9 (Timsit et al., 1995). Interestingly, in the forebrain, a study describes three distinct waves of OPCs in a ventral-to-dorsal progression (Kessaris et al., 2006). Specifically, the first wave of OPCs originates in the ventral forebrain at E12.5, followed by a second wave of OPCs from the lateral and/or caudal ganglionic eminences at around E14.5. Finally, a third wave of OPCs occurs within the postnatal cortex. OPCs are proliferative cells with highly migratory capabilities, allowing them to migrate along with vasculature (Tsai et al., 2016) to the whole CNS before maturation and myelination. Unlike neurons that proliferate and migrate in the embryonic stage, these two processes in OPCs occur mainly in postnatal phases in rodents (Snaidero & Simons, 2014). At about postnatal day (P) 2, late OPCs account for the majority of OL lineage cells, with a small amount of immature OLs existing. Mature OLs appear at P7 to initiate the CNS myelination (Figure 2), which is almost completed in most brain regions by P60 (Snaidero & Simons, 2014). In adults, OPCs and intermediately differentiated OLs are present across the entire CNS (Dawson et al., 2003). Of note, CNS myelination follows a specific time course and sequence rather than occurring simultaneously. Generally, in mammals, CNS myelination starts in the brainstem and then continues rostrally to the forebrain and caudally to the spinal cord. This pattern correlates very closely with developmental milestones (Dietrich et al., 1988). For instance, myelination occurs early from motor-sensory roots, special senses, and the brainstem (Kinney et al., 1988), which are the necessary structures for reflex behaviors and survival.
Figure 2.
Timeline of CNS Myelination Development in Humans, Rodents, and Zebrafish. The green box indicates the time of OPCs appearance. The orange box indicates the stage where OPCs and pre-myelinating OLs exist. The blue box indicates the time of myelination onset. g.w.= gestational weeks. E = embryonic day. OPCs, oligodendrocyte precursor cells. OLs, oligodendrocyte. hpf, hours post fertilization. dpf, days post fertilization. CNS, central nervous system.
Compared with rodents, only a few studies have addressed the OLs lineage in humans. OPCs in humans are first observed in the forebrain at 10 gestational weeks (g.w.). Immature OLs are observed between 18 and 28 g.w., although OPCs and late OPCs are still the major cell types during this stage (Barateiro et al., 2016). Thus, OLs lineage development during 18–28 g.w. of humans is similar to that of rodents at P2. Myelin basic protein (MBP) positive OLs are initially observed at around 28 g.w. during human development (Back et al., 2001; Barateiro et al., 2016; Hüppi et al., 1998), resembling the time point of P7 in rodents (Figure 2).
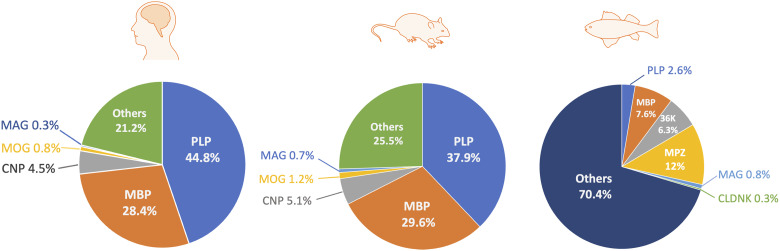
In zebrafish, another broadly used model for myelination study, OPCs are initially observed as early as 36 h post fertilization (hpf) (Kirby et al., 2006) and MBP, a marker of mature myelin, appears at about 60 hpf (Figure 2) (Almeida et al., 2011). Generally, CNS myelination is structurally and functionally conserved between zebrafish and mammals, although differences regarding the composition of myelin proteins are noticed (Figure 3) (Gargareta et al., 2022; Jahn et al., 2020; Siems et al., 2021). For example, myelin protein zero, a protein only found in the peripheral nervous system (PNS) of mammals, is present in the CNS of zebrafish (Brösamle & Halpern, 2002). Besides, zebrafish myelin also expresses some proteins that are absent in mammals, such as claudin K (Münzel et al., 2012), 36 K (Morris et al., 2004), and Zwilling-A/B (Schaefer & Brösamle, 2009). Moreover, a recent study shows that CD59 is the fourth most abundant (4.6%) myelin protein in zebrafish (Siems et al., 2021), while its abundance in mammals is comparatively low (0.01%) (Gargareta et al., 2022). Hence, more studies are required to explain the differences and determine the precise role of different myelin proteins in both zebrafish and mammals.
Figure 3.
Relative Abundance of CNS Myelin Proteins in Humans (Jahn et al., 2020), Rodents (Gargareta et al., 2022), and Zebrafish (Siems et al., 2021). OPCs, oligodendrocyte precursor cells. OLs, oligodendrocyte. CNP, CNPase, 2′,3′-cyclic nucleotide 3′-phosphodiesterase. MAG, myelin-associated glycoprotein. MBP, myelin basic protein. MOG, myelin oligodendrocyte glycoprotein. PLP, proteolipid protein. MPZ, myelin protein zero. CLDNK, claudin K. CNS, central nervous system.
Axon Targeting, Contact, and Process Polarization
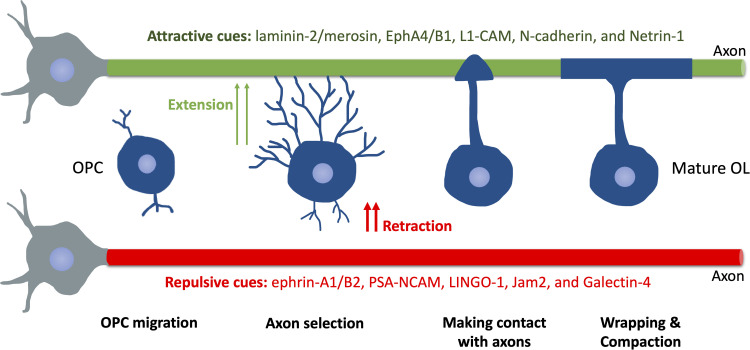
After ceasing migration, OPCs undergo continual process extension until making stable contact with the axonal membrane, subsequently forming an axon–glial interaction domain for further communication, namely, branching (Figure 4).
Figure 4.
Schematic of Myelination Progression in Mammals. After migrating to the final spot, OPCs extend or retract processes following attractive or repulsive cues on axons. OPC, oligodendrocyte precursor cell. OL, oligodendrocyte.
How OLs precisely target axons during initial myelination remain largely unknown. A recent study suggests that axon caliber alone, in the absence of axonal signals, allows myelination to initiate (Mayoral et al., 2018). Moreover, a study shows that OLs can sense the diameter of microfibers in vitro, thereby increasing sheath length with larger fibers (Bechler et al., 2015). These results are in line with the fact that nearly all axons with a diameter greater than 0.2 μm are myelinated (Goebbels et al., 2017), indicating an important role of axonal biophysical properties in axon selection during myelination. Presumably, this may involve a curvature-dependent mechanism, such as regulations of certain membrane-anchored proteins (Chang-Ileto et al., 2011; McMahon & Boucrot, 2015). Interestingly, similar to the “growth cone” model in axon outgrowth and pathfinding, a growing body of evidence also suggests that OLs processes extend or retract following permissive or repulsive cues on axons, respectively. In rodents, such permissive cues include laminin-2/merosin, EphA4/B1, L1-CAM, N-cadherin, and Netrin-1 (Figure 4) (Thomason et al., 2020). The negative/repulsive cues include axonal ephrin-A1/B2, PSA-NCAM, LINGO-1, Jam2, and Galectin-4 (Sherman & Brophy, 2005; Thomason et al., 2020). However, none of these cues are sufficient to instruct OLs to target axons in the CNS, which requires further research for a better understanding of this.
Interestingly, recent studies also support the role of neural activity in regulating axon targeting during initial myelination. For example, blocking action potentials in the zebrafish spinal cord results in axonal mistargeting and hypomyelination (Hines et al., 2015). However, some in vitro studies show that OLs can also myelinate paraformaldehyde (PFA)-fixed axons (Rosenberg et al., 2008) and nanofibers (Lee et al., 2013) in the absence of neural activity. Likely, correct axonal targeting depends on multiple cues, including both neural activity and OL-intrinsic signals.
After an initial axon–glia contact, a series of changes in OLs occur, including the downregulation of RhoA activity (Baer et al., 2009) and the local enrichment of phospholipid in OL membranes, such as PIP2 and PIP3 (Goebbels et al., 2010; Snaidero et al., 2014), eventually leading to a polarization of myelinating cells toward axons.
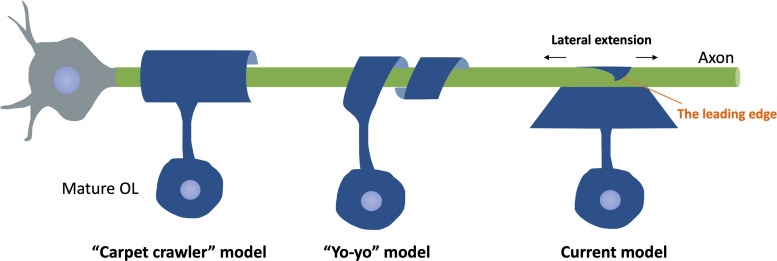
Myelin Wrapping
Once contact is formed and the polarization is finished, the wrapping process initiates. In the CNS, individual OL can myelinate up to 50 axons (Baumann & Pham-Dinh, 2001), with the number of membrane layers in one axon reaching up to 160 (Hildebrand et al., 1993). Nevertheless, the exact geometry of myelin wrapping is challenging to document due to the limitation of imaging technologies. Previously, two wrapping models are proposed, namely the “carpet crawler” model and the “yo-yo” model (Figure 5). The “carpet crawler” model suggests that, after axon–glia contact formation, the leading edge of the OLs membrane spreads and flattens into a broadsheet and then moves underneath itself to form the mature myelin sheath (Bunge et al., 1961). Alternatively, the “yo-yo” model proposes that the OL processes wrap around the axons until an appropriate number of layers are formed, then extend laterally into overlapping sheets (Pedraza et al., 2009). However, neither of these models provides the mechanisms of how synthesized molecules go through the compact myelin and reach the innermost layer, which cannot be easy as the inner layers become more and more distant to neural somas as the wrapping proceeds. With the advancement of imaging technologies, better visualization and understanding regarding the wrapping model are gained. By taking advantage of three-dimensional electron microscopy (3D EM), a newer model is proposed: the triangle-shaped leading edge moves underneath previously deposited layers, followed by lateral extension of each layer of myelin (Snaidero et al., 2014). This wrapping model allows close contact between each myelin layer and its underlying axon, thereby contributing to axon–glia communication. Notably, the authors also suggest that cytoplasmic channels appear transiently within compact myelin to provide a short connection between the outer and inner tongue, allowing the transportation of newly synthesized membrane components from the soma to the inner tongue. As myelination completes, these cytoplasmic channels resolve (Snaidero et al., 2014).
Figure 5.
Schematic of Different Wrapping Models Proposed. The “carpet crawler” model proposes that the process flattens into a broad sheet before wrapping. The “yo-yo” model suggests that the OL processes wrap around the axons until an appropriate number of layers are formed, then extend laterally into overlapping sheets. The current model suggests that the triangle-shaped leading edge moves underneath previously deposited layers, followed by lateral extension of each layer of myelin.
The molecular mechanisms of the wrapping process have also become a focus of many studies. Of note, OLs show a periodic actin pattern that is not seen in astrocytes or microglia, indicating tight cytoskeletal regulation during myelination (Brown & Macklin, 2019). In addition, one study indicates that actin polymerization in the leading edge is the main force driving the wrapping process, while actin depolymerization promotes membrane spreading by reducing surface tension (Nawaz et al., 2015). This is in line with another model proposing that actin polymerization powers OLs process extension in an Arp2/3-dependent manner, while actin depolymerization drives myelin wrapping (Zuchero et al., 2015). In this model, MBP prevents the actin disassembly factors cofilin and gelsolin from binding to PIP2, resulting in a release of cofilin/gelsolin to disassemble actin, thereby driving myelin wrapping.
Myelin Compaction
Myelin compaction coincides with wrapping. Initially, myelin membranes are negatively charged due to the abundance of phospholipids (PIP2, PIP3). Therefore, a mechanism of neutralizing these negative charges and pulling adjacent layers together is needed for compaction, which is now recognized as the key role of MBP. MBP shows a high affinity to negatively charged phospholipids, such as PIP2, thereby functioning as a “zipper” by pulling together two bilayers. Mice with MBP gene mutation (shiverer mice) fail to form compact myelin, resulting in severe dysmyelination with a characteristic “shivering” symptom (Readhead & Hood, 1990).
Of note, Snaidero et al. (2014) demonstrate that there is a delay of two or three wraps behind the growing tip during compaction, which makes room for newly formed layers. In this study, knockout of 2′,3′-Cyclic nucleotide 3′-phosphodiesterase (CNP), the third most abundant protein in CNS myelin in humans and rodents (Figure 3), results in a reduction of uncompacted wraps within the myelin sheath, indicating a potential role of CNP in preventing the myelin compaction in the innermost regions. It is likely that the leading edge grows together with the accumulation of CNP, while MBP appears two or three wraps behind to initiate myelin compaction. Therefore, proper myelin compaction may require equilibrium between MBP and CNP.
Regulation of the Initial CNS Myelination
Transcription Factors
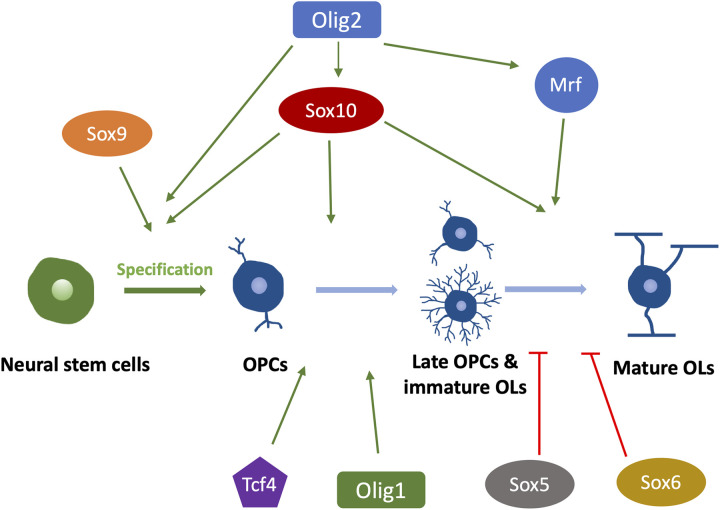
OL lineage development and myelination are under tight transcriptional control (Figure 6). The Olig family is one of the most studied transcriptional regulators for CNS myelination. Olig2 is required during OPCs specification and maintenance, as exemplified by the loss of OPCs in Olig2 null mice (Ligon et al., 2006; Lu et al., 2002). In addition, Olig2 also plays a prominent role in promoting OPCs migration and OLs differentiation (Maire et al., 2010; Wegener et al., 2015; Zhu et al., 2012). Therefore, Olig2 is considered a master transcriptional regulator for most stages of OL lineage development, from OPCs specification to myelin protein production (Zhang et al., 2022). In contrast, Olig1 seems to be more involved in OLs maturation. For example, Olig1 null mice exhibit severer dysmyelination due to the loss of mature/myelinating OLs, eventually leading to death within the third postnatal day (Xin et al., 2005). However, a subsequent study using new Olig1 knockout mice models shows that Olig1 null mice only exhibit a delay of OL differentiation but do not develop long-term myelin deficits (Paes de Faria et al., 2014), indicating a nonessential role of Olig1 in OL development. The possible reasons for the conflicting results may lie in how the mice lines were generated in these two studies. Paes de Faria et al. generated two independent Olig1 null mice models by different methods—one by homologous recombination in mouse embryonic stem cells, and the other one by transgenic rescue of an Olig1/Olig2 double-null mice line. Both lines showed consistent results. In contrast, Xin et al. crossed Olig1 null mice (with a Cre-Pgk-Neo cassette at the Olig1 locus) with FLP-expressing mice to remove the Pgk-Neo, which could potentially affect Olig2 expression considering that Olig1 and Olig2 locate close to each other. In zebrafish, both Olig1 and Olig2 are reported to control OL differentiation (Li et al., 2007; Park et al., 2002), indicating the similar role of the Olig family in regulating OL development.
Figure 6.
Schematic of Transcriptional Regulations in Different Stages of the OL Lineage Development. Arrows (→) indicate positive regulations, and the symbol (T) represents negative regulations. OPCs, oligodendrocyte precursor cells. OLs, oligodendrocyte. TFEB, transcription factor EB.
The Sox family is another major group of transcriptional regulators involved in myelination (Figure 6). Sox10 is required for terminal OLs differentiation, as exemplified by the complete loss of MBP + and PLP + OLs in Sox 10 null mice (Stolt et al., 2002). In zebrafish, the in vivo time-lapse imaging shows that the myelinating OLs in Sox 10 mutants can initiate axon ensheathment without expressing myelin proteins but soon die after the onset of myelination (Takada et al., 2010). This may suggest a more important role of Sox10 in myelin maintenance than during the onset of myelination. Like Olig2, some Sox factors, such as Sox8 and Sox9, are also required for OL specification and differentiation (Stolt et al., 2003, 2004). In contrast, other Sox members are found to inhibit OL differentiation by competing with Sox10, such as Sox5 and Sox6 (Emery & Lu, 2015).
In addition to the two major families of transcriptional regulators mentioned above, some new factors have been recently identified. For example, Tcf4 positively regulates OL differentiation in both myelination and remyelination without engaging its downstream Wnt/ β-catenin pathway (Hammond et al., 2015). Tcf4 null mice develop myelin deficits due to a lack of mature OLs (Phan et al., 2020). More recently, an elegant study shows that transcription factor EB induces selective OLs elimination in normally unmyelinated brain regions, thereby controlling when and where the initial myelination occurs (Sun et al., 2018). In addition, the myelin gene regulatory factor (Mrf) is also proven to crucially regulate CNS myelination by cooperating with Sox10. Mrf physically and functionally interacts with Sox10, synergistically promoting myelin gene expression (Hornig et al., 2013). Indeed, most transcriptional factors regulate myelination by directly binding to the promoter regions of myelin genes, thereby influencing myelin protein (MBP, PLP, and MAG) production (Tiane et al., 2019).
Extracellular Factors
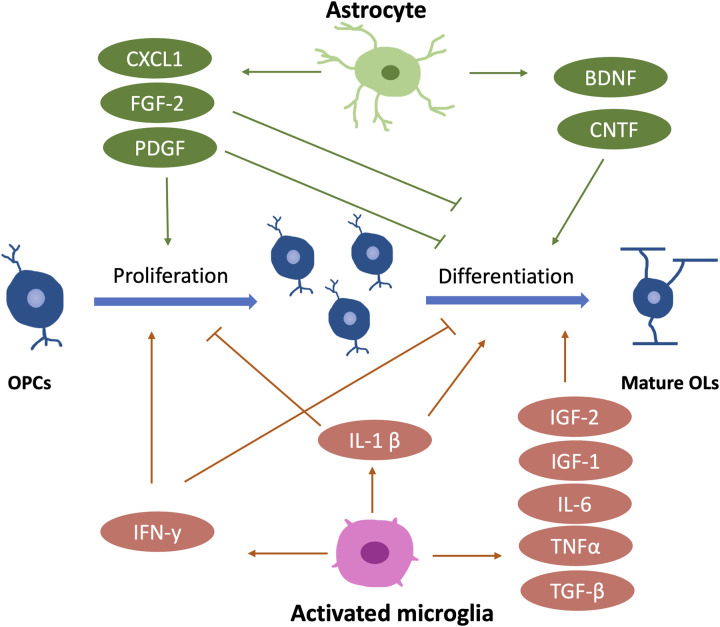
Many studies have highlighted the importance of some growth factors in CNS myelination. Notably, many of them are released by astrocytes and microglia, the other two major glial cell types in the CNS (Figure 7). As an example of astrocyte-derived factors, the platelet-derived growth factor-alpha significantly promotes OPC proliferation while inhibiting its maturation (Traiffort et al., 2020). This is considered an important mechanism for maintaining the OPC pool and preventing OPCs from premature differentiation (Noble et al., 1988; Raff et al., 1988). The brain-derived neurotrophic factor (BDNF), derived from astrocytes and neurons (Miranda et al., 2019), is required for myelin development, as exemplified by the delayed CNS myelination in BDNF null mice (Cellerino et al., 1997). Moreover, the fibroblast growth factor 2 (FGF-2), highly expressed in astrocytes (Newman et al., 2000), is suggested to promote OPCs proliferation and inhibit their differentiation into mature OLs (Bansal & Pfeiffer, 1994; McKinnon et al., 1990). In addition, astrocytes also secrete ciliary neurotrophic factor (CNTF) to benefit OL maturation through the Janus kinase (JAK) pathway (Stankoff et al., 2002). Some microglia-derived growth factors are also reported to regulate myelin development, such as insulin-like growth factor 1 (IGF-1, also produced by neurons) and insulin-like growth factor 2 (IGF-2). IGF-1 null mice exhibit reduced OL survival and maturation, suggesting an important role of IGF-1 in regulating OL lineage development (Ye et al., 2002). Interestingly, IGF-1 is expressed in the activated (ameboid-like) microglia within the corpus callosum until P7, the time point of myelination onset. After P7, IGF-1 expression levels decrease along with the morphological changes of microglia from ameboid to ramified shape (Traiffort et al., 2020). Therefore, IGF-1 may play a unique role during the initiation of myelination rather than after the myelination onset. Besides IGF-1, IGF-2 also promotes OL survival in vitro (Nicholas et al., 2002), although its role in myelination is less extensively studied.
Figure 7.
Major Growth Factors, Chemokines and Cytokines Released from Astrocytes and Microglia During Oligodendrocyte Lineage Development and Myelination. Arrows (→) indicate positive regulations, and the symbol (T) represents negative regulations. OPCs, oligodendrocyte precursor cells. OLs, oligodendrocyte. PDGF, platelet-derived growth factor. BDNF, brain-derived neurotrophic factor. CNTF, ciliary neurotrophic factor. IGF, insulin-like growth factor. FGF-2, fibroblast growth factor 2.
Interestingly, compared to astrocyte-derived factors, microglia-derived factors appear to play a more significant role in OL differentiation and myelination. A study shows that microglia-conditioned medium, but not astrocyte-conditioned medium, significantly enhances myelin protein expression and myelin sheath formation in the neuron-OL myelination coculture (Pang et al., 2013). However, the fundamental mechanism of how microglia regulate myelination through their secretome remains unclear.
Cytokines and chemokines are also important players in the regulation of CNS myelination, and most of them are microglia-derived (Figure 7). Activated microglia secrete TGF-β, IL-1β, TNF-α, IL-6, and IFN-γ to regulate OL development during development, and a blockade of these factors leads to impaired oligodendrogenesis (Shigemoto-Mogami et al., 2014). Specifically, IL-1β enhances OL maturation but negatively regulates OPC proliferation (Vela et al., 2002). In contrast, IFN-γ promotes OPC proliferation while inhibiting its differentiation (Baerwald & Popko, 1998; Chew et al., 2005), indicating that different microglia-derived cytokines may have opposite influences on OL development, and they work synergistically to determine OL fates. Besides microglia, astrocytes also produce chemokines to regulate OL development, such as CXCL-1. CXCL-1 may inhibit OPC migration while promoting its proliferation (Tsai et al., 2002).
Hormones, such as thyroid hormone (T3), are also closely involved in myelination. During early development, brain T3 is of maternal origin via placenta (Morreale de Escobar et al., 1987). T3 controls OL development in mammals by inhibiting OPC proliferation before promoting OL differentiation in vitro and in vivo (Almazan et al., 1985; Calza et al., 2002). Consistently, a lack of T3 in zebrafish results in hypomyelination, which is restored after T3 supplementation (Farías-Serratos et al., 2021). This sheds light on a well-conserved regulation mechanism of CNS myelination between mammals and zebrafish.
Axonal Regulations
Regarding axonal signaling during initial myelination, one interesting fact is that OLs can myelinate PFA-fixed axons and even nanofibers in vitro. This is agreed by a more recent study showing OLs derived from different areas (cortex and spinal cord) can myelinate microfibers in vitro with the same internode length as seen in corresponding regions in vivo (Bechler et al., 2015). However, there may be other explanations. Alternatively, it is very likely that some inhibitory molecules expressed on the surface of axons prevent myelination from happening until neutralized in vivo. Indeed, many studies have identified various axonal adhesion molecules that negatively regulate OLs development and myelination. PSA-NCAM and L1, the best-studied adhesion molecules, are expressed on nonmyelinated axons and are significantly downregulated during the onset of myelination (Charles et al., 2000; Jakovcevski et al., 2007). Presumably, these inhibitory adhesion molecules need to be neutralized before the wrapping process initiates, although a more direct correlation between these two events awaits testing. LINGO-1 is another inhibitor of myelination. In rats, downregulation of LINGO-1 promotes myelination, and conversely, overexpression of LINGO-1 inhibits myelin formation by activating RhoA signaling (Mi et al., 2005). Consistently, LINGO-1 knockdown in zebrafish enhances OL differentiation and promotes subsequent myelination (Yin & Hu, 2014). Together, these inhibitory molecules are closely involved in regulating myelination development, potentially the onset of myelination. If these inhibitors are the key factors to control the timing of myelination onset, some mechanisms must exist to regulate the persistence of the inhibitory signals during initial myelination, such as the “inhibitor of the inhibitor.”
In addition to adhesion molecules, neuronal activity is also suggested to profoundly regulate myelination in vivo. A study shows that pharmacogenetic stimulation of somatosensory axons in the mouse brain increases OPCs differentiation, resulting in thicker myelin in simulated axons compared to neighboring nonsimulated ones (Mitew et al., 2018). Moreover, using zebrafish, a recent study (Mensch et al., 2015) shows that a reduction in synaptic vesicle release results in a decrease in the axon numbers myelinated by one single OL. Furthermore, when increasing neural activity, 40% more axons are myelinated by a single OL, indicating an activity-dependent regulation during myelination. However, even without neural activity, an individual OL still myelinates around 60% axons of its full capacity. Therefore, it is reasonable to believe that neuronal activity is an essential modulator for myelin sheath development and refinement, although it may not be required for the ensheathment itself. Indeed, an emerging consensus nowadays is that the activity-driven myelination plasticity is essentially important for the myelination maintenance stage, during which learning and exercising occur frequently. Socially isolated mice develop hypomyelination in the prefrontal cortex (Liu et al., 2012; Makinodan et al., 2012). Consistently, an enriched environment increases myelination in rat corpus callosum (Zhao et al., 2012). Together, these studies highlight the important role of environmental input in myelin sheath formation. One can assume that the results are led by the changes in neuronal activity, although more straightforward evidence is needed in future studies.
The molecular mechanisms of this activity-dependent myelination remain unclear. Some studies propose that the glutamate released from axons may interact with myelinating OLs since both OPCs and OLs express the AMPA and NMDA receptors for glutamate (Bakiri et al., 2009; Butt, 2006; Káradóttir & Attwell, 2007). Alternatively, another study suggests that neuronal activity may control the release of neuregulin 1 from neural axons, thereby switching OLs between activity-dependent mode and activity-independent mode (Lundgaard et al., 2013).
Signaling Pathways
Many highly conserved intracellular signaling pathways, such as Wnt/β-catenin, PI3 K/AKT/mTOR, and ERK/MAPK signaling pathways, are suggested to tightly regulate CNS myelination. Initially, Wnt signaling is identified as a negative regulator of OL differentiation and myelination. Activation of Wnt signaling in mice leads to a delayed appearance of mature OLs and myelin proteins, while the number of OPCs remains unchanged (Feigenson et al., 2009), suggesting that the Wnt pathway is potentially involved in the OL differentiation stage. Similarly, upregulation of Wnt/β-catenin by deleting its antagonist in mice, such as APC and Axin2, results in impaired OL maturation, subsequently leading to hypomyelination (Fancy et al., 2011; Lang et al., 2013). However, conflicting results are obtained when inhibiting endogenous β-catenin in the conditional knockout mice. One study demonstrates that the Cre activity-induced β-catenin knockout does not influence OL differentiation (Lang et al., 2013), whereas another study shows that the conditional knockout of β-catenin causes a significant reduction in the numbers of mature OLs in mice brains from E18 to P15 (Dai et al., 2014). Of note, the latter study applies tamoxifen to delete the β-catenin gene much earlier than the former (E10.5 versus P6), potentially leading to conflicting results. Early loss of β-catenin may have more meaningful influences on OL development. However, in a zebrafish study, siRNA-induced knockdown of β-catenin inhibits OL differentiation and myelination in zebrafish larvae (Tawk et al., 2011), suggesting that the Wnt signaling pathway may also have promoting effects on myelination. Further studies are needed to clarify these seemingly conflicting conclusions.
PI3K/AKT/mTOR is another classic intracellular signaling pathway involved in many basic processes, such as cell proliferation and survival (Dudek et al., 1997; Franke et al., 1997). OL development is no exception. Overactivation of PI3 K/AKT/mTOR results in increased myelin thickness in mice (Flores et al., 2008; Harrington et al., 2010). Conversely, the downregulation of mTOR by its inhibitor rapamycin significantly impairs myelination in transgenic mice (Narayanan et al., 2009). Interestingly, in studies using knockout mice, the spinal cord seems more vulnerable to the deletion of mTOR than the brain. Mice with mTOR conditional ablation exhibit nearly unaffected myelination in the corpus callosum, while the spinal cord, in contrast, shows severe hypomyelination (Bercury et al., 2014; Wahl et al., 2014). Many studies show that mTOR deletion causes a reduction in MBP mRNA and MBP proteins in the spinal cord (Bercury et al., 2014; Wahl et al., 2014), highlighting the role of mTOR signaling in regulating MBP production at both transcription and translation levels.
ERK/MAPK is also considered a positive regulator of CNS myelination. Reduced myelin thickness is observed in Erk1/2 conditional knockout mice (Ishii et al., 2019). Furthermore, one study identifies the FGF-Receptor-type-2 (FGFR2) as a key upstream signal of ERK by showing that the myelin thickness reduction induced by conditional ablation of FGFR2 can be rescued by upregulating ERK signaling in transgenic mice (Furusho et al., 2017). ERK signaling also regulates adulthood myelination, as exemplified by the reinitiation of myelin growth in adult mice following ERK upregulation (Ishii et al., 2016; Jeffries et al., 2016). More recently, a study points out that the ERK/MAPK and the PI3 K/AKT/mTOR signaling pathways need to work both independently and cooperatively for a finely tuned myelination (Ishii et al., 2019), suggesting the presence of crosstalk among these key signaling pathways during myelination.
Some other signaling pathways also contribute to myelin development. For example, the bone morphogenetic protein (BMP) pathway is a potent inhibitor of OL differentiation and myelin protein expression (Grinspan, 2015). When treating rodent OPCs with BMP4 in vitro, their differentiation is significantly inhibited in a dose-dependent manner (Grinspan et al., 2000). Similarly, Notch signaling pathways are also suggested to inhibit OPCs differentiation during development (Genoud et al., 2002; Wang et al., 1998).
Remyelination Versus Initial Myelination
Remyelination refers to the adaptive responses to dys- and demyelination whereby the myelin sheath is structurally and functionally restored. Many similarities are shared between initial myelination and remyelination regarding the major steps and the regulating mechanisms. By analogy with initial myelination, remyelination starts with the recruitment of adult OPCs to the lesion sites, followed by morphological changes of OPCs, OPCs differentiation, wrapping, and myelin compaction. Both OPCs and adult OPCs proliferate and migrate to the spot where myelination is needed, although adult OPCs have a longer basal cell cycle time and slower migration speed (Wolswijk & Noble, 1989). As seen in initial myelination, an upregulation of several transcription factors (Olig2, Sox2) is also observed when remyelination initiates (Fancy et al., 2004; Shen et al., 2008; Watanabe et al., 2004). In addition, the inhibitory roles of LINGO-1 and Wnt signaling can also serve as important regulating mechanisms for remyelination (Fancy et al., 2009; Mi et al., 2007).
However, the differences between initial myelination and remyelination are documented. First, the relationship between the caliber of axons and the thickness of myelin sheath, namely g-ratio, is altered. The optimal g-ratio in initial myelination is 0.6, while remyelination only generates a thinner and shorter myelin sheath, resulting in a greater g-ratio than expected (Blakemore, 1974). Second, some molecular mechanisms vary between initial myelination and remyelination. On the one hand, some molecules are more involved in initial myelination than in remyelination. For example, a recent study shows that fatty acid–binding protein 7 (FABP7) is important in OPCs differentiation during development but not in remyelination (Foerster et al., 2020). On the other hand, some mechanisms are, at least for now, remyelination-specific. For example, CD47, a well-documented “don’t eat me signal,” may serve as a key mechanism of remyelination failure. CD47 tags myelin debris after demyelination and functions as a marker of “self,” thereby preventing their clearance by microglia (Gitik et al., 2011). However, the role of CD47 in initial myelination has not been addressed yet. Indeed, since initial myelination and remyelination have similarities but are not identical, it is reasonable that they share some signaling pathways but also own their unique way of orchestrating the processes.
Concluding Remarks
Research regarding initial CNS myelination has gained great progression due to researchers’ passion for this field. It is well recognized that myelination involves many steps regulated in time and space, during which various molecules and signaling pathways are responsible for the orchestration.
In this review, we have covered some mechanisms in initial CNS myelination. However, the picture is incomplete, with many challenging questions remaining to be answered. For example, the key signal that triggers the initiation of CNS myelination remains largely unknown. In contrast, a large body of evidence has suggested a key mechanism for PNS myelination. Specifically, the level of neuregulin 1 type III expressed on the PNS axons is a pivotal instructive cue for myelination. Myelinated axons express significantly higher levels of neuregulin 1 type III than unmyelinated ones, resulting in different ensheathment fates of PNS axons (Taveggia et al., 2005). This explains the fact that Schwann cells, the myelinating glia cells in the PNS, do not myelinate the neuregulin 1 type III-deficient nanofibers in vitro (Bechler et al., 2015). However, such key factors or mechanisms have yet to be identified in the CNS, which will become a focus in future studies. Fortunately, a diversity of animal models (zebrafish) and imaging techniques (3D EM) offer preeminent tools for in vivo myelination studies. Hopefully, researchers can take advantage of them to get a deeper insight into not only initial myelination but also myelin repair after injuries or demyelinating diseases, thereby helping identify the potential treatment targets for them.
Acknowledgments
The authors would like to thank China Scholarship Council as it provides financial support for author Q.Y's study in Canada.
Footnotes
Author contributions: J.K. selected the topic. Q.Y. wrote the paper; T.G. and Y.G. reviewed and edited the paper; J.K. critically revised the paper.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Q.Y. has received financial support from China Scholarship Council.
ORCID iD: Qiang Yu https://orcid.org/0000-0002-0094-0479
References
- Almazan G., Honegger P., Matthieu J. M. (1985). Triiodothyronine stimulation of oligodendroglial differentiation and myelination. A developmental study. Dev Neurosci, 7(1), 45–54. 10.1159/000112275 [DOI] [PubMed] [Google Scholar]
- Almeida R. G., Czopka T., Ffrench-Constant C., Lyons D. A. (2011). Individual axons regulate the myelinating potential of single oligodendrocytes in vivo. Development, 138(20), 4443–4450. 10.1242/dev.071001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back S. A., Luo N. L., Borenstein N. S., Levine J. M., Volpe J. J., Kinney H. C. (2001). Late oligodendrocyte progenitors coincide with the developmental window of vulnerability for human perinatal white matter injury. J Neurosci, 21(4), 1302–1312. 10.1523/jneurosci.21-04-01302.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer A. S., Syed Y. A., Kang S. U., Mitteregger D., Vig R., Ffrench-Constant C., Franklin R. J., Altmann F., Lubec G., Kotter M. R. (2009). Myelin-mediated inhibition of oligodendrocyte precursor differentiation can be overcome by pharmacological modulation of Fyn-RhoA and protein kinase C signalling. Brain, 132(Pt 2), 465–481. 10.1093/brain/awn334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerwald K. D., Popko B. (1998). Developing and mature oligodendrocytes respond differently to the immune cytokine interferon-gamma. J Neurosci Res, 52(2), 230–239. [DOI] [PubMed] [Google Scholar]
- Bakiri Y., Burzomato V., Frugier G., Hamilton N. B., Káradóttir R., Attwell D. (2009). Glutamatergic signaling in the brain’s white matter. Neuroscience, 158(1), 266–274. 10.1016/j.neuroscience.2008.01.015 [DOI] [PubMed] [Google Scholar]
- Bansal R., Pfeiffer S. E. (1994). Inhibition of protein and lipid sulfation in oligodendrocytes blocks biological responses to FGF-2 and retards cytoarchitectural maturation, but not developmental lineage progression. Dev Biol, 162(2), 511–524. 10.1006/dbio.1994.1105 [DOI] [PubMed] [Google Scholar]
- Barateiro A., Brites D., Fernandes A. (2016). Oligodendrocyte development and myelination in neurodevelopment: Molecular mechanisms in health and disease. Curr Pharm Des, 22(6), 656–679. 10.2174/1381612822666151204000636 [DOI] [PubMed] [Google Scholar]
- Barateiro A., Fernandes A. (2014). Temporal oligodendrocyte lineage progression: In vitro models of proliferation, differentiation and myelination. Biochim Biophys Acta, 1843(9), 1917–1929. 10.1016/j.bbamcr.2014.04.018 [DOI] [PubMed] [Google Scholar]
- Baumann N., Pham-Dinh D. (2001). Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev, 81(2), 871–927. 10.1152/physrev.2001.81.2.871 [DOI] [PubMed] [Google Scholar]
- Bechler M. E., Byrne L., Ffrench-Constant C. (2015). CNS Myelin sheath lengths are an intrinsic property of oligodendrocytes. Curr Biol, 25(18), 2411–2416. 10.1016/j.cub.2015.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercury K. K., Dai J., Sachs H. H., Ahrendsen J. T., Wood T. L., Macklin W. B. (2014). Conditional ablation of raptor or rictor has differential impact on oligodendrocyte differentiation and CNS myelination. J Neurosci, 34(13), 4466–4480. 10.1523/jneurosci.4314-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakemore W. F. (1974). Pattern of remyelination in the CNS. Nature, 249(457), 577–578. 10.1038/249577a0 [DOI] [PubMed] [Google Scholar]
- Brösamle C., Halpern M. E. (2002). Characterization of myelination in the developing zebrafish. Glia, 39(1), 47–57. 10.1002/glia.10088 [DOI] [PubMed] [Google Scholar]
- Brown T. L., Macklin W. B. (2019). The actin cytoskeleton in myelinating cells. Neurochem Res, 45(3), 684–693. 10.1007/s11064-019-02753-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge M. B., Bunge R. P., Ris H. (1961). Ultrastructural study of remyelination in an experimental lesion in adult cat spinal cord. J Biophys Biochem Cytol, 10(1), 67–94. 10.1083/jcb.10.1.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt A. M. (2006). Neurotransmitter-mediated calcium signalling in oligodendrocyte physiology and pathology. Glia, 54(7), 666–675. 10.1002/glia.20424 [DOI] [PubMed] [Google Scholar]
- Calza L., Fernandez M., Giuliani A., Aloe L., Giardino L. (2002). Thyroid hormone activates oligodendrocyte precursors and increases a myelin-forming protein and NGF content in the spinal cord during experimental allergic encephalomyelitis. Proc Natl Acad Sci U S A, 99(5), 3258–3263. 10.1073/pnas.052704499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellerino A., Carroll P., Thoenen H., Barde Y. A. (1997). Reduced size of retinal ganglion cell axons and hypomyelination in mice lacking brain-derived neurotrophic factor. Mol Cell Neurosci, 9(5–6), 397–408. 10.1006/mcne.1997.0641 [DOI] [PubMed] [Google Scholar]
- Chang-Ileto B., Frere S. G., Chan R. B., Voronov S. V., Roux A., Di Paolo G. (2011). Synaptojanin 1-mediated PI(4,5)P2 hydrolysis is modulated by membrane curvature and facilitates membrane fission. Dev Cell, 20(2), 206–218. 10.1016/j.devcel.2010.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles P., Hernandez M. P., Stankoff B., Aigrot M. S., Colin C., Rougon G., Zalc B., Lubetzki C. (2000). Negative regulation of central nervous system myelination by polysialylated-neural cell adhesion molecule. Proc Natl Acad Sci U S A, 97(13), 7585–7590. 10.1073/pnas.100076197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew L. J., King W. C., Kennedy A., Gallo V. (2005). Interferon-gamma inhibits cell cycle exit in differentiating oligodendrocyte progenitor cells. Glia, 52(2), 127–143. 10.1002/glia.20232 [DOI] [PubMed] [Google Scholar]
- Coman I., Aigrot M. S., Seilhean D., Reynolds R., Girault J. A., Zalc B., Lubetzki C. (2006). Nodal, paranodal and juxtaparanodal axonal proteins during demyelination and remyelination in multiple sclerosis. Brain, 129(12), 3186–3195. 10.1093/brain/awl144 [DOI] [PubMed] [Google Scholar]
- Dai Z. M., Sun S., Wang C., Huang H., Hu X., Zhang Z., Lu Q. R., Qiu M. (2014). Stage-specific regulation of oligodendrocyte development by Wnt/β-catenin signaling. J Neurosci, 34(25), 8467–8473. 10.1523/jneurosci.0311-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M. R., Polito A., Levine J. M., Reynolds R. (2003). NG2-expressing Glial progenitor cells: An abundant and widespread population of cycling cells in the adult rat CNS. Mol Cell Neurosci, 24(2), 476–488. 10.1016/s1044-7431(03)00210-0 [DOI] [PubMed] [Google Scholar]
- Dietrich R. B., Bradley W. G., Zaragoza E. J. t., Otto R. J., Taira R. K., Wilson G. H., Kangarloo H. (1988). MR evaluation of early myelination patterns in normal and developmentally delayed infants. AJR Am J Roentgenol, 150(4), 889–896. 10.2214/ajr.150.4.889 [DOI] [PubMed] [Google Scholar]
- Dudek H., Datta S. R., Franke T. F., Birnbaum M. J., Yao R., Cooper G. M., Segal R. A., Kaplan D. R., Greenberg M. E. (1997). Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science (New York, N.Y.), 275(5300), 661–665. 10.1126/science.275.5300.661 [DOI] [PubMed] [Google Scholar]
- Edgar J. M., Garbern J. (2004). The myelinated axon is dependent on the myelinating cell for support and maintenance: Molecules involved. J Neurosci Res, 76(5), 593–598. 10.1002/jnr.20063 [DOI] [PubMed] [Google Scholar]
- Emery B., Lu Q. R. (2015). Transcriptional and epigenetic regulation of oligodendrocyte development and myelination in the central nervous system. Cold Spring Harb Perspect Biol, 7(9), a020461. 10.1101/cshperspect.a020461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy S. P., Baranzini S. E., Zhao C., Yuk D. I., Irvine K. A., Kaing S., Sanai N., Franklin R. J., Rowitch D. H. (2009). Dysregulation of the wnt pathway inhibits timely myelination and remyelination in the mammalian CNS. Genes Dev, 23(13), 1571–1585. 10.1101/gad.1806309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy S. P., Harrington E. P., Yuen T. J., Silbereis J. C., Zhao C., Baranzini S. E., Bruce C. C., Otero J. J., Huang E. J., Nusse R., Franklin R. J., Rowitch D. H. (2011). Axin2 as regulatory and therapeutic target in newborn brain injury and remyelination. Nat Neurosci, 14(8), 1009–1016. 10.1038/nn.2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy S. P., Zhao C., Franklin R. J. (2004). Increased expression of Nkx2.2 and Olig2 identifies reactive oligodendrocyte progenitor cells responding to demyelination in the adult CNS. Mol Cell Neurosci, 27(3), 247–254. 10.1016/j.mcn.2004.06.015 [DOI] [PubMed] [Google Scholar]
- Farías-Serratos B. M., Lazcano I., Villalobos P., Darras V. M., Orozco A. (2021). Thyroid hormone deficiency during zebrafish development impairs central nervous system myelination. PLoS One, 16(8), e0256207. 10.1371/journal.pone.0256207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigenson K., Reid M., See J., Crenshaw E. B., III, Grinspan J. B. (2009). Wnt signaling is sufficient to perturb oligodendrocyte maturation. Mol Cell Neurosci, 42(3), 255–265. 10.1016/j.mcn.2009.07.010 [DOI] [PubMed] [Google Scholar]
- Flores A. I., Narayanan S. P., Morse E. N., Shick H. E., Yin X., Kidd G., Avila R. L., Kirschner D. A., Macklin W. B. (2008). Constitutively active akt induces enhanced myelination in the CNS. J Neurosci, 28(28), 7174–7183. 10.1523/jneurosci.0150-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerster S., Guzman de la Fuente A., Kagawa Y., Bartels T., Owada Y., Franklin R. J. M. (2020). The fatty acid binding protein FABP7 is required for optimal oligodendrocyte differentiation during myelination but not during remyelination. Glia, 68(7), 1410–1420. 10.1002/glia.23789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke T. F., Kaplan D. R., Cantley L. C. (1997). PI3K: Downstream AKTion blocks apoptosis. Cell, 88(4), 435–437. 10.1016/s0092-8674(00)81883-8 [DOI] [PubMed] [Google Scholar]
- Furusho M., Ishii A., Bansal R. (2017). Signaling by FGF receptor 2, not FGF receptor 1, regulates myelin thickness through activation of ERK1/2-MAPK, which promotes mTORC1 activity in an Akt-independent manner. J Neurosci, 37(11), 2931–2946. 10.1523/jneurosci.3316-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargareta V. I., Reuschenbach J., Siems S. B., Sun T., Piepkorn L., Mangana C., Späte E., Goebbels S., Huitinga I., Möbius W., Nave K. A., Jahn O., Werner H. B. (2022). Conservation and divergence of myelin proteome and oligodendrocyte transcriptome profiles between humans and mice. Elife, 11, e77019. 10.7554/eLife.77019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud S., Lappe-Siefke C., Goebbels S., Radtke F., Aguet M., Scherer S. S., Suter U., Nave K. A., Mantei N. (2002). Notch1 control of oligodendrocyte differentiation in the spinal cord. J Cell Biol, 158(4), 709–718. 10.1083/jcb.200202002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitik M., Liraz-Zaltsman S., Oldenborg P. A., Reichert F., Rotshenker S. (2011). Myelin down-regulates myelin phagocytosis by microglia and macrophages through interactions between CD47 on myelin and SIRPα (signal regulatory protein-α) on phagocytes. J Neuroinflammation, 8, 24. 10.1186/1742-2094-8-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebbels S., Oltrogge J. H., Kemper R., Heilmann I., Bormuth I., Wolfer S., Wichert S. P., Mobius W., Liu X., Lappe-Siefke C., Rossner M. J., Groszer M., Suter U., Frahm J., Boretius S., Nave K. A. (2010). Elevated phosphatidylinositol 3,4,5-trisphosphate in glia triggers cell-autonomous membrane wrapping and myelination. J Neurosci, 30(26), 8953–8964. 10.1523/jneurosci.0219-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebbels S., Wieser G. L., Pieper A., Spitzer S., Weege B., Yan K., Edgar J. M., Yagensky O., Wichert S. P., Agarwal A., Karram K., Renier N., Tessier-Lavigne M., Rossner M. J., Káradóttir R. T., Nave K. A. (2017). A neuronal PI(3,4,5)P(3)-dependent program of oligodendrocyte precursor recruitment and myelination. Nat Neurosci, 20(1), 10–15. 10.1038/nn.4425 [DOI] [PubMed] [Google Scholar]
- Grinspan J. B. (2015). Bone morphogenetic proteins: Inhibitors of myelination in development and disease. Vitam Horm, 99, 195–222. 10.1016/bs.vh.2015.05.005 [DOI] [PubMed] [Google Scholar]
- Grinspan J. B., Edell E., Carpio D. F., Beesley J. S., Lavy L., Pleasure D., Golden J. A. (2000). Stage-specific effects of bone morphogenetic proteins on the oligodendrocyte lineage. J Neurobiol, 43(1), 1–17. [DOI] [PubMed] [Google Scholar]
- Hammond E., Lang J., Maeda Y., Pleasure D., Angus-Hill M., Xu J., Horiuchi M., Deng W., Guo F. (2015). The wnt effector transcription factor 7-like 2 positively regulates oligodendrocyte differentiation in a manner independent of wnt/β-catenin signaling. J Neurosci, 35(12), 5007–5022. 10.1523/jneurosci.4787-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington E. P., Zhao C., Fancy S. P., Kaing S., Franklin R. J., Rowitch D. H. (2010). Oligodendrocyte PTEN is required for myelin and axonal integrity, not remyelination. Ann Neurol, 68(5), 703–716. 10.1002/ana.22090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand C., Remahl S., Persson H., Bjartmar C. (1993). Myelinated nerve fibres in the CNS. Prog Neurobiol, 40(3), 319–384. 10.1016/0301-0082(93)90015-k [DOI] [PubMed] [Google Scholar]
- Hines J. H., Ravanelli A. M., Schwindt R., Scott E. K., Appel B. (2015). Neuronal activity biases axon selection for myelination in vivo. Nat Neurosci, 18(5), 683–689. 10.1038/nn.3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig J., Fröb F., Vogl M. R., Hermans-Borgmeyer I., Tamm E. R., Wegner M. (2013). The transcription factors Sox0 and Myrf define an essential regulatory network module in differentiating oligodendrocytes. PLoS Genet, 9(10), e1003907. 10.1371/journal.pgen.1003907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüppi P. S., Warfield S., Kikinis R., Barnes P. D., Zientara G. P., Jolesz F. A., Tsuji M. K., Volpe J. J. (1998). Quantitative magnetic resonance imaging of brain development in premature and mature newborns. Ann Neurol, 43(2), 224–235. 10.1002/ana.410430213 [DOI] [PubMed] [Google Scholar]
- Ishii A., Furusho M., Dupree J. L., Bansal R. (2016). Strength of ERK1/2 MAPK activation determines its effect on myelin and axonal integrity in the adult CNS. J Neurosci, 36(24), 6471–6487. 10.1523/jneurosci.0299-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii A., Furusho M., Macklin W., Bansal R. (2019). Independent and cooperative roles of the Mek/ERK1/2-MAPK and PI3 K/Akt/mTOR pathways during developmental myelination and in adulthood. Glia, 67(7), 1277–1295. 10.1002/glia.23602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn O., Siems S. B., Kusch K., Hesse D., Jung R. B., Liepold T., Uecker M., Sun T., Werner H. B. (2020). The CNS myelin proteome: Deep profile and persistence after post-mortem delay. Front Cell Neurosci, 14, 239. 10.3389/fncel.2020.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovcevski I., Mo Z., Zecevic N. (2007). Down-regulation of the axonal polysialic acid-neural cell adhesion molecule expression coincides with the onset of myelination in the human fetal forebrain. Neuroscience, 149(2), 328–337. 10.1016/j.neuroscience.2007.07.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries M. A., Urbanek K., Torres L., Wendell S. G., Rubio M. E., Fyffe-Maricich S. L. (2016). ERK1/2 Activation in preexisting oligodendrocytes of adult mice drives new myelin synthesis and enhanced CNS function. J Neurosci, 36(35), 9186–9200. 10.1523/jneurosci.1444-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Káradóttir R., Attwell D. (2007). Neurotransmitter receptors in the life and death of oligodendrocytes. Neuroscience, 145(4), 1426–1438. 10.1016/j.neuroscience.2006.08.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessaris N., Fogarty M., Iannarelli P., Grist M., Wegner M., Richardson W. D. (2006). Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci, 9(2), 173–179. 10.1038/nn1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney H. C., Brody B. A., Kloman A. S., Gilles F. H. (1988). Sequence of central nervous system myelination in human infancy. II. Patterns of myelination in autopsied infants. J Neuropathol Exp Neurol, 47(3), 217–234. 10.1097/00005072-198805000-00003 [DOI] [PubMed] [Google Scholar]
- Kirby B. B., Takada N., Latimer A. J., Shin J., Carney T. J., Kelsh R. N., Appel B. (2006). In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat Neurosci, 9(12), 1506–1511. 10.1038/nn1803 [DOI] [PubMed] [Google Scholar]
- Kolodny E. H. (1993). Dysmyelinating and demyelinating conditions in infancy. Curr Opin Neurol Neurosurg, 6(3), 379–386. [PubMed] [Google Scholar]
- Lang J., Maeda Y., Bannerman P., Xu J., Horiuchi M., Pleasure D., Guo F. (2013). Adenomatous polyposis coli regulates oligodendroglial development. J Neurosci, 33(7), 3113–3130. 10.1523/jneurosci.3467-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Chong S. Y., Tuck S. J., Corey J. M., Chan J. R. (2013). A rapid and reproducible assay for modeling myelination by oligodendrocytes using engineered nanofibers. Nat Protoc, 8(4), 771–782. 10.1038/nprot.2013.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Lu Y., Smith H. K., Richardson W. D. (2007). Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J Neurosci, 27(52), 14375–14382. 10.1523/jneurosci.4456-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon K. L., Kesari S., Kitada M., Sun T., Arnett H. A., Alberta J. A., Anderson D. J., Stiles C. D., Rowitch D. H. (2006). Development of NG2 neural progenitor cells requires Olig gene function. Proc Natl Acad Sci U S A, 103(20), 7853–7858. 10.1073/pnas.0511001103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Dietz K., DeLoyht J. M., Pedre X., Kelkar D., Kaur J., Vialou V., Lobo M. K., Dietz D. M., Nestler E. J., Dupree J., Casaccia P. (2012). Impaired adult myelination in the prefrontal cortex of socially isolated mice. Nat Neurosci, 15(12), 1621–1623. 10.1038/nn.3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q. R., Sun T., Zhu Z., Ma N., Garcia M., Stiles C. D., Rowitch D. H. (2002). Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell, 109(1), 75–86. 10.1016/s0092-8674(02)00678-5 [DOI] [PubMed] [Google Scholar]
- Lundgaard I., Luzhynskaya A., Stockley J. H., Wang Z., Evans K. A., Swire M., Volbracht K., Gautier H. O., Franklin R. J., Attwell D., Káradóttir R. T. (2013). Neuregulin and BDNF induce a switch to NMDA receptor-dependent myelination by oligodendrocytes. PLoS Biol, 11(12), e1001743. 10.1371/journal.pbio.1001743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maire C. L., Wegener A., Kerninon C., Nait Oumesmar B. (2010). Gain-of-function of olig transcription factors enhances oligodendrogenesis and myelination. Stem Cells, 28(9), 1611–1622. 10.1002/stem.480 [DOI] [PubMed] [Google Scholar]
- Makinodan M., Rosen K. M., Ito S., Corfas G. (2012). A critical period for social experience-dependent oligodendrocyte maturation and myelination. Science, 337(6100), 1357–1360. 10.1126/science.1220845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayoral S. R., Etxeberria A., Shen Y. A., Chan J. R. (2018). Initiation of CNS myelination in the optic nerve is dependent on axon caliber. Cell Rep, 25(3), 544–550.e543. 10.1016/j.celrep.2018.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon R. D., Matsui T., Dubois-Dalcq M., Aaronsont S. A. (1990). FGF modulates the PDGF-driven pathway of oligodendrocyte development. Neuron, 5(5), 603–614. 10.1016/0896-6273(90)90215-2 [DOI] [PubMed] [Google Scholar]
- McMahon H. T., Boucrot E. (2015). Membrane curvature at a glance. J Cell Sci, 128(6), 1065–1070. 10.1242/jcs.114454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mensch S., Baraban M., Almeida R., Czopka T., Ausborn J., El Manira A., Lyons D. A. (2015). Synaptic vesicle release regulates myelin sheath number of individual oligodendrocytes in vivo. Nat Neurosci, 18(5), 628–630. 10.1038/nn.3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., Hu B., Hahm K., Luo Y., Kam Hui E. S., Yuan Q., Wong W. M., Wang L., Su H., Chu T. H., Guo J., Zhang W., So K. F., Pepinsky B., Shao Z., Graff C., Garber E., Jung V., Wu E. X., Wu W. (2007). LINGO-1 antagonist promotes spinal cord remyelination and axonal integrity in MOG-induced experimental autoimmune encephalomyelitis. Nat Med, 13(10), 1228–1233. 10.1038/nm1664 [DOI] [PubMed] [Google Scholar]
- Mi S., Miller R. H., Lee X., Scott M. L., Shulag-Morskaya S., Shao Z., Chang J., Thill G., Levesque M., Zhang M., Hession C., Sah D., Trapp B., He Z., Jung V., McCoy J. M., Pepinsky R. B. (2005). LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci, 8(6), 745–751. 10.1038/nn1460 [DOI] [PubMed] [Google Scholar]
- Miranda M., Morici J. F., Zanoni M. B., Bekinschtein P. (2019). Brain-derived neurotrophic factor: a key molecule for memory in the healthy and the pathological brain. Front Cell Neurosci, 13. 10.3389/fncel.2019.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitew S., Gobius I., Fenlon L. R., McDougall S. J., Hawkes D., Xing Y. L., Bujalka H., Gundlach A. L., Richards L. J., Kilpatrick T. J., Merson T. D., Emery B. (2018). Pharmacogenetic stimulation of neuronal activity increases myelination in an axon-specific manner. Nat Commun, 9(1), 306. 10.1038/s41467-017-02719-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje M. (2018). Myelin plasticity and nervous system function. Ann Rev Neurosci, 41, 61–76. 10.1146/annurev-neuro-080317-061853 [DOI] [PubMed] [Google Scholar]
- Morreale de Escobar G., Obregon M. J., Escobar del Rey F. (1987). Fetal and maternal thyroid hormones. Horm Res, 26(1–4), 12–27. 10.1159/000180681 [DOI] [PubMed] [Google Scholar]
- Morris J. K., Willard B. B., Yin X., Jeserich G., Kinter M., Trapp B. D. (2004). The 36 K protein of zebrafish CNS myelin is a short-chain dehydrogenase. Glia, 45(4), 378–391. 10.1002/glia.10338 [DOI] [PubMed] [Google Scholar]
- Münzel E. J., Schaefer K., Obirei B., Kremmer E., Burton E. A., Kuscha V., Becker C. G., Brösamle C., Williams A., Becker T. (2012). Claudin k is specifically expressed in cells that form myelin during development of the nervous system and regeneration of the optic nerve in adult zebrafish. Glia, 60(2), 253–270. 10.1002/glia.21260 [DOI] [PubMed] [Google Scholar]
- Narayanan S. P., Flores A. I., Wang F., Macklin W. B. (2009). Akt signals through the mammalian target of rapamycin pathway to regulate CNS myelination. J Neurosci, 29(21), 6860–6870. 10.1523/jneurosci.0232-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nave K. A., Werner H. B. (2014). Myelination of the nervous system: Mechanisms and functions. Ann Rev Cell Dev Biol, 30, 503–533. 10.1146/annurev-cellbio-100913-013101 [DOI] [PubMed] [Google Scholar]
- Nawaz S., Sanchez P., Schmitt S., Snaidero N., Mitkovski M., Velte C., Bruckner B. R., Alexopoulos I., Czopka T., Jung S. Y., Rhee J. S., Janshoff A., Witke W., Schaap I. A. T., Lyons D. A., Simons M. (2015). Actin filament turnover drives leading edge growth during myelin sheath formation in the central nervous system. Dev Cell, 34(2), 139–151. 10.1016/j.devcel.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M. P., Féron F., Mackay-Sim A. (2000). Growth factor regulation of neurogenesis in adult olfactory epithelium. Neuroscience, 99(2), 343–350. 10.1016/s0306-4522(00)00194-9 [DOI] [PubMed] [Google Scholar]
- Nicholas R. S., Stevens S., Wing M. G., Compston D. A. (2002). Microglia-derived IGF-2 prevents TNFalpha induced death of mature oligodendrocytes in vitro. J Neuroimmunol, 124(1–2), 36–44. 10.1016/s0165-5728(02)00011-5 [DOI] [PubMed] [Google Scholar]
- Noble M., Murray K., Stroobant P., Waterfield M. D., Riddle P. (1988). Platelet-derived growth factor promotes division and motility and inhibits premature differentiation of the oligodendrocyte/type-2 astrocyte progenitor cell. Nature, 333(6173), 560–562. 10.1038/333560a0 [DOI] [PubMed] [Google Scholar]
- Paes de Faria J., Kessaris N., Andrew P., Richardson W. D., Li H. (2014). New Olig1 null mice confirm a non-essential role for Olig1 in oligodendrocyte development. BMC Neurosci, 15, 12. 10.1186/1471-2202-15-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y., Fan L. W., Tien L. T., Dai X., Zheng B., Cai Z., Lin R. C., Bhatt A. (2013). Differential roles of astrocyte and microglia in supporting oligodendrocyte development and myelination in vitro. Brain Behav, 3(5), 503–514. 10.1002/brb3.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. C., Mehta A., Richardson J. S., Appel B. (2002). Olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol, 248(2), 356–368. 10.1006/dbio.2002.0738 [DOI] [PubMed] [Google Scholar]
- Pedraza L., Huang J. K., Colman D. (2009). Disposition of axonal caspr with respect to glial cell membranes: Implications for the process of myelination. J Neurosci Res, 87(15), 3480–3491. 10.1002/jnr.22004 [DOI] [PubMed] [Google Scholar]
- Phan B. N., Bohlen J. F., Davis B. A., Ye Z., Chen H. Y., Mayfield B., Sripathy S. R., Cerceo Page S., Campbell M. N., Smith H. L., Gallop D., Kim H., Thaxton C. L., Simon J. M., Burke E. E., Shin J. H., Kennedy A. J., Sweatt J. D., Philpot B. D., Jaffe A. E., Maher B. J. (2020). A myelin-related transcriptomic profile is shared by Pitt-Hopkins syndrome models and human autism spectrum disorder. Nat Neurosci, 23(3), 375–385. 10.1038/s41593-019-0578-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips T., Rothstein J. D. (2017). Oligodendroglia: Metabolic supporters of neurons. J Clin Invest, 127(9), 3271–3280. 10.1172/JCI90610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff M. C., Lillien L. E., Richardson W. D., Burne J. F., Noble M. D. (1988). Platelet-derived growth factor from astrocytes drives the clock that times oligodendrocyte development in culture. Nature, 333(6173), 562–565. 10.1038/333562a0 [DOI] [PubMed] [Google Scholar]
- Readhead C., Hood L. (1990). The dysmyelinating mouse mutations shiverer (Shi) and myelin deficient (shimld). Behav Genet, 20(2), 213–234. 10.1007/BF01067791 [DOI] [PubMed] [Google Scholar]
- Rosenberg S. S., Kelland E. E., Tokar E., De la Torre A. R., Chan J. R. (2008). The geometric and spatial constraints of the microenvironment induce oligodendrocyte differentiation. Proc Natl Acad Sci U S A, 105(38), 14662–14667. 10.1073/pnas.0805640105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer K., Brösamle C. (2009). Zwilling-A and -B, two related myelin proteins of teleosts, which originate from a single bicistronic transcript. Mol Biol Evol, 26(3), 495–499. 10.1093/molbev/msn298 [DOI] [PubMed] [Google Scholar]
- Shen S., Sandoval J., Swiss V. A., Li J., Dupree J., Franklin R. J., Casaccia-Bonnefil P. (2008). Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat Neurosci, 11(9), 1024–1034. 10.1038/nn.2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman D. L., Brophy P. J. (2005). Mechanisms of axon ensheathment and myelin growth. Nat Rev Neurosci, 6(9), 683–690. 10.1038/nrn1743 [DOI] [PubMed] [Google Scholar]
- Shigemoto-Mogami Y., Hoshikawa K., Goldman J. E., Sekino Y., Sato K. (2014). Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J Neurosci, 34(6), 2231–2243. 10.1523/jneurosci.1619-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siems S. B., Jahn O., Hoodless L. J., Jung R. B., Hesse D., Möbius W., Czopka T., Werner H. B. (2021). Proteome profile of myelin in the zebrafish brain. Front Cell Dev Biol, 9, 640169. 10.3389/fcell.2021.640169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M., Nave K. A. (2015). Oligodendrocytes: Myelination and axonal support. Cold Spring Harbor Perspect Biol, 8(1), a020479. 10.1101/cshperspect.a020479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaidero N., Mobius W., Czopka T., Hekking L. H., Mathisen C., Verkleij D., Goebbels S., Edgar J., Merkler D., Lyons D. A., Nave K. A., Simons M. (2014). Myelin membrane wrapping of CNS axons by PI(3,4,5)P3-dependent polarized growth at the inner tongue. Cell, 156(1–2), 277–290. 10.1016/j.cell.2013.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaidero N., Simons M. (2014). Myelination at a glance. J Cell Sci, 127(Pt 14), 2999–3004. 10.1242/jcs.151043 [DOI] [PubMed] [Google Scholar]
- Stankoff B., Aigrot M. S., Noël F., Wattilliaux A., Zalc B., Lubetzki C. (2002). Ciliary neurotrophic factor (CNTF) enhances myelin formation: A novel role for CNTF and CNTF-related molecules. J Neurosci, 22(21), 9221–9227. 10.1523/jneurosci.22-21-09221.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt C. C., Lommes P., Friedrich R. P., Wegner M. (2004). Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development, 131(10), 2349–2358. 10.1242/dev.01114 [DOI] [PubMed] [Google Scholar]
- Stolt C. C., Lommes P., Sock E., Chaboissier M. C., Schedl A., Wegner M. (2003). The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev, 17(13), 1677–1689. 10.1101/gad.259003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt C. C., Rehberg S., Ader M., Lommes P., Riethmacher D., Schachner M., Bartsch U., Wegner M. (2002). Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev, 16(2), 165–170. 10.1101/gad.215802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. O., Mulinyawe S. B., Collins H. Y., Ibrahim A., Li Q., Simon D. J., Tessier-Lavigne M., Barres B. A. (2018). Spatiotemporal control of CNS myelination by oligodendrocyte programmed cell death through the TFEB-PUMA axis. Cell, 175(7), 1811–1826.e1821. 10.1016/j.cell.2018.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada N., Kucenas S., Appel B. (2010). Sox10 is necessary for oligodendrocyte survival following axon wrapping. Glia, 58(8), 996–1006. 10.1002/glia.20981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taveggia C., Zanazzi G., Petrylak A., Yano H., Rosenbluth J., Einheber S., Xu X., Esper R. M., Loeb J. A., Shrager P., Chao M. V., Falls D. L., Role L., Salzer J. L. (2005). Neuregulin-1 type III determines the ensheathment fate of axons. Neuron, 47(5), 681–694. 10.1016/j.neuron.2005.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawk M., Makoukji J., Belle M., Fonte C., Trousson A., Hawkins T., Li H., Ghandour S., Schumacher M., Massaad C. (2011). Wnt/beta-catenin signaling is an essential and direct driver of myelin gene expression and myelinogenesis. J Neurosci, 31(10), 3729–3742. 10.1523/jneurosci.4270-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason E. J., Escalante M., Osterhout D. J., Fuss B. (2020). The oligodendrocyte growth cone and its actin cytoskeleton: A fundamental element for progenitor cell migration and CNS myelination. Glia, 68(7), 1329–1346. 10.1002/glia.23735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiane A., Schepers M., Rombaut B., Hupperts R., Prickaerts J., Hellings N., van den Hove D., Vanmierlo T. (2019). From OPC to oligodendrocyte: An epigenetic journey. Cells, 8(10), 1236. 10.3390/cells8101236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit S., Martinez S., Allinquant B., Peyron F., Puelles L., Zalc B. (1995). Oligodendrocytes originate in a restricted zone of the embryonic ventral neural tube defined by DM-20 mRNA expression. J Neurosci, 15(2), 1012–1024. 10.1523/jneurosci.15-02-01012.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traiffort E., Kassoussi A., Zahaf A., Laouarem Y. (2020). Astrocytes and microglia as major players of myelin production in normal and pathological conditions. Front Cell Neurosci, 14, 79. 10.3389/fncel.2020.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H. H., Frost E., To V., Robinson S., Ffrench-Constant C., Geertman R., Ransohoff R. M., Miller R. H. (2002). The chemokine receptor CXCR2 controls positioning of oligodendrocyte precursors in developing spinal cord by arresting their migration. Cell, 110(3), 373–383. 10.1016/s0092-8674(02)00838-3 [DOI] [PubMed] [Google Scholar]
- Tsai H. H., Niu J., Munji R., Davalos D., Chang J., Zhang H., Tien A. C., Kuo C. J., Chan J. R., Daneman R., Fancy S. P. (2016). Oligodendrocyte precursors migrate along vasculature in the developing nervous system. Science, 351(6271), 379–384. 10.1126/science.aad3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela J. M., Molina-Holgado E., Arévalo-Martín A., Almazán G., Guaza C. (2002). Interleukin-1 regulates proliferation and differentiation of oligodendrocyte progenitor cells. Mol Cell Neurosci, 20(3), 489–502. 10.1006/mcne.2002.1127 [DOI] [PubMed] [Google Scholar]
- Wahl S. E., McLane L. E., Bercury K. K., Macklin W. B., Wood T. L. (2014). Mammalian target of rapamycin promotes oligodendrocyte differentiation, initiation and extent of CNS myelination. J Neurosci, 34(13), 4453–4465. 10.1523/jneurosci.4311-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Sdrulla A. D., diSibio G., Bush G., Nofziger D., Hicks C., Weinmaster G., Barres B. A. (1998). Notch receptor activation inhibits oligodendrocyte differentiation. Neuron, 21(1), 63–75. 10.1016/s0896-6273(00)80515-2 [DOI] [PubMed] [Google Scholar]
- Watanabe M., Hadzic T., Nishiyama A. (2004). Transient upregulation of Nkx2.2 expression in oligodendrocyte lineage cells during remyelination. Glia, 46(3), 311–322. 10.1002/glia.20006 [DOI] [PubMed] [Google Scholar]
- Wegener A., Deboux C., Bachelin C., Frah M., Kerninon C., Seilhean D., Weider M., Wegner M., Nait-Oumesmar B. (2015). Gain of Olig2 function in oligodendrocyte progenitors promotes remyelination. Brain, 138(Pt 1), 120–135. 10.1093/brain/awu375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolswijk G., Noble M. (1989). Identification of an adult-specific glial progenitor cell. Development, 105(2), 387–400. 10.1242/dev.105.2.387 [DOI] [PubMed] [Google Scholar]
- Xin M., Yue T., Ma Z., Wu F. F., Gow A., Lu Q. R. (2005). Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci, 25(6), 1354–1365. 10.1523/jneurosci.3034-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P., Li L., Richards R. G., DiAugustine R. P., D’Ercole A. J. (2002). Myelination is altered in insulin-like growth factor-I null mutant mice. J Neurosci, 22(14), 6041–6051. 10.1523/jneurosci.22-14-06041.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Hu B. (2014). Knockdown of Lingo1b protein promotes myelination and oligodendrocyte differentiation in zebrafish. Exp Neurol, 251, 72–83. 10.1016/j.expneurol.2013.11.012 [DOI] [PubMed] [Google Scholar]
- Zhang K., Chen S., Yang Q., Guo S., Chen Q., Liu Z., Li L., Jiang M., Li H., Hu J., Pan X., Deng W., Xiao N., Wang B., Wang Z. X., Zhang L., Mo W. (2022). The oligodendrocyte transcription factor 2 OLIG2 regulates transcriptional repression during myelinogenesis in rodents. Nat Commun, 13(1), 1423. 10.1038/s41467-022-29068-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. Y., Shi X. Y., Qiu X., Lu W., Yang S., Li C., Chen L., Zhang L., Cheng G. H., Tang Y. (2012). Enriched environment increases the myelinated nerve fibers of aged rat corpus callosum. Anat Rec (Hoboken), 295(6), 999–1005. 10.1002/ar.22446 [DOI] [PubMed] [Google Scholar]
- Zhu X., Zuo H., Maher B. J., Serwanski D. R., LoTurco J. J., Lu Q. R., Nishiyama A. (2012). Olig2-dependent developmental fate switch of NG2 cells. Development, 139(13), 2299–2307. 10.1242/dev.078873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero J. B., Fu M. M., Sloan S. A., Ibrahim A., Olson A., Zaremba A., Dugas J. C., Wienbar S., Caprariello A. V., Kantor C., Leonoudakis D., Lariosa-Willingham K., Kronenberg G., Gertz K., Soderling S. H., Miller R. H., Barres B. A. (2015). CNS Myelin wrapping is driven by actin disassembly. Dev Cell, 34(2), 152–167. 10.1016/j.devcel.2015.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]