Abstract
Background
Despite well-known susceptibilities to other respiratory viral infections, individuals with allergic asthma have shown reduced susceptibility to severe coronavirus disease 2019 (COVID-19).
Objective
We sought to identify mechanisms whereby type 2 inflammation in the airway protects against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by using bronchial airway epithelial cells (AECs) from aeroallergen-sensitized children with asthma and healthy nonsensitized children.
Methods
We measured SARS-CoV-2 replication and ACE2 protein and performed bulk and single-cell RNA sequencing of ex vivo infected AEC samples with SARS-CoV-2 infection and with or without IL-13 treatment.
Results
We observed that viral replication was lower in AECs from children with allergic asthma than those from in healthy nonsensitized children and that IL-13 treatment reduced viral replication only in children with allergic asthma and not in healthy children. Lower viral transcript levels were associated with a downregulation of functional pathways of the ciliated epithelium related to differentiation as well as cilia and axoneme production and function, rather than lower ACE2 expression or increases in goblet cells or mucus secretion pathways. Moreover, single-cell RNA sequencing identified specific subsets of relatively undifferentiated ciliated epithelium (which are common in allergic asthma and highly responsive to IL-13) that directly accounted for impaired viral replication.
Conclusion
Our results identify a novel mechanism of innate protection against SARS-CoV-2 in allergic asthma that provides important molecular and clinical insights during the ongoing COVID-19 pandemic.
Key words: SARS-CoV-2, COVID-19, asthma, airway epithelial cells, epithelium, IL-13, children
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has rapidly infected humans across the globe, causing one of the most devastating pandemics in modern history, with 670 million confirmed cases and more than 6.7 million deaths worldwide by January 2023.1 Although most cases of the resulting coronavirus disease 2019 (COVID-19) are mild, some cases are severe and complicated by respiratory and multiorgan failure.2 , 3 As the pandemic has evolved, despite shifting risks with the arrival of new variants and reduced morbidity and mortality with the advent of vaccines and antiviral drugs, there has been marked heterogeneity among individuals with regard to the risk of infection with SARS-CoV-2 and/or severity of COVID-19 disease.4, 5, 6 Reported mortality rates have been as low as 0.2% and as high as 27% depending on patient age and underlying medical comorbidities,7 which have been identified as the main risk factors for more severe COVID-19.2 , 5 , 8, 9, 10, 11, 12, 13 Identifying mechanisms that explain disease heterogeneity with SARS-CoV-2 infection, in particular, those that may provide protection, has been a goal throughout the pandemic and can inform efforts to develop therapeutic interventions to prevent and treat COVID-19.
Higher airway viral loads14, 15, 16, 17, 18 and deficient antiviral interferon responses have been convincingly demonstrated to be associated with more severe disease9, 10, 11 , 13; however, the mechanisms of innate protection from SARS-CoV-2 remain poorly understood. Despite their well-known susceptibilities to other common respiratory viruses, especially human rhinoviruses,19 patients with respiratory allergy and allergic asthma, surprisingly, have shown reduced susceptibility to severe COVID-19.20, 21, 22, 23, 24, 25 These allergic diseases are characterized by chronic type 2 inflammation in the airways, driven in part through elevated expression of, and response to, IL-13.26 The hypothesized mechanisms by which type 2 inflammation may be protective against SARS-CoV-2 have included observed lower angiotensin-converting enzyme 2 (ACE2) expression in airway epithelium27, 28, 29 and greater airway mucus production,30 but prior studies have demonstrated that these factors cannot fully account for impaired viral entry or replication in airway epithelial cells (AECs) treated with IL-13.31 , 32
We sought to identify the mechanism(s) by which type 2 inflammation in the airway protects against SARS-CoV-2 infection. We used primary bronchial AECs from well-characterized aeroallergen-sensitized children with asthma (hereafter referred to as allergic asthma) and healthy nonsensitized children. AECs were differentiated ex vivo at an air-liquid interface (ALI) to generate organotypic cultures. Cultures were treated without or with IL-13 starting 1 week before SARS-CoV-2 infection and then infected with SARS-CoV-2. We measured SARS-CoV-2 replication and ACE2 protein expression and performed bulk and single-cell RNA sequencing (sc-RNAseq) of samples. Using combined bulk and single-cell transcriptomics, we investigated whether IL-13 induced type 2 inflammation would decrease SARS-CoV-2 replication, and if so, by what mechanisms.
Methods
ALI culture of bronchial AECs
Bronchial AECs from healthy nonsensitized children (n = 17) and children with allergic asthma (n = 15) aged 6 to 18 years (Tables I and II ) were obtained from subjects while under general anesthesia; the cells were obtained by using 4-mm Harrell unsheathed bronchoscope cytology brushes (ConMed, Utica, NY). As we and others have described33 , 34 an unprotected brush was inserted through an endotracheal tube, advanced until resistance was felt, and rubbed against the airway surface for 2 seconds. Cells were then seeded onto T-25 cell culture flasks precoated with type I collagen and proliferated under submerged culture conditions, as we have previously described.33 AECs from children were obtained and utilized in these experiments under studies no. 12490 and no. 00002603 approved by the Seattle Children’s Hospital institutional review board. Parents of subjects provided written consent and children older than 7 years provided assent. Passage 3 AECs were differentiated ex vivo for 21 days at an ALI on 12-well collagen-coated Corning plates with permeable transwells in PneumaCult ALI media (Stemcell Technologies, Vancouver, British Columbia, Canada) at 37˚C in an atmosphere of 5% CO2, as we have previously described, producing an organotypic differentiated epithelial culture with mucociliary morphology.33 , 35, 36, 37
Table I.
AEC donor characteristics
| Characteristic | Donors with allergic asthma (n = 15) | Healthy nonsensitized donors (n = 17) |
|---|---|---|
| Age (y), mean ± SD | 10.2 ± 3.6 | 11.3 ± 2.4 |
| Female sex, no. (%) | 8 (53%) | 9 (53%) |
| Serum IgE level, mean ± SD | 193 ± 229 | 91 ± 150 |
| Positive for ≥2 aeroallergens according to allergen-specific IgE, no. (%) | 9 (60%) | 0 (0%) |
| Positive for ≥3 aeroallergens according to allergen-specific IgE, no. (%) | 5 (33%) | 0 (0%) |
| Feno level (ppb), mean ± SD) | 18.4 ± 12.9 | 8.8 ± 2.1 |
| FEV1 % predicted, mean ± SD | 99.6 ± 9.8 | 102.2 ±11.6 |
| FEV1/FVC ratio, mean ± SD | 0.84 ± 0.07 | 0.88 ± 0.05 |
| History of severe asthma exacerbation, no. (%) | 12 (80%) | 0 (0%) |
Feno, Fraction of exhaled nitric oxide; FVC, forced vital capacity.
Table II.
Samples used for each condition across assays
| Sample | From patient with allergic asthma |
From healthy donor |
||||||
|---|---|---|---|---|---|---|---|---|
| qPCR | Protein | Bulk RNA-seq | scRNA-seq | qPCR | Protein | Bulk RNA-seq | scRNA-seq | |
| Uninfected | 15 | 15 | N/A | N/A | 17 | 17 | N/A | N/A |
| SARS-CoV-2 alone | 15 | 15 | 9 | 2 | 17 | 17 | 5 | 2 |
| IL-13 alone | N/A | 10 | N/A | N/A | N/A | 10 | N/A | N/A |
| SARS-CoV-2 + IL-13 | N/A | N/A | 9 | 2 | N/A | N/A | 5 | 2 |
N/A, Not available.
In a biologic safety level 3 (BSL3) facility, AEC cultures were infected with SARS-CoV-2 (USA-WA1/2020) at a multiplicity of infection (MOI) of 0.5. SARS-CoV-2 replication was assessed by quantitative PCR (qPCR) 96 hours post infection. ACE2 expression was assessed by qPCR, and protein abundance was assessed in AEC lysates by ELISA in parallel uninfected AEC cultures. In additional experiments, cultures of AECs from children with allergic asthma were exposed to IL-13 (10 ng/mL), with each medium change starting 1 week before and throughout SARS-CoV-2 infection.31 , 38 , 39 To determine the impact of IL-13 alone on ACE2 protein abundance in uninfected cultures, cultures of AECs from patients with allergic asthma (n = 10) and healthy donors (n = 10) were exposed to IL-13 (10 ng/mL) for 1 week before exposure to UV-inactivated SARS-CoV-2.40
At 96 hours post infection with SARS-CoV-2, RNA was isolated from cells by using Trizol and protein was isolated from cell lysates with radioimmunoprecipitation assay buffer (Sigma-Aldrich, St Louis, Mo) containing Triton X100 1% and SDS 0.1%, methods that we have demonstrated to fully inactivate SARS-CoV-2. Expression of ACE2, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), hypoxanthine-guanine phosphoryltransferase (HPRT), and peptidylprolyl isomerase A (PPIA) were measured by qPCR using Taqman probes (Thermo Fisher Scientific, Waltham, Mass). To measure SARS-CoV-2 replication in AEC cultures we used the Genesig Coronavirus Strain 2019-nCoV Advanced PCR Kit (Primerdesign, Chandler's Ford, United Kingdom), with duplicate assays of harvested RNA from each SARS-CoV-2–infected AEC experimental condition. The viral copy number used in analyses of each experimental condition was the mean of duplicate assays from each experimental condition.
To extract protein from the cell layer of SARS-CoV-2–infected AEC cultures, medium was first removed from the basolateral chamber of the transwells. Next, 100 μL of cold PBS was added to the apical surface of cultures and 1 mL was added to the basolateral chamber of cultures as a wash step. Then, 50μL of radioimmunoprecipitation assay buffer for protein extraction ready-to-use-solution (Sigma-Aldrich, product no. R0278) containing Triton X100 1% and SDS 0.1% was added to the apical surface of the AECs and incubated for 15 minutes on ice. A pipet tip was then used to gently scratch each apical well in a crosshatch pattern to loosen the AECs from the transwell membrane. Material was collected and centrifuged at 10,000 rpm at 4°C for 10 minutes, after which isolated protein was collected. ACE2 protein concentrations were measured in cell layer lysates via ELISA (R&D Systems, Minneapolis, Minn), with protein concentrations normalized to total protein levels in the lysate (bicinchoninic acid protein assay [Sigma-Aldrich]).
qPCR gene expression and protein levels are presented as means plus or minus the SD when data were normally distributed and as medians with interquartile ranges if 1 or more groups were not normally distributed. To determine whether data were normally distributed, the Kolmogorov-Smirnov test was used (α = 0.05). ACE2 qPCR relative expression was standardized by using GAPDH as a nonregulated reference gene. Additional analyses of ACE2 expression were also conducted by using HPRT and PPIA as reference genes. GenEx, version 5.0.1, was used to quantify gene expression from qPCR normalized to GAPDH (MultiD Analyses AB, Göteborg, Sweden) based on methods described by Pfaffl.41 Data on at least 1 group or condition in each experiment analyzed were determined to be nonnormally distributed; therefore, nonparametric tests were used for the analyses. To compare gene expression data and distributions of protein concentrations in cell lysates between paired groups, the Wilcoxon matched-pairs signed rank test was used. For unpaired data the Mann-Whitney test was used for analyses. For experiments with 3 or more conditions, the Kruskal-Wallis 1-way ANOVA on ranks test was used, and post hoc comparisons between pairs of subject groups were made by using the Dunn multiple comparisons test (significance level set at P <.05). Correlations were determined by using the Spearman's rank correlation coefficient. Data were analyzed by using Prism 9.0 software (GraphPad Software, Inc, San Diego, Calif.). Statistical significance was set at P less than .05.
Bulk RNA-seq sample preparation, sequencing, processing, and analysis
For bulk RNA sequencing (RNA-seq), bronchial AECs from a subset of the healthy nonsensitized children (n = 5) and children with allergic asthma (n = 9) were differentiated ex vivo at an ALI to generate organotypic cultures by the same methods as already mentioned. Cultures were stimulated without or with IL-13 (10 ng/mL), with each medium change starting 1 week before and throughout SARS-CoV-2 infection. In a BSL3 facility, AEC cultures were infected with SARS-CoV-2 (USA-WA1/2020) at an MOI of 0.5. RNA was isolated from AECs by using Trizol with the PureLink RNA Mini Kit (Invitrogen ThermoFisher) and Phasemaker Tubes (Invitrogen ThermoFisher) and then eluted into RNAse-free water. Total RNA was used to construct libraries by using the SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara, San Jose, Calif), with reverse transcription followed by PCR amplification to generate full-length amplified cDNA. Sequencing libraries were constructed by using the NexteraXT DNA sample preparation kit with unique dual indexes (Illumina, San Diego, Calif) to generate Illumina-compatible barcoded libraries. Libraries were pooled and quantified using a Qubit Fluorometer (Thermo Fisher Scientific, Waltham, Mass). Sequencing of pooled libraries was carried out on a NextSeq 2000 sequencer (Illumina) with paired-end 53-base reads, using NextSeq P2 and NextSeq P3 sequencing kits (Illumina) with a target depth of 5 million reads per sample. Base calls were processed to FASTQs on BaseSpace (Illumina), and a base call quality-trimming step was applied to remove low-confidence base calls from the ends of reads. Samples were sequenced in 2 batches by using repeated samples across batches to allow assessment for batch effects. Resulting bcl files were deconvoluted and converted to fastq format using Casava from Illumina. Fastq files were aligned to the Ensembl version of the human genome (GRCh38, Ensembl 91) by using STAR (version 2.4.2a). HTSeq-count (version 0.4.1) was used to generate gene counts with mode as “intersection (nonempty)” and minimum alignment quality set to 20 and otherwise set to default parameters. Quality metrics were compiled from PICAR (version 1.134), FASTQC (version 0.11.3), Samtools (version 1.2), and HTSeq-count (version 0.4.1). For quality control, samples that had human aligned counts greater than 1 million mapped reads and a median coefficient of variation coverage less than 0.9 were kept. Genes were filtered to include those that had a trimmed mean of M value normalization count of at least 1 in at least 10% of samples and further filtered for only protein coding genes. Normalized counts were transformed to log2 counts per million mapped reads along with observations level weights by using voomWithQualityWeights from the limma R package (version 3.5.1). We did not observe any noticeable batch effect assessed using principal component analysis, so for replicate samples, the replicate with the highest aligned counts was included for the analysis and other replicates were removed. The final data set included 28 samples composed of 14,019 genes. Differentially expressed genes (DEGs) were identified by using the limma R package42 by linear mixed effects model, ∼stimulation + (1|donor). For plotting purposes, log2 fold change (log2FC) values of more than 8 were truncated to log2FC = 8 . Gene ontology (GO) and pathway enrichment analyses were performed by using the enrichR R package (version 3.0).43
scRNA-seq sample preparation, sequencing, processing, and analysis
For scRNA-seq, bronchial AECs from a subset of the healthy nonsensitized children (n = 2) and children with allergic asthma (n = 2) were differentiated ex vivo at an ALI to generate organotypic cultures by the same methods as already mentioned. Cultures were stimulated without or with IL-13 (10 ng/mL), with each medium change starting 1 week before and throughout SARS-CoV-2 infection. In a BSL3 facility, AEC cultures were infected with SARS-CoV-2 (USA-WA1/2020) at an MOI of 0.5. To dissociate the differentiated AEC ALI cultures into single cells, the apical surface of the transwell was washed twice with 0.5 mL of PBS (Gibco, Billings, Mont), after which 300 μL of TrypLE Express (Gibco, 12604-013) warmed to 37°C was added to the apical surface and 600 μL was added to the basolateral chamber. The wells were incubated for 10 minutes at 37°C, after which 200 μL of Minimum Essential Medium, Spinner Modification (SMEM) (Quality Biological, VWR, Gaithersburg, Md) was added to the apical surface and gently pipetted 10 times by using a wide-orifice P1000 tip (Rainin, Oakland, Calif). The cell suspension was transferred to a 50-mL conic tube containing 10 mL of 37°C SMEM. The transwell was rinsed with 500 μL of PBS by using wide-orifice tips, and the rinse was added to the cell suspension. The cells were mixed by pipetting 20 times with a 5 mL serological pipet set to slow and then centrifuged at 250 g for 5 minutes with brake and acceleration set to slow. Next, the cell pellet was resuspended in 1 mL of TrypLE Express and incubated for 5 minutes at 37°C. Then, the cell suspension was diluted with 5 mL of SMEM and centrifuged at 250 g for 5 minutes with brake and acceleration set to slow. The cell pellet was resuspended with 1 mL of 1% BSA (Sigma-Aldrich, A7906-50G) in PBS by using a wide-orifice P1000 and gently pipetted 30 times maximum. An additional 2 mL of 1% BSA in PBS was added and mixed gently with a 5 mL serological pipet set to slow. The cell suspension was then filtered in a 2-step process, first using a 30-μm filter (pluriSelect, El Cajon, Calif) with a wide-orifice P1000 tip and then with a 40-μm Flowmi Cell Strainer (SP Bel-Art, Wayne, NJ). The 10× Genomics (Pleasanton, Calif) Chromium Next GEM Single Cell 3ʹ Reagent Kit v 3.1 was used to partition AECs into nanoliter-scale Gel Beads-in-EMulsion (GEMs). GEMs were generated by combining barcoded single-cell 3ʹ version 3.1 gel beads, master mix with 10× cell multiplexing oligo labeled cells, and partitioning oil onto a 10× Chromium Chip G. Incubation of the GEMs produced barcoded, full-length cDNA from polyadenylated mRNA and barcoded DNA from the Cell Multiplexing Oligo Feature Barcode. Following GEM generation, the gel beads were dissolved, releasing primers containing Illumina TruSeq Read 1 (read 1 sequencing primer), 10× Barcode, unique molecular identifier, and poly(dT) sequence or primers containing an Illumina Nextera Read 1 (read 1 sequencing primer), 10× Barcode, unique molecular identifier, and Capture Sequence 1 or 2, thereby generating cDNA from poly-adenylated mRNA and DNA from Cell Multiplexing Oligo Feature Barcode simultaneously from the same single cell inside the GEM. After incubation, the GEMs were broken and pooled fractions were recovered. Silane magnetic beads were used to purify the cell barcoded products from the post–GEM reverse transcription reaction mixture. The cell barcoded cDNA molecules were amplified via PCR to generate sufficient mass for library constructions. Size selection was used to separate the amplified cDNA molecules for 3ʹ Gene Expression and Cell Multiplexing library construction. Single-cell gene expression libraries were then constructed by using the 3’ version 3.1 Chromium Next GEM Single Cell Kit (10× Genomics). Libraries were pooled and treated with Illumina Free Adapter Blocking Reagent (Illumina) to block free adapters and reduce index hopping. Sequencing of pooled libraries was carried out on a NextSeq 2000 sequencer (Illumina) with use of NextSeq P3 flowcells (Illumina).
scRNA-seq (10×) data set preprocessing including cell demultiplexing and alignment were performed by using the Cell Ranger Single-Cell Software Suite (version 6.1.1, 10× Genomics, Inc). We prepared a custom reference genome with human reference genome (GRCh38, Ensembl 91) and SARS-CoV-2 genome (National Center for Biotechnology Information Refseq identifier NC_045512.2) as an additional chromosome and used it for alignment. First, we removed genes expressed in fewer than 4 cells. Next, we removed cells with fewer than 100 genes detected or cells with more than 20% mitochondrial reads. Further, to remove possible doublet cells, we removed cells with more than 2500 genes detected. Counts data were normalized to 10,000 reads per cell, after which samples were integrated by using a stepwise canonical correlation analysis (CCA) approach on 20 components and 2,000 variable genes. After integration, principal component analysis was run on integrated data from the samples of AECs from individuals with allergic asthma by using the first 30 principal components, followed by Uniform Manifold Approximation and Projection (UMAP) with 20 components and clustering with a resolution of 0.3 and 20 components to identify distinct cell clusters. Cluster-specific markers for each cluster were identified by using the FindAllMarkers function, and cell types were manually annotated by assessing the expression of cluster-specific markers to known cell type markers.44, 45, 46, 47, 48, 49 Cells from the healthy nonsensitized donors were then mapped to the identified cell clusters from the allergic asthma UMAP by utilizing the Seurat mapQuery function; this approach assigned cells from the healthy control samples to the cells with the greatest transcriptional similarity in the allergic asthma UMAP space. All these analyses were performed in the Seurat R package (version 4.1.0) and R (version 4.1.2).
Results
SARS-CoV-2 replication in AECs from healthy nonsensitized children and children with allergic asthma
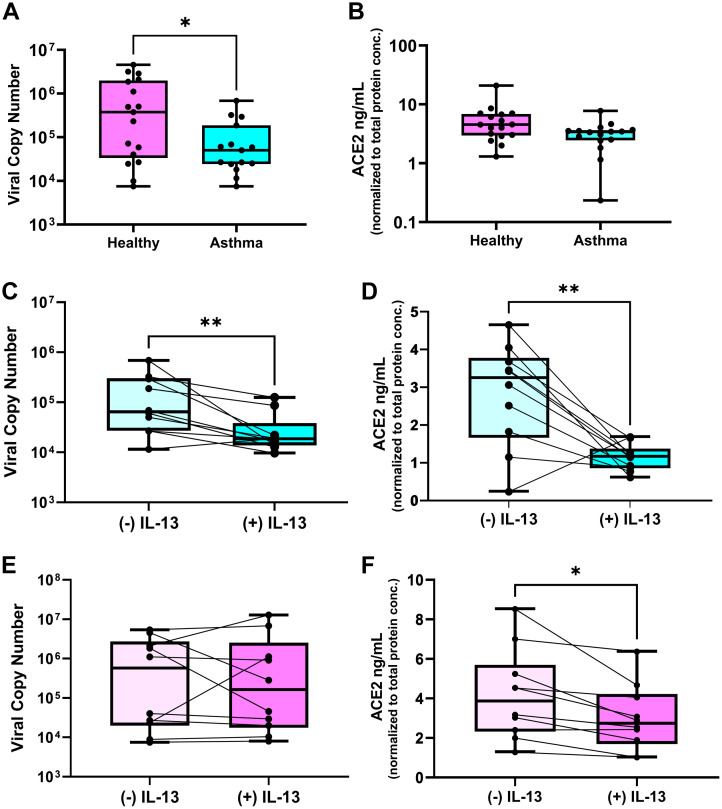
In the first phase of this study, we compared SARS-CoV-2 replication and ACE2 gene and protein expression in primary bronchial AECs from children with allergic asthma (n = 15) and healthy nonsensitized children (n = 17) (Tables I and II). SARS-CoV-2 replication was assessed by qPCR at 96 hours post infection. In parallel uninfected AEC cultures, ACE2 gene expression was assessed by qPCR and ACE2 protein abundance was assessed in AEC lysates by ELISA. We observed that SARS-CoV-2 replication was significantly lower in cultures of cells from children with allergic asthma than in cultures of AECs from healthy nonsensitized children (median viral copy 5 × 104 vs 3.8 × 105 [P = .04]) (Fig 1 , A). In uninfected cultures, ACE2 gene expression was similar between the cultures from children with allergic asthma and those from healthy nonsensitized children (see Fig E1 in the Online Repository at www.jacionline.org); ACE2 protein abundance was not significantly different but trended toward lower in AECs from children with allergic asthma than in AECs from healthy nonsensitized children (median ACE2 protein 3.4 ng/mL vs 4.5 ng/mL [P = .06]) (Fig 1, B). Among the primary AEC cultures from healthy children we observed a significant correlation between SARS-CoV-2 levels in infected cultures and the ACE2 protein abundance in the parallel uninfected cultures (Spearman r = 0.66; P = .005 [see Fig E2 in the Online Repository at www.jacionline.org]), whereas in the cultures of AECs from children with allergic asthma we did not observe a significant correlation between SARS-CoV-2 copy number and ACE2 protein abundance (Spearman r = 0.18; P = .52 [see Fig E2 in the Online Repository at www.jacionline.org]).
Fig 1.
SARS-CoV-2 replication in AECs from healthy nonsensitized children and children with allergic asthma and effect of IL-13 stimulation on ACE2 and SARS-CoV-2 replication. A, Boxplot showing SARS-CoV-2 copy number measured by qPCR 96 hours after infection in bronchial AEC cultures from healthy nonsensitized children (pink) and children with allergic asthma (light blue) (∗P = .04). B, Boxplot showing SARS-CoV-2 viral entry factor and ACE2 protein abundance in parallel uninfected AEC cultures from healthy nonsensitized children (pink) and children with allergic asthma (light blue) (P = .06). C, Boxplot showing viral copy number in cultures from children with allergic asthma infected with SARS-CoV-2 without and with IL-13 (10 ng/mL) treatment (∗∗P < .01). D, Boxplot showing ACE2 protein abundance in cultures from children with allergic asthma infected with SARS-CoV-2 without and with IL-13 (10 ng/mL) treatment (∗∗P < .01). E, Boxplot showing viral copy number in cultures from healthy nonsensitized infected with SARS-CoV-2 without and with IL-13 (10 ng/mL) treatment. F, Boxplot showing ACE2 protein abundance in cultures from healthy nonsensitized infected with SARS-CoV-2 without and with IL-13 (10 ng/mL) treatment (∗P < .05).
Effect of IL-13 stimulation on ACE2 and SARS-CoV-2 replication in AECs from healthy nonsensitized children and children with allergic asthma
In the second phase of this study, we observed that in experiments wherein AECs from children with allergic asthma (n = 10) were treated without or with IL-13 (10 ng/mL), ACE2 protein abundance decreased by 3-fold (mean ACE2 protein of 2.8 ng/mL in untreated cultures versus 1.1 ng/mL in IL-13 treated cultures [P = .006]) (Fig 1, D). Moreover, IL-13 stimulation significantly reduced SARS-CoV-2 replication in infected cultures from children with allergic asthma (median viral copy number of 6.4 × 104 in untreated AECs versus 1.9 × 104 in IL-13 treated AECs; P = .004) (Fig 1, C). When AECs from healthy nonsensitized children (n = 10) were treated with IL-13, ACE2 protein abundance decreased from a mean of 4.2 to 3 ng/mL (P = .01, Fig 1, F); however, IL-13 stimulation did not reduce SARS-CoV-2 replication in AECs from healthy nonsensitized children (Fig 1, E).
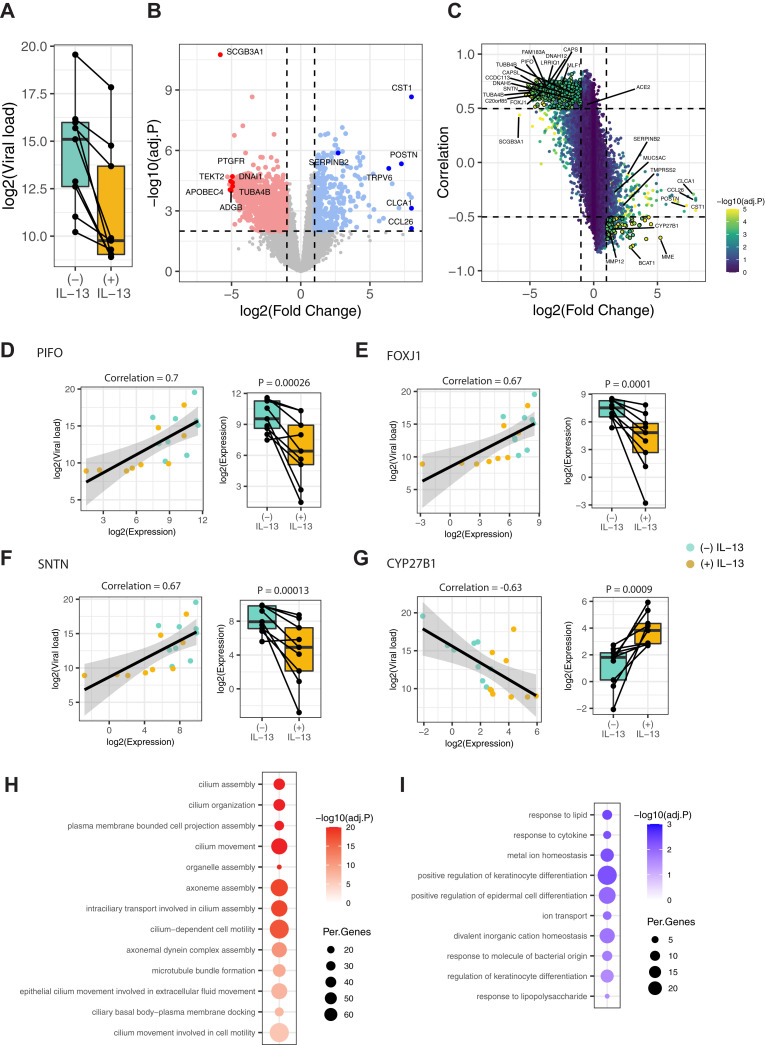
Bulk transcriptomics data demonstrate that IL-13 stimulation reduces SARS-CoV-2 replication in AECs from children with allergic asthma in parallel with downregulation of ciliated epithelial cell pathways
Next, we utilized bulk and single-cell transcriptomics approaches to identify potential mechanisms by which IL-13 stimulation reduces SARS-CoV-2 replication in the allergic asthma airway epithelium. RNA was isolated from AEC cultures 96 hours post infection for RNA-seq to comprehensively model cellular responses to SARS-CoV-2 and IL-13 (n = 9). We first quantified viral load from bulk RNA-seq data by counting the number of reads mapped to the SARS-CoV-2 genome normalized to total unmapped human reads. Consistent with the qPCR results shown earlier in this article, IL-13 stimulation significantly reduced viral load compared with unstimulated conditions in allergic asthma (log2FC = –5.34; P = .03, Fig 2, A). Next, we characterized the genes differentially expressed by IL-13 stimulation after infection; in total, we identified 1756 differentially expressed genes (absolute log2FC > 1; false discovery rate [FDR]-adjusted P < .05) composed of 534 IL-13–upregulated and 1222 IL-13–downregulated genes (Fig 2, B and see Table E1 in the Online Repository at www.jacionline.org). Consistent with the results from prior assays shown earlier in this article, ACE2 expression was relatively decreased by IL-13 stimulation (log2FC = –0.6; P = .014; FDR = 0.056). Next, we aimed to identify the genes likely mediating the reduction in SARS-CoV-2 replication by IL-13 stimulation in AECs from children with allergic asthma. We identified the genes with strong association between viral load and expression across samples and filtered to those genes differentially expressed by IL-13 stimulation. A total of 473 genes demonstrated a positive association between viral load and expression (Pearson correlation > 0.5) and a corresponding robust downregulation by IL-13 stimulation (log2FC < –1 and FDR < 0.001) (Fig 2, C and see Table E2 in the Online Repository at www.jacionline.org). This gene set included multiple airway ciliated epithelial cell–specific genes44 , 45 (Fig 2, C). For example, PIFO, FOXJ1, and SNTN, which are well-established ciliated cell marker genes, had strong positive associations with viral load (Pearson correlation for PIFO = 0.7; FOXJ1 = 0.67; SNTN = 0.67), and their expressions were decreased by IL-13 stimulation (log2FC for PIFO = –3.9; FOXJ1 = –4.1; SNTN = –4.7) (Fig 2, D-F and see Fig E4 in the Online Repository at www.jacionline.org). To understand the biologic functions represented by the 473 genes with this pattern, we performed GO and pathway enrichment analysis, which showed that these genes specifically represent functions of ciliated epithelial cells and in particular aspects of cilia axoneme assembly and movement, microtubule bundle formation, and intracellular transport along cilia (Fig 2, H). Conversely, 74 genes were negatively associated with viral load (Pearson correlation < –0.5) and were robustly upregulated by IL-13 stimulation (log2FC > 1; FDR < 0.001). For example, CYP27B1, a known gene in the keratinocyte differentiation process, showed a strong negative association with viral load (Pearson correlation = –0.63) and its expression was upregulated by IL-13 stimulation (log2FC = 2.8) (Fig 2, G). GO analysis of these 74 genes showed enrichment for keratinocyte differentiation pathways, response to lipids and cytokines, and ion transport (Fig 2, I). In contrast, a targeted analysis of type I IFN response genes using the MSigDB50 HALLMARK_INTERFERON_ALPHA_RESPONSE gene set showed modest differential expression with IL-13 but without a consistent relationship to viral load (see Fig E4).
Fig 2.
Bulk transcriptomics demonstrates that IL-13 stimulation reduces SARS-CoV-2 replication in AECs from children with allergic asthma in parallel with downregulation of ciliated epithelial cell pathways. A, Boxplot showing the viral load assessed by bulk mRNA sequencing in AECs from children with allergic asthma stimulated without or with IL-13 and infected with SARS-CoV-2. Lines connecting the dots denote samples from the same donor. B, Volcano plot showing the differentially upregulated and downregulated genes by IL-13 stimulation. Significantly (FDR < 0.01) upregulated (log2FC > 1) and downregulated genes (log2FC > –1) are denoted by blue and red color points, respectively. Select top-ranked genes by log2FC are highlighted. C, Scatter plot showing the association between viral load and IL-13–induced differential gene expression. The x-axis represents the change in expression due to IL-13 stimulation, and the y-axis represents the correlation to viral load. The color of the point shows the statistical significance of differential expression [–log10(adjusted P [adj.P])]. Genes significantly differentially expressed (FDR < 0.001) and highly correlated with viral load (Pearson correlation coefficient > 0.5 or < –0.5) are highlighted by larger point size and bolded border. Select ciliary and type 2 specific genes are highlighted. D-G, Examples of genes showing significant association of viral load and expression (scatter plots) and significant differential expression with IL-13 stimulation (box plots). H, Enriched GO terms for the 473 genes showing positive association between viral load and expression and downregulated by IL-13 stimulation. The size of each dot denotes the percentage of genes, and the intensity of color denotes statistical significance as –log10(adj.P). I, Enriched GO terms for the 74 genes showing negative association between viral load and expression and upregulated by IL-13 stimulation. The size of each dot denotes the percentage of genes, and the intensity of color denotes statistical significance as –log10(adj.P).
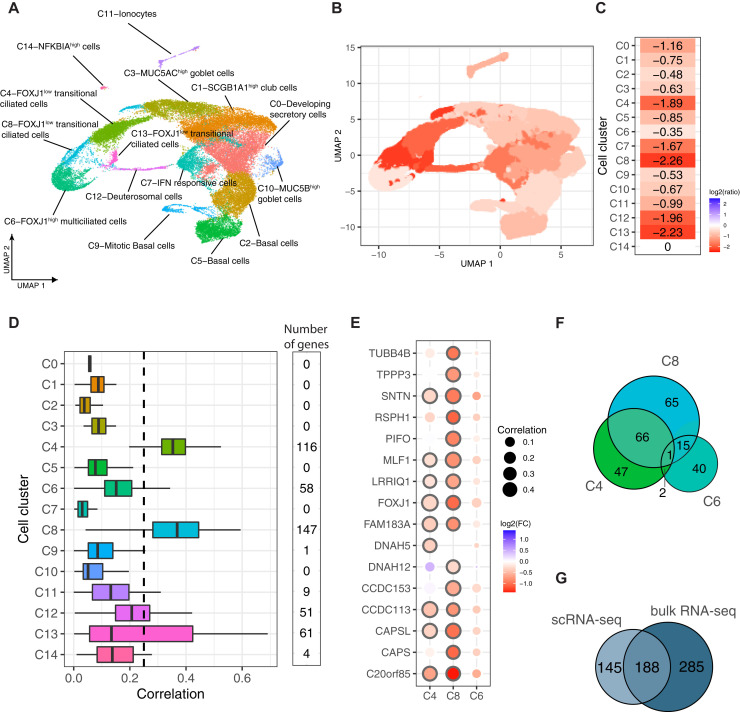
IL-13 stimulation decreases viral replication through specific effects on clusters of relatively undifferentiated ciliated epithelial cells common in AECs from children with allergic asthma
Next, we performed scRNA-seq on cultures of AECs from children with allergic asthma to understand how individual cell type–specific responses reduce SARS-CoV-2 replication with IL-13 stimulation. scRNA-seq libraries were generated from 2 donors with allergic asthma treated with or without IL-13 and then infected with SARS-CoV-2 by using the same methods as described earlier in this article. These yielded 37,828 individual cells for analysis after filtering and quality control (Fig 3 and see Fig E5 in the Online Repository at www.jacionline.org). Clustering analysis identified 15 cell clusters, which we annotated by using cell type–specific markers for known airway cell types (Fig 3, A and see Fig E5). This analysis revealed major epithelial cell types: basal cells (clusters C2 and C5), mitotic basal cells (cluster C9), developing secretory cells (C0), club cells (cluster C1), goblet cells (clusters C3 and C10), ciliated cells (clusters C4, C6, C8, and C13), deuterosomal cells (cluster C12), and ionocytes (cluster C11). In addition, we identified a cell cluster characterized primarily by high expression of interferon-stimulated genes (cluster C7). To measure the viral load on a per-cell basis, we mapped reads to the SARS-CoV-2 genome and quantified viral load as the number of SARS-CoV-2 reads normalized to the total unmapped human reads. We compared the differences in viral load as a ratio of viral load in IL-13–stimulated samples to that in unstimulated samples within each cell cluster and observed that IL-13 stimulation decreased SARS-CoV-2 replication across all cell populations, albeit to strikingly variable degrees of magnitude (Fig 3, B and C). The largest reductions in viral load occurred in 3 of the 4 ciliated cell clusters (ie, C4, C8, and C13) and also in the deuterosomal cells (C12), which are ciliated cell precursors (log2FC = –1.89 to –2.26). Interestingly, in contrast, the other ciliated cell cluster, C6, showed the least reduction in viral load among all of the clusters (log2FC = –0.35). We focused on identifying differences in expression among these ciliated cell clusters and their responses to IL-13 that account for these large differences in reduction in SARS-CoV-2 replication. We discovered that C4, C8, C13, and C12 had significantly lower expression of ciliated cell–specific marker genes than of C6 (Fig E5) and further that these marker genes were significantly decreased by IL-13 (see Fig E6 in the Online Repository at www.jacionline.org). The expression levels of many ciliated cell marker genes were correlated with viral load specifically in these clusters (Fig 3, D and see Table E2) but not in cluster C6. For example, PIFO was significantly downregulated by IL-13 and was correlated with viral load in C4 and C8 (Pearson correlations C4 = 0.26 and C8 = 0.35) but not in cluster C6 (Pearson correlation = 0.085) (Fig 3, E). A similar observation was true for FOXJ1 (Pearson correlations C4 = 0.47 and C8 = 0.32) and several other ciliary cell marker genes (Fig 3, E). Overall, we observed that many ciliated cell–specific genes were downregulated by IL-13 and had stronger correlations with viral load in clusters C4, C8, C13, and C12 than in C6. Furthermore, the majority of genes that were strongly associated with viral levels on a per-cluster basis were in these clusters rather than in cluster C6 (Fig 3, D and F). Additionally, 333 genes were associated with viral load on a per-cell basis among these ciliated cell clusters; these genes were significantly overlapping with the 473 genes previously identified in the bulk RNA-seq analysis already described (P < 1.0e–16; hypergeometric test) (Fig 3, G) and were similarly enriched for many of the same ciliated epithelial cell functions, including cilium assembly, axoneme assembly and movement, and intracellular transport along cilia (see Fig E6). Thus, although globally showing an expression pattern of ciliated cells, ciliated cell clusters C4, C8, and C13 demonstrated overall lower expression of genes of ciliary function than C6 did (Fig E5), and they were highly responsive to IL-13 in further downregulating these genes and reducing viral replication. Taken together, the data indicate that these clusters (C4, C8, and C13) are relatively undifferentiated and/or transitional ciliated epithelial cells when compared with C6, which are terminally differentiated multiciliated cells.
Fig 3.
IL-13 stimulation decreases viral replication through specific effects on clusters of relatively undifferentiated ciliated epithelial cells common in AECs from children with allergic asthma. A, UMAP clustering and visualization of scRNA-seq data from AECs from 2 children with allergic asthma stimulated with or without IL-13 and infected with SARS-CoV-2. Cell clusters are labeled based on highly expressed marker genes of airway epithelial cell types. B and C, Cell type–specific differences in viral load are presented as the ratio of viral load in the IL-13–stimulated condition to that in the unstimulated condition on a log2 scale shown in each cell cluster on the UMAP (B) and summary of the magnitude of change in viral load due to IL-13 stimulation in each cluster (C). D, Boxplot showing the distribution of single-cell–level correlations between viral load and expression of cell cluster genes; the numbers of significant genes with a Pearson correlation greater than 0.25 are noted. E, Dot plot showing the degree of differential expression in the IL-13–stimulated condition versus in the unstimulated condition and the correlation to viral load for ciliated cell marker genes in 3 ciliated cell clusters (C4, C8, and C13 represent relatively undifferentiated ciliated cell clusters, whereas C6 represents the terminally differentiated multiciliated cell cluster). F, Venn diagram showing the overlap of genes correlated with viral load within 3 ciliated cell clusters. G, Venn diagram showing the overlap of genes positively associated with viral load from bulk RNA-seq data and cell cluster–specific genes positively associated with viral load from scRNA-seq data.
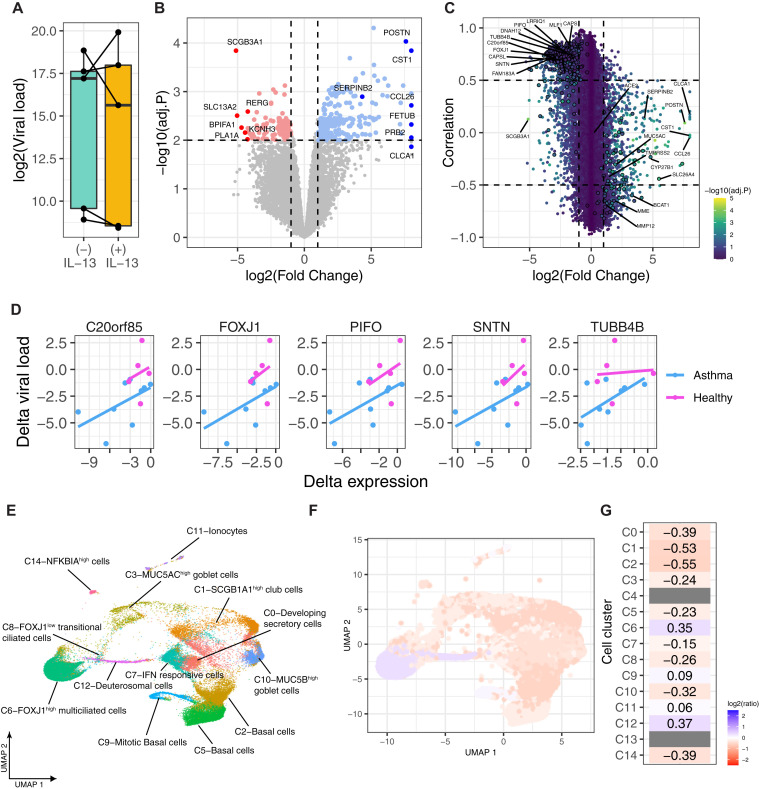
IL-13 stimulation has a variable effect on SARS-CoV-2 replication in healthy nonsensitized children
For comparison, we investigated the effect of IL-13 stimulation on SARS-CoV-2 replication and bulk gene expression in AECs from healthy nonsensitized children (n = 5). Interestingly, we did not see a significant change in viral load after IL-13 stimulation (P = .8); rather, there was a variable response among individuals (Fig 4, A). IL-13 stimulation had a lesser effect on gene expression in healthy nonsensitized than in children with allergic asthma, with relatively fewer differentially expressed genes (1179 differentially expressed genes [553 genes upregulated and 626 genes downregulated at an absolute log2FC > 1 and FDR < 0.05]) (Fig 4, B and see Table E1). We still observed a correlation among ciliary cell gene expression and viral load that is congruent with the observation in the group with allergic asthma (438 genes overlapping with the 473 genes from children with allergic asthma described earlier in this article), but these genes were not significantly decreased in expression by IL-13 stimulation in the nonsensitized healthy group (Fig 4, C and see Table E3 in the Online Repository at www.jacionline.org). The reason for this was that these genes generally showed larger and more consistent decreases with IL-13 stimulation in children with allergic asthma than in healthy nonsensitized children, resulting in larger and more significant effect sizes; this was in contrast to many other canonical IL-13 response genes (eg, POSTN and CLCA1) that showed similar changes in both groups (see Fig E7 in the Online Repository at www.jacionline.org). By examining the associations between the Δ expression (IL-13–stimulated minus unstimulated) of these genes and the Δ viral load, we demonstrated that the variable response of these genes to IL-13 corresponded closely to the variable change in viral load (Fig 4, D and see Fig E7).
Fig 4.
IL-13 stimulation has a variable effect on SARS-CoV-2 replication in healthy nonsensitized children. A, Boxplot showing the differences in viral load between healthy nonsensitized children infected with SARS-CoV-2 and those stimulated without or with IL-13. Lines connecting the dots denote samples from the same donor. B, Volcano plot showing the differentially upregulated and downregulated genes by IL-13 stimulation. Significantly (FDR < 0.01) upregulated (log2FC > 1) and downregulated genes (log2FC < –1) are denoted by blue and red color points, respectively. Select top-ranked genes by log2FC are highlighted. C, Scatter plot showing the association between viral load and expression of genes. Each point denotes a gene, and the color of the point shows the statistical significance from differential expression analysis in terms of –log10(adjusted P [adj.P]), and top-ranked genes are highlighted by larger point size and a circle around the points. Select ciliary and type 2–specific genes are highlighted in the same way as in Fig 2, C. D, Examples of select ciliated marker genes association between Δ viral load (difference in viral load between IL-13–stimulated and unstimulated) and Δ expression (difference in expression between IL-13–stimulated and unstimulated) in children with allergic asthma and healthy nonsensitized donors. E, Annotation of cells from scRNA-seq data generated from healthy nonsensitized children by using cells from children with allergic asthma as a reference to map cells onto the allergic asthma UMAP space. F and G, Cell type–specific differences in viral load as the ratio of viral load in the IL-13–stimulated to that in the unstimulated condition on a log2 scale are shown on the allergic asthma UMAP space (F) and cluster-level summary on the heatmap (G).
Next, we examined the cell-specific responses from scRNA-seq data generated from healthy nonsensitized children. To compare cell populations directly between healthy nonsensitized children and children with allergic asthma, we mapped and annotated individual cells from the healthy nonsensitized children to previously identified cell clusters from children with allergic asthma by utilizing the Seurat mapQuery function. This approach finds whether cells from the healthy nonsensitized children were more similar to one cluster than to the others and thereby superimposes these cells on the previously identified clusters from children with allergic asthma. Consistent with the bulk RNA-seq data, we observed only small and variable viral load differences due to IL-13 stimulation in the cell clusters in AECs from healthy nonsensitized donors (Fig 4, F and see Fig E8 in the Online Repository at www.jacionline.org). Most notably, almost all of the ciliated cells in healthy nonsensitized donors mapped to cluster C6 (99.3%), the terminally differentiated multiciliated cells (which had relatively minimal downregulation of ciliated cell genes with IL-13 and which we had seen previously) did not show a large drop in viral load with IL-13 in allergic asthma. A small number (0.7%) mapped to C8, but none mapped to C4 or C13 (Fig 4, E and see Table E4 in the Online Repository at www.jacionline.org). This means that most of the ciliated cells in healthy controls were more similar in gene expression to C6 (the terminally differentiated multiciliated cells) than to C4, C8, or C13 (the undifferentiated and/or transitional ciliated epithelial cells).
Discussion
We have demonstrated that in organotypic AEC cultures, SARS-CoV-2 replication was lower in epithelium from children with allergic asthma than healthy nonsensitized children. IL-13 stimulation further decreased viral replication in children with allergic asthma, but interestingly, it had a variable effect in healthy nonsensitized children. Bulk RNA-seq data demonstrated that lower viral transcript levels were closely associated with a downregulation of functional pathways of the ciliated epithelium, as opposed to lower ACE2 expression or increases in goblet cell counts or mucus secretion pathways. Furthermore, scRNA-seq data demonstrated a congruent result and identified specific subsets of relatively undifferentiated ciliated epithelial cells, common in cultures from donors with allergic asthma and highly responsive to IL-13, that show an impairment of viral replication. Specifically, these cells showed relative decreases in the key ciliary transcription factor FOXJ1 and corresponding downregulation of axoneme assembly and intraciliary transport machinery necessary for efficient viral replication,51 , 52 likely resulting in the lower viral levels within these individual cells. AECs from healthy nonsensitized children largely lacked these IL-13 responsive undifferentiated ciliated epithelial cells. Congruently, we did not observe a consistent decrease in viral replication in response to IL-13 in healthy nonsensitized AECs; rather, viral load remained closely associated with the relative expression of these ciliary cell–specific genes. In contrast, terminally differentiated multiciliated cells showed efficient viral replication. and their cilia specific genes were less affected by IL-13.
In the airway epithelium, ACE2 is relatively specifically localized to terminally differentiated multiciliated cells, which are the primary targets for SARS-CoV-2 replication in the early stages of disease; additionally, multiple studies have confirmed that viral tropism is quite specific to multiciliated cells.46 , 52, 53, 54, 55 The cilia on these cells contain the molecular components required for SARS-CoV-2 entry, and it has been proposed that the cilia specifically mediate both endocytosis and exocytosis of mature virions within multiciliated cells, initiating and spreading infection.51 Although cilia are generally understood to provide a protective role in the airways through their role in mucociliary clearance, in the case of SARS-CoV-2, they are exploited to propagate infection. SARS-CoV-2 does not infect epithelial basal cells; in fact, it is the renewal of differentiated multiciliated epithelial cells that has been shown to be necessary to maintain SARS-CoV-2 replication.56 Ciliated cell death and ciliary dysfunction result during SARS-CoV-2 infection, which is postulated to be due to cell damage from the virus; however, a decrease in FOXJ1 also occurs, resulting in a dedifferentiation of multiciliated cells.57 It has been hypothesized that the dedifferentiation of multiciliated cells may in fact serve as a protective host mechanism that prevents viral particles from interacting with cilia58; however, whether downregulation of FOXJ1 is a pathologic effect of the virus or a protective host response has remained unclear. Our data demonstrate that a naturally occurring downregulation of FOXJ1 owing to IL-13, and the associated functional alteration of ciliated epithelial cells toward an undifferentiated state, impairs viral replication. IL-13 has long been known to downregulate FOXJ1 and cause retraction of motile cilia,59 and in hamsters, IL-13 is known to be upregulated in the respiratory tract with SARS-CoV-2 infection,60 which is postulated as a protective response. Our data indicate that the chronic effects of type 2 inflammation in the airway of individuals with allergic asthma results in subsets of ciliated epithelial cells that have lower FOXJ1 expression and concomitant lower expression of key machinery of the cilia, axoneme, and cytoskeleton of ciliated cells. Moreover, we conclude that AECs from the airway in allergic asthma are highly sensitive to the effects of IL-13 to significantly impair SARS-CoV-2 replication. Although in the setting of allergic asthma these cells likely contribute to airway dysfunction, they also replicate SARS-CoV-2 less efficiently than do AECs from healthy nonsensitized children. Although type 2 inflammation can decrease interferon responses, this does not result in an increase in SARS-CoV-2 replication, and this effect of IL-13 is superseded by the identified effects on the ciliated epithelium that impair viral replication. We conclude that paradoxically, this “adverse” effect of type 2 inflammation on ciliated cells in the airway provides innate protection against SARS-CoV-2. We hypothesize that this innate cellular protection contributes significantly to the counterintuitive clinical protection against severe COVID-19 observed in individuals with allergic asthma.
Our analyses were limited in sample size to a small group of children with allergic asthma and healthy nonsensitized comparators. The high-quality data and large number of single-cell transcriptomes provided ample power to identify this signal, but they do limit our ability to generalize the relevance of our results more broadly, for instance, to allergen sensitized individuals without asthma,61 to adults with allergic asthma, or to children or adults with other asthma endotypes. Also, because our results derive from cell culture experiments, we cannot conclude the extent to which these effects of type 2 inflammation on SARS-CoV-2 would occur in vivo or the degree to which they might affect clinical severity of COVID-19. Additionally, because we did not examine uninfected epithelium, we cannot conclude that specific cell clusters of transitional/undifferentiated ciliated cells uniquely identified in allergic asthma already exist in asthma epithelium or are generated in response to SARS-CoV-2 infection specifically in asthma epithelium. Furthermore, our data do not allow us to identify underlying epigenetic or genetic features that mediate the observed differences in cellular expression whereby type 2 inflammation impairs SARS-CoV-2 replication; nor do the data identify the extent to which it is a feature of a chronic change in the epithelium in allergic asthma or an immunologic feature of allergic sensitization. Finally, it would be of significant value to contrast our data to data on the effects of type 2 inflammation on other respiratory viruses that drive more severe clinical disease in allergic asthma, such as rhinoviruses. Nevertheless, we believe that our data identify a highly significant and novel mechanism of innate protection against SARS-CoV-2 in allergic asthma that provides important molecular and clinical insights during the ongoing COVID-19 pandemic.
Clinical implications.
An important open question regarding the protective effects of type2 inflammation against SARS-CoV-2 infection with important mechanistic and clinical relevance has been addressed.
Footnotes
Supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases (to N.D.J., M.C.A, B.B., M.D., E.R.V., L.M.R., M.P.W., D.J.J, and J.S.D.), including grants K24AI150991-01S1 (to J.S.D.) and R01AI163160-01A1 (to J.S.D.).
Disclosure of potential conflict of interest: M. C. Altman reports personal fees from Sanofi-Regeneron and additional grant support from the National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases and the American Academy of Allergy, Asthma & Immunology outside the submitted work. D. J. Jackson reports personal fees from Novartis, Pfizer, Regeneron, AstraZeneca, Sanofi, and Vifor Pharma, as well as grants and personal fees from GlaxoSmithKline and grants from the NIH/National Heart, Lung and Blood Institute (NHLBI) outside the submitted work. J. S. Debley reports additional grant support from the NIH/NHLBI and the Bill and Melinda Gates Foundation outside the submitted work. The rest of the authors declare that they have no relevant conflicts of interest.
Supplementary data
References
- 1.Johns Hopkins Coronavirus Resource Center homepage Johns Hopkins University. https://coronavirus.jhu.edu/ Available at:
- 2.O’Driscoll M., Ribeiro Dos Santos G., Wang L., Cummings D.A.T., Azman A.S., Paireau J., et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590:140–145. doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- 3.Lai C.C., Liu Y.H., Wang C.Y., Wang Y.H., Hsueh S.C., Yen M.Y., et al. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): facts and myths. J Microbiol Immunol Infect. 2020;52:404–412. doi: 10.1016/j.jmii.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultze J.L., Aschenbrenner A.C. COVID-19 and the human innate immune system. Cell. 2021;184:1671–1692. doi: 10.1016/j.cell.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Degarege A., Naveed Z., Kabayundo J., Brett-Major D. Heterogeneity and risk of bias in studies examining risk factors for severe illness and death in COVID-19: a systematic review and meta-analysis. Pathogens. 2022;11:563. doi: 10.3390/pathogens11050563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borges do Nascimento I.J., von Groote T.C., O’Mathúna D.P., Abdulazeem H.M., Henderson C., Jayarajah U., et al. Clinical, laboratory and radiological characteristics and outcomes of novel coronavirus (SARS-CoV-2) infection in humans: a systematic review and series of meta-analyses. PLoS One. 2020;15 doi: 10.1371/journal.pone.0239235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CDC COVID-19 Response Team Severe outcomes among patients with coronavirus disease 2019 (COVID-19) - United States, February 12-March 16, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:343–346. doi: 10.15585/mmwr.mm6912e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levin A.T., Hanage W.P., Owusu-Boaitey N., Cochran K.B., Walsh S.P., Meyerowitz-Katz G. Assessing the age specificity of infection fatality rates for COVID-19: systematic review, meta-analysis, and public policy implications. Eur J Epidemiol. 2020;35:1123–1138. doi: 10.1007/s10654-020-00698-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor E.H., Marson E.J., Elhadi M., Macleod K.D.M., Yu Y.C., Davids R., et al. Factors associated with mortality in patients with COVID-19 admitted to intensive care: a systematic review and meta-analysis. Anaesthesia. 2021;76:1224–1232. doi: 10.1111/anae.15532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manry J., Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q., et al. The risk of COVID-19 death is much greater and age dependent with type I IFN autoantibodies. Proc Natl Acad Sci U S A. 2022;119 doi: 10.1073/pnas.2200413119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bermejo-Martin J.F., González-Rivera M., Almansa R., Micheloud D., Tedim A.P., Domínguez-Gil M., et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit Care. 2020;24:691. doi: 10.1186/s13054-020-03398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogan C.A., Stevens B.A., Sahoo M.K., Huang C., Garamani N., Gombar S., et al. High frequency of SARS-CoV-2 RNAemia and association with severe disease. Clin Infect Dis. 2021;72:e291–e295. doi: 10.1093/cid/ciaa1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miki S., Sasaki H., Horiuchi H., Miyata N., Yoshimura Y., Miyazaki K., et al. On-admission SARS-CoV-2 RNAemia as a single potent predictive marker of critical condition development and mortality in COVID-19. PLoS One. 2021;16 doi: 10.1371/journal.pone.0254640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawasuji H., Morinaga Y., Tani H., Yoshida Y., Takegoshi Y., Kaneda M., et al. SARS-CoV-2 RNAemia with a higher nasopharyngeal viral load is strongly associated with disease severity and mortality in patients with COVID-19. J Med Virol. 2022;94:147–153. doi: 10.1002/jmv.27282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozonoff A., Schaenman J., Jayavelu N.D., Milliren C.E., Calfee C.S., Cairns C.B., et al. Phenotypes of disease severity in a cohort of hospitalized COVID-19 patients: results from the IMPACC study. EBioMedicine. 2022;83 doi: 10.1016/j.ebiom.2022.104208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson D.J., Gern J.E. Rhinovirus infections and their roles in asthma: etiology and exacerbations. J Allergy Clin Immunol Pract. 2022;10:673–681. doi: 10.1016/j.jaip.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chhiba K.D., Patel G.B., Vu T.H.T., Chen M.M., Guo A., Kudlaty E., et al. Prevalence and characterization of asthma in hospitalized and nonhospitalized patients with COVID-19. J Allergy Clin Immunol. 2020;146:307–314.e4. doi: 10.1016/j.jaci.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eggert L.E., He Z., Collins W., Lee A.S., Dhondalay G., Jiang S.Y., et al. Asthma phenotypes, associated comorbidities, and long-term symptoms in COVID-19. Allergy. 2022;77:173–185. doi: 10.1111/all.14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S., Cao Y., Du T., Zhi Y. Prevalence of comorbid asthma and related outcomes in COVID-19: a systematic review and meta-analysis. J Allergy Clin Immunol Pract. 2021;9:693–701. doi: 10.1016/j.jaip.2020.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Y.D., Agache I., Akdis M., Nadeau K., Klimek L., Jutel M., et al. The effect of allergy and asthma as a comorbidity on the susceptibility and outcomes of COVID-19. Int Immunol. 2022;34:177–188. doi: 10.1093/intimm/dxab107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsson S.C., Gill D. Genetic predisposition to allergic diseases is inversely associated with risk of COVID-19. Allergy. 2021;76:1911–1913. doi: 10.1111/all.14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scala E., Abeni D., Tedeschi A., Manzotti G., Yang B., Borrelli P., et al. Atopic status protects from severe complications of COVID-19. Allergy. 2021;76:899–902. doi: 10.1111/all.14551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodruff P.G., Boushey H.A., Dolganov G.M., Barker C.S., Yang Y.H., Donnelly S., et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A. 2007;104:15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson D.J., Busse W.W., Bacharier L.B., Kattan M., O’Connor G.T., Wood R.A., et al. Association of respiratory allergy, asthma, and expression of the SARS-CoV-2 receptor ACE2. J Allergy Clin Immunol. 2020;146:203–206.e3. doi: 10.1016/j.jaci.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura H., Francisco D., Conway M., Martinez F.D., Vercelli D., Polverino F., et al. Type 2 inflammation modulates ACE2 and TMPRSS2 in airway epithelial cells. J Allergy Clin Immunol. 2020;146:80–88.e8. doi: 10.1016/j.jaci.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sajuthi S.P., DeFord P., Li Y., Jackson N.D., Montgomery M.T., Everman J.L., et al. Type 2 and interferon inflammation regulate SARS-CoV-2 entry factor expression in the airway epithelium. Nat Commun. 2020;11:5139. doi: 10.1038/s41467-020-18781-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farne H., Singanayagam A. Why asthma might surprisingly protect against poor outcomes in COVID-19. Eur Respir J. 2020;56 doi: 10.1183/13993003.03045-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonser L.R., Eckalbar W.L., Rodriguez L., Shen J., Koh K.D., Ghias K., et al. The type 2 asthma mediator IL-13 inhibits severe acute respiratory syndrome coronavirus 2 infection of bronchial epithelium. Am J Respir Cell Mol Biol. 2022;66:391–401. doi: 10.1165/rcmb.2021-0364OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison C.B., Edwards C.E., Shaffer K.M., Araba K.C., Wykoff J.A., Williams D.R., et al. SARS-CoV-2 infection of airway cells causes intense viral and cell shedding, two spreading mechanisms affected by IL-13. Proc Natl Acad Sci U S A. 2022;119 doi: 10.1073/pnas.2119680119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altman M.C., Reeves S.R., Parker A.R., Whalen E., Misura K.M., Barrow K.A., et al. Interferon response to respiratory syncytial virus by bronchial epithelium from children with asthma is inversely correlated with pulmonary function. J Allergy Clin Immunol. 2018;142:451–459. doi: 10.1016/j.jaci.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane C., Burgess S., Kicic A., Knight D., Stick S. The use of non-bronchoscopic brushings to study the paediatric airway. Respir Res. 2005;6:53. doi: 10.1186/1465-9921-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez-Guisa J.M., Powers C., File D., Cochrane E., Jimenez N., Debley J.S. Airway epithelial cells from asthmatic children differentially express proremodeling factors. J Allergy Clin Immunol. 2012;129:990–997.e6. doi: 10.1016/j.jaci.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reeves S.R., Kolstad T., Lien T.Y., Elliott M., Ziegler S.F., Wight T.N., et al. Asthmatic airway epithelial cells differentially regulate fibroblast expression of extracellular matrix components. J Allergy Clin Immunol. 2014;134:663–670.e1. doi: 10.1016/j.jaci.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.James R.G., Reeves S.R., Barrow K.A., White M.P., Glukhova V.A., Haghighi C., et al. Deficient follistatin-like 3 secretion by asthmatic airway epithelium impairs fibroblast regulation and fibroblast-to-myofibroblast transition. Am J Respir Cell Mol Biol. 2018;59:104–113. doi: 10.1165/rcmb.2017-0025OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonser L.R., Zlock L., Finkbeiner W., Erle D.J. Epithelial tethering of MUC5AC-rich mucus impairs mucociliary transport in asthma. J Clin Invest. 2016;126:2367–2371. doi: 10.1172/JCI84910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonser L.R., Koh K.D., Johansson K., Choksi S.P., Cheng D., Liu L., et al. Flow-cytometric analysis and purification of airway epithelial-cell subsets. Am J Respir Cell Mol Biol. 2021;64:308–317. doi: 10.1165/rcmb.2020-0149MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrow K.A., Rich L.M., Vanderwall E.R., Reeves S.R., Rathe J.A., White M.P., et al. Inactivation of material from SARS-CoV-2-infected primary airway epithelial cell cultures. Methods Protoc. 2021;4:7. doi: 10.3390/mps4010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziegler C.G.K., Miao V.N., Owings A.H., Navia A.W., Tang Y., Bromley J.D., et al. Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19. Cell. 2021;184:4713–4733.e22. doi: 10.1016/j.cell.2021.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hewitt R.J., Lloyd C.M. Regulation of immune responses by the airway epithelial cell landscape. Nat Rev Immunol. 2021;21:347–362. doi: 10.1038/s41577-020-00477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ravindra N.G., Alfajaro M.M., Gasque V., Huston N.C., Wan H., Szigeti-Buck K., et al. Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loske J., Röhmel J., Lukassen S., Stricker S., Magalhães V.G., Liebig J., et al. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat Biotechnol. 2022;40:319–324. doi: 10.1038/s41587-021-01037-9. [DOI] [PubMed] [Google Scholar]
- 48.Hatton C.F., Botting R.A., Dueñas M.E., Haq I.J., Verdon B., Thompson B.J., et al. Delayed induction of type I and III interferons mediates nasal epithelial cell permissiveness to SARS-CoV-2. Nat Commun. 2021;12:7092. doi: 10.1038/s41467-021-27318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nature Biotechnology. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 50.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buqaileh R., Saternos H., Ley S., Aranda A., Forero K., AbouAlaiwi W.A. Can cilia provide an entry gateway for SARS-CoV-2 to human ciliated cells? Physiol Genomics. 2021;53:249–258. doi: 10.1152/physiolgenomics.00015.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahn J.H., Kim J., Hong S.P., Choi S.Y., Yang M.J., Ju Y.S., et al. Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19. J Clin Invest. 2021;131 doi: 10.1172/JCI148517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee I.T., Nakayama T., Wu C.T., Goltsev Y., Jiang S., Gall P.A., et al. ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat Commun. 2020;11:5453. doi: 10.1038/s41467-020-19145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mulay A., Konda B., Garcia G., Jr., Yao C., Beil S., Villalba J.M., et al. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hao S., Ning K., Kuz C.A., Vorhies K., Yan Z., Qiu J. Long-term modeling of SARS-CoV-2 infection of cultured polarized human airway epithelium. MBio. 2020;11:e02852. doi: 10.1128/mBio.02852-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinot R., Hubert M., de Melo G.D., Lazarini F., Bruel T., Smith N., et al. SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nat Commun. 2021;12:4354. doi: 10.1038/s41467-021-24521-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schreiner T., Allnoch L., Beythien G., Marek K., Becker K., Schaudien D., et al. SARS-CoV-2 infection dysregulates cilia and basal cell homeostasis in the respiratory epithelium of hamsters. Int J Mol Sci. 2022;23:5124. doi: 10.3390/ijms23095124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gomperts B.N., Kim L.J., Flaherty S.A., Hackett B.P. IL-13 regulates cilia loss and foxj1 expression in human airway epithelium. Am J Respir Cell Mol Biol. 2007;37:339–346. doi: 10.1165/rcmb.2006-0400OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Francis M.E., Goncin U., Kroeker A., Swan C., Ralph R., Lu Y., et al. SARS-CoV-2 infection in the Syrian hamster model causes inflammation as well as type I interferon dysregulation in both respiratory and non-respiratory tissues including the heart and kidney. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seibold M.A., Moore C.M., Everman J.L., Williams B.J.M., Nolin J.D., Fairbanks-Mahnke A., et al. Risk factors for SARS-CoV-2 infection and transmission in households with children with asthma and allergy: a prospective surveillance study. J Allergy Clin Immunol. 2022;150:302–311. doi: 10.1016/j.jaci.2022.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.